Abstract
The multifunctional protein helper component proteinase (HC-Pro) is thought to interfere with the activity of the 20S proteasome; however, no sites of interaction have been identified for either protein. Here, we first show that the Potato virus Y (PVY) HC-Pro protein can interact with three Arabidopsis 20S proteasome subunits (PAA, PBB, and PBE), using a yeast two-hybrid system and the bimolecular fluorescence complement assay. In addition, yeast two-hybrid analysis of the interaction between several mutant subunits of the 20S proteasome and PVY HC-Pro confirmed that residues 81 to 140 of PAA, 1 to 80 of PBB, and 160 to 274 of PBE are necessary for binding PAA, PBB, and PBE to PVY HC-Pro, respectively. Deletion mutant analysis of PVY HC-Pro showed that the N terminus (residues 1 to 97) is necessary for its interaction with three Arabidopsis 20S proteasome subunits. The ability of HC-Pro to interact and interfere with the activity of the 20S proteasome may help explain the molecular basis of its multifunctional character.
The helper component proteinase (HC-Pro) of the genus Potyvirus is one of three viral proteinases that release the viral proteins needed for infection (27). Although HC-Pro has several functions in the viral cycle, including transmission via aphids, replication, and cell-to-cell and systemic virus movement (12, 32), little is known about the molecular mechanisms involved or the links between the protein's various activities (12). HC-Pro suppresses RNA silencing, which is regarded as an in vivo plant antiviral defense, but details of the molecular mechanism are lacking (1, 6, 23, 24, 28). Recently, it was discovered that HC-Pro of Lettuce mosaic virus (LMV) could interfere with the activity of the 20S proteasome, a key element of the ubiquitin/26S proteasome system (Ub/26S), which is also involved in the antiviral response (4).
Ub/26S is one of the most important proteolytic pathways in eukaryotes (11, 12, 15). The core element of this tightly regulated and highly specific system is the 26S proteasome, a high-molecular-mass complex consisting of a cylindrical 20S core protease capped on each end by a 19S regulatory particle (25, 26, 31, 33, 34). The central barrel-shaped 20S proteasome is composed of four stacked rings, including seven α subunits in the two outer rings and seven β subunits in the two inner rings, resulting in an overall configuration of α1-α7/β1-β7//β1-β7/α1-α7 (7, 30).
Ub/26S contributes significantly to plant development by affecting a wide range of processes, including embryogenesis, hormone signaling, and senescence (13, 25, 30). In addition, the proteasome can interfere with the translation of viral RNA and mRNA in virus-infected cells (3, 5, 8, 14, 16, 22). Ballut et al. (5) provided the first evidence of RNA endonuclease activity associated with the plant proteasome. Their results, which showed that the RNAs of two plant viruses, Tobacco mosaic virus and LMV, could be hydrolyzed by the proteasome, suggested that the endonuclease of the 20S proteasome might play an antiviral role in vivo (5).
Research on animal viral proteins has revealed that viruses can direct or subvert Ub/26S by interacting with the core of the 20S proteasome or with its 19S regulatory particle. For example, the human immunodeficiency virus type 1 Tat protein inhibits the peptidase activity and assembly of the 26S proteasome by interacting with distinct proteasome subunits (α and β) (2, 21, 29), while the hepatitis B virus X protein affects the 26S proteasome by interacting with the PMAS7 subunit of the 20S proteasome and PSMC1 subunit, which is a component of the 19S regulatory particle (19, 20, 40). Otherwise, the sequence of a viral protein can also interrupt proteasome function; for instance, Gly-Ala repeats act as stop-transfer signals during proteasome substrate processing (39).
Ballut et al. (4) were the first to describe the ability of a plant viral protein to interfere with the activity of the proteasome. They showed that HC-Pro from LMV could bind the proteasome and inhibit its 20S endonuclease activity in vitro, while its proteolytic activity was either unchanged or slightly stimulated (4). However, the interaction of a plant viral protein with the subunits of the 20S proteasome has not been described.
Using a yeast two-hybrid assay and bimolecular fluorescence complement (BiFC) assay, we found that potato virus Y (PVY) HC-Pro could interact with the PAA, PBB, and PBE subunits of the Arabidopsis 20S proteasome in yeast and in tobacco epidermal cells. Furthermore, we analyzed the domains necessary for these interactions in order to elucidate the mode of interaction between HC-Pro and the 20S proteasome.
MATERIALS AND METHODS
Plant materials and growth conditions.
Arabidopsis thaliana ecotype Columbia was used in this study. Surface-sterilized seeds were sown on Murashige-Skoug medium, placed at 4°C in darkness for 4 days, and then allowed to germinate. All seedlings transferred to soil were grown under standard greenhouse conditions.
Cloning and identification of the 20S proteasome cDNAs from A. thaliana.
The 14 cDNAs encoding the Arabidopsis 20S proteasome subunits included in this work are PAA2 (GenBank accession no. AF043519), PAB1 (AF043520), PAC1 (AF043521), PAD1 (AF043522), PAE1 (AF043524), PAF1 (AF043526), PAG1 (AF043526), PBA1 (AF043529), PBB2 (AF043531), PBC2 (AF043533), PBD1 (AF043537), PBE1 (AF043536), PBF1 (AF043537), and PBG1 (AF043538) (9); we designated the Arabidopsis 20S proteasome α and β subunits PAA-PAG and PBA-PBG as per Fu et al. (9). The primers used to clone these cDNAs are listed in Table 1. All cDNAs were cloned into pMD18-T (Takara) and verified by sequencing.
TABLE 1.
Primers used for cloning 14 Arabidopsis cDNAs
| Primer | Sequence |
|---|---|
| PAAF | 5′-CGATTTCGGAAGAGAGTTGTTC-3′ |
| PAAR | 5′-TCCCGACTGAATGTTCCA-3′ |
| PABF | 5′-AAGGAGCTATGGGAGAT-3′ |
| PABR | 5′-TGTAAATGCCACGAAA-3′ |
| PACF | 5′-AAGAAGAACCGAACGAG-3′ |
| PACR | 5′-TACCAGGCGAATAAGAC-3′ |
| PADF | 5′-AGATGGCGAGATACGAT-3′ |
| PADR | 5′-ATCTGAGACCGCTACTTT-3′ |
| PAEF | 5′-CGTTGTGCGAAGGAC-3′ |
| PAER | 5′-TCAAAGACGAGCGATG-3′ |
| PAFF | 5′-CGACGACGAGTGAAGAA-3′ |
| PAFR | 5′-CACAAGCCTTACATTTCC-3′ |
| PAGF | 5′-AATCAAGGAAGATGAGTAGC-3′ |
| PAGR | 5′-CATCAAACGACGCAAT-3′ |
| PBAF | 5′-CATCAGTCATCACCGGAATC-3′ |
| PBAR | 5′-GAAAGATCGGGTCATTCA-3′ |
| PBBF | 5′-AAAATGTCGCAGTCTTCC-3′ |
| PBBR | 5′-GAAATGGAGGCAACTACG-3′ |
| PBCF | 5′-CATCTTGCCTTGCGTCTT-3′ |
| PBCR | 5′-ATCAATCCATCCTTCCCT-3′ |
| PBDF | 5′-TCTGTAGACGGAAGATGGAGT-3′ |
| PBDR | 5′-ATCCTTTACGGACTGACGC-3′ |
| PBEF | 5′-TTTCTCCAGTAAGGGTTT-3′ |
| PBER | 5′-AAGGGAGTGAACGATTAG-3′ |
| PBFF | 5′-TAAGCCCTAGCTCAAA-3′ |
| PBFR | 5′-GATAAGGTTGGCTAAATG-3′ |
| PBGF | 5′-GAAGAACCTAAAGCTCACTACGATA-3′ |
| PBGR | 5′-AGCCGATGTGGTTGCTTGG-3′ |
Yeast two-hybrid assay.
The full-length coding sequence of PVY HC-Pro, which was fused to the GAL4 DNA-binding domain, was amplified by PCR from PVY N strain cDNA using the primers HC-Pro5 and HC-Pro3. The product was then cloned into pGBKT7 via BamHI/PstI digestion to form pGBKT7-HC-Pro. The full-length coding sequences of 14 Arabidopsis 20S proteasome subunits, which were fused to the GAL4 DNA-binding domain, were cloned into pGADT7. The primers and restriction sites used to create these constructs are listed in Table 2. Each of the 14 plasmids carrying a 20S proteasome subunit was separately cotransformed with pGBKT7-HC-Pro into Saccharomyces cerevisiae AH109 cells as described in the BD Matchmaker library construction and screening kit user manual. At the same time, the two plasmids supplied with the kits, pGBKT7-53 and pGADT7-RecT, were cotransformed as positive controls, while pGBKT7-HC-Pro and pGADT7 were cotransformed as negative controls.
TABLE 2.
Primers used in plasmid construction
| Primer | Sequence | Restriction enzyme | Construct |
|---|---|---|---|
| HC-Pro5 | 5′-TAGGATTCTGTCGAATGCCGACAATTTT-3′ | BamHI | pGBKT7-HC-Pro |
| HC-Pro3 | 5′-TACTGCAGACCAACTCTATAATGTTT-3′ | PstI | |
| PAA5 | 5′-ATGAATTCATGAGCAGAGGAAGCGGC-3′ | EcoRI | pGADT7-PAA |
| PAA3 | 5′-ATCTCGAGTCAGTCACGTTCACTAAT-3′ | XhoI | |
| PAB5 | 5′-ATGAATTCATGGGAGATAGTCAGTAC-3′ | EcoRI | pGADT7-PAB |
| PAB3 | 5′-ATCTCGAGTTACTCGACTTCAGCAAG-3′ | XhoI | |
| PAC5 | 5′-ATGAATTCATGTCTCGGAGATATGAT-3′ | EcoRI | pGADT7-PAC |
| PAC3 | 5′-ATGAGCTCTTAAGATGTTTCAGCAGC-3′ | SacI | |
| PAD5 | 5′-ATGAATTCATGGCGAGATACGATCGA-3′ | EcoRI | pGADT7-PAD |
| PAD3 | 5′-ATCTCGAGTCATGTTTCCTTCGCAGG-3′ | XhoI | |
| PAE5 | 5′-ATGAATTCATGTTTCTCACTAGAACT-3′ | EcoRI | pGADT7-PAE |
| PAE3 | 5′-ATCTCGAGTCAAAGACGAGCGATGAC-3′ | XhoI | |
| PAF5 | 5′-ATGAATTCATGTTCAGAAACCAATAC-3′ | EcoRI | pGADT7-PAF |
| PAF3 | 5′-ATCTCGAGTTACATTTCCATTGGAGC-3′ | XhoI | |
| PAG5 | 5′-ATGAATTCATGAGTAGCATTGGAACT-3′ | EcoRI | pGADT7-PAG |
| PAG3 | 5′-ATCTGCAGTTAGTCAGCATCCATCTC-3′ | XhoI | |
| PBA5 | 5′-ATGAATTCATGGATCTCAATCTCGAT-3′ | EcoRI | pGADT7-PBA |
| PBA3 | 5′-ATCTCGAGTCACATGGCCATTGGTTC-3′ | XhoI | |
| PBB5 | 5′-ATGAATTCATGTCGCAGTCTTCCGTC-3′ | EcoRI | pGADT7-PBB |
| PBB3 | 5′-ATCTGCAGTCATTCCTCCATAGCTTC-3′ | XhoI | |
| PBC5 | 5′-ATGAATTCATGTCGATCTTCGAGTAC-3′ | EcoRI | pGADT7-PBC |
| PBC3 | 5′-ATCTCGAGTCAATCCATCCTTCCCTT-3′ | XhoI | |
| PBD5 | 5′-ATGAATTCATGGAGTGTGTATTCGGT-3′ | EcoRI | pGADT7-PBD |
| PBD3 | 5′-ATCTCGAGTCAGACAACAGCAGTTGT-3′ | XhoI | |
| PBE5 | 5′-ATGAATTCATGAAGCTTGATACTAGT-3′ | EcoRI | pGADT7-PBE |
| PBE3 | 5′-ATCTGCAGTTATTCGGCTGTTGCTTC-3′ | SpeI | |
| PBF5 | 5′-ATGAATTCATGACTAAACAGCACGCG-3′ | EcoRI | pGADT7-PBF |
| PBF3 | 5′-ATCTCGAGTTAGTCTTTCCTCAGGTC-3′ | XhoI | |
| PBG5 | 5′-ATATCGATACATGACGACTTTTTCTG-3′ | ClaI | pGADT7-PBG |
| PBG3 | 5′-ATCTCGAGTTACCAGGAGCCTGCAGC-3′ | XhoI | |
| HcProBiFC5 | 5′-ATTCTAGAATGTCGAATGCCGACAATTT-3′ | XbaI | pUC-SPYCE-HC-Pro/pSPYNE-35S-HC-Pro |
| HcProBiFC3 | 5′-ATACTAGTACCAACTCTATAATGTTTTA-3′ | SpeI | |
| BIFCPAA5 | 5′-ATTCTAGAATGAGCAGAGGAAGCGGCGC-3′ | XbaI | |
| BIFCPAA3-1 | 5′-TAGGTACCGTCACGTTCACTAATGGCCG-3′ | KpnI | pUC-SPYCE-PAA/pSPYCE-35S-PAA |
| BIFCPAA3-2 | 5′-TACTCGAGGTCACGTTCACTAATGGCCG-3′ | XhoI | |
| BIFCPBB5 | 5′-ATTCTAGAATGTCGCAGTCTTCCGTC-3′ | XbaI | |
| BIFCPBB3-1 | 5′-ATGGTACCTTCCTCCATAGCTTCACC-3′ | KpnI | pUC-SPYCE-PBB/pSPYCE-35S-PBB |
| BIFCPBB3-2 | 5′-ATCCCGGGTTCCTCCATAGCTTCACC-3′ | XmaI | |
| BIFCPBE5 | 5′-ATTCTAGAATGAAGCTTGATACTAGTGG-3′ | XbaI | |
| BIFCPBE3-1 | 5′-TAGGTACCTTCGGCTGTTGCTTCCTCCA-3′ | KpnI | pUC-SPYCE-PBE/pSPYCE-35S-PBE |
| BIFCPBE3-2 | 5′-TAGGTACCTTCGGCTGTTGCTTCCTCCA-3′ | XmaI | |
| PAA1-5 | 5′-TAGAATTCATGACTGGCATGACAGCTGATTC-3′ | EcoRI | pGADT7-PAA1 |
| PAA1-3 | 5′-ATCTCGAGTCAGTCACGTTCACTAATGGCCG-3′ | XhoI | |
| PAA2-5 | 5′-ATGAATTCATGAGCAGAGGAAGCGGCGCTGG-3′ | EcoRI | pGADT7-PAA2 |
| PAA2-3 | 5′-ATCTCGAGTCAAACCATGGCAACTACTCCAA-3′ | XhoI | |
| PAA3-3 | 5′-TACTCGAGTTAGGCTAACAATCCAAGGT-3′ | XhoI | pGADT7-PAA3 |
| PBB1-5 | 5′-TAGAATTCATGTACTGTTGTGGTGCAGGA-3′ | EcoRI | pGADT7-PBB1 |
| PBB1-3 | 5′-TAGAGCTCTCATTCCTCCATAGCTTCACC-3′ | SacI | |
| PBB2-5 | 5′-TAGAATTCATGTCGCAGTCTTCCGTC-3′ | EcoRI | pGADT7-PBB2 |
| PBB2-3 | 5′-TAGAGCTCTCAGCTACCACTACCAAG-3′ | SacI | |
| PBE1-5 | 5′-TAGAATTCATGGGATATATCTCCTCACAATC-3′ | EcoRI | pGADT7-PBE1 |
| PBE1-3 | 5′-ATGAGCTCTTATTCGGCTGTTGCTTCCTCCA-3′ | SacI | |
| PBE2-5 | 5′-ATGAATTCATGAAGCTTGATACTAGTGGGTT-3′ | EcoRI | pGADT7-PBE2 |
| PBE2-3 | 5′-ATGAGCTCTTATCCAGCAATCATTGTACCAA-3′ | SacI | |
| HC-Pro1-5 | 5′-TAGAATTCATGATAGCGTTAGAGCATCTA-3′ | EcoRI | pGBKT7-HC-Pro1 |
| HC-Pro1-3 | 5′-TACTGCAGTTAACCAACTCTATAATGTTT-3′ | PstI | |
| HC-Pro2-5 | 5′-TAGAATTCATGTCGAATGCCGACAATTT-3′ | EcoRI | pGBKT7-HC-Pro2 |
| HC-Pro2-3 | 5′-TACTGCAGTTAACCATCATCAAGTGTTGT-3′ | PstI | |
| HC-Pro3-3 | 5′-TACTGCAGTTACAAGAACTTATTAACATG-3′ | PstI | pGBKT7-HC-Pro3 |
BiFC assay.
The vectors used in the BiFC assay (pUC-SPYNE, pUC-SPYCE, pSPYNE-35S, and pSPYCE-35S) were gifts from Klaus Harter and Jörg Kudla (36). The full-length coding sequence of PVY HC-Pro was cloned into pUC-SPYNE and pSPYNE-35S to create a fusion with the N-terminal fragment of yellow fluorescent protein (YFP) (pUC-SPYNE-HC-Pro and pSPYNE-35S-HC-Pro). The full-length coding sequences of three Arabidopsis 20S proteasome subunits (PAA, PBB, and PBE) were individually cloned into pUC-SPYCE and pSPYCE-35S to create a fusion with the C-terminal fragment of YFP. The primers and restriction sites used to construct these plasmids are listed in Table 2.
Two plasmids at a time were used to bombard onion (Allium cepa) epidermal cells by the method of Chen et al. (38), and YFP fluorescence was assessed 12 to 18 h later (38). The relevant negative controls were produced at the same time. For infiltration of Nicotiana benthamiana, Agrobacterium tumefaciens strain EHA105 was used to infiltrate the abaxial air space of 2- to 4-week-old plants; the epidermal cell layers of the leaves were assayed for fluorescence 2 days later (35, 37).
Fluorescence analysis was performed on a Nikon Eclipse TE2000-E inverted fluorescence microscope equipped with a Nikon D-Eclipse C1 spectral confocal laser scanning system. YFP fluorescence was examined at 514 nm (excitation) using an argon laser with an emission band of 515 to 530 nm and 650 nm (chlorophyll autofluorescence).
Construction of deletion mutants for PAA, PBB, PBE, and HC-Pro.
Deletion mutants were designed for each subunit that was determined to interact with the PVY HC-Pro protein: PAA1 (residues 81 to 246), PAA2 (residues 1 to 140), and PAA3 (residues 1 to 80) for PAA, PBB1 (residues 81 to 274), and PBB2 (residues 1 to 210) for PBB, and PBE1 (residues 81 to 274) and PBE2 (residues 1 to 159) for PBE. The coding sequences of these mutants were amplified individually by PCR using pGADT7-PAA, pGADT7-PBB, and pGADT7-PBE as the templates and the primers (PAA1-5, PAA1-3; PAA2-5, PAA2-3; PAA2-5, PAA3-3; PBB1-5, PBB1-3; PBB2-5, PBB2-3; PBE1-5, PBE1-3; and PBE2-5, PBE2-3) listed in Table 2. The products were then individually subcloned into pGADT7 and cotransformed with pGBKT7-HC-Pro into S. cerevisiae AH109 cells to determine which domains of the subunits were necessary for interacting with PVY HC-Pro.
Three deletion mutants were designed for PVY HC-Pro, HC-Pro1 (residues 98 to 456), HC-Pro2 (residues 1 to 298), and HC-Pro3 (residues 1 to 97). The coding sequences of these mutants were amplified individually by PCR using pGBKT7-HC-Pro as the templates and the primers (HC-Pro1-5, HC-Pro1-3; HC-Pro2-5, HC-Pro2-3; and HC-Pro2-5, HC-Pro3-3) listed in Table 2. The products were then individually subcloned into pGBKT7 and cotransformed with pGADT7-PAA, pGADT7-PBB, and pGADT7-PBE into S. cerevisiae AH109 cells to determine the necessary domain for interacting with PAA, PBB, and PBE.
RESULTS
Interaction of PVY HC-Pro with Arabidopsis 20S proteasome subunits in the yeast two-hybrid assay.
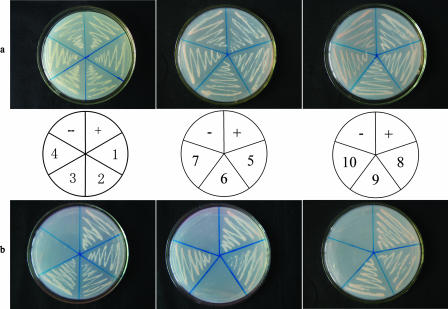
To test which subunit of the A. thaliana 20S proteasome could interact with PVY HC-Pro, we cloned 14 cDNAs encoding the subunits of the Arabidopsis 20S proteasome and subcloned them into pGADT7. All clones were verified by sequencing; the predicted primary sequences we obtained were consistent with those described by Fu et al. (9). Each of the 14 plasmids encoding a subunit of the 20S proteasome was then cotransformed with pGBKT7-HC-Pro into S. cerevisiae AH109 cells. The transformants were cultivated on synthetic dropout (SD) medium lacking Leu and Trp (SD/−Leu/−Trp), followed by selection on SD medium lacking Ade, His, Leu, and Trp (SD/−Ade/−His/−Leu/−Trp).
Our yeast two-hybrid results indicate that three Arabidopsis proteasome subunits, PAA, PBB, and PBE, are capable of interacting with HC-Pro in S. cerevisiae AH109 cells. In other words, the DNA-binding domain and activation domain were brought into sufficient proximity to drive the transcription of the reporter genes (ADE2, HIS3, lacZ, and MEL1) that allowed the yeast to grow on SD/−Ade/−His/−Leu/−Trp (Fig. 1). Subunits that did not interact with HC-Pro could not restore auxotrophy in the yeast cells (i.e., the transformants were able to grow on SD/−Leu/−Trp but not on SD/−Ade/−His/−Leu/−Trp) (Fig. 1). pGBKT7-53 and pGADT7-RecT were cotransformed as positive controls, while pGBKT7-HC-Pro and pGADT7 were cotransformed as negative controls; the transformants were selected on SD/−Ade/−His/−Leu/−Trp.
FIG. 1.
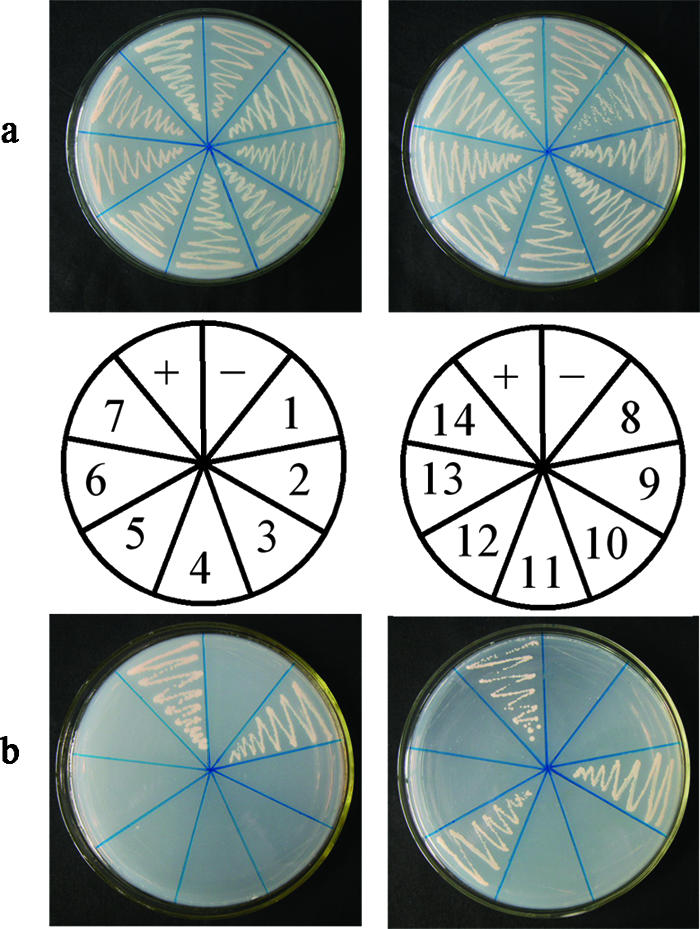
Interaction of 14 Arabidopsis 20S proteasome subunits and PVY HC-Pro in transformed S. cerevisiae AH109 cells grown on SD/−Leu/−Trp (a) and on SD/−Ade/−His/−Leu/−Trp (b). Cotransformants of seven a subunits (left) and seven β subunits (right) (PAA to PAE) and HC-Pro are shown. +, pGBKT7-53/pGADT7-RecT (positive control); −, pGBKT7-HC-Pro/pGADT7 (negative control); 1, pGBKT7-HC-Pro/pGADT7-PAA; 2, pGBKT7-HC-Pro/pGADT7-PAB; 3, pGBKT7-HC-Pro/pGADT7-PAC; 4, pGBKT7-HC-Pro/pGADT7-PAD; 5, pGBKT7-HC-Pro/pGADT7-PAE; 6, pGBKT7-HC-Pro/pGADT7-PAF; 7, pGBKT7-HC-Pro/pGADT7-PAG; 8, pGBKT7-HC-Pro/pGADT7-PBA; 9, pGBKT7-HC-Pro/pGADT7-PBB; 10, pGBKT7-HC-Pro/pGADT7-PBC; 11, pGBKT7-HC-Pro/pGADT7-PBD; 12, pGBKT7-HC-Pro/pGADT7-PBE; 13, pGBKT7-HC-Pro/pGADT7-PBF; 14, pGBKT7-HC-Pro/pGADT7-PBG.
Identification of the interaction between PVY HC-Pro and PAA, PBB, and PBE in living plant cells using BiFC.
To verify our yeast two-hybrid assay results, we analyzed the interaction between the three Arabidopsis 20S proteasome subunits (PAA, PBB, and PBE) and PVY HC-Pro by BiFC assay. In this approach, a fluorescent complex is formed when two nonfluorescent fragments of YFP are brought together by an interaction between the proteins fused to the fragments (17, 18). We constructed vectors expressing PAA, PBB, and PBE fused to the last 86 amino acids of YFP and a vector expressing HC-Pro fused to the first 155 amino acids of YFP (36). The two transient-expression vectors were then used to cobombard onion epidermal cells to test whether the proteins could interact, while the two constitutive vectors were individually transformed into A. tumefaciens strains EHA105 and EHA105 were used to coinfiltrate N. benthamiana.
As shown in Fig. 2, PAA, PBB, and PBE were able to interact with PVY HC-Pro in onion epidermal cells. YFP fluorescence was detected in the experimental samples 16 h after bombardment, whereas it was not detected in the negative control (i.e., pUC-SPYCE-HC-Pro and pUC-SPYNE [data not shown]). The assay in which A. tumefaciens was used to coinfiltrate N. benthamiana confirmed the interaction between PVY HC-Pro and PAA, PBB, and PBE in living plant cells (Fig. 3). Using the BiFC assay, we further confirmed that PVY HC-Pro could interact with these three Arabidopsis 20S proteasome subunits in living plant cells.
FIG. 2.
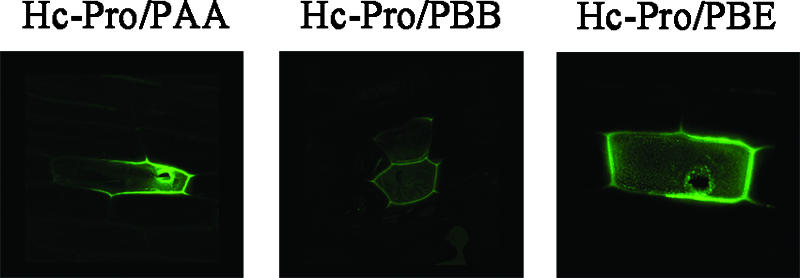
Fluorescence of reconstructed YFP complexes (green) in onion (A. cepa) epidermal cells.
FIG. 3.
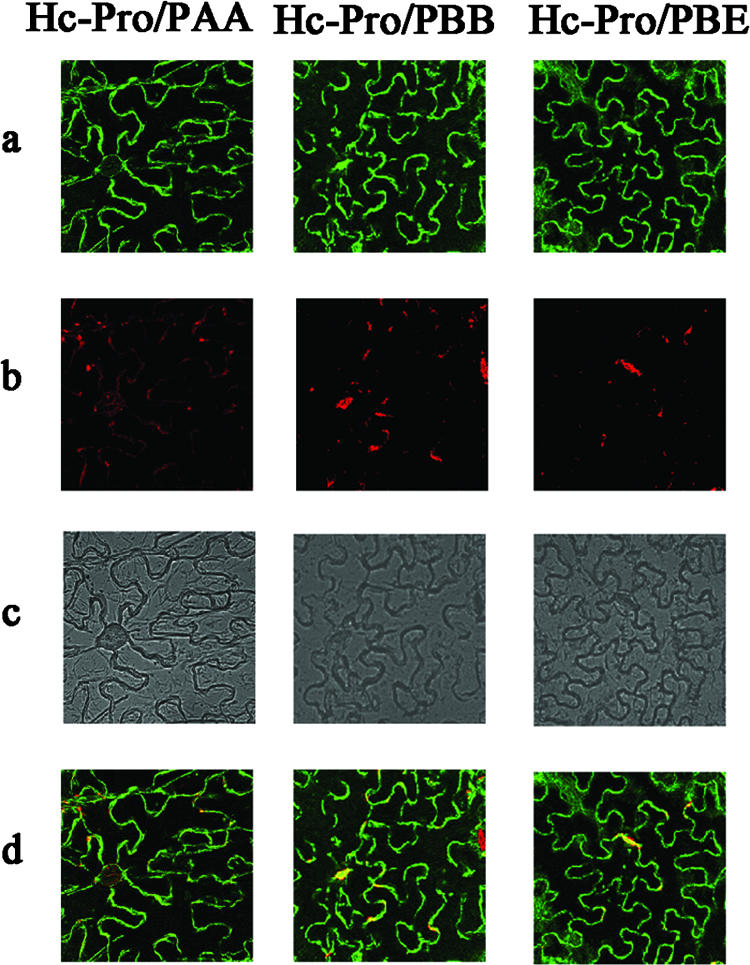
Subcellular localization of reconstructed YFP complexes determined in leaf epidermis of N. benthamiana. a, YFP fluorescence (green); b, chlorophyll autofluorescence (red); c, bright field; d, YFP-chlorophyll autofluorescence overlay.
Identification of the necessary domains of PAA, PBB, and PBE for interacting with PVY HC-Pro.
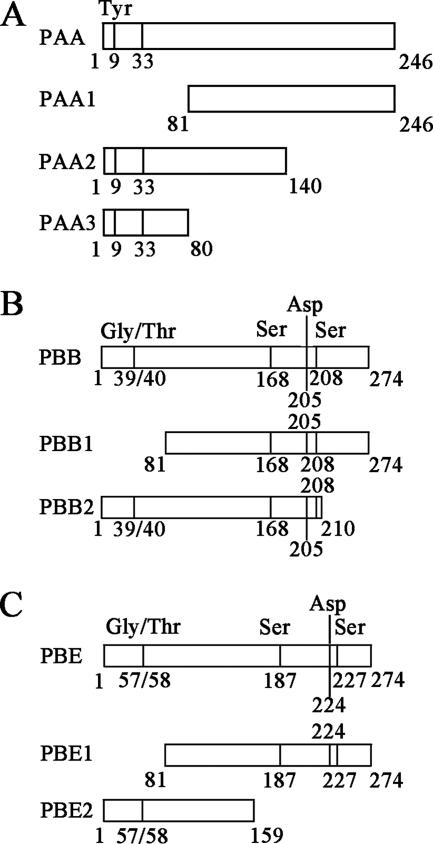
To determine the domains necessary for the interaction between HC-Pro and PAA, PBB, and PBE, we designed three deletion mutants for PAA and two deletion mutants for PBB and PBE (Fig. 4). The coding sequences of the mutants were subcloned into pGADT7 and separately cotransformed with pGBKT7-HC-Pro into S. cerevisiae AH109 cells. The domains required for the interaction were subsequently identified by a yeast two-hybrid assay.
FIG. 4.
Schematic overview of domains and deletion mutants of PAA (A), PBB (B), and PBE (C).
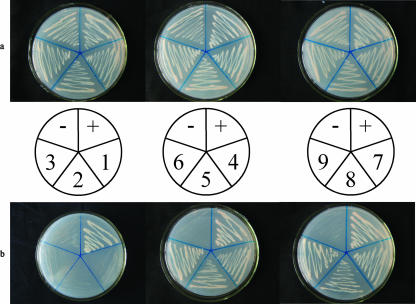
As shown in Fig. 5, the vectors expressing PAA1 (residues 81 to 246), PAA2 (residues 1 to 140), PBB2 (residues 1 to 210), and PBE1 (residues 81 to 274), which were each cotransformed with pGBKT7-HC-Pro into S. cerevisiae AH109 cells, restored the auxotrophy of the yeast cells, such that the transformants were able to grow on SD/−Ade/−His/−Leu/−Trp. In comparison, the transformants expressing PAA3 (residues 1 to 80), PBB1 (residues 81 to 274), or PBE2 (residues 1 to 159) with pGBKT7-HC-Pro were able to grow on SD/−Leu/−Trp but not on SD/−Ade/−His/−Leu/−Trp (Fig. 5).
FIG. 5.
Interaction of PVY HC-Pro and mutants of PAA (left), PBB (middle), and PBE (right) in transformed S. cerevisiae AH109 cells grown on SD/−Leu/−Trp (a) and on SD/−Ade/−His/−Leu/−Trp (b). +, pGBKT7-53/pGADT7-RecT (positive control); −, pGBKT7-HC-Pro/pGADT7 (negative control); 1, pGBKT7-HC-Pro/pGADT7-PAA; 2, pGBKT7-HC-Pro/pGADT7-PAA1; 3, pGBKT7-HC-Pro/pGADT7-PAA2; 4, pGBKT7-HC-Pro/pGADT7-PAA3; 5, pGBKT7-HC-Pro/pGADT7-PBB; 6, pGBKT7-HC-Pro/pGADT7-PBB1; 7, pGBKT7-HC-Pro/pGADT7-PBB2; 8, pGBKT7-HC-Pro/pGADT7-PBE; 9, pGBKT7-HC-Pro/pGADT7-PBE1; 10, pGBKT7-HC-Pro/pGADT7-PBE2.
These results indicate that the binding activities of the deletion mutants PAA1, PAA2, PBB2, and PBE1 were either similar to or greater than those of the wild-type proteins; in contrast, PAA3, PBB1, and PBE2 did not bind PVY HC-Pro. From these data, we conclude that residues 1 to 80 and 160 to 274 are necessary for the binding of PBB and PBE to PVY HC-Pro, respectively, because the deletion of these regions affected the interaction. Residues 81 to 140 of PAA may be the necessary domain for binding to PVY HC-Pro, because the deletion of the N terminus (residues 1 to 80) or the C terminus (residues 141 to 246) of PAA did not affect the interaction between PAA and PVY HC-Pro. In addition, the deletion mutant PAA1 (residues 1 to 80), which includes a conserved domain (residues 9 to 33), could not bind to PVY HC-Pro and bring the DNA-binding domain and activation domain into sufficient proximity to drive the transcription of the reporter genes (ADE2, HIS3, lacZ, and MEL1) that allowed the yeast to grow on SD/−Ade/−His/−Leu/−Trp (Fig. 5).
Identification of the necessary domain of PVY HC-Pro for interacting with PAA, PBB, and PBE.
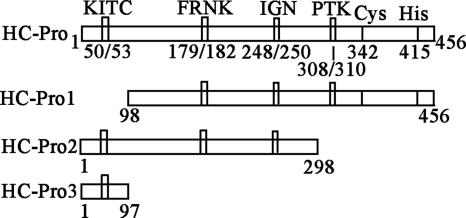
HC-Pro can be schematically divided into three regions: an N-terminal region, a C-terminal region, and a central region (27). In order to test which region is necessary for the binding to 20S proteasome subunits, we constructed three deletion mutants of PVY HC-Pro: HC-Pro1 (residues 98 to 456), HC-Pro2 (residues 1 to 298), and HC-Pro3 (residues 1 to 97) (Fig. 6). The domains required for the interaction were subsequently identified by a yeast two-hybrid assay.
FIG. 6.
Schematic overview of domains and deletion mutants of PVY HC-Pro.
As shown in Fig. 7, HC-Pro2 (residues 1 to 298) and HC-Pro3 (residues 1 to 97) could individually bind to PAA, PBB, and PBE and then drive the transcription of the reporter genes (ADE2, HIS3, lacZ, and MEL1) and restore the auxotrophy of the yeast cells, so that the transformants were able to grow on SD/−Ade/−His/−Leu/−Trp. However, HC-Pro1 (residues 98 to 456) did not interact with PAA, PBB, and PBE. From these results, we confirmed that the N terminus of PVY HC-Pro plays an important role in the binding to PAA, PBB, and PBE, because the deletion of this region affected the interaction between PVY HC-Pro and PAA, PBB, and PBE, and the N-terminal region of PVY HC-Pro (residues 1 to 97) itself interacted with PAA, PBB, and PBE.
FIG. 7.
Interaction of mutants of PVY HC-Pro and PAA, PBB, and PBE in transformed S. cerevisiae AH109 cells grown on SD/−Leu/−Trp (a) and on SD/−Ade/−His/−Leu/−Trp (b). Cotransformants of HC-Pro1 (left), HC-Pro2 (middle), and HC-Pro3 (right) and PAA, PBB, and PBE are shown. +, pGBKT7-53/pGADT7-RecT (positive control); −, pGBKT7-HC-Pro/pGADT7 (negative control); 1, pGBKT7-HC-Pro1/pGADT7-PAA; 2, pGBKT7-HC-Pro1/pGADT7-PBB; 3, pGBKT7-HC-Pro1/pGADT7-PBE; 4, pGBKT7-HC-Pro2/pGADT7-PAA; 5, pGBKT7-HC-Pro2/pGADT7-PBB; 6, pGBKT7-HC-Pro2/pGADT7-PBE; 7, pGBKT7-HC-Pro3/pGADT7-PAA; 8, pGBKT7-HC-Pro3/pGADT7-PBB; 9, pGBKT7-HC-Pro3/pGADT7-PBE.
DISCUSSION
HC-Pro is vital for the transmission of PVY, but the exact mechanisms underlying its multifunctional character are unclear. Recently, Ballut et al. (5) reported that HC-Pro from LMV was able bind to the 20S proteasome and affect its activity; however, exactly which subunits of the 20S proteasome are bound by HC-Pro is unknown. In this report, we present the first evidence that PVY HC-Pro interacts with three Arabidopsis 20S proteasome subunits, PAA, PBB, and PBE, using yeast two-hybrid and BiFC assays, which allow protein interactions to be examined under conditions that mimic the normal physiological environment (36). Our results demonstrate that PVY HC-Pro binds to distinct (α and β) subunits instead of binding one site on the Arabidopsis 20S proteasome.
The human proteasome 20S complex subunit Zeta harbors endonuclease activity (10); correspondingly, subunit PAE of the Arabidopsis 20S proteasome may also harbor this activity, especially since the Arabidopsis 20S proteasome α and β subunits have greater similarity to specific subunits in yeast and other organisms than with other α and β subunits in Arabidopsis (9). Recently, HC-Pro of LMV was reported to inhibit the endonuclease activity of the 20S proteasome in vitro, and the authors suggested that HC-Pro affects RNase activity directly rather than indirectly through protection of the template (4). According to our results, PVY HC-Pro did not directly bind to the subunit harboring the RNase activity; instead, it bound to another α subunit, PAA (Fig. 1, 2, and 3). We therefore conclude that HC-Pro may indirectly inhibit the endonuclease activity of the 20S proteasome by binding PAA. In addition, an analysis of the primary sequence of PAA revealed the existence of a conserved region near the N terminus (residues 9 to 33) (Fig. 4A). In the T. acidophilum-yeast complex, this region assumes an α-helical structure, which is necessary for the assembly and/or subsequent stabilization of specific α-subunit-α-subunit contacts, and the conserved Tyr9 residue in the N terminus of PAA plays a crucial role in this interaction (9). Using deletion mutants of PAA, we found that PVY HC-Pro might bind residues 81 to 140 of PAA but not the conserved region near the N terminus (Fig. 5); that is, the function of this conserved domain in PAA, like that in the T. acidophilum/yeast complex, may be either unaffected or indirectly affected.
We also found that PVY HC-Pro could interact with PBB and PBE of the Arabidopsis 20S proteasome (Fig. 1, Fig. 2, and Fig. 3). Based on sequence alignments with comparable T. acidophilum and yeast subunits, it has been suggested that PBB and PBE may be involved in forming protease active sites (9). Additional analysis of these two subunits revealed that Gly39/Thr40 in PBB and Gly57/Thr58 in PBE are cleaved to liberate the Thr active site (Fig. 4). In addition, Ser168, Asp205, and Ser208 in PBB (Fig. 4B) and Ser187, Asp224, and Ser227 in PBE (Fig. 4C) may be involved in forming the catalytic site, since in the three-dimensional complex they may be adjacent to the Thr in the active site. Our analysis of the interaction between PVY HC-Pro and several mutants of PBB and PBE indicates that HC-Pro binds residues 1 to 80 of PBB, which includes the conserved residues Gly39/Thr40, and residues 160 to 274 of PBE, which includes the conserved residues Ser187, Asp224, and Ser227 (Fig. 5). Thus, the domains of PBB and PBE that are necessary for binding to HC-Pro do not contain the same conserved residues. This indicates that HC-Pro does not directly bind these conserved residues, and it may not affect active-site formation in the protease.
We schematically divided PVY HC-Pro into three regions: an N-terminal region (residues 1 to 97), a central region (residues 98 to 298), and a C-terminal region (residues 299 to 456). Yeast two-hybrid analysis showed that the N-terminal region of PVY HC-Pro was necessary for the interaction with PAA, PBB, and PBE. The N-terminal region of HC-Pro was found to be involved only in the virus transmission process. Our results suggest that the N-terminal region of HC-Pro might have some other function in the infective process; for example, it may play an important role in the interaction with host proteins. However, our results are contrary to those reported by Ballut et al. (4), and there are some possible explanations for this. One may be that we use the yeast two-hybrid assay to confirm the interaction in vivo, while they analyzed the interaction in vitro. Another may be that we analyzed the exact subunit that can interact with PVY HC-Pro, while they studied the interaction between the whole 20S proteasome complex and LMV HC-Pro.
Based on our results, we propose that PVY HC-Pro binds the outside of the barrel of the Arabidopsis 20S proteasome by binding to several residues in the PAA, PBB, and PBE subunits. This would explain previous results showing that HC-Pro from LMV inhibits the endonuclease activity but not the proteolytic activity of the 20S proteasome in vitro (4). However, we have not yet identified the exact binding sites or the mode of interaction between HC-Pro and the 20S proteasome. Crystal structure analysis of the interaction may help resolve these issues.
Ub/26S plays a critical role in controlling development by affecting a wide range of processes, whereas viruses alter several functional systems, including Ub/26S, in order to facilitate their entry into cells (19, 20, 29). Some viral proteins can interfere with the function of Ub/26S by interacting with certain key elements. For example, several viral proteins, such as Tat and hepatitis B virus X protein, can interact with the 26S proteasome by binding to distinct subunits in the complex (2, 40). Here, we confirm that a plant virus protein, PVY HC-Pro, can interact with the PAA, PBB, and PBE subunits of the Arabidopsis 20S proteasome. Therefore, we presume that the ability to bind the proteasome may be a common feature of viral proteins that affect Ub/26S. Additional studies of the interaction between PVY HC-Pro and the Arabidopsis 20S proteasome will help us elucidate the molecular mechanisms underlying the multifunctional character of this protein as well as the relationship between viruses and cells.
Acknowledgments
This work was supported by the National Natural Science Foundation of China.
We thank Klaus Harter (Botanisches Institut, Universität zu Köln, Köln, Germany) and Jörg Kudla (Institut für Botanik und Botanischer Garten, Molekulare Entwichlungsbiologie der Pflanzen, Universität Münster, Münster, Germany) for the vectors used in the BiFC assay and Haiying Zhou and Liping Liang for help with the confocal microscope.
Footnotes
Published ahead of print on 26 September 2007.
REFERENCES
- 1.Anandalakshmi, R., G. J. Pruss, X. Ge, R. Marathe, A. C. Mallory, T. H. Smith, and V. B. Vance. 1998. A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 95:13079-13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apcher, G. S., S. Heink, D. Zantopf, P. M. Kloetzel, H. P. Schmid, R. J. Mayer, and E. Kruger. 2003. Human immunodeficiency virus-1 Tat protein interacts with distinct proteasomal alpha and beta subunits. FEBS Lett. 553:200-204. [DOI] [PubMed] [Google Scholar]
- 3.Aviel, S., G. Winberg, M. Massucci, and A. Ciechanover. 2000. Degradation of the Epstein-Barr virus latent membrane protein 1 (LMP1) by the ubiquitin-proteasome pathway. Targeting via ubiquitination of the N-terminal residue. J. Biol. Chem. 275:23491-23499. [DOI] [PubMed] [Google Scholar]
- 4.Ballut, L., M. Drucker, M. Pugniere, F. Cambon, S. Blanc, F. Roquet, T. Candresse, H. P. Schmid, P. Nicolas, O. L. Gall, and S. Badaoui. 2005. HcPro, a multifunctional protein encoded by a plant RNA virus, targets the 20S proteasome and affects its enzymic activities. J. Gen. Virol. 86:2595-2603. [DOI] [PubMed] [Google Scholar]
- 5.Ballut, L., F. Petit, S. Mouzeyar, O. Le Gall, T. Candresse, P. Schmid, P. Nicolas, and S. Badaoui. 2003. Biochemical identification of proteasome-associated endonuclease activity in sunflower. Biochim. Biophys. Acta 1645:30-39. [DOI] [PubMed] [Google Scholar]
- 6.Brigneti, G., O. Voinnet, W. X. Li, L. H. Ji, S. W. Ding, and D. C. Baulcombe. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17:6739-6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Coux, O., K. Tanaka, and A. L. Goldberg. 1996. Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 65:801-847. [DOI] [PubMed] [Google Scholar]
- 8.Drugeon, G., and I. Jupin. 2002. Stability in vitro of the 69K movement protein of Turnip yellow mosaic virus is regulated by the ubiquitin-mediated proteasome pathway. J. Gen. Virol. 83:3187-3197. [DOI] [PubMed] [Google Scholar]
- 9.Fu, H., J. H. Doelling, C. S. Arendt, M. Hochstrasser, and R. D. Vierstra. 1998. Molecular organization of the 20S proteasome gene family from Arabidopsis thaliana. Genetics 149:677-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gautier-Bert, K., B. Murol, A. S. Jarrousse, L. Ballut, S. Badaoui, F. Petit, and H. P. Schmid. 2003. Substrate affinity and substrate specificity of proteasomes with RNase activity. Mol. Biol. Rep. 30:1-7. [DOI] [PubMed] [Google Scholar]
- 11.Groll, M., and R. Huber. 2003. Substrate access and processing by the 20S proteasome core particle. Int. J. Biochem. Cell. Biol. 35:606-616. [DOI] [PubMed] [Google Scholar]
- 12.Harris, K., O. Smith, and J. Duffus. 2001. Virus-insect-plant interactions, p. 181-206. Academic Press, San Diego, CA.
- 13.Hellmann, H., and M. Estelle. 2002. Plant development: regulation by protein degradation. Science 297:793-797. [DOI] [PubMed] [Google Scholar]
- 14.Hericourt, F., S. Blanc, V. Redeker, and I. Jupin. 2000. Evidence for phosphorylation and ubiquitinylation of the turnip yellow mosaic virus RNA-dependent RNA polymerase domain expressed in a baculovirus-insect cell system. Biochem. J. 349:417-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hochstrasser, M. 1995. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr. Opin. Cell Biol. 7:215-223. [DOI] [PubMed] [Google Scholar]
- 16.Homma, S., A. Horsch, M. N. Pouch, F. Petit, Y. Briand, and H. P. Schmid. 1994. Proteasomes (prosomes) inhibit the translation of tobacco mosaic virus RNA by preventing the formation of initiation complexes. Mol. Biol. Rep. 20:57-61. [DOI] [PubMed] [Google Scholar]
- 17.Hu, C. D., Y. Chinenov, and T. K. Kerppola. 2002. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9:789-798. [DOI] [PubMed] [Google Scholar]
- 18.Hu, C. D., and T. K. Kerppola. 2003. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 21:539-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu, Z., Z. Zhang, E. Doo, O. Coux, A. L. Goldberg, and T. J. Liang. 1999. Hepatitis B virus X protein is both a substrate and a potential inhibitor of the proteasome complex. J. Virol. 73:7231-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, J., J. Kwong, E. C. Sun, and T. J. Liang. 1996. Proteasome complex as a potential cellular target of hepatitis B virus X protein. J. Virol. 70:5582-5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, X., U. Seifert, U. Salzmann, P. Henklein, R. Preissner, W. Henke, A. J. Sijts, P. M. Kloetzel, and W. Dubiel. 2002. The RTP site shared by the HIV-1 Tat protein and the 11S regulator subunit alpha is crucial for their effects on proteasome function including antigen processing. J. Mol. Biol. 323:771-782. [DOI] [PubMed] [Google Scholar]
- 22.Jockusch, H., and C. Wiegand. 2003. Misfolded plant virus proteins: elicitors and targets of ubiquitylation. FEBS Lett. 545:229-232. [DOI] [PubMed] [Google Scholar]
- 23.Kasschau, K. D., Z. Xie, E. Allen, C. Llave, E. J. Chapman, K. A. Krizan, and J. C. Carrington. 2003. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA unction. Dev. Cell 4:205-217. [DOI] [PubMed] [Google Scholar]
- 24.Mlotshwa, S., S. E. Schauer, T. H. Smith, A. C. Mallory, J. M. Herr, Jr., B. Roth, D. S. Merchant, A. Ray, L. H. Bowman, and V. B. Vance. 2005. Ectopic DICER-LIKE1 expression in P1/HC-Pro Arabidopsis rescues phenotypic anomalies but not defects in microRNA and silencing pathways. Plant Cell 17:2873-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moon, J., G. Parry, and M. Estelle. 2004. The ubiquitin-proteasome pathway and plant development. Plant Cell 16:3181-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 27.Plisson, C., M. Drucker, S. Blanc, S. German-Retana, O. Le Gall, D. Thomas, and P. Bron. 2003. Structural characterization of HC-Pro, a plant virus multifunctional protein. J. Biol. Chem. 278:23753-23761. [DOI] [PubMed] [Google Scholar]
- 28.Roth, B. M., G. J. Pruss, and V. B. Vance. 2004. Plant viral suppressors of RNA silencing. Virus Res. 102:97-108. [DOI] [PubMed] [Google Scholar]
- 29.Seeger, M., K. Ferrell, R. Frank, and W. Dubiel. 1997. HIV-1 tat inhibits the 20 S proteasome and its 11 S regulator-mediated activation. J. Biol. Chem. 272:8145-8148. [DOI] [PubMed] [Google Scholar]
- 30.Smalle, J., and R. D. Vierstra. 2004. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55:555-590. [DOI] [PubMed] [Google Scholar]
- 31.Sullivan, J. A., K. Shirasu, and X. W. Deng. 2003. The diverse roles of ubiquitin and the 26S proteasome in the life of plants. Nat. Rev. Genet. 4:948-958. [DOI] [PubMed] [Google Scholar]
- 32.Urcuqui-Inchima, S., A. L. Haenni, and F. Bernardi. 2001. Potyvirus proteins: a wealth of functions. Virus Res. 74:157-175. [DOI] [PubMed] [Google Scholar]
- 33.Vierstra, R. D. 1996. Proteolysis in plants: mechanisms and functions. Plant Mol. Biol. 32:275-302. [DOI] [PubMed] [Google Scholar]
- 34.Vierstra, R. D. 2003. The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 8:135-142. [DOI] [PubMed] [Google Scholar]
- 35.Voinnet, O., S. Rivas, P. Mestre, and D. Baulcombe. 2003. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33:949-956. [DOI] [PubMed] [Google Scholar]
- 36.Walter, M., C. Chaban, K. Schutze, O. Batistic, K. Weckermann, C. Nake, D. Blazevic, C. Grefen, K. Schumacher, C. Oecking, K. Harter, and J. Kudla. 2004. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40:428-438. [DOI] [PubMed] [Google Scholar]
- 37.Witte, C. P., L. D. Noel, J. Gielbert, J. E. Parker, and T. Romeis. 2004. Rapid one-step protein purification from plant material using the eight-amino acid StrepII epitope. Plant Mol. Biol. 55:135-147. [DOI] [PubMed] [Google Scholar]
- 38.Xu, X., C. Chen, B. Fan, and Z. Chen. 2006. Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18:1310-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, M., and P. Coffino. 2004. Repeat sequence of Epstein-Barr virus-encoded nuclear antigen 1 protein interrupts proteasome substrate processing. J. Biol. Chem. 279:8635-8641. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, Z., N. Torii, A. Furusaka, N. Malayaman, Z. Hu, and T. J. Liang. 2000. Structural and functional characterization of interaction between hepatitis B virus X protein and the proteasome complex. J. Biol. Chem. 275:15157-15165. [DOI] [PubMed] [Google Scholar]