Abstract
Neutralization of flaviviruses in vivo correlates with the development of an antibody response against the viral envelope (E) protein. Previous studies demonstrated that monoclonal antibodies (MAbs) against an epitope on the lateral ridge of domain III (DIII) of the West Nile virus (WNV) E protein strongly protect against infection in animals. Based on X-ray crystallography and sequence analysis, an analogous type-specific neutralizing epitope for individual serotypes of the related flavivirus dengue virus (DENV) was hypothesized. Using yeast surface display of DIII variants, we defined contact residues of a panel of type-specific, subcomplex-specific, and cross-reactive MAbs that recognize DIII of DENV type 2 (DENV-2) and have different neutralizing potentials. Type-specific MAbs with neutralizing activity against DENV-2 localized to a sequence-unique epitope on the lateral ridge of DIII, centered at the FG loop near residues E383 and P384, analogous in position to that observed with WNV-specific strongly neutralizing MAbs. Subcomplex-specific MAbs that bound some but not all DENV serotypes and neutralized DENV-2 infection recognized an adjacent epitope centered on the connecting A strand of DIII at residues K305, K307, and K310. In contrast, several MAbs that had poor neutralizing activity against DENV-2 and cross-reacted with all DENV serotypes and other flaviviruses recognized an epitope with residues in the AB loop of DIII, a conserved region that is predicted to have limited accessibility on the mature virion. Overall, our experiments define adjacent and structurally distinct epitopes on DIII of DENV-2 which elicit type-specific, subcomplex-specific, and cross-reactive antibodies with different neutralizing potentials.
Dengue fever (DF), the most prevalent arthropod-borne viral illness in humans, is caused by dengue virus (DENV). The four serotypes of DENV are transmitted to humans primarily by the mosquitoes Aedes aegypti and Aedes albopictus. DENV is a member of the Flaviviridae family and is related to the viruses that cause yellow fever and the Japanese, St. Louis, and West Nile encephalitides (8). Infection by DENV causes a spectrum of clinical disease, ranging from an acute, debilitating, self-limited febrile illness (DF) to a life-threatening hemorrhagic and capillary leak syndrome (dengue hemorrhagic fever/dengue shock syndrome). At present, no approved antiviral treatment or vaccine is available, and therapy is supportive in nature. DENV causes an estimated 25 to 100 million cases of DF and 250,000 cases of dengue hemorrhagic fever per year worldwide, with 2.5 billion people at risk for infection (27, 48).
DENV is an enveloped virus with a single-stranded, positive-sense RNA genome (11). The 10.7-kilobase genome is translated as a single polyprotein, which is then cleaved into three structural proteins (C, prM/M, and E) and seven nonstructural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) by virus- and host-encoded proteases. The 500-Å DENV mature virion has a well-organized outer protein shell, a 50-Å-thick lipid membrane bilayer, and a less-defined inner nucleocapsid core (37, 79). The icosahedral scaffold consists of 180 E and 180 M protein monomers arranged in a repeating pattern that lacks the predicted T=3 quasisymmetry (37, 78). The immature virion, which lacks cleavage of the prM protein, has a rough surface with 60 spikes (79), whereas the mature virion has a smooth surface. X-ray crystallographic analyses of the soluble ectodomains of the E proteins from tick-borne encephalitis virus and DENV demonstrated a dimeric assembly, with each subunit containing three domains (46, 59, 60). Domain III (DIII), which adopts an immunoglobulin-like fold, is believed by some to contain a cell surface receptor recognition site (3, 60, 74, 77). Recent structural results detailing the postfusion trimeric conformation of DENV type 2 (DENV-2) and tick-borne encephalitis virus E proteins has prompted a new model for type II viral fusion (7, 47). In the postfusion trimer, there is a reorganized E protein domain structure, with significant exposure of the hydrophobic fusion peptide in DII (47).
The majority of flavivirus-neutralizing antibodies recognize the structural E protein, although some also bind to the prM/M protein (16, 21, 58, 73). Serotype-specific epitopes elicit antibodies with the strongest neutralizing activities (62, 63), and protection in animals by antibodies correlates with neutralizing activity in vitro (6, 20, 25, 43, 53, 63). Based on epitope mapping data, many type-specific neutralizing antibodies against individual flaviviruses localize to DIII (1, 2, 10, 18, 39, 53, 61, 65, 66, 76), whereas neutralizing monoclonal antibodies (MAbs) that cross-react with other flaviviruses localize primarily to DII, near the fusion peptide (17, 23, 24, 54, 61, 68). Alteration of specific residues in DIII results in the loss of binding of neutralizing MAbs (2, 12, 32, 39, 40, 49). Recently, our group, along with others, localized individual contact residues of a large panel of anti-West Nile virus (anti-WNV) MAbs and defined a dominant neutralizing epitope on the lateral ridge of DIII (2, 53, 64). Crystallographic analysis indicated that a strongly neutralizing, DIII-specific anti-WNV MAb engaged four discontinuous segments, including the N-terminal linker region (residues 302 to 309) and three strand-connecting loops, namely, BC (residues 330 to 333), DE (residues 365 to 368), and FG (residues 389 to 391), which together form a single concave surface patch (51). Comparison of available WNV sequences revealed nearly complete conservation of the structurally defined epitope. However, sequence analysis of other flaviviruses revealed diversity in the four segments, with notable variation even between DENV serotypes. Interestingly, other groups have also identified individual flavivirus-specific neutralizing antibodies that localize to an analogous DIII binding region (32, 74, 75). Based on the coincident mapping of our and other neutralizing MAbs, we predicted that this structural epitope, although specific for individual flaviviruses, would have a dominant role in neutralization of all flaviviruses (51, 52).
In this study, we mapped the contact residues of a panel of anti-DENV-2, DIII-specific MAbs with distinct neutralizing potentials. Type-specific MAbs with the strongest neutralizing activities against DENV-2 localized to an epitope on the lateral ridge of DIII that was analogous in location to that seen with neutralizing WNV MAbs. Subcomplex-specific neutralizing MAbs that recognized several serotypes of DENV bound an adjacent epitope centered on the A strand of DIII. In contrast, several poorly neutralizing MAbs recognized conserved flavivirus residues within the AB loop that appear to have limited accessibility on the mature virion. Overall, in contrast to previous studies with WNV DIII, our data suggest the existence of two structurally distinct neutralizing epitopes on DIII of DENV-2 E protein, with a type-specific epitope on the lateral ridge of DIII centered at the unique FG loop and a subcomplex-specific epitope that binds the more conserved A strand.
MATERIALS AND METHODS
Cells and viruses.
Vero, BHK21-15, and Raji-DC-SIGN-R cells were maintained at 37°C in a 5% CO2 incubator in Dulbecco's modified essential medium or RPMI 1640 supplemented with 10% fetal bovine serum (Omega Scientific Inc., Tarzana, CA), 1% penicillin G, and 1% streptomycin. The DENV strains used were 16007 (DENV-1), 16681 (DENV-2), 16652 (DENV-3), and H241 (DENV-4). The viruses were amplified in C6/36 cells according to established protocols (19). Plaque reduction neutralization titer (PRNT) assays were performed on BHK21-15 cells with individual MAbs and quantitated as previously described (53).
MAbs.
The MAbs used in this study are presented in Table 1 and were purified from hybridoma culture supernatants or mouse ascites fluid by using an NAB protein A spin purification kit (Pierce, Rockford, IL). For sorting experiments, MAbs were labeled with Alexa Fluor 647 or Alexa Fluor 488, using a MAb labeling kit (Molecular Probes, Invitrogen, Carlsbad, CA).
TABLE 1.
Characteristics of DIII-specific MAbs used in this study
| MAb | Isotype | Neutralizing activity (PRNT50 [μg/ml])a | Reference |
|---|---|---|---|
| 1F1 | IgG2a | 0.01 | 44 |
| 3H5-1 | IgG1 | 0.18 | 22 |
| 6B6-10 | IgG2a | 0.07 | 34 |
| 9A3D-8 | IgG2a | 0.03 | 61 |
| M8051122 | IgG1 | 2 | Unpublished data |
| 9F16 | IgG1 | 0.8 | Unpublished data |
| 9F11 | IgG1 | 2 | Unpublished data |
| 2Q1899 | IgG1 | 5 | Unpublished data |
| 9D12 | IgG1 | 2 | 30 |
| 1A1D-2 | IgG1 | 0.3 | 61 |
| 5A2-7 | IgG2a | >100 | 44 |
| 13D4-1 | IgG2a | >100 | 42a |
| E111 | IgG2a | >100 | 54 |
| E114 | IgG1 | >100 | 54 |
Determined by PRNT assay on BHK21 cells with 102 PFU of DENV-2.
Expression of DENV-2 E protein DIII in yeast.
The DNA fragment encoding amino acid residues 294 to 409 (DIII) of the DENV-2 E protein was amplified from a DENV-2 strain 16681 infectious cDNA clone (35) by PCR, with BamHI and XhoI sites added at the 5′ and 3′ ends, respectively. The PCR products were digested and cloned as downstream fusions to Aga2 and Xpress epitope tag genes in the yeast surface display vector pYD1 (Invitrogen, Carlsbad, CA), under the control of an upstream GAL1 promoter. These constructs were transformed into Saccharomyces cerevisiae strain EBY100 (4, 5), using an S.c. EasyComp transformation kit (Invitrogen, Carlsbad, CA), to generate yeast that expressed DENV-2 DIII. Individual yeast colonies were grown to logarithmic phase at 30°C in tryptophan-free yeast medium containing 2% glucose. Fusion protein expression was induced on the surface by growing yeast cells for an additional 48 h in tryptophan-free medium containing 2% galactose at 20°C. Yeast cells were harvested, washed with phosphate-buffered saline (PBS) supplemented with bovine serum albumin (1 mg/ml), and stained with 50 μl of diluted MAbs. Purified antibodies were used at a concentration of 50 μg/ml, and MAbs from ascites fluid were used at a 1:100 dilution. After a 30-min incubation on ice, yeast cells were washed in PBS with bovine serum albumin and then stained with a goat anti-mouse immunoglobulin G (IgG) secondary antibody conjugated to Alexa Fluor 647 (Molecular Probes, Invitrogen, Carlsbad, CA). After fixation with 1% paraformaldehyde in PBS, yeast cells were analyzed on a FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ).
Library construction and screening.
To generate a random mutant library, DENV-2 DIII was mutated by error-prone PCR, using a GeneMorph II random mutagenesis kit (Stratagene). The mutant library was ligated into the pYD1 vector and transformed into XL2-Blue ultracompetent cells (Stratagene, La Jolla, CA). The colonies were pooled and transformed into yeast cells as described above.
For each individual MAb, the DENV-2 DIII mutant library was screened according to a previously described protocol (53). To identify yeasts that had selectively lost binding to a given MAb, the library was initially stained with this antibody conjugated to Alexa Fluor 647 for 30 min on ice. To control for the surface expression of DENV-2 DIII, yeast cells were subsequently stained for 30 min on ice with an Alexa Fluor 488-conjugated oligoclonal antibody that was derived from a pool of individual MAbs (1F1, 1A1D-2, 6B6-10, 9A3D-8, 13D4-1, and 5A2-7). After being stained, yeast cells were subjected to flow cytometry, and the population that was single MAb negative but oligoclonal antibody positive was identified. After three or four rounds of sorting, yeast cells were plated and individual colonies were tested for binding to individual MAbs by flow cytometry. For individual clones that had lost only the desired MAb binding epitope, the pYD1-DV2 DIII plasmid was recovered using a Zymoprep yeast miniprep kit (Zymo Research, Orange, CA). The plasmid was transformed into XL1-Blue competent cells (Stratagene, La Jolla, CA), purified using a QIAprep spin miniprep kit (QIAGEN), and sequenced. In some cases, DENV-2 DIII variants with multiple mutations were isolated. To determine which mutation conferred the phenotype, single independent mutations were engineered by site-directed mutagenesis, using a QuikChange II mutagenesis kit (Stratagene, La Jolla, CA), or by using splice-overlap PCR (45).
MAb staining of DENV-infected cells.
Raji-DC-SIGN-R cells were infected with DENV-1, DENV-2, DENV-3, or DENV-4 at a multiplicity of infection of 0.5 or 1. Depending on the strain, cells were harvested at 72 or 96 h, washed three times in PBS, and fixed in PBS with 1% paraformaldehyde. Cells were then washed twice in PBS and permeabilized in Hanks' balanced salt solution (Cellgro, Herndon, VA) containing 10 mM HEPES (pH 7.3), 0.1% saponin (Sigma, St. Louis, MO), and 0.02% NaN3 (HHSN). MAbs were bound to permeabilized virus-infected cells for 30 min on ice, washed three times in HHSN, and resuspended in 50 μl of a 1/500 dilution of Alexa Fluor-conjugated anti-mouse IgG (Molecular Probes, Invitrogen, Carlsbad, CA). After 15 min, cells were again washed in HHSN three times, fixed in PBS with 1% paraformaldehyde, and processed by flow cytometry.
Mapping of mutations onto the DENV-2 DIII crystal structure and virion.
Figures were prepared using the atomic coordinates of DENV-2 E (Research Collaboratory for Structural Bioinformatics [RCSB] accession number 1OAN) and the WNV E DIII/E16 Fab complex (RCSB accession number 1ZTX), using the programs Ribbons (9), MOLEMAN2 (36), Insight II (Accelrys, San Diego, CA), and PyMol (http://www.pymol.org). The representations of the DENV-2 virion were generated using the atomic coordinates and transformation matrices found in RCSB entry 1THD.
RESULTS
Strongly neutralizing MAbs against WNV localize to an epitope (see Fig. 2A) composed of the loops from three β-strands and an N-terminal region on the lateral ridge of DIII (2, 14, 51, 53, 64, 74). Sequence analysis indicated that this neutralizing epitope is highly conserved among WNV strains but divergent from those of other flaviviruses (38, 51). To test whether the analogous epitope on DIII of DENV-2 elicited strongly neutralizing antibodies, we screened a panel of 40 MAbs obtained from colleagues for immunoreactivity with DENV-2 strain 16681. Twenty-one of 40 MAbs exhibited significant reactivity with DENV-2-infected cells or purified recombinant soluble DENV-2 E protein derived from insect cells (data not shown). Of the MAbs that recognized DENV-2-infected cells, 14 bound strongly to yeast cells displaying DENV-2 DIII (data not shown). Of these, four had moderate (50% PRNT [PRNT50], 1 to 100 μg/ml) and six had strong (PRNT50 <1 μg/ml) neutralizing activities against strain 16681 in a standard PRNT assay (Table 1). Four others had weak or no appreciable inhibitory activity (PRNT50, >100 μg/ml). Notably, five of the six strongly neutralizing MAbs were type specific and showed no significant cross-reactivity with DENV-1-, DENV-3-, or DENV-4-infected cells (Table 2).
FIG. 2.
DIII lateral ridge antibody epitope. The structures of WNV and DENV-2 DIIIs are shown, with identification of binding sites of type-specific neutralizing antibodies. (A) Structure of WNV E16 neutralizing epitope determined by X-ray crystallography (16 residues in blue) or by using yeast display mapping (4 residues in orange). (B) Structure of DENV-2 DIII, with the corresponding 16 amino acids of the WNV E16 neutralizing antibody epitope highlighted. (C) Yeast display epitope residues (red) for the 1F1 MAb were mapped onto the pseudoatomic model of the mature DENV-2 virion (37). Virions are depicted as 2.0-Å-radius C-α atoms and are colored according to their E protein symmetry relationships, i.e., twofold (cyan), threefold (green), or fivefold (yellow) symmetry. (D to F) Structure of DENV DIII, with amino acid residues that significantly affect binding of type-specific neutralizing MAbs 3H5-1 (D), 6B6-10 (E), and 1F1 (F) marked in orange. The DIII disulfide bond is depicted in yellow.
TABLE 2.
Binding of MAbs to Raji-DC-SIGN-R cells infected with DENV-1, -2, -3, and -4
| Antibody | Neutralization strength | Specificity | Binding to virusa
|
|||
|---|---|---|---|---|---|---|
| DENV-1 16007 | DENV-2 16681 | DENV-3 16652 | DENV-4 H241 | |||
| 1F1 | Strong | Type | − | +++ | − | − |
| 3H5-1 | Strong | Type | − | +++ | − | − |
| 6B6-10 | Strong | Type | − | +++ | − | − |
| 9A3D-8 | Strong | Type | − | +++ | − | − |
| M8051122 | Moderate | Type | − | +++ | − | − |
| 9F16 | Strong | Type | − | +++ | − | − |
| 9F11 | Moderate | Type | − | +++ | − | − |
| 2Q1899 | Moderate | Type | − | +++ | − | − |
| 9D12 | Moderate | Subcomplex | + | +++ | − | +++ |
| 1A1D-2 | Strong | Subcomplex | +++ | +++ | +++ | − |
| 5A2-7 | Weak/none | Complex | +++ | +++ | +++ | +++ |
| 13D4-1 | Weak/none | Complex | +++ | +++ | +++ | +++ |
| E111 | Weak/none | Cross-reactive | +++ | +++ | +++ | +++ |
| E114 | Weak/none | Cross-reactive | +++ | +++ | +++ | +++ |
+++, strong binding (40 to 100% compared to control) to infected cells; +, weak binding (15 to 40% compared to control) to infected cells; −, no appreciable binding detected.
To map the amino acid contact residues of the DIII-specific DENV-2 MAbs with distinct neutralizing properties, we applied a high-throughput strategy that previously mapped 167 WNV E protein-specific MAbs (53, 54). Error-prone PCR mutagenesis introduced random point mutations within DIII of DENV-2 E. A library of ∼3.5 × 105 variants was pooled and used to create a mutant yeast expression library. Individual screens were performed to identify DIII mutants that lost binding selectively to strongly neutralizing MAbs. To eliminate mutants that abolished surface expression of DIII, yeast cells were stained sequentially with an Alexa Fluor 647-conjugated individual MAb and an Alexa Fluor 488-conjugated oligoclonal antibody derived from a pool of individual MAbs. After several rounds of cell sorting, yeast cells that selectively lost expression of an individual MAb epitope but retained expression of DIII on the surface were identified. Multiple independent yeast clones that selectively lost binding of individual MAbs were subjected to plasmid recovery and sequencing. Subsequently, mutant DIII expressed on the yeast surface was tested for MAb reactivity against the remainder of the panel of MAbs by flow cytometry (Fig. 1 and data not shown).
FIG. 1.
Flow cytometry histograms of loss-of-function DIII variants (G304Y, K307N, D329G, and P384N) selected by yeast surface display after being sorted with MAbs. Representative histograms are shown for MAbs 1F1, 3H5-1, and 13D4-1 with wild-type DIII (WT) and each of the DIII mutants. Data shown are representative of three independent experiments.
From each screen, we recovered 6 to 20 independent mutants that lost binding for an individual MAb. After sequencing them, we discovered that some of these mutants contained multiple mutations within the DIII region. In such cases, single mutations were engineered separately by site-directed mutagenesis to identify the change that caused the phenotype. Type-specific MAbs that localized to DIII and neutralized DENV-2 strongly (1F1, 3H5-1, 6B6-10, 9A3D-8, and 9F16) showed markedly reduced binding (>80% reduction) when residues G304 (five of five MAbs), E383 (three of five MAbs), and P384 (four of five MAbs) were altered (Fig. 1 and Table 3). Similarly, type-specific MAbs that neutralized moderately (PRNT50 of 2 to 5 μg/ml; M8051122, 9F11, and 2Q1899) also showed decreased binding when these three residues were changed. G304, E383, and P384 are located on adjacent loops and form a contiguous patch on the solvent-exposed surface at the lateral ridge of DIII (Fig. 2). As observed for DIII-specific neutralizing MAbs against WNV (53), the type-specific neutralizing DENV-2 MAbs bound overlapping epitopes. Consistent with this, the following additional point mutations caused significant loss of binding of individual type-specific neutralizing MAbs tested: T303Y (1F1), K307E (9A3D-8), E327R (9A3D-8), D329R (6B6-10), G330D (1F1 and 6B6-10), and S331Y (6B6-10). Mutation of G304, which appears to comprise part of a type-specific neutralizing epitope on DENV-2 DIII, also affected binding of both subcomplex-specific MAbs (9D12 and 1A1D-2) and one non- or weakly neutralizing MAb (E114). Because binding of several MAbs of different classes was affected by the G304 mutation, we cannot absolutely exclude perturbations of the structure of this mutant DIII. Against this, we did observe relatively wild-type levels of binding of two other nonneutralizing MAbs (13D4-1 and E111) to this mutant.
TABLE 3.
Summary of MAb binding to DENV-2 DIII mutants expressed on the surfaces of yeast cells
| MAb | MAb binding to mutanta
|
|||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T303Y | G304Y | K305E | K307N | K307Q | K307I | K307E | K310E | T315G | T315G and S331Y | H317Y | R323E | E327R | D329G | D329R | G330D | S331Y | E338D | T359I | E383G | P384A | P384N | N390H | N390Y | |
| Type-specific neutralizing MAbs | ||||||||||||||||||||||||
| 1F1 | <1 | <1 | 98 | 100 | 81 | 36 | 88 | 48 | 100 | 51 | 96 | 100 | 27 | 37 | 37 | 2 | 100 | 100 | 100 | 3 | <1 | <1 | 100 | 76 |
| 3H5-1 | 60 | 1 | 93 | 86 | 93 | 43 | 62 | 41 | 96 | 88 | 92 | 100 | 39 | 99 | 48 | 100 | 100 | 84 | 100 | 5 | <1 | <1 | 100 | 60 |
| 6B6-10 | 33 | 2 | 98 | 80 | 90 | 67 | 85 | 44 | 100 | <1 | 100 | 100 | 26 | <1 | 2 | <1 | <1 | 100 | 100 | 100 | 17 | 31 | 100 | 76 |
| 9A3D-8 | 88 | 19 | 76 | 89 | 67 | 29 | 5 | 63 | 83 | 67 | 100 | 100 | 17 | 100 | 100 | 87 | 100 | 100 | 100 | 87 | 59 | 60 | 100 | 75 |
| M8051122 | 68 | 1 | 67 | 92 | 90 | 67 | 48 | 17 | ND | ND | ND | 42 | 64 | 60 | 54 | 100 | 100 | 100 | 100 | 4 | 3 | 1 | 54 | 90 |
| 9F16 | 100 | 1 | 100 | 100 | 100 | 100 | 69 | 33 | ND | ND | ND | 59 | 83 | 100 | 80 | 100 | 89 | 100 | 100 | 1 | 2 | 2 | 78 | 59 |
| 9F11 | 89 | 2 | 100 | 89 | 100 | 60 | 60 | 23 | ND | ND | ND | 82 | 48 | 100 | 66 | 100 | 88 | 89 | 63 | <1 | 1 | <1 | 62 | 100 |
| 2Q1899 | 80.8 | 2 | 100 | 100 | 100 | 80 | 43 | 25 | 100 | 100 | 100 | 69 | 98 | 93 | 44 | 100 | 56 | 100 | 100 | 1 | 2 | <1 | 43 | 100 |
| Subcomplex-specific neutralizing MAbs | ||||||||||||||||||||||||
| 9D12 | 35 | 9 | 4 | 32 | 32 | 21 | 17 | <1 | 83 | 76 | 100 | 100 | 100 | 45 | 28 | 72 | 58 | 74 | 100 | 74 | 40 | 3 | 66 | 32 |
| 1A1D-2 | 100 | 4 | <1 | <1 | <1 | <1 | <1 | <1 | 67 | 73 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 90 | 68 | 47 | 100 | 74 |
| Nonneutralizing MAbs | ||||||||||||||||||||||||
| 5A2-7 | 100 | 40 | 100 | 100 | 98 | 62 | 100 | 27 | 29 | 5 | 17 | 100 | 60 | 66 | 86 | 100 | 89 | 78 | 100 | 38 | 100 | 83 | 84 | 89 |
| 13D4-1 | 74 | 100 | 94 | 99 | 59 | 51 | 100 | 44 | 100 | 3 | 2 | 100 | 67 | 93 | 87 | 100 | 93 | 100 | 8 | 69 | 100 | 76 | 90 | 90 |
| WN E111 | 100 | 92 | 61 | 93 | 100 | 70 | 86 | 35 | 100 | <1 | 14 | 61 | 76 | 17 | 85 | 100 | 100 | 74 | 100 | 100 | 93 | 74 | 100 | 86 |
| WN E114 | 100 | 2 | 60 | 78 | 61 | 48 | 45 | 4 | 100 | 100 | 100 | 72 | 37 | 47 | 46 | 100 | 91 | 42 | 100 | 90 | 49 | 48 | 75 | 56 |
Values shown were obtained by dividing the total fluorescence product (percent positive population × mean linear fluorescence intensity) for a mutant for each individual antibody by the total fluorescence product for wild-type DIII. Values in bold indicate reductions in MAb binding of >80% for a given mutation. The results are averages for three to five independent experiments for each mutant and each antibody. Since the G304Y mutant had slightly less surface expression on yeast cells, the values for this mutant only were normalized to the strongest binding antibody for that mutant × 100. ND, not determined.
In our panel of MAbs, we also identified two neutralizing MAbs (9D12 and 1A1D-2) that reacted with additional DENV serotypes. These subcomplex-specific MAbs showed moderate and strong inhibitory activities against DENV-2 infection, respectively (Tables 1 and 2). These MAbs, however, bound DIII somewhat distinctly. 1A1D-2, which binds DENV-1, DENV-2, and DENV-3, retained relatively normal binding with E383G and P384N mutations but showed markedly reduced binding with mutations in residues K305, K307, and K310. In comparison, 9D12, which binds DENV-2 and DENV-4 and weakly binds DENV-1, retained binding with the E383G and P384A mutations but had markedly reduced binding with K305E, K307E, K310E, and P384N mutations (Table 3).
Also in our panel were four MAbs (5A2-7, 13D4-1, E111, and E114) that had little or no neutralizing activity (Table 1). This group of MAbs recognized all four serotypes of DENV (Table 2). Two of them, E111 and E114, were cross-reactive and also recognized yeast expressing WNV DIII (data not shown). Independent yeast sorting for loss-of-binding mutations was performed with three of the four MAbs. Several mutations that specifically reduced binding of the poorly neutralizing MAbs but had little effect on most strongly neutralizing MAbs were identified. For example, H317Y mutation specifically reduced binding of the 5A2-7, 13D4-1, and E111 MAbs (Table 3), and a DIII variant with two mutations (T315G and S331Y) also diminished binding of these MAbs but did not affect any neutralizing MAbs, with the exception of 6B6-10. Interestingly, the T315G single mutant only modestly decreased 5A2-7 (71% reduction) binding, and the S331Y single mutant had no effect on binding of 5A2-7, 13D4-1, or E111.
To visualize spatially the different recognition patterns of MAbs that strongly, moderately, and weakly neutralized DENV-2 infection, we docked the loss-of-binding mutations (<20% of wild-type binding) defined by the yeast assay onto the existing crystallographic structure of DENV-2 DIII, the prefusion DENV-2 E protein dimer structure (46), and the pseudoatomic model of the mature DENV-2 virion (37). The DIII structures were compared to the previously defined crystallographic epitope (16 contact residues) of E16 on the lateral ridge of DIII of WNV (51) (Fig. 2A and B). Type-specific MAbs with the strongest neutralizing activities (3H5-1, 6B6-10, and 1F1) localized to amino acids on the analogous lateral ridge of DIII (Fig. 2D, E, and F). Nonetheless, the yeast mapping did suggest some subtle differences. Whereas the lateral ridge epitope of the strongly neutralizing anti-WNV MAb E16 was centered on the N-terminal region and the BC loop (K307, T330, and T332 of WNV), strongly neutralizing type-specific anti-DENV-2 MAbs localized more to the FG loop (E383 and P384 of DENV-2). The E383 and P384 residues in the FG loop are highly conserved among DENV-2 isolates but are not present in DENV-1, DENV-3, or DENV-4 strains (Fig. 3). The epitope of type-specific neutralizing MAbs against DENV-2 followed the same exposure pattern on the virion as that previously identified for E16 (34, 51): two of the three DIIIs per icosahedral asymmetric unit were exposed, with steric hindrance noted at the fivefold clustered DIII (Fig. 2C).
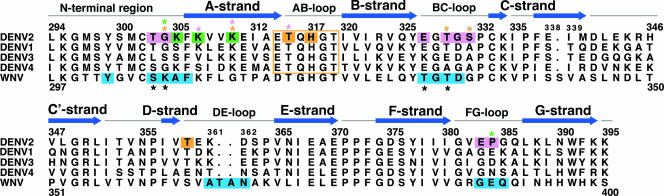
FIG. 3.
DIII amino acid sequence alignment. The sequence and secondary structure of DIII from the DENV-2 (strain 16681) E protein are aligned with those for DENV-1 (strain 16007), DENV-3 (strain 16652), DENV-4 (strain 1036), and WNV (New York 1999). The secondary structure of DENV-2 E DIII residues 294 to 395 (RCSB entry 1OAN) was predicted by DSSP (33). The results of yeast surface display epitope mapping are highlighted, with DENV-2 residues recognized primarily by type-specific MAbs colored magenta, subcomplex-specific residues colored green, and cross-reactive residues colored orange. The residues contacted by E16 on WNV DIII, as determined by crystallography, are colored blue (51), with black asterisks denoting residues identified by yeast display (53). Colored asterisks denote DENV-2 residues that are recognized by multiple classes of antibodies. For example, G304Y mutation resulted in a loss of binding of all type- and subcomplex-specific MAbs and a single cross-reactive MAb.
Subcomplex-specific MAbs (1A1D-2 and 9D12) that strongly and moderately neutralized DENV-2 infection recognized a flanking epitope (Fig. 4A and B). This epitope was centered on amino acids K305, K307, and K310 on the A strand. Consistent with the limited cross-reactivity of these subcomplex-specific MAbs, K310 is completely conserved among all four DENV serotypes, whereas K305 and K307 are conserved in DENV-4 and DENV-1 strains, respectively (Fig. 3). Although the 1A1D-2 and 9D12 MAb epitopes are reasonably exposed in all three symmetry environments on the mature virion (Fig. 4C and data not shown), some differences with the type-specific lateral ridge epitope were apparent, as follows: the 9D12 epitope appears predominantly exposed on the fivefold clustered DIII, and thus, one could speculate that a full complement of 180 Fabs could bind DENV-2 at saturation.
FIG. 4.
DIII A-strand epitope. The structure of DENV-2 DIII is shown, with identification of binding sites of subcomplex-specific neutralizing antibodies. (A and B) Structure of DENV DIII, with amino acid residues that significantly affect binding of subcomplex-specific neutralizing MAbs 1A1D-2 (A) and 9D12 (B) marked in orange. (C) Yeast display epitope residues (red) for the 9D12 MAb were mapped onto the pseudoatomic model of the mature DENV-2 virion. Virions are depicted as described in the legend to Fig. 2.
Several of the poorly neutralizing, cross-reactive MAbs (5A2-7, 13D4-1, and E111) mapped to an amino acid residue (e.g., H317) that was localized to the back side of DIII in the AB loop (Fig. 5A and B). Consistent with the cross-reactive nature of these MAbs, the entire AB loop sequence (E314, T315, Q316, H317, G318, and T319) is completely conserved among all four DENV serotypes. Moreover, the H317, G318, and T319 residues are present in virtually all WNV isolates and the H317 and T319 amino acids are conserved among flaviviruses (Fig. 3 and data not shown). Structurally, the AB loop has limited exposure on the surface of the E protein dimer and faces inward toward the nucleocapsid in all three symmetry environments of the mature virion (Fig. 5C and Fig. 6).
FIG. 5.
DIII AB loop epitope. The structure of DENV-2 DIII is shown, with identification of binding sites of poorly neutralizing antibodies. (A and B) Structure of DENV DIII, with amino acid residues that significantly affect binding of the poorly neutralizing MAbs E111 (A) and 13D4-1 (B) marked in orange. (C) Yeast display epitope residues (red) for the 13D4-1 MAb were mapped onto the pseudoatomic model of the mature DENV-2 virion. Virions are depicted as described in the legend to Fig. 2. Note that the 13D4-1 epitope is poorly accessible on the virion compared to the 1F1 (Fig. 2C) and 9D12 (Fig. 4C) epitopes.
FIG. 6.
Mapping of MAb epitopes onto the DENV-2 E protein dimer. The yeast display epitope residues (in red) for the type-specific 1F1 (A), subcomplex-specific 9D12 (B), and cross-reactive 13D4-1 (C) MAbs were rendered on the crystal structure of the DENV-2 E protein dimer (46) and are shown in side view, with the bottom side facing the viral lipid membrane.
DISCUSSION
The goal of this study was to characterize epitopes on DIII recognized by potent neutralizing antibodies against DENV-2. Previous studies had established a dominant type-specific epitope for eliciting protective antibodies in vitro and in vivo against the related flavivirus WNV. Here we tested a panel of MAbs against DIII of the DENV-2 E protein. Type-specific strongly neutralizing MAbs mapped to an analogous epitope centered on the FG loop of the lateral ridge region on DIII of DENV-2. Subcomplex-specific strongly neutralizing MAbs localized to a flanking epitope that was centered on three lysine residues in the A strand of DIII.
Extensive MAb competition binding studies have been performed by several groups to identify distinct antigenic and functional determinants on DENV-2 (18, 22, 29, 30, 61). Potently inhibitory type-specific MAbs were localized to domain B, which is now called DIII, based on structural analysis of the domain organization of flavivirus E proteins (46, 60). Nonetheless, amino acid contact residues of few neutralizing MAbs that react with DENV-2 have been established. For these (3H5-1, 4E11, G8D11, and 4G2), precise mapping data were obtained by analyzing neutralization escape mutants (39) and by differential recognition of chimeric DENV variants (32), site-specific DENV-2 mutants (17, 67), and E protein peptide sequences (44, 70, 72). We used a forward genetic strategy, error-prone PCR mutagenesis of DIII of DENV-2 E protein, and expression on yeast cells to map antibody contact residues in a nonbiased manner. By having a panel of DIII MAbs with differing neutralization potentials, we minimized the possibility that mutations would grossly affect folding. The validity of the yeast approach for identifying critical contact residues was confirmed by X-ray crystallographic studies that resolved the structural interface between DIII and a neutralizing anti-WNV Fab fragment (51). Of the DENV-2 MAbs that have been reported to contact specific amino acid residues in DIII, only the type-specific MAb 3H5-1 was available for our analysis. Prior fine mapping studies suggested that 3H5-1 recognized either a Glu-Pro-Gly motif centered at amino acids 383, 384, and 385 (32) or a linear peptide encompassing amino acids 386 to 397 (32, 72). Our yeast mapping experiments confirmed an essential role for residues E383 and P384 but also suggested an additional important contact residue (G304) located on an adjacent strand.
DIII of the E protein adopts an immunoglobulin-like fold (46, 60) that is significantly exposed on the surface of the mature virion (50, 78). The lateral ridge epitope on DIII was previously defined by X-ray crystallographic and nuclear magnetic resonance studies of Fab-DIII complexes of WNV and Japanese encephalitis virus and encompasses four discontinuous loops (51, 74, 75). In our study, we found six different strongly neutralizing MAbs against DENV-2 that localized to two overlapping structural epitopes on the lateral ridge and adjacent A strand of DIII.
For flaviviruses, virus type-specific epitopes generally elicit the most potent neutralizing antibodies (2, 28, 42, 53, 56, 63, 64). Of the DIII-specific MAbs against DENV-2 in our panel, in general, the ones with the strongest neutralizing activities were type specific and localized to the lateral ridge of DIII centered at the FG loop, near residues E383 and P384. Nonetheless, some of the type-specific neutralizing MAbs inhibited virus infectivity less strongly, although they recognized similar residues. Although further biophysical studies are needed, the affinities of binding of MAbs for a given DENV-2 DIII epitope may correlate with relative occupancy and may predict the strength of neutralization. Such a result was observed with less strongly neutralizing MAbs that recognized the lateral ridge epitope of DIII of WNV (57).
The two subcomplex-specific (1A1D-2 [PRNT50, 0.3 μg/ml] and 9D12 [PRNT50, 2 μg/ml]) MAbs that we tested had strong and moderate neutralizing activities, respectively, consistent with prior studies with the group-specific MAb 4E11 (PRNT50 values of 0.3 to 2.4 μg/ml for DENV-1 to DENV-4) (71). 1A1D-2 and 9D12 bind a more conserved epitope among DENV serotypes that partially overlaps with the type-specific lateral ridge neutralizing epitope but is centered on the A strand at residues K305, K307, and K310. Notably, several of these amino acids were recently identified as contact residues for the MAb 4E11, which neutralizes all four DENV serotypes (41, 70). Such broadly neutralizing subcomplex- or group-specific MAbs may have potential for development as antibody-based therapeutics against all serotypes of DENV.
An epitope map analysis of the different DENV-2 MAbs at the amino acid sequence level begins to explain their serotype-specific properties. Type-specific neutralizing MAbs against DENV-2 are centered on amino acids E383 and P384 in the FG loop, which are conserved among DENV-2 isolates but divergent in all other DENV serotypes. Subcomplex-specific neutralizing MAbs that bind some but not all DENV serotypes recognize a distinct epitope centered on three A-strand lysines of DIII. K310 is completely conserved among all four DENV serotypes, whereas K305 and K307 are conserved in DENV-4 and DENV-1 strains, respectively. The distinct binding specificities of 9D12 and 1A1D-2 for other DENV serotypes likely reflect differences in interactions with specific lysines on the A strand and the divergence of other additional contact residues in the FG loop and the G strand. Cross-reactive poorly neutralizing MAbs localized to the AB loop on DIII. Their lack of neutralizing activity is probably explained by the limited surface exposure of the AB loop. As seen for the DIII-specific poorly neutralizing anti-WNV MAb E9, a relative lack of accessibility directly affects the stoichiometry of binding, such that a threshold for antibody neutralization is rarely reached (57). The cross-reactivity of these AB loop MAbs with all DENV serotypes and distantly related flaviviruses is reasonably explained by sequence conservation: amino acids 314 to 319 are completely conserved among all four DENV serotypes, and residues 317 and 319 (which include the critical H317 amino acid) are conserved in virtually all flaviviruses. This analysis has implications for the development of novel DIII epitope-based diagnostic reagents for polyclonal antibodies, as mutation of AB loop sequences could abolish serotype and flavivirus nonneutralizing cross-reactive epitopes, making DIII-based immunologic assays more predictive of serotype-specific neutralizing antibodies.
Although the sequence of the lateral ridge epitope of DIII is variable among flaviviruses, the majority of mutations that abolish binding of virus-specific strongly neutralizing antibodies map there (2, 10, 12, 18, 53, 61, 63, 64, 67, 75), suggesting the existence of a type-specific neutralizing epitope for flaviviruses on DIII. It is important, however, that these epitope maps were derived using murine MAbs. Whether the human antibody response recognizes the same or different epitopes has not yet been determined in detail. Only 8% (4 of 51 antibodies) and 0% (0 of 11 antibodies) of human single-chain antibodies that were isolated from phage display libraries of WNV-infected or naïve patients reacted with DIII (26, 69). These results suggest that the human antibody repertoire against flaviviruses may actually be directed away from these DIII neutralizing epitopes and toward the inherently less neutralizing immunodominant epitopes on DI and DII (55). Since our results suggest that strongly neutralizing antibodies against DIII of DENV-2 map to the lateral ridge and A-strand epitopes, flavivirus DIII-based vaccines (13, 15, 31) that intentionally skew the humoral response to these epitopes and away from the cross-reactive, poorly accessible epitope in the AB loop could elicit a greater protective response.
Acknowledgments
We thank C. Nelson for help with structural analysis of DENV E proteins, members of our laboratories for critical reviews of the manuscript, and R. Putnak for providing several of the MAbs used in this study.
This work was supported by the Pediatric Dengue Vaccine Initiative (M.S.D., D.H.F., J.T.R., J.J.S., and A.D.B.) and by NIH grants AI061373 (M.S.D.) and U54 AI057160 (Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research).
Footnotes
Published ahead of print on 19 September 2007.
REFERENCES
- 1.Beasley, D. W., and J. G. Aaskov. 2001. Epitopes on the dengue 1 virus envelope protein recognized by neutralizing IgM monoclonal antibodies. Virology 279:447-458. [DOI] [PubMed] [Google Scholar]
- 2.Beasley, D. W., and A. D. Barrett. 2002. Identification of neutralizing epitopes within structural domain III of the West Nile virus envelope protein. J. Virol. 76:13097-13100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhardwaj, S., M. Holbrook, R. E. Shope, A. D. Barrett, and S. J. Watowich. 2001. Biophysical characterization and vector-specific antagonist activity of domain III of the tick-borne flavivirus envelope protein. J. Virol. 75:4002-4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boder, E. T., and K. D. Wittrup. 1998. Optimal screening of surface-displayed polypeptide libraries. Biotechnol. Prog. 14:55-62. [DOI] [PubMed] [Google Scholar]
- 5.Boder, E. T., and K. D. Wittrup. 1997. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15:553-557. [DOI] [PubMed] [Google Scholar]
- 6.Brandriss, M. W., J. J. Schlesinger, E. E. Walsh, and M. Briselli. 1986. Lethal 17D yellow fever encephalitis in mice. I. Passive protection by monoclonal antibodies to the envelope proteins of 17D yellow fever and dengue 2 viruses. J. Gen. Virol. 67:229-234. [DOI] [PubMed] [Google Scholar]
- 7.Bressanelli, S., K. Stiasny, S. L. Allison, E. A. Stura, S. Duquerroy, J. Lescar, F. X. Heinz, and F. A. Rey. 2004. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 23:728-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burke, D. S., and T. P. Monath. 2001. Flaviviruses, p. 1043-1125. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 9.Carson, M. 1987. Ribbon models of macromolecules. J. Mol. Graphics 5:103-106. [Google Scholar]
- 10.Cecilia, D., and E. A. Gould. 1991. Nucleotide changes responsible for loss of neuroinvasiveness in Japanese encephalitis virus neutralization-resistant mutants. Virology 181:70-77. [DOI] [PubMed] [Google Scholar]
- 11.Chambers, T. J., C. S. Hahn, R. Galler, and C. M. Rice. 1990. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 44:649-688. [DOI] [PubMed] [Google Scholar]
- 12.Chambers, T. J., M. Halevy, A. Nestorowicz, C. M. Rice, and S. Lustig. 1998. West Nile virus envelope proteins: nucleotide sequence analysis of strains differing in mouse neuroinvasiveness. J. Gen. Virol. 79:2375-2380. [DOI] [PubMed] [Google Scholar]
- 13.Chen, S., M. Yu, T. Jiang, Y. Deng, C. Qin, and E. Qin. 2007. Induction of tetravalent protective immunity against four dengue serotypes by the tandem domain III of the envelope protein. DNA Cell Biol. 26:361-367. [DOI] [PubMed] [Google Scholar]
- 14.Choi, K. S., J. J. Nah, Y. J. Ko, Y. J. Kim, and Y. S. Joo. 2007. The DE loop of the domain III of the envelope protein appears to be associated with West Nile virus neutralization. Virus Res. 123:216-218. [DOI] [PubMed] [Google Scholar]
- 15.Chu, J. H., C. C. Chiang, and M. L. Ng. 2007. Immunization of flavivirus West Nile recombinant envelope domain III protein induced specific immune response and protection against West Nile virus infection. J. Immunol. 178:2699-2705. [DOI] [PubMed] [Google Scholar]
- 16.Colombage, G., R. Hall, M. Pavy, and M. Lobigs. 1998. DNA-based and alphavirus-vectored immunisation with prM and E proteins elicits long-lived and protective immunity against the flavivirus, Murray Valley encephalitis virus. Virology 250:151-163. [DOI] [PubMed] [Google Scholar]
- 17.Crill, W. D., and G. J. Chang. 2004. Localization and characterization of flavivirus envelope glycoprotein cross-reactive epitopes. J. Virol. 78:13975-13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crill, W. D., and J. T. Roehrig. 2001. Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to Vero cells. J. Virol. 75:7769-7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diamond, M. S., D. Edgil, T. G. Roberts, B. Lu, and E. Harris. 2000. Infection of human cells by dengue virus is modulated by different cell types and viral strains. J. Virol. 74:7814-7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diamond, M. S., E. Sitati, L. Friend, B. Shrestha, S. Higgs, and M. Engle. 2003. Induced IgM protects against lethal West Nile virus infection. J. Exp. Med. 198:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falconar, A. K. 1999. Identification of an epitope on the dengue virus membrane (M) protein defined by cross-protective monoclonal antibodies: design of an improved epitope sequence based on common determinants present in both envelope (E and M) proteins. Arch. Virol. 144:2313-2330. [DOI] [PubMed] [Google Scholar]
- 22.Gentry, M. K., E. A. Henchal, J. M. McCown, W. E. Brandt, and J. M. Dalrymple. 1982. Identification of distinct antigenic determinants on dengue-2 virus using monoclonal antibodies. Am. J. Trop. Med. Hyg. 31:548-555. [DOI] [PubMed] [Google Scholar]
- 23.Goncalvez, A. P., R. Men, C. Wernly, R. H. Purcell, and C. J. Lai. 2004. Chimpanzee Fab fragments and a derived humanized immunoglobulin G1 antibody that efficiently cross-neutralize dengue type 1 and type 2 viruses. J. Virol. 78:12910-12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goncalvez, A. P., R. H. Purcell, and C. J. Lai. 2004. Epitope determinants of a chimpanzee Fab antibody that efficiently cross-neutralizes dengue type 1 and type 2 viruses map to inside and in close proximity to fusion loop of the dengue type 2 virus envelope glycoprotein. J. Virol. 78:12919-12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gould, E. A., A. Buckley, A. D. Barrett, and N. Cammack. 1986. Neutralizing (54K) and non-neutralizing (54K and 48K) monoclonal antibodies against structural and non-structural yellow fever virus proteins confer immunity in mice. J. Gen. Virol. 67:591-595. [DOI] [PubMed] [Google Scholar]
- 26.Gould, L. H., J. Sui, H. Foellmer, T. Oliphant, T. Wang, M. Ledizet, A. Murakami, K. Noonan, C. Lambeth, K. Kar, J. F. Anderson, A. M. de Silva, M. S. Diamond, R. A. Koski, W. A. Marasco, and E. Fikrig. 2005. Protective and therapeutic capacity of human single-chain Fv-Fc fusion proteins against West Nile virus. J. Virol. 79:14606-14613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halstead, S. B. 1988. Pathogenesis of dengue: challenges to molecular biology. Science 239:476-481. [DOI] [PubMed] [Google Scholar]
- 28.Heinz, F. X. 1986. Epitope mapping of flavivirus glycoproteins. Adv. Virus Res. 31:103-168. [DOI] [PubMed] [Google Scholar]
- 29.Henchal, E. A., M. K. Gentry, J. M. McCown, and W. E. Brandt. 1982. Dengue virus-specific and flavivirus group determinants identified with monoclonal antibodies by indirect immunofluorescence. Am. J. Trop. Med. Hyg. 31:830-836. [DOI] [PubMed] [Google Scholar]
- 30.Henchal, E. A., J. M. McCown, D. S. Burke, M. C. Seguin, and W. E. Brandt. 1985. Epitopic analysis of antigenic determinants on the surface of dengue-2 virions using monoclonal antibodies. Am. J. Trop. Med. Hyg. 34:162-169. [DOI] [PubMed] [Google Scholar]
- 31.Hermida, L., L. Bernardo, J. Martin, M. Alvarez, I. Prado, C. Lopez, L. Sierra Bde, R. Martinez, R. Rodriguez, A. Zulueta, A. B. Perez, L. Lazo, D. Rosario, G. Guillen, and M. G. Guzman. 2006. A recombinant fusion protein containing the domain III of the dengue-2 envelope protein is immunogenic and protective in nonhuman primates. Vaccine 24:3165-3171. [DOI] [PubMed] [Google Scholar]
- 32.Hiramatsu, K., M. Tadano, R. Men, and C. J. Lai. 1996. Mutational analysis of a neutralization epitope on the dengue type 2 virus (DEN2) envelope protein: monoclonal antibody resistant DEN2/DEN4 chimeras exhibit reduced mouse neurovirulence. Virology 224:437-445. [DOI] [PubMed] [Google Scholar]
- 33.Kabsch, W., and C. Sander. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577-2637. [DOI] [PubMed] [Google Scholar]
- 34.Kauffman, B., G. Nybakken, P. R. Chipman, W. Zhang, D. H. Fremont, M. S. Diamond, R. J. Kuhn, and M. G. Rossmann. 2006. West Nile virus in complex with a neutralizing monoclonal antibody. Proc. Natl. Acad. Sci. USA 103:12400-12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kinney, R. M., S. Butrapet, G. J. Chang, K. R. Tsuchiya, J. T. Roehrig, N. Bhamarapravati, and D. J. Gubler. 1997. Construction of infectious cDNA clones for dengue 2 virus: strain 16681 and its attenuated vaccine derivative, strain PDK-53. Virology 230:300-308. [DOI] [PubMed] [Google Scholar]
- 36.Kleywegt, G. J. 1997. Validation of protein models from Calpha coordinates alone. J. Mol. Biol. 273:371-376. [DOI] [PubMed] [Google Scholar]
- 37.Kuhn, R. J., W. Zhang, M. G. Rossmann, S. V. Pletnev, J. Corver, E. Lenches, C. T. Jones, S. Mukhopadhyay, P. R. Chipman, E. G. Strauss, T. S. Baker, and J. H. Strauss. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, L., A. D. Barrett, and D. W. Beasley. 2005. Differential expression of domain III neutralizing epitopes on the envelope proteins of West Nile virus strains. Virology 335:99-105. [DOI] [PubMed] [Google Scholar]
- 39.Lin, B., C. R. Parrish, J. M. Murray, and P. J. Wright. 1994. Localization of a neutralizing epitope on the envelope protein of dengue virus type 2. Virology 202:885-890. [DOI] [PubMed] [Google Scholar]
- 40.Lin, C. W., and S. C. Wu. 2003. A functional epitope determinant on domain III of the Japanese encephalitis virus envelope protein interacted with neutralizing-antibody combining sites. J. Virol. 77:2600-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lisova, O., F. Hardy, V. Petit, and H. Bedouelle. 2007. Mapping to completeness and transplantation of a group-specific, discontinuous, neutralizing epitope in the envelope protein of dengue virus. J. Gen. Virol. 88:2387-2397. [DOI] [PubMed] [Google Scholar]
- 42.Mandl, C. W., F. Guirakhoo, H. Holzmann, F. X. Heinz, and C. Kunz. 1989. Antigenic structure of the flavivirus envelope protein E at the molecular level, using tick-borne encephalitis virus as a model. J. Virol. 63:564-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42a.Mason, P. W., M. U. Zügel, A. R. Semproni, M. J. Fournier, and T. L. Mason. 1990. The antigenic structure of dengue type 1 virus envelope and NS1 proteins expressed in Escherichia coli. J. Gen. Virol. 71:2107-2114. [DOI] [PubMed] [Google Scholar]
- 43.Mathews, J. H., and J. T. Roehrig. 1984. Elucidation of the topography and determination of the protective epitopes on the E glycoprotein of Saint Louis encephalitis virus by passive transfer with monoclonal antibodies. J. Immunol. 132:1533-1537. [PubMed] [Google Scholar]
- 44.Megret, F., J. P. Hugnot, A. Falconar, M. K. Gentry, D. M. Morens, J. M. Murray, J. J. Schlesinger, P. J. Wright, P. Young, M. H. Van Regenmortel, et al. 1992. Use of recombinant fusion proteins and monoclonal antibodies to define linear and discontinuous antigenic sites on the dengue virus envelope glycoprotein. Virology 187:480-491. [DOI] [PubMed] [Google Scholar]
- 45.Misulovin, Z., X. W. Yang, W. Yu, N. Heintz, and E. Meffre. 2001. A rapid method for targeted modification and screening of recombinant bacterial artificial chromosome. J. Immunol. Methods 257:99-105. [DOI] [PubMed] [Google Scholar]
- 46.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2003. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. USA 100:6986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature 427:313-319. [DOI] [PubMed] [Google Scholar]
- 48.Monath, T. P. 1994. Dengue: the risk to developed and developing countries. Proc. Natl. Acad. Sci. USA 91:2395-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morita, K., M. Tadano, S. Nakaji, K. Kosai, E. G. Mathenge, B. D. Pandey, F. Hasebe, S. Inoue, and A. Igarashi. 2001. Locus of a virus neutralization epitope on the Japanese encephalitis virus envelope protein determined by use of long PCR-based region-specific random mutagenesis. Virology 287:417-426. [DOI] [PubMed] [Google Scholar]
- 50.Mukhopadhyay, S., B. S. Kim, P. R. Chipman, M. G. Rossmann, and R. J. Kuhn. 2003. Structure of West Nile virus. Science 302:248. [DOI] [PubMed] [Google Scholar]
- 51.Nybakken, G., T. Oliphant, S. Johnson, S. Burke, M. S. Diamond, and D. H. Fremont. 2005. Structural basis for neutralization of a therapeutic antibody against West Nile virus. Nature 437:764-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nybakken, G. E., C. A. Nelson, B. R. Chen, M. S. Diamond, and D. H. Fremont. 2006. Crystal structure of the West Nile virus envelope glycoprotein. J. Virol. 80:11467-11474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oliphant, T., M. Engle, G. Nybakken, C. Doane, S. Johnson, L. Huang, S. Gorlatov, E. Mehlhop, A. Marri, K. M. Chung, G. D. Ebel, L. D. Kramer, D. H. Fremont, and M. S. Diamond. 2005. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat. Med. 11:522-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oliphant, T., G. Nybakken, M. Engle, Q. Xu, C. A. Nelson, S. Sukupolvi-Petty, A. Marri, B. Lachmi, U. Olshevsky, D. H. Fremont, T. C. Pierson, and M. S. Diamond. 2006. Determinants of West Nile virus envelope protein domain I and II antibody recognition and neutralization. J. Virol. 80:12149-12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oliphant, T., G. E. Nybakken, S. K. Austin, Q. Xu, J. Bramson, M. Loeb, M. Throsby, D. H. Fremont, T. C. Pierson, and M. S. Diamond. 2007. Induction of epitope-specific neutralizing antibodies against West Nile virus. J. Virol. 81:11828-11839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peiris, J. S. M., J. S. Porterfield, and J. T. Roehrig. 1982. Monoclonal antibodies against the flavivirus West Nile. J. Gen. Virol. 58:283-289. [DOI] [PubMed] [Google Scholar]
- 57.Pierson, T. C., Q. Xu, S. Nelson, T. Oliphant, G. E. Nybakken, D. H. Fremont, and M. S. Diamond. 2007. The stoichiometry of antibody-mediated neutralization and enhancement of West Nile virus infection. Cell Host Microbe 1:135-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pincus, S., P. W. Mason, E. Konishi, B. A. Fonseca, R. E. Shope, C. M. Rice, and E. Paoletti. 1992. Recombinant vaccinia virus producing the prM and E proteins of yellow fever virus protects mice from lethal yellow fever encephalitis. Virology 187:290-297. [DOI] [PubMed] [Google Scholar]
- 59.Rey, F. A. 2003. Dengue virus envelope glycoprotein structure: new insight into its interactions during viral entry. Proc. Natl. Acad. Sci. USA 100:6899-6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 angstrom resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 61.Roehrig, J. T., R. A. Bolin, and R. G. Kelly. 1998. Monoclonal antibody mapping of the envelope glycoprotein of the dengue 2 virus, Jamaica. Virology 246:317-328. [DOI] [PubMed] [Google Scholar]
- 62.Roehrig, J. T., J. H. Mathews, and D. W. Trent. 1983. Identification of epitopes on the E glycoprotein of Saint Louis encephalitis virus using monoclonal antibodies. Virology 128:118-126. [DOI] [PubMed] [Google Scholar]
- 63.Roehrig, J. T., L. A. Staudinger, A. R. Hunt, J. H. Mathews, and C. D. Blair. 2001. Antibody prophylaxis and therapy for flaviviral encephalitis infections. Ann. N. Y. Acad. Sci. 951:286-297. [DOI] [PubMed] [Google Scholar]
- 64.Sanchez, M. D., T. C. Pierson, D. McAllister, S. L. Hanna, B. A. Puffer, L. E. Valentine, M. M. Murtadha, J. A. Hoxie, and R. W. Doms. 2005. Characterization of neutralizing antibodies to West Nile virus. Virology 336:70-82. [DOI] [PubMed] [Google Scholar]
- 65.Schlesinger, J. J., S. Chapman, A. Nestorowicz, C. M. Rice, T. E. Ginocchio, and T. J. Chambers. 1996. Replication of yellow fever virus in the mouse central nervous system: comparison of neuroadapted and non-neuroadapted virus and partial sequence analysis of the neuroadapted strain. J. Gen. Virol. 77:1277-1285. [DOI] [PubMed] [Google Scholar]
- 66.Seif, S. A., K. Morita, S. Matsuo, F. Hasebe, and A. Igarashi. 1995. Finer mapping of neutralizing epitope(s) on the C-terminal of Japanese encephalitis virus E-protein expressed in recombinant Escherichia coli system. Vaccine 13:1515-1521. [DOI] [PubMed] [Google Scholar]
- 67.Serafin, I. L., and J. G. Aaskov. 2001. Identification of epitopes on the envelope (E) protein of dengue 2 and dengue 3 viruses using monoclonal antibodies. Arch. Virol. 146:2469-2479. [DOI] [PubMed] [Google Scholar]
- 68.Stiasny, K., S. Kiermayr, H. Holzmann, and F. X. Heinz. 2006. Cryptic properties of a cluster of dominant flavivirus cross-reactive antigenic sites. J. Virol. 80:9557-9568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Throsby, M., C. Geuijen, J. Goudsmit, A. Q. Bakker, J. Korimbocus, R. A. Kramer, M. Clijsters-van der Horst, M. de Jong, M. Jongeneelen, S. Thijsse, R. Smit, T. J. Visser, N. Bijl, W. E. Marissen, M. Loeb, D. J. Kelvin, W. Preiser, J. ter Meulen, and J. de Kruif. 2006. Isolation and characterization of human monoclonal antibodies from individuals infected with West Nile virus. J. Virol. 80:6982-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thullier, P., C. Demangel, H. Bedouelle, F. Megret, A. Jouan, V. Deubel, J. C. Mazie, and P. Lafaye. 2001. Mapping of a dengue virus neutralizing epitope critical for the infectivity of all serotypes: insight into the neutralization mechanism. J. Gen. Virol. 82:1885-1892. [DOI] [PubMed] [Google Scholar]
- 71.Thullier, P., P. Lafaye, F. Megret, V. Deubel, A. Jouan, and J. C. Mazie. 1999. A recombinant Fab neutralizes dengue virus in vitro. J. Biotechnol. 69:183-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Trirawatanapong, T., B. Chandran, R. Putnak, and R. Padmanabhan. 1992. Mapping of a region of dengue virus type-2 glycoprotein required for binding by a neutralizing monoclonal antibody. Gene 116:139-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vazquez, S., M. G. Guzman, G. Guillen, G. Chinea, A. B. Perez, M. Pupo, R. Rodriguez, O. Reyes, H. E. Garay, I. Delgado, G. Garcia, and M. Alvarez. 2002. Immune response to synthetic peptides of dengue prM protein. Vaccine 20:1823-1830. [DOI] [PubMed] [Google Scholar]
- 74.Volk, D. E., D. W. Beasley, D. A. Kallick, M. R. Holbrook, A. D. Barrett, and D. G. Gorenstein. 2004. Solution structure and antibody binding studies of the envelope protein domain III from the New York strain of West Nile virus. J. Biol. Chem. 279:38755-38761. [DOI] [PubMed] [Google Scholar]
- 75.Wu, K. P., C. W. Wu, Y. P. Tsao, T. W. Kuo, Y. C. Lou, C. W. Lin, S. C. Wu, and J. W. Cheng. 2003. Structural basis of a flavivirus recognized by its neutralizing antibody: solution structure of the domain III of the Japanese encephalitis virus envelope protein. J. Biol. Chem. 278:46007-46013. [DOI] [PubMed] [Google Scholar]
- 76.Wu, S. C., W. C. Lian, L. C. Hsu, and M. Y. Liau. 1997. Japanese encephalitis virus antigenic variants with characteristic differences in neutralization resistance and mouse virulence. Virus Res. 51:173-181. [DOI] [PubMed] [Google Scholar]
- 77.Yu, S., A. Wuu, R. Basu, M. R. Holbrook, A. D. Barrett, and J. C. Lee. 2004. Solution structure and structural dynamics of envelope protein domain III of mosquito- and tick-borne flaviviruses. Biochemistry 43:9168-9176. [DOI] [PubMed] [Google Scholar]
- 78.Zhang, W., P. R. Chipman, J. Corver, P. R. Johnson, Y. Zhang, S. Mukhopadhyay, T. S. Baker, J. H. Strauss, M. G. Rossmann, and R. J. Kuhn. 2003. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat. Struct. Biol. 10:907-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang, Y., J. Corver, P. R. Chipman, W. Zhang, S. V. Pletnev, D. Sedlak, T. S. Baker, J. H. Strauss, R. J. Kuhn, and M. G. Rossmann. 2003. Structures of immature flavivirus particles. EMBO J. 22:2604-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]