Abstract
Whole genome phylogenetic analysis in this study resolved a total of five major genotypes among the 22 varicella-zoster virus (VZV) strains or isolates for which complete genomic sequences are available. Consistent with earlier publications we have designated these genotypes European 1 (E1), European 2 (E2), Japanese (J), mosaic 1 (M1), and mosaic 2 (M2). Single nucleotide polymorphism (SNP) analysis performed in a whole-genome alignment revealed that VZV isolates of all five genotypes can be accurately genotyped using SNPs from two amplicons: open reading frame 22 (ORF22) and either ORF21 or ORF50. This modified approach identifies all of the genotypes observed using any of the published genotyping protocols. Of 165 clinical varicella and zoster isolates from Australia and New Zealand typed using this approach, 67 of 127 eastern Australian isolates were E1, 30 were E2, 16 were J, 10 were M1, and 4 were M2; 25 of 38 New Zealand isolates were E1, 8 were E2, and 5 were M1. VZV strain diversity in eastern Australia is thus broader than has been described for any other region, including Europe, Africa, and North America. J strains were far more prevalent than previously observed in countries other than Japan. Two-amplicon typing was in complete accord with genotypes derived using SNP in multiple ORFs (ORFs 1, 21, 22, 38, 50, 54, and 62). Two additional minor genotypes, M3 and M4, could also be resolved using two-amplicon typing.
Varicella (chickenpox) results from primary infection with varicella-zoster virus (VZV), which characteristically occurs early in life and usually follows a benign course (1). A lifelong latent infection is established on first exposure and can reactivate, typically after age 50, to cause zoster (shingles). VZV can be transmitted to susceptible persons from either disease condition, although zoster carries a significantly lower risk of transmission. Transmission from zoster can reintroduce strains that were circulating decades earlier and, as such, likely contributes to the genetic stability of VZV. The epidemiology of VZV infection varies geographically. Varicella displays a marked seasonality (peaking in late winter or spring) in temperate climates, and infection is nearly ubiquitous by age 20. For reasons that remain unclear, seasonality does not occur in tropical countries, and a larger proportion of people enter adulthood uninfected by VZV (14, 15). In addition, several studies have demonstrated a distinctive geographic distribution of the major VZV genotypes aligning with cool versus warm global climates (2, 18, 28). It is unclear whether the strain distribution is actually driven primarily by climate or by other factors, such as immigration patterns.
A number of methods have been reported for identifying and genotyping VZV strains (4, 18, 21). Early VZV typing efforts relied on DNA restriction fragment length polymorphism (RFLP) analysis, an approach that first demonstrated the high degree of sequence conservation among VZV strains. Nevertheless, some intrastrain variation among wild-type isolates of VZV was observed (5, 31). RFLP has also been used to distinguish most wild-type VZV isolates from Oka vaccine preparations, using single nucleotide polymorphisms (SNPs) located in open reading frame 38 (ORF38; a PstI site present in many wild-type strains) and ORF54 (a BglI site found in Oka vaccine preparations) (11, 12, 17, 18). It was also determined that wild-type VZV strains isolated in the United States and Japan have distinctive PstI and BglI RFLP profiles. Japanese isolates are either PstI+ BglI+ or PstI− BglI+, while most isolates from the United States, United Kingdom, Europe, and eastern Australia isolates are PstI+ BglI− (11, 12, 13, 18, 19). BglI+ strains, apart from those isolated in Japan, are common in tropical regions such as equatorial Africa, India, Bangladesh, China, Central America, and northern Australia (18, 28). An unusual PstI− BglI− VZV strain was reported in Australia (3), a variation that could represent recombination between the dominant genotypes or point mutation at the ORF38 locus.
Whole-genome screening for SNPs using a heteroduplex mobility assay was used to identify variants of VZV circulating in the United Kingdom and elsewhere (2, 21), an approach that evaluated selected SNPs in ORFs 1, 21, 50, and 54. The method was modified to include more SNPs with the expectation of improving capacity to discriminate genotypes, but results were similar to those obtained using the original method (4). This technique distinguished at least three major genotypes among global VZV isolates (A, B, and C) (2), and the genotypes had distinctive geographic distributions. Genotype A strains formed an African-Asian grouping, whereas genotype B and C strains were found primarily in European populations. Genotype J was subsequently added to this genotyping scheme to accommodate Japanese strains (21). It was hypothesized that recombination between types A and C viruses resulted in genotype B viruses as well as unclassified A/C recombinants. This method was used by another laboratory to evaluate VZV variability in Ireland, where all four genotypes were detected (4).
Recent phylogenetic analyses of partial and whole VZV genome sequences suggest that VZV genomic variation can be categorized into specific genotypes and probable recombinant viruses (18, 22, 26). Loparev et al. (18) performed targeted, multilocus analysis of VZV polymorphisms for a large number of clinical isolates obtained from every populated continent, distinguishing three major genotypes: E (European), J (Japanese), and M (mosaic). The M strains were later subdivided into M1, M2, M3, and M4, which are each considered separate genotypes (16, 18, 30). Evaluation of the global distribution of VZV strains using sequence variation in a single amplicon from ORF22 sorted VZV isolates from anywhere in the world into four dominant VZV genotypes with a well-defined longitudinal distribution. Strains belonging to M1 and M2 genotypes were most common in tropical regions, E strains were most common in temperate longitudes, and J strains were most common in Japan. In separate studies an M3 genotype strain was isolated in the United States (30), and four M4 genotype strains were isolated in Spain and France (8). Complete genomic sequences for these two genotypes are not available; therefore, only E, J, M1, and M2 genotypes will be discussed in our comparison of ORF22 genotyping with other methods (21, 33).
A third genotyping strategy, based on complete DNA sequences for five glycoprotein genes (gH, gI, gL, gB, and gE) and the IE62 major transactivator gene, was used to classify VZV strains into three genotypes, also designated A, B, and C (7). This study revealed that VZV strains circulating in Japan, Iceland, and The Netherlands had uniform genotypes (A, B, and C, respectively), whereas genotypes circulating in different regions of the United States or even within the same state were diverse. These findings were recently extended to include viruses from Singapore and Thailand (33). Based on this approach viral isolates from the United States and Europe and from Singapore and Japan were segregated into four distinct genotypes arbitrarily designated A, B, C, and D. Viral isolates from Singapore and Japan were from genotypes B and C, and isolates from Western Europe and the United States were from genotypes A and D. Finally, in a study using complete genome sequence information available from a public database, VZV was segregated into four genotypes: A, B, C, and D (26). It is important to note that these similar nomenclature schemes do not correlate between methods (2, 4, 7, 18, 26, 33).
No consensus for strain nomenclature has been reached for VZV (2, 7, 21, 28, 33). For simplicity and convenience, we assigned VZV strains homologous to the Dumas reference strain to the European (E) genotype, strains homologous to the Oka parental strain to the Japanese (J) genotype, and strains displaying alternating J-like and E-like regions to the mosaic (M) genotypes.
We report here a direct comparison of most of the genotyping methods described above in a single study using clinical isolates from Australia and New Zealand. Australia, while somewhat geographically isolated, has a long history of European colonization and a more recent history of non-European immigration and includes climates ranging from tropical to temperate. Therefore, it is not surprising that the strains circulating in Australia would reflect that diverse history and climate. All of genotyping methods described above are capable of discriminating only four major VZV genotypes, and the four groups identified differ slightly among methods. By employing an approach that uses sequence data from our previously described 447-bp amplicon in ORF22 with data from one additional amplicon (either ORF21 or ORF 50), it was possible to allocate VZV isolates into five dominant VZV genotypes, each of which identifies a genotype described by one or more of the other genotyping protocols. Viruses from genotype B by the method of W. B. Muir et al. (21, 28) appeared to be recombinants between M and E viruses and were here designated E2 genotype strains since they were more closely related to the European genotype. Genotype E (Dumas-like strains) was changed to E1.
These data shed light on relationships between VZV genotypes and should help to monitor vaccine impact in countries with universal childhood varicella vaccination policies; vaccination might be expected to increase the frequency of VZV reinfection with local wild-type strains in persons vaccinated with J genotype Oka vaccine and possibly lead to an increase in recombination events. This study also describes a simple approach for broadening the ability to discriminate among the major circulating genotypes of VZV using sequence data from only two, rather than multiple, amplicons.
MATERIALS AND METHODS
Patients.
Ninety-nine specimens from 31 cases of varicella and 68 cases of zoster were collected in 2003 and 2004 from patients seen at tertiary care hospitals in eastern Australia. An additional 28 VZV isolates were obtained but lacked data regarding clinical presentation. Separately, 36 specimens from 24 cases of zoster and 12 cases of varicella (8 from pediatric patients) were isolated from patients seen in the greater Auckland region, New Zealand, between September 2004 and March 2005. Two additional specimens without data about clinical presentation were also collected. VZV DNA for all of the New Zealand isolates was obtained from infected cells that had been inoculated with virus recovered from vesicular swabs.
Molecular epidemiology.
DNA was purified, and PCR-based diagnostic assays were performed as previously described (16). Australian VZV DNA-positive specimens were placed on Whatman FTA filters and stored at room temperature. For specimens from New Zealand, 70 μl of suspended cells infected with single-passage VZV isolate were tested. Specimens were applied to FTA cards (Whatman, Inc., Florham Park, NJ) to inactivate virus and shipped to the CDC for VZV genotyping. One 1- to 3-mm-diameter punch from FTA cards was prepared for PCR according the manufacturer's instructions. In some instances, filters were washed with Whatman FTA washing solution and reused in several PCRs. Oka vaccine-associated SNPs were evaluated to confirm that isolates obtained in the study were wild-type strains and to validate the use of various strategies to discriminate the vaccine strain from wild type in the context of Australia and New Zealand. Vaccine-specific SNPs in ORF38, ORF54, and ORF62 were determined using fluorescent resonance energy transfer-based PCR performed on a LightCycler (Roche, Pleasanton, CA) as previously described (19), and additional testing for varicella vaccine-associated SNPs at positions 106710 and 107136 was performed as described in Loparev et al. (20). VZV genome loads were assessed using real-time quantitative PCR analysis with the minor groove binding (MGB) Eclipse probe system for allelic discrimination (Epoch Biosciences, Inc.). In this test additional VZV Oka vaccine-specific SNPs were evaluated (MspI/NaeI sites; VZV genome position 107252) using a modification of the method described by Campsall et al. (3). Briefly, modified MGB Eclipse primer mixes contained the forward (5′-GCCCAAAAACACTTTATCCTAC) and reverse (5′-GTTGTTGGAGAAGGGTGA) primers for amplification and the following MGB Dark Quencher Eclipse probes for detection and SNP discrimination: a wild-type probe (GCCTTTGCCAGC) labeled with 6-carboxyfluorescein and a mutant probe (5′-GAGCCTTTGCCGG) labeled with tetrachloro-6-carboxyfluorescein by using a controlled-pore glass column. Only samples with at least 1 ×106 VZV genome copies were used for this study to ensure sufficient material to generate between 10 and 20 amplicons for DNA sequencing. The PCR forward and reverse primers (p22R1f and p22R1r) were designed to amplify a 447-bp fragment (positions 37837 to 38264) of VZV ORF22, and sequence analysis within this amplicon was conducted as described previously (17). Multilocus DNA sequence analyses (see Table 3) at ORF1, ORF21, ORF50, ORF54, and ORF62 were performed as described previously (4). All VZV genomic locus numbers used in this study are based on the published nucleotide sequence for the Dumas strain.
TABLE 3.
Analysis of genomic variation using data from multiple VZV ORFs for VZV reference strains and clinical specimens isolated in Australia and New Zealanda
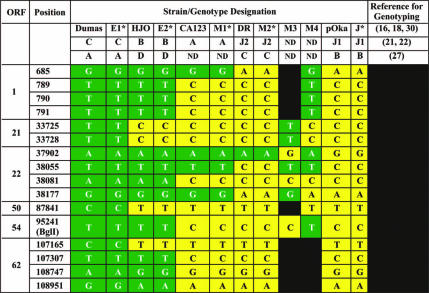
Genotypes were derived using the two-amplicon protocol introduced in this report. All clinical specimens from Australia and New Zealand allocated to the E1, E2, M1, M2, and J genotypes (Table 2) had the corresponding array of SNPs displayed in this table at ORFs 21, 22, and 50. Green cells represent the E1 (Dumas) strain marker, yellow cells represent the J (pOka) strain marker, and black indicates that the marker has not been determined for that strain. ND, not discriminated.
Bioinformatics.
CLUSTALW, version 1.83 (9, 32), was used to generate sequence alignments. Each alignment was inspected and optimized to remove short five-repeat regions in VZV genomes. Bayesian posterior probability inference of phylogeny used MrBayes, version 3.1 (10). MrBayes settings for the best-fit model (GTR+I+G) were selected by the hierarchical likelihood ratio test in MrModeltest, version 2.2 (23, 27). Phylogenetic trees were visualized using Treeview (24).
Nucleotide sequence accession numbers.
The accession numbers for the full genomic sequences for the 22 VZV isolates used for VZV genome genotyping are given in Table 1.
TABLE 1.
Strains used in the complete genome alignment and phylogenetic analysis
| VZV straina | Genotype by the indicated method:b
|
Diseasec | Year isolated | Location | Dominant regiond | Accession no.e | ||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | ||||||
| Dumas | E1 | C | A | V | 1970s | The Netherlands | Europe, North America, Australia | XO4370 |
| BC | E1 | C | A | Z | 1999 | Canada | AY548171 | |
| MSP | E1 | C | A | V | 1995 | United States | AY548170 | |
| SD | E1 | C | A | NA | 1980 | United States | DQ479953 | |
| KEL | E1 | C | A | Z | 2002 | United States | DQ479954 | |
| 36 | E1 | C | A | V | 1998 | Canada | DQ479958 | |
| 49 | E1 | C | A | V | 1999 | Canada | DQ479959 | |
| 32 P5 | E1 | C | A | V | 1976 | United States | DQ479961 | |
| 32 P22 | E1 | C | A | NA | DQ479962 | |||
| Russia | E1 | C | A | Z | Russia | NA | ||
| NH293 | E1 | C | A | V | 2000 | United States | DQ674250 | |
| HJO | E2 | B | D | Z | 1990s | Germany | Not dominant in any region | AJ871403 |
| 11 | E2 | B | D | Z | 1996 | Canada | DQ479955 | |
| 22 | E2 | B | D | Z | 1998 | Canada | DQ479956 | |
| 03-500 | E2 | B | D | NA | 2003 | Canada | DQ479957 | |
| CA123 | M1 | A | V | 1990s | United States | Central Africa | DQ457052 | |
| DR | M2 | J1 | C | Z | United States | Indochina | DQ452050 | |
| 8 | M2 | J1 | C | Z | 1995 | Canada | DQ479960 | |
| pOkaf | J | J1 | B | V | 1970 | Japan | Japan | AB097933 |
| vOka | AB097932 | |||||||
| VarilRix | DQ008354 | |||||||
| VariVax | DQ008355 | |||||||
Strains are grouped with reference strains (in boldface).
See text for a description of methods 1, 2, and 3.
V, varicella; Z, zoster; NA, disease condition of isolate is not available.
In the region(s), >50% of isolates are of the indicated genotype(s).
NA, not available.
vOka (Biken), VarilRix, and VariVax are all preparations of attenuated VZV derived from strain pOka.
RESULTS
Prediction of VZV genotypes using full genome sequence data.
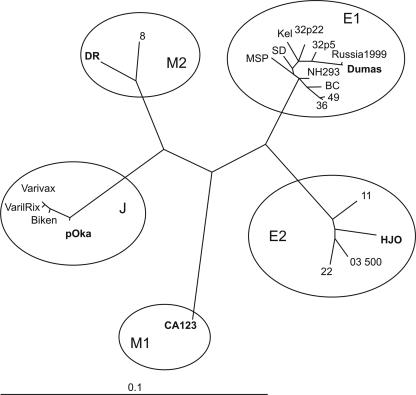
An optimal scheme for identifying the major circulating genotypes of VZV was established by analyzing the 22 complete genomic sequences available in the GenBank database. The outcome of phylogenetic analysis of these sequence data (shown in Table 3) clearly demonstrate the separation of VZV strains into five distinct genotypes. Henceforth, the two-amplicon method introduced here will be referred to as method 1, the scheme developed by Barrett-Muir et al. (2) as method 2, and the scheme of Peters et al. (26) as method 3. Among the complete published sequences, the largest number of strains were from genotype E1, including the first fully sequenced VZV isolate (Dumas); it is the most commonly circulating genotype in North America and Europe. E1 viruses belong to genotype B by method 2 and to genotype A by method 3. All 11 fully sequenced E1 isolates have >99.9% identity to the Dumas strain at the nucleotide level, excluding insertions and deletions, ranging between 33 and 47 total nucleotide differences.
A whole-genome sequence comparison placed four of the strains in a new genotype E2 (genotype B using method 2). These strains are not detected as a distinct genotype using ORF22-based genotyping alone but are clearly distinguished if SNPs from a targeted region of either ORF21 or ORF50 are included in the analysis (Fig. 1). Genotype E2 viruses comprise 15 to 40% of strains isolated from varicella cases in Europe and North America (2, 18). Strain HJO was isolated in Germany, and its sequence serves as the reference strain for genotype E2. All genotype E2 strains have the PstI+ BglI− markers in ORF38 and ORF54, in common with genotype E1.
FIG. 1.
Phylogenetic analysis based on the complete VZV genomes. Nucleotide substitutions supporting each genotype are shown. Reference strains for each genotype are shown in bold.
Genotype M1 strains are distinguished using ORF22 sequence information alone; the only fully sequenced representative of this genotype at present is strain CA123, isolated in California. M1 strains correspond to genotype A viruses using method 2, and the genotype has no analog in the method 3 genotyping scheme. Genotype M1 (A) strains are common in Central Africa and Indochina (17, 28). Genotype M1 strains have PstI+ BglI+ markers in ORF38 and ORF54.
The African strain DR is the reference strain for genotype M2, and a second fully sequenced isolate, strain 8, also grouped with M2 strains in the phylogenetic tree (Fig. 1). VZV strain DR had 181 single-base differences compared to Dumas, or 99.9% identity at the nucleotide level. Strain DR and North American strain 8 were among the isolates examined that were genotype C by method 3 (Table 1) and assigned to genotype M2 using ORF22 typing alone. Typing by method 2 assigned strains 8 and DR to genotype J1, the same classification to which the parental Oka (pOka) strain and the three vaccine preparations were assigned. Methods 1 and 3 both classified the M2 genotype viruses DR and VZV strain 8 into a distinct genotype, but method 2 cannot differentiate M2 genotype strains from Japanese (J) strains. Genotype M2 viruses are dominant in North Africa and Indochina and have been found occasionally in North America. All M2 strains have PstI+ BglI+ markers in ORF38 and ORF54 in common with most Japanese (genotype J) strains.
The only fully sequenced wild-type virus representing genotype J is the pOka strain from which all varicella vaccine preparations were derived. Again, excluding the deletions and insertions that are generally confined to the VZV genomic repeat elements, pOka had 99.82% identity with the Dumas strain at the nucleotide level, which translates to a total of 187 single nucleotide differences between these two viruses. About 95% of strains in Japan have the PstI+ BglI+ markers at ORF38 and ORF54, but 5% of these strains are PstI− BglI+ like the pOka reference strain (18). All Japanese isolates examined thus far belong exclusively to genotype J. The three varicella vaccine preparations vOka, VarilRix, and VariVax are also represented in the analysis shown in Fig. 1, and all of these vary slightly from one another. Genotype J strains are genotype J1 by method 2 and genotype B by method 3.
As shown in Fig. 1, genotypes E1 and J include the most divergent strains of VZV. Largely on the basis of providing the first complete sequences available, we propose here that the Dumas strain should be regarded as the reference for genotype E1, the HJO strain for genotype E2, the pOka strain for genotype J, the DR strain for genotype M1, and the CA123 strain for genotype M2. Each of these reference viruses is representative of the sequence variations characteristic of the five respective genotypes.
Characterization of isolates from Australia and New Zealand using SNPs associated with varicella vaccine.
We analyzed genetic variation among VZV isolates obtained from varicella and zoster patients in Australia and New Zealand. The 165 specimens were first confirmed VZV PCR positive, wild type, using PCR methods targeting VZV vaccine-associated SNPs in ORFs 38, 54, and 62. None of the VZV strains included in the study carried any of the vaccine-associated markers and, therefore, were wild-type strains. Single SNPs located in ORF38 and ORF54 have been used in combination to discriminate vaccine strains from wild-type strains. It has been recognized for some time that these markers fail to distinguish Oka vaccine from a subset of wild-type PstI− Japanese isolates and thus are not reliable for this purpose. Nonetheless, ORF38 (PstI site) and ORF54 (BglI site) data are included alongside the genotype determinations of isolates since these markers are also helpful for characterizing VZV strain variation (Table 2). All of the isolates examined were wild type based on markers analyzed in ORF62; 130 specimens were PstI+ BglI−, 34 were PstI+ BglI+, and one Australian isolate was PstI− BglI+ (the Oka vaccine profile for these markers) but was a wild-type strain using the more reliable markers in ORF62.
TABLE 2.
Allocation of Australian and New Zealand samples to five predicted VZV genotypes
| Genotype | No. of isolates (%) from:
|
ORF38/54 profilea | |
|---|---|---|---|
| Australia | New Zealand | ||
| E1 | 67 (52.8) | 25 (65.8) | PstI+ BglI− |
| E2 | 30 (23.6) | 8 (21.1) | PstI+ BglI− |
| J | 16 (12.6) | 0 (0) | PstI− BglI+ (1); PstI+ BglI+ (15) |
| M1 | 10 (7.9) | 5 (13.1) | PstI+ BglI+ |
| M2 | 4 (3.1) | 0 (0) | PstI+ BglI+ |
| M3 | 0 (0) | 0 (0) | PstI+ BglI+ |
| M4 | 0 (0) | 0 (0) | PstI+ BglI− |
| Total | 127 (100) | 38 (100) | |
Numbers in parentheses represent the numbers of isolates with the indicated profiles. No PstI− BglI− isolates were identified in this study.
Evaluation of genotyping method 1 using clinical specimens.
We evaluated the ability of genotyping method 1 (ORF22 SNPs plus variation in targeted SNPs in either ORF21 or ORF50) to categorize wild-type isolates collected in Australia and New Zealand into the five genotypes identified using published sequence data. These data are presented in Table 2.
Using this approach all of the clinical VZV isolates could be allocated to one of the five major VZV genotypes. Ninety-two isolates were E1 genotype strains (67 from Australia and 25 from New Zealand), 38 were genotype E2 (30 from Australia and 8 from New Zealand), 15 were genotype M1 (5 from Australia and 5 from New Zealand), 4 were genotype M2 (all from Australia), and 16 were genotype J (all from Australia). As expected, all 130 genotype E1 and E2 strains were PstI+ BglI−, and the genotype M1, M2, and 15/16 genotype J strains were PstI+ BglI+; the remaining genotype J strain was PstI− BglI+ (Table 2). No BglI− PstI− isolates were identified in this study, although a double-negative isolate was described in an earlier study from Australia (3). Other putative recombinant genotypes M3 (30) and M4 (15) were not observed in this study.
DISCUSSION
Refined strategy for VZV genotyping.
The VZV genome is highly conserved, and recombination between different VZV strains has also been hypothesized (22); both of these factors complicate strategies for genotypic analysis of VZV and for strain surveillance. Nonetheless, several reasonably robust methods for genotyping VZV have led to a wealth of useful information on VZV genotypic variation (18, 21, 22, 26) and were the driving impetus for this effort to develop a method that successfully identifies all of the genotypes described to date. The approach evaluated here requires the analysis of SNP information from only two PCR amplifications (ORF22 with four SNPs and either ORF21 with two SNPs or ORF50 with one SNP).
Several recently reported methods are capable of discriminating four major VZV genotypes; however, some discrepancies exist between these methods. For example, analysis based on ORF22 SNPs alone (18, 30) indicated that E strains constituted a uniform, single genotype and represented the dominant genotype in temperate climates. In contrast, the use genotyping method 2 (analysis of SNPs in at least four to five amplicons from ORFs 1, 21, 50, 54, and 68) revealed that the E-type strains were subdivided into two distinct genotypes (21). However, the ORF22-based strategy differentiated M2 and J as two distinct BglI+ genotypes, (18, 22), while method 2 resolved these strains as a single genotype J (4, 21). Yet another group that used whole-genome sequence analysis confirmed that there were four genotypes (26), but these investigators did not have access to complete sequence information for a genotype M1 strain as detected with ORF22 analysis (18) for their analysis. The current analysis of whole-genome sequence data from 22 published sequences has helped to clarify the clustering of VZV strains and provided the key to simplifying the strategy for genotyping VZV.
Comprehensive phylogenetic analysis of complete genome sequences resolved the separation of European, Japanese, and North American strains into five discrete genotypes, which we have designated E1, E2, J, M1, and M2. Strains allocated to any individual genotype display roughly a dozen coding differences in nonrepetitive genome sequences compared to the reference genotype. For example, the genotype E1 strains MSP and Dumas have 5 differences that lead to amino acid changes; BC and Dumas have 12 differences. Notably, this observation holds true for strains isolated on different continents; the genotype E2 strains HJO and 03-500, isolated in Germany and Canada, respectively, are nearly identical, with only a single SNP leading to an amino acid shift. Nearly complete identity was also observed in the coding regions between M2 strains DR and 8 (19, 26).
As of this time there is only one complete wild-type genomic sequence available for each representative strain of genotypes J and M1. However, extensive sequence analysis among isolates obtained from Japan, Central Africa, America, Asia, and Africa suggests that the genotype J and M1 strains are also closely similar (18, 21, 33). J and M1 viruses differ from the Dumas strain by 61 and 47 SNPs, respectively, for differences that confer an amino acid change. Genotype E and J strains are most divergent, whereas the M genotypes comprise VZV strains with variable assortments of genotype E (particularly E1) and genotype J SNPs. That said, HJO and related strains are actually M-type viruses since they display a combination of E and J SNPs through approximately one-third of their genomes. Nonetheless, we designated them E2 viruses because the first representative strains for this genotype were identified in the United Kingdom, and the reference strain HJO was isolated in Germany. In addition, only genotype E1 and E2 strains uniformly bear the BglI− marker in ORF54 and are identical across approximately two-thirds of their genomes, including the SNPs located in ORF22. Genotype E2 strains have been found dominant in some European countries and have also been isolated in the United State, Canada, and Australia, although these strains are also found all over the world. Three complete genome sequences for North American genotype E2 isolates (Table 1) were recently made available (strains 11, 22, and 03-500).
The detection of all five genotypes predicted by whole-genome phylogenetic analysis required the expansion of ORF22-based sequencing to include sequence data that reliably discriminate genotype E1 and E2 strains. Using ORF22-based sequencing as a foundation for the expanded technique made sense, since the protocol has successfully discriminated most of the major circulating genotypes on nearly 1,000 clinical isolates obtained from countries on every inhabited continent (6, 18, 30; also additional unreported observations). The approach was practical, parsimonious, and able to yield results from the limited amount of DNA typically recovered from clinical specimens, usually nonviable material collected as vesicular swabs or scabs. Bioinformatics analysis of all available whole-genome sequence data for VZV indicated that a genotyping scheme based on sequencing ORF22 and one additional amplicon would satisfy the requirement for detecting and discriminating all of the major genotypes even in specimens with limited template volume. We determined that a combination of the ORF22 sequence with a targeted region in either ORF21 or ORF50 would provide a robust, and still relatively straightforward, method. Finally, it should be noted that the other methods capable of resolving these genotypes require multiple amplification and sequencing steps.
We analyzed 12 well-characterized SNPs located in ORFs 1, 21, 22, 50, 54, and 62 (2, 4) as potential candidates for routinely discriminating all five VZV genotypes (Table). We determined that SNPs at positions 789, 790, 791, 95241, and 107307 discriminate genotype E strains from genotype M1, M2, and J strains. The SNP at position 685 differentiates genotype E and M1 strains from genotype M2 and J strains. Only analysis based on a combination of sequence data from ORF22 and from either ORF21 (SNPs in positions 33725 and 33728), ORF50 (SNP in position 87841), or ORF62 (SNP in position 107165, 108747, or 108951) facilitated the separation of genotypes E1 and E2 from genotype E, thus detecting all five genotypes using only sequence data from two amplicons. It should also be noted that the two-amplicon method reported here also discriminates two additional, previously described genotypes, M3 and M4, both of which were absent from the specimens collected for this study (16, 23).
Evaluation of genotyping method 1 using clinical specimens.
As expected based on a long history of European colonization, a significant proportion of VZV strains circulating in New Zealand and Australia are European (genotype E1 or E2) strains: 76.38% (52.76% E1 and 23.62% E2) of the isolates obtained in Australia and 86.85% (65.8% E1 and 21.05% E2) of the isolates obtained in New Zealand. In general, the proportion of E strains is similar to that observed in Europe and North America (18, 30).
Genotype E1 strains are more common than E2 strains in Europe and Australia. We speculate that genotype E2 strains may have arisen more than a century ago in remote European colonies such as Australia and New Zealand through recombination events between E1 and tropical genotype M strains and effectively competed with the original tropical and imported E1 strains, establishing themselves in the population. Recombination between E1 and J strains could also play a role in the appearance of genotype E2 strains. Alternatively, E2 strains might have emerged in Europe (16) in those countries with E1 and M strains in circulation, spreading afterwards to Australia and New Zealand; or recombinant variants of E2 could have been selected by temperate climates to which increasing numbers of people migrated from regions with warmer climates. The migration of persons native to tropical regions has been an ongoing and increasingly common occurrence in Australia and New Zealand for a number of decades. Strikingly, E2 strains currently circulating in the United Kingdom and Ireland (4, 21) are identical (based on limited SNP analysis) to Australian and New Zealand strains. It is possible that native Australians, native New Zealanders, or residents from tropical countries located to the north could be the source of the M-type strains that are circulating in these countries (18). It remains a possibility, given the very limited number of genetic changes involved, that these variant strains could have simply arisen through mutation, although recent analyses suggest otherwise (22). Another important finding was the observation of a large proportion of genotype J strains (12.6%) circulating in Australia. It appears from the data presented here that genotype J strains may be displacing genotype E1 strains in Australia. In previous studies we along with others were unable to find genotype J strains in Europe, Africa, and South America (18, 21, 30); however, a small number of genotype J strains have begun to circulate in regions of North America with substantial populations of Asian immigrants. These observations suggest that, in addition to the previously observed longitudinal distribution into tropical and temperate climate zones (18), VZV genotype distribution may also display a latitudinal distribution of temperate genotype E1 and J strains in the eastern and western hemispheres. Other putative VZV M3 (30) and M4 (16) genotype viruses were not detected in this study (Table 3).
The inclusion of large numbers of zoster cases in this study has the effect of broadening the time span of the survey, since the majority of those cases would reflect strains that were in circulation 5 to 8 decades ago. All five VZV genotypes were identified in senior Australian patients with zoster, indicating that all of the major VZV genotypes were in circulation in Australia during the first half of the 20th century. Also of interest is the correlation with other markers historically used to characterize VZV strains and to differentiate the Oka vaccine from wild type. All 97 E1 and E2 strains were PstI+ BglI−, 19 genotype M1 and M2 strains were PstI+ BglI+, and 15 genotype J isolates were PstI+ BglI+. However, one Australian genotype J strain was PstI− BglI+, resembling the vaccine strain and the wild-type pOka strain it was derived from. This strain, as with all wild-type genotype J isolates examined thus far (17, 19), was SmaI− and MspI− at ORF62 positions 106262 and 107252, respectively. This confirms that these ORF62 methods for discriminating the vaccine strain from wild type can be used reliably for that purpose in New Zealand and Australia (21, 25).
In this study we propose a new approach to VZV genotyping that effectively combines the genotype-discriminatory powers of all methods currently in use and that is based on the amplification and DNA sequence analysis of two short regions of the VZV genome. A method using amplification and sequencing of the previously reported 447-bp region of ORF22 (18, 30) together with the amplification and sequencing of either of two targeted regions in ORF21 or ORF50 was shown to effectively achieve that goal. Both versions of this protocol (ORF22/ORF21 and ORF22/ORF50) were able to clearly differentiate all five major VZV genotypes that were identified through phylogenetic analysis of the complete genomic sequences for 22 strains representing all of these major variants. Both versions of the protocol categorized wild-type strains of VZV into identical genotypes.
The study also confirmed the usefulness of ORF62 vaccine markers to discriminate wild-type varicella from Oka vaccine and simultaneously demonstrated that the method targeting SNPs in ORF38 and ORF54 would be unsuitable for that purpose in Australia. Genotype E1 strains represent >50% of the wild-type VZV isolates obtained from clinical cases of varicella and zoster in Europe and North America. All of those strains are PstI+ BglI−. Therefore, ORF38/ORF54-based SNP analysis formed a useful approach for discriminating vaccine from wild-type strains since few genotype J PstI− BglI+ strains were circulating in those countries. This is clearly not the case in Australia, where a significant fraction of isolated wild-type viruses belong to genotype J and would be mistaken for vaccine virus by this method.
The availability of technically practical, reliable methods for genotyping VZV strains will serve a critical function in countries with broad varicella vaccination policies, since tracking individual strains and identifying probable sources of infection are essential to effectively monitor vaccine impact. In addition, the study of global VZV genotype patterns will likely lead to a better understanding of global transmission patterns both before and after vaccination and of the evolutionary trends of this nearly ubiquitous virus.
Acknowledgments
We express our appreciation to all staff in the hospitals, health care centers, and laboratories that participated in skin lesion collection, and we thank them for excellent technical assistance with data entry. We also thank Olga Vankova (NPO Diagnostic Systems) for providing a complete genome sequence for VZV strain Russia 1999 before publication. We thank the CDC DNA Core Facility for oligonucleotide synthesis and DNA sequencing, which were invaluable for the completion of this project.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the funding agency.
All authors contributed equally to this work.
Footnotes
Published ahead of print on 26 September 2007.
REFERENCES
- 1.Arvin, A. M. 1996. Varicella-zoster virus: overview and clinical manifestations. Semin. Dermatol. 15:4-7. [PubMed] [Google Scholar]
- 2.Barrett-Muir, W., F. T. Scott, P. Aaby, J. John, P. Matondo, Q. L. Chaudhry, M. Siqueira, A. Poulsen, K. Yaminishi, and J. Breuer. 2003. Genetic variation of varicella-zoster virus: evidence for geographical separation of strains. J. Med. Virol. 70(Suppl. 1):S42—S47. [DOI] [PubMed] [Google Scholar]
- 3.Campsall, P. A., N. H. Au, J. S. Prendiville, D. P. Speert, R. Tan, and E. E. Thomas. 2004. Detection and genotyping of varicella-zoster virus by TaqMan allelic discrimination real-time PCR. J. Clin. Microbiol. 42:1409-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carr, M. J., G. P. McCormack, and B. Crowley. 2004. Genetic variation in clinical varicella-zoster virus isolates collected in Ireland between 2002 and 2003. J. Med. Virol. 73:131-136. [DOI] [PubMed] [Google Scholar]
- 5.Chow, V. T., S. S. Wan, S. Doraisingham, and A. E. Ling. 1993. Comparative analysis of the restriction endonuclease profiles of the Dumas and Singapore strains of varicella-zoster virus. J. Med. Virol. 40:339-342. [DOI] [PubMed] [Google Scholar]
- 6.Dayan, G. H., M. S. Panero, R. Debbag, A. Urquiza, M. Molina, S. Prieto, M. Del Carmen Perego, G. Scagliotti, D. Galimberti, G. Carroli, C. Wolff, D. S. Schmid, V. Loparev, D. Guris, and J. Seward. 2004. Varicella seroprevalence and molecular epidemiology of varicella-zoster virus in Argentina, 2002. J. Clin. Microbiol. 42:5698-5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faga, B., W. Maury, D. A. Bruckner, and C. Grose. 2001. Identification and mapping of single nucleotide polymorphisms in the varicella-zoster virus genome. Virology 280:1-6. [DOI] [PubMed] [Google Scholar]
- 8.Grossberg, R., R. Harpaz, E. Rubtcova, V. Loparev, J. F. Seward, and D. S. Schmid. 2006. Secondary transmission of varicella vaccine virus in a chronic care facility for children. J. Pediatr. 148:842-844. [DOI] [PubMed] [Google Scholar]
- 9.Higgins, D. G., and P. M. Sharp. 1988. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73:237-244. [DOI] [PubMed] [Google Scholar]
- 10.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 11.LaRussa, P., O. Lungu, I. Hardy, A. Gershon, S. P. Steinberg, and S. Silverstein. 1992. Restriction fragment length polymorphism of polymerase chain reaction products from vaccine and wild-type varicella-zoster virus isolates. J. Virol. 66:1016-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LaRussa, P., S. P. Steinberg, E. Shapiro, M. Vazquez, and A. A. Gershon. 2000. Viral strain identification in varicella vaccinees with disseminated rashes. Pediatr. Infect. Dis. J. 19:1037-1039. [DOI] [PubMed] [Google Scholar]
- 13.Larussa, P. S., and A. A. Gershon. 2001. Biologic and geographic differences between vaccine and clinical varicella-zoster virus isolates. Arch. Virol. Suppl. 2001:41-48. [DOI] [PubMed] [Google Scholar]
- 14.Lee, B. W. 1998. Review of varicella zoster seroepidemiology in India and Southeast Asia. Trop. Med. Int. Health 3:886-890. [DOI] [PubMed] [Google Scholar]
- 15.Lolekha, S., W. Tanthiphabha, P. Sornchai, P. Kosuwan, S. Sutra, B. Warachit, S. Chup-Upprakarn, Y. Hutagalung, J. Weil, and H. L. Bock. 2001. Effect of climatic factors and population density on varicella zoster virus epidemiology within a tropical country. Am. J. Trop. Med. Hyg. 64:131-136. [DOI] [PubMed] [Google Scholar]
- 16.Loparev, V., E. Martro, E. Rubtcova, C. Rodrigo, J. C. Piette, E. Caumes, J. P. Vernant, D. S. Schmid, and A. M. Fillet. 2006. Toward universal varicella-zoster virus genotyping: diversity of VZV strains from France and Spain. J. Clin. Microbiol. 45:559-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loparev, V. N., T. Argaw, P. R. Krause, M. Takayama, and D. S. Schmid. 2000. Improved identification and differentiation of varicella-zoster virus (VZV) wild-type strains and an attenuated varicella vaccine strain using a VZV open reading frame 62-based PCR. J. Clin. Microbiol. 38:3156-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loparev, V. N., A. Gonzalez, M. Deleon-Carnes, G. Tipples, H. Fickenscher, E. G. Torfason, and D. S. Schmid. 2004. Global identification of three major genotypes of varicella-zoster virus: longitudinal clustering and strategies for genotyping. J. Virol. 78:8349-8358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loparev, V. N., K. McCaustland, B. P. Holloway, P. R. Krause, M. Takayama, and D. S. Schmid. 2000. Rapid genotyping of varicella-zoster virus vaccine and wild-type strains with fluorophore-labeled hybridization probes. J. Clin. Microbiol. 38:4315-4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loparev, V. N., E. Rubtcova, J. Seward, M. Levin, and D. S. Schmid. 2007. DNA sequence variability in isolates recovered from patients with postvaccination rash or herpes zoster caused by Oka varicella vaccine. J. Infect. Dis. 195:502-510. [DOI] [PubMed] [Google Scholar]
- 21.Muir, W. B., R. Nichols, and J. Breuer. 2002. Phylogenetic analysis of varicella-zoster virus: evidence of intercontinental spread of genotypes and recombination. J. Virol. 76:1971-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Norberg, P., J. A. Liljeqvist, T. Bergstrom, S. Sammons, D. S. Schmid, and V. N. Loparev. 2006. Complete-genome phylogenetic approach to varicella-zoster virus evolution: genetic divergence and evidence for recombination. J. Virol. 80:9569-9576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nylander, J. A. A. 2004. MrModeltest 2.0 Program distributed by the author. Evolutionary Biology Centre, Uppsala University, Uppsala, Sweden.
- 24.Page, R. D. 1996. TreeView: an application to display Phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 25.Parker, S. P., M. Quinlivan, Y. Taha, and J. Breuer. 2006. Genotyping of varicella-zoster virus and the discrimination of Oka vaccine strains by TaqMan real-time PCR. J. Clin. Microbiol. 44:3911-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peters, G. A., S. D. Tyler, C. Grose, A. Severini, M. J. Gray, C. Upton, and G. A. Tipples. 2006. A full-genome phylogenetic analysis of varicella-zoster virus reveals a novel origin of replication-based genotyping scheme and evidence of recombination between major circulating genotypes. J. Virol. 80:9850-9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 28.Quinlivan, M., K. Hawrami, W. Barrett-Muir, P. Aaby, A. Arvin, V. T. Chow, T. J. John, P. Matondo, M. Peiris, A. Poulsen, M. Siqueira, M. Takahashi, Y. Talukder, K. Yamanishi, M. Leedham-Green, F. T. Scott, S. L. Thomas, and J. Breuer. 2002. The molecular epidemiology of varicella-zoster virus: evidence for geographic segregation. J. Infect. Dis. 186:888-894. [DOI] [PubMed] [Google Scholar]
- 29.Sauerbrei, A., E. Rubtcova, P. Wutzler, D. S. Schmid, and V. N. Loparev. 2004. Genetic profile of an Oka varicella vaccine virus variant isolated from an infant with zoster. J. Clin. Microbiol. 42:5604-5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sergeev, N., E. Rubtcova, V. Chizikov, D. S. Schmid, and V. N. Loparev. 2006. New mosaic subgenotype of varicella-zoster virus in the USA: VZV detection and genotyping by oligonucleotide-microarray. J. Virol. Methods 136:8-16. [DOI] [PubMed] [Google Scholar]
- 31.Takayama, M., and N. Takayama. 2002. Long PCR amplification of varicella-zoster virus DNA in clinical specimens from the patients with varicella and herpes zoster. Kansenshogaku Zasshi 76:347-354. [In Japanese.] [DOI] [PubMed] [Google Scholar]
- 32.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wagenaar, T. R., V. T. Chow, C. Buranathai, P. Thawatsupha, and C. Grose. 2003. The out of Africa model of varicella-zoster virus evolution: single nucleotide polymorphisms and private alleles distinguish Asian genotypes from European/North American genotypes. Vaccine 21:1072-1081. [DOI] [PubMed] [Google Scholar]