Abstract
During the late stages of adenovirus infection, the 100K protein (100K) inhibits the translation of cellular messages in the cytoplasm and regulates hexon trimerization and assembly in the nucleus. However, it is not known how it switches between these two functions. Here we show that 100K is methylated on arginine residues at its C terminus during infection and that this region is necessary for binding PRMT1 methylase. Methylated 100K is exclusively nuclear. Mutation of the third RGG motif (amino acids 741 to 743) prevents localization to the nucleus during infection, suggesting that methylation of that sequence is important for 100K shuttling. Treatment of infected cells with methylation inhibitors inhibits expression of late structural proteins. These data suggest that arginine methylation of 100K is necessary for its localization to the nucleus and is a critical cellular function necessary for productive adenovirus infection.
During a productive infection, adenovirus turns the infected cell into an efficient virion-producing factory. To do this, the virus must interfere with and manipulate several cellular processes to facilitate replication and packaging of its genome into newly synthesized virions. A characteristic of the late phase of infection is the severe inhibition of cellular protein synthesis (2) and the selective translation of late viral mRNAs (28, 29). At least one viral product has been implicated in this process: the 100K nonstructural protein (11). The adenovirus type 5 (Ad5) 100K protein (referred to here as 100K) is thought to inhibit host protein synthesis by binding to EIF4F on the EIF4F translation initiation complex in the cytoplasm and preventing its phosphorylation by mnk1 kinase (5, 6). Dephosphorylation of EIF4F is believed to lead to inhibition of cellular protein synthesis (26), whereas late viral messages are preferentially translated by a process known as ribosome shunting (8).
However, 100K is also involved in hexon trimerization and assembly, a part of the packaging process of the viral genomes into newly synthesized capsids that occurs in the nucleus (19), and cells infected with conditional temperature-sensitive (ts) 100K mutants show cytoplasmic accumulation of hexon monomers and fail to accumulate hexon trimers in the nucleus at the nonpermissive temperature (3, 20). Therefore, 100K must be able to localize in both subcellular compartments and may switch between these two functions during infection. Recently, a Rev-like nuclear export sequence and a nuclear localization sequence (NLS) were identified within the carboxy terminus of 100K (7). Deletion of the 100K C terminus resulted in its cytoplasmic accumulation, suggesting that the NLS is within that part of the molecule. However, the mechanism by which 100K subcellular localization and function is regulated during infection is currently unknown.
Posttranslational modification of proteins allows viruses to overcome the limitations imposed by the small size of their genomes, offering each viral product the capacity to expand its functional repertoire. 100K is known to be a phosphoprotein, although the function and the sites of phosphorylation remain to be elucidated. Moreover, 100K was recently reported to be methylated during infection of permissive tumor cells, and methylation is mediated by PRMT1, the most abundant methylase in the cell (15). However, the mechanism by which methylation controls 100K function and its importance during infection has not been determined. Many proteins are targets of arginine methylation, the majority of which are members of the family of heterogeneous nuclear ribonucleoproteins (16, 18), which play roles in pre-mRNA processing and nucleocytoplasmic RNA transport. Most of these proteins are methylated on arginine residues within RGG tripeptides (14), although several proteins have also been identified that are asymmetrically dimethylated at RXR, RG, and GRG motifs (17). Although arginine methylation does not alter the overall charge of the protein, the addition of methyl groups may alter the overall structure of the molecule due to increased steric hindrance and the reduction of hydrogen bonds as a result of the substitution of hydrogen atoms with methyl groups (17). Such alterations can affect protein-protein and protein-nucleic acid interactions, and several proteins have already been shown to be regulated at that level (1). Arginine methylation has also been shown to regulate nucleocytoplasmic localization. In many cases, methylation acts as an nuclear localization signal and results in the nuclear accumulation of the molecule (30), whereas in some others the opposite occurs (9).
The C terminus of 100K contains an RGG domain and several RXR and RG motifs which are well conserved among different adenovirus serotypes. It is therefore possible that part of the mechanism by which 100K switches between its two main known functions, namely, host protein synthesis inhibition and hexon trimerization, is by changing its subcellular localization during the course of infection, and methylation of the RGG domain may regulate this process.
The goal of the present study was to further define the mechanism by which 100K exerts its multicomponent function and to examine the role of posttranslational methylation in adenovirus-infected cells in the context of 100K regulation.
MATERIALS AND METHODS
Cell lines and culturing conditions.
U2OS cells were obtained from the American Type Culture Collection and grown at 37°C and 5% CO2 in Dulbecco modified Eagle medium (DMEM) supplemented with high glucose,10% fetal bovine serum (FBS), 100 μg of nonessential amino acids/ml, 4 mM l-glutamine,10 U of penicillin/ml, and 10 μg of streptomycin/ml. When indicated, cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol.
Plasmids and antibodies.
The plasmid pMYC-100K was generated by PCR amplification of the 100K sequence and cloning into pCAN-myc. Plasmids pMYC-100KΔC and pMYC-100KRGG3 were generated by PCR mutagenesis of the pMYC-100K (specific details for all cloning are available upon request). All constructs were confirmed by sequencing.
Mouse monoclonal antibodies to 100K were kindly provided by W. C. Russell (University of St. Andrews, Fife, United Kingdom) and were previously described (23). Mouse monoclonal α-myc 9E10 antibody was a generous gift from G. Evan. Other antibodies were from commercial sources: rabbit polyclonal α-myc (Cell Signaling Technology), mouse monoclonal α-PRMT-1 (Abcam catalog no. ab7027), mouse monoclonal α-hexon (Abcam catalog no. ab7428), mouse monoclonal α-fiber (Abcam catalog no. ab3233), mouse monoclonal α-E1A (Calbiochem catalog no. DP11), and mouse monoclonal asymmetric α-dimethyl-arginine (Upstate catalog no. 07414).
Immunofluorescence microscopy.
U2OS cells were transfected with plasmids expressing myc-100K, myc-100KΔC, or myc-100KRGG3 proteins and grown on coverslips. Cells were fixed for 15 min at room temperature with 4% paraformaldehyde freshly made in phosphate-buffered saline (PBS), permeabilized for 5 min with 0.1% Triton X-100, and blocked with 5% normal goat serum (NGS) for 20 min, followed by incubation with antibodies to myc tag epitope or to L4-100K in blocking buffer for 1 h at 37°C. Cells were then washed three times for 20 min each time with PBS, reacted with Alexα-488 anti-mouse fluorescently labeled antibody (Molecular Probes) for 30 min at 37°C, washed three times for 20 min each time with PBS, stained with Hoechst 33258, and mounted with Dako fluorescent mounting medium. Cells were visualized and photographed by using a Zeiss-LSM confocal microscope.
Immunoprecipitation and Western immunoblot analysis.
U2OS cells were lysed in NP-40 lysis buffer (1% NP-40, 150 mM NaCl, 10 mM HEPES [pH 7.4], 2 mM EDTA, 2 mM sodium orthovanadate, 0.1% β-mercaptoethanol, and protease inhibitor cocktail tablet [Roche]) and clarified by centrifugation for 20 min at 14,000 × g. Lysates were precleared for 20 min at 4°C with protein G-agarose and incubated with 9E10 agarose beads for 2 h at 4°C. Lysates were then washed five times with lysis buffer, and immunoprecipitated proteins were eluted off the agarose beads with sodium dodecyl sulfate (SDS) sample buffer. Samples were run on 4 to 20% or 6% Tris-glycine gels (Invitrogen) at 80 to 150 V and transferred at 300 mA for 90 min onto Immobilon polyvinylidene difluoride membranes.
S35 labeling.
U2OS cells were washed twice with PBS and supplemented with cysteine and methionine-free DMEM with 2% dialyzed FBS. Cells were starved for 2 h, 100 μCi of S35 was added to each well, and cells were incubated at 37°C for 2 h to label them. At the end of incubation the medium was removed, and the cells were washed twice with cold PBS before being harvested in the appropriate lysis buffer for subsequent analysis.
Nucleocytoplasmic fractionation.
Fractionation was performed by using a Sigma CelLytic nuclear extraction kit (Sigma product code N-Xtract) according to the manufacturer's protocol. Cells were treated with trypsin with 0.25 mM trypsin-EDTA solution (Gibco) and washed with PBS once, and pellets were incubated in hypotonic lysis buffer for 15 min at room temperature. Samples were centrifuged at 11,000 × g for 30 s at room temperature, and the supernatant was collected (cytoplasmic fraction). Nuclei were then incubated with extraction buffer, and samples were agitated for 30 min at room temperature. Samples were then centrifuged at 21,000 × g for 5 min, and the supernatant was collected (nuclear fraction).
Virus infections and quantification.
U2OS cells were infected in a low volume of DMEM with 2% FBS at 37°C and 5% CO2. Viruses were quantified in 293/E4/pIX cells by using an enzyme-linked immunosorbent assay as described previously (13). Wild-type Ad5 virus is dl309 (which has a partial deletion in the E3 region). Cells were infected at multiplicity of infection of 30.
Reversed-phase liquid chromatography-electrospray tandem mass spectrometry (LC-MS/MS) analysis.
Protein bands were excised from gels and digested in-gel with trypsin as described previously (22; donatello.ucsf.edu/ingel.html). The extracted digests were vacuum evaporated and resuspended in 1 μl of 0.1% formic acid in water. The digests were separated by nanoflow liquid chromatography using a 75-μm-by-150-mm reversed-phase C18 PepMap column (Dionex-LC-Packings, San Francisco, CA) at a flow rate of 350 nl/min in a NanoLC-1D proteomics high-performance liquid chromatography system (Eksigent Technologies, Livermore, CA) equipped with a FAMOS Autosampler (Dionex-LC-Packings, San Francisco, CA). Mobile phase A was 0.1% formic acid in water, and mobile phase B was 0.1% formic acid in acetonitrile. After equilibration of the column in 5% solvent B, approximately one-tenth of each digest (1 μl) was injected; the organic content of the mobile phase was increased linearly to 40% over 30 min and then to 50% in 1 min. The liquid chromatography eluate was coupled to a micro-ionspray source attached to a QSTAR XL or to a QSTAR Pulsar mass spectrometer (Applied Biosystems/MDS Sciex, South San Francisco, CA). Peptides were analyzed in positive ion mode. MS spectra were acquired for 1 s in the m/z range of 310 to 1,400. MS acquisitions were followed by 3-s collision-induced dissociation (CID) experiments in information-dependent acquisition mode. For each MS spectrum, the most intense multiple charged peaks over a threshold of 30 counts were selected for generation of CID mass spectra. The most common trypsin autolysis products were excluded (m/z = 523.280, 421.750, and 737.700). The CID collision energy was automatically set according to mass-to-charge (m/z) ratio and charge state of the precursor ion. A dynamic exclusion window was applied that prevented the same m/z from being selected for 1 min after its acquisition.
Data were analyzed with AnalystQS software (Applied Biosystems/MDS Sciex, South San Francisco, CA), and peak lists were generated by using the mascot.dll script. MS centroiding parameters were 50% peak height and a 0.02 amu merge distance. MS/MS centroiding parameters were 50% peak height and an 0.05 amu merge distance. The peak list was searched against the UniProt database as of 24 January 2006 by using ProteinProspector (4). Carbamidomethylation and carboxymethylation of cysteine, N acetylation of the N terminus of the protein, oxidation of methionine, phosphorylation of serine, threonine or tyrosine, mono-, di-, and trimethylation of lysine, and mono- and dimethylation of arginine were allowed as variable modifications. Peptide tolerance in MS and MS/MS modes was 150 ppm and 0.2 Da, respectively. The CID spectra with possible posttranslational modifications were further inspected manually.
Hits were considered significant when three or more peptide sequences matched a protein entry and the Prospector score was above the significant level (4). For identifications based on one or two peptide sequence with high MASCOT scores, the MS/MS spectrum was reinterpreted manually by matching all of the observed fragment ions to a theoretical fragmentation obtained by using MS Product (ProteinProspector) (4).
Analysis of relative abundance levels of methylated peptides.
Analysis by ProteinProspector of the fragment ions generated in CID experiments allowed the identification of several potentially modified peptides. These were confirmed by manual inspection. These peptides reveal the presence of mono- and dimethylation of R727, mono- and dimethylation of R741, and monomethylation of R736. In order to compare the abundance of these species between the different samples, MS data acquired from the tryptic digests of the protein purified from uninfected, 18-h-infected, or 36-h-infected cells were searched, looking for ions with the specific m/z values corresponding to the identified modified peptides (listed in the legend of fig 4). We plotted the extracted ion chromatograms (XIC) for ions of a given m/z and identified the chromatographic maximum of the peak that corresponds to the elution of the chemical species of interest. If several local maxima for a given m/z value were found on the ion current, ions with the same m/z values, with identical charge state and equivalent retention times in the chromatographic elution profiles (coelution with the same set of peptides according to the simultaneous presence on the MS spectra of ion peaks at defined m/z values) were chosen to be compared. The coincidence in these parameters suggests the identical chemical composition of these species found in the different samples. The number of ion counts for every ion at the maximum of the XIC peak was normalized against the maximum intensities reached in that particular sample by two reference ions at 633.0085+3 and 344.8776+3. This was done in order to be able to compare values between samples. These reference ions are very intense peaks corresponding to tryptic peptides from L4 100K from regions which have not been observed to present modifications (S259 to K275 and A668 to R675), so their intensities can be used as a reference to compensate for variations on the sensitivity of the instrument between different runs. Normalized ion counts are an average of the normalized values against both reference peaks for every ion.
FIG. 4.
Methylated 100K is exclusively nuclear and RGG mutants are defective for nuclear accumulation. (A) U2OS cells were transfected with myc-100K or myc-100KΔC; infected with dl309; and harvested at 18 and 36 h p.i. in a hypotonic lysis buffer. Lysates were then fractionated into nuclear and cytoplasmic fractions, and equal amounts were resolved by using an SDS-Tris-glycine 4 to 20% gradient gel and detected by immunoblotting with specific antibodies as indicated. N, nuclear fraction; C, cytoplasmic fraction; α-methyl-R, α-methyl arginine antibody. (B) Ionic currents detected in MS mode for selected ions at their maximum elution times. The sequences of the ions surveyed are EAAAAAATHGR*GGILGQSGR (ion at m/z = 467.0+4, containing monomethylated Arg727), EAAAAAATHGR**GGILGQSGR (ion at m/z = 470.5+4, dimethylated Arg727), GGFGR*GGGGHDGR (ion at m/z = 400.9+3, monomethylated Arg741), GGFGR**GGGGHDGR (ion at m/z = 405.5+3, dimethylated Arg741), and EAAAAAATHGR**GGILGQSGR* (ion at m/z = 474.0+4, simultaneously containing dimethylated R727 and monomethylated R736). R*, methylated arginine; R**, dimethylated arginine. The intensities of the different ions corresponding to species mono- or dimethylated at the same residue have been grouped and presented (as averaged values) in the graph. ColumnS: R727, ions at m/z = 467.0+4 and 470.5+4; R741, ions 400.9+3 and 405.5+3; R736R727, ion 474.0+4. M, monomethylated; DiM, dimethylated. Normalized ion counts are an average of the maximum intensity for every ion normalized against the maximum intensities of the reference peaks, as described in Materials and Methods. (C) Methylation inhibitors prevent 100K nuclear accumulation. Cells were infected with dl309, treated with 3 mM MTA at 12 h p.i., fixed with 4% PFA at 36 h p.i., and stained with a monoclonal antibody against 100K.
RESULTS
RGG boxes are well conserved in 100K and necessary for the nuclear accumulation of 100K during infection.
100K is a multifunctional protein critical for adenovirus functions during the late stages of infection. It is found both in the nucleus and the cytoplasm (10), but it is not clear how 100K switches between the different functions during infection. Since many multifunctional proteins are controlled by posttranslational modifications, we examined 100K for sequences that are potential sites for this type of regulation. The main structural characteristics of 100K include three coil-coil recognition motifs, which mediate protein-protein interactions and an RNA recognition motif at the central part of the protein which is involved in RNA binding (Fig. 1). A Rev-like nuclear export sequence was recently identified within this domain, and the EIF4G binding site was mapped to be between amino acids 280 to 345 (7). The C terminus of the protein contains RGG- and RGR-like sequences that are well conserved in 100K proteins from all sequenced serotypes (Fig. 1), and recent work suggested that this domain contains an NLS (7). Moreover, most adenovirus serotypes also contain several RG and RXR motifs upstream of the RGG domain within the C terminus of the protein (data not shown).
FIG. 1.
The RGG domain is well conserved at the C terminus of many adenovirus serotypes. The C terminus of Ad5 100K contains several RG and RGR motifs and an RGG domain between amino acids 727 and 743 with three RGG tripeptides. 100KΔC was constructed by deleting amino acids 651 to 805. We also constructed three mutant clones with each of the three RGG motifs with the RGG domain mutated to alanine residues.
In order to further study the function of this part of the protein in the context of a complete viral infection, we generated a C terminus truncated form of 100K (amino acids 651 to 805) lacking the RGG domain and the upstream RGR- and RG-containing region of the protein (myc-100KΔC), as well as three additional mutant clones, each of which has one of the three RGG sequences within the RGG box changed to alanine residues (100KRGG1, 100KRGG2, and 100KRGG3; Fig. 1). U2OS cells were transfected with wild-type 100K epitope tagged at the N terminus with the myc tag or with myc-tagged mutant clones and infected with wild-type Ad5. We then performed immunofluorescence studies to determine 100K localization during the course of infection. As shown in Fig. 2, nontagged, virus-coded 100K is in the cytoplasm during the earlier phase of infection, which is consistent with its initial involvement in host protein synthesis shutoff. During the later stages of infection 100K relocalizes to the nucleus (40 h postinfection [p.i.]), a finding consistent with the second 100K function in hexon trimerization and assembly, a process which is known to occur in the nucleus. In fact, 100K localization to the nucleus occurs at about the same time as hexon is first expressed, and the two proteins seem to move to the nucleus at the same time (data not shown), which also supports a previously suggested possible role of 100K in hexon nuclear transport. Transfected wild-type myc-100K localizes similarly to the 100K expressed from the virus: at 18 h and 40 h p.i. it localizes to the cytoplasm and nucleus, respectively. In contrast, myc-100KΔC remains cytoplasmic throughout infection. These data suggest that the C terminus, containing the RGG domain, is necessary for the nuclear accumulation of 100K. Therefore, we tested our three RGG mutants for their ability to relocalize to the nucleus after infection. myc-100KRGG1 and myc-100KRGG2 behaved like wild-type myc-100K, suggesting that these two boxes are not involved in the nuclear accumulation of the protein (data not shown). However, mutation of the third RGG tripeptide (amino acids 741 to 743) to alanine residues (myc-100KRGG3) resulted in a protein that failed to localize to the nucleus during the late phase of infection. These results suggest that the third RGG box within the carboxy terminus of 100K is the site that regulates 100K localization to the nucleus during the late phase of infection.
FIG. 2.
RGG mutants fail to localize to the nucleus during infection. U2OS cells were transfected with plasmids expressing myc-tagged wild-type 100K, a C terminus truncated 100K (100KΔC), or an RGG mutant with positions 741 to 743 changed to alanine residues. 100KΔC has a deletion of amino acids 680 to 805, which encompasses the arginine-glycine rich region with the RGG, RGR, and RG motifs. Untransfected and transfected cells were infected with dl309 (wild-type Ad5). Slides were fixed at 18 or 40 h p.i. and stained with α-100K (two leftmost panels) or α-myc antibodies as indicated.
100K is methylated and binds PRMT1 methylase through its RGG domain.
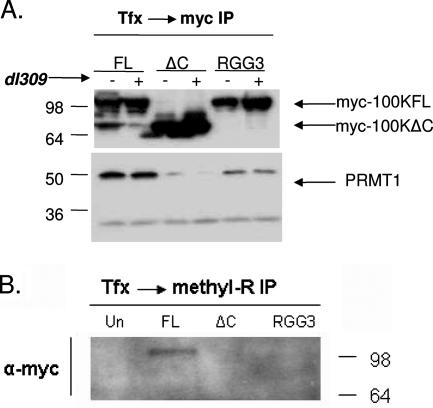
RGG boxes are sequences known in many cases to regulate nucleocytoplasmic localization of the protein, a function modulated by methylation on arginine residues adjacent to glycines. Recently, 100K was shown to be methylated during infection by PRMT1 methylase (11), which is the most predominant methylase in the cell. Therefore, we tested whether 100K is methylated at its RGG domain and whether this domain is necessary for the interaction with PRMT1. U2OS cells were transfected with wild-type myc-100K, myc-100KΔC, or myc-100KRGG3; infected with wild-type Ad5; and subjected to coimmunoprecipitation analysis with α-myc antibodies. Wild-type myc-100K coimmunoprecipitated with PRMT1 both in infected and uninfected cells (Fig. 3A). However, myc-100KΔC binding to PRMT1 is extremely reduced, showing that the C terminus of the protein is significant for PRMT1 binding. Moreover, binding of myc-100KRGG3 to PRMT1 was greatly reduced compared to the wild-type protein, implicating these particular residues as significant for the interaction between 100K and PRMT1.
FIG. 3.
100K is methylated and binds PRMT1 methylase through its RGG domain. (A and B) Cells were transfected with myc-100K, myc-100KΔC, or myc-100KRGG3; infected with dl309 when indicated; and harvested at 24 h p.i. Equal amounts of lysates were subjected to immunoprecipitation with α-myc (A) or α-methyl arginine (B) antibodies, and samples were resolved by using an SDS-Tris-glycine 4 to 20% gradient gel and detected by immunoblotting with specific antibodies as shown.
In order to show that 100K is methylated, myc-100KFL, myc-100KΔC, and myc-100KRGG3 were transfected in U2OS cells, immunoprecipitation was performed by using an antibody against dimethylated arginine residues, and samples were blotted with an α-myc antibody. As shown in Fig. 3B, myc-100KFL was pulled down using the α-dimethyl-arginine antibody. In contrast, myc-100KΔC and myc-100KRGG3 were not detected. These results show that the C terminus of 100K is the site of methylation and that removing this part of the molecule abolishes this type of posttranslational modification. More specifically, the third RGG box (amino acids 741 to 743) seems to be one of the 100K methylation sites since, similar to the complete C terminus deleted mutant, the myc-100KRGG3 shows reduced binding to PRMT1 (Fig. 3A) and does not immunoprecipitate with the α-dimethyl-arginine antibody (Fig. 3B).
Methylated 100K is exclusively nuclear and RGG mutants are defective for nuclear accumulation.
Because posttranslational methylation modulates subcellular localization of several RNA-binding proteins, we hypothesized that the C-terminal RGG containing region may be involved in 100K localization to the nucleus, potentially as a result of methylation of that domain. In support of this hypothesis, myc-100KRGG3 fails to localize to the nucleus (Fig. 2) and is not methylated (Fig. 3B). To further test this hypothesis, we first determined the subcellular localization of methylated myc-100KFL. U2OS cells were transfected with myc-100KFL, or myc-100KΔC as a control, and infected with wild-type Ad5, collected at 18 h and 36 h p.i., and lysates were fractionated to separate the nuclear and the cytoplasmic fractions. Nuclear and cytoplasmic fractions were then subjected to Western blot analysis with an α-methyl-arginine antibody. Figure 4A shows that methylated 100K is already present at 18 h p.i., and it increases at later stages of infection. More importantly, it remains exclusively nuclear. In contrast, in cells expressing the C terminus truncated protein, used here as a control, myc-100KΔC is not methylated, as expected from the fact that the 100K methylation site is within the C terminus of the molecule. The absence of a 100-kDa methylated band in the cells transfected with myc-100KΔC also confirms that the 100-kDa band present in the myc-100KFL transfected cells is the myc-100KFL protein and not a cellular protein that becomes methylated upon infection. Thus, our results from this experiment suggest that the function of methylated 100K must be in the nucleus.
To further define the role of arginine methylation on 100K, we searched for posttranslational modifications on the full-length protein by using reversed-phase LC-MS/MS analysis (Fig. 4B). Full-length 100K was purified from a stable cell line expressing TAP-tagged 100K and was subjected to LC-MS/MS analysis. To further characterize the role of methylation in the regulation of 100K localization during infection, we also purified TAP-tagged 100K from cells infected with wild-type adenovirus and harvested at 18 and 36 h p.i. Similar to the results shown in Fig. 2, 100K is cytoplasmic at 18 h p.i. and nuclear at 36 h p.i. in the TAP-100K stable cell line (data not shown). Analysis by ProteinProspector of the fragment ions generated in CID experiments allowed the identification of several potentially modified peptides. These were confirmed by manual inspection. These peptides reveal the presence of mono- and dimethylation of R727, mono- and dimethylation of R741, and monomethylation of R736 (Fig. 4B). Interestingly, the peptide GGFGR**GGGGHDGR containing methylated Arg741 (RGG3 box) was mostly identified in samples from uninfected cells and cells infected for 36 h and appeared at low levels in infected cells harvested at 18 h p.i. Thus, 100K methylation within the third RGG3 box is mostly detected when 100K is nuclear. The low levels of methylated 100KRGG3 in the cytoplasm (18-h sample) must be the 100K molecules methylated immediately before the cells were harvested for analysis and therefore had not localized to the nucleus yet. These results, suggest that methylation of 100K on R741 acts as a signal for the nuclear accumulation of the protein.
Finally, to further prove that arginine methylation is a posttranslational modification necessary for the nuclear accumulation of 100K, we treated Ad5-infected U2OS cells with methyl-thio-adenosine (MTA) a methylation inhibitor naturally generated in the cells as a metabolic intermediate in the conversion of putrescine to spermidine and of spermidine to spermine (27). Cells were infected with Ad5, treated with 0.3 mM MTA 18 h p.i., fixed with 4% paraformaldehyde (PFA) 36 h p.i., and stained with an α-100K antibody. Using confocal microscopy, we show that in adenovirus-infected cells treated with MTA, 100K fails to localize to the nucleus and is exclusively cytoplasmic, in contrast to untreated cells where 100K is already mostly nuclear at 36 h p.i. (Fig. 4C). Taken together, these results suggest that cellular arginine methylation is necessary for the nuclear accumulation of 100K.
100K-mediated host protein synthesis shutoff requires the C terminus.
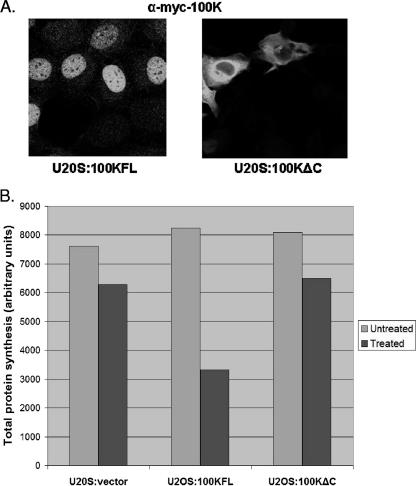
According to the current model of adenovirus-induced inhibition of cellular protein synthesis, 100K binds to the translation initiation complex EIF4F, the cap-binding complex required for cap-dependent translation, and displaces mnk1 kinase, which normally phosphorylates EIF4F (5, 7). Dephosphorylation of EIF4F is thought to inhibit the ability of the complex to bind to the cap structure on cellular RNA messages, and consequently cap-dependent translation is inhibited. The 100K binding site to EIF4F has been mapped to amino acids 280 to 365 (7), and this site was shown by others to be sufficient to displace mnk1. Thus, according to this model, a 100K stable cell line should be impossible to make, since inhibition of host protein synthesis would be toxic to the cell. However, we were able to generate such cell lines, by selecting U2OS cells transfected with myc-tagged 100KFL and 100KΔC. Since we already showed that arginine methylation of 100K results in the nuclear accumulation of this protein, we hypothesized that in our U2OS:100KFL stable cell line, myc-100KFL, may localize in the nucleus and therefore may not be available for binding to EIF4F in the cytoplasm. To test this hypothesis, U2OS:100KFL and U2OS:100KΔC cells were plated on chamber glass slides, fixed, and stained against the myc tag of the 100K clones in order to determine 100K subcellular localization. As shown in Fig. 5A, myc-100KFL is exclusively nuclear in the U2OS:100KFL stable cell line, which explains why 100K is not toxic to these cells, since nuclear 100K is not available to bind to the EIF4F complex in the cytoplasm. In contrast, myc-100KΔC was exclusively cytoplasmic, as expected from the results shown in Fig. 2, and the lack of the RGG domain which is necessary for nuclear accumulation. Nevertheless, this clone still contains the whole amino terminus of 100K, which includes the proposed binding site to EIF4F (amino acids 280 to 365) and should therefore be capable of binding to the translation initiation complex and induce shutoff of host protein synthesis. Thus, in order to further characterize 100K involvement in the inhibition of host protein synthesis and the role of arginine methylation in 100K function, we measured total protein synthesis in U2OS:vector (nonexpressing control), U2OS:100KFL, and U2OS:100KΔC cells in the presence or absence of the methylation inhibitor MTA (cells were treated with MTA for 12 h) by using S35 labeling. As shown in Fig. 5B, protein synthesis is not inhibited in the untreated U2OS cells expressing the FL or the ΔC 100K proteins, relative to the nonexpressing U2OS:vector control cells. In contrast, inhibiting methylation in U2OS:100KFL cells resulted in twofold-lower amounts of protein synthesized compared to the control cells. However, protein synthesis in U2OS:100KΔC cells remained unaffected. These results support our previous findings that arginine methylation is a posttranslational modification that controls 100K function by means of changing its localization during infection. Moreover, the fact that inhibiting methylation in the U2OS:100KΔC cell line did not have any effect on the overall protein synthesis suggests that the effects of inhibiting methylation in the U2OS:100KFL cell line are specific to 100K. In addition, these findings suggest that the carboxy terminus of 100K is necessary for host protein synthesis inhibition, in contrast to the current model of adenovirus-induced shutoff that proposes that amino acids 280 to 345 are sufficient for this function of the protein.
FIG. 5.
(A) U2OS:100KFL and U2OS:100KΔC cells were grown on glass chambers, fixed with 4% PFA, stained with an antibody against the myc tag and a fluorescently labeled secondary antibody, and visualized by confocal microscopy. In U2OS:100KFL cells, 100K is exclusively nuclear. However, the lack of the RGG domain in U2OS:100KΔC cells results in the cytoplasmic accumulation of 100K. (B) U2OS:vector, U2OS:100KFL, and U2OS:100KΔC cells were plated in the presence or absence of methylation inhibitor, labeled with S35 for 2 h, and harvested, and total lysates were run on a 4 to 20% gel, transferred to a polyvinylidene difluoride membrane, and visualized on PhosphorImager. Total protein synthesis was measured by using the ImageQuant software, as previously described (31), and the absolute value for each cell line is shown here (the results are the average value from two independent experiments).
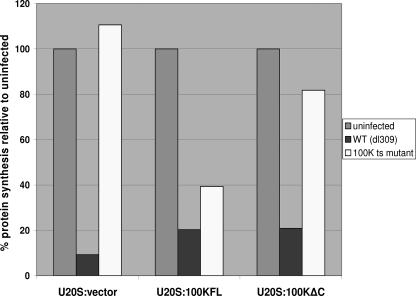
To further determine the importance of the C terminus of 100K in 100K-mediated shutoff, we used a 100K ts mutant virus (Ad2 ND1.ts4), which at the nonpermissive temperature (39.4°C) produces a defective 100K protein. Using this virus, and wild-type dl309 as a control, U2OS:vector, U2OS:100KFL, and U2OS:100KΔC cell lines were infected at the nonpermissive temperature; at 36 h p.i. the cells were S35 labeled for 1 h and harvested, and the total protein synthesis was measured, as described previously (21). The 100K ts mutant virus was defective for host protein synthesis shutoff, since in U2OS:control cells protein synthesis was not inhibited. In contrast, U2OS:100KFL cells complemented 100K-mediated shutoff, resulting in a 60% reduction in overall host protein synthesis (Fig. 6). The ability of stably expressed 100K to induce shutoff required, at least partially, the C terminus of the molecule, since U2OS:100KΔC cells were not able to support shutoff and showed only a partial reduction (18%) in the overall amounts of cellular proteins synthesized.
FIG. 6.
U2OS:vector, U2OS:100KFL, and U2OS:100KΔC cells were infected with wild type or with an 100K ts mutant adenovirus at the nonpermissive temperature (39.4°C). At 36 h p.i., cells were labeled with S35 and harvested, and lysates were run on a 4 to 20% Tris-glycine gel as described in Materials and Methods. Proteins were transferred onto a polyvinylidene difluoride membrane, and the total labeled protein was visualized by using PhosphorImager. The total protein synthesis was measured by using ImageQuant and is depicted on this graph as a percentage relative to the protein synthesis in uninfected cells. The results are the average values from two independent experiments.
Expression of late viral protein production is severely defective in MTA-treated cells.
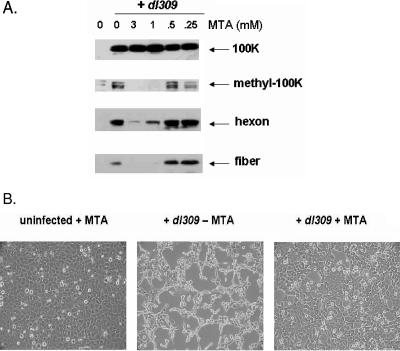
Cellular arginine methylation is necessary for 100K localization to the nucleus and may be an important type of posttranslational modification necessary for 100K function and vital to the virus life cycle. To test this hypothesis and to further define the role of arginine methylation during adenovirus infection, we investigated the late protein expression profile of U2OS cells infected with dl309 while treated with several concentrations of MTA at 12 h p.i. Total lysates were run on a 4 to 20% gradient gel and blotted with monoclonal antibodies against individual viral proteins (Fig. 7A). As shown in the figure, the expression of late adenovirus proteins in MTΑ-treated cells was severely reduced in a dose-dependent manner. The levels of methylated 100K were greatly reduced at higher concentrations of MTA, while the total 100K levels remained constant.
FIG. 7.
(A) Expression of late viral protein production is severely defective in MTA-treated cells. U2OS cells were infected with dl309 and treated with various concentrations of MTA at 12 h p.i.; lysates were normalized and resolved by using an SDS-Tris-glycine 4 to 20% gradient gel at 36 h p.i. and detected by immunoblot analysis using monoclonal antibodies against 100k, hexon, and fiber. Methyl-100K was detected by using an α-methyl arginine antibody. (B) U2OS cells were infected with dl309 in the presence or absence of the methylation inhibitor (MTA, added 12 h p.i.) and visualized under a light microscope at 48 h p.i. As a control, uninfected U2OS cells treated with the inhibitor were also included.
To further determine the importance of arginine methylation during adenovirus infection, we examined the effects of methylation inhibitors on the ability of wild-type adenovirus to induce cytopathic effect (Fig. 7B). U2OS cells were infected with dl309 and either treated with the methylation inhibitor 12 h p.i. or left untreated. As a control, we also included uninfected cells treated with the inhibitor to test the effects of inhibiting methylation on infected cells. As shown in Fig. 7B, inhibition of arginine methylation protects U2OS cells from adenovirus-induced cytopathic effect, further showing that arginine methylation is critical to the adenovirus life cycle. Finally, we measured the viral yield produced from infected U2OS cells treated with 3 mM MTA at 12 h p.i. by using an enzyme-linked immunosorbent assay as described previously (13). As expected from our results in Fig. 7, the viral yield produced from the MTA-treated cells was at least 1,000-fold lower than the yield produced from the untreated cells (data not shown).
DISCUSSION
100K is a multifunctional protein vital to the late phase of adenovirus infection. Its expression is necessary for adenovirus-induced inhibition of cellular protein synthesis and viral morphogenesis. These two processes occur at different subcellular compartments and at different times during infection. Thus, 100K function must be temporally and spatially regulated. However, the mechanism by which 100K switches between these functions remains unknown. In the present study, we demonstrate that Ad5 100K localization is controlled by posttranslational methylation within its RGG domain, and we show that arginine methylation is a necessary cellular component for a productive adenovirus infection.
Nuclear localization of 100K depends on arginine methylation.
Recently, it was shown that C-terminal truncation of 100K resulted in cytoplasmic accumulation of the protein, suggesting that this part of the molecule contains an NLS (7). However, cells transfected with wild-type 100K show both nuclear and cytoplasmic accumulation of the protein, suggesting that there is an additional level of complexity in the regulation of 100K localization. Using the methylation inhibitor MTA, we show here that in the absence of arginine methylation, 100K is exclusively cytoplasmic, suggesting that methylation may be critical for its nuclear accumulation. In support of this hypothesis, we show that methyl 100K is exclusively nuclear, along with most of the methylated proteins in the cell.
A critical RGG sequence necessary for methylation and nuclear import.
To date, several other cellular and viral proteins have been found to be controlled by posttranslational methylation, mostly at the level of subcellular localization and RNA-binding activity (see reference 17 for a review). Posttranslational methylation mainly occurs on arginine residues within RGG and RG motifs collectively called RGG boxes. The C terminus of Ad5 100K contains an arginine-glycine rich domain with three RGG tripeptides. As shown in Fig. 1, the RGG domain, as well as many upstream RG, RGR, and RXR motifs, are very well conserved across many adenovirus serotypes, which suggests that these sequences are functionally important. Using an antibody that recognizes asymmetrically dimethylated arginine residues, we show that 100K is methylated within its C terminus and that this part of the molecule is necessary for binding to PRMT1 methylase, since a C-terminal truncated 100K clone (myc-100KΔC) failed to bind to the methylase and was not methylated. Myc-100KΔC clone localized exclusively in the cytoplasm, further implicating the RGG domain in nuclear accumulation of the protein. Transfected wild-type myc-100K, as well as a mutant clone with the third RGG tripeptide (amino acids 741 to 743) changed to alanine residues (myc-100KRGG3), localized both in the nucleus and in the cytoplasm in uninfected cells. However, even though the wild-type myc-100K shuttled into the nucleus after infection, the myc-100KRGG3 mutant did not. Thus, our results suggest that even though the whole RGG domain may act as an NLS, the third RGG box (amino acids 741 to 743) is important in 100K transport from the cytoplasm to the nucleus during infection, and posttranslational methylation of arginine 741 may be a significant modification for this process.
Arginine methylation is a cellular function necessary for productive adenovirus infection.
Since nuclear localization of 100K is important for its function, and arginine methylation is necessary for nuclear accumulation of the protein, we hypothesized that arginine methylation must be vital to the late phase of adenovirus infection. Indeed, treatment of Ad5-infected U2OS cells with the methylation inhibitor MTA resulted in greatly reduced overall expression of late viral proteins in a dose-dependent manner. This result, along with the fact that the overall levels of 100K and total cellular protein synthesis also remained unaffected in MTA-treated cells, suggests that the effect of inhibition of arginine methylation in our cells is a result of inhibition of 100K methylation and not a nonspecific effect on the overall protein synthesis in the cell.
There are several possibilities as to how methylation may alter 100K function. One would be that by merely changing localization from the cytoplasm to the nucleus, 100K switches between its cytoplasmic function (host-protein synthesis shutoff) and its nuclear functions (hexon transport to the nucleus and hexon trimerization and assembly). It is also possible that methylation of the protein alters its binding properties, thereby affecting protein-protein and protein-RNA interactions, in a way that several other proteins are known to be regulated. For example, the RGG domain of herpes simplex virus ICP27 protein mediates binding to viral intronless RNAs (24, 25) and an ICP27-null virus is defective for nuclear export of these transcripts. ICP27 was also shown to be regulated by posttranslational arginine methylation (12), and removing the RGG domain abolishes its RNA-binding activity (18). It is therefore possible that adenovirus 100K is regulated in the same way. In support of this hypothesis, methyl-100K can be detected during the early phase of 100K expression and considerably earlier than when hexon protein can be detected (unpublished observations), suggesting that 100K methylation is needed before 100K is involved in hexon trimerization and assembly and may therefore also play a role in regulating RNA transport of late viral messages. Even though more experiments are needed in order to fully understand the multicomponent 100K function and the role that arginine methylation plays in regulating it, our study shows that posttranslational methylation of 100K is necessary for its nuclear accumulation and that the third RGG box (amino acids 741 to 743) is necessary for 100K shuttling from the cytoplasm to the nucleus during infection. Moreover, the dramatic effects of inhibition of protein arginine methylation on the late viral protein expression and overall viral yield suggests that this type of posttranslational modification is a critical cellular function necessary for a productive adenovirus infection.
Acknowledgments
We thank Mike Fried, Pablo Rodriguez, and Sang Lee for critical reading of the manuscript. We also thank W. C. Russell for kindly providing monoclonal antibodies to 100K and Hamish Young for kindly providing the 100K ts virus. We especially thank Conrado Soria, Serah Choi, and Jamie Smith for technical assistance and discussions. We also thank Abby Miller, Maria Christophorou, and all of the members of the McCormick lab for thoughtful discussions and comments. We also thank Jessie Castillo for administrative support. Confocal microscopy was supported by the UCSF Comprehensive Cancer Center Core facility, with thanks to Jane Gordon and Sarah Elmes.
The UCSF Mass Spectrometry Facility was supported by the Biomedical Research Technology Program of the National Center for Research Resources, NIH NCRR RR015804, NIH NCRR RR001614, and NIH NCRR RR012961. C.C.O. was supported by a Leukemia Society of America Fellowship. F.M. and D.C.I. were supported, in part, by ONYX Pharmaceuticals and UC Discovery grant bio-02-10242, and F.M. is a shareholder in ONYX Pharmaceuticals.
Footnotes
Published ahead of print on 8 August 2007.
REFERENCES
- 1.Bedford, M. T., A. Frankel, M. B. Yaffe, S. Clarke, P. Leder, and S. Richard. 2000. Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J. Biol. Chem. 275:16030-16036. [DOI] [PubMed] [Google Scholar]
- 2.Bello, L. J., and H. S. Ginsberg. 1967. Inhibition of host protein synthesis in type 5 adenovirus-infected cells. J. Virol. 1:843-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cepko, C. L., and P. A. Sharp. 1983. Analysis of Ad5 hexon and 100K ts mutants using conformation-specific monoclonal antibodies. Virology 129:137-154. [DOI] [PubMed] [Google Scholar]
- 4.Clauser, K. R., P. Baker, and A. L. Burlingame. 1999. Role of accurate mass measurement (±10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem. 71:2871-2882. [DOI] [PubMed] [Google Scholar]
- 5.Cuesta, R., Q. Xi, and R. J. Schneider. 2000. Adenovirus-specific translation by displacement of kinase Mnk1 from cap-initiation complex eIF4F. EMBO J. 19:3465-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuesta, R., Q. Xi, and R. J. Schneider. 2001. Preferential translation of adenovirus mRNAs in infected cells. Cold Spring Harbor Symp. Quant. Biol. 66:259-267. [DOI] [PubMed] [Google Scholar]
- 7.Cuesta, R., Q. Xi, and R. J. Schneider. 2004. Structural basis for competitive inhibition of eIF4G-Mnk1 interaction by the adenovirus 100-kilodalton protein. J. Virol. 78:7707-7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolph, P. J., V. Racaniello, A. Villamarin, F. Palladino, and R. J. Schneider. 1988. The adenovirus tripartite leader may eliminate the requirement for cap-binding protein complex during translation initiation. J. Virol. 62:2059-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gabler, S., H. Schutt, P. Groitl, H. Wolf, T. Shenk, and T. Dobner. 1998. E1B 55-kilodalton-associated protein: a cellular protein with RNA-binding activity implicated in nucleocytoplasmic transport of adenovirus and cellular mRNAs. J. Virol. 72:7960-7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gambke, C., and W. Deppert. 1981. Late nonstructural 100,000- and 33,000-dalton proteins of adenovirus type 2. I. Subcellular localization during the course of infection. J. Virol. 40:585-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayes, B. W., G. C. Telling, M. M. Myat, J. F. Williams, and S. J. Flint. 1990. The adenovirus L4 100-kilodalton protein is necessary for efficient translation of viral late mRNA species. J. Virol. 64:2732-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hibbard, M. K., and R. M. Sandri-Goldin. 1995. Arginine-rich regions succeeding the nuclear localization region of the herpes simplex virus type 1 regulatory protein ICP27 are required for efficient nuclear localization and late gene expression. J. Virol. 69:4656-4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson, L., A. Shen, L. Boyle, J. Kunich, K. Pandey, M. Lemmon, T. Hermiston, M. Giedlin, F. McCormick, and A. Fattaey. 2002. Selectively replicating adenoviruses targeting deregulated E2F activity are potent, systemic antitumor agents. Cancer Cell 1:325-337. [DOI] [PubMed] [Google Scholar]
- 14.Kiledjian, M., and G. Dreyfuss. 1992. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 11:2655-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kzhyshkowska, J., E. Kremmer, M. Hofmann, H. Wolf, and T. Dobner. 2004. Protein arginine methylation during lytic adenovirus infection. Biochem. J. 383:259-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lykke-Andersen, J., M. D. Shu, and J. A. Steitz. 2000. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103:1121-1131. [DOI] [PubMed] [Google Scholar]
- 17.McBride, A. E., and P. A. Silver. 2001. State of the arg: protein methylation at arginine comes of age. Cell 106:5-8. [DOI] [PubMed] [Google Scholar]
- 18.Mears, W. E., and S. A. Rice. 1996. The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA-binding activity and determines in vivo methylation. J. Virol. 70:7445-7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morin, N., and P. Boulanger. 1986. Hexon trimerization occurring in an assembly defective, 100K temperature-sensitive mutant of adenovirus 2. Virology 152:11-31. [DOI] [PubMed] [Google Scholar]
- 20.Oosterom-Dragon, E. A., and H. S. Ginsberg. 1981. Characterization of two temperature-sensitive mutants of type 5 adenovirus with mutations in the 100,000-dalton protein gene. J. Virol. 40:491-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Shea, C., L. Johnson, B. Bagus, S. Choi, C. Nicholas, A. Shen, L. Boyle, K. Pandey, C. Soria, J. Kunich, Y. Shen, G. Habets, D. Ginzinger, and F. McCormick. 2005. Late viral RNA export, rather than p53 inactivation, determines ONYX-015 tumor selectivity. Cancer Cell 8:61-74. [DOI] [PubMed] [Google Scholar]
- 22.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 23.Russell, W. C., G. Patel, B. Precious, I. Sharp, and P. S. Gardner. 1981. Monoclonal antibodies against adenovirus type 5: preparation and preliminary characterization. J. Gen. Virol. 56:393-408. [DOI] [PubMed] [Google Scholar]
- 24.Sandri-Goldin, R. M. 1998. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 12:868-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sandri-Goldin, R. M. 1998. Interactions between a herpes simplex virus regulatory protein and cellular mRNA processing pathways. Methods 16:95-104. [DOI] [PubMed] [Google Scholar]
- 26.Wang, X., A. Flynn, A. J. Waskiewicz, B. L. Webb, R. G. Vries, I. A. Baines, J. A. Cooper, and C. G. Proud. 1998. The phosphorylation of eukaryotic initiation factor eIF4E in response to phorbol esters, cell stresses, and cytokines is mediated by distinct MAP kinase pathways. J. Biol. Chem. 273:9373-9377. [DOI] [PubMed] [Google Scholar]
- 27.Williams-Ashman, H. G., J. Seidenfeld, and P. Galletti. 1982. Trends in the biochemical pharmacology of 5′-deoxy-5′-methylthioadenosine. Biochem. Pharmacol. 31:277-288. [DOI] [PubMed] [Google Scholar]
- 28.Yueh, A., and R. J. Schneider. 1996. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev. 10:1557-1567. [DOI] [PubMed] [Google Scholar]
- 29.Yueh, A., and R. J. Schneider. 2000. Translation by ribosome shunting on adenovirus and hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes Dev. 14:414-421. [PMC free article] [PubMed] [Google Scholar]
- 30.Yun, C. Y., and X. D. Fu. 2000. Conserved SR protein kinase functions in nuclear import and its action is counteracted by arginine methylation in Saccharomyces cerevisiae. J. Cell Biol. 150:707-718. [DOI] [PMC free article] [PubMed] [Google Scholar]