Abstract
A short open reading frame named the “U exon,” located on the adenovirus (Ad) l-strand (for leftward transcription) between the early E3 region and the fiber gene, is conserved in mastadenoviruses. We have observed that Ad5 mutants with large deletions in E3 that infringe on the U exon display a mild growth defect, as well as an aberrant Ad E2 DNA-binding protein (DBP) intranuclear localization pattern and an apparent failure to organize replication centers during late infection. Mutants in which the U exon DNA is reconstructed have a reversed phenotype. Chow et al. (L. T. Chow et al., J. Mol. Biol. 134:265-303, 1979) described mRNAs initiating in the region of the U exon and spliced to downstream sequences in the late DBP mRNA leader and the DBP-coding region. We have cloned this mRNA (as cDNA) from Ad5 late mRNA; the predicted protein is 217 amino acids, initiating in the U exon and continuing in frame in the DBP leader and in the DBP-coding region but in a different reading frame from DBP. Polyclonal and monoclonal antibodies generated against the predicted U exon protein (UXP) showed that UXP is ∼24K in size by immunoblot and is a late protein. At 18 to 24 h postinfection, UXP is strongly associated with nucleoli and is found throughout the nucleus; later, UXP is associated with the periphery of replication centers, suggesting a function relevant to Ad DNA replication or RNA transcription. UXP is expressed by all four species C Ads. When expressed in transient transfections, UXP complements the aberrant DBP localization pattern of UXP-negative Ad5 mutants. Our data indicate that UXP is a previously unrecognized protein derived from a novel late l-strand transcription unit.
An open reading frame (ORF) named the “U exon,” located on the l strand (for leftward transcription) between the early E3 region and the fiber gene, was first identified after the sequencing of Ad40 (9). Davison et al. (9) reported that the U exon that is conserved in mastadenoviruses would encode just over 50 amino acids (aa) for most mastadenoviruses and might encode the N terminus of a larger protein. U exon sequences are conserved within each of the four Ad genera but not between genera (10). Davison et al. (10) suggested that the U exon is an ancient feature in adenoviruses (Ads; all Ad genera genomes encode capsid proteins and the E2 proteins within the central region of the Ad genome; the U exon is encoded within this region).
In our work with a number of Ad E3 deletion mutants, we noted that mutants with deletions extending into the U exon have a mild growth defect and exhibit an altered Ad DNA-binding protein (DBP) (also known as E2 72K protein) intranuclear localization pattern when analyzed by indirect immunofluorescence. Reconstruction of the U exon DNA sequence reversed the phenotype of these mutants. Polyclonal and monoclonal antibodies were generated against peptides predicted from the putative U exon protein (UXP) ORF. We describe here the identification and characterization of UXP.
MATERIALS AND METHODS
Cell lines and adenoviruses.
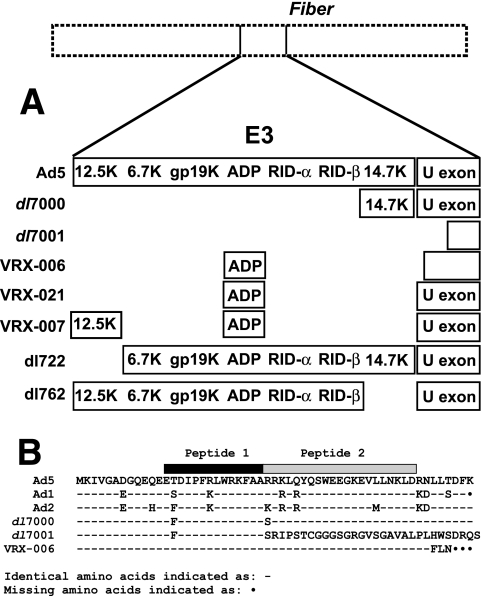
A549 cells (American Type Culture Collection [ATCC], Manassas, VA), HEK293 cells (Microbix, Toronto, Ontario, Canada), 21f1 cells (HeLa cells stably expressing DBP) (6), and A549E1 cells (A549 cells stably transfected with E1 sequences [bp 552 to 4090] under control of a mouse mammary tumor virus promoter; obtained from Genetic Therapy, Inc., Rockville, MD) were grown in Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS). HEL299 cells (human diploid embryonic lung cells; ATCC) were grown in DMEM with 10% FBS supplemented with nonessential amino acids and sodium pyruvate (1 mM). KB suspension culture cells were grown in Joklik's suspension culture medium containing 5% horse serum. Wild-type viruses used were Ad5 (ATCC VR-5), Ad1 (ATCC 1078-VR), Ad2, and Ad6 (ATCC 1083-VR). The Ad5-derived mutants used were dl327 (16), VRX-006, VRX-007 (13), VRX-021 (see below), dl7000 (42), and dl7001 (17, 42). The Ad E3 mutants dl722 (Δ12.5K) and dl762 (Δ14.7K) (5) were also used. Figure 1A is a schematic of the E3 genes present in these mutants, and Fig. 1B shows the U exon sequence present in these mutants.
FIG. 1.
Characteristics of the virus mutants used in the present study. (A) E3 regions of the mutants. Genes are indicated when present. Mutants dl7001 and VRX-006 have deletions that extend into the U exon region (depicted as smaller size of the U exon). Mutants dl722 and dl762 have deletions of the 12.5K and 14.7K genes, respectively. (B) U exon amino acid sequences of these viruses as deduced from the DNA sequence of Ad mutants through the U exon region.
Construction of VRX-006 and VRX-021.
The construction of mutants VRX-006 and VRX-007 has been described (13). In VRX-006 all E3 sequences between the L4 poly(A) site and a site (bp 30883) downstream of the E3B poly(A) site and upstream of the ORF for the fiber protein are deleted. The adp gene was reinserted into a newly created PacI site.
The virus mutant VRX-021 was constructed by using a shuttle plasmid encoding a version of the E3 region that is a hybrid between VRX-006 and VRX-007. Plasmid pA81 has a deletion of Ad5 bp 27858 to 27860 and an insert of TAA; deletion of Ad5 bp 27982 to 28134; deletion of Ad5 bp 28395 to 29397 and insert of CCTTAATTAAA; deletion of Ad5 bp 29783 to 30469. To construct VRX-021, pA81 was cotransfected into HEK293 cells along with dl327 DNA digested with EcoRI and SpeI. The resulting plaques were expanded on HEK293 cells and were analyzed by PCR and sequencing of the E3 region. A plaque of VRX-021 was plaque purified three times and expanded in A549 cell monolayers and KB spinner cells (45).
Antibodies.
Two peptides were synthesized and conjugated to keyhole limpet hemocyanin (Sigma-Geneosys; Sigma-Aldrich, St. Louis, MO). Peptide 1 (CETDIPFRLWRKFAA) corresponds to aa 13 to 26 of the Ad5 UXP and peptide 2 (CRRKLQYQSWEEGKEVLLNKLD) corresponds to aa 27 to 47 of the Ad5 UXP (Fig. 1B). These peptides were used for immunization of New Zealand White rabbits (Harlan, Indianapolis, IN) in the Saint Louis University Department of Comparative Medicine animal facilities and for generation of mouse monoclonal antibodies in the Saint Louis University Custom Hybridoma Development Service Facility (medschool.slu.edu/antibody/).
Infections.
Cells were infected with species C Ads (Ad1, Ad2, Ad5, and Ad6) or virus mutants at 10 to 50 PFU/cell as indicated in figure legends. Time points were taken at 18 to 48 h postinfection (p.i.) for immunoblots or 18 to 45 h p.i. for indirect immunofluorescence. 1-β-d-Arabinofuranosylcytosine (Ara-C) was added in some cases at a concentration of 20 μg/ml to inhibit Ad DNA replication and progression into the late stage of Ad infection; Ara-C was replenished at 10- to 12-h intervals. Actinomycin D was added at 1 μg/ml to some infections to inhibit RNA polymerase I.
Indirect immunofluorescence.
Cells were plated on #1 glass coverslips 1 to 2 days prior to infection. Cells were infected with 10 to 50 PFU/cell of virus as indicated in figure legends. At specific times p.i. (18 to 45 h), cells were fixed in 3.7% paraformaldehyde and subsequently permeabilized with methanol containing DAPI (4′,6′-diamidino-2-phenylindole) (48). Cells were incubated with anti-UXP primary antibodies. In some experiments, rabbit antiserum specific for the DBP C-terminal peptide (27) was used. The secondary antibodies were goat anti-rabbit immunoglobulin G (IgG) or goat anti-mouse IgG (Alexa Fluor 488 conjugates; Molecular Probes; Invitrogen Corp., Carlsbad, CA). When rabbit anti-DBP antiserum was used for costaining with the monoclonal anti-UXP antibodies, goat anti-rabbit IgG (rhodamine conjugate; Cappel, MP Biomedicals, Solon, OH) secondary antibody was used. Images were taken on a Nikon Optiphot microscope (Nikon, Melville, NY) equipped with a Nikon DXM1200 digital camera and ACT-1 software (Nikon) or on a Bio-Rad MRC 1024 confocal scanning microscope (Bio-Rad, Hercules, CA) with Confocal Assistant version 4.02 software.
Immunoblots.
A549 cells or KB cells were infected at 10 to 50 PFU/cell with Ad viruses or mutants or were mock infected. At the designated time points, cells were extracted in lysis buffer (10 mM Tris-HCl [pH 7.4], 66 mM EDTA, 1% Nonidet P-40 [NP-40], 0.4% deoxycholic acid) or Laemmli buffer. Samples were electrophoresed on 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to Immobilon-P membrane (Millipore, Billerica, MA). The blot was probed with rabbit antiserum (1:400 dilution) directed against UXP peptide 1 or peptide 2 and subsequently incubated with HRP-conjugated goat anti-rabbit IgG (1:4,000; Cappel, MP Biomedicals). When a pool of UXP-specific hybridoma supernatants was used as the primary antibody, the secondary antibody was HRP-conjugated goat anti-mouse IgG. The bands were visualized by using the LumiGLO peroxidase chemiluminescent substrate kit (KPL, Inc., Gaithersburg, MD).
Virus growth curves.
HEL299 cells were plated in six-well plates. When cells were nearly confluent (1.4 × 106 cells/well), cells were infected at 10 PFU/cell in serum-free DMEM. At 2.5 h p.i., cells were washed three times with DMEM containing 2% FBS to remove unattached virus. Initial samples were collected at 4 h p.i. to determine background virus levels. Daily samples were collected and processed by freeze-thawing and sonication. Virus titers were determined by plaque assay on A549 cells.
Plaque development assays.
Plaque assays were as described previously (45). Briefly, A549 cell monolayers were infected in triplicate with 10-fold serial dilutions of CsCl-banded Ad stocks. Ad5 was included in all assays as a control. Macroscopic plaques were counted at 1- to 2-day intervals for approximately 30 days p.i. The data were then plotted as the number of plaques at a given day as a percentage of the final plaque number to give the kinetics of plaque development. Typically, the results are based on 200 to 500 total plaques per virus.
Cloning the cDNA of UXP by reverse transcriptase PCR.
A549 cells were either mock infected or infected with Ad5 at 10 PFU/cell. At 42 h p.i., poly(A)+ RNA was isolated by using Oligotex Direct mRNA Micro kit (QIAGEN, Inc., Valencia, CA). For first-strand cDNA synthesis, 0.2 μg of poly(A)+ RNA in 8 μl of water was heated at 65°C for 10 min and then quenched on ice. Then, 5 μl of the bulk first-strand cDNA reaction mix (Amersham Biosciences, Piscataway, NJ), 1 μl of 200 mM dithiothreitol, and 1 μl of 20 μM oligo(dT)-anchor primer [5′-GAC CTC GAG ACT AGT GTC GAC (T17)-3′] were added. After incubation, 2 μl of the reverse transcriptase reaction was used for PCRs using primers F31038 (5′-ACG GAT CCA ACA ACA TGA AGA TAG TGG GT-3′) and R22393 (5′-ACA AGC TTG TAC ACT CTC GGG TGA TTA TT-3′) or R23593 (5′-ACA AGC TTA CTG GGT CGT CTT CAT TCA GC-3′). PCR products were analyzed by electrophoresis, extracted from the agarose gel, digested with BamHI/HindIII, and cloned into the expression plasmid pcDNA3.1/myc-His(−)A at the BamHI/HindIII sites to form plasmid pcDNA3.1-UXP. The insert was sequenced by using multiple primers, T7, F31038, R22393, or R23593 to sequence both strands.
Serotype alignments.
The CLUSTAL W program (44) was used in multiple sequence alignments. The EMBOSS pairwise alignment program was used in two sequence alignments. The GenBank sequences were used for Ad5 (accession no. AC_000008), Ad1 (accession no. AC_000017), and Ad2 (accession no. AC_000007). A portion of the Ad6 U exon sequence was available from Ad6 fiber gene sequencing (accession no. AB108424).
Transfection and immunoblotting.
HEK293 cells were grown in a six-well plate overnight and were transfected with 2 μg of the pcDNA3.1-UXP plasmid or empty plasmid by using the calcium phosphate method. At 48 h posttransfection, cells were washed twice with phosphate-buffered saline and lysed in 200 μl of Laemmli buffer. Then, 10 μl of each sample was subjected to immunoblot analysis as described above. The blot was probed with a mixture of two anti-UXP monoclonal antibodies diluted 1:10 in TBST (50 mM Tris-HCl [pH 7.6], 150 mM NaCl, 0.2% Tween 20) containing 5% nonfat dry milk for 1.5 h. The secondary antibody was horseradish peroxidase-conjugated goat anti-mouse IgG (1:4,000 dilution; Cappel, MP Biomedicals).
Transfection for immunofluorescence.
Cells were plated on #1 coverslips 1 day before transfection. Transfections were done on three cell lines: A549, 21f1, and A549E1. Cells were grown in six-well plates overnight and were transfected by using the calcium phosphate method with 2 μg of either the pcDNA3.1-UXP plasmid or the empty parental plasmid. For indirect immunofluorescence, cells were fixed at 48 h posttransfection (as described above). Primary antibody was anti-UXP (mixture of two mouse monoclonal antibodies). Secondary antibody was goat anti-mouse IgG (Alexa Fluor 488 conjugate; Molecular Probes).
Transfection and infection.
A549 and 21f1 cells, previously transfected as described above, were infected with VRX-006 at 5 × 107 PFU/35-mm well (approximately 50 PFU/cell) at 24 h posttransfection. After an additional 28 h, cells were fixed and permeabilized. Cells were incubated with primary antibodies: anti-UXP (as described above) and rabbit anti-DBP (1:800 dilution) (27). The secondary antibodies were anti-mouse IgG (Alexa Fluor 488 conjugate) and anti-rabbit IgG (rhodamine conjugate; Cappel). Images were acquired on a Nikon Optiphot microscope with Nikon DXM1200 digital camera and ACT-1 software.
Nucleotide sequence accession number.
The sequence determined in this study has been submitted to GenBank under accession number EU202643.
RESULTS
Our laboratory has constructed a large number of virus mutants with deletions within the E3 region of species C Ads. Figure 1A is a schematic of the E3 deletion mutants used here. Figure 1B shows the expected amino acid sequence of the U exon in Ad5, Ad1, Ad2, dl7000, dl7001, and VRX-006.
Altered DBP localization is associated with disruption of the U exon.
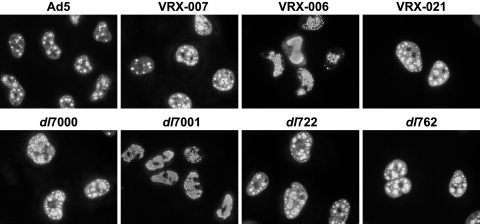
In characterization by indirect immunofluorescence of certain Ad E3 deletion mutants, we noted that the Ad E2 DBP pattern appeared different from the Ad5 DBP pattern (Fig. 2). In wild-type Ad5 infection, immunofluorescence shows uniform distribution of DBP throughout the nucleus (but excluded from nucleoli) during early infection. As infection progresses, DBP staining begins to appear in small dots that subsequently increase in size and appear as rings or spheres (Fig. 2). Later in infection, most DBP staining becomes localized to the central region of the nucleus. By comparison, the DBP pattern for dl7001 and VRX-006 (U exon-negative virus mutants) is initially similar to wild-type Ad5 with uniform nuclear staining, but, as infection progresses, DBP appears as a punctate pattern that fills the nucleus; organized replication centers do not form, and ring-like staining for DBP does not occur (Fig. 2). Mutant dl7000, which contains a large E3 deletion such that only the 14.7K gene is present and that retains the U exon, displayed a normal DBP distribution (Fig. 2). VRX-006 (U exon negative) shows the altered DBP staining pattern, but VRX-021 (in which the U exon is repaired) does not (Fig. 2).
FIG. 2.
The DBP immunofluorescence pattern is disrupted in cells infected with mutants with deletions in the UXP coding region. A549 cells were infected with the indicated viruses at 20 PFU/cell. The E3 genes present in each virus are shown in Fig. 1A. At 25 h p.i., cells were fixed and immunostained for DBP. The DBP is localized in distinct nuclear replication centers in all infected cells except dl7001- and VRX-006-infected cells, in which DBP is found in a uniform punctate pattern.
Mutants dl7001 and VRX-006 (14) have two of the largest E3 deletions that we have constructed. Both of these mutants lack parts of the left portion (3′ end) of the U exon, an ORF on the l strand. With VRX-006, the U exon ORF encodes 49 aa, followed by 3 missense aa's and a stop codon (Fig. 1B). With dl7001, the first 26 aa of UXP would be present followed by 30 missense aa's before a stop codon (Fig. 1B). Both deletions eliminate a potential consensus 5′-splice site. VRX-021 (sequence not shown) is exactly like VRX-006 except the small deletion in the U exon has been repaired; this results in a U exon amino acid sequence identical to Ad5.
Mutants with deletions within the 12.5K gene show normal DBP-defined replication centers (see dl722 in Fig. 2), indicating that deletion of the 12.5K gene is not responsible for the altered DBP immunofluorescence phenotype. Normal replication centers are also found with dl762, a 14.7K gene deletion mutant (Fig. 2). In addition, a series of mutants with deletion of each of the individual E3 genes was analyzed; none of these mutants had the altered DBP localization (data not shown). The altered DBP localization with UXP− mutants is evident on all human cell lines that we have tested. We conclude that the altered DBP pattern is associated with deletions affecting the U exon.
UXP is an ∼24K protein.
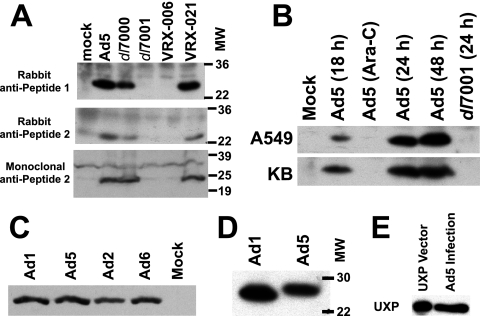
Two peptides from the translated Ad5 U exon ORF sequence (indicated in Fig. 1B) were synthesized and used to generate rabbit polyclonal antisera. Mouse monoclonal antibodies were generated against Peptide 2 (aa 27 to 47 of UXP). The specificity of the antibodies and UXP size were determined by immunoblotting. A549 cells were infected with Ad5, dl7000, dl7001, VRX-006, or VRX-021 at 10 PFU/cell or were mock infected. Rabbit antisera against either peptide detected the same Ad5 ∼24K band (Fig. 3A, the top and middle panels are antisera against peptides 1 and 2, respectively). Similar results were obtained with a pool of mouse monoclonal antibodies to peptide 2 (Fig. 3A, bottom panel). UXP was detected in extracts from cells infected with Ad5, dl7000, or VRX-021 (Fig. 3A), all of which have an intact U exon. The UXP band was not seen for dl7001- or VRX-006-infected cell extracts (Fig. 3A); these viruses have deletions in the U exon. Thus, immunoblots confirm that the U exon encodes a protein.
FIG. 3.
Characterization of UXP expression and size by immunoblotting. (A) UXP is an ∼24K protein on immunoblots. A549 cells were infected with Ad5, dl7000, dl7001, VRX-006, or VRX-021 at 10 PFU/cell or were mock infected. Infections were harvested at 45 h p.i. Immunoblots were incubated with rabbit serum generated against peptide 1 (top panel) or peptide 2 (middle panel) or with mouse monoclonal antibodies to peptide 2 (bottom panel). Mutants dl7001 and VRX-006 have deletions within the 3′ end of the U exon; VRX-021 is a mutant in which the U exon deletion of VRX-006 has been repaired. (B) UXP is a late protein which accumulates over the course of infection. A549 cells and KB cells were infected with Ad5 or dl7001 at 20 PFU/cell or were mock infected. Ara-C was added at 1 h p.i. to one infection to prevent the transition into late infection. Cells were harvested at the indicated time points. The primary antibody was rabbit antiserum against peptide 1. (C) UXP is expressed by all four species C Ads. A549 cells were infected at 50 PFU/cell and were harvested at ∼30 h p.i. The primary antibody was a pool of UXP-specific monoclonal antibodies. (D) The Ad1 UXP migrates faster on SDS-PAGE than the Ad5 UXP. The Ad1 and Ad5 UXPs are predicted to be 211 and 217 aa in length, respectively (see Fig. 8). Extracts and antibody were as in panel C, but the gel was run longer to better distinguish the size difference. (E) UXP expressed after transfection of the pcDNA3.1-UXP plasmid comigrates with UXP expressed during Ad5 infection. HEK293 cells transfected with the UXP expression plasmid were harvested at 48 h posttransfection. The Ad5 UXP is from the same extract as in panels C and D.
The size of ∼24K on SDS-PAGE suggests that UXP is generated from a spliced mRNA because the U exon ORF could only encode 55 aa. The ∼24K size indicates that the U exon is not spliced to the full-length versions of Ad polymerase (120K), preterminal protein (80K), or DBP (72K). In addition, we have not seen a ∼24K band on immunoblots with anti-DBP antiserum or antisera specific for terminal protein.
UXP is a late Ad protein.
The timeframe of Ad5 UXP expression was examined in both A549 and KB cells. UXP was not obtained from cells infected in the presence of Ara-C (Fig. 3B). Ara-C inhibits Ad DNA replication and therefore Ad infection remains in early stages. We conclude that UXP is not an early protein. UXP was detected at 18 h p.i. and in increasing amounts at 24 and 48 h p.i. (Fig. 3B). Thus, UXP is a late protein that accumulates in increasing amounts throughout infection. We have not detected UXP in immunoblots of CsCl-banded purified Ad5 virions (not shown), suggesting that UXP is not a virion protein. As was also shown in Fig. 3A, UXP was not obtained from mock-infected or dl7001-infected cells (Fig. 3B).
UXP is expressed by all Ad species C serotypes.
All four serotypes in species C exhibit a ∼24K band by immunoblot (Fig. 3C). The Ad6 genome has not been sequenced through all of the U exon, although a portion of the sequence has been submitted with the Ad6 fiber sequence. The Ad6 UXP migrated as a ∼24K band on SDS-PAGE. When the proteins were run further on the separating gel, the Ad1 UXP was slightly smaller than the Ad5 protein (Fig. 3D), as is expected after cloning of the entire UXP mRNA as cDNA (see below) because the predicted UXP from Ad1 lacks 6 aa at the C terminus compared to Ad5 (see Fig. 8). UXP from the Ad5 mutant in800 (49), which has a 4-aa insertion into the UXP and DBP coding sequence, is slightly larger by immunoblot (data not shown).
FIG. 8.
Predicted sequence of UXP in Ad5, Ad1, and Ad2 as deduced from a cDNA clone of the Ad5 UXP mRNA. A549 cells were infected with 10 PFU/cell of Ad5. At 42 h p.i., cytoplasmic mRNA was isolated. Primers within the U exon and within the DBP-coding region were used to generate the cDNA corresponding to the putative UXP mRNA. The clone was sequenced to determine the splice sites and the Ad5 UXP protein sequence. The genomic sequences of Ad1 and Ad2 were then used to predict the amino acid sequence for UXP for these species C Ads. The predicted molecular masses are: 23,567.1 Da (Ad5), 23,509.2 Da (Ad2), and 22,697.1 Da (Ad1).
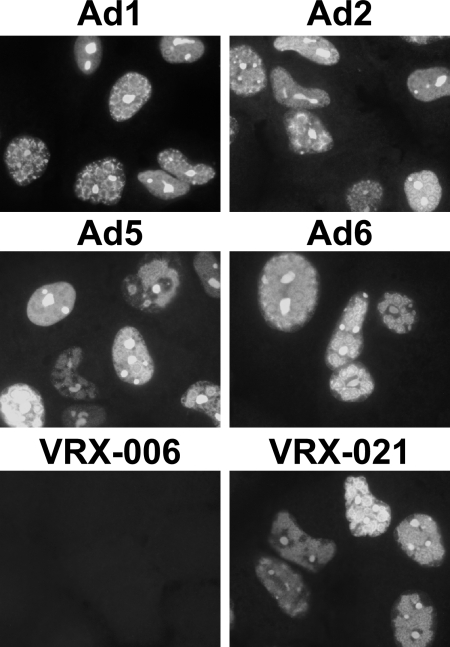
UXP expression by all species C Ads was also shown by indirect immunofluorescence (Fig. 4). UXP was not observed in cells infected with VRX-006 (U exon mutant), but UXP was seen in cells infected with VRX-021, in which the U exon was restored (Fig. 4).
FIG. 4.
Immunofluorescence of UXP in cells infected by all Ad species C serotypes (top four panels) and in cells infected with VRX-021 (bottom right panel). A549 cells were infected at 50 PFU/cell. Cells were fixed at 27 or 30 h p.i. and stained with monoclonal antibodies specific for UXP. UXP is not expressed after infection with VRX-006, a virus with a U exon deletion, but is present in VRX-021 in which the deletion has been repaired.
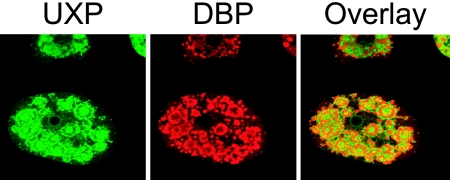
Replication centers show adjacent localization of UXP and DBP.
A549 cells were infected with Ad1 and were treated with actinomycin D at 27 h p.i. for 3 h. At 30 h p.i., cells were fixed and stained for UXP and DBP. Detection of UXP and DBP by confocal microscopy indicated that the two proteins are associated with Ad replication centers, but that the proteins are adjacent to each other rather than being coincident (Fig. 5). DBP staining defines Ad replication sites (ring-like structures); UXP surrounds these sites, as well as appearing to be within the DBP-defined replication centers. The combination of Ad1 and actinomycin D treatment was used because Ad1 appears to produce more abundant UXP by immunofluorescence and actinomycin D treatment results in better definition of replication centers for both UXP and DBP (the cell shown in Fig. 5 is very similar to cell “c” in the Ad5 infection shown in Fig. 6). In other published work on Ad infection, aspects of RNA transcription and processing occur at the edges of replication centers (see Discussion).
FIG. 5.
Comparison of DBP and UXP localization in Ad1-infected A549 cells by confocal microscopy. A549 cells were infected with Ad1 at 50 PFU/cell. At 27 h p.i., actinomycin D was added to the medium. At 30 h p.i., cells were fixed and stained for UXP (mouse monoclonal antibody; green) and DBP (rabbit polyclonal antibody; red). DBP rings are a marker for Ad replication centers. UXP staining is primarily adjacent to rather than coincident with DBP staining.
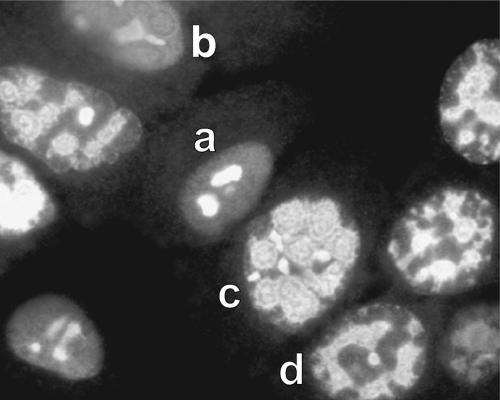
FIG. 6.
UXP localizes to the nucleus, nucleolus, and the periphery of viral replication centers. A549 cells were infected with Ad5 and immunostained with mouse monoclonal antibody. UXP is initially found in nucleoli and dispersed throughout the infected cell (cell “a”). UXP then becomes more abundant throughout the nucleus (cell “b”) and associates with replication centers (cell “c”). Subsequently, there appear to be connections between replication centers (cell “d”) and eventually localization to the center of the nucleus (not shown).
UXP localizes to the nucleus, nucleoli, and the periphery of Ad replication sites.
The monoclonal antibodies and rabbit antisera generated against either peptide display antinuclear and antinucleolar patterns and Ad replication center definition for Ad5-infected cells. UXP localization appears as several different patterns as infection progresses; a number of these appear in Fig. 6. Because the infections are not perfectly synchronous, it is possible to see multiple patterns at a single time point, but there is a general sequential progression of these patterns. When first detectable by indirect immunofluorescence (18 to 24 h p.i.), UXP is distributed throughout the nucleus, as well as being strongly associated with nucleoli (cell “a” in Fig. 6). At 26 to 32 h p.i., there is strong association with replication centers (as defined by coimmunostaining with rabbit polyclonal DBP antiserum) (cell “c” in Fig. 6). At 36 to 45 h p.i., UXP staining is more centrally located within nuclei as one large mass and is usually surrounded by DBP staining. At this very late time, the staining also localized with strong DAPI staining for DNA (presumably the Ad DNA; data not shown).
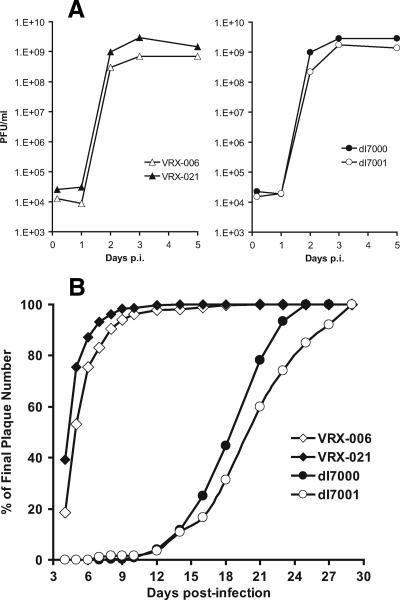
Virus mutants that lack UXP have a mild growth defect.
One of the reasons that we pursued the putative protein coded by the U exon is that E3 mutants whose deletions extend into U exon have a mild replication defect. CsCl-banded stocks of dl7001 or VRX-006 (which lack the U exon) prepared in KB cell spinner cultures are generally severalfold or even 1 log lower in titer than the corresponding dl7000, VRX-007, and VRX-021 mutants (which retain the U exon). The results of a single-step growth experiment in HEL299 (human diploid embryonic lung cells) with these mutants are shown in Fig. 7A. The yield of dl7001 was ∼4-fold lower than that of dl7000 at day 2 and ∼2-fold lower at days 3 and 5. The yield of VRX-006 was more than threefold (day 2) or fourfold (day 3) lower than that of VRX-021 (Fig. 7A), and it was more than sixfold lower than Ad5 at days 3 and 5 p.i. (data not shown).
FIG. 7.
Virus mutants in which the U exon is mutated have a mild growth defect. (A) The yield of UXP− mutants is slightly lower than the yield of UXP+ mutants. HEL299 cells were infected at 10 PFU/cell. At the indicated days p.i., cells and supernatants were harvested. Virus titers were determined by plaque assay on A549 cells. (B) UXP− mutants have slightly smaller plaques that take longer to develop than UXP+ mutants. The graph depicts the number of plaques seen during the plaque assay as a percentage of the total number of plaques seen at the end of the assay. Mutants dl7000 and dl7001 form plaques much more slowly than VRX-006 and VRX-021 because the former lack the gene for the adenovirus death protein.
The partial growth defect of U exon mutants is manifested in slightly smaller plaques that take longer to develop. For example, dl7001 plaques take longer to develop than dl7000 plaques (Fig. 7B); the same is true for VRX-006 compared to VRX-021 plaques (Fig. 7B). dl7001 and dl7000 plaques are much smaller than VRX-006, VRX-021, or Ad5 plaques (not shown) because dl7000 and dl7001 lack the adenovirus death protein, a key protein required for efficient plaque development (46, 47).
UXP is encoded by a novel l-strand transcription unit.
During late infection, the U exon is spliced to the DBP leader sequence and the DBP-encoding exon, but in a different reading frame from DBP. Guided by the work of Chow et al. (8), we have cloned the mRNA (as cDNA) that appears to encode UXP. Chow et al. (8) described an mRNA (“2c”), expressed at late stages of infection (16 and/or 22 h p.i. at an MOI of 100 to 200 PFU/cell), whose 5′ end maps to map unit (m.u.) 86.7 within the fiber gene but on the opposite stand (the l strand). The 5′ exon for this mRNA is spliced to a short exon at m.u. 68.6, and this exon is spliced to a larger 3′-terminal exon at m.u. 66.6. A good consensus sequence for 5′-splicing is found near the 3′ end of the U exon.
To determine whether the above mRNA encodes UXP, we isolated cytoplasmic mRNAs at 42 h p.i. from A549 cells infected at 10 PFU/cell. A cDNA was made and cloned into an expression plasmid by using primers within the U exon and the DBP-coding region. DNA sequencing suggested that the cloned cDNA encodes a protein with the sequence shown in Fig. 8. To determine whether the cloned cDNA produces a protein with the same characteristics as the UXP expressed during infections, the UXP expression plasmid or vector-only plasmid was used for transfections of HEK293 cells. Cells were extracted into Laemmli buffer at 48 h posttransfection. The HEK293 UXP extract was run adjacent to an extract from Ad5 infection. The blot was probed with UXP-specific monoclonal antibodies. UXP from Ad5-infected cells comigrated (or nearly comigrated) with UXP expressed after transient transfection of the cloned UXP cDNA (Fig. 3E); no band was seen for the vector-only plasmid transfection (data not shown). We conclude that the cloned cDNA encodes UXP.
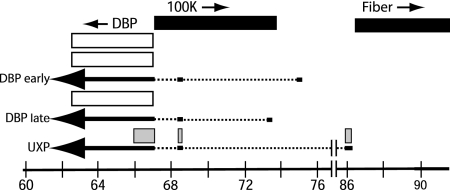
Figure 9 is a schematic of the UXP and DBP coding sequences as deduced from the work of Davison et al. (9, 10) and Chow et al. (8) and from our results as described here. There are three exons: Ad5 nucleotides 31038 to 30868, 24744 to 24668, and 24041 to 22393. The predicted amino acid sequence of the Ad5 UXP is shown in Fig. 8, as are the predicted sequences of UXP from Ad1 and Ad2. The predicted ATG initiation codon is at nucleotide 31031; this is the only ATG in the entire Ad5 protein. This ATG is predicted to be efficient by the Kozak rules (23). The 54 aa of the U exon continues in frame in the short second exon (27 aa), and this continues in frame in the third exon (136 aa) to yield a protein of 217 aa. Two of these amino acids are encoded across the two splice junctions. The second exon of UXP is embedded within the gene for the 100K protein, which is coded off the r strand. The second exon is also the DBP leader sequence; however, in contrast to UXP, this exon is not part of the coding sequences for DBP. The third exon of UXP is embedded within the gene for DBP, which, in common with UXP, is coded off the l strand, but UXP and DBP are in different reading frames.
FIG. 9.
Schematic of UXP and DBP transcripts and proteins. The Ad5 genomic m.u. are shown at the bottom. The leftward pointing arrows near the bottom are the spliced mRNAs for UXP and DBP; the solid parts are exons and dashed parts are introns. The bars above the arrows depict the UXP (□) and DBP (□) proteins. The UXP is expressed at late stages of infection. There are three exons in the UXP mRNA; the coding sequences for UXP protein are at Ad5 nucleotides 31031 to 30868, 24744 to 24668, and 24041 to 23629. The DBP early mRNA is expressed initially in early stages of infection, and the DBP late mRNA is expressed in late stages. The genes encoding DBP (nucleotides 22443 to 24032), the 100K protein (nucleotides 24061 to 26484), and fiber (nucleotides 31042 to 32787) are shown at the top; the arrows indicate the direction of transcription. The genes between the 100K protein and fiber genes are not shown. There are known promoters in the DNA coding sequences near the 5′ ends of the early and late DBP mRNAs, and there is a presumed promoter near the 5′ end of the UXP mRNA.
Examination of translated sequences from other human Ad serotypes (species A, B, D, E, and F) indicates that both the first exon (U exon) (10) and second exon (DBP leader sequence) sequences exhibit strong conservation. Although the third exon shows more divergence, the predicted proteins that would be encoded by other serotypes are of similar length, and all are very basic proteins with pI values of 11 or higher (data not shown).
UXP expressed after transient transfection shows nuclear and nucleolar localization.
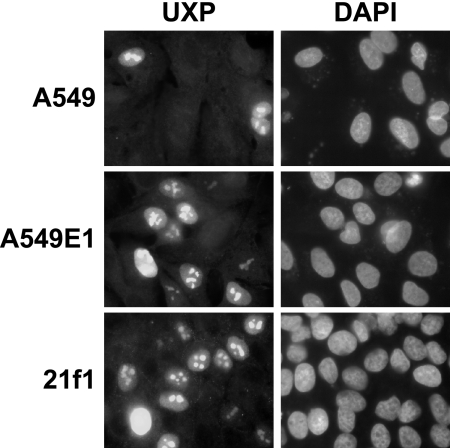
For immunofluorescence, A549, A549E1, or 21f1 cells were transfected with the pcDNA3.1-UXP plasmid that expresses UXP. Cells on coverslips were fixed at 48 h posttransfection. Individual cells were positive for UXP expression, and nuclei and nucleoli were stained in the transfected cells (Fig. 10). The same staining pattern was seen for cells that expressed no other Ad proteins (A549) or cells that were stably transfected with E1 (A549E1) or E2 DBP (21f1) plasmids.
FIG. 10.
UXP expressed after transfection localizes to nuclei and nucleoli. A549, A549E1 (A549 cells stably transfected with E1 genes), and 21f1 cells (HeLa cells stably transfected with DBP) were transfected with 2 μg of pcDNA3.1-UXP plasmid DNA per 35-mm well. At 48 h posttransfection cells were fixed, permeabilized, and immunostained using UXP-specific monoclonal antibodies. The secondary antibody was goat anti-mouse IgG (Alexa Fluor 488 conjugate).
UXP expression by transfection alters DBP localization in cells infected with a UXP mutant virus.
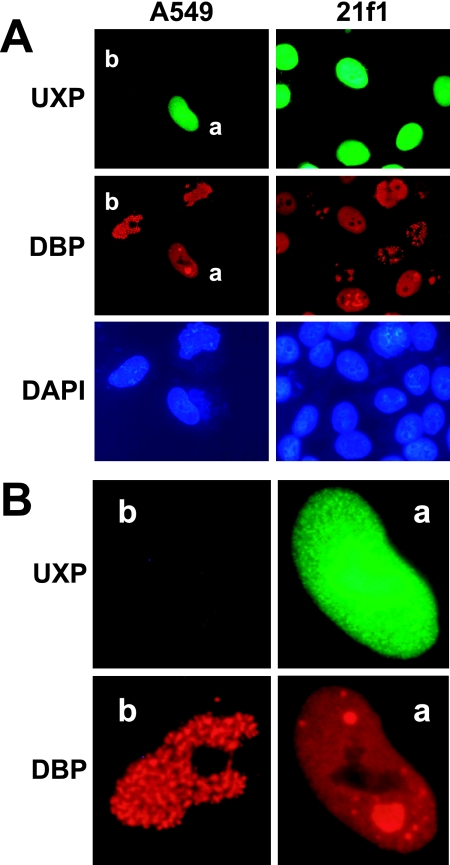
An experiment was conducted to determine whether UXP provided in trans would complement a virus with a UXP deletion. A549 or 21f1 cells were transfected with the UXP expression plasmid. After 24 h, the cells were infected with VRX-006, a U exon mutant, at ∼50 PFU/cell. After an additional 28 h, the cells were fixed and processed for immunostaining. Cells were stained for UXP by using a pool of two UXP-specific monoclonal antibodies and for DBP by using a rabbit DBP-specific antiserum. As shown in Fig. 11, nuclei positive for both UXP and DBP show uniform nuclear staining of DBP, as well as the formation of apparent replication centers as indicated by DBP staining. Only cells with UXP expression (e.g., the cell labeled “a”) show Ad5-like replication centers, while cells in which only DBP is present show the aberrant punctate DBP localization (e.g., the cell labeled “b”). Similar alteration of DBP localization was seen for both A549 and 21f1 cells. These results indicate that UXP can function in trans to complement the aberrant DBP localization pattern of UXP-negative virus mutants.
FIG. 11.
Transfected wild-type UXP acts in trans to facilitate the establishment of Ad replication centers after Ad infection with a UXP− mutant. HeLa 21f1 cells (expressing DBP) or A549 cells were transfected with the pcDNA3.1-UXP plasmid. At 24 h posttransfection, cells were infected at ∼50 PFU/cell with the UXP deletion mutant VRX-006. After an additional 28 h, cells were fixed and stained for DBP and UXP. (A) Cells expressing the transfected UXP show uniform anti-nuclear DBP staining, as well as the formation of replication centers. For A549 cells on the left, the top (UXP), middle (DBP), and bottom (DAPI) panels are the same field of cells. The same is true for 21f1 cells on the right. (B) A549 cells labeled cell “a” and cell “b” from panel A are shown at larger size. Cell “a” is positive for UXP, whereas cell “b” is not.
DISCUSSION
UXP appears to be encoded within a novel late l-strand transcription unit. UXP was not found in infections that were treated with Ara-C, a drug that inhibits the transition into late infection, indicating that UXP is a late Ad protein. This conclusion is consistent with the work of Chow et al. (8), who did not detect the putative UXP mRNA at early stages of infection. In addition, this mRNA was not seen by Chow et al. (8) when cycloheximide was added at 1 h p.i., suggesting that a new Ad early protein or a cellular protein must be synthesized in order for the promoter for mRNA “2c” to be used. When Ara-C was added to the Ad2 infections, a reduced amount of the mRNA “2c” was produced (8).
An unusual property of UXP is that it is encoded in an overlapping reading frame with DBP and that it also overlaps part of the gene for the 100K protein on the r strand. DBP has been reported to be associated with replication, transcription modulation, and host range of Ad (18). A number of Ad mutants have previously been generated within the region of DBP that also encodes the third exon of UXP (7, 43, 49). Some of these mutants failed to show complementation by DBP provided in trans by a HeLa cell line stably transfected with an inducible DBP expression plasmid (7). Any in-frame deletions or insertions in this portion of DBP would also mutate UXP. It would seem prudent to revisit the functions previously mapped to the N terminus of DBP (often described as the first one-third of DBP) since these functions could potentially be attributed to UXP function.
The first detectable UXP localization as shown by indirect immunofluorescence is to nucleoli, so the function of UXP in part probably involves nucleoli. Several other Ad proteins have been shown to localize to nucleoli, including IVa2 (29), the viral core proteins V (30, 31), Mu (26), and protein VII (25). The Ad E4orf4 protein is also targeted to nucleoli (33). Ad infection has been reported to disrupt nucleoli (37).
Viruses with nuclear replication and assembly alter the distribution of nuclear and nucleolar proteins in order to optimize conditions for virus replication and transcription. Most DNA viruses target and reorganize nucleolar proteins, PML/ND10 proteins, specific transcription factors, and DNA- and RNA-associated proteins (19). Virus replication typically occurs at discrete replication centers within the nucleus.
During Ad DNA synthesis, typical virus “replication centers” (also known as replication sites, replication compartments, or viral inclusion bodies) are found in the host nuclei. These replication centers are localized sites of Ad transcription and replication. As the infection proceeds, UXP becomes associated with replication centers, so UXP function likely involves these centers. Early Ad replicative centers contain single-stranded and double-stranded viral DNA and replication activity (40). At a later stage the replication centers enlarge and become sites of accumulation of large numbers of single-stranded DNA intermediates (40). Ad double-stranded DNA and its replication activity shift primarily into the immediately surrounding replicative zones (2). Ad DNA remains in the replicative foci to be replicated and transcribed prior to encapsulation, and nascent Ad RNA is found primarily within the peripheral replicative zone (the main replicative site of Ad5 genomes) (41). Late in infection, a single large centrally located mass is detected; this is the main site of storage of nonreplicating and nonencapsidated double-stranded Ad genomes (40).
The association of UXP with replication centers and the requirement for nuclease treatment prior to immunoprecipitation (data not shown) suggest that UXP is associating with nucleic acids, either RNA or DNA. Consistent with this idea, sequence analysis indicates that UXP is a very basic protein with a basic pI (11.78, 11.71, and 11.86 for the Ad5, Ad2, and Ad1 proteins, respectively). When Ad-infected cells were treated with actinomycin D, UXP association with replication centers was accentuated and UXP nucleolar staining decreased. Although actinomycin D treatment is used for inhibition of RNA polymerase activity, it has been reported that actinomycin D also inhibits Ad DNA replication at both initiation and elongation stages (36).
Aberrant DBP localization in UXP deletion mutants suggests that UXP is required for the establishment of Ad replication centers. UXP could be involved in DNA or RNA processing or could be a structural component of replication centers. Ad proteins that associate with replication centers include the E2 proteins DBP (39), preterminal protein, and polymerase, as well as E1B-55K/E4orf6 (35). The cellular proteins nuclear factor I (NF-I), NF-II, and NF-III are required for Ad replication (11); NF-I is associated with replication centers in Ad2-infected cells (4). Localization of E1B-55K/E4orf6 to the periphery of virus replication centers (35) is believed to be associated with their role in the export of viral RNA from the nucleus (12, 21). A pattern of protein localization similar to that seen with UXP has been reported for the L4 33K protein (15).
Other cellular proteins that have been reported to associate with Ad replication centers include: RNA splicing components (1), ASF (28), B23 (34), upstream-binding factor (24), YB-1 (20), BRCA1 (32), and ND10-associated proteins (especially Sp100) (22).
In other experiments, we expressed UXP in hamster and W162 cells (Vero cells stably transfected with the Ad E4 region [50]). UXP nuclear and nucleolar localization appear similar to that seen in infection of human cell lines (data not shown). Thus, if host proteins are involved in expression or localization of UXP, the host protein(s) must be well conserved across species.
Many Ad vectors have been built to maximize the deletion in the E3 region in order to increase the vector cloning capacity. Typically, sequences at the far right region of the E3 region are deleted (3). We have noticed a reduced yield of Ad E1-minus vectors when the U exon is disrupted. One such plasmid used by many research groups for vector construction has a large deletion that extends further into the U exon than either the VRX-006 or dl7001 deletions (the vectors would encode the first 10 aa of UXP, followed by 2 missense aa's and a stop codon). In addition, workers have modified the fiber gene or added additional fiber genes to change tissue tropism. In one study of a virus that lacks expression of fiber, major capsid proteins accumulated in amorphous nuclear inclusions that included PML and Sp100 (38). Because UXP affects formation of replication centers and the yields of virus are reduced for Ad viruses with deletions within this region, it would seem prudent to restore UXP function within the vector genome or by providing UXP in trans.
Acknowledgments
We thank Laura Gorges for contributions in the early stages of this project, Maurice Green for the rabbit anti-DBP antiserum, and Doug Brough for in800.
This study was supported by grant R01 CA118022 to W.S.M.W. from the National Institutes of Health.
Footnotes
Published ahead of print on 19 September 2007.
REFERENCES
- 1.Aspegren, A., and E. Bridge. 2002. Release of snRNP and RNA from transcription sites in adenovirus-infected cells. Exp. Cell Res. 276:273-283. [DOI] [PubMed] [Google Scholar]
- 2.Besse, S., and F. Puvion-Dutilleul. 1994. High resolution localization of replicating viral genome in adenovirus-infected HeLa cells. Eur. J. Cell Biol. 63:269-279. [PubMed] [Google Scholar]
- 3.Bett, A. J., W. Haddara, L. Prevec, and F. L. Graham. 1994. An efficient and flexible system for construction of adenovirus vectors with insertions or deletions in early regions 1 and 3. Proc. Natl. Acad. Sci. USA 91:8802-8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosher, J., A. Dawson, and R. T. Hay. 1992. Nuclear factor I is specifically targeted to discrete subnuclear sites in adenovirus type 2-infected cells. J. Virol. 66:3140-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brady, H. A., A. Scaria, and W. S. M. Wold. 1992. Map of cis-acting sequences that determine alternative pre-mRNA processing in the E3 complex transcription unit of adenovirus. J. Virol. 66:5914-5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brough, D. E., V. Cleghon, and D. F. Klessig. 1992. Construction, characterization, and utilization of cell lines which inducibly express the adenovirus DNA-binding protein. Virology 190:624-634. [DOI] [PubMed] [Google Scholar]
- 7.Brough, D. E., G. Droguett, M. S. Horwitz, and D. F. Klessig. 1993. Multiple functions of the adenovirus DNA-binding protein are required for efficient viral DNA synthesis. Virology 196:269-281. [DOI] [PubMed] [Google Scholar]
- 8.Chow, L. T., T. R. Broker, and J. B. Lewis. 1979. Complex splicing patterns of RNAs from the early regions of adenovirus-2. J. Mol. Biol. 134:265-303. [DOI] [PubMed] [Google Scholar]
- 9.Davison, A. J., E. A. Telford, M. S. Watson, K. McBride, and V. Mautner. 1993. The DNA sequence of adenovirus type 40. J. Mol. Biol. 234:1308-1316. [DOI] [PubMed] [Google Scholar]
- 10.Davison, A. J., K. M. Wright, and B. Harrach. 2000. DNA sequence of frog adenovirus. J. Gen. Virol. 81:2431-2439. [DOI] [PubMed] [Google Scholar]
- 11.de Jong, R. N., and P. C. van der Vliet. 1999. Mechanism of DNA replication in eukaryotic cells: cellular host factors stimulating adenovirus DNA replication. Gene 236:1-12. [DOI] [PubMed] [Google Scholar]
- 12.Dobner, T., and J. Kzhyshkowska. 2001. Nuclear export of adenovirus RNA. Curr. Top. Microbiol. Immunol. 259:25-54. [DOI] [PubMed] [Google Scholar]
- 13.Doronin, K., K. Toth, M. Kuppuswamy, P. Krajcsi, A. E. Tollefson, and W. S. M. Wold. 2003. Overexpression of the ADP (E3-11.6K) protein increases cell lysis and spread of adenovirus. Virology 305:378-387. [DOI] [PubMed] [Google Scholar]
- 14.Doronin, K., K. Toth, M. Kuppuswamy, P. Ward, A. E. Tollefson, and W. S. M. Wold. 2000. Tumor-specific, replication-competent adenovirus vectors overexpressing the adenovirus death protein. J. Virol. 74:6147-6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gambke, C., and W. Deppert. 1981. Late nonstructural 100,000- and 33,000-dalton proteins of adenovirus type 2. I. Subcellular localization during the course of infection. J. Virol. 40:585-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gooding, L. R., L. Aquino, P. J. Duerksen-Hughes, D. Day, T. M. Horton, S. P. Yei, and W. S. M. Wold. 1991. The E1B 19,000-molecular-weight protein of group C adenoviruses prevents tumor necrosis factor cytolysis of human cells but not of mouse cells. J. Virol. 65:3083-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gooding, L. R., T. S. Ranheim, A. E. Tollefson, L. Aquino, P. Duerksen-Hughes, T. M. Horton, and W. S. M. Wold. 1991. The 10,400- and 14,500-dalton proteins encoded by region E3 of adenovirus function together to protect many but not all mouse cell lines against lysis by tumor necrosis factor. J. Virol. 65:4114-4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hay, R. T., A. Freeman, I. Leith, A. Monaghan, and A. Webster. 1995. Molecular interactions during adenovirus DNA replication. Curr. Top. Microbiol. Immunol. 199(Pt. 2):31-48. [DOI] [PubMed] [Google Scholar]
- 19.Hiscox, J. A. 2002. The nucleolus: a gateway to viral infection? Arch. Virol. 147:1077-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holm, P. S., S. Bergmann, K. Jurchott, H. Lage, K. Brand, A. Ladhoff, K. Mantwill, D. T. Curiel, M. Dobbelstein, M. Dietel, B. Gansbacher, and H. D. Royer. 2002. YB-1 relocates to the nucleus in adenovirus-infected cells and facilitates viral replication by inducing E2 gene expression through the E2 late promoter. J. Biol. Chem. 277:10427-10434. [DOI] [PubMed] [Google Scholar]
- 21.Horridge, J. J., and K. N. Leppard. 1998. RNA-binding activity of the E1B 55-kilodalton protein from human adenovirus type 5. J. Virol. 72:9374-9379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishov, A. M., and G. G. Maul. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134:815-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozak, M. 1991. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 266:19867-19870. [PubMed] [Google Scholar]
- 24.Lawrence, F. J., B. McStay, and D. A. Matthews. 2006. Nucleolar protein upstream binding factor is sequestered into adenovirus DNA replication centres during infection without affecting RNA polymerase I location or ablating rRNA synthesis. J. Cell Sci. 119:2621-2631. [DOI] [PubMed] [Google Scholar]
- 25.Lee, T. W., G. E. Blair, and D. A. Matthews. 2003. Adenovirus core protein VII contains distinct sequences that mediate targeting to the nucleus and nucleolus, and colocalization with human chromosomes. J. Gen. Virol. 84:3423-3428. [DOI] [PubMed] [Google Scholar]
- 26.Lee, T. W., F. J. Lawrence, V. Dauksaite, G. Akusjarvi, G. E. Blair, and D. A. Matthews. 2004. Precursor of human adenovirus core polypeptide Mu targets the nucleolus and modulates the expression of E2 proteins. J. Gen. Virol. 85:185-196. [DOI] [PubMed] [Google Scholar]
- 27.Lillie, J. W., P. M. Loewenstein, M. R. Green, and M. Green. 1987. Functional domains of adenovirus type 5 E1a proteins. Cell 50:1091-1100. [DOI] [PubMed] [Google Scholar]
- 28.Lindberg, A., M. Gama-Carvalho, M. Carmo-Fonseca, and J. P. Kreivi. 2004. A single RNA recognition motif in splicing factor ASF/SF2 directs it to nuclear sites of adenovirus transcription. J. Gen. Virol. 85:603-608. [DOI] [PubMed] [Google Scholar]
- 29.Lutz, P., F. Puvion-Dutilleul, Y. Lutz, and C. Kedinger. 1996. Nucleoplasmic and nucleolar distribution of the adenovirus IVa2 gene product. J. Virol. 70:3449-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matthews, D. A. 2001. Adenovirus protein V induces redistribution of nucleolin and B23 from nucleolus to cytoplasm. J. Virol. 75:1031-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matthews, D. A., and W. C. Russell. 1998. Adenovirus core protein V is delivered by the invading virus to the nucleus of the infected cell and later in infection is associated with nucleoli. J. Gen. Virol. 79:1671-1675. [DOI] [PubMed] [Google Scholar]
- 32.Maul, G. G., D. E. Jensen, A. M. Ishov, M. Herlyn, and F. J. Rauscher, III. 1998. Nuclear redistribution of BRCA1 during viral infection. Cell Growth Differ. 9:743-755. [PubMed] [Google Scholar]
- 33.Miron, M. J., I. E. Gallouzi, J. N. Lavoie, and P. E. Branton. 2004. Nuclear localization of the adenovirus E4orf4 protein is mediated through an arginine-rich motif and correlates with cell death. Oncogene 23:7458-7468. [DOI] [PubMed] [Google Scholar]
- 34.Okuwaki, M., A. Iwamatsu, M. Tsujimoto, and K. Nagata. 2001. Identification of nucleophosmin/B23, an acidic nucleolar protein, as a stimulatory factor for in vitro replication of adenovirus DNA complexed with viral basic core proteins. J. Mol. Biol. 311:41-55. [DOI] [PubMed] [Google Scholar]
- 35.Ornelles, D. A., and T. Shenk. 1991. Localization of the adenovirus early region 1B 55-kilodalton protein during lytic infection: association with nuclear viral inclusions requires the early region 4 34-kilodalton protein. J. Virol. 65:424-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pincus, S., and D. Rekosh. 1984. The inhibition of both initiation and elongation of adenovirus DNA replication by actinomycin D. Biochem. Biophys. Res. Commun. 121:1033-1041. [DOI] [PubMed] [Google Scholar]
- 37.Puvion-Dutilleul, F., and M. E. Christensen. 1993. Alterations of fibrillarin distribution and nucleolar ultrastructure induced by adenovirus infection. Eur. J. Cell Biol. 61:168-176. [PubMed] [Google Scholar]
- 38.Puvion-Dutilleul, F., V. Legrand, M. Mehtali, M. K. Chelbi-Alix, H. de The, and E. Puvion. 1999. Deletion of the fiber gene induces the storage of hexon and penton base proteins in PML/Sp100-containing inclusions during adenovirus infection. Biol. Cell 91:617-628. [PubMed] [Google Scholar]
- 39.Puvion-Dutilleul, F., J. Pedron, and C. Cajean-Feroldi. 1984. Identification of intranuclear structures containing the 72K DNA-binding protein of human adenovirus type 5. Eur. J. Cell Biol. 34:313-322. [PubMed] [Google Scholar]
- 40.Puvion-Dutilleul, F., and E. Puvion. 1990. Analysis by in situ hybridization and autoradiography of sites of replication and storage of single- and double-stranded adenovirus type 5 DNA in lytically infected HeLa cells. J. Struct. Biol. 103:280-289. [DOI] [PubMed] [Google Scholar]
- 41.Puvion-Dutilleul, F., and E. Puvion. 1991. Sites of transcription of adenovirus type 5 genomes in relation to early viral DNA replication in infected HeLa cells. A high resolution in situ hybridization and autoradiographical study. Biol. Cell 71:135-147. [DOI] [PubMed] [Google Scholar]
- 42.Ranheim, T. S., J. Shisler, T. M. Horton, L. J. Wold, L. R. Gooding, and W. S. M. Wold. 1993. Characterization of mutants within the gene for the adenovirus E3 14.7-kilodalton protein which prevents cytolysis by tumor necrosis factor. J. Virol. 67:2159-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rice, S. A., and D. F. Klessig. 1985. Isolation and analysis of adenovirus type 5 mutants containing deletions in the gene encoding the DNA-binding protein. J. Virol. 56:767-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tollefson, A. E., M. Kuppuswamy, E. V. Shashkova, K. Doronin, and W. S. M. Wold. 2007. Preparation and titration of CsCl-banded adenovirus stocks. Methods Mol. Med. 130:223-235. [DOI] [PubMed] [Google Scholar]
- 46.Tollefson, A. E., J. S. Ryerse, A. Scaria, T. W. Hermiston, and W. S. M. Wold. 1996. The E3-11.6K adenovirus death protein (ADP) is required for efficient cell death: characterization of cells infected with adp mutants. Virology 220:152-162. [DOI] [PubMed] [Google Scholar]
- 47.Tollefson, A. E., A. Scaria, T. W. Hermiston, J. S. Ryerse, L. J. Wold, and W. S. M. Wold. 1996. The adenovirus death protein (E3-11.6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. J. Virol. 70:2296-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tollefson, A. E., K. Toth, K. Doronin, M. Kuppuswamy, O. A. Doronina, D. L. Lichtenstein, T. W. Hermiston, C. A. Smith, and W. S. M. Wold. 2001. Inhibition of TRAIL-induced apoptosis and forced internalization of TRAIL receptor 1 by adenovirus proteins. J. Virol. 75:8875-8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vos, H. L., D. E. Brough, F. M. Van der Lee, R. C. Hoeben, G. F. Verheijden, D. Dooijes, D. F. Klessig, and J. S. Sussenbach. 1989. Characterization of adenovirus type 5 insertion and deletion mutants encoding altered DNA binding proteins. Virology 172:634-642. [DOI] [PubMed] [Google Scholar]
- 50.Weinberg, D. H., and G. Ketner. 1983. A cell line that supports the growth of a defective early region 4 deletion mutant of human adenovirus type 2. Proc. Natl. Acad. Sci. USA 80:5383-5386. [DOI] [PMC free article] [PubMed] [Google Scholar]