Abstract
Serum response factor (SRF) was recently shown to bind and activate the human T-cell leukemia virus type 1 (HTLV-1) promoter at bases −116 to −125 relative to the transcription start site. In addition to the SRF binding site (CArG box), serum response elements (SRE) also typically contain a binding site for a member of the ternary complex factor (TCF) family. Here we demonstrate the presence of two TCF binding sites upstream of the viral CArG box. Binding of the TCF family member Elk-1 to these sites was shown to activate transcription of the promoter. Based on these results, the position of the previously described viral SRE (vSRE) within the HTLV-1 promoter can be extended from −116 to −157 to include the two newly identified TCF sites. Purified Elk-1 bound to a probe containing the vSRE, and this complex formed a ternary complex with SRF. In addition, the complex formed by nuclear extract on this probe contained Elk-1, as shown by electrophoretic mobility shift assay supershift. Both of the predicted TCF sites independently bound Elk-1. Elk-1 activated transcription of the HTLV-1 long terminal repeat (LTR), and mutations within either of the TCF sites or the CArG box reduced responsiveness of the LTR to Elk-1. Chromatin immunoprecipitation demonstrated that Elk-1 associates with the HTLV-1 LTR in vivo. These results identify a functional SRE within the HTLV-1 LTR and suggest that both Elk-1 and SRF play important roles in regulating basal HTLV-1 gene expression.
Elk-1 is a member of the Ets transcription factor family and the subfamily of ternary complex factor (TCF) proteins (19, 25). TCFs, in conjunction with serum response factor (SRF), regulate the expression of a class of cellular genes known as immediate-early genes. Immediate-early gene promoters contain serum response elements (SRE), which typically include an SRF binding site (CArG box) and a TCF binding site (consensus sequence, CCGGAA) (26). Although recruitment of SRF and TCF proteins to their respective binding sites can independently activate transcription, the interaction between TCF and SRF promotes the formation of a more stable ternary complex, allowing increased transcriptional activity.
The retrovirus human T-cell leukemia virus type 1 (HTLV-1) is the etiological agent of adult T-cell leukemia and the neurodegenerative disorder tropical spastic paraparesis/HTLV-1-associated myelopathy (16, 23, 33). Upon infection, the viral genome integrates into the host cell chromatin and viral gene expression is regulated by the viral promoter located in the U3 region of the 5′ long terminal repeat (LTR). Initial rounds of transcription result in expression of viral genes, including that encoding the viral oncoprotein Tax. Transcriptional activity of the LTR is required for viral genome production. Therefore, transcriptional regulation of the viral promoter is important both for disease progression and for viral replication.
Cellular transcription factors that are known to regulate the viral promoter include CREB/ATF family members, Ap-2, c-Myb, and SRF (1, 3, 10, 22, 30, 34). In addition to SRF and TCF binding sites, the viral promoter contains three 21-bp imperfect repeats, which are individually known as Tax-responsive element 1 (TRE1s) (6, 14, 24). CREB and other bZIP family members bind to the TRE1s and recruit Tax to activate transcription. The recently identified SRF binding site is located within a region of the viral promoter known as Tax-responsive element 2 (TRE2) (31). Other transcription factors known to bind this element include Sp-1, TIF-1, and c-Myb (32). The TRE2 binding site also overlaps an element that binds Ets1 and Ets2, termed the Ets responsive region 1 (ERR-1) (18). Therefore, a multitude of transcription factors regulate basal and activated transcription of the viral promoter.
Since a functional CArG box was identified in the viral LTR and Ets family members are known to bind this region, we investigated the possibility that the LTR contained a TCF binding site. Visual inspection of the viral promoter suggested two possible TCF binding sites, 23 and 33 bp upstream of the CArG box. The presence of these elements suggested that the 5′ end of the viral SRE (vSRE) should be expanded from the previously identified position −125 to −157 relative to the transcription start site (31). The predicted proximal TCF binding site contains a perfect consensus CCGGAA sequence and is located at bases −141 to −146, while the predicted distal TCF site has an additional G with the sequence CCGGGAA and is located at bases −151 to −157 upstream from the transcription start site. Therefore, we hypothesized that the vSRE contains both SRF and TCF binding sites. Here we report that Elk-1 and SRF bind to the vSRE. Specific binding of Elk-1 was observed on both of the predicted TCF binding sites. The wild-type LTR was activated by Elk-1 and SRF, while mutations in either the TCF site or the CArG box reduced activation. Finally, Elk-1 was shown to associate in vivo with the viral promoter in an HTLV-1-positive cell line. We conclude that the HTLV-1 LTR contains a functional SRE that is regulated by both Elk-1 and SRF.
MATERIALS AND METHODS
Recombinant protein purification.
Glutathione S-transferase (GST)-tagged SRF was purified according to the manufacturer's protocol (Amersham GE Healthcare, Piscataway, NJ). Briefly, GST-SRF fusion protein was expressed in Escherichia coli. Bacterial pellets were lysed in 100 mM Tris-HCl, pH 7.5, 200 mM KCl, 10 mM dithiothreitol, and 10 mM MgCl2, and GST-SRF protein was purified from the lysates in batch using glutathione beads. GST-SRF protein was eluted with glutathione elution buffer (25 mM reduced glutathione, 150 mM KCl, 50 mM Tris-HCl [pH 8.0]). His-tagged Elk-1 was also purified using nickel-nitrilotriacetic acid His Bind resin according to the manufacturer's instructions for batch purification under native conditions (Novagen, Madison, WI).
Cell lines.
CEM cells (a human T-lymphocyte cell line) were maintained in RPMI medium supplemented with 10% fetal bovine serum (FBS). MS9 cells (28) were maintained in RPMI supplemented with 10% FBS and 100 U/ml interleukin 2 (NCI Preclinical Repository, Frederick, MD). 293 cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% FBS.
Plasmids and oligonucleotides.
The sequences of the vSRE and mutant sense-strand oligonucleotides used in gel shift assays are shown in Table 1. The primers 5′-GAAGTCTGAGAAGGTCAGGG-3′ and 5′-CCACGCTTTTATAGACTCCTG-3′ were used to amplify the LTR in chromatin Immunoprecipitation (ChIP) assays. The primers 5′-ACCCTCGGTGTTGGCTG-3′ and 5′-TCCTAATCTCGTGAGCATTTCG-3′ were used to amplify the c-fos promoter in ChIP assays. The Elk-1 expression vector, pRSV-Elk-1-RSPA, was a gift from Maureen Shuh (Loyola University, New Orleans, LA). The SRF expression vector, pSG-SRF, was a gift from Robert Schwartz (Institute of Biotechnology, Texas A&M University, Houston, TX). The reporter constructs, pU3RLuc and pmvSRELuc, have been previously described (31). Other mutant constructs were made using a QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA). In this mutagenesis strategy, the parent plasmid was pU3RLuc or pmCArGLuc when the CArG box was to be wild type or mutant, respectively.
TABLE 1.
Sense sequences of oligonucleotides used in EMSAs
| Oligonucleotide | Sequencea |
|---|---|
| distproxCArG | |
| vSRE | CGGAGACCTCCGGGAAGCCACC-GGAACCACCCATTTCCTCCCCATGTTTGTCAAGCC |
| mCArG | CGGAGACCTCCGGGAAGCCACC-GGAACCACCCATTTCCTCCACATGTCTGTCAAGCC |
| mprox | CGGAGACCTCCGGGAAGCCACCAAGAACCACCCATTTCCTCCCCATGTTTGTCAAGCC |
| mprox/mCArG | CGGAGACCTCCGGGAAGCCACCAAGAACCACCCATTTCCTCCACATGTCTGTCAAGCC |
| mdist | CGGAGACCTCCGTCTCGCCACC-GGAACCACCCATTTCCTCCCCATGTTTGTCAAGCC |
| mdist/mCArG | CGGAGACCTCCGTCTCGCCACC-GGAACCACCCATTTCCTCCACATGTCTGTCAAGCC |
| mprox/mdist | CGGAGACCTCCGTCTCGCCACCAAGAACCACCCATTTCCTCCCCATGTTTGTCAAGCC |
| mvSRE | CGGAGACCTCCGTCTCGCCACCAAGAACCACCCATTTCCTCCACATGTCTGTCAAGCC |
dist, distal TCF site; prox, proximal TCF site.
EMSAs.
Double-stranded oligonucleotides were labeled with [α-32P]dCTP using Klenow enzyme. Electrophoretic mobility shift assays (EMSAs) using purified proteins included 1× EMSA buffer (20 mM HEPES, pH 7.4, 100 mM KCl, 0.5 mM EDTA, 1 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride, 10% glycerol), 1 μg sheared salmon sperm DNA (Sigma St. Louis, MO), 0.5 μg His-Elk-1, 2 μg GST-SRF, and 0.5 nM labeled probe in a 20-μl total reaction volume. The reaction mixtures were incubated at room temperature for 30 min. Complexes were resolved on a 4% nondenaturing polyacrylamide gel in 0.6× Tris-borate-EDTA. Antibodies (2 μg per 20-μl reaction mixture) used in supershift EMSA experiments (anti-SRF [SC-335], anti-Elk-1 [SC-355], and anti-AP-2 [SC25343] [Santa Cruz Biotechnology, Santa Cruz, CA]) were added 15 min after initiating the protein and DNA incubation. Reaction mixtures were incubated at room temperature for 30 min after addition of antibody. For EMSAs using nuclear extracts, 10 μg of CEM nuclear extract was incubated with the 32P-labeled probe. Unlabeled competitor oligonucleotides were added in a 200-fold excess over labeled probe.
Transfections.
293 cells were transfected using Fugene (Roche, Indianapolis, IN). Briefly, cells were distributed into six-well plates and allowed to adhere overnight. The Fugene transfection master mix was prepared by adding Fugene to serum-free Dulbecco's modified Eagle medium to create a final reaction mixture containing a ratio of 3 μl of Fugene to 2 μg of DNA. Aliquots of the Fugene-medium mixture were placed in individual tubes, and each reporter construct was added. Aliquots of each DNA mixture were placed in tubes for appropriate addition of an Elk-1 and/or SRF expression vector. After all components were added, reaction mixtures were incubated at room temperature for 30 min and then the transfection mixture (100 μl) was added to a well containing 293 cells. Each transfection was done in duplicate within each experiment, and each experiment was repeated three times.
Luciferase assays.
Cells were harvested 48 h after transfection by passive lysis (Promega, Madison, WI). Cells were washed once with phosphate-buffered saline, and then 250 μl of 1× passive lysis buffer was added to the wells and plates were rocked for 20 min. Relative luciferase activity was determined by adding 50 μl of a firefly luciferase substrate (Promega, Madison, WI) to 10 μl of lysate, and luminescence was detected using a Sirius luminometer (Berthold Detection Systems, Pforzheim, Germany). The degree of activation was determined by averaging the duplicate results in each experiment and normalizing them to the basal activity of each reporter. Results are displayed as the average n-fold activation from three independent experiments.
ChIP.
ChIP assays were performed as previously described (31). Briefly, lysates from 5 × 107 CEM or MS9 formaldehyde-cross-linked cells were precleared with protein-G beads (Upstate Biotechnology, Charlottesville, VA). Beads were prebound with an SRF-specific antibody (SC-335; Santa Cruz Biotechnology, Santa Cruz, CA) or Elk-1-specific antibody (SC-355; Santa Cruz Biotechnology, Santa Cruz, CA). Prebound beads were incubated with precleared lysates in immunoprecipitation (IP) buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 1 μg/ml leupeptin, 1 μg/ml aprotinin, 1 μg/ml pepstatin, 1 mM phenylmethylsulfonyl fluoride) (7). Precipitated beads were washed sequentially with IP buffer once, IP buffer with 0.5 M NaCl twice, LiCl wash buffer (20 mM Tris-HCl, pH 8.0, 250 mM LiCl, 2 mM EDTA, 0.5% Nonidet P-40) twice, and Tris-EDTA (pH 8.0) once (4, 5, 7). Elution buffer (0.1 M NaHCO3, 1% sodium dodecyl sulfate, 0.5 M NaCl) was added to the precipitated DNA-protein complexes, and the cross-links were reversed at 65°C overnight. Recovered DNA was analyzed by PCR with primers specific for the LTR or c-fos promoter that spanned the SRE in both promoters.
RESULTS
The HTLV-1 LTR can be activated by SRF, and SRF response elements typically contain binding sites for members of the TCF transcription factor family. Therefore, we investigated the possibility that Elk-1, a predominant TCF family member, would bind to the LTR and regulate transcription in combination with SRF. Visual examination of the LTR sequence upstream of the SRF binding site revealed two possible TCF binding sites at positions −141 to −146 and −151 to −157 relative to the transcription start site.
Elk-1 and SRF form a ternary complex on the vSRE.
EMSAs using a vSRE probe were used to determine whether Elk-1 bound to the predicted TCF sites and whether Elk-1 and SRF together could form a ternary complex on this element. Since SRF has previously been shown to bind the vSRE (31), it was used as a positive control in these experiments. Recombinant purified His-tagged Elk-1 and GST-tagged SRF were used to establish that these proteins can bind the vSRE independently (Fig. 1A, lanes 2 and 3), as well as in a ternary complex that migrates more slowly than either protein by itself (Fig. 1A, lane 4). As previously reported, SRF alone binds the vSRE weakly (31); therefore, approximately four times more SRF protein than Elk-1 protein was needed to detect a band by EMSA. Addition of Elk-1 or SRF antibody, but not a nonspecific antibody, produced a slower-migrating ternary complex in lanes containing both Elk-1 and SRF. The ability of either Elk-1- or SRF-specific antibody to shift virtually all of the ternary complex demonstrated that Elk-1 and SRF are both present in the complex (Fig. 1A, lanes 5 to 7). Thus, Elk-1 and SRF can independently bind the vSRE, and together they form a ternary complex on this element.
FIG. 1.
Elk-1 and SRF form a ternary complex on DNA. (A) EMSA using a 32P-labeled vSRE probe (lane 1) in the presence of recombinant SRF (rSRF) (lane 2), recombinant Elk-1 (rElk-1) (lane 3), or rElk-1 and rSRF (lanes 4 to 7). Specific antibody to Elk-1 (lane 5), SRF (lane 6), or AP-2 (lane 7) was added in the indicated reactions. The locations of supershifted bands are indicated on the right. (B) EMSA using 32P-labeled vSRE probe (lane 1) incubated with CEM nuclear extracts (lanes 2 to 5). Antibodies were added to the indicated lanes. The positions and identities of supershifted bands are indicated on the right. (C) EMSA using a vSRE probe (lane 1) incubated with CEM nuclear extracts (lanes 2 to 4) and a 25-fold excess of unlabeled vSRE (lane 3) or mvSRE (lane 4) used as specific or nonspecific competitors, respectively. Wild-type vSRE competes for the Elk-1 band (arrow). (D and E) Western blots of SRF and Elk-1 using recombinant proteins (D) or whole cell extracts (E) from MS9, 293, or CEM cells.
To determine whether Elk-1 can bind the vSRE in the presence of a complex protein mixture, antibody supershifts were used to detect Elk-1 and SRF binding from CEM nuclear extracts (Fig. 1B). In the presence of nuclear extract alone, we observed a band that migrated to a position in the gel similar to that of the recombinant SRF-Elk-1 complex. Other bands were also visible which were not observed when recombinant protein was used. The additional bands are likely due to the binding of as yet unidentified factors present in nuclear extracts, which contain a cadre of transcription factors, cofactors, and DNA binding proteins.
To determine whether Elk-1 can bind to the vSRE in the presence of a complex protein mixture, Elk-1 antibody was added to EMSA reaction mixtures containing nuclear extract. These reactions produced a band that migrated slightly faster in the gel than did recombinant Elk-1 (Fig. 1B, lane 3), indicating the presence of Elk-1 in these complexes. Binding of the Elk-1 antibody may displace one or more as yet unidentified cellular proteins from the complex, resulting in the faster-migrating band. The Elk-1 antibody supershift was specific because it was not observed with antibody to the unrelated transcription factor, AP-2. When the SRF antibody was used, a supershifted band was observed as expected (Fig. 1B, lane 4).
To determine whether the binding of Elk-1 from nuclear extracts to the vSRE was specific, unlabeled wild-type vSRE was used as a specific competitor and unlabeled mutant vSRE (mvSRE) was used as a nonspecific competitor (Fig. 1C, lanes 2 to 4). The mvSRE did not compete for any of the complexes, while the wild-type vSRE competed for the Elk-1 band. The ability of vSRE to compete for a slower-migrating band suggests that the vSRE binds different combinations of Elk-1, SRF, and possibly other transcription factors and cofactors. Taken together, these results establish that Elk-1 and SRF form a ternary complex on the vSRE.
Elk-1 binds to both TCF sites.
Since two putative TCF binding sites were identified within the vSRE, we next wanted to determine whether both of these sites could bind Elk-1. vSRE probes containing mutations in the proximal TCF site (mprox), distal TCF site (mdist), or the CArG box (mCArG) (Table 1) were used in EMSAs to examine their ability to bind purified Elk-1 or SRF. Elk-1 bound to each probe that contained at least one wild-type TCF site (Fig. 2A to F, lane 2) but not to probes containing mutations in both TCF sites (Fig. 2G to H, lane 2). Interestingly, the proximal TCF site appeared to bind Elk-1 more efficiently than the distal TCF site whether an SRF binding site was present (compare Fig. 2C with 2E) or not (compare Fig. 2D with 2F). Minor sequence differences between the two TCF sites or their positions within the SRE are most likely responsible for these differences in binding efficiency. The proximal TCF site has a perfect CCGGAA consensus sequence, while the distal TCF site (CCGGGAA) contains an additional G. Similar to Elk-1, SRF bound to each probe that contained a wild-type CArG box (Fig. 2A, C, E, and G, lane 3) but not to probes containing a mutation in the CArG box (Fig. 2B, D, F, and H, lane 3). Since the wild-type viral CArG box is 80% identical to a canonical CArG box, SRF alone binds to the viral CArG box with weak affinity (31).
FIG. 2.
Elk-1 binds to both TCF sites in the vSRE. 32P-labeled probes corresponding to wild-type vSRE (A), mutations in the CArG box (mCArG) (B), mutations in the proximal TCF (mprox) (C), mutations in the proximal TCF and CArG box (mprox/mCArG) (D), mutations in the distal TCF site (mdist) (E), mutations in the distal TCF site and CArG box (mdist/mCArG) (F), muations in the proximal and distal TCF sites (mprox/mdist) (G), or mutations in all three elements (mvSRE) (H) were incubated with rElk-1 alone (lane 2), rSRF alone (lane 3), or rElk-1 and rSRF (lane 4). A schematic diagram of each probe is shown below each panel. Samples were analyzed by EMSA. The sequence of each probe is shown in Table 1.
Incubation of Elk-1 and SRF together with a labeled probe containing at least one TCF site or CArG box produced a band that migrated more slowly than Elk-1 alone (compare Fig. 2A to G, lane 4, with Fig. 2H, lane 4). This slower-migrating band apparently reflects the binding of both Elk-1 and SRF to the probe. Even probes containing only a single binding site for TCF (Fig. 2D and F) or SRF (Fig. 2G) formed this slower-migrating band, suggesting that the ability of SRF and TCF to interact allows each of them to be recruited indirectly into the complex by the other. For example, mutation of the CArG box disables its ability to bind SRF alone (Fig. 2D, lane 3), but the interaction of Elk-1 with the distal TCF indirectly recruits SRF to the complex (lane 4), resulting in a slower-migrating ternary complex. These results confirm the ability of Elk-1 and SRF to form a ternary complex on the vSRE.
Elk-1 and SRF activate transcription from the vSRE.
Since Elk-1 and SRF are recruited to the vSRE, we next wanted to determine whether Elk-1 could activate transcription of the viral LTR. Each mutant analyzed in Fig. 2 was introduced into a reporter construct containing the viral LTR driving expression of firefly luciferase. Each reporter was transfected into 293 cells alone or together with an Elk-1 or SRF (Fig. 3A) expression vector. All mutants were activated less efficiently by Elk-1 than was the wild-type LTR (Fig. 3A). These results suggest that the CArG box and both TCF binding sites are necessary for full activation of the vSRE by Elk-1. Surprisingly, each mutant reporter retained some Elk-1 responsiveness, suggesting that additional unidentified Elk-1-responsive element(s) may exist elsewhere in the LTR. Although recombinant Elk-1 bound to the proximal TCF site more efficiently than to the distal TCF site (Fig. 2), constructs containing a wild-type proximal TCF binding site were not more responsive to Elk-1. This result suggests that Elk-1 binding affinity observed in in vitro EMSAs does not fully reflect in vivo functional activity.
FIG. 3.
Mutation of the vSRE reduces Elk-1 and SRF activation of the HTLV-1 LTR. (A) 293 cells were transfected with reporter plasmids containing either wild-type LTR (white bars) or LTR containing mutations within the three vSRE elements (patterned and shaded bars) alone (−) or with 2 μg of pRSV-Elk-1-RSPA expression plasmid (+). The mutant LTRs contained the same mutations shown in Table 1 and analyzed in Fig. 2. Results are presented as the average n-fold activation over the basal activity of each reporter from three experiments. (B) 293 cells were transfected as for panel A, but a pSG-SRF expression plasmid was used instead of Elk-1.
When an SRF expression vector was transfected together with a wild-type or mutant vSRE reporter construct, significant activation was observed only on the wild-type (vSRE), mdist, and mprox/mdist reporters (Fig. 3B). This was expected due to the presence of a wild-type CArG box in these vectors. However, the mprox reporter, which also contains a wild-type CArG box, was not activated by SRF. Thus, a factor that binds to the distal TCF site may repress SRF transactivation. This would explain why SRF activation was observed when the distal TCF site was mutated. These results also suggest that the proximal TCF binding site plays a positive role in SRE function, which is consistent with our finding that Elk-1 bound to the proximal TCF binding site more efficiently than to the distal TCF binding site.
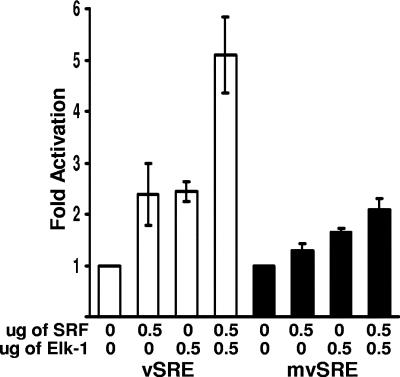
Since the TCF sites and CArG box were independently responsive to their respective binding factors, we wanted to determine whether these activities were additive or cooperative, Cotransfection of SRF and Elk-1 expression vectors into 293 cells together with a wild-type LTR resulted in twice as much reporter activity than was observed with either factor alone (Fig. 4). Transfection of a reporter containing mutations in the CArG box and both TCF binding sites (mvSRE) resulted in only a modest increase in activity with either factor alone or in combination. These results suggest that the cellular transcription factors SRF and Elk-1 induce additive transcriptional activation of the vSRE. Thus, although SRF can recruit Elk-1 to the vSRE when the TCF site(s) is mutated and visa versa, recruitment alone is not sufficient to fully activate transcription. It appears that both Elk-1 and SRF must bind DNA to fully activate transcription, probably because the binding of each protein to its respective DNA binding site and interactions between the proteins allow a more stable ternary complex to form and recruit necessary cofactors.
FIG. 4.
Elk-1 and SRF activate the HTLV-1 LTR. 293 cells were transfected with reporter plasmids containing either the wild-type LTR (white bars) or an LTR containing mutations in all three vSRE elements (mvSRE) (black bars) along with 0.5 μg of pRSV-Elk-1-RSPA, 0.5 μg of pSG-SRF, or 0.5 μg of Elk-1 and pSG-SRF expression plasmid. These results are the averages for three independent experiments.
Elk-1 associates with the HTLV-1 LTR in infected cells.
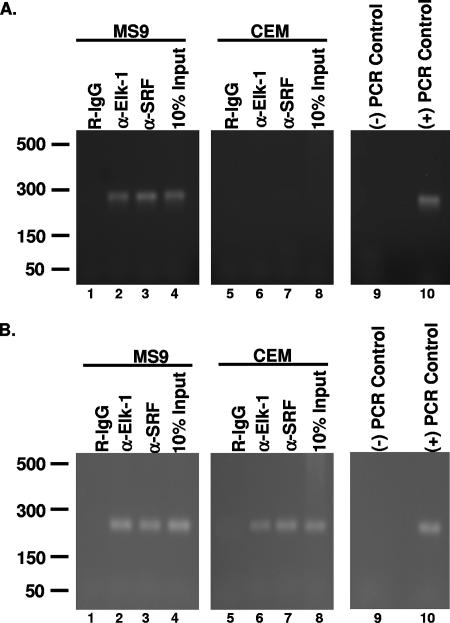
Since in vitro EMSA and in vivo transactivation studies discussed above suggest that Elk-1 associates with the HTLV-1 LTR in vivo, ChIP was performed to rigorously test this possibility. Extracts from formaldehyde-cross-linked MS9 (HTLV-1 positive) or CEM (HTLV-1 negative) human T-cell lines were precipitated with Elk-1-specific antibody, SRF-specific antibody, or normal rabbit immunoglobulin G (IgG), and PCR was performed using primers specific for the SRE regions of either the LTR or c-fos promoter (positive control). Since we had previously demonstrated that SRF could bind the LTR, an antibody to SRF was used as a positive control (31). As expected, the HTLV-1-positive MS9 cells produced a positive PCR signal for both the LTR (Fig. 5A) and c-fos promoter (Fig. 5B) when Elk-1 and SRF antibodies were used (lanes 2 and 3). These results demonstrate that SRF and ELK-1 associate both with the viral LTR and with the c-fos promoter in vivo. ChIP analysis of uninfected CEM cells demonstrated binding of Elk-1 and SRF to the c-fos promoter but not to the HTLV-1 LTR, which was expected since CEM cells do not contain the HTLV-1 LTR (Fig. 5A and B, lanes 6 and 7). The negative control rabbit IgG did not produce a positive PCR signal in any of our samples (Fig. 5A and B, lanes 1 and 5). The association of Elk-1 with the viral LTR in vivo further supports our hypothesis that Elk-1 is a transcriptional regulator of the HTLV-1 promoter.
FIG. 5.
Elk-1 associates with the vSRE in vivo. (A) Cross-linked MS9 (HTLV-1-positive) or CEM (HTLV-1-negative) nuclear lysates were immunoprecipitated using protein G beads prebound with anti-Elk-1 (α-Elk-1) or anti-SRF (α-SRF) antibody. Precipitated DNA was amplified using LTR-specific primers. Rabbit IgG was used as a negative control. Ten percent of the MS9 or CEM lysate used for IPs was PCR amplified as a positive control. Negative PCR control reactions were carried out in the absence of template. Positive PCR control reactions were carried out with a pU3RLuc plasmid. (B) PCR analysis of MS9 and CEM ChIPs with c-fos promoter-specific primers. DNA samples precipitated with anti-Elk-1 and anti-SRF antibodies as in panel A were amplified with c-fos promoter-specific primers. Negative PCR control reactions were carried out in the absence of template, and positive PCR control reactions used genomic CEM DNA. Each ChIP analysis was performed at least three times, and a representative result is shown.
DISCUSSION
The results of these experiments extend our definition of the vSRE to include the two predicted TCF sites and expand the repertoire of cellular transcription factors that regulate the HTLV-1 LTR to include Elk-1. Not only did Elk-1 bind the vSRE in vitro and in vivo through EMSAs and ChIPs, but we also demonstrated that Elk-1 forms a ternary complex with SRF and the vSRE. Functionally, Elk-1 activated the vSRE independently and in conjunction with SRF.
The Ets family members Ets1 and Ets2 are known to bind and activate the LTR at two locations, termed Ets responsive regions 1 and 2 (ERR-1 and ERR-2) (2). ERR-1 is located from bases −116 to −162 upstream of the transcription start site and therefore overlaps the vSRE (18). Within ERR-1, two Ets1 binding sites were identified, at bases −126 to −134 (ERE-A) and −149 to −157 (ERE-B). Interestingly, ERE-B corresponds to the distal TCF site, but ERE-A does not correspond to the proximal TCF site. Elk-1 and Ets1 are members of the same protein family and bind similar DNA sequences. While the core Ets binding sequence is GGAA, Elk-1 binding requires additional bases, optimized by the sequence CCGGAA (26). Slight differences in the sequence specificity of Ets1 and Elk-1 probably account for the identification of different binding sites for these factors within the LTR. The original characterizations of Ets binding sites in the LTR did not identify the proximal TCF site even though it contains the core GGAA sequence and was protected in footprinting analysis (2, 18). Therefore, the proximal TCF binding site was not previously recognized until this report demonstrated that Elk-1 can bind to, and activate through, the proximal TCF.
Although Elk-1 activated the vSRE, transcriptional activation was not completely abrogated by any of the vSRE mutations. Several possibilities may explain these results. First, SRF may indirectly recruit Elk-1 when the TCF sites are mutated and vice versa. Such indirect complexes may not be as stable as ternary complexes formed when both factors bind to adjacent sites, but the indirect complexes may promote some level of transcription. Second, an Elk-1 binding site, which may exist elsewhere in the viral promoter, could recruit Elk-1 to the promoter even when TCF binding sites within the vSRE are mutated. Third, since Elk-1 and SRF activate the expression of immediate-early genes, many of which are transcription factors, upregulation of one of these genes may encode a transcription factor that could activate the LTR through another element. Continued investigation of the complex regulation of this element will provide new insights into the transcriptional control of viral and cellular gene expression.
Our studies demonstrated that the proximal TCF site bound recombinant Elk-1 more efficiently than did the distal TCF site (Fig. 2C and E). However, Elk-1 did not preferentially activate the proximal TCF site in transfection experiments (Fig. 3). The difference in Elk-1 binding is probably due to slight variation of the distal TCF site sequence (CCGGGAA) from the canonical TCF site (CCGGAA), which is conserved in the proximal TCF site (26). Since ternary complexes containing Elk-1 and SRF formed regardless of TCF site status (Fig. 2C and E), the binding of SRF to the CArG box appears to recruit Elk-1 even if the TCF binding site is mutated. These interactions support equivalent activation from both the distal and proximal TCF reporter constructs (Fig. 3).
To date, an SRE that contains two TCF binding sites has not been described. However, both of the TCF binding sites in the HTLV-1 LTR reside within the reported tolerated distance of TCF binding sites (2 to 35 bases) from a CArG box within a functional SRE (29). Our previously published report characterizing the viral CArG box demonstrated that SRF has weak binding affinity for the vCArG box, probably because it shares only 80% identity with the canonical CArG box (31). Thus, redundancy of the TCF sites in the HTLV-1 promoter may increase the efficiency of SRF recruitment.
The viral promoter contains multiple regulatory elements that are responsive to the viral oncoprotein Tax. Three copies of TRE1 bind CREB/ATF family members and are potently activated in the presence of Tax (32). The other Tax-responsive element, TRE2, can be activated by Tax but not as strongly as the TRE1s (21). Interestingly, Tax is known to activate SRF transcription and to interact with Elk-1 (12, 13, 27). Since the vSRE is located in TRE2, it is possible that the vSRE contributes to transcriptional regulation in both the presence and absence of Tax. Fluctuation of Tax expression during viral infection, may allow the vSRE to modulate viral RNA expression when Tax expression is low.
Identification of an SRE regulated by SRF and Elk-1 within the HTLV-1 LTR establishes a new mechanism of HTLV-1 transcriptional regulation. Continued study of the role of this element during the viral life cycle will increase our understanding of viral gene expression and replication. Both SRF and Elk-1 are activated in response to mitogenic signals (8, 9, 17). Interestingly, HTLV-1 virions have been shown to induce the activation of resting T lymphocytes (11, 15), and SRF is known to be activated upon T-cell activation (9, 20). Thus, viral entry may activate mitogenic pathways, which could consequently result in activation of Elk-1 and SRF. The availability of these factors may allow the vSRE to play an important role in activation of viral gene expression from latency or during the initial phase of infection (20). We hypothesize that the vSRE could also play an important role in reactivation from latency when latently infected cells encounter mitogenic signals that activate Elk-1 and SRF. Continued study of these pathways will provide increased understanding of the mechanisms of HTLV-1 reactivation from latency and may reveal new therapeutic strategies for treatment of infected individuals.
Acknowledgments
We thank Robert Schwartz, the NCI Biological Resources Branch, and the NIH AIDS Research and Reference Reagent Program for providing reagents and members of the Marriott lab for helpful discussions and support. We also thank Diane Wycuff for her expertise and input during the initial stages of this work.
These studies were supported by Public Health Service grants CA-55684 and CA-77371, awarded to S.J.M., from the National Cancer Institute. H.Y.W. was supported in part by NIH training grant T32AI07471.
Footnotes
Published ahead of print on 26 September 2007.
REFERENCES
- 1.Barnhart, M. K., L. C. Connor, and S. J. Marriott. 1997. Function of the human T-cell leukemia-virus type 1 21-base-pair repeats in basal transcription. J. Virol. 71:337-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosselut, R., J. Duvall, A. Gegonne, M. Bailly, A. Hemar, J. N. Brady, and J. Ghysdael. 1990. The product of the c-ets-1 protooncogene and the related Ets2 protein act as transcriptional activators of the long terminal repeat of human T-cell leukemia virus, HTLV-I. EMBO J. 9:3137-3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosselut, R., F. Lim, P.-C. Romond, J. Frampton, J. N. Brady, and J. Ghysdael. 1992. Myb protein binds to multiple sites in the human T cell lymphotropic virus type I long terminal repeat and transactivates LTR mediated expression. Virology 186:764-769. [DOI] [PubMed] [Google Scholar]
- 4.Boyd, K. E., and P. J. Farnham. 1999. Coexamination of site-specific transcription factor binding and promoter activity in living cells. Mol. Cell. Biol. 19:8393-8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, K. E., J. Wells, J. Gutman, S. M. Bartley, and P. J. Farnham. 1998. c-Myc target gene specificity is determined by a post-DNA-binding mechanism. Proc. Natl. Acad. Sci. USA 95:13887-13892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brady, J. N., K.-T. Jeang, J. Duvall, and G. Khoury. 1987. Identification of p40x-responsive regulatory sequences within the human T-cell leukemia virus type I long terminal repeat. J. Virol. 61:2175-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braunstein, M., A. B. Rose, S. G. Holmes, C. D. Allis, and J. R. Broach. 1993. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 7:592-604. [DOI] [PubMed] [Google Scholar]
- 8.Buchwalter, G., C. Gross, and B. Wasylyk. 2004. Ets ternary complex transcription factors. Gene 324:1-14. [DOI] [PubMed] [Google Scholar]
- 9.Charvet, C., P. Auberger, S. Tartare-Decker, A. Bernard, and M. Deckert. 2002. Vav1 couples T cell receptor to serum response factor-dependent transcription via a MEK-dependent pathway. J. Biol. Chem. 277:15376-15384. [DOI] [PubMed] [Google Scholar]
- 10.Dasgupta, P., C. D. Reddy, P. Saikumar, and E. P. Reddy. 1992. The cellular proto-oncogene product Myb acts as transcriptional activator of the long terminal repeat of human T-lymphotropic virus type I. J. Virol. 66:270-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dodon, M. D., A. Bernard, and L. Gazzolo. 1989. Peripheral T-lymphocyte activation by human T-cell leukemia virus type I interferes with the CD2 but not with the CD3/TCR pathway. J. Virol. 63:5413-5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujii, M., T. Niki, T. Mori, T. Matsuda, M. Matsui, N. Nomura, and M. Seiki. 1991. HTLV-I Tax induces expression of various immediate early serum responsive genes. Oncogene 6:1023-1029. [PubMed] [Google Scholar]
- 13.Fujii, M., H. Tsuchiya, T. Chuhjo, T. Akizawa, and M. Seiki. 1992. Interaction of HTLV-I Tax1 with p67SRF causes the aberrant induction of cellular immediate early genes through CArG boxes. Genes Dev. 6:2066-2076. [DOI] [PubMed] [Google Scholar]
- 14.Fujisawa, J.-I., M. Seiki, M. Sato, and M. Yoshida. 1986. A transcriptional enhancer sequence of HTLV-I is responsible for transactivation mediated by p40x of HTLV-I. EMBO J. 5:713-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gazzolo, L., and M. DucDudon. 1987. Direct activation of resting T lymphocytes by human T-lymphotropic virus type I. Nature 326:714-717. [DOI] [PubMed] [Google Scholar]
- 16.Gessain, A., F. Barin, J. C. Vernant, O. Gout, L. Maurs, A. Calander, and G. DeThe. 1985. Antibodies to human T-lymphotropic virus type I in patients with tropical spastic paraparesis. Lancet ii:407-409. [DOI] [PubMed] [Google Scholar]
- 17.Gineitis, D., and R. Treisman. 2001. Differential usage of signal transduction pathways defines two types of serum response factor target gene. J. Biol. Chem. 276:24531-24539. [DOI] [PubMed] [Google Scholar]
- 18.Gitlin, S. D., R. Bosselut, A. Gegonne, J. Ghysdael, and J. N. Brady. 1991. Sequence-specific interaction of the Ets1 protein with the long terminal repeat of the human T-lymphotropic virus type I. J. Virol. 65:5513-5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hipskind, R. A., V. N. Rao, C. G. Mueller, E. S. Reddy, and A. Nordheim. 1991. Ets-related protein Elk-1 is homologous to the c-fos regulatory factor p62TCF. Nature 354:531-534. [DOI] [PubMed] [Google Scholar]
- 20.Lin, H. C., M. Hickey, L. Hsu, D. Medina, and A. B. Rabson. 2005. Activation of human T cell leukemia virus type 1 LTR promoter and cellular promoter elements by T cell receptor signaling and HTLV-1 Tax expression. Virology 339:1-11. [DOI] [PubMed] [Google Scholar]
- 21.Marriott, S. J., I. Boros, J. F. Duvall, and J. N. Brady. 1989. Indirect binding of human T-cell leukemia virus type I tax1 to a responsive element in the viral long terminal repeat. Mol. Cell. Biol. 9:4152-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matthews, M.-A. H., R.-B. Markowitz, and W. S. Dynan. 1992. In vitro activation of transcription by the human T-cell leukemia virus type I Tax protein. Mol. Cell. Biol. 12:1986-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osame, M., K. Usuku, S. Izumo, N. Ijichi, H. Amitani, A. Igata, M. Matsumoto, and M. Tara. 1986. HTLV-I associated myelopathy, a new clinical entity. Lancet i:1031-1032. [DOI] [PubMed] [Google Scholar]
- 24.Paskalis, H., B. K. Felber, and G. N. Pavlakis. 1986. cis-acting sequences responsible for the transcriptional activation of human T cell leukemia virus I constitute a conditional enhancer. Proc. Natl. Acad. Sci. USA 83:6558-6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao, V. N., K. Huebner, M. Isobe, A. ar-Rushdi, C. M. Croce, and E. S. Reddy. 1989. elk, tissue-specific ets-related genes on chromosomes X and 14 near translocation breakpoints. Science 244:66-70. [DOI] [PubMed] [Google Scholar]
- 26.Shore, P., A. J. Whitmarsh, R. Bhaskaran, R. J. Davis, J. P. Waltho, and A. D. Sharrocks. 1996. Determinants of DNA-binding specificity of ETS-domain transcription factors. Mol. Cell. Biol. 16:3338-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shuh, M., and D. Derse. 2000. Ternary complex factors and cofactors are essential for human T-cell leukemia virus type 1 Tax transactivation of the serum response element. J. Virol. 74:11394-11397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shuh, M., S. A. Hill, and D. Derse. 1999. Defective and wild-type human T-cell leukemia virus type I proviruses: characterization of gene products and trans-interactions between proviruses. Virology 262:442-451. [DOI] [PubMed] [Google Scholar]
- 29.Treisman, R., R. Marais, and J. Wynne. 1992. Spatial flexibility in ternary complexes between SRF and its accessory proteins. EMBO J. 11:4631-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wycuff, D. R., M. D. Goff, and S. J. Marriott. 2001. Identification of an Initiator-like element within the HTLV-I promoter. Virology 280:72-79. [DOI] [PubMed] [Google Scholar]
- 31.Wycuff, D. R., H. L. Yanites, and S. J. Marriott. 2004. Identification of a functional serum response element in the HTLV-I LTR. Virology 324:540-553. [DOI] [PubMed] [Google Scholar]
- 32.Yanites, H. L., K. K. Woodard, and S. J. Marriott. 2003. Modular control of HTLV-I transcription: a story of cooperation and competition. Curr. Top. Virol. 3:109-117. [Google Scholar]
- 33.Yoshida, M., I. Miyoshi, and Y. Hinuma. 1982. Isolation and characterization of retrovirus from cell lines of human adult T-cell leukemia and its importance in the disease. Proc. Natl. Acad. Sci. USA 79:2031-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao, L. J., and C. Z. Giam. 1992. Human T-cell lymphotropic virus type I (HTLV-1) transcriptional activator, Tax, enhances CREB binding to HTLV-1 21 base pair repeats by protein-protein interaction. Proc. Natl. Acad. Sci. USA 89:7070-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]