Abstract
The helper component-proteinase (HC-Pro) protein of potyviruses is a suppressor of gene silencing and has been shown to elicit plant developmental-defect-like symptoms. In Zucchini yellow mosaic virus (ZYMV), a mutation in the highly conserved FR180NK box of HC-Pro to FI180NK causes attenuation of these symptoms. At 5 days postinoculation and before symptoms appear, virus accumulation, HC-Pro protein levels, and viral short interfering RNA (siRNA) levels are similar for the severe (FRNK) and attenuated (FINK) strains. At this stage, ZYMVFRNK caused greater accumulation of most microRNAs (miRNAs), and especially of their complementary miRNA “passenger” strands (miRNA*s), in systemically infected leaves than the attenuated ZYMVFINK did. HC-ProFRNK specifically bound artificial siRNA and miRNA/miRNA* duplexes with a much higher affinity than the mutated HC-ProFINK. Further analysis of the mutant and wild-type HC-Pro proteins revealed that suppressor activity of the ZYMV HCFINK mutant was not diminished. However, the FINK mutation caused a loss of HC-Pro suppressor function in other potyviruses. Replacement of the second positively charged amino acid in the ZYMV FRNK box to result in FRNA also caused symptom attenuation and reduced small RNA duplex-binding affinity without loss of suppressor activity. Our data suggest that the highly conserved FRNK box in the HC-Pro of potyviruses is a probable point of contact with siRNA and miRNA duplexes. The interaction of the FRNK box with populations of miRNAs directly influences their accumulation levels and regulatory functions, resulting in symptom development.
Plants infected with viruses exhibit diverse symptoms, including pleiotropic developmental defects unique to virus infection (27, 71). To combat the damage caused by these intercellular parasites, plants possess an antiviral defense based on degradation of foreign RNA, known as RNA silencing or RNA interference (33, 65). In order to counteract RNA silencing within their hosts, plant viruses have acquired genes that encode suppressors that prevent degradation of their genomes and mRNAs (50, 65). RNA-silencing suppressors (RSS) in various plant virus families have been identified (67), and interestingly, they share no obvious similarities at either the nucleic acid level or the protein level, suggesting that they function through a variety of mechanisms.
The RSS of the Potyviridae family is a multifunctional protein named the helper component-proteinase (HC-Pro) (57). HC-Pro is essential for systemic infection in the genus Potyvirus, which is in contrast to the case for Wheat-streak mosaic virus (a tritimovirus), from which the entire HC-Pro can be removed without abolishing infectivity (55). Several different HC-Pro functions have been identified and mapped, including protease cleavage of the viral polyprotein (64), insect vector transmission (47), capsid binding (7), two single-stranded RNA-binding areas (58), viral movement and replication (11, 26), virus synergism (53, 68), and RNA-silencing suppression (3, 9, 25). Biochemical studies have shown that HC-Pro binds rgs-CaM (an endogenous suppressor of gene silencing) (2), inhibits the 20S proteosome endonuclease activity (6), and limits methylation of virus-derived small RNAs (smRNAs) (13, 74). HC-Pro forms dimers/multimers and is active in these forms (48, 51, 59). Schematically, the HC-Pro protein can be divided into three regions: an N-terminal region associated with aphid transmission, a C-terminal region associated with proteinase and RSS activity (62), and a central region implicated in many functions, including RSS activity (48). The central region contains several motifs that are highly conserved in all potyviruses, including the FRNK box at amino acid positions 179 to 182 in mature HC-Pro, which is associated with Zucchini yellow mosaic virus (ZYMV) symptom severity (18). The mutation of the FRNK box to FINK (R180I) causes a drastic reduction in symptom severity of the leaves of various cucurbit species without noticeably affecting virus accumulation or infectivity, and this mutation has been exploited for use in cross-protection (19). Other mutations in the central region of different potyviruses which are not located in the FRNK box also cause attenuation of symptoms without affecting virus accumulation (52).
Recently, a smRNA duplex-binding function has been discovered for HC-Pro (29, 40). This ability implies an effect on plant defense mechanisms and plant development, because short interfering RNAs (siRNAs) and microRNAs (miRNAs) are sequestered in an inactive duplex form. This smRNA binding strategy is similar to that of other RSS, such as the tombusvirus P19 protein and the P21 of closteroviruses (30, 54, 73).
RNA interference is based on smRNAs of 20 to 25 nucleotides (nts) which confer sequence specificity to regulatory or defensive pathways (22). Regulatory smRNAs include endogenous miRNAs (49) and trans-acting siRNAs (46) that originate from distinct loci and processing mechanisms. Other siRNAs are formed from double-stranded RNA (dsRNA) produced by RNA-dependent RNA polymerase (RDR) activity and derived from transposable elements and viral RNA. siRNAs can also originate from regions of viral RNA with intrinsic secondary structure (42). All smRNAs are processed by cleavage of dsRNA by one of several members of the DICER-like (DCL) enzyme family (23, 45). The cleavage products are 20- to 25-nt duplex dsRNA molecules that are bound by the RNA-induced silencing complex (RISC). From this duplex, the “passenger” or “star” strand (miRNA*) is released and rapidly degraded, while the remaining guide RNA molecule is incorporated into active RISC. This strand acts as a guide to bring the activated RISC complex into contact with complementary target RNAs, thereby mediating their cleavage and degradation (63).
Some symptoms of potyvirus infection can be phenocopied by the expression of HC-Pro in the absence of virus. Transgenic Arabidopsis thaliana and soybeans (Glycine max) with high and low HC-Pro expression levels exhibit severe and mild developmental abnormalities, respectively (35, 41). It has been hypothesized that these symptoms are caused by deregulation of the endogenous smRNA pathways (27). Because genes with mutations conferring defects in miRNA accumulation, such as dcl1, hen1, hyl1, ago1, and se1, have pleiotropic developmental phenotypes in arabidopsis (37), it was deduced that the similarity between virus symptoms and these phenotypes is caused by the disruption of shared components in these pathways. Indeed, miRNA and, strikingly, miRNA* levels rise upon viral infection (10, 12, 27, 39, 76). As a consequence of sequestration of miRNA duplexes by the viral suppressor, their mRNA targets initially accumulate to higher levels in virus-infected plants due to reduced cleavage by RISC (27, 76). Following the discovery that HC-Pro binds duplex smRNA (29, 40), we analyzed the effects of the expression of this protein in attenuated-symptom-causing forms of ZYMV. We present data indicating that the FRNK box of HC-Pro is required for smRNA binding and that this activity is correlated with differential miRNA sequestration and symptom severity.
MATERIALS AND METHODS
Plant material and virus inoculation.
Squash (Cucurbita pepo L. cv. Ma'ayan) seeds were sown in pots filled with a tuff-peat mixture amended with nutrients and placed in a growth chamber under continuous light at 23°C. Cotyledons of 5-day-old plants were inoculated with a plasmid containing an infectious cDNA encoding the full-length ZYMV under control of the 35S Cauliflower mosaic virus promoter (17). Inoculation was done by particle bombardment with a handheld device, the handgun (16). Squash leaves were collected on ice at different time points after inoculation for RNA and protein extraction. Leaves of plants that were not inoculated or were infected with either ZYMVFRNK (severe strain) or ZYMVFINK (attenuated strain) were collected at 5, 7, and 14 days postinoculation (dpi). Three independent groups of pooled plants were used as biological replicates with each pool consisting of partial leaves from seven to nine separately grown plants. Because symptoms were not clearly visible at 5 dpi, samples were not pooled until ZYMV infection was confirmed.
Viral constructs.
The ZYMV protein sequences used in this study were from the non-aphid-transmissible virus (ZYMVFRNK, GenBank accession no. EF062582) that induces severe symptoms and the attenuated ZYMV AGII (ZYMVFINK, GenBank accession no. EF062583) (15, 18), which differ by a single amino acid (R180I). To facilitate detection of HC-Pro by Western blotting, we added an N-terminal influenza virus hemagglutinin (HA) tag to HC-Pro. The wild-type and mutant versions of HC-Pro were PCR amplified by using a 5′ primer containing SalI and the HA tag and a 3′ primer containing BamHI. These PCR products were ligated into pKS35S-FLC-HIS, a His-tagged HC-Pro-containing version of ZYMV cited in reference 24 and digested with SalI and BamHI.
The attenuated ZYMVFINK infectious viral constructs expressing green fluorescent protein (GFP) or Cucumber mosaic virus (CMV) Fny 2b were designated ZYMV-AGII-GFP and ZYMV AGII-2b and were described in references 4 and 69. To insert the FRNK, FKNK, and FRNA HC-Pro mutations into ZYMV-AGII-GFP, we used splicing-overlap extension-PCR. Each of the resulting PCR products contained 5′ BstEII and 3′ BamHI restriction sites, which allowed them to be inserted into ZYMV-AGII-GFP digested with BstEII/BamHI (E. Haronsky, unpublished data).
Binary constructs.
To prepare binary constructs for the overexpression of HC-Pro in Nicotina benthamiana via infiltration with Agrobacterium tumefaciens (i.e., agroinfiltration), we used pBin19-GFP, which encodes GFP between the 35S promoter and a nopaline synthase terminator. The GFP coding region of this plasmid was excised with XbaI/NotI and replaced with an HA-tagged HC-Pro insert. This insert was prepared by PCR with a 5′ primer containing XbaI and the HA tag and a 3′ primer containing a stop codon and NotI (Haronsky, unpublished). For overexpression of Tobacco etch virus (TEV) HC-Pro in N. benthamiana, the GFP coding region of pBin19-GFP was excised with SalI/NotI and replaced with an HA-tagged TEV HC-Pro insert. This insert was prepared by PCR amplification with a 5′ primer containing SalI and the HA tag and a 3′ primer containing a stop codon and NotI. pGreen-TEV-HCFRNK and pGreen-TEV-HCFINK were used as templates for these PCRs (T. Arazi, unpublished data).
To prepare the GFP-silencing inverted-repeat construct (IR), pEGFP (Clontech) was PCR amplified by using primers designed according to the work of Wesley et al. (70). The PCR products were cloned into the appropriate restriction sites of the pHannibal vector (70). The resulting GFP-IR construct with accompanying regulatory sequences was then transferred to the binary vector pART27 (70) digested with NotI.
smRNA Northern blot hybridization analysis.
Total squash or N. benthamiana RNA was prepared from leaves by using TRI reagent (MRC). Two types of 15% acrylamide-8 M urea gels were used. Most gels were buffered, electrophoresed (by use of the Bio-Rad Mini-PROTEAN instrument with a 1.5-mm-thick comb), and electrotransferred (Bio-Rad semidry blotting system) in 0.5× Tris-borate-EDTA (TBE). smRNAs were transferred to a Nytran SPC membrane (Whatman) and cross-linked with 0.24 J UV light in a Stratalinker (Stratagene). Alternatively, a highly sensitive EDC [1-ethyl-3-(3-dimethylaminopropyl)carbodiimide] cross-linking protocol was performed exactly as described by Pall et al. (43). These gels were buffered and electrophoresed in 20 mM MOPS (3-[N-morpholino]propanesulfonic acid) at pH 7, and smRNAs were electrotransferred in water to a Hybond NX membrane (GE). For the TBE protocol, 20 μg of total RNA in 50% formamide per lane was loaded, while only 10 μg of the same was used in the EDC protocol. Despite the difference in sensitivity between these two protocols, they yielded identical conclusions.
For detection of miRNAs and miRNA* sequences, antisense oligonucleotide probes were synthesized to known (http://microrna.sanger.ac.uk/sequences/) or novel miRNAs discovered by bioinformatic analysis of Cucumis melo and Cucumis sativus expressed sequence tags (see Fig. S1 in the supplemental material). Probe sequences were chosen regardless of organism according to the highest signal obtained from the microarray described in the following section. Probe sequences were miR159 (5′-AAGAGCTCCCTTCAATCCAAA-3′), miR159* (5′-TATTGGACTTCAAGGAGCTCCA-3′), miR166 (5′-GGGAATGAAGCCTGGTCCGA-3′), miR166* (5′-CCTCGAGCCAGCCAACATTCC-3′), miR168 (5′-TTCCCGACCTGCACCAAGCGA-3′), and miR168* (5′-ATTCAGTTGATGCAAGGCGGG-3′). The source organisms for the probes and their full names are detailed in Fig. S1 in the supplemental material. Probes were radioactively end labeled with [γ-32P]ATP by using T4 polynucleotide kinase (New England Biolabs). End-labeled probes were hybridized overnight at 40°C in rapid hybridization buffer (Biological Industries, Beit Ha'emek, Israel), and blots were washed twice for 15 min in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.2% sodium dodecyl sulfate (SDS) at 50°C.
Probes for detecting GFP siRNAs were made by linearizing pBluescript-GFP with SalI followed by in vitro transcription using T7 RNA polymerase (Fermentas) in the presence of [α-32P]UTP. Hybridization was performed overnight in a solution of 2× SSC, 1% SDS, and 100 μg/ml herring sperm DNA (ICN) at 45°C, and the blots were washed twice for 15 min in 2× SSC-0.1% SDS at 40°C.
smRNA-specific microarray.
Microarrays were printed by LC Sciences (Houston, TX) with all known plant miRNA sequences from Sanger miRBase release 7.1 (http://microrna.sanger.ac.uk/sequences/), and corresponding probes were designed to detect miRNA*s. The third experiment utilized an updated microarray containing additional nonredundant miRNA sequences from Sanger miRBase release 10.0. The miRNA* sequences were bioinformatically extracted from the stem-loop sequences of the miRNA precursor and were not verified by cloning and sequencing. The microarray also included computationally discovered miRNAs and miRNA*s from several publications (1, 34, 38). For detection of viral siRNA, the complete genome of ZYMV, in both its sense and antisense orientations, was parsed in silico into 25-nt-long probes that were tiled with a 13-nt overlap and printed. Sense and antisense probe pairs were numbered according to their sense 5′ coordinates. Array hybridization was performed by LC Sciences with smRNA (from 20 μg of total RNA from squash leaves) fractionated with the mirVana miRNA isolation kit (Ambion). This fraction was tailed with poly(A) polymerase and ligated to one of two nucleotide tags (31). Following hybridization to the array, a second hybridization with tag-specific dendrimers labeled with Cy3 or Cy5 dyes was performed. RNA was spiked with several internal-control RNAs that were used for in-chip normalization.
EMSA.
In order to test the binding of HC-Pro to smRNA duplexes, electromobility shift assays (EMSAs) were conducted using a protocol modified from the method of Merai et al. (40). Extracts were prepared by grinding squash or N. benthamiana leaves 1/4 (wt/vol) in gel shift buffer (GSB; 30 mM HEPES [pH 7.3], 66 mM KCl, 100 mM NaCl, 10 mM dithiothreitol, 0.02% Tween 20, 0.8 mM MgCl2, and 4% glycerol). Samples were centrifuged twice for 10 min at 15,000 × g at 4°C, and the supernatant was frozen in aliquots at −80°C. Radiolabeled smRNA duplex probes were prepared using a protocol modified from Lakatos et al. (30). An RNA oligonucleotide (100 pmol) corresponding to human miR16 (5′-UAGCAGCACGUAAAUAUUGGCG-3′) was end labeled with [γ-32P]ATP by using T4 polynucleotide kinase (NEB). After Sepharose G25 gel filtration and ethanol precipitation with NaCl and glycogen, this labeled oligonucleotide was annealed to a 2× molar excess of (cold) phosphorylated human miR16* (5′-CCAGUAUUAACUGUGCUGCUGAA-3′). In addition to this synthetic miRNA/miRNA* duplex, a perfectly complementary artificial siRNA duplex was also created with 2-nt 5′-recessed and 2-nt 3′-overhang ends by using the oligonucleotide 5′-CCAAUAUUUACGUGCUGCUAAA-3′. Annealing was performed by resuspending the pellets in a double concentration (critical for miRNA/miRNA* duplex) of annealing buffer as described in reference 14, modified as follows: 40 mM K-acetate, 12 mM HEPES (pH 7.3), 0.8 mM Mg-acetate in 10 μl, heating to 97°C, and cooling slowly overnight to 4°C. Radiolabeled duplexes were then gel purified from a 15% acrylamide-0.5× TBE gel electrophoresed on ice, ethanol precipitated with NaCl and glycogen, and redissolved in GSB to a working stock of 100 nM. The concentrations of the labeled duplexes were approximated based on an unlabeled duplex gel purified alongside them and measured by use of a spectrophotometer (Nanodrop ND-1000). Unlabeled duplexes for competition assays were not gel purified but were directly quantified by spectrophotometer. EMSA was normally performed by adding 1 μl of radiolabeled probe (final concentration of 4.76 nM) to 20 μl of extract, followed by an incubation at room temperature for 15 min. In competition assays, an extra 1 μl of unlabeled probe brought the radiolabeled probe to 4.55 nM. Ten microliters of this reaction mixture was loaded on the gel with a dye composed of GSB with bromophenol blue and xylene cyanol. Polyacrylamide gel composition was found to be critical (0.5× Tris-borate buffer, 2.5% glycerol, 4% acrylamide-BIS [N,N-methylenebisacrylamide] [58:1], 1/2,000 TEMED [N,N,N′,N′′-tetramethylethylenediamine], 1/133 10% ammonium persulfate). Gels (1-mm thickness; Bio-Rad Mini-PROTEAN system) were polymerized for >3 h at 4°C, washed, and prerun before use. Electrophoresis was done on ice at 75 V for 75 min in 0.5× TBE. The gels were then lifted off the glass plates with dry Whatman 3MM paper, sealed in a polyethylene bag, and exposed to a phosphorimager screen (Fuji) for about 3 h. Images were quantified using ImageJ software.
HC-Pro quantification.
ZYMV HC-Pro was quantified by using Western blot analysis. Leaves from inoculated plants were weighed and homogenized in 3.3 volumes of denaturing ESB buffer (28). Alternatively, when samples were also used for binding assays, they were homogenized in GSB (see above) and treated as described above for the EMSA, after which an aliquot was denatured by the addition of SDS-polyacrylamide gel electrophoresis loading buffer and boiling. Samples (8 μl) were electrophoresed and electroblotted to a nitrocellulose membrane (Protran; Schleicher & Schuell) as described by Kimalov et al. (28). Blots were incubated with a rat monoclonal anti-HA antibody (Roche) at a 1:5,000 dilution in Tris-buffered saline-Tween overnight at 4°C followed by an alkaline phosphatase (AP)-immunoglobulin G (IgG) anti-rat conjugate (Sigma) at a 1:5,000 dilution for an hour at ambient temperature. In experiments with untagged HC-Pro, a polyclonal rabbit anti-ZYMV HC antiserum was used at a 1:4,000 dilution, followed by the AP-IgG anti-rabbit conjugate as described above. TEV HC-Pro was quantified using Western blot analysis with rabbit anti-TEV HC-Pro antiserum at 1:1,000 in Tris-buffered saline-Tween and a goat anti-rabbit AP-IgG as described above. Membranes were developed with BCIP (5-bromo-4-chloro-3-indolylphosphate)-nitroblue tetrazolium (Bio-Rad) according to the manufacturer's instructions, scanned on a tabletop scanner, and quantified using ImageJ software.
GFP suppression assay.
To test RSS activity, we employed a GFP suppression assay similar to that of Brigneti et al. (9). N. benthamiana leaves were agroinfiltrated with plasmids containing a constitutive enhanced GFP-expressing construct, an inverted-repeat GFP-silencing construct (IR), wild-type and mutant HC-Pro constructs, or combinations of these. pBin19-HC-Pro-expressing cultures were agroinfiltrated with A. tumefaciens strain EHA105, and pART-EGFP- and pART-IR-EGFP-expressing cultures were agroinfiltrated with the A. tumefaciens GV2260-C58 strain. In preparative agroinfiltration assays, equal concentrations of HC-Pro-expressing culture were diluted to a 1.5 optical density at 600 nm (OD600) in fresh LB-MES (morpholineethanesulfonic acid) medium and infiltrated into whole leaves. In coinfiltration assays, each culture was diluted to an 0.5 OD600 by the other cultures or a culture containing an insertless plasmid, empty vector (EV), and infiltrated into a restricted sector of the leaves. Protein samples were prepared from two or three leaves per plant as described above for squash leaves. GFP fluorescence was photographed in detached leaves with a digital camera using a yellow filter under a long-wave UV lamp (UVP model B100A).
RESULTS
A mutation in the FRNK box of ZYMV causes attenuation of symptoms.
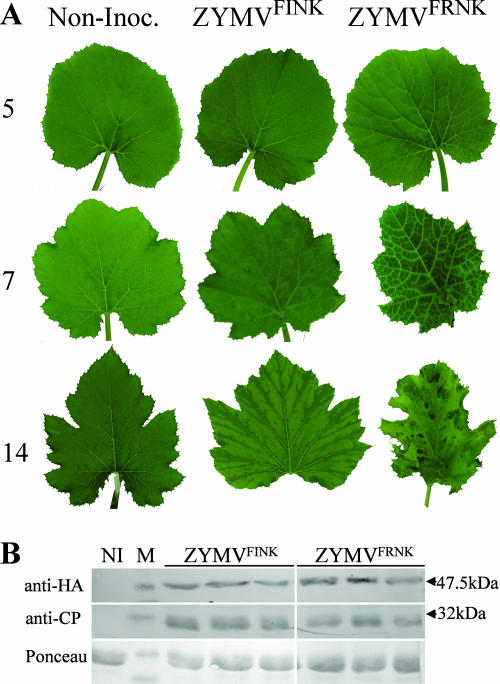
As has been previously reported (18), a point mutation in an infectious ZYMV cDNA clone that changes the HC-Pro FR180NK box to FI180NK causes symptom attenuation in cucurbits. In squash plants, symptoms of infection with both ZYMVFINK and ZYMVFRNK began with subtle vein clearing on the first true leaf at 5 dpi (Fig. 1). In ZYMVFRNK-infected plants, symptoms progressed to vein clearing in the second leaf at 7 dpi and to leaf deformation, dwarfing, and a green-island phenotype by 14 dpi. At later stages, ZYMVFRNK-infected plants became stunted and exhibited developmental abnormalities that were characterized by filamentous leaves displaying green islands (not shown). In contrast, the ZYMVFINK-infected plants appeared very similar to healthy plants and exhibited only a few discernible symptoms, including faint discoloration and very slight fanlike folding along veins (Fig. 1A).
FIG. 1.
A mutation in the FRNK motif of HC-Pro attenuates symptoms without reducing initial HC-Pro levels. (A) Squash (cv. Ma'ayan) leaves showing systemic symptoms at 5, 7, and 14 dpi with either severe (ZYMVFRNK) or attenuated (ZYMVFINK) ZYMV infection. Noninoculated leaves (Non-Inoc.), as a control, are shown. (B) The levels of HA-tagged HC-Pro and CP at 5 dpi were measured by Western blotting by using the antibodies indicated. A noninoculated leaf sample (NI) was loaded as a control. Arrows indicate the sizes of the nearest protein marker (M) bands. Ponceau staining was used to demonstrate equal protein loading.
Attenuation does not affect initial ZYMV HC-Pro levels and viral siRNA accumulation.
HC-Pro is associated with the elicitation of potyvirus-induced symptoms (5). To confirm that the observed symptom attenuation in ZYMVFINK-infected plants was not merely due to reduced HC-Pro concentrations or virus titers, we compared the HC-Pro and coat protein (CP) accumulations of these plants to those of plants infected with the severe ZYMVFRNK. Similar levels of HC-Pro protein and CP were detected in ZYMVFRNK- and ZYMVFINK-infected squash at 5 dpi (Fig. 1B). However, at 7 and 14 dpi, somewhat lower levels of HC-Pro were observed in plants infected with ZYMVFINK (not shown).
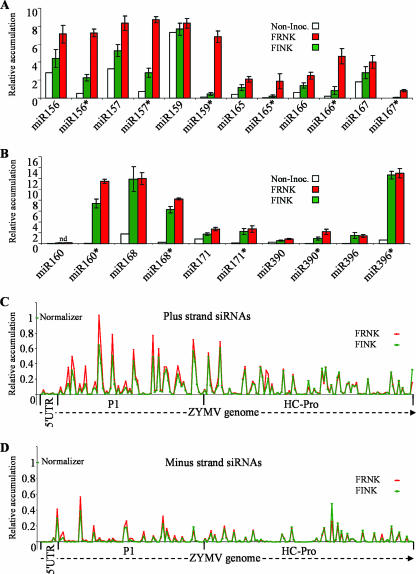
In addition to comparing HC-Pro protein levels, we examined the accumulation of virus-derived siRNA (vsiRNA) produced in planta following infection with ZYMVFRNK and ZYMVFINK. For this purpose, we used a microarray printed with oligonucleotide probes corresponding to the ZYMV genome, miRNA and miRNA* sequences. This microarray was hybridized with low-molecular-weight squash RNA fractions in three separate biological repeats (Fig. 2). ZYMV vsiRNA levels were detected by a series of probes that were 25 nt in length with 13-bp overlaps and covered the full ZYMV genome in both its sense (plus strand) (Fig. 2C) and antisense (minus strand) (Fig. 2D) orientations. Thus, 746 paired oligonucleotide probes were printed for each of the sense and antisense strands. ZYMV vsiRNA levels were assayed in RNA extracted at 5 dpi (Fig. 2C and D). An arbitrary 10% of normalizer signal threshold (0.1 relative accumulations) (Fig. 2) was used to compare the accumulated levels of the different siRNAs. In virus-free plants, no ZYMV probe hybridization signal came near the threshold level, and the signal was on average ∼850-fold lower than the normalizer signal (not shown). At 5 dpi, 21% of the probes for ZYMVFRNK and 26% of the probes for ZYMVFINK from the plus strand crossed the threshold, while only 5% and 10% of the minus-strand probes exceeded this threshold value for ZYMVFRNK and ZYMVFINK, respectively. This observation demonstrates that plus-strand siRNAs accumulate to higher levels than the minus-strand siRNAs. Importantly, the accumulating sense and antisense siRNAs did not match or pair up, demonstrating that there is no correlation between accumulation of the plus-strand vsiRNA and its minus-strand partner.
FIG. 2.
smRNAs of ZYMV-infected squash were quantified by microarray analysis at 5 dpi. The average expression levels of selected miRNAs (miR) and miRNA*s (miR*) that were quantified by using a comprehensive smRNA microarray assay (see Fig. S1 in the supplemental material for sequences shown in Fig. 2A and B). Noninoculated squash plants (Non-Inoc.) were compared to attenuated ZYMVFINK (FINK)- and severe ZYMVFRNK (FRNK)-infected plants. This graphic is arbitrarily divided to present two charts of miRNA/miRNA* pairs. For the pairs presented in panel A, the concentration of FRNK miRNA* is more than threefold greater than the concentration of FINK miRNA*. For the pairs presented in panel B, the concentration of FRNK miRNA* is less than threefold greater than the concentration of FINK miRNA*. The signals were normalized in-chip to a spike-in control RNA, and chip signals were subsequently normalized to each other. The smRNAs that were labeled and hybridized were from pools of seven to nine plants for each treatment. Error bars indicate the SEMs of three independent biological repeats. The relative accumulations of smRNAs derived from the plus (C) and minus (D) strands of the ZYMV genome in ZYMVFRNK (FRNK)- and ZYMVFINK (FINK)-infected leaves were measured. Viral siRNAs were assayed using tiled 25-nt probes on a representative microarray, as described above, that were tiled with a 13-bp overlap. Data shown are for 193 consecutive plus-strand probes and their complementary minus-strand probes that represent the first 2,437 nt, which encode the 5′ UTR protein, P1, and HC-Pro. For clarity in panels C and D, only one of three biological replicates is shown.
Levels of plus-strand ZYMVFRNK vsiRNA probes that exceeded the threshold were similar to those for ZYMVFINK; these probes had mean signals of 25% and 27% of the normalizer signal, respectively. The accumulation of minus-strand ZYMVFRNK vsiRNA was somewhat greater than the accumulation of ZYMVFINK vsiRNA; these probes had mean signals of 20% and 13% of the normalizer signal, respectively. Interestingly, probes producing significant signals were not distributed equally across the ZYMV genome. For the plus strand, no siRNA probes reacted in either the 5′ untranslated region (UTR) (Fig. 2C) or the 3′ UTR (not shown), and for the minus strand, none reacted in the 3′ UTR (not shown). These results were similar in all three replicates of the hybridization. These results demonstrate that symptoms, HC-Pro levels, and vsiRNA levels are similar at 5 dpi in the first true leaf for the two isolates. Thus, this time point and this tissue were used for all subsequent experiments.
miRNA* accumulation is increased in response to ZYMV infection and is considerably affected by viral attenuation.
miRNA and its complementary miRNA* strand have been shown to hyperaccumulate in potyvirus-infected plants (10). To test if miRNA and miRNA* levels are affected by the FRNK box of HC-Pro and are correlated with symptom expression, the levels of all highly conserved miRNAs (75) and their miRNA*s were quantified using the smRNA microarray described in the previous section (Fig. 2A and B). Arrays were hybridized to products of the smRNA fraction from the 5-dpi RNA samples of the three biological replicates. Analysis of the data demonstrates that many miRNAs that are predicted to be conserved are expressed at significant levels in squash (Fig. 2A and B). In addition, internal controls show that the array is very sensitive to single-base mismatches, which caused a >21-fold reduction in signal (not shown). This property allowed us to deduce a nearest estimate of the true miRNA and miRNA* sequences in squash by selecting only the highest average signal among the variants of the different miRNAs and miRNA*s that were available on the microarray (see Fig. S1 in the supplemental material).
In general, miRNA and miRNA* levels in virus-infected plants, and especially in ZYMVFRNK-infected plants, were higher than those in uninfected plants. miRNA* levels were especially elevated in comparison to those of miRNA (Fig. 2A and B). The miRNA* expression levels were usually very low in uninfected plants as expected due to their degradation in vivo. The accumulation of detectable miRNA* in ZYMV-infected plants was always much higher than that in uninfected plants, suggesting that viral infection prevents its degradation. Major differences in miRNA* expression were found between ZYMVFRNK- and ZYMVFINK-infected plants for the group of miRNA*s shown in Fig. 2A. In this group, miRNA* levels in ZYMVFRNK-infected plants were much higher than those in ZYMVFINK-infected plants. For example, the average difference (n-fold) in miR159* accumulations between ZYMVFRNK-infected plants and ZYMVFINK-infected plants was 19.7-fold (standard error of the mean [SEM], ±7.6), and for miR166* accumulation, it was 10.1-fold (SEM, ±4.8) (Fig. 2A). In a second group of miRNA*s shown in Fig. 2B, the differences were slighter. For example, the average difference between miR168* accumulations in ZYMVFRNK-infected plants and ZYMVFINK-infected plants was 1.3-fold (SEM, ±0.2) (Fig. 2B).
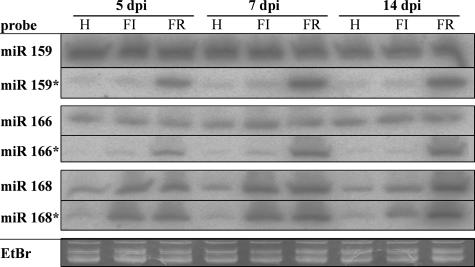
To better understand the kinetics of the expression of these highly and moderately different miRNA/miRNA* pairs in relation to ZYMV infection, the accumulations of miR159/miR159*, miR166/miR166*, and miR168/miR168* were analyzed over a 14-day time course by using smRNA Northern blot analysis (Fig. 3). RNA was extracted at 5 dpi from the first true leaf, at 7 dpi from the second true leaf, and at 14 dpi from the third or fourth leaf. Viral RNA levels, but not HC-Pro protein accumulation levels, were already at their peak at 5 dpi (data not shown). miR159 and miR166 were expressed at high levels in squash leaves (Fig. 3). The levels of both miRNAs were similar in ZYMV-infected plants and noninoculated plants. In contrast to these miRNAs, miR159* and miR166* were hardly detectable in healthy or ZYMVFINK-infected leaves, but they accumulated to very high levels in ZYMVFRNK-infected leaves. The accumulation of these miRNA*s continued to increase in ZYMVFRNK-infected leaves at 7 and 14 dpi.
FIG. 3.
Accumulations of miRNAs and miRNA*s 159, 166, and 168 in ZYMV-infected squash leaves are differently modulated over time by a mutation in the FRNK motif. RNA filter hybridization of noninoculated (H), attenuated ZYMVFINK-infected (FI), and severe ZYMVFRNK-infected (FR) squash RNA with antisense probes to the smRNAs indicated (see Fig. S1 in the supplemental material for the sequences). Ten or 20 μg of total RNA that was isolated from eight plants per treatment was loaded per lane. Days postinfection (dpi) are indicated at the top of the figure. Ethidium bromide (EtBr) staining in the tRNA zone is shown as an equal-loading control for one of the six similar gels.
miR168 was expressed in healthy squash leaves, and infection with either ZYMVFINK or ZYMVFRNK caused its increased accumulation. At 5 and 7 dpi, miR168 levels in ZYMVFRNK-infected leaves were initially similar to those seen in ZYMVFINK-infected leaves. However, at 14 dpi, the level in the ZYMVFRNK-infected leaves was higher than that observed in the ZYMVFINK-infected leaves (Fig. 3). Healthy leaves accumulated very low levels of miR168*, whereas ZYMV-infected leaves accumulated high levels of miR168*. In contrast to miR159* and miR166*, miR168* levels were initially quite similar for ZYMVFRNK- and ZYMVFINK-infected leaves at 5 and 7 dpi, and they decreased somewhat in ZYMVFINK at 14 dpi. This pattern of expression was very similar to that observed for the corresponding miRNA. This experiment shows that the majority of the effect of ZYMV infection on miRNA/miRNA* accumulation is already evident in the first leaf at 5 dpi.
The FRNK box is required for smRNA duplex binding.
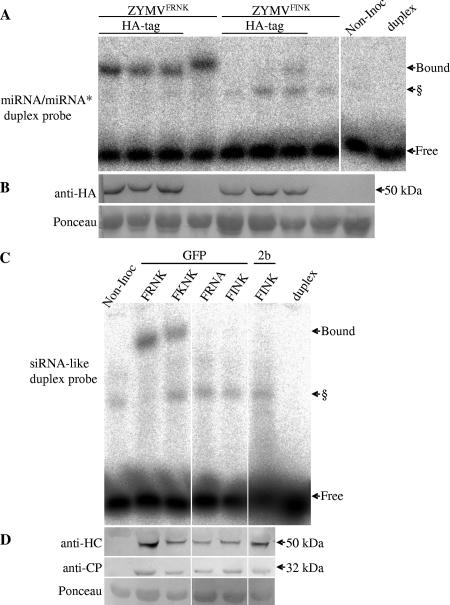
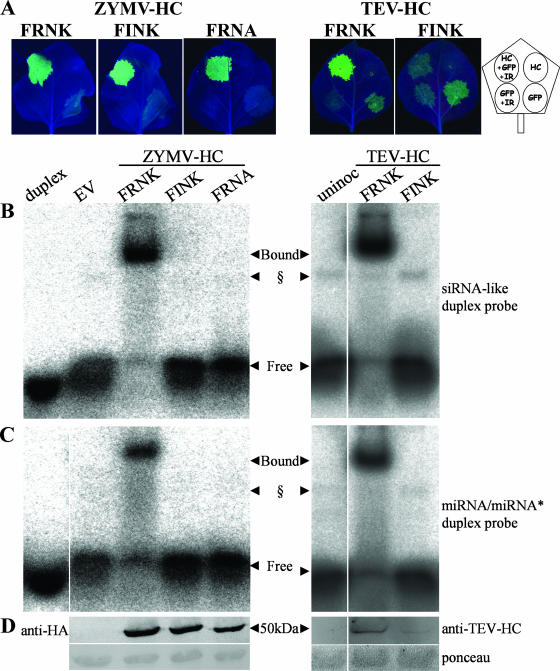
HC-Pro has previously been demonstrated to bind smRNA duplexes in vitro (29). To examine whether mutations in the FRNK box of HC-Pro influence miRNA/miRNA* duplex binding, an EMSA was conducted using extracts prepared from ZYMVFRNK- and ZYMVFINK-infected squash leaves. These extracts were incubated with an artificial miRNA/miRNA* duplex based on human miR16 that was radiolabeled and phosphorylated (Fig. 4A). To facilitate HC-Pro quantification, ZYMV constructs with an HA-tagged HC-Pro were used in three biological replicates per construct. The results show that the extract from ZYMVFRNK-infected leaves binds a fraction of the radiolabeled duplex that is not bound in healthy plants. In contrast, the ZYMVFINK extract binds much less of the duplex, frequently to an undetectable level. The untagged ZYMVFRNK HC-Pro protein also bound the duplex but migrated slightly more slowly than the HA-tagged protein. A putative plant-endogenous duplex-binding protein was often visible in both healthy and virus-infected samples (see also Fig. 6 and 7). Western blot analysis with anti-HA antibody detected similar levels of HC-Pro accumulation in ZYMVFRNK- and ZYMVFINK-infected plants (Fig. 4B). This result demonstrates that the reduced binding in the ZYMVFINK extracts was not due to lower HC-Pro levels.
FIG. 4.
The ZYMV HC-Pro FRNK motif is involved in double-stranded smRNA duplex binding. Squash leaf extracts were prepared for EMSA from ZYMV-infected (A and B) or foreign-gene-expressing, ZYMV-infected (C and D) plants. Panel A shows HC-Pro binding to an artificial miRNA/miRNA*-like duplex probe, and panel C shows HC-Pro binding to an artificial siRNA-like duplex probe. The corresponding protein-level controls for each experiment are shown in the Western blots in panels B and D. (A and B) Three leaf samples with HA-tagged HC-Pro-expressing virus and one sample with virus expressing untagged protein were collected at 5 dpi (for either severe ZYMVFRNK- or attenuated ZYMVFINK-infected plants), along with a noninoculated control sample (Non-Inoc.). (C and D) Representative samples from leaves inoculated with ZYMV clones that expressed GFP or CMV Fny 2b (2b) and had the indicated mutations in the FRNK motif. Bound and free radiolabeled duplexes are indicated by arrows. The radiolabeled probe alone was run as a control (duplex). § denotes a putative endogenous plant RNA duplex-binding protein that was sometimes observed in EMSA of infected and noninoculated samples. The miRNA/miRNA* duplex probe was based on the human precursor of miR16 and included mismatches and noncanonical base pairing, and the siRNA-like duplex probe had an artificial fully matching sequence with 3′ overhangs. Western blots were probed with monoclonal anti-HA antibody (B) or polyclonal anti-ZYMV HC-Pro and anti-ZYMV CP antibodies (D). Ponceau staining of the same membrane, as an equal loading control, is shown. The expected sizes of the HC-Pro monomer and the CP are marked by the arrows (50 kDa and 32 kDa, respectively).
FIG. 6.
Replacement of positively charged amino acids in two different locations of ZYMV HC-Pro reduces duplex RNA binding without abolishing RSS activity. (A) ZYMV HCFINK and ZYMV HCFRNA overexpressed in N. benthamiana are active as suppressors, while TEV HCFINK is inactive. N. benthamiana leaves were agrobacterium infiltrated with different combinations of binary constructs overexpressing the different HC-Pro constructs ZYMV HCFRNK, ZYMV HCFINK, ZYMV HCFRNA, TEV HCFRNK, and TEV HCFINK. ZYMV suppressors were HA tagged. Pictures of detached leaves were taken at 4 dpi (right panel) or 7 dpi (left panel) under UV illumination. The diagram (far right) shows the leaf infiltration pattern with binary constructs (HC-Pro construct [HC], GFP-expressing construct [GFP], GFP-silencing inverted-repeat construct [IR]). All cultures were diluted in each other or in culture with EV to an OD600 of 0.5. (B and C) Binding assay. Leaf extracts from a separate leaf from the same plants as those in panel A were collected at 3 dpi for EMSA, which was conducted with radioactive artificial siRNA (B) or miRNA (C) duplexes. (D) The same extracts were used for Western blot analysis with the antibodies shown. The amounts of HC-Pro protein loaded per lane were estimated to be 1.7 μg for ZYMV HCFRNK and 1.1 μg for ZYMV HCFINK. TEV HCFRNK protein levels were estimated to be 10-fold greater than TEV HCFINK levels. The concentration of each probe used in the binding assays was 5 nM. § marks a putative plant-endogenous duplex-binding protein that is sometimes observed. uninoc, uninoculated.
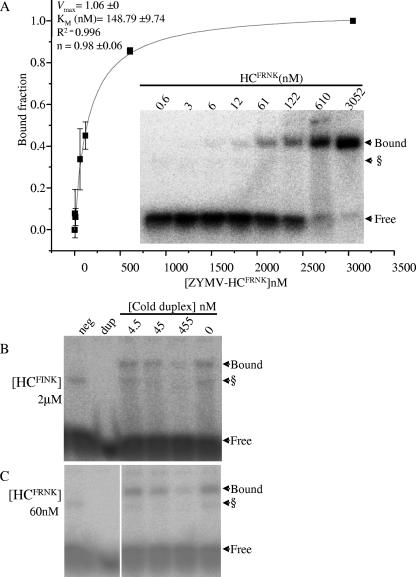
FIG. 7.
ZYMV HCFRNK binds an siRNA-like duplex with a much greater affinity than ZYMV HCFINK. (A) A dilution series of N. benthamiana leaf extract expressing ZYMV HCFRNK was assayed in an EMSA binding reaction with 4.76 nM siRNA-like duplex. A representative EMSA gel is shown (inset). Samples were diluted in EV-infiltrated leaf extracts to the HC-Pro concentrations indicated. The Hill plot (line graph) was derived from the averages of four such experiments. Signals were quantified from phosphorimager data using ImageJ software. The Hill plot and biochemical values (top left) were generated using Origin 7.5 software. Bars show standard deviations for the four replicates. n, apparent Hill coefficient. (B) Electromobility shift competition assay. γ-32P-radiolabeled siRNA-like duplex was premixed to 4.54 nM with different concentrations of nonradioactive phosphorylated competitor duplex (cold) and incubated with N. benthamiana extracts expressing HA-tagged ZYMV HCFRNK or HCFINK. Competition assays were conducted with undiluted (2 μM) HCFINK-expressing extract (B) and at a 50-fold dilution (in uninoculated leaf extract) for HCFRNK (60 nM) (C). Extract from noninoculated leaves (neg) and RNA duplex without extract (dup) were run as controls. § marks a putative plant endogenous duplex-binding protein sometimes seen in EMSA.
The R180-to-I180 mutation in ZYMV HC-Pro maintains suppressor activity.
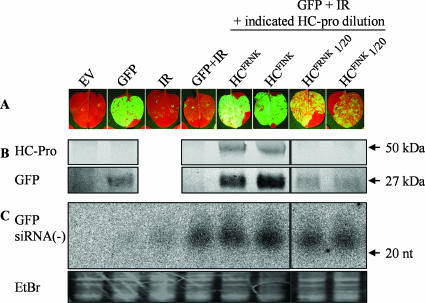
Since a major difference in ability to bind a smRNA duplex was observed between the mutant and wild-type ZYMV HC-Pro proteins, we examined their relative RSS activities in the absence of the virus by using a GFP-silencing assay. The wild-type HC-Pro gene of ZYMV and its R180-to-I180 mutant were cloned into a binary vector and were tested for RSS activity by agroinfiltration into N. benthamiana leaves (Fig. 5). A GFP-expressing construct and a GFP-IR silencing construct were used in conjunction with HA-tagged ZYMV HC-Pro constructs. GFP fluorescence could be detected at 2 dpi and was completely silenced by IR at this time (Fig. 5A and B). No differences in GFP fluorescence, GFP protein accumulation, or minus-strand GFP siRNA were observed between the wild-type and mutant HC-Pro constructs (Fig. 5). In order to test if the R180I mutation had more subtle effects on RSS activity, each of the HC-Pro-expressing cultures was diluted 1/20 in the EV-containing culture, while holding the GFP and IR dilutions constant. The dilution weakened their RSS activities equally, as elucidated from the similar reductions in GFP accumulation and levels of GFP siRNA that were observed (Fig. 5).
FIG. 5.
The suppressor activities of attenuated forms of HC-Pro are not compromised. N. benthamiana was agrobacterium infiltrated with equal culture concentrations with different combinations of binary constructs expressing GFP, a GFP inverted repeat (IR), the HA-tagged ZYMV HC-Pro constructs HCFRNK (wild type) and HCFINK (attenuated), or the EV. HC-Pro-expressing cultures were also diluted 20-fold (1/20) in EV-containing culture, while the GFP and IR concentrations were held constant. (A) Photographs were taken under UV light at 4 days postinfiltration, and protein and RNA samples were taken at the same time. (B) HC-Pro and GFP expression levels were detected by Western blotting in equal amounts of protein extract, probed with anti-HA or with anti-GFP monoclonal antibodies. The expected sizes of the HC-Pro monomer and GFP are marked by the arrows (50 kDa and 27 kDa, respectively). (C) GFP siRNA accumulation was detected by use of smRNA Northern blots containing 10 μg of total RNA per lane. The membrane was hybridized with radiolabeled sense-strand GFP RNA transcript. Size was assessed with DNA oligonucleotides stained with ethidium bromide (EtBr). The EtBr-stained gel for the tRNA zone is shown as an equal-loading control.
Suppressor activity and smRNA binding of HC-Pro in potyviruses.
To examine the general significance of the FR180NK box in potyviruses, we repeated the GFP-silencing assay with the FI180NK mutant by using ZYMV, TEV, Turnip mosaic virus (TuMV), and Potato virus Y (PVY) HC-Pro proteins. In this experiment, GFP could be detected at 4 dpi (Fig. 6A). GFP silencing also occurred without the inverted-repeat construct, as evident from the stronger GFP fluorescence in the presence of an active suppressor. The wild-type HCFRNK of each of the four potyviruses tested in this assay was a functional suppressor of GFP silencing (Fig. 6A; data for TuMV and PVY are not shown). Although the ZYMV HCFINK was an active RSS, the corresponding TEV, PVY, and TuMV mutants were not active, as demonstrated by their inability to suppress GFP silencing (Fig. 6A; data for PVY and TuMV are not shown).
Binding assays for the artificial siRNA (Fig. 6B) and miRNA/miRNA* (Fig. 6C) duplexes were performed with N. benthamiana leaf extracts. ZYMV and TEV HCFRNK leaf extracts bound both duplex types to nearly full depletion of the free duplex. Artificial siRNA duplex binding could also be detected for ZYMV HCFINK, although at a much lower level (see also Fig. 7B), but this could not be detected at all for its TEV counterpart. The difference in binding between ZYMV extracts was not due to differences in protein accumulation as determined by Western blot analysis (Fig. 6D). The ZYMV HCFINK level was estimated to be 1.1 μg/lane, compared to 1.7 μg/lane for ZYMV HCFRNK, by using a dilution series of purified HC-Pro. TEV HCFINK levels, on the other hand, were about 10-fold lower than those of TEV HCFRNK. We diluted TEV HCFRNK 100-fold, and it still bound siRNA-like duplex (data not shown), indicating that the lower expression level of TEV HCFINK protein is not sufficient to explain the abolished duplex binding. Consistent with the results described above, TEV and TuMV both lost their ability to infect their hosts when their HC-Pro proteins contained the mutation of R180 to I180 (data not shown).
Both positively charged amino acids of the FRNK box of ZYMV HC-Pro affect duplex binding and symptom severity.
A positive-charge-retaining mutant, the R180K mutant (ZYMVFKNK) had a smRNA duplex-binding affinity similar to that of the wild-type ZYMVFRNK virus (Fig. 4C) and caused severe squash leaf symptoms (Haronsky, unpublished). To investigate the importance of other positively charged amino acid residues in the FRNK box, the K182A mutation was introduced into ZYMV to make the ZYMVFRNA construct. ZYMVFRNA caused mild squash leaf symptoms, and its viral RNA levels were similar to those of ZYMVFINK (Haronsky, unpublished). As seen for ZYMVFINK-infected squash extract, ZYMVFRNA-infected squash extract did not bind detectable levels of siRNA duplex in an electromobility shift binding assay (Fig. 4C). Similarly, binding to artificial siRNA (Fig. 6B) and miRNA/miRNA* (Fig. 6C) duplexes was lost in N. benthamiana leaf extracts that were agroinfiltrated with an HA-tagged HCFRNA construct (Fig. 6B and C). However, despite this loss of smRNA binding, the RSS activity of HCFRNA was retained. Transient coexpression of GFP and GFP-IR with ZYMV HCFRNK, HCFINK, or HCFRNA resulted in sustained, high GFP expression levels, demonstrating their RSS activity (Fig. 6A). These results demonstrate that both positively charged amino acid residues of the FRNK motif are necessary for smRNA duplex binding.
Previously, we have shown that it is possible to alter the observed leaf symptoms from mild to severe by expression of the 2b RSS gene of CMV strain Fny in ZYMVFINK (69). Although symptoms caused by ZYMV 2b were very severe, the level of binding of its FINK-containing HC-Pro was below detection (Fig. 4C).
Mutation in the FRNK box greatly lowers HC-Pro's affinity for duplex RNA.
The expression of ZYMV HC-Pro protein in N. benthamiana leaves by agroinfiltration was much higher than that in ZYMV-infected squash, and thus, this system was utilized for biochemical studies. The N. benthamiana leaf extract was serially diluted to 0.6 nM from an ∼3 × 103 nM initial HC concentration, incubated with 4.8 nM siRNA-like duplex, and assayed (Fig. 7A). A Hill plot demonstrated that binding of the ZYMV HCFRNK to an siRNA-like duplex is noncooperative, with a Km value of about 150 nM (Fig. 7A). A Km value could not be calculated for the attenuated HCFINK, because binding at the initial concentration (∼2 × 103 nM) was much below saturation, and even a fivefold dilution caused a loss of detectable binding. To obtain purified proteins for continued biochemical studies of ZYMV HC-Pro, HA-tagged and His-tagged proteins were column purified or enriched by immunoprecipitation, but these proteins did not bind siRNA duplex in our assay. To demonstrate specificity and to accentuate the differences between the binding affinities of the HCFRNK and HCFINK proteins, a homologous competition assay was performed using phosphorylated, nonradiolabeled, artificial siRNA duplex as a competitor. HCFINK was assayed undiluted, at ∼2 μM (Fig. 7B), and HCFRNK was assayed in a 50-fold dilution of 60 nM (Fig. 7C) to achieve saturation of HC-Pro with the artificial siRNA duplex. Slightly reduced binding was observed for both HCFINK and HCFRNK in the 10 volumes of competitor-to-probe ratio (45 nM), declining further at the 100-volume ratio (455 nM). Thus, it was necessary to use at least 33 times the concentration of the HCFINK protein to observe a response comparable to that of the HCFRNK protein, demonstrating the importance of this charged residue to smRNA duplex binding.
DISCUSSION
We have shown that the FRNK box in the central domain of HC-Pro is required for binding double-stranded smRNA duplexes and that replacement of charged amino acids within this sequence has major effects on smRNA binding and elicitation of ZYMV infection symptoms. The FRNK box of potyviruses lies in the HC-Pro central domain and is highly conserved among all sequenced members of the Potyvirus genus (Fig. 8). This motif delineates the left border of a highly conserved trypsin-protected zone inside single-stranded RNA-binding domain A (48). Previously, we have shown that the HC-Pro mutation R180I is associated with very mild symptoms, compared to the severe wild-type symptoms in squash, without affecting the ZYMV life cycle (Fig. 1) (18). Two other ZYMV mutations also reduce symptoms, one occurring near the FRNK box, F205L, and the other near the C terminus, Q396D (36). A mutation in Plum pox potyvirus (PPV) in the HC-Pro central region can affect symptom severity without affecting virus accumulation (K107Q or S231G) (52). Loss of both Potato virus X/potyviral synergism and suppression of gene silencing were associated with the mutation L134H in PPV (21), and a loss of suppression together with loss of synergism (with a comovirus) was also found in Soybean mosaic virus (K142I) and TEV (K144I) mutants which are relatively conserved (20). Thus, the FRNK box is not the only motif governing HC-Pro RSS function and symptom expression.
FIG. 8.
The HC-Pro FRNK motif is highly conserved in potyviruses. Forty-four distinct potyvirus HC-Pro sequences are aligned to show a region in the central domain. The FRNK motif and two adjacent positively charged (+) and aromatic (a) residue-rich regions are underlined. The shading threshold is 80% amino acid identity. Numbering from the first amino acid of each HC-Pro is indicated. Virus proteins, extracted from viral sequences with the indicated GenBank accession numbers, are as follows: ZYMV gi 118566318 (ZYMV), Bean common mosaic necrosis virus gi 25013913 (BCMNV), Bean common mosaic virus gi 25013490 (BCMV), Bean yellow mosaic virus gi 25013500 (BYMV), Beet mosaic virus gi 40254030 (BtMV), Chilli veinal mottle virus gi 45004657 (ChiVMV), Clover yellow vein virus gi 25013510 (ClYVV), Cocksfoot streak virus gi 25014039 (CSV), Cowpea aphid-borne mosaic virus gi 25013520 (CABMV), Daphne virus Y gi 96980663 (DVY), Dasheen mosaic virus gi 25013784 (DsMV), East Asian passiflora virus gi 85539887 (EAPV), Japanese yam mosaic virus gi 25013883 (JYMV), Johnsongrass mosaic virus gi 25013809 (JGMV), Konjak mosaic virus gi 90093254 (KoMV), Leek yellow stripe virus gi 25013893 (LYSV), Lettuce mosaic virus gi 25013530 (LMV), Lily mottle virus gi 39163617 (LMoV), Maize dwarf mosaic virus gi 25013540 (MDMV), Onion yellow dwarf virus gi 32493289 (OYDV), Papaya leaf distortion mosaic potyvirus gi 32493279 (PLDMV), Papaya ringspot virus gi 25013550 (PRSV), Pea seed-borne mosaic virus gi 25013560 (PSbMV), Peanut mottle virus gi 25013833 (PeMoV), Pepper mottle virus gi 25013570 (PepMoV), Peru tomato mosaic virus gi 28519942 (PTV), PPV gi 25013580 (PPV), Potato virus A gi 25013590 (PVA), Potato virus V gi 25013850 (PVV), PVY gi 25013600 (PVY), Scallion mosaic virus gi 25013997 (SVY), Shallot yellow stripe virus gi 76803358 (SYSV), Sorghum mosaic virus gi 25013823 (SrMV), Soybean mosaic virus gi 25013610 (SMV), Sugarcane mosaic virus gi 25013620 (SCMV), Sweet potato feathery mottle virus gi 25013774 (SPFMV), Thunberg fritillary virus gi 68989219 (TFV), TEV gi 25013634 (TEV), Tobacco vein mottling virus gi 25013639 (TVMV), TuMV gi 25013650 (TuMV), Watermelon mosaic virus gi 51949948 (WMV), Wild potato mosaic virus gi 25141239 (WPMV), Wisteria vein mosaic virus gi 116723233 (WVMV), and Yam mosaic virus gi 48249198 (YMV).
HC-Pro is a strong suppressor of antiviral defense and gene silencing (3), and it alters smRNA accumulation and causes abnormal plant development (10, 27, 41). To examine the involvement of the FRNK box in symptom elicitation and RSS activity, we compared the differences in smRNA accumulation in plants infected with the wild-type severe ZYMVFRNK with that of plants infected with a symptom-attenuated viral strain, ZYMVFINK, differing in a single amino acid. We studied smRNA changes in the first true leaf at 5 dpi and before symptom appearance. At this stage, the accumulations of viral RNA and HC-Pro protein were similar for ZYMVFRNK and ZYMVFINK (Fig. 1). Thus, smRNA accumulation and subsequent symptom attenuation are not the result of differences in virus titer or HC-Pro concentration.
ZYMV infection causes increased accumulation of most detectable miRNAs along with their miRNA*s (Fig. 2A). miRNAs accumulated in a pattern similar to that of the miRNAs of transgenic arabidopsis leaves expressing TuMV P1-HC-Pro (27). miRNA*s are typically degraded to undetectable levels in uninfected plants, and levels of these were very low in virus-free squash. In contrast, virus-infected squash plants accumulated high levels of miRNA*, as observed for other potyviral infections (10). miRNA* levels in ZYMVFRNK-infected plants were higher than those of ZYMVFINK-infected plants, and the average differences ranged from less than 2-fold to 20-fold. We expect duplex stabilization alone might not account for the accumulation of some miRNAs being greater than that of others (Fig. 2). There may be secondary effects, such as transcriptional enhancement due to loss of feedback inhibition and up-regulation of DCL1 and AGO1 gene expression through loss of miR162 and miR168 control, as has been shown in HC-Pro-expressing arabidopsis (41, 76).
The accumulation of vsiRNA (plus and minus strands) in response to infection with ZYMVFINK was very similar to that seen in response to the wild-type ZYMVFRNK (Fig. 2C and D). The plus-strand vsiRNAs were more abundant than the minus-strand vsiRNAs. In addition, accumulation of a vsiRNA from one strand did not mimic the accumulation of its complementary opposing-strand vsiRNA. Thus, it is likely that at this early stage of infection (5 dpi), vsiRNA accumulation is primarily based on processing of single-strand self-folding viral RNA, as was found with Tombusvirus infection (42, 44, 56), and not from cleavage of viral dsRNA derived from RDR amplification.
The fact that vsiRNA levels and ZYMV viability are not primarily affected by the HCFINK mutation (Fig. 2C and D) indicates that RSS activity is not disrupted in vivo. Indeed, RSS assays with ZYMV HC-Pro in N. benthamiana (Fig. 5) show that there is no difference between the wild type and mutant in this respect. In our RSS assay, we observed that siRNA derived from GFP antisense transcripts accumulated in the presence of high levels of HC-Pro. However, no difference in siRNA accumulation between the mutant and wild-type HC-Pro was observed. We hypothesize that HC-Pro does not negatively affect DCL or RDR activity but might stabilize DCL products by binding them in vivo, thus preventing their incorporation into RISC.
High accumulation of miRNA/miRNA*, but not of vsiRNA, in severe ZYMV-infected plants relative to attenuated ZYMV-infected plants led us to examine differential smRNA binding by ZYMV HC-Pro mutant proteins. Wild-type ZYMV HC-Pro binds miRNA/miRNA* duplexes in vitro (Fig. 4 and 6), as has been shown for TEV HC-Pro and other viral suppressors (29, 40). This binding presumably stabilizes such duplexes in vivo. EMSAs demonstrated substantial binding of both miRNA/miRNA*-like and siRNA-like duplexes in extracts of ZYMVFRNK-infected squash plants and in agroinfiltrated N. benthamiana leaf extracts expressing ZYMV HCFRNK. Even though the ZYMV HCFINK and HCFRNA mutant proteins had greatly reduced affinities for artificial miRNA/miRNA* and siRNA duplexes in vitro compared to that of wild-type HCFRNK (Fig. 4, 6, and 7), both of these charge-reduced mutants maintained their RSS activities. We did not observe smRNA binding by HCFRNA, although it may occur below our assay detection limits.
We found that HC-Pro bound miRNA/miRNA* duplexes less efficiently than it bound siRNA duplexes (Fig. 6B and C) and propose that this might have implications in vivo. Each of the six conserved miRNAs whose complementary miRNA*s accumulated to levels threefold or higher during ZYMVFRNK infection (Fig. 2A) was found to have at least one bulge resulting from an insertion or deletion or a stretch of two mismatches when aligned by mfold (see Fig. S1 in the supplemental material) (77). None of the five miRNA/miRNA* pairs in which the difference in expression between the viruses was threefold or less (Fig. 2B) had any bulges or more than one mismatch in a row. Based on this observation, we hypothesize that bulges or stretches of mismatches in smRNA duplexes can interfere with HC-Pro binding and that this interference is more pronounced for the weaker-binding HCFINK. The implication is that miRNAs that have less homology with their respective miRNA*s are less sensitive to viral interference (Fig. 9; see Fig. S1 in the supplemental material). However, there is ample silencing suppression by HCFINK (Fig. 5 and 6), possibly due to the fact that siRNA and highly complementary duplexes can still be bound by the HC-Pro mutant (Fig. 4A).
FIG. 9.
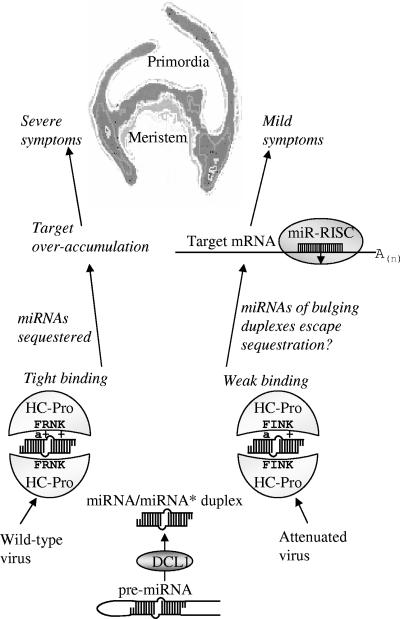
Model of HC-Pro influence on symptom expression. miRNAs are processed from a hairpin precursor (pre-miRNA) by a DICER-like enzyme (DCL1) to make duplex miRNA/miRNA*. In uninfected plants, miRNA is incorporated into the RISC, and miRNA* is degraded. RISC then down-regulates the expression of a target mRNA, complementary to the miRNA, through cleavage. In potyvirus-infected plants, HC-Pro binds the miRNA/miRNA* duplex, effectively sequestering miRNA and causing ectopic expression of target mRNAs. In wild-type HC-Pro, the FRNK motif contains an aromatic amino acid residue (a) and two positively charged amino acid residues (+) which are involved in duplex binding. Charge reduction in the ZYMVFINK-attenuated virus causes lower affinity for miRNA/miRNA* duplexes, especially for those which contain bulges or multiple mismatches, allowing adequate RISC function and, thus, mild symptom expression.
While the lower affinity of HCFINK to smRNA duplexes had no effect on viral siRNA levels at an early stage of infection, a large effect on miRNA/miRNA* duplexes was observed (Fig. 2). The contrast between the similar vsiRNA accumulations and the altered miRNA profiles probably reflects the difference in their biogeneses. miRNA/miRNA* biogenesis is regulated and affected by levels of other miRNAs, while vsiRNA levels are presumably a function of viral genomic RNA levels and their processing by DCL2 and DCL4 (8). However, the weaker binding of these vsiRNAs by HCFINK might explain the reduction in viral genomic RNA levels compared to those observed for wild-type ZYMV at later stages of infection.
The importance of the charge, not sequence, in the FRNK box was demonstrated by the result that the charge-preserving ZYMV HCFKNK mutant caused severe symptoms and bound smRNA with an affinity similar to ZYMV HCFRNK. In contrast, ZYMV HCFRNA and ZYMV HCFINK mutants were permissive for virus replication and caused mild symptoms, but they had weaker binding activities (Fig. 4C and 6B). This finding is consistent with the roles of aromatic (F, Y, W, and H) and positively charged (K, R, and H) residues in RNA binding (60). Indeed, only conservative amino acid variations are naturally found in the FRNK box of the potyvirus HC-Pro (Fig. 8) (e.g., three potyviruses have a positively charged amino acid R180K exchange and two have an aromatic F179Y replacement). Further inspection of the central domain shows that two distinct aromatic and positively charged amino acid residue-rich regions flank the FRNK box (Fig. 8). In other RSS acting through RNA binding, such as P19 (61) and P21 (72), aromatic and positively charged residues play a direct role in binding. Thus, the FRNK box is a potential point of contact with smRNA duplexes, and it contributes to the ability of HC-Pro to sequester miRNA and directly influence symptom development (Fig. 9).
The 2b protein of CMV Fny was recently shown to interfere with plant defenses and to affect plant development by disrupting the expression of miRNA targets (32) through its interaction with AGO1 (slicer) (76). Therefore, CMV 2b and potyvirus HC-Pro interfere with miRNA function by different mechanisms, and as such, their concerted action is synergistic (68). Their abilities to interfere with smRNA function in different ways explain the enhanced symptoms when CMV Fny 2b was expressed in the attenuated ZYMVFINK virus vector (Fig. 4C) (69).
We found that maintenance of RSS activity (and resulting virus viability) of the HC-Pro R180I mutation is specific to ZYMV and not to the host (Fig. 6). HCFINK mutants of TEV, which did not bind detectable smRNA duplex, had reduced expression of HC-Pro (Fig. 6D). Thus, no binding or undetectable binding may lead to silencing of the HC-Pro construct itself following agroinfiltration. This outcome is expected when the innate silencing activity is not suppressed, as with the GFP construct alone (Fig. 6A) and as has been previously shown (66). The FINK mutation causes TEV (Arazi, unpublished) and TuMV (S. A. Whitham, unpublished data) infectious clones to lose their infectivity, further supporting the idea that RSS and smRNA binding activities are coupled, at least in some potyviruses. The reason for survival of ZYMV bearing the naturally occurring HCFINK mutation is unclear, but it may be related to differences in the HC-Pro protein structures of ZYMV and other potyviruses. It is conceivable that HC-Pro is able to suppress silencing in more than one way, for example, through interaction with an endogenous RSS, such as rgs-CaM (2), or through some other protein-protein interaction. This issue may be resolved by addressing the issue of how RSS activity is preserved in the HCFINK mutant in contrast to the reduced binding of siRNA in vitro and of miRNA/miRNA* duplexes in vivo.
In summary, as levels of ZYMV and HC-Pro are already high in the first true leaf at 5 dpi but leaf deformations appear only later, we propose that these symptoms are a result of viral interference with miRNA-mediated gene regulation in young leaves just past their primordial stage. The first and second leaves, which develop normally and remain symptomless, may have already crossed a certain developmental threshold by the time ZYMV and HC-Pro arrive. However, subsequent leaves are increasingly symptomatic. We propose that in very young leaves, the sequestration of miRNA/miRNA* duplexes by HC-Pro renders most of the miRNAs inactive, which causes levels of critical miRNA or trans-acting siRNA target mRNAs to rise until loss of equilibrium occurs (Fig. 9). The lower affinity of the HCFINK mutants for miRNA/miRNA* duplexes allows sufficient populations of free miRNAs to be maintained in young leaves, permitting normal development. Thus, the decreased accumulation of miRNA and miRNA* levels observed for ZYMVFINK compared to ZYMVFRNK may account for the abrogation of symptoms (Fig. 9).
Supplementary Material
Acknowledgments
We are grateful to O. Ostersetzer (Volcani Center, Israel) for advice on binding assays, to N. Katzir (Neve-Yaar, Volcani Center, Israel) for providing us with melon-expressed sequence tags, to V. Dolja (Oregon State University) for providing us with anti-TEV HC antiserum, to S.D. Yeh (National Chung Hsing University, Taichung, Taiwan) for providing us with anti-ZYMV HC-Pro antiserum, and to A. Palmenberg (University of Wisconsin—Madison) for performing RNA folding.
This research was supported by grant USA-3623-04 from the United States-Israel Binational Agricultural Research and Development Fund, grant 460/04 from the Israel Science Foundation, and grant WA1019/5-1 from the German Research Council's trilateral (German-Israeli-Palestinian) project.
Footnotes
Published ahead of print on 26 September 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Adai, A., C. Johnson, S. Mlotshwa, S. Archer-Evans, V. Manocha, V. Vance, and V. Sundaresan. 2005. Computational prediction of miRNAs in Arabidopsis thaliana. Genome Res. 15:78-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anandalakshmi, R., R. Marathe, X. Ge, J. M. Herr, Jr., C. Mau, A. Mallory, G. Pruss, L. Bowman, and V. B. Vance. 2000. A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science 290:142-144. [DOI] [PubMed] [Google Scholar]
- 3.Anandalakshmi, R., G. J. Pruss, X. Ge, R. Marathe, A. C. Mallory, T. H. Smith, and V. B. Vance. 1998. A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 95:13079-13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arazi, T., S. G. Slutsky, Y. M. Shiboleth, Y. Wang, M. Rubinstein, S. Barak, J. Yang, and A. Gal-On. 2001. Engineering zucchini yellow mosaic potyvirus as a non-pathogenic vector for expression of heterologous proteins in cucurbits. J. Biotechnol. 87:67-82. [DOI] [PubMed] [Google Scholar]
- 5.Atreya, C. D., P. L. Atreya, D. W. Thornbury, and T. P. Pirone. 1992. Site-directed mutations in the potyvirus HC-Pro gene affect helper component activity, virus accumulation, and symptom expression in infected tobacco plants. Virology 191:106-111. [DOI] [PubMed] [Google Scholar]
- 6.Ballut, L., M. Drucker, M. Pugniere, F. Cambon, S. Blanc, F. Roquet, T. Candresse, H. P. Schmid, P. Nicolas, O. L. Gall, and S. Badaoui. 2005. HcPro, a multifunctional protein encoded by a plant RNA virus, targets the 20S proteasome and affects its enzymic activities. J. Gen. Virol. 86:2595-2603. [DOI] [PubMed] [Google Scholar]
- 7.Blanc, S., J. J. Lopez-Moya, R. Wang, S. Garcia-Lampasona, D. W. Thornbury, and T. P. Pirone. 1997. A specific interaction between coat protein and helper component correlates with aphid transmission of a potyvirus. Virology 231:141-147. [DOI] [PubMed] [Google Scholar]
- 8.Bouché, N., D. Lauressergues, V. Gasciolli, and H. Vaucheret. 2006. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 25:3347-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brigneti, G., O. Voinnet, W. X. Li, L. H. Ji, S. W. Ding, and D. C. Baulcombe. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17:6739-6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Chapman, E. J., A. I. Prokhnevsky, K. Gopinath, V. V. Dolja, and J. C. Carrington. 2004. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 18:1179-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cronin, S., J. Verchot, R. Haldeman-Cahill, M. C. Schaad, and J. C. Carrington. 1995. Long-distance movement factor: a transport function of the potyvirus helper component proteinase. Plant Cell 7:549-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunoyer, P., C. H. Lecellier, E. A. Parizotto, C. Himber, and O. Voinnet. 2004. Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell 16:1235-1250. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Ebhardt, H. A., E. P. Thi, M. B. Wang, and P. J. Unrau. 2005. Extensive 3′ modification of plant small RNAs is modulated by helper component-proteinase expression. Proc. Natl. Acad. Sci. USA 102:13398-13403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elbashir, S. M., W. Lendeckel, and T. Tuschl. 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15:188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gal-On, A., Y. Antignus, A. Rosner, and B. Raccah. 1990. Nucleotide sequence of the zucchini yellow mosaic virus capsid-encoding gene and its expression in Escherichia coli. Gene 87:273-277. [DOI] [PubMed] [Google Scholar]
- 16.Gal-On, A., E. Meiri, C. Elman, D. J. Gray, and V. Gaba. 1997. Simple hand-held devices for the efficient infection of plants with viral-encoding constructs by particle bombardment. J. Virol. Methods 64:103-110. [DOI] [PubMed] [Google Scholar]
- 17.Gal-On, A., E. Meiri, H. Huet, W. J. Hua, B. Raccah, and V. Gaba. 1995. Particle bombardment drastically increases the infectivity of cloned DNA of zucchini yellow mosaic potyvirus. J. Gen. Virol. 76:3223-3227. [DOI] [PubMed] [Google Scholar]
- 18.Gal-On, A., and B. Raccah. 2000. A point mutation in the FRNK motif of the potyvirus helper component-protease gene alters symptom expression in cucurbits and elicits protection against the severe homologous virus. Phytopathology 90:467-473. [DOI] [PubMed] [Google Scholar]
- 19.Gal-On, A., and Y. M. Shiboleth. 2005. Cross protection, p. 261-288. In G. Loebenstein and J. P. Carr (ed.), Natural resistance mechanisms of plants to viruses. Springer, Dordrecht, The Netherlands.
- 20.Ghabrial, S., and C. Zhang. 2005. A single amino acid change in potyvirus HcPro abolishes its function as a suppressor of RNA silencing. Abstr. XIII Int. Congr. Virol. V- 554, p. 21182. [Google Scholar]
- 21.González-Jara, P., F. A. Atencio, B. Martínez-García, D. Barajas, F. Tenllado, and J. R. Díaz-Ruíz. 2005. A single amino acid mutation in the plum pox virus helper component-proteinase gene abolishes both synergistic and RNA silencing suppression activities. Phytopathology 95:894-901. [DOI] [PubMed] [Google Scholar]
- 22.Hamilton, A. J., and D. C. Baulcombe. 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286:950-952. [DOI] [PubMed] [Google Scholar]
- 23.Hutvágner, G., J. McLachlan, A. E. Pasquinelli, E. Balint, T. Tuschl, and P. D. Zamore. 2001. A cellular function for the RNA-interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science 293:834-838. [DOI] [PubMed] [Google Scholar]
- 24.Kadouri, D., Y. Peng, Y. Wang, S. Singer, H. Huet, B. Raccah, and A. Gal-On. 1998. Affinity purification of HC-Pro of potyviruses with Ni2+-NTA resin. J. Virol. Methods 76:19-29. [DOI] [PubMed] [Google Scholar]
- 25.Kasschau, K. D., and J. C. Carrington. 1998. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95:461-470. [DOI] [PubMed] [Google Scholar]
- 26.Kasschau, K. D., and J. C. Carrington. 1995. Requirement for HC-Pro processing during genome amplification of tobacco etch potyvirus. Virology 209:268-273. [DOI] [PubMed] [Google Scholar]
- 27.Kasschau, K. D., Z. Xie, E. Allen, C. Llave, E. J. Chapman, K. A. Krizan, and J. C. Carrington. 2003. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4:205-217. [DOI] [PubMed] [Google Scholar]
- 28.Kimalov, B., A. Gal-On, R. Stav, E. Belausov, and T. Arazi. 2004. Maintenance of coat protein N-terminal net charge and not primary sequence is essential for zucchini yellow mosaic virus systemic infectivity. J. Gen. Virol. 85:3421-3430. [DOI] [PubMed] [Google Scholar]
- 29.Lakatos, L., T. Csorba, V. Pantaleo, E. J. Chapman, J. C. Carrington, Y. P. Liu, V. V. Dolja, L. F. Calvino, J. J. Lopez-Moya, and J. Burgyan. 2006. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 25:2768-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lakatos, L., G. Szittya, D. Silhavy, and J. Burgyan. 2004. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 23:876-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, H.-J., M. Palkovits, and W. S. Young III. 2006. From the cover: miR-7b, a microRNA up-regulated in the hypothalamus after chronic hyperosmolar stimulation, inhibits Fos translation. Proc. Natl. Acad. Sci. USA 103:15669-15674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewsey, M., F. C. Robertson, T. Canto, P. Palukaitis, and J. P. Carr. 2007. Selective targeting of miRNA-regulated plant development by a viral counter-silencing protein. Plant J. 50:240-252. [DOI] [PubMed] [Google Scholar]
- 33.Li, F., and S. W. Ding. 2006. Virus counterdefense: diverse strategies for evading the RNA silencing immunity. Annu. Rev. Microbiol. 60:503-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, X., and Y. Z. Zhang. 2005. Computational detection of microRNAs targeting transcription factor genes in Arabidopsis thaliana. Comput. Biol. Chem. 29:360-367. [DOI] [PubMed] [Google Scholar]
- 35.Lim, H. S., T. S. Ko, H. A. Hobbs, K. N. Lambert, J. M. Yu, N. K. McCoppin, S. S. Korban, G. L. Hartman, and L. L. Domier. 2007. Soybean mosaic virus helper component-protease alters leaf morphology and reduces seed production in transgenic soybean plants. Phytopathology 97:366-372. [DOI] [PubMed] [Google Scholar]
- 36.Lin, S. S., H. W. Wu, F. J. Jan, R. F. Hou, and S. D. Yeh. 2007. Modifications of the helper component-protease of zucchini yellow mosaic for generation of attenuated mutants for cross protection against severe infection. Phytopathology 97:287-296. [DOI] [PubMed] [Google Scholar]
- 37.Lobbes, D., G. Rallapalli, D. D. Schmidt, C. Martin, and J. Clarke. 2006. SERRATE: a new player on the plant microRNA scene. EMBO Rep. 7:1052-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu, C., S. S. Tej, S. Luo, C. D. Haudenschild, B. C. Meyers, and P. J. Green. 2005. Elucidation of the small RNA component of the transcriptome. Science 309:1567-1569. [DOI] [PubMed] [Google Scholar]
- 39.Mallory, A. C., B. J. Reinhart, D. Bartel, V. B. Vance, and L. H. Bowman. 2002. A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro-RNAs in tobacco. Proc. Natl. Acad. Sci. USA 99:15228-15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mérai, Z., Z. Kerenyi, S. Kertesz, M. Magna, L. Lakatos, and D. Silhavy. 2006. Double-stranded RNA binding may be a general plant RNA viral strategy to suppress RNA silencing. J. Virol. 80:5747-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mlotshwa, S., S. E. Schauer, T. H. Smith, A. C. Mallory, J. M. Herr, Jr., B. Roth, D. S. Merchant, A. Ray, L. H. Bowman, and V. B. Vance. 2005. Ectopic DICER-LIKE1 expression in P1/HC-Pro Arabidopsis rescues phenotypic anomalies but not defects in microRNA and silencing pathways. Plant Cell 17:2873-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Molnár, A., T. Csorba, L. Lakatos, E. Varallyay, C. Lacomme, and J. Burgyan. 2005. Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J. Virol. 79:7812-7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pall, G. S., C. Codony-Servat, J. Byrne, L. Ritchie, and A. Hamilton. 2007. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res. 35:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pantaleo, V., G. Szittya, and J. Burgyan. 2007. Molecular bases of viral RNA targeting by viral small interfering RNA-programmed RISC. J. Virol. 81:3797-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Papp, I., M. F. Mette, W. Aufsatz, L. Daxinger, S. E. Schauer, A. Ray, J. van der Winden, M. Matzke, and A. J. Matzke. 2003. Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol. 132:1382-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peragine, A., M. Yoshikawa, G. Wu, H. L. Albrecht, and R. S. Poethig. 2004. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in arabidopsis. Genes Dev. 18:2368-2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pirone, T., and S. Blanc. 1996. Helper-dependent vector transmission of plant viruses. Annu. Rev. Phytopathol. 34:227-247. [DOI] [PubMed] [Google Scholar]
- 48.Plisson, C., M. Drucker, S. Blanc, S. German-Retana, O. Le Gall, D. Thomas, and P. Bron. 2003. Structural characterization of HC-Pro, a plant virus multifunctional protein. J. Biol. Chem. 278:23753-23761. [DOI] [PubMed] [Google Scholar]
- 49.Reinhart, B. J., E. G. Weinstein, M. W. Rhoades, B. Bartel, and D. P. Bartel. 2002. MicroRNAs in plants. Genes Dev. 16:1616-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roth, B. M., G. J. Pruss, and V. B. Vance. 2004. Plant viral suppressors of RNA silencing. Virus Res. 102:97-108. [DOI] [PubMed] [Google Scholar]
- 51.Ruiz-Ferrer, V., J. Boskovic, C. Alfonso, G. Rivas, O. Llorca, D. Lopez-Abella, and J. J. Lopez-Moya. 2005. Structural analysis of tobacco etch potyvirus HC-pro oligomers involved in aphid transmission. J. Virol. 79:3758-3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sáenz, P., L. Quiot, J. B. Quiot, T. Candresse, and J. A. Garcia. 2001. Pathogenicity determinants in the complex virus population of a plum pox virus isolate. Mol. Plant-Microbe Interact. 14:278-287. [DOI] [PubMed] [Google Scholar]
- 53.Shi, X. M., H. Miller, J. Verchot, J. C. Carrington, and V. B. Vance. 1997. Mutations in the region encoding the central domain of helper component-proteinase (HC-Pro) eliminate potato virus X/potyviral synergism. Virology 231:35-42. [DOI] [PubMed] [Google Scholar]
- 54.Silhavy, D., A. Molnar, A. Lucioli, G. Szittya, C. Hornyik, M. Tavazza, and J. Burgyan. 2002. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 21:3070-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stenger, D. C., R. French, and F. E. Gildow. 2005. Complete deletion of wheat streak mosaic virus HC-Pro: a null mutant is viable for systemic infection. J. Virol. 79:12077-12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Szittya, G., A. Molnar, D. Silhavy, C. Hornyik, and J. Burgyan. 2002. Short defective interfering RNAs of tombusviruses are not targeted but trigger post-transcriptional gene silencing against their helper virus. Plant Cell 14:359-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Urcuqui-Inchima, S., A. L. Haenni, and F. Bernardi. 2001. Potyvirus proteins: a wealth of functions. Virus Res. 74:157-175. [DOI] [PubMed] [Google Scholar]
- 58.Urcuqui-Inchima, S., I. G. Maia, P. Arruda, A. L. Haenni, and F. Bernardi. 2000. Deletion mapping of the potyviral helper component-proteinase reveals two regions involved in RNA binding. Virology 268:104-111. [DOI] [PubMed] [Google Scholar]
- 59.Urcuqui-Inchima, S., J. Walter, G. Drugeon, S. German-Retana, A. L. Haenni, T. Candresse, F. Bernardi, and O. Le Gall. 1999. Potyvirus helper component-proteinase self-interaction in the yeast two-hybrid system and delineation of the interaction domain involved. Virology 258:95-99. [DOI] [PubMed] [Google Scholar]
- 60.van Tilbeurgh, H., X. Manival, S. Aymerich, J. M. Lhoste, C. Dumas, and M. Kochoyan. 1997. Crystal structure of a new RNA-binding domain from the antiterminator protein SacY of Bacillus subtilis. EMBO J. 16:5030-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vargason, J. M., G. Szittya, J. Burgyan, and T. M. Tanaka Hall. 2003. Size selective recognition of siRNA by an RNA silencing suppressor. Cell 115:799-811. [DOI] [PubMed] [Google Scholar]
- 62.Varrelmann, M., E. Maiss, R. Pilot, and L. Palkovics. 2007. Use of pentapeptide-insertion scanning mutagenesis for functional mapping of the plum pox virus helper component proteinase suppressor of gene silencing. J. Gen. Virol. 88:1005-1015. [DOI] [PubMed] [Google Scholar]
- 63.Vaucheret, H., A. C. Mallory, and D. P. Bartel. 2006. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell 22:129-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Verchot, J., K. L. Herndon, and J. C. Carrington. 1992. Mutational analysis of the tobacco etch potyviral 35-kDa proteinase: identification of essential residues and requirements for autoproteolysis. Virology 190:298-306. [DOI] [PubMed] [Google Scholar]
- 65.Voinnet, O. 2005. Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 6:206-220. [DOI] [PubMed] [Google Scholar]
- 66.Voinnet, O., S. Rivas, P. Mestre, and D. Baulcombe. 2003. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33:949-956. [DOI] [PubMed] [Google Scholar]
- 67.Wang, M. B., and M. Metzlaff. 2005. RNA silencing and antiviral defense in plants. Curr. Opin. Plant Biol. 8:216-222. [DOI] [PubMed] [Google Scholar]
- 68.Wang, Y., V. Gaba, J. Yang, P. Palukaitis, and A. Gal-On. 2002. Characterization of synergy between cucumber mosaic virus and potyviruses in cucurbit hosts. Phytopathology 92:51-58. [DOI] [PubMed] [Google Scholar]
- 69.Wang, Y., T. Tzfira, V. Gaba, V. Citovsky, P. Palukaitis, and A. Gal-On. 2004. Functional analysis of the cucumber mosaic virus 2b protein: pathogenicity and nuclear localization. J. Gen. Virol. 85:3135-3147. [DOI] [PubMed] [Google Scholar]
- 70.Wesley, S. V., C. A. Helliwell, N. A. Smith, M. B. Wang, D. T. Rouse, Q. Liu, P. S. Gooding, S. P. Singh, D. Abbott, P. A. Stoutjesdijk, S. P. Robinson, A. P. Gleave, A. G. Green, and P. M. Waterhouse. 2001. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27:581-590. [DOI] [PubMed] [Google Scholar]
- 71.Whitham, S. A., C. Yang, and M. M. Goodin. 2006. Global impact: elucidating plant responses to viral infection. Mol. Plant-Microbe Interact. 19:1207-1215. [DOI] [PubMed] [Google Scholar]
- 72.Ye, K., L. Malinina, and D. J. Patel. 2003. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature 426:874-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ye, K., and D. J. Patel. 2005. RNA silencing suppressor p21 of beet yellows virus forms an RNA binding octameric ring structure. Structure 13:1375-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu, B., E. J. Chapman, Z. Yang, J. C. Carrington, and X. Chen. 2006. Transgenically expressed viral RNA silencing suppressors interfere with microRNA methylation in arabidopsis. FEBS Lett. 580:3117-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang, B. H., X. P. Pan, Q. L. Wang, G. P. Cobb, and T. A. Anderson. 2005. Identification and characterization of new plant microRNAs using EST analysis. Cell Res. 15:336-360. [DOI] [PubMed] [Google Scholar]
- 76.Zhang, X., Y. R. Yuan, Y. Pei, S. S. Lin, T. Tuschl, D. J. Patel, and N. H. Chua. 2006. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 20:3255-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.