Abstract
A monoclonal Fab (Fab 3674) selected from a human nonimmune phage library by panning against the chimeric construct NCCG-gp41 (which comprises an exposed coiled-coil trimer of gp41 N helices fused in the helical phase onto the minimal thermostable ectodomain of gp41) is described. Fab 3674 is shown to neutralize diverse laboratory-adapted B strains of human immunodeficiency virus type 1 (HIV-1) and primary isolates of subtypes A, B, and C in an Env-pseudotyped-virus neutralization assay, albeit with reduced potency (approximately 25-fold) compared to that of 2F5 and 4E10. Alanine scanning mutagenesis maps a novel epitope to a shallow groove on the N helices of gp41 that is exposed between two C helices in the fusogenic six-helix bundle conformation of gp41. Bivalent Fab 3674 and the C34 peptide (a potent fusion inhibitor derived from the C helix of gp41) are shown to act at similar stages of the fusion reaction and to neutralize HIV-1 synergistically, providing additional evidence that the epitope of Fab 3674 is new and distinct from the binding site of C34.
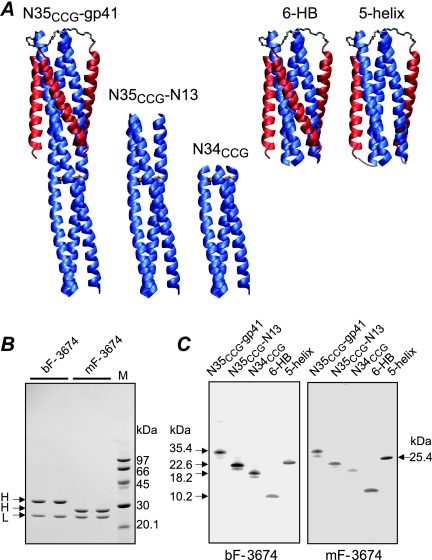
The initial steps of human immunodeficiency virus (HIV) infection require fusion of viral and host cell membranes (1, 11). Membrane fusion is mediated by the surface envelope (Env) glycoproteins gp120 and gp41, the latter of which possesses both ecto- and transmembrane domains. gp120 governs attachment and the first steps of the fusion reaction by binding to the primary receptor CD4 and to coreceptor CXCR4 or CCR5. These interactions initiate a series of conformational changes in gp120 and gp41 that lead to formation of a prehairpin intermediate (PHI) of the ectodomain of gp41 in which its N-heptad repeat (N-HR; residues 542 to 591) and C-heptad repeat (C-HR; residues 623 to 663) become accessible and are present in an overall extended conformation (5, 13, 14). The PHI bridges the viral and target cell membranes through the N and C termini of gp41: the PHI is tethered to the viral membrane by the transmembrane domain located C terminal to the C-HR, and a fusion peptide located N terminal to the N-HR is inserted into the target cell membrane (5, 11, 30). The subsequent formation of a six-helix bundle (6-HB) with the N-HR trimer surrounded by three C-HR helices drives apposition of the viral and target cell membranes, which ultimately leads to fusion (6, 42). Prior to fusion, the N-HR forms a fully accessible trimeric, helical coiled-coil that is targeted by a number of fusion inhibitors, including peptides derived from the C-HR (such as DP178 or enfuvirtide and C34) (18, 29, 43) and those derived from the N-HR that can form mixed N-HR trimers (2). Because the C-HR and N-HR are distal to one another in the PHI, the C-HR can also be targeted by constructs that present fully (NCCG-gp41 [23], N35CCG-N13, or N34CCG [24]) or partially (5-helix [36]) exposed N-HR trimers, constructs that are depicted in Fig. 1A.
FIG. 1.
Characterization of bivalent (bF) and monovalent (mF) Fab 3674 antibodies. (A) Ribbon diagrams of the engineered gp41-derived constructs used in this study. The N-HR and C-HR helices are displayed in blue and red, respectively, and disulfide bridges linking the N-HR helices (NCCG constructs) are shown as gold rods. (B) SDS-PAGE of reduced bF-3674 and mF-3674 stained with Coomassie. H and L denote heavy and light chains, respectively. Molecular masses of protein standards are indicated. The larger mass of the heavy chain of bF-3674 relative to that for mF-3674 is due to the helix-loop-helix dimerization domain. (C) Western blot analysis under nonreducing conditions of gp41-derived constructs reacting with bF-3674 and mF-3674. Molecular masses are indicated. Disulfide-linked NCCG-gp41, N35CCG-N13, and N34CCG refold spontaneously, just like the 6-HB. Binding is conformation dependent as reduced forms of NCCG-gp41, N35CCG-N13, and N34CCG, which do not refold spontaneously but form heterogeneous aggregates due to the exposed N-HR region (23, 24) not conformationally restricted by disulfide bridges, do not interact with the Fabs.
In a recent paper, we described a set of eight monoclonal Fabs obtained from a human nonimmune phage library by panning against two constructs, namely, NCCG-gp41 and N35CCG-N13 (25). Both constructs present the N-HR as a stable, helical, disulfide-linked trimer, and in NCCG-gp41, the helical trimer extends in helical phase from the 6-HB core (Fig. 1A) (23, 24). Those eight monoclonal Fabs were shown to recognize three broad epitopes on gp41 that could be grouped as follows: class A recognized the exposed surface of the C-HR helices of the 6-HB, class B recognized a surface comprising the exposed regions of both the N-HR and C-HR helices of the 6-HB, and class C was specific for the trimeric coiled-coil of N-HR helices. Screening of that group of antibodies in a vaccinia virus-based HIV-1 fusion assay using Env from HIV-1 LAV showed that the most potent fusion inhibitors belonged to class B. Unfortunately, none of the eight Fabs were found to be inhibitory in Env-pseudotyped-virus neutralization assays. Contemporaneously, Miller et al. reported the discovery of a monoclonal antibody, D5, derived from a naive scFv library selected against the gp41-derived construct 5-helix (31) (Fig. 1A). Using biochemical techniques and crystallography, D5 was shown to bind to 5-helix in the hydrophobic pocket that comprises a portion of the interaction surface for the C-HR and is critical for 6-HB formation (27, 31). D5 and the C-HR-derived peptide C34 compete for the same binding site on the PHI, and the mode of action of D5, like that of the C34 peptide, is to block the formation of the 6-HB.
Here, we report the discovery of a broadly HIV-1 neutralizing monoclonal antibody (Fab 3674) from a human nonimmune phage library obtained by selection against NCCG-gp41. Neutralization activity for Fab 3674 is demonstrated for laboratory-adapted B strains of HIV-1 and diverse primary isolates belonging to subtypes A, B, and C. The epitope for Fab 3674 is unique and comprises a shallow groove at the C-terminal end of the N-HR helices that is exposed between two C-HR helices in the fusogenic 6-HB. Viral neutralization assays performed using synchronized infection demonstrate that C34 and Fab 3674 act at the same stage in the fusion process, although Fab 3674 binds tightly not only to the N-HR trimer but also to the 6-HB. In addition, Fab 3674 neutralizes HIV-1 synergistically with the C34 peptide, providing further evidence that the epitope for Fab 3674 is adjacent to but does not obstruct the interaction surface for the C-HR.
MATERIALS AND METHODS
gp41-derived proteins.
Protocols for the expression, purification, and folding of NCCG-gp41 and N-terminal His-tagged N35CCG-N13, N34CCG, and 6-HB have been described previously (23, 24). (The His tag comprises 20 residues.) NCCG-gp41 is a chimeric protein comprising N35CCG (residues 546 to 580 of gp41 HIV-1 Env with Leu576, Gln577, and Ala578 replaced by Cys, Cys, and Gly, respectively) fused onto the minimal thermostable ectodomain core of gp41. Thus, each chain of NCCG-gp41 comprises N35CCG-N34-(L6)-C28, where N34 and C28 represent portions of the N-HR and C-HR regions of gp41 (residues 546 to 579 and 628 to 655, respectively) and L6 is a six-residue linker (SGGRGG). N35CCG-N13 is a 48-residue polypeptide that comprises N35CCG, immediately followed by N13 (residues 546 to 548). Three chains of NCCG-gp41, N35CCG-N13, and N34CCG are linked covalently via three intermolecular disulfide bridges to form stable trimers which refold spontaneously from a denatured state. The 6-HB is the minimal thermostable ectodomain core of gp41, comprising N34-(L6)-C28. Single (m81 to m89) and double (m90 to m97) His-tagged 6-HB alanine mutants were made using standard procedures. C-terminal His-tagged 5-helix was expressed and purified as described previously for the His-tagged 6-HB (25). All proteins underwent a final purification step using size exclusion chromatography and were characterized by gel electrophoresis and electrospray mass spectrometry. The masses of the monomeric His-tagged 6-HB and 5-helix were 10,033 and 25,395 Da, respectively; the masses of the three other gp41-derived constructs have been reported previously (23, 24). 6-HB constructs bearing double mutations were characterized additionally by circular dichroism (CD), and double mutants found to react weakly with Fabs in a Western blot format were verified to be trimeric by multiangle light scattering (see below).
Recombinant mini-antibodies.
Recombinant mini-antibodies were obtained from the HuCAL GOLD collection of human antibody genes after three rounds of selection against immobilized NCCG-gp41 and His-tagged 6-HB as described previously (25). The 18 positive clones were isolated and purified for use in subsequent studies. Bivalent and monovalent Fab 3674 antibodies were further purified by size exclusion chromatography eluting with phosphate-buffered saline, and peak fractions were pooled and concentrated. The masses of the heavy and light chains of mF-3674, determined by electrospray mass spectrometry, were 26,240 and 22,615 Da, respectively.
SDS-PAGE.
The composition and purity of antibodies were confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Solutions containing ∼1 mg/ml of reduced bF-3674 and mF-3674 (0.2 mM dithiothreitol) were electrophoresed on a 20% homogeneous PhastGel. Proteins were stained with Coomassie R350. The heavy chain of bF-3674 is larger than that of mF-3674, since the former is linked to the helix-loop-helix dimerization domain.
CD measurements.
CD spectra of His-tagged 6-HB and the double mutants m90 to m97 (Figure 2) were recorded at 25°C in 20 mM sodium formate buffer, pH 3, on a JASCO J-720 spectropolarimeter, using a 0.1-cm-path-length cell. Evaluation of secondary structure was carried out using the program CDNN (3). Reversible temperature denaturation was also followed by CD, and all mutants refolded spontaneously.
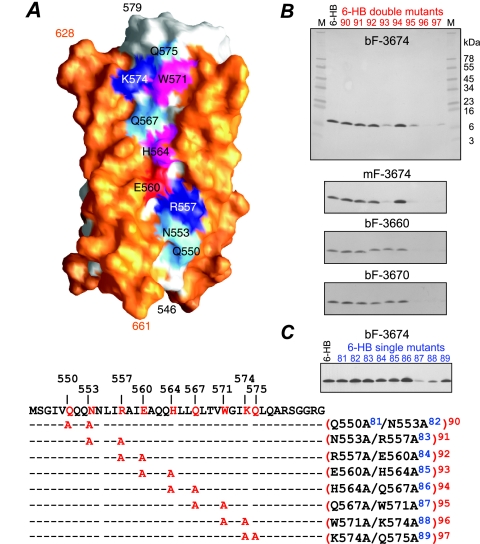
FIG. 2.
Epitope mapping of neutralizing Fab 3674 and nonneutralizing Fabs 3663 and 3670 by alanine scanning mutagenesis on the 6-HB. (A) Surface representation of the 6-HB core, with the C-HR helices in orange, the N-HR helices in white, and the solvent exposed N-HR residues that lie in a shallow groove between two C-HR helices in cyan for neutral hydrophilic residues (Q550, N553, Q567, and Q575), magenta for aromatic residues (H564 and W571), dark blue for positively charged residues (R557 and K574), and red for negatively charged residues (E560). The single (m81 to m89)- and double (m90 to m97)-alanine mutants are listed below the sequence of the N-HR. (B and C) Western blots of bF-3674, mF-3674, bF-3663, and bF-3670 against the 6-HB double-alanine mutants (M denotes SeeBlue-prestained standards) (B) and bF-3674 against the 6-HB single-alanine mutants (C). All 6-HB variants refold spontaneously.
Western blot analyses.
gp41-derived proteins (500 ng/lane) in nonreduced form were electrophoresed in 10 to 20% Tris-Tricine gradient gels, transferred to a membrane, and blocked with Tris-buffered saline containing 0.05% Tween 20 (TBST) and 10% bovine serum albumin (BSA) for 1 h at room temperature. Following three 5-min washes in TBST, membranes were incubated for 1 h in TBST, 1% BSA, and 5 μg/ml antibody. After being washed in TBST (three times), membranes were incubated for 1 h in TBST containing 1% BSA and 0.3 μg goat anti-human anti-Fab antibody-alkaline phosphatase conjugate (Jackson Immunoresearch) and then washed in TBST (three times) and developed in Fast 5-bromo-4-chloro-3-indolyl phosphate-nitro blue tetrazolium (one tablet dissolved in 20 ml TBST-1% BSA; Sigma) solution. Upon acquisition of the desired signal, the enzymatic reaction was terminated by rinsing the blot extensively in deionized water.
Surface plasmon resonance.
Surface plasmon resonance studies were conducted with a Biacore T100 system. Sensor chips, reagents, and supplies were purchased from Biacore International (Piscataway, NJ). Samples for kinetics experiments were prepared in a formate running buffer (25 mM formate, 100 mM NaCl, and 0.005% [vol/vol] detergent P20, pH 5.0), unless specified. Fab mF-3674 (500 nM in 10 mM acetate buffer, pH 4.5) was immobilized to a level of 100 RU on a series S 1 sensor chip activated with two consecutive 600-s injections of EDC-NHS [N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide-N-hydroxysuccinimide], using a flow rate of 10 ml/min. The remaining activated esters on the surface were quenched by an injection of ethanolamine (1 M, pH 8.5, for 300 s). Binding to mF-3674 was analyzed by running solutions of various concentrations of the 6-HB (twofold serial dilutions starting at 100 nM) sequentially through control and active flow cells at a flow rate of 50 μl/min, with association and dissociation phases of 60 s and 180 s, respectively. The surface was regenerated with a 15-s pulse of regeneration solution (0.2% SDS and 0.5 M NaCl) at a flow rate of 60 ml/min. No mass transport effects were observed under these conditions. Injection of gp41 (2.5 nM) over immobilized Fab at flow rates ranging from 20 to 100 μl/min (10-μl intervals) did not produce detectable changes in Rmax. Reference-subtracted and zero-concentration-corrected curves were fit using steady-state and/or heterogeneous-analyte models as part of the BIAevaluation package.
Multiangle light scattering.
The concentration of the 6-HB variants on the nitrocellulose membrane used in Western blot analysis during reaction with the Fabs is expected to be ∼2.5 μM based on band area and loading concentration. To verify that the 6-HB proteins are trimeric at this concentration, the double-alanine mutants (m93, m95, and m96) that reacted poorly with Fab 3674 were subject to size exclusion chromatography coupled with multiangle light scattering and refractive index measurements. The experimental masses of 30,220 ± 302, 30,030 ± 240, and 27,600 ± 276 Da for m93, m95, and m96, respectively, indicate that the double mutations borne in these constructs do not affect the monomer-trimer equilibrium under these conditions. More than 80% of the injected protein (150 μg/125 μl) can be accounted for in the ∼1-ml volume of the peak fraction after the column, indicative of a concentration of ∼5 μM during light-scattering and refractive index measurements. Under the same conditions, the His-tagged 6-HB construct yields a similar trimer molecular mass of 30,460 ± 913 Da.
Recombinant vaccinia viruses, cell lines, molecular clones, and antibodies.
The following vaccinia virus Env-expressing and reporter gene-expressing plasmids, cell lines, and clones were obtained from the NIH AIDS Research and Reference Reagent Program: recombinant vaccinia viruses vSC60 (encoding the HIV-1 IIIB Env protein; catalogue no. 3377), vCB41 (encoding the HIV-1 LAV Env protein; catalogue no. 3375), vCB43 (encoding the HIV-1 BaL Env protein; catalogue no. 3376), vCB28 (encoding the HIV-1 JR FL Env protein; catalogue no. 3368), vCB39 (encoding the HIV-1 ADA Env protein; catalogue no. 3373), vCB34 (encoding the HIV-1 SF2 Env protein; catalogue no. 3371), vBD3 (encoding the HIV-1 89.6 Env protein; catalogue no. 4082), vCB21R-LacZ (catalogue no. 3365), and vCBYF1-fusin (encoding the CXCR4 coreceptor; catalogue no. 3364); the HIV-1 expression plasmid SG3Δenv (catalogue no. 11051); the standard reference panel of subtype B HIV-1 Env clones (catalogue no. 11227); HIV-1 subtype C Env clones ZM233.PB6 and DU172.17 as part of a standard reference panel of subtype C HIV-1 Env clones (catalogue no. 11326); the HIV-1 Env molecular clone pCAGGS SF162 gp160 (catalogue no. 10463); and TZM-b1 (or JC53BL-13; catalogue no. 8129) indicator cells. 293T cells were obtained from the American Type Culture Collection. B-SC-1 cells and recombinant vaccinia viruses vP11T7gene1 (encoding the phage T7 polymerase), vCB32 (encoding the HIV-1 SF162 Env protein), and vCCR5 (encoding the CCR5 coreceptor) were provided by C. Broder, P. Kennedy, and E. Berger; gp160 expression plasmids pSVIII HXBc2, 89.6, JR CSF, and YU2 were provided by J. Sodroski (39, 40); and HIV-1 subtype A Env clones pSVIII RW020 (15) and pSVIII DJ263.8 (26) and HIV-1 subtype B gp160 expression plasmids pSVIII BaL01 and BaL26 (22) were provided by J. Mascola. Monoclonal antibodies 2F5 (32) and 4E10 (38) were provided by H. Katinger.
Fusion assay.
Inhibition of HIV Env-mediated cell fusion was examined using a vaccinia virus-based gene reporter assay as described previously (17).
Virus neutralization assay.
Pseudovirus stocks were prepared as described previously (21). Exponentially dividing 293T cells were cotransfected (Fugene6 transfection kit; Roche) with the Env-deficient HIV-1 SG3Δenv expression plasmid and an Env-expressing plasmid in ratios proportionate to their individual sizes (approximately 16 μg total DNA per 50 to 80% confluent T-150 culture flask). Culture supernatants were collected at 2 days posttransfection, filtered through a 0.45-μm filter, and stored at −86°C until further use. Neutralization assays were performed as described previously (21, 22). Serial dilutions of Fabs were added to Env-pseudotyped viruses, followed by addition of TZM-bl indicator cells (JC53BL-13). After overnight incubation at 37°C, fresh medium was added to each well, and luciferase activity was measured at ∼48 h postinfection (BrightGlo; Promega). Fifty percent inhibitory concentration (IC50) values were calculated using the following activity relationship: percent activity = 100/(1 + KA[I]), where KA is the equilibrium association constant and [I] is the concentration of the inhibitor.
Synergy experiments and analyses.
Env-pseudotyped-HXB2-virus neutralization assays were carried out using a range of fixed combination ratios of C34 and Fab 3674 (both monovalent and bivalent) as described previously (17). Analysis of combination effects followed that of Chou and Talalay (8, 9), where a dose reduction index (DRI) of inhibitor x in combination with inhibitor y is given by DRIx = (IC50)x/(IC50)x,y, where (IC50)x is the activity of inhibitor x alone and (IC50)x,y is the activity of inhibitor x in combination with inhibitor y. The summation of the effects of the two inhibitors in combination is characterized by the combination index (CI), given by CI = (DRI)x−1 + (DRI)y−1 + (DRI)x(DRI)y−1, where (DRI)x(DRI)y−1 represents the state in which both inhibitors are bound (9, 16). CI values equal to, greater than, or less than 1 are indicative of additive, antagonistic, and synergistic effects, respectively (9). DRI and CI values were calculated on the basis of individual IC50 values for Fabs bF-3674 and mF-3674 and C34 alone versus in combination.
Synchronized viral infection assays.
Synchronized viral infection assays in the context of the Env-pseudotyped-HXB2-virus neutralization assay were performed using spinoculation (34). In brief, following overnight incubation at 37°C, plated cells were cooled and centrifuged at 4°C (1,200 × g), and the medium was removed and cold virus (4°C) added to the wells. Following centrifugation for 2 h (1,100 × g) at 4°C, cells were gently washed twice with cold media (to remove any remaining virus that is not bound to CD4-expressing cells) before transferring the plates to 30°C or 37°C. While the plates were maintained at these temperatures, fully inhibitory concentrations of Fab or C34 were added at increasing times. After 2 h, all plates were incubated at 37°C for 48 h. The data were analyzed as described previously (35).
RESULTS
Antibody selection.
Recombinant monoclonal Fabs were selected from the HuCal Gold nonimmune phage library of 109 human antibody genes (20), using immobilized NCCG-gp41 (23) and the minimal thermostable ectodomain core (40) of gp41, the 6-HB (Fig. 1A). NCCG-gp41 consists of residues 546 to 580 of the N-HR of gp41 HIV-1 Env (HXB2) fused in helical phase to the minimal thermostable ectodomain core of gp41 (residues 546 to 579 of the N-HR and 628 to 655 of the C-HR gp41 connected by a six-residue linker). The three chains of NCCG-gp41 are linked covalently via three intermolecular disulfide bridges (arising from mutations L576C, Q577C, and A587C) to form a stable trimer. Thus, the minimal thermostable ectodomain core presents only the 6-HB of gp41 as a target for antibody selection, while NCCG-gp41 presents in addition a stably folded 3-HB of the N-HR.
Twelve Fabs were isolated from the NCCG-gp41 selection (clones 3663 to 3674) and 6 from the 6-HB selection (clones 3675 to 3680). The 18 Fabs were expressed as monovalent and bivalent mini-antibodies (the latter bearing a dimerization helix-loop-helix domain at the C terminus of the antibody heavy chain) (Fig. 1B). Hereafter, we use the notation mF or bF followed by the clone number to denote monovalent or bivalent Fabs, respectively. Fusion inhibitory activity was initially assessed for these 18 Fabs in an HIV-1 Env-mediated cell fusion assay (23, 33) using Env from HIV-1 LAV and an HXB2 Env-pseudotyped-virus neutralization assay (21). Only three clones, 3663, 3670, and 3674, were inhibitory in the fusion assay (Table 1), and only one clone, 3674, displayed antiviral activity in the neutralization assay (Table 2).
TABLE 1.
Fabs 3674, 3670, and 3663 inhibit HIV-1 Env-mediated fusiona
| Env strain | IC50 (μg/ml)b for:
|
|||
|---|---|---|---|---|
| bF-3674 | mF-3674 | bF-3670 | bF-3663 | |
| LAV | 8 ± 1 | 34 ± 6 | 20 ± 6 | 22 ± 8 |
| IIIB | 14 ± 3 | 23 ± 4 | 29 ± 6 | 81 ± 8 |
| BaL | 35 ± 9 | 68 ± 20 | 227 ± 138 | 253 ± 120 |
| SF162 | 191 ± 51 | 159 ± 26 | NA | NA |
| SF2 | 68 ± 13 | 103 ± 33 | NA | NA |
The fusion assays were carried out as described (17) using diverse lab-adapted subtype B viruses.
NA, no activity: in the range of concentrations tested, inhibition of fusion was too weak to reliably determine an IC50 value.
TABLE 2.
Fab 3674 antibodies neutralize diverse HIV-1 subtype B pseudotypes
| HIV-1 Env strain | IC50 (μg/ml)a for:
|
|||
|---|---|---|---|---|
| bF-3674 | mF-3674 | 2F5 | 4E10 | |
| Laboratory adapted | ||||
| HXB2 | 9 ± 1 | 30 ± 3 | 0.4 ± 0.1 | 0.3 ± 0.1 |
| YU2 | 114 ± 18 | 203 ± 25 | 18.0 ± 5 | 24.8 ± 7 |
| JR CSF | 78 ± 20 | 106 ± 30 | 5.3 ± 1.5 | 8.6 ± 2.3 |
| SF162 | 218 ± 62 | NA | 8.4 ± 1.5 | 6.6 ± 0.1 |
| 89.6 | 94 ± 18 | NA | ||
| Primary | ||||
| Bal01 | 29 ± 9 | 89 ± 9 | ||
| BaL26 | 294 ± 75 | 113 ± 20 | ||
NA, no activity: in the range of Fab concentrations tested, neutralization was too weak to reliably determine an IC50 value.
Fabs 3663, 3670, and 3674 bind to an exposed, polar face of the N-HR helices of gp41.
To determine the specificities of Fabs 3663, 3670, and 3674 toward the N-HR trimer, the 6-HB, or both, five distinct but complementary gp41 constructs were used for Western blot analyses. These included NCCG-gp41 (23); the 6-HB; two stable, disulfide-linked, trimeric N-HR constructs, N35CCG-N13 and N34CCG (24); and the single-chain construct 5-helix, in which one C-HR helix of the 6-HB is absent (36). All four Fabs showed similar specificities toward gp41 reacting with the 6-HB as well as constructs presenting N-HR trimers alone or a combination of both (displayed in Fig. 1C for bF-3674 and mF-3674). Since these Fabs recognize both the 6-HB and the N-HR trimers, the epitope(s) must comprise those residues of the N-HR that are not buried by the C-HR helix in the 6-HB (Fig. 2A).
To more precisely map the epitope recognized by Fabs 3663, 3670, and 3674, we mutated to alanine the nine N-HR residues that are solvent exposed in the 6-HB and located in a shallow groove between two C-HR helices (Fig. 2A). Mutations were introduced as a series of eight overlapping double mutations (m90 to m97) and nine single mutations (m81 to m89). CD studies verified that all nine 6-HB double mutants were helical (90 to 100%, taking into account the disordered His tag and linker connecting the N-HR and C-HR), with midpoint melting temperatures between 55 and 70°C, consistent with values observed for the wild type, and underwent reversible temperature denaturation (unpublished data). Western blots of the double mutants (m90 to m97) incubated with bF-3674, mF-3674, bF-3663, and bF-3670 Fabs are shown in Fig. 2B. Bands for both bF-3674 and mF-3674 show diminished binding to double mutants m93 (E650A/H564A), m95 (Q567A/W571A), m96 (W571A/K574A), and m97 (K574A/Q575A) in the order m96 < m97 < m93 < m95. Interestingly, the intensity of the band corresponding to the intervening double mutant m94 (H564A/Q567A) is the same as that for the wild-type 6-HB. In contrast, the Western blots for bF-3663 and bF-3670 show that these Fabs bind equivalently to double mutants m90 to m94 and the 6-HB, but binding to double mutants m95 to m97 is greatly reduced, with binding in the order m97 < m96 < m95. Fab 3674 antibodies versus Fabs 3663 and 3670 therefore recognize overlapping epitopes, but the binding site for 3674 is more extensive and includes residues of the N-HR displaced by approximately one helical turn: the epitope for Fab 3674 comprises E560, H564, W571, K574, and Q575, while that for Fabs 3663 and 3670 consists of W571, K574, and Q575, with a likely contribution from Q567.
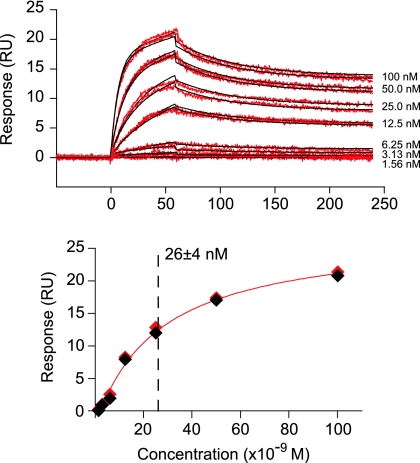
The epitope for bF-3674 was further defined by Western blot analysis of the series of single-mutant 6-HBs, which showed the largest decrease in band intensity for mutant m87 (W571A), followed by that for mutant m88 (K574A), with smaller decreases in band intensities for mutants m84 (E560A), m85 (H564A), and m89 (Q575A) (Fig. 2C). Consistent with the epitopes revealed by the double mutants, these results suggest that W571 and K574 contribute strongly to recognition of all four Fabs reported here. Last, the 6-HB binds immobilized mF-3674 with high affinity, with an equilibrium dissociation constant (KD) of ∼26 ± 4 nM, as determined by surface plasmon resonance (Fig. 3).
FIG. 3.
Surface plasmon resonance of gp41 6-HB binding to mF-3674. (A) Sensorgrams (red) of wild-type gp41 6-HB binding to immobilized mF-3674 Fab. The association [ka = (1.07 ± 0.01) × 106 M−1 s−1] and dissociation [kd = (1.93 ± 0.01) × 10−2 s−1] phases obtained at all concentrations of the 6-HB were fit simultaneously with a heterogeneous analyte model using BIAevaluation software. All data were reference subtracted and zero-concentration corrected prior to fitting. Nonlinear least-squares best fits to the data are shown as black lines. (B) Plot of maximum response units (RU) as a function of concentration yields an equilibrium dissociation constant (KD) of 26 ± 4 nM, in good agreement with the value of ∼18 nM derived from the association and dissociation rate constants. (The diamonds are the experimental data, and the red line represents the best fit to a simple binding isotherm.)
Monoclonal Fabs inhibit HIV-1 Env-mediated fusion.
Bivalent Fabs 3663, 3670, and 3674 were tested for their inhibitory effects on HIV-1 Env-mediated fusion, using Env from diverse laboratory-adapted subtype B HIV-1 strains in a quantitative vaccinia virus-based cell fusion assay (17, 33) (Table 1). bF- and mF-3674 showed the most potent inhibition as well as the largest breadth of activity, followed by bF-3670, while bF-3363 was the least active. In this assay, the overall fusion inhibitory activity of mF-3674 was only slightly reduced relative to that of bF-3674. All three Fabs showed the strongest inhibitory activity against X4 Envs derived from LAV and IIIB viruses, followed by BaL (R5 virus). Whereas bF- and mF-3674 moderately inhibited fusion of the dual-tropic strain SF2 (X4/R5) and SF162 (R5), no fusion inhibitory activity was observed for either bF-3670 or bF-3663. None of the Fabs inhibited fusion mediated by R5 Env JRFL or dual-tropic 89.6 at concentrations as high as 100 μg per ml (data not shown).
Bivalent and monovalent Fab 3674 antibodies neutralize diverse HIV-1 strains.
The antiviral activities of bF- and mF-3674 against diverse laboratory-adapted subtype B strains of HIV-1, as well as the primary B subtype Bal01 and Bal26 strains, were further examined using an Env-pseudotyped-virus neutralization assay (21, 22) (Table 2). The neutralizing activities of bF-3674 and mF-3674 were comparable to the activities observed in the fusion assays. The bivalent Fab 3674 was ∼2 to 4 times more potent than the monovalent form against all the viruses tested in this panel, with the exception of Bal26 Env.
To further examine the breadth of antiviral activity of Fab 3674, we carried out Env-pseudotyped-virus neutralization assays using a standard reference panel of subtype B Env clones derived from acute and early subtype B infections (Table 3) (28). bF- and mF-3674 were active against most of the panel B pseudoviruses, with the exception of viruses pseudotyped with Env from B.6535.3 and B.PVO.4 isolates. Again, bF-3674 was slightly more active than mF-3674 against most of the pseudoviruses, with the exception of B.WIT04160.33 and B.REJ04541.67. bF-3674 also showed measurable neutralization activity against B.TRJ04551.58 and B.TRO.11, while mF-3674 displayed no activity.
TABLE 3.
Fab 3674 neutralizes the diverse primary isolatesa
| HIV-1 Env subtype and strain | IC50 (μg/ml)b for:
|
|
|---|---|---|
| bF-3674 | mF-3674 | |
| Subtype B | ||
| B.AC10.0.29 | 28 ± 8 | 88 ± 15 |
| B.CAAN5342.A2 | 28 ± 6 | 94 ± 11 |
| B.SC422661.8 | 30 ± 7 | 185 ± 36 |
| B.QH0692.42 | 71 ± 12 | 209 ± 32 |
| B.WITO4160.33 | 104 ± 35 | 76 ± 29 |
| B.REJO4541.67 | 128 ± 66 | 106 ± 16 |
| B.RHPA4259.7 | 150 ± 54 | 296 ± 72 |
| B.THRO4156.18 | 231 ± 82 | 373 ± 135 |
| B.TRJO4551.58 | 82 ± 28 | NA |
| B.TRO.11 | 86 ± 27 | NA |
| B.PVO.4 | NA | NA |
| B.6535.3 | NA | NA |
| Subtype A | ||
| A.DJ263.8 | 158 ± 29 | 210 ± 68 |
| A.RW020 | 74 ± 21 | 145 ± 46 |
| Subtype C | ||
| C.ZM233M.PB6 | 6 ± 2 | 10 ± 3 |
| C.DU172.17 | 24 ± 3 | 157 ± 44 |
The viruses were pseudotyped with Envs from the standard reference panel of subtype B (28) and C (44) HIV-1 clones and with Env from subtype A HIV-1 strains derived from primary isolates.
NA, no activity: in the range of concentrations of Fabs tested, viral neutralization was too weak to reliably determine an IC50 value.
Finally, cross-clade antiviral activity was examined using Env clones derived from primary isolates of subtypes A (A.DJ263.8 and A.RW020) (15, 26) and C (C.ZM233 and C.DU172) (44) HIV-1 (Table 3). Both bF- and mF-3674 Fabs showed neutralizing activity toward the clade A and C pseudoviruses tested, with the bivalent form being more active than the monovalent one in all cases. BF-3674 displayed quite potent activity toward clade C pseudoviruses, with IC50 values ranging from 6 to 24 μg/ml.
Fab 3674 and C34 do not compete and are synergistic in a neutralization assay.
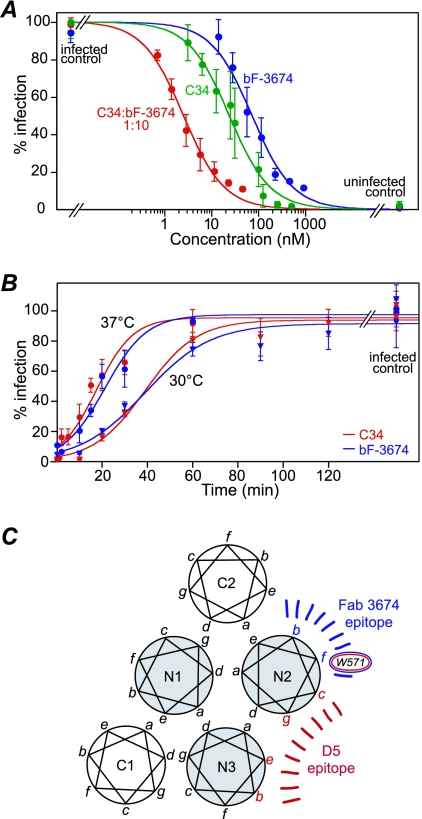
The gp41 C-HR-derived peptide C34 (residues 628 to 661) is a highly potent inhibitor of HIV-1 fusion. By targeting the exposed N-HR trimer of the PHI, C34 prevents the formation of the fusogenic 6-HB by competing with the intramolecular interaction between the N-HR and C-HR of gp41. Because Fab 3674 is small relative to immunoglobulin G and recognizes an epitope on the N-HR helix that does not overlap with the interface for the C-HR helix, the viral neutralizing activity of Fab 3674 and C34 together should be either synergistic or additive but not antagonistic. Analysis of the effects on virus neutralization of C34 and Fab 3674 antibodies (monovalent and bivalent) applied over a range of fixed combination ratios (9, 17) to Env-pseudotyped HXB2 (Table 4 and Fig. 4A) demonstrates that mF-3674 and C34 are additive and bF-3674 and C34 are synergistic, with average respective CIs of 1.0 ± 0.2 and 0.6 ± 0.1. To further probe the mechanism of inhibition of bF-3674, neutralization assays where infection was synchronized (34) and viruses were subjected to single-point, fully inhibitory concentrations of C34 or bF-3674 at increasing points in time were carried out. Plots showing percent infection as a function of time (Fig. 4B) reveal that C34 and bF-3674 act at very similar stages of the entry process. Together, these data are consistent with the notion that Fab 3674 antibodies and C34 both target the N-HR trimer yet are not mutually competitive as inhibitors of fusion and must therefore target distinct regions of the N-HR trimer.
TABLE 4.
Combined treatment of C34 and Fab 3674 in HXB2 Env-pseudotyped virus neutralization assaysa
| Fab and C34/Fab 3674 ratio | Dose reduction indexb for:
|
CIc | |
|---|---|---|---|
| C34 | Fab-3674 | ||
| bF-3674 | |||
| 1:5 | 4.4 ± 0.6 | 2.7 ± 0.4 | 0.7 ± 0.2 |
| 1:7 | 6.7 ± 1.0 | 2.8 ± 0.5 | 0.6 ± 0.1 |
| 1:10 | 10.2 ± 1.6 | 2.8 ± 0.5 | 0.5 ± 0.1 |
| 1:15 | 9.5 ± 1.4 | 2.0 ± 0.3 | 0.7 ± 0.2 |
| 1:20 | 18.4 ± 3.1 | 2.9 ± 0.6 | 0.4 ± 0.1 |
| 1:25 | 16.2 ± 2.6 | 2.0 ± 0.3 | 0.6 ± 0.2 |
| mF-3674 | |||
| 1:20 | 2.0 ± 0.4 | 2.1 ± 0.5 | 1.2 ± 0.4 |
| 1:30 | 3.0 ± 0.6 | 2.1 ± 0.6 | 1.0 ± 0.4 |
| 1:40 | 4.3 ± 1.1 | 2.3 ± 0.7 | 0.8 ± 0.3 |
| 1:50 | 5.1 ± 1.0 | 2.1 ± 0.5 | 0.8 ± 0.3 |
The experiments were carried out as described previously (17).
The DRI is the ratio of IC50s in the absence and presence of the second inhibitor.
The CI is the summation of the effects of the two inhibitors in combination (see Materials and Methods). The synergistic, additive, and antagonistic effects of the combination of two inhibitors are defined by CI values of <1, 1, and >1, respectively (9). The IC50 values for C34, bF3674, and mF3674 alone are 28 ± 3, 78 ± 11, and 582 ± 112 nM, respectively.
FIG. 4.
Synergistic and synchronized antiviral activities of Fab bF-3674 and C34 further define the Fab 3674 epitope. (A) Dose response curves for antiviral activity against HXB2 Env-pseudotyped virus of C34 (green), bF-3674 Fab (blue), and a typical fixed combination ratio (1:10) of C34/bF-3674 (red). The experimental data (closed circles) are the averages for 6 to 10 independent experiments, with error bars indicating standard deviations and solid lines representing best-fit curves using the following simple activity relationship: percent fusion = 100/(1 + [Fab]/IC50). The concentrations for the combination curve refer to the molar concentration of the C34 peptide. Dose reduction indices and CIs for different ratio combinations are provided in Table 4. (B) Synchronized viral infection assays using fully inhibitory concentrations of C34 or bF-3674 at increasing time points. Assays were run at 30°C (triangles) and 37°C (circles); data were fit to a sigmoidal curve as described previously (34). (C) Comparison of epitopes for neutralizing anti-gp41 antibodies Fab 3674 and D5. Mutagenesis and Western blot analyses show that Fab 3674-N-HR binding is governed by residues located at positions b and f of the heptad repeat (indicated in blue), while crystallographic analysis (27) shows that D5 binding to the N-HR occurs in a hydrophobic pocket located between two N helices. W571, located at position f, is critical for recognition of both antibodies.
DISCUSSION
The ability of HIV to escape a neutralizing immune response has been a major challenge of the vaccine development effort. To date, only four potent and broadly neutralizing monoclonal antibodies have been reported. 2G12 and b12 target gp120 (4, 41), and 2F5 and 4E10 target the membrane-proximal region of gp41 (32, 38, 45) that is located C terminal to the C-HR. The accessibility of the PHI, coupled with the highly conserved sequence of gp41, has long rendered gp41 as an attractive target for inhibitors and antibodies with anti-HIV activity. Antibodies and Fabs raised or selected against gp41-derived constructs such as the 6-HB and the N-HR and C-HR peptides have been described. However, with the exception of D5 (28) and most recently m46 (10), all have been only weakly inhibitory or nonneutralizing (7, 16, 25). This shortcoming has generally been attributed to the difficulty of inhibiting a transient intermediate, namely, the PHI, or the sheer sizes of immunoglobulin Gs, which were perceived as being too bulky to reach a target whose structure and accessibility in the context of virions are largely unknown.
Using the chimeric construct NCCG-gp41 for panning, we have succeeded in selecting from a human nonimmune phage library a broadly neutralizing Fab (3674) that inhibits infection by HIV-1 pseudotyped with Envs of primary and laboratory-adapted strains belonging to subtypes A, B, and C. Fab 3674 targets a structurally contiguous epitope on the N-HR of gp41 that is solvent accessible and located in a shallow groove between two C-HR helices in the 6-HB. This epitope is displaced by approximately one helical turn from the N-HR epitopes for Fabs 3663 and 3670, which are also inhibitory in a fusion assay but are nonneutralizing.
The binding site recognized by Fab 3674, defined by mutagenesis and Western blot analyses using multiple well-characterized gp41 constructs, partially overlaps with that of monoclonal antibody 5H/I1-BMV-D5 (D5) (31). D5 was selected from a scFv library, using 5-helix and the N-HR mimetic IZN36 as targets. Like Fab 3674, D5 is also broadly neutralizing, although bF-3674 appears to be two to three times more potent against strains for which both antibodies were tested (specifically HXB2, JR CSF, SF162, and 89.6). The epitopes of Fab 3674 and D5, however, are clearly distinct. D5 binds to the hydrophobic pocket situated between and formed by two N-HR helices and interacts with both (27, 31) (Fig. 1A). With the exception of W571 (position f), the D5-gp41 interaction surface is composed of residues at positions b, e, g, and c of the heptad repeat (Fig. 4C). Because this pocket normally accommodates the C-HR helix, D5 blocks binding of the C34 peptide to 5-helix. Likewise, a cyclic D peptide that binds specifically to this hydrophobic pocket (12) blocks binding of D5 to 5-helix. Thus, D5 inhibits fusion in a manner analogous to that of C34 by binding to the exposed N-HR trimer in the PHI conformation of gp41, thereby preventing formation of the fusogenic 6-HB. Fab 3674, however, binds tightly to the 6-HB (Fig. 1C, 3, and 4) as well as stable N-HR trimers, and residues at positions b and f of the N-HR mediate its binding to gp41. Moreover, C34 and Fab 3674 do not compete with one another and neutralize HIV-1 (HXB2) either synergistically (for bF-3674) or additively (for mF-3674). Thus, in addition to the distinct binding sites, we can surmise that the mode of action of Fab 3674 may also differ from that of D5, with Fab 3674 having the potential to act on multiple targets during or after 6-HB formation. In addition to the PHI, these may include the proposed pre-6-HB intermediate (30) or the 6-HB itself, interactions with which may prevent formation of an effective fusion pore comprising multiple gp41 molecules (16, 30).
The epitope for Fab 3674, comprising E560, H564, W571, K574, and Q575, is fully conserved throughout the Envs of the clade A, B, and C viruses tested, with one exception: H564 is replaced by an Arg in JRFL. Interestingly, Fab 3674 did not inhibit JRFL env-mediated cell fusion in our preliminary screen, a cell-cell fusion assay. In addition to being consistent with H564 contributing to the epitope of Fab 3674 antibodies, this observation suggests that the presence of a large and/or positively charged residue in this region abrogates Fab binding. For the other viruses tested, no correlations were observed between virus subtype or coreceptor usage and degree of inhibition of cell fusion or virus neutralization. Together, these results suggest that the neutralizing activity of Fab 3674 toward a particular HIV-1 strain may be determined not only by affinity to gp41, which should in this case be fairly constant, with the exception of JRFL, but also by kinetic restriction. In particular, the effectiveness of gp41-directed inhibitors has been shown to be dependent on the ratio of the apparent unimolecular-rate constants for the formation of an irreversible fusogenic state versus a dead-end-inhibited state (37).
This study constitutes the first example of a 6-HB-directed antibody that can neutralize diverse HIV-1 strains. In addition to Fab 3674, several other antibodies that target the ectodomain of gp41 have been described very recently and include D5 (28) and m46 (10), each of which exhibits various degrees of potency, depending on the virus strain and assay used. These results provide additional support for ongoing vaccine design efforts aimed at targeting the surface envelope glycoproteins gp41 and gp120.
Acknowledgments
This work was supported by the Intramural AIDS Targeted Antiviral Program of the Office of the Director of the National Institutes of Health and by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (to G.M.C. and C.A.B.).
We thank E. Berger, R. Brundiers, C. Frisch, L. Laugenaur, J. Mascola, and N. Ray for useful discussions and reagents; A. Aniana for technical assistance; and the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, for many of the reagents used in this study.
Footnotes
Published ahead of print on 26 September 2007.
REFERENCES
- 1.Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism and disease. Annu. Rev. Immun. 17:657-700. [DOI] [PubMed] [Google Scholar]
- 2.Bewley, C. A., J. M. Louis, R. Ghirlando, and G. M. Clore. 2002. Design of a novel peptide inhibitor of HIV fusion that disrupts the internal trimeric coiled-coil of gp41. J. Biol. Chem. 277:14238-14245. [DOI] [PubMed] [Google Scholar]
- 3.Bohm, G., R. Muhr, and R. Jaenicke. 1992. Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng. 5:191-195. [DOI] [PubMed] [Google Scholar]
- 4.Burton, D. R., P. Jayashree, R. Kodury, S. J. Sharp, G. B. Thornton, P. W. H. I. Parren, L. S. W. Sawyer, R. M. Hendry, N. Dunlop, P. Nara, M. Lamacchia, E. Garrati, E. R. Stiehm, Y. J. Bryson, Y. Cao, J. P. Moore, D. D. Ho, and C. F. Barbas III. 1994. Efficient neutralization of primary isolates of HIV 1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 5.Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93:681-684. [DOI] [PubMed] [Google Scholar]
- 6.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C. H., M. L. Greenberg, D. P. Bolognest, and T. J. Matthews. 2000. Monoclonal antibodies that bind to the core of fusion-active glycoprotein 41. AIDS Res. Hum. Retrovir. 16:2037-2041. [DOI] [PubMed] [Google Scholar]
- 8.Chou, T. C., and P. Talalay. 1981. Generalized equations for the analysis of inhibitions of Michaelis-Menten and higher-order kinetic systems for two or more mutually exclusive and non-exclusive inhibitors. Eur. J. Biochem. 115:207-216. [DOI] [PubMed] [Google Scholar]
- 9.Chou, T. C., and P. Talalay. 1984. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 22:27-55. [DOI] [PubMed] [Google Scholar]
- 10.Choudry, V., M.-Y. Zhang, I. A. Sidorov, J. M. Louis, I. Harris, A. S. Dimitrov, P. Bouma, F. Cham, A. Choudhary, S. M. Rybak, T. Fouts, D. C. Montefiori, C. C. Broder, G. V. Quinnan, Jr., and D. S. Dimitrov. 2007. Cross-reactive HIV-1 neutralizing monoclonal antibodies selected by screening of an immune human phage library against and envelope glycoprotein (gp140) isolated from a patient (R2) with broadly HIV-1 neutralizing antibodies. Virology 363:79-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 12.Eckert, D. M., V. N. Malashkevich, L. H. Hong, P. A. Carr, and P. S. Kim. 1999. Inhibiting HIV-1 entry: discovery of D-peptide inhibitors that target the gp41 coiled-coil pocket. Cell 99:103-115. [DOI] [PubMed] [Google Scholar]
- 13.Furata, R. A., C. T. Wild, Y. Weng, and C. D. Weiss. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat. Struct. Biol. 5:276-279. [DOI] [PubMed] [Google Scholar]
- 14.Gallo, S. A., C. M. Finnegan, M. Viard, Y. Raviv, A. Dimitrov, S. S. Rawat, A. Puri, S. Durell, and R. Blumenthal. 2003. The HIV Env-mediated fusion reaction. Biochim. Biophys. Acta 1614:36-50. [DOI] [PubMed] [Google Scholar]
- 15.Gao, F., S. G. Morrison, D. L. Robertson, C. L. Thornton, S. Craig, G. Karlsson, J. Sodroski, M. Morgado, B. Galvao-Castro, H. von Briesen, S. Beddows, J. Weber, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 1996. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G: the WHO and NIAID networks for HIV isolation and characterization. J. Virol. 70:1651-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Golding, H., M. Zaitseva, E. de Rosny, L. R. King, J. Manischewitz, I. Sidorov, M. K. Gorny, S. Zolla-Pazner, D. S. Dimitrov, and C. D. Weiss. 2002. Dissection of human immunodeficiency virus type 1 entry with neutralizing antibodies to gp41 fusion intermediates. J. Virol. 76:6780-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gustchina, E., J. M. Louis, C. A. Bewley, and G. M. Clore. 2006. Synergistic inhibition of HIV-1 envelope-mediated membrane fusion by inhibitors targeting the N- and C-terminal heptad repeats of gp41. J. Mol. Biol. 364:283-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang, S., K. Lin, N. Strick, and A. R. Neurath. 1993. HIV-1 inhibition by a peptide. Nature 365:113. [DOI] [PubMed] [Google Scholar]
- 19.Karlsson, G. B., F. Gao, J. Robinson, B. Hahn, and J. Sodroski. 1996. Increased envelope spike intensity and stability are not required for the neutralization of primary human immunodeficiency viruses. J. Virol. 70:6136-6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knappik, A., L. Ge, A. Honegger, P. Pack, M. Fisher, G. Wellnhofer, A. Hoess, J. Wölle, A. Plückthun, and B. Virnekäs. 2000. Fully synthetic human combinatorial antibody libraries (HuCAL) based on molecular consensus frameworks and CDRs randomized with trinucleotides. J. Mol. Biol. 296:57-86. [DOI] [PubMed] [Google Scholar]
- 21.Li, M., F. Gao, J. R. Mascola, L. Stamatatos, V. R. Polonis, M. Koutsoukos, G. Voss, P. Goepfert, P. Gilbert, K. M. Greene, M. Bilska, D. L. Kothe, J. F. Salazar-Gonzalez, X. Wei, J. M. Decker, B. H. Hahn, and D. C. Montefiori. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108-10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, Y., K. Svehla, N. L. Mathy, G. Voss, J. R. Mascola, and R. Wyatt. 2006. Characterization of antibody responses elicited by human immunodeficiency virus type 1 primary isolate trimeric and monomeric envelope glycoproteins in selected adjuvants. J. Virol. 80:1414-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Louis, J. M., C. A. Bewley, and G. M. Clore. 2001. Design and properties of NCCG-gp41, a chimeric gp41 molecule with nanomolar HIV-1 fusion inhibitory activity. J. Biol. Chem. 276:29485-29489. [DOI] [PubMed] [Google Scholar]
- 24.Louis, J. M., G. M. Clore, and C. A. Bewley. 2003. Covalent trimers of the internal N-terminal trimeric coiled-coil of gp41 and antibodies directed against them are potent inhibitors of HIV envelope-mediated cell fusion. J. Biol. Chem. 278:20278-20285. [DOI] [PubMed] [Google Scholar]
- 25.Louis, J. M., C. A. Bewley, E. Gustchina, A. Aniana, and G. M. Clore. 2005. Characterization and HIV-1 fusion inhibitory properties of monoclonal Fabs obtained from a human non-immune phage library selected against diverse epitopes of the ectodomain of HIV-1 gp41. J. Mol. Biol. 353:945-951. [DOI] [PubMed] [Google Scholar]
- 26.Louwagie, J., W. Janssens, J. Mascola, L. Heyndrickx, P. Hegerich, G. van der Groen, F. E. McCutchan, and D. S. Burke. 1995. Genetic diversity of the envelope glycoprotein from human immunodeficiency virus type 1 isolates of African origin. J. Virol. 69:263-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luftig, M. A., M. Mattu, P. Di Giovine, R. Geleziunas, R. Hrin, G. Barbato, E. Bianchi, M. D. Miller, A. Pessi, and A. Carfi. 2006. Structural basis for HIV-1 neutralization by a gp41 fusion intermediate-directed antibody. Nat. Struct. Mol. Biol. 13:740-747. [DOI] [PubMed] [Google Scholar]
- 28.Mascola, J. R., P. D'Souza, P. Gilbert, B. H. Hahn, N. L. Haigwood, L. Morri, C. J. Petropoulos, V. R. Polonis, M. Sarzotti, and D. C. Montefiori. 2005. Recommendations for the design and use of standard virus panels to assess neutralizing antibody responses elicited by candidate human immunodeficiency virus type 1 vaccines. J. Virol. 79:10103-10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matthews, T., M. Salgo, M. Greenberg, J. Chung, R. DeMasi, and C. Bolognesi. 2004. Enfuvirtide: the first therapy to inhibit the entry of HIV-1 into host CD4 lymphocytes. Nat. Rev. Drug Discov. 3:215-225. [DOI] [PubMed] [Google Scholar]
- 30.Melikyan, G. B., M. Egelhofer, and D. von Laer. 2006. Membrane-anchored inhibitory peptides capture human immunodeficiency virus type 1 gp41 conformations that engage the target membrane prior to fusion. J. Virol. 80:3249-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, M. D., R. Geleziunas, E. Bianchi, S. Lennard, R. Hrin, H. Zhang, M. Lu, Z. An, P. Ingallinella, M. Finotto, M. Mattu, A. C. Finnefrock, D. Bramhill, J. Cook, D. M. Eckert, R. Hampton, M. Patel, S. Janatow, J. Joyce, G. Ciliberto, R. Cortese, P. Lu, W. Strohl, W. Schleif, M. McElhaugh, S. Lane, C. Lloyd, D. Lowe, J. Osbourn, T. Vaughan, E. Emini, G. Barbato, P. S. Kim, D. J. Hazua, J. W. Shiver, and A. Pessi. 2005. A human monoclonal antibody neutralizes diverse HIV-1 isolates by binding a critical gp41 epitope. Proc. Natl. Acad. Sci. USA 102:14759-14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muster, T., F. Steindl, M. Purtscher, A. Trkola, A. Klima, G. Himmler, F. Ruker, and H. Katinger. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67:6642-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nussbaum, O., C. C. Broder, and E. A. Berger. 1994. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J. Virol. 68:5411-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Doherty, U., W. J. Swiggard, and M. H. Malim. 2000. A sensitive quantitative assay for human immunodeficiency virus type 1 integration. J. Virol. 74:10074-10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reeves, J. D., S. A. Gallo, N. Ahmad, J. L. Miamidian, P. E. Harvey, M. Sharron, S. Pohlmann, J. N. Sfakianos, C. A. Derdeyn, R. Blumenthal, E. Hunter, and R. W. Doms. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density and fusion kinetics. Proc. Natl. Acad. Sci. USA 99:16249-16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Root, M. J., M. S. Kay, and P. S. Kim. 2001. Protein design of an HIV-1 entry inhibitor. Science 291:884-888. [DOI] [PubMed] [Google Scholar]
- 37.Steger, H. K., and M. J. Root. 2006. Kinetic dependence to HIV-1 entry inhibition. J. Biol. Chem. 281:25813-25821. [DOI] [PubMed] [Google Scholar]
- 38.Stiegler, G., R. Kunert, M. Purtscher, S. Wolbank, R. Voglauer, F. Steindl, and H. Katinger. 2001. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 17:1757-1765. [DOI] [PubMed] [Google Scholar]
- 39.Sullivan, N., Y. Sun, J. Li, W. Hofmann, and J. Sodroski. 1995. Replicative function and neutralization sensitivity of envelope glycoproteins from primary and T-cell line-passaged human immunodeficiency virus type 1 isolates. J. Virol. 69:4413-4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan, K., J. Liu, S. Wang, S. Sgen, and M. Lu. 1997. Atomic structure of a thermostable subdomain of gp41. Proc. Natl. Acad. Sci. USA 94:12303-12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 43.Wild, C., T. Oas, C. McDanal, D. Bolognesi, and T. Matthews. 1992. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc. Natl. Acad. Sci. USA 89:10537-10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williamson, C., L. Morris, M. F. Maughan, L. H. Ping, S. A. Dryga, R. Thomas, E. A. Reap, T. Cilliers, J. van Harmelen, A. Pascual, G. Ramjee, G. Fray, R. Johnston, S. A. Karim, and R. Swanström. 2003. Characterization and selection of HIV-1 subtype C isolates for use in vaccine development. AIDS Res. Hum. Retrovir. 19:133-144. [DOI] [PubMed] [Google Scholar]
- 45.Zwick, M. B., A. F. Labrijn, M. Wang, C. Spenlehauer, E. O. Saphire, J. M. Binley, J. P. Moore, G. Stiegler, H. Katinger, D. R. Burton, and P. W. Parren. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]