Abstract
The APOBEC3 cytidine deaminases are potent antiviral factors that restrict replication of human immunodeficiency virus type 1 (HIV-1). HIV-1 Vif binds APOBEC3G and APOBEC3F and targets these proteins for ubiquitination by forming an E3 ubiquitin ligase with cullin 5 and elongins B and C. The N-terminal region of Vif is required for APOBEC3G binding, but the binding site(s) is unknown. To identify the APOBEC3G binding site in Vif, we established a scalable binding assay in a format compatible with development of high-throughput screens. In vitro binding assays using recombinant proteins identified Vif peptides and monoclonal antibodies that inhibit Vif-APOBEC3G binding and suggested involvement of Vif residues 33 to 83 in APOBEC3G binding. Cell-based binding assays confirmed these results and demonstrated that residues 40 to 71 in the N terminus of Vif contain a nonlinear binding site for APOBEC3G. Mutation of the highly conserved residues His42/43 but not other charged residues in this region inhibited Vif-APOBEC3G binding, Vif-mediated degradation of APOBEC3G, and viral infectivity. In contrast, mutation of these residues had no significant effect on Vif binding and degradation of APOBEC3F, suggesting a differential requirement for His42/43 in Vif binding to APOBEC3G and APOBEC3F. These results identify a nonlinear APOBEC3 binding site in the N terminus of Vif and demonstrate that peptides or antibodies directed against this region can inhibit Vif-APOBEC3G binding, validating the Vif-APOBEC3 interface as a potential drug target.
The human immunodeficiency virus type 1 (HIV-1) Vif protein is required during virus replication to overcome the antiviral activity of the cytidine deaminases APOBEC3G and APOBEC3F. In the absence of Vif, APOBEC3G and APOBEC3F are packaged into HIV-1 virions and deaminate cytidines in viral minus-strand DNA during reverse transcription, resulting in G-to-A hypermutation and premature degradation of newly synthesized viral DNA (7, 14, 16, 41). APOBEC3G and APOBEC3F are associated with high-molecular-weight ribonucleoprotein complexes in the cytoplasm, possibly localizing to P bodies and stress granules (3, 6, 12, 36), and may also inhibit viral replication via deamination-independent mechanisms (1, 8, 22, 29). Vif neutralizes the antiviral activity of APOBEC3G and APOBEC3F predominantly by forming an E3 ubiquitin ligase with cullin 5 (Cul5), elongin B (EloB), and elongin C (EloC) that targets these proteins for degradation by the ubiquitin-proteasome pathway (4, 13, 18-20, 27, 33, 38, 39). Vif may also inhibit APOBEC3 activity through mechanisms independent of proteasomal degradation (10, 11, 20, 24, 33). Vif associates with the Cul5-EloB-EloC complex by binding directly to EloC via a BC box motif at positions 144 to 153 and to Cul5 via hydrophobic residues at positions 120, 123, and 124 within a zinc-binding region (residues 100 to 142) formed by a conserved H-X5-C-X17-18C-X3-5-H (HCCH) motif (19, 21). Binding of Vif to APOBEC3G and APOBEC3F is essential for their degradation by the Vif-Cul5 E3 ligase (18). The Vif-APOBEC3G interaction is species specific; HIV-1 Vif binds to and inactivates human APOBEC3G and APOBEC3F but not APOBEC3 proteins derived from African green monkeys (AGM) and rhesus macaques (2, 15, 17, 25, 37). Conversely, simian immunodeficiency virus SIV(AGM) Vif inactivates AGM and rhesus macaque but not human APOBEC3G. A single amino acid difference in APOBEC3G, aspartic acid at position 128 in human APOBEC3G versus lysine in AGM APOBEC3G, controls species specificity by influencing Vif-APOBEC3G binding (2, 15, 25, 37). The N-terminal region of HIV-1 Vif is important for binding and neutralization of APOBEC3G and APOBEC3F and also contributes to species-specific recognition (18, 26, 32, 34), but the specific binding site(s) for APOBEC3 proteins has not been determined.
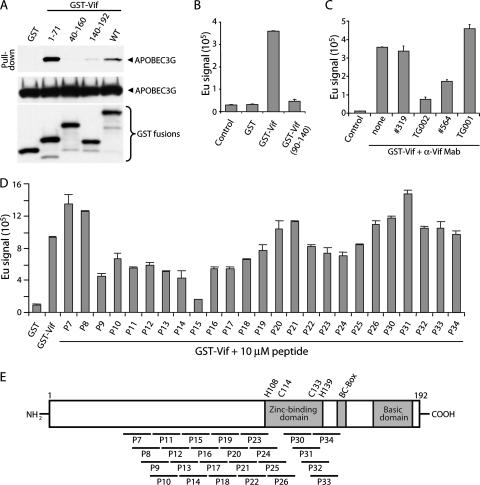
To investigate the APOBEC3G binding site in HIV-1 Vif, we performed coprecipitation experiments using full-length and truncated recombinant GST-Vif fusion proteins. Glutathione S-transferase (GST) and GST-Vif proteins (>95% pure) were purified from bacteria and incubated with lysate prepared from 293T cells expressing APOBEC3G-hemagglutinin (HA) as described previously (20). GST proteins were precipitated with glutathione-Sepharose and washed extensively, and bound proteins were detected by Western blotting (Fig. 1A, top). APOBEC3G was coprecipitated by full-length GST-Vif and GST-Vif(1-71), whereas GST-Vif(40-160) and GST-Vif(140-192) precipitated APOBEC3G at background levels similar to those of the GST control. Western blots demonstrated similar amounts of GST proteins and APOBEC3G in each sample (Fig. 1A, middle and bottom). These data suggest that Vif residues 1 to 71 are sufficient for APOBEC3G binding.
FIG. 1.
APOBEC3G binds the N-terminal region of Vif. (A) Recombinant GST and GST-Vif fusion proteins (2 μg) prebound to glutathione-Sepharose were incubated with lysate prepared from 293T cells expressing APOBEC3G-HA. APOBEC3G coprecipitating with GST proteins was detected by anti-HA western blotting (top). Equivalent levels of input APOBEC3G (middle) and GST (bottom) proteins were confirmed by western blotting with anti-HA or anti-GST, respectively. (B) Recombinant His-APOBEC3G (100 ng) (Immunodiagnostics, Woburn, MA) was incubated with no GST protein, GST, GST-Vif, or GST-Vif(90-140) (100 ng) bound to glutathione-coated 96-well plates for 1 h at room temperature. Bound APOBEC3G was detected with Eu-anti-His in a heterogenous TRF assay (Delfia) using a Wallac Victor II Multiplate reader. (C) Inhibition of APOBEC3G binding by anti-Vif monoclonal antibody TG002. Binding reactions were carried out as described above in the presence of 1 μg of the indicated antibody. (D) Peptide competition of Vif-APOBEC3G binding. Binding assays were performed in the presence of 10 μM Vif peptides. All data are presented as means of duplicate samples ± standard deviations. (E) Diagram of HIV-1 Vif and overlapping 15-mer Vif peptides used to test for competition of Vif-APOBEC3G binding.
To further characterize the APOBEC3G binding site in Vif and take initial steps toward establishing a screen for compounds that inhibit Vif-APOBEC3G binding, we developed a scalable binding assay in a format compatible with development of high-throughput screens. Binding of recombinant GST-Vif to recombinant His-APOBEC3G (Immunodiagnostics) was measured by a heterogenous time-resolved fluorometry (TRF) assay (Delfia; Perkin-Elmer). GST or GST-Vif (100 ng) was bound to glutathione-coated 96-well plates and incubated with His-APOBEC3G (100 ng). Plates were washed, and bound APOBEC3G was detected with Europium (Eu)-labeled anti-His using a Wallac Victor II Multiplate reader. The Eu signal increased approximately 20-fold above background levels following binding of APOBEC3G to GST-Vif, whereas the signal obtained with GST-Vif(90-140) was similar to that of the GST control (Fig. 1B). We next used this assay to identify Vif monoclonal antibodies (MAbs) that inhibit Vif-APOBEC3G binding (Fig. 1C). Competition with MAb TG002 (NIH AIDS Reagent Program), which maps to Vif residues 34 to 47, reduced Vif-APOBEC3G binding by approximately 80% (Fig. 1C). MAb #564 (31) reduced Vif-APOBEC3G binding by ∼50%, whereas MAbs #319 (31) and TG001, which maps to residues 176 to 192, had no inhibitory effect. We screened overlapping Vif peptides corresponding to the clade B consensus sequence (15-mers at 10 μM, overlapping by 11 amino acids) (NIH AIDS Reagent Program) for their ability to inhibit Vif-APOBEC3G binding. Peptides P9 to P18, derived from sequences spanning Vif residues 33 to 83, decreased the Eu signal induced by Vif-APOBEC3G binding by ∼40 to 80% (Fig. 1D). P15 (derived from residues 57 to 71) was the most potent inhibitory peptide, reducing the Eu signal to levels near background; whether its increased potency relative to those of the other inhibitory peptides reflects enhanced competition or an intrinsic property of this peptide (e.g., improved solubility and/or folding) is unclear. Peptides derived from the central and C-terminal regions of Vif did not inhibit Vif-APOBEC3G binding (Fig. 1D; also data not shown). Peptides 7, 8, and 31 slightly enhanced the TRF signal. The explanation for this finding is unclear; one possibility is that these peptides may interact with Vif and/or APOBEC3G and induce a conformational change that enhances Vif-APOBEC3G association. These results suggest that the N-terminal region of Vif is involved in APOBEC3G binding.
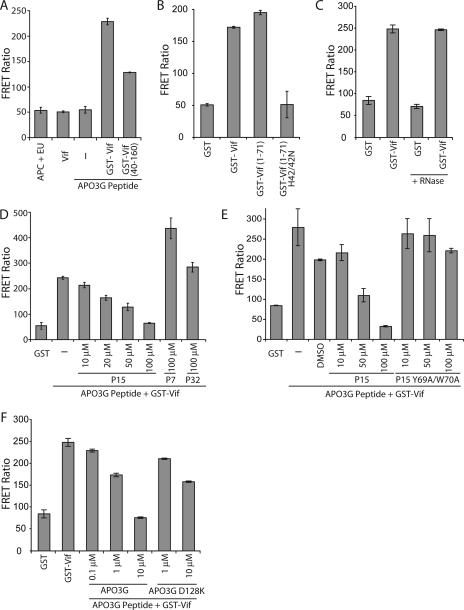
We confirmed the TRF binding results and simplified the binding assay format by developing a homogenous time-resolved fluorescence resonance energy transfer (FRET) assay (Lance; Perkin-Elmer) using recombinant Vif and a synthetic APOBEC3G peptide. The APOBEC3G peptide sequence (residues 110 to 148) was based on modeling of the APOBEC3G sequence onto the B. subtilis cytidine deaminase (ljkt) structure (data not shown) and is predicted to be composed of two alpha-helices flanking a loop containing the D128 residue that determines species-specific Vif-APOBEC3G binding (2, 15, 25, 37). Circular dichroism spectral analysis of the APOBEC3G peptide confirmed the presence of a helical secondary structure (data not shown). Binding assays were performed according to the manufacturer's protocol using GST or GST-Vif proteins (15 nM) incubated with the biotinylated APOBEC3G peptide (500 nM) in a 384-well format. Vif and APOBEC3G were labeled with anti-GST-Eu (2 nM) and streptavidin-allophycocyanin (APC) (25 nM), respectively, and the FRET signal was measured as fluorescence emission intensity. GST-Vif binding to the APOBEC3G peptide produced a significant increase in the FRET ratio over background levels; GST-Vif(40-160) bound to the APOBEC3G peptide but less efficiently than full-length GST-Vif (Fig. 2A). The low dynamic range of this assay compared to that of the TRF assay was expected, since the FRET signal is dependent on the proximity of the Eu and APC labels and both proteins are labeled indirectly. An important advantage over the TRF assay, however, is the homogeneous format of the FRET assay, which is better suited for development of high-throughput screens. GST-Vif(1-71) bound to the APOBEC3G peptide at levels similar to those of full-length GST-Vif, providing further evidence that the APOBEC3G binding site is located within the N terminus of Vif (Fig. 2A and B). Mutation of the conserved histidines at positions 42 and 43, which are important for Vif-APOBEC3G binding (see below), disrupted GST-Vif(1-71) binding to the APOBEC3G peptide (Fig. 2B). Vif and APOBEC3G are RNA-binding proteins (5, 9, 40). We therefore investigated whether the observed binding was direct or dependent upon RNA. The efficiencies of GST-Vif-APOBEC3G peptide binding in the presence and absence of RNase A were similar (Fig. 2C), consistent with results in previous studies demonstrating that Vif binds directly to APOBEC3G (20).
FIG. 2.
Peptide inhibition of Vif-APOBEC3G binding in a homogenous FRET assay (Lance). (A to C) Recombinant GST-Vif protein (15 nM) was incubated with biotinylated APOBEC3G peptide (aa 110 to 148) (500 nM) in a 384-well plate. Binding was detected using APC-streptavidin and Eu-anti-GST and expressed as a ratio [(fluorescence emission intensity at 665 nm/intensity at 615 nm) × 104]. RNase A (10 μg/ml) was added to reaction mixtures where indicated. (D to F) Peptide competition of Vif-APOBEC3G binding. Binding reactions were carried out as for panel A with Vif peptides 7 (aa 25 to 39), 9 (aa 33 to 47), 15 (aa 57 to 71), mutant 15 (aa 57 to 71; Y69A/W70A), and 32 (aa 125 to 139) or wild-type or D128K APOBEC3G peptide (aa 110 to 148) added at the indicated concentrations. Data are presented as means for duplicate samples ± standard deviations.
To further demonstrate the specificity of binding in the FRET assay, we performed peptide competition assays using peptides derived from regions of Vif and APOBEC3G that were previously shown to be important for function in vivo. Binding of Vif to the APOBEC3G peptide was competed by P15 (residues 57 to 71). Similar to the TRF assay, binding was inhibited by increasing amounts of P15, whereas P7 (residues 25 to 39) and P32 (residues 125 to 139) failed to inhibit binding at the highest peptide concentration (Fig. 2D). Peptide P7 enhanced the FRET signal, similar to results in the TRF assay (Fig. 1D). The requirement for a large molar excess of P15 and other Vif peptides for inhibition in the FRET and TRF assays might be due to the fact that 15-amino-acid (aa) peptides represent only a small fragment of the larger region involved in APOBEC3G binding (see below). The MAb TG002 inhibited Vif-APOBEC3G binding in the FRET assay, similar to results in the TRF assay (data not shown). Previous mutagenic analysis of Vif showed that mutation of the conserved residues Tyr69 and Trp70 present in P15 inhibits Vif function in H9 cells, which express APOBEC3G endogenously (30). A mutant P15 containing Y69A and W70A failed to inhibit Vif-APOBEC3G binding, even at the highest concentration tested (Fig. 2E); FRET signals with mutant P15 were similar to those obtained in the presence of dimethyl sulfoxide alone. Binding reactions were also performed in the presence of a competitor APOBEC3G peptide (residues 110 to 148) lacking a biotin moiety. The nonbiotinylated APOBEC3G peptide inhibited Vif-APOBEC3G binding, whereas the peptide containing the D128K mutation, which impairs species-specific Vif-APOBEC3G interactions, had weaker inhibitory activity (Fig. 2F). These data confirm the specificity of the FRET binding assay and demonstrate that the in vitro binding assay recapitulates physiologically relevant interactions. Together, results from coprecipitation experiments and the TRF and FRET assays demonstrate that the N-terminal region of Vif is required for APOBEC3G binding and contains the binding site for APOBEC3G.
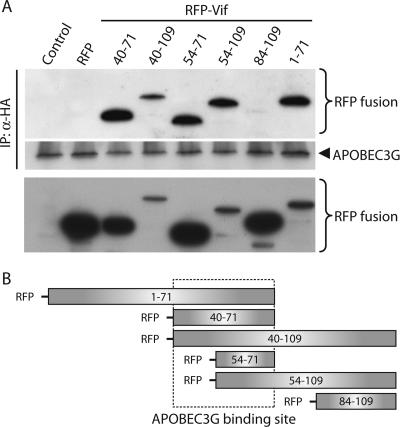
The in vitro binding assays suggested that the N-terminal region of Vif binds to APOBEC3G. Cell-based binding assays using Vif mutants have been complicated by the relative instability and/or aggregation of Vif deletion mutants, possibly due to improper folding (18). To identify the N-terminal region(s) of Vif required for APOBEC3G binding in cells, we prepared a series of red fluorescent protein (RFP) fusion proteins (Fig. 3), since this approach was expected to improve the stability of Vif deletion mutants. Vif fragments were fused to the C terminus of monomeric RFP (28) and coexpressed in 293T cells with APOBEC3G-HA. RFP-Vif fusion proteins demonstrating a punctate expression pattern by fluorescence microscopy [i.e., RFP-Vif(1-109)], indicative of protein aggregation, were excluded from further analysis (data not shown). Cells were transfected with identical amounts of each RFP-Vif plasmid; differences in expression levels (Fig. 3A) might therefore result from intrinsic differences in protein folding or stability (data not shown). APOBEC3G was immunoprecipitated with anti-HA (Roche), and coprecipitating RFP-Vif fusions were detected by Western blotting with anti-RFP (Stratagene). RFP-Vif(1-71) was coprecipitated by APOBEC3G, suggesting that Vif residues 1 to 71 are sufficient for APOBEC3G binding (Fig. 3A). In contrast, residues 84 to 109 failed to coprecipitate with APOBEC3G despite high levels of expression detected by Western blotting. RFP alone did not bind APOBEC3G. A minimal fragment containing Vif residues 54 to 71 bound APOBEC3G, but Vif-APOBEC3G binding was enhanced by the presence of flanking residues based on the levels of coprecipitated full-length RFP-Vif, RFP-Vif(54-71), RFP-Vif(40-71), and RFP-Vif(54-109) compared to their relative expression in cell lysate (Fig. 3A; also data not shown). RFP-Vif(40-71) was more efficiently coprecipitated by APOBEC3G than RFP-Vif(54-71), even though RFP-Vif(54-71) was expressed at higher levels. Binding assays with full-length GST-Vif and GST-Vif(40-160) proteins suggested that Vif residues 1 to 40 are important for APOBEC3G binding (Fig. 1A) (20). In contrast, results from cell-based binding assays (Fig. 3) suggested these residues are dispensable for APOBEC3G binding by RFP-Vif fusion proteins, consistent with results of the peptide inhibition studies (Fig. 1D). This discrepancy, which might result from the use of different fusion tags and/or intrinsic differences in protein folding or differences between cell-based and in vitro assays, raises the possibility that residues upstream of position 40 might contribute to APOBEC3G binding. Nonetheless, binding experiments using RFP-Vif fusions suggest that amino acids 40 to 71 comprise the major region in Vif important for APOBEC3G binding in cells (Fig. 3B).
FIG. 3.
Vif residues 40 to 71 are important for APOBEC3G binding. (A) Lysates prepared from 293T cells coexpressing APOBEC3G-HA and RFP-Vif fusions were subjected to anti-HA immunoprecipitation. Proteins were detected by Western blotting using anti-RFP (top) or anti-HA (α-HA) (middle). Expression of RFP fusions was confirmed by anti-RFP Western blotting of cell lysates (bottom). (B) Diagram of RFP-Vif fusions.
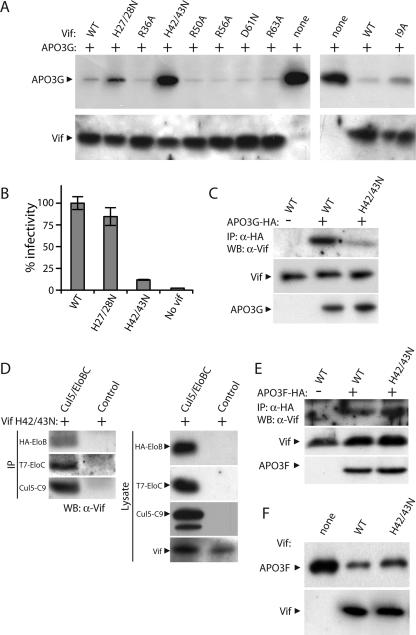
We next used Vif point mutants to investigate the Vif-APOBEC3G interaction and determine functional consequences of binding mutations on the anti-APOBEC3 activity of Vif. The charged residue at position 128 in human APOBEC3G regulates species-specific interaction with Vif, suggesting the presence of a complementary charge on Vif (25). Therefore, we analyzed the importance of conserved charged residues in the N-terminal region of Vif for APOBEC3G degradation (Fig. 4A). Vif and APOBEC3G-HA were coexpressed in 293T cells, and the ability of Vif to induce APOBEC3G degradation was determined by Western blotting of cell lysate. Mutation of the histidines at positions 42 and 43, which are conserved in >99% of HIV-1 Vif sequences, prevented Vif-mediated degradation of APOBEC3G; APOBEC3G levels were similar to those present in the absence of Vif. Notably, these residues are located within the APOBEC3G binding region and the inhibitory peptide P9 identified in the preceding experiments. Furthermore, histidine 43 was previously identified as being important for Vif function during virus production in H9 cells (30). Mutation of the conserved histidines at positions 27 and 28 resulted in only a minor reduction in Vif-mediated degradation of APOBEC3G, whereas mutation of other charged residues within the N terminus, R36, R50, R56, D61, and R63, had no significant effect. Residue I9 has been suggested to be important for APOBEC3G binding based on results obtained with YFP-Vif fusions (35). However, in the context of Vif alone, the I9A mutant functioned similar to the wild type for Vif-mediated degradation of APOBEC3G (Fig. 4A).
FIG. 4.
Histidines 42/43 in Vif are important for binding and neutralization of APOBEC3G but not APOBEC3F. (A) APOBEC3G-HA was expressed in 293T cells with wild-type (WT) and mutant Vif proteins. Protein levels were determined by Western blotting. (B) Virus was produced from 293/APOBEC3G cells following transfection with a vif-deleted proviral plasmid and WT or mutant pCDNA3.Vif. Infectivity of normalized virus was measured in Cf2-luc cells. Shown are means ± standard deviations (n = 3). (C) Vif residues His42/43 are important for APOBEC3G binding. 293T cells were cotransfected with APOBEC3G-HA and WT or H42/43N Vif expression plasmids. Lysates were immunoprecipitated (IP) with anti-HA (α-HA) and probed by Western blotting (WB). Equivalent levels of expression were confirmed by Western blotting of cell lysates. α-Vif, anti-Vif. (D) Vif H42/43N binds to Cul5-EloBC. 293T cells were cotransfected with Vif expression vectors and pCDNA3.HA-Cul5, pCDNA3.HA-EloB, or pCDNA3.T7-EloC. Lysates were immunoprecipitated (IP) with antibodies recognizing epitope tags on the indicated proteins and probed by Western blotting (WB). Protein expression was confirmed by Western blotting of cell lysates. α-Vif, anti-Vif. (E) Vif residues His42/43 are not important for APOBEC3F binding. Vif and APOBEC3F-HA were expressed in 293T cells, and immunoprecipitations (IP) and blotting (WB) were performed as for panel C. α-HA, anti-HA; α-Vif, anti-Vif. (F) Vif residues His42/43 are dispensible for Vif-mediated degradation of APOBEC3F. APOBEC3F-V5 was expressed in 293T cells with WT and mutant Vif proteins. Protein expression was detected by Western blotting.
To confirm the biological relevance of these observations, we tested the ability of mutant Vif proteins to support the production of infectious virus in the presence of APOBEC3G. Vif H42/43N was severely impaired for the production of infectious HIV-1 (Fig. 4B), consistent with the reduced capacity of this mutant to induce APOBEC3G degradation (Fig. 4A). In contrast, Vif H27/28N supported the production of infectious virus at levels near those of wild-type Vif. Coprecipitation experiments demonstrated that Vif H42/43N is defective for APOBEC3G binding (Fig. 4C), suggesting that decreased Vif-APOBEC3G binding is responsible for the reduced capacity to enhance viral infectivity. Vif H42/43N retained the ability to bind to EloB, EloC, and Cul5, suggesting that the loss of APOBEC3G binding did not result from a global defect in protein folding (Fig. 4D). Finally, we tested whether His42/43 were important for APOBEC3F binding and degradation. Analysis of naturally occurring Vif variants demonstrated the differential requirement for certain residues to overcome APOBEC3G versus APOBEC3F (32). Vif H42/43N bound to and induced degradation of APOBEC3F at levels similar to those of wild-type Vif (Fig. 4E and F), indicating that these residues are not important for APOBEC3F binding or degradation. These results are in agreement with those recently reported by Russell and Pathak, which demonstrated a requirement for Vif residues 40 to 44 (and the importance of the individual histidine residues at positions 42/43) to counteract APOBEC3G but not APOBEC3F (23). Together, these data suggest that H42/43 are selectively required for counteracting APOBEC3G but not APOBEC3F.
In summary, we demonstrated that residues 40 to 71 in the N terminus of Vif contain a nonlinear binding site for APOBEC3G. Vif residues 54 to 71 are sufficient for APOBEC3G binding, but amino acids 40 to 71 bind APOBEC3G more efficiently and His42/43 are important for Vif-APOBEC3G binding and Vif-mediated APOBEC3G degradation in vivo. Functional analysis of Vif variants isolated from patients demonstrated that mutations within the APOBEC3G binding region (Y40H and E45G) compromise the ability of Vif to overcome APOBEC3G (32). These data from patient isolates suggest that the region we identified as important for APOBEC3G binding is likely to be important for HIV replication in vivo, but they do not exclude possible involvement of other regions of Vif in binding to APOBEC3 proteins. For example, RFP-Vif(54-71) and RFP-Vif(54-109) bound to APOBEC3G in cells (Fig. 3A) even though they lack His42/43, which appear to be important for APOBEC3G binding by full-length Vif (without RFP) in cells (Fig. 4A). The conserved tryptophan residues at positions 11 and 79 are important for Vif-mediated suppression of APOBEC3F but not APOBEC3G. In addition, residues 14 to 17 (DRMR), which are not essential for Vif binding to APOBEC3G, facilitate species-specific recognition of APOBEC3G (23, 26). Together these results suggest that the N terminus of Vif contains a nonlinear structure that forms the APOBEC3 protein binding site and binds to APOBEC3G and APOBEC3F through distinct interfaces (23, 25, 34). Vif may function like a molecular scaffold, since it contains at least four sites that mediate protein-protein interactions—Vif-APOBEC3G (Fig. 1 to 4) (23), Vif-APOBEC3F (23, 34), Vif-EloC (19, 39), and Vif-Cul5 (21). The APOBEC3G and APOBEC3F binding sites likely overlap. Importantly, results from the Vif-APOBEC3G TRF and FRET binding assays establish a format for high-throughput screens to identify small-molecule inhibitors that disrupt this critical interaction and validate the Vif-APOBEC3 interface as a potential drug target.
Acknowledgments
We thank M. Malim and K. Strebel for reagents and R. Ptak and M. Glicksman for helpful advice. Vif monoclonal antibodies, peptides, and 293/APOBEC3G cells were obtained from the AIDS Research and Reference Reagent Program (donated by M. Malim, Transgene, and NIAID).
A.M. was supported in part by an NSF Predoctoral Fellowship. This work was supported by NIH grants AI67032 and AI62555. Core facilities were supported by Harvard Center for AIDS Research (CFAR) and DF/HCC Cancer Center grants.
Footnotes
Published ahead of print on 26 September 2007.
REFERENCES
- 1.Bishop, K. N., R. K. Holmes, and M. H. Malim. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 80:8450-8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogerd, H. P., B. P. Doehle, H. L. Wiegand, and B. R. Cullen. 2004. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc. Natl. Acad. Sci. USA 101:3770-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiu, Y. L., V. B. Soros, J. F. Kreisberg, K. Stopak, W. Yonemoto, and W. C. Greene. 2005. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature 435:108-114. [DOI] [PubMed] [Google Scholar]
- 4.Conticello, S. G., R. S. Harris, and M. S. Neuberger. 2003. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 13:2009-2013. [DOI] [PubMed] [Google Scholar]
- 5.Dettenhofer, M., S. Cen, B. A. Carlson, L. Kleiman, and X. F. Yu. 2000. Association of human immunodeficiency virus type 1 Vif with RNA and its role in reverse transcription. J. Virol. 74:8938-8945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallois-Montbrun, S., B. Kramer, C. M. Swanson, H. Byers, S. Lynham, M. Ward, and M. H. Malim. 2007. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J. Virol. 81:2165-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 8.Holmes, R. K., F. A. Koning, K. N. Bishop, and M. H. Malim. 2007. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation: comparisons with APOBEC3G. J. Biol. Chem. 282:2587-2595. [DOI] [PubMed] [Google Scholar]
- 9.Jarmuz, A., A. Chester, J. Bayliss, J. Gisbourne, I. Dunham, J. Scott, and N. Navaratnam. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79:285-296. [DOI] [PubMed] [Google Scholar]
- 10.Kao, S., M. A. Khan, E. Miyagi, R. Plishka, A. Buckler-White, and K. Strebel. 2003. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J. Virol. 77:11398-11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kao, S., E. Miyagi, M. A. Khan, H. Takeuchi, S. Opi, R. Goila-Gaur, and K. Strebel. 2004. Production of infectious human immunodeficiency virus type 1 does not require depletion of APOBEC3G from virus-producing cells. Retrovirology 1:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozak, S. L., M. Marin, K. M. Rose, C. Bystrom, and D. Kabat. 2006. The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J. Biol. Chem. 281:29105-29119. [DOI] [PubMed] [Google Scholar]
- 13.Liu, B., P. T. Sarkis, K. Luo, Y. Yu, and X. F. Yu. 2005. Regulation of Apobec3F and human immunodeficiency virus type 1 Vif by Vif-Cul5-ElonB/C E3 ubiquitin ligase. J. Virol. 79:9579-9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 15.Mangeat, B., P. Turelli, S. Liao, and D. Trono. 2004. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J. Biol. Chem. 279:14481-14483. [DOI] [PubMed] [Google Scholar]
- 16.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 17.Mariani, R., G. Rutter, M. E. Harris, T. J. Hope, H. G. Krausslich, and N. R. Landau. 2000. A block to human immunodeficiency virus type 1 assembly in murine cells. J. Virol. 74:3859-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398-1403. [DOI] [PubMed] [Google Scholar]
- 19.Mehle, A., J. Goncalves, M. Santa-Marta, M. McPike, and D. Gabuzda. 2004. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev. 18:2861-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mehle, A., B. Strack, P. Ancuta, C. Zhang, M. McPike, and D. Gabuzda. 2004. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 279:7792-7798. [DOI] [PubMed] [Google Scholar]
- 21.Mehle, A., E. R. Thomas, K. S. Rajendran, and D. Gabuzda. 2006. A zinc-binding region in Vif binds Cul5 and determines cullin selection. J. Biol. Chem. 281:17259-17265. [DOI] [PubMed] [Google Scholar]
- 22.Newman, E. N., R. K. Holmes, H. M. Craig, K. C. Klein, J. R. Lingappa, M. H. Malim, and A. M. Sheehy. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15:166-170. [DOI] [PubMed] [Google Scholar]
- 23.Russell, R. A., and V. K. Pathak. 2007. Identification of two distinct HIV-1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J. Virol. 81:8201-8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santa-Marta, M., F. A. da Silva, A. M. Fonseca, and J. Goncalves. 2005. HIV-1 Vif can directly inhibit apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G-mediated cytidine deamination by using a single amino acid interaction and without protein degradation. J. Biol. Chem. 280:8765-8775. [DOI] [PubMed] [Google Scholar]
- 25.Schrofelbauer, B., D. Chen, and N. R. Landau. 2004. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc. Natl. Acad. Sci. USA 101:3927-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schrofelbauer, B., T. Senger, G. Manning, and N. R. Landau. 2006. Mutational alteration of human immunodeficiency virus type 1 Vif allows for functional interaction with nonhuman primate APOBEC3G. J. Virol. 80:5984-5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404-1407. [DOI] [PubMed] [Google Scholar]
- 28.Sherer, N. M., M. J. Lehmann, L. F. Jimenez-Soto, A. Ingmundson, S. M. Horner, G. Cicchetti, P. G. Allen, M. Pypaert, J. M. Cunningham, and W. Mothes. 2003. Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic (Copenhagen) 4:785-801. [DOI] [PubMed] [Google Scholar]
- 29.Shindo, K., A. Takaori-Kondo, M. Kobayashi, A. Abudu, K. Fukunaga, and T. Uchiyama. 2003. The enzymatic activity of CEM15/Apobec-3G is essential for the regulation of the infectivity of HIV-1 virion but not a sole determinant of its antiviral activity. J. Biol. Chem. 278:44412-44416. [DOI] [PubMed] [Google Scholar]
- 30.Simon, J. H., A. M. Sheehy, E. A. Carpenter, R. A. Fouchier, and M. H. Malim. 1999. Mutational analysis of the human immunodeficiency virus type 1 Vif protein. J. Virol. 73:2675-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simon, J. H., T. E. Southerling, J. C. Peterson, B. E. Meyer, and M. H. Malim. 1995. Complementation of vif-defective human immunodeficiency virus type 1 by primate, but not nonprimate, lentivirus vif genes. J. Virol. 69:4166-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simon, V., V. Zennou, D. Murray, Y. Huang, D. D. Ho, and P. D. Bieniasz. 2005. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 1:e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stopak, K., C. de Noronha, W. Yonemoto, and W. C. Greene. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12:591-601. [DOI] [PubMed] [Google Scholar]
- 34.Tian, C., X. Yu, W. Zhang, T. Wang, R. Xu, and X. F. Yu. 2006. Differential requirement for conserved tryptophans in human immunodeficiency virus type 1 Vif for the selective suppression of APOBEC3G and APOBEC3F. J. Virol. 80:3112-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wichroski, M. J., K. Ichiyama, and T. M. Rana. 2005. Analysis of HIV-1 viral infectivity factor-mediated proteasome-dependent depletion of APOBEC3G: correlating function and subcellular localization. J. Biol. Chem. 280:8387-8396. [DOI] [PubMed] [Google Scholar]
- 36.Wichroski, M. J., G. B. Robb, and T. M. Rana. 2006. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog. 2:e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu, H., E. S. Svarovskaia, R. Barr, Y. Zhang, M. A. Khan, K. Strebel, and V. K. Pathak. 2004. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc. Natl. Acad. Sci. USA 101:5652-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed] [Google Scholar]
- 39.Yu, Y., Z. Xiao, E. S. Ehrlich, X. Yu, and X. F. Yu. 2004. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 18:2867-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, H., R. J. Pomerantz, G. Dornadula, and Y. Sun. 2000. Human immunodeficiency virus type 1 Vif protein is an integral component of an mRNP complex of viral RNA and could be involved in the viral RNA folding and packaging process. J. Virol. 74:8252-8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]