Abstract
The salt overly sensitive (SOS) pathway is critical for plant salt stress tolerance and has a key role in regulating ion transport under salt stress. To further investigate salt tolerance factors regulated by the SOS pathway, we expressed an N-terminal fusion of the improved tandem affinity purification tag to SOS2 (NTAP-SOS2) in sos2-2 mutant plants. Expression of NTAP-SOS2 rescued the salt tolerance defect of sos2-2 plants, indicating that the fusion protein was functional in vivo. Tandem affinity purification of NTAP-SOS2-containing protein complexes and subsequent liquid chromatography-tandem mass spectrometry analysis indicated that subunits A, B, C, E, and G of the peripheral cytoplasmic domain of the vacuolar H+-ATPase (V-ATPase) were present in a SOS2-containing protein complex. Parallel purification of samples from control and salt-stressed NTAP-SOS2/sos2-2 plants demonstrated that each of these V-ATPase subunits was more abundant in NTAP-SOS2 complexes isolated from salt-stressed plants, suggesting that the interaction may be enhanced by salt stress. Yeast two-hybrid analysis showed that SOS2 interacted directly with V-ATPase regulatory subunits B1 and B2. The importance of the SOS2 interaction with the V-ATPase was shown at the cellular level by reduced H+ transport activity of tonoplast vesicles isolated from sos2-2 cells relative to vesicles from wild-type cells. In addition, seedlings of the det3 mutant, which has reduced V-ATPase activity, were found to be severely salt sensitive. Our results suggest that regulation of V-ATPase activity is an additional key function of SOS2 in coordinating changes in ion transport during salt stress and in promoting salt tolerance.
To cope with salt stress, plants have evolved strategies to maintain low Na+ concentrations in the cytoplasm. The salt overly sensitive (SOS) pathway, identified through isolation and study of the sos1, sos2, and sos3 mutants, is essential for maintaining favorable ion ratios in the cytoplasm and for tolerance of salt stress (63, 64). SOS1 is a Na+/H+ exchanger located on the plasma membrane (39, 53); SOS3 is a myristoylated EF hand-type Ca2+-binding protein able to sense specific salt stress-induced calcium signals (19), and SOS2 is a Ser/Thr kinase with a C-terminal regulatory domain and an N-terminal catalytic domain (24). During salt stress conditions, the SOS2-SOS3 complex phosphorylates and activates the transport activity of the SOS1 antiporter (42).
The function of the SOS2-SOS3 regulatory complex depends on interaction of SOS2 and regulatory proteins, including SOS3. The C-terminal regulatory domain of SOS2 consists of an autoinhibitory FISL motif that binds to SOS3 (13, 24) and a PPI motif that binds to type 2C protein phosphatase abcisic acid (ABA)-insensitive 2 (ABI2) (33). Yeast two-hybrid experiments have shown that the SOS2 protein physically interacts with SOS3, and in vitro phosphorylation assays have shown that Ca2+ is required to activate the kinase activity of the SOS2-SOS3 complex (16). SOS3 binding also recruits SOS2 to the plasma membrane (13, 16). Activated SOS2 is then able to phosphorylate SOS1, enhancing its activity during salt stress (39, 42). SOS2 and other SnRK3 class kinases closely related to SOS2 have also been shown to interact with ABI2 or ABI1 (15, 33), thus forming a point of potential connection between ABA signaling and salt stress responses. Additional work in our laboratory has sought to identify other SOS2-interacting proteins.
The most studied mechanism, although not the only mechanism, by which the SOS pathway can promote salt tolerance is through the regulation of ion transport processes. A number of lines of evidence suggest that this includes not only regulation of SOS1 activity but also the regulation of other transporters. Tonoplast vesicles isolated from sos2-2, but not sos3-1, cells had a greatly reduced Na+/H+ exchange activity compared to the wild type (40). Furthermore, in the presence of 100 mM NaCl, this activity could be stimulated by adding constitutively active SOS2 to the vesicles. This indicated that the SOS2 protein kinase is required for activation of tonoplast-localized Na+/H+ exchangers (NHXs) during salt stress, and this activation likely occurs independently of SOS3 (40). Similarly, SOS2 was shown to interact with and activate the vacuolar H+/Ca2+ antiporter CAX1, independently of SOS3 (7). Thus, a number of transport activities may be regulated by SOS2, and these may occur by mechanisms distinct from the SOS2-SOS3 interaction that is required for the regulation of SOS1.
The driving force for SOS1 and the tonoplast-located NHXs, as well as many other transport activities, is the proton motive force generated by H+-pumping ATPases, such as the plasma membrane H+-ATPase and the tonoplast H+-pyrophosphatase and H+-ATPase. The vacuolar H+-ATPase (V-ATPase) is the major proton pump that establishes and maintains an electrochemical proton gradient across the tonoplast, thus providing the driving force for the secondary active transport of metabolites and ions (27). The V-ATPase is a complex multisubunit enzyme, composed of a V1 peripheral sector (subunits A to H), which binds and hydrolyzes ATP, and a V0 membrane sector (subunits a to e), which provides the pathway for the entry of protons into the vacuolar lumen (54). In the genome of Arabidopsis thaliana, genes encoding at least 12 V-ATPase subunits have been identified. While the majority of the V0 subunits appear to be encoded by multiple genes, the subunits that constitute the V1 peripheral stalk are mainly encoded by single genes, with the exceptions of the subunits B, E, and G. Each of these subunits has three different isoforms: AtVHA-B1, -B2, and -B3; AtVHA-E1, -E2, and -E3; and AtVHA-G1, -G2, and -G3 (54, 59).
Because the V-ATPase has such a key function in cellular transport processes, it plays an important role in growth and development and in the responses to external stimuli. This is illustrated by the phenotype of the deetiolated 3 (det3) mutant (4). The det3 mutant has a twofold reduction in levels of the V-ATPase subunit C (AtVHA-C), leading to a conditional lack of V1 peripheral sector assembly and V-ATPase activity (51). It is also defective in cell expansion and morphogenesis, as det3 seedlings have decreased cell expansion in the hypocotyl, petioles, and inflorescence stems and fail to arrest shoot development when seedlings are grown in the dark. AtVHA-C has also been shown to bind to and be phosphorylated by the protein kinase AtWITH NO K8 (AtWNK8; K refers to lysine), but the significance of this phosphorylation remains to be determined (18).
In addition to the general importance of V-ATPase in cell expansion and development, a number of studies have reported an increase in mRNA, protein levels, or activity of V-ATPase in response to salt stress. In Mesembryanthemum crystallinum, a coordinate up-regulation of gene expression in response to NaCl treatment was observed for V-ATPase subunits A, B, E, and F of the peripheral domain and subunit c of the membrane domain (11, 12, 25, 55) and a 2.5-fold increase in the total amount of the V-ATPase complex was also observed (43). In the halophyte Suaeda salsa, an increase in protein amount and in V-ATPase activity was found in salt-stressed, but not osmotically stressed, plants, suggesting that the ionic component of salt stress is responsible for the induction of V-ATPase expression and activity (57). Similarly, it has been shown that NaCl but not ABA can stimulate V-ATPase activity (20). Salt-induced transcript up-regulation of one or more V-ATPase subunits or an increase in V-ATPase activity has also been observed in a number of glycophytic species (3, 4, 22, 30, 45, 61). This includes A. thaliana, where an increase in the transcript levels of AtVHA-C (36) but not of subunit D (AtVHA-D) (23) was found upon salt stress. These studies suggest that increased V-ATPase level and/or activity may be required to drive Na+ sequestration under salt stress. Because V-ATPase can provide the driving force only for ion transport into the vacuole, any effect of the V-ATPase on salt tolerance depends on the activity of a number of other transporters needed to sequester ions into the vacuole. However, it has also been recently observed that Arabidopsis mutants carrying transferred DNA insertions in the coding sequence of the vacuolar Ca2+/H+ antiporters CAX1 and CAX2 had reduced V-ATPase activity (6, 37). This suggests coordinate regulation of tonoplast ion transport processes. The mechanism underlying this coordination, however, remains unclear. Based on the studies described above, SOS2 is one candidate for a protein that may coordinate various ion transport processes during salt stress.
In this study, we used tandem affinity purification (TAP) tagging to isolate proteins interacting with SOS2 in vivo. We found that SOS2 interacts with subunits forming the cytoplasmic sector of the V-ATPase and that this interaction is enhanced under salt stress conditions. Parallel experiments using the yeast two-hybrid system showed that SOS2 interacts directly with at least two of the three VHA-B subunit isoforms present in Arabidopsis. In addition, the V-ATPase-deficient det3 mutant was found to be extremely salt sensitive and this, along with altered V-ATPase activity in both sos2-2 and sos3-1 mutants, demonstrates the importance of SOS2 regulation of V-ATPase activity in salt tolerance. The results define a new function of SOS2 in interacting with cytoplasmic VHA-B subunits and stimulating H+ transport activity to provide an increased driving force for compartmentation of Na+ into the vacuole.
MATERIALS AND METHODS
Plant materials and salt tolerance assay.
Arabidopsis thaliana ecotype Columbia was used in all experiments. The sos2-2 and sos3-1 mutants have been previously described (64). The det3 mutant was obtained from the Arabidopsis Biological Resource Center. Soil-grown plants were kept under continuous light (70 mmol m−2 s−1) at 23°C. Seedlings were grown under the same conditions. For seedling salt tolerance assays, sterilized seed was plated on half-strength MS media without sucrose or other sugars (unless otherwise noted in text or figure legends) and stratified for 3 days at 4°C. Four-day-old seedlings were transferred to fresh plates of either the same media (control) or media containing NaCl at concentrations indicated in the text or figures. Root length increase was monitored over the following 7 days, and seedlings were photographed at the end of the experiment. For experiments involving dark-grown seedlings, seed was plated on control or salt-containing media with 0.5% sucrose, stratified, and either placed under the same growth conditions or kept in darkness by wrapping the plates in foil.
Construction of NTAP-SOS2 plants.
To make TAP-tagged SOS2, SOS2 cDNA was amplified by PCR with KOD polymerase (Takara) using SOS2-pGEX (13) as the template. The primers used amplified a 1,207-bp fragment encoding SOS2 with flanking BP clonase recognition sites. Primer sequences were 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCAACCATGACAAAGAAAATGAGAAGAG-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTCCTCAAAACGTGATTGTTCTGAGA-3′. The PCR product was first cloned into pDONOR207 and then transferred to the pNTAPi vector (48) using the Gateway system (Invitrogen). The recombinant plasmid was introduced into Agrobacterium tumefaciens strain GV3101 by electroporation and used to transform sos2-2 plants by the floral dip method. Transgenic plants were selected on MS agar plates containing 25 mg liter−1 glufosinate ammonium.
TAP and mass spectrometry analysis.
TAP was carried out using NTAP-SOS2 plants grown in soil for 20 days and either harvested directly (control) or irrigated with 150 mM NaCl for 24 h (salt stressed). For both control and salt-stressed samples, entire plants (30 g of tissue) were collected from pots, briefly washed with water to remove soil from the roots, ground in liquid nitrogen, and used for TAP as previously described (48). After the final elution step, the isolated protein was precipitated using STRATARESIN (Stratagene) and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 8% polyacrylamide gels, and proteins were visualized by Coomassie staining. The gel was cut into horizontal slices, and corresponding slices (containing proteins of the same molecular weight) from control and salt-treated samples were processed in parallel. In each slice, proteins were subject to in-gel trypsin digestion and processed essentially as described previously (58). The trypsin-digested peptides were analyzed by a micro-liquid chromatography (μLC) nano-electrospray ionization tandem mass spectrometry (MS/MS) with a Micromass quadrupole time-of-flight API US instrument (Waters, Milford, MA). For protein identification, the resulting MS/MS spectra were then searched against the NCBInr Arabidopsis database using the MASCOT program. For the relative quantification of proteins purified from salt-treated and control plants, samples were analyzed in a pairwise manner to minimize variation and an LC-mass spectrometry (LC-MS)-based label-free quantification strategy (49) was used. Briefly, the LC-MS spectral intensity (counts) of an individually extracted ion chromatographic peak that was located by the m/z ratio, charge state, and retention time identified in the preceding LC-MS/MS experiment was used to represent the relative amount of the corresponding peptide ion. The total ion counts of several selected peptides were calculated for both control and salt-stressed samples to obtain relative quantitative ratios. The values were normalized by ion counts of autodigested tryptic peptides that were common to all samples. An average ratio ± standard deviation of all the peptides belonging to a single protein was calculated to estimate relative abundances of SOS2-interacting proteins for salt stress and control conditions.
Immunoblotting.
Immunoblotting was performed both to check the levels of NTAP-SOS2 expression and to check the levels of NTAP-SOS2 present before and after the TAP procedure was performed. For detection of NTAP-SOS2 expression, 2-week-old seedlings grown on MS agar plates were homogenized in liquid nitrogen and suspended in extraction buffer (50 mM HEPES-KOH, pH 7.5, 5 mM EDTA, 5 mM EGTA, 20% glycerol, 2 mM dithiothreitol, 2 mM phenylmethylsulfonyl fluoride). An equal volume of 2× SDS-containing sample buffer was added, and the samples were heated at 95°C for 5 min and subjected to SDS-PAGE. Western blotting was performed using standard protocols. Briefly, proteins were electrophoretically transferred to a nitrocellulose membrane (Amersham) and blocked with 5% milk in phosphate-buffered saline containing 0.1% Tween 20. Blots were incubated with either SOS2 or VHA-B antibody, washed, and probed with horseradish peroxidase-conjugated anti-rabbit immunoglobulin G antibody (Bio-Rad), and immunoreactive bands were detected with the ECL Western blotting detection system (Amersham). The SOS2 antibody has been previously described by our laboratory (14). The VHA-B antibody was a generous gift from Heven Sze (University of Maryland) and has also been previously described (56). Immunoblotting was also used to check the levels of NTAP-SOS2 and VHA-B present either before and after TAP in extracts from 20-day-old NTAP-SOS2 plants.
Yeast two-hybrid assay.
The entire coding sequence of VHA-B1 (At1g76030) was amplified by PCR from the cDNA clone U21927, obtained from the Arabidopsis Biological Resource Center, with primers containing restriction sites and cloned in frame between NcoI and XmaI sites of pACT2. Primer sequences were 5′-CATGCCATGGGGACGAATGATCTCGA-3′, 5′-TCCCCCCGGGTTAACTGGTTGAGTCGCGGCT-3′, and 5′-CATGCCATGGAGAGAACCTATCCTGAAGAGATGAT-3′. Similarly, the full-length coding sequence of VHA-B2 (At4g38510) was cloned in frame in pACT2 using NcoI and PstI and the cDNA clone U12748, obtained from the Arabidopsis Biological Resource Center, as a template. Primers sequences were 5′-CATGCCATGGGTGCTGCTGAAAACAACCTT-3′ and 5′-AAAACTGCAGTCAGTTGGTGGTATCGCGACTGTA-3′. For cloning of VHA-B1 and VHA-B2 Δ140, the same forward primer was used; the sequence was 5′-CATGCCATGGAGAGAACCTATCCTGAAGAGATGAT-3′. pAS-SOS2, pAS-SOS1, and pAS-SOS2K40N were used as previously described (33). Plasmid DNA of bait and prey constructs was cotransformed (2) into the Saccharomyces cerevisiae strain Y190. Yeast growth and the β-galactosidase assay were performed as previously described (16, 33).
Proton transport assays.
Transport assays were conducted using membranes isolated from suspension-cultured cells originated from wild-type, sos2-2, or sos3-1 calli. Calli were induced by transferring 2-week-old seedlings to plates containing callus initiation medium (4.3 g/liter MS salts, 30 g/liter sucrose, 1× MS vitamins, 3 mg/liter 2,4-dichlorophenoxyacetic acid, 0.05 mg/liter kinetin, 1 g/liter casein hydrolysate, and 7 g/liter agar at pH 5.7). The plates were then placed in the dark and subcultured onto new medium every 2 weeks. After three to five passages, friable callus formed and was transferred from plates to liquid culture (4.3 g/liter MS salts, 30 g/liter sucrose, 1× MS vitamins, 3 mg/liter 2,4-dichlorophenoxyacetic acid, 0.05 mg/liter kinetin, and 1 g/liter casein hydrolysate at pH 5.0). Cells were cultured in the dark at 24°C with shaking at 130 rpm and were subcultured every 5 to 7 days to generate sufficient suspension cells for the vesicle isolation and transport assays. Cells were harvested and used for membrane isolation at 7 days after subculturing.
Tonoplast vesicles were isolated using dextran gradients, and H+ transport assays were conducted as previously described (39, 40). The proton transport activity of the tonoplast H+-ATPase was measured as a decrease (quench) in the fluorescence of the pH-sensitive fluorescent probe quinacrine (39, 40). For V-ATPase H+ transport activity, the assay medium (1 ml) contained 5 μM quinacrine, 3 mM ATP, 100 mM 1,3-bis-Tris propane (BTP) chloride, 25 mM BTP-HEPES (pH 7.5), 250 mM mannitol, and 50 μg tonoplast membrane protein. Glutathione S-transferase-tagged recombinant, constitutively active T/DSOS2DF protein was expressed in Escherichia coli and purified as previously described (13, 39). When used in transport assays, 0.2 μg of T/DSOS2DF was incubated with membrane vesicles for 7 min at room temperature before pH gradient formation across the membrane (ΔpH) was initiated with the addition of MgSO4.
RESULTS
Analysis of TAP-tagged SOS2 identifies V-ATPase subunits as SOS2-interacting proteins.
To identify proteins interacting with the protein kinase SOS2, we used the TAP method (38, 47, 48, 50). The coding sequence of SOS2 was fused to that of the C terminus of the TAP tag sequence in the binary plasmid pNTAPi (48). This plasmid (encoding NTAP-SOS2) was used for transformation of sos2-2 mutant plants, and, among the different lines generated, a line that expressed TAP-tagged SOS2 and rescued the salt sensitivity phenotype of the sos2-2 mutant (Fig. 1A and B) was used for purification of SOS2-containing complexes.
FIG. 1.
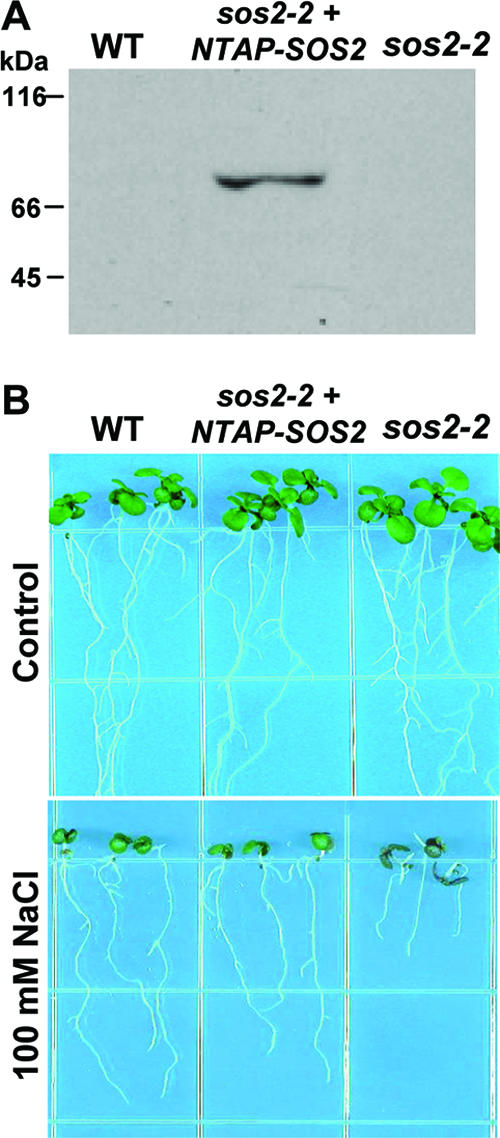
Expression of a functional TAP-tagged SOS2 complements the salt sensitivity of a sos2-2 mutant. (A) Western blot analysis with SOS2-specific antibody detected NTAP-SOS2 expressed in the sos2-2 mutant at the molecular mass expected for a fusion of the TAPi tag with SOS2 (molecular mass of SOS2 alone is 50.6 kDa). SOS2 was not detected in the wild type (WT) because of its relatively low level of expression compared to the expression of NTAP-SOS2 driven by the 35S promoter. (B) Complementation of the salt sensitivity phenotype of the sos2-2 mutant by NTAP-SOS2. Photographs were taken 10 days after the transfer of seedlings to control or salt-containing media.
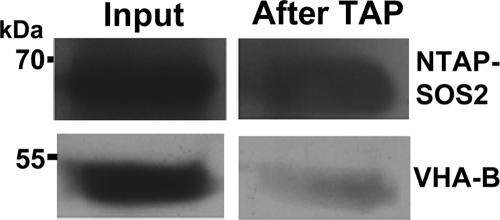
TAP was performed essentially as described previously (48) using 20-day-old NTAP-SOS2 (sos2-2) plants that had been exposed to 150 mM NaCl for 24 h. Salt-treated plants were used to maximize the chances of identifying interactions that are important in the function of SOS2 in salt stress tolerance. The proteins were then separated by SDS-PAGE, and the individual bands were isolated and digested in situ with trypsin for analysis using LC-MS/MS. Using this strategy, we identified several subunits constituting the peripheral stalk of the V-ATPase (Table 1). We also confirmed the presence of both NTAP-SOS2 and VHA-B in the purified protein complexes by performing Western blot analysis (Fig. 2).
TABLE 1.
V-ATPase proteins identified by LC-MS/MS in NTAP-SOS2 complexes
| Protein | Locus in A. thaliana | No. of peptides identified | Protein sequence coverage (%) |
|---|---|---|---|
| AtVHA-A | At1g78900 | 7 | 14 |
| AtVHA-B1/B2/B3 | At1g76030/At4g38510/At1g20260 | 5 | 12 |
| AtVHA-C | At1g12840 | 4 | 16 |
| AtVHA-E1 | At4g11150 | 2 | 10 |
| AtVHA-G1 | At3g01390 | 1 | 15 |
FIG. 2.
Both NTAP-SOS2 and VHA-B are present after TAP. Western blot analysis using SOS2- or VHA-B-specific antibodies detected both proteins in samples before (input) and after TAP.
Salt treatment enhances interaction of SOS2 with V-ATPase peripheral subunits.
After our initial identification of proteins from the V-ATPase peripheral domain as SOS2-interacting proteins, we performed additional experiments to determine whether the interaction is affected by salt stress and whether the interaction may be affected by altered abundance of either SOS2 or VHA-B in salt-stressed tissue. NTAP-SOS2 complexes were purified in parallel from untreated plants and from plants exposed to salt stress (150 mM NaCl) for 24 h. To estimate the relative levels of various proteins in the purified complexes from control versus salt-stressed plants, the arbitrary total ion current counts of all the peptides representing the SOS2-interacting proteins including VHA-B were summed and expressed relative to the trypsin enzyme used for peptide digestion (49).
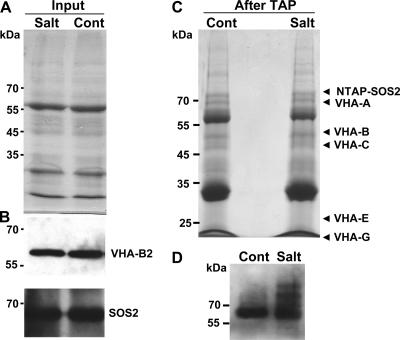
Extracts from control and salt-treated plants had similar total protein levels both before and after TAP (Fig. 3A and C). Likewise, the amounts of NTAP-SOS2 recovered from the two samples were not significantly different in either the protein extract before purification (input) or after TAP (Fig. 3B). This was expected since the expression of NTAP-SOS2 was driven by the constitutive 35S promoter. Western blot analysis also showed similar total amounts of VHA-B in the extracts from control and salt-treated plants before TAP (Fig. 3B), indicating that the total protein abundance of VHA-B was not altered by the salt treatment imposed. However, LC-MS/MS analysis of the purified complexes showed that greater amounts of several V-ATPase peripheral subunits copurified with NTAP-SOS2 in samples from salt-treated plants than in control samples (Table 2). In particular, in the NTAP-SOS2 complexes purified from salt-stressed plants we observed a 4.1-fold increase in the relative amount of VHA-C, a 3.6-fold increase in VHA-A, and a 2.5-fold increase in VHA-B compared to the control. The consistently higher levels of several V-ATPase peripheral subunits associated with SOS2 in salt-stressed plants suggested that interaction of SOS2 with the V-ATPase complex is enhanced by salt stress.
FIG. 3.
Purification of NTAP-SOS2 protein complexes from control and salt-treated plants. (A) Coomassie-stained SDS-PAGE gel showing protein extracts from control (Cont) and salt-stressed (Salt) NTAP-SOS2 sos2-2 plants. Similar amounts of total protein were present in both extracts. (B) Western blot analysis showing similar amounts of NTAP-SOS2 and VHA-B in control and salt-stressed samples. (C) Coomassie-stained SDS-PAGE gel of protein isolated from control and salt-stressed plants after TAP. Positions of the VHA subunits identified by LC-MS/MS analysis are indicated on the right side of the gel. Note the similar amounts of total protein in the control versus salt-stressed samples. (D) Western blot analysis with anti-SOS2 antibody of TAP-purified samples. While the amounts of N-TAPiSOS2 recovered are similar in both samples, in salt-stressed plants the antibody recognizes additional bands, perhaps indicating posttranslational modifications of SOS2 occurring only under salt stress conditions.
TABLE 2.
Relative quantification of V-ATPase proteins found in NTAP-SOS2 protein complexes isolated from control or salt-stressed plants
| Protein | No. of peptides | Salt/control ratioa |
|---|---|---|
| VHA-A | 4 | 3.6 ± 1.2 |
| VHA-B1/B2/B3 | 3 | 2.5 ± 0.2 |
| VHA-C | 2 | 4.1 ± 2.0 |
| VHA-E1 | 2 | 2.2 ± 0.3 |
| VHA-G1 | 1 | 2.3 |
Ratios are the means ± standard deviations of the ratios of individual peptides from each protein.
Yeast two-hybrid analysis identifies VHA-B subunits as SOS2-interacting proteins.
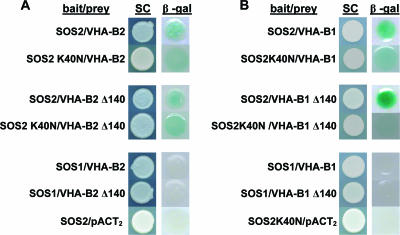
While successful in identifying a salt-inducible interaction between SOS2 and the V-ATPase complex as a whole, the TAP-tagging experiments could not establish which of the V-ATPase peripheral subunits interacted directly with SOS2 and which were simply copurified as part of the V-ATPase complex. Our previous yeast two-hybrid screen using SOS2 as the bait protein identified not only ABI2 (33) but also VHA-B and several other clones as putative interactors with SOS2 (data not shown). VHA-B, together with VHA-A, controls the binding of ATP, whose hydrolysis is then catalyzed by VHA-A alone while VHA-B, it has been suggested, has a role in regulating the activity of the V-ATPase complex (29, 44, 54). To confirm the interaction between VHA-B proteins and SOS2, the entire open reading frames of VHA-B1 and VHA-B2 were fused with the GAL4 activation domain in the prey plasmid pACT2 and cotransformed with the bait plasmid pAS-SOS2, pAS-SOS2 K40N, or pAS-SOS1 (33) into the yeast strain Y190. SOS2 interacted with both of the VHA-B subunits, as indicated by the β-galactosidase activity produced when pAS-SOS2 or pAS-SOS2K40N was cotransformed with pACT-VHA-B1 or pACT-VHA-B2 (Fig. 4A and B). The positive interaction observed using the catalytically inactive SOS2-K40N mutant, indicated that the interaction did not depend on SOS2 kinase activity (Fig. 4B); however, the interaction with SOS2-K40N did appear somewhat weaker than the interaction with wild-type SOS2. As expected, a SOS1 C-terminal fragment, used as negative control, showed no interaction with VHA-B1 or -B2 (SOS1 is a plasma membrane protein and so would not be expected to interact with the V-ATPase complex on the tonoplast).
FIG. 4.
Yeast two-hybrid analysis detects interaction of SOS2 with VHA-B1 and -B2. VHA-B2 (A) and VHA-B1 (B) were cloned into the prey plasmid pACT2 and cotransformed with the bait plasmid encoding SOS2, SOS2-K40N, or SOS1 (a negative control) or the empty pACT2 vector into the yeast strain Y190. Yeast grown on synthetic complete (SC) medium and the β-galactosidase filter assay (β- gal) are shown. SOS2-K40N is a catalytically inactive mutant of SOS2. Middle sections of both panels A and B show the interaction of SOS2 or SOS2-K40N with VHA-B2 Δ140 or VHA-B1 Δ140, which are lacking the N-terminal 140 amino acids. Bottom sections of panels A and B show negative controls.
We also investigated the interaction of SOS2 with VHA-B1 and -B2 constructs with the N-terminal 140 amino acids deleted. This was because previous yeast two-hybrid screening using SOS2 as the bait protein had identified VHA-B clones with this N-terminal portion deleted (data not shown). Also, the N-terminal 140 amino acids of the VHA-B subunits contain a domain called the ATP-synt_ab_N domain (amino acids 1 to 90) and part of the ATP binding domain (amino acids 90 to 380). The ATP-synt_ab_N domain forms a closed beta-barrel with Greek-key topology. Although the Greek-key motifs do not have an assigned function, computational analysis suggests that they mediate protein-protein interactions (10). Interestingly, SOS2 could still interact with VHA-B1 or -B2 Δ140 constructs (Fig. 4). These results indicate that the N-terminal 140 amino acids of VHA-B did not participate in SOS2 binding.
The V-ATPase-impaired det3-1 mutant is hypersensitive to salt stress.
Although a number of studies report an increase in V-ATPase activity and/or amounts in response to salt stress, to our knowledge, the effect of reduced V-ATPase activity on salt tolerance has not been directly tested. To address the physiological importance of V-ATPase activity in salt tolerance, we tested the salt response of the det3 mutant. The det3 mutant was isolated by screening for mutants that had a light-grown phenotype, most notably a reduction in hypocotyl length, even when grown in the absence of light (5). The det3 mutant was subsequently shown to have a twofold reduction in levels of VHA-C and a conditional lack of V-ATPase peripheral sector assembly and V-ATPase activity, which were more pronounced in etiolated seedlings (51).
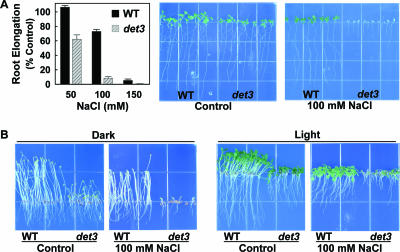
det3 seedlings grown on control media and then transferred to salt-containing media exhibited a severe salt sensitivity phenotype (Fig. 5A; no sucrose or other sugars were added to the media in these experiments). Root elongation of the det3 mutant was inhibited by more than 40% on 50 mM NaCl, a concentration that caused no root growth inhibition in the wild type. On 100 mM NaCl, root elongation was almost completely inhibited and over the course of 7 days the det3 seedlings became completely bleached (Fig. 5A). To determine whether this increased sensitivity was also true in dark-grown seedlings, where the reduction in V-ATPase activity should be the greatest, wild-type and det3 seedlings were sown directly on control or 100 mM NaCl media and kept either in darkness or standard growth conditions. In these experiments, 0.5% sucrose was added to all the treatments to support seedling growth in the dark. Dark-grown det3 seedlings on control media exhibited the characteristic reduction in hypocotyl length that was used to isolate the mutation (Fig. 5B). det3 seedlings on 100 mM NaCl in the dark again had greatly reduced growth. Light-grown det3 seedlings on sucrose-containing media were also more salt sensitive, although the addition of sucrose seemed to partially alleviate the salt sensitivity of the det3 mutant. Overall, the severe salt sensitivity of the det3 mutant is consistent with a key role of V-ATPase activity in salt tolerance.
FIG. 5.
The det3 mutant, which is impaired in V-ATPase activity, is salt sensitive. (A, left) Effect of NaCl on root elongation in the wild type (WT; ecotype Columbia) and det3 mutant. Root elongation is presented as a percentage of the elongation on control media for each genotype. Data are means ± standard errors (n = 14 to 16). (A, right) Pictures of representative seedlings 6 days after transfer to either control media (half-strength MS without sugar) or the same media with the addition of 100 mM NaCl. (B) Phenotypes of 6-day-old wild-type and det3 seedlings germinated and grown on control media (half-strength MS, 0.5% sucrose) in either light or dark.
H+ transport activity of the V-ATPase is reduced in the sos2-2 mutant but enhanced in the sos3-1 mutant compared to the wild type.
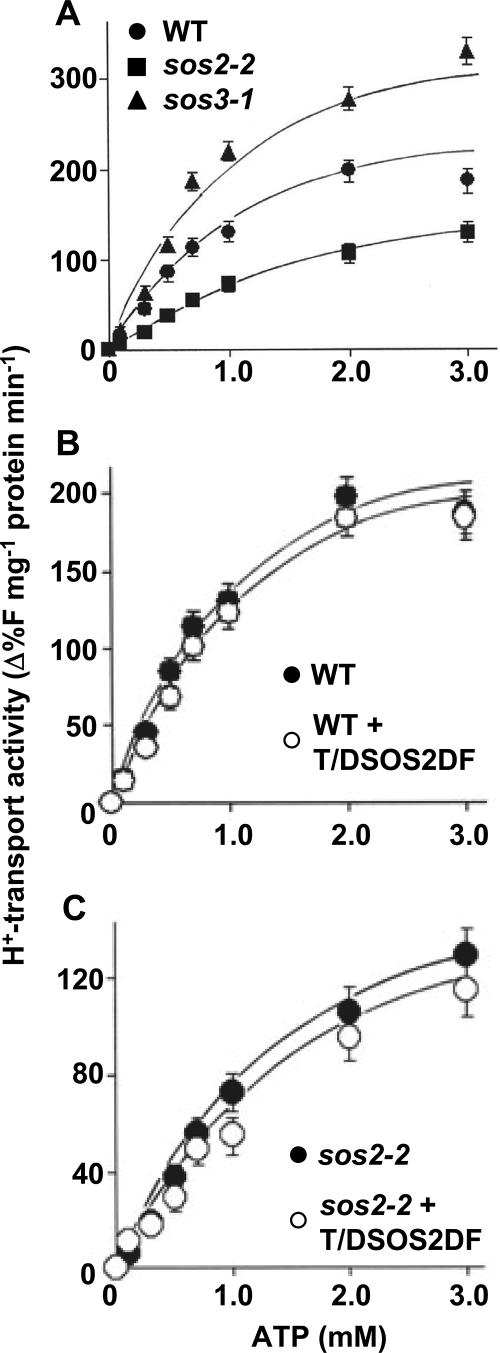
The interaction of SOS2 with VHA-B1 and -B2 along with the salt sensitivity of the det3 mutant suggested that one mechanism by which SOS2 promotes salt tolerance may be by stimulation of V-ATPase activity. To test this possibility, we measured the H+ transport activity of vacuolar membrane vesicles purified from wild-type, sos2-2, and sos3-1 vesicles. In wild-type vesicles the H+ transport activity of the V-ATPase increased with increased ATP concentration (Fig. 6A) and kinetic analyses indicated a Km for ATP of 0.75 mM and Vmax of 220 units (Table 3). In contrast, the H+ transport activity of tonoplast vesicles isolated from sos2-2 cells was reduced significantly relative to that of the wild-type vesicles. At 3 mM ATP, sos2-2 mutant transport activity was reduced by 30.7% (Fig. 6A) and kinetic analysis showed a reduced Vmax and a higher Km for ATP (Table 3). The results suggest that SOS2 is required for maximal activity of the V-ATPase.
FIG. 6.
Effect of sos2 and sos3 mutants on V-ATPase activity. (A) V-ATPase activity of tonoplast vesicles isolated from the wild type (WT) and sos2-2 and sos3-1 mutants. Tonoplast vesicles were isolated from cell cultures of each genotype by dextran gradient purification, and transport assays were performed as described in Materials and Methods. Initial rates of fluorescence quench were calculated over a range of ATP concentrations from 0 to 3 mM. All data represent means ± standard errors of at least three replicate experiments. Each replicate experiment was performed using independent membrane preparations. (B) V-ATPase activity of vesicles from wild-type cells with or without the addition of recombinant T/DSOS2DF. Assay conditions are as described for panel A, and wild-type data are the same as in panel A and are reproduced in this panel for ease of comparison. (C) V-ATPase activity of vesicles from sos2-2 cells with or with the addition of T/DSOS2DF. sos2-2 cell data are the same as in panel A and are reproduced here for ease of comparison.
TABLE 3.
Kinetic analysis of the proton transport activity of the vacuolar H+-ATPasea
| Genotype | Km (mM ATP) | Vmax |
|---|---|---|
| WTb | 0.75 | 220 |
| sos2-2 | 1.35 | 140 |
| sos3-1 | 0.85 | 320 |
| WT + T/DSOS2DF | 0.71 | 215 |
| sos2-2 + T/DSOS2DF | 1.30 | 134 |
Data shown in Fig. 6 were transformed using a Hanes-Woolf plot in order to determine kinetic parameters (x intercept = Km; Km/y intercept = Vmax). Units for Vmax are Δ%F mg protein[minus]1 min−1.
WT, wild type.
In previous studies addition of mutant forms of SOS2 with constitutively active phosphorylation activity could directly stimulate ion transport activity of either plasma membrane Na+ transport mediated by SOS1 or tonoplast Na+/H+ exchange mediated by NHX transporters (14, 39, 40). Thus, we attempted a similar experiment by adding constitutively active SOS2 (T/DSOS2DF) which has a T168D mutation combined with deletion of the FISL domain (14). Interestingly, we observed that T/DSOS2DF could not stimulate the V-ATPase activity of wild-type tonoplast vesicles (Fig. 6B; Table 3), nor could it rescue the reduced V-ATPase activity of vesicles from sos2-2 cells (Fig. 6C; Table 3) in the in vitro assay. Together, these V-ATPase assays indicated that SOS2 may be required for V-ATPase activation although SOS2 kinase activity itself cannot stimulate V-ATPase activity. It is possible that other factors not present in these in vitro assays, such as additional proteins, the FISL domain of SOS2, or posttranslational modification of SOS2, may be required to stimulate V-ATPase activity in vivo.
Another interesting result was that the V-ATPase activity of vesicles obtained from the sos3-1 mutant was significantly enhanced relative to that of the wild type (activity of sos3-1 vesicles was 77% higher than that of the wild type in the presence of 3 mM ATP; Fig. 6A and Table 3). While the exact mechanism behind this increased activity is not known, one possibility is that SOS3 competes with the V-ATPase for SOS2 binding, possibly through the FISL domain of SOS2.
DISCUSSION
Previous studies in our laboratory have identified the components of the SOS pathway as key elements in coordinating salt stress responses, particularly ion transport on both the plasma membrane and tonoplast, and several ion transport processes known to be regulated by SOS2 are diagrammed in Fig. 7. The SOS pathway, and SOS2 in particular, is also a point of cross talk between salt stress and other stress signals and stress responses (8, 35, 62). We have continued to search for additional factors that interact with SOS2 using both yeast two-hybrid and proteomic approaches and here report identification of the V-ATPase as a target of SOS2 interaction and regulation. The importance of V-ATPase and its potential regulation by SOS2 in salt stress resistance at the cellular and physiological levels is indicated by both the severe salt sensitivity of the V-ATPase-deficient det3 mutant and direct observation of altered tonoplast H+ transport in the sos2-2 mutant.
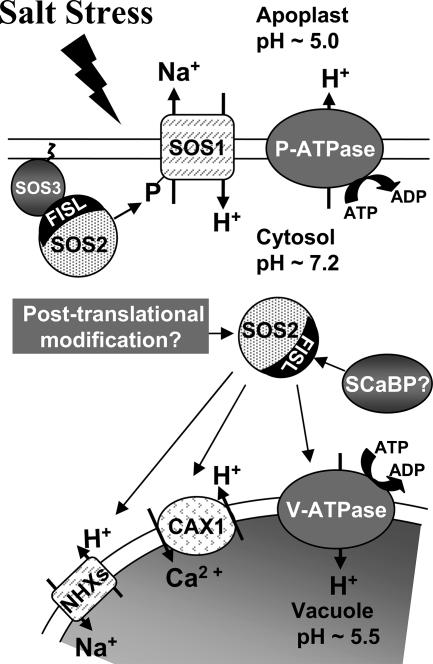
FIG. 7.
Diagram of SOS2 regulation of ion transport. In response to salt stress, SOS2 is activated and is recruited to the plasma membrane by binding to SOS3. The activated SOS3-SOS2 regulatory complex activates the Na+/H+ antiporter SOS1, which reduces Na+ concentration in the cytosol by extrusion to the apoplast. On the plasma membrane, the H+ gradient needed to drive Na+ extrusion is maintained by the plasma membrane H+-ATPase (P-ATPase). SOS2 also activates tonoplast-located NHX antiporters to compartmentalize Na+ ions to the vacuole and the H+/Ca2+ transporter CAX1. SOS2 has been shown to regulate these tonoplast transporters independently of SOS3, perhaps by mechanisms other than phosphorylation of the target transporter. Instead, an SCaBP may be responsible for targeting SOS2 to the tonoplast. Posttranslational modification of SOS2 may also be important for these interactions. The H+ gradient needed to drive Na+ transport across the tonoplast is maintained by the tonoplast H+-pyrophosphatase (not shown) and the V-ATPase. The data reported here indicate that SOS2 interacts with and regulates the V-ATPase and that V-ATPase activity is required for salt tolerance. The FISL domain of SOS2 is known to be required for SOS3 binding and could also have a role in the interaction of SOS2 with the V-ATPase either directly or through SCaBPs.
SOS2 interaction with and regulation of V-ATPase.
NTAP-SOS2 purification identified five subunits (A, B1/2/3, C, E1, and G1) of the V-ATPase peripheral (cytoplasmic) sector as being components of a SOS2-containing protein complex. Of these subunits, VHA-A is responsible for the binding and hydrolysis of ATP, VHA-B is involved in noncatalytic ATP binding and, it has also been suggested, has a regulatory role, and VHA-C, VHA-E, and VHA-G are involved in the assembly and coupling of the peripheral and integral membrane sectors of the V-ATPase complex (29, 34, 44, 51, 54).
Comparison of protein complexes isolated from unstressed and salt-stressed plants indicated that the association of the V-ATPase peripheral sector with SOS2 was enhanced by salt stress. At least in the case of VHA-B, this appeared to be caused not by an increased amount of SOS2 or VHA-B under salt stress but rather another mechanism that altered binding affinity or stability of the SOS2 interaction with the V-ATPase. This is consistent with other reports showing that the amount of V-ATPase does not change in short-term salt treatments (26, 28, 46). While it is not known what modification could have altered the binding affinity of SOS2 for the V-ATPase complex, one possibility is posttranslational modification of SOS2 itself. The multiple bands of SOS2 from salt-stressed, but not control plants seen in Fig. 3D provide an indication of such posttranslational modifications and evidence of SOS2 autophosphorylation, and phosphorylation by other kinases has also been observed in our laboratory (16; H. Fujii and J.-K. Zhu, unpublished observations). This possibility is also consistent with our observation that it was not possible to activate tonoplast H+ transport activity in vitro by adding recombinant T/DSOS2DF alone, which presumably is not posttranslationally modified in the same manner as SOS2 from salt-stressed plants. Overall, the enhanced association of SOS2 with the V-ATPase complex in salt-stressed plants indicates that this interaction likely has a specific role in regulating V-ATPase activity under high salt conditions.
While the identification of multiple V-ATPase subunits in a complex with SOS2 and their increased abundance in the complex after salt stress confirm that an interaction did take place and was influenced by salt stress, it does not imply that SOS2 interacts directly with each of the identified subunits. Our yeast two-hybrid results revealed that SOS2 interacts directly with VHA-B proteins (Fig. 4). VHA-B has been previously shown to interact with VHA-A, VHA-C, VHA-E, and possibly VHA-G (31, 54). Therefore, it appears that the other VHA proteins copurify with NTAP-SOS2 because of their interaction with VHA-B. This provides an indication that SOS2 interacts with VHA-B proteins after their incorporation into a functional V-ATPase complex on the tonoplast rather than interacting with VHA-B alone in another cellular compartment, such as the recently reported interaction of VHA-B1 and hexokinase in the nucleus (9). However, the possibility that SOS2 might also directly interact with other V-ATPase subunits cannot be ruled out.
The SOS2/VHA-B interaction, along with our direct observation of an increased Km for ATP and decreased Vmax of the V-ATPase in the absence of SOS2, raises the question of how SOS2 may affect V-ATPase activity. The most obvious possibility is through direct phosphorylation of VHA-B by SOS2. We investigated this by conducting in vitro phosphorylation assays using the T/DSOS2/308 constitutively active form of SOS2 (13, 14) with VHA-B1 or VHA-B2 as substrates. Our previous observations showed that T/DSOS2/308 has a high kinase activity (13, 14); however, we failed to detect any VHA-B phosphorylation despite repeated attempts carried out using conditions similar to those previously described for the in vitro phosphorylation of SOS1 (data not shown). This observation taken together with the observations that addition of T/DSOS2DF was not able to stimulate V-ATPase activity in vitro and that the catalytically inactive SOS2-K40N mutant was still capable of interaction with VHA-B1 and -B2 in the yeast two-hybrid system raises the possibility that SOS2 regulates the activity of the V-ATPase through a physical interaction that does not involve phosphorylation. Previous reports describing SOS2 interaction with and regulation of the tonoplast transporters CAX1 and NHX1 also showed SOS2 effects on transport activity (7, 40). For CAX1, at least partial stimulation of transport activity could be achieved by SOS2-K40N (7). For NHX1, it was not possible to detect in vitro phosphorylation using the same types of active SOS2 that could stimulate transport activity (40). Thus, as was the case in the V-ATPase experiments reported here, it was also not possible to associate stimulation of transport activity with SOS2 phosphorylation of CAX1 or NHX1. While this may indicate a phosphorylation-independent mechanism by which SOS2 can stimulate transport activity, it is also possible that conditions required for the phosphorylation of tonoplast-located transporters are different than those for SOS1 and thus could not be reproduced in our in vitro assays.
SOS2 regulation of the V-ATPase might instead require the presence of the FISL domain which is deleted in both T/DSOS2DF and T/DSOS2/308. The FISL domain might be necessary for the binding of SOS2 to VHA-B, which by itself alters the transport activity of the V-ATPase complex, or the FISL domain might mediate binding of SOS2 prior to phosphorylation. A role for the FISL domain may also explain the higher Vmax of the V-ATPase in the absence of SOS3. SOS3 also binds to the FISL domain of SOS2 (13), and the absence of SOS3 may result in more SOS2 being bound to the V-ATPase, thus stimulating H+ transport. The involvement of other proteins in this interaction should also be considered. SOS2 has previously been shown to interact with other members of the family of SOS3-like Ca2+ binding proteins (SCaBP1, SCaBP3, SCaBP5, SCaBP6, and SCaBP8) (13, 41), and it is possible that interaction with one of these SCaBPs, possibly through the FISL domain, could target SOS2 to the tonoplast for regulation of the V-ATPase (Fig. 7).
Role of the V-ATPase and its interaction with SOS2 in salt tolerance.
SOS2 not only enhances, together with SOS3, Na+ extrusion into the apoplast by regulation of the plasma membrane Na+/H+ antiporter SOS1 but has also been shown to increase the activity of tonoplast Na+/H+ antiporters (Fig. 7) to increase sodium transport into the vacuole in response to salt stress (39, 40, 42). Both of these transport activities depend on the maintenance of H+ gradients across the membranes to provide the driving force for Na+ transport, and it is therefore necessary to increase the activities of the H+-pumping transporters located on the plasma membrane (P type H+ ATPase) and on the tonoplast (V-ATPase or tonoplast H+-pyrophosphatase) during salt stress. Indeed, it has been shown that the expression of genes encoding plasma membrane ATPases is upregulated during salt stress (32, 60). In addition, it has been shown that transferred-DNA knockout mutants of AHA4, which encodes one isoform of the plasma membrane P-type H+-ATPase, is more salt sensitive than the wild type (56). A recent study has also used direct measurements of H+ fluxes in roots of the wild type and sos mutants to directly show that disruption of the SOS pathway causes altered H+ transport at the plasma membrane (52).
On the tonoplast, the V-ATPase, together with the H+-translocating pyrophosphatase, is the major enzyme responsible for maintaining a high concentration of H+ inside the vacuole relative to the cytoplasm. As described in the introduction, a number of studies have found increased expression or activity of the V-ATPase during salt stress; however, the significance of this up-regulation for salt tolerance has not been examined. The severe salt sensitivity we observed in the det3 mutant directly demonstrated the importance of the V-ATPase in plant salt tolerance at the physiological level. It also implies that, in terms of promoting salt tolerance, the loss of V-ATPase activity could not be compensated for by other activities, for example, the tonoplast H+-pyrophosphatase. Thus, the interaction of SOS2 with the V-ATPase complex is consistent with the central role of SOS2 in regulating ion transport processes critical for salt tolerance and also with a previous report of the importance of tonoplast sodium transport mediated by NHX1 in salt tolerance (1).
Despite the many indications of the importance of V-ATPase activity, the physiological factors and signaling mechanisms underlying its regulation are largely unknown. Our present results indicate that SOS2 functions in regulating the V-ATPase and coordinating its activity with those of other ion transporters regulated by the SOS pathway (Fig. 7). There is also evidence that increased tonoplast H+ transport activity may be Ca2+ dependent (17). Another study has suggested that nitric oxide (NO) is involved in the induction of tonoplast H+ transport under salt stress (61). This same study suggested that NO treatment could improve salt tolerance of maize plants. This result is interesting in the context of several recent results from our laboratory which suggest an interaction of the SOS pathway with reactive oxygen detoxification. This includes the interaction of SOS1 with RCD1 (21) as well as additional data from our study of SOS2-interacting proteins that indicate that SOS2 is also a point of interaction between salt and reactive oxygen signaling through its interaction with NDPK2 and catalases.
Acknowledgments
We thank Heven Sze (University of Maryland) for the VHA-B antibody, the Arabidopsis Biological Resource Center for distributing the det3 seed, and Rebecca Stevenson for technical assistance.
This work is contribution no. 91 of CNR-IGV, Italy.
This work was supported by a National Institutes of Health grant R01GM59138 to J.-K.Z and a Department of Energy grant (DE-FG02-04ER15616) to K.S.S. P.E.V. was supported by an NIH postdoctoral fellowship (F32GM074445).
Footnotes
Published ahead of print on 17 September 2007.
REFERENCES
- 1.Apse, M. P., J. B. Sottosanto, and E. Blumwald. 2003. Vacuolar cation/H+ exchange, ion homeostasis, and leaf development are altered in a T-DNA insertional mutant of AtNHX1, the Arabidopsis vacuolar Na+/H+ antiporter. Plant J. 36:229-239. [DOI] [PubMed] [Google Scholar]
- 2.Bai, C., and S. J. Elledge. 1997. Gene identification using the yeast two-hybrid system. Methods Enzymol. 283:141-156. [DOI] [PubMed] [Google Scholar]
- 3.Binzel, M. L., and J. R. Dunlap. 1995. Abscisic acid does not mediate NaCl-induced accumulation of 70-kDa subunit tonoplast H+-ATPase message in tomato. Planta 197:563-568. [Google Scholar]
- 4.Binzel, M. L., and R. Ratajczak. 2001. Function of membrane transport systems under salinity: tonoplast, p. 423-450. In A. Läuchli and U. Luttge (ed.), Salinity: environments-plants-molecules. Kluver, Dordrecht, The Netherlands.
- 5.Cabrera y Poch, H., C. Peto, and J. Chory. 1993. A mutation in the Arabidopsis DET3 gene uncouples photoregulated leaf development from gene expression and chloroplast biogenesis. Plant J. 4:6671-6682. [Google Scholar]
- 6.Cheng, N. H., J. K. Pittman, B. J. Barkla, T. Shigaki, and K. D. Hirschi. 2003. The Arabidopsis cax1 mutant exhibits impaired ion homeostasis, development, and hormonal responses and reveals interplay among vacuolar transporters. Plant Cell 15:347-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, N. H., J. K. Pittman, J.-K. Zhu, and K. D. Hirschi. 2004. The protein kinase SOS2 activates the Arabidopsis H+/Ca2+ antiporter CAX1 to integrate calcium transport and salt tolerance. J. Biol. Chem. 279:2922-2926. [DOI] [PubMed] [Google Scholar]
- 8.Chinnusamy, V., A. Jagendorf, and J.-K. Zhu. 2005. Understanding and improving salt tolerance in plants. Crop Sci. 45:437-448. [Google Scholar]
- 9.Cho, Y.-H., S. H. Yoo, and J. Sheen. 2006. Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 127:579-589. [DOI] [PubMed] [Google Scholar]
- 10.Crabbe, M. J. C., and D. Goode. 1995. Protein folds and functional similarity; Greek key/immunoglobulin fold. Comput. Chem. 19:343-349. [PubMed] [Google Scholar]
- 11.Dietz, K. J., and B. Arbinger. 1996. cDNA sequence and expression of subunit E of the vacuolar H+-ATPase in the inducible Crassulacean acid metabolism plant Mesembryanthemum crystallinum. Biochim. Biophys. Acta 1281:134-138. [DOI] [PubMed] [Google Scholar]
- 12.Golldack, D., and K. J. Dietz. 2001. Salt-induced expression of the vacuolar H+-ATPase in the common ice plant is developmentally controlled and tissue specific. Plant Physiol. 125:1643-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo, Y., U. Halfter, M. Ishitani, and J.-K. Zhu. 2001. Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 13:1383-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo, Y., Q. Qiu, F. J. Quintero, J. M. Pardo, M. Ohta, C. Zhang, K. S. Schumaker, and J.-K. Zhu. 2004. Transgenic evaluation of activated mutant alleles of SOS2 reveals a critical requirement for its kinase activity and C-terminal regulatory domain for salt tolerance in Arabidopsis thaliana. Plant Cell 16:435-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo, Y., L. Xiong, C.-P. Song, D. Gong, U. Halfter, and J.-K. Zhu. 2002. A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev. Cell 3:233-244. [DOI] [PubMed] [Google Scholar]
- 16.Halfter, U., M. Ishitani, and J.-K. Zhu. 2000. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. USA 97:3735-3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han, N., Q. Shao, C. M. Lu, and B. S. Wang. 2005. The leaf tonoplast V-H+-ATPase activity of a C3 halophyte Suaeda salsa is enhanced by salt stress in a Ca-dependent mode. J. Plant Physiol. 162:267-274. [DOI] [PubMed] [Google Scholar]
- 18.Hong-Hermesdorf, A., A. Brüx, A. Grüber, G. Grüber, and K. Schumacher. 2006. A WNK kinase binds and phosphorylates V-ATPase subunit C. FEBS Lett. 580:932-939. [DOI] [PubMed] [Google Scholar]
- 19.Ishitani, M., J. Liu, U. Halfter, C.-S. Kim, W. Shi, and J.-K. Zhu. 2000. SOS3 function in plant salt tolerance requires N-myristoylation and calcium-binding. Plant Cell 12:1667-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janicka-Russak, M., and G. Klobus. 2006. Modification of plasma membrane and vacuolar H+-ATPases in response to NaCL and ABA. J. Plant Physiol. 141:97-107. [DOI] [PubMed] [Google Scholar]
- 21.Katiyar-Agarwal, S., J. Zhu, K. Kim, X. Fu, A. Huang, and J.-K. Zhu. 2006. The plasma membrane Na+/H+ antiporter SOS1 interacts with RCD1 and functions in oxidative stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 103:18002-18007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirsch, M., Z. An, R. Viereck, R. Low, and T. Rausch. 1996. Salt stress induces an increased expression of V-type Hg+-ATPase in mature sugar beet leaves. Plant Mol. Biol. 32:543-547. [DOI] [PubMed] [Google Scholar]
- 23.Kluge, K., D. Golldack, and K. J. Dietz. 1999. Subunit D of the vacuolar H+-ATPase of Arabidopsis thaliana. Biochim. Biophys. Acta 1419:105-110. [DOI] [PubMed] [Google Scholar]
- 24.Liu, J., M. Ishitani, U. Halfter, C.-S. Kim, and J.-K. Zhu. 2000. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. USA 97:3730-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Löw, R., B. Rockel, M. Kirsch, R. Ratajczak, S. Hortensteiner, E. Martinoia, U. Lüttge, and T. Rausch. 1996. Early salt stress effects on the differential expression of vacuolar H+-ATPase genes in roots and leaves of Mesembryanthemum crystallinum. Plant Physiol. 110:259-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Löw, R., and T. Rausch. 1996. In suspension-cultured Daucus carota cells salt stress stimulates H+-transport but not ATP hydrolysis of the V-ATPase. J. Exp. Bot. 47:1725-1732. [Google Scholar]
- 27.Lüttge, U., E. Fischer-Schliebs, and R. Ratajczak. 2001. The H+ pumping V-ATPase of higher plants: a versatile “eco-enzyme” in response to environmental stress. Cell. Biol. Mol. Lett. 6:356-361. [Google Scholar]
- 28.Mariaux, J. B., E. Fischer-Schliebs, U. Lüttge, and R. Ratajczak. 1997. Dynamics of activity and structure of the tonoplast vacuolar-type H+-ATPase in plants with different CAM expression and in a C3 plant under salt stress. Protoplasma 196:181-189. [Google Scholar]
- 29.Martiny-Baron, G., M. F. Manolson, R. J. Poole, D. Hecker, and G. F. E. Scherer. 1992. Proton transport and phosphorylation of tonoplast polypeptides from zucchini are stimulated by the phospholipid platelet-activating factor. Plant Physiol. 99:1635-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narasimhan, M. L., M. L. Binzel, E. Perez-Prat, Z. Chen, D. E. Nelson, N. K. Singh, R. A. Bressan, and P. M. Hasegawa. 1991. NaCl regulation of tonoplast ATPase 70-kilodalton subunit mRNA in tobacco cells. Plant Physiol. 97:562-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishi, T., and M. Forgac. 2002. The vacuolar (H+)-ATPases—nature's most versatile proton pumps. Nat. Rev. Mol. Cell Biol. 3:94-103. [DOI] [PubMed] [Google Scholar]
- 32.Niu, X., B. Damsz, A. K. Kononowicz, R. A. Bressan, and P. M. Hasegawa. 1996. NaCl-induced alterations in both cell structure and tissue-specific plasma membrane H+-ATPase gene expression. Plant Physiol. 111:679-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohta, M., Y. Guo, U. Halfter, and J.-K. Zhu. 2003. A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc. Natl. Acad. Sci. USA 100:11771-11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Owegi, M. A., A. L. Carenbauer, N. M. Wick, J. F. Brown, K. L. Terhune, S. A. Bilbo, R. S. Weaver, R. Shircliff, N. Newcomb, and K. J. Parra-Belky. 2005. Mutational analysis of the stator subunit E of the yeast V-ATPase. J. Biol. Chem. 280:18393-18402. [DOI] [PubMed] [Google Scholar]
- 35.Pardo, J. M., B. Cubero, E. O. Leidi, and F. J. Quintero. 2006. Alkali cation exchangers: roles in cellular homeostasis and stress tolerance. J. Exp. Bot. 57:1181-1199. [DOI] [PubMed] [Google Scholar]
- 36.Perera, I. Y., X. Li, and H. Sze. 1995. Several distinct genes encode nearly identical to 16 kDa proteolipids of the vacuolar H+-ATPase from Arabidopsis thaliana. Plant Mol. Biol. 29:227-244. [DOI] [PubMed] [Google Scholar]
- 37.Pittman, J. K., T. Shigaki, J. L. Marshall, J. L. Morris, N. H. Cheng, and K. D. Hirschi. 2004. Functional and regulatory analysis of the Arabidopsis thaliana CAX2 cation transporter. Plant Mol. Biol. 56:959-971. [DOI] [PubMed] [Google Scholar]
- 38.Puig, O., F. Caspary, G. Rigaut, B. Ruitz, E. Bouvert, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complexes purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 39.Qiu, Q. S., Y. Guo, M. A. Dietrich, K. S. Schumaker, and J.-K. Zhu. 2002. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 99:8436-8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qiu, Q. S., Y. Guo, F. J. Quintero, J. M. Pardo, K. S. Schumaker, and J.-K. Zhu. 2004. Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt-overly-sensitive (SOS) pathway. J. Biol. Chem. 279:207-215. [DOI] [PubMed] [Google Scholar]
- 41.Quan, R., H. Lin, I. Mendoza, Y. Zhang, W. Cao, Y. Yang, M. Shang, S. Chen, J. M. Pardo, and Y. Guo. 2007. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 19:1415-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quintero, F. J., M. Ohta, H. Shi, J.-K. Zhu, and J. M. Pardo. 2002. Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Nat1. Acad. Sci. USA 99:9061-9066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ratajczak, R., J. Richter, and U. Lüttge. 1994. Adaptation of the tonoplast V-type H+-ATPase of Mesembryanthemum crystallinum to salt stress, C3-CAM transition and plant age. Plant Cell Environ. 17:1101-1112. [Google Scholar]
- 44.Ratajczak, R. 2000. Structure, function and regulation of the plant vacuolar H+-translocating ATPase. Biochim. Biophys. Acta 1465:17-36. [DOI] [PubMed] [Google Scholar]
- 45.Rausch, T., M. Kirsch, R. Low, A. Lehr, R. Viereck, and A. Zhigang. 1996. Salt stress responses of higher plants: the role of proton pumps and Na+/H+-antiporters. J. Plant Physiol. 148:425-433. [Google Scholar]
- 46.Reuveni, M., A. B. Bennett, R. A. Bressan, and P. M. Hasegawa. 1990. Enhanced H+-transport capacity and ATP hydrolysis activity of the tonoplast H+-ATPase after NaCl adaptation. Plant Physiol. 94:524-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 48.Rohila, J. S., M. Chen, R. Cerny, and M. E. Fromm. 2004. Improved tandem affinity purification tag and methods for isolation of protein heterocomplexes from plants. Plant J. 38:172-181. [DOI] [PubMed] [Google Scholar]
- 49.Rojo, E., R. Martin, C. Carter, J. Zouhar, S. Pan, J. Plotnikova, H. Jin, M. Paneque, J. J. Sanchez-Serrano, B. Baker, F. Ausubel, and N. Raikhel. 2004. VPEγ exhibits a caspase-like activity that contributes to defense against pathogens. Curr. Biol. 21:1897-1906. [DOI] [PubMed] [Google Scholar]
- 50.Rubio, V., Y. Shen, Y. Liu, G. Gusmaroli, S. P. Dinesh-Kumar, and X. W. Deng. 2005. An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. 41:767-778. [DOI] [PubMed] [Google Scholar]
- 51.Schumacher, K., D. Vafeados, M. McCarthy, H. Sze, T. Wilkins, and J. Chory. 1999. The Arabidopsis det3 mutant reveals a central role for the vacuolar H+-ATPase in plant growth and development. Genes Dev. 13:3259-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shabala, L., T. A. Cuin, I. A. Newman, and S. Shabala. 2005. Salinity-induced ion flux patterns from the excised roots of Arabidopsis sos mutants. Planta 222:1041-1050. [DOI] [PubMed] [Google Scholar]
- 53.Shi, H., M. Ishitani, C.-S. Kim, and J.-K. Zhu. 2000. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 97:6896-6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sze, H., K. Schumacher, M. L. Muller, S. Padmanaban, and L. Taiz. 2002. A simple nomenclature for a complex proton pump: VHA genes encode the vacuolar H+-ATPase. Trends Plant Sci. 7:157-161. [DOI] [PubMed] [Google Scholar]
- 55.Tsiantis, M. S., D. M. Bartholomew, and J. A. C. Smith. 1996. Salt regulation of transcript levels for the c subunit of a leaf vacuolar H+-ATPase in the halophyte Mesembryanthemum crystallinum. Plant J. 9:729-736. [DOI] [PubMed] [Google Scholar]
- 56.Vitart, V., I. Baxter, P. Doerner, and J. F. Harper. 2001. Evidence for a role in growth and salt resistance of a plasma membrane H+-ATPase in the root endodermis. Plant J. 27:191-201. [DOI] [PubMed] [Google Scholar]
- 57.Wang, B., U. Lüttge, and R. Ratajczak. 2001. Effects of salt treatment and osmotic stress on V-ATPase and V-PPase in leaves of the halophyte Suaeda salsa. J. Exp. Bot. 52:2355-2365. [DOI] [PubMed] [Google Scholar]
- 58.Wang, Y., J. Sun, and P. R. Chitnis. 2000. Proteomic study of the peripheral proteins from thylakoid membranes of the cyanobacterium Synechocystis sp. Electrophoresis 21:1746-1754. [DOI] [PubMed] [Google Scholar]
- 59.Ward, J. M., and H. Sze. 1992. Subunit composition and organization of the vacuolar H+-ATPase from oat roots. Plant Physiol. 99:170-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, J.-S., C. Xie, Z.-Y. Li, and S.-Y. Chen. 1999. Expression of the plasma membrane H+-ATPase gene in response to salt stress in a rice salt-tolerant mutant and its original variety. Theoret. Appl. Genet. 99:1006-1011. [Google Scholar]
- 61.Zhang, Y. Y., L. L. Wang, Y. L. Liu, Q. Zhang, Q. P. Wei, and W. H. Zhang. 2006. Nitric oxide enhances salt tolerance in maize seedlings through increasing activities of proton-pump and Na+/H+ antiport in the tonoplast. Planta 224:545-555. [DOI] [PubMed] [Google Scholar]
- 62.Zhu, J.-K. 2003. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 6:441-445. [DOI] [PubMed] [Google Scholar]
- 63.Zhu, J.-K. 2002. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53:247-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu, J.-K., J. Liu, and L. Xiong. 1998. Genetic analysis of salt tolerance in Arabidopsis thaliana: evidence of a critical role for potassium nutrition. Plant Cell 10:1181-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]