Abstract
Ribosome biogenesis requires equimolar amounts of four rRNAs and all 79 ribosomal proteins (RP). Coordinated regulation of rRNA and RP synthesis by eukaryotic RNA polymerases (Pol) I, III, and II is a key requirement for growth control. Using a novel global genetic approach, we showed that the absence of Hmo1 becomes lethal when combined with mutations of components of either the RNA Pol II or Pol I transcription machineries, of specific RP, or of the TOR pathway. Hmo1 directly interacts with both the region transcribed by Pol I and a subset of RP gene promoters. Down-regulation of Hmo1 expression affects RP gene expression. Upon TORC1 inhibition, Hmo1 dissociates from ribosomal DNA (rDNA) and some RP gene promoters simultaneously. Finally, in the absence of Hmo1, TOR-dependent repression of RP genes is alleviated. Therefore, we show here that Saccharomyces cerevisiae Hmo1 is directly involved in coordinating rDNA transcription by Pol I and RP gene expression by Pol II under the control of the TOR pathway.
Ribosome biogenesis on its own consumes up to 80% of a proliferating cell's energy and represents about 95% of total transcription (44). Recent evidence suggests that ribosome biogenesis is required not only for growth but also for regulation of cell growth (51). 25S, 18S, and 5.8S ribosomal RNAs are synthesized by RNA polymerase I (Pol I), while the 5S rRNA is transcribed by RNA Pol III. RNA Pol II produces messenger RNAs encoding 79 ribosomal proteins (RP), expressed from 138 genes in budding yeast (62). To generate mature ribosomal subunits, ribosomal components have to be transported, processed, and assembled, using more than 200 different trans-acting factors (70). The second group of Pol II-transcribed genes, called Ribi, is coregulated with the RP genes (21, 29). Global coordination of ribosome component transcription by all three transcriptional machineries remains to be understood.
In Saccharomyces cerevisiae, ribosome biosynthesis is primarily regulated at the level of transcription and under the control of the conserved TORC1 (target of rapamycin complex 1) pathway (discussed in reference 45). Recent studies have identified essential transcription factors controlled by the TORC1 pathway involved specifically in transcription by either Pol I, Pol III, or Pol II. These factors seem to act on single transcription apparatus, such as Rrn3/TIFIA (14, 19) and UBF (24) for Pol I, Maf1 for Pol III (17, 57, 71), and the Forkhead-like transcription factor Fhl1 and two cofactors, Ifh1 and Crf1, for Pol II (46, 62, 63, 72). However, cross talk between Pols is well documented, and recent findings argue for an upstream function of Pol I in this cross-regulation (9, 37).
Transcription by Pol I is known to be the major rate-limiting step in ribosome biogenesis (52). TOR complex 1 (TORC1) binds directly to the ribosomal DNA (rDNA) and activates Pol I (38). A subpopulation of Pol I in complex with Rrn3 is competent for initiation (48, 49). An essential Pol I subunit, Rpa43, interacts with Rrn3 (56). Inhibition of TORC1 by rapamycin decreased the amount of the Rrn3-Pol I complex (14). An overexpressed Rrn3-Rpa43 fusion protein can substitute for both essential proteins Rrn3 and Rpa43. If this fusion protein, named CARA (constitutive association of Rrn3 and RpA43), is overexpressed in a mutant strain lacking both Rpa43 and Rrn3, Pol I remains competent for initiation even when TORC1 is inactivated (37). Interestingly, the CARA mutant can alleviate TORC1-dependent regulation of RP genes (37). This finding could suggest that a Pol I factor contributes to TORC1 regulation of RP gene transcription.
We have previously shown that Hmo1 is a bona fide Pol I transcription factor (20). Recently, it has been shown that Hmo1 is bound to both rDNA and RP gene promoters (23). Hmo1 binding to RP promoters requires Rap1 and is required for the assembly of Fhl1 and Ifh1 onto RP promoters. However, Hmo1 appears not to be required for global RP gene expression (23).
To better understand which role Hmo1 fulfills in vivo, we tried to identify new genetic partners of Hmo1. Using a novel systematic genetic approach, we established that Hmo1 is genetically linked to Pol I, to specific RP genes, and to TORC1 in vivo. Like the CARA strain, Hmo1 is hypersensitive to TORC1 inhibition. In the absence of Hmo1, Ifh1 is still essential for RP gene expression but some specific RP genes are deregulated. We established that in the absence of Hmo1, the cross-regulation of Pol I-RP gene transcription is alleviated. Therefore, Hmo1 is required for the TORC1-regulatable expression of RP genes.
MATERIALS AND METHODS
Yeast and plasmid constructions.
Yeast media and genetic techniques were described previously (26, 67). Yeast strains are described in Table 1. Oligonucleotides are described in Table S1 in the supplemental material. Plasmids pFS-Tap-Kan (75), pFS-Tap-TRP (75), pENTR3C (Invitrogen), pDONR201 (Invitrogen), and pFA6-13myc-TRP1 (41) were previously described. Plasmid pRC1 (10), containing the TOR1-1 (or DRR1-1) allele, was subcloned using the BamHI-ClaI fragment (3.6 kb) in pRS305 to generate pRS305-TOR1-1, which was cut with AgeI for integration. pGID1, pGID2, pRS316-CII, pFL36-CII, and pAG32-ttt, used in the genetic-interaction-with-gene-deletion (GID) screen, are described in detailed GID protocols. pUC19-HPHMX4 was constructed by ligating the HindIII-XbaI fragment of pAG32 (22) into the HindIII-SpeI-digested vector pUC19 (74). The following plasmids were constructed using Gateway technology (Invitrogen): pDONR201-HMO1 (by a BP recombinase enzyme mix with pDONR201 and HMO1 [oligonucleotides 1529 and 1530], using strain BY4741 as the template) and pDONR-FPR1 (using the directional TOPO cloning strategy [Stratagene] with pENTR/D-TOPO and FPR1 [oligonucleotides 1547 and 1548], with strain BY4741 as the template). pGID-HMO1, pRS316-HMO1, and pFL36-HMO1 were obtained by LR recombinase enzyme mix between pDONR201-HMO1 and pGID, pRS316-CII, and pFL36-CII, respectively. Similarly, pGID-FPR1 and pRS316-FPR1 were obtained by LR reactions between pDONR-FPR1 and pGID and pRS316-CII, respectively.
TABLE 1.
Yeast strains
| Strain | Genotype | Source or reference |
|---|---|---|
| BMA64-1a | MATaleu2-3,112 his3-11,15 trp1Δ ade2-1 ura3-1 | 2 |
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Euroscarf |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Euroscarf |
| YO6969 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hmo1-Δ::KAN-MX4 | Euroscarf |
| YO2941 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 fpr1-Δ::KAN-MX4 | Euroscarf |
| OGP126-1a | MATaleu2-3,112 his3-11,15 trp1Δ can1-100 ade2-1 ura3-52 HMO1::TAP-KAN | This work |
| YAB2-2a | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hmo1Δ::PrαNAT-TetR-VP16 + pGID-HMO1 (TetO-CEN URA3 MET15 HMO1) | This work |
| YAB11-1a | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hmo1Δ::PrαNAT-TetR-VP16 rpa34-Δ::KANMX4 + pGID-HMO1 (TetO-CEN URA3 MET15 HMO1) | This work |
| YAB9-1a | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hmo1Δ::PrαNAT-TetR-VP16 rpa49-Δ::KANMX4 + pGID-HMO1 (TetO-CEN URA3 MET15 HMO1) | This work |
| YAB4-1a | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hmo1Δ::PrαNAT-TetR-VP16 yjl075c-Δ::KANMX4 (or net1-Δc) + pGID-HMO1 (TetO-CEN URA3 MET15 HMO1) | This work |
| YAB5-1a | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hmo1Δ::PrαNAT-TetR-VP16 rps23a-Δ::KANMX4 + pGID-HMO1 (TetO-CEN URA3 MET15 HMO1) | This work |
| YAB6-1a | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hmo1Δ::PrαNAT-TetR-VP16 fpr1-Δ::KANMX4 + pGID-HMO1 (TetO-CEN URA3 MET15 HMO1) | This work |
| YAB7-1a | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hmo1Δ::PrαNAT-TetR-VP16 tom1-Δ::KANMX4 + pGID-HMO1 (TetO-CEN URA3 MET15 HMO1) | This work |
| YAB8-1a | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hmo1Δ::PrαNAT-TetR-VP16 ncs2-Δ::KANMX4 + pGID-HMO1 (TetO-CEN URA3 MET15 HMO1) | This work |
| YAB100-1a | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hmo1Δ::PrαNAT-TetR-VP16 HPH-pTET-RPA190 + pGID-HMO1 (TetO-CEN URA3 MET15 HMO1) | This work |
| YAB101-1a | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hmo1Δ::PrαNAT-TetR-VP16 HPH-pTET-RPB1 + pGID-HMO1 (TetO-CEN URA3 MET15 HMO1) | This work |
| YAB102-1a | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hmo1Δ::PrαNAT-TetR-VP16 HPH-pTET-KOG1 + pGID-HMO1 (TetO-CEN URA3 MET15 HMO1) | This work |
| YAB103-1a | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hmo1Δ::PrαNAT-TetR-VP16 HPH-pTET-IFH1 + pGID-HMO1 (TetO-CEN URA3 MET15 HMO1) | This work |
| YAB104-1a | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 fpr1Δ::PrαNAT-TetR-VP16 HPH-pTET-RPA190 + pGID-FPR1 (TetO-CEN URA3 MET15 FPR1) | This work |
| YAB105-1a | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 fpr1Δ::PrαNAT-TetR-VP16 HPH-pTET-RPB1 + pGID-FPR1 (TetO-CEN URA3 MET15 FPR1) | This work |
| YAB106-1a | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 fpr1Δ::PrαNAT-TetR-VP16 HPH-pTET-IFH1 + pGID-FPR1 (TetO-CEN URA3 MET15 FPR1) | This work |
| YAB107-1a | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 fpr1Δ::PrαNAT-TetR-VP16 HPH-pTET-KOG1 + pGID-FPR1 (TetO-CEN URA3 MET15 FPR1) | This work |
| YAB108-1a | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 fpr1Δ::PrαNAT-TetR-VP16 HPH-pTET-HMO1 + pGID-FPR1 (TetO-CEN URA3 MET15 FPR1) | This work |
| YPH500 | MATα leu2Δ1 his3Δ200 trp1Δ63 lys2-801 ade2-101 ura3-52 | 37 |
| CARA | MATα leu2Δ1 his3Δ200 trp1Δ63 lys2-801 ade2-101 ura3-52 ccn3Δ::HIS5 rpa43Δ::KANMX4 + pGEN-RRN3-RPA43 (2μm TRP1 RRN3::RPA43) | 37 |
| YAB73-1a | MATα leu2Δ1 his3Δ200 trp1Δ63 lys2-801 ade2-101 ura3-52 rrn3Δ::HIS5 rpa43Δ::KANMX4 hmo1Δ::HPH + pGEN-RRN3-RPA43 (2μm TRP1 RRN3::RPA43) | This work |
| YAB72-1a | MATα leu2Δ1 his3Δ200 trp1Δ63 lys2-801 ade2-101 ura3-52 hmo1Δ::HPH | This work |
| YAB82-1a | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 TOR1-1::LEU2 | This work |
Strains were generated by genetic crosses with indicated parental strains (Table 1). The strategy for the modification of strains used in the GID screen is described in the supplemental material. C-terminal fusion proteins were generated using the F2 or R1 oligonucleotide, with pFA6-13myc-TRP1, pFS-Tap-Kan, or pFS-Tap-TRP as the template (41). Insertion of the regulatable promoter was achieved using the PCR product from pAG32-ttt and oligonucleotides 1836 to 1837 (RPA190), 1841 to 1842 (RPB1), 1806 to 1807 (IFH1), 1597 to 1598 (KOG1), and 1856 to 1857 (HMO1). Strains YAB72 and YAB73 were generated by using oligonucleotides 887 and 888 on plasmid pUC19-HPHMX4.
Immunoblotting.
Exponentially growing cells from strain OGP126-1a were untreated or treated for 60 min with 400 ng/ml rapamycin. Cells were collected, and proteins were extracted by NaOH-loading buffer treatment (36). Cell extracts were loaded on 10% polyacrylamide sodium dodecyl sulfate gels, and proteins were separated by electrophoresis and transferred onto nitrocellulose membranes. Western blotting was performed with anti-protein A-peroxidase mouse antiperoxidase antibodies (DAKO) and antiactin antibodies (AC-40; Sigma). As secondary antibodies, anti-mouse antibody-horseradish peroxidase (Jackson Laboratories) conjugates were used, followed by detection with chemiluminescence (Pierce).
ChIP.
Hmo1 chromatin immunoprecipitation (ChIP) was performed using strains OG126-1a and BMA64-1a as an untagged control. Tandem affinity purification was performed according to Rigaut et al. (60). Increased specificity was achieved using three washes, each three times: with lysis buffer (50 mM HEPES [pH 7.5], 1% Triton X-100, 0.1% Na deoxycholate, 1 mM EDTA), washing buffer 1 (50 mM HEPES [pH 7.5], 500 mM NaCl, 1% Triton X-100, 0.1% Na deoxycholate, 1 mM EDTA), and then washing buffer 2 (10 mM Tris-HCl [pH 8], 250 mM LiCl, 0.5% NP-40, 1 mM EDTA, 0.1% Na deoxycholate) as described in Bier et al. (8). Next, tobacco etch virus Nia protease cleavage was done for 2 h at 16°C. The amount of immunoprecipitated DNA was expressed as a value relative to the amount of input DNA, where 1 unit represents 0.005% of input DNA.
Systematic SL screen: GID.
Methods used for the systematic synthetic lethal (SL) approach are explained in detail as a supplemental material protocol.
Transcription analysis.
For depletion experiments, strains bearing pTET promoters were treated for 6 h with 5 μg/ml of doxycycline prior to harvesting. Total RNA was extracted from yeast using the hot phenol procedure (9).
For quantitative reverse transcription-PCR, total RNA was directly reverse transcribed using an oligonucleotide deoxyribosylthymine primer and Superscript II reverse transcriptase (Invitrogen). cDNAs were RNase H treated and used in a 1:5 dilution to perform a quantitative PCR with primers 2011 and 2012, 2025 and 2026, 2029 and 2032, and 1122 and 1123. RNA levels were normalized relative to ACT1 mRNA levels.
Microarray data accession number.
Microarray protocols and analysis have been deposited in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) under accession no. E-MEXP-651.
RESULTS
Systematic SL screen using an hmo1Δ mutant: the GID approach.
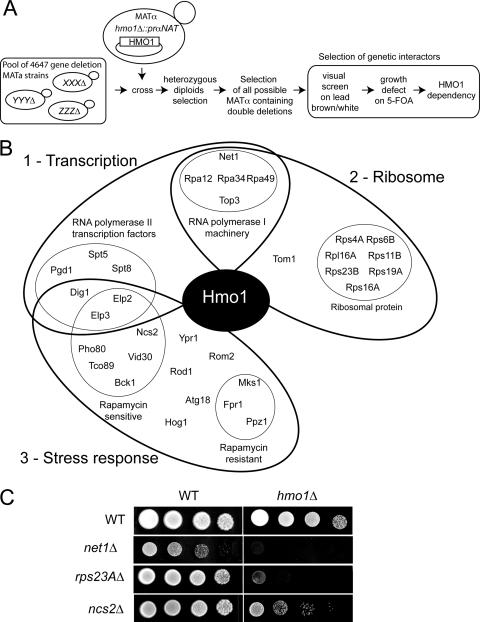
We have previously shown that Hmo1 is genetically linked to the Pol I apparatus (20). Interestingly, Hmo1 is not essential (42) but is implicated in the control of mutagenesis (1), interacts with long CAG repeat tracts (33), and is bound to both rDNA and RP gene promoters and may contribute to rRNA processing (23). The precise function of Hmo1 remains elusive, but the available data suggest that Hmo1 couples Pol I transcription to other cellular processes. To identify putative other functions of Hmo1 in vivo, we performed a systematic SL screen with hmo1Δ as the bait, using a new plasmid-based approach named GID (Fig. 1A). Our method combines the simplicity of classical SL screens, using a plasmid dependency assay (6), and makes use of the available collection of all nonessential yeast gene deletions (73). The construction of the test strain and the detailed steps of the screen are described in Fig. S1 and S2 in the supplemental material. In brief, in the test strain, the HMO1 gene is replaced by a nourseothricin resistance marker (NAT) placed under the control of a MAT-alpha-specific promoter. This test strain is transformed with a pGID-HMO1 complementing plasmid which carries both a MET15 and a URA3 marker and a hygromycin resistance marker. It is first mated with pools of all viable haploid deletion strains of the a mating type marked by a kanamycin resistance marker (KAN). Diploids were selected on G418 and hygromycin-containing medium and sporulated. After sporulation, alpha-haploids bearing double deletions were selected on nourseothricin and G418 double-selective medium. The identification of genetic partners was performed by selection of mutant cells from the library requiring HMO1 for growth in three successive steps (Fig. 1A). We began with a color assay using lead-containing medium. In lead-containing medium, H2S-producing colonies develop a dark brown color due to formation of PbS (53). Strains bearing mutations such as met15− or met2− are overproducing H2S (53). Therefore, colonies that can lose the pGID-HMO1 plasmid carrying the MET15 marker develop on lead-containing medium a brown color (73, 15). A white color could indicate a requirement for HMO1 for survival. Next, we scored for the ability of cells to grow without this complementing plasmid by direct counterselection of pGID-HMO1 (using the URA3 marker) on 5-fluoroorotic acid (5-FOA)-containing medium. Finally, we reversed those phenotypes by providing another plasmid bearing HMO1. Using three selection steps, we established the requirement for Hmo1 in some double-deletion strains. The gene deletion responsible for the SL phenotype was then identified by using the “molecular bar codes” associated with each deletion mutant (see the supplemental material).
FIG. 1.
Genetic interactions of Hmo1. (A) Flow diagram of the GID screen. (B) Schematic representation of identified genetic interactors with HMO1. The interactors can be grouped roughly into three different classes of function that are partially overlapping: (i) transcription, (ii) ribosome, and (iii) stress response. (C) Growth comparison of wild-type, single-mutant, and double-deletion strains. The single-deletion hmo1Δ strain (YAB2-2a) and double-deletion hmo1Δ/net1Δc (YAB4-1a), hmo1Δ/rps23aΔ (YAB5-1a), and hmo1Δ/nsc2Δ (YAB8-1a) strains containing plasmid pGID-HMO1 were transformed with pFL36-HMO1 (left; WT) or an empty vector (right; hmo1Δ). Tenfold serial dilution series were spotted on 5-FOA-containing plates. Growth was scored after 4 days at 30°C. On 5-FOA medium, the pGID-HMO1 plasmid is lost. In the left panel, HMO1 is still present on plasmid pFL-HMO1, scoring the growth of the single-deletion mutant; in the right panel, HMO1 is absent, scoring the growth of the double-deletion mutant hmo1Δ plus the indicated deletion.
We performed the screen for 120,000 individual colonies, resulting in 15-fold library coverage of each possible double mutant. With the color assay, 922 candidates were selected. Of these, 123 double mutants grew poorly or not at all without plasmid on 5-FOA-containing medium. In 63 clones out of the 123, representing 28 different double mutants, the growth defect was fully restored by providing an HMO1-containing plasmid (see Table S2 in the supplemental material). Surprisingly, rpa12Δ, rpa34Δ, top3Δ, and rpa49Δ, previously identified as genetically interacting with hmo1Δ (20), have not been isolated. The corresponding double mutants, generated by individual crosses, were not white on lead-containing medium, probably due to H2S formation even in the presence of MET15, resulting in brown colonies (53). Since the white color is required for the first selection step, our screening procedure does not allow the identification of all possible genetic interactions (data not shown). However, we confirmed our previously published interactions with hmo1Δ on 5-FOA medium (e.g., rpa12Δ, rpa34Δ, top3Δ, and rpa49Δ) (20). Furthermore, we reproduced the strong SL interaction of Hmo1 with Fpr1 (18).
In summary, out of 32 gene deletions, 7 were SL and 25 caused synthetic slow growth to various extents in combination with hmo1Δ (see Table S2 in the supplemental material). From these 32 genes, we defined three putative functional groups, according to known phenotypes: genes involved in ribosome biogenesis (13 genes), genes involved in transcription of Pol I (5 genes) or Pol II (6 genes), and a group of genes involved in very distinct functions in vivo but all connected to stress response pathways (16 genes) (Fig. 1B). The growth defects of three double mutants are shown in Fig. 1C. We observed that using this novel strategy (GID) to identify SL candidates, we are able to select a panel of candidates having various growth defects (see Table S2 in the supplemental material).
In conclusion, using this systematic screen, we confirmed that Hmo1 becomes essential when Pol I transcription is defective and new links to two functional pathways were unveiled: Hmo1 becomes essential when (i) stress response pathways such as TORC1 are affected and (ii) specific RP are down-regulated.
Hmo1 is enriched on a subset of the rDNA unit.
We have previously established the direct involvement of Hmo1 in Pol I transcription (20). From our genetic data, we established that Hmo1 is linked to Pol I initiation (rpa43-24 or net1) (20, 65), termination mutant rpa12Δ (58), or the newly defined Pol I elongation factor Spt5 (64) (Fig. 1B).
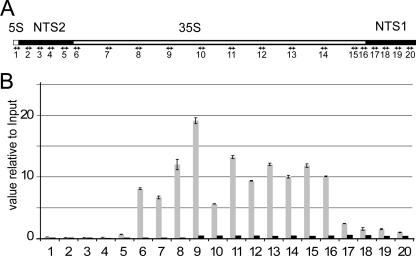
To determine at which step of the transcription process the Hmo1 protein acts, we mapped its association within the rDNA unit. Because rDNA is a highly repeated genomic locus, we used a ChIP protocol with extensive and stringent washing optimized to detect specific associations within the rDNA unit (8). Furthermore, we chose not to normalize our ChIP results using a region outside rDNA, which may lack comparability because of the repeated nature of rDNA and a difference in extractability, but to compare using an untagged strain (Fig. 2). Using a total of 20 primer pairs covering the rDNA unit (Fig. 2A), we observed a roughly 100-fold enrichment of the region transcribed by Pol I (35S; primer pairs 6 to 16) (Fig. 2B, compare gray bar [Hmo1] to black bar [control]) compared to the untagged-control-strain level. We reproducibly observed a low binding efficiency for Hmo1 within the NTS2 region (primer pairs 1 to 5) and mild binding to the terminator region (primer pairs 17 to 20). We observed drastic increases between amplicons 5 and 6, at positions −271 to −131 and positions +6 to +108, respectively, relative to the Pol I transcription start site (13). This result demonstrates no enrichment upstream of promoter-bound elements at positions −146 to −100 for UAF and −28 to +8 for CF relative to the Pol I transcription start site (32, 39). We detected no greater enrichment near the CF binding site (amplicon 6) than for amplicons 7 to 16, which were totally devoid of any promoter elements. This result is different from the previously described association throughout the rRNA gene (23). We interpret this discrepancy as an effect of the normalization procedure and our stringent washing protocol. We conclude that Hmo1 is specifically enriched within the Pol I-transcribed region of the rDNA unit, suggesting that it may function during elongation.
FIG. 2.
Hmo1 binds preferentially to the Pol I-transcribed region of the rDNA. ChIP experiments were performed using strain HMO1-TAP (OGP126-1a) or the untagged BMA64-1a control strain, followed by a two-step purification. (A) Schematic representation of an rDNA unit. Sequence elements within the rDNA (nontranscribed spacers 1 and 2 [NTS1 and -2], the 5S RNA Pol III transcript, and the 35S RNA Pol I transcript) are noted. The oligonucleotides used to analyze the immunoprecipitates via quantitative PCR are indicated by arrows (1 to 20). (B) Hmo1 ChIP. Relative values of immunoprecipitated DNA and the whole-cell extract (Input) are shown. The amount of immunoprecipitated DNA was expressed as a value relative to that for input DNA, where 1 unit represents 0.005% of input DNA. DNAs immunoprecipitated from the Hmo1-TAP strain (OGP126-1a) and from the untagged BMA64-1a control strain are shown in gray and in black, respectively. Standard deviations for three different measurements are indicated.
Hmo1 is connected to the TORC1 pathway.
Hmo1 absence requires an optimal Pol I transcription for viability (20). We found here that mutation of some specific RP genes and of stress response pathways is also genetically linked to Hmo1. Several lines of evidence link Hmo1 more specifically to the TORC1 stress response. One of the major regulators of RP gene expression is the TORC1 complex. Among the genetic interactors of Hmo1, Fpr1 was the most frequently found gene, confirming an earlier report (18). Fpr1 is one of the four peptidyl-prolyl cis-trans isomerases of the FK506 binding protein family (FKBPs) and is the specific cytoplasmic receptor of rapamycin. An fpr1Δ strain is resistant to rapamycin treatment (25). In complex with rapamycin, Fpr1 binds TORC1 and inhibits its function (40). TORC1 is made of two exchangeable kinases, Tor1 and Tor2; two essential proteins, Lst8 and Kog1 (40); and one nonessential factor, Tco89 (59). Essential genes are not tested in our GID screen, but Tco89 had been identified (Fig. 1B).
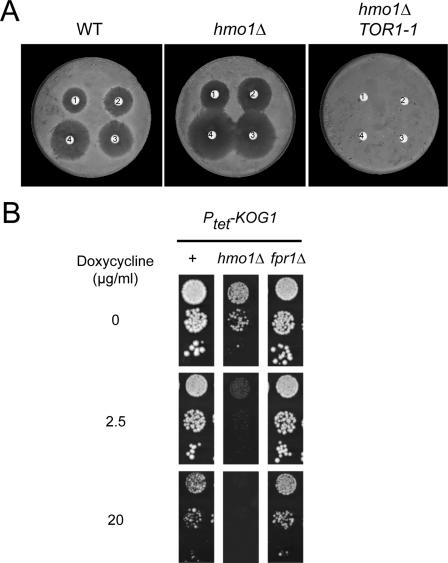
We checked the sensitivity of hmo1Δ to TORC1 inhibition using rapamycin treatment. As a control, we introduced in the hmo1Δ strain a dominant mutant of TOR1 (DRR1dom, or TOR1-1), which makes cells resistant to rapamycin (10). As shown in Fig. 3A, the hmo1Δ strain is more sensitive to rapamycin than the corresponding wild type. This sensitivity is fully rescued by the TOR1-1 mutation.
FIG. 3.
Inhibition of the TORC1-pathway is lethal in an hmo1Δ background. (A) An hmo1Δ mutant is hypersensitive to rapamycin. BY4741 (WT), hmo1Δ (Y16969), and hmo1Δ TOR1-1 strains were spread homogenously onto yeast extract-peptone-dextrose plates by using glass beads. TOR1-1 is generated by a dominant mutation in the TORC1-kinase TOR1, making TORC1 rapamycin insensitive. Sterile Whatman 3MM filter paper containing different amounts of rapamycin (1, 0.01 μg; 2, 0.02 μg; 3, 0.05 μg; 4, 2 μg) were deposited on top of these plates. The halo around each filter is indicative of the growth inhibition. (B) Hmo1 is genetically linked to the TOR pathway. KOG1 was placed under the control of a tetracycline-regulatable promoter (Ptet-KOG1). Growth was scored without (0 μg/ml), with mild (2.5 μg/ml) or with strong (20 μg/ml) repression of KOG1 transcription. Kog1 depletion was performed in wild-type (+; YAB102-1a), hmo1Δ (YAB102-1a cured of pRS316-HMO1), and fpr1Δ (YAB107-1a cured of pRS316-FPR1) strains. Tenfold serial dilution series were spotted on rich medium in the presence of increasing concentrations of doxycycline. Growth was observed after 5 days of incubation at 30°C.
To further confirm the connection between Hmo1 and the TORC1 pathway, we next assayed the consequences of combining hmo1Δ with depletion of essential genes of the TORC1 complex. The replacement of the native promoter by the tetracycline-regulatable promoter was initially developed by Herrero's group (4) and has recently been used for large-scale studies (50). We inserted a tetracycline-regulatable promoter upstream of three essential genes, RPA190 (the largest subunit of Pol I), RPB1 (the largest subunit of Pol II), and KOG1 (a TORC1-specific member) (27, 40, 47). To increase promoter shutoff, the repression in the presence of doxycycline was increased by introduction of the repressor tetR′-SSN6 (tTA′) (5). We have previously shown that specific mutations of Pol I are SL with hmo1Δ but that Pol II mutations such as rpb1-1 are not (20). Consistently, we observed that Hmo1 becomes fully essential for growth when Pol I (ptet-RPA190) is inhibited by mild depletion but not when Pol II (ptet-RPB1) is (data not shown). Using a doxycycline concentration allowing depletion of Kog1 without a detectable growth defect in a wild-type background (2.5 μg/ml), we observed no detectable growth in an hmo1Δ mutant (Fig. 3B). As a control, we tested the same four depletions in combination with the fpr1Δ mutation. None of the depletions were synergistically affected by the fpr1 deletion, showing the specificity of the Hmo1-Kog1 interaction. Therefore, Hmo1 becomes essential when KOG1 is depleted. In conclusion, hmo1Δ is sensitive to both TORC1 inhibition and depletion of an essential TORC1 component.
Hmo1 is a nonessential regulator of RP gene expression.
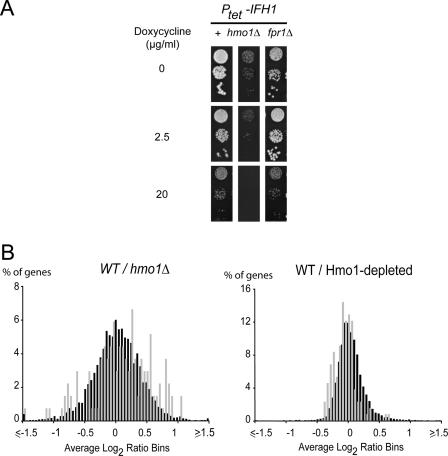
We have established a genetic link between hmo1Δ and some specific RP genes. To identify a link between Hmo1 and transcription of RP genes, we focused on Ifh1, an essential activator of RP gene expression, regulated by TORC1 (12, 46, 63, 72). We inserted the tetracycline-regulatable promoter upstream of the IFH1 gene in a wild-type strain in the absence of Hmo1 or in the absence of Fpr1 (Fig. 4A). Note that the introduction of the promoter is well tolerated in the wild type but seems to be toxic in the absence of Hmo1. Interestingly, mild depletion of Ifh1 results in no detectable growth defect in wild-type cells or in an FPR1 deletion background but is fully lethal in the absence of Hmo1 (Fig. 4A). Therefore, Ifh1 and Hmo1 are strongly linked, suggesting a role for Hmo1 in RP gene expression.
FIG. 4.
Hmo1 is a nonessential activator of RP gene expression. (A) Ifh1 depletion is lethal in the absence of Hmo1. IFH1 was placed under the control of a tetracycline-regulatable promoter (Ptet-IFH1). Growth was scored without (0 μg/ml), with mild (2.5 μg/ml), or with strong (20 μg/ml) repression of IFH1 transcription. Ifh1 depletion was performed in wild-type (+; YAB103-1a), hmo1Δ (YAB103-1a cured of pRS316-HMO1), and fpr1Δ (YAB106-1a cured of pRS316-FPR1) strains. Tenfold serial dilution series were spotted on rich medium in the presence of increasing concentrations of doxycycline. Growth was observed after 5 days of incubation at 30°C. (B) Transcriptome analysis of hmo1 deletion and of Hmo1 depletion. Total RNA was extracted and reverse transcribed from a wild-type strain (BY4741) or from an hmo1-Δ strain (Y06969). Depletion of Hmo1 (Hmo1-depleted) was performed for 6 h by adding 5 μg/ml doxycycline to YAB108-1a; untreated cells were used as a control (WT). The histograms represent the percentages of total transcript (black) and RP gene mRNAs (gray) with given variations compared to the wild-type levels (x axis).
We then investigated RP gene expression in an hmo1Δ background or the effect of Hmo1 and Ifh1 depletion for 6 h using the regulatable tetracycline promoter. Transcriptome analysis was done using microarrays consisting of long oligonucleotides representing all 6,000 yeast genes (see Materials and Methods). Our depletion approach makes use of doxycycline, which has virtually no effect on global gene expression at concentrations used for promoter shutoff (5 μg/ml). Interestingly, hmo1 deletion (Fig. 4B, left) or 6 h of depletion (Fig. 4B, right) have very different consequences. More than 500 genes are twofold up- or down-regulated in the hmo1Δ strain. As previously reported by Hall et al., no significant difference was observed between the mean distribution of RP genes in the hmo1Δ strain and that in the wild type (23) (Fig. 4B, left). Moreover, the Ribi regulon, which has been reported to show transcriptional responses identical to those of RP genes (21, 29), is significantly up-regulated in the hmo1Δ mutant (Kolmogorov-Smirnov test comparing the Ribi regulon to the total population; P = 3.8 × 10−17; data not shown). Conversely, after 6 h of Hmo1 depletion, a very limited number of mRNAs are affected. Taken individually, none of the RP mRNAs is significantly affected (more than twofold). However, when we considered all detected RP mRNAs one class, we observed a mild but significant decrease of RP mRNAs compared to the total mRNA level (Kolmogorov-Smirnov test between RP genes for general response with Hmo1 depletion; P = 3.2 × 10−9). Under these conditions, the Ribi regulon is not affected, showing that we could separate the direct functions of Hmo1 from secondary adaptation mechanisms. We then compared depletion of Ifh1 with that of Hmo1 (see Fig. S4 in the supplemental material). Ifh1 depletion also shows a very significant decrease of RP mRNAs (P = 1.8 × 10−66), with no significant effect on the Ribi regulon. Therefore, we showed that Hmo1 depletion leads to a mild repression of RP gene expression. Due to the limited repression level observed, we cannot distinguish between a general effect on all RP genes and a specific effect on a subset of RP genes.
Hmo1 is required for coordinated expression of RP genes.
The function of Hmo1 in RP gene expression is nonessential. We detect a defect in RP gene expression before cells can adapt to the absence of Hmo1 or by combining an hmo1Δ mutation with Ifh1 depletion. However, we have observed lethality for HMO1 deletion combined with specific RP gene deletions. Most RP genes in yeast are duplicated. Despite the lack of effect on the mean expression levels, we noticed that in the hmo1Δ background, the distribution of RP gene expression levels is significantly broader than in the total mRNA population (Kolmogorov-Smirnov testing for equal distribution; P = 0.009), resulting in mild down- or up-regulation of a limited number of RP genes. For example, we observed a more-than-twofold decrease of three RP gene mRNAs (RPL16B, RPS5, and RPS15). In several instances, when one of the two copies of a given RP gene is down-regulated in the absence of Hmo1, the other copy is genetically linked to the HMO1 deletion. RPL16B is down-regulated in the absence of Hmo1, and the double mutant rpl16AΔ hmo1Δ is lethal, most likely because the essential L16 protein is not produced to a sufficient level.
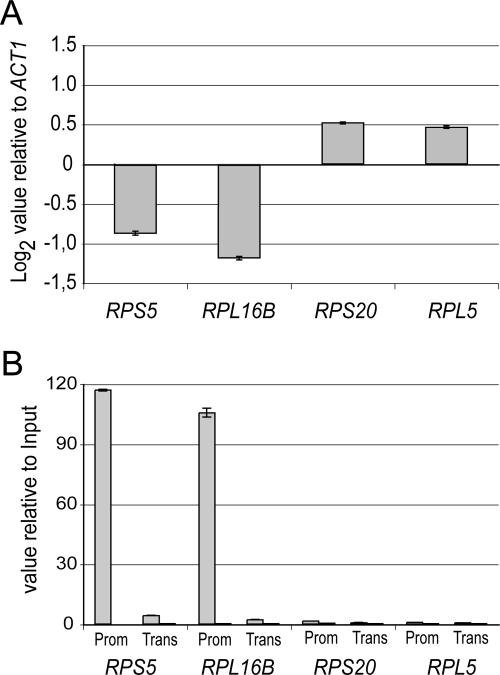
To understand the criterion under which a given RP gene will be up- or down-regulated in the absence of Hmo1, we focused on four RP genes which respond differentially to the absence of Hmo1 (Fig. 5). RPS5 and RPL16B are fourfold down-regulated in the absence of Hmo1, while RPL5 and RPS20 are up-regulated in the hmo1Δ strain, as shown by quantitative PCR after reverse transcription of total cellular mRNAs (Fig. 5A). To investigate promoter occupancy by Hmo1 upstream of these genes, we performed ChIP assays with an Hmo1-TAP strain (Fig. 5B). We observed a highly significant enrichment of Hmo1 on the RPS5 and RPL16B gene promoters and no interaction with RPL5 and RPS20 gene promoters (Fig. 5B). Therefore, we unveiled a correlation between Hmo1 binding to the promoter region and Hmo1-dependent expression on four individual RP genes. Our results suggest that under exponential growth conditions, Hmo1 is activating expression of at least a subset of RP genes, such as RPS5 or RPL16B. In an hmo1Δ background, cells adapt to the absence of Hmo1, resulting in an unbalanced expression of RP genes.
FIG. 5.
Hmo1 is binding to specific RP gene promoters. (A) Effect of an hmo1Δ mutation on the steady-state mRNA levels of RPS5, RPL16B, RPS20, and RPL5. Quantitative PCR was performed on reverse-transcribed total mRNA extracted from hmo1Δ (Y16969) and BY4741 strains. mRNA levels were normalized against ACT1 transcript levels. Log2 values for ratios of mutant versus wild-type levels are represented with standard errors for three independent experiments. (B) Hmo1 binds specifically to the promoter regions of RPS5 and RPL16B. A ChIP experiment was performed using strain HMO1-TAP (OGP126-1a), followed by a two-step purification (see Materials and Methods). Amplicons targeting the promoter of the indicated genes (Prom) contain the putative Fhl1 binding site. Amplicons marked “Trans” target the transcribed regions of the indicated gene.
Hmo1 is implicated in coordinated expression of rRNA and RP gene expression.
Hmo1 is required for Pol I-dependent rRNA expression (20) and for expression of at least a subset of RP genes and directly interacts with both transcription machineries. A recent report has established that deregulated Pol I activity (the “CARA mutant,” bearing an overexpressed Rrn3-Rpa43 fusion protein) can alleviate TORC1-dependent regulation of RP genes (37). This finding suggests that a bona fide Pol I transcription factor might also interact with the Pol II-dependent RP gene expression system. Hmo1 appeared a good candidate for participation in this regulatory pathway.
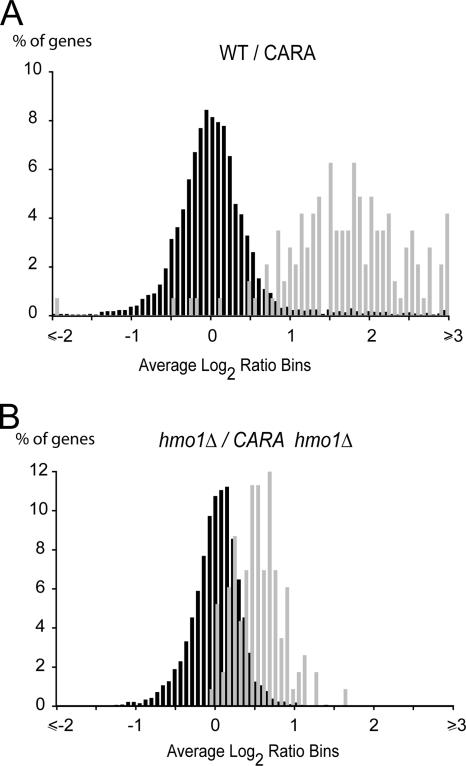
We investigated the global transcriptional response following TORC1 inhibition by rapamycin in the CARA mutant versus that in its wild type (Fig. 6A) or in CARA combined with the absence of Hmo1 versus that in a sole hmo1Δ mutant (Fig. 6B). Confirming published data, we detected 107 RP genes more than twofold up-regulated in the CARA mutant in the presence of rapamycin. Therefore, most if not all RP genes appear up-regulated in the CARA mutant compared to the wild-type levels (11, 37). Interestingly, in the absence of Hmo1, the effect of CARA on global RP gene expression after rapamycin treatment is largely alleviated. We observed only eight genes more than twofold up-regulated (RPP0, RPS0A, RPS9A, RPS22B, RPL18A, RPL26A, RPL31B, and RPL36B). This result demonstrates that Hmo1 is involved in coupling Pol I transcription to RP gene expression after TORC1 inhibition.
FIG. 6.
Hmo1 is involved in TORC1-dependent coregulation of rDNA and RP gene transcription. (A) The CARA mutant results in an up-regulation of RP genes upon TORC1 inhibition. (B) Hmo1 contributes to the coupling of Pol I and RP gene expression. The histograms represent the percentages of total transcript (black) and RP gene mRNAs (gray) with given variations compared to the wild-type (WT) levels (x axis). Total RNA was extracted and reverse transcribed from strains YPH500, CARA, YPH500/hmo1Δ (YAB72), and CARA/hmo1Δ (YAB73) treated for 60 min with 400 ng/ml rapamycin.
In conclusion, we have shown that Hmo1 contributes to the CARA-dependent up-regulation of RP genes under TORC1 inhibition.
DNA binding of Hmo1 to rDNA and RP promoters is abolished by TORC1 inhibition, and Hmo1 is required for TORC1-dependent inhibition of RP gene expression.
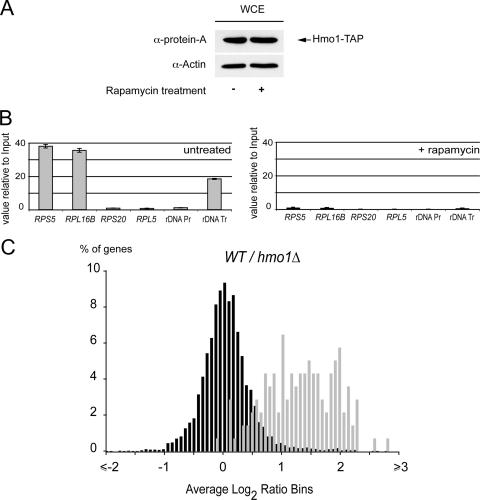
We have shown that Hmo1 contributes to the coupling between Pol I transcription and the transcription of a subset of RP genes by Pol II. To understand how Hmo1 contributes to TORC1 regulation, we investigated the consequence of TORC1 inhibition on the DNA binding properties of Hmo1. Following rapamycin treatment, Hmo1 is not degraded (Fig. 7A) but dissociates from both the RP gene promoters and the rDNA, as shown by Hmo1 ChIP experiments (Fig. 7B). Since the depletion of Hmo1 has a mild inhibitory effect on the expression of the RP genes class, and since Hmo1 dissociates from a subset of RP promoters upon TORC1 inhibition, we speculated that Hmo1 could participate in the inactivation of RP gene transcription after TORC1 inhibition.
FIG. 7.
Hmo1 is required for TORC1-dependent inhibition of RP gene expression. (A) The Hmo1 protein steady-state level is unaffected after 60 min of TORC1 inhibition. Total proteins were extracted from strain HMO1-TAP (OGP126-1a) without or after treatment for 60 min with 400 ng/ml rapamycin. Actin was used as a loading control. (B) Hmo1 DNA binding is rapamycin dependent. ChIP experiments were performed using strain HMO1-TAP (OGP126-1a) without (left) or after treatment with rapamycin (A). (C) RP genes are up-regulated in an hmo1Δ strain compared to wild-type (WT) levels upon TORC1 inhibition. The histogram represents the percentages of total transcript (black) and RP gene mRNAs (gray) with given variations compared to the wild-type levels (x axis). Total RNA was extracted and reverse transcribed from strains YPH500 hmo1Δ (YAB72) and YPH500 treated for 60 min with 400 ng/ml rapamycin.
We investigated RP gene expression levels following TORC1 inhibition by rapamycin treatment in an hmo1Δ background (Fig. 7C). This treatment produces on a wild-type strain a well-defined transcriptional response: down-regulation of ribosome biogenesis (Pol I, II, and III) and up-regulation of defined stress response genes (69). Strikingly, when treated with rapamycin, an hmo1Δ mutant causes a threefold up-regulation of the mean expression level of the RP genes compared to the wild-type level (Fig. 7C). We observed 87 RP genes up-regulated more than twofold compared to the wild-type levels. Therefore, under stress conditions, Hmo1 is required for the repression of the RP gene class. The expression of RP genes in the absence of Hmo1 is largely insensitive to TORC1 inhibition. In conclusion, we have shown that Hmo1 is required for RP gene expression in a TORC1-regulatable manner.
DISCUSSION
In this work, we unveiled a link between Hmo1, the TORC1 pathway, and RP gene expression. Hmo1 binds specific RP gene promoters and activates a subset of RP gene expression in exponentially growing cells. The absence of Hmo1 is viable but results in an unbalanced expression of RP genes. Following TORC1 inhibition, we observed an attenuated down-regulation of expression of RP genes in an hmo1Δ background. Conversely, RP genes are partly insensitive to TORC1 inhibition in a deregulated Pol I mutant (37). This positive regulatory effect of Pol I on RP gene expression is largely lost in the absence of Hmo1. Therefore, Hmo1 contributes, both positively and negatively, to the TORC1-regulatable expression of RP genes.
DNA binding properties of Hmo1.
Hmo1 is a member of the high-mobility-group (HMG) protein family. Detailed biochemical studies of Hmo1 established the presence of two DNA binding motifs and a lysine-rich C-terminal extension (3, 30). Like HMGB1 and -2 in mammals, Hmo1 belongs to the non-sequence-specific group of HMG proteins, but it preferentially binds DNA with altered conformations (30). A recent study by Hall et al. demonstrated an association with RP gene promoters which depends on Rap1 binding (23), and we report here a specific enrichment over the transcribed regions of the rDNA. This specific association may suggest a common topological arrangement between these two sites. The Rap1 consensus site in RP genes is associated with nucleosome depletion (7). Similarly, even if nucleosomes are essential for Pol I transcription (68), the transcribed region of the rDNA was initially described as depleted of nucleosomes (16). A recent study has elegantly demonstrated that nucleosomes are present but are highly dynamic (28). This common feature could suggest a preferential association between Hmo1 and domains with dynamic nucleosomal architectures.
Hmo1 and Pol I transcription.
We have previously demonstrated that Hmo1 is a Pol I transcription factor (20). Here, we now show that Hmo1 interacts strongly with the region of the rDNA transcribed by RNA Pol I. The enrichment at the transcribed region suggests a function in transcription elongation rather than in initiation only. Abnormal accumulation of rRNA precursors in the absence of Hmo1 was observed by members of the Struhl laboratory (23). The imbalanced amounts of pre-rRNAs versus rRNA observed in the hmo1-Δ strain could simply result from the slow-growth phenotype of this strain, since a similar imbalance is also observed in an unrelated Pol II mutant, rpb9-Δ (20). Such an imbalance could also be more directly linked to a Pol I elongation defect since it was found in a mutant affecting the elongation rate of Pol I (64). Therefore, the precise function of Hmo1 on the Pol transcription cycle remains to be elucidated but could involve Pol I elongation. A putative function of Hmo1 during elongation brings into question our assumption that Hmo1 might be the ortholog of animal UBF (20), which is primarily described to be a key transcription factor in Pol I initiation (55). Recent work has, however, established a function of UBF during Pol I elongation (66). hUBF1 acts at multiple steps of the Pol I transcription cycle. With respect to rRNA synthesis, the function of UBF in animals may be recapitulated in yeast by more than one protein: while UAF performs a UBF-like function in initiation, either by stabilization of SL1/CF binding on the Pol I promoter as previously hypothesized (55) or by a function in promoter escape as more recently demonstrated (54), the second function of UBF in elongation may in yeast be fulfilled by Hmo1. Interestingly, the properties of both UBF and Hmo1 are regulated by TOR (66). In yeast, following TORC1 inhibition, Pol I transcription is abolished primarily by dissociation of Rrn3 from Pol I. However, when Rrn3 and Pol I are constitutively associated, a TORC1 inhibition of 35S rRNA production in vivo is still detected (37). This result may suggest a TORC1-dependent regulation of Pol I elongation. Interestingly, hmo1Δ and CARA mutants (11) are both hypersensitive to TORC1 inhibition by rapamycin. We suggest that Hmo1 could be involved in a Pol I-regulatable elongation process.
The function of Hmo1 in RP gene expression is revealed by specific interactions with RP genes.
Recent work has demonstrated that the binding of Hmo1 to most RP gene promoters is not required for global RP gene expression (23). Most RP genes are duplicated and are thought to be redundant in their function (34). We identified some genetic interactions with one of the two RP genes coding for an essential RP. For example, Hmo1 is fully essential when RPS19A or RPS4A is absent. These specific interactions could result from either a divergent function of each of the proteins produced or a distinct transcriptional regulation of each RP gene (34). In our screen, we propose that specific interactions result from a transcriptional defect of the remaining copy in the absence of Hmo1. Note that genetic links could have been missed due to duplication of the remaining copy of the RP gene in the deletion collection (35).
Interplay between RP promoter-bound factors.
RP gene transcription is mainly controlled by the interplay of Rap1, Fhl1, and Ifh1 at RP promoters (46, 62, 63, 72). In the absence of Hmo1, no binding of Fhl1, a functional platform for its associated coactivator Ifh1, is detected on a reporter construct (23). Importantly, we show here that in the absence of Hmo1, Ifh1 is still essential (Fig. 4A), suggesting that RP gene promoters are still dependent on Fhl1/Ifh1 even in an hmo1Δ mutant. Other factors are also bound to RP promoters in exponentially growing cells, such as Esa1 (61) or Sfp1 (29, 43). When TORC1 is inhibited, Ifh1 dissociates from Fhl1 and recruits Crf1, a corepressor (46) which seems to be strain specific (76). Furthermore, Esa1 and Sfp1 are released, and the Rpd3-Sin3 histone deacetylase complex is recruited to the promoters (43, 61). Importantly, like Hmo1, both Sfp1 and the Rpd3-Sin3 complex are required for RP repression during TORC1 inhibition. Sfp1 and Hmo1 are released from a specific subset of RP promoters after TORC1 inhibition and are required for TORC1 repression of RP genes. However, genetic evidence argues against the involvement of Hmo1 and Sfp1 in the same pathway. An sfp1Δ mutant is epistatic to an fhl1Δ mutant, suggesting that Sfp1 acts via Fhl1 and Ifh1 (29). Our result demonstrates a clear synergy between the hmo1Δ mutation and Ifh1 depletion, suggesting that Hmo1 and Sfp1 behave differently on RP genes. We show here that RP genes are expressed in an hmo1Δ mutant but not regulated by TORC1. We propose that Hmo1 can bend DNA of a subset of RP gene promoters via its HMG-B domain and contributes to the recruitment of specific factors. A study just published, focusing on the assembly of regulatory factors on RP gene promoters in the presence or absence of Hmo1, is in full agreement with our conclusion (31). In the absence of Hmo1, Fhl1 recruitment is impaired (23) but not abolished, and other factors which are TORC1 independent in their activity are recruited.
Hmo1 is involved in coupling Pol I-dependent rRNA and Pol II-dependent RP gene transcription.
It has been previously observed that cells are able to adjust transcription of rRNA in response to reduced production of RP (62). Recent work has shown that constitutive Pol I activation serves as an activating signal for RP expression during TORC1 inhibition (37). In both studies, Pol I and Pol II can adjust their respective activities but the molecular mechanism remains unclear. Hmo1 appears to be a good candidate for contribution to this cross talk between the Pol I and the Pol II transcription apparatus, since it is binding to both the rDNA transcribed region and RP gene promoters and since this binding is regulated by the TORC1 complex. Additionally, our data suggest that Hmo1 is directly involved in Pol I and RP gene expression in vivo. In conclusion, we suggest that Hmo1 contributes to the TORC1-regulatable coexpression of ribosomal components in yeast.
Supplementary Material
Acknowledgments
We thank Christophe Charles for strains YPH500 and CARA and for sharing unpublished data, George P. Livi for the plasmid bearing the TOR1-1 allele, Yoshiko Kikuchi for the plasmid bearing TOM1, Frédéric Devaux for his help in the transcriptome study, Florence Hantraye for the transcriptome normalization procedure, A. Reimann for his help concerning the quantitative PCR, and Y. Henry, M. Mhlanga, K. Bystrincky, and F. Feueurbach-Fournier for critical reading of the manuscript.
A.B.B. was supported by fellowships from the French Ministry of Research and Technology (MNRT) and the German Academic Exchange Service (DAAD). This work was supported by an ATIP grant from CNRS and the Association Nationale pour la Recherche.
Footnotes
Published ahead of print on 17 September 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alekseev, S. Y., S. V. Kovaltsova, I. V. Fedorova, L. M. Gracheva, T. A. Evstukhina, V. T. Peshekhonov, and V. G. Korolev. 2002. HSM2 (HMO1) gene participates in mutagenesis control in yeast Saccharomyces cerevisiae. DNA Repair 1:287-297. [DOI] [PubMed] [Google Scholar]
- 2.Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauerle, K. T., E. Kamau, and A. Grove. 2006. Interactions between N- and C-terminal domains of the Saccharomyces cerevisiae high-mobility group protein HMO1 are required for DNA bending. Biochemistry 45:3635-3645. [DOI] [PubMed] [Google Scholar]
- 4.Belli, G., E. Gari, M. Aldea, and E. Herrero. 1998. Functional analysis of yeast essential genes using a promoter-substitution cassette and the tetracycline-regulatable dual expression system. Yeast 14:1127-1138. [DOI] [PubMed] [Google Scholar]
- 5.Belli, G., E. Gari, L. Piedrafita, M. Aldea, and E. Herrero. 1998. An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res. 26:942-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bender, A., and J. R. Pringle. 1991. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:1295-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein, B. E., C. L. Liu, E. L. Humphrey, E. O. Perlstein, and S. L. Schreiber. 2004. Global nucleosome occupancy in yeast. Genome Biol. 5:R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bier, M., S. Fath, and H. Tschochner. 2004. The composition of the RNA polymerase I transcription machinery switches from initiation to elongation mode. FEBS Lett. 564:41-46. [DOI] [PubMed] [Google Scholar]
- 9.Briand, J. F., F. Navarro, O. Gadal, and P. Thuriaux. 2001. Cross talk between tRNA and rRNA synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:189-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cafferkey, R., P. R. Young, M. M. McLaughlin, D. J. Bergsma, Y. Koltin, G. M. Sathe, L. Faucette, W. K. Eng, R. K. Johnson, and G. P. Livi. 1993. Dominant missense mutations in a novel yeast protein related to mammalian phosphatidylinositol 3-kinase and VPS34 abrogate rapamycin cytotoxicity. Mol. Cell. Biol. 13:6012-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chedin, S., A. Laferte, T. Hoang, D. L. Lafontaine, M. Riva, and C. Carles. 2007. Is ribosome synthesis controlled by pol I transcription? Cell Cycle 6:11-15. [DOI] [PubMed] [Google Scholar]
- 12.Cherel, I., and P. Thuriaux. 1995. The IFH1 gene product interacts with a fork head protein in Saccharomyces cerevisiae. Yeast 11:261-270. [DOI] [PubMed] [Google Scholar]
- 13.Choe, S. Y., M. C. Schultz, and R. H. Reeder. 1992. In vitro definition of the yeast RNA polymerase I promoter. Nucleic Acids Res. 20:279-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Claypool, J. A., S. L. French, K. Johzuka, K. Eliason, L. Vu, J. A. Dodd, A. L. Beyer, and M. Nomura. 2004. Tor pathway regulates Rrn3p-dependent recruitment of yeast RNA polymerase I to the promoter but does not participate in alteration of the number of active genes. Mol. Biol. Cell 15:946-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cost, G. J., and J. D. Boeke. 1996. A useful colony colour phenotype associated with the yeast selectable/counter-selectable marker MET15. Yeast 12:939-941. [DOI] [PubMed] [Google Scholar]
- 16.Dammann, R., R. Lucchini, T. Koller, and J. M. Sogo. 1993. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 21:2331-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desai, N., J. Lee, R. Upadhya, Y. Chu, R. D. Moir, and I. M. Willis. 2005. Two steps in Maf1-dependent repression of transcription by RNA polymerase III. J. Biol. Chem. 280:6455-6462. [DOI] [PubMed] [Google Scholar]
- 18.Dolinski, K. J., and J. Heitman. 1999. Hmo1p, a high mobility group 1/2 homolog, genetically and physically interacts with the yeast FKBP12 prolyl isomerase. Genetics 151:935-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fath, S., P. Milkereit, G. Peyroche, M. Riva, C. Carles, and H. Tschochner. 2001. Differential roles of phosphorylation in the formation of transcriptional active RNA polymerase I. Proc. Natl. Acad. Sci. USA 98:14334-14339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gadal, O., S. Labarre, C. Boschiero, and P. Thuriaux. 2002. Hmo1, an HMG-box protein, belongs to the yeast ribosomal DNA transcription system. EMBO J. 21:5498-5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldstein, A. L., and J. H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541-1553. [DOI] [PubMed] [Google Scholar]
- 23.Hall, D. B., J. T. Wade, and K. Struhl. 2006. An HMG protein, Hmo1, associates with promoters of many ribosomal protein genes and throughout rRNA gene locus in Saccharomyces cerevisiae. Mol. Cell. Biol. 26:3672-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hannan, K. M., Y. Brandenburger, A. Jenkins, K. Sharkey, A. Cavanaugh, L. Rothblum, T. Moss, G. Poortinga, G. A. McArthur, R. B. Pearson, and R. D. Hannan. 2003. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol. Cell. Biol. 23:8862-8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heitman, J., N. R. Movva, and M. N. Hall. 1991. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253:905-909. [DOI] [PubMed] [Google Scholar]
- 26.Hermann-Le Denmat, S., M. Werner, A. Sentenac, and P. Thuriaux. 1994. Suppression of yeast RNA polymerase III mutations by FHL1, a gene coding for a fork head protein involved in rRNA processing. Mol. Cell. Biol. 14:2905-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ingles, C. J., H. J. Himmelfarb, M. Shales, A. L. Greenleaf, and J. D. Friesen. 1984. Identification, molecular cloning, and mutagenesis of Saccharomyces cerevisiae RNA polymerase genes. Proc. Natl. Acad. Sci. USA 81:2157-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones, H. S., J. Kawauchi, P. Braglia, C. M. Alen, N. A. Kent, and N. J. Proudfoot. 2007. RNA polymerase I in yeast transcribes dynamic nucleosomal rDNA. Nat. Struct. Mol. Biol. 14:123-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jorgensen, P., I. Rupes, J. R. Sharom, L. Schneper, J. R. Broach, and M. Tyers. 2004. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 18:2491-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamau, E., K. T. Bauerle, and A. Grove. 2004. The Saccharomyces cerevisiae high mobility group box protein HMO1 contains two functional DNA binding domains. J. Biol. Chem. 279:55234-55240. [DOI] [PubMed] [Google Scholar]
- 31.Kasahara, K., K. Ohtsuki, S. Ki, K. Aoyama, H. Takahashi, T. Kobayashi, K. Shirahige, and T. Kokubo. 2007. Assembly of regulatory factors on rRNA and ribosomal protein genes in Saccharomyces cerevisiae. Mol. Cell. Biol. [DOI] [PMC free article] [PubMed]
- 32.Keys, D. A., B. S. Lee, J. A. Dodd, T. T. Nguyen, L. Vu, E. Fantino, L. M. Burson, Y. Nogi, and M. Nomura. 1996. Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev. 10:887-903. [DOI] [PubMed] [Google Scholar]
- 33.Kim, H., and D. M. Livingston. 2006. A high mobility group protein binds to long CAG repeat tracts and establishes their chromatin organization in Saccharomyces cerevisiae. J. Biol. Chem. 281:15735-15740. [DOI] [PubMed] [Google Scholar]
- 34.Komili, S., and F. P. Roth. 2007. Genetic interaction screens advance in reverse. Genes Dev. 21:137-142. [DOI] [PubMed] [Google Scholar]
- 35.Koszul, R., S. Caburet, B. Dujon, and G. Fischer. 2004. Eucaryotic genome evolution through the spontaneous duplication of large chromosomal segments. EMBO J. 23:234-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kushnirov, V. V. 2000. Rapid and reliable protein extraction from yeast. Yeast 16:857-860. [DOI] [PubMed] [Google Scholar]
- 37.Laferté, A., E. Favry, A. Sentenac, M. Riva, C. Carles, and S. Chedin. 2006. The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes Dev. 20:2030-2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, H., C. K. Tsang, M. Watkins, P. G. Bertram, and X. F. Zheng. 2006. Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature 442:1058-1061. [DOI] [PubMed] [Google Scholar]
- 39.Lin, C. W., B. Moorefield, J. Payne, P. Aprikian, K. Mitomo, and R. H. Reeder. 1996. A novel 66-kilodalton protein complexes with Rrn6, Rrn7, and TATA-binding protein to promote polymerase I transcription initiation in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:6436-6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loewith, R., E. Jacinto, S. Wullschleger, A. Lorberg, J. L. Crespo, D. Bonenfant, W. Oppliger, P. Jenoe, and M. N. Hall. 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10:457-468. [DOI] [PubMed] [Google Scholar]
- 41.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 42.Lu, J., R. Kobayashi, and S. J. Brill. 1996. Characterization of a high mobility group 1/2 homolog in yeast. J. Biol. Chem. 271:33678-33685. [DOI] [PubMed] [Google Scholar]
- 43.Marion, R. M., A. Regev, E. Segal, Y. Barash, D. Koller, N. Friedman, and E. K. O'Shea. 2004. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc. Natl. Acad. Sci. USA 101:14315-14322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin, D. E., and M. N. Hall. 2005. The expanding TOR signaling network. Curr. Opin. Cell Biol. 17:158-166. [DOI] [PubMed] [Google Scholar]
- 45.Martin, D. E., T. Powers, and M. N. Hall. 2006. Regulation of ribosome biogenesis: where is TOR? Cell Metab. 4:259-260. [DOI] [PubMed] [Google Scholar]
- 46.Martin, D. E., A. Soulard, and M. N. Hall. 2004. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 119:969-979. [DOI] [PubMed] [Google Scholar]
- 47.Memet, S., M. Gouy, C. Marck, A. Sentenac, and J. M. Buhler. 1988. RPA190, the gene coding for the largest subunit of yeast RNA polymerase A. J. Biol. Chem. 263:2830-2839. [PubMed] [Google Scholar]
- 48.Milkereit, P., and H. Tschochner. 1998. A specialized form of RNA polymerase I, essential for initiation and growth-dependent regulation of rRNA synthesis, is disrupted during transcription. EMBO J. 17:3692-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller, G., K. I. Panov, J. K. Friedrich, L. Trinkle-Mulcahy, A. I. Lamond, and J. C. Zomerdijk. 2001. hRRN3 is essential in the SL1-mediated recruitment of RNA Polymerase I to rRNA gene promoters. EMBO J. 20:1373-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mnaimneh, S., A. P. Davierwala, J. Haynes, J. Moffat, W. T. Peng, W. Zhang, X. Yang, J. Pootoolal, G. Chua, A. Lopez, M. Trochesset, D. Morse, N. J. Krogan, S. L. Hiley, Z. Li, Q. Morris, J. Grigull, N. Mitsakakis, C. J. Roberts, J. F. Greenblatt, C. Boone, C. A. Kaiser, B. J. Andrews, and T. R. Hughes. 2004. Exploration of essential gene functions via titratable promoter alleles. Cell 118:31-44. [DOI] [PubMed] [Google Scholar]
- 51.Moss, T. 2004. At the crossroads of growth control; making ribosomal RNA. Curr. Opin. Genet. Dev. 14:210-217. [DOI] [PubMed] [Google Scholar]
- 52.Moss, T., and V. Y. Stefanovsky. 2002. At the center of eukaryotic life. Cell 109:545-548. [DOI] [PubMed] [Google Scholar]
- 53.Ono, B., N. Ishii, S. Fujino, and I. Aoyama. 1991. Role of hydrosulfide ions (HS-) in methylmercury resistance in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 57:3183-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Panov, K. I., J. K. Friedrich, J. Russell, and J. C. Zomerdijk. 2006. UBF activates RNA polymerase I transcription by stimulating promoter escape. EMBO J. 25:3310-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paule, M. R. 1998. Transcription of ribosomal genes by eukaryotic RNA polymerase I. Landes Bioscience, Austin, TX.
- 56.Peyroche, G., P. Milkereit, N. Bischler, H. Tschochner, P. Schultz, A. Sentenac, C. Carles, and M. Riva. 2000. The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. EMBO J. 19:5473-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pluta, K., O. Lefebvre, N. C. Martin, W. J. Smagowicz, D. R. Stanford, S. R. Ellis, A. K. Hopper, A. Sentenac, and M. Boguta. 2001. Maf1p, a negative effector of RNA polymerase III in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:5031-5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prescott, E. M., Y. N. Osheim, H. S. Jones, C. M. Alen, J. G. Roan, R. H. Reeder, A. L. Beyer, and N. J. Proudfoot. 2004. Transcriptional termination by RNA polymerase I requires the small subunit Rpa12p. Proc. Natl. Acad. Sci. USA 101:6068-6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reinke, A., S. Anderson, J. M. McCaffery, J. Yates III, S. Aronova, S. Chu, S. Fairclough, C. Iverson, K. P. Wedaman, and T. Powers. 2004. TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J. Biol. Chem. 279:14752-14762. [DOI] [PubMed] [Google Scholar]
- 60.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 61.Rohde, J. R., and M. E. Cardenas. 2003. The Tor pathway regulates gene expression by linking nutrient sensing to histone acetylation. Mol. Cell. Biol. 23:629-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rudra, D., Y. Zhao, and J. R. Warner. 2005. Central role of Ifh1p-Fhl1p interaction in the synthesis of yeast ribosomal proteins. EMBO J. 24:533-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schawalder, S. B., M. Kabani, I. Howald, U. Choudhury, M. Werner, and D. Shore. 2004. Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature 432:1058-1061. [DOI] [PubMed] [Google Scholar]
- 64.Schneider, D. A., S. L. French, Y. N. Osheim, A. O. Bailey, L. Vu, J. Dodd, J. R. Yates, A. L. Beyer, and M. Nomura. 2006. RNA polymerase II elongation factors Spt4p and Spt5p play roles in transcription elongation by RNA polymerase I and rRNA processing. Proc. Natl. Acad. Sci. USA 103:12707-12712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shou, W., K. M. Sakamoto, J. Keener, K. W. Morimoto, E. E. Traverso, R. Azzam, G. J. Hoppe, R. M. Feldman, J. DeModena, D. Moazed, H. Charbonneau, M. Nomura, and R. J. Deshaies. 2001. Net1 stimulates RNA polymerase I transcription and regulates nucleolar structure independently of controlling mitotic exit. Mol. Cell 8:45-55. [DOI] [PubMed] [Google Scholar]
- 66.Stefanovsky, V., F. Langlois, T. Gagnon-Kugler, L. I. Rothblum, and T. Moss. 2006. Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling. Mol. Cell 21:629-639. [DOI] [PubMed] [Google Scholar]
- 67.Stettler, S., N. Chiannilkulchai, S. Hermann-Le Denmat, D. Lalo, F. Lacroute, A. Sentenac, and P. Thuriaux. 1993. A general suppressor of RNA polymerase I, II and III mutations in Saccharomyces cerevisiae. Mol. Gen. Genet. 239:169-176. [DOI] [PubMed] [Google Scholar]
- 68.Tongaonkar, P., S. L. French, M. L. Oakes, L. Vu, D. A. Schneider, A. L. Beyer, and M. Nomura. 2005. Histones are required for transcription of yeast rRNA genes by RNA polymerase I. Proc. Natl. Acad. Sci. USA 102:10129-10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsang, C. K., and X. F. Zheng. 2007. TOR-in(g) the nucleus. Cell Cycle 6:25-29. [DOI] [PubMed] [Google Scholar]
- 70.Tschochner, H., and E. Hurt. 2003. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 13:255-263. [DOI] [PubMed] [Google Scholar]
- 71.Upadhya, R., J. Lee, and I. M. Willis. 2002. Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Mol. Cell 10:1489-1494. [DOI] [PubMed] [Google Scholar]
- 72.Wade, J. T., D. B. Hall, and K. Struhl. 2004. The transcription factor Ifh1 is a key regulator of yeast ribosomal protein genes. Nature 432:1054-1058. [DOI] [PubMed] [Google Scholar]
- 73.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, N. Liebundguth, D. J. Lockhart, A. Lucau-Danila, M. Lussier, N. M'Rabet, P. Menard, M. Mittmann, C. Pai, C. Rebischung, J. L. Revuelta, L. Riles, C. J. Roberts, P. Ross-MacDonald, B. Scherens, M. Snyder, S. Sookhai-Mahadeo, R. K. Storms, S. Veronneau, M. Voet, G. Volckaert, T. R. Ward, R. Wysocki, G. S. Yen, K. Yu, K. Zimmermann, P. Philippsen, M. Johnston, and R. W. Davis. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 74.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 75.Zenklusen, D., P. Vinciguerra, Y. Strahm, and F. Stutz. 2001. The yeast hnRNP-like proteins Yra1p and Yra2p participate in mRNA export through interaction with Mex67p. Mol. Cell. Biol. 21:4219-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao, Y., K. B. McIntosh, D. Rudra, S. Schawalder, D. Shore, and J. R. Warner. 2006. Fine-structure analysis of ribosomal protein gene transcription. Mol. Cell. Biol. 26:4853-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.