Abstract
The insulator element at the 5′ end of the chicken β-globin locus acts as a barrier, protecting transgenes against silencing effects of adjacent heterochromatin. We showed earlier that the transcription factor USF1 binds within the insulator and that this site is important for generating in adjacent nucleosomes histone modifications associated with active chromatin and, by inference, with barrier function. To understand the mechanism of USF1 action, we have characterized USF1-containing complexes. USF1 interacts directly with the histone H4R3-specific methyltransferase PRMT1. USF1, PRMT1, and the histone acetyltransferases (HATs) PCAF and SRC-1 form a complex with both H4R3 histone methyltransferase and HAT activities. Small interfering RNA downregulation of USF1 results in localized loss of H4R3 methylation, and other histone modifications associated with euchromatin, at the insulator. A dominant negative peptide that interferes with USF1 binding to DNA causes silencing of an insulated reporter construct, indicating abolition of barrier function. These results show that USF1 plays a direct role in maintaining the barrier, supporting a model in which the insulator works as a barrier by maintaining a local environment of active chromatin.
Within the nucleus, heterochromatic and euchromatic domains may lie next to one another. In the absence of constraints, a variety of mechanisms may allow the extension of repressive heterochromatic structures into adjacent euchromatin (14, 20, 28). Barrier insulators are capable of preventing this heterochromatic encroachment. They are distinct in properties and composition from enhancer-blocking insulators, which prevent inappropriate interactions of neighboring gene systems (6, 41).
Although there may be more than one way to block the propagation of condensed chromatin structures into an active chromatin domain, experiments both in yeast (30) and in vertebrates (42) have suggested that elements which recruit high levels of histone modifications associated with transcriptional activation may establish such a barrier. The 5′HS4 insulator at the 5′ end of the chicken β-globin locus lies immediately downstream of an ∼16-kb condensed chromatin domain and is thus in a position to protect the globin locus against the extension downstream of this heterochromatic region (24, 25, 34). The nucleosomes adjacent to the insulator site are highly enriched in active histone modifications, including acetylation of histones H3 and H4 and methylation of lys4 on histone H3 and of arg3 on H4 (15, 24, 25). Consistent with barrier function, we have shown that a 250-bp core sequence, derived from the β-globin insulator, can protect a stably integrated transgene from silencing by endogenous heterochromatic sequences at the site of integration (32, 36). This barrier assay enabled us to show that a single factor binding site within the core was required to maintain high levels of histone acetylation as well as H3K4 methylation over the protected sequences and that this site was essential for insulation. We identified the factor as the regulatory protein USF1 and showed that it bound to the site as a heterodimer with USF2 and recruited histone acetyltransferases (HATs) as well as the H3K4 methyltransferase SET 7/9 (42).
As a next step, we therefore sought both to confirm the role of USF1 in barrier function and to identify the native USF1 complexes that are involved. A combination of gel filtration chromatography, coimmunoprecipitation, and immunopurification assays allowed us to demonstrate a direct interaction between USF1 and the arginine methyltransferase PRMT1. The ∼400-kDa complex that contains USF1 and PRMT1 also contains USF2 as well as the HATs PCAF and SRC-1. We had previously shown that PRMT1 plays a major role in H4R3 methylation, which in turn is necessary for a number of histone acetylation events over the entire folate receptor/globin domain (15).
We also provide both indirect and direct evidence for the role of USF1 in barrier insulator function. We show first that RNA interference-mediated downregulation of USF1 results in the appearance at sites downstream over the endogenous β-globin locus of H3K27 trimethylation, a mark of heterochromatin formation. These sites in wild-type cells exhibit unusually low levels of this modification. This is consistent with the view that the wild-type insulator protects against such incursions and that USF1, together with PRMT1 and other factors recruited by USF1, is essential to such protection. We have also used our standard barrier assay to study directly the effect of depletion of USF1 binding at the insulator. We find that expression of a dominant negative protein that interferes with binding results in complete loss of ability of the insulator element to protect against heterochromatic silencing.
MATERIALS AND METHODS
Cell lines and small interfering RNA (siRNA) knockdown.
6C2 cells were maintained as described previously (34). Ten-day chicken embryonic erythrocytes were harvested from 10-day embryos from fertilized White Leghorn chicken eggs (Truslow Farms, Chestertown, MD). Five 65-bp oligonucleotide duplexes that specifically target different portions of the chicken USF1 were cloned into a pSilencer3.1-H1 hygro vector, following the manufacturer's instructions (Ambion, Austin, TX). The vectors were transfected into 6C2 cells by electroporation. Stable transfectants were selected in the presence of 1 mg/ml of hygromycin B. The stable hairpin-expressing cell lines were analyzed for USF expression by Western blotting.
Purification of USF1 complexes and its associated HMTs.
Twenty g of nuclear protein was extracted from 2 liters of whole chicken blood (Pelfreez Biologicals) in 20 mM HEPES (pH 7.9), 420 mM NaCl, 0.2 mM EDTA, and 1 mM dithiothreitol (DTT) as described previously (5). The nuclear extract was adjusted to 20 mM HEPES (pH 7.9), 125 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, and 10% glycerol. Ten-ml aliquots of extracts were fractionated on a Sephacryl S-300 HR column (Amersham, Piscataway, NJ). The eluted fractions were analyzed for DNA site footprint IV (FIV) DNA binding activity and histone methyltransferase (HMT) activities.
Purification of a USF1 complex was done in a stably transduced HeLa S3 cell line with an N-terminal FLAG-hemagglutinin (HA) double-tagged chicken USF1 construct using a system previously described (29). Positive expressing cells also coexpress the interleukin-2 receptor (IL-2R), and multiple rounds of selection by anti-IL-2R magnetic sorting were conducted to obtain >95% stably expressing cells. To make soluble nuclear extract, cells were washed once and swollen in hypotonic buffer (10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, 0.2 mM phenylmethylsulfonyl fluoride [PMSF], and protease inhibitors) for 10 min on ice followed by homogenization 10 times with a “loose” pestle. Nuclei were centrifuged at 2,000 × g for 10 min at 4°C, and nuclear pellets were resuspended in 0.5 volumes low-salt buffer (20 mM HEPES, pH 7.9, 20 mM KCl, 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.2 mM PMSF, and protease inhibitors). High-salt buffer (20 mM HEPES, pH 7.9, 1.2 M KCl, 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM DTT, protease inhibitors) was slowly added (0.5 volume; ∼0.42 M KCl final), nuclei were rotated for 30 min at 4°C, and insoluble material was removed by centrifugation at 14,000 × g for 15 min. Soluble extract was then dialyzed in buffer containing 20 mM HEPES, pH 7.9, 100 mM KCl, 20% glycerol, 0.2 mM EDTA, 0.5 mM DTT, 0.2 mM PMSF, and protease inhibitors. Complexes were first purified with M2-anti-FLAG resin (Sigma) by incubating 300 μl packed resin with the nuclear extracts for 4 hours at 4°C, which were then washed three times with 10 ml wash buffer (20 mM Tris, pH 7.9, 0.1 M KCl, 5 mM MgCl2, 10% glycerol, 0.1% Tween 20, 1 mM DTT, protease inhibitors) in a 10-ml disposable column followed by two elutions with 300 μl of wash buffer plus 0.16 μg/ml FLAG peptide. The eluted material was resolved on a 4 to 12% NuPAGE gel in morpholineethanesulfonic acid buffer (Invitrogen) and Coomassie stained. Bands were identified by liquid chromatography-tandem mass spectrometry identification at the Harvard University Taplin protein sequencing facility.
Plasmids.
cDNA encoding human and chicken USF1 was isolated by reverse transcription-PCR using gene-specific primers. The PCR product was then cloned into a pCRII-TOPO vector (Invitrogen) and subjected to sequencing. Full-length cDNA and its truncated fragments were then transferred into a pGEX-5X-1 vector. PRMT1 cDNA was a kind gift from Harvey R. Herschman. The plasmid pCMV-AUSF1 was obtained from Charles Vinson (NIH).
HMT, gel retardation, and GST pull-down assays.
The column fractions were incubated with chicken core histones or recombinant H4 in a total volume of 30 μl containing 20 mM Tris-HCl (pH 8.0), 4 mM EDTA, 1 mM PMSF, 0.5 mM DTT, and 1 μl [3H]S-adenosylmethionine (15 Ci/mmol; NEN Life Science Products) at 30°C for 1 h. Gel retardation assays were carried out as described previously (42). 35S-labeled PRMT1 was synthesized using a coupled in vitro transcription-translation system in reticulocyte lysates (Promega) in the presence of translation-grade [35S]methionine (Amersham). The glutathione S-transferase (GST) pull-down assay was performed as described elsewhere (16).
Immunoprecipitation and Western blot analysis.
Nuclear extracts were prepared as previously described (5). Anti-USF1 (H-86; a kind gift from Emery H. Bresnick, University of Wisconsin) and anti-PRMT1 (G-13) antibodies were purchased from Santa Cruz Biotechnology, Santa Cruz, CA. Immunoprecipitations were performed using a Seize X protein A immunoprecipitation kit following the manufacturer's instructions (Pierce, Rockford, IL). One ml of nuclear extract (from 5 × 107 to 1 × 108 adult chicken erythrocytes) dialyzed into immunoprecipitation (IP) buffer (20 mM HEPES, pH 7.9, 125 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 10% glycerol) was used per IP mixture. Nuclear extract aliquots were precleared with 5 μg of normal rabbit immunoglobulin G (IgG) for 2 h at 4°C. A 200-μg aliquot of anti-USF1 antibody or normal rabbit IgG (Santa Cruz Biotechnology) was immobilized and cross-linked to protein A beads (Invitrogen). Nuclear extracts were incubated with antibody-immobilized protein A and then washed briefly four times with phosphate-buffered saline (PBS) followed by a wash with PBS for 30 min at 4°C with agitation. Immunoprecipitated proteins were eluted with 200 μl of elution buffer (containing primary amine, pH 2.8). A 15-μl volume of this sample was fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electrotransferred to polyvinylidene difluoride membranes. Blots were incubated with the indicated primary antibodies and then with horseradish peroxidase-conjugated secondary antibodies. Proteins were visualized by enhanced chemiluminescence (Pierce, Rockford, IL).
ChIP assay and barrier analysis.
Chromatin immunoprecipitation (ChIP) assays for the modifications of core histones were performed in the absence of formaldehyde cross-linking as described previously (25) using antibodies specific for various posttranslational modifications of histone tails (anti-histone H4 dimethyl R3 [07-213], anti-histone H3 acetyl K9/acetyl K14 [06-599], anti-histone H4 tetra-acetyl K5/K8/K12/K16 [06-598], and anti-histone H3 dimethyl K4 [07-030] [all from Upstate Biotechnology, Lake Placid, NY]; anti-histone H3 trimethyl K27 was a kind gift from Thomas Jenuwein). The difference was determined by the following equation: [(PCR signal in the IP)/(DNA concentration in the IP)]/[(PCR signal in the input)/(DNA concentration in the input)]. The polymerase II (Pol II) ChIP assays were performed with formaldehyde cross-linked chromatin using antibodies specific for Ser 2 (H5) or Ser 5 (H14) phosphorylated RNA polymerase II (Covance). The difference was determined by measuring the PCR signal of the specific IP and preimmune IP and comparing their ratio to that for a site within the 16-kb condensed chromatin region.
Cells were stained with fluorescein isothiocyanate (FITC)-anti-human CD25 (eBioscience, San Diego, CA). Immunostained cells were analyzed on a flow cytometer. Fluorescence-activated cell sorting (FACS) was performed on days 0 and 15 in the absence of selection.
RESULTS
Analysis of HMT activities associated with USF1 in erythroid cells.
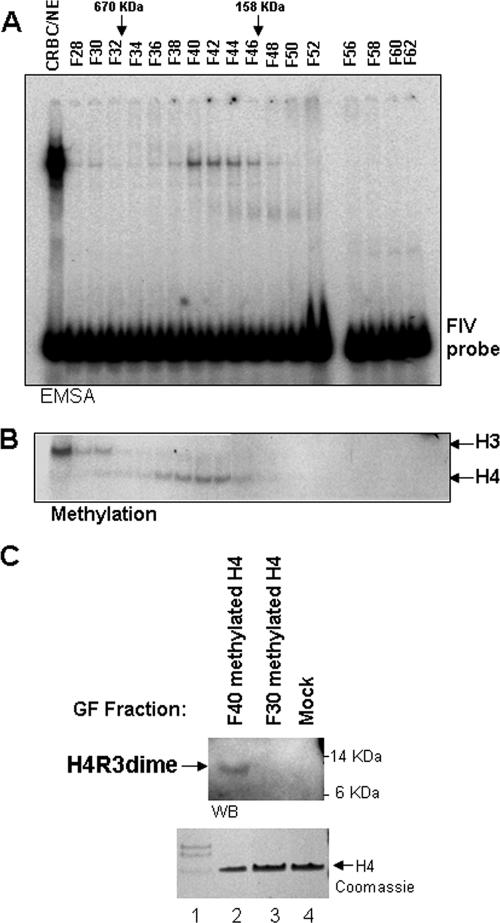
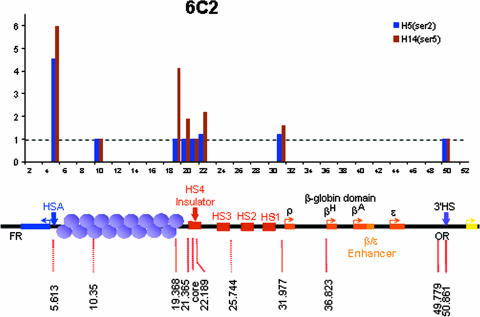
We had shown earlier that USF1 is a critical component of the 5′HS4 β-globin insulator. It binds as a heterodimer with USF2 to DNA site FIV within the insulator element (42). Either deletion of this site or downregulation of USF1 protein abolishes the ability of the insulator element to modify histones in the immediately surrounding nucleosomes. The data suggest that USF1 orchestrates recruitment of multiple histone modification activities at the insulator. Consistent with this view, ChIP studies showed that the histone acetyltransferases pCAF, CBP, and p300 were localized at the insulator (42). To understand further the molecular mechanism by which USF1 mediates formation of a chromatin barrier, we searched for USF1-containing nuclear complexes that might include histone-modifying enzymes. Nuclear extract from chicken erythrocytes was fractionated using a Sephacryl S-300 HR gel filtration column (Amersham Biosciences), and electrophoretic mobility shift assays were carried out to detect complexes that contained USF1, using a DNA probe (FIV) from 5′HS4 that carried the USF1 binding site. The distribution of USF activity suggested the existence of at least two discrete complexes that migrated on the column at approximately 1.8 MDa and 400 kDa (Fig. 1A).
FIG. 1.
Histone methyltransferases associate with the FIV DNA-binding complex. (A) Fractionation of nuclear extract from chicken erythrocytes (CRBC/NE). Column fractions derived from the Sephacryl S-300 HR column were analyzed. An electrophoretic mobility shift assay with column fractions using 32P-labeled FIV oligonucleotide duplexes shows that USF1 DNA-binding activity is present in two complexes of approximately 1.8 MDa and 180 to 400 kDa by gel filtration. (B) Fluorogram of the same column fractions used in the HMT assay. Each USF1 complex coelutes with different HMT activities with specificity for H3 or H4 separately. (C) Association of histone H4R3 methylation with USF1 activity. Recombinant histone H4 was subjected to methylation with column fractions number 30 and 40, representing two different USF1 DNA-binding complexes. (Top) Western blotting analysis with a dimethyl H4R3-specific antibody. (Bottom) Coomassie stain of recombinant histone H4 and core histone isolated from chicken used in the Western blot assay.
We began by assaying the fractions for HMTs and found that each complex cofractionated with a distinct HMT activity. H3-specific histone methyltransferase activity was found specifically in fractions containing the 1.8-MDa complex (Fig. 1B). In contrast, the 400-kDa complex overlapped with a histone H4-specific methyltransferase activity (Fig. 1B). To identify positively the USF1-associated H4-specific modifications involved in the latter complex, core histones incubated with peak fraction 40 were tested for reactivity with antibody specific for asymmetrically methylated dimethyl H4R3. A positive reaction was detected with this fraction, but not with fraction 30 (Fig. 1C, compare lanes 2 and 3). Under the same conditions, we did not detect methylation from a fraction of mock-methylated H4 substrates (Fig. 1C, lane 4). In addition, the antibody specific to methyl-H4K20 did not detect methylated K20 in either fraction (data not shown). We conclude that the small USF1 complex migrating in the 400-kDa range specifically cofractionates with H4R3-specific methyltransferase activity.
USF1 interacts with PRMT1 in vitro and in vivo.
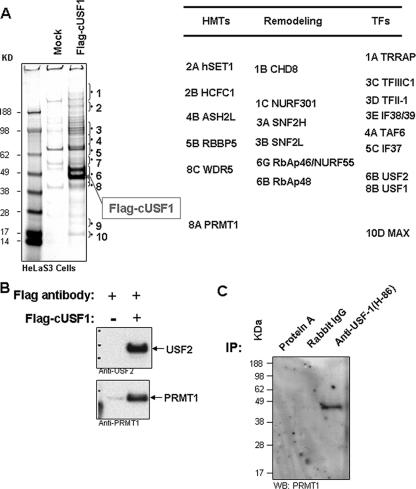
To further purify and characterize the USF1-containing chromatin-modifying complexes, we expressed FLAG/HA epitope-tagged chicken USF1 in HeLa cells. The FLAG/HA-tagged USF1 and its associated complexes were purified through a FLAG epitope-specific immuno-affinity column followed by FLAG peptide elution. In order to monitor the specificity of purification, nuclear extracts from HeLa cells without expression of fusion protein were used in a parallel mock purification. The resulting polypeptides were resolved by SDS-PAGE and identified by mass spectrometry (Fig. 2A). The majority of polypeptides identified are involved in the pathways that regulate transcription as well as chromatin structure and function. They fall into the following categories: (i) the USF1/USF2 heterodimer; (ii) PRMT1, the only HMT known to target H4R3; (iii) MRE11, involved in DNA double-stranded break repair; (iv) transcription factors; (v) chromatin remodeling complexes; (vi) an H3K4-specific HMT complex; (vii) histone deacetylase complexes. In this report, we will focus on the PRMT1- and HAT-containing complexes because of their relevance to the establishment of active chromatin structure within the globin locus and the function of barrier insulators.
FIG. 2.
Identification of the USF1-interacting proteins. (A) USF1-interacting polypeptides were purified from 1 × 109 FLAG-HA-tagged USF1-transduced and mock-transduced HeLa cells by using a FLAG antibody-conjugated column. The polypeptides were eluted by the FLAG peptide and were resolved by SDS-PAGE. The proteins were visualized with Coomassie blue staining and were analyzed by mass spectrometry. A partial list of polypeptides identified is listed on the right. (B) The FLAG-purified samples from FLAG-USF1-transduced and mock-transduced cells were analyzed by immunoblotting with antibodies against USF1, USF2, and PRMT1. (C) USF1 interacts with PRMT1 in chicken erythroleukemia 6C2 cells. Nuclear extracts from 6C2 cells were immunoprecipitated with protein A beads, rabbit IgG, and USF1 antibody. The precipitated samples were analyzed by immunoblotting with PRMT1 antibody.
We confirmed the mass spectrometry results by Western blot analysis using the FLAG-purified material. In most cases tested, proteins identified by mass spectrometry were present in the FLAG-USF1 purification but absent in the mock purification (data not shown). Figure 2B shows that both USF2 and PRMT1 were enriched in the FLAG-USF1 purification, which suggests that USF1 associates with PRMT1 in vivo (Fig. 2B). To confirm this interaction, we performed an immunoprecipitation in which nuclear extracts were precipitated with USF1-specific antibody and immunoblotted with PRMT1 antibody. Compared to protein A beads and rabbit IgG, USF1 again specifically interacted with PRMT1 (Fig. 2C).
These data suggest that there might be a direct interaction between USF1 and PRMT1. To establish this and to characterize the domains involved in this interaction, USF1 and the truncated mutants containing or lacking the helix-loop-helix (HLH) domain were expressed as GST fusion proteins (Fig. 3A). These fusion proteins were used in a GST pull-down assay by incubation with [35S]methionine-labeled PRMT1, which confirmed that USF1 and PRMT1 directly interact. The basic helix-loop-helix (bHLH) domain of USF1 is essential for this interaction (Fig. 3B, compare lanes 2, 3, 4, and 5), but these data do not exclude the possibility of contributions from the more-N-terminal part of the protein. The results indicate that USF1 directly recruits the histone H4R3-specific methyltransferase PRMT1 and that the interaction is mediated by the bHLH domain of USF1.
FIG. 3.
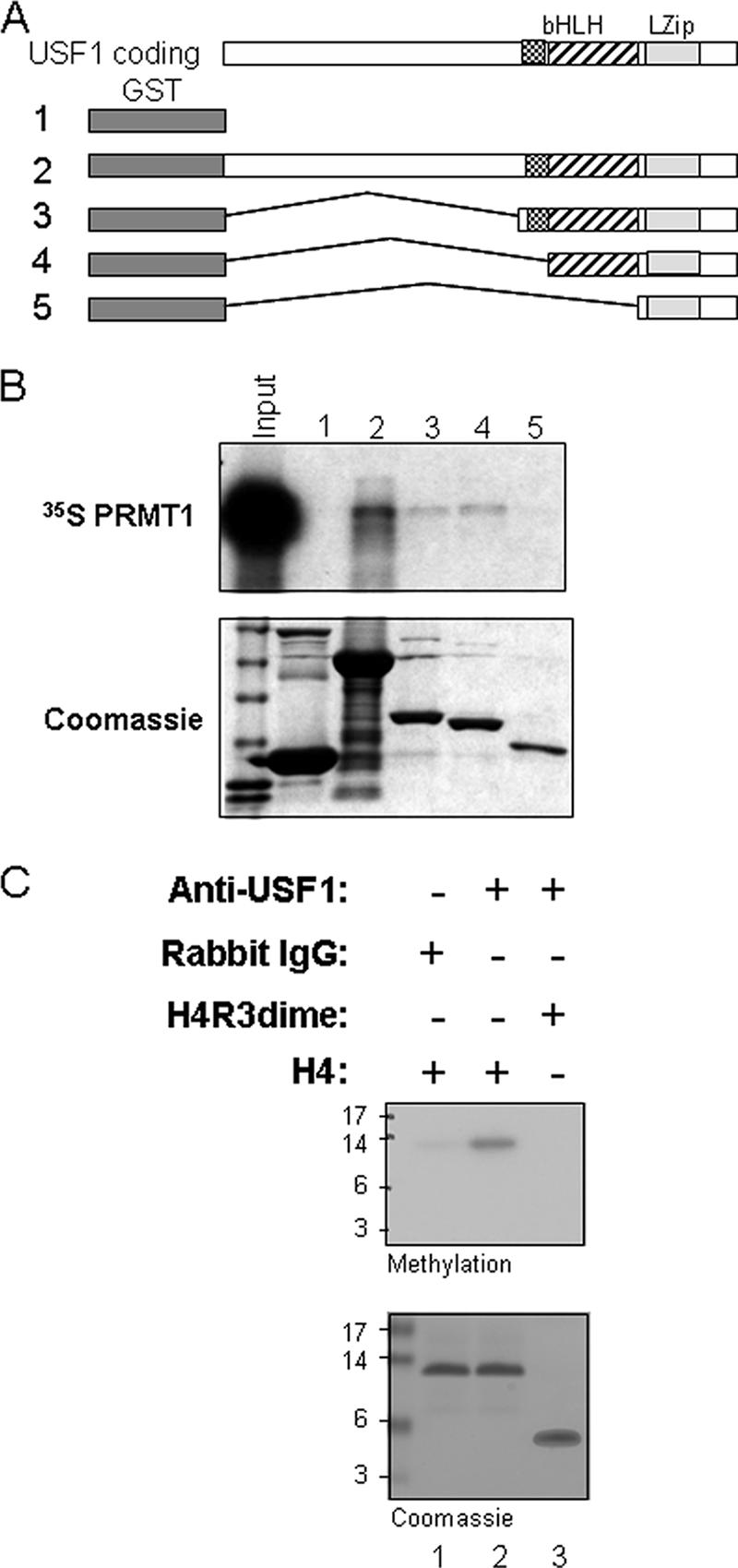
USF1 directly recruits PRMT1 and H4R3 HMT activity. (A) The HLH domain of USF1 interacts with Arg 3-specific methyltransferase PRMT1 directly. Shown is a schematic representation of the GST-USF1 fusion proteins used in the GST pull-down assay. (B) 35S-labeled PRMT1 was incubated with GST and GST-USF1 fusion proteins preadsorbed to glutathione-Sepharose beads. (Top) Bound PRMT1 was eluted and visualized by fluorography following SDS-PAGE. (Bottom) A Coomassie-stained gel shows the relative amounts of fusion proteins used in the assay. (C) USF1 recruits H4R3-specific HMT. Nuclear extracts from HeLa cells were immunoprecipitated with rabbit IgG or USF1 antibody. The precipitates were incubated with unmethylated H4 peptide (lane 1 and 2) or R3 dimethylated H4 peptide (lane 3) in the presence of [3H]AdoMet as cofactor. The proteins were resolved by SDS-PAGE and were visualized by fluorography (top). The loading amounts of H4 used in the assay were demonstrated by Coomassie blue staining (bottom).
USF1-containing complexes exhibit HMT and HAT activities.
It has been reported that PRMT1 is the major enzyme asymmetrically methylating H4R3 in vitro and in vivo (15, 39, 40). We therefore asked whether the USF1-containing complex exhibited H4R3 methyltransferase activity. Nuclear extracts were immunoprecipitated with USF1 antibody, and the immunoprecipitates were assayed for methylation activity. The USF1-containing complex showed robust HMT activity on an unmodified histone H4 substrate (Fig. 3C, lane 2). However, the activity was completely abolished when we used a histone H4 substrate dimethylated at the R3 residue (Fig. 3C, lane 3). We conclude that USF1 functionally associates with PRMT1.
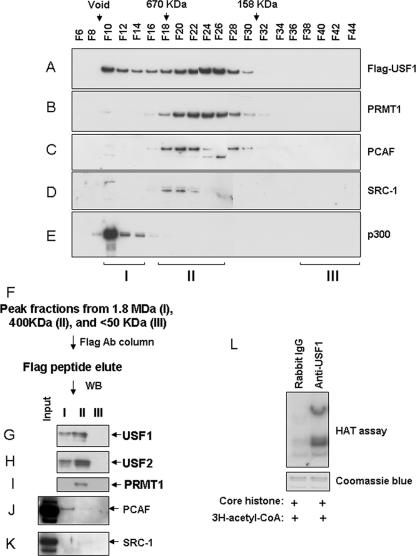
USF1 has been shown to colocalize in vivo with HATs p300 and PCAF at the 5′HS4 insulator (42), and the complexes contain TRRAP, which is a PCAF coregulator (Fig. 2A), suggesting that they may be present in the same complex with PRMT1. To test this possibility, nuclear extracts from transformed HeLa cells expressing FLAG/HA-USF1 were fractionated through a Sephacryl S-300 HR column and subjected to Western blot analysis. USF1 and PRMT1 comigrate in the ∼400-kDa fractions (Fig. 4A and B). The peak fractions containing the histone acetyltransferases PCAF and SRC1 overlap those containing USF1 and PRMT1, but at positions corresponding to a slightly higher molecular weight than the peaks for the latter. This is consistent with the formation of two complexes, both containing USF1 and PRMT1 but only a subset containing the HATs (Fig. 4C and D). In contrast, p300, as well as PCAF, appear in early fractions that contain 1.8-MDa complexes close to the void volume in this kind of column (Fig. 4E).
FIG. 4.
USF1 forms a complex with PRMT1 and HATs. Nuclear extracts from HeLa cells expressing FLAG-USF1 were fractionated through a Sephacryl S-300 HR column. (A to E) Fractions were collected and were analyzed by immunoblotting with antibodies against USF1 (A), PRMT1 (B), PCAF (C), SRC-1 (D), and p300 (E). (F to K) The peak fractions from 1.8 MDa (I), 400 kDa (II), and <50 kDa (III) were collected. The pooled fractions were immunoprecipitated with FLAG antibody. The precipitates were then analyzed by Western blot analysis using anti-USF1, anti-USF2, anti-PRMT1, anti-PCAF, and anti-SRC-1 antibodies. (L) HeLa nuclear extracts were immunoprecipitated with rabbit IgG or USF1 antibody and then were incubated with chicken core histones in the presence of [3H]acetyl coenzyme A (CoA). Histones were resolved by SDS-PAGE and were visualized by fluorography (top) or Coomassie blue staining (bottom).
To provide further evidence that USF1, PRMT1, and histone acetyltransferases form a multiprotein complex in the 400-kDa size range, the peak fractions from the Sephacryl column corresponding to sizes of 1.8 MDa (I), 400 kDa (II), and less than 50 kDa (III; negative control) were collected separately and immunoprecipitated with anti-FLAG-conjugated agarose beads. The immunoprecipitates from these fractions were analyzed for the presence of USF1, USF2, PRMT1, PCAF, and SRC-1 by Western blotting analysis (Fig. 4F). Both 1.8-MDa and 400-kDa complexes contained USF1 and USF2 heterodimer, consistent with the formation of two distinct USF1-containing complexes (Fig. 4G and H). The histone acetyltransferase PCAF associates with USF in both complexes (Fig. 4J). However, PRMT1 and SRC-1 are exclusively present with USF1 in the 400-kDa complex (Fig. 4I and K). Together with the data presented above (Fig. 2A and B), these results suggest that USF1 can recruit both HMTs and HATs, although not in every case into the same complex. The ability of USF1 to interact with HATs was confirmed by measuring HAT activity in fractions immunoprecipitated from HeLa nuclear extracts with an antibody to USF1. Chicken core histones incubated with these immunocomplexes showed strong HAT activities toward histones H3, H2A, H2B, and H4 (Fig. 4L). It should be noted that we had shown earlier that such immunocomplexes contain PCAF (42). Coactivator SRC1 has been shown to be present in nuclear receptor coactivator complexes that include PCAF and PRMT1 (9, 19), and SRC1 also possesses HAT activity (38).
Role of USF1 in the chromatin insulator.
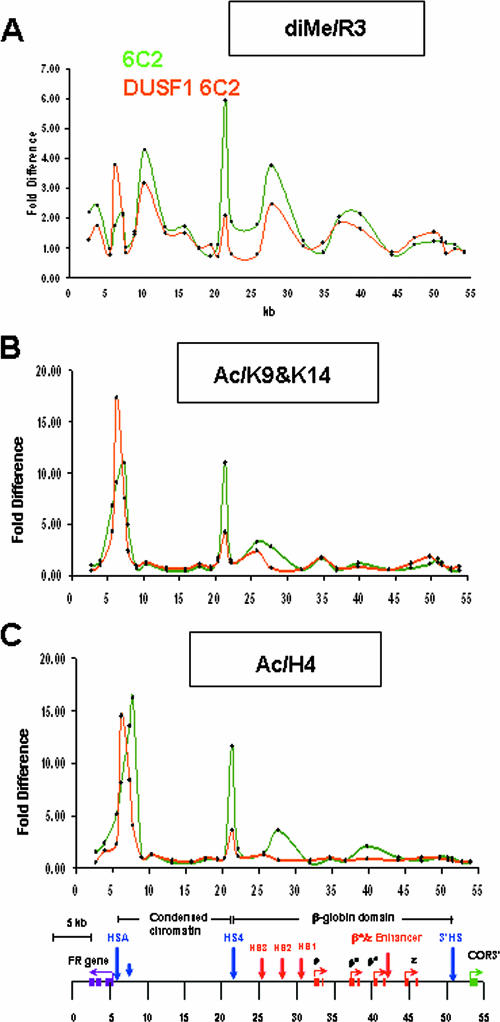
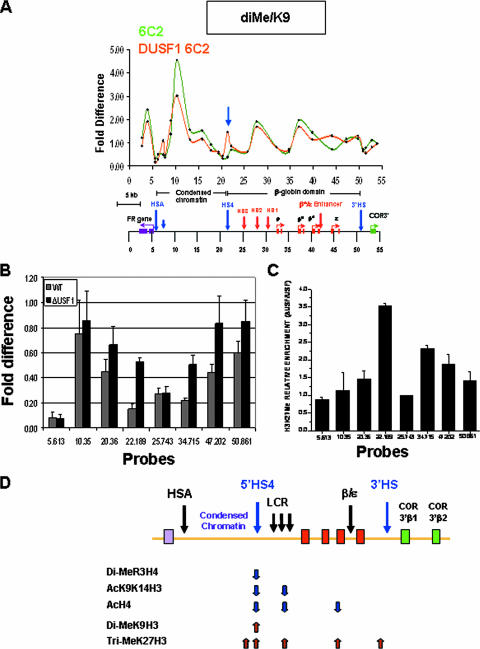
Given that a plausible model of barrier insulator function is that it maintains high levels of “positive” histone modifications on adjacent nucleosomes, we wished to examine the possible role of histone methylation and acetylation at the HS4 β-globin insulator. Our earlier studies demonstrated the localization of USF1 (complexed to USF2) at the 5′HS4 insulator and described the effects of deletion of its binding site or USF1 knockdown on local histone modifications around the insulator (42). ChIP studies also show that PRMT1 colocalizes with USF1 at the 5′HS4 insulator element (A. G. West, G. Felsenfeld, et al., unpublished data). In an attempt to better characterize USF1 function at the chromatin barrier, and its relationship with a variety of histone modifications that mark the insulator element, we extended the ChIP analysis over the entire 54-kb globin locus, comparing the wild type with USF1 knockdown 6C2 cells, in which the expression of USF1 is reduced 80 to 90% by siRNA-mediated knockdown (42). In addition, we used antibodies directed against histone modifications not reported in our earlier paper: acetyl H4, methyl H4R3, and trimethyl H3K27. In these studies, we employed the method we have previously described (25), in which formaldehyde cross-linking is not introduced. Nuclei are directly digested with micrococcal nuclease and then fractionated on a sucrose gradient. The mono- and di-nucleosome fractions are retained for immunoprecipitation, followed by measurement using real-time PCR, permitting analysis at high resolution.
In the USF1 knockdown cells, there was a 65% decrease in H4R3 methylation specifically localized at the 5′HS4 insulator element (Fig. 5A). Concomitant with the reduction in this modification, the acetylation of H3 and H4 tails at the insulator was reduced 62% and 70%, respectively (Fig. 5B and C). There was a notable reduction in both H3 and H4 acetylation over the locus control region HS2 site (site at 27.729) (Fig. 5B and C). The drop of acetylation in this region may reflect the loss of insulator protection from invasion of the 16-kb condensed chromatin (see Discussion, below). It should be noted that the histone modifications measured at the insulator actually reflect values at the nucleosomes adjacent to the insulator itself. Results from our laboratory (V. Calhoun, C. Jin, and G. Felsenfeld, unpublished data) for the endogenous site and from Zhao et al. (44) for an insulator element carried on an episome show that the insulator domain itself is depleted of nucleosomes, which led the latter authors to suggest that this could contribute to barrier function by preventing propagation of a silencing signal from one nucleosome to the next.
FIG. 5.
Knockdown of USF1 leads to decreased methylation at H4R3 as well as acetylation of H3 and H4. USF1 expression was reduced 80 to 90% in 6C2 cells based on RNA interference technology (42). The results of ChIP assays with antibodies specific to dimethylated H4R3 (A), acetylated H3K9/K14 (B), and acetylated H4 (C) from wild-type (green) or USF1 knockdown (orange) 6C2 cell line are shown. See Materials and Methods for a description of the measurements and calculations of the difference.
The distribution of dimethyl H3K9 in the USF1 knockdown line is similar to that of wild-type 6C2 cells throughout the region, except for the insulator element at 5′HS4, where there is a notable appearance of H3K9 dimethylation (site 21.365), which is completely absent from wild-type 6C2 cells (Fig. 6A). More extensive changes were seen in levels of trimethyl H3K27, a modification associated with repressive chromatin (8, 33). In wild-type cells (Fig. 6B), there is a high level of H3K27 trimethyl over the 16-kb condensed chromatin region, terminating at the 3′ end of this region at the nucleosomes just 5′ of the insulator. The probe situated just 3′ of the insulator (at 22.189 kb) detects only low levels of this modification, and these low levels extend 3′ for at least another 10 kb. The pattern is different in the USF1-depleted line (Fig. 6B and C). The site at 22.189 is now highly modified by H3K27 trimethylation, increased by at least threefold. Modification at downstream sites up to about 35 kb is increased 1.5- to 2-fold, except for site 25.743, which marks a locus control region element and may be partially depleted of histones (Fig. 6B and C). These results reflect the high overall genomic levels of enrichment for H3K27Me3. A recent genome-wide study (4) showed that there is only a ∼2- to 3-fold increase in the level of K27 trimethylation associated with inactive compared to active genes. The observed change over the β-globin locus that accompanies USF1 depletion (Fig. 6D) is entirely consistent with these measurements and supports the idea that the globin locus is inactivated when USF1 is depleted.
FIG. 6.
USF1 is required for histone modifications at the 5′HS4 insulator element. (A) Results of chromatin IP with antibodies specific for dimethyl H3K9 from wild-type (green) and USF1 knockdown (orange) 6C2 cell lines across 54 kb of the chicken β-globin locus. (B) Focusing on histone H3K27 trimethylation at the chicken β-globin domain following USF1 knockdown. ChIP of trimethylated H3K27 at endogenous sequences in wild-type (WT; gray bars) or USF1 knockdown (black bars) cells. (C) The relative enrichment of histone H3 K27 trimethylation is shown across the β-globin domain by comparing the USF1 knockdown to wild-type 6C2 cells. (D) Summary of the ChIP data across the chicken β-globin locus, showing the results of the USF1 knockdown. The arrows pointing up and down indicate increases or decreases in the modification, respectively.
USF1 is responsible for chromatin barrier function.
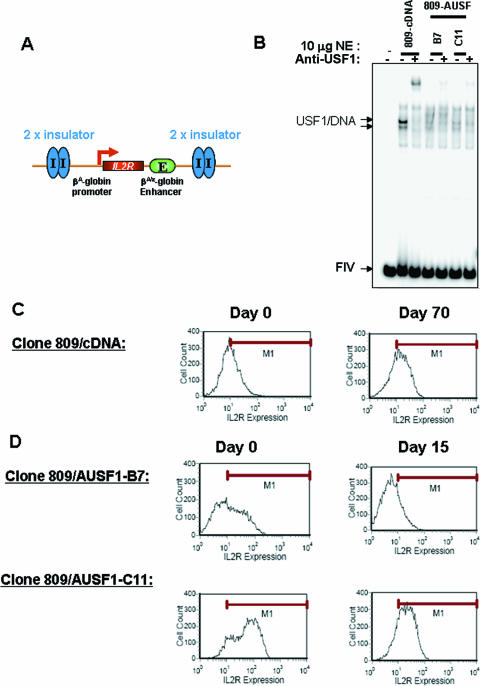
The experiments described so far show that USF1 is responsible for recruiting histone-modifying enzymes, and their associated modifications, to the nucleosomes surrounding the insulator. They do not show that USF1 protein is essential to barrier activity. We have previously described an assay for chromatin barrier activity that uses a human IL-2R reporter driven by the chicken βA-globin promoter and the β/ɛ enhancer (32). This construct is flanked by two copies of the chicken 5′HS4 core insulator sequence and integrated stably into the genome of the chicken erythroid 6C2 cell line (Fig. 7A) (32). After withdrawal of hygromycin selection, the insulated reporter continues to be expressed for at least 100 days in culture (cell line 809) (32). In contrast, expression of an uninsulated transgene, in a parallel experiment, was rapidly silenced by chromosomal position effects.
FIG. 7.
USF1 is responsible for preventing chromosomal position-effect silencing. (A) Schematic representation of the insulated IL-2R transgene, which was amplified four times (four copies) in the 6C2 genome (line 809). (B) Line 809 was transfected with vector containing AUSF1 cDNA, which lacks the basic DNA-binding region and inhibits both USF1 and USF2 activities. The single clones (B7 and C11) were selected in the presence of 100 μg/ml zeocin (Invitrogen). Gel mobility shift assays of nuclear extracts from the control and AUSF1 clones were carried out using 32P-labeled FIV oligonucleotide duplexes. (C) Results of the FACS analysis for IL-2R expression at 0 and 70 days after removal of hygromycin selection. M1 designates IL-2R-positive cells. On these histograms, cell number is indicated on the y axis and the logarithm of fluorescein isothiocyanate fluorescence intensity is on the x axis. (D) Results of the FACS analysis for IL-2R expression of clone B7 (top) and clone C11 (bottom) at 0 and 15 days after removal of hygromycin selection.
To test directly the role of USF1 in maintaining an active chromatin environment, we made use of 6C2 line 809, which contains four copies of the IL-2R transgene flanked with the 5′HS4 insulators; expression of IL-2R is maintained in this fully insulated line during 70 days in culture (Fig. 7C). We stably expressed in this line a USF1 mutant gene, AUSF1, which lacks the basic DNA-binding region and inhibits both the endogenous USF1 and USF2 activities (35). Two clones, B7 and C11, exhibited high AUSF1 expression by Western blot analysis (data not shown) and strongly inhibited endogenous USF1 DNA-binding activity in gel shift assays (Fig. 7B, compare lanes 4 and 6 to lane 2). In these lines, we observed already at day 15 a marked reduction in the IL-2R expression level (overall intensity) by 54% and 64% in clones B7 and C11, respectively (Fig. 7D). In addition, the number of IL-2R-positive cells decreased 53% in clone B7 and 13% in clone C11 (Fig. 7D). These results demonstrate that the USF1 protein is directly involved in the protection of transgenes against chromosomal position-effect silencing in the chromatin barrier assay. They support the model in which USF1 establishes a chromatin barrier by maintaining a local environment of active chromatin.
The 5′HS4 insulator element does not recruit Pol II.
Both enhancer and insulator elements have the ability to establish an active chromatin domain, yet the insulator element does not activate transcription of genes. In the human and mouse globin loci, USF1 has been shown to recognize E-box elements located at the enhancer HS2 and the β-globin promoter (7, 21). We explored further the differences between the USF1 site at the insulator and those at the mouse β-globin locus. At the mouse locus during erythropoiesis, the enhancer HS2 and β-major promoter recruit several active histone modifications, including H3 and H4 acetylation and H3K4 methylation (18, 23); RNA Pol II is recruited to these sites in MEL cells (17). We asked whether Pol II also was recruited to the chicken β-globin 5′HS4 insulator. In the 6C2 cell line the folate receptor gene is transcriptionally active, and the globin genes are largely inactive. Immunoprecipitation with antibodies against the Ser 2- and Ser 5-phosphorylated forms of Pol II shows that there are high concentrations of both species at the folate receptor promoter (Fig. 8). Neither species is detected within the 16-kb condensed chromatin region or in the globin locus, but there is an accumulation of the Ser 5-P form 5′ of the insulator, and some is also detectable about 2 kb away on the 3′ side (Fig. 8). We did not look for the unmodified form of Pol II in 6C2 cells, but in separate experiments in 10-day embryonic erythrocytes we detected all three forms. These were all present only at low levels in the neighborhood of the insulator (data not shown). Consistent with this, we do not detect the recruitment of TAFII 250, the largest component of the TFIID complex, at the endogenous 5′HS4 insulator in the chicken β-globin locus (data not shown). The concentration of polymerase on the 5′ side is similar to that reported by Zhao et al. for a minichromosome construct in which the 5′HS4 insulator is inserted between the HS2 enhancer and ɛ-globin gene and which they concluded is the result of the insulator's ability to block the passage of polymerase (43). Our data are consistent with this result and also show that the insulator itself does not seem to recruit polymerase in the same way as an active promoter and enhancer, such as that found at the folate receptor. The data suggest that at the insulator element USF and its cofactors recruit a large array of histone modification activities associated with chromatin opening but that this has only a local effect on modifying the neighboring nucleosomes, rather than functioning as a classical activating element.
FIG. 8.
USF1 does not recruit RNA polymerase II to the 5′HS4 insulator at the chicken β-globin locus. A ChIP analysis of Pol II localization at the 54-kb chicken β-globin locus in 6C2 cells is shown. The IPs using anti-Pol II phosphor Ser 2 are indicated by blue bars, and those that used anti-Pol II phosphor Ser 5 are indicated by red bars. RNA polymerase levels were compared to those for a site within the 16-kb condensed chromatin region, which was assumed to be low (see Materials and Methods).
DISCUSSION
USF1 as a critical component of a barrier insulator.
We recently identified USF1 and USF2 as essential components of the vertebrate barrier insulator located within 5′HS4 at the 5′ end of the chicken β-globin locus. This insulator confers on reporter constructs the ability to resist the encroachment of transcriptionally repressive chromosomal condensation processes (42). In this report, we purified USF1-containing multiple protein complexes with the aim of further understanding the mechanism by which this barrier works. We found that USF1 associates with two distinct histone methyltransferase activities. One of them is PRMT1, which catalyzes the methylation of histone H4R3. USF1 and PRMT1 interact with each other, and it is likely that PCAF and SRC-1, which both interact with USF1 and comigrate with USF1 and PRMT1 on a gel filtration column, are part of the same complex, which displays their corresponding HAT activities (Fig. 4L). Downregulation of USF1 by siRNA in erythroid cells not only results in local loss of H4R3 methylation in 5′HS4 (Fig. 5A) but also reduces acetylation of H3 and H4 at the same location (Fig. 5B and C). The decrease in H4R3 methylation is largely confined to the insulator region, reflecting the loss of tethering of PRMT1 through its interaction with USF1 (Fig. 6D). This can be compared with the much more widely disseminated loss of methylated H4R3 when PRMT1 itself is downregulated (15). Loss of USF1 also leads to an increase of repressive modifications, such as methylation of H3K9 and H3K27 (Fig. 6A and B). However, in contrast to the localized changes, confined to the insulator, seen with other modifications, the absence of USF1 affects H3K27 methylation at sites downstream over the β-globin locus. Of particular interest is the very large increase seen at kb 22.189, immediately downstream of the insulator, as well as at other downstream sites, 34.715 and 47.202 (Fig. 6B and C). As noted above, the magnitude of these increases is entirely consistent with the changes expected in H3K27 trimethylation when an active gene becomes inactive (4). Our results indicate that the overall genomic levels of this enrichment are rather high. Thus, as in the case of H3K9 dimethylation (24), the most significant sites are those with low levels of methylation, such as the β-globin locus in wild-type cells. The increases seen over this locus in the USF1-depleted cells indicate that its H2K27 modification levels are approaching those of the 16-kb condensed chromatin region, consistent with the loss of insulator barrier function.
Taken together, our data indicate that PRMT1 is recruited to the 5′HS4 insulator by USF1 and that both USF1 and PRMT1 are required to maintain activating histone modifications localized over the insulator element. Consistent with this conclusion, although downregulation of PRMT1 has been shown to decrease levels of dimethylated H4R3 at the 5′HS4 insulator (15), it does not affect USF1 association with the 5′HS4 insulator (data not shown). In addition, the observation that loss of USF1 from the insulator results in propagation of silencing modifications (particularly H3K27Me3) over the globin locus is consistent with a model in which USF1 works with histone-modifying enzymes to establish a barrier against the encroachment of adjacent heterochromatin into the globin locus.
This view of the central role of USF1 in barrier activity was confirmed by the experiments shown in Fig. 7. The transformed 6C2 cell line clone 809 carries four copies of a fragment of the IL-2R gene, driven by erythroid-specific regulatory elements, each flanked by two copies of the core insulator. As shown earlier (32, 36) and confirmed in Fig. 7C, the IL-2R gene is protected in these cells from extinction of expression over very long times in culture, whereas loss of expression is usually rapid in uninsulated lines (32, 36). The protection conferred by the insulators can be overcome, however, by expression of a dominant negative mutant USF protein which prevents binding of USF1 to its site on the insulator. Under those conditions, silencing of IL-2R expression is rapid (Fig. 7D). Thus, USF1 and by inference its associated cofactors are essential to the maintenance of insulator barrier function.
USF1 as a transcription factor.
Although this paper focuses on the role of USF1 at the chicken β-globin 5′HS4 insulator, USF1 is also a ubiquitous transcription factor that is normally dimerized with USF2 and typically recognizes the canonical E box sequence, CACGTG. This is distinct from the degenerate E box, CACGGG, to which the USF1/USF2 heterodimer binds within the insulator at 5′HS4 (42). USF1 protein functions as a transcriptional regulator involved in a variety of cellular growth and differentiation pathways, including lipid and glucose metabolism and cell cycle and growth control (3, 11, 31, 42, 46). During hematopoiesis, USF1 plays an essential role in regulating expression of the HOXB4 gene, which is preferentially transcribed in immature hematopoietic cells and implicated in the transition from primitive to definitive hematopoiesis as well as in hematopoietic stem cell self-renewal and expansion (1, 13, 45). It has been suggested that USF1 may be directly responsible for activating HOXB4 transcription. USF proteins have also been shown to regulate human β-globin HS2 enhancer and promoter activities (7, 21). However, the underlying mechanisms by which USF1 activates these genes are unclear.
In agreement with the role of USF1 in activation of β-globin gene transcription (12), we found that USF1 colocalizes with the arginine methyltransferase PRMT1 and the modification it catalyzes, at the mouse β-globin HS2 enhancer and β-major promoter in cells where the β-globin is actively transcribed (S. Huang et al., unpublished data). But these sites differ from the insulator, because they recruit RNA Pol II while the insulator does not (Fig. 8). We suggest that USF1 is playing a similar roles at both insulator and enhancer: modifying adjacent nucleosomes to make the region more accessible to the other factors binding to neighboring sites. When these factors are transcriptional activators, such as EKLF or GATA-1, the region has enhancer activity. When the nearby factor (such as CTCF) is not an activator of transcription, the region may function as a barrier insulator because of the strong histone-modifying activity of USF and its cofactors, but it will not be an enhancer.
PRMT1 as a coactivator.
PRMT1 was originally shown to be an H4R3-specific methyltransferase that stimulates HAT activities and acts as a coactivator for nuclear receptor-mediated transcription (9, 22, 40). In the context of nuclear receptor-mediated transcription, the function of PRMT1 as coactivator is strictly dependent on its ability to interact with the p160 family of coactivators (19, 22). Consistent with these reports, we found that one of the p160 family of coactivators, SRC-1, forms a multiprotein complex with USF1 and PRMT1 in a gel filtration column, suggesting that they are present in the same complex (Fig. 4). We do not know, however, whether SRC-1 is important in insulator function or only for these other activities that involve the USF1/PRMT1 complex. We note that SRC-1 HAT activity is relatively weak (38). PRMT1 can also be targeted onto chromatin by YY1 and p53 and activates sequence-specific transcription mediated by these factors. Methylation of H4R3 by PRMT1 is required for this activation (2, 37).
Mechanisms of barrier insulation.
The barrier function of the chicken β-globin 5′HS4 insulator was originally detected through its ability to protect randomly integrated transgenes from chromosomal position-effect silencing (32, 36). In the absence of this insulator, the transgene used in the barrier assay is silenced rapidly (36). Silencing is characterized by early loss of histone acetylation in parallel with loss of transcription, followed by an accumulation of H3K9 methylation and DNA methylation at the transgene promoter (26, 27). This activity is distinct in properties and mechanism from the enhancer-blocking activity mediated by CTCF (5, 10). Unlike the enhancer-blocking activity, the barrier function of the 5′HS4 insulator requires binding of USF1 and the ability of USF1 to recruit histone modification activities to the insulator (42; this paper).
In an earlier study, we showed that downregulation of PRMT1 in 6C2 cells has a global effect on methylation of Arg 3 on histone H4, resulting in greatly decreased methylation over the folate receptor gene and the globin genes as well (15). This also affects H4R3 methylation at the 5′HS4 insulator, which normally displays high levels of this modification. Reduction in PRMT1 expression disrupts other activating histone modifications and impairs barrier activity of the chicken 5′HS4 insulator (15).
We have proposed (24, 25) that the recruitment to the globin insulator of enzymes associated with activating histone modifications serves to block the processive mechanisms that could otherwise propagate silencing modifications into active chromatin domains. Such mechanisms have already been demonstrated in yeast (30) and involve maintenance of high levels of histone acetylation at the boundary. In the case of 5′HS4, more elaborate and possibly redundant mechanisms appear to be involved. Consistent with this view, we showed that USF1 is essential to the protection of the endogenous β-globin gene against silencing histone modifications and that in its absence the core insulator element fails completely to protect a transgene against silencing. This insulator function is mediated by the numerous histone-modifying enzymes recruited to the insulator site by the USF1/USF2 heterodimer. It is clear that USF1 forms at least two distinct high-molecular-weight complexes that carry out this function. The extent to which each complex contributes to USF1 function may depend on the local environment. It seems likely that at sites throughout the genome a major role of USF1 is to establish a region of histone modification conducive to the action of other regulatory factors that are bound nearby. This property also forms the basis of USF1 function as a barrier insulator element.
Acknowledgments
We thank Yoshihiro Nakatani, Emery H. Bresnick, Charles Vinson, and Harvey R. Herschman for generously providing reagents and Rodolfo Ghirlando for helping with gel filtration. We are grateful to Jörg Bungert and members of the Felsenfeld laboratory for their helpful suggestions and comments on the manuscript. The FACS analysis was carried out in the Shands Cancer Center Flow Cytometry Core Facility.
The research is in part supported by an American Cancer Society Institutional research grant and the Bankhead-Coley Cancer Research Grant (S.H.). It was also supported by the Intramural Research Program, NIDDK, NIH.
Footnotes
Published ahead of print on 10 September 2007.
REFERENCES
- 1.Abramovich, C., and R. K. Humphries. 2005. HOX regulation of normal and leukemic hematopoietic stem cells. Curr. Opin. Hematol. 12:210-216. [DOI] [PubMed] [Google Scholar]
- 2.An, W., J. Kim, and R. G. Roeder. 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117:735-748. [DOI] [PubMed] [Google Scholar]
- 3.Andrews, G. K., D. K. Lee, R. Ravindra, P. Lichtlen, M. Sirito, M. Sawadogo, and W. Schaffner. 2001. The transcription factors MTF-1 and USF1 cooperate to regulate mouse metallothionein-1 expression in response to the essential metal zinc in visceral endoderm cells during early development. EMBO J. 20:1114-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barski, A., S. Cuddapah, K. Cui, T.-Y. Ron, D. E. Schones, Z. Wang, G. Wei, L. Chepelev, and K. Zhao. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823-837. [DOI] [PubMed] [Google Scholar]
- 5.Bell, A. C., A. G. West, and G. Felsenfeld. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98:387-396. [DOI] [PubMed] [Google Scholar]
- 6.Bell, A. C., A. G. West, and G. Felsenfeld. 2001. Insulators and boundaries: versatile regulatory element in the eukaryotic genome. Science 291:447-450. [DOI] [PubMed] [Google Scholar]
- 7.Bresnick, E. H., and G. Felsenfeld. 1993. Evidence that the transcription factor USF is a component of the humen beta-globin locus control region heteromeric protein complex. J. Biol. Chem. 268:18824-18834. [PubMed] [Google Scholar]
- 8.Cao, R., L. Wang, H. Wang, L. Xia, H. Erdjument-Bromage, P. Tempst, R. S. Jones, and Y. Zhang. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298:1039-1043. [DOI] [PubMed] [Google Scholar]
- 9.Chen, D., S. M. Huang, and M. R. Stallcup. 2000. Synergistic, p160 coactivator-dependent enhancement of estrogen receptor function by CARM1 and p300. J. Biol. Chem. 275:40810-40816. [DOI] [PubMed] [Google Scholar]
- 10.Chung, J. H., M. Whiteley, and G. Felsenfeld. 1993. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell 74:505-514. [DOI] [PubMed] [Google Scholar]
- 11.Coon, H., Y. Xin, P. N. Hopkins, R. M. Cawthon, S. J. Hasstedt, and S. C. Hunt. 2005. Upstream stimulatory factor 1 associated with familial combined hyperlipidemia, LDL cholesterol, and triglycerides. Hum. Genet. 117:444-451. [DOI] [PubMed] [Google Scholar]
- 12.Crusselle-Davis, V. J., K. F. Vieira, Z. Zhou, A. Anantharaman, and J. Bungert. 2006. Antagonistic regulation of β-globin gene expression by helix-loop-helix proteins USF and TFII-I. Mol. Cell. Biol. 26:6832-6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giannola, D. M., W. D. Shlomchic, M. Jegathesan, D. Liebowitz, C. S. Abrams, T. Kadesch, A. Dancis, and S. G. Emerson. 2000. Hematopoietic expression of HOXB4 is regulated in normal and leukemic stem cells through transcriptional activation of the HOXB4 promoter by upstream stimulating factor (USF)-1 and USF-2. J. Exp. Med. 192:1479-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grewal, S. I., and D. Moazed. 2003. Heterochromatin and epigenetic control of gene expression. Science 301:798-802. [DOI] [PubMed] [Google Scholar]
- 15.Huang, S., M. Litt, and G. Felsenfeld. 2005. Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications. Genes Dev. 19:1885-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, S., Y. Qiu, R. W. Stein, and S. J. Brandt. 1999. P300 functions as a transcriptional coactivator for the TAL1/SCL oncoprotein. Oncogene 18:4958-4967. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, K. D., J. A. Grass, M. E. Boyer, G. M. Kiekhaefer, G. A. Blobel, M. J. Weiss, and E. H. Bresnick. 2002. Cooperative activities of hematopoietic regulators recruits RNA polymerase II to a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. USA 99:11760-11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiekhaefer, G. M., J. A. Grass, K. D. Johnson, M. E. Boyer, and E. H. Bresnick. 2002. Hematopoietic-specific activators establish an overlapping pattern of histone acetylation and methylation within a mammalian chromatin domain. Proc. Natl. Acad. Sci. USA 99:14309-14314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koh, S. S., D. Chen, Y. H. Lee, and M. R. Stallcup. 2001. Synergistic enhancement of nuclear receptor function by p160 coactivators and two coactivators with protein methyltransferase activities. J. Biol. Chem. 276:1089-1098. [DOI] [PubMed] [Google Scholar]
- 20.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 21.Leach, K. M., K. F. Vieira, S. H. Kang, A. Aslanian, M. Teichmann, R. G. Roeder, and J. Bungert. 2003. Characterization of the human beta-globin downstream promoter region. Nucleic Acids Res. 31:1292-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, Y.-H., S. S. Koh, X. Zhang, X. Cheng, and M. R. Stallcup. 2002. Synergy among nuclear receptor coactivators: selective requirement for protein methyltransferase and acetyltransferase activities. Mol. Cell. Biol. 22:3621-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Letting, D. L., C. Rakowski, M. J. Weiss, and G. A. Blobel. 2003. Formation of a tissue-specific histone acetylation pattern by the hematopoietic transcription factor GATA-1. Mol. Cell. Biol. 23:1334-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Litt, M. D., M. Simpson, M. Gaszner, C. D. Allis, and G. Felsenfeld. 2001. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science 293:2453-2455. [DOI] [PubMed] [Google Scholar]
- 25.Litt, M. D., M. Simpson, F. Recillas-Targa, M. N. Prioleau, and G. Felsenfeld. 2001. Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J. 20:2224-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mutskov, V., and G. Felsenfeld. 2004. Silencing of transgene transcription precedes methylation of promoter DNA and histone H3 lysine 9. EMBO J. 23:138-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mutskov, V. J., C. M. Farrell, P. A. Wade, A. P. Wolffe, and G. Felsenfeld. 2002. The barrier function of an insulator couples high histone acetylation levels with specific protection of promoter DNA from methylation. Genes Dev. 16:1540-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakayama, J., J. C. Rice, B. D. Strahl, C. D. Allis, and S. I. Grewal. 2001. Role of histone lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110-113. [DOI] [PubMed] [Google Scholar]
- 29.Ogawa, H., K. Ishiguro, S. Gaubatz, D. M. Livingston, and Y. Nakatani. 2002. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296:1132-1136. [DOI] [PubMed] [Google Scholar]
- 30.Oki, M., L. Valenzuela, T. Chiba, T. Ito, and R. T. Kamakaka. 2004. Barrier proteins remodel and modify chromatin to restrict silenced domain. Mol. Cell. Biol. 24:1956-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pajukanta, P., H. E. Lilja, J. S. Sinsheimer, R. M. Cantor, A. J. Lusis, M. Gentile, X. J. Duan, A. Soro-Paavonen, J. Naukkarinen, J. Saarela, M. Laakso, C. Ehnholm, M.-R. Taskinen, and L. Peltonen. 2004. Familial combined hyperlipidemia is associated with upstream transcription factor 1 (USF1). Nat. Genet. 36:371-376. [DOI] [PubMed] [Google Scholar]
- 32.Pikaart, M. J., F. Recillas-Targa, and G. Felsenfeld. 1998. Loss of transcriptional activity of a transgene is accompanied by DNA methylation anf histone deacetylation and is prevented by insulators. Genes Dev. 12:2852-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plath, K., J. Fang, S. K. Mlynarczyk-Evans, R. Cao, K. A. Worringer, H. Wang, C. C. La Cruz, A. P. Otte, B. Panning, and Y. Zhang. 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300:131-135. [DOI] [PubMed] [Google Scholar]
- 34.Prioleau, M. N., P. Nony, M. Simpson, and G. Felsenfeld. 1999. An insulator element and condensed chromatin region separate the chicken beta-globin locus from an independently regulated erythroid-specific folate receptor gene. EMBO J. 18:4035-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qyang, Y., X. Luo, T. Lu, P. M. Ismail, D. Krylov, C. Vinson, and M. Sawadogo. 1999. Cell-type-dependent activity of the ubiquitous transcription factor USF in cellular proliferation and transcriptional activation. Mol. Cell. Biol. 19:1508-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Recillas-Targa, F., M. J. Pikaart, B. Burgess-Beusse, A. C. Bell, M. D. Litt, A. G. West, M. Gaszner, and G. Felsenfeld. 2002. Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. Proc. Natl. Acad. Sci. USA 99:6883-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rezai-Zadeh, N., X. Zhang, F. Namour, G. Fejer, Y. Wen, Y. Yao, I. Gyory, K. Wright, and E. Seto. 2003. Targeted recruitment of a histone H4-specific methyltransferase by the transcription factor YY1. Genes Dev. 17:1019-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spencer, T. E., G. Jenster, M. M. Burcin, et al. 1997. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature 389:194-198. [DOI] [PubMed] [Google Scholar]
- 39.Strahl, B. D., S. D. Briggs, C. J. Brame, J. A. Caldwell, S. S. Koh, H. Ma, R. G. Cook, J. Shabanowitz, D. F. Hunt, M. R. Stallcup, and C. D. Allis. 2001. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr. Biol. 11:996-1000. [DOI] [PubMed] [Google Scholar]
- 40.Wang, H., Z. Huang, L. Xia, Q. Feng, H. Erdjument-Bromage, B. D. Strahl, S. D. Briggs, C. D. Allis, J. Wong, P. Tempst, and Y. Zhang. 2001. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 293:853-857. [DOI] [PubMed] [Google Scholar]
- 41.West, A. G., M. Gaszner, and G. Felsenfeld. 2002. Insulator: many functions, many mechanisms. Genes Dev. 16:271-288. [DOI] [PubMed] [Google Scholar]
- 42.West, A. G., S. Huang, M. Gaszner, M. D. Litt, and G. Felsenfeld. 2004. Recruitment of histone modifications by USF proteins at a vertebrate barrier element. Mol. Cell 16:453-463. [DOI] [PubMed] [Google Scholar]
- 43.Zhao, H., and A. Dean. 2004. An insulator blocks spreading of histone acetylation and interferes with RNA polymerase II transfer between an enhancer and gene. Nucleic Acids Res. 32:4903-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou, H., S. H. Song, and A. Dean. 2006. Enhancer blocking by chicken beta-globin 5′-HS4: role of enhancer strength and insulator nucleosome depletion. J. Biol. Chem. 281:30572-30580. [DOI] [PubMed] [Google Scholar]
- 45.Zhu, J., D. M. Giannola, Y. Zhang, A. J. Rivera, and S. G. Emerson. 2003. NF-Y cooperates with USF1/2 to induce the hematopoietic expression of HOXB4. Blood 102:2420-2427. [DOI] [PubMed] [Google Scholar]
- 46.Zhu, Y., M. Casado, S. Vaulont, and K. Sharma. 2005. Role of upstream stimulatory factors in regulation of renal transforming growth factor-β1. Diabetes 54:1976-1984. [DOI] [PubMed] [Google Scholar]