Abstract
Prep1 is known to interact in vivo with Pbx1 to regulate development and organogenesis. We have identified a novel Prep1-interacting protein, p160 c-Myb binding protein (p160). p160 and Pbx1 compete for Prep1 in vitro, and p160 inhibits Prep1-dependent HoxB2 expression in retinoic acid-treated NT2-D1 cells. The N-terminal physiologically truncated form of p160, p67, binds the sequence 63LFPLL67 in the HR1 domain of Prep1. Mutation of both L63 and L66 impairs the binding of Prep1 to both p160/p67 and Pbx1. The sequences required to bind Prep1 are mainly located in residues 51 to 151. Immunofluorescence colocalization and coimmunoprecipitation of endogenous p160 and Prep1 are induced by ActD, which translocates p160 from the nucleolus to the nucleoplasm. These data therefore show that p160 is a novel regulator of Prep1-Pbx1 transcriptional activity.
Meinox and Pbx interaction (6, 44) controls the expression and activity of Hox gene products (8, 10, 17, 18). These proteins are expressed well before any Hox gene and hence must have additional functions (10). Pbx1-deficient mice show an embryonic-lethal phenotype characterized by homeotic transformations of the cranial and neck cartilages, hypoplastic or absent organs, deficient pancreas development, and altered hematopoietic development (13, 28, 34, 44). Meis1-null and Prep1 hypomorphic Prep1i/i (“i” means hypomorphic) mutants both exhibit an embryonic-lethal phenotype with major defects in early hematopoiesis, angiogenesis, and oculogenesis (1, 19, 23). A Prep1-null mutation causes early lethality (embryonic day 7.5) (L. C. Fernandez-Diaz and F. Blasi, unpublished data).
An important property of Prep1 is its ability to control the levels of all Pbx and Meis proteins, which is at least in part responsible for the Prep1i/i phenotype (10, 19, 33). Therefore, the activity of Prep1 is expected to be tightly regulated. Pbx-interacting partners have been described, such as Pdx in the somatostatin and the elastase enhancers (20, 46), Pax6 in the glucagon promoter (22), Oct-1 in GnRH (42), Cdx-2 in the proglucagon gene (32), and MyoD during myogenic differentiation (3, 30). Smads-2, -3, and -4 also interact with both Prep1 and Pbx1 in the FSH promoter (2). However, no unique direct interactor with Prep1 capable of preventing binding to Pbx and hence inhibiting Prep1-Pbx DNA-binding and transcriptional activities has been described.
We have searched for specific Prep1-interacting proteins that might compete with Pbx. Among the recently purified new Prep1-interacting proteins isolated by tandem-affinity purification (11), we found p160 c-Myb binding protein (p160), a mainly nucleolar protein known to bind the negative domains of the nuclear transcription factors c-Myb (16, 47) and PPAR-γ coactivator 1α (PGC1-α) (15). An endogenous, proteolytically generated amino-terminal fragment of p160, p67, is found in myeloid cells (25). This posttranslational modification is functionally relevant, since p67 inhibits the activities of c-Myb and PGC1-α (16, 47).
Here, we characterize the novel Prep1-p160 (and p67) interaction and show that p160 (and p67) is a direct Prep1-binding protein. The essential p160 (and p67)-binding residues are found in the first homology domain (HR1) of Prep1 in the 63LFPLL67 sequence. Alanine substitution for both L63 and L67 prevents binding of Prep1 to both p160 (and p67) and Pbx1. In p160, Prep1-binding sequences are located in the 51-to-151 region. In vitro, p160 (and p67) competes with Pbx1 for Prep1 binding and ectopic p160 expression inhibits the binding of Prep1/Pbx1 to the enhancer of HoxB2 and the retinoic acid (RA) induction of HoxB2 transcription. In the cell, p160 and Prep1 are found in the nucleolus (26) and nucleus (4), respectively. Interestingly, treatment of cells with actinomycin D (ActD), which extrudes p160 from the nucleolus, induces colocalization and coimmunoprecipitation of the endogenous proteins.
MATERIALS AND METHODS
Cell culture and reagents.
NIH 3T3, F9, NT2-D1, and COS-7 cells (ATCC, Manassas, VA) were maintained in Dulbecco's modified Eagle's medium (GIBCO-BRL, Gaithersburg, MD) with 10% heat-inactivated fetal bovine serum (GIBCO-BRL) at 37°C in 5% CO2. Differentiation of NT2-D1 cells was carried out as described previously (33, 45) with 10 μM RA (with dimethyl sulfoxide as a control). The nonoverexpressing and the Prep1-overexpressing clones of F9 (A2 and 2a18, respectively) have been described previously (33).
Construction of expression vectors.
PSG5-Prep1, pSG5-Pbx1a/b, pSG5-Hoxb1, pADML-R3 (HoxB1 enhancer), and pADML-R4 (HoxB2 enhancer) were described previously (6, 7, 14, 24). Pact-c-p160-FLAG and pact-c-p67*-FLAG vectors (47) were used for in vivo expression. pGEM3Z-p160, pGEM3Z-p67*, and p67 deletion constructs (47) were used for in vitro translation studies. Deletions of p160, generated in B. Spiegelmann's laboratory (Dana Farber Institute for Cancer Research, Boston, MA) (15), were provided by Addgene (Boston MA). pRUFneo-p160-FLAG was provided by T. J. Gonda (Hanson Centre for Cancer Research, Australia) (47) and used to generate p160 retrovirus. The bacterial expression plasmid for Prep1-glutathione S-transferase (GST) has been described previously (7).
Prep1-GST deletion and point mutants.
Bacterial expression plasmids for Prep1 deletion mutants were PCR amplified from pSG5-Prep1. Fragments were cloned into pGEX-4T1 (Stratagene, La Jolla, CA). The primers were F1-R4 for HR1, F4-R3 for HR2, F1-R3 for HR1 plus HR2, F1-R2 for HR1 plus HR2*, and F5-R1 for the homeodomain plus the C terminus. The primers used to create the Prep1 deletion mutants were as follows: F1(BamHI), 5′CCGGGATCCATGATGGCTACACAG-3′; F2(XhoI), 5′-CCGCTCGAGATGAACAGTGAAACTCTG-3′; F4(BamHI), 5′-CCGGGATCCACAACTTCTGCCAG-3′; F5(HindIII), 5′-CCGCTCGAGGGCCAAGTGGTCACACAG-3′; R1(NotI), 5′-TCGCGGCCGCTGCAGGGAGTCACTGTTCGC-3′; R2(EcoRI), 5′-TCCGAATTCTTGGGGAGTGACGAC-3′; R3(EcoRI), 5′-TCCGAATTCTTCTGTTTTCAGACAAGC-3′; and R4(EcoRI), 5′-TCCGAATTCGCCTTCAGAGCCCTG-3′.
The Prep1(HR1) point mutants L63-A, L66-A, L63/66-zThe Prep1(HR1) point mutants L63-A, L66-A, L63/66-A, and L69-A were generated using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and the following primers: L63A(sense), CAGGCCATTTATAGGCATCCAGCATTTCCATTATTAGCTTTGTTG; L63A(antisense), CAACAAAGCTAATAATGGAAATGCTGGATGCCTATAAATGGCCTG; L66A(sense), GCCATTTATAGGCATCCACTATTTCCAGCATTAGCTTTGTTG; L66A(antisense), CAACAAAGCTAATGCTGGAAATAGTGGATGCCTATAAATGGC; L63/66A(sense), CAAGCAGGCCATTTATAGGCATCCAGCATTTCCAGCATTAGCTTTGTTG; L63/66A(antisense), CAACAAAGCTAATGCTGGAAATGCTGGATGCCTATAAATGGCCTGCTTG; L69A(sense), GGCATCCACTATTTCCATTATTAGCTGCGTTGTTTGAAAAATGTGAAC; and L69A(antisense), GTTCACATTTTTCAAACAACGCAGCTAATAATGGAAATAGTGGATGCC. All constructs were verified by sequencing them in both directions.
Generation of retroviruses and infection of NIH 3T3 cells.
Phoenix ecotropic packaging cells (29) were transfected with 10 μg of retroviral plasmid using the calcium phosphate protocol (38) and incubated overnight. NIH 3T3 cells were infected with the filtered viral supernatant supplemented with 8 μg/ml Polybrene (Sigma, St. Louis, MO) for 4 h at 37°C.
Transfection, immunoprecipitation, and Western blotting.
Cells were plated on 10-cm plates for 24 h, transfected with 4 μg of plasmids using FuGene 6 (Roche, Nutley, NJ), and harvested after 48 h, and nuclear extracts were obtained as described before (13). The nuclear extracts were adjusted to IBB buffer (10 mM Tris-HCl, pH 8, 0.2% NP-40, 150 mM NaCl) and precleared with protein G-Sepharose beads for 1 h at 4°C. The clarified supernatants were incubated with 5 μg of the indicated antibodies and recovered on protein G-Sepharose beads or on M2-anti-FLAG affinity resin (Sigma, St. Louis, MO) overnight at 4°C. The beads were rinsed several times with IBB buffer, resuspended in Laemmli buffer twice, heated at 85°C, and centrifuged at 10,000 × g. In the coimmunoprecipitation experiment with F9 cells, the immunoprecipitating anti-Prep1 antibody was the B2 monoclonal antibody from Santa Cruz Biotech (Santa Cruz, CA).
RNA extraction and real-time PCR.
Total RNA was extracted from transfected cells with an RNeasy minikit (QIAGEN) and quantitated by spectrophotometry (Nanodrop). Five micrograms of total RNA was reverse transcribed using a SuperScript First-Strand kit with random primers (Invitrogen) according to the manufacturer's instructions. For quantitative real-time PCR, 5 ng of reverse-transcribed RNA was amplified in a light cycler instrument (Roche) using a FastStart DNA mix SYBR Green I kit (Roche). The PCR conditions for HoxB2 mRNA were as follows: first denaturation and DNA polymerase activation step, 95°C for 10 min; second denaturation step, 95°C for 15 seconds; annealing step, 56°C for 6 seconds; extension step, 72°C for 20 seconds. For actin, the conditions were as follows: first denaturation and DNA polymerase activation step, 95°C for 10 min; second denaturation step, 95°C for 15 seconds; annealing step, 58°C for 6 seconds; extension step, 72°C for 20 seconds. The amount of HoxB2 mRNA was normalized to actin mRNA. The following primers were used: HoxB2 fw, TCCTCCTTTCGAGCAAACCTTCC; HoxB2 rev, AGTGGAATTCCTTCTCCAGTTCC; Actin fw, GGCATCCTGACCCTGAAGT; and Actin rev, CGGATGTCAACGTCACACTT.
Immunocytochemical staining and confocal microscopy.
Cells were grown on sterile coverslips, treated with the indicated drugs, and fixed with phosphate-buffered saline (PBS)-3% paraformaldehyde for 15 min at room temperature. After being washed with PBS, the cells were permeabilized with 0.2% PBS-Triton X-100 [vol/vol] and blocked with PBS-1% bovine serum albumin for 1 h at room temperature. Coverslips were incubated with the indicated antibodies in blocking buffer (0.1% PBS-bovine serum albumin), rinsed with PBS, incubated with purified Alexa 488- and Alexa 586-conjugated goat anti-mouse or anti-rabbit immunoglobulin Gs (IgGs), rinsed, and mounted with Immuno-Fluore Mounting Medium (ICN, Costa Mesa, CA). Nuclei were visualized by Hoeschst 33258 (Sigma) staining. Fluorescence was visualized with an inverted fluorescence microscope (DM IRBE; Leica, Wetzlar, Germany) and captured with a TCS-NT argon/krypton confocal laser microscope (Leica, Wetzlar, Germany). Incubations with matched mouse isotype IgGs, irrelevant rabbit IgGs, or secondary antibodies were always negative.
In vitro transcription-translation.
Proteins were in vitro synthesized using the TNT coupled transcription-translation kit (Promega, Madison, WI) from 1 μg of plasmid and 2 μl of translation grade [35S]methionine (Amersham Biosciences, Amersham Place, United Kingdom). Protein synthesis was evaluated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the gels were fixed, dried, and exposed to Amersham films.
Pull-down assays with GST-tagged protein beads.
GST fusion proteins were purified from Escherichia coli BL21 cells grown at 37°C to an optical density at 600 nm of 1, and protein expression was induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside for 3 h, as described previously (7). The pellets were sonicated in PBS with Complete protease inhibitor cocktail (Roche) supplemented with 1% (vol/vol) Triton X-100 and centrifuged, and the supernatants were collected and stored at −80°C. Lysates containing the GST proteins were incubated with glutathione-Sepharose (Amersham) beads for 30 min at 4°C.
For pull-down assays, 10 μl of in vitro-translated proteins precleared with 20 μl of glutathione beads in 200 μl of NET-N buffer (10 mM Tris-HCl, pH 8, 150 mM NaCl, 0.2% NP-40 [vol/vol]) supplemented with the Complete protease inhibitor cocktail were incubated for 1 h at 4°C with 1 to 3 μg of the GST protein beads, washed five times in NET-N buffer, and eluted twice for 15 min each time at room temperature with 10 mM glutathione. The quantity of eluted GST fusion proteins was evaluated by digital densitometry (ImageQuant 5.2; Molecular Dynamics) of Coomassie-stained gels. Nonspecific binding to GST was subtracted, and the data were normalized to the input.
EMSA.
Electrophoretic mobility shift assays (EMSA) with b2-PP2 and Sp1 oligonucleotides were done as described previously (5, 7). The sequence of oligonucleotide b2-PP2 (5′-GGAGCTGTCAGGGGGCTAAGATTGATCGCCTCA-3′) contains both the Prep1-Pbx1 and the Pbx1-HoxB1 binding sites of the HoxB2 r4 enhancer (underlined) (17). The sequence of the Sp1-binding oligonucleotide is 5′-GATCCGATCGGGGCGGGGCGATC-3′ (19). Extracts were prepared as described previously (12).
Antibody and reagent sources.
Polyclonal antibodies against Prep1 were previously described (6); monoclonal CH12.2 recognizing only human Prep1 was obtained by immunization with a bacterially expressed amino terminus of human Prep1 (unpublished data). Anti-p160 and anti-Pbx1b antibodies were kindly donated by S. Ishi (RIKEN Tsukuba Institute, Japan) and Michael L. Cleary (Stanford University Medical Center, Stanford, CA), respectively. The commercial antibodies were Pbx2 and nucleolin antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) and Prep1 (Meis4) (Upstate Biotechnology, Upstate House, Dundee, United Kingdom); the immunoprecipitating anti-Prep1 monoclonal antibody B-2 obtained from Santa Cruz Biotechnology (Santa Cruz, CA); anti-FLAG antibodies and goat anti-mouse/rabbit IgG Alexa 488 or Alexa 586 from Sigma, St. Louis, MO; and mouse nonimmune IgGs (DAKO, Glostrup, Denmark). Protein G-agarose beads and the Complete EDTA-free protease inhibitor cocktail were from Roche (Nutley, NJ).
Luciferase assays.
A total of 4 × 104 cells were plated in 24-well plates and transiently transfected with 25 ng/well of pAMDL-R3Hoxb1, R4-HoxB2 luciferase, or pSG5-Hoxb1 vector; 50 ng of Pbx1a or Prep1 vector; 20 ng β-galactosidase (β-Gal) plasmid; and 50 to 500 ng of pact-c-p160FLAG or pact-c-p67 FLAG vector, using FuGene 6 (Roche). Total DNA was held constant with pSG5 or pact-c empty vector. After 24 h, the cells were lysed with reporter lysis buffer, and the luciferase activity was determined using the Promega Luciferase Kit (Promega) in a Mithras Luminometer (Berthold, Oak Ridge, TN). The luciferase data were normalized by β-Gal assays as described previously (8).
RESULTS
p160 copurifies and coprecipitates with Prep1.
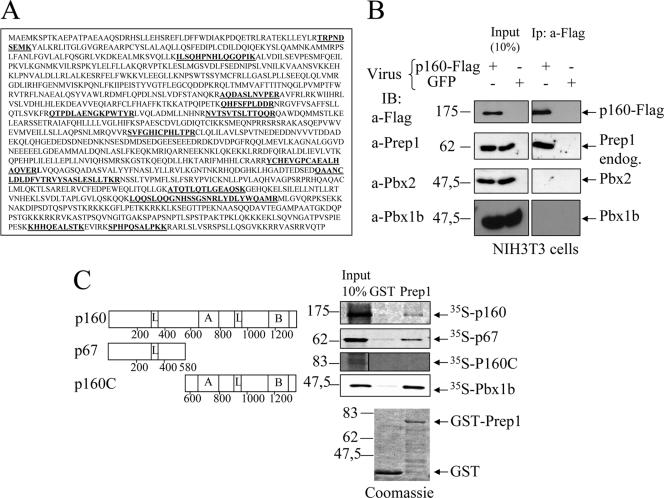
Using tandem-affinity purification (43) and matrix-assisted laser desorption ionization-time of flight protein sequencing, we isolated proteins that specifically interact with Prep1 in NIH 3T3 cells (11). One of these proteins was p160 (25, 47), which was identified by the sequence of 13 different peptides in two independent preparations (Fig. 1A). We reproduced the Prep1-p160 interaction by infecting NIH 3T3 cells with either p160-FLAG or control green fluorescent protein (GFP) retrovirus and immunoprecipitation with an M2 anti-FLAG affinity resin (Fig. 1B). p160-FLAG overexpression did not affect the nuclear level of Prep1, Pbx1b, or Pbx2 (Fig. 1B, input panel). Prep1, but not Pbx1b or Pbx2, specifically coprecipitated with p160-FLAG (Fig. 1B). We also carried out transient-cotransfection experiments with both Prep1 and p160-FLAG, immunoprecipitating the extracts with monoclonal anti-Prep1 and immunoblotting them with anti-FLAG antibodies (see Fig. S1 in the supplemental material) or immunoprecipitating them with anti-FLAG (p160) antibodies and blotting them with anti-Prep1 (not shown). Overall, these experiments indicated that Prep1 and p160 can form a stable complex in vivo that does not contain Pbx and, hence, that p160 may be a specific Prep1-interacting protein.
FIG. 1.
In vivo and in vitro interactions of Prep1 and p160. (A) The sequence of mouse p160. In boldface are the peptides identified by matrix-assisted laser desorption ionization-time of flight after tandem-affinity purification (11). (B) Immunoprecipitation with M2 anti-FLAG antibody and immunoblotting with the indicated antibodies of nuclear extracts of NIH 3T3 cells infected with p160-FLAG or a control GFP retrovirus as indicated. Input and immunoprecipitates (Ip) are compared (see Materials and Methods). (C) Schematic representation of p160 deletion mutants (left) and binding to recombinant GST-Prep1 (right). The leucine zipper-like motifs (L), the acidic domain (A), and the basic carboxyl-terminal region (B) are indicated. Binding was performed with in vitro-translated 35S-p160, 35S-p67, 35S-p160C, and 35S-Pbx1b. On the upper right is shown autoradiography of different pull downs performed with either GST (control) or GST-Prep1 (Prep1) and the 35S-labeled proteins. On the bottom are the Coomassie-stained gels of GST and GST-Prep1.
The same sequence (63LFPLLALL70) of Prep1 is required for binding both p160 and Pbx1.
To confirm the interaction between Prep1 and p160, we used pull-down assays with GST-Prep1 and in vitro-translated 35S-p160 or 35S-p67 (which is a physiological cleavage product of p160 found in myeloid cells) (16, 25, 47). As shown in Fig. 1C, 35S-p160 specifically associated with Prep1-GST but not with GST, indicating a direct interaction. Interestingly, 35S-p67 also bound strongly to Prep1-GST. However, the C-terminal fragment, p160C (amino acids 580 to 1344), did not associate with Prep1-GST. Recombinant Prep1-GST, as expected, strongly bound 35S-Pbx1b (Fig. 1C). Figure 1C (bottom) also shows the Coomassie staining of the GST and Prep1-GST preparations. These experiments suggest that the interaction between p160 and Prep1 is direct and mainly involves the N-terminal p67 fragment.
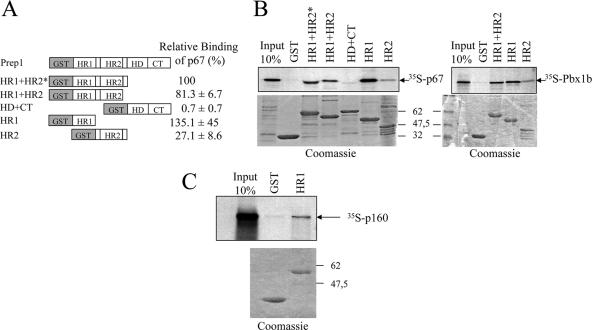
To determine the domain of Prep1 involved in the interaction, we generated GST-Prep1 deletion mutants (Fig. 2A) and performed pull-down assays with in vitro-translated 35S-p67 and 35S-p160. As a positive control we used 35S-Pbx1b, the binding region for which also resides in the N-terminal part of Prep1 (HR1 plus HR2) (6). As shown in Fig. 2B, a specific interaction with both p67 and Pbx1b was detected with the N-terminal part of Prep1 (constructs HR1, HR2, HR1 plus HR2, and HR1-HR2*), but not with a construct containing the homeodomain and C terminus. The isolated HR1 domain displayed full binding to both p67 and full-length p160 (Fig. 2B and C). Thus, p160 and Pbx1 bind the same domain of Prep1, HR1.
FIG. 2.
Mapping of the Prep1 domains required for p67 interaction. Shown is the binding of in vitro-translated 35S-p67 or 35S-Pbx1b to Prep1-GST deletion mutant beads. (A) Schematic representation of Prep1 mutants and quantitation of binding of 35S-p67 (the average of three determinations). (B) Representative pull downs of 35S-p67 (left) and 35S-Pbx1b (right). (C) Representative pull downs of 35S-p160 with GST-HR1. (Top) Autoradiography. (Bottom) Coomassie staining of the gel.
We also found that the salt sensitivity of the p67 and Pbx1 interactions with Prep1 were comparable, both resisting high-salt conditions but not sodium dodecyl sulfate. Prep1-Pbx1 salt resistance was slightly higher than that of Prep1-p67 (see Fig. S2 in the supplemental material).
We observed in the HR1 domain the conserved sequence 63LFPLLALL70, composed of two overlapping LXXLL motifs, 63LFPLL67 and 66LLALL70 (Fig. 3A). Similar sequences are also present in other p160-binding proteins (15, 36, 47). Pull-down experiments with the GST-HR1 construct showed that mutagenesis of L63 (L63A) or L66 (L66A) decreased binding to in vitro-translated 35S-p67 and 35S-Pbx1b (53 and 72%, respectively) (Fig. 3B and C). Similar results were obtained with Pbx1a (not shown). Interestingly, the double mutant L63/L66A completely lost binding to both p67 and Pbx1b, indicating that this sequence is fundamental for the binding of both proteins (Fig. 3B and C). Thus, p67 and Pbx1 bind strongly to the very same region of Prep1. Pull-down experiments with GST-Prep1 and GST-Prep1L63AL66A demonstrated the relevance of these mutations in the full-length protein. As shown in Fig. 3D, while Pbx1, p67, and p160 were pulled down by wild-type GST-Prep1, the mutant Prep1 was much less efficient. Quantitation of the gels showed that the mutation decreased the binding of Pbx1a by 47% and the binding of p67 by 67%. Mutant L69A also decreased the binding of p67 by 70%, suggesting that the second LXXLL motif also collaborates in p67 binding (not shown).
FIG. 3.
An LFPLL sequence of the HR1 domain of Prep1 is required to interact with both p67 and Pbx1. (A) Alignment of the p160-binding sequences in Prep1 and its Meinox homologs, highlighting the conservation of the leucine-rich region (shaded). The asterisks indicate conserved amino acids. (B) Binding of in vitro-translated 35S-p67 or 35S-Pbx1b to wild-type (WT) and mutant (L63A, L66A, and L63/L66-A) GST-HR1 beads. Control GST beads are included. (Top) Autoradiography. (Bottom) Representative Coomassie staining of GST fusion proteins. (C) Quantitation of binding after densitometric analysis. The error bars indicate standard deviations. (D) Pull downs of 35S-p67 and 35S-Pbx1a by GST-Prep1 and GST-Prep1L63/L66A mutants. (Top) Autoradiography. (Bottom) Coomassie staining. (E) Schematic representation of p67 and its mutants highlighting the leucine-rich sequences. L, leucine zipper domain. On the right is shown the quantification by densitometry expressed as relative binding of each mutant to GST-HR1 (the average of at least three determinations). Wild-type p67 was given the arbitrary value of 100%. (F) Representative pull down of GST-HR1 with the indicated in vitro-translated 35S-p67 mutants. (Left) Input. (Right) Pull down.
We conclude that p160 and its proteolytic fragment p67 specifically interact with Prep1 and use the same binding domain as Pbx1b.
Residues 51 to 151 of p160 (containing the first three LXXLL motifs) participate in binding to Prep1.
We also investigated the roles of the amino-terminal leucine-rich motifs of p67 using in vitro-translated deletion mutants. The p67 fragment has a leucine zipper domain at positions 307 to 335 and four leucine-rich motifs (LXXLL) at positions 94, 135, 160, and 245 (47) (Fig. 3E). Deletion of the C-terminal region up to residue 228 (p52, p38, and p25 in Fig. 3F) barely affected the binding of p67 to GST-HR1. Thus, the leucine zipper is not involved in binding. Deletion of the first 50 amino-terminal residues (p61) also had no effect (Fig. 3E and F). However, decreased binding was observed when the central portion of p67 was deleted, in particular the 51-to-151 region (p12 and p49; 38 and 42% binding, respectively). This region contains two leucine-rich motifs at positions 94 (LXXLL) and 135 (LXXVL). We did not observe a complete loss of binding activity with any of the deletion mutants, suggesting that other regions may also contribute to the overall binding.
p160 competes with Pbx1 and inhibits Prep1 transcriptional activity.
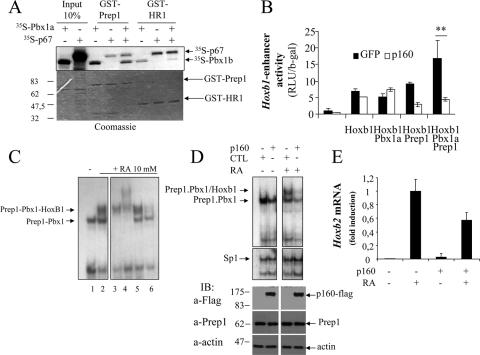
p160 may regulate Prep1 transcriptional activity by preventing the formation or promoting the dissociation of Prep1-containing complexes. To directly test the competition between Pbx1 and p160 for Prep1 binding, we performed pull-down assays with GST-Prep1 and in vitro-translated p67 and Pbx1b. As shown in Fig. 4A, in the presence of p67, the amount of Pbx1b pulled down by GST-Prep1 decreased, demonstrating a competitive interaction. Interestingly, using GST-HR1 and in the presence of a 10-fold excess of p67, a major Pbx1a binding reduction was observed. These results reinforce the hypothesis that p160(p67) competes with Pbx1 to bind Prep1.
FIG. 4.
p160 competes with Pbx1 and decreases Prep1-dependent DNA binding and transcription. (A) Competitive pull down of 35S-p67 and 35S-Pbx1a with GST-Prep1 and GST-HR1 beads. (Top) Autoradiography. (Bottom) Coomassie staining showing equal loading of GST-Prep1 and GST-HR1 in the various reactions. The proteins tested in the pull downs are indicated on top. (B) Transient transfection of a luciferase reporter plasmid (pADML-R3) containing the R3 sequence of the HoxB1 enhancer into NIH 3T3 cells stably expressing p160-FLAG (white) or GFP (black), with the indicated expression plasmids (see Materials and Methods). Activity (RLU/β-Gal) was measured 24 h later and divided by the expression of the internal transfection control β-Gal. The values represent the average plus standard error of at least three independent experiments performed in triplicate. **, P < 0.001; paired t test. (C) EMSA analysis of the Prep1-Pbx binding activity of dimethyl sulfoxide-treated and 10 μM RA-treated NT2-D1 nuclear extracts to the 32P-labeled b2-PP2 oligonucleotide. Identification of the shifted bands in untreated (−) or 48-h RA-treated (+) empty-vector-transfected cells, using inhibition by specific antibodies, is shown. Lane 1, EMSA with nuclear extracts from untreated cells; lanes 2 to 6, nuclear extracts of RA-treated cells. Lanes 1 and 2, no addition; lane 3, anti-Prep1 antibodies; lane 4, anti-Pbx1 antibodies; lane 5, anti-Pbx2 antibodies; lane 6, anti-HoxB1 antibodies. (D) Effects of p160 overexpression on the EMSA of NT2-D1 nuclear extracts with the b2-PP2 oligonucleotide. Nuclear extracts from cells stably transfected with p160-FLAG (p160) or control (CTL) vectors, treated (+) or not (−) with 10 μM RA for 48 h, were incubated with 32P-b2-PP2 (see Materials and Methods). Below is a control EMSA performed with an oligonucleotide carrying the sequence of the SP1 binding site (see Materials and Methods). At the bottom, the presence of p160 in the nuclear extract is shown by immunoblotting (IB) with anti-FLAG antibodies; actin antibodies were used as loading controls. (E) Real-time PCR measurement of the HoxB2 mRNA level in NT2-D1 cells expressing or not p160-FLAG and treated or not with 10 μM RA. The data were from two independent experiments performed in triplicate.
To test whether p160 modifies Prep1 transcriptional activity, we carried out transient-transfection experiments in NIH 3T3 cells with a luciferase reporter driven by the HoxB1 enhancer. As described previously (6, 17, 18, 24), the highest transcriptional activation was obtained in the presence of cotransfected Prep1, Pbx1, and HoxB1 expression plasmids. This activity was totally lost when the experiment was carried out in NIH 3T3 cells stably expressing p160 (Fig. 4B). We also tested whether p160 inhibited the DNA-binding activity of Prep1-Pbx1, using EMSA with in vitro-translated proteins and the b2-PP2 oligonucleotide from the HoxB2 enhancer (see Materials and Methods). In vitro-translated Prep1 and Pbx1a formed a retarded complex that was not observed with Prep1 or Pbx1 alone, as expected (18). No binding was observed with p160 alone. Preincubation with in vitro-translated p160 specifically decreased the binding of Prep1-Pbx1a complexes (see Fig. S5 in the supplemental material). Finally, transient-transfection experiments showed that expression of either p160 or p67 also inhibited the transcription-activating effect of Prep1, Pbx1, and HoxB1 on the luciferase activity of the HoxB1 and HoxB2 enhancer constructs (see Fig. S4 in the supplemental material). These data are consistent with a model in which p160 and/or p67 interacts with Prep1, preventing the binding of Prep1 to Pbx1 and to DNA and thus the Prep1-Pbx1 transcriptional activity.
To explore whether p160 overexpression inhibited endogenous Prep1 activity, we turned to NT2-D1 teratocarcinoma cells, which can be induced by RA to differentiate and to transcribe the HoxB genes (45). We first used EMSA and inhibition by specific antibodies to determine the nature of the proteins binding the enhancer of the HoxB2 gene (oligonucleotide b2-PP2; see Materials and Methods). As shown in Fig. 4C, control cell extracts (lane 1) produced a single band. In cells treated with 10 μM RA, two major bands appeared (lane 2), one comigrating with the single band in control cells and an upper band. The lower band was inhibited by Prep1 and Pbx1 antibodies (lanes 3 and 4) and hence corresponds mostly to a Prep1-Pbx1 dimer. The upper band was inhibited by Prep1, Pbx1 (not Pbx2), and HoxB1 antibodies and hence was mostly represented by the Prep1-Pbx1-HoxB1 trimer, as expected (18). We then tested pools of cells stably expressing p160 or empty vector to compare their DNA-binding activities (with or without treatment with 10 μM RA) and their RA-induced expression of HoxB2. As shown in the EMSA in Fig. 4D, overexpression of p160 decreased the intensity of the shifted band in uninduced cells and even more in RA-treated cells (i.e., the binding due to Prep1-Pbx1 dimers and Prep1-Pbx1-HoxB1 trimers). Expression of p160 had no effect on the level of endogenous Prep1 (Fig. 4D) or Pbx1b (not shown) and did not change the DNA-binding activity of the unrelated SP1 transcription factor.
The functional significance of the effect observed by EMSA was seen when we determined by real-time PCR the level of HoxB2 mRNA induced by RA. As shown in Fig. 4E, HoxB2 mRNA induction was decreased by about 45% in the cells expressing p160.
Overall, these experiments allowed us to conclude that the ectopic expression of p160 inhibits Prep1 DNA binding and transcriptional activities both in vitro and in vivo, in agreement with the hypothesis that p160 competes with Pbx1 for Prep1 binding.
p160 shuttles between nucleoli and the nucleoplasm but interacts with Prep1 only in the nucleus.
We studied the subcellular localization of endogenous p160 by confocal microscopy using polyclonal antibodies against the C terminus of p160. These antibodies preferentially stained few nuclear organelles, possibly nucleoli, in exponentially growing NIH 3T3 cells (Fig. 5A), as expected (26, 47). The nucleolar nature of the p160-positive nuclear bodies was confirmed by costaining them with antibodies to nucleolin, a specific marker of nucleoli (not shown). In some cells, staining of p160 was also detected in the nucleoplasm (Fig. 5A), as also described by others (26, 47). Under basal conditions, less than 5% of the cells also presented nucleoplasmic staining (see the histogram in Fig. S3 in the supplemental material). Transient transfection of a p160-FLAG construct also gave (mainly) nucleolar FLAG staining, but cells in mitosis displayed clear nucleoplasmic staining (Fig. 5B). Conversely, a p67-FLAG construct was also detected in the nucleoplasm, with total nucleolar exclusion (Fig. 5C), in agreement with the C-terminal region of p160 being required to direct the protein to nucleoli (26). The nucleolar/nucleoplasmic localization suggests that the protein, in addition to nucleocytoplasmic shuttling (26), also shuttles between the subnuclear compartments.
FIG. 5.
Subcellular localization of p160/p67 is modulated by ActD. (A) Confocal immunofluorescence microscopy of NIH 3T3 cells showing staining of endogenous p160 with polyclonal anti-C terminus antibodies (left). The nuclei were stained with Hoechst stain (right). The arrow points to a cell with both nucleolar and nucleoplasmic staining. (B) Confocal immunofluorescence of NIH 3T3 cells transiently transfected with a p160-FLAG construct. Shown are two cells positive for the anti-FLAG antibodies, one with nucleolar staining (arrowhead) and the other, which is dividing, with nuclear staining (arrow). (C) Confocal immunofluorescence localization of transiently transfected p67-FLAG using polyclonal anti-FLAG antibodies (left) and nuclear Hoechst staining (right). (D) Confocal immunofluorescence of endogenous p160 detected with anti-C terminus polyclonal antibodies in control (CTL)- or ActD (0.1 μM; 2 h)-stimulated NIH 3T3 cells. ActD + 24 FBS refers to cells treated with ActD as described above, washed, and left in complete medium for 24 h before microscopy. (E) Confocal immunofluorescence localization of endogenous Prep1 (red) and p160 (green) under basal conditions (left) or after ActD treatment (0.2 μM; 1 h) (right). The antibodies used are indicated. Colocalization is shown in yellow (merge).
Since the main role of Prep1 is in RNA polymerase II-dependent transcription, its interaction with p160 is expected to take place in the nucleoplasm. We tested whether translocation of p160 from the nucleolus to the nucleoplasm could be induced using agents known to do the same for other proteins (39). Neither UV at high doses (20 J/m2) or for prolonged times, known to redistribute nucleophosmin (31), nor the JNK pathway activator anisomycin, which stimulates the nucleolar translocation of RHII/Gu helicase (48), had any effect on p160 in NIH 3T3 cells (data not shown). Likewise, no effect was observed by inhibiting RNA polymerase II by protein synthesis or by DNA replication with alpha-amanitin, cycloheximide, or hydroxyurea (data not shown). However, ActD, an inhibitor of RNA polymerase I and of DNA replication, clearly induced complete, dose-dependent translocation of endogenous p160 to the nucleoplasm (Fig. 5D; see the histogram in Fig. S3 in the supplemental material). The effect was already visible after 0.5 h (not shown). After 1 h at 0.05 μM ActD, 20% of the cells lost nucleolar staining, and at 0.2 μM, the proportion of nucleoplasmic staining increased to 80% (see the histogram in Fig. S3 in the supplemental material). ActD-induced translocation was reversible: after withdrawal of the drug and in the presence of cycloheximide, the proportion of cells with exclusive nucleolar staining increased from 0 to 30% (Fig. 5D; see the histogram in Fig. S3 in the supplemental material).
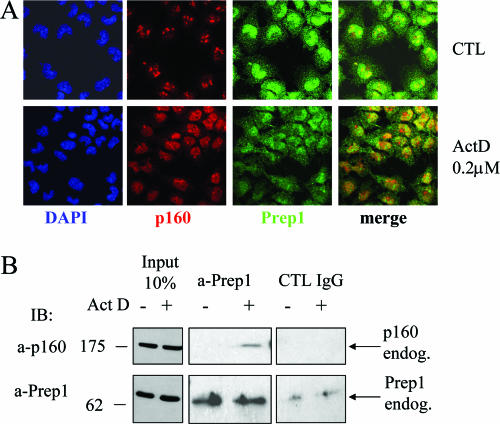
Overall, these results show that p160 can shuttle between nucleoli and the nucleoplasm and that the equilibrium is shifted during mitosis and under conditions inhibiting nucleolar function. We have therefore compared p160/Prep1 colocalization before and after treatment with ActD using confocal fluorescence microscopy. Under basal conditions, endogenous Prep1 of NIH 3T3 cells was diffusely distributed in the nucleus with minimal costaining with p160 (Fig. 5E). After brief ActD stimulation, a significant increase in colocalization with endogenous Prep1 was observed (Fig. 5E). Interestingly, in Prep1-overexpressing cells, nucleoplasmic p160 staining was significantly increased (not shown), suggesting that Prep1 may shift p160 from the nucleolus to the nucleoplasm.
To further demonstrate the interaction between endogenous Prep1 and p160, we performed coimmunoprecipitation experiments in the presence or in the absence of ActD. As we could not use NT2-D1 cells because they do not contain endogenous p160 (not shown) or NIH 3T3 cells because they express very low levels of Prep1, we used murine F9 teratocarcinoma cells, which contain immunoprecipitable p160 (not shown) and which are similar to NT2-D1 cells because they can be induced to express HoxB genes by RA treatment (33, 37). These cells contain nucleolar p160 and respond to ActD by translocating nucleolar p160 to the nucleoplasm, where it colocalizes with Prep1 (Fig. 6A). Under these conditions, ActD treatment also induced coimmunoprecipitation of endogenous Prep1 and p160 (Fig. 6B).
FIG. 6.
ActD-induced colocalization and coimmunoprecipitation of Prep1 and p160 in F9 teratocarcinoma cells. (A) Confocal immunofluorescence analysis with the indicated antibodies of cells treated with 0.2 μM ActD for 2 h (bottom) or of untreated control (CTL) cells (top). (B) Coimmunoprecipitation of Prep1 and p160 F9 cells treated (+) or not (−) with 0.2 μM ActD for 2 h. The cell extracts were immunoprecipitated with Prep1 monoclonal antibody or control IgG and blotted with polyclonal anti-Prep1 (a-Prep1) or anti-p160 (a-p160) antibodies. IB, immunoblot; endog., endogenous.
In conclusion, p160 may function in both nucleoli and the nucleoplasm; inhibition of RNA polymerase I favors p160 nucleoplasmic relocation; and nucleoplasmic, but not nucleolar, p160 interacts with Prep1.
DISCUSSION
In this paper, we identify an inhibitor of Prep1-Pbx1 function, p160/p67. p160/p67 competes with Pbx1 for Prep1 and inhibits its DNA-binding and transcriptional activities. This is the first Prep1 inhibitor identified. Moreover, we show that the interaction between p160/p67 and Prep1 is functionally relevant, as it inhibits RA-induced HoxB2 transcription. Finally, the interaction can be boosted by ActD, which forces p160 out of the nucleolus into the nucleoplasm (Fig. 7). The p160-binding region in Prep1 resides in the sequence 63LFPLLALL70 in HR1 and is essential for both p160 and Pbx1 interactions. Mutations of residues 63, 66, and 69 strongly inhibit the binding of Prep1 to both (Fig. 3).
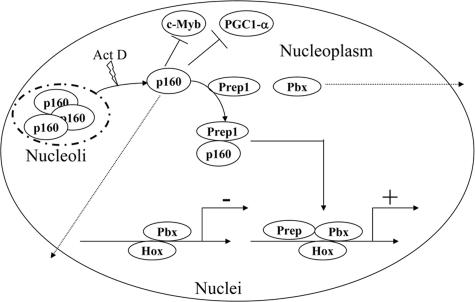
FIG. 7.
Scheme outlining the putative effects of the competition for Prep1 between Pbx1 and p160. p160 inhibits c-Myb and PGC1-α activities, but Prep1 binding might prevent these interactions. At the same time, the binding of Prep1 to p160 would inhibit or modify Pbx1 transcriptional activity, for example, on [i]Hox[r] genes. Dimerization with Prep1 may help to keep both p160 and Pbx1 in the nucleus and increase their half-lives. The absence of Prep1 might decrease the half-lives of both Pbx1 and p160 (dotted arrows). +, transcription activation; −, transcription inhibition.
The use of the same binding site for p160 and Pbx1 has important implications for Prep1. As Pbx1 and p160 compete for Prep1 (Fig. 4A and D), p160 can regulate Prep1 activity (Fig. 7). Moreover, since the p160-binding motif is conserved in all Prep1 orthologs (Fig. 3A), p160 may bind not only Prep1, but also Prep2 and all Meis proteins. Indeed, preliminary experiments have shown that in vitro-translated Meis1 also interacts with p67 (not shown).
LXXLL motifs (40) are present in many transcription factors and cofactors, like SRC-1-p160, TIF-2-GRIP-1, and CBP/p300, where they mediate interactions with hormone-activated nuclear receptors that can activate or repress transcription (21). Indeed, while p160 was originally demonstrated to bind the leucine zipper domain of c-Myb (16), the same group later reported that binding involved the leucine-rich domains rather than the leucine zipper, pointing out the presence of the LXXLL motifs (36). As deletions of the 51-to-151 sequence of p67, containing two LXXLL motifs, inhibits Prep1 binding, the Prep1-binding sequence may be represented by such motifs (Fig. 3F). However, specific experimental evidence to demonstrate this point is still not available. Moreover, although the 51-to-151 region appears to be important functionally, other binding sequences may be required for high-affinity interaction, since deletion of this region does not totally abolish binding (Fig. 3E and F).
An intriguing coincidence is the fact that p160 also binds to and inhibits the activity of PGC1-α, an essential regulator of glucose and energy metabolism (41) in which binding is also mediated by the LXXLL motifs (15). Indeed, another phenotype of the few surviving adult Prep1i/i mice is insulin hypersensitivity, which correlates with increased PGC1-α and decreased p160 (F. Oriente, L. C. Fernandez-Diaz, C. Miele, S. Iovimo, S. Mori, V. M. Draz, G. Tromcome, P. Formisano, F. Blasi, and F. Beguimot, unpublished data). In muscle cells, overexpression of Prep1, but not of the L63AL66A mutant, induces an opposite phenotype (i.e., reduced Glut4 and PGC1-α and increased p160). The fact that the same LXXLL motifs may be involved in both PGC1-α and Prep1 binding suggests that the p160-Prep1 interaction (or lack thereof) may be the basis of these phenotypes. Indeed, ectopic expression of Prep1, but not of the Prep1 L63AL66A mutant, increases the half-life of p160 and its steady-state level. In extracts of mouse muscle, Prep1/p160 coimmunoprecipitation is readily detected, and its efficiency is directly related to the level of Prep1 expression (i.e., wild type > Prep1i/+ > Prep1i/i) (Oriente et al., not shown).
Where does the interaction between Prep1 and p160 take place, i.e., the nucleolus or the nucleoplasm? Prep1 has no nuclear localization signal but is driven to the nucleus by Pbx1 (4), while p160 has mostly a nucleolar localization (26, 47). In NIH 3T3 cells, about 5% of the cells show a nucleoplasmic localization of both p160 and Prep1 (see Fig. S3 in the supplemental material). Increased localization of p160 in the nucleoplasm is found in dividing cells and under conditions in which p160 is translocated from the nucleolus to the nucleoplasm by ActD. This is supported by the coimmunoprecipitation of endogenous Prep1 and p160 (Fig. 6).
Prep1 protects Pbx1 from nuclear export (4, 27). It is possible that Prep1 also protects p160 from being exported from the nucleus, as both p160 and Pbx1 shuttle from the nucleus through a CRM-1-dependent export pathway (26, 27). However, additional experiments are required to demonstrate this hypothesis.
While the present work shows that p160 regulates Prep1-Pbx, other data show that Prep1 can in turn regulate p160. Indeed, as p160-dependent inhibition of PGC1-α affects energy metabolism, mitochondrial biogenesis, and respiration in muscles and gluconeogenesis in the liver (15, 41), the reduced availability of Prep1 in the livers and muscles of Prep1i/i mice affects the extent of PGC1-α inhibition, affecting insulin sensitivity. Indeed, heterozygous (and even more the rare homozygous) Prep1i/i hypomorphic mice display gene dosage-dependent increased insulin sensitivity and increases in glucose uptake and muscle Glut4 transporter and PGC1-α expression. Since Glut4 expression is dependent on PGC1-α (35), and since p160 inhibits PGC1-α (15), the absence of Prep1 might either activate p160, which would further inhibit PGC1-α, or inactivate p160 by inducing faster degradation, in analogy with Pbx proteins (33). In fact, the decrease in p160 due to a shorter half-life in the absence of Prep1 appears to be the cause of the insulin hypersensitivity in Prep1i/i mice (Oriente et al., not shown). Likewise, as the short form of p160, p67, inhibits c-Myb and PGC-1α activities (15, 47), the reduction of Prep1 in Prep1i/i embryos might be involved in the observed deficient hematopoiesis of these embryos (14a, 19).
In conclusion, the discovery of p160 and the identification of Prep1-binding residues considerably expands our views of the functions of this homeodomain protein and of p160 itself.
Supplementary Material
Acknowledgments
This work was supported by grants from Telethon Foundation Onlus, AIRC (Italian Association for Cancer Research), and the Italian Ministry of Education (PRIN) to F.B. V.M.D. was the recipient of a Marie Curie fellowship from the EU (contract number HPMF-CT-2002-01875).
We are grateful to S. Ishi for providing antibodies to p160 and to Tom Gonda for expression and retroviral constructs of p160.
Footnotes
Published ahead of print on 17 September 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Azcoitia, V., M. Aracil, A. C. Martinez, and M. Torres. 2005. The homeodomain protein Meis1 is essential for definitive hematopoiesis and vascular patterning in the mouse embryo. Dev. Biol. 280:307-320. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, J. S., N. Rave-Harel, S. M. McGillivray, D. Coss, and P. L. Mellon. 2004. Activin regulation of the follicle-stimulating hormone beta-subunit gene involves Smads and the TALE homeodomain proteins Pbx1 and Prep1. Mol. Endocrinol. 18:1158-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkes, C. A., D. A. Bergstrom, B. H. Penn, K. J. Seaver, P. S. Knoepfler, and S. J. Tapscott. 2004. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell 14:465-477. [DOI] [PubMed] [Google Scholar]
- 4.Berthelsen, J., C. Kilstrup-Nielsen, F. Blasi, F. Mavilio, and V. Zappavigna. 1999. The subcellular localization of PBX1 and EXD proteins depends on nuclear import and export signals and is modulated by association with PREP1 and HTH. Genes Dev. 13:946-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berthelsen, J., J. Vandekerkhove, and F. Blasi. 1996. Purification and characterization of UEF3, a novel factor involved in the regulation of the urokinase and other AP-1 controlled promoters. J. Biol. Chem. 271:3822-3830. [DOI] [PubMed] [Google Scholar]
- 6.Berthelsen, J., V. Zappavigna, E. Ferretti, F. Mavilio, and F. Blasi. 1998. The novel homeoprotein Prep1 modulates Pbx-Hox protein cooperativity. EMBO J. 17:1434-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berthelsen, J., V. Zappavigna, F. Mavilio, and F. Blasi. 1998. Prep1, a novel functional partner of Pbx proteins. EMBO J. 17:1423-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brendolan, A., E. Ferretti, V. Salsi, K. Moses, S. Quaggin, F. Blasi, M. L. Cleary, and L. Selleri. 2005. A Pbx1-dependent genetic and transcriptional network regulates spleen ontogeny. Development 132:3113-3126. [DOI] [PubMed] [Google Scholar]
- 9.Chang, C. P., Y. Jacobs, T. Nakamura, N. Jenkins, N. G. Copeland, and M. L. Cleary. 1997. Meis proteins are major in vivo DNA binding partners for wild-type but not chimeric Pbx proteins. Mol. Cell. Biol. 17:5679-5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deflorian, G., N. Tiso, E. Ferretti, D. Meyer, F. Blasi, M. Bortolussi, and F. Argenton. 2004. Prep1.1 has essential genetic functions in hindbrain development and cranial neural crest cell differentiation. Development 131:613-627. [DOI] [PubMed] [Google Scholar]
- 11.Díaz, V. M., A. Bachi, and F. Blasi. 2007. Purification of the Prep1 interactome identifies novel pathways regulated by Prep1. Proteomics 7:2617-2623. [DOI] [PubMed] [Google Scholar]
- 12.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DiMartino, J. F., L. Selleri, D. Traver, M. T. Firpo, J. Rhee, R. Warnke, S. O'Gorman, I. L. Weissman, and M. L. Cleary. 2001. The Hox cofactor and proto-oncogene Pbx1 is required for maintenance of definitive hematopoiesis in the fetal liver. Blood 98:618-626. [DOI] [PubMed] [Google Scholar]
- 14.Di Rocco, G., F. Mavilio, and V. Zappavigna. 1997. Functional dissection of a transcriptionally active, target-specific Hox-Pbx complex. EMBO J. 16:3644-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Di Rosa, P., J. C. Villaescusa, E. Longobardi, G. Iotti, E. Ferretti, V. M. Diaz, A. Micco, G. Ferrari, and F. Blasi. 24 August 2007. The homeodomain transcription factor Prep1 (pKnox1) is required for hematopoietic stem and progenitor cell activity. Dev. Biol. doi: 10.1016/j.ydbio.2007.08.031. [DOI] [PubMed]
- 15.Fan, M., J. Rhee, J. St-Pierre, C. Handschin, P. Puigserver, J. Lin, S. Jaeger, H. Erdjument-Bromage, P. Tempst, and B. M. Spiegelman. 2004. Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to PGC-1alpha: modulation by p38 MAPK. Genes Dev. 18:278-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Favier, D., and T. J. Gonda. 1994. Detection of proteins that bind to the leucine zipper motif of c-Myb. Oncogene 9:305-311. [PubMed] [Google Scholar]
- 17.Ferretti, E., F. Cambronero, S. Tumpel, E. Longobardi, L. M. Wiedemann, F. Blasi, and R. Krumlauf. 2005. Hoxb1 enhancer and control of rhombomere 4 expression: complex interplay between PREP1-PBX1-HOXB1 binding sites. Mol. Cell. Biol. 25:8541-8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferretti, E., H. Marshall, H. Popperl, M. Maconochie, R. Krumlauf, and F. Blasi. 2000. Segmental expression of Hoxb2 in r4 requires two separate sites that integrate cooperative interactions between Prep1, Pbx and Hox proteins. Development 127:155-166. [DOI] [PubMed] [Google Scholar]
- 19.Ferretti, E., J. C. Villaescusa, P. Di Rosa, L. C. Fernandez-Diaz, E. Longobardi, R. Mazzieri, A. Miccio, N. Micali, L. Selleri, G. Ferrari, and F. Blasi. 2006. Hypomorphic mutation of the TALE gene Prep1 (pKnox1) causes a major reduction of Pbx and Meis proteins and a pleiotropic embryonic phenotype. Mol. Cell. Biol. 26:5650-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goudet, G., S. Delhalle, F. Biemar, J. A. Martial, and B. Peers. 1999. Functional and cooperative interactions between the homeodomain PDX1, Pbx, and Prep1 factors on the somatostatin promoter. J. Biol. Chem. 274:4067-4073. [DOI] [PubMed] [Google Scholar]
- 21.Heery, D. M., E. Kalkhoven, S. Hoare, and M. G. Parker. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733-736. [DOI] [PubMed] [Google Scholar]
- 22.Herzig, S., L. Fuzesi, and W. Knepel. 2000. Heterodimeric Pbx-Prep1 homeodomain protein binding to the glucagon gene restricting transcription in a cell type-dependent manner. J. Biol. Chem. 275:27989-27999. [DOI] [PubMed] [Google Scholar]
- 23.Hisa, T., S. E. Spence, R. A. Rachel, M. Fujita, T. Nakamura, J. M. Ward, D. E. Devor-Henneman, Y. Saiki, H. Kutsuna, L. Tessarollo, N. A. Jenkins, and N. G. Copeland. 2004. Hematopoietic, angiogenic and eye defects in Meis1 mutant animals. EMBO J. 23:450-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobs, Y., C. A. Schnabel, and M. L. Cleary. 1999. Trimeric association of Hox and TALE homeodomain proteins mediates Hoxb2 hindbrain enhancer activity. Mol. Cell. Biol. 19:5134-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keough, R., E. Woollatt, J. Crawford, G. R. Sutherland, S. Plummer, G. Casey, and T. J. Gonda. 1999. Molecular cloning and chromosomal mapping of the human homologue of MYB binding protein (P160) 1A (MYBBP1A) to 17p13.3. Genomics 62:483-489. [DOI] [PubMed] [Google Scholar]
- 26.Keough, R. A., E. M. Macmillan, J. K. Lutwyche, J. M. Gardner, F. J. Tavner, D. A. Jans, B. R. Henderson, and T. J. Gonda. 2003. Myb-binding protein 1a is a nucleocytoplasmic shuttling protein that utilizes CRM1-dependent and independent nuclear export pathways. Exp. Cell Res. 289:108-123. [DOI] [PubMed] [Google Scholar]
- 27.Kilstrup-Nielsen, C., M. Alessio, and V. Zappavigna. 2003. Pbx1 nuclear export is regulated independently of Pbx-Meinox interaction by PKA phosphorylation of the PBC-B domain. EMBO J. 22:89-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, S. K., L. Selleri, J. S. Lee, A. Y. Zhang, X. Gu, Y. Jacobs, and M. L. Cleary. 2002. Pbx1 inactivation disrupts pancreas development and in Ipf1-deficient mice promotes diabetes mellitus. Nat. Genet. 30:430-435. [DOI] [PubMed] [Google Scholar]
- 29.Kinsella, T. M., and G. P. Nolan. 1996. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene Ther. 7:1405-1413. [DOI] [PubMed] [Google Scholar]
- 30.Knoepfler, P. S., D. A. Bergstrom, T. Uetsuki, I. Dac-Korytko, Y. H. Sun, W. E. Wright, S. J. Tapscott, and M. P. Kamps. 1999. A conserved motif N-terminal to the DNA-binding domains of myogenic bHLH transcription factors mediates cooperative DNA binding with pbx-Meis1/Prep1. Nucleic Acids Res. 27:3752-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurki, S., K. Peltonen, L. Latonen, T. M. Kiviharju, P. M. Ojala, D. Meek, and M. Laiho. 2004. Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell 5:465-475. [DOI] [PubMed] [Google Scholar]
- 32.Liu, T., D. R. Branch, and T. Jin. 2006. Pbx1 is a co-factor for Cdx-2 in regulating proglucagon gene expression in pancreatic A cells. Mol. Cell Endocrinol. 249:140-149. [DOI] [PubMed] [Google Scholar]
- 33.Longobardi, E., and F. Blasi. 2003. Overexpression of PREP-1 in F9 teratocarcinoma cells leads to a functionally relevant increase of PBX-2 by preventing its degradation. J. Biol. Chem. 278:39235-39241. [DOI] [PubMed] [Google Scholar]
- 34.Manley, N. R., L. Selleri, A. Brendolan, J. Gordon, and M. L. Cleary. 2004. Abnormalities of caudal pharyngeal pouch development in Pbx1 knockout mice mimic loss of Hox3 paralogs. Dev. Biol. 276:301-312. [DOI] [PubMed] [Google Scholar]
- 35.Michael, L. F., Z. Wu, R. B. Cheatham, P. Puigserver, G. Adelmant, J. J. Lehman, D. P. Kelly, and B. M. Spiegelman. 2001. Restoration of insulin-sensitive glucose transporter (GLUT4) gene expression in muscle cells by the transcriptional coactivator PGC-1. Proc. Natl. Acad. Sci. USA 98:3820-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nomura, T., J. Tanikawa, H. Akimaru, C. Kanei-Ishii, E. Ichikawa-Iwata, M. M. Khan, H. Ito, and S. Ishii. 2004. Oncogenic activation of c-Myb correlates with a loss of negative regulation by TIF1β and Ski. J. Biol. Chem. 279:16715-16726. [DOI] [PubMed] [Google Scholar]
- 37.Papalopulu, N., R. Lovell-Badge, and R. Krumlauf. 1991. The expression of murine Hox-2 genes is dependent on the differentiation pathway and displays a colinear sensitivity to retinoic acid in F9 cells and Xenopus embryos. Nucleic Acids Res. 19:5497-5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perlaky, L., B. C. Valdez, and H. Busch. 1997. Effects of cytotoxic drugs on translocation of nucleolar RNA helicase RH-II/Gu. Exp. Cell Res. 235:413-420. [DOI] [PubMed] [Google Scholar]
- 40.Plevin, M. J., M. M. Mills, and M. Ikura. 2005. The LxxLL motif: a multifunctional binding sequence in transcriptional regulation. Trends Biochem. Sci. 30:66-69. [DOI] [PubMed] [Google Scholar]
- 41.Puigserver, P., and B. M. Spiegelman. 2003. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocr. Rev. 24:78-90. [DOI] [PubMed] [Google Scholar]
- 42.Rave-Harel, N., M. L. Givens, S. B. Nelson, H. A. Duong, D. Coss, M. E. Clark, S. B. Hall, M. P. Kamps, and P. L. Mellon. 2004. TALE homeodomain proteins regulate gonadotropin-releasing hormone gene expression independently and via interactions with Oct-1. J. Biol. Chem. 279:30287-30297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 44.Selleri, L., M. J. Depew, Y. Jacobs, S. K. Chanda, K. Y. Tsang, K. S. Cheah, J. L. Rubenstein, S. O'Gorman, and M. L. Cleary. 2001. Requirement for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development 128:3543-3547. [DOI] [PubMed] [Google Scholar]
- 45.Simeone, A., D. Acampora, L. Arcioni, P. Andrews, E. Boncinelli, and F. Mavilio. 1990. Sequential activation of HOX2 homeobox genes by retinoic acid in human embryonic carcinoma cells. Nature 346:763-766. [DOI] [PubMed] [Google Scholar]
- 46.Swift, G. H., Y. Liu, S. D. Rose, L. J. Bischof, S. Steelman, A. M. Buchberg, C. V. Wright, and R. J. MacDonald. 1998. An endocrine-exocrine switch in the activity of the pancreatic homeodomain protein PDX1 through formation of a trimeric complex with PBX1b and MRG1 (MEIS2). Mol. Cell. Biol. 18:5109-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tavner, F. J., R. Simpson, S. Tashiro, D. Favier, N. A. Jenkins, D. J. Gilbert, N. G. Copeland, E. M. Macmillan, J. Lutwyche, R. A. Keough, S. Ishii, and T. J. Gonda. 1998. Molecular cloning reveals that the p160 Myb-binding protein is a novel, predominantly nucleolar protein which may play a role in transactivation by Myb. Mol. Cell. Biol. 18:989-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Westermarck, J., C. Weiss, R. Saffrich, J. Kast, A. M. Musti, M. Wessely, W. Ansorge, B. Seraphin, M. Wilm, B. C. Valdez, and D. Bohmann. 2002. The DEXD/H-box RNA helicase RHII/Gu is a co-factor for c-Jun-activated transcription. EMBO J. 21:451-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.