Abstract
Selenoprotein mRNAs are potential targets for degradation via nonsense-mediated decay due to the presence of in-frame UGA codons that can be decoded as either selenocysteine or termination codons. When UGA decoding is inefficient, as occurs when selenium is limiting, termination occurs at these positions. Based on the predicted exon-intron structure, 14 of the 25 human selenoprotein mRNAs are predicted to be sensitive to nonsense-mediated decay. Among these, sensitivity varies widely, resulting in a hierarchy of preservation or degradation of selenoprotein mRNAs and, thus, of selenoprotein synthesis. Potential factors in dictating the hierarchy of selenoprotein synthesis are the Sec insertion sequence RNA-binding proteins, SBP2 and nucleolin. To investigate the mechanistic basis for this hierarchy and the role of these two proteins, we carried out knockdowns of SBP2 expression and assessed the effects on selenoprotein mRNA levels. We also investigated in vivo binding of selenoprotein mRNAs by SBP2 and nucleolin via immunoprecipitation of the proteins and quantitation of bound mRNAs. We report that SBP2 exhibits strong preferential binding to some selenoprotein mRNAs over others, whereas nucleolin exhibits minimal differences in binding. Thus, SBP2 is a major determinant in dictating the hierarchy of selenoprotein synthesis via differential selenoprotein mRNA translation and sensitivity to nonsense-mediated decay.
Selenoproteins contain the trace element selenium in the form of the unusual amino acid selenocysteine. Selenocysteine is incorporated into selenoproteins via recoding of UGA codons that would otherwise function as termination codons (10, 16). Early studies on the first identified eukaryotic selenoprotein, cytoplasmic glutathione peroxidase (Gpx1), showed that dietary selenium status influenced Gpx1 enzyme activity levels in rat liver and that levels of the corresponding mRNA exhibited dependence on dietary selenium (27). This effect was shown to occur not at the level of transcription of the Gpx1 gene but, rather, via changes in RNA turnover (4). The mechanism by which selenium status influences Gpx1 mRNA turnover bears the hallmarks of nonsense-mediated decay (NMD), a pathway that targets mRNAs containing premature termination codons for degradation. The presence of both a UGA codon and an intron downstream of the UGA was shown to be required for selenium-dependent regulation of mRNA turnover (24, 31).
Studies from several laboratories have shown that selenoprotein mRNAs exhibit differential tissue and selenoprotein-specific dependence on dietary selenium status. Whereas the mRNA for Gpx1 is highly sensitive to changes in selenium status, other selenoprotein mRNAs such as those encoding type 1 iodothyronine deiodinase (Dio1) and selenoprotein P (SelP) exhibit intermediate sensitivity, while Gpx4 and thioredoxin reductase 1 (Trxr1) mRNA levels exhibit minimal changes in response to selenium deprivation (2, 15, 17, 20).
It is well documented that retention of selenium stores differs widely in different tissues (1) and that this is a likely factor in some of the reported differences in selenoprotein mRNA responses. Strikingly, however, even within a given tissue, the levels of some selenoproteins decrease with selenium depletion whereas others are preserved. This observation suggests that other factors may differentiate between the different selenoprotein mRNAs to elicit various expression levels of the corresponding proteins.
We previously suggested the Sec insertion sequence (SECIS)-binding protein, SBP2, as a candidate for establishing or contributing to the hierarchy of selenoprotein synthesis (21). SBP2 binds SECIS elements, the secondary structures in the 3′ untranslated regions (UTRs) of selenoproteins, and results in recoding UGA codons as selenocysteine instead of stop (5). Using a transient transfection system in which constructs encoded a selenoenzyme, Dio1, linked to different SECIS elements, we showed that different SECIS elements exhibited different responses to SBP2 cotransfection, presumably due to their respective interactions with SBP2 (21). A recent report by Dumitrescu et al. (11) demonstrated that mutations in SBP2 result in differential effects on expression levels of different selenoproteins. SelP levels and plasma glutathione peroxidase (Gpx3) activity in plasma from patients bearing the SBP2 mutation were ∼4- and ∼7.5-fold lower, respectively, than in unaffected siblings. Gpx1 and Dio2 activities in skin fibroblasts of the patients were ∼3- and 10-fold lower, respectively, relative to unaffected siblings. Binding of SECIS elements by other factors, including nucleolin and ribosomal protein L30 (3, 33), may also contribute to the hierarchy effect.
The goal of the present study was to gain insight into the factors and mechanism dictating the differential sensitivity of different selenoprotein mRNAs to degradation. We investigated the effects of SBP2 limitation via transient and stable RNA interference (RNAi) on selenoprotein mRNA levels. We show that SBP2 knockdown exerts differential effects on different selenoprotein mRNAs. We carried out immunoprecipitations of wild-type and mutant SBP2 and of nucleolin and assessed the levels of selenoprotein mRNAs bound by these factors. We report that SBP2 exhibits widely differing binding levels to different selenoprotein mRNAs, whereas nucleolin appears to bind most selenoprotein mRNAs at similar levels. We examined the in vivo binding of these factors to the two SECIS elements of SelP, as we have shown that the two elements function differently in SelP translation (29). The implications for elucidating how the biosynthesis of crucial selenoproteins is maintained at the expense of more expendable ones are discussed.
MATERIALS AND METHODS
Constructs and antibodies.
SBP2 short hairpin RNA (shRNA) and nonsilencing control shRNA gene constructs cloned into pSM2 retroviral vectors were purchased from Open Biosystems. SBP2 small interfering RNAs (siRNAs) and nontargeting control siRNAs were purchased from Dharmacon. Full-length human SBP2 cDNA was a generous gift from Paul Copeland. The region of human SBP2 (amino acids 408 to 783) corresponding to the “minimal functional domain” defined for rat SBP2 (7) was subcloned into the pDONR221 vector and recombined into pcDNA-DEST40, a C-terminal V5 tag vector for mammalian expression, using Gateway technology (Invitrogen). Inverse PCR was utilized to generate a missense mutation within the putative nucleolar localization signal and RNA binding domain (R540Q mutation) that has been implicated in Sec incorporation defects (11). Full-length murine EFsec (where EF is elongation factor) in pUHD10-3 has been described previously (30). Human SelP DNA was a generous gift from Kristina Hill and Ray Burk. SelP was subcloned into pcDNA 3.1 and linked to three copies of the lambda phage boxB sequence. This was used for derivatives that contain the coding region and either the first SECIS element plus flanking 3′ UTR sequences with the second SECIS element deleted or the second SECIS element plus flanking 3′ UTR sequences with the first SECIS element deleted. Each of these constructs contains three copies of the lambda boxB sequence. A lambda N peptide-glutathione S-transferase (GST) fusion was generated by subcloning the 22-amino-acid lambda boxB N protein-binding domain into Gateway pDEST15 vector (Invitrogen). Primary antibodies included rabbit anti-human SBP2 polyclonal antiserum (generous gift of Laura Papp and Kum Kum Khanna), mouse monoclonal anti-V5 (Invitrogen), mouse monoclonal nucleolin (Zymed), mouse monoclonal β-actin (Sigma), and mouse immunoglobulin G isotype control (Pharmingen).
Cell culture and transfections.
MSTO-211H human mesothelioma cells were cultured in RPMI 1640 medium (Gibco-Invitrogen) supplemented with 10% fetal bovine serum and incubated with 5% CO2 at 37°C. Cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Stable shRNA transfectants derived from single cells were selected and maintained in puromycin-supplemented medium (2 μg/ml). Transient siRNA transfectants were harvested 96 h posttransfection. HEK-293 cells were cultured and transfected, and 75Se labeling was carried out as described previously (29).
Immunoprecipitations.
MSTO-211H cells were harvested in lysis solution (0.5% Triton X-100, 25 mM Tris-HCl, 300 mM NaCl, 1 mM CaCl2) and subjected to freeze-thaw cycles to enhance rupture. MSTO-211H cells overexpressing recombinant V5-tagged SBP2 were lysed 48 h posttransfection. Lysate was precleared and incubated with 5 μg of anti-V5 antibody, antinucleolin antibody, or mouse immunoglobulin G isotype control antibody for 4 h followed by incubation with protein G-Sepharose (Zymed) overnight. The resin was washed three times (25 mM Tris-HCl, 140 mM NaCl, 1 mM CaCl2) followed by RNA isolation with Trizol reagent (Invitrogen) according to the manufacturer's instructions. RNA was resuspended in water for subsequent cDNA synthesis.
MSTO-211H cells were transiently transfected (Lipofectamine 2000; Invitrogen) with the human SelP-lambda boxB constructs containing either the first or second SECIS element, along with a construct consisting of the minimal functional domain of SBP2 (amino acids 408 to 783) bearing a V5 tag. Cells were lysed 48 h posttransfection and incubated with bacterially expressed lambda N peptide-GST protein fusion for 1 h at 4°C, followed by pull-down with glutathione-agarose beads, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and Western blotting with anti-V5 antibody (Invitrogen) or anti-nucleolin antibody (Zymed-Invitrogen). Proteins were detected by enhanced chemiluminescence (ECL Plus; GE Healthcare). Densitometric quantitation of bands was carried out using a Kodak Gel Logic 200 imager analyzed with Kodak molecular imaging software. RNA recovery was quantitated by real-time reverse transcription-PCR (RT-PCR) and was 0.010 and 0.009 pg/150 μl of eluate from immunoprecipitates for SECIS 1 and SECIS 2 mRNAs, respectively. RNA recovery for the control minus the boxB value was 3 orders of magnitude lower.
Western blotting.
Protein was harvested from MSTO-211H cells transfected with SBP2-targeted siRNA or nonsilencing siRNA 96 h posttransfection using CellLytic MT buffer (Sigma) containing 1 mM dithiothreitol, 5 mM EDTA, and 1× protease inhibitor cocktail. The cell suspension was sonicated (Fisher Scientific Sonic Dismembrator) and then centrifuged at 13,000 × g (Beckman Coulter microcentrifuge) for 10 min. A Bradford assay was performed on the supernatant using Bradford reagent (Bio-Rad, Hercules, CA). SelP-lambda boxB-associated protein was eluted in 10 mM reduced glutathione solution. Protein was mixed with reduced Laemmli buffer, boiled at 95°C for 10 min, and loaded onto a 10 to 14.5% polyacrylamide gel (Bio-Rad). Protein aliquots of 20, 10, 5, and 2.5 μg were loaded to analyze knockdown of SBP2 by siRNA. Protein was transferred onto polyvinylidene difluoride membranes, blocked for 1 h with 5% milk solution, and probed for 1 h using primary antibodies. Membranes were incubated with the appropriate horseradish peroxidase-conjugated secondary antibody for 45 min and detected using ECL Plus (GE Healthcare). Goat anti-mouse secondary antibodies (Jackson Immunolabs, West Grove, PA) were used for V5 and actin blots. Goat anti-rabbit horseradish peroxidase-conjugated antibodies (Jackson Immunolabs, West Grove, PA) were used for the SBP2 blots. Densitometric analysis of film was carried out using a Gel Logic 200 imager and Kodak molecular imaging software (Kodak Scientific Imaging Systems, Rochester, NY).
RNA isolation, cDNA synthesis, and real-time qPCR analysis.
RNA was isolated from the SBP2 knockdown cells using RNeasy spin columns and treated with RNase-free DNase I (QIAGEN). Concentration and purity of the extracted RNA were determined using the A260/A280 value measured on an ND1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE). One microgram of the RNA was used for cDNA synthesis. All cDNA was synthesized using the Applied Biosystems high-capacity cDNA synthesis kit. Real-time PCR was performed using Platinum SYBR Green quantitative PCR (qPCR) SuperMix (Invitrogen) in a Light Cycler 2.0 (Roche). Cycling conditions were used as suggested in the SYBR Green kit instructions, and results were analyzed using relative quantification software (Roche). Oligonucleotides used for qPCR along with predicted product sizes and amplification efficiencies are listed in Table S1 in the supplemental material. Hypoxanthine phosphoribosyltransferase (HPRT) was used as an internal standard, as we have found that the levels of this mRNA do not vary significantly in response to most changes in conditions. Student's paired t test was used to determine statistical significance for changes in mRNA levels following RNAi.
RESULTS
Expression levels of selenoprotein synthesis factors in cell lines.
In a recent analysis of the expression of 24 selenoprotein genes, three nonselenoprotein factors involved in their biosynthesis (SBP2, SECp43, and Sps1), and four housekeeping genes in eight murine tissues, SBP2 mRNA levels ranked among the lowest of any of the mRNAs measured in brain, heart, lung, and liver and in the low to middle range in kidney, intestine, and spleen. Only testes exhibited high levels of SBP2 mRNA relative to the majority of other mRNAs examined (19), in agreement with published findings (6).
To identify appropriate cell lines for knockdown or overexpression of SBP2 and nucleolin, we carried out real-time RT-PCR analysis of the mRNAs for these two factors and for SECp43, Sps1 and Sps2, soluble liver antigen/liver protein (SLA/LP), and EFsec in four mammalian cell lines, HEK-293, HepG2, MSTO-211H, and WISH cells. The levels of mRNA for the factors varied over a wide range, with nucleolin mRNA being the most abundant, followed by Sps1 and Sps2, SBP2, and SECp43 mRNAs (data not shown). EFsec and SLA/LP mRNA levels were the lowest in all four cell lines. SBP2 mRNA levels varied over a 20-fold range, with the highest level in MSTO-211H cells and the lowest in HEK-293 cells. This is consistent with our recent report that SBP2 protein is detectable by antibody staining in MSTO-211H cells but not in HEK-293 cells (9). Based on these findings, we chose to utilize HEK-293 cells for SBP2 overexpression studies and MSTO-211H cells for RNAi studies.
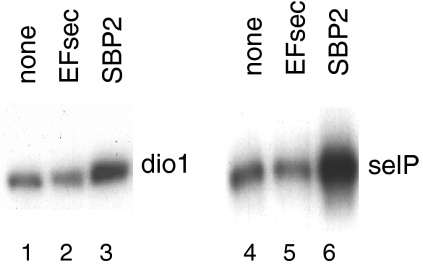
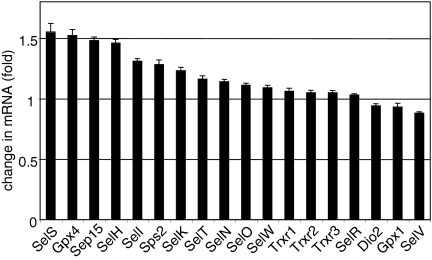
Transient transfection of SBP2 into HEK-293 cells stimulates selenocysteine incorporation, as assessed by 75Se labeling of two selenoproteins, Dio1 and SelP (Fig. 1, lane 3 versus lane 1 and lane 6 versus lane 4). These findings are consistent with our previous report that transfection of SBP2 overcomes competition among SECIS elements in an in vivo competition assay (20). In contrast, EFsec was not limiting in HEK-293 cells in spite of the low abundance of its mRNA, as transfection of an EFsec expression plasmid had no apparent effect on selenoprotein synthesis (Fig. 1, lane 2 versus lane 1 and lane 5 versus lane 4). SBP2 protein has been reported to be translated inefficiently both in vitro and in vivo (6). This is supported by the fact that although EFsec mRNA levels are lower than SBP2 mRNA levels, SBP2 is clearly limiting for selenoprotein synthesis in HEK-293 cells. Real-time RT-PCR quantitation of the effects of SBP2 overexpression on endogenous selenoprotein mRNA levels in HEK-293 cells revealed various but generally modest effects in response to SBP2 transfection, with the largest increases in the range of ∼50% (Fig. 2).
FIG. 1.
SBP2 cotransfection stimulates selenium labeling of Dio1 and SelP, but EFsec does not. SBP2 or EFsec expression constructs were cotransfected in HEK-293 cells with Dio1 or SelP expression plasmids. Cells were labeled with 75Se, and labeled protein in the cell lysate (dio1) or medium (selP) was analyzed by SDS-PAGE and autoradiography.
FIG. 2.
SBP2 overexpression differentially affects the levels of selenoprotein mRNAs. HEK-293 cells were transiently transfected with SBP2 or empty vector, and selenoprotein mRNA levels were quantitated by real-time PCR and normalized to HPRT (n = 3).
Transient and stable knockdown of SBP2 expression.
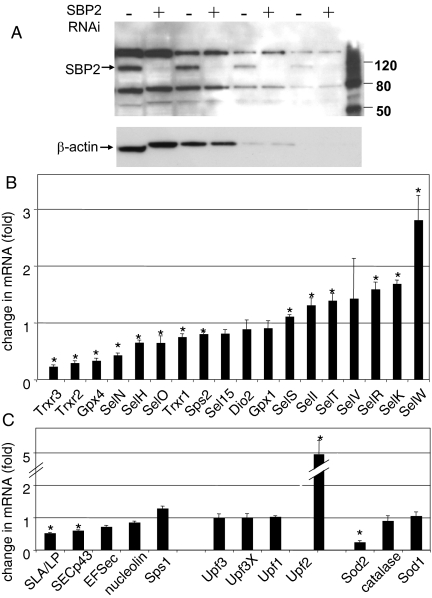
To assess the effects of competition for limiting SBP2, we carried out RNAi-mediated knockdown of SBP2 expression in MSTO-211H cells, followed by real-time RT-PCR to assess SBP2 and selenoprotein mRNA levels. Transient knockdown using siRNAs resulted in a decrease in SBP2 mRNA levels to approximately 25% of control levels. The effects of SBP2 knockdown on selenoprotein mRNAs ranged from 50% decreases in SelH and Gpx1 mRNAs and 40% and 30% decreases in Trxr1 and SelT mRNAs, respectively, to modest or minimal decreases in the majority of selenoprotein mRNAs (Fig. 3A). This finding is in accord with our previous report that SECIS elements from different selenoproteins exhibited different abilities to direct selenocysteine incorporation and to compete for available SBP2 (20). The SelW, Dio2, and SelV mRNAs increased slightly in the SBP2 knockdown cells compared to control cells, but these changes were not statistically significant. The effects on the mRNAs for the selenoprotein synthesis factors (EFsec, SECp43, SLA/LP, and Sps1) and for the NMD factors Upf1, Upf2, Upf3, and Upf3X were also examined. None of these was significantly affected by transient SBP2 knockdown (Fig. 3B).
FIG. 3.
Effects of transient knockdown of SBP2 in MSTO-211H cells on selenoprotein mRNA levels. MSTO-211H cells were transfected with SBP2 siRNAs or nontargeting siRNAs. RNA was harvested from the cells at 96 h posttransfection and used for cDNA synthesis. The levels of mRNAs for selenoproteins (A) and for selenoprotein synthesis factors, NMD factors, and antioxidant enzymes (grouped respectively) (B) relative to HPRT were analyzed by real-time PCR (n = 3). *, P ≤ 0.05, as determined by a Student's paired t test.
Stable knockdown of SBP2 was also carried out in MSTO-211H cells using an shRNA targeting the N-terminal region. SBP2 mRNA levels were knocked down to approximately 30% of control levels. Western analysis confirmed that SBP2 protein was undetectable following knockdown (Fig. 4A). Assessment of selenoprotein mRNA levels revealed similar patterns in response to both the transient and stable knockdowns for some selenoproteins, while others differed in their responses (Fig. 4B). The greatest decreases, in the range of 60 to 75%, were seen with Trxr2, Trxr3, Gpx4, and SelN mRNAs, all of which were minimally affected or unaffected in the transient knockdowns. SelV and SelW mRNA levels increased in the stable knockdowns, the latter by nearly threefold. SelK, SelR, and SelT mRNA levels increased by 40 to 70% in the stable line; but in the transient knockdowns there was no effect on SelK or SelR mRNAs, and SelT mRNA decreased 30%. SelH and Trxr1 mRNAs showed modest decreases in both the transient and stable knockdowns. The mRNAs for the selenoprotein synthesis factors EFsec, SECp43, and SLA/LP decreased ∼30, 40, and 50%, respectively, while Sps1 mRNA increased by ∼30% (Fig. 4C). The most dramatic effect on any mRNA examined in the stable knockdowns was the fivefold increase in Upf2 mRNA levels. This is particularly striking, given the lack of effect on the other NMD factors, Upf1, Upf3, and Upf3X.
FIG. 4.
Effects of stable knockdown of SBP2 in MSTO-211H cells on selenoprotein mRNA levels. MSTO-211H cells were stably transfected with SBP2-targeting shRNA or nontargeting control shRNA. Aliquots of 20, 10, 5, and 2.5 μg (left to right) of total cell lysates from control and stable knockdown cells were analyzed by SDS-PAGE and Western blotting with anti-SBP2 antibody, followed by stripping and reprobing for β-actin (A). The levels of selenoprotein mRNAs (B) and of mRNAs for selenoprotein synthesis factors, NMD factors, and antioxidant enzymes (grouped respectively) (C) relative to HPRT were analyzed by real-time PCR (n = 3). *, P ≤ 0.05, as determined by a Student's paired t test.
The differential effects of stable versus transient knockdown may be due to chronic versus acute effects of suppressing selenoprotein synthesis, including long-term effects on antioxidant and cellular redox status. We thus examined the mRNA levels for three factors involved in antioxidant defense, Sod1, Sod2, and catalase. In the transient knockdowns, none of these mRNAs were affected, whereas in the stable knockdowns, the mRNA for Sod2 decreased to ∼25% of control levels. Sod2 expression is regulated by the redox-responsive transcription factors NF-κB, Sp1, and early growth response (EGR) factor. Intriguingly, the four selenoprotein mRNAs that exhibited the greatest decreases in the stable knockdown cells and minimal responses in the transient knockdowns (Trxr2, Trxr3, Gpx4, and SelN mRNAs) all contain stress-responsive promoter elements. These include multiple NF-κB (3 to 8), Sp1 (9 to 27), Egr (10 to 24), and Ets (8 to 17) sites and Egr/Ets modules. Further analysis will be required to determine to what extent the decreases in the levels of these mRNAs are mediated by effects on transcription as a result of chronic oxidative stress versus effects on mRNA stability.
Predicted susceptibility of selenoprotein mRNAs to NMD.
NMD has been implicated in the changes in Gpx1 mRNA levels in response to selenium deficiency (24, 31). In mammals, recognition of a nonsense codon as premature typically occurs when the codon is located at least 50 to 55 nucleotides upstream of an intron in the pre-mRNA (25). Deposition of an exon junction complex downstream of a termination codon results in recruitment of the Upf factors and other factors eliciting mRNA degradation. We examined the genome structure of the 25 human selenoproteins to determine which mRNAs would be predicted to be sensitive to NMD due to the presence of introns more than 50 to 55 nucleotides downstream of the UGA codons. Fourteen of the selenoprotein mRNAs are predicted to be sensitive to NMD, and 11 are predicted to be resistant (Table 1). Most of the mRNAs predicted to be resistant, including those of Dio2, SelI, SelK, SelO, SelR, SelS, and Sps2, were either minimally affected or increased upon SBP2 knockdown. Likewise, several of the mRNAs that are predicted to be sensitive to NMD decreased in abundance in response to either transient (Gpx1, SelH, and SelT mRNAs), or stable (Gpx4, SelN, and Trxr2 mRNAs) SBP2 knockdown. However, the mRNAs for SelV, SelW, and others (e.g., SelT mRNA in the stable knockdown) increased in abundance, despite predicted sensitivity to NMD. Conversely, two mRNAs predicted to be resistant nonetheless decreased in abundance: Trxr1 mRNA in the transient knockdown and Trxr3 mRNA in the stable knockdown. The levels of some mRNAs, including those of Dio1, Dio3, Gpx2, Gpx3, Gpx6, SelM, and SelP, were too low to be reliably quantitated. To elucidate possible mechanistic explanations for these effects, we investigated the in vivo selenoprotein mRNA binding specificity of SBP2, as well as that of an additional factor implicated in SECIS binding, nucleolin.
TABLE 1.
Predicted NMD sensitivity and SECIS forms
| Selenoprotein | No. of exons | Position of intron relative to UGA codon
|
Subject to NMD | SECIS form(s) | |
|---|---|---|---|---|---|
| Distance from codon (bp)a | Direction | ||||
| Dio1 | 4 | 105 | Downstream | Yes | 1 |
| Dio2 | 2 | 174 | Upstream | No | 2 |
| Dio3 | 1 | No EJC | No | 2 | |
| Gpx1 | 2 | 105 | Downstream | Yes | 1 |
| Gpx2 | 2 | 105 | Downstream | Yes | 1 |
| Gpx3 | 5 | 141 | Downstream | Yes | 2 |
| Gpx4 | 7 | 10 | Downstream | Yes | 2 |
| Gpx6 | 5 | 141 | Downstream | Yes | 2 |
| SelH | 4 | 135 | Downstream | Yes | 2 |
| SelI | 10 | 66 | Upstream | No | 2 |
| SelK | 5 | 6 | Downstream | No | 2 |
| SelM | 7 | 138 | Downstream | Yes | 2 |
| SelN | 13 | 162 | Downstream | Yes | 1 |
| SelO | 10 | 330 | Upstream | No | 2 |
| SelP | 5 | 240 | Downstream | Yes | 1, 2 |
| SelR | 4 | 36 | Downstream | No | 2 |
| SelS | 7 | 6 | Downstream | No | 2 |
| SelT | 6 | 141 | Downstream | Yes | 2 |
| SelV | 6 | 72 | Downstream | Yes | 1 |
| SelW | 6 | 72 | Downstream | Yes | 2 |
| Sel15 | 5 | 78 | Downstream | Yes | 2 |
| Sps2 | 1 | No EJC | No | 2 | |
| Trxr1 | 15 | 63 | Upstream | No | 2 |
| Trxr2 | 18 | 65 | Downstream | Yes | 2 |
| Trxr3 | 16 | 69 | Upstream | No | 2 |
EJC, exon junction complex.
SBP2 exhibits widely differing levels of binding to different selenoprotein mRNAs.
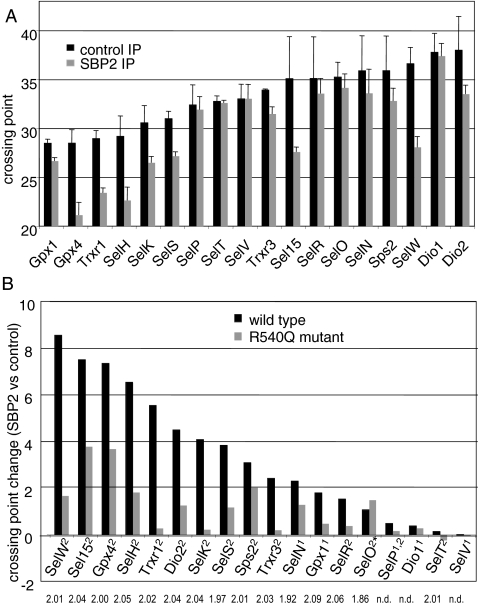
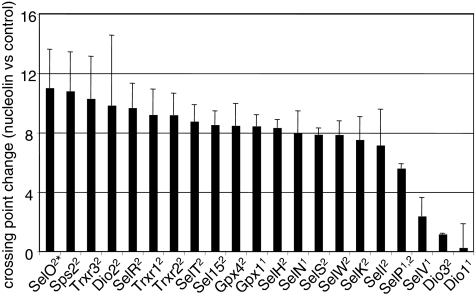
The range of responses of selenoprotein mRNAs to SBP2 knockdown suggests that SBP2 may exhibit differential binding to the SECIS elements of these mRNAs. To investigate the in vivo binding of SBP2 to these mRNAs, we carried out transient expression of an epitope-tagged SBP2 minimal functional domain, followed by immunoprecipitation and real time RT-PCR of mRNAs in the immunoprecipitates. An isotype control antibody was used for nonspecific immunoprecipitations, and mRNA levels in the SBP2 and isotype control pull-downs were compared. PCR crossing points were calculated for products amplified from the isotype control and SBP2 immunoprecipitations and plotted as the mean crossing point from three immunoprecipitations (Fig. 5A). The most abundant mRNAs were those of Gpx1, Gpx4, Trxr1, and SelH (Fig. 5A). The least abundant detectable mRNAs were those of Dio1 and Dio2. Changes in the crossing points for SBP2 versus control immunoprecipitations were also plotted for each selenoprotein mRNA (Fig. 5B, black bars) as an indication of the enrichment of individual mRNAs in the SBP2 immunoprecipitation. SelW mRNA consistently displayed the largest increase in crossing point, ∼8.5 (Fig. 5B). Intriguingly, SelW mRNA levels increased in the transient and stable SBP2 knockdowns, the latter by nearly threefold. Sel15, Gpx4, and SelH mRNAs exhibited crossing point increases greater than 6 in the SBP2 immunoprecipitations. All of these selenoprotein mRNAs are predicted to be sensitive to NMD. Intriguingly, the mRNAs exhibiting the largest increases in the SBP2 immunoprecipitations from MSTO-211H cells generally were also the mRNAs exhibiting the largest increases in levels in response to SBP2 transfection in HEK-293 cells (Fig. 2). Several selenoprotein mRNAs (Dio2, SelK, SelS, Sps2, and Trxr3 mRNAs) exhibited crossing point changes ranging from ∼2 to 4. The smallest changes were seen with Dio1, Gpx1, SelN, SelO, SelP, SelR, SelT, and SelV mRNAs. Of these, Dio1, Gpx1, SelN, SelP, and SelV mRNAs all contain form 1 SECIS elements (Table 1). It should be noted that the mRNA levels for Dio1 and Dio2 were at the limits of detection in the control immunoprecipitations but increased enough to be reliably quantitated in the SBP2 immunoprecipitations.
FIG. 5.
Coimmunoprecipitation of selenoprotein mRNAs with SBP2 wild-type protein and R540Q mutant. SBP2 was immunoprecipitated from MSTO-211H cells overexpressing wild-type or mutant V5-tagged SBP2 using anti-V5 antibody. Mouse isotype control antibody was used for control immunoprecipitations (IP) of the wild-type and R540Q SBP2-overexpressing cells. RNA was extracted from the immunoprecipitates and analyzed by real-time PCR analysis to determine relative quantities of selenoprotein mRNAs. (A) Crossing points of mRNAs in SBP2 and isotype control pull-downs. (B) Changes in crossing point are given for SBP2 versus isotype control and R540Q mutant versus isotype control pull-downs. The type of SECIS element is indicated in superscript after each selenoprotein name. The asterisk indicates the noncanonical form 2 element of SelO. Amplification efficiencies per cycle are given under each selenoprotein name. n.d., not done.
A mutation in the RNA binding domain of SBP2, R540Q, identified in patients with defects in thyroid hormone metabolism (11), was generated and tested for its ability to immunoprecipitate selenoprotein mRNAs. The R540Q mutant exhibited significant differences in RNA binding relative to wild-type SBP2 (Fig. 5B). Sel15 and Gpx4 mRNAs showed the highest crossing point changes, in the range of ∼4, whereas the changes for most selenoprotein mRNAs were lower, in the range of 0 to 2. Of note, the mRNA level of SelO, a selenoprotein with a noncanonical SECIS element, exhibited a greater increase in crossing point with the R540Q mutant than in the wild-type SBP2 immunoprecipitation. Thus, this mutation may alter the binding specificity of SBP2 such that a noncanonical SECIS is better recognized than canonical SECIS elements. The only other selenoprotein mRNA with a noncanonical SECIS, SelM mRNA, was below the limits of detection.
Nucleolin binds most selenoprotein mRNAs efficiently.
Immunoprecipitation of endogenous nucleolin was carried out with a nucleolin-specific antibody, followed by real time RT-PCR as above. Changes in crossing points were considerably higher for most selenoprotein mRNAs than with the SBP2 immunoprecipitations, but the variation between selenoprotein mRNAs was less marked, with a few exceptions (Fig. 6). This may in part be due to the abundance of nucleolin relative to SBP2. SelO mRNA, with its noncanonical SECIS element, exhibited the highest increase in crossing point, consistent with a report that nucleolin binding requires the conserved AUGA motif but does not require the AA motif in the apical loop (33). Dio1 and Dio3 mRNAs exhibited the lowest increases in crossing points, followed by SelV and then SelP mRNAs. High variability was seen with some mRNAs, such as Dio1 mRNA, presumably due to low mRNA abundance.
FIG. 6.
Coimmunoprecipitation of selenoprotein mRNAs with nucleolin. Nucleolin was immunoprecipitated from MSTO-211H cell lysates using antinucleolin antibody. Mouse isotype control antibody was used for control immunoprecipitations. RNA was extracted from the immunoprecipitate and analyzed by real-time PCR analysis to determine relative quantities of selenoprotein mRNAs.
SBP2 preferentially binds to the first versus the second SelP SECIS element.
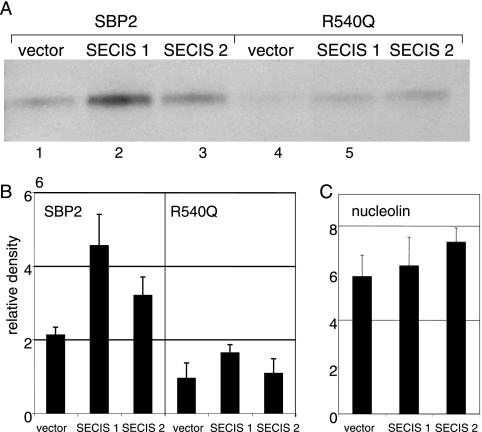
We have recently reported functional differences between the two SECIS elements of SelP, the only selenoprotein mRNA containing more than one SECIS element (29). We showed that the first SECIS element is essential for producing full-length protein and likely functions in decoding all but the first UGA codon, which appears to be decoded by the second SECIS element. To investigate the reasons for this functional difference, we used an in vivo affinity approach, termed glutathione RNA affinity chromatography (8, 12), to examine protein binding to the two SECIS elements of SelP. The 19-nucleotide RNA binding site of lambda bacteriophage antiterminator protein N, termed lambda boxB, was inserted in three copies in tandem into the 3′ UTR of SelP expression constructs comprised of the coding region and 3′ UTR but with either the first or second SECIS element deleted. As these deletions are the only difference between the constructs, differential binding patterns can be attributed to the SECIS elements. The constructs were cotransfected into HEK-293 cells with the epitope-tagged wild-type or mutant SBP2 constructs. Cell lysates were incubated with the RNA-binding domain of the lambda N peptide fused to GST, followed by pull-down with glutathione-agarose beads, SDS-PAGE, and Western blotting. Detection of SBP2 using anti-V5 antibody revealed ∼2.4-fold higher binding of SBP2 to SECIS 1 than SECIS 2 (Fig. 7A, lanes 1 to 3, and B) after the background binding to empty vector was subtracted. Analysis of binding with the R540Q mutant revealed minor differences in binding to the two SECIS elements and less total binding (Fig. 7A, lanes 4 to 6, and B). Nucleolin appeared to bind both SelP constructs comparably (Fig. 7C) and at levels not significantly above binding to empty vector.
FIG. 7.
SelP SECIS 1 and SECIS 2 exhibit differential binding to SBP2 in vivo. (A) Wild-type SBP2 or mutant R540Q SBP2 was cotransfected in HEK-293 cells with SelP-lambda boxB constructs containing either the SECIS 1(S1) or SECIS 2 (S2) sequence. The recombinant SelP was pulled down with lambda boxB N-protein-linked glutathione-Sepharose. Coelution of V5-tagged SBP2 was analyzed by Western blotting. (B) SBP2 and R540Q bands were scanned and quantitated from three independent experiments. Mean densities and standard deviations are shown. (C) Blots were stripped and reprobed with anti-nucleolin antibody, and bands were scanned and quantitated.
DISCUSSION
Differential responses of selenoprotein mRNAs to selenium limitation have long been known, but the reasons for these differences have remained elusive. Numerous studies have focused on the relatively large decreases in Gpx1 mRNA versus the minimal changes in Gpx4 mRNA (17). A few studies have also reported various effects on Dio1, SelP, SelW, and Trxr1 mRNAs (2, 15, 17, 20). The present study was aimed at investigating the reasons for these differences and provides evidence for a mechanism whereby some selenoprotein mRNAs, including those of Gpx4, SelH, and SelW, would be preferentially translated, thereby eluding NMD, at the expense of others such as Gpx1 and Dio1 mRNAs. In addition, our findings suggest a mechanistic basis for the differential functions of the two SECIS elements of SelP. Our finding that SBP2 immunoprecipitation pulls down widely varying levels of different selenoprotein mRNAs suggests that SBP2 is a major determinant in establishing the hierarchy of selenoprotein synthesis and selenoprotein mRNA turnover. This idea is supported by the various effects of SBP2 overexpression and transient and stable SBP2 knockdowns on different selenoprotein mRNAs. A prior study assessing in vitro binding of purified recombinant SBP2 to in vitro transcribed SECIS elements reported dissociation constants over only a twofold range for Dio1, Gpx1, Gpx4, SelN, SelP, and Sep15 mRNAs (13). The differences observed in vitro versus in vivo may be due to intracellular factors that influence binding, including other proteins, small molecules, and the microenvironment around the complexes, e.g., ions, salts, and pH.
In contrast to SBP2, nucleolin appears to be much less discriminatory in its RNA binding, with a few exceptions such as the Dio1 and Dio3 mRNAs. Among the reported functions for nucleolin are interactions with rRNA and proteins and roles in rRNA maturation and ribosome assembly (14). In addition, a small fraction of nucleolin has been shown in heterokaryon assays to shuttle between the nucleus and the cytoplasm. Thus, it is reasonable to speculate that nucleolin may be involved in promoting interactions between the SECIS elements in the 3′ UTR and ribosomal proteins in the nucleolus or perhaps even the ribosome at the decoding site.
It is notable that SelP mRNA is low in the hierarchy for SBP2 binding, suggesting that synthesis of the corresponding protein would be at risk when selenium is in short supply. However, SelP mRNA is highly abundant in liver, being present at levels ∼7-fold higher than the second most abundant liver selenoprotein mRNA, that of Gpx4, and ∼10-fold higher than the next two mRNAs, encoding Gpx1 and SelR (2, 15, 17, 20).
Targeted disruption of the SelP gene has shed some light on the mechanism of the tissue-specific differences in selenium supply via this selenium transport protein, revealing that brain and testes were particularly dependent on SelP for their selenium supply (18, 28). To further investigate this function in other tissues as well as the effects on additional selenoproteins, we recently examined the tissue-specific effects of SelP knockout on the levels of all 24 murine selenoprotein mRNAs in four tissues (19). While most selenoprotein mRNA levels either decreased or were unaffected in response to SelP knockout, the levels of four selenoprotein mRNAs, those of Gpx4, SelH, SelW, and Sel15, were increased in heart and lung in the knockout animals relative to wild type. Comparing the results of the previous study to the present one, we find that these four selenoprotein mRNAs showed the greatest increases in crossing point in the SBP2 immunoprecipitation. This supports our interpretation of the present study that SBP2 may play a major role in determining selenoprotein mRNA hierarchy.
The fact that SBP2 mRNA levels are typically lower than the other selenoprotein synthesis factors and most selenoprotein mRNAs, along with the reported inefficient translation of SBP2, suggests that the protein may be limiting in complexes catalyzing selenocysteine incorporation, at least under certain circumstances. Conditions that might decrease the ratio of SBP2 to selenoprotein mRNAs include genetic polymorphisms; disease states; cellular or environmental stresses (redox status, cytokines, or toxins) or developmental, metabolic, or hormonal changes (e.g., thyroid hormone levels) that alter transcription of selenoprotein genes; turnover of selenoprotein mRNAs; or expression, localization, or activity of factors necessary for selenocysteine incorporation (26).
The finding that stable knockdown of SBP2 resulted in a fivefold increase in the level of Upf2 mRNA is intriguing, although the reasons for this are unknown. Quantitation of Upf proteins in HeLa cells indicated that the ratios of Upf1:Upf2:Upf3 are approximately 40:2:1 (22). However, real-time RT-PCR analysis of the mRNA levels of these factors in MSTO-211H cells revealed that the Upf2 mRNA amplification curves were ∼5 crossing points higher than those of Upfs 1, 3, and 3X, corresponding to a decrease in mRNA on the order of ∼25-fold. Thus, it is feasible that Upf2 may be limiting in these cells. In accord with this, our recent attempts to knock down Upf2 expression in MSTO-211H cells have been challenging, resulting in either small decreases in expression or loss of viability. Transient Upf1 knockdown in HeLa cells resulted in seven- and twofold increases in Trxr2 and SelW mRNA levels, respectively (23). No changes in selenoprotein mRNAs were reported following stable Upf2 knockdown in HeLa cells, although the authors indicate that Trxr2 mRNA levels were below detection (32). No other selenoprotein mRNAs were discussed in either of these studies. Further investigations will be required to determine the mechanism of Upf2 upregulation in the present study and whether increases in the levels of NMD substrates might affect the levels of NMD-specific factors and, if so, how this might occur.
Supplementary Material
Acknowledgments
This work was supported by NIH grants DK52963 and DK47320 to M.J.B.
Footnotes
Published ahead of print on 10 September 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Behne, D., H. Hilmert, S. Scheid, H. Gessner, and W. Elger. 1988. Evidence for specific selenium target tissues and new biologically important selenoproteins. Biochim. Biophys. Acta 966:12-21. [DOI] [PubMed] [Google Scholar]
- 2.Bermano, G., J. R. Arthur, and J. E. Hesketh. 1996. Role of the 3′ untranslated region in the regulation of cytosolic glutathione peroxidase and phospholipid-hydroperoxide glutathione peroxidase gene expression by selenium supply. Biochem. J. 320:891-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chavatte, L., B. A. Brown, and D. M. Driscoll. 2005. Ribosomal protein L30 is a component of the UGA-selenocysteine recoding machinery in eukaryotes. Nat. Struct. Mol. Biol. 12:408-416. [DOI] [PubMed] [Google Scholar]
- 4.Christensen, M. J., and K. W. Burgener. 1992. Dietary selenium stabilizes glutathione peroxidase mRNA in rat liver. J. Nutr. 122:1620-1626. [DOI] [PubMed] [Google Scholar]
- 5.Copeland, P. R., and D. M. Driscoll. 2002. Purification and analysis of selenocysteine insertion sequence-binding protein 2. Methods Enzymol. 347:40-49. [DOI] [PubMed] [Google Scholar]
- 6.Copeland, P. R., and D. M. Driscoll. 1999. Purification, redox sensitivity, and RNA binding properties of SECIS-binding protein 2, a protein involved in selenoprotein biosynthesis. J. Biol. Chem. 274:25447-25454. [DOI] [PubMed] [Google Scholar]
- 7.Copeland, P. R., V. A. Stepanik, and D. M. Driscoll. 2001. Insight into mammalian selenocysteine insertion: domain structure and ribosome binding properties of Sec insertion sequence binding protein 2. Mol. Cell. Biol. 21:1491-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Czaplinski, K., T. Kocher, M. Schelder, A. Segref, M. Wilm, and I. W. Mattaj. 2005. Identification of 40LoVe, a Xenopus hnRNP D family protein involved in localizing a TGF-beta-related mRNA during oogenesis. Dev. Cell. 8:505-515. [DOI] [PubMed] [Google Scholar]
- 9.de Jesus, L. A., P. R. Hoffmann, T. Michaud, E. P. Forry, A. Small-Howard, R. J. Stillwell, N. Morozova, J. W. Harney, and M. J. Berry. 2006. Nuclear assembly of UGA decoding complexes on selenoprotein mRNAs: a mechanism for eluding nonsense mediated decay? Mol. Cell. Biol. 26:1795-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Driscoll, D. M., and P. R. Copeland. 2003. Mechanism and regulation of selenoprotein synthesis. Annu. Rev. Nutr. 23:17-40. [DOI] [PubMed] [Google Scholar]
- 11.Dumitrescu, A. M., X. H. Liao, M. S. Abdullah, J. Lado-Abeal, F. A. Majed, L. C. Moeller, G. Boran, L. Schomburg, R. E. Weiss, and S. Refetoff. 2005. Mutations in SECISBP2 result in abnormal thyroid hormone metabolism. Nat. Genet. 37:1247-1252. [DOI] [PubMed] [Google Scholar]
- 12.Duncan, K., M. Grskovic, C. Strein, K. Beckmann, R. Niggeweg, I. Abaza, F. Gebauer, M. Wilm, and M. W. Hentze. 2006. Sex-lethal imparts a sex-specific function to UNR by recruiting it to the msl-2 mRNA 3′ UTR: translational repression for dosage compensation. Genes Dev. 20:368-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fletcher, J. E., P. R. Copeland, D. M. Driscoll, and A. Krol. 2001. The selenocysteine incorporation machinery: interactions between the SECIS RNA and the SECIS-binding protein SBP2. RNA 7:1442-1453. [PMC free article] [PubMed] [Google Scholar]
- 14.Ginisty, H., H. Sicard, B. Roger, and P. Bouvet. 1999. Structure and Functions of Nucleolin. J. Cell Sci. 112:761-772. [DOI] [PubMed] [Google Scholar]
- 15.Hadley, K. B., and R. A. Sunde. 2001. Selenium regulation of thioredoxin reductase activity and mRNA levels in rat liver. J. Nutr. Biochem. 12:693-702. [DOI] [PubMed] [Google Scholar]
- 16.Hatfield, D. L., and V. N. Gladyshev. 2002. How selenium has altered our understanding of the genetic code. Mol. Cell. Biol. 22:3565-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill, K. E., P. R. Lyons, and R. F. Burk. 1992. Differential regulation of rat liver selenoprotein mRNAs in selenium deficiency. Biochem. Biophys. Res. Commun. 185:260-263. [DOI] [PubMed] [Google Scholar]
- 18.Hill, K. E., J. Zhou, W. J. McMahan, A. K. Motley, J. F. Atkins, R. F. Gesteland, and R. F. Burk. 2003. Deletion of selenoprotein P alters distribution of selenium in the mouse. J. Biol. Chem. 278:13640-13646. [DOI] [PubMed] [Google Scholar]
- 19.Hoffmann, P. R., S. Hoge, P. A. Li, F. Hoffmann, A. Hashimoto, and M. J. Berry. 2007. The selenoproteome exhibits widely varying, tissue-specific dependence on selenoprotein P for selenium supply. Nucleic Acids Res. 35:3963-3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lei, X. G., J. K. Evenson, K. M. Thompson, and R. A. Sunde. 1995. Glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase are differentially regulated in rats by dietary selenium. J. Nutr. 125:1438-1446. [DOI] [PubMed] [Google Scholar]
- 21.Low, S. C., E. Grundner-Culemann, J. W. Harney, and M. J. Berry. 2000. SECIS-SBP2 interactions dictate selenocysteine incorporation efficiency and selenoprotein hierarchy. EMBO J. 19:6882-6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maquat, L. E., and G. Serin. 2001. Nonsense-mediated mRNA decay: insights into mechanism from the cellular abundance of human Upf1, Upf2, Upf3, and Upf3X proteins. Cold Spring Harb. Symp. Quant. Biol. 66:313-320. [DOI] [PubMed] [Google Scholar]
- 23.Mendell, J. T., N. A. Sharifi, J. L. Meyers, F. Martinez-Murillo, and H. C. Dietz. 2004. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 36:1073-1078. [DOI] [PubMed] [Google Scholar]
- 24.Moriarty, P. M., C. C. Reddy, and L. E. Maquat. 1998. Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol. Cell. Biol. 18:2932-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagy, E., and L. E. Maquat. 1998. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 23:198-199. [DOI] [PubMed] [Google Scholar]
- 26.Papp, L. V., J. Lu, F. Striebel, D. Kennedy, A. Holmgren, and K. K. Khanna. 2006. The redox state of SECIS binding protein 2 controls its localization and selenocysteine incorporation function. Mol. Cell. Biol. 26:4895-4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saedi, M. S., C. G. Smith, J. Frampton, I. Chambers, P. R. Harrison, and R. A. Sunde. 1988. Effect of selenium status on mRNA levels for glutathione peroxidase in rat liver. Biochem. Biophys. Res. Commun. 153:855-861. [DOI] [PubMed] [Google Scholar]
- 28.Schomburg, L., U. Schweizer, B. Holtmann, L. Flohe, M. Sendtner, and J. Kohrle. 2003. Gene disruption discloses role of selenoprotein P in selenium delivery to target tissues. Biochem. J. 370:397-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoytcheva, Z., R. M. Tujebajeva, J. W. Harney, and M. J. Berry. 2006. Efficient incorporation of multiple selenocysteines involves an inefficient decoding step serving as a potential translational checkpoint and ribosome bottleneck. Mol. Cell. Biol. 26:9177-9184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tujebajeva, R. M., P. R. Copeland, X. M. Xu, B. A. Carlson, J. W. Harney, D. M. Driscoll, D. L. Hatfield, and M. J. Berry. 2000. Decoding apparatus for eukaryotic selenocysteine incorporation. EMBO Rep. 2:158-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiss, S. L., and R. A. Sunde. 1998. Cis-acting elements are required for selenium regulation of glutathione peroxidase-1 mRNA levels. RNA. 4:816-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wittmann, J., E. M. Hol, and H. M. Jack. 2006. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol. Cell. Biol. 26:1272-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu, R., Q. Shen, and P. E. Newburger. 2000. Recognition and binding of the human selenocysteine insertion sequence by nucleolin. J. Cell Biochem. 77:507-516. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.