Abstract
Transcription of the testis-specific Pgk2 gene is selectively activated in primary spermatocytes to provide a source of phosphoglycerate kinase that is critical to normal motility and fertility of mammalian spermatozoa. We examined dynamic changes in protein-DNA interactions at the Pgk2 gene promoter during murine spermatogenesis in vivo by performing genomic footprinting and chromatin immunoprecipitation assays with enriched populations of murine spermatogenic cells at stages prior to, during, and following transcription of this gene. We found that genes encoding the testis-specific homeodomain factor PBX4 and its coactivator, PREP1, are expressed in patterns that mirror expression of the Pgk2 gene and that these factors become bound to the Pgk2 enhancer in cells in which this gene is actively expressed. We therefore suggest that these factors, along with CREM and SP3, direct stage- and cell type-specific transcription of the Pgk2 gene during spermatogenesis. We propose that binding of PBX4, plus its coactivator PREP1, is a rate-limiting step leading to the initiation of tissue-specific transcription of the Pgk2 gene. This study provides insight into the developmentally dynamic establishment of tissue-specific protein-DNA interactions in vivo. It also allows us to speculate about the events that led to tissue-specific regulation of the Pgk2 gene during mammalian evolution.
Spermatogenesis is a highly ordered process by which the germ cell lineage gives rise to functional gametes in the male. Development of the spermatogenic lineage involves unique events, including meiosis and genetic recombination, as well as dramatic morphological changes during spermiogenesis that give rise to the uniquely structured spermatozoon (7). As with other specific lineages in mammalian organisms, the characteristic differentiation of male germ cells proceeds as a result of expression of unique combinations of genes (17). Recent studies have revealed that spermatogenic cells express a surprisingly large proportion of genes in the murine genome. Microarray studies show that the majority of differential gene expression observed in the developing testis occurs in spermatogenic cells and that dynamic changes in gene expression accompany transitions from premeiotic to meiotic and from meiotic to postmeiotic stages of spermatogenesis (2, 52, 53). The same microarray studies have also confirmed that a large number of genes are expressed exclusively in spermatogenic cells. Thus, spermatogenesis is characterized by a unique transcriptome that undergoes dynamic, stage-specific changes, and this implies the ongoing function of mechanisms regulating tissue- and stage-specific transcription (37, 39).
Among the many germ cell-specific genes expressed during spermatogenesis is a set encoding isozymes that function during glycolysis, primarily in spermatozoa. These include the Ldhc, Gapdhs, Hk1s, Pgm2, Pdha2, and Pgk2 genes, which encode sperm-specific isozymes of lactate dehydrogenase, glyceraldehyde-3-phosphate dehydrogenase, hexokinase, phosphoglycerate mutase, pyruvate dehydrogenase E1 alpha, and phosphoglycerate kinase, respectively (13, 17, 20, 42). In mammals, two genes encode the glycolytic enzyme phosphoglycerate kinase, the X-linked Pgk1 gene and the autosomal Pgk2 gene (58). The Pgk1 gene is widely expressed in all somatic cells, oogenic cells, and premeiotic spermatogenic cells, whereas the Pgk2 gene is expressed exclusively in meiotic spermatocytes and postmeiotic spermatids, where expression of the Pgk1 gene is repressed by meiotic sex chromosome inactivation (MSCI) (57) and by the subsequent repressive effects of postmeiotic sex chromatin (PMSC) (38, 45, 60).
The promoter of the Pgk2 gene is believed to have derived from that of the Pgk1 gene as part of a retroposition event that led to the genesis of the Pgk2 gene early during mammalian evolution (12, 40). Thus, although the Pgk2 promoter was likely initially identical to the Pgk1 promoter, it appears to have diverged, especially in eutherian mammals, to facilitate tissue-specific expression. Evidence favoring a common ancestry of these two promoters includes the absence of a TATA box and the presence of GC and CAAT box elements in both (34, 35). Although this “core promoter” region from the Pgk2 gene is able to direct transcription of a downstream reporter gene in transient transfection assays (12), this portion alone is insufficient to direct expression of a reporter gene in vivo in transgenic mice (48). Indeed, a minimum of an additional 42 bp of enhancer sequence immediately upstream of the Pgk2 core promoter, along with the core promoter itself, is required to direct testis-specific expression in vivo (62). The acquisition of tissue-specific function at this enhancer region, along with the loss in the Pgk2 promoter of the CpG island present in the Pgk1 promoter, represents key differences that we previously suggested may have facilitated the evolution of testis-specific regulation of the Pgk2 gene (34, 35). We previously used a combination of analyses in vitro and in vivo to determine that spermatogenesis-specific expression of the Pgk2 gene is regulated by a 5′-flanking region that includes enhancer and core promoter elements that bind testis-specific and ubiquitous transcription factors, respectively (21, 48, 62). We also showed that demethylation of a domain encompassing this regulatory region precedes decondensation of chromatin, factor binding, and initiation of transcription by several days (3, 4, 22, 28, 29, 63).
The mammalian spermatogenic cell lineage is well characterized with respect to the sequential appearance of specific spermatogenic cell types, permitting a detailed study of developmental processes (37, 39). Here we have taken advantage of this system to examine in vivo the developmental dynamics of protein-DNA interactions that regulate the activation of the Pgk2 gene and to identify the key factors involved. By characterizing expression patterns during spermatogenesis of the genes encoding these factors, we can suggest those most likely to be the key regulators of tissue-specific, cell type-specific, and developmental stage-specific transcription of the Pgk2 gene during spermatogenesis. Our results provide the first demonstration of a spermatogenesis-specific gene that is regulated by the testis-specific homeodomain factor PBX4 and form the basis for a model of gene activation whereby this “pioneer transcription factor” (8, 15, 16, 50) initiates an ordered cascade of events that includes remodeling of chromatin and binding of other transcription factors to initiate transcription of the Pgk2 gene. Finally, our results facilitate a functional comparison of the promoter of the testis-specific Pgk2 gene with that of the ubiquitously expressed Pgk1 gene that allows us to suggest key molecular changes that accompanied the evolution of tissue-specific regulation of the Pgk2 gene.
MATERIALS AND METHODS
Preparation of cells.
All mice were obtained from Charles River Laboratories (Wilmington, MA), and all procedures involving animals were approved in advance by the University of Texas at San Antonio Institutional Animal Care and Use committee. We isolated specific spermatogenic cell types using a 2 to 4% bovine serum albumin “Sta Put” gradient with sedimentation velocity at unit gravity as previously described (7, 38). Primitive type A spermatogonia were isolated from the testes of male CD-1 mice at 6 days postpartum (dpp). Type A and type B spermatogonia were isolated from testes of eight dpp CD-1 mice. Preleptotene spermatocytes, leptotene-plus-zygotene spermatocytes, and early (juvenile) pachytene spermatocytes were isolated from the testes of 18 dpp mice. Pachytene spermatocytes, round spermatids, and elongated spermatids plus residual bodies were isolated from the testes of adult (≥60 dpp) mice. Testicular spermatozoa were isolated from the testes of adult mice by dissociation of tissue and sonication to lyse all other cells (38). The purity of each cell population was determined by morphological examination under phase optics. Purities of spermatogonial cell types and juvenile spermatocytes were ≥85%, while those of adult spermatocytes, spermatids, and spermatozoa were ≥95%. Spleen cells from adult CD-1 mice were used as a somatic control. Spleens were mechanically teased open with hypodermic needles in phosphate-buffered saline, and the resulting cell suspensions were filtered through 100-μm nylon mesh (Millipore, Billerica, MA) and then treated with Tris-buffered ammonium chloride (0.017 M Tris-HCl, pH 7.6, 0.84% NH4Cl) to lyse contaminating erythrocytes.
Dimethyl sulfate and piperidine treatments of DNA for in vivo footprinting and ligation-mediated PCR (LM-PCR).
In vivo dimethyl sulfate (DMS) treatment was performed with tissues or enriched populations of specific cell types, as previously described by Hornstra and Yang (24). The double-stranded linker (made from oligonucleotides LP1 and LP2 [Table 1]) used for annealing to the piperidine-cleaved blunt ends of DMS-treated genomic DNA has been described previously (43, 44). One pmol of gene-specific primer 1 (Table 1) was annealed to 1 μg of treated DNA and extended using Vent (exo−) DNA polymerase (New England Biolabs, Ipswich, MA). The reaction was performed for 1 cycle at 94°C for 5 min followed by 1 min at each of the following temperatures, in succession: 57°C, 55°C, 54°C, 53°C, 52°C, 51°C, and 50°C. Following the annealing steps, extension was performed at 72°C for 20 min. The double-stranded linker (150 pmol) was ligated to the genomic DNA fragments in an overnight ligation at 16°C with T4 DNA ligase (New England Biolabs). Following ligation, genomic DNA fragments were purified by ethanol precipitation and then amplified by using primer LP2 and a gene-specific primer 2 (Table 1). The PCR was performed as follows: 94°C for 12 min, followed by 20 cycles of 94°C for 30 s, 61°C for 2 min, and 76°C for 30 s. Following this amplification, 0.1 pmol of radiolabeled gene-specific primer 3 (Table 1) was added to each tube, and an additional five cycles were run, consisting of 94°C for 1 min, 69°C for 1 min, and 76°C for 2 min. The amplification products were purified by ethanol precipitation, resolubilized in formamide loading buffer, and denatured by boiling for 5 min. They were then loaded, along with an aliquot of treated naked DNA (G ladder), onto a 6% polyacrylamide gel and subjected to electrophoresis at 55 W of constant power for 3 h. Each gel was then dried and exposed overnight to an imaging screen (Bio-Rad, Hercules, CA) and visualized on a Molecular Imager FX (Bio-Rad).
TABLE 1.
Primers for in vivo footprinting/ligation-mediated PCR
| Function and primer name | Sequence |
|---|---|
| Double stranded-linker | |
| LP1 | 5′-GAATTCAGATC-3′ |
| LP2 | 5′-GTGACCCGGGAGATCTGAATTC-3′ |
| Pgk2 core promoter upper-strand primer set | |
| Pgk2 promoter upper-1 | 5′-TCCACTTTGTCCAGAGTC-3′ |
| Pgk2 promoter upper-2 | 5′-CAGAAAGAGCCATCTTGATGGTATGC-3′ |
| Pgk2 promoter upper-3 | 5′-CTTGATGGTATGCACAACAGCCTCTTTACTGC-3′ |
| Pgk2 core promoter lower-strand primer set | |
| Pgk2 promoter lower-1 | 5′-AACTCTAACCCCAAGCTC-3′ |
| Pgk2 promoter lower-2 | 5′-CTGTTTAGTATTCTAGGCTGGCAGTTTG-3′ |
| Pgk2 promoter lower-3 | 5′-CTAGGCTGGCAGTTTGAGTCTGTACCAGGC-3′ |
| Pgk2 upstream enhancer upper-strand primer set | |
| Pgk2 enhancer upper-1 | 5′-GGTGATTTTTAACGCTTAAG-3′ |
| Pgk2 enhancer upper-2 | 5′-CCTTTCTTTGTGAGTTCAAAGTCTC-3′ |
| Pgk2 enhancer upper-3 | 5′-GTGAGTTCAAAGTCTCCTGCACAATAGCCATTGG-3′ |
Chromatin immunoprecipitation assay.
Chromatin immunoprecipitation (ChIP) assays were performed using an immunoprecipitation assay kit (Upstate, Charlottesville, VA) according to the manufacturer's instructions, with minor modifications. Cells were fixed in a final concentration of 1% formaldehyde for 10 min at room temperature, and then the reaction was stopped by addition of 125 mM glycine. After cells were washed once with phosphate-buffered saline, they were resuspended in sodium dodecyl sulfate (SDS) lysis buffer (1% SDS, 50 mM Tris-HCl, pH 8.1, 10 mM EDTA) containing protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO), 1 mM phenylmethylsulfonyl fluoride, and 5 mM sodium butyrate and incubated for 10 min on ice. The resulting lysate was sonicated five times for 10 s each at 50% amplitude (Branson Digital Sonifier model 450; Branson Ultrasonics, Danbury, CT) to reduce the size of DNA fragments to 100 to 1,000 bp. The sample was centrifuged to remove cell debris and diluted 10-fold in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.1, 167 mM NaCl, protease inhibitor cocktail, 1 mM phenylmethylsulfonyl fluoride, 5 mM sodium butyrate). After the chromatin solution was precleared, it was subjected to immunoprecipitation with antibodies against SP1, SP3, PBX1/2/3/4, PBX1/2/3, PREP1, CREB-1, CREM-1, and CBF-B/NF-YA (Santa Cruz Biotechnology, Santa Cruz, CA). Normal rabbit antiserum (Santa Cruz Biotechnology) was used as a nonspecific, negative control for binding of transcription factors. After samples were incubated overnight with antibody at 4°C, the resulting immune complexes were collected by addition of 60 μl of a salmon sperm DNA-protein A agarose slurry (Upstate) and incubated at 4°C with agitation for 1 h. The beads were washed several times, and the attached immune complexes were eluted with a buffer containing 1% SDS and 0.1 M NaHCO3. Cross-links were reversed by the addition of 5 M NaCl and incubation at 65°C for 5 h. The samples were then treated with RNase A for 30 min and proteinase K for 1 h, and DNA was purified by phenol-chloroform extraction and ethanol precipitation. After immune complexes were eluted, DNA was quantified using PicoGreen (Invitrogen, Carlsbad, CA) and analyzed by quantitative PCR (qPCR). Samples from at least two independent immunoprecipitations were analyzed for each transcription factor examined. In addition to providing a normalization standard, analysis of rabbit immunoglobulin G (IgG) also provided a control for nonspecific binding of antibodies by establishing a threshold value for such nonspecific binding at 1.0. In addition, the Pgk1 and Pgk2 genes were analyzed in the immunoprecipitated DNA samples. Thus, each analysis also served as a control for the other.
Real-time qPCR for ChIP assay.
qPCR analysis was carried out with DNA precipitated by ChIP using SYBR Premix Ex Taq (Takara Bio USA, Madison, WI) on a Chromo4 real-time PCR detection system (Bio-Rad). Each PCR assay was performed in triplicate. PCR primers used are listed in Table 2. Relative amounts of DNA in the input and bound fractions were determined from cycle threshold (CT) values, using a standard curve generated from DNA of known concentrations. Values for enrichment were calculated as the average from at least two independent ChIP experiments and multiple independent PCR analyses with each antibody. Results were normalized to control values from rabbit IgG arbitrarily set at 1.0.
TABLE 2.
Primers for PCRs used with ChIP assays
| PCR primer | Sequence (forward and reverse) | Annealing temp (°C) |
|---|---|---|
| Pgk1 CAAT box | 5′-ACTAGTCTCGTGCAGATGGACAGC-3′ | 62 |
| 5′-CTCTGAGCCCAGAAAGCGAAG-3′ | ||
| Pgk2 E1-E4 | 5′-AACCCCAAGCTCTCCGTTTA-3′ | 62 |
| 5′-TCTGTGATTGGTTCTCCATCC-3′ | ||
| Pgk2 CAAT box | 5′-GGCTGGCAGTTTGAGTCTGT-3′ | 62 |
| 5′-TTGAAAGGAAATCCAGCTCTGT-3′ | ||
| Pgk2 GC box | 5′-CAATGGCTATTGTGCAGGAG-3′ | 62 |
| 5′-AGGCTGGCTTGGTGGTGA-3′ |
Real-time quantitative reverse transcription-PCR analysis.
Total RNA was isolated from purified cell populations and tissues, using the guanidine thiocyanate/CsCl method as described previously (62). Before reverse transcription, total RNA was treated with RNase-free DNase I (Invitrogen) to remove any DNA contamination. Five hundred nanograms of total RNA were used as template to synthesize first-strand cDNA using the GeneAmp RNA PCR kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. To control for the possibility of genomic DNA contamination, first-strand cDNA synthesis was conducted without reverse transcriptase and exposed to PCR amplification. cDNAs were then amplified by PCR using SYBR Premix Ex Taq (Takara Bio USA) on a Chromo4 real-time PCR detection system (Bio-Rad). The PCR primers used in this analysis are listed in Table 3. The relative expression of target mRNAs was calculated from target CT values and β-actin or 18S rRNA CT values using the standard curve method.
TABLE 3.
Primers for quantitative real-time RT-PCRs
| Gene-specific PCR primer | Sequence (forward and reverse) | Annealing temp (°C) |
|---|---|---|
| Pgk1 | 5′-CAGTTGCACAGCATCTCAGC-3′ | 58 |
| 5′-AGACGCCCTCTACAATGCAC-3′ | ||
| Pgk2 | 5′-TGGTGGTGGAATGGCTTACACCTTCCTG-3′ | 68 |
| 5′-TCGTGGCTCCCTCTTCATCAAACAAGG-3′ | ||
| Pbx1a | 5′-AGCCCTCATCCCTATGTTGAC-3′ | 60 |
| 5′-CAGTGTGGTGTATTCCTGCTAAGA-3′ | ||
| Pbx1b | 5′-TCTTGTGCCATCTTTGGTGA-3′ | 60 |
| 5′-GCCTGCTTTATTGTAGACCTTGTT-3′ | ||
| Pbx2 | 5′-TGACCCCCTCAGACATCCTC-3′ | 60 |
| 5′-CCACCCTCTTCACTCCCTCA-3′ | ||
| Pbx3 | 5′-CAAGGTGTGCTCTTCCTCTGG-3′ | 64 |
| 5′-TGTGTTTTGATTGGTGGGATG-3′ | ||
| Pbx4 | 5′-TGCGGCTTTGCCCTTTTTCA-3′ | 64 |
| 5′-TCGGGTGCTCAGAATGAACAGTCC-3′ | ||
| Meis1 | 5′-TCGGGAAGGGTGGGAAAAACTG-3′ | 64 |
| 5′-ACACGGCCTTTGGGTTTCCA-3′ | ||
| Meis2 | 5′-ATGGGTATGGCCCAGCCAAGTT-3′ | 64 |
| 5′-GTGGGGATGGCTTGGCAAAT-3′ | ||
| Meis3 | 5′-TGACCACCAGCCTTGCACCTAA-3′ | 64 |
| 5′-CGTTGAATGGGGTGGGAGCTTT-3′ | ||
| Prep1 | 5′-CCCCGTGCACAGAGATGCTTTT-3′ | 64 |
| 5′-ATGCCATTGTGCCCAGCAAG-3′ | ||
| Prep2 | 5′-CCATGGCTCCGCATGGAAAA-3′ | 64 |
| 5′-TCCCAAGTCTGCCCCTTAGCAA-3′ | ||
| β-actin | 5′-CCGTGAAAAGATGACCCAG-3′ | 60 |
| 5′-TAGCCACGCTCGGTCAGG-3′ | ||
| 18S rRNA | 5′-GTAACCCGTTGAACCCCATT-3′ | 60 |
| 5′-CCATCCAATCGGTAGTAGCG-3′ |
RESULTS
Transcription kinetics of Pgk genes during spermatogenesis.
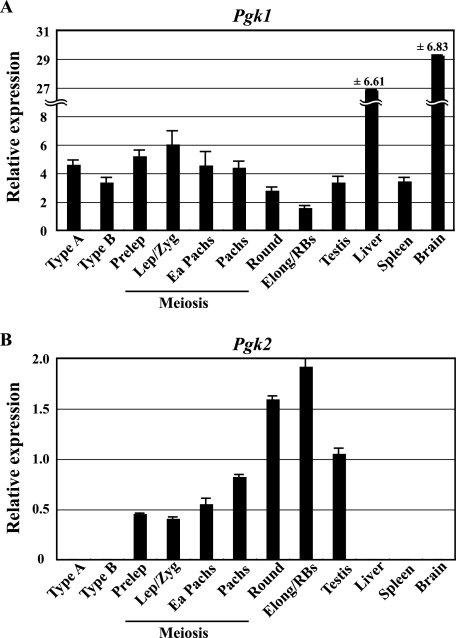
Pgk2 transcripts are first detectable at 10 to 12 dpp in mice, which correlates with the appearance of the first primary spermatocytes (38). This was confirmed using purified spermatogenic cell preparations, as Pgk2 transcripts were found in preleptotene spermatocytes but not in the immediately preceding cell type in the spermatogenic lineage, type B spermatogonia (38). Therefore, unlike the promoter-directing ubiquitous expression of the X-linked Pgk1 gene, the promoter of the autosomal Pgk2 gene directs highly regulated expression that is characterized by tissue-, cell type-, and stage-specific transcription. To more accurately characterize the kinetics of transcription of the Pgk genes during spermatogenesis, we performed real-time qRT-PCR with RNA samples from purified populations of spermatogonia, spermatocytes, and spermatids, as well as from whole adult testis, liver, spleen, and brain tissues (Fig. 1). As shown in Fig. 1A, moderate levels of transcript from the Pgk1 gene are present in premeiotic spermatogonia (type A and type B) but then show a steady decline during the latter half of the first meiotic prophase and continue to decline into the postmeiotic spermatid stage. This reflects the effects of MSCI and PMSC on expression of X-linked housekeeping genes during mammalian spermatogenesis (45, 57, 60). In contrast, the autosomal Pgk2 gene shows the opposite pattern of expression during spermatogenesis, with no expression in premeiotic spermatogonia and then a steady increase in expression during first meiotic prophase (Fig. 1B). Surprisingly, levels of Pgk2 transcript continued to show dramatic increases in round and elongated spermatids relative to those seen in primary spermatocytes, despite the fact that previous studies suggest that ongoing transcription of this gene in pachytene spermatocytes is as active as or more active than in round spermatids (29). It is possible that the relatively higher levels of Pgk2 transcripts detected in our qRT-PCR analysis of spermatids are due in part to an overall decrease in complexity of the RNA pool in these cells such that persistent levels of Pgk2 transcripts then represent a greater proportion of the total RNA pool. Nevertheless, the key point is the tightly regulated, tissue-, cell type-, and developmental stage-specific initiation of transcription of this gene in primary spermatocytes that is coincident with the onset of meiosis.
FIG. 1.
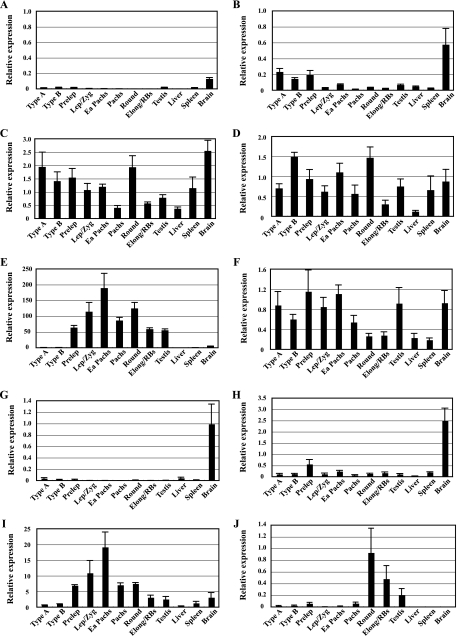
Quantitative RT-PCR analysis of Pgk gene expression during spermatogenesis. Expression of the Pgk1 (A) and Pgk2 (B) genes during spermatogenesis and in representative somatic tissues was quantified by real-time RT-PCR. The relative expression of target mRNA was calculated from the target CT values and β-actin mRNA CT values, using the standard curve method. Each error bar indicates standard error of the mean. Populations of the specific spermatogenic cell types examined included the following: Type A, type A spermatogonia; Type B, type B spermatogonia; Prelep, preleptotene spermatocytes; Lep/Zyg, leptotene plus zygotene spermatocytes; Ea Pachs, early (juvenile) pachytene spermatocytes; Pachs, pachytene spermatocytes; Round, round spermatids; Elong/RBs, elongated spermatids plus residual bodies.
Analysis of protein-DNA interactions at the Pgk2 promoter by DMS footprinting in vivo.
Previous electrophoretic mobility shift assays (EMSAs) demonstrated the capacity in vitro for both the CAAT and the GC box elements in the Pgk2 core promoter to bind ubiquitous factors found in nuclear extracts from either somatic cells or testicular cells (21). In addition, within the Pgk2 promoter region (Fig. 2A), two putative enhancer sequences (E1/E2 and E3/E4) located immediately upstream of the core promoter were shown to bind up to three different putative activator proteins in vitro from nuclear extracts of mouse testicular cells. Specifically, two binding activities were detected in the E1/E2 region, and a third binding activity was detected in the E3/E4 region. The E3/E4 region was also found to bind a putative repressor protein from nuclear extracts of nonexpressing somatic cells (21).
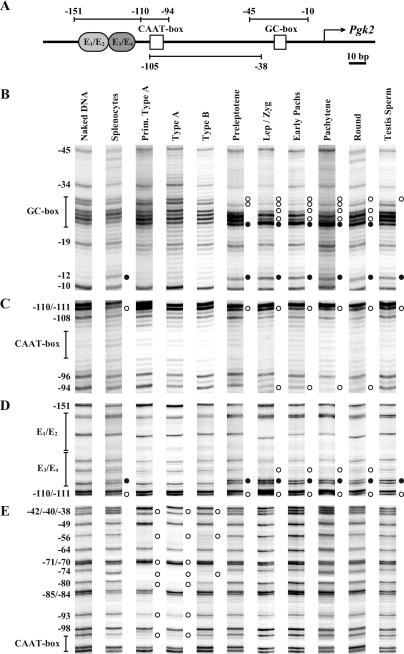
FIG. 2.
In vivo footprinting of the Pgk2 promoter during spermatogenesis in the mouse. (A) Schematic representation of putative regulatory elements in the Pgk2 gene promoter. The transcription start site is represented as a bent arrow. The core promoter includes a GC box and a CAAT box (open boxes). Immediately upstream from the core promoter is enhancer region E1/E2 and the E3/E4 subregions that have been shown to bind up to four anonymous factors. These sites are represented, respectively, as oval light and dark gray boxes. The location of amplimers analyzed by LM-PCR for in vivo footprinting are depicted as horizontal lines with numbers representing nucleotide positions relative to the transcription start site. Amplimers shown above and below the line represent analysis of upper and lower DNA strands, respectively. (B through E) DMS modification and LM-PCR were used to in vivo footprint the core promoter and enhancer regions of the Pgk2 promoter in purified populations of mouse spermatogenic cells. Open circles adjacent to bands represent protected cleavage sites, whereas filled circles represent enhanced cleavage sites. Numbers represent nucleotide positions relative to the transcriptional initiation site. Vertical lines represent the location of known or putative factor-binding elements. Prim. type A, primitive type A spermatogonia; type A, type A spermatogonia; type B, type B spermatogonia; Lep/Zyg, leptotene plus zygotene spermatocytes; early pachs, early (juvenile) pachytene spermatocytes; pachytene, pachytene spermatocytes; round, round spermatids; testis sperm, testicular sperm.
To determine which of the protein-DNA interactions detected in vitro actually occur in vivo, we performed in vivo genomic footprinting by treating freshly isolated cells with DMS, followed by LM-PCR-aided genomic sequencing. We purified populations of mouse spermatogenic cell types prior to, during, and following the period of active transcription of the Pgk2 gene and analyzed protein-DNA interactions in vivo in the core promoter and enhancer region (Fig. 2). We performed a similar analysis using somatic (spleen) cells to determine the status in vivo of protein-DNA interactions at the Pgk2 promoter when this gene is terminally repressed.
Because transcription of the Pgk2 gene is repressed in somatic cells, we expected to detect few protein-DNA interactions at the Pgk2 promoter in splenocytes other than those representing putative repressors. Indeed, we detected a binding activity at position −115 that extends to −110 and that overlaps the E3/E4 region, consistent with our previous EMSA data indicating binding of a single factor at the E3/E4 region in nonexpressing somatic cells (Fig. 2D) (21). This suggests that a repressor is constitutively bound to the Pgk2 enhancer in nonexpressing somatic cells; however, the identity of this repressor remains unknown. We also detected an enhancement at −12 in splenocytes, but we suspect that this represents nonspecific binding since it was detected in splenocytes, spermatocytes, and spermatids but not in spermatogonia.
Active transcription of the Pgk2 gene was previously demonstrated in primary spermatocytes and round spermatids by nuclear runoff experiments (29). We detected a variety of distinct footprints at the Pgk2 promoter site in vivo in spermatogenic cells. In the core promoter, a faint footprint first became evident at the GC box in preleptotene spermatocytes (extending from position −30 to −24) and became much more distinct in later spermatocytes (Fig. 2B). This footprint consisted of an enhancement at position −24 and several protections at −25, −27, −29, and −30 and developed gradually throughout the progression of first meiotic prophase, coinciding with the increase in Pgk2 transcriptional activity shown in Fig. 1B. In round spermatids, the footprint at the GC box diminished slightly (the protection at −29 disappeared) (Fig. 2B). This may indicate the beginning of a gradual unloading of a GC box-bound factor associated with changes in chromosomal architecture as histones are replaced by transition proteins and subsequently protamines during spermiogenesis (49).
The CAAT box (from position −105 to −100) lacks any guanosines within the putative binding sequence on the upper strand of the Pgk2 core promoter, such that factor binding in vivo cannot be assessed directly on this strand by using DMS as the footprinting agent (Fig. 2C). However, the guanosines (−105, −104, and −100) on the bottom strand also showed no significant footprint in spermatocytes and spermatids (Fig. 2E). Therefore, although a binding activity at the CAAT box was detected in vitro in EMSA experiments (21), we were not able to confirm a protein-DNA interaction at this site in vivo, even in cell types that were actively transcribing the Pgk2 gene.
In spermatocytes and round spermatids, protein-DNA interactions were also detected in the Pgk2 enhancer region, specifically in the E3/E4 region at about the same stage as factor binding developed in the core promoter (Fig. 2D). This was indicated by the appearance, initially in preleptotene spermatocytes, of an enhancement at position −115 and a protection at −110. Subsequently, in a mixture of leptotene and zygotene spermatocytes, an additional protection became visible at −119. These factor-binding activities persisted through the round spermatid stage. We conclude that this footprint is distinct from that seen in this same region in somatic splenocytes for two reasons. First, the footprint detectable in spermatocytes extends from −110 to −119 and, as such, is more expansive than the footprint detected in splenocytes that extends from −110 to only −115. Second, the footprint detected in splenocytes indicative of a putative repressor is not observed in spermatogonia, indicating that the putative repressor becomes dissociated from the E3/E4 region prior to activation and is subsequently replaced by an activator that binds to this same region in spermatocytes and spermatids.
While we were able to detect a protein-DNA interaction at the E3/E4 region of the Pgk2 enhancer in expressing spermatocytes and spermatids in vivo, we detected no binding activity at the E1/E2 region in these same cell types (Fig. 2D). Thus, as with the CAAT box, we found that certain sequence elements that demonstrated the capacity to bind nuclear factors in reconstitution EMSA experiments in vitro are, in fact, not bound by proteins in vivo. This demonstrates the need to confirm results obtained in vitro with analyses performed in vivo.
Interestingly, even though the GC box is bound in spermatocytes and spermatids by a ubiquitously expressed factor, we detected no evidence of protein binding at this site in any of the spermatogonial cell types we examined (Fig. 2B). This suggests that the Pgk2 promoter exists in a condensed (repressed) chromatin state in spermatogonia, such that factors that are already present at this stage are prevented from accessing their cognate binding sites. Similarly, we observed no evidence of factor binding in the Pgk2 enhancer region in spermatogonia (Fig. 2D). However, we did detect several apparent protein-DNA interactions in the form of protections in the region between the CAAT box and the GC box, specifically on the lower strand in primitive type A and type A spermatogonia (at positions −100, −93, −80, −74, −70, −56, and −40) and, to a lesser extent, in type B spermatogonia (only at positions −74, −56, and −40) (Fig. 2E). The pattern of these footprints suggests a periodicity of about 20 bp that could represent a phased nucleosome, but we have no other data to confirm this. If a phased nucleosome is present in this position in spermatogonia, then it apparently becomes displaced coincident with the subsequent initiation of active transcription of the Pgk2 gene in spermatocytes and spermatids.
Postmeiotic spermatogenic cells undergo the unique differentiative process of spermiogenesis that includes, among other things, significant chromatin condensation as histones associated with DNA in earlier spermatogenic cells are ultimately replaced by protamines (49). As a result, global transcription ceases, presumably due to restricted access of the transcriptional machinery to promoter sequences. Therefore, we expected to observe a reduction in protein-DNA interactions associated with the Pgk2 promoter in spermatozoa. Indeed, the protections and enhancements seen at the GC box and enhancer region in expressing spermatocytes and spermatids largely disappeared in testicular spermatozoa, wherein we observed a footprint pattern similar to that seen in nonexpressing somatic cells (Fig. 2B to D).
Identification by ChIP analysis of factors regulating Pgk gene transcription in vivo.
The GC box is well characterized as an SP/KLF family binding site and is a common cis regulatory element present in the promoters of several genes expressed in a developmentally specific manner in differentiating mouse germ cells (11, 25, 54, 56, 61). The SP/KLF family of transcription factors is united by a particular combination of three conserved C2H2 zinc fingers that form the sequence-specific DNA-binding domain (31). Within the SP/KLF family of transcription factors, SP1 and SP3 are ubiquitously expressed in mammalian cells, including spermatogenic cells (56) and so were primary candidates for the factors responsible for the footprint at the Pgk2 GC box in spermatocytes and spermatids.
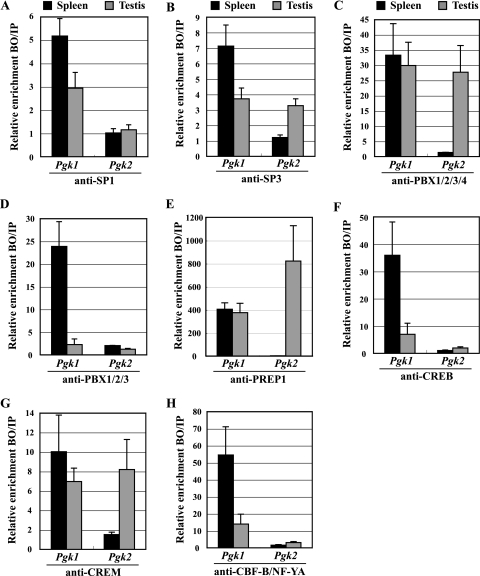
To examine the association of the SP1 and SP3 factors with GC boxes in the Pgk1 and Pgk2 promoters in vivo, we conducted ChIP experiments on cells from adult mouse testis and spleen. In contrast to the Pgk2 promoter, which contains a single GC box, the murine Pgk1 promoter has five GC box elements located within a CpG island (as revealed by TFSEARCH [http://www.cbrc.jp/research/db/TFSEARCH.html]). We observed an association of both SP1 and SP3 with the Pgk1 promoter in somatic splenocytes (Fig. 3A and B). Similarly, in adult mouse testis, which contains both somatic and germ cells, we also detected both SP1 and SP3 in association with the Pgk1 promoter (Fig. 3A and B). In contrast, the Pgk2 promoter showed only background levels of association with either factor in splenocytes (where this gene is not expressed) but showed a significant association with SP3, but not SP1, in the testis (where this gene is expressed) (Fig. 3A and B).
FIG. 3.
In vivo binding of transcription factors to the Pgk1 and Pgk2 promoters. ChIP analyses were performed with testicular germ cells and splenocytes, using antibodies specific for SP1 (A), SP3 (B), PBX1/2/3/4 (C), PBX1/2/3 (D), PREP1 (E), CREB (F), CREM (G), and CBF-B/NF-YA (H). Precipitated DNA was subjected to real-time PCR with the use of specific primers for the Pgk1 and Pgk2 promoters (see below), and relative enrichment of amplified regions in immunoprecipitated DNA (bound fractions; BO) was determined and compared to that in input (IP) DNA. All results were normalized to values for normal rabbit IgG, with that level arbitrarily set to 1.0. Each error bar indicates standard error of the mean. All PCR primers used are listed in Table 2. The PCR primer set named “Pgk1 CAAT box” was used to analyze the binding of all transcription factors examined in the Pgk1 promoter. This primer set amplified the “NF1-like region” (from position −180 to −77) (14, 47), including the consensus binding site for PBX factors and the CAAT box element (the binding site for CBF-B/NF-YA) in the Pgk1 promoter. Because of the size of the sonication fragments, amplification of this region was also used to examine the binding of SP factors to the nearby GC boxes at positions −66 and −254 in the Pgk1 promoter. Similarly, the PCR primer sets named “Pgk2 E1-E4,” “Pgk2 CAAT box,” and “Pgk2 GC box” (Table 2) were used to selectively amplify specific regions in the Pgk2 promoter, including a region containing the E1/E2-plus-E3/E4 enhancer sites, a region containing the CAAT box, and a region containing the GC box, respectively. The “Pgk2 GC box” primer set was also used to analyze the binding of CREB and/or CREM to the putative CRE site (+333). Deduction of specificity of binding of each factor to each element was based on the fact that, except where noted otherwise, each putative binding element represented the only candidate consensus binding sequence for each factor within at least 1 kb in each direction in each promoter.
A portion of the E3/E4 enhancer element (5′-AGATAGACAG-3′) in the Pgk2 gene shows consensus homology with a bipartite PBX-HOX binding site (5′-AGATTGATCG-3′) that is essential for tissue-specific enhancer activity in the Hoxb2 gene (18). Heterodimerization of PBX and PREP1 (also known as PKNOX1) yields a complex with strong binding affinity for the sequence 5′-TGACAG-3′ (9, 10, 19), and this site is also found within the E3/E4 enhancer element in the Pgk2 gene. Furthermore, a recently reported, expanded consensus MEIS1A-HOXA9 heterodimeric binding sequence (5′-TGACAGKTTWAYGA-3′; http://www.cbil.upenn.edu/cgi-bin/tess/tess) matches the 3′ half of the E3/E4 enhancer element as noted above plus an additional 8 bp that match the sequence in the Pgk2 promoter immediately 3′ to the E3/E4 enhancer element, yielding a total potential binding site in the Pgk2 enhancer for the PBX-MEIS, the PBX-HOX, or the HOX-MEIS complex of 18 bp (5′-AGATAGACAGGATGGAGA-3′). PBX and PREP1 are members of the TALE homeodomain family, with homeodomains most closely related to the PBC and MEIS subfamilies, respectively (41). Wagner et al. (59) identified PBX4 as a novel testis-specific member of the PBX family. Note that the mammalian Pbx4 gene expressed exclusively in the mammalian testis is not orthologous to the zebra fish pbx4 gene required for the development of the hindbrain (41). Expression of the mammalian Pbx4 gene is first detectable in the prepuberal testis coincident with the onset of the first wave of spermatogenesis (59). Therefore, PBX4 represented a strong candidate for the factor responsible for the footprint at the Pgk2 E3/E4 enhancer element in spermatocytes and spermatids. The Prep1 gene is also expressed predominantly in the testis (19) and so represented a candidate for a coactivator that could function in a complex with PBX4 to regulate testis-specific activation of the Pgk2 gene.
We used ChIP to investigate the interactions in vivo between PBX4 and PREP1 and the Pgk2 enhancer elements E3/E4. Because an antibody that solely recognizes PBX4 was not commercially available, we performed ChIP assays using an anti-PBX1/2/3/4 antibody, which recognizes all four isoforms of the PBX family, and an anti-PBX1/2/3 antibody that recognizes the first three but not the fourth isoform (Fig. 3C and D). The anti-PBX1/2/3/4 antibody precipitated Pgk1 sequences from both spleen and testis and Pgk2 sequences from testis only. The anti-PBX1/2/3 antibody also precipitated Pgk1 sequences at high levels from spleen but only at very low levels from testis. Pgk2 sequences were not precipitated from either spleen or testis, using the anti-PBX1/2/3 antibody. This suggests that PBX4 is the sole PBX isoform bound to the Pgk2 gene and is the predominant isoform bound to the Pgk1 gene in the testis. We also confirmed that PREP1 localizes to the Pgk2 promoter in the testis, but not in spleen, and to the Pgk1 gene in both spleen and testis (Fig. 3E). These associations support the contention that PBX factors function as activators in conjunction with the coactivator PREP1 to stimulate transcription from Pgk promoters.
Although they were not indicated by our footprinting experiments, two other candidate regulators of Pgk transcription were the cyclic AMP-responsive element modulator (CREM) and its family member the cyclic AMP-responsive element binding protein (CREB). CREM is a transcription factor known to be critically required for spermatogenesis (26). A microarray study of gene expression in testes of CREM-deficient mice revealed a 2.8-fold decrease in the level of Pgk2 transcripts (http://www.dkfz.de/tbi_old/crem/index.html) (6). Indeed, half CRE sites (5′-TGACG-3′ and/or 5′-CGTCA-3′) are found at −1632 and +333 of the murine Pgk2 gene (http://natural.salk.edu/CREB/) (64). CREB is known to function in glucose homeostasis, for which the glycolytic enzyme phosphoglycerate kinase, encoded by the Pgk1 and Pgk2 genes, is crucial (33). Previous studies showed that the human PGK1 promoter is occupied by CREB in primary hepatocytes and in the human embryonic kidney cell line HEK293T (64).
We examined the association of CREM and CREB with the Pgk1 and Pgk2 promoters in testis and spleen. We confirmed the binding of CREB to the Pgk1 promoter in both spleen and testis, but no significant association of CREB with the Pgk2 promoter was observed in either tissue (Fig. 3F). However, CREM, a master regulator of spermatogenesis (26), was found to be associated with both the Pgk1 and Pgk2 promoters in the testis (Fig. 3G). Thus, both CREB and CREM are bound to the Pgk1 gene in the testis, but only CREM is bound to the Pgk2 gene in this same tissue. There are both activating and repressing isoforms of CREM (26). Because a microarray study of gene expression during spermatogenesis in a CREM-deficient mouse revealed a 2.8-fold decrease in expression of the Pgk2 gene (6), we assume there are normally activating isoforms of CREM bound to this gene in spermatocytes and spermatids in vivo.
Finally, we examined the association of CBF-B/NF-YA in both the Pgk1 and Pgk2 promoters, because it is the most likely factor to bind to a CAAT box (55). Pfeifer et al. (46) and Dai et al. (14) used genomic sequencing to detect a footprint in vivo at the CAAT box in the actively expressed human PGK1 gene, and our ChIP analysis revealed an association of CBF-B/NF-YA with the mouse Pgk1 promoter in somatic (spleen) cells where the Pgk1 gene is actively expressed (Fig. 3H). However, as noted above, our in vivo footprinting analysis of the Pgk2 gene in spermatocytes and spermatids revealed no protein-DNA interaction at the Pgk2 CAAT box (Fig. 2C and E). This is consistent with our ChIP analysis that revealed no association of CBF-B/NF-YA with the Pgk2 promoter in testicular cells (Fig. 3H).
Spermatogenic stage-specific binding of transcription factors to the Pgk2 gene.
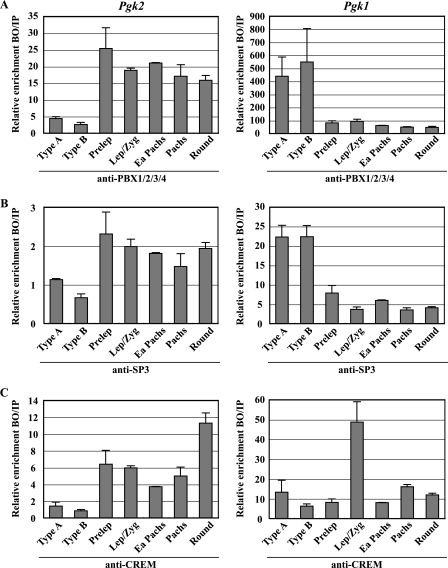
Having identified the key transcriptional regulators of the Pgk2 gene, we next sought to determine the developmental kinetics by which these factors interact with the Pgk2 promoter during spermatogenesis in vivo. We performed ChIP assays for PBX4, SP3, and CREM on highly enriched populations of specific mouse spermatogenic cell types at stages prior to, during, and following the period of active transcription of the Pgk2 gene. As shown in Fig. 1B, transcription of the Pgk2 gene begins at the onset of meiosis, and levels of Pgk2 transcript then increase steadily throughout spermatogenesis. On the other hand, the X-linked Pgk1 gene is expressed in all somatic cells and in premeiotic spermatogonia, but transcription of this gene ceases in primary spermatocytes due to MSCI (57) and continues to be repressed in spermatids due to the effects of PMSC (Fig. 1A) (45, 60). We found that PBX4, SP3, and CREM are all recruited to the Pgk2 promoter predominantly at the onset of meiosis and then remain present throughout meiosis and the subsequent differentiation of spermatogenic cells to the round spermatid stage (Fig. 4A to C, left side), coinciding exactly with the pattern of active transcription of the Pgk2 gene during spermatogenesis.
FIG. 4.
Developmental stage- and cell type-specific recruitment of transcription factors to the Pgk1 and Pgk2 promoters. ChIP analyses were performed with relatively pure populations of specific spermatogenic cell types prior to, during, and following the period of active transcription of the Pgk2 gene, using antibodies specific for PBX1/2/3/4 (A), SP3 (B), and CREM (C). Precipitated DNA was subjected to real-time PCR with the use of specific primers for the Pgk2 (left side panels) and Pgk1 (right side panels) promoters, and enrichment of immunoprecipitated DNA (bound fractions, BO) relative to input (IP) DNA was calculated in each case. All results were normalized to values for normal rabbit IgG, with that level arbitrarily set to 1.0. Each error bar indicates standard error of the mean. Type A, type A spermatogonia; Type B, type B spermatogonia; Prelep, preleptotene spermatocytes; Lep/Zyg, leptotene plus zygotene spermatocytes; Ea Pachs, early (juvenile) pachytene spermatocytes; Pachs, pachytene spermatocytes; Round, round spermatids.
In contrast, the binding of PBX factors and SP3 to the Pgk1 promoter was prominent in premeiotic type A and type B spermatogonia but then declined rapidly at the onset of meiosis, correlating with the cessation of transcription of the Pgk1 gene due to MSCI (Fig. 4A and B, right side). CREM remained associated with the Pgk1 promoter at relatively low levels throughout spermatogenesis, except in leptotene-plus-zygotene spermatocytes, which showed a striking transient increase (Fig. 4C, right side), possibly indicating a role for this factor in initiating MSCI.
Expression profile of transcriptional activators of the Pgk2 gene during spermatogenesis.
Our factor- and stage-specific ChIP assays revealed the identities of key factors regulating transcription of the Pgk2 gene but did not reveal which of these factors might be the primary regulator(s) of the tissue- and stage-specific transcription of this gene during spermatogenesis. To gain further insight into this aspect, we analyzed expression profiles during spermatogenesis of Pbx4 and Prep1, along with other members of the Pbx (Pbx1, Pbx2, Pbx3) and Meis (Meis1, Meis2, Meis3, and Prep2) families of genes encoding transcription factors. Using real-time RT-PCR, we examined total RNA from highly enriched populations of specific spermatogenic cell types, as well as from whole adult testis, liver, spleen, and brain tissues (Fig. 5). The relative expression levels of Pbx1a and Pbx1b, alternative splicing variants of transcripts from the Pbx1 gene, were consistently low at all stages of spermatogenesis and in all tissues examined, except for the expression level of Pbx1b in the brain (Fig. 5A and B), while the Pbx2 and Pbx3 genes were expressed at various levels throughout spermatogenesis, with the Pbx2 expression level showing a decrease during meiosis (Fig. 5C and D). However, the testis-specific Pbx4 gene showed a tightly regulated expression pattern during spermatogenesis, initiating transcription in meiotic spermatocytes and continuing in postmeiotic spermatids (Fig. 5E), mirroring the initial expression profile of the Pgk2 gene (Fig. 1B). Meis1 was also expressed at various levels during spermatogenesis (Fig. 5F), while the Meis2 and Meis3 genes were repressed in male germ cells, although these genes were expressed at high levels in the brain (Fig. 5G and H). Expression of the Prep1 and Prep2 genes was upregulated in the testis. Prep1 showed increasing expression beginning at the onset of meiosis in a pattern similar to that of the Pbx4 gene (Fig. 5I), whereas Prep2 was expressed almost exclusively in postmeiotic spermatids (Fig. 5J). These findings are complemented by the results of previous studies in which PBX4 protein was detected in a subnuclear domain in pachytene spermatocytes (59) and the PREP1 protein was detected in the adult testis (19). Because the expression patterns of the Pbx4 and Prep1 genes mirror that of Pgk2 and because our ChIP analysis demonstrates that these factors associate with the Pgk2 promoter in an appropriate stage-specific pattern, we suggest that a complex of PBX4 and PREP1 is likely to be the primary factor involved in initiating stage- and cell type-specific transcription of the Pgk2 gene during spermatogenesis.
FIG. 5.
qRT-PCR analysis of expression during spermatogenesis of genes encoding PBX and MEIS factors. Expression of different members of the Pbx family of genes, Pbx1a (A), Pbx1b (B), Pbx2 (C), Pbx3 (D), and Pbx4 (E), and the Meis family of genes, Meis1 (F), Meis2 (G), Meis3 (H), Prep1 (I), and Prep2 (J), was quantitated by real-time RT-PCR. The relative expression of each target mRNA was calculated from the target CT values and the 18S rRNA CT values, using the standard curve method. Each error bar indicates standard error of the mean. Type A, type A spermatogonia; Type B, type B spermatogonia; Prelep, preleptotene spermatocytes; Lep/Zyg, leptotene plus zygotene spermatocytes; Ea Pachs, early (juvenile) pachytene spermatocytes; Pachs, pachytene spermatocytes; Round, round spermatids; Elong/RBs, elongated spermatids plus residual bodies.
DISCUSSION
By taking advantage of the uniquely accessible spermatogenic cell lineage, we were able to capture a rare glimpse into the developmental dynamics of the establishment of key protein-DNA interactions associated with transcriptional activation of the testis-specific Pgk2 gene in vivo. A single footprint was detected in the E3/E4 enhancer region in somatic cells, where the Pgk2 gene is terminally repressed (Fig. 2D). We assume this represents binding of a repressor, although the identity of this repressor remains unknown. Interestingly, this interaction was not observed in spermatogonia, suggesting that the somatic repressor is either not expressed in spermatogenic cells or is present but no longer bound to the Pgk2 gene in these cells. Previous EMSA studies with nuclear extracts from prepuberal testes did show a band attributable to the E3/E4 site, favoring the latter alternative (21). We previously chronicled demethylation of DNA in a domain that encompasses the Pgk2 promoter (22). This demethylation takes place in fetal (type T1) and neonatal (type T2) prospermatogonia and therefore precedes the spermatogonial stage of spermatogenesis. Thus, it is possible that one direct or indirect effect of this spermatogenesis-specific demethylation event is to release or prevent binding of this repressor. The fact that the footprint of this putative repressor overlaps with that of the tissue-specific activator (PBX4) suggests a possible switch mechanism operating at the E3/E4 element based on differential binding of these two factors (see below).
Although transcription of the Pgk2 gene does not begin until the meiotic spermatocyte stage (Fig. 1B), we observed unique footprints within the Pgk2 promoter region in premeiotic spermatogonia (Fig. 2E). These footprints, located between the CAAT and GC boxes in the Pgk2 core promoter, were observed for premeiotic spermatogonia but not for subsequent spermatocytes or spermatids and were most prominent in primitive type A spermatogonia isolated from prepuberal males at 6 dpp and in type A spermatogonia isolated at 8 dpp and less prevalent in differentiating type B spermatogonia, also isolated from males at 8 dpp. Because these footprints were observed prior to but not during active transcription of the Pgk2 gene, we envision two potential explanations for what they represent. One possibility, supported by the ∼20-bp periodicity of these footprints, is that they represent the presence of a single, phased nucleosome positioned in this region just prior to the activation of transcription that then becomes displaced to facilitate factor binding required for the initiation of transcription. A second possibility is that these footprints could represent the presence of chromatin remodeling complexes involved in initiating critical changes in chromatin structure to potentiate the initiation of transcription of this gene in spermatocytes, although detectable decondensation of chromatin and formation of a DNase I hypersensitive site in the Pgk2 promoter do not occur until late in the spermatogonia type B stage (28, 29).
A distinct set of footprints was detected uniquely in those spermatogenic cell types in which the Pgk2 gene is known to be actively transcribed (Fig. 1B) (29, 38). Thus, in meiotic spermatocytes and postmeiotic round spermatids, we observed unique footprints at the GC box and in the E3/E4 enhancer region (Fig. 2B and D). Based on our ChIP assays, we report here, for the first time, the putative identity of the transcription factors responsible for initiation of transcription of the Pgk2 gene during spermatogenesis. We find that the SP3 factor is preferentially bound to the GC box in the Pgk2 core promoter in spermatocytes and spermatids and that the PBX4 factor is bound to the E3/E4 region in the Pgk2 enhancer in these same cells (Fig. 3 and 4). Mammalian PBX factors have been shown to independently dimerize with either HOX proteins or MEIS/PREP proteins (41). Trimeric complexes encompassing all three homeoproteins, PBX-HOX-MEIS/PREP, have also been characterized (41). Our ChIP assays detected PREP1 at the Pgk2 promoter in the testis (Fig. 3). Therefore, initiation of Pgk2 transcription appears to be driven by a complex involving the PBX4 and PREP1 factors bound to the E3/E4 enhancer region, and this complex presumably interacts with other factors bound to the Pgk2 promoter, including SP3 and CREM, to form an enhanceosome-like structure that directs the initiation of transcription of the Pgk2 gene (62). Interestingly, the activity of certain tissue-specific genes activated by PBX-containing complexes was previously shown to be regulated by a balance, or switch, between activator and repressor complexes (5, 51). The alternate protein-DNA interactions we observed at the E3/E4 enhancer in somatic cells, where the Pgk2 gene is repressed, and spermatocytes and spermatids, where the Pgk2 gene is actively transcribed, could represent the function of such a switch mechanism.
Our developmental analysis demonstrated that occupation of the Pgk2 enhancer site by the PBX4-PREP1 complex first occurs coincidentally with the onset of meiosis in primary spermatocytes and that the initial occupation of the GC box by SP3 in the Pgk2 core promoter and the association of CREM with the Pgk2 gene also occur at approximately this same time. This is despite the fact that SP3 and CREM are present in earlier, premeiotic spermatogonia, as are other members of the PBX and MEIS transcription factor families. This suggests that prior to the early primary spermatocyte stage, the Pgk2 promoter is inaccessible to binding by most transcription factors, consistent with our previous observations that chromatin in this region remains relatively condensed in spermatogonia and then decondenses in spermatocytes (28, 29). The stage-specific expression patterns during spermatogenesis of the genes encoding the PBX4 and PREP1 factors suggest that the initial formation of this complex directly regulates the coincident, stage-specific transcriptional activation of the Pgk2 gene.
In this regard, it is interesting to note that somatic isoforms of the PBX factor family have been implicated in the activation of muscle-specific gene expression (8, 15, 27). In muscle, PBX forms a complex with MEIS and penetrates repressive chromatin in the myogenin promoter to recruit MYOD, which then interacts with histone acetyltransferases to promote the acetylation of histones and the association of SWI/SNF complexes that then initiate chromatin remodeling, and this, in turn, facilitates access for stable binding of additional activator complexes to the myogenin promoter (8, 15). Indeed, it has been suggested that PBX-related complexes act as “pioneer” transcription factors to enter repressed (condensed) chromatin and initiate remodeling to facilitate initiation of transcription of tissue-specific genes (8, 15, 16, 50). Our observations of molecular changes in the Pgk2 promoter at the beginning of the spermatocyte stage, including the disappearance of a potential phased nucleosome (Fig. 2E), decondensation of chromatin (28, 29), and changes in histone modifications (H. Yoshioka, J. L. Hornecker, and J. R. McCarrey, unpublished data), all support the contention that chromatin remodeling is indeed induced in the Pgk2 promoter at this stage. The contention that binding of the PBX4-PREP1 complex to the Pgk2 enhancer is critical for the activation of Pgk2 transcription is further supported by our earlier observations that the E3/E4 region is required to direct the testis-specific expression in vivo of Pgk2 promoter/CAT reporter transgenes (48, 62) and that this region is also required to direct changes in histone modifications in the Pgk2 promoter (H. Yoshioka et al., unpublished data). Thus, these observations are consistent with a model in which the PBX4-PREP1 complex binds to the Pgk2 promoter and recruits histone-modifying enzymes and chromatin remodeling factors to facilitate decondensation of chromatin, reconfiguration of nucleosomes, and binding of transcription factors including SP3 and CREM to initiate spermatogenesis-specific transcription of the Pgk2 gene.
Our in vivo analysis of protein-DNA interactions in specific spermatogenic cell types allowed us to examine the developmental sequence of events leading to the initiation of transcription of the Pgk2 gene. Previous developmental studies of molecular events at the promoters of the HNF-4α (23), the collagenase (32), the IFN-β (1), and the chicken lysozyme (30) genes suggest that the order in which histone modifications, chromatin remodeling, and factor binding normally occur varies among different tissue-specific genes undergoing transcriptional activation. Our results indicate that recruitment of the PBX4-PREP1 complex, SP3, and CREM to the Pgk2 promoter occurs at approximately the same time as chromatin decondensation (28) and the formation of a DNase I hypersensitive site (29), as well as changes in histone modifications (H. Yoshioka et al., unpublished data), all of which occur coincident with the spermatogonium-spermatocyte transition. Importantly, however, the resolution of our developmental analyses in vivo was limited to days rather than minutes or hours. Thus, we cannot distinguish between truly simultaneous events and sequential events that occur in rapid succession. Nevertheless, we have shown that these parameters all undergo significant changes at this stage and none (other than demethylation of DNA [22]) is preset significantly prior to the stage at which transcription of the Pgk2 gene is first initiated.
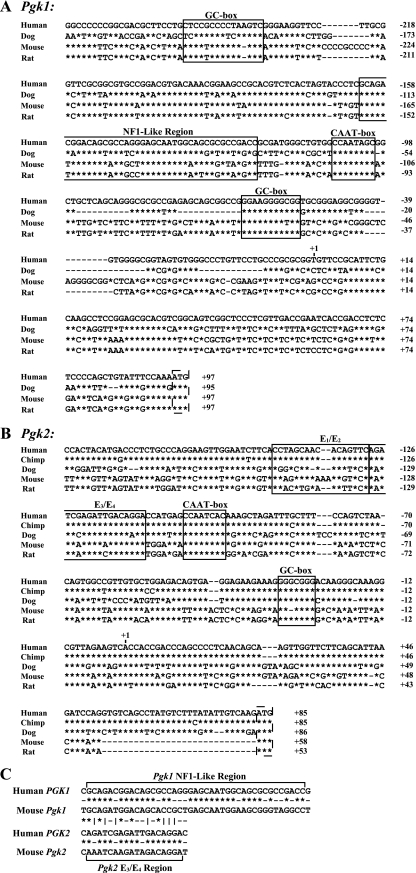
As noted in the introduction, the intronless Pgk2 gene is believed to have arisen in the mammalian genome as a retroposed copy of an intron-containing Pgk1-like progenitor gene early during mammalian evolution (12, 40). Transcription of the newly formed Pgk2 gene was most likely facilitated by the inclusion of a portion of the promoter from the Pgk1-like progenitor gene as part of the retroposed sequence (34-36). Thus, we previously speculated that the Pgk2 gene initially carried a housekeeping promoter sequence that directed ubiquitous low-level transcription and that this regulatory sequence has subsequently undergone a transition to direct, high-level, testis-specific transcription, at least in eutherian mammals (34-36). A comparison of mammalian Pgk1 and Pgk2 promoter sequences is shown in Fig. 6. One potentially important distinction not specifically shown in this figure is that the Pgk1 promoter is embedded in a CpG island, whereas the Pgk2 promoter has lost the density of CpG dinucleotides required to be considered an island and has retained only dispersed CpG dinucleotides (35). This difference is likely responsible in part for the fact that the Pgk1 promoter region typically remains constitutively hypomethylated (except on the inactive X chromosome) (47), whereas the Pgk2 promoter undergoes highly regulated demethylation in a tissue-, stage-, and cell type-specific manner uniquely in spermatogenic cells (22).
FIG. 6.
Pgk promoter sequences in mammals. Comparisons are shown among promoter sequences for the Pgk1 (A) and Pgk2 (B) genes from various mammalian species. Alignment of sequences upstream from the translational start codon (ATG, dashed rectangle) of each Pgk gene was done using CLUSTALW (http://align.genome.jp/). Identical nucleotides are represented by asterisks, and dashes indicate a gap. The numbers to the right of each sequence are relative to the transcription start site, which is designated +1. Highly conserved sequences for which potential binding functions are known are surrounded by rectangles. (C) An alignment is shown of the NF1-like region from the human and mouse Pgk1 genes and the E3/E4 region from the human and mouse Pgk2 genes and the potential relationship among these regions. Asterisks indicate sequence identity, dashes indicate a lack of identity, and vertical lines indicate partial identity.
Multiple conserved regions are evident within each Pgk promoter (Fig. 6). As confirmed by the results presented here, differential expression of the mammalian Pgk genes is clearly related to unique protein-DNA interactions associated with each Pgk gene. Surprisingly, CREM appears to bind both the Pgk1 and the Pgk2 genes and so is not likely to be primarily responsible for their differential expression. Although a GC box is common to both the Pgk1 and the Pgk2 promoters and binds SP transcription factors in both cases, we observed preferential binding of SP1 to the Pgk1 promoter in somatic cells and SP3 to the Pgk2 promoter in spermatogenic cells. All other protein-DNA interactions we observed were unique to one or the other of these genes. Not surprisingly, the GC box element is well conserved in both promoters (Fig. 6A and B). Somewhat more surprising is the similar conservation of the CAAT box element in each promoter, given the fact that we detected a bound protein at this site in the Pgk1 gene but not in the Pgk2 gene. However, this element may play a role in directing gene- and tissue-specific demethylation of the Pgk2 gene in prospermatogonia during fetal and perinatal stages (22), and that could be the reason for its conservation in this gene.
A key distinction between the Pgk1 and Pgk2 promoters is found in the region just upstream from the CAAT box in each. In the Pgk1 promoter, there is a region of 41 bp that has been termed an “NF1-like” binding sequence and has been shown to bind factors found in somatic cells (14, 46). This sequence is relatively well conserved among the four mammalian Pgk1 promoters, as shown in Fig. 6A, but is not found intact in the Pgk2 promoter. Rather, in the equivalent position, the Pgk2 gene shows a different conserved element of 18 bp that we previously termed the E3/E4 region (Fig. 6B) and showed by EMSA to have the capacity to bind one or more testis-specific factors (21). The results presented in this study indicate that this is the site of binding of the testis-specific PBX4-PREP1 complex. Interestingly, the E3/E4 enhancer element in the Pgk2 promoter shows some similarity to the 5′ half of the NF1-like element in the Pgk1 promoter (Fig. 6C). We assume that this portion of the Pgk1 promoter is responsible for binding somatic members of the PBX family, as indicated by precipitation of the Pgk1 promoter by the PBX1/2/3 antibody in splenocytes (Fig. 3C and D). However, the extensive conservation of sequence in the E3/E4 region among mammalian Pgk2 genes suggests that it has undergone selection for optimal binding of the PBX4-PREP1 complex.
This information allows us to speculate that the Pgk1-like promoter included in the original Pgk2 retroposon may have diverged to facilitate its transition from a housekeeping promoter to its present status as a tissue-specific promoter which functions uniquely in spermatogenic cells. It is noteworthy that the differences in protein-DNA interactions associated with each of the Pgk promoters are based largely on the preferential binding of different members of similar families of transcription factors. In particular, it would appear that the evolution of testis-specific members of the PBX activator and MEIS/PREP coactivator families, along with the optimization of a binding sequence to preferentially bind the testis-specific isoform of the PBX activator family (PBX4), have combined to impose testis-specific regulation onto the Pgk2 retroposon in eutherian mammals. As genomic sequences for additional mammalian species become available, it will be of interest to recapitulate this evolutionary process to determine if, as we suspect, the appearance of testis-specific members of the PBX and MEIS factor families coevolved in mammals with the appearance of an optimal binding site for this complex in the Pgk2 promoter.
In conclusion, we have identified key transcription factors that are associated with the actively transcribed Pgk2 gene in spermatocytes and spermatids, including CREM, SP3, and the testis-specific PBX4 activator and PREP1 coactivator. We have exploited the highly accessible spermatogenic cell lineage to characterize the developmental kinetics of associations between these factors and the Pgk2 promoter. Finally, we have suggested how the Pgk2 promoter may have evolved from a Pgk1-like housekeeping promoter to the tightly regulated, testis-specific promoter now associated with this gene in eutherian mammals.
Acknowledgments
We are grateful to Thomas P. Yang and Christine Mione (University of Florida College of Medicine, Gainesville) for technical advice on in vivo footprint analysis and Julia Atencio for assistance with procedures.
This work was supported by NIH grant HD46637 to J.R.M. K.T.P. was supported by NIH MBRS-RISE grant GM60655.
Footnotes
Published ahead of print on 17 September 2007.
REFERENCES
- 1.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 2.Almstrup, K., J. E. Nielsen, M. A. Hansen, M. Tanaka, N. E. Skakkebaek, and H. Leffers. 2004. Analysis of cell-type-specific gene expression during mouse spermatogenesis. Biol. Reprod. 70:1751-1761. [DOI] [PubMed] [Google Scholar]
- 3.Ariel, M., H. Cedar, and J. McCarrey. 1994. Developmental changes in methylation of spermatogenesis-specific genes include reprogramming in the epididymis. Nat. Genet. 7:59-63. [DOI] [PubMed] [Google Scholar]
- 4.Ariel, M., J. McCarrey, and H. Cedar. 1991. Methylation patterns of testis-specific genes. Proc. Natl. Acad. Sci. USA 88:2317-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asahara, H., S. Dutta, H. Y. Kao, R. M. Evans, and M. Montminy. 1999. Pbx-Hox heterodimers recruit coactivator-corepressor complexes in an isoform-specific manner. Mol. Cell. Biol. 19:8219-8225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beißbarth, T., I. Borisevich, A. Horlein, M. Kenzelmann, M. Hergenhahn, A. Klewe-Nebenius, R. Klaren, B. Korn, W. Schmid, M. Vingron, and G. Schutz. 2003. Analysis of CREM-dependent gene expression during mouse spermatogenesis. Mol. Cell. Endocrinol. 212:29-39. [DOI] [PubMed] [Google Scholar]
- 7.Bellvé, A. R. 1993. Purification, culture, and fractionation of spermatogenic cells. Methods Enzymol. 225:84-113. [DOI] [PubMed] [Google Scholar]
- 8.Berkes, C. A., D. A. Bergstrom, B. H. Penn, K. J. Seaver, P. S. Knoepfler, and S. J. Tapscott. 2004. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell 14:465-477. [DOI] [PubMed] [Google Scholar]
- 9.Berthelsen, J., V. Zappavigna, E. Ferretti, F. Mavilio, and F. Blasi. 1998. The novel homeoprotein Prep1 modulates Pbx-Hox protein cooperativity. EMBO J. 17:1434-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berthelsen, J., V. Zappavigna, F. Mavilio, and F. Blasi. 1998. Prep1, a novel functional partner of Pbx proteins. EMBO J. 17:1423-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blake, M. C., R. C. Jambou, A. G. Swick, J. W. Kahn, and J. C. Azizkhan. 1990. Transcriptional initiation is controlled by upstream GC-box interactions in a TATAA-less promoter. Mol. Cell. Biol. 10:6632-6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boer, P. H., C. N. Adra, Y. F. Lau, and M. W. McBurney. 1987. The testis-specific phosphoglycerate kinase gene pgk-2 is a recruited retroposon. Mol. Cell. Biol. 7:3107-3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boussouar, F., and M. Benahmed. 2004. Lactate and energy metabolism in male germ cells. Trends Endocrinol. Metab. 15:345-350. [DOI] [PubMed] [Google Scholar]
- 14.Dai, S. M., H. H. Chen, C. Chang, A. D. Riggs, and S. D. Flanagan. 2000. Ligation-mediated PCR for quantitative in vivo footprinting. Nat. Biotechnol. 18:1108-1111. [DOI] [PubMed] [Google Scholar]
- 15.de la Serna, I. L., Y. Ohkawa, C. A. Berkes, D. A. Bergstrom, C. S. Dacwag, S. J. Tapscott, and A. N. Imbalzano. 2005. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol. Cell. Biol. 25:3997-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de la Serna, I. L., Y. Ohkawa, and A. N. Imbalzano. 2006. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat. Rev. Genet. 7:461-473. [DOI] [PubMed] [Google Scholar]
- 17.Eddy, E. M. 2002. Male germ cell gene expression. Recent Prog. Horm. Res. 57:103-128. [DOI] [PubMed] [Google Scholar]
- 18.Ferretti, E., H. Marshall, H. Popperl, M. Maconochie, R. Krumlauf, and F. Blasi. 2000. Segmental expression of Hoxb2 in r4 requires two separate sites that integrate cooperative interactions between Prep1, Pbx and Hox proteins. Development 127:155-166. [DOI] [PubMed] [Google Scholar]
- 19.Ferretti, E., H. Schulz, D. Talarico, F. Blasi, and J. Berthelsen. 1999. The PBX-regulating protein PREP1 is present in different PBX-complexed forms in mouse. Mech. Dev. 83:53-64. [DOI] [PubMed] [Google Scholar]
- 20.Fitzgerald, J., W. M. Hutchison, and H. H. Dahl. 1992. Isolation and characterisation of the mouse pyruvate dehydrogenase E1 alpha genes. Biochim. Biophys. Acta 1131:83-90. [DOI] [PubMed] [Google Scholar]
- 21.Gebara, M. M., and J. R. McCarrey. 1992. Protein-DNA interactions associated with the onset of testis-specific expression of the mammalian Pgk-2 gene. Mol. Cell. Biol. 12:1422-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geyer, C. B., C. M. Kiefer, T. P. Yang, and J. R. McCarrey. 2004. Ontogeny of a demethylation domain and its relationship to activation of tissue-specific transcription. Biol. Reprod. 71:837-844. [DOI] [PubMed] [Google Scholar]
- 23.Hatzis, P., and I. Talianidis. 2002. Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Mol. Cell 10:1467-1477. [DOI] [PubMed] [Google Scholar]
- 24.Hornstra, I. K., and T. P. Yang. 1992. Multiple in vivo footprints are specific to the active allele of the X-linked human hypoxanthine phosphoribosyltransferase gene 5′ region: implications for X chromosome inactivation. Mol. Cell. Biol. 12:5345-5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iannello, R. C., J. Young, S. Sumarsono, M. J. Tymms, H. H. Dahl, J. Gould, M. Hedger, and I. Kola. 1997. Regulation of Pdha-2 expression is mediated by proximal promoter sequences and CpG methylation. Mol. Cell. Biol. 17:612-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimmins, S., N. Kotaja, I. Davidson, and P. Sassone-Corsi. 2004. Testis-specific transcription mechanisms promoting male germ-cell differentiation. Reproduction 128:5-12. [DOI] [PubMed] [Google Scholar]
- 27.Knoepfler, P. S., D. A. Bergstrom, T. Uetsuki, I. Dac-Korytko, Y. H. Sun, W. E. Wright, S. J. Tapscott, and M. P. Kamps. 1999. A conserved motif N-terminal to the DNA-binding domains of myogenic bHLH transcription factors mediates cooperative DNA binding with Pbx-Meis1/Prep1. Nucleic Acids Res. 27:3752-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kramer, J. A., J. R. McCarrey, D. Djakiew, and S. A. Krawetz. 1998. Differentiation: the selective potentiation of chromatin domains. Development 125:4749-4755. [DOI] [PubMed] [Google Scholar]
- 29.Kumari, M., J. C. Stroud, A. Anji, and J. R. McCarrey. 1996. Differential appearance of DNase I-hypersensitive sites correlates with differential transcription of Pgk genes during spermatogenesis in the mouse. J. Biol. Chem. 271:14390-14397. [DOI] [PubMed] [Google Scholar]
- 30.Lefevre, P., C. Lacroix, H. Tagoh, M. Hoogenkamp, S. Melnik, R. Ingram, and C. Bonifer. 2005. Differentiation-dependent alterations in histone methylation and chromatin architecture at the inducible chicken lysozyme gene. J. Biol. Chem. 280:27552-27560. [DOI] [PubMed] [Google Scholar]
- 31.Lomberk, G., and R. Urrutia. 2005. The family feud: turning off Sp1 by Sp1-like KLF proteins. Biochem. J. 392:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martens, J. H., M. Verlaan, E. Kalkhoven, and A. Zantema. 2003. Cascade of distinct histone modifications during collagenase gene activation. Mol. Cell. Biol. 23:1808-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayr, B., and M. Montminy. 2001. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2:599-609. [DOI] [PubMed] [Google Scholar]
- 34.McCarrey, J. R. 1987. Nucleotide sequence of the promoter region of a tissue-specific human retroposon: comparison with its housekeeping progenitor. Gene 61:291-298. [DOI] [PubMed] [Google Scholar]
- 35.McCarrey, J. R. 1990. Molecular evolution of the human Pgk-2 retroposon. Nucleic Acids Res. 18:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCarrey, J. R. 1994. Evolution of tissue-specific gene expression in mammals: a new gene is formed and refined. Bioscience 44:20-27. [Google Scholar]
- 37.McCarrey, J. R. 1998. Spermatogenesis as a model system for developmental analysis of regulatory mechanisms associated with tissue-specific gene expression. Semin. Cell Dev. Biol. 9:459-466. [DOI] [PubMed] [Google Scholar]
- 38.McCarrey, J. R., W. M. Berg, S. J. Paragioudakis, P. L. Zhang, D. D. Dilworth, B. L. Arnold, and J. J. Rossi. 1992. Differential transcription of Pgk genes during spermatogenesis in the mouse. Dev. Biol. 154:160-168. [DOI] [PubMed] [Google Scholar]
- 39.McCarrey, J. R., C. B. Geyer, and H. Yoshioka. 2005. Epigenetic regulation of testis-specific gene expression. Ann. N. Y. Acad. Sci. 1061:226-242. [DOI] [PubMed] [Google Scholar]
- 40.McCarrey, J. R., and K. Thomas. 1987. Human testis-specific PGK gene lacks introns and possesses characteristics of a processed gene. Nature 326:501-505. [DOI] [PubMed] [Google Scholar]
- 41.Moens, C. B., and L. Selleri. 2006. Hox cofactors in vertebrate development. Dev. Biol. 291:193-206. [DOI] [PubMed] [Google Scholar]
- 42.Mori, C., J. E. Welch, K. D. Fulcher, D. A. O'Brien, and E. M. Eddy. 1993. Unique hexokinase messenger ribonucleic acids lacking the porin-binding domain are developmentally expressed in mouse spermatogenic cells. Biol. Reprod. 49:191-203. [DOI] [PubMed] [Google Scholar]
- 43.Mueller, P. R., and B. Wold. 1989. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science 246:780-786. [DOI] [PubMed] [Google Scholar]
- 44.Mueller, P. R., and B. Wold. 1991. Ligation-mediated PCR: applications to genomic footprinting. Methods 2:20-31. [Google Scholar]
- 45.Namekawa, S. H., P. J. Park, L. F. Zhang, J. E. Shima, J. R. McCarrey, M. D. Griswold, and J. T. Lee. 2006. Postmeiotic sex chromatin in the male germline of mice. Curr. Biol. 16:660-667. [DOI] [PubMed] [Google Scholar]
- 46.Pfeifer, G. P., R. Drouin, A. D. Riggs, and G. P. Holmquist. 1992. Binding of transcription factors creates hot spots for UV photoproducts in vivo. Mol. Cell. Biol. 12:1798-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pfeifer, G. P., R. L. Tanguay, S. D. Steigerwald, and A. D. Riggs. 1990. In vivo footprint and methylation analysis by PCR-aided genomic sequencing: comparison of active and inactive X chromosomal DNA at the CpG island and promoter of human PGK-1. Genes Dev. 4:1277-1287. [DOI] [PubMed] [Google Scholar]
- 48.Robinson, M. O., J. R. McCarrey, and M. I. Simon. 1989. Transcriptional regulatory regions of testis-specific PGK2 defined in transgenic mice. Proc. Natl. Acad. Sci. USA 86:8437-8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rousseaux, S., C. Caron, J. Govin, C. Lestrat, A. K. Faure, and S. Khochbin. 2005. Establishment of male-specific epigenetic information. Gene 345:139-153. [DOI] [PubMed] [Google Scholar]
- 50.Sagerstrom, C. G. 2004. PbX marks the spot. Dev. Cell 6:737-738. [DOI] [PubMed] [Google Scholar]
- 51.Saleh, M., I. Rambaldi, X. J. Yang, and M. S. Featherstone. 2000. Cell signaling switches HOX-PBX complexes from repressors to activators of transcription mediated by histone deacetylases and histone acetyltransferases. Mol. Cell. Biol. 20:8623-8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schultz, N., F. K. Hamra, and D. L. Garbers. 2003. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc. Natl. Acad. Sci. USA 100:12201-12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shima, J. E., D. J. McLean, J. R. McCarrey, and M. D. Griswold. 2004. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol. Reprod. 71:319-330. [DOI] [PubMed] [Google Scholar]
- 54.Short, S., M. L. Short, D. M. Milkowski, and R. A. Jungmann. 1991. Functional analysis of cis- and trans-regulatory elements of the lactate dehydrogenase A subunit promoter by in vitro transcription. J. Biol. Chem. 266:22164-22172. [PubMed] [Google Scholar]
- 55.Testa, A., G. Donati, P. Yan, F. Romani, T. H. Huang, M. A. Vigano, and R. Mantovani. 2005. Chromatin immunoprecipitation (ChIP) on chip experiments uncover a widespread distribution of NF-Y binding CCAAT sites outside of core promoters. J. Biol. Chem. 280:13606-13615. [DOI] [PubMed] [Google Scholar]
- 56.Thomas, K., D. Y. Sung, J. Yang, K. Johnson, W. Thompson, C. Millette, J. McCarrey, A. Breitberg, R. Gibbs, and W. Walker. 2005. Identification, characterization, and functional analysis of Sp1 transcript variants expressed in germ cells during mouse spermatogenesis. Biol. Reprod. 72:898-907. [DOI] [PubMed] [Google Scholar]
- 57.Turner, J. M. 2007. Meiotic sex chromosome inactivation. Development 134:1823-1831. [DOI] [PubMed] [Google Scholar]
- 58.VandeBerg, J. L., D. W. Cooper, and P. J. Close. 1973. Mammalian testis phosphoglycerate kinase. Nat. New Biol. 243:48-50. [PubMed] [Google Scholar]
- 59.Wagner, K., A. Mincheva, B. Korn, P. Lichter, and H. Popperl. 2001. Pbx4, a new Pbx family member on mouse chromosome 8, is expressed during spermatogenesis. Mech. Dev. 103:127-131. [DOI] [PubMed] [Google Scholar]
- 60.Wang, P. J., D. C. Page, and J. R. McCarrey. 2005. Differential expression of sex-linked and autosomal germ-cell-specific genes during spermatogenesis in the mouse. Hum. Mol. Genet. 14:2911-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilkerson, D. C., S. A. Wolfe, and S. R. Grimes. 2002. H1t/GC-box and H1t/TE1 element are essential for promoter activity of the testis-specific histone H1t gene. Biol. Reprod. 67:1157-1164. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, L. P., J. Stroud, C. A. Eddy, C. A. Walter, and J. R. McCarrey. 1999. Multiple elements influence transcriptional regulation from the human testis-specific PGK2 promoter in transgenic mice. Biol. Reprod. 60:1329-1337. [DOI] [PubMed] [Google Scholar]
- 63.Zhang, L. P., J. C. Stroud, C. A. Walter, G. S. Adrian, and J. R. McCarrey. 1998. A gene-specific promoter in transgenic mice directs testis-specific demethylation prior to transcriptional activation in vivo. Biol. Reprod. 59:284-292. [DOI] [PubMed] [Google Scholar]
- 64.Zhang, X., D. T. Odom, S. H. Koo, M. D. Conkright, G. Canettieri, J. Best, H. Chen, R. Jenner, E. Herbolsheimer, E. Jacobsen, S. Kadam, J. R. Ecker, B. Emerson, J. B. Hogenesch, T. Unterman, R. A. Young, and M. Montminy. 2005. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl. Acad. Sci. USA 102:4459-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]