Abstract
Germ line missense mutations in HRAS and KRAS and in genes encoding molecules that function up- or downstream of Ras in cellular signaling networks cause a group of related developmental disorders that includes Costello syndrome, Noonan syndrome, and cardiofaciocutaneous syndrome. We performed detailed biochemical and functional studies of three mutant K-Ras proteins (P34R, D153V, and F156L) found in individuals with Noonan syndrome and cardiofaciocutaneous syndrome. Mutant K-Ras proteins demonstrate a range of gain-of-function effects in different cell types, and biochemical analysis supports the idea that the intrinsic Ras guanosine nucleotide triphosphatase (GTPase) activity, the responsiveness of these proteins to GTPase-activating proteins, and guanine nucleotide dissociation all regulate developmental programs in vivo.
Ras proteins are signal switch molecules that cycle between active GTP-bound and inactive GDP-bound conformations (Ras-GTP and Ras-GDP) (reviewed in references 10 and 31). The counterbalancing activities of guanine nucleotide exchange factors and GTPase-activating proteins (GAPs) control Ras-GTP levels in vivo (reviewed in reference 10). SOS1, the major guanine nucleotide exchange factor in many mammalian cells, is recruited to protein complexes that assemble on activated growth factor receptors. SOS1 binds to Ras to displace bound guanine nucleotides, and Ras then passively rebinds to either GDP or GTP. Because GTP is much more abundant in the cytosol (20), nucleotide exchange increases intracellular Ras-GTP levels. GTP binding induces a conformational shift in the switch I and II domains of Ras that allows Ras-GTP to interact productively with effectors such as Raf family members, phosphatidylinositol 3 kinase, and Ral-GDS. Signaling is terminated when Ras-GTP is hydrolyzed to Ras-GDP. This reaction is catalyzed by an inefficient intrinsic Ras GTPase activity that is markedly accelerated by binding to GAPs. Neurofibromin and p120 GAP are the predominant GAPs in most mammalian cells (2, 4, 10).
Somatic missense KRAS mutations that introduce amino acid substitutions at positions 12, 13, and 61 are among the most common molecular lesions found in human cancer. Oncogenic K-Ras proteins accumulate in the GTP-bound conformation due to defective intrinsic GTPase activity and resistance to GAPs (29). Surprisingly, germ line KRAS mutations that encode novel amino acid substitutions not found in cancer were recently discovered in 2 to 4% of individuals with Noonan syndrome (NS) as well as in some persons with cardiofaciocutaneous (CFC) syndrome (7, 24, 25, 32). We found that two NS-associated K-Ras proteins (V14I and T58I K-Ras) are gain-of-function alleles that are less activated than oncogenic G12D K-Ras by a variety of biochemical and functional criteria (24). V14 is located within the K-Ras phosphate-binding loop (P-loop), whereas T58 is near the switch II domain. Figure 1A shows the locations of amino acid substitutions found for persons with NS and CFC syndrome, which include alterations within the K-Ras switch I domain (P34L, P34Q, P34R, and I36M) and in the α-5 helix of the 4B isoform (V152G, D153V, F156I, and F156L) (7, 24, 25, 32). Here we describe a comprehensive biochemical and functional analysis of three mutant K-Ras proteins that cause NS and CFC syndrome: P34R, D153V, and F156L K-Ras. The phenotypic features of persons with each mutation have been described previously (7, 24, 32), including those of an individual with the F156L substitution who is one of two siblings with independent germ line RAS gene mutations (25).
FIG. 1.
Biochemical analysis of WT and mutant K-Ras proteins. (A) Schematic representation of K-Ras4B showing the distribution of the amino acid substitutions encoded by germ line mutations found for developmental disorders (above) and the three amino acids that are commonly altered by cancer-associated somatic mutations (below). The P-loop, switch I (Sw I), and switch II (Sw II) domains are conserved among all Ras isoforms (H-Ras, N-Ras, K-Ras4A, and K-Ras4B). These isoforms vary considerably in the hypervariable region. The germ line substitutions characterized in this study are shown in red. (B) Intrinsic GTP hydrolysis measured as the number of cpm released over time. (C and D) GTP hydrolysis stimulated by various concentrations of the GRD of neurofibromin (C) or p120 GAP (D). GTP hydrolysis was measured after 8 min. (E and F) Dissociation of bound GTP (E) and GDP (F) from WT and mutant K-Ras proteins over time.
MATERIALS AND METHODS
KRAS expression constructs.
Wild-type (WT) KRAS human cDNA was cloned into the pENTR/D-TOPO vector (Invitrogen, Carlsbad, CA). A QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) was used to introduce point mutations, which were verified by sequencing. Gateway technology (Invitrogen) was used to clone WT and mutant KRAS cDNAs into the pDEST12.2 vector (Invitrogen) and into the murine stem cell virus (MSCV) backbone containing a green fluorescent protein (GFP) cassette driven by an internal ribosome entry site (IRES) downstream of the KRAS sequence. In addition, KRAS cDNA encoding the first 166 amino acids of WT and mutant proteins was cloned into the pGEX-4T-2 vector (Amersham, Piscataway, NJ) to generate recombinant N-terminal glutathione S-transferase fusion proteins.
Biochemical analysis of recombinant proteins.
Intrinsic and GAP-stimulated GTP hydrolysis assays have been described in detail and were performed as described previously (4, 5, 24). Recombinant GAP-related domain (GRD) proteins from p120 GAP and neurofibromin were produced in Escherichia coli. We incubated 200 nM of each recombinant K-Ras protein that had been preloaded with [γ-32P]GTP without (intrinsic GTPase activity assay) or with (GAP assays) GRD proteins at room temperature. To measure guanine nucleotide dissociation, recombinant K-Ras proteins (500 nM) were loaded with either [α-32P]GTP or [8,5′-3H]GDP in 5 mM EDTA at room temperature. Aliquots were removed from the loading reaction and placed into 20 mM HEPES, pH 7.3, 50 mM NaCl2, 2 mM MgCl2, 2 mM dithiothreitol, 0.2 mg/ml bovine serum albumin, and 0.2 mM (excess) unlabeled GTP to allow guanine nucleotide exchange for the designated time points. The amount of labeled guanine nucleotide bound to K-Ras was determined by vacuum filtration through nitrocellulose filters (0.22-μm pore size) and liquid scintillation counting. Each data point was derived from duplicate samples, and the recombinant proteins were investigated in multiple independent experiments.
Retroviral transduction and hematopoietic progenitor assays.
All experimental procedures involving mice were reviewed and approved by the UCSF Committee on Animal Research. These assays were performed as described elsewhere on cells that were first infected with MSCV-KRAS-IRES-GFP retroviruses engineered to express WT or mutant K-Ras proteins and then sorted to isolate GFP-positive cells for culture in methylcellulose medium (M3231 for CFU-granulocyte-macrophage [CFU-GM] assays and M3234 for burst-forming unit-erythroid [BFU-E] assays; both from StemCell Technologies, Vancouver BC) (24). CFU-GM and BFU-E colonies were grown over a range of recombinant murine GM colony-stimulating factor (GM-CSF) (Peprotech, Rocky Hill, NJ) and recombinant murine erythropoietin (R&D Technologies, Minneapolis, MN) concentrations, respectively, and were counted by indirect microscopy. Macrophage progenitors were grown by culturing transduced GFP-positive fetal liver cells in 50 ng/ml macrophage CSF (Peprotech) as described by Chan et al. (8).
Ras signaling and Western blot analysis.
COS-7 cells were transiently transfected using Lipofectamine 2000 (Invitrogen) with pDEST12.2 vectors encoding WT, P34R, D153V, F156L, and G12D K-Ras mutant proteins. The medium was changed on cells 24 h after transfection into Dulbecco's modified Eagle medium-H21 containing 0.1% fetal bovine serum, and cells were collected after 6 h. COS-7 cells were lysed and Ras-GTP levels were measured as described previously (9) using Raf-1 RBD agarose (Upstate, Lake Placid, NY). Macrophage progenitors were collected using cell dissociation buffer (Invitrogen) and lysed similarly to COS-7 cells. The antibodies used for immunoblotting included anti-K-Ras (F234) (Santa Cruz Biotechnology); anti-phospho-MEK1/2 (Ser217/221), anti-MEK1/2, anti-phospho-p44/42 mitogen-activated protein kinase/extracellular signal-regulated kinase 1/2 (ERK1/2) (Thr202/Tyr204), anti-β-actin, and anti-phospho-S6 (Ser235/236) (all from Cell Signaling, Beverly, MA); and anti-phospho-AKT (a generous gift of David Stokoe, UCSF).
RESULTS AND DISCUSSION
We first loaded N-terminal glutathione S-transferase fusion proteins containing amino acids 1 to 166 of WT, G12D, P34R, D153V, and F156L K-Ras proteins with [γ-32P]GTP and measured intrinsic GTPase activities. P34R and D153V K-Ras both displayed normal intrinsic rates of GTP hydrolysis (Fig. 1B). In contrast, the F156L mutant protein demonstrated impaired intrinsic GTPase activity that was similar to that of oncogenic G12D K-Ras (Fig. 1B). We next assayed the ability of the GRDs of neurofibromin and p120 GAP to stimulate K-Ras GTPase activity. As expected (3, 5), both GAPs markedly enhanced the GTPase activity of WT K-Ras, whereas G12D K-Ras was resistant (Fig. 1C and D). Remarkably, the P34R K-Ras GTPase was insensitive to both GAPs. F156L K-Ras showed an intermediate level of responsiveness, whereas D153V showed a GTP hydrolysis level equivalent to that of WT K-Ras (Fig. 1C and D).
Based on structural modeling, Carta and colleagues (7) proposed that the D153V substitution destabilizes regions of K-Ras that contribute to guanine nucleotide binding, and they predicted that this destabilization would result in increased guanine nucleotide dissociation. To test this hypothesis directly, we loaded recombinant K-Ras proteins with either [α-32P]GTP or [8,5′-3H]GDP to assay guanine nucleotide dissociation. D153V K-Ras showed a normal rate of GTP and GDP dissociation over a 2- to 120-min time course (Fig. 1E and F and data not shown). By contrast, F156L K-Ras displayed a markedly increased rate of guanine nucleotide dissociation, which is consistent with a previous study of F156L H-Ras (21).
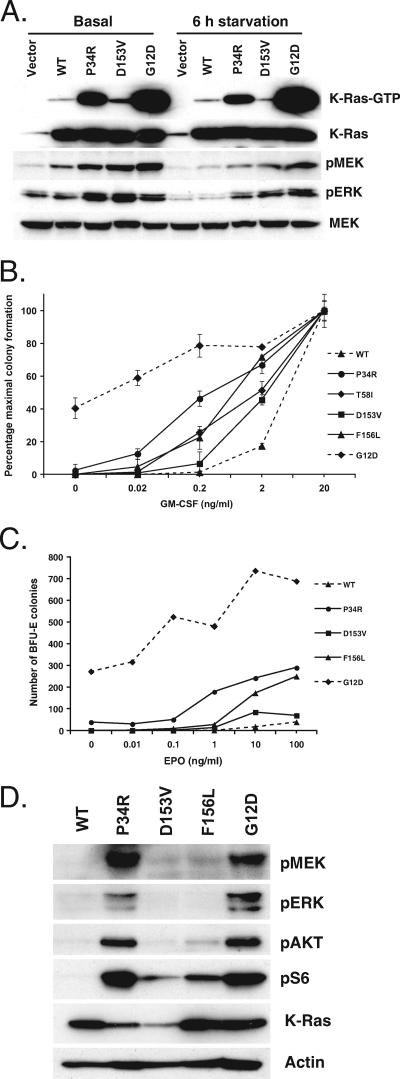
We next expressed mutant K-Ras proteins in COS-7 monkey kidney cells and measured Ras-GTP levels and the phosphorylation of downstream effectors. Cells expressing P34R K-Ras displayed elevated levels of Ras-GTP under basal growth conditions in 10% fetal calf serum and after serum deprivation (Fig. 2A). Ras-GTP levels were modestly elevated in D153V-expressing cells in basal growth conditions but similar to those seen for WT K-Ras after 6 and 12 h of serum deprivation (Fig. 2A and data not shown). P34R and D153V K-Ras expression also consistently induced higher levels of phosphorylated MEK and ERK (pMEK and pERK) in COS-7 cells (Fig. 2A). Cells expressing F156L K-Ras displayed elevated levels of Ras-GTP and pMEK that were similar to what we observed with the P34R mutant protein (25).
FIG. 2.
Functional characterization of P34R, D153V, and F156L K-Ras proteins. (A) Ras signaling in transiently transfected COS-7 cells under basal growth conditions or after 6 h in 0.1% fetal bovine serum (starvation). (B) CFU-GM colony formation of fetal liver cells expressing WT or mutant K-Ras proteins over a range of GM-CSF concentrations. (C) BFU-E colony formation of fetal liver cells expressing WT and mutant K-Ras proteins over a range of erythropoietin (EPO) concentrations. Data show number of colonies per 200,000 GFP-positive fetal liver cells. (D) Phosphorylation of signaling proteins downstream of Ras in macrophage progenitors expressing WT or mutant K-Ras proteins. The data shown in panels A to D are representative of at least three independent experiments.
Infants with NS show a spectrum of hematologic abnormalities and are predisposed to juvenile myelomonocytic leukemia (JMML) (18). We have previously shown that the mutant SHP-2 and K-Ras proteins identified in cases of NS and JMML have gain-of-function effects in primary hematopoietic progenitors, which correlate with the degree of biochemical activation (22, 24). We therefore infected mouse fetal liver cells with MSCV vectors encoding full-length WT or mutant K-Ras proteins and a GFP gene downstream of an IRES. GFP-positive cells were isolated by cell sorting and were plated in methylcellulose medium over a range of GM-CSF concentrations to enumerate CFU-GM colonies. P34R, D153V, and F156L K-Ras proteins all induced a hypersensitive pattern of CFU-GM colony formation that was most pronounced in cells expressing P34R K-Ras (Fig. 2B). The D153V mutant protein displayed the weakest effects with respect to both the number of colonies and their size and morphology. Importantly, only oncogenic G12D K-Ras induced cytokine-independent CFU-GM colony growth (Fig. 2B). We also investigated the effects of this K-Ras allele series on BFU-E colony formation. As in myeloid progenitors, P34R, D153V, and F156L K-Ras all potentiated BFU-E colony growth with the same relative potency as in the CFU-GM progenitor assay (Fig. 2C).
We previously characterized a germ line KRAS mutation encoding a T58I substitution in an infant with NS who presented with a JMML-like disorder (24). Interestingly, P34R K-Ras induced a more hypersensitive pattern of hematopoietic progenitor colony growth than the T58I mutant protein (Fig. 2B and data not shown).
To investigate Ras signaling in a disease-relevant primary cell type, we infected fetal liver cells with MSCV-KRAS-IRES-GFP viruses and cultured them in macrophage CSF to generate macrophage progenitors. Cells expressing mutant K-Ras proteins grew more rapidly (data not shown) and showed differing levels of pMEK, pERK, pAkt, and pS6 that correlated with their effects on CFU-GM and BFU-E colony growth (Fig. 2B and C). Notably, macrophage progenitors expressing P34R or G12D K-Ras showed markedly increased levels of pMEK, pERK, pAkt, and pS6 (Fig. 2D). F156L K-Ras induced a modest increase in both pAkt and pS6, but pMEK and pERK levels were normal. As in the colony assays and in COS-7 cells, D153V K-Ras was the least activated allele in macrophage progenitors, with cells expressing this mutant protein showing elevated levels only of pS6 (Fig. 2D). These data may underestimate the relative potency of D153V K-Ras, because macrophage progenitors grown in culture for 1 to 2 weeks tend to express somewhat lower levels of this protein (Fig. 2D).
The patterns of somatic and germ line KRAS, HRAS, and NRAS mutations observed for cancer and developmental disorders infer distinct biologic functions for specific Ras isoforms. Germ line NRAS mutations have not been reported to date, and almost all of the germ line HRAS mutations found for Costello syndrome introduce amino acid substitutions at positions that are also commonly altered in cancer (codons 12 and 13). By contrast, the KRAS mutations discovered in persons with NS and CFC syndrome are not found in cancer. Kras is the only Ras gene that is essential for murine embryogenesis (14, 16), and KRAS is by far the most commonly mutated RAS gene in human tumors (23). Thus, fully characterizing these novel K-Ras mutant proteins in vitro and in cells is an important priority. We find that they display a complex pattern of intrinsic biochemical properties (Table 1) and have variable effects on cellular signaling and hematopoietic progenitor colony growth (Fig. 2).
TABLE 1.
Summary of biochemical properties of mutant K-Ras proteinsa
| K-Ras | Clinical phenotypeb | Intrinsic GTPase activity | Response to neurofibromin | Response to p120 GAP | Nucleotide exchange |
|---|---|---|---|---|---|
| WT | Normal | Normal | Normal | Normal | Normal |
| G12D | Common somatic mutation in cancer | ↓↓↓↓↓ | ↓↓↓↓↓ | ↓↓↓↓↓ | Normal |
| V14I | NS | ↓↓ | ↓↓↓ | ↓↓↓ | Normal |
| P34R | CFC syndrome | Normal | ↓↓↓↓↓ | ↓↓↓↓↓ | Normal |
| T58I | NS | ↓↓↓ | ↓↓↓ | ↓ | Normal |
| D153V | NS, CFC syndrome, NS/CFC | Normal | Normal | Normal | Normal |
| F156L | CS, CFC syndrome | ↓↓↓↓↓ | ↓↓↓ | ↓↓↓ | ↑↑↑ |
Specific characteristics of each mutant are displayed according to a scoring system that relates the biochemical activities of each mutant protein to those of WT and G12D K-Ras.
Abbreviations: CS, Costello syndrome; NS/CFC, overlapping clinical features of NS and CFC syndrome.
P34R and G12D K-Ras are insensitive to p120 GAP and neurofibromin; however, P34R K-Ras retains normal intrinsic GTPase activity. P34R K-Ras is more potent than other proteins encoded by germ line KRAS mutations by many criteria; it accumulates in the GTP-bound conformation and induces high levels of pERK, pMEK, pAkt, and pS6 in COS-7 cells and primary macrophages and has dramatic effects on hematopoietic progenitor colony growth. These data as well as the disease phenotypes observed for individuals with neurofibromatosis type 1, NS, and CFC syndrome support an essential role of GAPs as negative regulators of Ras signaling in normal development. It is surprising that the P34R mutation is highly resistant to GAPs, because homozygous inactivation of either Gap or Nf1 is lethal in mouse embryos (6, 12, 13). Possible explanations for this apparent paradox include (i) the differential requirements for Ras signaling in murine and human development, (ii) the existence of essential Ras- or GAP-independent functions of neurofibromin and p120 GAP, (iii) the compromised affinity of P34R K-Ras for effectors, and (iv) the expression of normal Ras proteins in the tissues of individuals with germ line KRAS mutations, which can be regulated by GAPs. Our studies of P34R K-Ras also suggest that levels of intrinsic K-Ras GTPase activity modulate cell fates in specific tissues, as only G12D K-Ras induces cytokine-independent CFU-GM and BFU-E colony growth. However, the strong effects of P34R K-Ras could be explained in part by overexpression, which might have overcome reduced affinity for effectors that could be relevant when the protein is expressed at endogenous levels. We are generating KrasP34R “knock-in” mice to address this question and to determine the developmental consequences of expressing P34R K-Ras from the Kras locus (D. A. Tuveson and K. Shannon, unpublished data). Interestingly, Stone and coworkers (26) isolated P34R in a screen for activating substitutions in the c-H-Ras effector domain that could transform fibroblasts. They found that P34R H-Ras was insensitive to GAPs, and a competitive binding assay suggested that this resistance might be due to decreased affinity of the mutant protein for p120 GAP.
In contrast to P34R K-Ras, the F156L mutant protein has defective intrinsic GTPase activity but is partially responsive to GAPs. F156L K-Ras also shows a rapid rate of nucleotide exchange and accumulates in the GTP-bound conformation in cells. These biochemical data are consistent with a previous study of F156L H-Ras, which also reported that the H-Ras mutant protein showed modest transforming potential in fibroblasts (21). We believe the rapid rate of nucleotide dissociation did not significantly affect our ability to assay intrinsic and GAP-stimulated GTP hydrolysis of F156L K-Ras because the experimental conditions allowed for efficient rebinding of labeled nucleotide.
We have not uncovered aberrant biochemical properties of the recombinant D153V mutant protein. D153V K-Ras had the weakest effects on signaling in COS-7 cells and primary macrophages and induced modest, but reproducible, GM-CSF hypersensitivity in hematopoietic progenitors that is similar to the effects of germ line SHP-2 mutant proteins in these cells (19, 22). Primary hematopoietic stem/progenitor cells from Mx1-Cre; KrasG12D compound mutant mice show elevated levels of pS6, which are due to inputs from both the phosphatidylinositol 3 kinase/Akt and Raf/MEK/ERK cascades (30). Similarly, pS6 showed the greatest difference between WT macrophage progenitors and cells expressing each germ line K-Ras mutant protein, including D153V. It is possible that our in vitro assays are not sufficiently sensitive to detect subtle biochemical alterations in the recombinant D153V K-Ras protein or that it is deregulated by a novel mechanism such as increased effector affinity or enhanced sensitivity to SOS1 or another exchange factor. Further investigation of this mutation, which is associated with a range of clinical manifestations, may uncover a novel mechanism of regulating Ras output in vivo.
Many of the mutations detected in persons with disorders of the NS spectrum introduce novel amino acid substitutions in SHP-2, SOS1, H-Ras, K-Ras, B-Raf, MEK1, and MEK2. Characterizing the biochemical and functional properties of these mutant proteins can provide unexpected insights into mechanisms of normal growth control. This principle is perhaps best illustrated by in-depth studies of mutant SHP-2 proteins resulting from somatic and germ line PTPN11 mutations (1, 8, 15, 19, 22, 27, 28). Somatic leukemia-associated PTPN11 alleles encode strong gain-of-function SHP-2 proteins with elevated phosphatase activity, which is essential for aberrant hematopoietic growth (15, 19, 22, 27, 28). However, a thorough biochemical analysis of the germ line PTPN11 mutations found for NS uncovered a more complex picture, with mutant SHP-2 proteins displaying variable effects on phosphatase activity, the affinity of the SH2 domains for phosphotyrosyl ligands, and substrate specificities (15). Most surprisingly, the PTPN11 mutations that cause LEOPARD syndrome, a disorder that shares some clinical features with NS, abrogate or markedly diminish SHP-2 phosphatase activity (11, 17, 27).
Our studies of mutant K-Ras proteins found in NS and CFC syndrome support the idea that the intrinsic Ras GTPase activity, the responsiveness of these proteins to GAPs, and guanine nucleotide dissociation all regulate developmental programs in vivo. Analysis of α-5 helix mutations highlights the importance of regions outside the P-loop and switch domains in Ras regulation. As the molecular causes of developmental disorders are discovered, it is worth considering if the terms that are used to describe these diseases should be replaced by a diagnosis that is based on the affected gene. The highly variable and cell-context-specific properties of K-Ras mutant proteins found for human developmental disorders argue for establishing a hybrid classification system that includes both the clinical syndrome and the identity of the underlying mutation (e.g., NS due to a K-Ras D153 substitution). This will provide the most accurate information for performing genotype/phenotype correlations. Furthermore, the distinct biochemical properties of mutant proteins identified for patients who share the same clinical diagnosis will likely affect responses to molecularly targeted therapeutics.
Acknowledgments
We are indebted to Oddmund Sovik for referring the patient with the F156L mutation for study. We are grateful to David Tuveson for critical comments. We also thank Robert Hawley for providing the MSCV vector and the Laboratory for Cell Analysis Shared resource of the UCSF Comprehensive Cancer Center for assistance with cell sorting.
This work was supported, in part, by NIH grants R37 CA72614 and R01 CA104282.
Footnotes
Published ahead of print on 17 September 2007.
REFERENCES
- 1.Araki, T., M. G. Mohi, F. A. Ismat, R. T. Bronson, I. R. Williams, J. L. Kutok, W. Yang, L. I. Pao, D. G. Gilliland, J. A. Epstein, and B. G. Neel. 2004. Mouse model of Noonan syndrome reveals cell type- and gene dosage-dependent effects of Ptpn11 mutation. Nat. Med. 10:849-857. [DOI] [PubMed] [Google Scholar]
- 2.Boguski, M., and F. McCormick. 1993. Proteins regulating Ras and its relatives. Nature 366:643-653. [DOI] [PubMed] [Google Scholar]
- 3.Bollag, G., F. Adler, N. elMasry, P. C. McCabe, E. Conner, P. Thompson, F. McCormick, and K. Shannon. 1996. Biochemical characterization of a novel KRAS insertional mutation from a human leukemia. J. Biol. Chem. 271:32491-32494. [DOI] [PubMed] [Google Scholar]
- 4.Bollag, G., D. W. Clapp, S. Shih, F. Adler, Y. Zhang, P. Thompson, B. J. Lange, M. H. Freedman, F. McCormick, T. Jacks, and K. Shannon. 1996. Loss of NF1 results in activation of the Ras signaling pathway and leads to aberrant growth in murine and human hematopoietic cells. Nat. Genet. 12:144-148. [DOI] [PubMed] [Google Scholar]
- 5.Bollag, G., and F. McCormick. 1991. Differential regulation of rasGAP and neurofibromatosis gene product activities. Nature 351:576-579. [DOI] [PubMed] [Google Scholar]
- 6.Brannan, C. I., A. S. Perkins, K. S. Vogel, N. Ratner, M. L. Nordlund, S. W. Reid, A. M. Buchberg, N. Jenkins, L. Parada, and N. Copeland. 1994. Targeted disruption of the neurofibromatosis type 1 gene leads to developmental abnormalities in heart and various neural crest-derived tissues. Genes Dev. 8:1019-1029. [DOI] [PubMed] [Google Scholar]
- 7.Carta, C., F. Pantaleoni, G. Bocchinfuso, L. Stella, I. Vasta, A. Sarkozy, C. Digilio, A. Palleschi, A. Pizzuti, P. Grammatico, G. Zampino, B. Dallapiccola, B. D. Gelb, and M. Tartaglia. 2006. Germline missense mutations affecting KRAS isoform B are associated with a severe Noonan syndrome phenotype. Am. J. Hum. Genet. 79:129-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan, R. J., M. B. Leedy, V. Munugalavadla, C. S. Voorhorst, Y. Li, M. Yu, and R. Kapur. 2005. Human somatic PTPN11 mutations induce hematopoietic-cell hypersensitivity to granulocyte-macrophage colony-stimulating factor. Blood 105:3737-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donovan, S., W. See, J. Bonifas, D. Stokoe, and K. M. Shannon. 2002. Hyperactivation of protein kinase B and ERK have discrete effects on survival, proliferation, and cytokine expression in Nf1-deficient myeloid cells. Cancer Cell 2:507-514. [DOI] [PubMed] [Google Scholar]
- 10.Donovan, S., K. M. Shannon, and G. Bollag. 2002. GTPase activating proteins: critical regulators of intracellular signaling. BBA Rev. Cancer 1602:23-45. [DOI] [PubMed] [Google Scholar]
- 11.Hanna, N., A. Montagner, W. H. Lee, M. Miteva, M. Vidal, M. Vidaud, B. Parfait, and P. Raynal. 2006. Reduced phosphatase activity of SHP-2 in LEOPARD syndrome: consequences for PI3K binding on Gab1. FEBS Lett. 580:2477-2482. [DOI] [PubMed] [Google Scholar]
- 12.Henkemeyer, M., D. Rossi, D. Holmyard, M. Puri, G. Mbamalu, K. Harpal, T. S. Shih, T. Jacks, and T. Pawson. 1995. Vascular system defects and neuronal apoptosis in mice lacking Ras GTPase activating protein. Nature 377:695-701. [DOI] [PubMed] [Google Scholar]
- 13.Jacks, T., S. Shih, E. M. Schmitt, R. T. Bronson, A. Bernards, and R. A. Weinberg. 1994. Tumorigenic and developmental consequences of a targeted Nf1 mutation in the mouse. Nat. Genet. 7:353-361. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, L., D. Greenbaum, K. Cichowski, K. Mercer, E. Murphy, E. Schmitt, R. T. Bronson, H. Umanoff, W. Edelmann, R. Kucherlapati, and T. Jacks. 1997. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev. 11:2468-2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keilhack, H., F. S. David, M. McGregor, L. C. Cantley, and B. G. Neel. 2005. Diverse biochemical properties of Shp2 mutants: implications for disease phenotypes. J. Biol. Chem. 280:30984-30993. [DOI] [PubMed] [Google Scholar]
- 16.Koera, K., K. Nakamura, K. Nakao, J. Miyoshi, K. Toyoshima, T. Hatta, H. Otani, A. Aiba, and M. Katsuki. 1997. K-ras is essential for the development of the mouse embryo. Oncogene 15:1151-1159. [DOI] [PubMed] [Google Scholar]
- 17.Kontaridis, M. I., K. D. Swanson, F. S. David, D. Barford, and B. G. Neel. 2006. PTPN11 (Shp2) mutations in LEOPARD syndrome have dominant negative, not activating, effects. J. Biol. Chem. 281:6785-6792. [DOI] [PubMed] [Google Scholar]
- 18.Lauchle, J. O., B. S. Braun, M. L. Loh, and K. Shannon. 2006. Inherited predispositions and hyperactive Ras in myeloid leukemogenesis. Pediatr. Blood Cancer 46:579-585. [DOI] [PubMed] [Google Scholar]
- 19.Mohi, M. G., I. R. Williams, C. R. Dearolf, G. Chan, J. L. Kutok, S. Cohen, K. Morgan, C. Boulton, H. Shigematsu, H. Keilhack, K. Akashi, D. G. Gilliland, and B. G. Neel. 2005. Prognostic, therapeutic, and mechanistic implications of a mouse model of leukemia evoked by Shp2 (PTPN11) mutations. Cancer Cell 7:179-191. [DOI] [PubMed] [Google Scholar]
- 20.Proud, C. 1986. Guanine nucleotides, protein phosphorylation and the control of translation. Trends Biochem. Sci. 12:73-77. [Google Scholar]
- 21.Quilliam, L. A., S. Zhong, K. M. Rabun, J. W. Carpenter, T. L. South, C. J. Der, and S. Campbell-Burk. 1995. Biological and structural characterization of a Ras transforming mutation at the phenylalanine-156 residue, which is conserved in all members of the Ras superfamily. Proc. Natl. Acad. Sci. USA 92:1272-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schubbert, S., K. Lieuw, S. L. Rowe, C. M. Lee, X. Li, M. L. Loh, D. W. Clapp, and K. M. Shannon. 2005. Functional analysis of leukemia-associated PTPN11 mutations in primary hematopoietic cells. Blood 106:311-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schubbert, S., K. Shannon, and G. Bollag. 2007. Hyperactive Ras in developmental disorders and cancer. Nat. Rev. Cancer 7:295-308. [DOI] [PubMed] [Google Scholar]
- 24.Schubbert, S., M. Zenker, S. L. Rowe, S. Boll, C. Klein, G. Bollag, I. van der Burgt, L. Musante, V. Kalscheuer, L. E. Wehner, H. Nguyen, B. West, K. Y. Zhang, E. Sistermans, A. Rauch, C. M. Niemeyer, K. Shannon, and C. P. Kratz. 2006. Germline KRAS mutations cause Noonan syndrome. Nat. Genet. 38:331-336. [DOI] [PubMed] [Google Scholar]
- 25.Sovik, O., S. Schubbert, G. Houge, S. J. Steine, G. Norgard, B. Engelsen, P. R. Njolstad, K. Shannon, and A. Molven. 2007. De novo HRAS and KRAS mutations in two siblings with short stature and neuro-cardio-facio-cutaneous features. J. Med. Genet. 44:e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stone, J. C., M. Colleton, and D. Bottorff. 1993. Effector domain mutations dissociate p21ras effector function and GTPase-activating protein interaction. Mol. Cell. Biol. 13:7311-7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tartaglia, M., S. Martinelli, L. Stella, G. Bocchinfuso, E. Flex, V. Cordeddu, G. Zampino, I. Burgt, A. Palleschi, T. C. Petrucci, M. Sorcini, C. Schoch, R. Foa, P. D. Emanuel, and B. D. Gelb. 2006. Diversity and functional consequences of germline and somatic PTPN11 mutations in human disease. Am. J. Hum. Genet. 78:279-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tartaglia, M., C. M. Niemeyer, A. Fragale, X. Song, J. Buechner, A. Jung, K. Hahlen, H. Hasle, J. D. Licht, and B. D. Gelb. 2003. Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. Nat. Genet. 34:148-150. [DOI] [PubMed] [Google Scholar]
- 29.Trahey, M., and F. McCormick. 1987. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science 238:542-545. [DOI] [PubMed] [Google Scholar]
- 30.Van Meter, M. E., E. Diaz-Flores, J. A. Archard, E. Passegue, J. M. Irish, N. Kotecha, G. P. Nolan, K. Shannon, and B. S. Braun. 2007. K-RasG12D expression induces hyperproliferation and aberrant signaling in primary hematopoietic stem/progenitor cells. Blood 109:3945-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vetter, I. R., and A. Wittinghofer. 2001. The guanine nucleotide-binding switch in three dimensions. Science 294:1299-1304. [DOI] [PubMed] [Google Scholar]
- 32.Zenker, M., K. Lehmann, A. L. Schulz, H. Barth, D. Hansmann, R. Koenig, R. Korinthenberg, M. Kreiss-Nachtsheim, P. Meinecke, S. Morlot, S. Mundlos, A. S. Quante, S. Raskin, D. Schnabel, L. E. Wehner, C. P. Kratz, D. Horn, and K. Kutsche. 2007. Expansion of the genotypic and phenotypic spectrum in patients with KRAS germline mutations. J. Med. Genet. 44:131-135. [DOI] [PMC free article] [PubMed] [Google Scholar]