Abstract
While endocytosis attenuates signals from plasma membrane receptors, recent studies suggest that endocytosis also serves as a platform for the compartmentalized activation of cellular signaling pathways. Intersectin (ITSN) is a multidomain scaffolding protein that regulates endocytosis and has the potential to regulate various biochemical pathways through its multiple, modular domains. To address the biological importance of ITSN in regulating cellular signaling pathways versus in endocytosis, we have stably silenced ITSN expression in neuronal cells by using short hairpin RNAs. Decreasing ITSN expression dramatically increased apoptosis in both neuroblastoma cells and primary cortical neurons. Surprisingly, the loss of ITSN did not lead to major defects in the endocytic pathway. Yeast two-hybrid analysis identified class II phosphoinositide 3′-kinase C2β (PI3K-C2β) as an ITSN binding protein, suggesting that ITSN may regulate a PI3K-C2β-AKT survival pathway. ITSN associated with PI3K-C2β on a subset of endomembrane vesicles and enhanced both basal and growth factor-stimulated PI3K-C2β activity, resulting in AKT activation. The use of pharmacological inhibitors, dominant negatives, and rescue experiments revealed that PI3K-C2β and AKT were epistatic to ITSN. This study represents the first demonstration that ITSN, independent of its role in endocytosis, regulates a critical cellular signaling pathway necessary for cell survival.
Intersectin (ITSN) is a modular scaffold with multiple protein interaction domains that is conserved among metazoa. At the amino terminus are two Eps15 homology (EH) domains that bind NPF motifs on proteins such as epsin (36). The EH domains are followed by a coiled-coil domain that enables ITSN to homo- and heterodimerize with proteins such as Eps15 (24). The carboxy terminus consists of five Src homology 3 (SH3) domains that interact with Pro-rich motifs on a variety of proteins, several of which are involved in regulating endocytosis. Indeed, a subset of ITSN′s SH3 domains are potent inhibitors of clathrin-coated pit formation (26). Recent studies on the Drosophila melanogaster ortholog of ITSN, Dap160, indicate that this scaffold functions as a stabilizing or recruitment factor for components of the clathrin-coated pit (14, 17). The loss of Dap160 function results in fewer coated vesicles, as well as enlarged vesicles, indicating that ITSN functions in both the formation and maturation of endocytic vesicles. Consistent with this role in Drosophila, silencing ITSN expression in mammalian cells results in defects in epidermal growth factor receptor (EGFR) internalization (18).
Although ITSN is clearly linked with endocytosis, increasing evidence suggests that ITSN′s role within the cell is not limited to this process (1, 10, 18, 19, 27-29, 32). The cloning of mammalian ITSN revealed a longer, spliced product containing a guanine nucleotide exchange factor domain specific for Cdc42 (10, 27). Through the EH domains, ITSN activates a Jun N-terminal protein kinase-dependent pathway that cooperates with growth factors and Ras to stimulate transcription and the oncogenic transformation of cells (1, 19, 32). In addition to interacting with endocytic proteins, the SH3 region of ITSN also interacts with several proteins that regulate cellular signaling pathways. For example, ITSN forms complexes with the guanine nucleotide exchange factor Sos to stimulate Ras activation specifically on endomembrane compartments (19). More recently, it has been shown that ITSN associates with the Cbl E3 ubiquitin ligase to regulate EGFR ubiquitylation, trafficking, signaling, and degradation (18).
While loss-of-function mutations in ITSN result in synaptic vesicle defects and lethality in Drosophila (14, 17), these mutant flies possess only mild endocytic defects, raising the possibility that the loss of ITSN may result in additional deficits, particularly in signaling pathways. To address this possibility, we have stably silenced ITSN expression in neuronal cells to determine the importance of this scaffold in neuron function. We demonstrate that ITSN directly interacts with a novel isoform of phosphoinositide 3′-kinase (PI3K) to regulate the survival of neuronal cells through the activation of a PI3K-AKT pathway. This effect is distinct from ITSN′s involvement in endocytosis and indicates that ITSN function in the cell is pleiotrophic and not limited to regulation of the endocytic pathway.
MATERIALS AND METHODS
Cells and reagents.
HEK 293T, N1E-115, A431, and COS cells were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum at 37°C. The medium for A431 cells stably transfected with ITSN was supplemented with 100 μg/ml hygromycin B. Geneticin was purchased from Gibco, and puromycin was purchased from BD Biosciences. Human recombinant epidermal growth factor was purchased from Upstate Biotechnology. Monoclonal antihemagglutinin (anti-HA) antibody was purchased from Covance. Antibodies to Akt and phospho-Akt (pAKT) (pSer473) were purchased from Cell Signaling Technology. Antibodies to Cbl were purchased from Santa Cruz Biotechnology, Inc. Polyclonal antibodies to ITSN and PI3K-C2β have been described previously (2, 18). The PI3K inhibitor LY294002 was purchased from Calbiochem. Primary cortical neurons from day 18 rat embryos were purchased from Gelantis and cultured as indicated by Gelantis's protocol.
DNA constructs.
The yellow fluorescent protein (YFP)-tagged mouse ITSN (short isoform) and the constructs expressing HA-tagged ITSN and the EH, coiled-coil, and SH3 domains have been previously described (19). Glutathione S-transferase (GST) fusions with the individual SH3 domains were made by PCR of the following amino acids from mouse ITSN: SH3A, amino acids (aa) 738 to 801; SH3B, aa 909 to 963; SH3C, aa 998 to 1052; SH3D, aa 1070 to 1130; and SH3D, aa 1151 to 1209. Each fragment was designed with a BamHI site at the 5′ end and an EcoRI site at the 3′ end to clone into pGEX4T2. All clones were verified by DNA sequence analysis. Cyan fluorescent protein (CFP)-tagged PI3K-C2β was subcloned from Glu-tagged PI3K-C2β (2). Note that this PI3K-C2β clone was N-terminally truncated due to an internal methionine. Thus, aa 1 corresponds to aa 26 in full-length PI3K-C2β. The amino acid numbers of PI3K-C2β refer to sequence CAA72168 (EMBL database). The insert was excised by EcoRI-XhoI digestion, and the ends were filled in to generate blunt ends. Plasmid pECFP was digested with EcoRI, and the ends were filled in and then ligated to the insert. The ends were sequenced to confirm that the insert was in the correct orientation and in frame. The N terminus of PI3K-C2β (PI3K NT) fused to CFP (CFP-PI3K NT) was subcloned from the clone obtained from the yeast two-hybrid screen. A NotI fragment containing amino acids G18 to V475 was excised, and the ends were filled and then ligated to pECFP that had been previously digested with SmaI and dephosphorylated. The sequence of inserts was confirmed by DNA sequence analysis. The GST fusion proteins with PI3K NT (aa 27 to 356 as noted above) and the C2 domains of PI3K-C2α were as described previously (2). HA-Akt, Myr-Akt-HA, and HA-Akt-K179A were a gift from Al Baldwin, University of North Carolina, Chapel Hill, NC.
Construction of silencing vectors.
Pairs of oligonucleotides were annealed and then ligated into pSuperRetro.puro, referred to as pSR (Oligoengine), and digested with BglII and HindIII. The pLVTH lentiviral vector (35) was derived by subcloning the EcoRI-ClaI fragment of pSR-M1635 into pLVTH digested with EcoRI and ClaI. The oligonucleotides used to generate the short hairpin RNA (shRNA) constructs used in this study are as follows: R111-5, 5′GATCCCCAGCATGACCAGCAGTTCCATTCAAGAGATGGAACTGCTGGT CATGCTTTTTT; R111-3, 5′AGCTAAAAAAGCATGACCAGCAGTTCCATCTCTTGAATGGAACTGCTGGTCATGCTGGG; M207-5, 5′GATCCCCCGATGGAACGATGGATCAATTCAAGAGATTGATCCATCCTTCCA TCGTTTTT; M207-3, 5′AGCTAAAAACGATGGAAGGATGGATCAATCTCTTGAATTGATCCATCGTTCCATCGGGG; M268-5, 5′GATCCCCCAAGGATATCAGCTCCCCTTTCAAGAGAAGGGGAGCTGATATCCTTG TTTTT; M268-3, 5′AGCTAAAAACAAGGATATCAGCTCCCCTTCTCTTGAAAGGGGAGCTGATATCCTTGGGG; M599-5, 5′GATCCCCAGGCACAATCATTCGATGTTTCAAGAGAACATCGAATGATTGTGCCTTT TTT; M599-3, 5′AGCTAAAAAAGGCACAATCATTCGATGTTCTCTTGAAACATCGAATGATTGTGCCTGGG; M739-5, 5′GATCCCCACTATTCTCATGCAATCAATTCAAGAGATTGATTGCATGAGAATAGTTTTTT; M739-3, 5′AGCTAAAAAACTATTCTCATGCAATCAATCTCTTGAATTGATTGCATGAGAATAGTGGG; M1635-5, 5′GTCCCCGATACTCAGTGACCAGTTATTCAAGAGATAACTGGTCACTGAGTATCTTTTT; M1635-3, 5′AGCTAAAAAGATACTCAGTGACCAGTTATCTCTTGAATAACTGGTCACTG AGTATCGGG; miRNA-5, 5′GATCCCCGGATATCAGCTCCCCTCCATTCAAGAGATGGAGGGGAGCTGATATCCTTTTTGGAAA; and miRNA-3, 5′AGCTTTTCCAAAAAGGATATCAGCTCCCCTCCATCTCTTGAATGGAGGGGAGCTGATATCCGGG.
Description of PI3K Pro-rich mutants.
Mutation of Pro-rich domain 1 (PRD1) (aa 156 to 161; PPPLPP to AAALAA) and PRD2 (aa 169 to173; PPLPP to AALAA) in full-length pECFP-PI3K-C2β was carried out using a two-step site-directed PCR mutagenesis approach. Two pairs of oligonucleotides were used for generating the first-step PCR products—a 0.6-kb PRD1 fragment using primers pECFP5′621 (5′ CTGCCCGACAACCACTACCTG 3′) and PI3KPR1-3′PA (5′ AGCGGTATCCCAGATAGAAGCTCGGGCAGCCAGAGCAGCTGCGGATAGTTTC 3′) and a 0.5-kb PRD2 fragment using primers PI3KPR2-5′PA (5′ CGAGCTTCTATCTGGGATACCGCTGCCCTAGCTGCCAGAAAGGGGTC 3′) and PI3KPR2-3′ (5′ AGAGCCAGATGCAAGGATATCCAGCA 3′). These two PCR fragments were mixed, reamplified using pECFP5′621 and PI3KPR2-3′ as primer pairs, subcloned into pCR4-topo-TA, and subjected to DNA sequence analysis. This construct was digested with XhoI and EcoRV to release the PI3K fragment, which was subcloned into a pECFP Xho-EcoRV fragment produced by digestion of the pECFP-PI3K-C2β construct above with XhoI and EcoRV that releases the wild-type N-terminal fragment, the XhoI-EcoRV fragment (aa 27 to 345), and a 4-kb EcoRV PI3K fragment (aa 346 to 1634). The resulting construct encodes the Pro-to-Ala (PA) mutant CFP-PI3K NT PA. We then digested this construct with EcoRV and subcloned the 4-kb EcoRV PI3K fragment to generate the full-length PI3K PA mutant. The wild-type PI3K NT construct was obtained by subcloning the N-terminal XhoI-EcoRV fragment of PI3K (aa 27 to 345) into the pECFP Xho-EcoRV fragment above. For both the wild type and the PA mutant of PI3K NT, the cloning strategy results in an additional 25 amino acids on the C terminus (KLIDTVDLEILQSTVPRARDPPDLI), derived from the vector.
Differentiation of N1E-115 cells.
N1E-115 cells or stable subclones were plated at a density of 20,000 cells per well in six-well plates or 200,000 cells per 10-cm dish in complete medium (DMEM with 10% fetal bovine serum). On the following day, the medium was replaced with DMEM containing 0.1% bovine serum albumin (BSA) and no serum (differentiation medium). Cells were examined for differentiation as evidenced by changes in cell morphology and the production of neurites for 4 days following serum withdrawal. The images were recorded by using a Nikon digital camera attached to a Nikon inverted microscope. Each plate of cells in differentiation medium was paired with a plate of cells in complete medium for comparison. The experiments were done in duplicate and repeated at least three times. To quantify the cell death during differentiation, we used a CellTiter-Glo luminescent cell viability assay kit (Promega).
Yeast two-hybrid assay.
Analysis was performed through a contract with Myriad Genetics essentially as described previously (31), except that the various individual domains of mouse or human ITSN were used as bait. To confirm the yeast two-hybrid assay results with ITSN and PI3K, the Saccharomyces cerevisiae yeast strain AH109 (MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1UAS-GAL1TATA-HIS3 GAL2UAS-GAL2TATA-ADE2 URA3::MEL1UAS-MEL1TATA-lacZ) was transformed with ITSN:pGBT.Q/bait (aa 730 to 816 of ITSN) and PI3K-C2β:pGAD.PN2/prey (aa 18 to 475 of PI3K-C2β). Additionally, each plasmid, with corresponding empty vectors or both empty vectors, was also tested for growth on medium lacking histidine.
Phosphoinositide kinase assays.
Confluent cell cultures were incubated in the absence or presence of EGF at 37°C for the times indicated and then lysed at 4°C in cell lysis buffer (10 mM Tris-HCl [pH 7.6], 5 mM EDTA, 50 mM NaCl, 30 mM sodium pyrophosphate, 50 mM NaF, 100 μM Na3VO4, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride). The lysates were clarified by centrifugation (13,000 × g, 20 min), and the supernatants were transferred to a fresh tube and used for immunoprecipitation as indicated below. Kinase assays were performed in a total volume of 30 μl containing 20 mM HEPES, pH 7.4, 100 mM NaCl, 0.1 mM EGTA, 0.1 mM EDTA, and 200 μg/ml phosphatidylinositol (Sigma). After sonicated lipid was preincubated with the sample for 10 min, the reaction was initiated by adding divalent cation (6 mM) and 100 μM ATP (0.2 μCi [γ-32P]ATP). The assay mixtures were incubated at 30°C for 20 min and then terminated with acidified chloroform/methanol (1:1, vol/vol). The extracted phosphoinositide products were fractionated by thin-layer chromatography and visualized by autoradiography. All assays were linear with respect to time and enzyme addition.
AKT activation assays.
COS cells were transiently cotransfected with the expression constructs indicated in Fig. 4A along with an HA epitope-tagged AKT expression construct. Following an overnight incubation in serum-free medium, the cells were stimulated with EGF, the lysates harvested, and Western blot assays performed to assess the expression of HA-AKT. The lysates were normalized so that equal amounts of HA-AKT were immunoprecipitated with an HA monoclonal antibody. The precipitates were washed three times with ice-cold phospholipase C (PLC)-LB containing inhibitors and then resuspended in 25 μl of 4× NuPAGE sample buffer supplemented with 5% β-mercaptoethanol. After being heated to 70°C for 10 min, equivalent amounts of samples were fractionated on duplicate 4 to 12% NuPAGE gels (Invitrogen), transferred to Immobilon-P membranes, and probed with antibodies to the HA epitope to determine the total levels of AKT or antibodies to activated AKT (pSer473; New England Biolabs). The activation levels were determined by densitometric analysis of Western blots to determine the pAKT and total AKT levels. The activation (n-fold) was determined by dividing the level of pAKT by that of total AKT and normalizing to unstimulated, vector control sample (YFP or LVTH alone). The experiments were performed in triplicate.
FIG. 4.
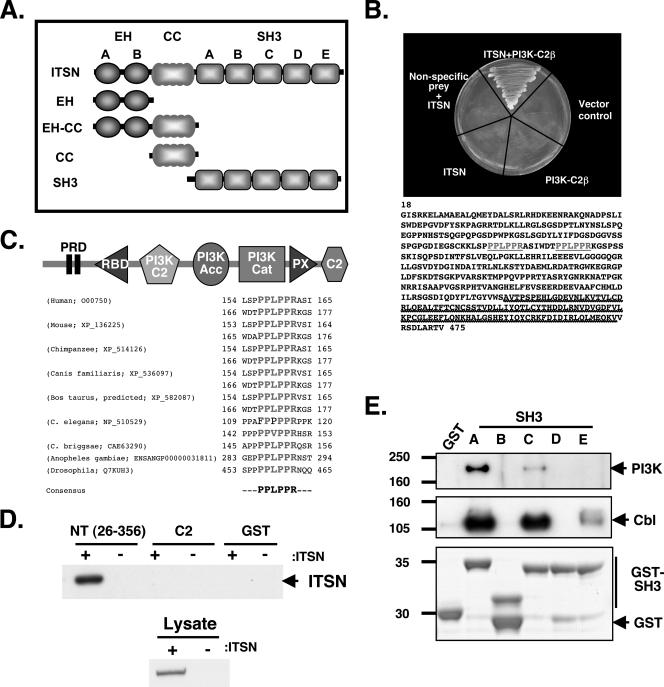
ITSN interacts with PI3K-C2β. (A) ITSN expression constructs. At the NH2 terminus are two EH domains, followed by a coiled-coil (CC) region and five SH3 domains, A through E. Each of the indicated proteins possesses an HA epitope tag at the NH2 terminus. (B) Yeast two-hybrid screening identified PI3K-C2β as an ITSN binding protein. Only the coexpression of ITSN and PI3K-C2β was sufficient to activate HIS3 expression, allowing growth on medium lacking histidine. Putative ITSN-binding sequences are highlighted in gray and underlined. Heavy underlining dentoes the putative Ras-binding region. (C) The modular structure of PI3K-C2β is shown at the top of the panel (15). RBD, putative PI3K Ras binding domain; PI3K C2, outlier PKC-related Ca+2 binding domain specific to PI3Ks; PI3K Acc, PI3K accessory domain; PI3K Cat, PI3K catalytic domain; PX, PhoX homology domain; C2, PKC-related Ca+2 binding domain. Shown below the diagram is an alignment of the PRDs from PI3K-C2β orthologs, with a consensus sequence at the bottom. C. elegans, Caenorhabditis elegans; C. briggsae, Caenorhabditis briggsae. (D) GST, GST-PI3K NT (aa 26 to 356 of PI3K-C2β), or GST-PI3K-C2α C2 was incubated with lysates from 293T cells transfected with either ITSN (+) or empty vector (−). PI3K-C2β (NT) but not the C2 domain of PI3K-C2α (C2) or GST by itself bound ITSN (top panel). Expression of HA-ITSN is shown in the lower panel. (E) PI3K-C2β binds specifically to SH3A of ITSN. GST fusions of the individual SH3 domains were used to precipitate endogenous PI3K-C2β from lysates of 293T cells. SH3A and, to a lesser extent, SH3C bound PI3K, while SH3B, -D, and -E and GST alone did not (top panel). In contrast, Cbl bound SH3A, -C, and -E (middle panel). The purified SH3 domains are shown in the bottom panel. GST-SH3B of ITSN runs anomalously on gels. The gels shown are representative of identical data from three separate experiments. Molecular masses (kDa) are shown on the left.
GST pull-downs.
GST fusion proteins with PI3K-C2β, ITSN, and GST alone were purified from Escherichia coli DH5α. Briefly, a 50-ml culture was grown at 37°C until the cell density reached 1 as measured by absorbance at 600 nm. The cultures were then induced with isopropyl-β-d-thiogalactopyranoside (IPTG) (0.1 mM), grown for an additional 3 h, and spun down. The cell pellet was lysed in 5 ml of B-PER solution (Pierce) supplemented with protease inhibitors and incubated at 4°C for 20 min on a nutator. The debris was pelleted, and the supernatant was placed in a new tube. A total of 200 μl of washed glutathione-agarose beads was added to the supernatant, and the mixture was incubated at 4°C for 2 h on the nutator, after which the beads were pelleted, washed three times with incubation buffer (25 mM HEPES, pH 7.2, 0.5% Triton X-100, 125 mM potassium acetate, 2.5 mM magnesium acetate, 5 mM EGTA, supplemented with 1 mM dithiothreitol and protease inhibitors), and resuspended in a final volume of 250 μl (including beads). A 10-μl amount of the slurry was run on a gel to estimate the concentration of the purified proteins.
Cell lysates were prepared from HA-tagged ITSN or mock-transfected HEK 293T cells. Forty-eight hours after transfection, the cells were washed with warm phosphate-buffered saline (PBS) and lysed in PLC lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 10% glycerol, 1% Triton X-100, 1 mM EGTA, 1.5 mM MgCl2, and 100 mM NaF) supplemented with protease inhibitors. After 30 min at 4°C on a nutator, the soluble lysates were obtained by centrifugation at 14,000 rpm for 10 min and used for the GST pull-down assay. One milligram of protein was added to 0.5 μg of the purified PI3K-C2α C2 or PI3K NT fusion protein or GST as a control and mixed on the nutator at 4°C for 2 h. At the end of the incubation, 20 μl of washed glutathione-agarose beads were added to the mixture so that the beads would be more visible. The beads were spun down and washed three times with the incubation buffer described above and then resuspended in 30 μl of LDS sample buffer (Invitrogen). The samples were fractionated on a gel, transferred to an Immobilon-P membrane, and probed with antibodies against HA. The blots were developed with SuperSignal chemiluminescence reagent (Pierce).
Confocal microscopy.
Confocal images were acquired using a Carl Zeiss LSM 510 mounted on an Axiovert 100 M microscope. Images were obtained using a 514 nm argon laser for YFP and 458 nm for CFP with a Plan-Apochromat 63×/1.4 oil immersion objective lens. A 531- to 595-nm-wavelength bandpass filter was used for YFP emission, and a 470- to 500-nm-wavelength bandpass filter was used for CFP, with a pinhole of 0.7 Airy units, which provides a z resolution of ∼0.6 um.
BiFC assays.
Bimolecular fluorescence complementation (BiFC) expression constructs consisting of the NH2 terminus or COOH terminus of Venus (VN and VC, respectively) were kindly provided by Chang-Deng Hu (25). ITSN or PI3K was cloned into these vectors such that the Venus fragments were fused to the NH2 terminus of the respective protein. COS cells were transiently transfected late in the day and then imaged the following morning so that the various proteins were not vastly overexpressed. CFP was cotransfected along with the BiFC constructs, but at a fivefold-lower amount, to mark transfected cells.
Endocytosis assays.
For analysis of fluorescent transferrin internalization, cells were serum starved for 60 min in the presence of 2% BSA and then incubated with Alexa 488-conjugated transferrin (50 μg/ml; Molecular Probes) for 15 min at 37°C. The cells were then washed in cold PBS, acid washed (0.2 N acetic acid-0.5 M NaCl) one time for 8 min, rinsed in PBS, and then fixed in 4% paraformaldehyde. Cells were analyzed by using a Zeiss LSM 510 META confocal microscope (excitation, 488 nm; filters, 505 to 530 nm), and z-stacks were collected for 20 to 25 cells per cell line for two different sets of cells. The images were imported into the three-dimensional-imaging software Volocity 3.70, and the total fluorescence was quantified for the entire z-stack. For analysis of the internalization of biotinylated transferrin, cells were treated with biotinylated transferrin (50 μg/ml; Sigma) as described above and then lysed in PLC-LB buffer. The biotinylated transferrin levels in the lysates were quantified by Western blotting with a streptavidin antibody. Changes in fluid-phase endocytosis were quantified by measurement of horseradish peroxidase (HRP) uptake according to previously published methods (16). Briefly, cells were serum starved, incubated with HRP (5 mg/ml; Sigma) for 1 h at 37°C, washed five times with cold PBS-1% BSA, and lysed in PBS-0.1% Triton X-100. The HRP internalization was then quantified by using 3,3-diaminobenzidine tablets (Sigma) and reading the absorbance at 490 nm.
TUNEL staining.
Apoptotic cells were identified by terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) staining using an Apo-Direct kit from Calbiochemical. Cells were counterstained with Hoechst 33258 (Molecular Probes), and the percent TUNEL-positive cells was determined by counting the number of TUNEL-Hoechst double-positive cells and dividing by the total number of Hoechst-positive cells. More than 250 total Hoechst-positive cells were scored for each time point.
RESULTS
ITSN regulates cell survival.
To determine the physiological importance of ITSN in mammalian cells, we silenced ITSN expression using shRNAs. We designed several shRNA vectors to different regions of mouse ITSN and generated stable polyclonal, as well as clonal, lines of N1E-115 mouse neuroblastoma cells with each of these vectors. These vectors inhibit the expression of both short and long isoforms of ITSN, although N1E-115 cells only express the ITSN short isoform (Fig. 1) (20). Western blot analysis revealed that each of these vectors silenced ITSN to different degrees (Fig. 1; see also Fig. S1 in the supplemental material). However, M1635 resulted in a >90% reduction in ITSN protein levels. When grown in the absence of serum, N1E-115 cells differentiate into neuron-like cells that are electrically excitable (13). Silencing ITSN expression had no appreciable effect on the growth of cells in complete medium (data not shown). However, upon shifting cells to serum-free conditions, we observed a precipitous decrease in the survival of ITSN-silenced cells (Fig. 1B and C). Similar results were obtained by differentiating with dimethyl sulfoxide (DMSO) (data not shown). A comparison of the various silenced cell lines revealed that a decrease in survival was only apparent when ITSN levels were reduced by >70% of the wild-type levels. These results indicate that ITSN is necessary for the survival of differentiating neuronal cells.
FIG. 1.
Silencing ITSN in N1E-115 neuroblastoma cells. (A) Stable shRNAs were transfected into N1E-115 cells and either clonal (designated with a number) or polyclonal (P) cell lines were selected. ITSN expression was determined by Western blot analysis of cell lysates. The relative ITSN expression was determined by dividing the ITSN signal by the calreticulin signal and normalizing to the ratio from the N1E-115 sample. CALR, calreticulin as a normalization control for loading. (B) N1E-115-derived lines were examined for their differentiation potential by removal of serum. Similar results were obtained by differentiation with DMSO (data not shown). (C) Cell survival was quantified using a CellTiterGlo kit for measuring total ATP levels. Silencing ITSN resulted in dramatically reduced survival following serum withdraw but did not affect the growth of cells in the presence of serum (data not shown). N1E, N1E-115 cells; pSR, N1E-115 cells stably transfected with empty pSuperRetro vector.
To determine whether this result was specific to N1E-115 cells or more generally applicable to primary neurons, we infected rat cortical neurons with lentivirus (35) expressing ITSN shRNA (M1635), which targets rat, as well as mouse, ITSN (Fig. 2). Infection of primary neurons resulted in a dramatic decrease in ITSN protein levels (Fig. 2C, bottom panel), as well as a gradual loss of neuron survival (Fig. 2A and B). Following plating, the neurons formed a dense network of neurites. However, ITSN silencing resulted in a gradual disintegration of the neurites, which appeared more granular with time, followed by the eventual death of the cells. This was accompanied by a dramatic increase in the number of apoptotic cells in the ITSN-silenced samples (Fig. 2C). Furthermore, ITSN silencing resulted in decreased activation of the prosurvival kinase AKT (Fig. 2D). These data demonstrate that ITSN is necessary for the survival of neurons and suggest a potential role for ITSN in the activation of AKT.
FIG. 2.
Silencing ITSN decreases survival of primary cortical neurons through an apoptotic pathway. (A) Primary rat cortical neurons were infected with lentivirus expressing green fluorescent protein alone (LVTH) or green fluorescent protein and shRNA to ITSN (M1635) and observed for differences at the indicated days postinfection. (B) Survival of neurons was quantified by CellTiterGlo ATP assay. (C) Presence of apoptotic cells was determined by TUNEL staining. At least 250 cells per condition were scored for TUNEL staining. ITSN expression in the individual samples is shown below the graph. Equal protein amounts were loaded in each lane. L, LVTH-infected cultures; M, M1635-infected cultures; ITSN-L, ITSN long isoform; ITSN-S, ITSN short isoform; Tuj1, neuron-specific tubulin. (D) Silencing ITSN results in decreased pAKT levels. The lysates whose results are shown in panel C were analyzed for the levels of pAKT versus total AKT. The numbers below the panels represent the percent reduction in AKT activation, i.e., pAKT/total AKT, compared to the ratio in LVTH-infected cells for that day. DPI, days postinfection.
ITSN interacts with PI3K-C2β.
Although ITSN regulates the endocytic pathway, we did not observe any significant decrease in clathrin-dependent internalization (transferrin receptor) or fluid-phase endocytosis (HRP) in the parental or ITSN-silenced N1E-115 cells (Fig. 3). This finding suggests that the decrease in neuron survival upon the silencing of ITSN may be due to defects in a signaling pathway critical for the survival of differentiating neuronal cells. ITSN functions as a molecular scaffold that associates with a number of endocytic and signal transduction proteins and regulates both of these processes (21). Given the multiplicity of domains in ITSN (Fig. 4A), we utilized a high-throughput yeast two-hybrid screen to identify novel binding partners. We identified a fragment of class II PI3K, specifically, PI3K-C2β, encompassing the N terminus (aa 18 to 475), as a binding partner for the first SH3 domain of ITSN (aa 730 to 816) (Fig. 4B). PI3K-C2β possesses two highly conserved Pro-rich sequences at the N terminus that are nearly identical to the ligand specificity of the first SH3 domain of ITSN (Fig. 4C) (29). However, these Pro-rich motifs are not present in other PI3K isoforms, including class I, class II α and γ isoforms, and class III PI3Ks. Using GST pull-down experiments, we found that the N terminus of PI3K-C2β containing the two Pro-rich sequences was sufficient to bind ITSN, whereas neither GST nor GST-PI3K-C2α C2 domain interacted with ITSN (Fig. 4D).
FIG. 3.
Stable silencing of ITSN does not affect constitutive endocytosis. (A) N1E-115 cells transfected with the indicated pSR construct were incubated with biotinylated transferrin (bTfn). Uptake of transferrin was quantified by Western blot analysis with an HRP-linked streptavidin antibody. Calr, calreticulin as control for loading. (B) N1E-115 cells stably silenced for ITSN (M1635) or vector control cells (pSR) were incubated with HRP and then assayed for uptake as previously described (16).
ITSN has five SH3 domains that each interact with different proteins, such as Sos (19), Cbl (18), Numb (20), and CdGAP (12). Using GST fusions to the individual SH3 domains, we demonstrated that PI3K-C2β bound almost exclusively to SH3A, with SH3C binding to a lesser extent (Fig. 4E). In contrast, Cbl, another ITSN binding protein, bound to SH3C and SH3E in addition to SH3A. These results demonstrate that the interaction of ITSN and PI3K-C2β is mediated predominantly by the SH3A domain of ITSN binding the Pro-rich N terminus of PI3K-C2β.
Immunocytochemical staining of endogenous ITSN and PI3K-C2β revealed that a portion of these two proteins colocalized in cells (Fig. 5A). We were unable to coprecipitate endogenous ITSN and PI3K-C2β, possibly due to the fact that the antibodies target epitopes in the regions of interaction between ITSN and PI3K-C2β. Therefore, we analyzed the interaction of epitope-tagged versions of the two proteins. This approach also allowed us to determine whether both isoforms of ITSN interacted with PI3K-C2β. Both the HA-tagged short and long ITSN isoforms associated with PI3K-C2β (Fig. 5B). The data from the yeast two-hybrid assay and cell-free experiments demonstrated that the SH3 domains of ITSN, specifically, SH3A, bound PI3K-C2β. In support of these results, we observed that only the full-length ITSN and the isolated SH3 domains associated with endogenous PI3K-C2β from cell lysates, whereas the remaining regions were not sufficient for stable interaction (Fig. 5C). PI3K-C2β was not present in the HA immunoprecipitates from vector-, EH-, EH-coil-, or coil-transfected cell lysates, indicating that this interaction was specific to the SH3 domains of ITSN. To determine the importance of the Pro-rich motifs in PI3K-C2β for interaction with ITSN, we mutated the Pro to Ala in both motifs. These mutations abolished the interaction of ITSN with both full-length PI3K-C2β and PI3K NT (Fig. 5D). However, deletion of SH3A from ITSN did not inhibit the interaction of ITSN with PI3K-C2β, suggesting that these two proteins interact in a multivalent fashion. Similar results were reported for the interaction of ITSN with Cbl (18). These data demonstrate that ITSN requires the Pro-rich motifs of PI3K-C2β for stable association.
FIG. 5.
ITSN associates with PI3K-C2β in vivo. (A) Immunocytochemical staining of endogenous ITSN (green) and endogenous PI3K-C2β (red) from LAN-1 human neuroblastoma cells. ITSN staining is shown in panels a and d. PI3K-C2β staining is shown in panels b and e. Overlap of ITSN and PI3K-C2β is shown in panels c and f. Panels g and h show LAN-1 cells stained with anti-rabbit or anti-mouse secondary antibody, respectively, as negative controls. Scale bars, 20 μm. (B) HA epitope-tagged ITSN proteins were expressed in 293T cells, immunoprecipitated with anti-HA, and then examined for association with endogenous PI3K-C2β (top panel). The bottom panel shows the expression levels for the HA-tagged ITSN proteins. Arrows indicate the positions of PI3K-C2β and the ITSN long (ITSN-L) and ITSN short (ITSN-S) isoforms. In similar experiments, antibodies to PI3K-C2α did not interact with ITSN (data not shown). (C) HA epitope-tagged ITSN truncation mutants were expressed in cells, immunoprecipitated as described for panel B, and then examined for association with endogenous PI3K-C2β. The position of PI3K-C2β is indicated by the arrow. The band above this represents a nonspecific contaminating band. The data shown are representative of the results of three independent experiments. Molecular masses (kDa) are shown on the left. (D) Mutation of the PRD of PI3K-C2β inhibits the interaction with ITSN. Full-length fragments of PI3K or PI3K NT fused to CFP were coexpressed with HA-ITSN. ITSN was immunopurified using HA antibodies (top and middle panels), and the association of PI3K was determined by Western blot analysis with green fluorescent protein antibodies (top panel). The middle panel represents a Western blot of the HA immunoprecipitates with HA antibody, demonstrating the level of HA-ITSN in each sample. The PA mutants of PI3K and PI3K NT did not interact with ITSN. The expression of each of the proteins in the cell lysates is shown in the lower panels. Molecular masses (kDa) are shown on the left. α, anti; IP, immunoprecipation; IB, immunoblot.
ITSN and PI3K-C2β colocalize in cells.
Given the association of ITSN with endocytic vesicles, we next evaluated the spatial interaction between ITSN and PI3K-C2β using BiFC (9). Coexpression of ITSN fused to VN along with PI3K-C2β fused to VC resulted in a BiFC signal on vesicles (Fig. 6A, top panels). This result is consistent with the overlap of endogenous ITSN and PI3K-C2β from the immunocytochemical staining (Fig. 5A) and the colocalization of YFP-ITSN with CFP-PI3K-C2β (see Fig. S2 in the supplemental material). The BiFC signal between ITSN and PI3K-C2β was specific, as mutation of the PRD abolished the BiFC signal (Fig. 6A, middle panels) without affecting the expression of PI3K-C2β (Fig. 6B). In addition, coexpression of VN-ITSN with VC fused to a nonspecific peptide did not result in the reconstitution of Venus fluorescence (Fig. 6A, bottom panels). VN-ITSN and VC-PI3K were expressed at nearly endogenous levels (Fig. 6C). These results demonstrate that ITSN interacts with PI3K on a subset of vesicles and that this association requires the PRD of PI3K-C2β for stable interaction with ITSN.
FIG. 6.
ITSN colocalizes with PI3K-C2β in vivo. (A) BiFC analysis. VN-ITSN was coexpressed with either VC-tagged PI3K-C2β (wild type or PRD mutant) or VC fused to a nonspecific peptide (VC-pep). Mutation of the PRD (PA mutant, middle panels) of PI3K dramatically reduced interaction with ITSN as measured by BiFC. Confocal settings were identical for each condition. Cells were transfected in the early evening and analyzed the following morning to minimize the expression of the transfected constructs. CFP was cotransfected with the BiFC constructs to mark transfected cells. The scale bar represents 20 μm. (B) BiFC-tagged proteins are expressed at near endogenous levels. The first lane corresponds to lysates from untransfected cells. The remaining lanes correspond to the samples shown in panel A. Endogenous ITSN is visible as a doublet in COS cell lysates. Calr, calreticulin as a loading control for the Western blots. The VC-nonspecific peptide fusion protein (VC-pep) was visible as a peptide of ∼16 kDa on the lower portion of the HA blot (not shown). WT, wild type; α, anti.
ITSN increases the lipid kinase activity of PI3K-C2β, resulting in AKT activation.
We next tested whether the interaction of ITSN with PI3K was regulated by growth factors. Although PI3K-C2β and ITSN were associated basally, EGF stimulation enhanced this interaction in a time-dependent manner (Fig. 7A). Furthermore, ITSN enhanced the lipid kinase activity of PI3K-C2β upon EGF stimulation, which peaked at 10 min and then returned to basal levels (Fig. 7B, top panel). In contrast, we did not observe an increase in PI3K activity over the basal levels in vector-transfected cells until later times following EGF stimulation, i.e., 30 to 60 min (Fig. 7B, bottom panel). In complementary experiments, PI3K-C2β activity was present in HA-ITSN immunoprecipitates, and EGF treatment resulted in a sixfold increase in this activity (Fig. 7C). However, PI3K-C2β activity was not present in HA-immunoprecipitates from the control, vector-transfected cells (Fig. 7C).
FIG. 7.
ITSN association with PI3K-C2β is enhanced upon growth factor stimulation and results in AKT activation. (A) A431 cells were stably transfected with either empty vector (−) or ITSN (+). Following EGF stimulation (100 ng/ml) for the indicated times, PI3K-C2β was immunoprecipitated and examined for association with ITSN by Western blot analysis. Molecular masses (kDa) are shown on the left. (B) Lipid kinase activity in PI3K-C2β immunoprecipitates from ITSN-expressing cells was enhanced following EGF stimulation. A431 cells transfected with either empty vector or HA-tagged ITSN were stimulated with EGF for the indicated times. PI3K-C2β was immunoprecipitated, and the Ca+2-dependent lipid kinase activity was measured. (C) EGF enhanced ITSN-associated PI3K-C2β activity. HA immunoprecipitates from A431 cells whose results are shown in panel B were stimulated without or with EGF (100 ng/ml, 10 min) and then subjected to a PI3K assay as described in Materials and Methods. Experiments were performed in triplicate, and the data represent the means ± standard errors of the means (SEM) of the results. (D) ITSN overexpression enhances AKT activation. COS cells were transfected with the indicated expression construct along with HA-tagged AKT. Cells were serum-starved and then stimulated without (−) or with (+) EGF (100 ng/ml, 10 min), lysed, and AKT immunoprecipitated with an anti-HA antibody. The immunoprecipitates were analyzed for activated AKT using a phosphospecific antibody (pSer473) and total AKT (anti-HA). The relative AKT activation was determined by dividing the level of pAKT by total AKT and normalizing to the level in the unstimulated vector control sample (YFP alone). The graph represents the means ± SEM of the results of three independent experiments. (E) ITSN activation of AKT is PI3K-dependent. AKT activation was determined as described for panel D, except that cells were treated overnight with LY294002 or control vehicle (DMSO). Results are the means ± SEM of the results of three independent experiments.
Given the role of 3′-phosphoinositides in the activation of AKT, we measured the effect of ITSN-PI3K on AKT activation. Transfection of cells with YFP-ITSN resulted in an increase in total pAKT levels compared to the levels with transfection of YFP alone or YFP-EH (Fig. 7D). Furthermore, this increase was enhanced by EGF stimulation and blocked by the PI3K inhibitor LY294002 (Fig. 7E). Together, these data demonstrate that ITSN enhances PI3K-C2β activity in cells, as well as increasing the EGF induction of lipid kinase activity of PI3K-C2β, resulting in AKT activation.
The ITSN-PI3K-AKT pathway regulates neuron cell survival.
To determine whether ITSN′s role in cell survival in neuronal cells (Fig. 1 and 2) was related to its ability to stimulate PI3K activity in epithelial cells (Fig. 7), we treated N1E-115 cells with LY294002. As with the silencing of ITSN, PI3K inhibition decreased cell survival during differentiation (Fig. 8A). Similarly, the stable expression of dominant-negative AKT (AKT DN) decreased the survival of differentiating N1E-115 cells (Fig. 8B and C). Thus, inhibition of the PI3K-AKT pathway phenocopied loss of ITSN. Conversely, the overexpression of either PI3K-C2β (Fig. 8D) or AKT (Fig. 8E and F) in ITSN-silenced N1E-115 cells rescued the survival of these cells during differentiation; however, neither the expression of CFP alone (Fig. 8D), PI3K NT (data not shown), nor AKT DN (Fig. 8E and F) rescued ITSN-silenced cells. Although ITSN regulates the endocytic pathway, these results appear to be independent of this activity, as neither transferrin internalization nor uptake of HRP were inhibited in ITSN-silenced cells compared to rescued ITSN-silenced cells, e.g., M1635 plus CFP-PI3K, M1635 plus AKT, or control cells (Fig. 3 and 9). These results demonstrate that ITSN regulates neuron survival through the activation of a PI3K-C2β-AKT signaling pathway.
FIG. 8.
PI3K-C2β and AKT are epistatic to ITSN. (A) Wild-type N1E-115 cells were differentiated in the presence of PI3K inhibitor (LY294002) or control vehicle (DMSO), and cell survival was quantified using an ATP assay. (B) N1E-115 cells were stably transfected with either empty vector or a kinase-dead AKT construct (AKT-DN). Cells were grown in the presence of serum (top panels) or differentiated by removal of serum (bottom panels). (C) Parallel cultures from the samples shown in panel B were incubated with CellTiterGlo reagent to quantify cell survival. Expression of kinase-dead AKT (AKT DN) resulted in increased cell death. (D) ITSN-silenced cells (M1635) were stably transfected with either CFP or CFP-PI3K-C2β. Cells were then grown in the presence of serum or differentiated by removal of serum. (E) ITSN-silenced cells (M1635) were stably transfected with either empty vector or HA-tagged wild-type (AKT-WT), activated (Myr-AKT), or kinase-dead (AKT-DN) AKT. Cells were then grown in the presence of serum (top panels) or differentiated by removal of serum (bottom panels). (F) Survival of cells shown in panel E was determined using CellTiterGlo reagent as described for panel C. (G) Expression of various HA-tagged AKT proteins was determined by Western blot analysis of cell lysates with anti-HA antibodies (top panel). These same lysates were also analyzed for expression of ITSN. pSR, N1E-115 cells stably transfected with empty pSuperRetro vector; M1635, N1E-115 cells stably transfected with the pSR-M1635 shRNA construct that silences ITSN expression. The M1635 cells were also stably transfected with AKT expression constructs (wild type [WT], kinase dead [DN], or activated [Myr]) or empty vector (vector). α, anti.
FIG. 9.
Endocytosis in ITSN-silenced cells. Internalization of Alexa 488-transferrin was measured in the various N1E-115 cells. (A) Confocal image analysis revealed no difference in the uptake of transferrin in the various N1E-115 clones. pcDNA, vector alone; WT, wild type. (B) Transferrin (Tf) internalization was quantified for each of the cell lines. The results shown are the means ± SEM of the results of two independent experiments in which relative levels of transferrin internalization were determined for the panel of cell lines. The differences between samples were not statistically significant.
DISCUSSION
ITSN is a multidomain scaffolding protein initially described as an endocytic regulatory protein (21). Indeed, ITSN associates with a number of endocytic accessory proteins, including epsin, Eps15, dynamin, synaptojanin, and SNAP-23/25 (21), and loss-of-function alleles in the Drosophila ortholog, Dap-160, result in defects in both the size and number of clathrin-coated vesicles (14, 17), consistent with a role for this unique scaffold in regulating endocytosis. However, several studies have revealed a more complex role for ITSN in regulating cell function. Through its interaction with Sos and CdGAP, ITSN regulates the activation of several Ras-like GTPases, including Ras (19) and Rac (12). In conjunction with Numb, ITSN regulates the activation of Cdc42 by the EphB2 receptor (11, 20). Furthermore, ITSN regulates receptor tyrosine kinase (RTK) ubiquitylation and trafficking (18) and cooperates with these receptors to activate the Jun N-terminal protein kinase-mitogen-activated protein kinase pathway, leading to changes in gene expression and oncogenic transformation of cells (1, 19). Thus, ITSN is involved in the regulation of cellular signaling pathways, as well as the endocytic pathway, although the physiological importance of this ITSN regulation is not clear.
This current study reveals a critical role for ITSN in regulating the survival of differentiating neurons through the regulation of a PI3K-C2β-AKT pathway. We demonstrate that ITSN and PI3K-C2β are associated and that growth factor enhances both the interaction of these two proteins and the activation of PI3K-C2β by ITSN. While PI3K activation and PtdIns(3,4,5)P3 levels have been thought to occur mainly at the plasma membrane, recent studies indicate that RTKs stimulate phosphatidylinositol-3,4,5-trisphosphate [PtdIns(3,4,5)P3] production on endocytic vesicles (23) and that these PtdIns(3,4,5)P3-positive vesicles are trafficked to the plasma membrane through a kinesin-dependent process (8). In neurons, this transport is necessary for determining polarity and specifying axon development (8). Our data suggest that ITSN-PI3K-C2β may contribute to PtdIns(3,4,5)P3 generation on intracellular vesicles. Although this interaction of ITSN was specific to PI3K-C2β, both PI3K-C2α and PI3K-C2β interact with clathrin, resulting in enhanced lipid kinase activity and PtdIns(3,4,5)P3 production (6, 33). This interaction appears to be important for the dynamic cycling of clathrin from vesicles at the plasma membrane, as well as in the trans-Golgi network (6). The activation of AKT in ITSN-overexpressing cells is consistent with the model that ITSN stimulates PI3K-C2β, resulting in increased PtdIns(3,4,5)P3 synthesis and activation of AKT.
PI3K-C2β is a member of the larger family of PI3Ks that consist of three distinct classes, each of which phosphorylates the 3′ hydroxyl position of the inositol ring of phosphatidylinositol-derived lipids (30). Class II PI3Ks have been implicated in trafficking events due to their association with the trans-Golgi network and clathrin-coated vesicles (6) and their ability to regulate both clathrin-dependent endocytosis and protein transport (5, 6). In addition, class II PI3Ks are downstream targets of activated receptors, resulting in the activation of AKT (3, 33, 34). Given the cooperativity between ITSN and RTKs (1, 11, 20), the association of ITSN with PI3K-C2β may represent an important step in the regulation of PI3K-C2β activity by growth factor receptors.
These studies demonstrate an important role for ITSN in cell survival through the regulation of phosphoinositide metabolism. The association of ITSN with synaptojanin I, an endocytic protein containing two phosphoinositide phosphatase domains (7), suggests that this multidomain scaffold may regulate both the production and the turnover of multiple phosphoinositides in vivo. Deletion of synaptojanin I results in altered phosphoinositide levels in the brain, with an increase in PtdIns(4,5)P2 levels (4). Interestingly, an increase in clathrin-coated vesicles and a decrease in the recycling of these vesicles was observed upon the deletion of synaptojanin I, consistent with its proposed role in vesicle trafficking. Synaptojanin I mutant animals exhibited enhanced synaptic depression, as well as neurologic defects, and died soon after birth (4). Thus, the association of ITSN with both PI3K-C2β and synaptojanin I suggests that this scaffolding molecule may play a central role in the dynamic regulation of phosphoinositides in vivo.
Our previous results indicated that transient silencing of ITSN resulted in decreased internalization of the EGFR, whereas the stably silenced N1E-115 cells did not exhibit defects in internalization. The lack of a gross endocytic defect in the stably silenced cells is not surprising, since the Dap160 loss-of-function mutants exhibit very mild endocytic defects which only become apparent during high-frequency stimulation of the synapse (14, 17). Thus, these mild defects may not be detectable under the conditions used in these studies. Furthermore, it is possible that the remaining ITSN in the cells is sufficient for maintaining endocytosis but not for the regulation of PI3K activity. Regardless, it is clear that the survival defect in the ITSN-silenced cells is rescued by the expression of either PI3K-C2β or AKT, both of which are activated downstream of ITSN.
Although class I PI3Ks regulate cell survival through the activation of AKT, our data demonstrate that PI3K-C2β plays a critical role in this process as well. Furthermore, our results reveal an important role for ITSN in regulating this PI3K-C2β-AKT survival pathway. Indeed, loss-of-function alleles of Drosophila ITSN resulted in larval lethality (14, 17), consistent with this possibility. In addition, a recent study by Predescu and colleagues (22) demonstrated that transient silencing of ITSN in human microvascular endothelial cells activated a mitochondrial apoptotic pathway, leading to death of the ITSN-silenced cells. However, the mechanism by which ITSN protects cells from apoptosis was not determined. Our silencing experiments demonstrate that the specific depletion of ITSN from neuronal cells decreased the survival both of differentiating N1E-115 cells and primary cortical neurons, which could be rescued by the overexpression of either PI3K-C2β or AKT. Interestingly, constitutive endocytosis was unaffected in the ITSN-silenced cells, rescued cells, or cells expressing AKT DN (Fig. 8). Thus, ITSN regulates cell survival, independent of its role in endocytosis, through the activation of a novel PI3K-C2β-AKT pathway.
Supplementary Material
Acknowledgments
The authors declare that there are no competing financial interests.
We thank Albert Baldwin for providing the ATK constructs, Chang-Deng Hu for the BiFC constructs, and Dieder Trono for the pLVTH lentiviral system. We thank Jeff Reese and Yawer Husain for assistance with the confocal imaging and quantification of transferrin fluorescence and Michael Stewart for quantification of N1E-115 cell survival. We also thank Athar Chishti, David Armstrong, and Fernando Ribeiro-Neto for helpful comments on the manuscript and members of the O'Bryan lab for many helpful discussions.
This work was funded in part by the intramural research program of the NIH (J.P.O.), support from a Concern Foundation grant (J.P.O.), and start-up funds from the University of Illinois at Chicago (J.P.O.).
Footnotes
Published ahead of print on 17 September 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adams, A., J. M. Thorn, M. Yamabhai, B. K. Kay, and J. P. O'Bryan. 2000. Intersectin, an adaptor protein involved in clathrin-mediated endocytosis, activates mitogenic signaling pathways. J. Biol. Chem. 275:27414-27420. [DOI] [PubMed] [Google Scholar]
- 2.Arcaro, A., S. Volinia, M. J. Zvelebil, R. Stein, S. J. Watton, M. J. Layton, I. Gout, K. Ahmadi, J. Downward, and M. D. Waterfield. 1998. Human phosphoinositide 3-kinase C2beta, the role of calcium and the C2 domain in enzyme activity. J. Biol. Chem. 273:33082-33090. [DOI] [PubMed] [Google Scholar]
- 3.Arcaro, A., M. J. Zvelebil, C. Wallasch, A. Ullrich, M. D. Waterfield, and J. Domin. 2000. Class II phosphoinositide 3-kinases are downstream targets of activated polypeptide growth factor receptors. Mol. Cell. Biol. 20:3817-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cremona, O., G. Di Paolo, M. R. Wenk, A. Luthi, W. T. Kim, K. Takei, L. Daniell, Y. Nemoto, S. B. Shears, R. A. Flavell, D. A. McCormick, and P. De Camilli. 1999. Essential role of phosphoinositide metabolism in synaptic vesicle recycling. Cell 99:179-188. [DOI] [PubMed] [Google Scholar]
- 5.Domin, J., I. Gaidarov, M. E. Smith, J. H. Keen, and M. D. Waterfield. 2000. The class II phosphoinositide 3-kinase PI3K-C2alpha is concentrated in the trans-Golgi network and present in clathrin-coated vesicles. J. Biol. Chem. 275:11943-11950. [DOI] [PubMed] [Google Scholar]
- 6.Gaidarov, I., M. E. Smith, J. Domin, and J. H. Keen. 2001. The class II phosphoinositide 3-kinase C2alpha is activated by clathrin and regulates clathrin-mediated membrane trafficking. Mol. Cell 7:443-449. [DOI] [PubMed] [Google Scholar]
- 7.Guo, S., L. E. Stolz, S. M. Lemrow, and J. D. York. 1999. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J. Biol. Chem. 274:12990-12995. [DOI] [PubMed] [Google Scholar]
- 8.Horiguchi, K., T. Hanada, Y. Fukui, and A. H. Chishti. 2006. Transport of PIP3 by GAKIN, a kinesin-3 family protein, regulates neuronal cell polarity. J. Cell Biol. 174:425-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu, C. D., Y. Chinenov, and T. K. Kerppola. 2002. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9:789-798. [DOI] [PubMed] [Google Scholar]
- 10.Hussain, N. K., S. Jenna, M. Glogauer, C. C. Quinn, S. Wasiak, M. Guipponi, S. E. Antonarakis, B. K. Kay, T. P. Stossel, N. Lamarche-Vane, and P. S. McPherson. 2001. Endocytic protein intersectin-l regulates actin assembly via Cdc42 and N-WASP. Nat. Cell Biol. 3:927-932. [DOI] [PubMed] [Google Scholar]
- 11.Irie, F., and Y. Yamaguchi. 2002. EphB receptors regulate dendritic spine development via intersectin, Cdc42 and N-WASP. Nat. Neurosci. 5:1117-1118. [DOI] [PubMed] [Google Scholar]
- 12.Jenna, S., N. K. Hussain, E. I. Danek, I. Triki, S. Wasiak, P. S. McPherson, and N. Lamarche-Vane. 2002. The activity of the GTPase-activating protein CdGAP is regulated by the endocytic protein intersectin. J. Biol. Chem. 277:6366-6373. [DOI] [PubMed] [Google Scholar]
- 13.Kimhi, Y., C. Palfrey, I. Spector, Y. Barak, and U. Z. Littauer. 1976. Maturation of neuroblastoma cells in the presence of dimethylsulfoxide. Proc. Natl. Acad. Sci. USA 73:462-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koh, T. W., P. Verstreken, and H. J. Bellen. 2004. Dap160/intersectin acts as a stabilizing scaffold required for synaptic development and vesicle endocytosis. Neuron 43:193-205. [DOI] [PubMed] [Google Scholar]
- 15.Letunic, I., R. R. Copley, S. Schmidt, F. D. Ciccarelli, T. Doerks, J. Schultz, C. P. Ponting, and P. Bork. 2004. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 32:D142-D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, G., and P. D. Stahl. 1993. Structure-function relationship of the small GTPase rab5. J. Biol. Chem. 268:24475-24480. [PubMed] [Google Scholar]
- 17.Marie, B., S. T. Sweeney, K. E. Poskanzer, J. Roos, R. B. Kelly, and G. W. Davis. 2004. Dap160/intersectin scaffolds the periactive zone to achieve high-fidelity endocytosis and normal synaptic growth. Neuron 43:207-219. [DOI] [PubMed] [Google Scholar]
- 18.Martin, N. P., R. P. Mohney, S. Dunn, M. Das, E. Scappini, and J. P. O'Bryan. 2006. Intersectin regulates epidermal growth factor receptor endocytosis, ubiquitylation, and signaling. Mol. Pharmacol. 70:1643-1653. [DOI] [PubMed] [Google Scholar]
- 19.Mohney, R. P., M. Das, T. G. Bivona, R. Hanes, A. G. Adams, M. R. Philips, and J. P. O'Bryan. 2003. Intersectin activates Ras but stimulates transcription through an independent pathway involving JNK. J. Biol. Chem. 278:47038-47045. [DOI] [PubMed] [Google Scholar]
- 20.Nishimura, T., T. Yamaguchi, A. Tokunaga, A. Hara, T. Hamaguchi, K. Kato, A. Iwamatsu, H. Okano, and K. Kaibuchi. 2006. Role of numb in dendritic spine development with a Cdc42 GEF intersectin and EphB2. Mol. Biol. Cell 17:1273-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Bryan, J. P., R. P. Mohney, and C. E. Oldham. 2001. Mitogenesis and endocytosis: what's at the INTERSECTIoN? Oncogene 20:6300-6308. [DOI] [PubMed] [Google Scholar]
- 22.Predescu, S. A., D. N. Predescu, I. Knezevic, I. K. Klein, and A. B. Malik. 2007. Intersectin-1s regulates the mitochondrial apoptotic pathway in endothelial cells. J. Biol. Chem. 282:17166-17178. [DOI] [PubMed] [Google Scholar]
- 23.Sato, M., Y. Ueda, T. Takagi, and Y. Umezawa. 2003. Production of PtdInsP3 at endomembranes is triggered by receptor endocytosis. Nat. Cell Biol. 5:1016-1022. [DOI] [PubMed] [Google Scholar]
- 24.Sengar, A. S., W. Wang, J. Bishay, S. Cohen, and S. E. Egan. 1999. The EH and SH3 domain Ese proteins regulate endocytosis by linking to dynamin and Eps15. EMBO J. 18:1159-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shyu, Y. J., H. Liu, X. Deng, and C. D. Hu. 2006. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. BioTechniques 40:61-66. [DOI] [PubMed] [Google Scholar]
- 26.Simpson, F., N. K. Hussain, B. Qualmann, R. B. Kelly, B. K. Kay, P. S. McPherson, and S. L. Schmid. 1999. SH3-domain-containing proteins function at distinct steps in clathrin-coated vesicle formation. Nat. Cell Biol. 1:119-124. [DOI] [PubMed] [Google Scholar]
- 27.Snyder, J. T., D. K. Worthylake, K. L. Rossman, L. Betts, W. M. Pruitt, D. P. Siderovski, C. J. Der, and J. Sondek. 2002. Structural basis for the selective activation of Rho GTPases by Dbl exchange factors. Nat. Struct. Biol. 9:468-475. [DOI] [PubMed] [Google Scholar]
- 28.Tong, X. K., N. K. Hussain, A. G. Adams, J. P. O'Bryan, and P. S. McPherson. 2000. Intersectin can regulate the Ras/MAP kinase pathway independent of its role in endocytosis. J. Biol. Chem. 275:29894-29899. [DOI] [PubMed] [Google Scholar]
- 29.Tong, X. K., N. K. Hussain, E. de Heuvel, A. Kurakin, E. Abi-Jaoude, C. C. Quinn, M. F. Olson, R. Marais, D. Baranes, B. K. Kay, and P. S. McPherson. 2000. The endocytic protein intersectin is a major binding partner for the Ras exchange factor mSos1 in rat brain. EMBO J. 19:1263-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vanhaesebroeck, B., and M. D. Waterfield. 1999. Signaling by distinct classes of phosphoinositide 3-kinases. Exp. Cell Res. 253:239-254. [DOI] [PubMed] [Google Scholar]
- 31.von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H. Y. Chung, E. Morita, H. E. Wang, T. Davis, G. P. He, D. M. Cimbora, A. Scott, H. G. Krausslich, J. Kaplan, S. G. Morham, and W. I. Sundquist. 2003. The protein network of HIV budding. Cell 114:701-713. [DOI] [PubMed] [Google Scholar]
- 32.Wang, J. B., W. J. Wu, and R. A. Cerione. 2005. Cdc42 and Ras cooperate to mediate cellular transformation by intersectin-L. J. Biol. Chem. 280:22883-22891. [DOI] [PubMed] [Google Scholar]
- 33.Wheeler, M., and J. Domin. 2006. The N-terminus of phosphoinositide 3-kinase-C2beta regulates lipid kinase activity and binding to clathrin. J. Cell. Physiol. 206:586-593. [DOI] [PubMed] [Google Scholar]
- 34.Wheeler, M., and J. Domin. 2001. Recruitment of the class II phosphoinositide 3-kinase C2β to the epidermal growth factor receptor: role of Grb2. Mol. Cell. Biol. 21:6660-6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiznerowicz, M., and D. Trono. 2003. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J. Virol. 77:8957-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamabhai, M., N. G. Hoffman, N. L. Hardison, P. S. McPherson, L. Castagnoli, G. Cesareni, and B. K. Kay. 1998. Intersectin, a novel adaptor protein with two Eps15 homology and five Src homology 3 domains. J. Biol. Chem. 273:31401-31407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.