Abstract
Activation-induced cytidine deaminase (AID) initiates antibody diversification processes by deaminating immunoglobulin sequences. Since transcription of target genes is required for deamination in vivo and AID exclusively mutates single-stranded DNA (ssDNA) in vitro, AID has been postulated to mutate transcription bubbles. However, since ssDNA generated by transcription can assume multiple structures, it is unknown which of these are targeted in vivo. Here we examine the enzymatic and binding properties of AID for different DNA structures. We report that AID has minimal activity on stem-loop structures and preferentially deaminates five-nucleotide bubbles. We compared AID activity on cytidines placed at various distances from the single-stranded/double-stranded DNA junction of bubble substrates and found that the optimal target consists of a single-stranded NWRCN motif. We also show that high-affinity binding is required for but does not necessarily lead to efficient deamination. Using nucleotide analogues, we show that AID's WRC preference (W = A or T; R = A or G) involves the recognition of a purine in the R position and that the carbonyl or amino side chains of guanosine negatively influence specificity at the W position. Our results indicate that AID is likely to target short-tract regions of ssDNA produced by transcription elongation and that it requires a fully single-stranded WRC motif.
Somatic hypermutation and class switch recombination (CSR) are characteristic of the secondary immune response (21, 27). Somatic hypermutation, CSR, and immunoglobulin gene conversion all require the enzyme activation-induced cytidine deaminase (AID) (1, 20, 30, 36, 37, 46). AID likely initiates these processes by deaminating cytidine to uridine exclusively within single-stranded DNA (ssDNA) regions (7, 16, 17, 24, 40, 41, 43, 51). Despite the many advances made over the last few years in the field of antibody diversification, many questions still exist regarding the regulation and biochemical mechanism of AID, in particular its target selection.
AID activity is believed to be regulated through interaction with cofactors (8) and through posttranslational modification (3, 33, 38). On the other hand, the basic enzymatic properties of AID itself are also crucial determinants of its activity. First, we and others found that purified AID preferentially mutates cytidines in WRC motifs (W is A or T; R is A or G) (24, 25, 41, 58). Findings that CSR breakpoints occur at WRC motifs (22, 61) and that this specificity is highly conserved (53) argue that the sequence preference inherent to AID plays a significant biological role. Second, AID deaminates ssDNA processively in vitro (41), and evidence for this type of activity has been documented for mice (55). Third, we recently showed that AID binds single-stranded or bubble-type DNA substrates with high affinity and a long half-life, irrespective of nucleotide sequence (25).
The mechanism that targets AID to immunoglobulin (Ig) genes is not known. Although cis-acting sequences that reside within the Ig locus have long been suspected to exist, data in support of this notion are currently incomplete. Recent developments have suggested that proteins that bind to e-box motifs (34) or the Ig promoter itself (56) help localize AID to Ig sequences. Indeed, it has been well documented that AID-generated mutation rates correlate with the transcriptional rate of target genes (2, 39) and that transcribed double-stranded DNA (dsDNA) is subject to AID activity in Escherichia coli, in mammalian cell lines, and in vitro (9, 21, 27, 29, 45). Despite this strong correlation with transcription, the precise nature of the in vivo AID substrate is unknown. It has been postulated that AID acts directly on the ssDNA of transcription bubbles (8, 31, 45, 51) or on R-loops formed between long-lived RNA-DNA hybrids (57, 59). In support of this notion, deoxyuridines have been reported to be generated preferentially in the nontranscribed strand (32, 45, 62), as would be predicted due to the “protection” offered by the nascent RNA strand. However, other groups find that both strands are more or less mutated equally in vivo (55) and in vitro (5, 49, 50). Alternatively, positive supercoiling induced downstream of the transcriptional machinery can generate stem-loop or G4 DNA structures (18, 44, 54), while negative supercoiling induced upstream of the RNA polymerase can also cause local unwinding of the DNA, creating ssDNA structures (15, 28). Thus, since transcription can generate multiple DNA structures, it is not clear which of these are the in vivo targets of AID. Furthermore, the architectural requirements of WRC specificity have not been examined in detail. For instance, while it is known that AID mutates cytidines in ssDNA (7), it is not known whether only the cytidine or the entire WRC motif is required to be single-stranded in order to be deaminated by AID. This distinction is particularly important, since some DNA binding proteins exhibit nucleotide sequence specificity in the context of dsDNA (e.g., DNA-binding transcription factors) (48) while others do so on ssDNA (e.g., NF-GMb and H16) (14, 19). To address these issues, we measured the catalytic preference and binding kinetics of AID for various DNA structures.
MATERIALS AND METHODS
AID purification.
The purification of glutathione S-transferase (GST)-AID has been previously described (25). Briefly, GST-AID expression was induced in E. coli Bl21(DE3), followed by incubation for 16 h at 16°C. GST-AID was purified from the supernatant of lysed Cells using a column of Glutathione Sepharose high-performance beads (Amersham) as per manufacturer's recommendations. GST-AID was purified on Glutathione Sepharose beads (Amersham) as per manufacturer's recommendations and stored in 20 mM Tris-Cl pH 7.5, 100 mM NaCl, 1 mM dithiothreitol.
Substrate preparation.
The bubble substrates are similar to those previously described (25) (Fig. 1A). 5′ labeling was done using [γ-32P]dATP and polynucleotide kinase (New England Biolabs [NEB]) followed by purification through mini-Quick spin DNA columns (Roche). A 2.5-pmol amount of the labeled strand was mixed with at least a twofold excess of the cold strand in a volume of 25 μl and annealed by slow cooling from 94°C. The stem-loop substrates were labeled and purified in a similar manner, but self-annealing was done in a 125-μl volume by heating to 95°C and snap-cooling. In order to verify their structure, 20 fmol of stem-loop substrate was digested with 1 U of BamHI in a volume of 20 μl for 1 h at 37°C (Fig. 2B). A single-stranded oligonucleotide containing a BamHI site was used to ensure that any BamHI cleavage was due to the presence of a double-stranded recognition sequence.
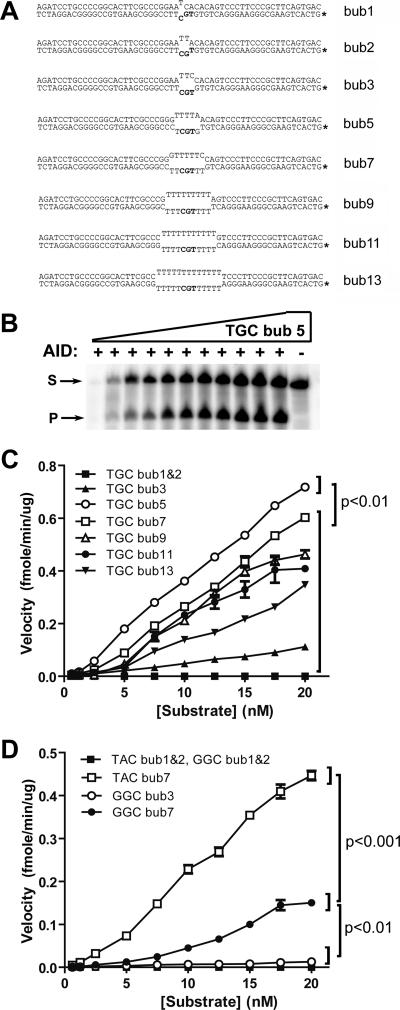
FIG. 1.
Minimal mismatch bubble substrates. (A) DNA substrates containing the mismatch sequences ranging from 1 to 13 nt. The dinucleotide 5′ of the target cytidine is TG, creating a TGC hot spot (i.e., WRC) motif for AID deamination. The asterisk indicates the 32P-labeled strand. The total substrate is 56 nt long, and the cleaved product of deamination is 28 nt long. (B) Typical denaturing polyacrylamide gel electrophoresis of a deamination assay showing the activity of AID on 0.125 to 200 fmol (0.0125 to 20 nM) of TGC bub5 (5-nt-long bubble shown in panel A). “S” indicates the substrate band, while “P” indicates the band corresponding to the cleavage product as a result of deamination. A control reaction without AID is shown on the right, and UDG was added to all reactions. (C) Deamination assays with each substrate shown in panel A. Product/substrate bands were quantitated and divided by the incubation time (min) and AID amount (μg) to calculate the reaction velocity at each substrate concentration (ranging from 0.0125 to 20 nM) for each of the substrates shown in panel A. The first part of the substrate designation indicates the WRC motif sequence, and the last number indicates the length of the bubble. P values were <0.01 between TGC bub5 plots at substrate concentrations of >5 nM and plots for all other substrates. (D) Same as panel C, expect with substrates that contain a different WRC motif (TAC) and a non-WRC motif (GGC). P values were <0.001 between TAC bub7 and GGC bub7 plots, and P values were <0.01 between 7-nt bubbles and 1-, 2-, or 3-nt bubbles at substrate concentrations of >7.5 nM.
FIG. 2.
Deamination kinetics on stem-loop and bubble substrates. (A) The stem-loop substrates containing ssDNA regions of 1 to 13 nt. All stem-loop substrates contain a WRC motif (AGC), with the exception of GGC S1, which contains a non-WRC motif (GGC) due to its structure. The substrates were 5′ labeled and self-annealed. A BamHI site was engineered in the stem to allow for the conformation of the proper dsDNA structure. (B) BamHI digestion of GGC S1, AGC S3, and AGC S7. The denaturing polyacrylamide gel shows, from left to right, 56-nt and 28-nt size markers and “C,” which is a 35-nt non-self-complementary oligonucleotide containing a single-stranded version of a BamHI site. (C) Native gel electrophoresis in order to confirm that the stem-loop structures did not form concatemers due to self-complementarity. A single band indicated a uniform single-molecule structure. (D) Deamination kinetics comparing the activity of AID on the stem-loop substrates to that on AGC bub7, which is identical to the 7-nt bubble shown in Fig. 1A except for the dinucleotide 5′ of the target C (AG instead of TG). Deamination assays similar to that shown in Fig. 1B were carried out over a range of substrate concentration from 0.0125 to 8 nM, and the velocity of product generation by AID was calculated at each substrate concentration. P values were <0.01 between the AGC bub7 plot and all stem-loop plots at substrate concentrations of >3 nM.
AID deamination assay.
The UDG-based deamination assay has been described previously (25, 51). Briefly, 0.1 to 500 fmol of labeled substrate was incubated with 0.3 to 0.9 μg GST-AID for 30 to 90 min at 37°C in 50 mM Tris (pH 7.5), 100 mM NaCl, and 2 μM MgCl in a volume of 10 μl. AID was then deactivated for 15 min at 75°C. The volume was then increased to 20 μl, and 1 U uracil DNA glycosylase and buffer (NEB) was added for a 60-min incubation period of 37°C in order to excise the uracil, followed by incubation at 95°C for 8 min in a final [NaOH] of 100 mM to cleave the alkali-labile abasic site. Samples were electrophoresed on 14% denaturing acrylamide gels, visualized using a PhosphorImager (Molecular Dynamics). ImageQuant 5.0 (Molecular Dynamics) was used for band quantitation. Product formation velocities were calculated at each input substrate concentration for a given amount of AID in a unit of time. Each line on a typical graph was obtained from at least two independent experiments, for each of which at least two independent gels were quantitated.
EMSA.
Detection of AID binding using an electrophoretic mobility shift assay (EMSA) has been previously described (25). Briefly, labeled substrate was incubated with 0.3 to 1 μg GST-AID in 50 mM Tris (pH 7.5), 2.0 μM MgCl, 50 mM NaCl, and 1 mM dithiothreitol in a final volume of 10 μl at 25°C for 45 min. Samples were then UV cross-linked (Stratagene) on ice at a distance of 2 cm from the UV source with 100 mJ and an irradiation time of 50 s. Samples were electrophoresed at 4°C on 8% native gels and visualized using a PhosphorImager (Molecular Dynamics). Gel quantitation was done using ImageQuant 5.0 software (Molecular Dynamics). Duplicate gels were used to obtain accurate average values of bound and free fractions at each substrate concentration. Data were plotted as bound and free fractions of the substrate. Graphpad Prism 5.0 software was used to fit the data to the equation derived from the law of mass action, [bound] = ([boundmax] × [free])/(Kd + [free]) (where [bound] is the concentration of bound fraction, [free] is the concentration of free fraction, [boundmax] is the maximum concentration of bound fraction, and Kd is the binding affinity of AID for the substrate) for the determination of approximate half-saturation values. The determination of complex half-life values was performed as previously mentioned (25). Briefly, binding reactions were set up using 10 fmol of radioactively labeled substrate and incubated for 45 min to allow for complex formation. A thousandfold excess (10 pmol) of unlabeled substrate was added to the binding mixture, followed by incubation for various lengths of time to allow for dissociation of AID from the labeled substrate prior to UV cross-linking of the reaction. Graphpad Prism 5.0 was used to fit the data to an exponential-decay model for the determination of complex half-life values.
MBN treatment of bubble substrates.
Labeled bubble-type substrate (50 fmol) was incubated with 0.1 or 1 U of mung bean nuclease (MBN) (NEB) in a buffer containing 50 mM sodium acetate, 30 mM NaCl, 1 mM ZnSO4 (pH 5.0) for 2 to 5 min at 37°C in a final volume of 10 μl. Reactions were stopped by the addition of 200 mM EDTA-10 mM ATP, pH 9.0 (4), electrophoresed on denaturing acrylamide gels, and visualized using a PhosphorImager (Molecular Dynamics).
Statistical analysis.
Each point on a velocity-versus-substrate-concentration plot was obtained from at least two independent experiments. Data were analyzed by using GraphPad Prism version 5.0. A two-way analysis-of-variance test was used for statistical analysis of the deamination assays, and one-way analysis-of-variance and Tukey's multiple comparisons tests were used for the EMSA data.
RESULTS
Optimal bubble size required for AID activity.
Because transcription can induce regions of ssDNA in different ways, we sought to determine the substrate preferences of AID in vitro to gain insight into its natural targets in vivo. First, we examined the optimal DNA bubble size for AID-mediated deamination. Previously we found that AID preferentially deaminates 5-nucleotide (nt) bubble substrates as compared to larger bubbles. However, since the 5-nt bubble was the smallest bubble examined, we extended our analysis to include smaller-bubble substrates to determine the optimal bubble size for AID deamination. We examined the enzymatic activity of AID using the UDG-based AID deamination assay, previously described by us and others (25, 58), to obtain enzyme kinetic data for AID. Figure 1B shows a typical deamination assay gel showing the activity of AID over a range of substrate concentrations. Enzyme velocity plots were generated by determining the amount of substrate deaminated by a unit of AID in a unit of time at each substrate concentration. Using substrates that contained bubbles ranging from 1 to 13 nt with identical WRC motifs (i.e., TGC), we found that AID preferentially mutated bubbles of 5 nt (Fig. 1C). For both WRC (Fig. 1C and D) and non-WRC (Fig. 1D) motifs, we failed to detect deamination activity on 1- and 2-nt bubble substrates, with only residual activity on 3-nt bubble substrates.
AID activity is reduced on stem-loop structures.
The preference of AID for small bubble-type structures as well as the high frequency of inverted repeat sequences in the immunoglobulin genes (21) raise the possibility that stem-loop structures generated through DNA unwinding during transcription may be the in vivo targets of AID rather than the transcription bubble itself (6, 9, 50). In order to test this possibility, we generated stem-loop structures of 1 (GGC S1), 3 (AGC S3), 7 (AGC S7), 9 (AGC S9), and 13 (AGC S13) nt, with the latter four containing WRC motifs (Fig. 2A). In order to verify that the proper stem-loop structure was achieved, the annealed stem-loop structures were designed with a BamHI restriction site in the stem region (Fig. 2A). Complete BamHI digestion indicated the formation of a double-stranded stem (Fig. 2B). Furthermore, the presence of a single band of the appropriate size upon native electrophoresis indicated that the annealed stem-loops were of a uniform monomolecular, nonconcatamerized structure (Fig. 2C; also data not shown). As shown in Fig. 2D, AID activity was significantly diminished on the stem-loop substrates compared to that on AGC bub7, indicating that AID prefers to deaminate bubble substrates.
High-affinity binding is required for AID activity.
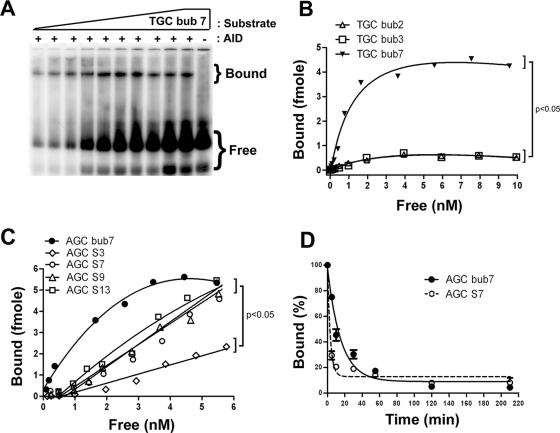
We previously showed that AID bound to bubble substrates with high affinity (i.e., nanomolar range) (25). To determine whether high-affinity binding is necessary for deamination, we used EMSAs to measure the bound and free fractions of substrate over a range of substrate concentrations in order to calculate the binding affinity (i.e., Kd) of AID for substrates that were poorly deaminated (i.e., bub2, bub3, S3, S7, S9, and S13). A typical EMSA experiment is shown in Fig. 3A. While the half-saturation (Kd) values obtained for binding to 2- and 3-nt bubbles (Fig. 3B) were approximately threefold higher than those measured on a 7-nt bubble (i.e., 4.51 ± 1.84 nM, 4.55 ± 3.90 nM, and 1.41 ± 0.58 nM, respectively), the amount of bound substrate was 5- to 10-fold lower than that with a 7-nt bubble (a point which will be addressed below; see Discussion). We compared AID binding to bubble and stem-loop substrates by measuring half-saturation values and the time of complex half-life. As shown in Fig. 3C, higher levels of complex were generated on the AGC bub7, AGC S7, S9, and S13 substrates than on AGC S3. However, binding saturation was not achieved with the stem-loop substrates (i.e., Kd of >10 nM for AGC S3 and S7, S9, and S13), indicating that AID binds stem-loop substrates with at least a 10-fold-lower binding affinity than that for AGC bub7. Complex decay studies indicated that the half-life of AID bound to AGC bub7 was approximately 10 min (Fig. 3D), in agreement with our previous findings (25). In contrast, AID bound to AGC S7 with a half-life of approximately 2 min (Fig. 3D). Taken together, these results indicate that lower binding affinities and lower levels of bound complex result in decreased AID activity.
FIG. 3.
Determination of AID binding parameters to bubble and stem-loop substrates. (A) Typical native EMSA gel showing the binding of AID to TGC bub7 over a range of substrate concentrations from 0.0125 to 10 nM. Bound and free substrate are shown. −, GST alone as a control for AID-specific complexes. (B) EMSA was carried out in the presence of increasing substrate concentrations ranging from 0.0125 to 10 nM for TGC bub1, TGC bub2, TGC bub3, and TGC bub7. In order to obtain half-saturation (Kd) values of interaction, fractions of bound and free substrate were quantitated at each substrate concentration and a bound-versus-free plot was generated. P values were <0.05 between TGC bub7 and other substrates. (C) Same as panel B, but comparing the binding of AID to AGC bub7, AGC S3, AGC S7, AGC S9, and AGC S13 over a substrate concentration range of 0.0125 to 6 nM. Binding saturation kinetics were not observed for either of the stem-loop substrates, indicating a significantly lower affinity of AID binding. P values were <0.05 in a comparison of AGC bub7 results to those for AGC S3. (D) EMSA comparing the complex half-life of GST-AID bound to AGC bub7 and AGC S7. All reactions were carried out in duplicate, and the average values at each time point (as indicated on the x axis) are shown. The amount of bound substrate after each incubation time with cold competitor is expressed as a percentage of the total bound substrate at time zero (no incubation with competitor) and is shown on the y axis. Complex half-life was determined to be 10 min for AGC bub7 and 2 min for AGC S7.
AID prefers to deaminate a WRC motif flanked by ssDNA.
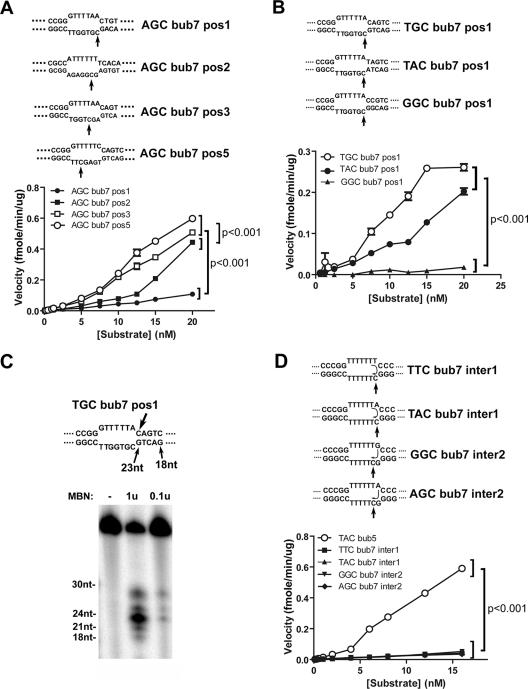
A possible explanation for the preference of AID for the 5-nt bubble substrate over the 1- to 3-nt bubble substrates could be that AID prefers to target cytidines that are surrounded by ssDNA rather than those located proximal to a single-stranded/double-stranded DNA (ss/dsDNA) junction. In order to test this possibility, we generated 7-nt bubble substrates containing the same WRC motif (AGC) and varied the position of the target cytidine (Fig. 4A, upper panel). Although weak deamination was observed when the target cytidine was next to the ss/dsDNA junction, AID preferentially deaminated the more central cytidines of AGC bub7 pos3 and pos5 compared to the AGC bub7 pos1 and pos2 substrates (Fig. 4A, lower panel). This indicates that the WR dinucleotide and the target cytidine must be fully single stranded for efficient deamination by AID. The preferential deamination of a cytidine located at positions 5 and 4 (in the case of the optimal bub5 substrate) suggested that poor deamination of larger bubble substrates (e.g., bub13) (Fig. 1) may be due to a preference of AID for deaminating cytidines in close proximity to an ss/dsDNA junction. However, we found that 17-nt bubble substrates with the target cytidine located at either 10 or 4 nt from the bubble junction were deaminated equally by AID (data not shown). Thus, weak deamination of larger bubble substrates (e.g., 13 nt and above) compared to that of optimal bubble substrates (5- and 7-nt bubbles) is not due to differences in the position of the target cytidine.
FIG. 4.
Deamination kinetics on 7-nt bubble substrates. (A) Upper panel shows substrates with the target cytidine in the context of the WRC motif (AGC) placed in different positions (pos1, -2, -3, or -5) in the bubble region of the substrate. The bubble and surrounding sequences are shown, and the substrates are otherwise identical to those shown in Fig. 1A. The velocity of deamination by AID was measured for each substrate over a range of substrate concentrations from 0.0125 to 20 nM and is shown in the lower panel. P values were <0.001 for a comparison between AGC bub7 pos5 and pos3 to AGC bub7 pos2 and pos 1 results at a substrate concentration of >5 nM. (B) Upper panel shows substrates with a first position target cytidine and the −1 and −2 nucleotides placed in the stem region of the bubble substrate and varied to form WRC (TGC or TAC) or non-WRC (GGC) motifs. The lower panel shows deamination kinetics similar to those described above over a substrate concentration of 0.0125 to 20 nM. P values were <0.001 between the WRC (TGC and TAC) and the non-WRC (GGC) motifs at a substrate concentration of >7.5 nM. (C) MBN digestion of TGC bub7 pos1, which is shown in panel C. The upper panel shows the bubble and surrounding regions of the bubble and the expected sizes of digestion products at various positions. Substrate (50 fmol) was digested with 0.1 U or 1 U of MBN for 5 min at 37°C, and the mixture was electrophoresed on a 14% denaturing gel along with labeled oligonucleotide size markers ranging from 18 to 30 nt. (D) The upper panel shows substrates with the cytidine in the first (TTC bub7 inter1 and TAC bub7 inter1) or second (GGC bub7 inter2 and AGC bub7 inter2) position in the context of a non-WRC motif (GGC). Top strands were varied such that if the WRC motif were read in trans (as indicated by the arrows inside the bubble), the target cytidine would be place in either a WRC or non-WRC motif (TTC versus TAC or GGC versus AGC). The lower panel shows deamination kinetics determined as described for part A over a substrate concentration of 0.0125 to 15 nM along with a 5-nt bubble containing a WRC motif (TAC bub5) as a positive control for the deamination activity of AID. P values were <0.001 between results for TAC bub7 and all other substrates at substrate concentrations of >5 nM.
The WRC motif may be recognized in transiently ssDNA but only in cis.
Since we detected low but measurable levels of AID activity on the bubble substrate with the target cytidine placed next to the ss/dsDNA junction (i.e., AGC bub7 pos1) (Fig. 4A), we sought to determine whether the activity of AID on junctional cytidines could be modulated by the upstream dinucleotide in the double-stranded region of the substrate. We generated bubble substrates where the dinucleotide preceding the target cytidine was placed in the double-stranded region of the bubble and was varied to form WRC (TGC bub7 pos1 or TAC bub7 pos1) or non-WRC (GGC bub7 pos1) motifs (Fig. 4B, upper panel). The results show that AID preferentially deaminated WRC over non-WRC motifs even when the WR was placed in the double-stranded region (Fig. 4B, lower panel). This suggests either that AID has sufficient DNA melting activity to expose the WR motif, that it recognizes the WR in dsDNA (albeit to a lesser degree than in ssDNA based on results shown in Fig. 4A), or that the WR sequence can be exposed by transient denaturation (“breathing”) of the dsDNA region. To distinguish between these possibilities, we used two approaches. First, digestion of TGC bub7 pos1 by MBN, an ssDNA endonuclease (4), revealed a major expected product of 23 nt corresponding to complete digestion of ssDNA up to the edge of the double-stranded bubble stem (Fig. 4C, 0.1u lane). However, in the presence of larger amounts of MBN, weaker products of 21 and 19 nt corresponding to digestion products inside the double-stranded region of the substrate were also detected (Fig. 4C, 1u lane). Second, with substrates labeled on the top strand, we were able to detect weak AID activity on the cytidines located within the dsDNA region of bubble substrates (e.g., Fig. 4D, top panel, arrow pointing to top-strand cytidine; data not shown), confirming that these positions were at least transiently single-stranded since the primary amino group on cytidine must be accessible for AID deamination. Taken together, these results indicate that the weak activity observed when the WR motif was in the double-stranded region was due to transient ssDNA formation as a result of “breathing” allowing for the recognition of the WR dinucleotide features by AID. Thus, these data show that a fully single-stranded WRC motif is required for deamination by AID.
Since we found that the deamination was strongly inhibited if the WR motif was placed within the dsDNA region of a bubble, we tested whether the WR motif could be placed in ssDNA but on the opposite strand of the bubble (Fig. 4D, upper panel). We designed target bubble structures in which the sequence of the top (cold) strand was varied to form WRC or non-WRC motifs in trans with respect to the target cytidine located on the bottom strand (Fig. 4D, upper panel). We found that AID activity on the target cytidine was not affected by the WR sequence of the top strand (Fig. 4D, lower panel), indicating that WRC preference must be enforced 5′ to 3′ in the context of the same DNA strand.
Recognition of specific nucleotide side chain groups by AID.
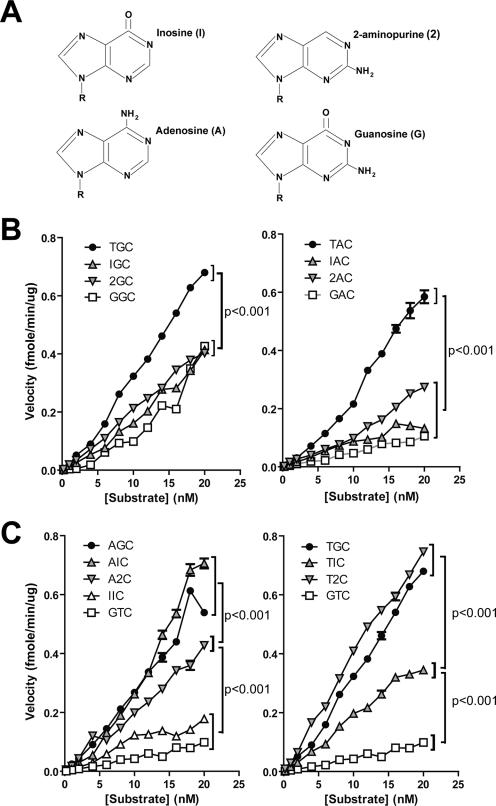
Although we have previously shown that the WRC specificity of AID represents a catalytic rather than a binding preference (25), the reason for this specificity is unknown. AID's preference for R nucleotides (i.e., A or G) in the −1 position relative to the cytidine could simply reflect a requirement for a purine rather than a pyrimidine at this position. However, the reasons for AID's preference of a W nucleotide (i.e., A or T) in the −2 position relative to the cytidine are less clear, since one purine (A) or pyrimidine (T) generates a hot spot while the other purine (G) or pyrimidine (C) results in a cold spot. In order to gain insight into the WRC specificity of AID, we utilized the purine analogues inosine and 2-aminopurine in catalytic assays described above. As shown in Fig. 5A, inosine and 2-aminopurine resemble G, with the former harboring the carbonyl group and the latter the amino group of G. We reasoned that substitution of A or G by either of these purine analogues would facilitate the identification of the purine features that are important for AID activity in the −1 and −2 positions. Thus, we created 7-nt bubble substrates (similar to TGC bub7, shown in Fig. 1A) in which we inserted inosine or 2-aminopurine in the −1 or −2 positions relative to the cytidine in various combinations and assessed AID deamination in parallel with conventional WRC and non-WRC substrates.
FIG. 5.
Importance of nucleotide side chains in WRC specificity. (A) The nucleotide analogues inosine and 2-aminopurine are shown with adenosine and guanosine. Inosine and 2-aminopurine harbor, respectively, only the carbonyl or amino side chain of G. Seven-nucleotide bubble substrates similar to TGC bub7 shown in Fig. 1A were constructed with different combinations of A, G, inosine, and 2-aminopurine in either the −2 or −1 position. (B) Deamination kinetics comparing the activities of AID on a range of substrate concentrations for substrates containing inosine or 2-aminopurine in the −2 position along with a natural purine in the −1 position, which is either A (right plot) or G (left plot). Each velocity value is the average for two independent experiments, and error bars are shown. P values were <0.001 between results for the natural hot-spot motifs TGC, TAC, and AGC (shown in panel C, left) and those for all other plots at substrate concentrations of >6 nM. (C) Deamination kinetics comparing the activities of AID on a range of substrate concentrations for substrates where either inosine or 2-aminopurine was placed in the −1 position in combination with a natural W nucleotide in the −2 position, which is either T (right plot) or A (left plot). The non-WRC substrate GTC was used as a typical cold spot to highlight the range of AID activity. For the left panel, P values were <0.001 in a comparison of AGC and AIC results to those for A2C at substrate concentrations of >8 nM and comparing A2C results with those for IIC and GTC at substrate concentrations of >4 nM. For the right panel, P values were <0.001 in a comparison of TGC and T2C results to those for TIC at substrate concentrations of >6 nM and comparing TIC results to those for GTC at substrate concentrations of >4 nM.
First, we examined the effect of either purine analogue in the −2 position (W), while the −1 position (R) was either G (Fig. 5B, left panel) or A (Fig. 5B, right panel). Neither inosine nor 2-aminopurine, like guanosine, was able to substitute for a W (A or T) in the −2 position (i.e., AID deamination was greater with TGC or AGC than with IGC, 2GC, or GGC and greater with TAC than with IAC, 2AC, or GAC) (Fig. 5B). We conclude that the carbonyl and amino moieties of guanosine have a negative effect on AID deamination in the −2 position relative to the cytidine. We next examined the effects of either purine analogue in the −1 position relative to the cytidine in the AID deamination assays while the −2 position was either an A (Fig. 5C, left panel) or T (Fig. 5C, right panel). In contrast to the results described above, here either inosine or 2-aminopurine was able to support efficient AID deamination. These results show that AID prefers a purine at the −1 position relative to the cytidine. Interestingly, if the −2 position was an A, inosine supported better deamination than 2-aminopurine in the −1 position (i.e., greater deamination with AIC than with A2C) (Fig. 5C). In contrast, if the −2 position was a T, 2-aminopurine supported better deamination than inosine in the −1 position (i.e., deamination was greater with T2C than with TIC) (Fig. 5C). This result reflects an interdependence of the −1 and −2 positions on each other (see Discussion).
DISCUSSION
Preferential deamination of small-bubble substrates.
It has been proposed that AID might target the transcription bubble itself or other ssDNA structures generated upstream or downstream of the elongation complex (6, 9, 21, 31, 45, 50). Thus, to gain insight into the in vivo substrate of AID, we examined its requirements in vitro. We reasoned that AID is likely to have an in vitro pattern of activity that is reflective of its in vivo targets, a correlation that is observed for many enzymes. In support of this rationale, we and others have shown that other properties of AID, such as WRC specificity, when measured in vitro, intimately match the pattern of hot-spot mutations in vivo (24, 25, 41). As another well-studied example of immunological relevance, the pattern of RAG1/2 cleavage and binding to different immunoglobulin recombination signal sequences in vitro correlates with the usage of gene segments in vivo (23, 26, 35, 60).
We found that deamination was optimal on 5-nt bubble substrates, undetectable or very weak on 1- to 3-nt bubble substrates or stem-loop substrates of 3 to 13 nt, and reduced on larger bubbles of 9 to 13 nt. Our finding that 7-nt bubbles contained transiently ssDNA at the dsDNA junctions (Fig. 4D) suggests that 3-nt bubbles would also be subject to this effect and thus the poor but measurable activity on 3-nt bubbles could simply reflect AID's deamination of “breathing” 3-nt bubbles. This would indicate that the minimal DNA binding/catalytic domain for AID is likely greater than 3 nt in length. Although our work argues against AID deaminating stem-loop structures in vivo, we cannot exclude the possibility that larger stem-loops or R-loops may be acted on by AID. However, based on the results shown here, bubble, stem-loop, or R-loop structures greater than 7 nt in length are probably recognized by AID as less efficiently ssDNA and than small bubbles, possibly due to the more rigid structure of the latter. An earlier report showed that the optimal substrate for AID activity was a 9-nt bubble substrate (7), a discrepancy that we propose might be due to different preparations of AID or various reaction conditions. Additionally, we found that AID activity levels may appear similar at low substrate concentrations and differences may become apparent only when enzymatic velocities are examined over a wide range of substrate concentrations, which can also explain this discrepancy. We suggest that since transcription bubbles are proposed to be ∼18 nt long (12), with the majority of their length occupied by the elongation complex, the preference of AID for deamination of WRC motifs in 5-nt-long bubbles reflects its optimization for targeting the accessible regions of the transcription bubble. Alternatively, AID may have evolved to react with small regions of transcription-induced denatured DNA, as was recently observed for hypermutating genes (47).
As mentioned in the introduction, current literature has not addressed whether the WR motif must also be fully single stranded. That is, significant data show that AID can react with WRC motifs in ssDNA but don't rule out that AID can react with a WRC motif which is only partially located in ssDNA, such as would occur at the edges of DNA gaps or transcription bubble junctions in vivo. To this end, we examined whether the WR motif can be recognized in dsDNA as well as ssDNA. By using bubble substrates in which the position of the target cytidine was varied as well as substrates in which the WR motif was placed in the stem rather than the bubble region, we showed that the most efficient deamination target consisted of a WRC motif flanked by ssDNA. This result indicates that in contrast to sequence-specific dsDNA recognition enzymes (e.g., restriction endonucleases and transcription factors), AID is not capable of discerning nucleotide features in dsDNA.
The significance of binding affinity for AID activity.
To understand why some structures are preferentially deaminated over other substrates, we examined whether AID binding affinity correlates with deamination activity. First, we found that stem-loop substrates, which were poorly deaminated by AID, were also bound by AID with lower affinity than was the case with optimal substrates (i.e., 5- to 7-nt bubble substrates). The notable difference in AID binding between a 7-nt bubble and a 7-nt stem-loop substrate suggests that AID can discern specific shape differences, possibly due to DNA bending. Second, we found that 2- or 3-nt bubbles, which were poorly deaminated by AID, exhibit highly diminished boundmax values despite only slightly reduced affinity compared to that for bub7. This indicates heterogeneity in the substrate or the enzyme pool, such that a smaller fraction of the substrate is bound. As discussed above, since the bubble substrates contain ss/dsDNA junctions which are “breathing,” we believe that the lower boundmax values obtained with 3-nt bubbles are due to AID interacting with “opened-up” bubbles that exist at a constant but low concentration relative to the “closed” 3-nt length bubbles. Collectively, our data suggest that high-affinity binding of AID to DNA substrates is required for efficient deamination in vitro. Definitive confirmation of this notion would require in vitro and in vivo data correlating substrate affinity with activity using mutants of AID.
Our finding that AID binds ssDNA with high affinity provides an explanation for AID processivity (41). That is, because AID binds rapidly and dissociates slowly, it is more likely to bind the same substrate molecule after dissociation (25, 42), thus accounting for the short-track processivity of AID (13). Substrate “jumping” has also been shown to account for the processive-like behavior of some restriction endonucleases (52), in contrast to substrate “sliding,” which accounts for the processivity of polymerases (10). Nonetheless, formal proof is lacking that AID processivity is necessary for biological function, and this might be satisfied by the experiments mentioned in the previous paragraph.
Target requirements for WRC preference.
Since direct analysis of the AID catalytic site is not yet possible due to the lack of an X-ray crystal structure, we aimed to gain insight into the mechanism of WRC specificity by understanding the specific nucleotide side chains that are of importance for the recognition of a WRC by AID. By using the purine analogues inosine and 2-aminopurine in the −1 position in combination with a natural W (A or T) in the −2 position relative to the target cytidine, we showed that both analogues support efficient deamination. This indicates that AID prefers any purine at the −1 position. However, as noted in Results, the nucleotide preference of AID at the −2 position is paradoxical. We found that when placed in the −2 position in combination with natural R nucleotides (i.e., A or G) in the −1 position, both inosine and 2-aminopurine functioned similarly to a G, rather than A. This indicates either that the amino moiety of A is recognized by the catalytic site of AID in this position or that the carbonyl and amino moieties of G are a hindrance to recognition. The former model would predict that G, inosine, and 2-aminopurine would all be equally inefficient at supporting deamination. However, our observation that in the −2 position G is slightly less efficient than either inosine or 2-aminopurine, each of which carries only one of its side chains, supports the latter model. Overall, the result that no trinucleotide combination with an analogue was a significantly better deamination substrate than natural WRC motifs indicates that the catalytic site of AID is optimized for its natural targets in vivo. We propose that the catalytic site of AID possesses two recognition pockets for the −2 position, one that fits the purine A and one that fits the pyrimidine T. Since any purine could support efficient deamination in the −1 position, this suggests that interactions between AID and DNA might occur on shared groups between purines via involvement of amino acids such as Asn, Gln, Ser, Thr, Tyr, Arg, and Lys, all of which are thought to form hydrogen bonds with the pentamer or hexamer backbone of purines (11). However, our finding that AID's preference for a nucleotide in the −1 position depended on which nucleotide was present in the −2 position (i.e., preference for T2C over TIC but preference for AIC over A2C) suggests that different amino acid side chains of the −1 pocket of AID prefer different purine side chains, such that depending on which −2 pocket is occupied, either a carbonyl or an amino side chain bearing purine is preferred in the −1 position. We note that this model represents the simplest scenario for an AID catalytic site compatible with the current in vitro data but that alternative models are possible, since stacking interactions between adjacent nucleotides and the recognition of a dinucleotides motif by a single amino acid may be important.
Acknowledgments
We are grateful to members of the Martin lab and Marc Shulman, Gillian Wu, and David Pulleyblank for helpful discussions and to Oxana Kolenchenko, Maribel Berru, and Yin Ying Zhang for technical help.
This research was funded by a grant from the Canadian Institute of Health Research to A.M., who is supported by a Canada Research Chair award. M.L. was supported by the David Rae Memorial Award from the Leukemia and Lymphoma Society of Canada and is presently supported by a Terry Fox Foundation award from the National Cancer Institute of Canada.
Footnotes
Published ahead of print on 24 September 2007.
REFERENCES
- 1.Arakawa, H., J. Hauschild, and J. M. Buerstedde. 2002. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science 295:1301-1306. [DOI] [PubMed] [Google Scholar]
- 2.Bachl, J., C. Carlson, V. Gray-Schopfer, M. Dessing, and C. Olsson. 2001. Increased transcription levels induce higher mutation rates in a hypermutating cell line. J. Immunol. 166:5051-5057. [DOI] [PubMed] [Google Scholar]
- 3.Basu, U., J. Chaudhuri, C. Alpert, S. Dutt, S. Ranganath, G. Li, J. P. Schrum, J. P. Manis, and F. W. Alt. 2005. The AID antibody diversification enzyme is regulated by protein kinase A phosphorylation. Nature 438:508-511. [DOI] [PubMed] [Google Scholar]
- 4.Baumann, U., and S. Chang. 1995. Asymmetric structure of five and six membered DNA hairpin loops. Mol. Biol. Rep. 22:25-31. [DOI] [PubMed] [Google Scholar]
- 5.Besmer, E., E. Market, and F. N. Papavasiliou. 2006. The transcription elongation complex directs activation-induced cytidine deaminase-mediated DNA deamination. Mol. Cell. Biol. 26:4378-4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bransteitter, R., P. Pham, P. Calabrese, and M. F. Goodman. 2004. Biochemical analysis of hypermutational targeting by wild type and mutant activation-induced cytidine deaminase. J. Biol. Chem. 279:51612-51621. [DOI] [PubMed] [Google Scholar]
- 7.Bransteitter, R., P. Pham, M. D. Scharff, and M. F. Goodman. 2003. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl. Acad. Sci. USA 100:4102-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaudhuri, J., C. Khuong, and F. W. Alt. 2004. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature 430:992-998. [DOI] [PubMed] [Google Scholar]
- 9.Chaudhuri, J., M. Tian, C. Khuong, K. Chua, E. Pinaud, and F. W. Alt. 2003. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature 422:726-730. [DOI] [PubMed] [Google Scholar]
- 10.Chen, X., S. Zuo, Z. Kelman, M. O'Donnell, J. Hurwitz, and M. F. Goodman. 2000. Fidelity of eucaryotic DNA polymerase delta holoenzyme from Schizosaccharomyces pombe. J. Biol. Chem. 275:17677-17682. [DOI] [PubMed] [Google Scholar]
- 11.Cheng, A. C., W. W. Chen, C. N. Fuhrmann, and A. D. Frankel. 2003. Recognition of nucleic acid bases and base-pairs by hydrogen bonding to amino acid side-chains. J. Mol. Biol. 327:781-796. [DOI] [PubMed] [Google Scholar]
- 12.Choder, M., and Y. Aloni. 1988. RNA polymerase II allows unwinding and rewinding of the DNA and thus maintains a constant length of the transcription bubble. J. Biol. Chem. 263:12994-13002. [PubMed] [Google Scholar]
- 13.Coker, H. A., and S. K. Petersen-Mahrt. 2007. AID's distributive mode of action: a definition. DNA Repair (Amsterdam) 6:693-694. [DOI] [PubMed] [Google Scholar]
- 14.Coles, L. S., F. Occhiodoro, M. A. Vadas, and M. F. Shannon. 1994. A sequence-specific single-strand DNA binding protein that contacts repressor sequences in the human GM-CSF promoter. Nucleic Acids Res. 22:4276-4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins, I., A. Weber, and D. Levens. 2001. Transcriptional consequences of topoisomerase inhibition. Mol. Cell. Biol. 21:8437-8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickerson, S. K., E. Market, E. Besmer, and F. N. Papavasiliou. 2003. AID mediates hypermutation by deaminating single stranded DNA. J. Exp. Med. 197:1291-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Noia, J., and M. S. Neuberger. 2002. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature 419:43-48. [DOI] [PubMed] [Google Scholar]
- 18.Duquette, M. L., P. Handa, J. A. Vincent, A. F. Taylor, and N. Maizels. 2004. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 18:1618-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaillard, C., and F. Strauss. 1990. Sequence-specific single-strand-binding protein for the simian virus 40 early promoter stimulates transcription in vitro. J. Mol. Biol. 215:245-255. [DOI] [PubMed] [Google Scholar]
- 20.Harris, R. S., J. E. Sale, S. K. Petersen-Mahrt, and M. S. Neuberger. 2002. AID is essential for immunoglobulin V gene conversion in a cultured B cell line. Curr. Biol. 12:435-438. [DOI] [PubMed] [Google Scholar]
- 21.Honjo, T., K. Kinoshita, and M. Muramatsu. 2002. Molecular mechanism of class switch recombination: linkage with somatic hypermutation. Annu. Rev. Immunol. 20:165-196. [DOI] [PubMed] [Google Scholar]
- 22.Huang, F. T., K. Yu, C. L. Hsieh, and M. R. Lieber. 2006. Downstream boundary of chromosomal R-loops at murine switch regions: implications for the mechanism of class switch recombination. Proc. Natl. Acad. Sci. USA 103:5030-5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larijani, M., S. Chen, L. A. Cunningham, J. M. Volpe, L. G. Cowell, S. M. Lewis, and G. E. Wu. 2006. The recombination difference between mouse kappa and lambda segments is mediated by a pair-wise regulation mechanism. Mol. Immunol. 43:870-881. [DOI] [PubMed] [Google Scholar]
- 24.Larijani, M., D. Frieder, W. Basit, and A. Martin. 2005. The mutation spectrum of purified AID is similar to the mutability index in Ramos cells and in ung(−/−)msh2(−/−) mice. Immunogenetics 56:840-845. [DOI] [PubMed] [Google Scholar]
- 25.Larijani, M., A. P. Petrov, O. Kolenchenko, M. Berru, S. N. Krylov, and A. Martin. 2007. AID associates with single-stranded DNA with high affinity and a long complex half-life in a sequence-independent manner. Mol. Cell. Biol. 27:20-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, A. I., S. D. Fugmann, L. G. Cowell, L. M. Ptaszek, G. Kelsoe, and D. G. Schatz. 2003. A functional analysis of the spacer of V(D)J recombination signal sequences. PLoS Biol. 1:E1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, Z., C. J. Woo, M. D. Iglesias-Ussel, D. Ronai, and M. D. Scharff. 2004. The generation of antibody diversity through somatic hypermutation and class switch recombination. Genes Dev. 18:1-11. [DOI] [PubMed] [Google Scholar]
- 28.Liu, L. F., and J. C. Wang. 1987. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. USA 84:7024-7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Longerich, S., A. Tanaka, G. Bozek, D. Nicolae, and U. Storb. 2005. The very 5′ end and the constant region of Ig genes are spared from somatic mutation because AID does not access these regions. J. Exp. Med. 202:1443-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin, A., P. D. Bardwell, C. J. Woo, M. Fan, M. J. Shulman, and M. D. Scharff. 2002. Activation-induced cytidine deaminase turns on somatic hypermutation in hybridomas. Nature 415:802-806. [DOI] [PubMed] [Google Scholar]
- 31.Martin, A., and M. D. Scharff. 2002. AID and mismatch repair in antibody diversification. Nat. Rev. Immunol. 2:605-614. [DOI] [PubMed] [Google Scholar]
- 32.Martomo, S. A., D. Fu, W. W. Yang, N. S. Joshi, and P. J. Gearhart. 2005. Deoxyuridine is generated preferentially in the nontranscribed strand of DNA from cells expressing activation-induced cytidine deaminase. J. Immunol. 174:7787-7791. [DOI] [PubMed] [Google Scholar]
- 33.McBride, K. M., A. Gazumyan, E. M. Woo, V. M. Barreto, D. F. Robbiani, B. T. Chait, and M. C. Nussenzweig. 2006. Regulation of hypermutation by activation-induced cytidine deaminase phosphorylation. Proc. Natl. Acad. Sci. USA 103:8798-8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michael, N., H. M. Shen, S. Longerich, N. Kim, A. Longacre, and U. Storb. 2003. The E box motif CAGGTG enhances somatic hypermutation without enhancing transcription. Immunity 19:235-242. [DOI] [PubMed] [Google Scholar]
- 35.Montalbano, A., K. M. Ogwaro, A. Tang, A. G. Matthews, M. Larijani, M. A. Oettinger, and A. J. Feeney. 2003. V(D)J recombination frequencies can be profoundly affected by changes in the spacer sequence. J. Immunol. 171:5296-5304. [DOI] [PubMed] [Google Scholar]
- 36.Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102:553-563. [DOI] [PubMed] [Google Scholar]
- 37.Okazaki, I. M., K. Kinoshita, M. Muramatsu, K. Yoshikawa, and T. Honjo. 2002. The AID enzyme induces class switch recombination in fibroblasts. Nature 416:340-345. [DOI] [PubMed] [Google Scholar]
- 38.Pasqualucci, L., Y. Kitaura, H. Gu, and R. Dalla-Favera. 2006. PKA-mediated phosphorylation regulates the function of activation-induced deaminase (AID) in B cells. Proc. Natl. Acad. Sci. USA 103:395-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peters, A., and U. Storb. 1996. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity 4:57-65. [DOI] [PubMed] [Google Scholar]
- 40.Petersen-Mahrt, S. K., R. S. Harris, and M. S. Neuberger. 2002. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature 418:99-104. [DOI] [PubMed] [Google Scholar]
- 41.Pham, P., R. Bransteitter, J. Petruska, and M. F. Goodman. 2003. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature 424:103-107. [DOI] [PubMed] [Google Scholar]
- 42.Pham, P., L. Chelico, and M. F. Goodman. 2007. DNA deaminases AID and APOBEC3G act processively on single-stranded DNA. DNA Repair (Amsterdam) 6:689-692. [DOI] [PubMed] [Google Scholar]
- 43.Rada, C., G. T. Williams, H. Nilsen, D. E. Barnes, T. Lindahl, and M. S. Neuberger. 2002. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 12:1748-1755. [DOI] [PubMed] [Google Scholar]
- 44.Rahmouni, A. R., and R. D. Wells. 1992. Direct evidence for the effect of transcription on local DNA supercoiling in vivo. J. Mol. Biol. 223:131-144. [DOI] [PubMed] [Google Scholar]
- 45.Ramiro, A. R., P. Stavropoulos, M. Jankovic, and M. C. Nussenzweig. 2003. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat. Immunol. 14:14. [DOI] [PubMed] [Google Scholar]
- 46.Revy, P., T. Muto, Y. Levy, F. Geissmann, A. Plebani, O. Sanal, N. Catalan, M. Forveille, R. Dufourcq-Labelouse, A. Gennery, I. Tezcan, F. Ersoy, H. Kayserili, A. G. Ugazio, N. Brousse, M. Muramatsu, L. D. Notarangelo, K. Kinoshita, T. Honjo, A. Fischer, and A. Durandy. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the hyper-IgM syndrome (HIGM2). Cell 102:565-575. [DOI] [PubMed] [Google Scholar]
- 47.Ronai, D., M. D. Iglesias-Ussel, M. Fan, Z. Li, A. Martin, and M. D. Scharff. 2007. Detection of chromatin-associated single-stranded DNA in regions targeted for somatic hypermutation. J. Exp. Med. 204:181-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosenberg, J. M., H. W. Boyer, and P. Greene. 1981. The structure and function of the EcoRI restriction endonuclease. Gene Amplif. Anal. 1:131-164. [PubMed] [Google Scholar]
- 49.Shen, H. M., S. Ratnam, and U. Storb. 2005. Targeting of the activation-induced cytosine deaminase is strongly influenced by the sequence and structure of the targeted DNA. Mol. Cell. Biol. 25:10815-10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen, H. M., and U. Storb. 2004. Activation-induced cytidine deaminase (AID) can target both DNA strands when the DNA is supercoiled. Proc. Natl. Acad. Sci. USA 101:12997-13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sohail, A., J. Klapacz, M. Samaranayake, A. Ullah, and A. S. Bhagwat. 2003. Human activation-induced cytidine deaminase causes transcription-dependent, strand-biased C to U deaminations. Nucleic Acids Res. 31:2990-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stanford, N. P., M. D. Szczelkun, J. F. Marko, and S. E. Halford. 2000. One- and three-dimensional pathways for proteins to reach specific DNA sites. EMBO J. 19:6546-6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wakae, K., B. G. Magor, H. Saunders, H. Nagaoka, A. Kawamura, K. Kinoshita, T. Honjo, and M. Muramatsu. 2006. Evolution of class switch recombination function in fish activation-induced cytidine deaminase, AID. Int Immunol. 18:41-47. [DOI] [PubMed] [Google Scholar]
- 54.Wang, Z., and P. Droge. 1997. Long-range effects in a supercoiled DNA domain generated by transcription in vitro. J. Mol. Biol. 271:499-510. [DOI] [PubMed] [Google Scholar]
- 55.Xue, K., C. Rada, and M. S. Neuberger. 2006. The in vivo pattern of AID targeting to immunoglobulin switch regions deduced from mutation spectra in msh2-/- ung-/- mice. J. Exp. Med. 203:2085-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang, S. Y., S. D. Fugmann, and D. G. Schatz. 2006. Control of gene conversion and somatic hypermutation by immunoglobulin promoter and enhancer sequences. J. Exp. Med. 203:2919-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu, K., F. Chedin, C. L. Hsieh, T. E. Wilson, and M. R. Lieber. 2003. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 4:442-451. [DOI] [PubMed] [Google Scholar]
- 58.Yu, K., F. T. Huang, and M. R. Lieber. 2004. DNA substrate length and surrounding sequence affect the activation-induced deaminase activity at cytidine. J. Biol. Chem. 279:6496-6500. [DOI] [PubMed] [Google Scholar]
- 59.Yu, K., D. Roy, M. Bayramyan, I. S. Haworth, and M. R. Lieber. 2005. Fine-structure analysis of activation-induced deaminase accessibility to class switch region R-loops. Mol. Cell. Biol. 25:1730-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu, K., A. Taghva, and M. R. Lieber. 2002. The cleavage efficiency of the human immunoglobulin heavy chain VH elements by the RAG complex: implications for the immune repertoire. J. Biol. Chem. 277:5040-5046. [DOI] [PubMed] [Google Scholar]
- 61.Zarrin, A. A., F. W. Alt, J. Chaudhuri, N. Stokes, D. Kaushal, L. Du Pasquier, and M. Tian. 2004. An evolutionarily conserved target motif for immunoglobulin class-switch recombination. Nat. Immunol. 5:1275-1281. [DOI] [PubMed] [Google Scholar]
- 62.Zeng, X., G. A. Negrete, C. Kasmer, W. W. Yang, and P. J. Gearhart. 2004. Absence of DNA polymerase eta reveals targeting of C mutations on the nontranscribed strand in immunoglobulin switch regions. J. Exp. Med. 199:917-924. [DOI] [PMC free article] [PubMed] [Google Scholar]