Abstract
DNA methyltransferase 1 (DNMT1) plays an important role in the inheritance of genomic DNA methylation, which is coupled to the DNA replication process. Early embryonic lethality in DNMT1-null mutant (Dnmt1c) mice indicates that DNA methylation is essential for mammalian development. DNMT1, however, interacts with a number of transcriptional regulators and has a transcriptional repressor activity independent of its catalytic activity. To examine the roles of the catalytic activity of DNMT1 in vivo, we generated a Dnmt1ps allele that expresses a point-mutated protein that lacks catalytic activity (DNMT1-C1229S). Dnmt1ps mutant mice showed developmental arrest shortly after gastrulation, near-complete loss of DNA methylation, and an altered distribution of repressive chromatin markers in the nuclei; these phenotypes are quite similar to those of the Dnmt1c mutant. The mutant DNMT1 protein failed to associate with replication foci in Dnmt1ps cells. Reconstitution experiments and replication labeling in Dnmt1−/− Dnmt3a−/− Dnmt3b−/− (i.e., unmethylated) embryonic stem cells revealed that preexisting DNA methylation is a major determinant for the cell cycle-dependent localization of DNMT1. The C-terminal catalytic domain of DNMT1 inhibited its stable association with unmethylated chromatin. Our results reveal essential roles for the DNA methylation mark in mammalian development and in DNMT1 localization.
The methylation of DNA at the C-5 position of cytosine residues is a heritable epigenetic mechanism that is involved in a broad range of biological processes in vertebrates, plants, and fungi (3). In mammals, DNA methylation is coordinately regulated by three DNA methyltransferases, DNMT1, DNMT3A, and DNMT3B (6), and plays a crucial role in the regulation of gene expression, the silencing of parasitic elements, genomic imprinting, and embryogenesis (3). The hypermethylation of promoter CpG islands in tumor suppressor genes is a well-recognized feature of many cancers (28). Mammalian genomes are mostly methylated at symmetrical CpG sites, and the pattern of methylated CpGs is thought to propagate through the copying of a template parental strand (3). After semiconservative DNA replication, hemi-methylated CpG sites are recognized and preferentially methylated at cytosine on the opposite, nascent DNA strand.
DNMT1 is the major enzyme responsible for the stable inheritance of DNA methylation patterns after DNA replication (21, 27). Inactivation of DNMT1 results in extensive loss of DNA methylation in the mouse (32, 34). DNMT1 has a strong preference for hemi-methylated DNA as a substrate. The major isoform of mouse DNMT1 is a 1,620-amino-acid protein that has a large N-terminal regulatory domain, which contains several functional subdomains, and a C-terminal catalytic domain, separated by a linker region, a Lys-Gly (KG) repeat. The N-terminal regulatory domain and the C-terminal catalytic domain can interact directly with each other (17). This intramolecular interaction seems to be required for the enzymatic function of DNMT1 (17, 38). DNMT1 contains DNA-binding domains in both the N- and C-terminal regions that preferentially bind methylated DNA (17). Enzymological kinetic studies of DNMT1 suggest that the binding of methylated DNA at a site other than its catalytic center increases the enzymatic activity by a conformational change; this is known as an allosteric activation (2). DNMT1 methylates its hemi-methylated DNA substrate in a processive manner, in which it moves along a DNA strand, methylating multiple hemi-methylated CpG sites during one DNA-binding cycle (22, 56).
Cytological studies have revealed that DNMT1 dynamically changes its intranuclear localization in a cell cycle-dependent manner (12, 33). During S phase, DNMT1 is largely localized to the site of DNA replication (33). A replication focus target sequence (RFT), also known as the targeting sequence (TS) (33), and a proliferating cell nuclear antigen (PCNA)-binding domain (PBD) (8, 12) in DNMT1's N-terminal region are reported to be critical for DNMT1's association with replication foci. The PBD is also important for DNMT1's localization to DNA repair sites, following UV irradiation (39). In addition, some DNMT1 remains associated with heterochromatin in the late S and G2 phases, even after heterochromatin replication (12). Fluorescence-recovery-after-photobleaching analysis showed that DNMT1 is loaded de novo onto heterochromatin during these phases and thus via a replication-independent mechanism (12). Such cell cycle-dependent changes in DNMT1 localization probably contribute to the stable inheritance of DNA methylation patterns, although their precise roles and mechanisms remain elusive.
A number of studies suggest that DNMT1 has nonenzymatic functions. Its N-terminal domain interacts with histone-modifying enzymes, transcriptional repressors, and cell cycle/checkpoint-regulatory molecules (15, 16, 19, 20, 45, 48, 49, 57). DNMT1 can mediate transcriptional repression (19, 48, 49) or cell cycle progression (55) in a catalytic activity-independent manner in mammalian cell culture. A loss or decrease of DNMT1 causes early embryonic lethality in mouse, frog, and zebrafish, probably due to multiple defects (32, 34, 46, 51). Inactivation of DNMT1 in mouse embryonic fibroblast cells causes defective growth and cell death, accompanied by the dysregulated transcription of numerous genes (26). Its inactivation in human cancer cells leads to activation of the G2/M checkpoint and mitotic catastrophe (5). Because the previous Dnmt1 null allele in mice disrupts the protein structure and results in loss of the DNMT1 protein (32), it is possible that the reported phenotype of Dnmt1 knockout mice reflects the loss of both its enzymatic function and its other, nonenzymatic functions. Thus, it remains unclear to what extent the catalytic activity contributes to the major defects induced by Dnmt1 inactivation during embryogenesis.
Here, to reevaluate the role of DNA methylation during embryogenesis, we generated embryonic stem (ES) cells and mice expressing a protein with a subtle mutation in the catalytic center of DNMT1 (Dnmt1ps allele). We found that embryos carrying either the null mutation (Dnmt1c) or Dnmt1ps showed developmental arrest around embryonic day 8.5 (E8.5), accompanied by multiple defects, suggesting that the loss of catalytic activity is mainly responsible for these phenotypes. Cytological analysis using Dnmt1ps mutant cells and ES cells deficient for DNA methyltransferases demonstrated that a preexisting DNA methylation mark in the genome is crucial for regulating DNMT1's localization within the cell nucleus. Our results reveal an importance of the DNA methylation mark in vivo that could not be fully evaluated for Dnmt1 null mutant mice and provide a novel contribution to our understanding of the complex regulation of DNMT1 localization.
MATERIALS AND METHODS
ES cell culture.
ES cells were maintained in Glasgow modified Eagle's medium (Sigma) supplemented with 15% fetal bovine serum, 0.1 mM nonessential amino acids (Invitrogen), 1 mM sodium pyruvate, 0.1 mM 2-mercaptoethanol, and 1,000 U/ml leukemia inhibitory factor and grown on a gelatinized culture dish with feeder cells.
Generation of Dnmt1-ps mutant ES cells and mice.
To construct the Dnmt1 targeting vector for a point mutation, pTS015, the mouse Dnmt1 genomic DNA fragment containing exons 31 to 33 was amplified by PCR, and a C1229S point mutation in the PC motif was introduced using a QuikChange site-directed mutagenesis kit (Stratagene). The fragment was ligated into the other Dnmt1 targeting vector for null mutation, pTA010, in which exons 31 to 33 are replaced with an IRES-β geo cassette with flanking loxP sites (54). pTS015 was introduced by electroporation into J1 mouse ES cells. One targeted ES cell line was injected into blastocysts to generate chimeric mice. Offspring of these mice were crossed with β-actin Cre mice to remove the IRES-β geo cassette. A different targeted ES cell line was transiently transfected with a Cre expression vector to remove the IRES-β geo cassette, and the remaining wild-type locus was targeted by pTA010. For the PCR identification of the Dnmt1ps allele in mice, we used the primer sequences AAGCGTGTGCCGTCTAGGCA and GCAGTCCTCTGTGAACACTG, which amplify a 600-bp fragment from the wild-type allele and an 820-bp fragment from the Dnmt1ps allele, respectively. For the Dnmt1c allele (32), we used primer sequences GTGTTCACAGAGGACTGCAACG and CCAGCCTCTGAGCCCAGAAAGCGA. To confirm that the Dnmt1ps mutants did not have any coding-region sequence mutations besides C1229S in the PC motif, the full-length Dnmt1 cDNA was PCR amplified from Dnmt1−/ps ES cells and Dnmt1c/ps mice by using the forward primer GGGGTACCATGCCAGCGCGAACAGCTCC and the reverse primer GGGGTACCCTAGTCCTTGGTAGCAGCCT, each of which contains a KpnI site. The PCR fragments were digested with KpnI and cloned into pBluescript M13 SK+ (Stratagene). At least three independent clones for each mutant cDNA were sequenced and compared with the sequence of wild-type Dnmt1 cDNA derived from J1 ES cells.
DNA methylation analysis.
Bisulfite sequence analysis was carried out as described previously (9). Bisulfite-treated genomic DNA was PCR amplified using specific primers (IAP, LINE1, major satellite repeats) (7, 30) with HotStar Taq (Qiagen) for 30 cycles. The size-fractionated PCR products were subcloned into a TA-cloning vector. We sequenced 12 to 32 clones for each sample.
For Southern blot analysis, genomic DNA was digested with CpG methylation-sensitive restriction enzymes (HpaII or MaeII), blotted, and hybridized with IAP (Fusd) or major satellite (pSAT) probes (42).
For immunostaining 5-meC, cells were incubated in a hypotonic solution (0.075 M KCl), fixed in methanol-acetic acid (3:1), and dropped onto a glass slide. After air drying, the slides were irradiated with UV light for 8 h under a germicidal lamp. Detection of 5-meC was carried out according to standard immunostaining protocols. The anti-MeC monoclonal antibody (kindly provided by Hiroshi Sano) was used at a 1:500 dilution (41).
RT-PCR.
RNA was isolated from ES cells with TRIzol extraction reagent (Invitrogen). RNA was isolated from embryos with an RNA extraction kit (Qiagen), according to the manufacturer's instructions. First-strand cDNA was synthesized with the SuperScript first-strand synthesis system for reverse transcription-PCR (RT-PCR). The following primer sets were used: p21-f (AGTGTGCCGTTGTCTCTTCG), p21-r (ACACCAGAGTGCAAGACAGC), p27-f (TCGCAGAACTTCGAAGAGG), p27-r (TGACTCGCTTCTTCCATATCC), p53-f (CCATGGAGGAGTCACAGTCGG), p53-r (TGTCAGGAGCTCCTGCAGCAC), Igf2-f (GACGTGTCTACCTCTCAGGCCGTACTT), Igf2-r (GGGTGTCAATTGGGTTGTTTAGAGCCA), H19-f (CCACTACACTACCTGCCTCAGAATCTGC), H19-r (GGTGGGTACTGGGGCAGCATTG), IAP-typeI-f (GAGCAGGTGAAACCACTGGTCAAT), IAP-typeI-r (ACCCTTGCCACACTTAGAGCAAAG), GAPDH-f (ATGGTGAAGGTCGGTGTGAACGGATTTGGC), and GAPDH-r (GCATCGAAGGTGGAAGAGTGGGAGTTGCTG).
Construction of expression plasmids.
Full-length and truncated forms of mouse Dnmt1 cDNA were ligated in-frame with the 3′ end of the yellow fluorescence protein (YFP) gene, whose expression is regulated by the cytomegalovirus promoter of pcDNA (Invitrogen).
Site-directed mutagenesis was performed with a QuikChange site-directed mutagenesis kit (Stratagene), according to the manufacturer's instructions. The primers used for each point mutation were as follows: DNMT1C1229S-f (GGTGGGCCACCCAGCCAGGGCTTCAG), DNMT1C1229S-r (CTGAAGCCCTGGCTGGGTGGCCCACC), DNMT1H168R-f (GACCACCATCACGGCTCGCTTCACGAAGGGCCCC), DNMT1H168R-r (GGGGCCCTTCGTGAAGCGAGCCGTGATGGTGGTC), DNMT1E1171A-f (ACGCTGTGGGCCATCGCCATGTGGGACCCGGCAG), DNMT1E1171A-r (CTGCCGGGTCCCACATGGCGATGGCCCACAGCGT), DNMT3AC706S-f (GATTGGAGGCAGTCCCAGCAATGACCTCTCC), DNMT3AC706S-r (GGAGAGGTCATTGCTGGGACTGCCTCCAATC), DNMT1C1229A-f (GGTGGGCCACCCGCCCAGGGCTTCAG), DNMT1C1229A-r (CTGAAGCCCTGGGCGGGTGGCCCACC), DNMT1E1269D-f (TCTTCCTTCTGGACAACGTCAGGAAC), DNMT1E1269D-r (GTTCCTGACGTTGTCCAGAAGGAAGA).
ES cells were transfected with each construct by Lipofectamine 2000 (Invitrogen). After 24 h in culture, the transfected cells were collected and fixed on glass slides for microscopic evaluation.
Immunostaining.
ES cells grown on culture dishes were collected by trypsinization, cytospun onto glass slides, fixed with 4% paraformaldehyde in phosphate-buffered saline (10 min, 25°C), washed, and then permeabilized with 0.2% Triton X-100 in phosphate-buffered saline (10 min at 25°C). For immunostaining, the samples were incubated in blocking solution (3% bovine serum albumin, 0.1% Tween 20, 4× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) for 30 min at 37°C to reduce nonspecific binding and then in detection solution containing primary antibodies (1% bovine serum albumin, 0.1% Tween 20, 4× SSC) for 1 h at 37°C. After three washes with 4× SSC, the samples were incubated in detection solution containing secondary antibodies. Wild-type and mutant cells were obtained by trypsinizing embryos. The cells were fixed, permeabilized, and immunostained as described above. For the PCNA immunostaining, the cells were treated with 0.5% Triton X-100 in CSK buffer {100 mM NaCl, 300 mM sucrose, 10 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 6.8, 3 mM MgCl2, 1 mM EGTA} for 30 seconds at 4°C, fixed with paraformaldehyde, and then treated with methanol for 20 min at −20°C.
The primary antibodies were anti-DNMT1 rabbit polyclonal antibody (sc-20701; Santa Cruz Biotechnology) diluted 1:20, anti-PCNA mouse monoclonal antibody (sc-56; Santa Cruz Biotechnology) diluted 1:20, anti-acetylated histone H4 rabbit polyclonal antibody (06-866; Upstate) diluted 1:200, anti-HP1β mouse monoclonal antibody (MAB3448; Chemicon) diluted 1:500, anti-H3K9me3 rabbit polyclonal antibody (07-442; Upstate) diluted 1:500, and anti-H4K20me3 rabbit polyclonal antibody (07-463; Upstate) diluted 1:500. Alexa Fluor 488 goat anti-mouse immunoglobulin G (IgG) (A11017; Molecular Probes) and Alexa Fluor 555 goat anti-rabbit IgG (A21430; Molecular Probes) were the secondary antibodies.
Replication labeling.
Nucleotide analogs were introduced into cells by “bead loading” as described previously (37, 52), with slight modifications. One-hundred micromolar digoxigenin-11-dUTP (Roche Applied Science) was added to serum-free medium containing 50 μM dATP, dGTP, and dCTP. This mixture was added to cells growing in a 35-mm culture dish, and glass beads (106 μm; Sigma) were added. The dish was shaken vigorously, the glass beads were washed away, and the cells were grown for a further 30 min before sample preparation. Dig-labeled DNA was detected using anti-digoxigenin conjugated with rhodamine. To introduce methylated or unmethylated dCTP into the cell nucleus, we used the hypotonic shift method described previously, with slight modifications (29). One millimolar 5-me-dCTP (Roche Applied Science) or dCTP was added simultaneously with 100 μM bio-dUTP (Roche Applied Science) to KHB buffer (10 mM HEPES, pH 7.4, 30 mM KCl). The mixture was added to cells growing in a 48-well culture dish. After incubation at 37°C with 5% CO2 for 5 min, the cells were washed and cultured in normal medium for 1 h to introduce the nucleotides into nascent DNA. Bio-labeled DNA was detected using Alexa Fluor 647 streptavidin (S32357; Molecular Probes).
Immunoblotting.
Cell extracts lysed with sample buffer (50 mM Tris-HCl, pH 6.8, 2% sodium dodecyl sulfate, 6% β-mercaptoethanol, 10% glycerol, 0.05% bromophenol blue) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (8%) and transferred to nitrocellulose membranes. Anti-DNMT1 rabbit polyclonal antibody (a gift from F. Ishikawa) diluted 1:500, anti-phospho-p53 (Ser15) rabbit polyclonal antibody (9284; Cell Signaling Technology) diluted 1:1,000, and anti-α-tubulin mouse monoclonal antibody (CP06; Oncogene) were used as primary antibodies. Cell extraction with Triton X-100 was performed as described previously (47).
Imaging system and measurement.
Images were collected using a Leica DM RA2 fluorescence microscope equipped with a cooled charge-coupled-device camera (C4742-95-12ER; Hamamatsu Photonics, Inc.), controlled by a Macintosh G4 computer running the software program IPLab (Signal Analytics Co.). The images were captured at different stage positions and processed using deconvolution software (Scientific Volume Imaging).
Accession number.
The RIKEN accession number for the Dnmt1ps mice is CDB0500K.
RESULTS
Generation of a catalytic-defective Dnmt1 allele.
To examine the in vivo contribution of DNMT1's catalytic activity, we generated both ES cells and mice bearing a point mutation in the Dnmt1 gene in the Pro-Cys (PC) motif of its catalytic center, in which Cys1229 was replaced with Ser (Fig. 1A and B) (59). In ES cells, the point mutation was introduced by homologous recombination (termed the Dnmt1ps allele), and the remaining wild-type allele in the Dnmt1ps heterozygous ES cells was deleted by a second gene targeting, which removed the exons encoding the PC motif and the Glu-Asn-Val (ENV) motif of the catalytic domain, to obtain a compound heterozygous Dnmt1−/ps ES cell line (Fig. 1B). We confirmed the correct targeting by Southern blot analysis (Fig. 1C). We did not observe any sequence mutations except for the Cys1229-to-Ser substitution in the coding region of the Dnmt1 transcript in the Dnmt1−/ps cells (Fig. 1D and data not shown) (see Materials and Methods). The Dnmt1−/ps cells expressed a mutant DNMT1 protein with almost the same molecular size as the wild-type protein at a level comparable to that of the endogenous protein (Fig. 1E).
FIG. 1.
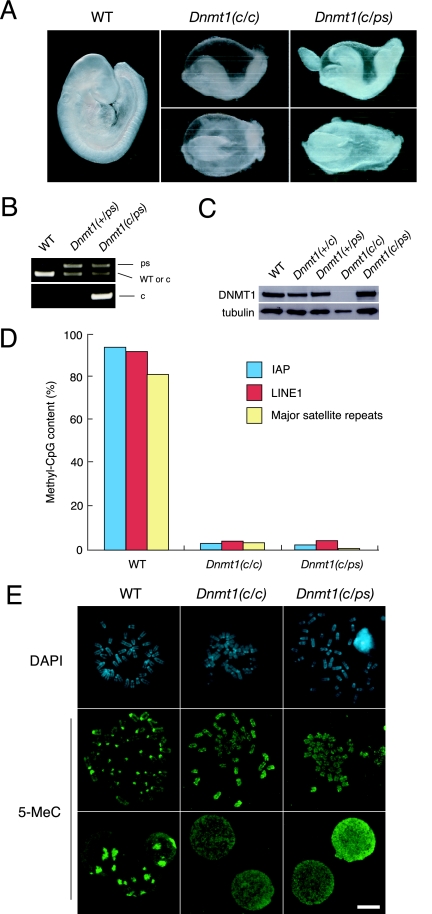
Generation of the Dnmt1ps allele by homologous recombination in ES cells. (A) Schematic illustration of the DNMT1 structure. (B) Generation of the Dnmt1-ps allele. The wild-type Dnmt1 locus was targeted with pTS015 carrying a mutated exon 32 and the IRES-βgeo cassette with flanking loxP sites (Dnmt1-neo-ps allele). After homologous recombination, the cassette was removed by Cre recombinase, resulting in the Dnmt1-ps allele. Cysteine 1229 of PC residues (box shade) in motif IV was changed to serine. The remaining wild-type locus was targeted by pTA010, in which exons 31 to 33 were deleted, to create a null mutation. Filled boxes indicate exons, a red star indicates a mutated exon, and arrowheads indicate loxP sites. Locations of the 5′ and 3′ probes for genomic Southern analysis are shown. (C) Genomic Southern analysis of the targeted ES cells digested with EcoRV for the 5′ probe (left) or XmaI for the 3′ probe (right). The 5′ probe detects a >20-kb band (wild type, ps) or an 8.8-kb band (neo-ps, null), and the 3′ probe detects a 20-kb band (wild-type, ps), a 14-kb band (neo-ps), or a 12.5-kb band (null). WT, wild-type. (D) Sequences of Dnmt1 transcripts in wild-type and Dnmt1−/ps ES cells. The red star indicates the mutated base. (E) Immunoblot analysis of ES cell extracts by use of an anti-DNMT1 antibody. (F) DNA methylation of repetitive sequences (IAP and pericentromeric major satellite repeats), analyzed by Southern blotting. Genomic DNA from each cell line was digested with methylation-sensitive (HpaII or MaeII) or methylation-insensitive (MspI) restriction enzymes and analyzed with the probe indicated at the bottom of each blot. (G) Quantitative analysis of the methyl-CpG content of repetitive sequences (IAP, LINE1, and pericentromeric major satellite repeats) by bisulfite sequencing analysis. The percentages of methyl-CpG in the total CpG sites analyzed in wild-type and mutant ES cells are indicated. (H) Anti-methylcytosine antibody staining in wild-type, Dnmt1c/c, Dnmt1−/ps, and Dnmt1−/− Dnmt3a−/− Dnmt3b−/− (TKO) ES cells. Mitotic chromosomes were stained with DAPI (blue). Distribution of methyl-cytosine (green) was detected in mitotic chromosomes and interphase nuclei. Scale bar, 10 μm.
To evaluate the effect of the C1229S mutation on DNMT1's catalytic activity, we examined the global DNA methylation level of the Dnmt1−/ps ES cells. First, we analyzed the DNA methylation of a dispersed repeat, the intracisternal A particle (IAP), and a tandem repeat, a major satellite sequence, by Southern blot analysis using methylation-sensitive restriction enzymes. We found that the DNA methylation of both IAP and the major satellite repeats in the Dnmt1−/ps ES cells decreased greatly, as reported previously for mutant ES cells (Dnmt1c/c) (Fig. 1F) carrying a null allele (32). Using the bisulfite sequencing method, we next quantified the amounts of methylated CpG in dispersed repeats (IAP and LINE1) and a tandem repeat (pericentromeric major satellite repeats) (Fig. 1G). We found that the DNA methylation of the Dnmt1−/ps ES cells decreased globally to the same level as that seen in the Dnmt1c/c null mutant ES cells. Notably, both the Dnmt1c/c and Dnmt1−/ps ES cells retained significant amounts of DNA methylation compared with Dnmt1−/− Dnmt3a−/− Dnmt3b−/− triple-knockout (TKO) ES cells, in which CpG methylation is nearly absent (54). Consistent with this, mitotic chromosomes or interphase nuclei in the Dnmt1−/ps and Dnmt1c/c mutant ES cells showed punctate staining with an anti-5-methylcytosine antibody, which corresponded to the pericentromeric heterochromatin regions that were strongly stained by 4′,6′-diamino-2-phenylindole (DAPI); this 5-methylcytosine staining pattern was not observed in TKO ES cells (Fig. 1H). The residual methylation in the Dnmt1−/ps and Dnmt1c/c mutant ES cells is probably attributable to the relatively high expression levels of DNMT3A and DNMT3B seen in undifferentiated ES cells (43). These results indicate that the C1229S substitution in the PC motif abolished the catalytic activity of DNMT1.
Loss of DNA methylation and developmental arrest in Dnmt1ps mice.
To evaluate the role of DNMT1 catalytic activity in mouse embryogenesis, we examined the phenotypes of Dnmt1ps mutant mice and compared them with those of Dnmt1c/c null mutant mice. The Dnmt1ps/+ heterozygous mice grew normally and were fertile. To obtain compound heterozygous or homozygous Dnmt1ps mutants, we crossed Dnmt1ps/+ mice with Dnmt1c/+ mice or intercrossed Dnmt1ps/+ mice. From these crosses, we recovered embryos with the corresponding genotypes at close to the expected ratios at E9.5 (Table 1) . All the recovered Dnmt1c/ps and Dnmt1ps/ps embryos were developmentally arrested at similar embryonic stages; few or no somites were formed, and embryonic turning and neural tube closure had not occurred (Fig. 2A and B). These gross morphological phenotypes of the Dnmt1c/ps and Dnmt1ps/ps embryos were indistinguishable from those of the Dnmt1c/c null mutant embryos (Fig. 2A and data not shown). Immunoblot analysis showed that the Dnmt1c/ps embryos expressed the mutant DNMT1 protein with an appropriate molecular size at a level comparable to that of wild-type DNMT1 (Fig. 2C). Thus, our results suggest that the loss of DNMT1's catalytic activity is the main reason for the embryonic lethal phenotype of the Dnmt1 null mutant embryos.
TABLE 1.
Genotypes of offspring derived from intercrosses between Dnmt1c/+ and Dnmt1ps/+ mice
| Genotypes of interbred micea | Stage | No. of mice
|
No. of embryos
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| In litter | Total | Resorbed and not genotyped | Genotyped (resorbed and genotyped using yolk sac only) as:
|
|||||||
| +/+ | +/ps | +/c | c/c | c/ps | ps/ps | |||||
| F, Dnmt1+/c; M, Dnmt1+/c | E10.5 | 2 | 7 | 0 | 4 | 0 | 3 (1) | |||
| E9.5 | 18 | 137 | 8 | 35 | 58 (1) | 36 (6) | ||||
| F, Dnmt1+/ps; M, Dnmt1+/c | E10.5 | 3 | 21 | 0 | 6 | 7 | 2 | 6 (3) | ||
| E9.5 | 15 | 114 | 4 | 27 (1) | 26 (3) | 24 (1) | 33 (4) | |||
| F, Dnmt1+/c; M, Dnmt1+/ps | E10.5 | 2 | 13 | 1 | 2 | 3 | 4 | 3 (1) | ||
| E9.5 | 4 | 32 | 8 | 8 (1) | 5 (1) | 4 | 7 (2) | |||
| F, Dnmt1+/ps; M, Dnmt1+/ps | E9.5 | 7 | 53 | 5 | 5 (1) | 30 (2) | 13 (2) | |||
F, female; M, male.
FIG. 2.
Early embryonic lethality of Dnmt1c/ps embryos. (A) Gross morphology of wild-type (WT), Dnmt1c/c, and Dnmt1c/ps embryos dissected at E9.5. Growth of Dnmt1c/c and Dnmt1c/ps embryos was retarded at similar embryonic stages. Embryos of both mutant genotypes formed no or few (up to seven) somites and had distorted neural tubes. (B) PCR genotype analysis of genomic DNA isolated from wild-type, Dnmt1ps/+, and Dnmt1c/ps embryos. The upper panel shows results for a primer set for the Dnmt1-ps allele, and the lower panel shows results for a primer set for the Dnmt1-c allele (see Materials and Methods). (C) Immunoblot analysis of embryonic cell extracts (E9.5) probed with an anti-DNMT1 antibody. For the loading control, the same membrane was stripped and reprobed with an antitubulin antibody. (D) Bisulfite sequencing analysis of repetitive sequences (IAP, LINE1, and pericentromeric major satellite repeats) as described for Fig. 1G. Genomic DNA was isolated from E9.5 embryos. (E) Anti-methylcytosine antibody staining in wild-type, Dnmt1c/c, and Dnmt1c/ps cells from E9.5 embryos. Mitotic chromosomes were visualized by DAPI staining (blue). Distribution of methyl-cytosine (green) was detected in mitotic chromosomes and interphase nuclei. Scale bar, 10 μm.
We next examined the DNA methylation of the Dnmt1c/ps embryos by using bisulfite sequencing analysis and anti-5-meC immunostaining. The bisulfite sequencing showed a severe loss of DNA methylation in the Dnmt1c/ps embryos (Fig. 2D) at a level similar to that seen in the Dnmt1c/c null mutant embryos. Notably, the DNA methylation levels of the Dnmt1c/ps and Dnmt1c/c embryos were significantly lower than those of the ES cells with the same genotypes and were comparable to those of the TKO ES cells (Fig. 1G and 2D). Similarly, the Dnmt1c/ps and Dnmt1c/c embryonic cells did not show punctate 5-meC staining patterns at heterochromatin regions (Fig. 2E), similar to the TKO ES cells (Fig. 1H). These results suggested that the loss of DNMT1's catalytic activity resulted in the near absence of DNA methylation in the embryos, probably because DNMT3A and DNMT3B are expressed at low levels at this embryonic stage.
Transcriptional derepression, p53-activation, and partial cell growth defects in Dnmt1ps mutant mice.
The inactivation of DNMT1 dysregulates the transcription of many genes, including imprinted genes and retrotransposons (26). We found the transcriptional dysregulation of Igf2 and H19 (imprinted genes) and IAP (a retrotransposon) in both the Dnmt1c/c and Dnmt1c/ps embryos by RT-PCR analysis (Fig. 3A), indicating that the loss of DNMT1's catalytic activity was sufficient to cause the transcriptional dysregulation of these genes in mouse embryos.
FIG. 3.
Growth defect and damage response in Dnmt1c/ps embryonic cells. (A) RT-PCR expression analysis of cell cycle-related (p21, p27, and p53) and DNA methylation-sensitive (H19, Igf2, and IAP) genes in wild-type (WT), Dnmt1c/c, and Dnmt1c/ps embryos at E9.5. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as the loading control. (B) Immunoblot analysis of p53 phosphorylated on Ser15 (p53-P). The results for the loading control, β-tubulin, were the same as those in Fig. 2C. (C) Immunofluorescence staining of PCNA in wild-type, Dnmt1c/c, and Dnmt1c/ps embryonic cells. Nuclei were visualized by DAPI staining. Conventional microscopic images are shown. Scale bar, 10 μm. Average percentages of PCNA-positive cells, with standard deviations, are shown at right (n > 300, triplicate). The difference between the wild-type and Dnmt1c/c or Dnmt1c/ps embryonic cells was statistically significant (P < 0.01, chi-square analysis).
Previous studies showed that the loss of DNMT1 in mouse somatic cells causes growth arrest and p53-dependent cell death (26). In both the Dnmt1c/c and Dnmt1c/ps embryonic extracts, we detected phosphorylation on Ser15 of the p53 protein, one of the major cellular responses to DNA damage (Fig. 3B). This result was consistent with the increased expression of p21, a direct target of activated p53, in these mutants (Fig. 3A).
To examine cell growth in Dnmt1 mutant embryos, we scored proliferating cells by staining embryonic cells with an anti-PCNA antibody. We found that, although both Dnmt1c/c and Dnmt1c/ps mutants included fewer PCNA-positive cells than did wild-type embryos (Fig. 3C), a substantial percentage of the cells (42 to 45%) were still proliferating in the Dnmt1c/c and Dnmt1c/ps mutant embryos at E9.5 (Fig. 3C). Immunofluorescence analyses of the E9.5 Dnmt1c/ps mutant embryos suggested that the G2/M population (cells that were PCNA negative and contained histone H3 phosphorylated on Ser10) was not noticeably enriched in the mutants compared with wild-type embryos and that p53 was activated (phosphorylation on Ser15) in a small fraction of the mutant cell population (data not shown). These results suggested that a growth defect and DNA damage response occurred in a small cell population of the Dnmt1 mutant embryos, while much of the embryonic cell population was still alive and growing in the absence of DNA methylation and DNMT1, at least at E9.5. The lack of DNMT1 activity may differentially affect certain cell types within embryos or may lead to a cellular growth defect and damage response in either a progressive or stochastic manner at this embryonic stage.
Altered distribution of chromatin marks and effectors in Dnmt1ps mutant mice.
A number of chromatin-regulatory molecules interact with DNMT1, implicating DNMT1 in the direct control of chromatin properties through its nonenzymatic functions, in vivo. We first examined the distribution of repressive chromatin markers, i.e., heterochromatin protein 1β (HP1β), histone H3 trimethylated at Lys9 (H3K9me3), and histone H4 trimethylated at Lys20 (H4K20me3), in the Dnmt1 mutant embryos by immunofluorescence staining (Fig. 4A and B and data not shown). In wild-type embryos, HP1β, H3K9 trimethylation, and H4K20 trimethylation signals were detected strongly and specifically in the DAPI-dense pericentromeric heterochromatin regions. In contrast, in the Dnmt1c/c and Dnmt1c/ps mutant embryos, the distribution of these repressive chromatin markers was globally affected; their staining intensities in the pericentromeric heterochromatin relative to the euchromatic signals were decreased in the entire cell population. Since the growth defect was observed in only a portion of the cell population (Fig. 3C), it seems unlikely that secondary effects of the growth defects solely account for these changes in chromatin properties. We did not detect a difference in the resistance of HP1β to Triton X-100 extraction between wild-type embryos and either Dnmt1 mutant, suggesting that the association of HP1β with chromatin or nuclear structures is not perturbed in these mutant cells (Fig. 4C). Our results suggest that the loss of DNMT1's catalytic activity alters the repressive chromatin properties in embryonic somatic cells.
FIG. 4.
Dysregulation of chromatin structure in Dnmt1c/ps embryos. (A and B) Immunofluorescence staining of embryonic cells isolated from wild-type (WT) and mutant embryos at E9.5 with specific antibodies to HP1β (A) and H3K9me3 (B). Nuclei were stained with DAPI. The upper panels are low-magnification images (conventional microscopic images), and the lower panels are high-magnification images (deconvolution images). Percentages of cells showing the decreased heterochromatin signal (left) and diffuse staining pattern (right) of HP1β are shown (A). For H3K9me3 staining, interphase (left) and mitotic (right) nuclei are shown (B). (C) Immunoblot analysis of HP1β in the Triton-resistant and Triton-extracted fractions of embryonic cells. PCNA was used as the control for the Triton-extracted fraction. (D, E) Immunostaining analysis of acetylated H4 in embryonic cells. A small population of cells in both the Dnmt1c/c and Dnmt1c/ps embryos had abnormally hyperacetylated histone H4 (D). An abnormal association of HP1β proteins with mitotic chromosomes with hyperacetylated histone H4 was observed (E). Scale bars, 10 μm.
We next examined the distribution of an active chromatin marker, acetylated histone H4. In wild-type animals, acetylated histone H4 was distributed throughout the nucleus but absent from pericentromeric heterochromatin (Fig. 4D, WT). For most cells, the histone H4 staining patterns were essentially the same in the wild-type and the Dnmt1 mutant embryos (data not shown). However, a small population of cells (∼1%) in the Dnmt1 mutants contained unusually hyperacetylated histone H4 (Fig. 4D, Dnmt1c/c and Dnmt1c/ps). This population was never detected in the wild-type embryos. We often observed hyperacetylated cells in mitosis (Fig. 4E), suggesting that it was unlikely that the growth defects in the Dnmt1 mutant embryos caused this hyperacetylation. Interestingly, these hyperacetylated cells retained HP1β in their mitotic chromosomes (Fig. 4E, HP1β), even though HP1 normally dissociates from chromatin in the M phase (18, 24). DNMT1 may be involved in maintaining the global histone acetylation profiles in certain cell types, as observed in human cancer cells (14).
Failure of DNMT1 to associate with replication foci in Dnmt1ps embryos and ES cells.
It is well established that DNMT1 associates with replication foci during the S phase (33). To determine whether mutant DNMT1 localized properly in the nuclei of Dnmt1ps embryos and ES cells, we performed an immunofluorescence analysis of DNMT1 and the replication foci marker PCNA. In mouse cells, genomic DNA is replicated initially at euchromatic regions inside the nucleus in the early S phase, at DAPI-dense heterochromatin regions in the middle S phase, and finally at regions close to the nuclear membrane in the late S phase (Fig. 5A to F, PCNA) (40, 44). We confirmed that during the S phase, DNMT1 associated with the replication foci, as revealed by its colocalization with PCNA (Fig. 5A to F, DNMT1 and Merge) (33). Also, as expected, in the late-S and G2 phases, DNMT1 was retained in the heterochromatin regions after their replication (Fig. 5C and F, showing DNMT1 signals in DAPI-dense heterochromatin without overlap with PCNA) (12). These cell cycle-dependent localizations of DNMT1 occurred in the nuclei of both embryonic and ES cells (Fig. 5A to C and D to F).
FIG. 5.
Perturbed localization of DNMT1 in Dnmt1c/ps embryos and Dnmt1−/ps ES cells. Localization of endogenous DNMT1 proteins in wild-type embryos (A to C), wild-type ES cells (D to F), Dnmt1c/ps embryos (G to I), and Dnmt1−/ps ES cells (J to L) is shown. Colocalization of DNMT1 (green) with PCNA (red) was visualized in the early S (A, D, G, and J), middle S (B, E, H, and K), and late S (C, F, I, and L) phases. Merged images at right show overlays of PCNA and DNMT1 staining (yellow indicates overlap). Nuclei were stained with DAPI (blue). (A, D, G, and J) Magnified views of the regions of the early-S-phase cells in white squares are shown. Scale bars, 10 μm.
In the cells from the Dnmt1c/ps embryos, however, the endogenous DNMT1-C1229S protein showed diffuse localization patterns in all cells, regardless of their point in the cell cycle (Fig. 5G to I and data not shown). We did not observe a clear colocalization of DNMT1 and PCNA signals in these cells (Fig. 5G to I), indicating that DNMT1-C1229S failed to associate with replication foci. No stable association of the mutant DNMT1 with the heterochromatic region was found in the late S or G2 phase either. We observed similar diffuse patterns of DNMT1 distribution in most of the cells from the Dnmt1ps/ps homozygous embryos (data not shown).
In the Dnmt1−/ps ES cells, we also found that the endogenous DNMT1-C1229S protein was abnormally distributed. However, the distribution patterns in the Dnmt1−/ps ES cells were distinct from those in the somatic cells from the Dnmt1−/ps embryos. In most of the Dnmt1−/ps ES cells (>80%), DNMT1-C1229S localized aberrantly to the pericentromeric region, independent of the cell cycle (Fig. 5J to L). This focal pericentromeric localization of DNMT1 was observed even during euchromatic replication in the early S phase (Fig. 5J). We observed hardly any overlaps between DNMT1 and PCNA signals during the early- and late-S phases in the Dnmt1−/ps ES cells (Fig. 5J and L), indicating that DNMT1-C1229S was not enriched at the replication foci, at least in these cell cycle phases. We found no discernible difference in DNMT1 localization between the wild-type and Dnmt1ps/+ ES cells (data not shown), suggesting that DNMT1-C1229S localized properly in the Dnmt1ps/+ ES cells.
Essential role for the genomic DNA methylation mark in DNMT1's association with replication foci.
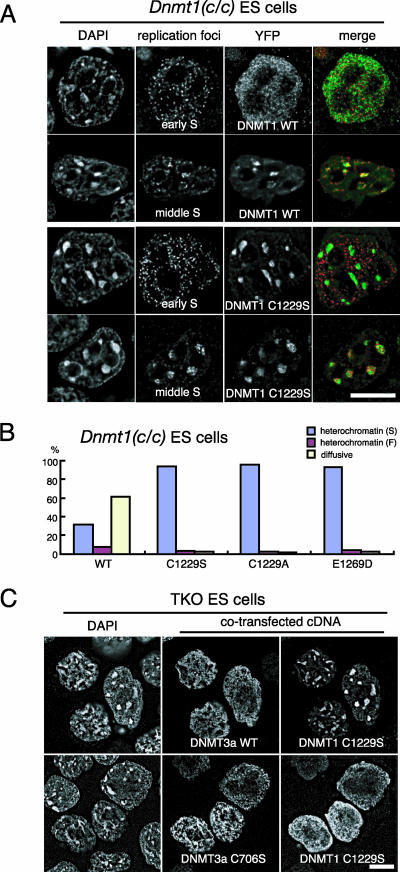
We first asked how the catalytic activity of DNMT1 affects its association with replication foci. Our results raised two possibilities. The C1229S mutation might directly affect DNMT1's localization because of a structural change or a loss of molecular interactions. Alternatively, the loss of DNMT1's catalytic activity might indirectly lead to its localization change through decreased DNA methylation of the genome. To examine these possibilities, DNMT1 proteins with various mutations were expressed as fusion proteins with YFP-DNMT1 in ES cells with various Dnmt mutant backgrounds. Replication foci were simultaneously visualized by the incorporation of digoxigenin-dUTP. In wild-type ES cells, the YFP-fused wild-type DNMT1 protein (YFP-DNMT1-WT) was diffuse in the nuclei during euchromatic replication in the early S phase (data not shown), whereas the focal YFP-DNMT1-WT pattern showing colocalization with the replication foci was observed in the DAPI-dense heterochromatin in the middle S phase (Fig. 6A). These YFP-DNMT1-WT localizations were comparable to those of the endogenous DNMT1 protein (Fig. 5D to F), indicating that the localization of YFP-DNMT1 was properly regulated in the ES cells. We found that the YFP-fused DNMT1-C1229S protein (YFP-DNMT1-C1229S) showed cell cycle-dependent localization similar to that of YFP-DNMT1-WT (Fig. 6B), indicating that the catalytic-defective C1229S mutation itself is not sufficient to alter DNMT1's localization in wild-type ES cells. This result is consistent with our observation that the endogenous DNMT1-C1229S protein in Dnmt1ps/+ ES cells was distributed properly.
FIG. 6.
Essential role for DNA methylation mark in DNMT1 localization. (A to H) Localization of YFP-DNMT1 fusion proteins during replication of the pericentromeric heterochromatin. YFP fusions with wild-type (A and E) and mutant (C1229S [B and F], H168R [C and G], and ΔRFT [D and H]) DNMT1 proteins were transiently expressed in wild-type (A to D) and TKO (E to H) ES cells. Replication sites were visualized by the incorporation of digoxigenin-dUTP. Nuclei were visualized by DAPI staining. Merged images of YFP-DNMT1 (green) and replication foci (red) are shown. (I to L) YFP fusions with wild-type (I and J) and mutant (H168R [K] and ΔRFT [L]) DNMT1 proteins were transiently coexpressed with wild-type (I, K, and L) or catalytic-defective (J) DNMT3A proteins in TKO ES cells, and the recovery of the cell cycle-dependent localization of DNMT1 was assessed. DNMT3A proteins were detected by immunofluorescence staining. (M) Localization of DNMT1 at replicated regions in Dnmt3a−/− Dnmt3b−/− ES cells. Results are shown for immunofluorescence staining of endogenous DNMT1 and PCNA proteins in low-passaged Dnmt3a−/− Dnmt3b−/− ES cells. Nuclei were visualized by DAPI staining. A merged image of DNMT1 (green) and PCNA (red) is shown. Scale bars, 10 μm.
We also examined the effects of the DNMT1 mutations in the PBD and the RFT/TS domain (8, 12, 33) on the association of YFP-DNMT1 with replication foci. Interestingly, we found that the PBD mutant forms of YFP-DNMT1 with amino acid substitutions (YFP-DNMT1-H168R, -H168V, and -R161A) (8, 25) and a small deletion (YFP-DNMT1-delPBD) were all able to localize to the replication foci in a large percentage of the cell population (Fig. 6C and data not shown), although in some cells no or only partial association with the replication foci was observed. These results indicate that the PCNA binding is beneficial but not essential for the association of DNMT1 with replication foci in ES cells. On the other hand, a YFP-fused DNMT1 protein with a small deletion in the RFT/TS domain (YFP-DNMT1-delRFT) showed diffuse nuclear staining in the entire cell population, independent of the cell cycle (Fig. 6D), indicating that, as described previously (33), the RFT/TS domain plays a critical role in DNMT1's association with replication foci, at least in the middle S phase in ES cells.
To examine the roles of preexisting DNA methylation marks on DNMT1 localization, we introduced the same wild-type and mutant YFP-DNMT1 proteins into TKO ES cells, which lack DNA methylation. Strikingly, all of the YFP-DNMT1 constructs (YFP-DNMT1-WT, -C1229S, -H168R, and -delRFT) showed diffuse nuclear distribution in the entire cell population, independent of the cell cycle, and did not show stable association with the replication foci, even in the middle S phase (Fig. 6E to H). A small percentage of cells showed a faint enrichment of the signal in the heterochromatin region, with mostly diffuse patterns (data not shown), although the significance of this observation is currently unknown. Importantly, even YFP-DNMT1-WT, which had intact PBD and RFT/TS domains, could not localize properly in TKO ES cells (Fig. 6E), indicating that the DNA methylation state plays an essential role in DNMT1 localization.
Previous studies have shown that DNMT1 is poor at introducing new global DNA methylation into an extensively hypomethylated genome in the absence of DNMT3 (7). We found that the coexpression of YFP-DNMT1-WT with the wild-type DNMT3A protein (DNMT3A-WT) restored the stable association of YFP-DNMT1-WT with replication foci in TKO ES cells in a cell cycle-dependent manner (Fig. 6I). In contrast, the coexpression of a catalytic-defective DNMT3A protein, DNMT3A-C706S, did not restore the cell cycle-dependent localization of YFP-DNMT1-WT in TKO ES cells (Fig. 6J), indicating that the catalytic activity of DNMT3A is required for this rescue. The distributions of the YFP-DNMT1-WT and DNMT3A proteins were mostly different (Fig. 6I and J). We also observed a similar rescue of the YFP-DNMT1-WT localization by coexpression of the wild-type DNMT3B protein, but not a catalytic-defective DNMT3B protein, in TKO ES cells (data not shown). Furthermore, we found that the endogenous DNMT1 protein correctly associated with replication foci in a cell cycle-dependent manner in low-passage Dnmt3a−/− Dnmt3b−/− ES cells (Fig. 6M), which retain significant genomic DNA methylation in euchromatic and heterochromatic regions (data not shown) (7, 42). From these results, we conclude that DNA methylation marks, but not the DNMT3A and DNMT3B proteins, are required for the cell cycle-dependent DNMT1 localization to replication foci.
We found that the coexpression of DNMT3A-WT restored the stable association of the PBD mutant YFP-DNMT1-H168R with replication foci in TKO ES cells, but the RFT/TS mutant YFP-DNMT1-delRFT remained diffuse under the same conditions (Fig. 6K and L). These results indicate that an RFT/TS-dependent mechanism, but not PCNA binding, is required for the DNA methylation-dependent localization of DNMT1.
Mislocalization of catalytic-defective DNMT1 to heterochromatin in a methylation mark-dependent manner.
We next asked how the endogenous DNMT1-C1229S protein is retained in the pericentromeric heterochromatin in Dnmt1−/ps ES cells, independent of their point in the cell cycle (Fig. 5J to L). To examine whether this mislocalization by the C1229S mutation is caused by the loss of catalytic activity, YFP-DNMT1 fusion proteins with various mutations in the catalytic center were expressed in Dnmt1c/c ES cells. The expression of YFP-DNMT1-C1229S in Dnmt1c/c ES cells (Fig. 7A) recapitulated the mislocalization of the endogenous DNMT1-C1229S protein to the heterochromatin in Dnmt1−/ps ES cells (Fig. 5J to L), although it localized properly in wild-type ES cells (Fig. 6B). We found that the two catalytic-defective YFP-DNMT1s with independent mutations at the PC motif (YFP-DNMT1-C1229A) and the ENV motif (YFP-DNMT1-E1269D) behaved similarly to YFP-DNMT1-C1229S in the wild-type and Dnmt1c/c ES cells (Fig. 7B and data not shown). These results suggest that the loss of catalytic activity caused the mislocalization of DNMT1 to heterochromatin in the Dnmt1c/c ES cells, although we cannot rule out the possibility that these subtle mutations caused overall structural changes unrelated to the catalytic activity. It is also possible that the catalytic-defective DNMT1 protein is tightly bound to heterochromatin in the absence of catalytically active DNMT1. However, a photobleaching analysis suggested that most (>80%) of the YFP-DNMT1-C1229S protein exchanged within minutes, comparable to what was found for YFP-DNMT1-WT, in the heterochromatin of Dnmt1c/c ES cells (data not shown), indicating that DNMT1-C1229S is able to dissociate from its binding sites in the absence of wild-type DNMT1 protein.
FIG. 7.
Mislocalization of catalytic-defective DNMT1 to heterochromatin in Dnmt1-deficient ES cells in a methylation mark-dependent manner. (A) Localization of YFP-fused wild-type and C1229S DNMT1 proteins during the replication of euchromatin (early S) and pericentromeric heterochromatin (middle S) in Dnmt1c/c ES cells. Replication sites were visualized by the incorporation of digoxigenin-dUTP. Nuclei were visualized by DAPI staining. Merged images of YFP-DNMT1 (green) and replication foci (red) are shown. (B) Localization analysis of YFP-fused wild-type or catalytic-defective DNMT1 in Dnmt1c/c ES cells. DNMT1 localization was classified into three patterns: focal heterochromatin (S; blue), characterized by strong localization to pericentromeric heterochromatin; faint heterochromatin (F; red), characterized by localization to both pericentromeric heterochromatin and euchromatin; and diffuse (yellow), charac-terized by diffuse localization throughout the euchromatin. The percentage of cells showing each staining pattern is shown (n > 180). The catalytic-defective YFP-DNMT1 mutants were retained in the heterochromatin regions in the Dnmt1c/c ES cells. (C) YFP-DNMT1-C1229S fusion protein was transiently coexpressed with the wild-type (WT) or catalytic-defective (C706S) DNMT3A protein in TKO ES cells. DNMT3A proteins were detected by immunofluorescence staining. Scale bars, 10 μm.
We observed that considerable DNA methylation was retained in the heterochromatin regions of both Dnmt1c/c and Dnmt1−/ps ES cells (Fig. 1H), probably because DNMT3A and DNMT3B are expressed at relatively high levels in ES cells. Based on our finding that DNA methylation marks are essential for proper DNMT1 localization (Fig. 6), it is possible that DNA methylation marks remaining on the heterochromatin direct the mislocalization of DNMT1 to the heterochromatin. To test this possibility, we cointroduced YFP-DNMT1-C1229S and DNMT3 into TKO ES cells. When coexpressed with wild-type DNMT3A or DNMT3B, YFP-DNMT1-C1229S preferentially localized to heterochromatin in almost all the cells (Fig. 7C and data not shown), as observed in the Dnmt1c/c ES cells (Fig. 7A). In contrast, when coexpressed with the catalytic-defective DNMT3A or DNMT3B protein, YFP-DNMT1-C1229S failed to localize to the heterochromatin and showed diffuse staining patterns in all of the TKO ES cells (Fig. 7C and data not shown). These reconstitution experiments indicated that the residual DNA methylation in the heterochromatin directed the ectopic localization of the catalytic-defective DNMT1 protein in the Dnmt1c/c and Dnmt1−/ps ES cells.
However, these results raised the question of why the catalytic-defective DNMT1 protein was distributed appropriately in the wild-type (Fig. 6B) and Dnmt1ps/+ (data not shown) ES cells without localizing to the even more heavily methylated heterochromatin of these ES cells. Considering that the enzymatic function of DNMT1 converts hemi-methylation to full methylation, we hypothesized that DNMT1 may recognize the state of a DNA methylation mark, hemi-methylation, which is enriched in newly replicated regions and thus distributed in a cell cycle-dependent manner.
To test this hypothesis, we introduced hemi-methylated sites into the unmethylated genome of TKO ES cells by replication labeling with 5-methylcytosine and examined their effects on the localization of YFP-DNMT1 (Fig. 8A). We found that YFP-DNMT1-C1229S clearly associated with replication foci within 1 h when 5-methylcytosine, but not cytosine, was incorporated in TKO ES cells (Fig. 8B). YFP-DNMT1-C1229S associated only with replicating heterochromatin that incorporated 5-methylcytosine (DAPI dense, dUTP positive), not with the juxtaposed nonreplicating heterochromatin region (DAPI dense, dUTP negative) (Fig. 8B, middle, inset). These results indicate that a DNA methylation mark is sufficient to direct DNMT1 association to the unmethylated chromatin of TKO ES cells.
FIG. 8.
Prompt and stable association of DNMT1-C1229S with hemi-methylated sites introduced by 5-methylcytosine incorporation in TKO ES cells. (A) Summary of the experimental procedure. YFP-DNMT1-C1229S fusion protein was transiently expressed in TKO ES cells, which were then treated with hypotonic buffer for 5 min to introduce methylated or unmethylated dCTP exogenously, cultured for 1 h, and then fixed for immunofluorescence detection. Biotin-dUTP was introduced simultaneously with dCTP and detected with avidin-Cy3 to visualize replication sites in the nucleus. (B) Cell nuclei that incorporated methylated dCTP during euchromatic replication (top) and pericentromeric replication (middle) or incorporated unmethylated dCTP (bottom) during pericentromeric replication are shown. Magnified views (insets) are shown for the sites of pericentromeric replication marked with white squares. Scale bar, 10 μm. (C) YFP-DNMT1-C1229S fusion protein was transiently coexpressed with untagged wild-type (WT) or catalytic-defective DNMT1 in TKO ES cells before methylated-dCTP incorporation. The percentages of YFP-DNMT1-C1229S enrichment at pericentromeric replication sites (biotin-dUTP positive) are shown (n = 50). The number of cells that were labeled with biotin-dUTP at the pericentromeric heterochromatin regions is considered 100%.
Under our experimental conditions, we did not observe a clear enrichment of YFP-DNMT1-WT association with replication foci with 5-methylcytosine (data not shown). It is possible that the formation of hemi-methylated CpG sites in TKO ES cells by 5-methylcytosine incorporation is not as efficient as that at newly replicated regions of wild-type ES cells. Our results suggest that the catalytically active DNMT1 protein promptly eliminates the sites that would prompt further DNMT1 loading by converting hemi-methylation to full methylation. Consistently, when coexpressed with the wild-type DNMT1 protein, the association of YFP-DNMT1-C1229S with the 5-methylcytosine-incorporated heterochromatin in TKO ES cells was dramatically reduced (Fig. 8C). These results are consistent with the model in which DNMT1 associates stably with hemi-methylated regions.
Negative regulatory role of the C-terminal catalytic domain of DNMT1.
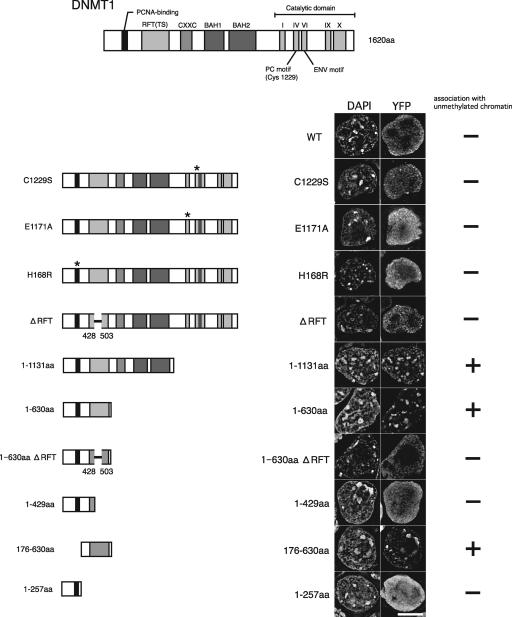
During a search for a regulatory domain for the methylation-dependent localization of DNMT1, we found that the N-terminal region of DNMT1 without the C-terminal catalytic domain (YFP-DNMT1-[1-1131]) could associate with unmethylated heterochromatin in TKO ES cells (Fig. 9). This was in clear contrast to the diffuse distribution of the full-length YFP-DNMT1-WT and YFP-DNMT1-C1229S proteins in TKO ES cells (Fig. 6E and F and 9, YFP-DNMT1-WT and -C1229S). Further analyses using YFP-DNMT1 proteins with various deletions indicated that the RFT/TS domain was required and sufficient for the N-terminal region of DNMT1 to localize to unmethylated heterochromatin (Fig. 9). Because the N-terminal region and the C-terminal catalytic domain, separated by the KG-repeat linker region, have been shown to interact with each other, these results suggest that the C-terminal catalytic domain suppresses the association of DNMT1 with unmethylated chromatin by a mechanism that is independent of its catalytic activity.
FIG. 9.
Inhibitory effect of the C-terminal catalytic domain on pericentromeric heterochromatin localization in TKO ES cells. Maps of YFP-DNMT1 deletion mutant constructs are shown at left. Representative images of TKO ES cells expressing each construct are shown. Association with pericentromeric heterochromatin was assessed for each mutant protein. Nuclei were visualized by DAPI staining. WT, wild type. Scale bar, 10 μm.
DISCUSSION
In this study, we generated a novel Dnmt1 mutant allele, Dnmt1ps, that bears a C1229S mutation in the PC motif in its catalytic center and has no catalytic activity. Through our analysis of Dnmt1ps mutant embryos and ES cells, we came to two major conclusions. First, the catalytic activity of DNMT1 makes a major contribution to the in vivo functions of DNMT1 during mouse embryogenesis. Second, a preexisting DNA methylation mark in the genome is a major determinant for the appropriate localization of DNMT1 to newly replicated regions.
Roles of the catalytic activity of DNMT1 in mouse embryogenesis.
Previous studies suggested the involvement of DNMT1's nonenzymatic functions in the cell cycle regulation of human cancer cells (55) and vertebrate embryogenesis (23). On the other hand, the catalytic activity of DNMT1 is required for some organ-specific gene regulation in the late development of zebrafish (46) and for retrotransposon silencing and cell survival in in vitro-differentiated mouse ES cells (10). In this study, we showed that the phenotypes of Dnmt1ps embryos were quite similar to those of Dnmt1 null mutant embryos (Fig. 2) (32). These results suggested that the loss of catalytic activity is the main reason for the embryonic lethality of Dnmt1-deficient mice and that CpG methylation is essential, at least for mouse early embryogenesis, in vivo. We also observed a damage response and perturbed cell growth, as revealed by the upregulation of p21 gene expression, phosphorylation of p53, and a slight decrease in the population of proliferating (PCNA-positive) cells, in Dnmt1c null and Dnmt1ps mutant embryonic somatic cells (Fig. 3). In the embryos of both mutants, substantial cell populations still proliferated in the near absence of DNA methylation (Fig. 2 and 3), consistent with the observation that mouse embryonic fibroblasts divide a few times upon the inactivation of DNMT1 (26). In contrast, the complete inactivation of DNMT1 in HCT116 cells leads to rapid mitotic arrest before extensive passive demethylation occurs (5). Undifferentiated mouse ES cells grow actively in the absence of DNMT1 (32). These results suggest that various cell types respond differently to the loss of DNMT1 protein.
The biochemical interactions of DNA methyltransferases and methyl-CpG-binding proteins with histone modification enzymes suggest that DNA methylation contributes to the properties of global chromatin structures such as heterochromatin in vivo. Inactivation of DNMT1 in plants and zebrafish results in a decreased level of a repressive chromatin marker, trimethylation of histone H3 at lysine9 (H3K9me3) (46, 53). The global distribution of heterochromatic markers H3K9me3 and HP1 in pericentromeric heterochromatin is generally retained in extensively hypomethylated Dnmt-deficient mouse ES cells (31, 54), whereas its level is decreased in human cancer cells (14). In this study, we found a global change in the distribution of heterochromatin markers (H3K9me3, H4K20me3, HP1) in most cells and abnormal histone H4 hyperacetylation in a small cell population in Dnmt1-deficient embryos (Fig. 4). Because both Dnmt1c-null and Dnmt1ps mutant embryonic cells showed the same chromatin defects, our results clearly indicate that the enzymatic function of DNMT1 is required for the integrity of the chromatin structure in embryonic somatic cells. These previous studies and ours also suggest that the contributions of DNA methylation to chromatin structure and histone modification may be different for ES cells and somatic cells.
Role of DNA methylation state in cell cycle-dependent DNMT1 localization.
One of the major cytological features of DNMT1 is its association with replication foci during the S phase, which is believed to be important for inheritance of the DNA methylation profile (33). Our results obtained using catalytic-defective DNMT1 support a model in which the state of a DNA methylation mark, hemi-methylated CpG, plays an active role in the cell cycle-dependent localization of DNMT1. Normally, hemi-methylated sites are enriched at newly replicated regions or repaired sites where DNMT1 is colocalized. We found that hemi-methylated sites introduced by 5-methylcytosine incorporation were sufficient to direct the association of the catalytic-defective DNMT1 with the chromatin of TKO ES cells (Fig. 8). We also observed that the catalytic-defective DNMT1 protein mislocalized to the heterochromatin in a cell cycle-independent manner in a Dnmt1-deficient background (Fig. 7A and B). A similar mislocalization to heterochromatin was recently reported for Dnmt1 knockout ES cells expressing DNMT1-C1229S, suggesting the involvement of DNA methylation in DNMT1 localization (10). Our further mechanistic analysis by reconstitution experiments using TKO ES cells revealed that the residual DNA methylation marks created by DNMT3A and DNMT3B attracted the catalytic-defective DNMT1 protein to the heterochromatin under conditions of Dnmt1 deficiency (Fig. 7C). It is likely that the heterochromatin in Dnmt1-deficient ES cells retains hemi-methylated sites due to the absence of the hemi-methyltransferase activity of DNMT1. In a similar way, it is possible to explain the physiological association of DNMT1 with the heterochromatin in the late S and G2 phases (12) as reflecting the retention of hemi-methylated sites due to delayed maintenance methylation in the heterochromatin, which is replicated in the middle S phase. Previous studies showed that a substantial amount of the methylation of newly synthesized DNA is significantly delayed after replication (35). The diffuse distribution of DNMT1 in TKO ES cells suggests that the association of DNMT1 with heterochromatin in the late S and G2 phases also requires the methylation mark (Fig. 6E, I, and J).
The hemi-methylation association model was considered previously when DNMT1 localization at replication foci was reported, although the replication-targeting mechanism was proposed as a preferable model in that study (33). However, the replication-targeting model alone cannot fully explain our finding that the intact DNMT1 protein was unable to associate stably with replication foci in TKO ES cells. Thus, we propose that the cell cycle-dependent localization of DNMT1 may be regulated, at least in part, by hemi-methylated CpG as an epigenetic mark of newly replicated regions.
Roles of regulatory domains in DNMT1 localization.
A number of biochemical studies suggested that conformational change and the interaction between the N-terminal region and the C-terminal catalytic domain play important roles in DNMT1 function (1, 2, 17, 25, 38). Our results suggested that, unlike full-length DNMT1, the N-terminal domain of DNMT1 associated with unmethylated chromatin, and the RFT/TS domain was responsible for this function (Fig. 9). These results suggest that the RFT/TS domain promotes DNMT1's association with chromatin independent of its methylation status and that the C-terminal catalytic domain suppresses the RFT/TS domain function by a mechanism that is independent of its catalytic activity. Based on our results, we propose the following model as a mechanism for DNMT1 localization. The N-terminal domain of DNMT1 associates with chromatin through its RFT/TS domain, and the C-terminal catalytic domain blocks the RFT/TS function as a default state. Upon sensing DNA methylation marks, possibly hemi-methylated sites, the C-terminal domain releases its suppression activity through a conformational change in DNMT1, allowing a stable association between DNMT1 and chromatin through RFT/TS. While DNMT1's intrinsic abilities of methylation recognition may contribute the control of its localization (17), it is possible that molecules other than DNMT1 are involved in its methylation recognition and chromatin association processes. Genetic studies with mice have shown that several chromatin regulators are required for maintenance of the proper global DNA methylation level (4, 11). In Arabidopsis, a molecule possessing methyl-CpG-binding activity is involved in the methylation inheritance of centromeric repeats (58).
Roles of DNMT1 localization in DNA methylation inheritance.
Recent studies using human HCT116 cells show that DNMT1's interaction with the replication machinery through its PDB improves the efficiency of, but is not essential for, postreplicative methylation (13, 50). The hemi-methylation association model does not absolutely require DNMT1's direct coupling with the replication machinery, although it would be beneficial to increase the frequency of DNMT1's association with methylated sites. These two mechanisms may cooperatively control DNMT1's localization to replication foci. The relative contribution of these mechanisms may depend on the cell type or developmental or pathological state. In addition to the cellular localization control, DNMT1 distinguishes unmethylated from hemi-methylated CpG sites for its enzymatic reaction as it moves along a DNA strand (22, 56). Thus, multiple mechanisms ensure the fidelity of methylation inheritance.
For the proper inheritance of DNA methylation patterns, it is important that unmethylated regions in the parental genome remain unmethylated in the daughter cells. Although DNMT1 has a higher enzymatic activity for unmethylated DNA substrate than do DNMT3A and DNMT3B in vitro (43), previous studies using Dnmt knockout ES cells (7, 42) or transgene expression in Drosophila (36) have shown that, in the absence of DNMT3A and DNMT3B, DNMT1 has a poor de novo methylation activity in vivo. Since the localization of DNMT1 depends largely on preexisting genomic DNA methylation, DNMT1 may not be fully functional with unmethylated genomic regions as its substrate in vivo.
Roles of nonenzymatic functions of DNMT1 in mouse embryogenesis.
The contributions of the nonenzymatic functions of DNMT1 in mouse embryogenesis remain elusive. First, major defects caused by the loss of the catalytic activity may mask the nonenzymatic functions. In addition, these functions may be more important in later developmental stages. To address this issue, further studies using a conditional mutant allele of Dnmt1 will be necessary. Second, nonenzymatic functions of DNMT1 may be coupled with DNMT1's localization. DNMT1 and histone deacetylase HDAC2 are reported to colocalize only at late replication sites, suggesting that DNMT1 cooperates with histone deacetylation in replicating inactive chromatin (49). The histone methyltransferase G9a also associates with DNMT1 at replication foci (16). It is possible that the nonenzymatic functions of DNMT1 contribute to the phenotype of Dnmt1ps mutant embryos owing to the failure of DNMT1 to localize to the proper nuclear sites. However, our results suggest that the loss of DNA methylation causes the mislocalization of DNMT1 protein, indicating that such nonenzymatic functions of DNMT1 are dependent on its enzymatic function and on a DNA methylation mark in vivo.
In conclusion, our results extend the understanding of in vivo DNMT1 function by demonstrating an essential and major role for the catalytic activity of DNMT1 in mouse embryogenesis. Our findings also provide firm evidence that preexisting DNA methylation is required for the stable association of DNMT1 with chromatin in vivo. This concept forms the basis for understanding the propagation mechanism of DNA methylation. Further mechanistic analyses may also be useful for developing novel DNMT1-targeted drugs.
Acknowledgments
We thank A. Yamagiwa for help with the bisulfite sequencing analysis; A. Tsumura for the Dnmt1 targeting cassette; E. Li for the mouse Dnmt1 cDNA, Dnmt1c mutant mice, and ES cells; H. Sano for the antimethylcytosine antibody; F. Ishikawa for the anti-DNMT1 antibody; H. Enomoto for the β-actin Cre transgenic mice; and S. Hayashi for critical reading. We are grateful to the Laboratory for Animal Resources and Genetic Engineering at RIKEN CDB for the blastocyst injection of ES cells and for the housing and reproduction support of the mice and to S. Hayashi, H. Wada, and K. Kato for the equipment and consultation for the photobleaching analysis.
This work was supported in part by grants in aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to M.O.
Footnotes
Published ahead of print on 24 September 2007.
REFERENCES
- 1.Araujo, F. D., S. Croteau, A. D. Slack, S. Milutinovic, P. Bigey, G. B. Price, M. Zannis-Hajopoulos, and M. Szyf. 2001. The DNMT1 target recognition domain resides in the N terminus. J. Biol. Chem. 276:6930-6936. [DOI] [PubMed] [Google Scholar]
- 2.Bacolla, A., S. Pradhan, R. J. Roberts, and R. D. Wells. 1999. Recombinant human DNA (cytosine-5) methyltransferase. II. Steady-state kinetics reveal allosteric activation by methylated dna. J. Biol. Chem. 274:33011-33019. [DOI] [PubMed] [Google Scholar]
- 3.Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6-21. [DOI] [PubMed] [Google Scholar]
- 4.Carlone, D. L., J. H. Lee, S. R. Young, E. Dobrota, J. S. Butler, J. Ruiz, and D. G. Skalnik. 2005. Reduced genomic cytosine methylation and defective cellular differentiation in embryonic stem cells lacking CpG binding protein. Mol. Cell. Biol. 25:4881-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, T., S. Hevi, F. Gay, N. Tsujimoto, T. He, B. Zhang, Y. Ueda, and E. Li. 2007. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat. Genet. 39:391-396. [DOI] [PubMed] [Google Scholar]
- 6.Chen, T., and E. Li. 2004. Structure and function of eukaryotic DNA methyltransferases. Curr. Top. Dev. Biol. 60:55-89. [DOI] [PubMed] [Google Scholar]
- 7.Chen, T., Y. Ueda, J. E. Dodge, Z. Wang, and E. Li. 2003. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell. Biol. 23:5594-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chuang, L. S., H. I. Ian, T. W. Koh, H. H. Ng, G. Xu, and B. F. Li. 1997. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science 277:1996-2000. [DOI] [PubMed] [Google Scholar]
- 9.Clark, S. J., J. Harrison, C. L. Paul, and M. Frommer. 1994. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 22:2990-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damelin, M., and T. H. Bestor. 2007. Biological functions of DNA methyltransferase 1 require its methyltransferase activity. Mol. Cell. Biol. 27:3891-3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dennis, K., T. Fan, T. Geiman, Q. Yan, and K. Muegge. 2001. Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev. 15:2940-2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Easwaran, H. P., L. Schermelleh, H. Leonhardt, and M. C. Cardoso. 2004. Replication-independent chromatin loading of Dnmt1 during G2 and M phases. EMBO Rep. 5:1181-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egger, G., S. Jeong, S. G. Escobar, C. C. Cortez, T. W. Li, Y. Saito, C. B. Yoo, P. A. Jones, and G. Liang. 2006. Identification of DNMT1 (DNA methyltransferase 1) hypomorphs in somatic knockouts suggests an essential role for DNMT1 in cell survival. Proc. Natl. Acad. Sci. USA 103:14080-14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Espada, J., E. Ballestar, M. F. Fraga, A. Villar-Garea, A. Juarranz, J. C. Stockert, K. D. Robertson, F. Fuks, and M. Esteller. 2004. Human DNA methyltransferase 1 is required for maintenance of the histone H3 modification pattern. J. Biol. Chem. 279:37175-37184. [DOI] [PubMed] [Google Scholar]
- 15.Esteve, P. O., H. G. Chin, and S. Pradhan. 2005. Human maintenance DNA (cytosine-5)-methyltransferase and p53 modulate expression of p53-repressed promoters. Proc. Natl. Acad. Sci. USA 102:1000-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esteve, P. O., H. G. Chin, A. Smallwood, G. R. Feehery, O. Gangisetty, A. R. Karpf, M. F. Carey, and S. Pradhan. 2006. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 20:3089-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fatemi, M., A. Hermann, S. Pradhan, and A. Jeltsch. 2001. The activity of the murine DNA methyltransferase Dnmt1 is controlled by interaction of the catalytic domain with the N-terminal part of the enzyme leading to an allosteric activation of the enzyme after binding to methylated DNA. J. Mol. Biol. 309:1189-1199. [DOI] [PubMed] [Google Scholar]
- 18.Fischle, W., B. S. Tseng, H. L. Dormann, B. M. Ueberheide, B. A. Garcia, J. Shabanowitz, D. F. Hunt, H. Funabiki, and C. D. Allis. 2005. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438:1116-1122. [DOI] [PubMed] [Google Scholar]
- 19.Fuks, F., W. A. Burgers, A. Brehm, L. Hughes-Davies, and T. Kouzarides. 2000. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat. Genet. 24:88-91. [DOI] [PubMed] [Google Scholar]
- 20.Fuks, F., P. J. Hurd, R. Deplus, and T. Kouzarides. 2003. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 31:2305-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goll, M. G., and T. H. Bestor. 2005. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74:481-514. [DOI] [PubMed] [Google Scholar]
- 22.Goyal, R., R. Reinhardt, and A. Jeltsch. 2006. Accuracy of DNA methylation pattern preservation by the Dnmt1 methyltransferase. Nucleic Acids Res. 34:1182-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hashimoto, H., I. Suetake, and S. Tajima. 2003. Monoclonal antibody against dnmt1 arrests the cell division of xenopus early-stage embryos. Exp. Cell Res. 286:252-262. [DOI] [PubMed] [Google Scholar]
- 24.Hirota, T., J. J. Lipp, B. H. Toh, and J. M. Peters. 2005. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 438:1176-1180. [DOI] [PubMed] [Google Scholar]
- 25.Iida, T., I. Suetake, S. Tajima, H. Morioka, S. Ohta, C. Obuse, and T. Tsurimoto. 2002. PCNA clamp facilitates action of DNA cytosine methyltransferase 1 on hemimethylated DNA. Genes Cells 7:997-1007. [DOI] [PubMed] [Google Scholar]
- 26.Jackson-Grusby, L., C. Beard, R. Possemato, M. Tudor, D. Fambrough, G. Csankovszki, J. Dausman, P. Lee, C. Wilson, E. Lander, and R. Jaenisch. 2001. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat. Genet. 27:31-39. [DOI] [PubMed] [Google Scholar]
- 27.Jeltsch, A. 2006. Molecular enzymology of mammalian DNA methyltransferases. Curr. Top. Microbiol. Immunol. 301:203-225. [DOI] [PubMed] [Google Scholar]
- 28.Jones, P. A., and S. B. Baylin. 2007. The epigenomics of cancer. Cell 128:683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koberna, K., D. Stanek, J. Malinsky, M. Eltsov, A. Pliss, V. Ctrnacta, S. Cermanova, and I. Raska. 1999. Nuclear organization studied with the help of a hypotonic shift: its use permits hydrophilic molecules to enter into living cells. Chromosoma 108:325-335. [DOI] [PubMed] [Google Scholar]
- 30.Lane, N., W. Dean, S. Erhardt, P. Hajkova, A. Surani, J. Walter, and W. Reik. 2003. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis 35:88-93. [DOI] [PubMed] [Google Scholar]
- 31.Lehnertz, B., Y. Ueda, A. A. Derijck, U. Braunschweig, L. Perez-Burgos, S. Kubicek, T. Chen, E. Li, T. Jenuwein, and A. H. Peters. 2003. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13:1192-1200. [DOI] [PubMed] [Google Scholar]
- 32.Lei, H., S. P. Oh, M. Okano, R. Juttermann, K. A. Goss, R. Jaenisch, and E. Li. 1996. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 122:3195-3205. [DOI] [PubMed] [Google Scholar]
- 33.Leonhardt, H., A. W. Page, H. U. Weier, and T. H. Bestor. 1992. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71:865-873. [DOI] [PubMed] [Google Scholar]
- 34.Li, E., T. H. Bestor, and R. Jaenisch. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69:915-926. [DOI] [PubMed] [Google Scholar]
- 35.Liang, G., M. F. Chan, Y. Tomigahara, Y. C. Tsai, F. A. Gonzales, E. Li, P. W. Laird, and P. A. Jones. 2002. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol. Cell. Biol. 22:480-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lyko, F., B. H. Ramsahoye, H. Kashevsky, M. Tudor, M. A. Mastrangelo, T. L. Orr-Weaver, and R. Jaenisch. 1999. Mammalian (cytosine-5) methyltransferases cause genomic DNA methylation and lethality in Drosophila. Nat. Genet. 23:363-366. [DOI] [PubMed] [Google Scholar]
- 37.Manders, E. M., H. Kimura, and P. R. Cook. 1999. Direct imaging of DNA in living cells reveals the dynamics of chromosome formation. J. Cell Biol. 144:813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Margot, J. B., A. M. Aguirre-Arteta, B. V. Di Giacco, S. Pradhan, R. J. Roberts, M. C. Cardoso, and H. Leonhardt. 2000. Structure and function of the mouse DNA methyltransferase gene: Dnmt1 shows a tripartite structure. J. Mol. Biol. 297:293-300. [DOI] [PubMed] [Google Scholar]
- 39.Mortusewicz, O., L. Schermelleh, J. Walter, M. C. Cardoso, and H. Leonhardt. 2005. Recruitment of DNA methyltransferase I to DNA repair sites. Proc. Natl. Acad. Sci. USA 102:8905-8909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakayasu, H., and R. Berezney. 1989. Mapping replicational sites in the eucaryotic cell nucleus. J. Cell Biol. 108:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishiyama, R., M. Ito, Y. Yamaguchi, N. Koizumi, and H. Sano. 2002. A chloroplast-resident DNA methyltransferase is responsible for hypermethylation of chloroplast genes in Chlamydomonas maternal gametes. Proc. Natl. Acad. Sci. USA 99:5925-5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okano, M., D. W. Bell, D. A. Haber, and E. Li. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247-257. [DOI] [PubMed] [Google Scholar]
- 43.Okano, M., S. Xie, and E. Li. 1998. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 19:219-220. [DOI] [PubMed] [Google Scholar]
- 44.Panning, M. M., and D. M. Gilbert. 2005. Spatio-temporal organization of DNA replication in murine embryonic stem, primary, and immortalized cells. J. Cell. Biochem. 95:74-82. [DOI] [PubMed] [Google Scholar]
- 45.Pradhan, S., and G. D. Kim. 2002. The retinoblastoma gene product interacts with maintenance human DNA (cytosine-5) methyltransferase and modulates its activity. EMBO J. 21:779-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rai, K., L. D. Nadauld, S. Chidester, E. J. Manos, S. R. James, A. R. Karpf, B. R. Cairns, and D. A. Jones. 2006. Zebra fish Dnmt1 and Suv39h1 regulate organ-specific terminal differentiation during development. Mol. Cell. Biol. 26:7077-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reyes, J. C., C. Muchardt, and M. Yaniv. 1997. Components of the human SWI/SNF complex are enriched in active chromatin and are associated with the nuclear matrix. J. Cell Biol. 137:263-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robertson, K. D., S. Ait-Si-Ali, T. Yokochi, P. A. Wade, P. L. Jones, and A. P. Wolffe. 2000. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 25:338-342. [DOI] [PubMed] [Google Scholar]
- 49.Rountree, M. R., K. E. Bachman, and S. B. Baylin. 2000. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 25:269-277. [DOI] [PubMed] [Google Scholar]
- 50.Spada, F., A. Haemmer, D. Kuch, U. Rothbauer, L. Schermelleh, E. Kremmer, T. Carell, G. Langst, and H. Leonhardt. 2007. DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells. J. Cell Biol. 176:565-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stancheva, I., and R. R. Meehan. 2000. Transient depletion of xDnmt1 leads to premature gene activation in Xenopus embryos. Genes Dev. 14:313-327. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Takebayashi, S., K. Sugimura, T. Saito, C. Sato, Y. Fukushima, H. Taguchi, and K. Okumura. 2005. Regulation of replication at the R/G chromosomal band boundary and pericentromeric heterochromatin of mammalian cells. Exp. Cell Res. 304:162-174. [DOI] [PubMed] [Google Scholar]
- 53.Tariq, M., H. Saze, A. V. Probst, J. Lichota, Y. Habu, and J. Paszkowski. 2003. Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc. Natl. Acad. Sci. USA 100:8823-8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsumura, A., T. Hayakawa, Y. Kumaki, S. Takebayashi, M. Sakaue, C. Matsuoka, K. Shimotohno, F. Ishikawa, E. Li, H. R. Ueda, J. Nakayama, and M. Okano. 2006. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 11:805-814. [DOI] [PubMed] [Google Scholar]
- 55.Unterberger, A., S. D. Andrews, I. C. Weaver, and M. Szyf. 2006. DNA methyltransferase 1 knockdown activates a replication stress checkpoint. Mol. Cell. Biol. 26:7575-7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vilkaitis, G., I. Suetake, S. Klimasauskas, and S. Tajima. 2005. Processive methylation of hemimethylated CpG sites by mouse Dnmt1 DNA methyltransferase. J. Biol. Chem. 280:64-72. [DOI] [PubMed] [Google Scholar]
- 57.Vire, E., C. Brenner, R. Deplus, L. Blanchon, M. Fraga, C. Didelot, L. Morey, A. Van Eynde, D. Bernard, J. M. Vanderwinden, M. Bollen, M. Esteller, L. Di Croce, Y. de Launoit, and F. Fuks. 2006. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439:871-874. [DOI] [PubMed] [Google Scholar]
- 58.Woo, H. R., O. Pontes, C. S. Pikaard, and E. J. Richards. 2007. VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev. 21:267-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wyszynski, M. W., S. Gabbara, E. A. Kubareva, E. A. Romanova, T. S. Oretskaya, E. S. Gromova, Z. A. Shabarova, and A. S. Bhagwat. 1993. The cysteine conserved among DNA cytosine methylases is required for methyl transfer, but not for specific DNA binding. Nucleic Acids Res. 21:295-301. [DOI] [PMC free article] [PubMed] [Google Scholar]