Abstract
Ku70 forms a heterodimer with Ku80, called Ku, that is critical for repairing DNA double-stand breaks by nonhomologous end joining and for maintaining telomeres. Mice with either gene mutated exhibit similar phenotypes that include increased sensitivity to ionizing radiation and severe combined immunodeficiency. However, there are also differences in the reported phenotypes. For example, only Ku70 mutants are reported to exhibit a high incidence of thymic lymphomas while only Ku80 mutants are reported to exhibit early aging with very low cancer levels. There are two explanations for these differences. First, either Ku70 or Ku80 functions outside the Ku heterodimer such that deletion of one is not identical to deletion of the other. Second, divergent genetic backgrounds or environments influence the phenotype. To distinguish between these possibilities, the Ku70 and Ku80 mutations were crossed together to generate Ku70, Ku80, and double-mutant mice in the same genetic background raised in the same environment. We show that these three cohorts have similar phenotypes that most resemble the previous report for Ku80 mutant mice, i.e., early aging without substantially increased cancer levels. Thus, our observations suggest that the Ku heterodimer is important for longevity assurance in mice since divergent genetic backgrounds and/or environments likely account for these previously reported differences.
Ku70 and Ku80 form a heterodimer called Ku that is well known for its role in repairing DNA double-strand breaks by nonhomologous end joining (NHEJ) (33). The other known components of mammalian NHEJ include DNA-PKCS, Artemis, Xrcc4, DNA ligase IV, and Xrcc4-like factor (34, 50). Ku and the 460-kDa catalytic subunit DNA-PKCS form a holoenzyme referred to as DNA-PK (DNA-dependent protein kinase). Artemis and DNA-PKCS form a complex that opens hairpins and processes overhangs (36). These ends are then ligated by the Xrcc4-DNA ligase IV heterodimer in a complex with Xrcc4-like factor (1, 6, 21). Cells with any of these proteins deleted are defective in assembling V(D)J [variable (diverse) joining] segments of antigen receptor genes, thus causing failed lymphocyte development, resulting in severe combined immunodeficiency (SCID) (3, 14, 18, 22, 49, 55). In addition, these cells are hypersensitive to agents that cause DNA double-strand breaks (DSBs) and are genetically unstable (2, 9, 17, 19, 31). Thus, mutating any of these genes causes similar phenotypes that reflect defective DNA end joining that includes defective repair of nonspecific DSBs and defective repair of the coding ends associated with V(D)J recombination. However, there are also important differences in the phenotypes. For example deletion of Ku, Xrcc4, and DNA ligase IV, but not DNA-PKCS, results in small mice, prohibits repair of signal ends associated with V(D)J recombination, and causes neuronal apoptosis. The latter phenotype manifests itself to various degrees, depending on the component being deleted (24).
There are also striking differences reported for the Ku70 and Ku80 mutant phenotypes. This is surprising since they form a heterodimer. ku80−/− mutant mice are reported to exhibit early aging without increased cancer (26, 41, 56, 58). On the other hand, ku70−/− mutant mice are reported to exhibit a high incidence of thymic lymphoma (23, 32). These differences may be due to additional functions for either Ku70 or Ku80 outside the Ku heterodimer. This is possible even though most of Ku70 is degraded in the absence of Ku80 (41) and vice versa (22) since some protein remains. In addition, evidence supports Ku70 function outside the Ku heterodimer since it associates with a variety of proteins, including Bax (7a, 20a, 32a, 53a), cyclin E (38), Hp1α (53), TRF2 (52), MRE11 (20), p95vav (42), apolipoprotein J, and unidentified proteins (57). Thus, unregulated Ku70 function may accelerate aging in ku80−/− mice. Alternatively, these different phenotypes may be due to differences in the genetic background and/or the housing environment. Even though the Ku70 and Ku80 mutant mice are in the same genetic background (129 × C57BL/6 cross), genetic drift is rapid, particularly for 129 mice (48). In this context, different breeding strategies could inadvertently select for different groups of genetic modifiers. Environmental differences are also possible since housing impacts physiological factors, including lymphocyte homeostasis (13). To support these possibilities, the genetic background and environment influence adulthood diseases, including cancer, for mice and humans (5, 12, 51).
In order to determine whether there are different roles for Ku and its subunits in the aging process and cancer development, Ku70 and Ku80 mutant mice were crossed to generate ku70−/−, ku80−/−, and double-mutant mice. These three cohorts are in the same genetic background raised in the same environment. Here we show that all three cohorts exhibit lower cancer levels than previously reported for ku70−/− mice. These mutant cohorts also exhibit an early-aging phenotype, as previously reported for ku80−/− mice. Therefore, the previously reported phenotypic differences between ku70−/− and ku80−/− mice are likely due to differences in genetic background and/or environment and not to Ku-independent functions for either Ku70 or Ku80.
MATERIALS AND METHODS
Mouse genotyping.
Total DNA was isolated from mouse tails with tail lysis buffer (100 mM Tris-Cl [pH 8.5], 5 mM EDTA, 0.2% sodium dodecyl sulfate, 200 mM NaCl), followed by ethanol precipitation. The DNA was resuspended in water. PCRs were carried out with Taq polymerase (PGC Scientific, Frederick, MD) on an Eppendorf Mastercycler Gradient (Eppendorf, Westbury, NY). For the Ku80 wild-type allele, we used sense primer 5′GAGAGTCTACGACAACTGTGC 3′ and antisense primer 5′-AGAGGGACTGCAGCCATATTA-3′. For the Ku80 mutant allele, we used sense primer 5′-GGTTGCCAGTCATGCTACGGT-3′ and antisense primer 5′-CCAAAGGCCTACCCGCTTCCATT-3′. The conditions were 29 cycles of 94°C for 30 s, 59°C for 1 min, and 72°C for 30 s. For the Ku70 wild-type and mutant alleles, we used antisense primer 5′-GGCTGGCTTTAGCACTGTCA-3′. For the Ku70 wild-type allele, we used sense primer 5′-ACACGGCTTCCTTAATGTGA-3′. For the Ku70 mutant allele, we used sense primer 5′-ACGTAAACTCCTCTTCAGACCT-3′. The conditions used were 35 cycles of 94°C for 45 s, 56°C for 45 s, and 72°C for 1.5 min.
Phenotypic observation and mouse husbandry.
Mice were housed in a specific-pathogen-free environment in microisolator cages. They were observed five or six times a week for the entire course of their life spans. The person identifying aging characteristics was blind to the genotype. Moribund mice (losing weight and responsiveness) were observed multiple times a day, and all mice were euthanatized when they were immobile and could no longer reach the water source. Morbidities were scored by Kaplan-Meier analysis and measured for statistical significance by the Wilcoxon rank sum test. Euthanatized mice were observed by necropsy, and organs were removed and fixed for histology. Mice were housed in microisolator cages in a specific-pathogen-free environment. Serum samples from sentinel mice were tested twice a year for murine rotavirus strain EDIM, mouse hepatitis virus, minute virus of mice, Mycoplasma pulmonis, mouse parvovirus, parvovirus NS-1, polyomavirus, pneumonia virus of mice, reovirus type 3, Sendai virus, and Theiler's murine encephalomyelitis virus. Pinworms were detected by the anal tape test and cecum examination. The fur was microscopically examined for mites. The rodent diet was irradiated, the water was acidified to pH 2.5 to 3.0, and the bedding and the whole cage setup were autoclaved, including the wire top, isolator cage, cardholders, water, and water bottles. All mouse procedures were done in accordance with The Guide for the Care and Use of Laboratory Animals and approved by the institutional IACUC.
Histology.
Tissues were fixed in 10% neutral buffered formalin for 24 h and then in 70% ethanol until embedded in paraffin, cut into sections, and stained with hematoxylin and eosin by standard procedures.
Histology quantification.
Mouse femurs were photographed at ×40 magnification and printed. In order to evaluate the changes in cortical wall surface area between the control mice and the mutant mice, 2-in. lengthwise sections of cortical wall (from the growth plate toward the metaphysis) were cut from the bone pictures and weighed in grams. Averages were taken for mice with like genotypes, and percent change was calculated by dividing the average cortical wall weight of each mutant cohort by the control average at the young (first) time point and then multiplying by 100%. The percent change for the control cohorts was determined by dividing the average cortical wall weight at the middle-age and old time points by the control average at the young time point and then multiplying by 100%. In order to evaluate the changes in trabecular surface area, all trabeculae, from the growth plate to the articular surface, were cut from the bone printout and weighed in grams. Percent change in the trabecular surface area for the mutant cohorts was calculated by dividing the average trabecular weight by the control average at the young time point times 100%. The percent change for the control cohorts was determined by dividing the average trabecular weight at the middle-age and old time points by the control average at the young time point and then multiplying by 100%. The growth plate of the femur was analyzed at ×200 magnification in order to visualize the chondrocytes. The entire lengths of the growth plates were cut from a printout and weighed in grams. Percent change in growth plate weight for the mutant cohorts was calculated by dividing their average growth plate weight by the control average at the young time point times 100%. The percent change for the control cohorts was determined by dividing the average growth plate weight at the middle-age and old time points by the control average at the young time point then multiplying by 100%.
Two-color fluorescence in situ hybridization (FISH).
To generate mouse skin fibroblasts (MSFs), both ears were removed from the mice of all of the cohorts at a variety of ages throughout their life spans. These ears were cut into small millimeter-sized pieces and seeded onto a 3.5-cm plate in M15 with fresh antibiotics at 20% O2 (Dulbecco's modified Eagle's medium with 15% fetal bovine serum, 2 mM glutamine, 30 mg penicillin/ml, 50 mg streptomycin/ml). This was passage 0. MSFs at passage 2 were treated with 10 mg Colcemid for 4 h and then trypsinized to release them from the plate. For slide preparation, cells were spun (1,000 rpm) and then washed twice for 10 min each time in phosphate-buffered saline (all phosphate-buffered saline washes were at pH 7.4 unless otherwise noted). The pellet was resuspended in 300 μl 75 mM KCl dropwise by flicking the tube and then incubated in a 37°C water bath for 15 min. Three hundred microliters of methanol-acetic acid (3:1) fixative was added dropwise by flicking the tube, and the mixture was spun at 3,000 rpm for 30 min. Cells were washed in 300 μl of 3:1 fixative added dropwise by flicking the tube, spun at 3,000 rpm for 30 min, and then washed again. For hybridization, slides were denatured in a 70% formamide-2× SSC solution (1× SSC is 0.15 M sodium chloride plus 0.015 M sodium citrate, pH 7.0) at 70°C for 4 min. Slides were immediately transferred to 30% formamide-2× SSC in 1 μl of 1-mg/ml telomere probe [6-carboxyfluorescein 5′ (CCCTAA)7 3′] per slide and incubated at 37°C for 5′. Slides were washed by being dipped in 2× SSC 10 times, air dried, mounted with Vectashield containing 4′,6′-diamidino-2-phenylindole dihydrochloride (DAPI; Vector Laboratories) at a 0.6-μg/ml final concentration, and imaged with a Zeiss Axioplan 2 microscope.
RESULTS
In order to determine if the reported differences in the Ku70 and Ku80 mutant phenotypes are due to either Ku70 or Ku80 independent functions or to the genetic background and/or environmental differences, we crossed Ku70+/− mice to Ku80+/− mice to generate three mutant cohorts [ku70−/− Ku80(+/+)+/−, Ku70(+/+)+/− ku80−/−, and ku70−/− ku80−/−]. There was also one control cohort [Ku70(+/+)+/− Ku80(+/+)+/−]. Previous reports described two different strains of Ku70 mutant mice (22, 32) and two different strains of Ku80 mutant mice (41, 58). We used the Ku70 mutant strain previously reported to show neuronal apoptosis (22-24) and the Ku80 mutant strain previously reported to exhibit early aging (56, 58). Both are defective for V(D)J recombination and repair of ionizing radiation-induced damage; however, only the Ku70 mutant strain is reported to exhibit a high incidence of lymphoma while only the Ku80 mutant strain is reported to exhibit early aging without increased cancer.
All of the mice in this study were generated from 57 different Ku80+/− Ku70+/− breeding pairs; therefore, these cohorts are generation 1 brothers, sisters, and cousins. This large number of breeding pairs eliminates overrepresentation of genetic outliers that may arise because of a crossbred background or random genetic changes. In addition, generation 1 mice are not subject to the gradual degenerative defects that occur with progressive generations of homozygous mutant crosses such as mice with a deletion of the telomerase RNA component (mTR) that is essential for telomere length maintenance; mtr−/− mice exhibit progressive telomere erosion with each generation (4). These generational crosses would be extremely difficult to perform since Ku70 mutant (32) and Ku80 mutant (P.H., personal observation) breeding pairs rarely produce offspring.
Life span.
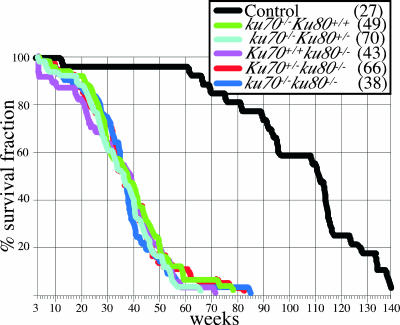
The life spans of the control and mutant cohorts were determined (Fig. 1). All of the mutant cohorts had a shortened life span compared to that of the control cohort (P < 0.0001 [Wilcoxon test] for all mutant cohorts compared to the control cohort). However, the life spans of these three mutant cohorts were very similar and not statistically significantly different (Wilcoxon test P values ranged from 0.3552 to 0.8973). The mean life span was ∼37 weeks for these mutant cohorts, compared to ∼108 weeks for the control cohort. Thus, deletion of Ku70, Ku80, or both reduces the life span by ∼66%. Since there was no observable difference in the homozygous wild-type and heterozygous genotypes for Ku70 and Ku80, these genotypes were combined for simplicity for the remainder of the study.
FIG. 1.
Life span analysis. Survival curves (100% × number of mice alive after each week/total number of mice at 3 weeks) are shown. Mice in the first 3 weeks were not included since Ku mutant mice often die before weaning because they are unable to compete with their littermates. Which line colors represent which mouse strains is shown in the inset (numbers of mice observed are in parentheses).
Tumors.
Mice were observed for tumors throughout the entirety of their life spans. Twenty-six percent (7/27) of the control mice had tumors, including lymphoma (n = 4), adenocarcinoma (n = 1), and hemangioma, a benign tumor (n = 2). All of the mutant cohorts had fewer tumors than control mice, i.e., 6% of the ku70−/− mutant cohort (7/119, six lymphomas and one hepatocellular carcinoma; P < 0.005), 2% of the ku80−/− mutant cohort (2/109, one lymphoma and one hemangioma; P < 0.001), and 0% of the ku70−/− ku80−/− double-mutant cohort (0/38; P < 0.0002). With the numbers observed, there was no statistically significantly difference in cancer incidence between the mutant cohorts (ku70−/− versus ku80−/−, P = 0.175; ku70−/− versus ku70−/− ku80−/−, P = 0.197; ku80−/− versus ku70−/− ku80−/−, P = 1.0).
Even though these mutant cohorts exhibited fewer tumors than the control cohort, their onset was much earlier. The Ku70 mutant mice exhibited lymphoma at the ages of 7, 29, 30, 40, 42, and 47 weeks and hepatocellular carcinoma at 40 weeks. The Ku80 mutant mice exhibited lymphoma at 34 weeks and hemangioma at 78 weeks, while the control mice exhibited lymphoma at 65, 94, 96, and 98 weeks; hemangioma at 114 and 135 weeks; and adenocarcinoma at 117 weeks. Thus, mutant mice exhibited more tumors within their first year compared to control mice.
Aging.
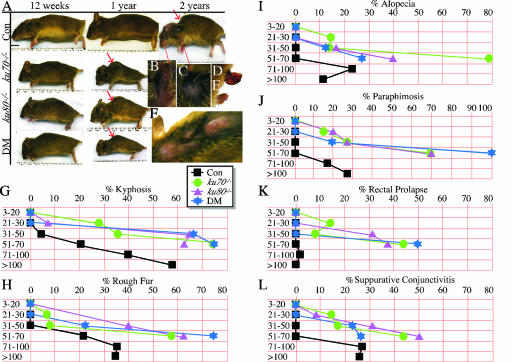
Control and mutant mice were observed for gross signs of aging by a cross-sectional analysis, with particular attention given to the following external characteristics, as shown in old control mice: kyphosis (Fig. 2A, red arrow), suppurative conjunctivitis (Fig. 2B), a rough fur coat (Fig. 2C), rectal prolapse (Fig. 2D), paraphimosis (Fig. 2E, penis permanently extended beyond the prepuce), and alopecia (Fig. 2F). A series of time frames were used to measure phenotype onset and incidence (Table 1). These time frames span between 10 and 20 weeks and are necessary to allow the aging characteristics to become obvious since aging is a highly stochastic process with wide variations in both onset and incidence (26). In addition, long intervals are needed to fully realize each phenotype since it takes time to fully develop.
FIG. 2.
External ageing characteristics. (A) Gross aging seen in control (Con), ku70−/−, ku80−/−, and ku80−/− ku70−/− (double mutant, DM) littermates. Red arrows point to kyphosis. Increased magnifications of an old control mouse (>120 weeks old) show suppurative conjunctivitis (B), rough fur coat (C), rectal prolapse (D), paraphimosis (E), and alopecia (F) (dorsal view of cranium, neck, and thorax). The graphs show the onset and incidence of kyphosis (G), rough fur coat (H), alopecia (I), paraphimosis (J), rectal prolapse (K), and suppurative conjunctivitis (L). Statistical analysis based on a t test comparing the groups listed in Table 1: 1 versus 5 and 1 versus 9, P = 1.0 (all phenotypes); 1 versus 13, P = 0.06 (kyphosis, rough fur, and alopecia) and P = 1.0 (the remaining phenotypes); 1 versus 17, <0.001 (kyphosis and rough fur), P = 0.02 (alopecia), P = 0.006 (suppurative conjunctivitis), P = 0.11 (paraphimosis), and P = 1.0 (rectal prolapse); 1 versus 18, P < 0.001 (kyphosis), P < 0.002 (rough fur), P = 0.31 (alopecia), P = 0.01 (suppurative conjunctivitis), and P = 1.0 (rectal prolapse); 2, 3, and 4 versus 6, 7, and 8, P = 0.02 (kyphosis), P = 0.09 (rough fur), P = 0.21 (alopecia), P = 0.09 (suppurative conjunctivitis), P = 0.23 (paraphimosis), and P = 0.21 (rectal prolapse); 2, 3, and 4 versus 10, 11, and 12, P < 0.001 (kyphosis, rough fur, and suppurative conjunctivitis), P = 0.02 (alopecia), P = 0.002 (paraphimosis), and P = 0.008 (rectal prolapse); 2, 3, and 4 versus 14, 15, and 16, P < 0.001 (all phenotypes).
TABLE 1.
Summary of gross age-related characteristics
| Mouse age (wk), group no. | Genotype | No. of mice/total
|
|||||
|---|---|---|---|---|---|---|---|
| Kyphosis | Rough fur | Alopecia | Suppurative conjunctivitis | Paraphimosis | Rectal prolapse | ||
| 3-20 | |||||||
| 1 | Wild type (control) | 0/21 | 0/21 | 0/21 | 0/21 | 0/13 | 0/21 |
| 2 | ku70−/− | 0/14 | 0/14 | 0/14 | 0/14 | 0/7 | 0/14 |
| 3 | ku80−/− | 0/15 | 0/15 | 0/15 | 0/15 | 0/6 | 0/15 |
| 4 | DMa | 0/14 | 0/14 | 0/14 | 0/14 | 0/11 | 0/14 |
| 21-30 | |||||||
| 5 | Wild type (control) | 0/16 | 0/16 | 0/16 | 0/16 | 0/11 | 0/16 |
| 6 | ku70−/− | 4/14 | 1/14 | 2/14 | 2/14 | 1/7 | 2/14 |
| 7 | ku80−/− | 1/13 | 2/13 | 0/13 | 1/13 | 2/10 | 0/13 |
| 8 | DM | 0/10 | 0/10 | 0/10 | 0/10 | 0/7 | 0/10 |
| 31-50 | |||||||
| 9 | Wild type (control) | 1/24 | 0/24 | 0/24 | 0/24 | 0/14 | 0/24 |
| 10 | ku70−/− | 9/25 | 2/25 | 3/25 | 4/25 | 4/11 | 2/25 |
| 11 | ku80−/− | 13/20 | 8/20 | 3/20 | 6/20 | 4/11 | 6/20 |
| 12 | DM | 6/9 | 2/9 | 1/9 | 2/9 | 1/5 | 0/9 |
| 51-70 | |||||||
| 13 | Wild type (control) | 3/13 | 3/13 | 3/13 | 0/13 | 0/8 | 0/13 |
| 14 | ku70−/− | 9/12 | 7/12 | 9/12 | 9/12 | 3/5 | 5/12 |
| 15 | ku80−/− | 5/8 | 5/8 | 3/8 | 5/8 | 3/5 | 3/8 |
| 16 | DM | 3/4 | 3/4 | 1/4 | 3/4 | 3/3 | 2/4 |
| 71-100, 17 | Wild type (control) | 39/98 | 34/98 | 21/98 | 25/98 | 12/63 | 2/98 |
| >100, 18 | Wild type (control) | 29/50 | 17/50 | 5/50 | 12/50 | 8/27 | 0/50 |
DM, ku70−/− ku80−/− double mutant.
We measured the onset and incidence of kyphosis (Fig. 2G), rough fur coat (Fig. 2H), alopecia (Fig. 2I), paraphimosis (Fig. 2J), and prolapsed rectum (Fig. 2K). No mice presented with these characteristics for the first 20 weeks. However, mice from all of the mutant cohorts commonly exhibited these characteristics within a year while mice from the control cohort commonly exhibited most of these characteristics 20 to 40 weeks later. Thus, these mutant cohorts exhibited an early onset of the same external age-related characteristics as the control cohort.
We also observed mice for inflammatory responses. Previously we showed that control and ku80−/− mice presented with external signs of inflammation as they aged; however, these signs were observed earlier in ku80−/− mutant mice (56). These signs typically include suppurative conjunctivitis with skin ulcerations at the mucocutaneous junction. Here we quantitated suppurative conjunctivitis since this phenotype is commonly observed as 129 mice age (54). No mice presented with suppurative conjunctivitis for the first 20 weeks. However, the mutant cohorts exhibited suppurative conjunctivitis by 21 to 50 weeks while the control cohort exhibited suppurative conjunctivitis by 71 to 100 weeks. Pasteurella multocida was isolated from periorbital exudates, suggesting that suppurative conjunctivitis was caused by opportunistic bacterial infections. In all of the cohorts, including the control, many of the mice likely died from sepsis since they often exhibited signs of severe inflammatory responses that included large abscesses located in the pleural and peritoneal cavities.
It is possible that the early onset of these inflammatory responses was caused by SCID in the Ku mutant mice since Ku is required for the assembly of the V(D)J segments of antigen receptor genes that is requisite for lymphocyte development (22, 23, 58). Additionally, when challenged, ku80−/− mice are susceptible to infectious disease, which greatly reduces their life span (56). However, the suppurative conjunctivitis shown here was likely age related and not caused by SCID for the following reasons. (i) These mice were house in a specific-pathogen-free colony (see Materials and Methods). Ku80 mutant mice that succumbed to infectious disease were housed in a conventional colony contaminated with mouse hepatitis virus and Theiler's murine encephalomyelitis virus. These challenged ku80−/− mice exhibited a mean life span of only 17 weeks and showed clear signs of infection that included liver necrosis (56). By contrast, the Ku mutant mice presented here lived much longer and did not show these signs. (ii) Suppurative conjunctivitis is commonly observed as control 129 mice age (54), and the mice in this study were in a 129 × C57BL/6 background. Therefore, this is an expected age-related observation for this mouse strain. Thus, suppurative conjunctivitis is dependent not on SCID but instead on age and is a typical sign of aging in 129 mice.
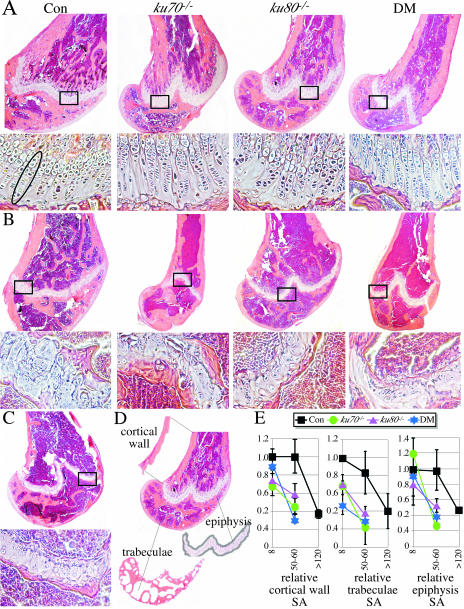
We investigated bones for age-related changes by histology since kyphosis may be caused by osteoporosis in aged individuals (40). The surface areas of the bone cortical wall and trabeculae were quantitated for three mice in each cohort at three ages, 8 weeks (Fig. 3A), 50 to 60 weeks (Fig. 3B), and >120 weeks (Fig. 3C; only control mice remained at this late time point). To evaluate the cortical wall surface area, the distal part of the femur was observed at the point proximal to the epiphysis (Fig. 3D). To evaluate the trabecular surface area, the distal part of the femur was observed from the border of the epiphysis distal to the articular surface of the knee joint (Fig. 3D). Mice from all cohorts at all time points were compared to the control cohort at the 8-week time point. Both the control and mutant cohorts exhibited a decrease in cortical wall and trabecular surface areas with age, but this reduction occurred earlier in the mutant cohorts than in the control cohort (Fig. 3E).
FIG. 3.
Microscopic analysis. (A) Eight-week-old control (Con) and mutant femurs. Double mutant, DM. Top images, distal femur; bottom images, section of epiphysis from the boxed insets. Note that the chondrocytes are aligned in a series of columns, as highlighted by the oval for the control. (B) Fifty- to 60-week-old control and mutant mice. Note that the columns of chondrocytes are diminished in the control and mostly gone in the mutants. Also note that the cortical wall is thinner with fewer trabeculae in the mutants but not in the control. (C) Control at >120 weeks. The columns of chondrocytes are mostly gone. The cortical wall is thinner with fewer trabeculae compared to earlier time points. (D) Diagram showing the section of cortical wall (proximal to the epiphysis), trabeculae (between the epiphysis and articular surface), and epiphysis (the entire length) used to measure age-related changes. See Materials and Methods for the quantification methods used. (E) Graphs displaying the age-related changes in surface area for the cortical wall (left), trabeculae (middle), and epiphysis (right). There are three mice in each group, and shown is the surface area (SA) relative to the mean of the 8-week-old control (hence, the 8-week-old control is always 1.0). Statistical analysis based on a t test: 8-week-old control versus 50- to 60-week-old control, P = 0.9624 (cortical wall), P = 0.2707 (trabeculae), and P = 0.9251 (epiphysis); 8-week-old control versus >120-week-old control, P = 0.0036 (cortical wall), P = 0.0621 (trabeculae), and P = 0.0003 (epiphysis); 8-week-old mutants combined versus 50- to 60-week-old mutants combined, P = 0.00081 (cortical wall), P = 0.00739 (trabeculae), and P = 0.00158 (epiphysis).
We also investigated the epiphysis for age-related changes by histology (the same femurs were observed). The epiphysis is a cartilaginous plate located at the ends of long bones that contain matrix-producing cells called chondrocytes. These chondrocytes are important for bone growth and are arranged in a columnar pattern (Fig. 3A); however, with age they diminish in number and lose their columnar organization. A qualitative examination for chondrocyte number and columnar organization showed a reduction with age in both the control and mutant cohorts, but this reduction occurred earlier in the mutant cohorts (Fig. 3A to C). In addition, with age, cartilage is replaced by bone, effectively reducing the epiphyseal surface area. Therefore, we measured the surface area of the entire length of the epiphysis and found epiphyseal surface area decreases in both the control and mutant cohorts with an earlier onset in the mutant cohorts (Fig. 3E).
Chromosomal instability.
Ku70 and Ku80 maintain genomic stability by repairing DNA DSBs through NHEJ to suppress an accumulation of breaks (31). In addition, Ku70 and Ku80 maintain telomeres since they associate with telomeres (8, 29), suppress chromosomal fusions either with (2, 30, 43) or without (8) telomeres, and impact telomere length maintenance (8, 15). NHEJ may be important during the aging process since chromosomal rearrangements accumulate as mice age (10) while telomere maintenance may impact aging since telomere erosion exacerbates aging for both Ku80 and DNA-PKCS mutant mice (16). In order to observe genomic instability, we analyzed metaphase spreads generated from primary MSFs (passage 2) derived from the ears of control and mutant mice at a variety of ages (4 to 9 weeks, 32 to 62 weeks, and 132 to 140 weeks). These metaphase spreads were observed by two-color FISH with a telomere probe and DAPI. We observe a variety of chromosomal changes that are related to either age or Ku deletion (Table 2).
TABLE 2.
Summary of FISH analysis
| Mouse age (wk), group no. | Genotype | No. of MPSsc | No. of events/metaphase spreadb
|
|||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Single-chromatid telomere loss
|
Fragments/breaks
|
Telomere association
|
Telomere fusion
|
|||||||||||||||||||||||||||
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 15 | Total | 0 | 1 | 2 | 3 | 4 | 5 | 6 | Total | 0 | 1 | 2 | Total | 0 | 1 | 2 | 3 | Total | |||
| 4-9 | ||||||||||||||||||||||||||||||
| 1 | Wild type (control) | 207 | 158 | 29 | 10 | 7 | 2 | 0 | 1 | 0 | 0 | 0 | 84 | 190 | 14 | 2 | 0 | 1 | 0 | 0 | 22 | 203 | 4 | 0 | 4 | 205 | 2 | 0 | 0 | 2 |
| 2 | ku70−/− | 90 | 59 | 15 | 10 | 3 | 2 | 0 | 1 | 0 | 0 | 0 | 58 | 66 | 17 | 5 | 2 | 0 | 0 | 0 | 33 | 89 | 1 | 0 | 1 | 89 | 1 | 0 | 0 | 1 |
| 3 | ku80−/− | 136 | 106 | 17 | 9 | 0 | 3 | 0 | 0 | 1 | 0 | 0 | 54 | 95 | 23 | 13 | 4 | 0 | 1 | 0 | 66 | 133 | 2 | 1 | 4 | 128 | 7 | 1 | 0 | 9 |
| 4 | DMa | 238 | 154 | 50 | 16 | 9 | 4 | 2 | 2 | 0 | 0 | 1 | 162 | 176 | 47 | 10 | 4 | 1 | 0 | 0 | 83 | 228 | 10 | 0 | 10 | 206 | 27 | 4 | 1 | 38 |
| 32-62 | ||||||||||||||||||||||||||||||
| 5 | Wild type (control) | 215 | 151 | 39 | 17 | 4 | 3 | 0 | 0 | 1 | 0 | 0 | 104 | 187 | 26 | 2 | 0 | 0 | 0 | 0 | 30 | 212 | 3 | 0 | 3 | 214 | 1 | 0 | 0 | 1 |
| 6 | ku70−/− | 78 | 52 | 12 | 4 | 1 | 3 | 1 | 2 | 1 | 1 | 1 | 82 | 55 | 13 | 6 | 4 | 0 | 0 | 0 | 37 | 73 | 5 | 0 | 5 | 77 | 1 | 0 | 0 | 1 |
| 7 | ku80−/− | 144 | 71 | 29 | 18 | 10 | 9 | 3 | 2 | 2 | 0 | 0 | 172 | 98 | 27 | 16 | 2 | 0 | 0 | 1 | 71 | 138 | 5 | 1 | 7 | 124 | 18 | 1 | 1 | 23 |
| 8 | DM | 31 | 13 | 4 | 6 | 7 | 0 | 1 | 0 | 0 | 0 | 0 | 42 | 19 | 7 | 4 | 1 | 0 | 0 | 0 | 18 | 27 | 4 | 0 | 4 | 29 | 1 | 1 | 0 | 3 |
| 132-140, 9 | Wild type (control) | 116 | 43 | 33 | 19 | 11 | 4 | 4 | 1 | 1 | 0 | 0 | 153 | 92 | 20 | 3 | 1 | 0 | 0 | 0 | 29 | 116 | 0 | 0 | 0 | 114 | 2 | 0 | 0 | 2 |
DM, ku70−/− ku80−/− double mutant.
The total number of events per metaphase spread is shown as 0 to as many as 15.
MPSs, metaphase spreads.
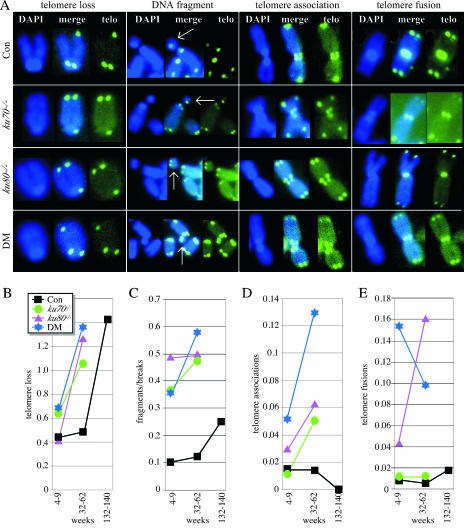
Both control and Ku mutant spreads exhibited single-chromatid telomere loss (on either the short or the long arm) (Fig. 4A and B) and chromosomal fragments and breaks (Fig. 4A and C). These abnormalities were commonly observed and progressively increased with age. Thus, these events are age-related phenotypes similar to those presented in Fig. 2 and 3.
FIG. 4.
Chromosomal abnormalities observed by two-color FISH on metaphase spreads. (A) Representative examples of chromosomal abnormalities. Shown are the DAPI stain, telomere probe (telo), and merge. The white arrow points to a chromosomal fragment. Con, control. (B to E) Graphs showing the total number of events per total number of metaphase spreads for single-chromatid telomere loss (B), fragments/breaks (C), telomere associations (D), and telomere fusions (E). The number may be greater than 1 if enough metaphase spreads have multiple events (see Table 2). Statistical analysis based on the likelihood ratio test comparing groups listed in Table 2: 1 versus 2, 3, and 4, P = 0.0513 (single-chromatid telomere loss), P < 0.0001 (fragments/breaks), P = 0.4469 (telomere associations), and P < 0.0001 (telomere fusions); 1 versus 5, P = 0.3570 (single-chromatid telomere loss), P < 0.3141 (fragments/breaks), P = 0.3338 (telomere associations), and P = 0.2814 (telomere fusions); 1 versus 9, P < 0.0001 (single-chromatid telomere loss), P = 0.0017 (fragments/breaks), P < 0.0001 (telomere associations), and P = 0.2959 (telomere fusions); 2, 3, and 4 versus 6, 7, and 8, P < 0.0001 (single-chromatid telomere loss), P < 0.0419 (fragments/breaks), P < 0.0001 (telomere associations), and P = 0.5530 (telomere fusions).
Ku deletion increases the level of telomere associations and telomere fusions (Fig. 4A, D, and E), as previously reported by three groups (2, 30, 43); however, Ku deletion does not increase the level of chromosomal fusions without telomeres, as reported by another group (8). The latter abnormality was observed in only two ku70−/− ku80−/− metaphase spreads from the 4- to 9-week age group and one ku80−/− and two control metaphase spreads from the 32- to 62-week age group (not shown). Telomere associations, but not telomere fusions, increased with age for the Ku mutant spreads. Both of these events were rarely observed in the control spreads at any age. Thus, these abnormalities are specific to Ku deletion and not to aging.
DISCUSSION
The Ku heterodimer is an essential component of NHEJ. However, the phenotypic differences among various strains of Ku70 and Ku80 mutant mice imply an independent role for each subunit. To address this issue, we crossed Ku70 and Ku80 mutant mice to generate Ku70, Ku80, and double-mutant mice in the same genetic background and housed them in the same cages. We found that mice with a deletion of Ku70, Ku80, or both had similar phenotypes, including early aging without substantially increased cancer.
Do Ku mutant mice age like control mice?
Accelerated-aging models are often called “segmental progerias” since they frequently display only a subset of age-related symptoms naturally found in the entire population. This is certainly true for many of the models of accelerated aging in humans like Werner's syndrome and Hutchinson-Gilford syndrome (25, 37). Thus, some have questioned the relevance of these progerias to normal aging (39). To address this controversial issue, three broad criteria were established to maximize the odds that any particular aging model actually reflects normal aging (26, 27). These criteria are as follows. (i) Aging signs occur after development, preferably after reproductive maturation. (ii) The same signs occur in the control population at a later age but at a similar point in the life span. (iii) A diverse set of tissues and organs are affected. These Ku mutant mice fit all three of these criteria. Here we show that Ku mutant mice exhibit the same aging signs as controls, meaning that all of the aging signs observed in our control mice were also observed in our mutant mice. Therefore, these mice do not display a “segmental” phenotype. However, we have not performed an exhaustive analysis, leaving open the possibility that certain age-related characteristics differ between Ku mutant and control mice. In addition, the incidence of aging signs seems to increase for Ku mutant mice compared to control mice. This is visible in all of phenotypes shown in Fig. 2G to L but is most obvious for rectal prolapse. Thus, Ku mutant mice exhibit an array of aging signs remarkably similar to those of control mice but there are some differences, in particular, increased incidence, suggesting that Ku deletion actually accentuates aging as measured by both onset and incidence.
How does Ku deletion impact cancer incidence?
One interesting reversal from the above trend is that control mice show more cancer than Ku mutant mice. Perhaps Ku mutant mice exhibit less cancer because they have less time to develop it. This is the logical conclusion when cancer incidence is viewed from the perspective of chronological time since most Ku mutant mice die before the control mice begin to show cancer. However, this conclusion does not correlate with the other aging signs that show an increased incidence within the same time frame. It is also possible that Ku deletion reduces cancer in real time. This is the logical conclusion when aging signs are viewed from the perspective of biological time (a measurement relative to the life span) since all cohorts exhibit these aging characteristics at approximately the same point within their life span. Previously, we proposed that Ku deletion indirectly suppresses cancer through persistent anticancer responses to inefficiently repaired DNA damage, resulting in excessive apoptosis or cellular senescence (35, 56). We have begun to address this issue by crossing the Ku80 mutation into a p53 mutant background (11). The p53 mutant mice obtained were defective for cellular responses to DNA damage and showed very high levels of spontaneous cancer in less than 1 year. We have reported that simultaneous deletion of Ku80 and p53 greatly shortens the latency and increases the incidence of two cancer types, i.e., pro-B-cell lymphoma (35) and medulloblastoma (28). These observations support the possibility that persistent anticancer responses influence the Ku mutant phenotype by reducing cancer. This possibility is in line with the hypothesis that anticancer responses contribute to aging, a concept termed antagonistic pleiotropy (7). In order to fully evaluate the possibility that antitumor responses are elevated in Ku mutant mice, the role that Ku deletion plays during oncogenesis must be evaluated in cancer models that maintain normal DNA damage responses.
The cancer incidence in Ku mutant mice may be strongly influenced by the genetic background and environment. Previously, two papers showed that Ku70 mutant mice have cancer levels higher than those reported here. The first report states that “we have observed six cases of thymic tumors among approximately 40 Ku70−/− mice ranging in age from 2 to 7 months” (23). Those mice were of the same strain as those used for this study. Comparing the lymphoma incidence in our present study to that in the previous study suggests that a lower lymphoma level is described here, although the number is at the margin of significance (6/119 versus 6/40, P = 0.076 [Fisher's exact test]). Even though the first report shows 6 of 40 mice with lymphoma within 7 months, it must be noted that a life span study was not preformed and lymphoma was an incidental finding as mice were sacrificed for B- and T-cell analysis. Therefore, taking this into account, there is a significant difference in cancer levels between the previous and present studies since only 3/119 Ku70 mutant mice observed in this study showed signs of cancer by 7 months (P = 0.0084). The second report states that >50% and perhaps has high as 85% of their Ku70 mutant mice died from T-cell lymphoma (32). Even though exact numbers are not given for the second report, it is clear that our ku70−/− mice had much less lymphoma. It is possible that the Ku70 mutant mice in our study had low cancer levels because of early death from another illness; however, this is unlikely since our Ku70 mutants actually lived longer than those in the second study (median life span of 37 versus 28 weeks). Thus, there is a clear divergence in cancer incidence between the Ku70 mutant mice in this study and those in the previous two studies, suggesting that divergent genetic backgrounds or environments impact cancer incidence in mice with Ku70 deleted.
How do Ku70 and Ku80 impact telomere maintenance?
Several groups have shown that either Ku70 or Ku80 associates with telomeres (8, 29), suggesting that Ku is important for telomere maintenance; however, their exact role is controversial. One report shows that Ku80 suppresses telomerase-mediated telomere lengthening since Ku80 mutant cells exhibit telomerase-dependent telomere elongation (15). In addition, Ku80 mutant cells have been reported by this group and several others (2, 30, 43) to exhibit telomere-telomere fusions, resulting in joined chromosomes. This observation suggests that Ku caps telomeres to prevent telomere-telomere joining. However, another report shows that Ku70 and Ku80 maintain telomere length since cells with Ku deleted display telomere shortening (8). Furthermore, this report shows that deletion of Ku70 or Ku80 results in chromosomal fusions without telomeres. Our results are consistent with the former reports suggesting Ku caps telomeres since telomere fusions and associations, but not chromosomal fusions without telomeres, are clearly increased in Ku mutant cells. The authors in the latter group suggested that the divergent results were due to genetic background and/or environmental differences, similar to our conclusion about the different cancer incidences reported for Ku70 mutant mice.
Are defects in NHEJ or telomere maintenance critical for aging in Ku mutant mice?
We show a variety of chromosomal abnormalities in MSFs derived from control and Ku mutant mice as they age. These abnormalities could be placed into the following three broad categories: (i) abnormalities caused by defective DSB repair (DNA breaks and fragments), (ii) abnormalities caused by defective telomere maintenance (telomere-telomere fusions and associations), and (iii) abnormities caused by either defective DSB repair or telomere maintenance (single-chromatid telomere loss). Chromosomal abnormalities from the second category are clearly increased in Ku mutant spreads but are not observed in control spreads. Therefore, these abnormalities are not likely a part of the normal aging process. However, telomere associations increase with age for the Ku mutant spreads; therefore, this event is an age-related phenotype unique to Ku deletion. Chromosomal abnormalities from the first and third categories increase with age for both controls and Ku mutants, with an earlier onset in the Ku mutants. These results suggest that these events are a part of normal aging. It is difficult to know if single-chromatid telomere loss is due to selective telomere erosion or to a chromatid break; therefore, the question of whether this is a telomere maintenance defect or a DSB repair defect remains open. However, chromosomal fragments and breaks suggest defective DSB repair.
Conclusion.
We show that deletion of Ku70, Ku80, or both results in identical aging phenotypes without substantially increasing the incidence of cancer. This report suggests that the Ku70 mutant phenotype is sensitive to the genetic background and environment since two previous studies reported a higher incidence of lymphoma. In addition, this report suggests that defective NHEJ and/or telomere maintenance contributes to normal aging since Ku70 and Ku80 mutant mice display a wide range of normal age-related phenotypes. Our observations corroborate previous reports that end-joining capacity and fidelity decline as normal human fibroblasts age (46, 47), suggesting that Ku function influences normal aging and establishes Ku70 and Ku80 as important longevity assurance proteins.
Acknowledgments
This work was supported by grants UO1 ES11044, R01 CA76317-05A1, and P01 AG17242 to P.H.; by American Cancer Society grant RSG-04-019-01-CNE to Y.G.; and by DOD grant W81XWH-04-1-0325 to V.B.H.
Footnotes
Published ahead of print on 17 September 2007.
REFERENCES
- 1.Ahnesorg, P., P. Smith, and S. P. Jackson. 2006. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell 124:301-313. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, S. M., J. Meyne, D. J. Chen, A. Kurimasa, G. C. Li, B. E. Lehnert, and E. H. Goodwin. 1999. DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc. Natl. Acad. Sci. USA 96:14899-14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biedermann, K. A., J. R. Sun, A. J. Giaccia, L. M. Tosto, and J. M. Brown. 1991. scid mutation in mice confers hypersensitivity to ionizing radiation and a deficiency in DNA double-strand break repair. Proc. Natl. Acad. Sci. USA 88:1394-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blasco, M. A., H. W. Lee, M. P. Hande, E. Samper, P. M. Lansdorp, R. A. DePinho, and C. W. Greider. 1997. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91:25-34. [DOI] [PubMed] [Google Scholar]
- 5.Bond, G. L., and A. J. Levine. 2007. A single nucleotide polymorphism in the p53 pathway interacts with gender, environmental stresses and tumor genetics to influence cancer in humans. Oncogene 26:1317-1323. [DOI] [PubMed] [Google Scholar]
- 6.Buck, D., L. Malivert, R. de Chasseval, A. Barraud, M. C. Fondaneche, O. Sanal, A. Plebani, J. L. Stephan, M. Hufnagel, F. le Deist, A. Fischer, A. Durandy, J. P. de Villartay, and P. Revy. 2006. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell 124:287-299. [DOI] [PubMed] [Google Scholar]
- 7.Campisi, J. 2005. Aging, tumor suppression and cancer: high wire-act! Mech. Ageing Dev. 126:51-58. [DOI] [PubMed] [Google Scholar]
- 7a.Cohen, H. Y., S. Lavu, K. J. Bitterman, B. Hekking, T. A. Imahiyerobo, C. Miller, R. Frye, H. Ploegh, B. M. Kessler, and D. A. Sinclair. 2004. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol. Cell 13:627-638. [DOI] [PubMed] [Google Scholar]
- 8.d'Adda di Fagagna, F., M. P. Hande, W. M. Tong, D. Roth, P. M. Lansdorp, Z. Q. Wang, and S. P. Jackson. 2001. Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr. Biol. 11:1192-1196. [DOI] [PubMed] [Google Scholar]
- 9.Difilippantonio, M. J., J. Zhu, H. T. Chen, E. Meffre, M. C. Nussenzweig, E. E. Max, T. Ried, and A. Nussenzweig. 2000. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature 404:510-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dollé, M. E., H. Giese, C. L. Hopkins, H. J. Martus, J. M. Hausdorff, and J. Vijg. 1997. Rapid accumulation of genome rearrangements in liver but not in brain of old mice. Nat. Genet. 17:431-434. [DOI] [PubMed] [Google Scholar]
- 11.Donehower, L. A., M. Harvey, B. L. Slagle, M. J. McArthur, C. A. Montgomery, Jr., J. S. Butel, and A. Bradley. 1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356:215-221. [DOI] [PubMed] [Google Scholar]
- 12.Donehower, L. A., M. Harvey, H. Vogel, M. J. McArthur, C. A. Montgomery, Jr., S. H. Park, T. Thompson, R. J. Ford, and A. Bradley. 1995. Effects of genetic background on tumorigenesis in p53-deficient mice. Mol. Carcinog. 14:16-22. [DOI] [PubMed] [Google Scholar]
- 13.Dorshkind, K., L. Welniak, R. A. Gault, J. Hixon, E. Montecino-Rodriguez, N. D. Horseman, J. M. Gertner, and W. J. Murphy. 2003. Effects of housing on the thymic deficiency in dwarf mice and its reversal by growth hormone administration. Clin. Immunol. 109:197-202. [DOI] [PubMed] [Google Scholar]
- 14.Errami, A., V. Smider, W. K. Rathmell, D. M. He, E. A. Hendrickson, M. Z. Zdzienicka, and G. Chu. 1996. Ku86 defines the genetic defect and restores X-ray resistance and V(D)J recombination to complementation group 5 hamster cell mutants. Mol. Cell. Biol. 16:1519-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Espejel, S., S. Franco, S. Rodriguez-Perales, S. D. Bouffler, J. C. Cigudosa, and M. A. Blasco. 2002. Mammalian Ku86 mediates chromosomal fusions and apoptosis caused by critically short telomeres. EMBO J. 21:2207-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Espejel, S., P. Klatt, J. Menissier-de Murcia, J. Martin-Caballero, J. M. Flores, G. Taccioli, G. de Murcia, and M. A. Blasco. 2004. Impact of telomerase ablation on organismal viability, aging, and tumorigenesis in mice lacking the DNA repair proteins PARP-1, Ku86, or DNA-PKcs. J. Cell Biol. 167:627-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferguson, D. O., J. M. Sekiguchi, S. Chang, K. M. Frank, Y. Gao, R. A. DePinho, and F. W. Alt. 2000. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc. Natl. Acad. Sci. USA 97:6630-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fulop, G. M., and R. A. Phillips. 1990. The scid mutation in mice causes a general defect in DNA repair. Nature 347:479-482. [DOI] [PubMed] [Google Scholar]
- 19.Gao, Y., D. O. Ferguson, W. Xie, J. P. Manis, J. Sekiguchi, K. M. Frank, J. Chaudhuri, J. Horner, R. A. DePinho, and F. W. Alt. 2000. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature 404:897-900. [DOI] [PubMed] [Google Scholar]
- 20.Goedecke, W., M. Eijpe, H. H. Offenberg, M. van Aalderen, and C. Heyting. 1999. Mre11 and Ku70 interact in somatic cells, but are differentially expressed in early meiosis. Nat. Genet. 23:194-198. [DOI] [PubMed] [Google Scholar]
- 20a.Gomez, J. A., V. Gama, T. Yoshida, W. Sun, P. Hayes, K. Leskov, D. Boothman, and S. Matsuyama. 2007. Bax-inhibiting peptides derived from Ku70 and cell-penetrating pentapeptides. Biochem. Soc. Trans. 35:797-801. [DOI] [PubMed] [Google Scholar]
- 21.Grawunder, U., M. Wilm, X. Wu, P. Kulesza, T. E. Wilson, M. Mann, and M. R. Lieber. 1997. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature 388:492-495. [DOI] [PubMed] [Google Scholar]
- 22.Gu, Y., S. Jin, Y. Gao, D. T. Weaver, and F. W. Alt. 1997. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc. Natl. Acad. Sci. USA 94:8076-8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu, Y., K. J. Seidl, G. A. Rathbun, C. Zhu, J. P. Manis, N. van der Stoep, L. Davidson, H. L. Cheng, J. M. Sekiguchi, K. Frank, P. Stanhope-Baker, M. S. Schlissel, D. B. Roth, and F. W. Alt. 1997. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity 7:653-665. [DOI] [PubMed] [Google Scholar]
- 24.Gu, Y., J. Sekiguchi, Y. Gao, P. Dikkes, K. Frank, D. Ferguson, P. Hasty, J. Chun, and F. W. Alt. 2000. Defective embryonic neurogenesis in Ku-deficient but not DNA-dependent protein kinase catalytic subunit-deficient mice. Proc. Natl. Acad. Sci. USA 97:2668-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hasty, P., J. Campisi, J. Hoeijmakers, H. van Steeg, and J. Vijg. 2003. Aging and genome maintenance: lessons from the mouse? Science 299:1355-1359. [DOI] [PubMed] [Google Scholar]
- 26.Hasty, P., and J. Vijg. 2004. Accelerating aging by mouse reverse genetics: a rational approach to understanding longevity. Aging Cell 3:55-65. [DOI] [PubMed] [Google Scholar]
- 27.Hasty, P., and J. Vijg. 2004. Rebuttal to Miller: ‘Accelerated aging’: a primrose path to insight. Aging Cell 3:67-69. [DOI] [PubMed] [Google Scholar]
- 28.Holcomb, V. B., H. Vogel, T. Marple, R. W. Kornegay, and P. Hasty. 2006. Ku80 and p53 suppress medulloblastoma that arise independent of Rag-1-induced DSBs. Oncogene 25:7159-7165. [DOI] [PubMed] [Google Scholar]
- 29.Hsu, H. L., D. Gilley, E. H. Blackburn, and D. J. Chen. 1999. Ku is associated with the telomere in mammals. Proc. Natl. Acad. Sci. USA 96:12454-12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu, H. L., D. Gilley, S. A. Galande, M. P. Hande, B. Allen, S. H. Kim, G. C. Li, J. Campisi, T. Kohwi-Shigematsu, and D. J. Chen. 2000. Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev. 14:2807-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karanjawala, Z. E., U. Grawunder, C. L. Hsieh, and M. R. Lieber. 1999. The nonhomologous DNA end joining pathway is important for chromosome stability in primary fibroblasts. Curr. Biol. 9:1501-1504. [DOI] [PubMed] [Google Scholar]
- 32.Li, G. C., H. Ouyang, X. Li, H. Nagasawa, J. B. Little, D. J. Chen, C. C. Ling, Z. Fuks, and C. Cordon-Cardo. 1998. Ku70: a candidate tumor suppressor gene for murine T cell lymphoma. Mol. Cell 2:1-8. [DOI] [PubMed] [Google Scholar]
- 32a.Li, Y., T. Yokota, V. Gama, T. Yoshida, J. A. Gomez, K. Ishikawa, H. Sasaguri, H. Y. Cohen, D. A. Sinclair, H. Mizusawa, and S. Matsuyama. 2007. Bax-inhibiting peptide protects cells from polyglutamine toxicity caused by Ku70 acetylation. Cell Death Differ. [Epub ahead of print.] doi: 10.1038/sj.cdd.4402219. [DOI] [PubMed]
- 33.Liang, F., P. J. Romanienko, D. T. Weaver, P. A. Jeggo, and M. Jasin. 1996. Chromosomal double-strand break repair in Ku80-deficient cells. Proc. Natl. Acad. Sci. USA 93:8929-8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lieber, M. R. 1998. Warner-Lambert/Parke-Davis Award Lecture. Pathological and physiological double-strand breaks: roles in cancer, aging, and the immune system. Am. J. Pathol. 153:1323-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim, D. S., H. Vogel, D. M. Willerford, A. T. Sands, K. A. Platt, and P. Hasty. 2000. Analysis of ku80-mutant mice and cells with deficient levels of p53. Mol. Cell. Biol. 20:3772-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma, Y., U. Pannicke, K. Schwarz, and M. R. Lieber. 2002. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 108:781-794. [DOI] [PubMed] [Google Scholar]
- 37.Martin, G. M., and M. S. Turker. 1988. Model systems for the genetic analysis of mechanisms of aging. J. Gerontol. 43:B33-B39. [DOI] [PubMed] [Google Scholar]
- 38.Mazumder, S., D. Plesca, M. Kinter, and A. Almasan. 2007. Interaction of a cyclin E fragment with Ku70 regulates Bax-mediated apoptosis. Mol. Cell. Biol. 27:3511-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller, R. A. 2004. ‘Accelerated aging’: a primrose path to insight? Aging Cell 3:47-51. [DOI] [PubMed] [Google Scholar]
- 40.Moro Alvarez, M. J., and M. Diaz-Curiel. 2007. Pharmacological treatment of osteoporosis for people over 70. Aging Clin. Exp. Res. 19:246-254. [DOI] [PubMed] [Google Scholar]
- 41.Nussenzweig, A., C. Chen, V. da Costa Soares, M. Sanchez, K. Sokol, M. C. Nussenzweig, and G. C. Li. 1996. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature 382:551-555. [DOI] [PubMed] [Google Scholar]
- 42.Romero, F., C. Dargemont, F. Pozo, W. H. Reeves, J. Camonis, S. Gisselbrecht, and S. Fischer. 1996. p95vav associates with the nuclear protein Ku-70. Mol. Cell. Biol. 16:37-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samper, E., F. A. Goytisolo, P. Slijepcevic, P. P. van Buul, and M. A. Blasco. 2000. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 1:244-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reference deleted.
- 45.Reference deleted.
- 46.Seluanov, A., J. Danek, N. Hause, and V. Gorbunova. 6 August 2007, posting date. Changes in the level and distribution of Ku proteins during cellular senescence. DNA Repair (Amsterdam) doi: 10.1016/j.dnarep.2007.06.010. [DOI] [PMC free article] [PubMed]
- 47.Seluanov, A., D. Mittelman, O. M. Pereira-Smith, J. H. Wilson, and V. Gorbunova. 2004. DNA end joining becomes less efficient and more error-prone during cellular senescence. Proc. Natl. Acad. Sci. USA 101:7624-7629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simpson, E. M., C. C. Linder, E. E. Sargent, M. T. Davisson, L. E. Mobraaten, and J. J. Sharp. 1997. Genetic variation among 129 substrains and its importance for targeted mutagenesis in mice. Nat. Genet. 16:19-27. [DOI] [PubMed] [Google Scholar]
- 49.Smider, V., W. K. Rathmell, M. R. Lieber, and G. Chu. 1994. Restoration of X-ray resistance and V(D)J recombination in mutant cells by Ku cDNA. Science 266:288-291. [DOI] [PubMed] [Google Scholar]
- 50.Smith, G. C. M., and S. P. Jackson. 1999. The DNA-dependent protein kinase. Genes Dev. 266:288-290. [DOI] [PubMed] [Google Scholar]
- 51.Smith, G. D. 2007. Life-course approaches to inequalities in adult chronic disease risk. Proc. Nutr. Soc. 66:216-236. [DOI] [PubMed] [Google Scholar]
- 52.Song, K., D. Jung, Y. Jung, S. G. Lee, and I. Lee. 2000. Interaction of human Ku70 with TRF2. FEBS Lett. 481:81-85. [DOI] [PubMed] [Google Scholar]
- 53.Song, K., Y. Jung, D. Jung, and I. Lee. 2001. Human ku70 interacts with heterochromatin protein 1α. J. Biol. Chem. 276:8321-8327. [DOI] [PubMed] [Google Scholar]
- 53a.Subramanian, C., A. W. Opipari, Jr., X. Bian, V. P. Castle, and R. P. Kwok. 2005. Ku70 acetylation mediates neuroblastoma cell death induced by histone deacetylase inhibitors. Proc. Natl. Acad. Sci. USA 102:4842-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sundberg, J. P., K. S. Brown, R. Bates, T. L. Cunliffe-Beamer, and H. Bedigian. 1991. Suppurative conjunctivitis and ulcerative blepharitis in 129/J mice. Lab. Anim. Sci. 41:516-518. [PubMed] [Google Scholar]
- 55.Taccioli, G. E., T. M. Gottlieb, T. Blunt, A. Priestley, J. Demengeot, R. Mizuta, A. R. Lehmann, F. W. Alt, S. P. Jackson, and P. A. Jeggo. 1994. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science 265:1442-1445. [DOI] [PubMed] [Google Scholar]
- 56.Vogel, H., D. S. Lim, G. Karsenty, M. Finegold, and P. Hasty. 1999. Deletion of Ku86 causes early onset of senescence in mice. Proc. Natl. Acad. Sci. USA 96:10770-10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang, C. R., S. Yeh, K. Leskov, E. Odegaard, H. L. Hsu, C. Chang, T. J. Kinsella, D. J. Chen, and D. A. Boothman. 1999. Isolation of Ku70-binding proteins (KUBs). Nucleic Acids Res. 27:2165-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu, C., M. A. Bogue, D. S. Lim, P. Hasty, and D. B. Roth. 1996. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell 86:379-389. [DOI] [PubMed] [Google Scholar]