Abstract
Canonical Wnt signaling and its nuclear effectors, β-catenin and the family of T-cell factor (TCF) DNA-binding proteins, belong to the small number of regulatory systems which are repeatedly used for context-dependent control of distinct genetic programs. The apparent ability to elicit a large variety of transcriptional responses necessitates that β-catenin and TCFs distinguish precisely between genes to be activated and genes to remain silent in a specific context. How this is achieved is unclear. Here, we examined patterns of Wnt target gene activation and promoter occupancy by TCFs in different mouse cell culture models. Remarkably, within a given cell type only Wnt-responsive promoters are bound by specific subsets of TCFs, whereas nonresponsive Wnt target promoters remain unoccupied. Wnt-responsive, TCF-bound states correlate with DNA hypomethylation, histone H3 hyperacetylation, and H3K4 trimethylation. Inactive, nonresponsive promoter chromatin shows DNA hypermethylation, is devoid of active histone marks, and additionally can show repressive H3K27 trimethylation. Furthermore, chromatin structural states appear to be independent of Wnt pathway activity. Apparently, cell-type-specific regulation of Wnt target genes comprises multilayered control systems. These involve epigenetic modifications of promoter chromatin and differential promoter occupancy by functionally distinct TCF proteins, which together determine susceptibility to Wnt signaling.
During embryonic development, multicellular organisms employ only a few families of growth factors and signal transduction pathways to precisely orchestrate the cell-type- and stage-specific activation of distinct genetic programs (9). The recurrent use of a limited number of signaling systems and signal transducers poses the fundamental question of how the same effector molecules can selectively control different sets of target genes in a tissue-specific manner. This problem can be exemplified by canonical Wnt signaling, which is involved in many aspects of embryonic development as well as the homeostasis of adult tissues (18, 50). In canonical Wnt signaling, secreted glycoproteins of the family of Wnt growth factors bind to LRP/Frizzled receptor complexes, thereby triggering a signaling cascade which leads to the activation of β-catenin (18, 50). In the absence of a Wnt stimulus, β-catenin becomes phosphorylated upon recruitment to a multimeric protein complex containing the scaffolding proteins Axin1 or Axin2, adenomatous polyposis coli (APC), and the Ser/Thr kinases casein kinase I and glycogen synthase kinase 3 (GSK3). Phosphorylation marks β-catenin for proteasomal degradation. Wnt pathway activation disables the destruction machinery and allows β-catenin to translocate to the nucleus, where it interacts with one of the four members of the lymphoid enhancer factor (LEF)/T-cell factor (TCF) family of DNA-binding proteins (3, 71). Although β-catenin can additionally interact with a range of other DNA-binding transcription factors (1, 24, 39, 40, 57, 67, 82), it appears that β-catenin/TCF complexes are the prime nuclear effectors of canonical Wnt signaling (26) and in conjunction with diverse corepressors and coactivators catalyze both repression and Wnt-induced activation, respectively, of their target genes (reviewed in references 71 and 79).
Wnt target genes can be differentially regulated in a tissue-specific and developmentally controlled manner such that in a given cell type Wnt-responsive and -nonresponsive genes coexist. How the separation of the two groups of genes is brought about mechanistically is currently not known. Recent reports indicate that TCF family members perform distinct and nonoverlapping functions during embryogenesis and skin development, although phenotypic analyses of mutant animals suggest a certain degree of redundancy (26, 30, 49, 53, 58, 63, 72). Moreover, the E-type splice variants of TCF1 and TCF4 exhibit promoter-specific activities and are not interchangeable with other isoforms or TCF family members (6, 31). Gene-specific activities of the TCFs thus may contribute to differential Wnt target gene regulation. A certain complication, though, arises from the fact that all TCF family members appear to uniformly interact with β-catenin and to recognize the same DNA elements. Furthermore, genetic evidence from Drosophila melanogaster as well as promoter studies with Xenopus laevis suggest that TCFs contribute to both repression and activation of Wnt target genes (15, 16). Accordingly, TCFs are thought to bind to promoter regions of their targets irrespective of Wnt pathway activity in order to provide essential chromosomal docking sites for their diverse interaction partners (21, 40, 41, 66). However, the apparent promiscuity with respect to promoter recognition and binding to β-catenin, as well as constant promoter occupancy, is difficult to reconcile with the observed selective responsiveness of disparate subgroups of Wnt targets. Thus, current models for gene regulation by canonical Wnt signaling do not provide a satisfactory mechanistic framework to explain how an incoming activating signal provided by β-catenin would be enabled to precisely distinguish between genes to be activated and genes to remain silent in a specific context.
Aside from specific functional properties of TCFs, could additional mechanisms be involved in the differential regulation of Wnt target gene transcription? Epigenetic factors are known to establish and maintain specific patterns of transcription during development (52, 65), and key developmental regulators and components of signaling pathways are subject to epigenetic control (13, 14, 44). Epigenetic determinants control the accessibility of promoter chromatin and establish lineage-specific heritable states of gene expression through modulation of DNA methylation and posttranslational modifications of core histones (36, 52). A tight link between chromatin structure and transcriptional regulation of Wnt target genes is provided by multiple histone-modifying and chromatin-remodeling enzyme complexes among the cofactors of β-catenin and TCFs (8, 11, 17, 28, 32, 56, 74). Also, Wnt-induced transcription of the c-Myc gene is accompanied by cyclic changes in the methylation of lysine 4 of histone H3 (H3K4) (66). Therefore, it is tempting to speculate that epigenetic mechanisms additionally contribute to setting up distinct functional states of Wnt-regulated genes. They could, for example, act upstream of TCFs and constitute a preselection mechanism for potentially Wnt-responsive genes by allowing or preventing promoter binding of TCF proteins.
In this study we have used different cellular models and Wnt-inducible genes to systematically investigate the regulatory properties of TCFs and the contribution of epigenetic mechanisms to differential Wnt target gene control, which had not been tested in a rigorous and comprehensive manner before. Our findings confirm and extend previous reports of cell-type- and promoter-specific functional differences among TCF family members. Notably, TCFs are not present at promoter regions of nonresponding genes. Rather, Wnt responsiveness correlates with differential promoter occupancy by selected TCFs. Furthermore, Wnt-inducible promoters are distinguished by DNA hypomethylation and a Wnt-independent occurrence of active histone marks. Thus, we hypothesize that cell-type-specific inducibility of Wnt target genes is based on complex regulatory systems involving epigenetic factors and distinct promoter occupancy by TCFs.
(S. Wöhrle performed this work in partial fulfillment of the requirements of his Ph.D. studies in the Faculty of Biology of the University of Freiburg, Freiburg, Germany.)
MATERIALS AND METHODS
Cell culture and generation of stable cell lines.
C17.2 cells, NIH 3T3 fibroblasts expressing murine Wnt3a (NIH 3T3-Wnt3a), and E14 embryonic stem (ES) cells were cultured as described previously (2, 5, 69, 80). C2C12 cells (ECACC 91031101) were cultured in Dulbecco's modified Eagle's medium (PAN-Biotech) containing 10% fetal calf serum and 100 units/ml penicillin-streptomycin. To generate cell lines stably expressing hemagglutinin (HA)-tagged TCFs, the cDNAs of the different TCF/LEF constructs were cloned upstream of the internal ribosome entry site/enhanced green fluorescent protein cassette in pSF91, a vector derived from murine ES cell virus (34). Phoenix Eco cells (61) were transfected with the retroviral constructs, and the viral supernatants were used for infection of C17.2, C2C12, and E14 cells. Cells expressing enhanced green fluorescent protein were sorted using a MoFlo high-speed cell sorter (Dako) into 96-well plates by the core facility for flow cytometry and cell sorting at the University of Freiburg Medical Center. Single-cell-derived clones with similar TCF expression levels were chosen for further analyses.
Wnt pathway activation and purification of recombinant Wnt3a.
For activation of canonical Wnt signaling, the GSK3 inhibitor SB-216763 (Sigma), recombinant Wnt3a (R&D Systems), or Wnt3a partially purified from conditioned media was used. Partial purification was achieved following published procedures (42, 78) with the following modifications. NIH 3T3-Wnt3a cells were passaged 1:20 in 15-cm culture dishes, and conditioned medium was collected after 4 days. Fresh medium was added, and a second batch of medium was collected after 3 days. Both batches were combined, and a total of 500 ml conditioned medium was used for the purification. Detergent was omitted from the conditioned medium prior to application onto the Blue Sepharose column as well as from the washing buffer. Proteins bound to the Sepharose column were eluted with elution buffer containing CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} in two steps (at 20% and 40% elution buffer for 10 column volumes each). Fractions were analyzed for Wnt activity by a β-catenin stabilization assay (35) in C17.2 cells. Wnt-containing fractions were pooled for further purification by gel filtration as described previously. Due to the weak binding of Wnt3a to heparin Sepharose, subsequent cation-exchange chromatography was omitted from the purification protocol and pooled fractions of the gel filtration column containing Wnt activity were directly used for stimulation of the Wnt pathway. Typically, Wnt3a activity was enriched about 150-fold with respect to the starting material and contained an equivalent of 500 ng/ml recombinant Wnt3a (R&D Systems).
RNA isolation, RT-PCR, and quantitative real-time RT-PCR.
Cells were plated in 6-well or 12-well plates, and total cellular RNA was isolated using the NucleoSpin RNA II kit (Macherey and Nagel). Oligo(dT)-primed cDNA was synthesized with 0.5 to 2 μg RNA and Superscript II reverse transcriptase (RT) (Invitrogen). An equivalent of 50 ng total cellular RNA served as the template in PCRs. PCR products were analyzed by gel electrophoresis and staining with ethidium bromide. Real-time PCR was performed in a MyiQ single-color real-time PCR detection system (Bio-Rad) using SYBR green reaction mix (Invitrogen) and an equivalent of 40 or 80 ng RNA of each sample. The data were normalized to GAPDH expression. All experiments were performed in duplicate at least three separate times. Primer sequences and details of PCR conditions for each primer set are available upon request.
ChIP.
Chromatin immunoprecipitation (ChIP) was performed as previously described (12) with modifications. Cells were plated in 15-cm tissue culture dishes and treated where appropriate with SB-216763, trichostatin A (TSA), or with dimethyl sulfoxide (DMSO) only as the solvent control. Formaldehyde was added directly to the cell culture medium at a final concentration of 0.5%. Fixation was stopped after 10 min at room temperature by the addition of glycine to a final concentration of 0.125 M. Cells were rinsed twice with ice-cold phosphate-buffered saline and incubated with 2 ml trypsin-EDTA for 10 min at room temperature. Trypsin was inactivated by adding fetal calf serum (final concentration 10%), and cells were scraped off. Cells were collected by centrifugation and washed once with ice-cold phosphate-buffered saline. The pellet was resuspended in NP-40 lysis buffer (5 mM HEPES-KOH [pH 7.9], 85 mM KCl, 0.5% NP-40, and protease inhibitors [Roche]) and incubated for 10 min. The cells were disrupted by Dounce homogenization and nuclei were collected by centrifugation at 4,500 × g, resuspended in nucleus lysis buffer (50 mM Tris-HCl [pH 8.1], 10 mM EDTA, 1% sodium dodecyl sulfate [SDS], and protease inhibitors), and incubated for 5 min on ice. Samples were sonicated on ice at 40% amplitude 10 times for 10 s each by use of a Branson sonifier model W-450D. The average DNA fragment size was 250 to 500 bp. The lysate was microcentrifuged at 16,100 × g for 10 min and the supernatant was transferred to a clean tube. The optical density at 260 nm was determined, and aliquots corresponding to 60 optical density units were used for each IP. The lysates were diluted with two sample volumes of IP dilution buffer (16.7 mM Tris-HCl [pH 8.0], 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 167 mM NaCl, and protease inhibitors), and the appropriate antibodies or isotype controls were added. Antibodies used include goat anti-TCF4 (sc-8631; Santa Cruz), rat anti-HA (3F10; Roche), rabbit anti-acetyl histone H3 (06-599; Upstate), rabbit anti-trimethyl histone H3K4 (ab8580; Abcam), rabbit anti-trimethyl histone H3K27 (07-449; Upstate), and isotype control immunoglobulin Gs (IgGs) (Santa Cruz). Thirty microliters protein A- or G-Dynabeads (Invitrogen), preblocked with 250 μg/ml bovine serum albumin and 250 μg/ml salmon sperm DNA, were added, and the samples were incubated overnight at 4°C. The next day, beads were washed twice with dialysis buffer (50 mM Tris-HCl [pH 8.0], 2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride) and four times with IP wash buffer (100 mM Tris-HCl [pH 9.0], 500 mM LiCl, 1% NP-40, and 1% deoxycholic acid). For each wash, beads were resuspended in 1.2 ml buffer and incubated at 4°C for 10 min with constant agitation. Before the first wash, 20% of the supernatant of the antibody isotype control was saved as reference material for quantification. Immunoprecipitates were eluted from the beads by two consecutive elution steps with 150 μl elution buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA, 1% SDS) at 67°C for 10 min on a thermomixer. Eluates were combined, and 10 μg RNase (Roche) and NaCl (final concentration, 300 mM) were added to the immunoprecipitate and the reference material. Cross-links were reversed at 67°C for 4 h. Proteinase K (80 μg) was added and the samples were incubated for 1 h at 45°C. DNA was then purified using QIAquick spin columns (Qiagen) as described by the manufacturer by use of 75 μl elution buffer. PCR was performed using 5 μl of the eluted DNA. For real-time PCR, 4 μl of the immunoprecipitated DNA and 2% of the reference material were used as templates. Sequence information for the primers used and details of PCR conditions for each primer set are available upon request. The data were normalized to unspecific GAPDH amplification and are presented as enrichment (n-fold) with standard errors of the mean (SEM) with respect to isotype control samples. For normalization of data obtained after the precipitation of trimethylated H3K4, values derived from the amplification of a distal Axin2 promoter region (nucleotide positions −11202 to −11033 relative to the transcriptional start site) were used instead of GAPDH. All experiments were performed in duplicate at least three separate times.
Immunoprecipitation and Western blotting.
Nuclear lysates for immunoprecipitation and Western blot analyses of the acetylation state of histone H3 were prepared as for ChIP except that formaldehyde fixation and two of the washing steps with IP wash buffer were omitted from the protocol. The protein concentration of the nuclear lysates was determined by the DC protein assay (Bio-Rad), and 300 μg of total protein was used for immunoprecipitations with anti-acetylated H3 antibodies (06-599; Upstate) or rabbit control IgGs (Santa Cruz). Immunoprecipitates and a fraction of the nuclear lysates (2.5%) were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose membranes, and analyzed with antibodies to acetylated H3, appropriate horseradish peroxidase-labeled secondary antibodies (Dianova), and a chemiluminescence detection reagent. Equal loading was confirmed by Coomassie brilliant blue staining of fractions of nuclear lysates separated by SDS-PAGE (7.5%).
Sodium bisulfite genomic sequencing.
Genomic DNA, isolated from cells plated in 10-cm culture dishes by use of a DNeasy blood and tissue kit (Qiagen) according to the manufacturer's protocol, was digested to completion with BglII, and purified using QIAquick spin columns (Qiagen). Bisulfite conversion was performed using an EZ DNA methylation-gold kit (Zymo Research) with 1 μg of BglII-digested genomic DNA. For PCR amplification, 100 to 200 ng bisulfite-treated DNA was used in a 50-μl reaction mix containing 200 μM of each of the four dinucleoside triphosphates, 30 pmol of each primer, 1 × PCR buffer, and 2.5 units of Taq polymerase (Q-Biogene). Primers specific to bisulfite-converted DNA were designed using the MethPrimer program (46) containing 5′ extensions for BamHI and HindIII restriction endonucleases. Primer sequences and PCR conditions are available upon request. PCR products were purified using QIAquick spin columns, digested with BamHI and HindIII restriction endonucleases, and subsequently cloned into pBS(+KS) vector (Stratagene). Clones were sequenced by the Sequencing Core Facility, University of Freiburg Medical Center.
RESULTS
Differential expression and inducibility of Wnt target genes in neural, myogenic, and embryonic cell lines.
To systematically compare Wnt target gene activation levels and to gain insight into the mechanisms underlying differential responsiveness, we chose the murine cell lines C17.2, C2C12, and E14 as model systems. C17.2 cells are derived from neonatal cerebellum and possess neural stem cell-like properties. C2C12 are myogenic cells, and E14 is an ES cell line. As a representative set of Wnt target genes with distinct activation patterns in embryonic development, we selected Axin2, Cdx1, and T/Brachyury (T/Bra) (7, 22, 54, 73, 77). The genomic organization of these genes is partially depicted in Fig. 1. Examination of the DNA sequences revealed that in all three cases the promoter regions are associated with CpG islands. Moreover, multiple sequence motifs matching the consensus binding site for TCF family members are present distal and proximal to their transcriptional start sites. While all of these elements appear to contribute to Wnt inducibility of the Axin2 gene, only the promoter-proximal TCF-binding elements (TBEs) 3 to 5 of the Cdx1 and T/Bra genes appear to be functionally relevant (5, 7, 29, 37, 45, 47, 48, 81). The expression and Wnt inducibility of Axin2, Cdx1, and T/Bra were analyzed by semiquantitative and quantitative RT-PCR in C17.2, C2C12, and E14 cells. Activation of the canonical Wnt pathway by recombinant Wnt3a or the GSK3 inhibitor SB-216763 led to an upregulation of Axin2 expression within 3 h after stimulation (Fig. 2). Axin2 expression responds to Wnt pathway activation in all three cell lines, which is consistent with the view that Axin2 is a ubiquitous Wnt target (18). Moreover, this indicates that all of the cell lines analyzed are able to sense and process a Wnt signal. Induction of Axin2 occurs with similar kinetics in each of the cell types analyzed, reaching its maximum between 6 and 10 h (Fig. 2B). In contrast to Axin2, Cdx1 and T/Bra can be induced by Wnt pathway activation only in E14 cells but not in C17.2 or C2C12 cells (Fig. 2A). Compared to the activation of Axin2, the induction of Cdx1 and T/Bra expression is delayed and reaches maximal levels at 24 h of stimulation (Fig. 2B). The fact that Cdx1 and T/Bra are not Wnt inducible in C2C12 and C17.2 cells corresponds very well to their expression patterns during embryonic development (22, 54, 73, 77) and shows that our cellular models faithfully recapitulate their differential regulation in vivo.
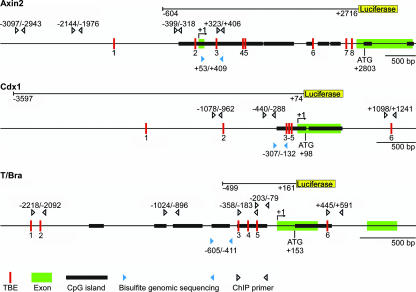
FIG. 1.
Schematic representation of regulatory regions of Wnt target genes. Shown are parts of the murine Axin2, Cdx1, and T/Bra gene loci. Thin lines represent DNA, TBEs are indicated by red bars and numbered, exons are highlighted in green, and CpG islands are in black. Blue arrowheads mark the location of primer pairs used for genomic bisulfite sequencing. White arrowheads indicate primer pairs used for the analyses of chromatin immunoprecipitates. Numbers refer to nucleotide positions relative to the transcriptional start sites (arrows, +1). For each gene, DNA fragments used to drive the expression of a luciferase reporter gene (yellow bar) are displayed and nucleotide coordinates are shown. In each case, reporter gene expression driven by the promoter fragments used recapitulates important aspects of Axin2, Cdx1, and T/Bra embryonic expression patterns in transgenic animals and mutational inactivation of TCF binding elements contained within the promoter regions abrogates transgene expression in cognate T/Bra, Cdx1, and Axin2 expression domains (7, 48, 81).
FIG. 2.
Wnt target genes are differentially activated in neural, myogenic, and embryonic cell lines. (A) Wnt target gene activation in C17.2 neural stem-cell like cells, C2C12 myogenic cells, and E14 embryonic stem cells. Total RNA was isolated from cells treated with 25 μM (C17.2, C2C12) or 5 μM (E14) SB-216763 (lanes 1 to 3) and 200 ng/ml recombinant Wnt3a (lanes 5 to 10) as indicated. Activation of the Wnt target genes Axin2, Cdx1, and T/Bra was analyzed by RT-PCR. Amplification of GAPDH was used to control sample integrity and loading. Control reactions received no template (no templ.), cDNA samples which were mock processed in the absence of RT (−RT), or plasmid DNA or a pretested cDNA as the template (pos. control). n.a.: not analyzed. (B) Quantitative real-time PCR of Wnt target gene activation. C17.2, C2C12, and E14 cells were treated with recombinant Wnt3a for the times indicated. Activation of the Wnt target genes Axin2, Cdx1, and T/Bra is shown as induction (n-fold) compared to transcript levels in untreated cells. Data given are the average and SEM from at least three experiments.
Only promoters of Wnt-inducible target genes are occupied by endogenous TCF4.
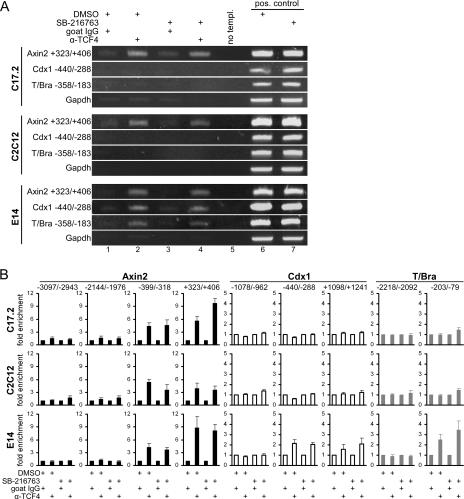
TCF family members exhibit cell-type- and promoter-specific functional differences (3, 6, 31). A simple explanation for the restricted inducibility of Cdx1 and T/Bra therefore was that C17.2 and C2C12 cells did not express a TCF family member specifically required for their activation. However, all four TCF factors are expressed in C17.2, C2C12, and E14 cells (see Fig. S1 in the supplemental material), arguing that the differential responsiveness of Wnt target genes is not due to the selective expression of TCFs. To examine whether the differential Wnt responsiveness of Cdx1 and T/Bra is linked to distinct functional properties of TCFs, we analyzed the abilities of the different TCF family members to induce the expression of luciferase reporter genes harboring genomic DNA fragments from the Axin2, Cdx1, and T/Bra genes (Fig. 1). The results of these experiments revealed that cellular background and promoter-specific determinants appear to influence the transactivation capacities of TCF family members and further confirm that TCF family members are functionally distinct (see Fig. S1 in the supplemental material). However, we observed that, in contrast to the endogenous genes which were refractory to Wnt pathway activation, the transiently transfected Cdx1 and T/Bra reporter genes were responsive to the expression of β-catenin and TCF family members. In view of this diverging behavior, we therefore wished to determine whether TCFs were actually present at the promoter regions of the endogenous Axin2, Cdx1, and T/Bra genes in our cellular models. To this end, we initially performed ChIP with antibodies against TCF4 (Fig. 3). Semiquantitative or quantitative real-time PCR was used to monitor the enrichment of specific promoter sequences in precipitated DNA and revealed that the Axin2 gene is occupied by TCF4 in both SB-216763-treated and control C17.2, C2C12, and E14 cells (Fig. 3; Fig. 1 shows primer locations). In E14 cells, where the Cdx1 and T/Bra genes are Wnt responsive, TCF4 also occupies the Cdx1 and T/Bra promoters, again in a stimulus-independent manner. TCF4 is also present at TBE6 of the intronic enhancer of the Cdx1 gene (29). Even more importantly, TCF4 is not found at functionally irrelevant locations around −3 kb and −2 kb upstream of the Axin2 transcriptional start site and at −2 kb and −1 kb, respectively, upstream of the Cdx1 and T/Bra promoter regions. Furthermore, TCF4 is not associated with the nonresponsive Cdx1 and T/Bra promoters in C17.2 and C2C12 cells. Quantitative analysis revealed stronger enrichment of the Axin2 promoter DNA relative to Cdx1 and T/Bra in E14 cells (Fig. 3B). This could indicate that TCF4 is bound more frequently to the Axin2 promoter than to the Cdx1 and T/Bra promoter regions. Interestingly, the inhibition of GSK3 appeared to increase the binding of TCF4 to the TBE3 region of the Axin2 gene, but this was observed for C17.2 cells only.
FIG. 3.
Promoters of Wnt-inducible target genes are occupied by endogenous TCF4 irrespective of pathway activity. (A) ChIP analysis for TCF4 in C17.2, C2C12, or E14 cells. Cells were stimulated for 24 h with 25 μM (C17.2, C2C12) or 5 μM (E14) SB-216763 or with DMSO as the solvent control. Formaldehyde-fixed chromatin was precipitated using 2 μg of a TCF4-specific antibody or 2 μg of a nonspecific goat IgG as control. Precipitated DNA was analyzed by PCR using primer sets indicated in Fig. 1 with close proximity to TCF-binding elements in the Axin2, Cdx1, and T/Bra promoters. Primers within the coding region of the GAPDH gene, which is not regulated by TCFs, were used to monitor nonspecific precipitation. Control PCRs received no template (no templ.) or genomic DNA as the positive control. (B) Quantitative real-time PCR of ChIP DNA from C17.2, C2C12, and E14 cells as described above. For each gene, precipitated DNA was analyzed using multiple primer sets. Numbers refer to nucleotide positions of the amplicons relative to the transcriptional start site as indicated in Fig. 1. Bars represent relative enrichment compared to that of the nonspecific isotype control (relative enrichment of 1). Data given are the average and SEM from at least three experiments. α-, anti-.
Promoter occupation by the different TCF family members is limited to Wnt-responsive target genes.
Unfortunately, we were limited to TCF4 in the analysis of promoter occupation by endogenous TCF factors due to lack of antibodies which do not cross-react among TCF family members and which are functional in ChIP (S. Wöhrle and A. Hecht, unpublished observation). Nevertheless, to be able to comparatively analyze the bindings of different TCF family members to target gene promoters, we generated stable cell lines expressing HA-tagged TCFs. These cell lines showed low to moderate increases in the overall levels of the respective TCF family members (see Fig. S2 in the supplemental material). Individual cell clones were selected for further analyses based on similar expression levels of ectopic TCF1E, LEF1, TCF3, and TCF4E (see Fig. S2 in the supplemental material). To validate our strategy, we carried out ChIP analyses with TCF4-HA cells derived from C17.2, C2C12, and E14 ES cells using both anti-TCF4 and anti-HA antibodies. In each case, cell-type-specific and differential promoter occupation were preserved, and similar results were obtained with the anti-TCF4 and anti-HA antibodies (data not shown). Similarly, the cell-type-specific Wnt responsiveness of Axin2, Cdx1, and T/Bra was maintained in the presence of ectopic TCFs, even though expression levels and degrees of inducibility under Wnt-responsive conditions varied, perhaps due to clonal variation (see Fig. S3 in the supplemental material). Having validated the experimental approach, we extended the analyses to cell lines expressing HA-tagged versions of the other TCF family members. As it turns out, just like TCF4, LEF1, TCF1, and TCF3 also occupy target genes only when they are in a Wnt-responsive state (Axin2 in all cell clones, Cdx1 and T/Bra in E14 cell clones) (Fig. 4). Notably, none of the TCFs associated with nonresponsive promoter regions such as Cdx1 or T/Bra in C17.2 and C2C12 cells. Moreover, TCF family members differ in their abilities to associate even with active target promoters. LEF1 and TCF3 showed weak promoter binding compared to TCF1 and TCF4, which had exceeded LEF1 and TCF3 also with respect to their transactivation capacities in transient reporter gene assays (see Fig. S1 in the supplemental material). Particularly striking is the apparent absence of TCF3 from the Axin2 promoter in C17.2 cells and the T/Bra promoter in C17.2, C2C12, and E14 cells. Similarly, LEF1 displayed comparatively weaker Axin2 promoter association in C17.2 cells and was not present at the T/Bra promoter in C17.2, C2C12, and E14 cells. In summary, these results further support cell-type- and promoter-specific functional differences among TCFs and reveal TCF-specific profiles of promoter occupancy. Furthermore, TCF family members are not constantly bound to promoter regions of their target genes and are not able to access nonresponsive genes.
FIG. 4.
Promoter occupancy by different TCF family members is limited to Wnt-responsive target genes. ChIP analysis was performed in C17.2, C2C12, or E14 cells stably transfected with expression vectors for HA-tagged TCFs. Chromatin was precipitated using 2 μg of a rat anti-HA (α-HA) antibody or equal amounts of the corresponding isotype control. Precipitated DNA was analyzed by quantitative real-time PCR using primer sets within the Axin2, Cdx1, and T/Bra promoter regions. Nucleotide positions of the amplicons relative to the transcriptional start site are indicated (Fig. 1). Bars represent relative enrichment compared to that of the nonspecific isotype control (relative enrichment of 1). Data given are the average and SEM from at least four experiments.
Differential Wnt inducibility correlates with distinct epigenetic states.
Contrasting inducibilities of episomal and endogenous Cdx1 and T/Bra promoters in C17.2 and C2C12 cells and conditional promoter occupancy by TCFs hint that a chromosomal context and epigenetic modifications are additional critical determinants of Wnt target gene control. As mentioned above (Fig. 1), the binding sites for TCF factors and the transcriptional start sites of the Axin2, Cdx1, and T/Bra genes lie within or in close proximity to CpG islands and thus could potentially be subject to regulation by DNA methylation. Bisulfite conversion and sequencing of PCR-amplified genomic DNA were used to analyze the DNA methylation patterns of the Axin2, Cdx1, and T/Bra promoters in C17.2, C2C12, and E14 cells treated with or without the GSK3 inhibitor to activate canonical Wnt signaling (Fig. 5A). In agreement with its ubiquitous inducibility, the Axin2 promoter is only marginally methylated in all three cell lines. Likewise, we observed little or no methylation of the Cdx1 and T/Bra promoters in E14 cells. In contrast, both promoters show a high or intermediate degree of methylation in C17.2 and C2C12 cells, in which they are refractory to Wnt stimulation. Regardless of which gene was analyzed, the induction of the Wnt pathway by the inhibition of GSK3 had no effect on the methylation status in all three cell lines under investigation.
FIG. 5.
Differential Wnt inducibility correlates with distinct epigenetic states of the corresponding target genes. (A) Analysis of DNA methylation patterns at the Axin2, Cdx1, and T/Bra promoters. Genomic DNA was isolated from C17.2, C2C12, or E14 cells after treatment for 24 h with 25 μM (C17.2, C2C12) or 5 μM (E14) SB-216763 or with DMSO as the solvent control. Bisulfite-treated DNA fragments of the promoter regions were PCR amplified with primer pairs shown in Fig. 1, subcloned, and analyzed by sequencing. Each line represents the results from a single sequence and each circle denotes a CpG dinucleotide (filled circles, methylated CpGs; open circles, nonmethylated CpGs). (B) ChIP analysis for acetylated histone H3 in C17.2, C2C12, or E14 cells. Formaldehyde-fixed chromatin was prepared after treatment with 25 μM (C17.2, C2C12) or 5 μM (E14) SB-216763 or with DMSO as the solvent control for 24 h. Chromatin was precipitated with 2 μg of an anti-acetylated histone H3 (α-H3ac) antibody or equal amounts of a nonspecific rabbit isotype control. ChIP DNA was analyzed by quantitative real-time PCR with primer pairs within the Axin2, Cdx1, and T/Bra promoter regions. Nucleotide positions of the amplicons relative to the transcriptional start site are indicated (Fig. 1). Bars represent relative enrichment compared to that of the nonspecific isotype control (relative enrichment of 1). Data given are the average and SEM from at least three experiments.
Histone modifications are further epigenetic determinants of gene activity (52). We therefore examined whether differential Wnt inducibility of the Axin2, Cdx1, and T/Bra genes is accompanied by the presence of active or repressive histone marks by use of ChIP with antibodies directed against histone H3 acetylated at lysines 9 and 14 (H3K9/14) or trimethylated at lysines 4, 9, or 27 (H3K4, H3K9, or K3K27, respectively). DNA precipitated from SB-216763-treated or control cells was analyzed by quantitative real-time PCR. Figure 5B shows the results of the analyses for histone H3 acetylation. This modification is associated with sequences immediately upstream and downstream of the transcriptional start site of the Axin2 gene in all cell lines tested. Similarly, T/Bra promoter regions are enriched in material precipitated with antibodies against acetylated H3 from E14 ES cells, whereas distal Axin2 and T/Bra sequences appear to be devoid of acetylated H3. Furthermore, the active mark is absent from the inactive T/Bra locus in C17.2 and C2C12 cells. There is also a weak association of acetylated H3 with different regions around the Cdx1 promoter which is most pronounced in E14 ES cells and less evident in C2C12 and C17.2 cells (Fig. 5B). Interestingly, acetylated H3 is present at transcriptionally poised Axin2, Cdx1, and T/Bra promoters even in unstimulated cells, and the association shows no or at best a slight increase upon Wnt pathway activation.
Trimethylated H3K4 is another histone modification characteristic of chromatin of transcriptionally poised or active genes (52, 65). Consistent with this, we find high levels of trimethylated H3K4 around the promoter but not at far upstream regions of the Axin2 gene in E14 ES cells, C2C12 cells, and C17.2 cells (Fig. 6). Trimethylated H3K4 is also present in the immediate vicinity of the transcriptional start sites of the Wnt-responsive Cdx1 and T/Bra genes in ES cells (Fig. 6). In contrast, trimethylated H3K4 shows only much-reduced or no association with Cdx1 and T/Bra chromatin when the genes are in a transcriptionally inactive and nonresponsive state as in C2C12 and C17.2 cells. Like H3 acetylation, methylation of H3K4 is largely stimulus independent and appears to be affected by Wnt pathway activation only at the Cdx1 gene in ES cells, where its levels increase approximately twofold at the promoter and within the transcribed region. To further extend our analyses of epigenetic modifications at Wnt target genes, we monitored the occurrence of trimethylated H3K9 (H3K9me3) and H3K27 (H3K27me3) at the Axin2, Cdx1, and T/Bra genes in our cellular model systems. However, ChIP experiments with antibodies against H3K9me3 were not informative. Also, little or no association of Axin2 and Cdx1 sequences with H3K27me3 was found in C2C12 and C17.2 cells, and only low levels of H3K27me3 were observed at the Cdx1 and T/Bra genes in ES cells (see Fig. S4 in the supplemental material). In contrast, T/Bra chromatin is strongly enriched for H3K27me3 in C17.2 and C2C12 cells (see Fig. S4 in the supplemental material). Apparently, H3K27me3 is specific for the nonresponsive state of the T/Bra locus. Taken together, the Wnt-responsive state appears to be characterized by a stimulus-independent presence of acetylated histone H3 and methylated H3K4 and DNA hypomethylation of the corresponding promoter regions. Conversely, when found in the nonresponsive state, Wnt target genes display DNA hypermethylation, and active histone marks are largely absent.
FIG. 6.
Wnt responsiveness is paralleled by the cell-type-specific occurrence of methylated H3K4 at target gene promoters. ChIP analysis for methylated histone H3K4 in C17.2, C2C12, or E14 cells. Formaldehyde-fixed chromatin was prepared after treatment with 25 μM (C17.2, C2C12) or 5 μM (E14) SB-216763 or with DMSO as the solvent control for 24 h. Chromatin was precipitated with 2 μg of an antibody recognizing trimethylated histone H3K4 (α-H3K4me3) or equal amounts of a nonspecific rabbit isotype control. ChIP DNA was analyzed by quantitative real-time PCR with primer pairs within the Axin2, Cdx1, and T/Bra promoter regions. Nucleotide positions of the amplicons relative to the transcriptional start site are indicated (Fig. 1). Bars represent relative enrichment compared to that of the nonspecific isotype control (relative enrichment of 1). Data given are the average and SEM from at least three experiments.
The nonresponsive state of Wnt target genes is unaffected by the inhibition of histone deacetylases and DNA methylases.
DNA methylation and histone modifications correlate with differential Wnt inducibility and occupation of the Axin2, Cdx1, and T/Bra promoters by TCFs and thus may be involved in setting up responsive and nonresponsive states of the Cdx1 and T/Bra genes in C17.2 and C2C12 cells. If so, interfering with the epigenetic status of target gene promoters might establish Wnt inducibility. To test this, we analyzed expression and Wnt inducibility as well as TCF promoter occupancy upon the inhibition of histone deacetylases by TSA and DNA methyltransferases by 5-aza-2′-deoxycytidine (Aza) in C17.2 cells (Fig. 7). Treatment with TSA or Aza had no effect on Axin2 expression and did not interfere with its Wnt inducibility (Fig. 7A). Cdx1 expression was also not affected and remained refractory to Wnt3a in the presence of TSA or Aza. Similarly, Aza had no effect on T/Bra expression. Therefore, to demonstrate the efficacy of the Aza treatment, we monitored levels of DNA methylation by bisulfite conversion and genomic sequencing. Furthermore, because DNA methyltransferases and histone-modifying enzymes form an intricate network of epigenetic regulators (65, 75, 76), ChIP experiments were performed to analyze potential changes in acetylation of H3K9/14 and methylation of H3K27 upon Aza treatment. However, despite efficiently lowering DNA methylation levels (compare Fig. 7B, right, to 5A), Aza did not affect the various histone modifications present at the Axin2, Cdx1, and T/Bra genes in C17.2 cells (data not shown). Similarly, Aza treatment did not allow for promoter occupancy by TCF4 (data not shown).
FIG. 7.
Nonresponsive Wnt target genes remain refractory to Wnt induction upon inhibition of histone deacetylases or DNA methyltransferases. (A) TSA increases the basal expression level but does not restore the Wnt inducibility of the T/Bra gene. C17.2 cells were treated for 24 h with 0.5 μM TSA or 5 μM Aza in combination with 50 ng/ml Wnt3a purified from conditioned medium (lanes 1 to 6) as indicated. Total RNA was isolated, and the activation of the Wnt target genes Axin2, Cdx1, and T/Bra was analyzed by RT-PCR. Amplification of GAPDH was used to control sample integrity and loading. Control reactions were done as described for Fig. 2A. (B) DNA methylation analysis of the T/Bra promoter in C17.2 cells upon TSA and Aza treatment. Bisulfite-treated DNA was amplified by PCR with primers indicated in Fig. 1 and analyzed as described before (Fig. 5). (C) Treatment with TSA increases levels of acetylated histone H3. Immunoprecipitations with anti-acetylated H3 (α-acH3) antibodies were performed with nuclear lysates from C17.2 cells treated for 24 h with 0.5 μM TSA, 5 μM Aza, or DMSO as the solvent control. A fraction of the nuclear (nucl.) lysates (2.5%) and the immunoprecipitates were analyzed by immunoblotting with anti-acetylated H3 antibodies. Coomassie brilliant blue staining of nuclear lysates separated by SDS-PAGE was used to confirm equal loading. (D) Inhibition of histone deacetylases does not alter the epigenetic states and TCF promoter occupancy of Wnt target genes. ChIP analysis for TCF4, acetylated histone H3, and methylated histone H3K27 in TSA-treated C17.2 cells. Formaldehyde-fixed chromatin was prepared from C17.2 cells after treatment with 1 μM TSA or DMSO as the solvent control for 24 h. Chromatin was precipitated using 2 μg of a TCF4-specific antibody, antibodies to acetylated histone H3 (2 μg) or trimethylated histone H3K27 (α-H3K27me3) (4 μg), or equivalent amounts of the corresponding isotype controls. Precipitated DNA was analyzed by quantitative real-time PCR using primer sets within the promoter regions of the Axin2, Cdx1, and T/Bra genes. Nucleotide positions of the amplicons relative to the transcriptional start site are indicated (Fig. 1). Bars represent relative enrichment compared to that of the nonspecific isotype controls (relative enrichment of 1). Data given are the average and SEM from at least three experiments.
Whereas Aza proved to be ineffective, TSA increased the basal expression of T/Bra (Fig. 7A, compare lanes 1 and 3), albeit this did not render the promoter Wnt inducible (Fig. 7A, compare lanes 3 and 4). The efficiency of TSA treatment was confirmed by Western blotting and immunoprecipitation (Fig. 7C). Since histone-modifying enzymes can interact with DNA methyltransferases (75, 76), we additionally analyzed whether TSA altered T/Bra promoter methylation in C17.2 cells. This was not the case (compare Fig. 7B, left, to 5A). These findings rule out that decreased promoter methylation is responsible for the enhanced expression of T/Bra upon TSA treatment and together with the results described above indicate that low levels of DNA methylation per se are not sufficient to alleviate the transcriptional repression of Wnt target genes.
ChIP analysis was performed to test whether the acetylation or methylation of H3 and the binding of TCF4 to the Axin2, Cdx1, and T/Bra promoter regions were altered in C17.2 cells treated with TSA (Fig. 7D). Although global levels of H3 acetylation were clearly elevated (Fig. 7C), TSA did not increase the levels of acetylated H3 at the Cdx1 promoter. Irrespective of its effects on the basal transcription of the T/Bra gene, TSA also failed to alter the acetylation of H3 at multiple locations surrounding this gene (Fig. 7D). In contrast, we observed a twofold decrease in methylated H3K27 at −2 kb and −1 kb upstream of the transcriptional start site of the T/Bra gene (Fig. 7D). However, this effect appears to be locally restricted and was not seen at the promoter-proximal TBEs or within the transcribed region of the gene (Fig. 7D). In agreement with this, TSA treatment did not lead to the occupancy of the Cdx1 and T/Bra promoters by TCF4 (Fig. 7D). This is consistent with our observation that TSA-mediated expression of T/Bra remains resistant to Wnt stimulation. On the other hand, the occupancy of the Axin2 promoter by TCF4 and its association with acetylated H3 (Fig. 7D) remained unchanged under these conditions, showing that TSA and Aza do not generally impair TCF4 activity. Thus, Aza and TSA treatment appear to be accompanied by a certain reorganization of promoter chromatin which allows basal level expression of T/Bra, while the resulting structure is still not permissive for TCF binding and Wnt-inducible transcription of nonresponsive promoters. Nonetheless, epigenetic factors and chromatin structure appear to contribute to Wnt target gene control, as indicated by the TSA effects on T/Bra expression and H3K27 trimethylation.
DISCUSSION
The molecular basis of stage- and tissue-specific transcriptional control under the influence of a limited number of signal transduction pathways and their nuclear effectors is only poorly understood. Here, we have used canonical Wnt signaling as a paradigm to investigate mechanisms of differential gene expression. One of our key findings is that TCF family members differ dramatically with respect to their transactivation potentials and to their abilities to bind to target gene promoters in vivo. Two developmentally regulated Wnt target genes are occupied by TCFs only when they are in an active and Wnt-responsive state. In cell types where expression is uninducible, their promoter regions are devoid of bound TCFs. This result differs from what might have been expected according to current literature models (3, 18, 71, 79). On the basis of genetic analyses in Drosophila melanogaster and Xenopus laevis which had revealed a dual function of TCFs in both the activation and the repression of Wnt target genes (15, 16, 64) and the results of ChIP experiments showing Wnt-independent occupancy of the c-Myc, cyclin D1, and Suz12 promoters by TCFs in different cellular backgrounds (41, 66), these models imply constant promoter occupancy by TCFs. However, the present conceptual framework for canonical Wnt signaling does not exclude the possibility that promoter occupancy by TCFs is restricted to specific functional states of their target genes, and at present it is not known whether TCFs are permanently present at all target genes and in all tissues. Our findings show that this is not the case and suggest that developmentally regulated Wnt target genes can be temporarily or permanently withdrawn from Wnt signaling in a cell-type-specific way by excluding TCFs from their DNA-binding elements.
Due to largely identical DNA-binding specificities and their uniform interaction with β-catenin, for a long time the prevailing view was that TCF family members are functionally interchangeable effectors of Wnt signaling. However, evidence is accumulating that TCFs perform divergent functions (6, 26, 30, 31, 49, 53, 58, 63, 72). Our results further extend these findings. Interestingly, the functional properties of TCF family members vary considerably with cellular background and promoter context. TCF1E and TCF4E contain context-dependent transactivation domains which influence DNA binding and help to recruit coactivators (3, 31). Physical interactions enable LEF1 to synergize with DNA-binding proteins of the Smad family at promoters with binding sites for both LEF1 and Smads (43, 59). Cell-specific expression of cofactors and promoter-specific configurations of DNA-binding elements thus might explain some of the observed functional specificities. The finding that T/Bra is apparently not a preferred target of LEF1 was unexpected because only LEF1/TCF1 doubly homozygous mutant animals show a Wnt3a−/−-like phenotype (26). However, instead of interchangeably controlling the same genes, LEF1 and TCF1 may attend to different sets of targets, and phenotypic changes upon combined deletion of TCF1 and LEF1 could be due to cumulative effects. Alternatively, they could additionally be backed up by TCF4, which would also be consistent with our results. Regardless of the actual relationships, it will be interesting to determine the exact range of target genes for individual TCFs.
We find that the transcriptionally active and Wnt-responsive states of Axin2, Cdx1, and T/Bra promoters are characterized by low or absent methylation of CpG islands and stimulus-independent association of promoter DNA with acetylated H3K9/14 and methylated H3K4, as well as selective occupancy with activation-competent TCF family members. In contrast, TCFs are not associated with nonresponsive targets which additionally are characterized by DNA hypermethylation and the lack of active chromatin marks. Unlike the constant levels of H3 acetylation and H3K4 methylation at the Axin2, Cdx1, and T/Bra genes which we observed and which apparently are not affected by Wnt pathway activity, alterations of the H3K4 methylation state have previously been described at the cell cycle control gene c-Myc in response to alterations of APC and β-catenin activity (66). However, this study by Sierra and colleagues differs from ours with respect to the experimental settings, the biological function of the gene investigated, and the time course of its transcriptional induction, which may explain the divergent chromatin structural dynamics at the c-Myc gene. A certain degree of variability of epigenetic features which accompany distinct functional states of Wnt target genes is additionally revealed by repressive H3K27 trimethylation. H3K27me3 is present at low levels in chromatin of the Cdx1 and T/Bra genes in ES cells. However, only the T/Bra locus shows association with high levels of H3K27me3 in C17.2 and C2C12 cells. Also, the functional significance of H3K27me3 is unclear. In ES cells, the Cdx1 and T/Bra genes appear to reside in bivalent chromatin domains characterized by the simultaneous presence of methylated H3K4 and H3K27 (this study and references 10 and 55). However, the concomitant presence of H3K27me3 does not interfere with Wnt inducibility of the T/Bra and Cdx1 genes in ES cells. Furthermore, ES cells with lowered levels of H3K27me3 show reduced expression of T/Bra during differentiation (60), which is the opposite of what one would expect if H3K27me3 served to repress T/Bra expression. In addition, despite different states of H3K27 methylation, both the T/Bra and the Cdx1 gene are refractory to Wnt stimulation in C17.2 and C2C12 cells. Clearly, further experimentation is needed to clarify the role of H3K27 methylation in the differential regulation of Wnt target genes.
In case of β-catenin/TCF-regulated genes, Wnt responsiveness, i.e., the ability of a Wnt growth factor to elicit an acute transcriptional response, appears to critically depend on the ability of TCFs to access and occupy corresponding promoter elements. This is suggested by the lack of Wnt inducibility of the Cdx1 and T/Bra genes in the absence of promoter-bound TCFs. The observation that the Cdx1 and T/Bra promoters respond to TCFs and β-catenin in transient transfections argues that indeed the limiting factor in order to confer Wnt responsiveness is promoter occupancy by TCFs. Apparently, all trans-acting factors required for their transcriptional induction are present even in cells in which the endogenous Cdx1 and T/Bra promoters are unavailable for the Wnt pathway. On the other hand, the differential occupation of the Axin2, Cdx1, and T/Bra promoters by TCFs within the same cell type indicates that promoter accessibility for TCFs is not controlled in a global but rather in a gene-specific manner. The molecular mechanisms underlying this are not known. Based on our findings, local chromatin structure appears to play a role in these processes, but the precise causal relationships between different levels of DNA methylation, histone modification, and TCF promoter occupancy remain to be established. Alterations in epigenetic modifications may be a consequence of the absence or presence of TCFs from respective promoter regions. Alternatively, the acquisition of different chromatin structural features may be independent of promoter occupancy by TCFs. Support for the latter hypothesis comes from the observation that TCF family members are unable to access the nonresponsive Cdx1 and T/Bra genes even when overexpressed, and the cell type specificity of these genes is maintained. Moreover, the chromatin states of Cdx1 and T/Bra are similar in E14 ES cells, but they differ with respect to H3K27 trimethylation in C17.2 and C2C12 cells. This gene-specific divergence suggests that the absence of TCFs does not by default lead to the uniform appearance of H3K27 trimethylation and that distinct chromatin structural states are not merely a consequence of the presence or absence of TCFs. In fact, epigenetic determinants of chromatin structure might establish conditions which are either permissive or nonpermissive for the binding of selected TCF family members, although this idea needs further investigation. If able to access a promoter, TCF family members, based on their specific functional properties, confer Wnt inducibility and take over the fine-tuning of Wnt target gene control. In support of such a multilayered model of transcriptional control, it is known that canonical Wnt signaling does not in all cases appear to initiate the transcription of its targets from scratch but rather maintains expression after a priming event (4, 19, 27, 33). In this sense, the observation that H3 acetylation and H3K4 trimethylation at the Axin2, Cdx1, and T/Bra genes remained unchanged upon Wnt pathway actuation may simply reflect a preceding and Wnt-independent activation step leading to the opening of chromatin structure and thereby enable TCF association. A precedent for the activation of a gene by the sequential action of signaling cascades involving the restructuring of chromatin is provided by the GFAP locus. Here, fibroblast growth factor signaling leads to a locally confined switch from H3K9 to H3K4 methylation and thereby facilitates the binding of STAT transcription factors in response to CNTF stimulation (70). As an alternative, a priming event may also involve the action of a locus control region and bring about promoter accessibility for TCFs by promoting widespread changes of chromatin structure over extended regions at Wnt target loci (20). Interestingly, scenarios like these may be specific for Wnt targets involving TCF family members as binding partners of β-catenin, such as Axin2, Cdx1, T/Bra, and Survivin (51). An example diverging from the situation at these genes is provided by cyclin D2, where transcriptional activation following the inhibition of GSK3 is accompanied by increased acetylation of histones H3 and H4 (40). However, the induction of cyclin D2 expression occurs independently of TCF family members and instead is based on an interaction between β-catenin and PitX2, both of which are loaded simultaneously onto the cyclin D2 promoter upon Wnt stimulation (40). It thus appears that PitX2, unlike TCFs, does not require prior chromatin opening and is able to invade an inactive promoter region on its own.
Using inhibitors of histone deacetylases and DNA methyltransferases, we have attempted to impart Wnt responsiveness on the Cdx1 and T/Bra genes in C2C12 and C17.2 cells (B. Wallmen and A. Hecht, unpublished observation, and this study). However, in contrast to certain tumor suppressor genes which are inactivated by DNA methylation (36), Cdx1 and T/Bra could not be reactivated by Aza treatment. Unlike Aza treatment, which proved to be ineffective, the inhibition of histone deacetylases led to elevated basal level transcription of T/Bra and altered H3K27 trimethylation at regions distal to the promoter, although it did not restore Wnt inducibility, TCF binding, and H3 acetylation at the promoter. Combined exposure to TSA and Aza resulted in the same phenotype of T/Bra expression as TSA treatment alone (data not shown). The lack of Suz12 did not activate T/Bra expression during ES cell differentiation (60), and knockdown of Suz12 or treatment with the general methylase inhibitor MTA (70) did not lead to Wnt inducibility of T/Bra in neural cells (Wöhrle and Hecht, unpublished observation). However, epigenetic modifiers form complex and intricate regulatory networks (25, 68, 75, 76). For example, the histone methyltransferase complex PRC2 cooperates with DNA methyltransferases to promote de novo methylation of CpG dinucleotides (76). Similarly, in differentiating ES cells, the Oct3/4 locus is permanently inactivated by the sequential action of transcriptional repressors, histone-modifying enzymes, and DNA methylation (25). Because so far we have been able to target only a subset of epigenetic modifications and have not achieved their complete erasure, nonresponsive chromatin at Wnt target genes may not have been sufficiently destabilized to allow TCF binding. On the other hand, it cannot be excluded that the actual loading of TCFs still depends on the initial activation of target genes by an appropriate priming event, even under conditions where chromatin structure has become permissive.
The need to selectively target only a subset of all potential target genes in a particular cell type and at a specific developmental stage is not a problem unique to Wnt signaling. The Notch and Hedgehog signal cascades are widely used in developmental processes and elicit distinct transcriptional responses in a context-dependent manner (23, 38). Target genes of both pathways toggle between repressed and activated states in ways very similar to those seen for Wnt target genes. To some extent, mechanisms underlying tissue- and stage-specific transcriptional control may be inherent properties of signal transduction processes and pathway components. A single type of activator usually is insufficient to induce gene expression, and typically the functionality of regulatory elements depends on the combined and cooperative input of multiple regulators (9). To sharpen transcriptional responses and to prevent leaky expression due to the activity of a single activator default repression in the absence of the full activator complement appear to be another common feature in transcriptional control by extracellular signaling pathways and their effectors (9). In the case of canonical Wnt signaling, these principles can be illustrated by the dual activator/repressor activity of TCFs (15, 16, 64) and their ability to cooperate with other types of proteins, for instance, Smad proteins (43, 59, 62). Beyond this, our findings add chromatin structural changes as an additional control layer operating in conjunction with signal transducers in order to generate selective transcriptional responses. The principle of multilayered transcriptional control as exemplified by Wnt/β-catenin targets may serve as a paradigm for several other signaling pathways. Thus, epigenetic mechanisms may turn out to be even more widely used to endow separate groups of genes with differential capacities to respond to developmentally relevant signal transduction pathways and their nuclear effectors.
Supplementary Material
Acknowledgments
We are grateful to K. Bruser and M. Schrempp for excellent technical assistance and to R. Cassada for critical reading of the manuscript.
This work was supported by DFG research grants He 2004/4 and He 2004/6 to A. Hecht.
Footnotes
Published ahead of print on 8 October 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Akiyama, H., J. P. Lyons, Y. Mori-Akiyama, X. Yang, R. Zhang, Z. Zhang, J. M. Deng, M. M. Taketo, T. Nakamura, R. R. Behringer, P. D. McCrea, and B. de Crombrugghe. 2004. Interactions between Sox9 and beta-catenin control chondrocyte differentiation. Genes Dev. 18:1072-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoki, M., A. Hecht, U. Kruse, R. Kemler, and P. K. Vogt. 1999. Nuclear endpoint of Wnt signaling: neoplastic transformation induced by transactivating lymphoid-enhancing factor 1. Proc. Natl. Acad. Sci. USA 96:139-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arce, L., N. N. Yokoyama, and M. L. Waterman. 2006. Diversity of LEF/TCF action in development and disease. Oncogene 25:7492-7504. [DOI] [PubMed] [Google Scholar]
- 4.Arias, A. M., and P. Hayward. 2006. Filtering transcriptional noise during development: concepts and mechanisms. Nat. Rev. Genet. 7:34-44. [DOI] [PubMed] [Google Scholar]
- 5.Arnold, S. J., J. Stappert, A. Bauer, A. Kispert, B. G. Herrmann, and R. Kemler. 2000. Brachyury is a target gene of the Wnt/beta-catenin signaling pathway. Mech. Dev. 91:249-258. [DOI] [PubMed] [Google Scholar]
- 6.Atcha, F. A., J. E. Munguia, T. W. Li, K. Hovanes, and M. L. Waterman. 2003. A new beta-catenin-dependent activation domain in T cell factor. J. Biol. Chem. 278:16169-16175. [DOI] [PubMed] [Google Scholar]
- 7.Aulehla, A., C. Wehrle, B. Brand-Saberi, R. Kemler, A. Gossler, B. Kanzler, and B. G. Herrmann. 2003. Wnt3a plays a major role in the segmentation clock controlling somitogenesis. Dev. Cell 4:395-406. [DOI] [PubMed] [Google Scholar]
- 8.Barker, N., A. Hurlstone, H. Musisi, A. Miles, M. Bienz, and H. Clevers. 2001. The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation. EMBO J. 20:4935-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barolo, S., and J. W. Posakony. 2002. Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev. 16:1167-1181. [DOI] [PubMed] [Google Scholar]
- 10.Bernstein, B. E., T. S. Mikkelsen, X. Xie, M. Kamal, D. J. Huebert, J. Cuff, B. Fry, A. Meissner, M. Wernig, K. Plath, R. Jaenisch, A. Wagschal, R. Feil, S. L. Schreiber, and E. S. Lander. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125:315-326. [DOI] [PubMed] [Google Scholar]
- 11.Billin, A. N., H. Thirlwell, and D. E. Ayer. 2000. β-Catenin-histone deacetylase interactions regulate the transition of LEF1 from a transcriptional repressor to an activator. Mol. Cell. Biol. 20:6882-6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyd, K. E., J. Wells, J. Gutman, S. M. Bartley, and P. J. Farnham. 1998. c-Myc target gene specificity is determined by a post-DNA binding mechanism. Proc. Natl. Acad. Sci. USA 95:13887-13892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyer, L. A., K. Plath, J. Zeitlinger, T. Brambrink, L. A. Medeiros, T. I. Lee, S. S. Levine, M. Wernig, A. Tajonar, M. K. Ray, G. W. Bell, A. P. Otte, M. Vidal, D. K. Gifford, R. A. Young, and R. Jaenisch. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441:349-353. [DOI] [PubMed] [Google Scholar]
- 14.Bracken, A. P., N. Dietrich, D. Pasini, K. H. Hansen, and K. Helin. 2006. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 20:1123-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brannon, M., M. Gomperts, L. Sumoy, R. T. Moon, and D. Kimelman. 1997. A beta-catenin/XTcf-3 complex binds to the siamois promoter to regulate dorsal axis specification in Xenopus. Genes Dev. 11:2359-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cavallo, R. A., R. T. Cox, M. M. Moline, J. Roose, G. A. Polevoy, H. Clevers, M. Peifer, and A. Bejsovec. 1998. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature 395:604-608. [DOI] [PubMed] [Google Scholar]
- 17.Chen, G., J. Fernandez, S. Mische, and A. J. Courey. 1999. A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev. 13:2218-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clevers, H. 2006. Wnt/beta-catenin signaling in development and disease. Cell 127:469-480. [DOI] [PubMed] [Google Scholar]
- 19.Danielian, P. S., and A. P. McMahon. 1996. Engrailed-1 as a target of the Wnt-1 signalling pathway in vertebrate midbrain development. Nature 383:332-334. [DOI] [PubMed] [Google Scholar]
- 20.Dean, A. 2006. On a chromosome far, far away: LCRs and gene expression. Trends Genet. 22:38-45. [DOI] [PubMed] [Google Scholar]
- 21.de la Roche, M., and M. Bienz. 2007. Wingless-independent association of Pygopus with dTCF target genes. Curr. Biol. 17:556-561. [DOI] [PubMed] [Google Scholar]
- 22.Duprey, P., K. Chowdhury, G. R. Dressler, R. Balling, D. Simon, J. L. Guenet, and P. Gruss. 1988. A mouse gene homologous to the Drosophila gene caudal is expressed in epithelial cells from the embryonic intestine. Genes Dev. 2:1647-1654. [DOI] [PubMed] [Google Scholar]
- 23.Ehebauer, M., P. Hayward, and A. Martinez-Arias. 2006. Notch signaling pathway. Sci. STKE 2006:cm7. [DOI] [PubMed] [Google Scholar]
- 24.Essers, M. A., L. M. de Vries-Smits, N. Barker, P. E. Polderman, B. M. Burgering, and H. C. Korswagen. 2005. Functional interaction between beta-catenin and FOXO in oxidative stress signaling. Science 308:1181-1184. [DOI] [PubMed] [Google Scholar]
- 25.Feldman, N., A. Gerson, J. Fang, E. Li, Y. Zhang, Y. Shinkai, H. Cedar, and Y. Bergman. 2006. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat. Cell Biol. 8:188-194. [DOI] [PubMed] [Google Scholar]
- 26.Galceran, J., I. Farinas, M. J. Depew, H. Clevers, and R. Grosschedl. 1999. Wnt3a−/−-like phenotype and limb deficiency in Lef1(−/−)Tcf1(−/−) mice. Genes Dev. 13:709-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galceran, J., S. C. Hsu, and R. Grosschedl. 2001. Rescue of a Wnt mutation by an activated form of LEF-1: regulation of maintenance but not initiation of Brachyury expression. Proc. Natl. Acad. Sci. USA 98:8668-8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallant, P. 2007. Control of transcription by Pontin and Reptin. Trends Cell Biol. 17:187-192. [DOI] [PubMed] [Google Scholar]
- 29.Gaunt, S. J., D. Drage, and A. Cockley. 2003. Vertebrate caudal gene expression gradients investigated by use of chick cdx-A/lacZ and mouse cdx-1/lacZ reporters in transgenic mouse embryos: evidence for an intron enhancer. Mech. Dev. 120:573-586. [DOI] [PubMed] [Google Scholar]
- 30.Gregorieff, A., R. Grosschedl, and H. Clevers. 2004. Hindgut defects and transformation of the gastro-intestinal tract in Tcf4(-/-)/Tcf1(-/-) embryos. EMBO J. 23:1825-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hecht, A., and M. P. Stemmler. 2003. Identification of a promoter-specific transcriptional activation domain at the C terminus of the Wnt effector protein T-cell factor 4. J. Biol. Chem. 278:3776-3785. [DOI] [PubMed] [Google Scholar]
- 32.Hecht, A., K. Vleminckx, M. P. Stemmler, F. van Roy, and R. Kemler. 2000. The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J. 19:1839-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heemskerk, J., S. DiNardo, R. Kostriken, and P. H. O'Farrell. 1991. Multiple modes of engrailed regulation in the progression towards cell fate determination. Nature 352:404-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hildinger, M., K. L. Abel, W. Ostertag, and C. Baum. 1999. Design of 5′ untranslated sequences in retroviral vectors developed for medical use. J. Virol. 73:4083-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirsch, C., L. M. Campano, S. Wohrle, and A. Hecht. 2007. Canonical Wnt signaling transiently stimulates proliferation and enhances neurogenesis in neonatal neural progenitor cultures. Exp. Cell Res. 313:572-587. [DOI] [PubMed] [Google Scholar]
- 36.Jaenisch, R., and A. Bird. 2003. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33:245-254. [DOI] [PubMed] [Google Scholar]
- 37.Jho, E. H., T. Zhang, C. Domon, C. K. Joo, J. N. Freund, and F. Costantini. 2002. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell. Biol. 22:1172-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jia, J., and J. Jiang. 2006. Decoding the Hedgehog signal in animal development. Cell. Mol. Life Sci. 63:1249-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kan, L., N. Israsena, Z. Zhang, M. Hu, L. R. Zhao, A. Jalali, V. Sahni, and J. A. Kessler. 2004. Sox1 acts through multiple independent pathways to promote neurogenesis. Dev. Biol. 269:580-594. [DOI] [PubMed] [Google Scholar]
- 40.Kioussi, C., P. Briata, S. H. Baek, D. W. Rose, N. S. Hamblet, T. Herman, K. A. Ohgi, C. Lin, A. Gleiberman, J. Wang, V. Brault, P. Ruiz-Lozano, H. D. Nguyen, R. Kemler, C. K. Glass, A. Wynshaw-Boris, and M. G. Rosenfeld. 2002. Identification of a Wnt/Dvl/beta-Catenin →Pitx2 pathway mediating cell-type-specific proliferation during development. Cell 111:673-685. [DOI] [PubMed] [Google Scholar]
- 41.Kirmizis, A., S. M. Bartley, and P. J. Farnham. 2003. Identification of the polycomb group protein SU(Z)12 as a potential molecular target for human cancer therapy. Mol. Cancer Ther. 2:113-121. [PubMed] [Google Scholar]
- 42.Kishida, S., H. Yamamoto, and A. Kikuchi. 2004. Wnt-3a and Dvl induce neurite retraction by activating Rho-associated kinase. Mol. Cell. Biol. 24:4487-4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Labbe, E., A. Letamendia, and L. Attisano. 2000. Association of Smads with lymphoid enhancer binding factor 1/T cell-specific factor mediates cooperative signaling by the transforming growth factor-beta and wnt pathways. Proc. Natl. Acad. Sci. USA 97:8358-8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee, T. I., R. G. Jenner, L. A. Boyer, M. G. Guenther, S. S. Levine, R. M. Kumar, B. Chevalier, S. E. Johnstone, M. F. Cole, K. Isono, H. Koseki, T. Fuchikami, K. Abe, H. L. Murray, J. P. Zucker, B. Yuan, G. W. Bell, E. Herbolsheimer, N. M. Hannett, K. Sun, D. T. Odom, A. P. Otte, T. L. Volkert, D. P. Bartel, D. A. Melton, D. K. Gifford, R. Jaenisch, and R. A. Young. 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125:301-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leung, J. Y., F. T. Kolligs, R. Wu, Y. Zhai, R. Kuick, S. Hanash, K. R. Cho, and E. R. Fearon. 2002. Activation of AXIN2 expression by beta-catenin-T cell factor. A feedback repressor pathway regulating Wnt signaling. J. Biol. Chem. 277:21657-21665. [DOI] [PubMed] [Google Scholar]
- 46.Li, L. C., and R. Dahiya. 2002. MethPrimer: designing primers for methylation PCRs. Bioinformatics 18:1427-1431. [DOI] [PubMed] [Google Scholar]
- 47.Lickert, H., C. Domon, G. Huls, C. Wehrle, I. Duluc, H. Clevers, B. I. Meyer, J. N. Freund, and R. Kemler. 2000. Wnt/(beta)-catenin signaling regulates the expression of the homeobox gene Cdx1 in embryonic intestine. Development 127:3805-3813. [DOI] [PubMed] [Google Scholar]
- 48.Lickert, H., and R. Kemler. 2002. Functional analysis of cis-regulatory elements controlling initiation and maintenance of early Cdx1 gene expression in the mouse. Dev. Dyn. 225:216-220. [DOI] [PubMed] [Google Scholar]
- 49.Liu, F., O. van den Broek, O. Destree, and S. Hoppler. 2005. Distinct roles for Xenopus Tcf/Lef genes in mediating specific responses to Wnt/{beta}-catenin signalling in mesoderm development. Development 132:5375-5385. [DOI] [PubMed] [Google Scholar]
- 50.Logan, C. Y., and R. Nusse. 2004. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 20:781-810. [DOI] [PubMed] [Google Scholar]
- 51.Ma, H., C. Nguyen, K. S. Lee, and M. Kahn. 2005. Differential roles for the coactivators CBP and p300 on TCF/beta-catenin-mediated survivin gene expression. Oncogene 24:3619-3631. [DOI] [PubMed] [Google Scholar]
- 52.Margueron, R., P. Trojer, and D. Reinberg. 2005. The key to development: interpreting the histone code? Curr. Opin. Genet. Dev. 15:163-176. [DOI] [PubMed] [Google Scholar]
- 53.Merrill, B. J., U. Gat, R. DasGupta, and E. Fuchs. 2001. Tcf3 and Lef1 regulate lineage differentiation of multipotent stem cells in skin. Genes Dev. 15:1688-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meyer, B. I., and P. Gruss. 1993. Mouse Cdx-1 expression during gastrulation. Development 117:191-203. [DOI] [PubMed] [Google Scholar]
- 55.Mikkelsen, T. S., M. Ku, D. B. Jaffe, B. Issac, E. Lieberman, G. Giannoukos, P. Alvarez, W. Brockman, T.-K. Kim, R. P. Koche, W. Lee, E. Mendenhall, A. O'Donovan, A. Presser, C. Russ, X. Xie, A. Meissner, M. Wernig, R. Jaenisch, C. Nusbaum, E. S. Lander, and B. E. Bernstein. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448:553-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mosimann, C., G. Hausmann, and K. Basler. 2006. Parafibromin/Hyrax activates Wnt/Wg target gene transcription by direct association with beta-catenin/Armadillo. Cell 125:327-341. [DOI] [PubMed] [Google Scholar]
- 57.Mulholland, D. J., S. Dedhar, G. A. Coetzee, and C. C. Nelson. 2005. Interaction of nuclear receptors with the Wnt/beta-catenin/Tcf signaling axis: Wnt you like to know? Endocr. Rev. 26:898-915. [DOI] [PubMed] [Google Scholar]
- 58.Nguyen, H., M. Rendl, and E. Fuchs. 2006. Tcf3 governs stem cell features and represses cell fate determination in skin. Cell 127:171-183. [DOI] [PubMed] [Google Scholar]
- 59.Nishita, M., M. K. Hashimoto, S. Ogata, M. N. Laurent, N. Ueno, H. Shibuya, and K. W. Cho. 2000. Interaction between Wnt and TGF-beta signalling pathways during formation of Spemann's organizer. Nature 403:781-785. [DOI] [PubMed] [Google Scholar]
- 60.Pasini, D., A. P. Bracken, J. B. Hansen, M. Capillo, and K. Helin. 2007. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell. Biol. 27:3769-3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Riese, J., X. Yu, A. Munnerlyn, S. Eresh, S. C. Hsu, R. Grosschedl, and M. Bienz. 1997. LEF-1, a nuclear factor coordinating signaling inputs from wingless and decapentaplegic. Cell 88:777-787. [DOI] [PubMed] [Google Scholar]
- 63.Roel, G., F. S. Hamilton, Y. Gent, A. A. Bain, O. Destree, and S. Hoppler. 2002. Lef-1 and Tcf-3 transcription factors mediate tissue-specific Wnt signaling during Xenopus development. Curr. Biol. 12:1941-1945. [DOI] [PubMed] [Google Scholar]
- 64.Roose, J., M. Molenaar, J. Peterson, J. Hurenkamp, H. Brantjes, P. Moerer, M. van de Wetering, O. Destree, and H. Clevers. 1998. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature 395:608-612. [DOI] [PubMed] [Google Scholar]
- 65.Schuettengruber, B., D. Chourrout, M. Vervoort, B. Leblanc, and G. Cavalli. 2007. Genome regulation by polycomb and trithorax proteins. Cell 128:735-745. [DOI] [PubMed] [Google Scholar]
- 66.Sierra, J., T. Yoshida, C. A. Joazeiro, and K. A. Jones. 2006. The APC tumor suppressor counteracts beta-catenin activation and H3K4 methylation at Wnt target genes. Genes Dev. 20:586-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sinner, D., S. Rankin, M. Lee, and A. M. Zorn. 2004. Sox17 and beta-catenin cooperate to regulate the transcription of endodermal genes. Development 131:3069-3080. [DOI] [PubMed] [Google Scholar]
- 68.Smallwood, A., P. O. Esteve, S. Pradhan, and M. Carey. 2007. Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev. 21:1169-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Snyder, E. Y., D. L. Deitcher, C. Walsh, S. Arnold-Aldea, E. A. Hartwieg, and C. L. Cepko. 1992. Multipotent neural cell lines can engraft and participate in development of mouse cerebellum. Cell 68:33-51. [DOI] [PubMed] [Google Scholar]
- 70.Song, M. R., and A. Ghosh. 2004. FGF2-induced chromatin remodeling regulates CNTF-mediated gene expression and astrocyte differentiation. Nat. Neurosci. 7:229-235. [DOI] [PubMed] [Google Scholar]
- 71.Stadeli, R., R. Hoffmans, and K. Basler. 2006. Transcription under the control of nuclear Arm/beta-catenin. Curr. Biol. 16:R378-R385. [DOI] [PubMed] [Google Scholar]
- 72.Standley, H. J., O. Destree, M. Kofron, C. Wylie, and J. Heasman. 2006. Maternal XTcf1 and XTcf4 have distinct roles in regulating Wnt target genes. Dev. Biol. 289:318-328. [DOI] [PubMed] [Google Scholar]
- 73.Subramanian, V., B. Meyer, and G. S. Evans. 1998. The murine Cdx1 gene product localises to the proliferative compartment in the developing and regenerating intestinal epithelium. Differentiation 64:11-18. [DOI] [PubMed] [Google Scholar]
- 74.Takemaru, K. I., and R. T. Moon. 2000. The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression. J. Cell Biol. 149:249-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van der Vlag, J., and A. P. Otte. 1999. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat. Genet. 23:474-478. [DOI] [PubMed] [Google Scholar]
- 76.Vire, E., C. Brenner, R. Deplus, L. Blanchon, M. Fraga, C. Didelot, L. Morey, A. Van Eynde, D. Bernard, J. M. Vanderwinden, M. Bollen, M. Esteller, L. Di Croce, Y. de Launoit, and F. Fuks. 2006. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439:871-874. [DOI] [PubMed] [Google Scholar]
- 77.Wilkinson, D. G., S. Bhatt, and B. G. Herrmann. 1990. Expression pattern of the mouse T gene and its role in mesoderm formation. Nature 343:657-659. [DOI] [PubMed] [Google Scholar]
- 78.Willert, K., J. D. Brown, E. Danenberg, A. W. Duncan, I. L. Weissman, T. Reya, J. R. Yates III, and R. Nusse. 2003. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature 423:448-452. [DOI] [PubMed] [Google Scholar]
- 79.Willert, K., and K. A. Jones. 2006. Wnt signaling: is the party in the nucleus? Genes Dev. 20:1394-1404. [DOI] [PubMed] [Google Scholar]
- 80.Williams, R. L., D. J. Hilton, S. Pease, T. A. Willson, C. L. Stewart, D. P. Gearing, E. F. Wagner, D. Metcalf, N. A. Nicola, and N. M. Gough. 1988. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature 336:684-687. [DOI] [PubMed] [Google Scholar]
- 81.Yamaguchi, T. P., S. Takada, Y. Yoshikawa, N. Wu, and A. P. McMahon. 1999. T (Brachyury) is a direct target of Wnt3a during paraxial mesoderm specification. Genes Dev. 13:3185-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zorn, A. M., G. D. Barish, B. O. Williams, P. Lavender, M. W. Klymkowsky, and H. E. Varmus. 1999. Regulation of Wnt signaling by Sox proteins: XSox17 alpha/beta and XSox3 physically interact with beta-catenin. Mol. Cell 4:487-498. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.