Abstract
The forkhead transcription factor FoxM1 has been reported to regulate, variously, proliferation and/or spindle formation during the G2/M transition of the cell cycle. Here we define specific functions of FoxM1 during brain development by the investigation of FoxM1 loss-of-function mutations in the context of Sonic hedgehog (Shh)-induced neuroproliferation in cerebellar granule neuron precursors (CGNP). We show that FoxM1 is expressed in the cerebellar anlagen as well as in postnatal proliferating CGNP and that it is upregulated in response to activated Shh signaling. To determine the requirements for FoxM1 function, we used transgenic mice carrying conventional null alleles or conditionally targeted alleles in conjunction with specific Cre recombinase expression in CGNP or early neural precursors driven by Math1 or Nestin enhancers. Although the overall cerebellar morphology was grossly normal, we observed that the entry into mitosis was postponed both in vivo and in Shh-treated CGNP cultures. Cell cycle analysis and immunohistochemistry with antibodies against phosphorylated histone H3 indicated a significant delay in the G2/M transition. Consistent with this, FoxM1-deficient CGNP showed decreased levels of the cyclin B1 and Cdc25b proteins. Furthermore, the loss of FoxM1 resulted in spindle defects and centrosome amplification. These findings indicate that the functions of FoxM1 in Shh-induced neuroproliferation are restricted to the regulation of the G2/M transition in CGNP, most probably through transcriptional effects on target genes such as those coding for B-type cyclins.
The cerebellum, derived from the embryonic midhindbrain region, serves critical roles in the coordination of movement and some higher-order functions. Cerebellar granule neuron precursors (CGNP) are initially formed in rhombomere 1 of the embryonic neural tube. They then migrate dorsally where a second phase of postnatal proliferation takes place in the external granular cell layer (EGL), which in mice lasts for approximately 2 weeks. Subsequently, CGNP differentiate and migrate from the EGL into deeper parts of the cortex (1). While granule neurons represent the most abundant neuronal cell type in the entire nervous system, relatively little is known about the mediators and mechanisms that regulate the proliferation and differentiation of these neurons.
Activation of the Sonic hedgehog (Shh) pathway is required for the expansion of specific neural precursor populations during normal development of the central nervous system (CNS). In granule cells, Shh acts on the transmembrane protein Patched (Ptc) to relieve inhibition of the Smoothened receptor and to upregulate target genes (13). Shh has furthermore been implicated in a variety of human cancers including cerebellar medulloblastoma, the most frequently found malignant brain tumor of childhood, which is believed to derive from granule cell precursors (34, 41). Proliferative effects of Shh have been especially well characterized in the EGL of the cerebellum, where the expansion of CGNP is dependent upon Shh signaling (42). The transcription factors known to mediate Shh mitogenic signaling within this system include the Gli proteins and the proto-oncogene product N-myc (5, 15). Indirect transcriptional targets of Shh signaling that are integral to the cell cycle machinery include the cyclins A2, B1, B2, D1, and D2 (16, 36, 45).
A novel candidate that has recently been implicated as a transcriptional target of hedgehog signaling in neural precursors (33) as well as in human colorectal cancer (7) and basal cell carcinomas (38) is the forkhead transcription factor FoxM1 (also known as Trident, HFH-11, and WIN). The role of FoxM1 in normal development was first reported in the myocardium. FoxM1 null mice die around birth and are found with disorganized and polyploid cardiomyocytes that lead to circulatory failure (20). A similar significance has been reported for mice with FoxM1-deficient liver, where diminished DNA replication and a failure to enter mitosis result in a reduced number of hepatoblasts and severe developmental abnormalities (21). Finally, mice with FoxM1-deficient lungs exhibit severe hypertrophy of arteriolar smooth muscle cells and a significant reduction of lung mesenchyme proliferation (18). However, the function of FoxM1 in the CNS and especially in the cerebellum, a model system used to study Shh mitogenic effects, has not been characterized. In particular, we asked whether FoxM1 function is required for (i) overall rates of Shh-induced neuroproliferation, (ii) mitotic entry of CGNP, and/or (iii) mitotic spindle formation during the G2/M transition.
We show that FoxM1 is specifically expressed in granule cell precursors of the developing cerebellar cortex and that it is upregulated after the activation of the Shh pathway. While FoxM1-deficient CGNP from conditional knockout mice maintain their responsiveness to Shh with respect to DNA synthesis during S phase, they express decreased levels of cyclin B1 and Cdc25b protein expression, and their ability to enter mitosis is disturbed, as shown by a significantly elevated number of cells in G2 or prophase. We show, moreover, that FoxM1 is crucial for the spindle apparatus and centrosome duplication in CGNP. Together, these findings suggest a restricted role for FoxM1 in the G2/M transition in CGNP.
MATERIALS AND METHODS
Transgenic animals and genotyping.
Generation and characterization of Nestin-cre [B6.Cg-Tg(Nes-cre)1Kln/J], FoxM1−/−, FoxM1Fl/Fl, and Wnt1/GAL4 UAS-Shh animals have been described previously (20, 32, 39, 40). Math1-cre transgenic animals, which carry bacteriophage P1 cre recombinase under the control of a 1.4-kb upstream Math1 enhancer element (11), have also been described previously (26). Embryos were obtained from timed pregnancies, with the plug date defined as embryonic day 0.5 (E0.5). For all animals, genotyping was done by PCR using genomic DNA which was extracted from mouse tail tissue. Primers have been published previously. Cre expression was further verified by immunohistochemistry, and global or cell-type-specific knockout of FoxM1 was verified by Western blotting or by in situ hybridization.
Histology.
For hematoxylin and eosin (H&E) staining and for immunohistochemical procedures, mouse embryos and brains were fixed in a 4% paraformaldehyde-phosphate-buffered saline (PBS) solution overnight at 4°C. Tissue for frozen sections was equilibrated in 20% glucose-PBS (pH 7.4) and embedded in optimal-cutting-temperature compound. Twelve-micrometer parasagittal sections were prepared on Superfrost Plus slides (Fisher). Tissue for paraffin wax-embedded sections was dehydrated, embedded, and cut into 5-μm-thick sections, according to standard protocols. Overall morphology was assessed by H&E staining. All photomicrographs, including those from immunohistochemical experiments, were taken digitally, using a Zeiss Axioskop microscope and an Axiocam imaging system.
Immunohistochemistry.
Tissue sections were subjected to heat-induced antigen retrieval at 99°C in a 10 mM sodium citrate buffer for 20 min for all antibodies. Staining was performed using an HRP (horseradish peroxidase)-DAB (3,3′-diaminobenzidine)-based Envision+ staining system (Dako) according to the manufacturer's specifications, except that incubation times were increased for primary antibodies (overnight at 4°C) and secondary antibodies (1 h at room temperature). Counterstaining was performed using Harris hematoxylin (Sigma). Cultured CGNP were immunolabeled using fluorescence-coupled secondary antibodies (Alexa 488 and Alexa 546, 1:500; Molecular Probes) and counterstained with DAPI (4′,6′-diamidino-2-phenylindole; Sigma). The primary antibodies used were anti-phospho-histone H3 (6G3 and polyclonal, 1:100; Cell Signaling), anti-8-bromodeoxyuridine (BrdU) (B44, 1:100; Becton-Dickinson), anti-cleaved caspase 3 (Asp175, 1:100; Cell Signaling), anti-FoxM1 (polyclonal, 1:50; Santa Cruz Biotechnology), anti-Cre (polyclonal, 1:1,500; Covance), anti-green fluorescent protein (anti-GFP) (polyclonal, 1:500; Abcam), anti-α-tubulin (B-5-1-2, 1:1,000; Sigma), and anti-γ-tubulin (GTU88, 1:500; Sigma).
Preparation of protein extracts and Western blotting.
Protein lysates were prepared and quantified as described previously (16). One hundred micrograms of each sample was separated on 10% sodium dodecyl sulfate-polyacrylamide gels and then transferred to Immobilon polyvinylidene difluoride (Millipore) membranes. The primary antibodies used were anti-FoxM1 (polyclonal, 1:500; Santa Cruz Biotechnology), anti-cyclin B1 (polyclonal, 1:200; Santa Cruz Biotechnology), anti-Cdc25b (polyclonal, 1:200; Santa Cruz Biotechnology), anti-N-myc (monoclonal, 1:500; BD; a kind gift from W. Weiss, San Francisco, CA), and anti-β-tubulin (TUB 2.1, 1:5,000; Sigma). Peroxidase-conjugated secondary antibodies included donkey anti-mouse (1:5,000; Jackson Immunoresearch Laboratories) and goat anti-rabbit (1:10,000; Pierce) antibodies. Blots were developed using enhanced chemiluminescence (Amersham-Pharmacia), according to the manufacturer's instructions. Chemiluminescent immunoreactivity was detected using Kodak X-Omat X-ray film.
Probe synthesis and in situ hybridization.
Gene fragments were amplified directly from plasmids, using ready-made T7, T3, or SP6 primers (Integrated DNA Technologies, Coralville, IA). Digoxigenin-labeled antisense RNA probes were made by using amplified DNA as the template and SP6, T3, or T7 as the RNA polymerase (Roche). In situ hybridization with frozen sections was performed according to standard protocols. Briefly, brain sections were hybridized overnight with labeled RNA probes at 65°C, washed twice in 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), pH 4.5, and 0.1% Tween 20 at 65°C, washed twice in MBST buffer, pH 7.5, containing 100 mM maleic acid, 150 mM NaCl2, 2 mM levamisole, and 0.1% Tween 20, blocked in MBST with 2% BM blocking agent (Roche) and 20% lamb serum, and incubated with alkaline phosphatase-labeled antidigoxigenin antibodies (1:2,500 in 2% serum; Roche) for 2 h. Sections were washed, and color was visualized using BM purple (Roche) (a detailed protocol is available from the author upon request).
Primary cell culture and virus production.
Primary cultures of P5/P6 mouse granule cell precursors were established as previously described (15). For immunohistochemical experiments, cells were plated on polyornithine-plated glass coverslips. For experiments including virus infection of cultured granule cell precursors, cells were maintained in serum-free medium containing Shh (3 μg/ml) for 24 h prior to virus infection. For virus preparation, 293 EBNA (Invitrogen) packaging cells were cotransfected with the gag-pol gene fusion, with vesicular stomatitis virus G glycoprotein plasmids, and with retroviral constructs carrying either internal ribosome entry site (IRES)-GFP alone, Cre-IRES-GFP, or FoxM1-IRES-GFP sequences, using Fugene 6 transfection reagent (Roche). Packaging cells were re-fed at 12 h after transfection. Retroviral supernatants (4 ml) were harvested every 12 h for 72 h and kept at 4°C until they were pooled, filtered through 0.45-mm syringe filters, aliquoted, and stored at −80°C until use. For the infection of granule cell precursors, conditioned medium was removed and saved, and cells were exposed to freshly thawed retroviral supernatants for 2 to 3 h. Supernatants were removed, and conditioned medium was replaced.
For proliferation assays, granule cell precursors plated on coverslips were pulsed with 25 mg/ml BrdU for 2 h prior to fixation in 4% paraformaldehyde. Cells were washed in PBS, treated for 10 min with 4 N HCl, and neutralized for 5 min with 0.1 M sodium borate buffer (pH 8.5) before being processed for immunocytochemistry.
Cell cycle analysis.
For flow cytometry, cells were fixed in ethanol, immunostained with antibodies against phospho-histone H3, treated with 500 units/ml RNase (Sigma) for 15 min at 37°C, and afterwards stained with propidium iodine (25 μg/ml; Sigma). Cell cycle analysis and immunofluorescence detection were performed using a FACScan (Becton Dickinson) and CellQuest software (Becton Dickinson) for data acquisition. Data shown in Fig. 6 and described in the accompanying text are representative of results from experiments performed in triplicate.
FIG. 6.
Delay in G2/M transition in Math1-cre FoxM1Fl/Fl granule cells. Immunohistochemistry of sections of P7 cerebella, using antibodies against phospho-histone H3, reveals a significantly elevated number of labeled cells in the EGL of Math1-cre FoxM1Fl/Fl animals (A). Cell cycle analysis and immunolabeling of freshly dissected granule cell precursors show a higher proportion of cells in G2/M phase, as well as more phospho-histone H3-positive cells in the mutant as opposed to the wild-type mice (B). Immunohistochemistry of tissue cultures from dissociated granule cell precursors indicates that mutant cells accumulate in late G2, where they express low levels of phospho-histone H3 (panel C, arrows). Asterisks mark granule cells in M phase, which show strong expression of phosphorylated histone H3.
Quantitation and statistics.
All in situ hybridizations, as well as conventional and immunohistochemical staining, were carried out at least three times with at least two different wild-type and mutant mice from separate litters. In vivo counting of phospho-histone H3-positive granule cells was performed by averaging the total number of immunolabeled cells per section by the total length of the EGL. Counts were taken from three parasagittal sections of two wild-type and two mutant animals by an unbiased observer. To assess the percentage of cultured granule cells that incorporated BrdU, 10 fields of 100 cells were analyzed per condition, using DAPI staining to select homogeneously distributed and dispersed (as opposed to clumped) cell fields. M-phase analysis of cultured granule cell precursors was performed by counting at least 100 cells in M phase from at least three independent experiments. Statistical significance was calculated using a two-tailed t test (Excel software) in which P values of <0.05 were considered significant. Error bars represent standard deviations of the means.
RESULTS
FoxM1 is expressed in proliferating granule cell precursors and is responsive to Sonic hedgehog signaling.
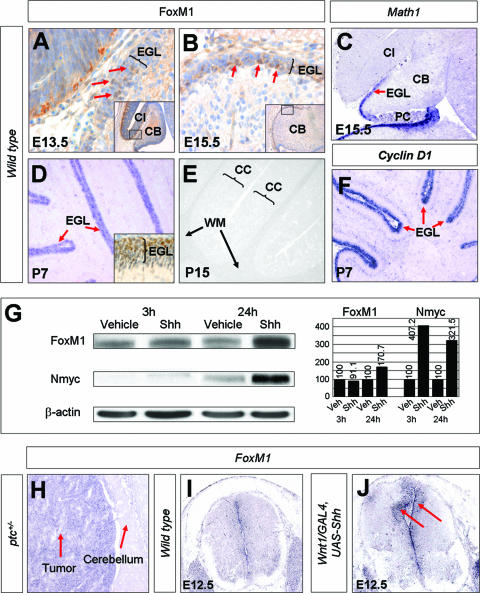
We first characterized FoxM1 expression in the developing mouse midbrain-hindbrain region during embryonic development. As shown in Fig. 1, the expression of FoxM1 is detected in the cerebellar anlagen as early as E13.5 in a population of cells that also expresses Math1, a marker of granule cell precursors. Although FoxM1 continues to show expression on into the postnatal stages of cerebellar development, with a peak at P7 (Fig. 1D), its expression is restricted to a rapidly proliferating population of EGL cells that also expresses the cyclin genes such as cyclin D1 (Fig. 1F), a regulator of G1 progression, and cyclin B1 (not shown), which regulates the G2/M transition (15, 45). FoxM1 is rapidly downregulated thereafter and is undetectable at P15 (Fig. 1E).
FIG. 1.
Expression of FoxM1 during normal cerebellar development and in response to activation of the Shh signaling pathway. Panels A and B and the inset in panel D show results from immunohistochemistry analysis, while all other images are from in situ hybridizations. Expression of FoxM1 in the cerebellum is detectable at E13.5 (panel A, arrows) and covers the entire EGL from E15.5 onward (panel B, arrows). It is restricted to granule cell precursors in the EGL as shown in sagittal sections of P7 cerebella (D) and is downregulated when granule cell precursors start to differentiate. No more expression is observable at P15 (E). Note that FoxM1 is expressed in the same regions as Math1 (C) and cyclin D1 (F), as shown in E15.5 and P7 cerebella, respectively. Western blotting analysis from protein extracts of cultured granule cell precursors shows an upregulation of FoxM1 levels after a 24-h treatment with Shh (G). In situ hybridization for FoxM1 in a cerebellar tumor from an adult Ptc+/− animal with a strong signal in the tumor tissue, but not in adjacent cerebellar tissue (H). FoxM1 is expressed as a consequence of ectopic Shh expression in the dorsal neural tube of E12.5 Wnt1/GAL4 UAS-Shh animals (I and J). CB, cerebellum; CC, cerebellar cortex; CI, colliculus inferior; PC, plexus choroideus; WM, white matter.
The temporal and spatial expression of FoxM1 in the EGL suggested that FoxM1 might be responsive to the activation of Shh signaling. We therefore treated CGNP in vitro with recombinant Shh protein or vehicle and analyzed FoxM1 expression by Western blotting. As shown in Fig. 1G, FoxM1 protein expression levels are upregulated at 24 h posttreatment with Shh, similar to the upregulation of N-myc protein expression levels. Since the genes directly targeted by Hedgehog signaling, such as N-myc (15), usually show robust upregulation after 3 h, we also assessed FoxM1 expression levels at the corresponding early time points. However, the absence of FoxM1 upregulation at that time suggests that FoxM1 is probably an indirect Shh target.
Further evidence for the upregulation by Shh in vivo is provided by strong expression in cerebellar tumors that form in Ptc+/− mice due to constitutive activation of Hedgehog signaling (8) (Fig. 1H) and by analysis of Wnt1/GAL4 UAS-Shh mice, in which Shh is ectopically expressed in the dorsal spinal cord (32) (Fig. 1I and J). While these data collectively show that Shh can upregulate FoxM1 expression, they do not show that FoxM1 is a specific target of Shh signaling. Indeed, the expression of FoxM1 is widespread in precursor cells during development, clearly indicating that its activity can be recruited by additional mitogens.
FoxM1 function is dispensable for overall cerebellar morphogenesis.
To analyze roles for FoxM1 in cerebellar development, we first studied mice carrying a conventional null allele of FoxM1 (20). As shown in Fig. 2, we did not detect any obvious abnormalities in cerebellar development at E17.5, just prior to birth, the latest time point we could study. Mice that are homozygous for null alleles of FoxM1 have limited viability due to cardiac defects (20). Indeed, we failed to detect any surviving FoxM1 null pups out of 69 tested.
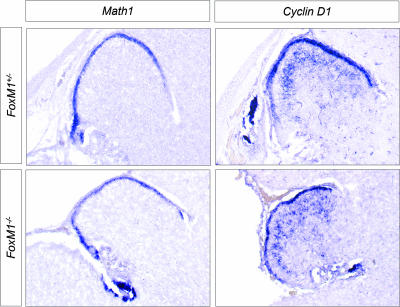
FIG. 2.
FoxM1−/− animals die around birth but develop cerebella that are indistinguishable from those of the wild type at E18.5. Sagittal sections from E18.5 cerebella show a normal overall morphology and expression of granule cell markers Math1 and cyclin D1.
To circumvent this problem and to study FoxM1 functions at postnatal stages of cerebellar development, we employed a conditional allele of FoxM1 (39). In order to achieve conditional knockout of FoxM1 specifically in granule cell precursors, we used transgenic animals that expressed Cre recombinase under the control of Math1 regulatory sequences (26). The specificity of such regulatory sequences within the cerebellar cortex has been shown previously (11, 25).
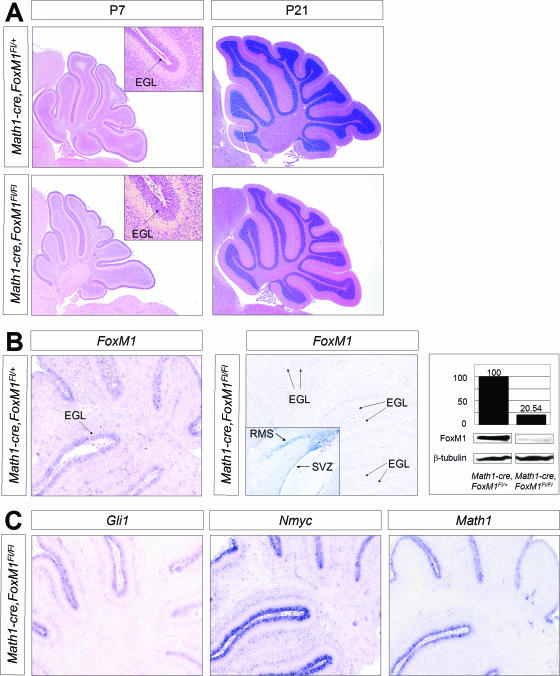
We then crossed the Math1-cre mice with the FoxM1Fl/Fl mice and analyzed conditional knockout animals at postnatal stages. As shown in Fig. 3, we found that the deletion of FoxM1 in granule cell precursors failed to produce a gross morphological phenotype at P7 and P21, and we did not observe neurological abnormalities. Analysis by in situ hybridization and Western blotting confirmed significant downregulation of FoxM1 expression in the conditional knockout animals (Fig. 3B). As expected, expression of the Hedgehog transcriptional targets Gli1 and N-myc as well as Math1, a marker for granule cell precursor cells, was unaffected (Fig. 3C), indicating that the FoxM1 function is dispensable for Shh signal transduction. Furthermore, apoptosis was measured by immunohistochemistry and Western blotting by using antibodies against cleaved caspase 3, but no significant differences were detectable between the wild-type and the mutant mice (see Fig. S1 in the supplemental material).
FIG. 3.
Cerebella from Math1-cre FoxM1Fl/Fl animals appear grossly normal at P7 and P21 (A). Insets of H&E-stained sections show parts of the cerebellar EGL with proliferating granule cell precursors that, in P7 wild-type mice, express FoxM1, as shown in panel B. As expected, FoxM1 is lost in cerebella from P7 Math1-cre FoxM1Fl/Fl mice (B). Western blots show the levels of FoxM1 expression, and normalized quantities are given in percentages. In situ hybridization for mutant mice reveals no FoxM1 expression in the cerebellum but clear signals in the forebrain subventricular zone (SVZ) and rostral migratory stream (RMS), where Math1-cre is not expressed (inset in panel B, arrows). The Sonic hedgehog target genes Gli1 and Nmyc as well as Math1 are properly expressed in FoxM1-deficient granule cell precursors (C).
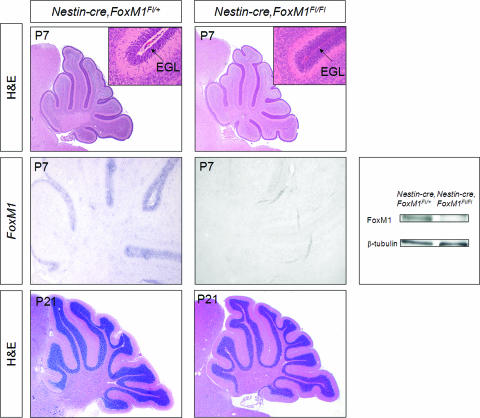
To rule out the possibility that Math1 gene-driven Cre activity might have failed to target granule cell precursors at a key stage in the developmental sequence, we sought to confirm our findings by using an independent conditional knockout strategy. Because Nestin-cre has been used previously to show requirements for N-myc during granule cell and cerebellar development (19), we crossed Nestin-cre animals with FoxM1Fl/Fl mice to generate conditional null mice and heterozygous controls. As shown in Fig. 4, the cerebellar phenotype of the Nestin-cre FoxM1Fl/Fl mice was indistinguishable from that of the Math1-cre FoxM1Fl/Fl animals. Moreover, we did not detect any obvious histological phenotype in the Nestin-cre FoxM1Fl/Fl animals (U. Scüller. and D. H. Rowitch, unpublished data), although FoxM1 was expressed in the forebrain subventricular zone and rostral migratory stream (Fig. 3B, inset).
FIG. 4.
Morphology of Nestin-cre FoxM1Fl/Fl cerebella appears normal in H&E-stained sagittal sections of P7 and P21 cerebella. Insets in P7 images show higher magnifications of the cerebellar EGL, which is morphologically indistinguishable in wild-type and mutant mice. FoxM1 is strongly reduced in P7 cerebella from Nestin-cre FoxM1Fl/Fl animals, as shown by in situ hybridization and Western blotting. Despite the broad expression of Nestin-cre outside the cerebellum, gross examination of Nestin-cre FoxM1Fl/Fl mice did not reveal any obvious developmental defects in other parts of the CNS (data not shown).
FoxM1-deficient CGNP show a normal S-phase response to Shh.
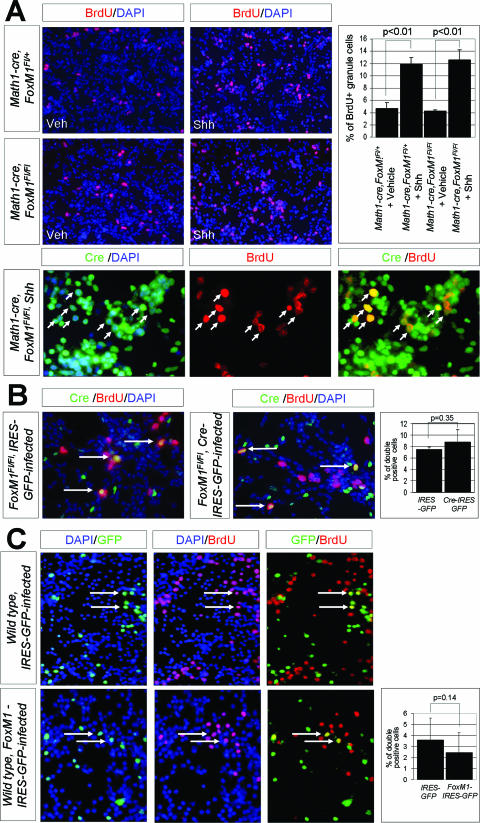
Although the results described above indicated that FoxM1 function was not required for achieving the ultimate cerebellar size, they did not rule out more subtle roles for FoxM1 in CGNP cell cycle regulation. To more precisely characterize this, we first turned to an in vitro culture system that has been established to show mitogenic responses to Shh signaling. As shown in Fig. 5A, the treatment of P5 to P6 wild-type CGNP with Shh produces a threefold increase in proliferation as measured by BrdU incorporation during DNA synthesis (42). Identical results were found with FoxM1-deficient cells. The absence of abnormalities with respect to BrdU incorporation in FoxM1 knockout granule cells raised the possibility that Cre recombinase activity was affecting only a fraction of the present granule cell neurons. Therefore, we performed colabeling with antibodies against Cre recombinase and BrdU, which is incorporated into DNA during S phase. As shown in Fig. 5A, we detected normal BrdU incorporation levels in the Cre-expressing cells themselves that are very probably deleted the FoxM1Fl/Fl mutant alleles. In addition, we infected the FoxM1Fl/Fl mutant CGNP with retroviruses carrying Cre recombinase and counted infected proliferating cells. As shown in Fig. 5B, Cre recombinase-carrying retroviruses had only insignificant effects on BrdU incorporation compared to that of GFP retrovirus controls. Together, these data show that FoxM1 is not required for DNA synthesis of CGNP in response to Shh signaling in vitro.
FIG. 5.
(A) S-phase response to Shh, as measured by BrdU uptake, is not affected in CGNP from Math1-cre FoxM1Fl/Fl animals. The CGNP from both the wild-type and the mutant animals showed an approximately threefold increase of BrdU uptake after treatment with Shh. Double labeling of CGNP from Math1-cre FoxM1Fl/Fl animals using antibodies against BrdU and Cre proved that the response to Shh was indeed occurringin mutant CGNP. (B) Acute removal of FoxM1 in CGNP from FoxM1Fl/Fl animals by adding retroviruses carrying Cre-IRES-GFP sequences did not result in significant alterations of BrdU uptake compared to that of cells transduced with IRES-GFP sequences only. This was shown by determining the percentage of cells positive for BrdU and GFP within the population of GFP-positive cells. (C) Overexpression of FoxM1 using retroviruses carrying FoxM1-IRES-GFP sequences in wild-type CGNP does not significantly alter BrdU-uptake, compared to wild-type cells transduced with IRES-GFP sequences only. The histogram shows percentages of cells positive for BrdU and GFP within the population of GFP-positive cells.
We next tested whether the forced expression of FoxM1 alters BrdU uptake in cultured wild-type CGNP. However, as shown in Fig. 5C, we failed to detect any positive impact of FoxM1 overexpression in this regard. Thus, FoxM1 is neither necessary nor sufficient for the regulation of S phase in response to Shh treatment.
FoxM1 is required for the regulation of G2/M transition in CGNP.
The results described above were consistent with previous analyses of FoxM1, which is a known regulator of a later phase of the cell cycle, the G2/M transition (6). In order to analyze the G2/M transition in FoxM1-deficient CGNP, we used antibodies against phosphorylated histone H3. Phosphorylation of histone H3 at its amino terminus (serine 10), which is tightly correlated with expression of the FoxM1 target Aurora B (37) is initiated by the Msk1 and Rsk2 kinases in late G2 interphase (31), and the expression of phosphorylated histone H3 is maximal in M phase (12). We observed a significant increase for the number of phospho-histone H3-positive CGNP in FoxM1-deficient cerebella in vivo as well as in vitro, using immunohistochemistry and flow cytometry (Fig. 6A and B), indicating an increased population of mutant cells in late G2 interphase and/or M phase.
To define the cell cycle alterations more precisely, we performed immunohistochemistry with cultured granule cells and found that the proportion of CGNP in late G2 interphase was significantly increased. Such late G2 cells are characterized by a weak immunopositivity for phospho-histone H3 (12) and account for some 2.7% of the cells in wild-type animals as opposed to 6.7% of the cells in Math1-cre FoxM1Fl/Fl CGNP (P < 0.01; Fig. 6C). These findings indicate that FoxM1 is crucial for timely mitotic entry.
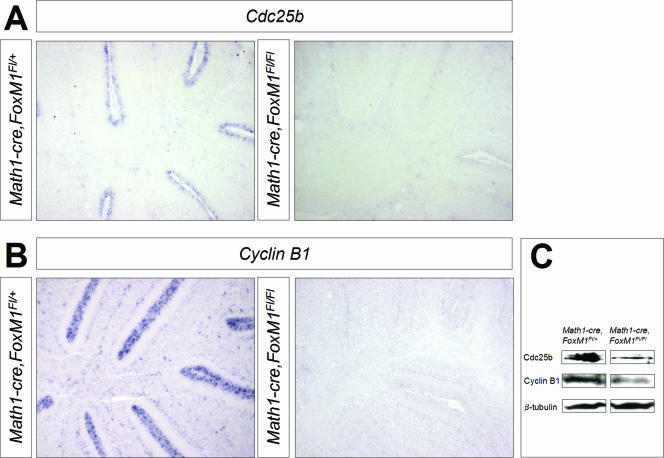
In order to identify mechanisms that may lead to the G2/M defects described above, we focused on cell cycle-regulating genes that have been described to have roles downstream of FoxM1. Among the previously published FoxM1 target genes, we found that cyclin A2, cyclin B1, cyclin B2, and Cdc25b were expressed by CGNP in the cerebellar EGL. While we did not detect any significant differences between the levels of cyclin A2 and cyclin B2 protein expression in the wild-type and that of the mutant mice (data not shown), Western blotting analysis and in situ hybridization showed a clear decrease in the cyclin B1 and Cdc25b expression levels of FoxM1-deficient granule cells compared to that of wild-type cells (Fig. 7). This is consistent with the role of cyclin B1 and Cdc25b for normal mitotic entry that has been described for other systems (for a review, see reference 3).
FIG. 7.
In situ hybridization (A and B) and Western blotting analysis (C) reveal that the expression of Cdc25b (A and C) and cyclin B1 (B and C) is decreased in P7 Math1-cre FoxM1Fl/Fl cerebella compared to those of wild-type littermates.
Formation of the normal mitotic spindle is regulated by FoxM1.
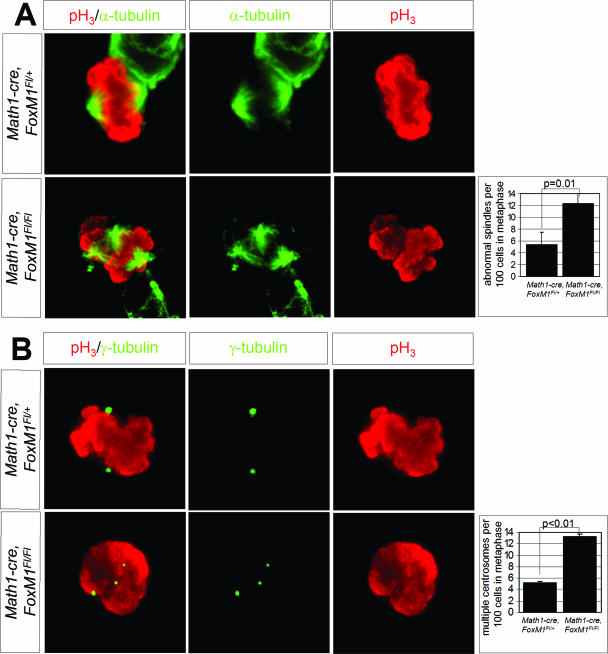
To further characterize the phenotype caused by the loss of FoxM1, we analyzed the formation of the spindle apparatus and the separation of the centrosomes, both of which are mechanisms that have been associated with the function of cyclin B1 (4, 43). Thus, we first performed double labeling using antibodies against phospho-histone H3 and α-tubulin, which labels the spindle apparatus of CGNP (36). The distribution of mutant cells in prophase, metaphase, anaphase, or telophase was not obviously different from that of wild-type cells (data not shown). However, as shown in Fig. 8A, 12.3% of the mutant cells in prometaphase/metaphase showed an abnormal formation of the spindle apparatus, a highly significant increase compared to that of the wild-type cells (P = 0.01). Additionally, immunostaining using antibodies against γ-tubulin, a core component of the centrosome, revealed significantly more mutant cells with multiple centrosomes than wild-type cells (13.2% versus 5.2%, respectively; P < 0.01; Fig. 8B). These findings suggest that the loss of FoxM1 results in defects of the spindle apparatus and centrosome amplification in cerebellar granule cell precursors.
FIG. 8.
Defects of the spindle apparatus and centrosome amplification in Math1-cre FoxM1Fl/Fl CGNP. (A) Representative mitotic CGNP cultured from Math1-cre FoxM1Fl/+ and Math1-cre FoxM1Fl/Fl animals are labeled with antibodies against phospho-histone H3 (M-phase marker) and against α-tubulin which marks the microtubules of the cell. While the wild-type cell shows regular alignment of the chromosomes, the mutant cell reveals clear abnormalities of the spindle apparatus with multiple apparent spindle poles throughout the cell. Such spindle cell aberrations occurred significantly more often in mutant cells than in wild-type CGNP. (B) Multiple centrosomes, as visualized by using antibodies against the centrosome core component γ-tubulin, are visible significantly more often in cultured Math1-cre FoxM1Fl/Fl CGNP than in wild-type cells.
DISCUSSION
Recent reports have suggested roles for FoxM1 in development and tumorigenesis downstream of Shh signaling. Here we have analyzed the expression and function of FoxM1 in developing CGNP, a population that depends on the mitogenic activity of Shh. We provide evidence that FoxM1 is needed for efficient mitotic entry, both in vivo and in vitro, and that in its absence, there are abnormalities in spindle assembly which likely engender aneuploidy. However, ultimate cerebellar morphogenesis is grossly unaffected, and rates of Shh-induced neuroproliferation are normal in FoxM1 mutants, suggesting that its functions in the cell cycle are limited to regulation of the G2/M transition. The implications of these findings and the regulation of aneuploidy during normal development and tumorigenesis are discussed further.
FoxM1 is an indirect downstream transcriptional target of Hedgehog signaling in proliferating CGNP.
A recent screening study identified FoxM1 among a number of transcription factors that are expressed at postnatal stages of cerebellar granule neuron development (9). We extended this analysis to antenatal stages and tested more directly the role of Shh signaling in FoxM1 regulation. While we gathered clear evidence that FoxM1 expression is indeed controlled by Shh signaling in CGNP, FoxM1 expression levels were elevated only at 24 h after treatment. This is in contrast to the direct targets of Shh signaling, such as D-type cyclins and N-myc, that show upregulation after a short 3-h treatment with Shh. We conclude from this that FoxM1 is most probably an indirect transcriptional target of Shh.
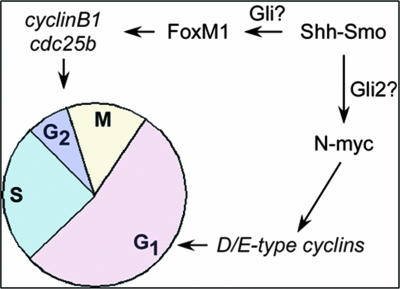
Previous work shows that Shh treatment of CGNP results in the upregulation of G1 targets such as cyclin D1, cyclin D2, and N-myc, as well as G2/M targets such as A- and B-type cyclins (16, 36, 45). Based on our observations of decreased cyclin B1 expression levels, we propose that FoxM1 acts downstream of Shh primarily by regulating gene targets that control the G2/M transition such as Cdc25b and cyclin B1 (Fig. 9). Consistent with this proposed model, the expression of G1 effectors of Shh signaling (e.g., cyclin D1 and N-myc) is unperturbed in FoxM1-deficient animals.
FIG. 9.
A model for the possible regulatory mechanisms of Shh signaling on cell cycle progression in CGNP, based on present and published results. Previous work suggests that N-myc is a direct target of Shh signaling and the activity of Gli transcription factors (most probably Gli2), which regulates G1-phase cell cycle progression via D/E-type cyclins (5, 10, 15, 16, 28). In contrast, FoxM1 activates gene targets such as the cyclin B1 and Cdc25b genes encoding regulators of G2/M progression. While our findings suggest that FoxM1 upregulation by Shh in CGNP may be indirect, previous studies have suggested this could occur through Gli proteins (38).
Gross cerebellar morphogenesis is unaffected in FoxM1 mutants.
Granule cells are the most abundant cell type in the cerebellum, and adequate proliferation of those cells takes place over a protracted period in the mammalian brain (e.g., about 3 weeks in rodents and up to 18 months in humans) (1). FoxM1 is required for the proliferation of hepatocytes in the liver (40), mesenchyme cells in the lung (18), and cardiac precursors (20). The lack of obvious behavioral deficits and detectable gross abnormalities at various stages of cerebellar morphogenesis in the FoxM1−/−, Math1-cre FoxM1Fl/Fl, and Nestin-cre FoxM1Fl/Fl mice in vivo provides strong evidence that FoxM1 function is dispensable for the overall rates of CGNP proliferation. However, our data do show delayed mitotic entry and spindle abnormalities. Despite abnormalities in the G2/M transition, proliferation rates are unaffected, a situation characteristic of many cancers that demonstrate high rates of cell division and aneuploidy (27).
Restricted roles for FoxM1 in the G2/M transition of the cell cycle regulation in CGNP.
We set out to resolve specific roles for FoxM1 during the cell cycle, with a well-characterized primary CGNP model system of Shh-induced proliferation. FoxM1 function has been shown in other organ systems to be a transcriptional regulator of both G1/S progression and G2/M transition (6). G1/S progression is thought to be regulated by diminishing nuclear levels of the inhibitory cell-cycle regulators p21Cip1 and p27Kip1 and by activating Cdc25a transcription (40). In distinction, G2/M transition is mediated by the upregulation of cyclin B1 and Cdc25b (22). Our results show that in the cerebellum, FoxM1 functions are restricted to the G2/M transition. First, BrdU uptake studies indicated that G1/S progression is not significantly affected by a deficiency of FoxM1 in CGNP (Fig. 5). Second, Cdc25a protein levels are not altered in Math1-cre FoxM1Fl/Fl cerebella (data not shown). Third, overall cerebellar morphogenesis is unaffected (see description above), which also argues against widespread mitotic catastrophe or apoptosis. In contrast, mitotic entry is affected in FoxM1-deficient granule cells. We found a significantly elevated number of cells in late stages of G2. Those cells are characterized by a weak but clearly detectable immunoreactivity for phospho-histone H3, which reflects the initiation of histone H3 phosphorylation in late G2 (31) (Fig. 6).
One possible explanation for this phenomenon is the downregulation of cyclin B1 and Cdc25b that we observed in FoxM1-deficient CGNP (Fig. 7). Cyclin B1 null mice are not viable, and it is therefore unclear whether the growth of the cerebellum is dependent on cyclin B1 (3). Expression of Cdc25b phosphatase has recently been tightly linked to Hedgehog activity (2), in keeping with the possibility of roles for FoxM1 in this pathway (Fig. 9). However, Cdc25b null mice are viable and did not display any obvious cerebellar phenotype (23), indicating that this axis is not required for overall cerebellar morphogenesis. Taken together, these findings suggest the existence of a synergistic and/or compensatory pathway in proliferating CGNP that is FoxM1 independent.
Although our results are remarkably similar to those described by Wonsey and Follettie (44), the frequency of mitotic defects was much lower. Therefore, a compensatory mechanism in FoxM1 mutant CGNP might exist that maintains normal mitosis in most of the cells. Further work is needed to understand precisely the relationship between the delay in mitotic entry and the spindle abnormalities that we observed. However, one possibility is that cells entering mitosis with compromised cyclin B/cyclin-dependent kinase 1 (CDK1) activity might have chromosome segregation defects. Chromosome segregation defects can lead to the failure of cytokinesis, polyploidy, and cells with multiple centrosomes (35). In this context, it seems noteworthy that, as we know from cancer biology, aneuploidy is not necessarily coupled to apoptosis which is unaffected in FoxM1 mutant mice (see Fig. S1 in the supplemental material) (29).
Roles for FoxM1 in brain cancer.
A variety of human cancers, such as prostate, lung, and breast cancer, require FoxM1 for growth and tumor progression both in vitro (44) and in vivo (14, 17). Intriguingly, this is also true for gliomas (24), although we have learned from the Nestin-cre FoxM1Fl/Fl mice analyzed in this study that FoxM1 is dispensable for the overall growth of the CNS, including the glial compartment (U. Schüller and D. H. Rowitch, unpublished observations).
The loss of FoxM1 in breast cancer cells has a dramatic effect on mitosis, leading to mitotic catastrophe due to an increase of multipolar spindles and overduplicated centrosomes (44). There are several potential mechanisms by which the loss of FoxM1 could promote cell division failure and centrosome amplification. One likely mechanism is through failed cytokinesis. Failed cytokinesis produces tetraploid cells with extra centrosomes. At least in the absence of p53, this leads to aneuploidy and promotes tumorigenesis. One FoxM1 transcriptional target is the spindle midzone kinesin Kif20a. Kif20a is required for normal cytokinesis, and diminished expression is thus expected to increase the rate at which cytokinesis errors occur. Furthermore, the reduced levels of cyclin B/Cdk1 activity in FoxM1 cells are expected to impair chromosome segregation. Chromosome segregation errors, especially if they lead to chromatin blocking the cleavage furrow, can inhibit cytokinesis. Additionally, the loss of FoxM1 through undefined mechanisms could cause centrosome overreplication or centriole splitting, contributing to spindle multipolarity. Medulloblastoma, a highly malignant cerebellar childhood tumor that is thought to arise from granule cell precursors (34), expresses high levels of FoxM1 (30). Thus, it is possible that FoxM1 has critical roles in the context of medulloblastoma genesis, and research is under way to explore such potential functions.
Supplementary Material
Acknowledgments
We thank Jason Ling, Dongin Yuk, and Sovann Kaing for excellent technical assistance. We also thank the late Robert Costa (University of Illinois, Chicago, IL) for contributing FoxM1Fl/Fl mice, Jane Johnson (University of Texas, Dallas, TX) for providing the Math1 plasmid, and Fabienne Pituello (Université P. Sabatier, Toulouse, France) for providing the Cdc25b plasmid.
U.S. is a scholar of the Mildred-Scheel-Stiftung für Krebsforschung. This work was supported by grants (to D.H.R.) from the NINDS (NS 047527), the James S. McDonnell Foundation, and the Pediatric Brain Tumor Foundation of the United States.
Footnotes
Published ahead of print on 24 September 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Altmann, J., and S. A. Bayer. 1997. Development of the cerebellar system in relation to its evolution, structure and functions. CRC Press, New York, NY.
- 2.Benazeraf, B., Q. Chen, E. Peco, V. Lobjois, F. Medevielle, B. Ducommun, and F. Pituello. 2006. Identification of an unexpected link between the Shh pathway and a G2/M regulator, the phosphatase CDC25B. Dev. Biol. 294:133-147. [DOI] [PubMed] [Google Scholar]
- 3.Ciemerych, M. A., and P. Sicinski. 2000. Cell cycle in mouse development. Oncogene 24:2877-2898. [DOI] [PubMed] [Google Scholar]
- 4.Clute, P., and J. Pines. 1999. Temporal and spatial control of cyclin B1 destruction in metaphase. Nat. Cell Biol. 1:82-87. [DOI] [PubMed] [Google Scholar]
- 5.Corrales, J. D., G. L. Rocco, S. Blaess, Q. Guo, and A. L. Joyner. 2004. Spatial pattern of sonic hedgehog signaling through Gli genes during cerebellum development. Development 131:5581-5590. [DOI] [PubMed] [Google Scholar]
- 6.Costa, R. H. 2005. FoxM1 dances with mitosis. Nat. Cell Biol. 7:108-110. [DOI] [PubMed] [Google Scholar]
- 7.Douard, R., S. Moutereau, P. Pernet, M. Chimingqi, Y. Allory, P. Manivet, M. Conti, M. Vaubourdolle, P. H. Cugnenc, and S. Loric. 2006. Sonic hedgehog-dependent proliferation in a series of patients with colorectal cancer. Surgery 139:665-670. [DOI] [PubMed] [Google Scholar]
- 8.Goodrich, L. V., L. Milenkovic, K. M. Higgins, and M. P. Scott. 1997. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 277:1109-1113. [DOI] [PubMed] [Google Scholar]
- 9.Gray, P. A., H. Fu, P. Luo, Q. Zhao, J. Yu, A. Ferrari, T. Tenzen, D. i. Yuk, E. F. Tsung, Z. Cai, J. A. Alberta, L. p. Cheng, Y. Liu, J. M. Stenman, M. T. Valerius, N. Billings, H. A. Kim, M. E. Greenberg, A. P. McMahon, D. H. Rowitch, C. D. Stiles, and Q. Ma. 2004. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science 306:2255-2257. [DOI] [PubMed] [Google Scholar]
- 10.Hallikas, O., K. Palin, N. Sinjushina, R. Rautiainen, J. Partanen, E. Ukkonen, and J. Taipale. 2006. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell 124:47-59. [DOI] [PubMed] [Google Scholar]
- 11.Helms, A. W., A. L. Abney, N. Ben Arie, H. Y. Zoghbi, and J. E. Johnson. 2000. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development 127:1185-1196. [DOI] [PubMed] [Google Scholar]
- 12.Hendzel, M. J., Y. Wei, M. A. Mancini, A. Van Hooser, T. Ranalli, B. R. Brinkley, D. P. Bazett-Jones, and C. D. Allis. 1997. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106:348-360. [DOI] [PubMed] [Google Scholar]
- 13.Ho, K. S., and M. P. Scott. 2002. Sonic hedgehog in the nervous system: functions, modifications and mechanisms. Curr. Opin. Neurobiol. 12:57-63. [DOI] [PubMed] [Google Scholar]
- 14.Kalin, T. V., I. C. Wang, T. J. Ackerson, M. L. Major, C. J. Detrisac, V. V. Kalinichenko, A. Lyubimov, and R. H. Costa. 2006. Increased levels of the FoxM1 transcription factor accelerate development and progression of prostate carcinomas in both TRAMP and LADY transgenic mice. Cancer Res. 66:1712-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kenney, A. M., M. D. Cole, and D. H. Rowitch. 2003. Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development 130:15-28. [DOI] [PubMed] [Google Scholar]
- 16.Kenney, A. M., and D. H. Rowitch. 2000. Sonic hedgehog promotes G1 cyclin expression and sustained cell cycle progression in mammalian neuronal precursors. Mol. Cell. Biol. 20:9055-9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, I. M., T. Ackerson, S. Ramakrishna, M. Tretiakova, I. C. Wang, T. V. Kalin, M. L. Major, G. A. Gusarova, H. M. Yoder, R. H. Costa, and V. V. Kalinichenko. 2006. The forkhead box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res. 66:2153-2161. [DOI] [PubMed] [Google Scholar]
- 18.Kim, I. M., S. Ramakrishna, G. A. Gusarova, H. M. Yoder, R. H. Costa, and V. V. Kalinichenko. 2005. The forkhead Box M1 transcription factor is essential for embryonic development of pulmonary vasculature. J. Biol. Chem. 280:22278-22286. [DOI] [PubMed] [Google Scholar]
- 19.Knoepfler, P. S., P. F. Cheng, and R. N. Eisenman. 2002. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 16:2699-2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korver, W., M. W. Schilham, P. Moerer, M. J. van den Hoff, K. Dam, W. H. Lamers, R. H. Medema, and H. Clevers. 1998. Uncoupling of S phase and mitosis in cardiomyocytes and hepatocytes lacking the winged-helix transcription factor Trident. Curr. Biol. 8:1327-1330. [DOI] [PubMed] [Google Scholar]
- 21.Krupczak-Hollis, K., X. Wang, V. V. Kalinichenko, G. A. Gusarova, I. C. Wang, M. B. Dennewitz, H. M. Yoder, H. Kiyokawa, K. H. Kaestner, and R. H. Costa. 2004. The mouse forkhead Box m1 transcription factor is essential for hepatoblast mitosis and development of intrahepatic bile ducts and vessels during liver morphogenesis. Dev. Biol. 276:74-88. [DOI] [PubMed] [Google Scholar]
- 22.Laoukili, J., M. R. H. Kooistra, A. Bras, J. Kauw, R. M. Kerkhoven, A. Morrison, H. Clevers, and R. H. Medema. 2005. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat. Cell Biol. 7:126-136. [DOI] [PubMed] [Google Scholar]
- 23.Lincoln, A. J., D. Wickramasinghe, P. Stein, R. M. Schultz, M. E. Palko, M. P. D. De Miguel, L. Tessarollo, and P. J. Donovan. 2002. Cdc25b phosphatase is required for resumption of meiosis during oocyte maturation. Nat. Genet. 30:446-449. [DOI] [PubMed] [Google Scholar]
- 24.Liu, M., B. Dai, S. H. Kang, K. Ban, F. J. Huang, F. F. Lang, K. D. Aldape, T. X. Xie, C. E. Pelloski, K. Xie, R. Sawaya, and S. Huang. 2006. FoxM1B is overexpressed in human glioblastomas and critically regulates the tumorigenicity of glioma cells. Cancer Res. 66:3593-3602. [DOI] [PubMed] [Google Scholar]
- 25.Machold, R., and G. Fishell. 2005. Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron 48:17-24. [DOI] [PubMed] [Google Scholar]
- 26.Matei, V., S. Pauley, S. Kaing, D. Rowitch, K. W. Beisel, K. Morris, F. Feng, K. Jones, J. Lee, and B. Fritzsch. 2005. Smaller inner ear sensory epithelia in Neurog1 null mice are related to earlier hair cell cycle exit. Dev. Dyn. 234:633-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakagawa, S., M. Watanabe, and Y. Inoue. 1996. Altered gene expression of the N-methyl-d-aspartate receptor channel subunits in Purkinje cells of the staggerer mutant mouse. Eur. J. Neurosci. 8:2644-2651. [DOI] [PubMed] [Google Scholar]
- 28.Oliver, T. G., L. L. Grasfeder, A. L. Carroll, C. Kaiser, C. L. Gillingham, S. M. Lin, R. Wickramasinghe, M. P. Scott, and R. Wechsler-Reya. 2003. Transcriptional profiling of the Sonic hedgehog response: a critical role for N-myc in proliferation of neuronal precursors. Proc. Natl. Acad. Sci. USA 100:7331-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pellman, D. 2007. Aneuploidy and cancer. Nature 446:38-39. [DOI] [PubMed] [Google Scholar]
- 30.Pomeroy, S. L., P. Tamayo, M. Gaasenbeek, L. M. Sturla, M. Angelo, M. E. McLaughlin, J. Y. Kim, L. C. Goumnerova, P. M. Black, C. Lau, J. C. Allen, D. Zagzag, J. M. Olson, T. Curran, C. Wetmore, J. A. Biegel, T. Poggio, S. Mukherjee, R. Rifkin, A. Califano, G. Stolovitzky, D. N. Louis, J. P. Mesirov, E. S. Lander, and T. R. Golub. 2002. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature 415:436-442. [DOI] [PubMed] [Google Scholar]
- 31.Prigent, C., and S. Dimitrov. 2003. Phosphorylation of serine 10 in histone H3, what for? J. Cell Sci. 116:3677-3685. [DOI] [PubMed] [Google Scholar]
- 32.Rowitch, D. H., B. St.Jacques, S. M. K. Lee, J. D. Flax, E. Y. Snyder, and A. P. McMahon. 1999. Sonic hedgehog regulates proliferation and inhibits differentiation of CNS precursor cells. J. Neurosci. 19:8954-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanai, N., A. Alvarez-Buylla, and M. S. Berger. 2005. Neural stem cells and the origin of gliomas. N. Engl. J. Med. 353:811-822. [DOI] [PubMed] [Google Scholar]
- 34.Schüller, U., A. T. Kho, Q. Zhao, Q. Ma, and D. H. Rowitch. 2006. Cerebellar “transcriptome” reveals cell-type and stage-specific expression during postnatal development and tumorigenesis. Mol. Cell Neurosci. 33:247-259. [DOI] [PubMed] [Google Scholar]
- 35.Shi, Q., and R. W. King. 2005. Chromosome nondisjunction yields tetraploid rather than aneuploid cells in human cell lines. Nature 437:1038-1042. [DOI] [PubMed] [Google Scholar]
- 36.Sjoström, S. K., G. Finn, W. C. Hahn, D. H. Rowitch, and A. M. Kenney. 2005. The Cdk1 complex plays a prime role in regulating N-Myc phosphorylation and turnover in neural precursors. Dev. Cell 9:327-338. [DOI] [PubMed] [Google Scholar]
- 37.Tanaka, J., S. Nakagawa, E. Kushiya, M. Yamasaki, M. Fukaya, T. Iwanaga, M. I. Simon, K. Sakimura, M. Kano, and M. Watanabe. 2000. Gq protein alpha subunits galphaq and galpha11 are localized at postsynaptic extra-junctional membrane of cerebellar Purkinje cells and hippocampal pyramidal cells. Eur. J. Neurosci. 12:781-792. [DOI] [PubMed] [Google Scholar]
- 38.Teh, M. T., S. T. Wong, G. W. Neill, L. R. Ghali, M. P. Philpott, and A. G. Quinn. 2002. FOXM1 is a downstream target of Gli1 in basal cell carcinomas. Cancer Res. 62:4773-4780. [PubMed] [Google Scholar]
- 39.Tronche, F., C. Kellendonk, O. Kretz, P. Gass, K. Anlag, P. C. Orban, R. Bock, R. Klein, and G. Schütz. 1999. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 23:99-103. [DOI] [PubMed] [Google Scholar]
- 40.Wang, X., H. Kiyokawa, M. B. Dennewitz, and R. H. Costa. 2002. The forkhead box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proc. Natl. Acad. Sci. USA 99:16881-16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wechsler-Reya, R., and M. P. Scott. 2001. The developmental biology of brain tumors. Annu. Rev. Neurosci. 24:385-428. [DOI] [PubMed] [Google Scholar]
- 42.Wechsler-Reya, R. J., and M. P. Scott. 1999. Control of neuronal precursor proliferation in the cerebellum by sonic hedgehog. Neuron 22:103-114. [DOI] [PubMed] [Google Scholar]
- 43.Wolf, F., C. Wandke, N. Isenberg, and S. Geley. 2006. Dose-dependent effects of stable cyclin B1 on progression through mitosis in human cells. EMBO J. 25:2802-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wonsey, D. R., and M. T. Follettie. 2005. Loss of the forkhead transcription factor FoxM1 causes centrosome amplification and mitotic catastrophe. Cancer Res. 65:5181-5189. [DOI] [PubMed] [Google Scholar]
- 45.Zhao, Q., A. Kho, A. M. Kenney, D. D. Yuk, I. Kohane, and D. H. Rowitch. 2002. Identification of genes expressed with temporal-spatial restriction to developing cerebellar neuron precursors by a functional genomic approach. Proc. Natl. Acad. Sci. USA 99:5704-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.