Abstract
Invadopodia are Src-induced cellular structures that are thought to mediate tumor invasion. ASAP1, an Arf GTPase-activating protein (GAP) containing Src homology 3 (SH3) and Bin, amphiphysin, and RVS161/167 (BAR) domains, is a substrate of Src that controls invadopodia. We have examined the structural requirements for ASAP1-dependent formation of invadopodia and related structures in NIH 3T3 fibroblasts called podosomes. We found that both predominant splice variants of ASAP1 (ASAP1a and ASAP1b) associated with invadopodia and podosomes. Podosomes were highly dynamic, with rapid turnover of both ASAP1 and actin. Reduction of ASAP1 levels by small interfering RNA blocked formation of invadopodia and podosomes. Podosomes were formed in NIH 3T3 fibroblasts in which endogenous ASAP1 was replaced with either recombinant ASAP1a or ASAP1b. ASAP1 mutants that lacked the Src binding site or GAP activity functioned as well as wild-type ASAP1 in the formation of podosomes. Recombinant ASAP1 lacking the BAR domain, the SH3 domain, or the Src phosphorylation site did not support podosome formation. Based on these results, we conclude that ASAP1 is a critical target of tyrosine kinase signaling involved in the regulation of podosomes and invadopodia and speculate that ASAP1 may function as a coincidence detector of simultaneous protein association through the ASAP1 SH3 domain and phosphorylation by Src.
The ability of malignant cells to invade normal tissues underlies much of the pathology of cancer. Invasion depends on adhesion, cell movement, and penetration into the extracellular matrix. Cellular structures that mediate these behaviors include invadopodia (4-6, 15, 24, 36, 37). Invadopodia are similar in appearance and composition to structures called podosomes, which are found in osteoclasts and macrophages (5). (We refer to similar structures found in the fibroblastic cell line NIH 3T3 after Src transformation as podosomes.) Both invadopodia and podosomes are large macromolecular complexes that connect the extracellular matrix with the intracellular actin cytoskeleton. They are highly dynamic and coordinately turn over to elicit cell movement and invasion. Proteins controlling invadopodia include nonreceptor tyrosine kinases (Src and FAK), Rho and Arf family GTP-binding proteins, and regulators of Rho and Arf proteins. ASAP1, a GTPase-activating protein (GAP) for Arf1, has also been implicated in the control of cell adhesive structures, including invadopodia (10, 21, 22, 29, 33).
ASAP1 is a phosphatidylinositol 4,5-bisphosphate-dependent Arf GAP (34) specific for class 1 and 2 Arfs (3, 10, 21, 22). ASAP1 contains, from the N to C termini, BAR (Bin, amphiphysin, and RVS161/167), pleckstrin homology, Arf GAP, ankyrin repeat, proline-rich, and Src homology 3 (SH3) domains. Two predominant splice variants, ASAP1a and ASAP1b, differ by 57 amino acids within the proline-rich region (3). ASAP1 binds a number of proteins that regulate focal adhesions (FAs) and invadopodia. ASAP1 was initially identified on the basis of GAP activity and in screens for proteins that bind to the SH3 domain of Src (3, 16), an oncogene that is a nonreceptor tyrosine kinase and induces the formation of invadopodia and podosomes (5). ASAP1 has also been found to bind to and be phosphorylated by Pyk2, a FA kinase (FAK)-type protein. FAK is a nonreceptor tyrosine kinase that mediates signaling from FAs and controls the turnover of the same structures (30, 31, 35). Additional proteins reported to bind ASAP1 include CrkL, CD2AP, CIN85, and cortactin (17, 21, 22, 28, 29).
The first reported evidence for a role for ASAP1 in cytoskeletal regulation was the finding that it associates with FAs (33). This localization is dependent on binding to CrkL (28) and FAK (21). Either overexpression or reduced expression of ASAP1 decreases the levels of paxillin in FAs (21, 33). This effect is dependent on Arf GAP activity (21) and on ASAP1 interaction with FAK (21) and CD2AP (22). ASAP1a has also been implicated in the formation of invadopodia. ASAP1a, but not ASAP1b, was found to bind to cortactin, a protein found in invadopodia but not FAs (5, 29). Either reducing ASAP1a levels with small interfering RNA (siRNA) or blocking the complex of ASAP1 with cortactin abrogates the formation of invadopodia and cell invasion (29). The gene for ASAP1 is located on chromosome 8q in a region that is amplified in a number of malignancies (7). In uveal melanoma and mammary carcinoma, ASAP1 expression levels have been found to correlate with invasive potential, consistent with ASAP1-dependent regulation of cell adhesive structures (7, 29).
Here we have determined the structural requirements for the ASAP1-dependent formation of invadopodia and podosomes. Contrary to expectations based on previous work (12, 29), both major ASAP1 isoforms bound cortactin, associated with invadopodia and podosomes, and supported podosome formation. Neither direct interaction with Src nor GAP activity was necessary for ASAP1 to support podosome formation. The BAR and SH3 domains of ASAP1 were necessary for podosome formation. Src-dependent phosphorylation of ASAP1 was also necessary. Based on these results, we conclude that protein association with the SH3 domain of ASAP1 together with ASAP1 phosphorylation leads to the formation of podosomes. We speculate that the function of the BAR domain is related to the tubulation of membranes within podosomes.
MATERIALS AND METHODS
Plasmids.
An expression vector for ASAP1b (GenBank accession no. AF075462) was previously described (3). The open reading frame for mouse ASAP1a (GenBank accession no. AF075461) that was fused to the sequence encoding an N-terminal Flag tag was subcloned into pCI (Promega Biosciences, Inc., San Luis Obispo, CA) as described previously (3, 33). Mouse ASAP1b was also subcloned into pSI (Promega Biosciences, Inc., San Luis Obispo, CA). [ΔBAR]ASAP1b has previously been described (27). A reading frame for Flag-[1-1022]ASAP1b, which consists of amino acids 1 to 1022 of ASAP1b ([ΔSH3]ASAP1b), was generated by PCR and subcloned into pCI. Point mutations in ASAP1b cDNA (R497K, R811A, Y782E, and Y782F) and in ASAP1a cDNA (Y1094A, E1103A, and W1122A) were generated by site-directed mutagenesis using sequence-specific oligonucleotides and the QuikChange mutagenesis kit (Stratagene, La Jolla, CA). The open reading frame for ASAP1b (GenBank accession no. AF075462) was cloned into pEGFPC1 using the EcoRI site. Mammalian expression vectors of constitutively active chicken Src (c-Src) (pCEFL-[Y527F]c-Src), dominant negative c-Src (pCEFL-[R295M,Y527F]c-Src), and focal adhesion kinase (FAK) were kind gifts from J. Silvio Gutkind (OPCB, NIDCR, National Institutes of Health, Bethesda, MD). A mammalian expression vector for dominant negative FAK (FRNK) (13) was a kind gift from J. Thomas Parsons (University of Virginia at Charlottesville). An expression vector for the expression of green fluorescent protein (GFP)-cortactin was a kind gift from Takehito Uruno, Laboratory of Cell Biology, NHLBI.
Cell lines and transfections.
NIH 3T3 fibroblasts and NIH 3T3 fibroblasts stably transfected with a plasmid directing expression of a fusion protein of red fluorescent protein (RFP) and actin were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 2 mM l-glutamine and 10% (vol/vol) heat-inactivated fetal bovine serum (FBS). MDA-MB-231 breast cancer cells stably expressing [Y527F]c-Src ([Y527F]c-Src-MDA-MB-231) were maintained in phenol red-free DMEM supplemented with 10% (vol/vol) heat-inactivated FBS, 2 mM l-glutamine, and 100 units/ml penicillin-100 μg/ml streptomycin. NIH 3T3 fibroblasts were seeded at 5 × 104 cells/cm2 overnight either in tissue culture dishes or in 12-well tissue culture plates containing fibronectin-coated (25 μg·ml−1) (Sigma, St. Louis, MO) coverslips. Cells were transfected for 18 to 24 h using FuGENE 6 per the manufacturer's instructions (Roche Diagnostics Corp., Indianapolis, IN). For cotransfections, equal amounts of each plasmid DNA were used. 293T cells were handled similarly, but Lipofectamine 2000 (Invitrogen) was used for transfections.
Antibodies.
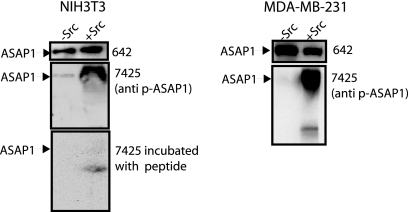
Rabbit polyclonal anti-ASAP1 serum (antibody 642) has previously been described (33). Mouse anti-c-Src immunoglobulin G (IgG) (monoclonal antibody 327) used for immunofluorescence was a kind gift of Joan Brugge (Department of Cell Biology, Harvard Medical School, Boston, MA). Monoclonal anti-c-Src antibody (clone EC10) from Upstate Biotech (Lake Placid, NY) was used for immunoblotting. Polyclonal and monoclonal (clone M5) antibodies recognizing the Flag epitope were from Sigma. Rabbit anti-actin IgG was from Molecular Probes (Eugene, OR). A rabbit polyclonal antibody for GFP was from Invitrogen, and a monoclonal antibody was from Covance. Antibodies against cortactin (clone 4F11), FAK (clone 2A7), phosphotyrosine (clone 4G10), and the myc epitope were from Upstate Biotech (Lake Placid, NY). An antibody for paxillin was from BD Transduction Laboratories (San Diego, CA). Rockland Antibody Company (Gilbertsville, PA) raised antisera against the phosphopeptide acetyl-KQRLS[pY]GAFTN-Ahx-C-amide for the detection of ASAP1 phosphorylated on Tyr-782. The antibody, named pAB7425, was affinity purified and used for immunoblotting. The antibody did not detect protein in NIH 3T3 fibroblasts or MDA-MB-231 cells without active Src. In cells expressing [Y527F]c-Src, the antibody detected a protein migrating with an Mr of 125,000 by immunoblotting (Fig. 1). The signal was blocked by the peptide to which the antibody was raised.
FIG. 1.
Characterization of an antibody raised against a phosphopeptide from ASAP1. An antibody (642) raised against ASAP1 and an affinity-purified antibody (pAB7425) from a serum raised against a phosphopeptide from ASAP1, containing phospho-Y782 (P-Y782), were used to immunoblot lysates from NIH 3T3 and MDA-MB-231 cells expressing [Y527F]c-Src, as indicated. In the lower left panel, pAB7425 was mixed with the peptide against which it was raised prior to immunoblotting.
Immunofluorescence and microscopy.
Cells were fixed, permeabilized, and stained for immunofluorescence microscopy as described previously (33). Rhodamine-conjugated phalloidin and Alexa 647-conjugated phalloidin (Molecular Probes) were used to detect F-actin. Rabbit primary antibodies were detected with goat anti-rabbit IgG conjugated to Alexa 488 (Molecular Probes). Mouse monoclonal antibodies were detected using Alexa 594 goat anti-mouse IgG (Molecular Probes). Confocal microscopy was performed using an LSM510 or an LSM510 Meta confocal microscope attached to a Zeiss Axiovert 100 M microscope equipped with a 63× 1.4-numerical-aperture (NA) Plan-Neofluar oil immersion lens (Carl Zeiss, Thornwood, NY).
Live-cell imaging.
NIH 3T3 cells stably transfected with RFP-actin were transiently transfected with enhanced GFP (eGFP)-ASAP1b and [Y527F]c-Src. After 24 h, 4 x 105 cells were seeded on chambered glass slides coated with fibronectin. The movements of fluorescent proteins in live cells were observed with a Zeiss Axiovert 200 microscope equipped with a PerkinElmer Ultraview spinning disc confocal system (PerkinElmer, Boston MA), using a 63× 1.4 NA Plan-Neofluar oil immersion objective. Images were captured with an Orca-ERII charge-coupled-device camera (Hamamatsu, Bridgewater, NJ). Photobleaching studies were performed using the PhotoKinesis attachment, using both the 488- and 568-nm laser lines for bleaching.
Movies were produced from a series of TIFF images of single confocal sections using IPLab3.6 (Scanalytics, Inc., Fairfax, VA).
Protein extraction, immunoprecipitation, and immunoblotting.
Proteins were extracted in 20 mM Tris-Cl (pH 8.0), 100 mM NaCl, 10% glycerol as described previously (26). Protease activity was inhibited using Roche Complete mini protease inhibitor cocktail tablets (Roche, Mannheim, Germany). For experiments examining phosphotyrosine levels, 1 mM Na3VO4 was included in the extraction buffer. Ectopically expressed protein levels and actin protein levels were determined by immunoblotting whole-cell lysates. For immunoprecipitation, cell lysates were incubated overnight with 15 μl of anti-Flag M2-agarose affinity gel (Sigma). Samples were centrifuged and rinsed three times with lysis buffer. Bound proteins were eluted with 30 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer. Eluted proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose. Horseradish peroxidase-conjugated secondary antibodies were detected with the ECL Plus Western blotting detection system (Amersham Biosciences, Piscataway, NJ).
Gelatin matrix degradation assay.
Gelatin Oregon Green-Alexa 488 conjugate was purchased from Molecular Probes and resuspended according to the manufacturer's instructions. Poly-d-lysine, sodium borohydride (NaBH4), and porcine type A gelatin were from Sigma; glutaraldehyde was purchased from Ted Pella, Inc. (Redding, CA). Gelatin-coated coverslips were prepared as described previously (2) with the following modifications. Coverslips were first incubated at room temperature for 20 min in 50 mg/ml poly-d-lysine and then rinsed with phosphate-buffered saline (PBS). Coverslips were then treated with 0.5% (vol/vol) glutaraldehyde in PBS for 15 min and washed with PBS. Gelatin Oregon Green-Alexa 488 was diluted 1:30 in 0.2% (wt/vol) unlabeled gelatin and preheated to 37°C prior to incubation on coverslips; coverslips were rinsed and then used. NIH 3T3 fibroblasts were seeded in 10% FBS-DMEM and incubated for 18 to 24 h on gelatin-coated coverslips prior to fixation and staining. [Y527F]c-Src-MDA-MB-231 breast cancer cells were analyzed following a 5-h incubation on gelatin coverslips. Cells were counted as having a degraded matrix when black regions, indicative of the loss of fluorescently labeled gelatin, were observed beneath the cell.
ASAP1 siRNA.
SMARTpool siRNA from Dharmacon was used to target ASAP1 in NIH 3T3 fibroblasts (GenBank accession no. NM_010026) and MDA-MB-231 cells (GenBank accession no. NM_018482). HiPerfect transfection reagent from Qiagen was used for this purpose. For siRNA rescue experiments, custom-made ASAP1 siRNA against the 3′ untranscribed region (UTR) was designed by Dharmacon and was electroporated using an Amaxa kit and instrument (Amaxa, Gaithersburg, MD) according to the manufacturer's protocol. GCAUGCAUCUAGACGCUUAUU was the sense sequence and phospho-UAAGCGUCUAGAUGCAUGCUU was the antisense sequence used. Cells were used 72 h posttransfection. A siCONTROL nontargeting siRNA pool (product no. D-001206-13-05; Dharmacon) was used as a control in some experiments. It had no effect on podosomes (not shown).
RESULTS
Isoform specificity of ASAP1 association with invadopodia.
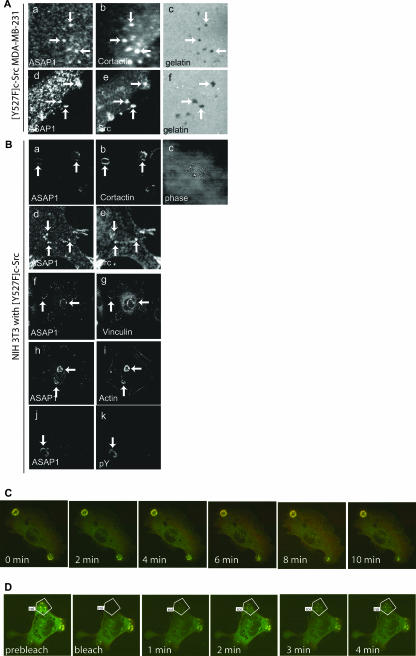
To study the function of ASAP1 in invadopodium formation, we set out to determine the structural requirements, starting with an examination of two ASAP1 isoforms. In previous work, ASAP1b was found to associate with and regulate FAs in a number of cell lines (21, 22, 33). ASAP1a associates with invadopodia in a breast cancer cell line, MDA-MB-231 (29). Invadopodia and analogous structures called podosomes in NIH 3T3 fibroblasts are induced by Src activation. Both invadopodia and podosomes are found on the ventral surfaces of cells and contain a complex of proteins, including actin, cortactin, vinculin, and Src. The structures are rich in protein tyrosine phosphate. We examined the localization of endogenous ASAP1 in a cell line derived from MDA-MB-231, called [Y527F]c-Src-MDA-MB-231, which stably expresses activated Src, and in NIH 3T3 fibroblasts that transiently express the active Src mutant, [Y527F]c-Src. ASAP1 associated with punctate structures on the ventral surface of MDA-MB-231 cells (Fig. 2A) and NIH 3T3 fibroblasts (Fig. 2B). In the latter case, the puncta often associated in circular patterns termed rosettes (Fig. 2B). These structures contained cortactin (Fig. 2A and B, panels b), Src (Fig. 2A and B, panels e), vinculin (Fig. 2B, panel g), actin (Fig. 2B, panel i), and phosphotyrosine (Fig. 2B, panel k). Invadopodia can proteolytically digest cross-linked gelatin from surfaces on which cells are grown. We found that in MDA-MB-231 cells, the gelatin matrix was degraded directly under the structures containing ASAP1, cortactin, and Src (Fig. 2A, panels c and f). Using live-cell imaging of cells stably expressing RFP-actin and transiently expressing eGFP-ASAP1b and [Y527F]c-Src, we found that some podosomes turned over on a minute time scale (Fig. 2C). ASAP1b association with podosomes paralleled their formation as detected by actin. In podosomes that appeared to be stable, photobleaching experiments revealed that actin turnover rates were on the time scale of minutes. The rates of ASAP1b and actin association were indistinguishable (Fig. 2D). Based on these criteria, we conclude that ASAP1b is an integral component of invadopodia in MDA-MB-231 cells as previously reported (29) and podosomes in NIH 3T3 fibroblasts.
FIG. 2.
Localization of ASAP1. (A) MDA-MB-231 cells. MDA-MB-231 cells stably expressing [Y527F]c-Src were grown on Alexa 488-conjugated gelatin and stained for ASAP1 (a and d) and either cortactin (b) or Src (e). The fluorescently labeled gelatin matrix is visualized (c and f). Black areas (arrows) are regions where gelatin matrix degradation has occurred. (B) NIH 3T3 fibroblasts. NIH 3T3 fibroblasts transiently transfected with plasmids directing the expression of [Y527F]c-Src were grown on a gelatin matrix, fixed, and stained for ASAP1 (a, d, f, h, and j) and either cortactin (b), Src (e), vinculin (g), actin (i), or phosphotyrosine (pY) (k). A phase-contrast image is shown (c). (C) Live-cell imaging of podosomes. NIH 3T3 fibroblasts stably expressing RFP-actin were transfected with plasmids directing the expression of eGFP-ASAP1b and [Y527F]c-Src. The cells were imaged for 10 min. (D) ASAP1 and actin turnover in podosomes. The indicated region of NIH 3T3 fibroblasts were photobleached (area outlined in white), and the recovery of red (RFP-actin) and green (GFP-ASAP1b) fluorescence was determined. roi, region of interest.
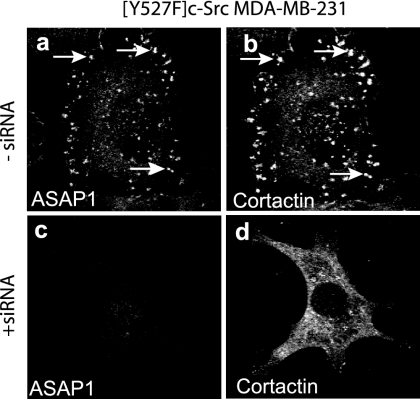
The reduction of ASAP1 expression levels has been reported to diminish the formation of invadopodia in mammary carcinoma cells (29). We obtained similar results. ASAP1 levels were reduced through the use of siRNA (siRNA targeted both isoforms) in the mammary carcinoma cell line [Y527F]c-Src-MDA-MB-231 (Fig. 3) and in NIH 3T3 fibroblasts that expressed [Y527F]c-Src (see Fig. 5A, panels a through f). In cells with reduced ASAP1 expression, cortactin did not target invadopodia or podosomes nor did the cells contain polymerized actin typical of these structures (see Fig. 5).
FIG. 3.
Effect of reduction of ASAP1 levels on [Y527F]c-Src-MDA-MB-231 cells. Cells grown on fibronectin-coated coverslips were treated with siRNA targeting ASAP1 and stained for ASAP1 (a and c) and cortactin (b and d).
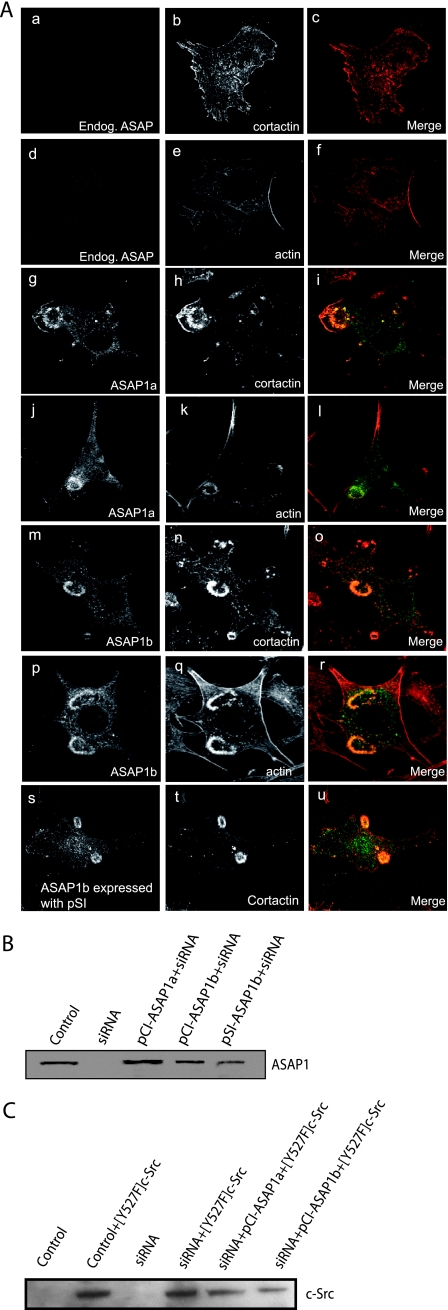
FIG. 5.
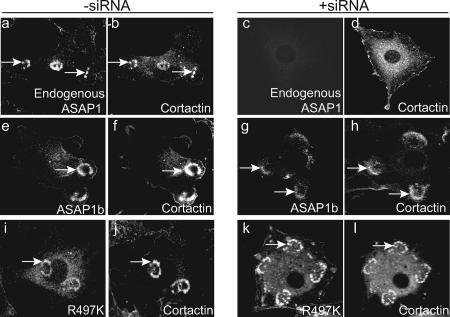
Effect of ASAP1a and ASAP1b on the localization of cortactin in NIH 3T3 fibroblasts transformed with [Y527F]c-Src. NIH 3T3 fibroblasts were transfected with a plasmid directing the expression of [Y527F]c-Src and siRNA to the 3′UTR of ASAP1 to reduce the levels of endogenous ASAP1. As indicated, the cells were also transfected with plasmids with the cytomegalovirus promoter directing the expression of Flag-ASAP1a (pCI-ASAP1a), Flag-ASAP1b (pCI-ASAP1b), or a plasmid with a simian virus 40 promoter directing the expression of Flag-ASAP1b (pSI-ASAP1b); these cDNAs are resistant to the siRNA. (A) Representative micrographs. Podosomes were visualized using an antibody to cortactin or rhodamine-phalloidin. Quantitation is provided in Table 1. (B) Relative expression levels of ASAP1. ASAP1 was detected in the indicated cell lysates by immunoblotting with antibody 642 for ASAP1. (C) Relative expression levels of [Y527F]c-Src. Src in the indicated cell lysates was detected by immunoblotting.
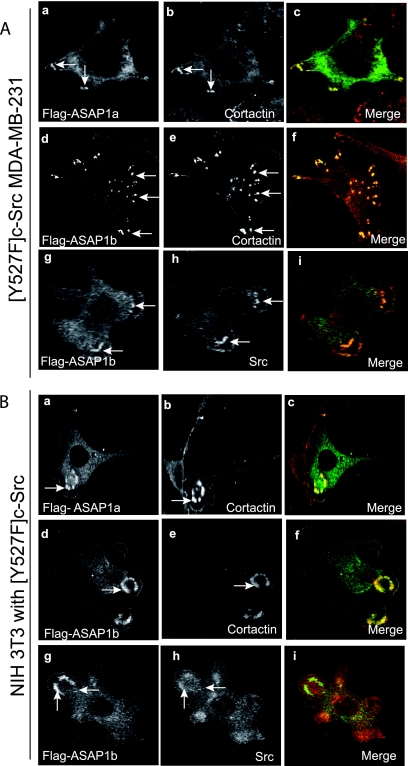
Using recombinant ASAP1, we determined whether the association with podosomes and invadopodia was specific for ASAP1 isoforms. Both ASAP1a and ASAP1b associated with invadopodia in [Y527F]c-Src-MDA-MB-231 cells (Fig. 4A). Our live-cell imaging experiments supported the idea that ASAP1b is in podosomes (Fig. 2C and D). To further examine podosomes, NIH 3T3 fibroblasts were cotransfected with plasmids for expression of [Y527F]c-Src and either epitope-tagged ASAP1a or ASAP1b (Fig. 4B). Podosomes were detected with antibodies for cortactin. Both ASAP1 isoforms associated with podosomes with the same efficiency. Because ASAP1b was previously reported to not associate with invadopodia (29), we examined Src as an additional marker. Src colocalized with ASAP1b in both cell types.
FIG. 4.
Comparison of ASAP1a and ASAP1b. (A) Localization of ASAP1a and ASAP1b in [Y527F]c-Src-MDA-MB-231 cells. Cells were transfected with plasmids directing expression of Flag-ASAP1a (a to c) and Flag-ASAP1b (d to i). ASAP1 was detected with antibody to the Flag epitope (a, d, and g) and either cortactin (b and e) or Src (h). (B) Localization of ASAP1a and ASAP1b in Src-transformed NIH 3T3 fibroblasts. NIH 3T3 fibroblasts expressing [Y527F]c-Src and either Flag-ASAP1a (a to c) or Flag-ASAP1b (d to i) were stained for ASAP1 using an antibody to the Flag epitope (a, d, and g) and either cortactin (b and e) or Src (h). Panels c, f, and i are the merged images.
Previous reports indicate that ASAP1a associates with invadopodia but that ASAP1b does not because ASAP1b lacks the cortactin binding site. We considered the possibility that our results differed because ASAP1 forms dimers (27). A heterodimer of ASAP1a/ASAP1b might associate with invadopodia since one subunit would have the putative cortactin binding site. To test this idea, we examined the individual isoforms expressed as recombinant proteins in NIH 3T3 fibroblasts in which expression of ASAP1 was reduced with siRNA that targeted both isoforms. NIH 3T3 fibroblasts with reduced ASAP1 expression did not form podosomes when [Y527F]c-Src was expressed (Fig. 5A and Table 1). Podosome formation, detected by staining for cortactin or actin, could be restored by expression of either recombinant ASAP1a or ASAP1b (Fig. 5A and Table 1). Both forms of ASAP1 associated with podosomes. In these experiments, recombinant ASAP1 was expressed using a plasmid (pCI-ASAP1) containing a cytomegalovirus promoter resulting in relatively high expression levels. We also examined the effect of recombinant ASAP1b expressed from a plasmid containing the weaker simian virus 40 promoter (pSI-ASAP1b) (relative expression levels shown in Fig. 5B). Even at lower levels of expression, ASAP1b had an efficiency for supporting podosome formation similar to that of ASAP1a (Table 1). [Y527F]c-Src was expressed to similar levels under each condition (Fig. 5C).
TABLE 1.
Effect of ASAP1 mutants on podosome formation
| Recombinant protein | % of cells with podosomes (no. of expts)a
|
|
|---|---|---|
| −siRNA | +siRNA | |
| None | 38 ± 5 (7) | 6 ± 4 (6)** |
| ASAP1a | 35 ± 4 (7) | 31 ± 3 (6) |
| ASAP1b | 46 ± 10 (5) | 38 ± 7 (4) |
| ASAP1bb | 30 ± 3 (5) | 30 ± 2 (4) |
| [R497K]ASAP1b | 42 ± 10 (5) | 33 ± 5 (4) |
| [Y782F]ASAP1b | 2 ± 3 (5)** | 4 ± 5 (4)** |
| [R811A]ASAP1b | 29 ± 3 (5) | 32 ± 5 (3) |
| [R1021A]ASAP1b | 38 ± 7 (5) | 29 ± 8 (4) |
| [Y1094A]ASAP1a | 24 ± 4 (7)** | 22 ± 3 (6)* |
| [E1103A]ASAP1a | 23 ± 7 (7)** | 22 ± 10 (6)* |
| [W1122A]ASAP1a | 13 ± 1 (2)** | 12 ± 1 (2)** |
| [ΔBAR]ASAP1b | 31 ± 4 (5) | 15 ± 6 (4)** |
| [ΔSH3]ASAP1b | 4 ± 3 (5)** | 7 ± 3 (4)** |
| [817-873]ASAP1a | 25 ± 5 (3)* | ND |
Podosomes were visualized in NIH 3T3 fibroblasts expressing [Y527F]c-Src and the indicated recombinant ASAP1 protein. Where indicated, the fibroblasts were left untreated (−siRNA) or were treated with siRNA (+siRNA) targeting endogenous ASAP1 but not the RNA for the recombinant proteins. Cells were scored for the presence of podosomes. At least 30 cells were counted for each experiment. The values presented are means ± standard deviations. The data were analyzed by analysis of variance, followed by Dunnett's multiple-comparison test. For the first column, significant differences from the value for no recombinant ASAP1 are indicated by asterisks; for the second column, significant differences from the value for cells expressing ASAP1a are indicated by asterisks (*, P < 0.05; **, P < 0.01). ND, not determined.
Expressed using the pSI plasmid resulting in levels less than those achieved with the pCI plasmid used for all other constructs presented in this table (Fig. 5B).
Role of Arf GAP activity for invadopodium formation.
ASAP1 could contribute to the formation of podosomes or invadopodia in several ways. We first considered the contribution of Arf GAP activity of ASAP1. [R497K]ASAP1b, a mutant with 10−5-fold the activity of wild-type ASAP1 when measured in vitro (23), was compared to the wild-type protein. In NIH 3T3 fibroblasts expressing [Y527F]c-Src, Flag-[R497K]ASAP1b associated with podosomes to the same extent as did wild-type ASAP1b (Fig. 6). Replacing endogenous ASAP1 with Flag-[R497K]ASAP1b restored podosome formation to the same extent as that of wild-type ASAP1b (Fig. 6 and Table 1), leading us to conclude that GAP activity was not critical for forming podosomes.
FIG. 6.
Effect of ASAP1 Arf GAP activity on podosome formation. The left micrographs (−siRNA) are NIH 3T3 fibroblasts either transfected with a plasmid directing expression of [Y527F]c-Src (a and b) or cotransfected with the plasmid for [Y527F]c-Src and plasmids directing expression of Flag-ASAP1b (e and f) or Flag-[R497K]ASAP1b (i and j). On the right (+siRNA), NIH 3T3 fibroblasts were transfected with plasmids for [Y527F]c-Src and siRNA targeted to the 3′UTR of mouse ASAP1 mRNA and, for some panels, ASAP1b (g and h) or [R497K]ASAP1b (k and l) plasmids resistant to this siRNA. Podosomes were detected with an antibody to cortactin.
Role of the BAR domain for invadopodium formation.
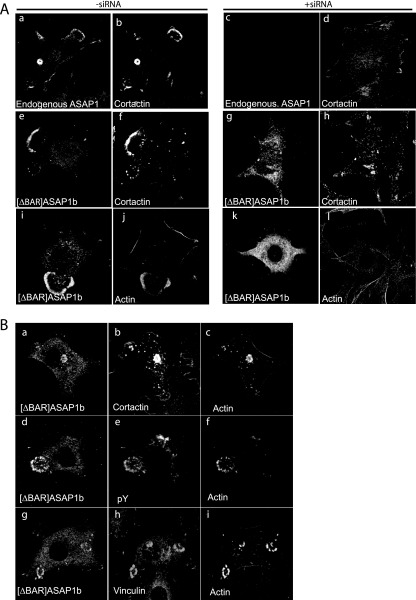
The BAR domain is a recently identified structure within ASAP1 (27, 32) that causes membrane tubulation and may mediate protein-protein binding. We tested for a function in invadopodium formation by examining a recombinant ASAP1 lacking the BAR domain, [ΔBAR]ASAP1b (Flag-[331-1091]ASAP1b). [ΔBAR]ASAP1b had no detectable effect on the formation of podosomes when endogenous ASAP1 was present (Fig. 7A and B and Table 1). To confirm that podosomes were formed, cells were triple stained to examine ASAP1 and actin together with cortactin, phosphotyrosine, or vinculin as additional markers of podosomes. The structures contained all markers that were examined. We concluded that the structures were either podosomes or podosome-like and that [ΔBAR]ASAP1 did not affect podosome formation when endogenous ASAP1 was present. However, cells in which endogenous ASAP1 was replaced with [ΔBAR]ASAP1b formed few podosomes when expressing [Y527F]c-Src (Fig. 7 and Table 1), leading us to further conclude that the BAR domain was important for this function of ASAP1.
FIG. 7.
(A) Role of the BAR domain of ASAP1 in podosome formation. The experiment was performed as described for Fig. 6 but using [ΔBAR]ASAP1b to replace endogenous ASAP1. Podosomes were detected with antibodies to cortactin (b, f, d, and h). Cells were also stained for polymerized actin (j and l). (B) Analysis of podosome-like structures formed in the presence of [ΔBAR]ASAP1 and endogenous ASAP1. NIH 3T3 fibroblasts forming podosomes were triple labeled for the indicated proteins or the protein phosphotyrosine (pY).
Role of ASAP1/protein association in invadopodium formation.
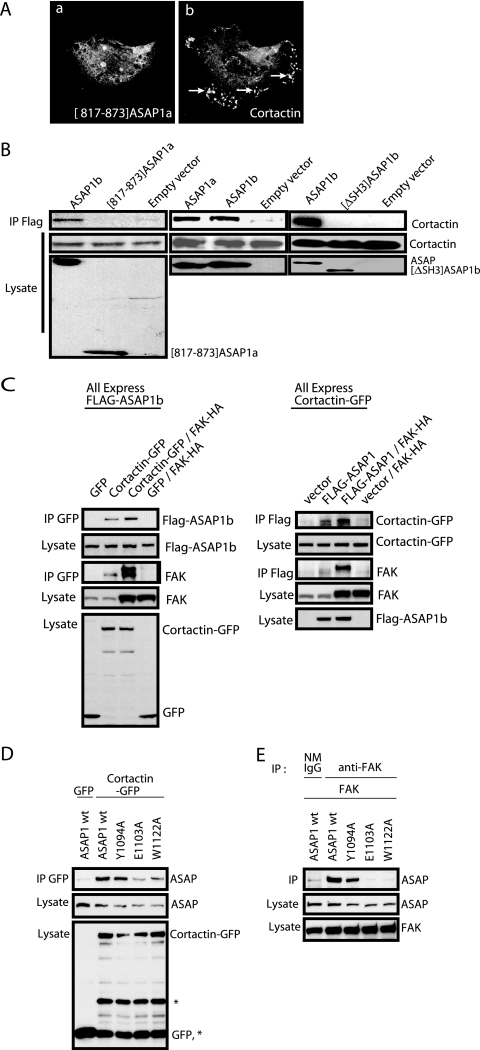
We next considered the role of three proteins that associate with ASAP1 for the formation of invadopodia: cortactin, Src, and FAK. All three are involved in invadopodium and podosome formation, and the last two are also involved in FA dynamics. We started by examining cortactin binding, since this was found in previous work to be critical (12, 29). We found that the natural splice variant ASAP1b that lacked the putative cortactin binding site was as efficient as ASAP1a in supporting podosome formation (Fig. 5 and Table 1). Residues 817 to 873 of ASAP1a (these residues, which contain a PPPPPP motif, are not present in ASAP1b) have been reported to directly bind cortactin (29). To test this assertion, NIH 3T3 fibroblasts coexpressing [Y527F]c-Src and Flag-[817-873]ASAP1a were examined by immunofluorescence (Fig. 8A). Flag-[817-873]ASAP1a did not colocalize with cortactin. It slightly decreased the number of cells forming podosomes (Table 1). When proteins were immunoprecipitated through the Flag epitope, cortactin was not detected with [817-873]ASAP1a. In contrast, both ASAP1a and ASAP1b associated with cortactin in immunoprecipitates (Fig. 8B). These results support the idea that the PPPPPP motif within this region of ASAP1a does not mediate binding to cortactin.
FIG. 8.
ASAP1-cortactin association. (A) Cellular distribution and effects of [817-873]ASAP1a. Flag-[817-873]ASAP1 was expressed in NIH 3T3 fibroblasts with [Y527F]c-Src. Podosomes were detected with an antibody to cortactin. Arrows indicate podosomes. (B) Association of ASAP1 with cortactin. NIH 3T3 fibroblasts were transfected with plasmids directing the expression of the indicated proteins. Association of recombinant ASAP1 with cortactin was determined by immunoprecipitation (IP) through the Flag tag and immunoblotting for both the Flag tag and cortactin. (C) Effect of FAK on ASAP1-cortactin association. Flag-ASAP1b, GFP-cortactin, and HA-FAK were expressed in HEK 293T cells as indicated. In the experimental results shown on the left, proteins were immunoprecipitated with an antibody for GFP. The levels of Flag-ASAP1, GFP-cortactin, and HA-FAK were determined in the lysates and precipitates. In the experimental results shown on the right, proteins were immunoprecipitated with an antibody to the Flag tag. (D) Effect of point mutations within the SH3 domain of ASAP1 on cortactin binding. NIH 3T3 fibroblasts were transfected with expression vectors for Flag-ASAP1 wild type (wt) or the indicated SH3 mutants (W1122A, Y1094A, and E1103A) with GFP-tagged cortactin. Immunoprecipitation was done with anti-GFP polyclonal antibody, and the precipitated materials were analyzed by immunoblotting with monoclonal anti-Flag antibody (M5) or monoclonal anti-GFP antibody. *, GFP-cortactin degradation products. (E) Effect of point mutations within the SH3 domain of ASAP1 on FAK binding. NIH 3T3 fibroblasts were transfected with expression vectors for FLAG-tagged wild-type ASAP1 or the indicated ASAP1 mutants with FAK. Cells were lysed and proteins immunoprecipitated as described for panel D, except that anti-FAK 2A7 monoclonal antibody and normal mouse IgG (NMIgG) (as the negative control) were used instead of anti-GFP for immunoprecipitation.
We considered the possibility that the SH3 domain of ASAP1 mediates binding to the proline-rich domain of cortactin. To test this idea, Flag-[ΔSH3]ASAP1b (Flag-[1-1022]ASAP1b) or Flag-tagged wild-type ASAP1b were coexpressed with [Y527F]c-Src in NIH 3T3 fibroblasts. The cells were lysed, and proteins were immunoprecipitated from the lysates with an antibody to the Flag epitope. Similar levels of Flag-[ΔSH3]ASAP1b and Flag-ASAP1b were detected in the precipitates (Fig. 8B). As previously observed, precipitates with Flag-ASAP1b contained cortactin. In contrast, cortactin was not detected in precipitates with Flag-[ΔSH3]ASAP1b (Fig. 8B). These results are consistent with cortactin binding being dependent on the SH3 domain.
The SH3 domain of ASAP1 also binds to FAK (21). We tested for competitive binding of FAK and cortactin to the SH3 domain. Flag-tagged ASAP1b, hemagglutinin (HA)-tagged FAK, and GFP-tagged cortactin were expressed in combination as indicated in Fig. 8C. We immunoprecipitated cortactin through the GFP tag and examined precipitates for Flag-tagged ASAP1b (Fig. 8C, left). We found no detectable ASAP1b signal if cortactin was not expressed. More ASAP1b was detected in precipitates from cells expressing HA-FAK than in those that did not. We also immunoprecipitated through the Flag tag and examined the precipitates for the presence of GFP-cortactin. More cortactin was detected with the expression of HA-FAK than in the cells not expressing HA-FAK. Furthermore, in both experiments, FAK was found in the precipitates with ASAP1 and cortactin. Based on these combined results, we concluded that FAK and cortactin binding to ASAP1 are codependent.
Several amino acids within the SH3 domain of ASAP1 align with highly conserved residues in other SH3 domains that mediate binding to PXXP motifs (1, 19, 20). To further characterize the role of the SH3 domain in cortactin binding and the relationship to FAK binding, we mutated three residues in ASAP1a—Tyr-1094, Glu-1103, and Trp-1122—to Ala. Using immunoprecipitation, we determined the binding levels of the mutant proteins to cortactin and FAK (Fig. 8D and E). The three mutants were able to bind to cortactin, but [W1122A] ASAP1a and [E1103A]ASAP1a did not bind cortactin as efficiently as wild-type ASAP1b or [Y1094A]ASAP1a (Fig. 8D). [Y1094A]ASAP1a bound effectively to FAK, whereas both [W1122A]ASAP1a and [E1103A]ASAP1a did not bind FAK. Thus, although FAK enhances the cortactin-ASAP1 association, cortactin can bind ASAP1 without FAK.
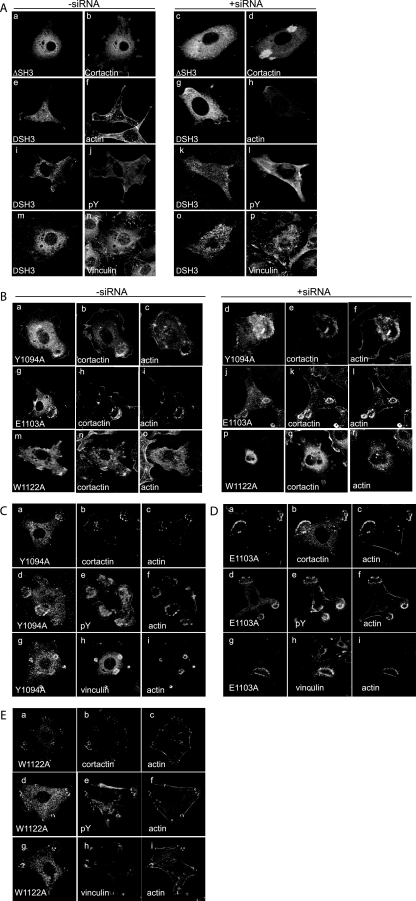
The SH3 domain of ASAP1 is critical for the ability of ASAP1 to support podosome formation. As a first test of the function of the SH3 domain, the effects of expressing ASAP1 and [ΔSH3]ASAP1b in NIH 3T3 fibroblasts were compared. We found that [ΔSH3]ASAP1b disrupted podosome formation (Fig. 9 and Table 1). In cells with reduced levels of endogenous ASAP1, expression of [ΔSH3]ASAP1 together with [Y527F]c-Src did not induce the formation of podosomes. Instead, cortactin was present in amorphous structures in some cells (Fig. 9A, panel d). Cortactin and [ΔSH3]ASAP1 did not colocalize. Polymerized actin was present in the cell cortex; some puncta of actin were observed, but these were not on the ventral surface of the cell nor did they colocalize with ASAP1 (Fig. 9A, panels f and h). [ΔSH3]ASAP1 did not colocalize with phosphotyrosine (Fig. 9A, panels j and l) or vinculin (Fig. 9A, panels n and p), two other markers of podosomes. To further characterize the role of the SH3 domain, the effect of expressing [W1122A]ASAP1a, [Y1094A]ASAP1a, and [E1103A]ASAP1a on the formation of podosomes was determined (Fig. 9B and Table 1). Each blocked the formation of podosomes in the presence of endogenous ASAP1. The effects of [Y1094A]ASAP1a and [E1103A]ASAP1a were modest. [W1122A]ASAP1a blocked to the greatest extent. The podosomes or podosome-like structures that formed contained cortactin, actin, phosphotyrosine, and vinculin (Fig. 9C, D, and E). After depletion of endogenous ASAP1 with siRNA, [Y1094A]ASAP1a and [E1103A]ASAP1a supported podosomes or podosome-like structures (Fig. 9B) but less efficiently than did wild-type ASAP1a or ASAP1b (Table 1). [W1122A]ASAP1a poorly supported the formation of podosomes or podosome-like structures (Fig. 9B and Table 1).
FIG. 9.
(A) Role of SH3 domain of ASAP1 for podosome formation. The experiment was performed as described in the legend to Fig. 6. Representative micrographs are shown, with the quantitation presented in Table 1. Flag-[ΔSH3]ASAP1 transfected cells were detected with polyclonal anti-Flag antibody (a, e, c, and g). Podosomes were detected with cortactin (b and d), actin (f and h), phosphotyrosine (pY) (j and l), and vinculin (n and p). (B) Effect of point mutations within the SH3 domain on podosome formation. NIH 3T3 cells were transfected with the W1122A, Y1094A, and E1103A point mutants and [Y527F]c-Src in the presence (−siRNA) or absence (+siRNA) of endogenous ASAP1. Cells were stained with polyclonal anti-Flag antibody (a, g, m, d, j, and p) and anticortactin antibody (b, h, n, e, k, and q) and for polymerized actin (c, i, o, f, l, and r). (C, D, and E) Analysis of podosome-like structures formed in cells expressing ASAP1 with mutations in the SH3 domain. Cells expressing [Y1094A]ASAP1 (C), [E1103A]ASAP1 (D), or [W1122A]ASAP1 (E) were triple stained for the indicated proteins.
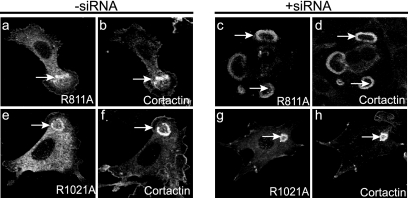
A PXXP motif at residues 806 to 811 of ASAP1 is highly conserved and binds to Src (3). We used ASAP1b with a mutation in this motif ([R811A]ASAP1b) to determine if Src binding was necessary for ASAP1 function. [R811A]ASAP1b associated with podosomes as efficiently as wild-type recombinant protein in the presence of endogenous protein (Fig. 10). We also examined cells in which endogenous ASAP1 levels were reduced with siRNA. [R811A]ASAP1b was as efficient as wild-type ASAP1 in restoring podosomes (Fig. 10 and Table 1). We used the same approach to examine the role of the atypical PXPXP motif at residues 1016 to 1021 (ASAP1b, mouse sequence), which binds CIN85 (17). ASAP1 with this motif mutated, [R1021A]ASAP1b, was indistinguishable from wild-type Flag-ASAP1b in associating with podosomes and supporting their formation when endogenous ASAP1 was reduced (Fig. 10 and Table 1).
FIG. 10.
Role of SH3-binding proline-rich motifs in ASAP1 for podosome formation. The experiment was performed as described in the legend to Fig. 6 using plasmids directing expression of Flag-[R811A]ASAP1b or Flag-[R1021A]ASAP1b. Quantitation is provided in Table 1. Transfected cells were visualized using polyclonal anti-Flag antibody (a, e, c, and g). Podosomes were visualized using cortactin (b, f, d, and h).
The role of ASAP1 phosphorylation in podosome formation.
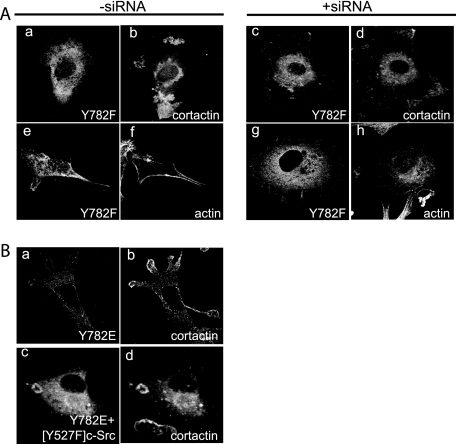
ASAP1 has been found to bind to both Src and FAK and to be phosphorylated by Src and the FAK homolog Pyk2. Because of the importance of Src- and FAK-dependent phosphorylation for podosome and invadopodium formation (5, 11), we considered the possibility that phosphorylation of ASAP1 may have a function in podosome formation. ASAP1 is phosphorylated on Tyr-782 (18). As an initial test of the idea, we mutated Tyr-782 to Phe in ASAP1b. Without [Y527F]c-Src, [Y782F]ASAP1b was diffusively distributed, with a small amount concentrated at the cell edge (data not shown). In cells expressing [Y527F]c-Src, coexpression of [Y782F]ASAP1b prevented the formation of podosomes, which were detected by staining for either cortactin or actin (Fig. 11A). Instead, cortactin was found in amorphous aggregates. Similarly, in cells with reduced levels of endogenous ASAP1, [Y782F]ASAP1b did not support the formation of Src-dependent podosomes. Based on these results, we concluded that Tyr-782 was necessary for podosome formation, presumably functioning as a phosphate acceptor for a kinase. We next considered whether ASAP1 phosphorylation is sufficient for podosome formation. As a test, we generated a mutant cDNA encoding ASAP1b with a Glu in place of Tyr-782, reasoning that the negative charge of the amino acid side group would mimic a phosphate (Fig. 11B). [Y782E]ASAP1 did target podosomes like wild-type protein. However, the expression of [Y782E]ASAP1 was not sufficient to induce podosomes.
FIG. 11.
Role of Tyr-782 of ASAP1 for podosome formation. (A) [Y782F]ASAP1. This experiment was performed as described in the legend to Fig. 6 using a plasmid directing expression of [Y782F]ASAP1. Quantitation is provided in Table 1. (B) Localization of [Y782E]ASAP1. Flag-[Y782E]ASAP1 was expressed in NIH 3T3 fibroblasts that also expressed [Y527F]c-Src where indicated (c). The cells were immunostained with antibodies against the Flag epitope (a and c) and cortactin (b and d).
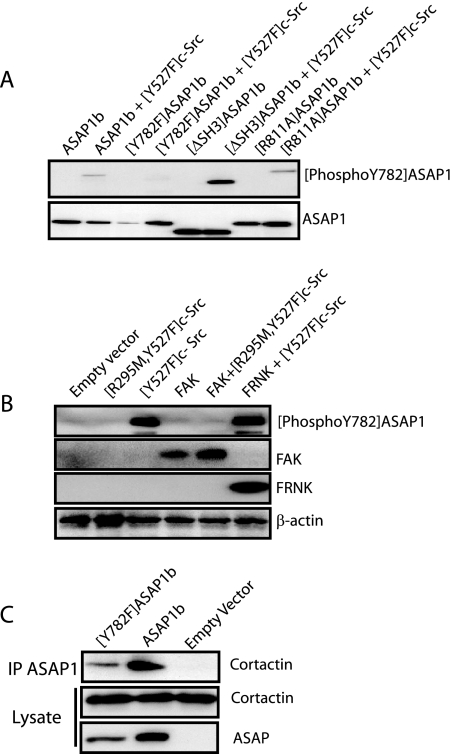
Mutants lacking the Src phosphorylation site or the FAK binding site blocked podosome formation. Both mutations might disrupt podosome formation secondary to disrupting phosphorylation of ASAP1. To test this idea, phosphorylation of ASAP1 was monitored using a specific antibody raised against a phosphopeptide as described in Materials and Methods. We found that expression of [Y527F]c-Src resulted in the phosphorylation of both [R811A]ASAP1, which does not bind Src efficiently, and [ΔSH3]ASAP1, which does not bind FAK (Fig. 12A). As anticipated, expression of [Y527F]c-Src did not result in phosphorylation of [Y782F]ASAP1. Overexpression of FAK did not result in the phosphorylation of ASAP1b as previously described (18) (Fig. 12B). Furthermore, ASAP1b was as efficiently phosphorylated by [Y527F]c-Src in the presence of FRNK, a dominant-negative form of FAK, as in its absence (Fig. 12B). Based on these results, we conclude that Src induces phosphorylation of ASAP1 on Tyr-782 independent of the function of FAK. As a test of a possible regulatory relationship between phosphorylation and protein binding to the SH3 domain of ASAP1, cortactin binding levels to wild-type and [Y782F]ASAP1 were compared in cells expressing [Y527F]c-Src (Fig. 12C). Cortactin associated with both proteins.
FIG. 12.
Src-dependent phosphorylation of ASAP1. (A) Phosphorylation of ASAP1 mutants. Phosphorylation of Flag-ASAP1, Flag-[Y782F]ASAP1, Flag-[ΔSH3]ASAP1, and Flag-[R811A]ASAP1 expressed in NIH 3T3 fibroblasts with [Y527F]c-Src, as indicated, was determined by immunoblotting with anti-[P-Y782]ASAP1 antibody, pAB7425 (see Materials and Methods for a description of this antibody). Total cell lysates were also probed with monoclonal Flag antibody to control for transfection efficiency (lower panel). (B) Role of Src and FAK in phosphorylation of ASAP1. [P-Y782]ASAP1 was determined in lysates of NIH 3T3 fibroblasts expressing [R295M, Y527F]c-Src, [Y527F]c-Src, FAK, FAK with [R295M, Y527F]c-Src, or [Y527F]c-Src with FRNK. Total cell lysates were prepared and probed with anti-[P-Y782]ASAP1 (antibody pAB7425), anti-FAK, anti-myc for detection of myc-FRNK, and an antibody against actin to control for loading. (C) Role of phosphorylation in cortactin binding. The indicated recombinant Flag-ASAP1b protein was expressed in NIH 3T3 fibroblasts. Proteins were immunoprecipitated through the Flag epitope. Immunoblottings of the immunoprecipitated (IP) material and total cell lysates were performed as indicated.
Taken together, our results support a model in which the function of ASAP1 in invadopodia is regulated through protein interaction mediated by the SH3 domain of ASAP1 and phosphorylation downstream of Src activation.
DISCUSSION
ASAP1 is an Arf GAP and a target of Src and FAK signaling that regulates FAs, podosomes, and invadopodia. These cytoskeletal structures are coordinately remodeled for cell movement and, in cancer cells, invasion. Our results indicate that podosomes are highly dynamic. Some podosomes turned over on a minute time scale. For those podosomes that did not, the two molecular components that we examined, actin and ASAP1, were in rapid flux. The rate of ASAP1 association was indistinguishable from that for actin. This result, together with the results of experiments in which reduced expression of ASAP1 blocked podosome formation, supported the idea that ASAP1 is an integral component of these structures. With the complex domain structure of ASAP1, we hypothesized that it could have a regulatory function. Therefore, to understand the basis for the regulation of podosomes and invadopodia, we examined the structural determinants of ASAP1 function in podosomes. We found that neither ASAP1 Arf GAP activity nor the Src binding site was important for the formation of podosomes. Phosphorylation of ASAP1 by Src and an intact SH3 domain on ASAP1 were required for podosome formation. Phosphorylation of ASAP1 was independent of the presence of the SH3 domain. Based on these results, we conclude that simultaneous association of protein with the ASAP1 SH3 domain and phosphorylation of ASAP1 are necessary to form podosomes.
Our work is the first examination of the role of phosphorylation of ASAP1 for cytoskeleton remodeling. Previous examination of Src- and Pyk2-dependent phosphorylation of ASAP1 focused on GAP activity (18). Both Src and Pyk2 were found to phosphorylate two Tyrs, one at position 308 (mouse sequence, accession no. AF 075462) and the other at 782. Phosphorylation of ASAP1 inhibited GAP activity. Tyr-308 is not present in the identified splice variants of the human protein so we did not examine the effect of mutating this residue. Tyr-782 and the surrounding 10 amino acids are identical in chimpanzee, chicken, dog, mouse, rat, cow, and human ASAP1 proteins. Consistent with the high degree of conservation of this sequence, Tyr-782 was critical to ASAP1 function. ASAP1 with Tyr-782 changed to phenylalanine functioned as a dominant negative, blocking podosome formation.
Though necessary for the formation of podosomes, phosphorylation of ASAP1 on Tyr-782 may not be sufficient for podosome formation. [Y782E]ASAP1 expression did not induce podosomes. In this mutant, a glutamate provides the negative charge at position 782, mimicking phosphotyrosine in the wild-type protein. Glutamate is not an ideal phosphomimetic. Nevertheless, it is possible that a second Src-dependent event may be necessary for podosome/invadopodium formation. Thus, ASAP1 may mediate one of several parallel events initiated by activated Src that lead to the formation of podosomes and invadopodia.
Src activity is necessary for ASAP1 phosphorylation, but the kinase that directly phosphorylates ASAP1 has not been identified. Both Src and Pyk2 have previously been implicated in ASAP1 phosphorylation (18). The Src binding site was not necessary for either Src-dependent podosome formation or phosphorylation of Tyr-782. We considered that Src might work through FAK or Pyk2; however, FRNK, a dominant negative form of FAK which should bind and thereby sequester the SH3 domain of ASAP1 from both FAK and Pyk2, had no effect on Src-dependent phosphorylation. Three explanations for the results are as follows: (i) Src bound the mutant with sufficient affinity so that, at high concentrations, it was able to directly phosphorylate ASAP1; (ii) Src or Pyk2 can phosphorylate without binding to a site distinct from the phosphorylation site; or (iii) another kinase in the Src cascade is responsible for directly phosphorylating ASAP1.
We found that the SH3 domain of ASAP1 was important for both podosome formation and cortactin binding. ASAP1 binding to cortactin has previously been reported to be important for invadopodium formation (12, 29). Our results differ from the previous reports in two ways. First, we found that ASAP1 associates with cortactin in cells that are not forming invasive structures in addition to those that do form invadopodia; previously ASAP1 was found to associate with cortactin only in invasive cells (9, 24). Second, the molecular basis for the interaction between ASAP1 and cortactin that we identified is different from previous work, which identified a PXXP motif that distinguishes ASAP1a from ASAP1b as the cortactin binding site (12, 29); ASAP1b could not be shown to interact with cortactin (11, 28). In contrast, we found that ASAP1a and ASAP1b bound cortactin to the same extent. Furthermore, a recombinant protein comprised of the PXXP motif identified as the binding site in the previous work neither bound cortactin, as determined by immunoprecipitation, nor colocalized with cortactin, as determined by immunofluorescence. This recombinant protein had little effect on the cellular localization of cortactin. Instead of a PXXP motif, the SH3 domain of ASAP1 contributes to binding cortactin. A recombinant ASAP1 mutant lacking the SH3 domain was unable to bind cortactin as determined by immunoprecipitation, did not colocalize with cortactin, and blocked podosome formation.
We are currently examining the molecular mechanism by which the SH3 domain of ASAP1 contributes to podosome formation and the binding of cortactin. Our result that FAK increased the interaction between ASAP1 and cortactin has led us to consider two mechanisms by which the SH3 domain may mediate binding to cortactin. One is that FAK acts as a bridge between the proteins. The results of experiments using SH3 domain point mutants exclude this possibility. The other is that FAK phosphorylates ASAP1 and/or cortactin directly or indirectly. The phosphorylation may stimulate the interaction of ASAP1 and cortactin. The data available from the point mutants did not provide a test of the second possibility but do indicate that FAK binding and cortactin binding were at least partly independent. A related unresolved question is whether cortactin, FAK, or both bind to ASAP1 to induce podosomes.
Another difference between our results and those previously reported (25) is related to the role of CIN85 binding to ASAP1. The interaction is mediated by the SH3 domains of CIN85 and an atypical PXPXPR motif in ASAP1. We found that a mutant ASAP1 that does not bind CIN85 supported podosome formation as well as did wild-type ASAP1. Dominant negative CIN85 has been reported to block invadopodium formation (25), which was interpreted as a requirement for the CIN85-ASAP1 complex. The apparent discrepancy between the results could be due to differences between cell types or between podosomes and invadopodia. Furthermore, CIN85 could function independently of ASAP1 to induce invadopodia, which would explain the results that a mutant of ASAP1 that does not bind CIN85 supported podosome formation but that dominant negative CIN85 blocked the formation.
We found that the BAR domain of ASAP1 contributes to podosome formation. In previous work, we found that the BAR domain induces tubulation of lipid bilayers (27). Given that podosomes are comprised of labyrinths of tubulated membranes (5), the BAR domain could contribute by inducing the observed tubulation of membranes. Alternatively, the BAR domain could function as a sensor of tubulation (8, 9, 14, 32), targeting ASAP1 to the site of imminent podosome formation, or the BAR domain could bind other proteins involved in forming podosomes.
With its multiple functional domains, ASAP1 may be a coincidence detector that regulates formation of podosomes/invadopodia. The result that both phosphorylation of ASAP1 and the presence of the SH3 domain were required for podosome formation has at least two possible explanations. One is that the events are codependent: phosphorylation is necessary for protein binding to the SH3 domain or vice versa. However, [ΔSH3]ASAP1 was efficiently phosphorylated; [Y782F]ASAP1, which cannot be phosphorylated, bound cortactin as efficiently as wild-type ASAP1; and the expression of [Y527F]c-Src did not affect ASAP1-cortactin association. The remaining explanation is that the two signals are independently generated but impinge on a single target, ASAP1. The simultaneous presence of the two signals leads to the cellular response.
Acknowledgments
We thank Joan Brugge (Harvard), J. Silvio Gutkind (NIDCR), Takehito Uruno (NHLBI), and J. Thomas Parsons (University of Virginia at Charlottesville) for reagents and J. Thomas Parsons for insightful discussions. We also thank Susan Garfield (NCI, Confocal Microscope Core) for assistance.
This work was supported by the Intramural Research Program of the National Cancer Institute, Department of Health and Human Services.
Footnotes
Published ahead of print on 24 September 2007.
REFERENCES
- 1.Arold, S., P. Franken, M. P. Strub, F. Hoh, S. Benichou, R. Benarous, and C. Dumas. 1997. The crystal structure of HIV-1 Nef protein bound to the Fyn kinase SH3 domain suggests a role for this complex in altered T cell receptor signaling. Structure 5:1361-1372. [DOI] [PubMed] [Google Scholar]
- 2.Bowden, E. T., M. Barth, D. Thomas, R. I. Glazer, and S. C. Mueller. 1999. An invasion-related complex of cortactin, paxillin and PKCμ associates with invadopodia at sites of extracellular matrix degradation. Oncogene 18:4440-4449. [DOI] [PubMed] [Google Scholar]
- 3.Brown, M. T., J. Andrade, H. Radhakrishna, J. G. Donaldson, J. A. Cooper, and P. A. Randazzo. 1998. ASAP1, a phospholipid-dependent Arf GTPase-activating protein that associates with and is phosphorylated by Src. Mol. Cell. Biol. 18:7038-7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunton, V. G., I. R. J. MacPherson, and M. C. Frame. 2004. Cell adhesion receptors, tyrosine kinases and actin modulators: a complex three-way circuitry. Biochim. Biophys. Acta 1692:121-144. [DOI] [PubMed] [Google Scholar]
- 5.Buccione, R., J. D. Orth, and M. A. McNiven. 2004. Foot and mouth: podosomes, invadopodia and circular dorsal ruffles. Nat. Rev. Mol. Cell Biol. 5:647-657. [DOI] [PubMed] [Google Scholar]
- 6.Carragher, N. O., and M. C. Frame. 2004. Focal adhesion and actin dynamics: a place where kinases and proteases meet to promote invasion. Trends Cell Biol. 14:241-249. [DOI] [PubMed] [Google Scholar]
- 7.Ehlers, J. P., L. Worley, M. D. Onken, and J. W. Harbour. 2005. DDEF1 is located in an amplified region of chromosome 8q and is overexpressed in uveal melanoma. Clin. Cancer Res. 11:3609-3613. [DOI] [PubMed] [Google Scholar]
- 8.Farsad, K., and P. De Camilli. 2003. Mechanisms of membrane deformation. Curr. Opin. Cell Biol. 15:372-381. [DOI] [PubMed] [Google Scholar]
- 9.Farsad, K., N. Ringstad, K. Takei, S. R. Floyd, K. Rose, and P. De Camilli. 2001. Generation of high curvature membranes mediated by direct endophilin bilayer interactions. J. Cell Biol. 155:193-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furman, C., S. M. Short, R. R. Subramanian, B. R. Zetter, and T. M. Roberts. 2002. DEF-1/ASAP1 is a GTPase-activating protein (GAP) for ARF1 that enhances cell motility through a GAP-dependent mechanism. J. Biol. Chem. 277:7962-7969. [DOI] [PubMed] [Google Scholar]
- 11.Gimona, M., and R. Buccione. 2006. Adhesions that mediate invasion. Int. J. Biochem. Cell Biol. 38:1875-1892. [DOI] [PubMed] [Google Scholar]
- 12.Hashimoto, S., M. Hirose, A. Hashimoto, M. Morishige, A. Yamada, H. Hosaka, K. I. Akagi, E. Ogawa, C. Oneyama, T. Agatsuma, M. Okada, H. Kobayashi, H. Wada, H. Nakano, T. Ikegami, A. Nakagawa, and H. Sabe. 2006. Targeting AMAP1 and cortactin binding bearing an atypical Src homology 3/proline interface for prevention of breast cancer invasion and metastasis. Proc. Natl. Acad. Sci. USA 103:7036-7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayasaka, H., K. Simon, E. D. Hershey, K. Masumoto, and J. T. Parsons. 2005. FRNK, the autonomously expressed C-terminal region of focal adhesion kinase, is uniquely regulated in vascular smooth muscle: analysis of expression in transgenic mice. J. Cell. Biochem. 95:1248-1263. [DOI] [PubMed] [Google Scholar]
- 14.Itoh, T., and P. De Camilli. 2006. BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim. Biophys. Acta 1761:897-912. [DOI] [PubMed] [Google Scholar]
- 15.Juliano, R. L. 2002. Signal transduction by cell adhesion receptors and the cytoskeleton: functions of integrins, cadherins, selectins, and immunoglobulin-superfamily members. Annu. Rev. Pharmacol. Toxicol. 42:283-323. [DOI] [PubMed] [Google Scholar]
- 16.King, F. J., E. D. Hu, D. F. Harris, P. Sarraf, B. M. Spiegelman, and T. M. Roberts. 1999. DEF-1, a novel Src SH3 binding protein that promotes adipogenesis in fibroblastic cell lines. Mol. Cell. Biol. 19:2330-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kowanetz, K., K. Husnjak, D. Holler, M. Kowanetz, P. Soubeyran, D. Hirsch, M. H. H. Schmidt, K. Pavelic, P. De Camilli, P. A. Randazzo, and I. Dikic. 2004. CIN85 associates with multiple effectors controlling intracellular trafficking of epidermal growth factor receptors. Mol. Biol. Cell 15:3155-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kruljac-Letunic, A., J. Moelleken, A. Kallin, F. Wieland, and A. Blaukat. 2003. The tyrosine kinase Pyk2 regulates Arf1 activity by phosphorylation and inhibition of the Arf-GTPase-activating protein ASAP1. J. Biol. Chem. 278:29560-29570. [DOI] [PubMed] [Google Scholar]
- 19.Lee, C. H., B. Leung, M. A. Lemmon, J. Zheng, D. Cowburn, J. Kuriyan, and K. Saksela. 1995. A single amino-acid in the SH3 domain of Hck determines its high-affinity and specificity in binding to HIV-1 Nef protein. EMBO J. 14:5006-5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, C. H., K. Saksela, U. A. Mirza, B. T. Chait, and J. Kuriyan. 1996. Crystal structure of the conserved core of HIV-1 Nef complexed with a Src family SH3 domain. Cell 85:931-942. [DOI] [PubMed] [Google Scholar]
- 21.Liu, Y., J. C. Loijens, K. H. Martin, A. V. Karginov, and J. T. Parsons. 2002. The association of ASAP1, an ADP ribosylation factor-GTPase activating protein, with focal adhesion kinase contributes to the process of focal adhesion assembly. Mol. Biol. Cell 13:2147-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, Y., G. M. Yerushalmi, P. R. Grigera, and J. T. Parsons. 2005. Mislocalization or reduced expression of Arf GTPase-activating protein ASAP1 inhibits cell spreading and migration by influencing Arf1 GTPase cycling. J. Biol. Chem. 280:8884-8892. [DOI] [PubMed] [Google Scholar]
- 23.Luo, R., B. Ahvazi, D. Amariei, D. Shroder, B. Burrola, W. Losert, and P. A. Randazzo. 2007. Kinetic analysis of GTP hydrolysis catalysed by the Arf1-GTP-ASAP1 complex. Biochem. J. 402:439-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mclean, G. W., N. O. Carragher, E. Avizienyte, J. Evans, V. G. Brunton, and M. C. Frame. 2005. The role of focal-adhesion kinase in cancer—a new therapeutic opportunity. Nat. Rev. Cancer 5:505-515. [DOI] [PubMed] [Google Scholar]
- 25.Nam, J. M., Y. Onodera, Y. Mazaki, H. Miyoshi, S. Hashimoto, and H. Sabe. 2007. CIN85, a Cbl-interacting protein, is a component of AMAP1-mediated breast cancer invasion machinery. EMBO J. 26:647-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nie, Z., M. Boehm, E. Boja, W. Vass, J. Bonifacino, H. Fales, and P. A. Randazzo. 2003. Specific regulation of the adaptor protein complex AP-3 by the Arf GAP AGAP1. Dev. Cell 5:513-521. [DOI] [PubMed] [Google Scholar]
- 27.Nie, Z., D. S. Hirsch, R. Luo, X. Jian, S. Stauffer, A. Cremesti, J. Andrade, J. Lebowitz, M. Marino, B. Ahvazi, J. E. Hinshaw, and P. A. Randazzo. 2006. A BAR domain in the N terminus of the Arf GAP ASAP1 affects membrane structure and trafficking of epidermal growth factor receptor. Curr. Biol. 16:130-139. [DOI] [PubMed] [Google Scholar]
- 28.Oda, A., I. Wada, K. Miura, K. Okawa, T. Kadoya, T. Kato, H. Nishihara, M. Maeda, S. Tanaka, K. Nagashima, C. Nishitani, K. Matsuno, M. Ishino, L. M. Machesky, H. Fujita, and P. Randazzo. 2003. CrkL directs ASAP1 to peripheral focal adhesions. J. Biol. Chem. 278:6456-6460. [DOI] [PubMed] [Google Scholar]
- 29.Onodera, Y., S. Hashimoto, A. Hashimoto, M. Morishige, A. Yamada, E. Ogawa, M. Adachi, T. Sakurai, T. Manabe, H. Wada, N. Matsuura, and H. Sabe. 2005. Expression of AMAP1, an ArfGAP, provides novel targets to inhibit breast cancer invasive activities. EMBO J. 24:963-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parsons, J. T. 2003. Focal adhesion kinase: the first ten years. J. Cell Sci. 116:1409-1416. [DOI] [PubMed] [Google Scholar]
- 31.Parsons, J. T., K. H. Martin, J. K. Slack, J. M. Taylor, and S. A. Weed. 2000. Focal adhesion kinase: a regulator of focal adhesion dynamics and cell movement. Oncogene 19:5606-5613. [DOI] [PubMed] [Google Scholar]
- 32.Peter, B. J., H. M. Kent, I. G. Mills, Y. Vallis, P. J. G. Butler, P. R. Evans, and H. T. McMahon. 2004. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303:495-499. [DOI] [PubMed] [Google Scholar]
- 33.Randazzo, P. A., J. Andrade, K. Miura, M. T. Brown, Y. Q. Long, S. Stauffer, P. Roller, and J. A. Cooper. 2000. The Arf GTPase-activating protein ASAP1 regulates the actin cytoskeleton. Proc. Natl. Acad. Sci. USA 97:4011-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Randazzo, P. A., and D. S. Hirsch. 2004. Arf GAPs: multifunctional proteins that regulate membrane traffic and actin remodelling. Cell. Signal. 16:401-413. [DOI] [PubMed] [Google Scholar]
- 35.Turner, C. E., M. D. Schaller, and J. T. Parsons. 1993. Tyrosine phosphorylation of the focal adhesion kinase pp125FAK during development relation to paxillin. J. Cell Sci. 105:637-645. [DOI] [PubMed] [Google Scholar]
- 36.Webb, D. J., J. T. Parsons, and A. F. Horwitz. 2002. Adhesion assembly, disassembly and turnover in migrating cells—over and over and over again. Nat. Cell Biol. 4:E97-E100. [DOI] [PubMed] [Google Scholar]
- 37.Wehrle-Haller, B., and B. A. Imhof. 2002. The inner lives of focal adhesions. Trends Cell Biol. 12:382-389. [DOI] [PubMed] [Google Scholar]