Abstract
mRNA stability is a major determinant of inflammatory gene expression. Rapid degradation of interleukin-8 (IL-8) mRNA is imposed by a bipartite AU-rich element (ARE) in the 3′ untranslated region (R. Winzen et al., Mol. Cell. Biol. 24:4835-4847, 2004). Small interfering RNA-mediated knockdown of the ARE-binding protein KSRP resulted in stabilization of IL-8 mRNA or of a β-globin reporter mRNA containing the IL-8 ARE. Rapid deadenylation was impaired, indicating a crucial role for KSRP in this step of mRNA degradation. The two IL-8 ARE domains both contribute to interaction with KSRP, corresponding to the importance of both domains for rapid degradation. Exposure to the inflammatory cytokine IL-1 has been shown to stabilize IL-8 mRNA through p38 mitogen-activated protein (MAP) kinase and MK2. IL-1 treatment impaired the interaction of KSRP with the IL-8 ARE in a manner dependent on p38 MAP kinase but apparently independent of MK2. Instead, evidence that TTP, a target of MK2, can also destabilize the IL-8 ARE reporter mRNA is presented. In a comprehensive approach to identify mRNAs controlled by KSRP, two criteria were evaluated by microarray analysis of (i) association of mRNAs with KSRP in pulldown assays and (ii) increased amounts in KSRP knockdown cells. According to both criteria, a group of 100 mRNAs is controlled by KSRP, many of which are unstable and encode proteins involved in inflammation. These results indicate that KSRP functions as a limiting factor in inflammatory gene expression.
Rapid degradation controls the levels of many mRNAs that are translated into transiently expressed proteins. These include cytokines, growth factors, proto-oncogene products, and other proteins participating in acute reactions. The short half-lives of their mRNAs depend on regulatory RNA sequences, the most widely distributed being AU-rich elements (AREs) located in their 3′ untranslated regions (UTRs) (8, 39). AREs have been divided into three classes, differing in the sequences and modes of degradation imposed by them (8). Class I AREs contain one to three scattered AUUUA motifs, and class II AREs contain multiple overlapping AUUUA motifs. Class III AREs are less well defined and lack an AUUUA motif. With the search pattern WWWU(AUUUA)UUUW, 4,000 human mRNAs have been reported to contain AREs and grouped into the ARED database (1), where the class II AREs are further subdivided into different groups, depending on the number of AUUUA motifs present in an ARE.
Interleukin-8 (IL-8) is a member of the CXC chemokine family, released from different types of cells in response to direct cell stress, pathogens, or the proinflammatory cytokines tumor necrosis factor (TNF) and IL-1 (reference 25 and references therein). It attracts and activates leukocytes and also plays a role in angiogenesis. Studying its induction in response to IL-1, we previously observed that in addition to transcriptional activation of the IL-8 gene, its mRNA is stabilized (26, 46). The latter response involves the activation of p38 mitogen-activated protein (MAP) kinase and its substrate kinase MK2. Stabilization of IL-8 mRNA can contribute to enhanced IL-8 expression, e.g., in viral infection (22). Our recent studies showed that the IL-8 mRNA contains an ARE which consists of two functionally distinct domains. They cooperate for maximal destabilization and interaction with cytoplasmic proteins in vitro (44).
Control of mRNA degradation by AREs involves the function of proteins binding to them. ARE-binding proteins include destabilizing factors such as TTP, BRF1, or KSRP, which recruit RNA degrading enzymes, as well as stabilizing factors like HuR (12, 17). AUF1/hnRNP D has been shown to function in both ways.
The KH-type splicing regulatory protein (KSRP) was originally identified as a factor involved in regulated splicing of c-src (35). It contains four hnRNP K homology domains and is a member of the family of far upstream sequence binding proteins (FUBP) (11), also named FUBP2 accordingly. KSRP has been shown to play a role in rapid degradation of ARE-containing transcripts (3, 7, 9, 15, 18, 19, 32, 38). Other functions which depend on interactions with mRNA sequences distinct from AREs have been ascribed to KSRP or its homologs. A chicken homolog interacts with the zipcode sequence that controls β-actin mRNA localization (23). In rat, KSRP also binds to the β-actin zipcode sequence (40) and to a region determining localization of microtubule-associated protein 2 mRNA (36). In Xenopus oocytes, a member of the FUBP family stimulates removal of a translational repressor element from the Vg1 mRNA (30). Despite these observations, more comprehensive information on target mRNAs directly regulated by KSRP is largely missing.
We now report that in HeLa cells suppression of KSRP expression by small interfering RNA (siRNA) indicates an essential role for rapid degradation of IL-8 mRNA and its deadenylation as the initiating step in it. The two domains of the IL-8 ARE are involved in interaction with KSRP in vitro and in intact cells. Interaction with the IL-8 ARE is impaired by IL-1 in a manner dependent on p38 MAP kinase but not on MK2, explaining in part the stabilization induced by the p38/MK2 pathway. Microarray experiments indicate that KSRP interacts with numerous ARE-containing mRNAs but also with transcripts that lack AREs, including its own mRNA. By comparison of mRNAs associated with KSRP and upregulated by KSRP knockdown, using IL-1 stimulation as in vitro model of an inflammatory response, a group of bona fide targets of KSRP is defined. Their regulation of expression by KSRP is expected to have significant impact on inflammatory gene expression in vivo.
MATERIALS AND METHODS
Plasmids.
The expression vectors for β-globin (ptetBBB) carrying insertions of the IL-8 3′ UTR, for constitutively active MAP kinase kinase 6 (MKK62E) and MK2 (MK2EE), and for negative interfering MK2 (MK2K76R) have been described in detail previously (44, 46). ptetBBB-TNF1315-1350 expresses an mRNA that contains the ARE of human TNF-α in its 3′ UTR. To express KSRP with an N-terminal Strep tag (stKSRP), a sequence encoding the Strep-tactin target peptide Strep tag III (29) was inserted into pcDNA3.1-HisC-KSRP (a kind gift from Ching-Yi Chen). The KSRP sequence in the resulting plasmid was replaced by the coding region of green fluorescent protein (GFP) to express Strep-tagged GFP (stGFP) or of human TTP to express Strep-tagged TTP (stTTP).
Transfection and determination of mRNA degradation.
HeLa cells constitutively expressing the tetracycline-controlled transactivator protein were transfected by the calcium phosphate method, and the degradation kinetics of plasmid-expressed mRNA was determined using the tet-off system as detailed earlier (44). Cells were stimulated with human recombinant IL-1α (a gift from A. Stern and P. T. Lomedico, Hoffman-La Roche) where indicated. The degradation kinetics for endogenous mRNA was determined by inhibiting transcription with actinomycin D (5 μg/ml). Total RNA was isolated and Northern blot analysis performed using digoxigenin-labeled antisense RNA probes. RNA half-lives were determined as described in reference 46, using a video imaging system and the Molecular Analyst program (Bio-Rad). siRNA duplexes (Qiagen) specifically interfering with expression of KSRP (18) or GFP (47) were transfected by the calcium phosphate method. Transfection was repeated 2 days later with the respective siRNA together with plasmids for reporter mRNA expression. Samples for determining KSRP protein levels and mRNA degradation were prepared the following day. Essentially similar increases in mRNA stability in KSRP knockdown cells were observed with a second siRNA duplex (modified from reference 3).
Analysis of poly(A) tail length.
To resolve heterogeneity in poly(A) tail length, RNA samples were separated on denaturating polyacrylamide gels (4% [wt/vol] acrylamide, 7 M urea) as described previously (44). Running in 1× Tris-borate-EDTA was performed at about 10 V/cm for variable times, depending on the size of the mRNA monitored. Deadenylated reference RNAs were prepared by RNase H digestion in the presence of oligo(dT) (44).
Strep tag affinity chromatography.
All steps were carried out in the cold. Cells were pelleted, resuspended at 107 cells per 125 μl in lysis buffer (20 mM β-glycerophosphate [pH 7.4], 150 mM NaCl, 7.5% [vol/vol] glycerol, 1 mM EDTA, 1 mM sodium orthovanadate, 0.5% [vol/vol] Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 200 units/ml RNase inhibitor [MBI Fermentas]) and kept on ice for 15 min. After centrifugation at 16,000 × g, the supernatant was collected (cytoplasmic extract). Aliquots were frozen for determination of stKSRP protein and input mRNAs. Strep-tactin agarose beads (IBA) preincubated for 30 min with tRNA from Escherichia coli (50 μg/ml) in 500 μl wash buffer (20 mM β-glycerophosphate [pH 7.4], 150 mM NaCl, 1 mM EDTA) were washed twice in the same buffer and added to the cytoplasmic extract. After 4 h with constant mixing, the beads were spun down and washed three times with wash buffer. Aliquots of the beads were heated to 95°C with sample buffer for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) for subsequent analysis of stKSRP amounts or were supplied with 10 μg of bacterial rRNA as carrier RNA and subjected to total RNA isolation (RNA isolation kit; Macherey & Nagel).
Reverse transcription-PCR (RT-PCR).
Total RNA was reverse transcribed with Moloney murine leukemia virus reverse transcriptase and amplified with Taq polymerase (both from MBI Fermentas) and specific primers (MWG Biotech) for CXCL3 (sense, 5′TCTCGCACAGCTTCCCGA; antisense, 5′CTAGAAAGCTGCTGTTCTC), 14-3-3ζ (sense, 5′AGGTCATCTTGGAGGGTC; antisense, 5′CTCCTTGGGTATCCGATG), IκBζ (sense, 5′CTCCTGTTGCACATCCGA; antisense, 5′CAGCTAACTTGAACTGTGTT), IL-6 (sense, 5′CTGGGCACAGAACTTATGTTG; antisense, 5′GGTAAGCCTACACTTTCCA), IL-8 (sense, ATCAAATATTTGTGCAAGAATTTGG; antisense, TTTCAGATAAACAATAATGT); KSRP (sense, 5′GAGAAGATAGCGAGATCTAA; antisense, 5′GAATGTTCCACCTCTAACTA); FUBP3 (sense, 5′AATCAAGCAGTTGCAGGAG; antisense, 5′CCGTGTATGTTATCTCCTG), and TTP (sense, 5′GATCTGACTGCCATCTACG; antisense, TCACTCAGAAACAGAGATGC).
Western blot analysis.
Proteins were separated by SDS-PAGE (10% acrylamide) and blotted onto polyvinylidene difluoride membranes (Millipore). Equal loading and protein transfer were controlled by staining with Coomassie brilliant blue. KSRP was detected by chemiluminescence using a specific monoclonal antibody (kindly provided by C.-Y. Chen) and alkaline-phosphatase-labeled secondary antibody.
Microarray analysis.
RNA from pulldown or siRNA-mediated knockdown experiments was purified and Cy3-labeled cRNA prepared as described previously (21). For analysis of KSRP-associated mRNAs on high-density arrays, RNA was isolated from Strep-tactin pulldown in four independent experiments which yielded essentially similar sets of mRNAs enriched by stKSRP compared to stGFP. Global mRNA expression of input samples was analyzed in parallel. Table S1 in the supplemental material shows results from one experiment where stringency was increased by raising the NaCl concentration to 700 mM in the first two washes of Strep-tactin agarose beads after incubation with cytoplasmic extract (see above). Cy3-labeled cRNA was generated using an Amino Allyl MessageAmp II with Cy3 kit according to the manufacturer's recommendations (AM1795; Applied Biosystems). Even amplification efficiency was controlled by spiking reverse transcription reactions with a mixture of 10 different E1A mRNA species (5188-5977; Agilent Technologies) that were each detected by 32 probes on the microarray. cRNA quality, yield, and labeling efficiency were analyzed by an Agilent 2100 bioanalyzer. To minimize technically caused variations, cRNA for each RNA sample was generated in duplicate and pooled prior to labeling and hybridizing. Labeled cRNAs were hybridized to the Whole Human Genome Microarray (G4112F, ID 014850; Agilent Technologies) containing 45,015 probes for 15,713 annotated genes and 15,269 uncharacterized transcripts. cRNA fragmentation, hybridization, and washing steps were performed exactly as directed by the manufacturer's One-Color Microarray-Based Gene Expression Analysis Protocol V5.0.1 manual. Slides were scanned on an Agilent G2505 B microarray scanner at high (100%) and low (5%) photomultiplier tube settings. Data extraction and intra-array normalization were performed with Feature Extraction V9.1.3.1 software (Agilent protocol file GE1-v5_91_0806.xml). Flagged spots were excluded from further analysis. Interarray normalization was performed according to E1A signals. Intensity values close to background levels were replaced by surrogate values, calculated from E1A signals, to compensate for elevated bias at the low end of the dynamic intensity range. Ratio data of probes directed against the same RNA were averaged by calculating the geometric mean.
In vitro transcription and electrophoretic mobility shift assays.
Radiolabeled RNA fragments corresponding to the IL-8 ARE and mutants thereof were synthesized with T7 RNA polymerase from linearized plasmids as described recently (44). The labeled RNA probes were purified using mini Quick Spin RNA columns (Roche Diagnostics) and incubated with cytoplasmic extract (prepared by Nonidet P-40 lysis as in reference 45) or with affinity-purified stKSRP eluted from the Strep-tactin agarose with desthiobiotin (10 mM; IBA). After digestion with RNase T1, samples were separated on nondenaturing polyacrylamide gels (44) or UV cross-linked with 900 mJ/cm2 and separated by SDS-PAGE (45).
RESULTS
KSRP is essential for rapid degradation of IL-8 mRNA.
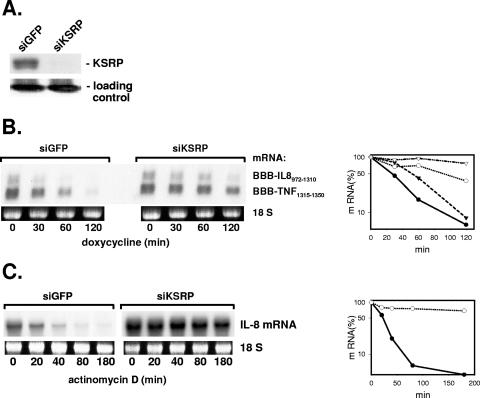
Rapid turnover of IL-8 mRNA is largely dependent on an ARE characterized in detail recently (44). To find out if the ARE-binding protein KSRP is involved in the function of the IL-8 ARE, HeLa cells were transfected with siRNAs specific for KSRP mRNA. Amounts of KSRP were strongly diminished compared to those in cells transfected with siRNA against GFP as a control (Fig. 1A). Degradation of β-globin reporter mRNAs carrying the IL-8 ARE as well as the TNF ARE was slowed down in the KSRP knockdown cells compared to that in the control cells (Fig. 1B). Endogenous IL-8 mRNA is barely detectable in unstimulated HeLa cells but strongly and rapidly induced by the proinflammatory cytokine IL-1. Two hours after addition of IL-1, the stability of endogenous IL-8 mRNA was strongly increased when KSRP expression was suppressed by siRNA (Fig. 1C).
FIG. 1.
Evidence for a crucial role for KSRP in the control of IL-8 mRNA decay. HeLa cells were transfected with siRNA specific for KSRP or for GFP as a control (siKSRP or siGFP, respectively). (A) Western blot with antibodies against KSRP to control knockdown efficiency. Coomassie brilliant blue staining of an irrelevant protein is shown as a loading control. (B) Cells were cotransfected with plasmids for β-globin mRNAs containing destabilizing elements of IL-8 or of TNF-α mRNA. At the indicated times after stopping transcription with doxycycline (3 μg/ml), total RNA was isolated and analyzed by Northern blotting with a β-globin antisense probe. Ethidium bromide staining of the 18S rRNA is shown to allow comparison of the RNA amounts loaded. Results were quantified by a video analyzer system (amount of mRNA at the time of doxycycline addition [0 min] = 100%; circles, BBB-IL-8972-1310; triangles, BBB-TNF1315-1350; closed symbols, siGFP; open symbols, siKSRP). (C) Endogenous IL-8 mRNA was induced by incubating the cells for 2 h with IL-1α (2 ng/ml). mRNA half-life was determined after stopping transcription with actinomycin D (5 μg/ml). Northern blots were hybridized to an IL-8 antisense probe and results quantified as described for panel B.
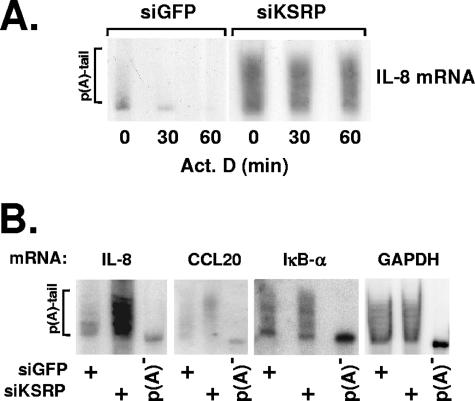
Suppression of KSRP impairs deadenylation of IL-8 mRNA.
Evidence has been presented for recruitment of the deadenylating enzyme PARN by KSRP (18). To compare the poly(A) tail lengths of IL-8 mRNA in control and KSRP knockdown cells, the RNA samples were separated by PAGE. This revealed primarily short poly(A) tails in control cells after 2 h of stimulation with IL-1, as expected from previous work in which we showed that IL-1 induces rapid and transient IL-8 mRNA expression with long poly(A) tails at the 1-h maximum, with rapid poly(A) tail shortening and decrease in RNA amounts ensuing thereafter (20). In the cells treated with siRNA against KSRP, IL-8 transcripts with intermediate and long poly(A) tails were prominent (Fig. 2A). The weighted arithmetic mean length of poly(A) tails increased from 58 nucleotides (nt) in control cells to 120 nt in KSRP knockdown cells. Furthermore, in the latter cells amounts and poly(A) tail lengths remained fairly constant for 1 hour after inhibiting transcription. Similarly, steady-state levels and poly(A) tail lengths of CCL20 mRNA were also increased in the KSRP knockdown cells (Fig. 2B) (mean length 136 nt versus 96 nt in the control cells). The mRNA of IκBα is short lived but does not contain a typical ARE with overlapping AUUUA motifs in its 3′ UTR. Amounts and poly(A) tail lengths are found to be hardly affected when cells treated with control or KSRP-specific siRNA are compared (Fig. 2B) (mean lengths of 97 and 110 nt, respectively), indicating that KSRP knockdown selectively affects only certain mRNAs. A stable mRNA for a housekeeping enzyme, GAPDH (glyceraldehyde-3-phosphate dehydrogenase), also remains unaffected by suppressing KSRP levels (mean lengths of 107 and 104 nt, respectively, in control and KSRP knockdown). These results provide evidence for a role for KSRP in the rapid and specific deadenylation and degradation of endogenous ARE-containing mRNAs.
FIG. 2.
Effect of KSRP knockdown on the poly(A) [p(A)] lengths of mRNAs. IL-1α was added to HeLa cells transfected with siRNA specific for KSRP or for GFP (siKSRP or siGFP, respectively). (A) After 2 h, transcription was inhibited by addition of actinomycin D (Act. D; 5 μg/ml). Total RNA was isolated at the indicated times thereafter, separated on a polyacrylamide gel, and analyzed by Northern blotting with an IL-8 antisense probe. (B) Polyacrylamide gel analysis of the indicated mRNAs from cells transfected with siRNAs and stimulated with IL-1α as described for panel A. Aliquots of the RNAs from cells transfected with siRNA against KSRP were subjected to in vitro deadenylation [p(A)−].
Both domains of the IL-8 ARE are involved in its interaction with KSRP.
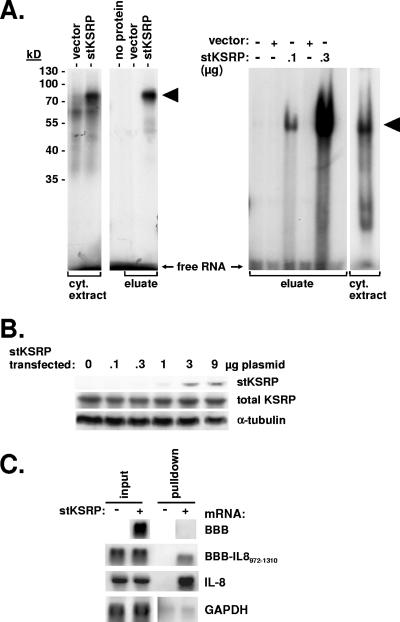
In UV cross-linking experiments, radiolabeled RNA corresponding to the IL-8 ARE forms protein-RNA complexes with cytoplasmic extract (Fig. 3A). Formation of the highest-molecular-weight complex is increased with cytoplasmic extract from cells expressing stKSRP. Furthermore, IL-8 ARE RNA can be UV cross-linked to purified stKSRP, resulting in a product of the expected molecular weight in SDS-PAGE. Complex formation of stKSRP and the IL-8 ARE is also detected in nondenaturing gels (Fig. 3A, right).
FIG. 3.
Interaction of the IL-8 ARE with KSRP. (A) Radiolabeled in vitro-transcribed RNA corresponding to the IL-8 ARE was incubated alone (no protein), with cytoplasmic (cyt.) extracts from cells transfected with empty vector or with an stKSRP expression plasmid, or with eluate from Strep-tactin-agarose beads after incubation with the cytoplasmic extracts. Complexes were either separated by SDS-PAGE after UV cross-linking in vitro (left) or separated by nondenaturing gel electrophoresis (right). KSRP-RNA complexes are marked by arrowheads. (B) stKSRP expression plasmid was transfected in increasing amounts. Western blot detection of stKSRP was done with horseradish peroxidase-labeled Strep-tactin, and total KSRP and α-tubulin were detected with specific antibodies. (C) β-Globin reporter mRNAs without insertion (BBB) or with an IL-8 RNA fragment containing the ARE (BBB-IL-8972-1310) were coexpressed with stKSRP where indicated and the cells stimulated with IL-1α. Total RNA was prepared from cytoplasmic extract (input) and from eluate of Strep-tactin beads (pulldown). β-Globin and endogenous IL-8 and GAPDH mRNAs were detected by Northern blot analysis.
Interaction of KSRP with the IL-8 ARE was confirmed in pulldown assays with stKSRP. First, stKSRP plasmid was transfected in different amounts and stKSRP and total KSRP were detected by Western blot analysis (transfected and endogenous KSRP could not be separated in SDS-PAGE). In the range of plasmid amounts tested, strong increases in total KSRP were not observed (Fig. 3B). Amounts that corresponded to 7 μg in the setting of Fig. 3B were chosen for further experiments. In Northern analysis of mRNAs associated with stKSRP after pulldown from cytoplasmic extract with Strep-tactin agarose beads, β-globin mRNA without an ARE was not detected (Fig. 3C). β-Globin mRNA with the ARE-containing region of IL-8 mRNA was selectively detected in pulldown from cells expressing stKSRP but not in pulldown from control cells. Importantly, endogenous IL-8 mRNA was also found to associate with stKSRP. GAPDH mRNA, which exhibits very high signal intensity in total RNA, was detected in spurious and not significantly differing amounts in both the specific and the control pulldown preparations.
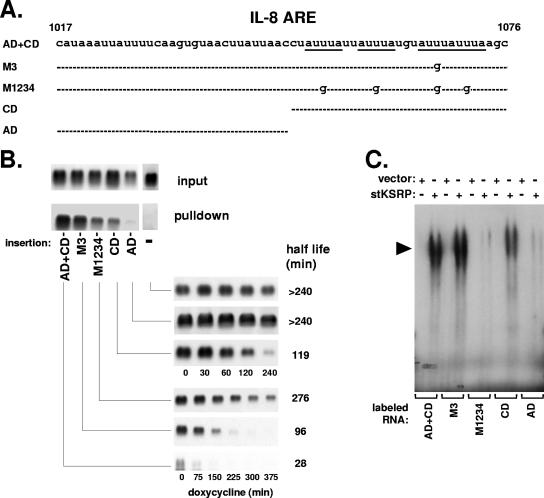
We further studied the ARE-KSRP interaction by investigating the importance of the four AUUUA motifs and of the two functionally distinct domains in the ARE that we had previously defined (44). β-Globin reporter RNAs containing the ARE or derivatives thereof in the 3′ UTR were expressed (Fig. 4A). Compared to the wild-type ARE with an intact auxiliary domain and core domain (AD+CD), a mutant in which the third AUUUA motif of the core domain was changed into AUGUA (M3) showed slightly diminished interaction with KSRP in pulldown assays (Fig. 4B). Interaction was strongly reduced for a mutant in which all four AUUUA motifs were destroyed (M1234). Comparably strong impairment of KSRP interaction was observed for an mRNA that contained only the core domain. An RNA containing only the auxiliary domain showed the weakest interaction. The results revealed a close correlation between the interaction of the mRNAs with KSRP and their half-lives in intact cells (Fig. 4B) (see also reference 44). Importantly, the auxiliary domain strongly contributes to interaction of KSRP with the ARE, suggesting that this is the cause for its enhancing effect on the moderate destabilization exerted by the core domain alone.
FIG. 4.
Participation of both domains of the IL-8 ARE in KSRP interaction and destabilization. (A) Scheme of the IL-8 ARE and derivatives assayed (CD, core domain; AD, auxiliary domain). (B) RNA isolated from cytoplasmic extract (input) and stKSRP pulldown was subjected to Northern blot analysis of β-globin reporter mRNAs without insertion (−), containing the complete IL-8 ARE (AD+CD) or mutants in which the third AUUUA motif (M3) or all four motifs (M1234) were changed into AUGUA, or containing a single domain as indicated. Also shown are the corresponding degradation kinetics of these mRNAs in HeLa cells (taken from reference 44 with permission). (C) Complexes formed between purified stKSRP (0.4 μg) and radiolabeled RNA fragments corresponding to the complete IL-8 ARE or its derivatives were analyzed by nondenaturing gel electrophoresis. KSRP-RNA complexes are marked by arrowheads.
The relationship between the destabilizing function of the IL-8 ARE and its interaction with KSRP was further probed in in vitro binding experiments. Complexes were analyzed in nondenaturing gels to avoid the problem of different cross-link efficiency rather than affinity when derivatives of the ARE were compared in their abilities to interact with KSRP. As shown in Fig. 4C, interaction was hardly affected if one of the four AUUUA motifs was mutated (M3). Strong decrease in interaction was observed for the mutant in which all motifs were destroyed (M1234). The core domain alone, which contains all four AUUUA motifs, showed markedly reduced interaction, and for the auxiliary domain alone, the interaction was reduced even stronger. Taken together, these data show a close correlation between the importance of the two domains for ARE function and interaction with KSRP.
Interaction of KSRP with the IL-8 ARE is impaired by p38 MAP kinase activation without involvement of MK2.
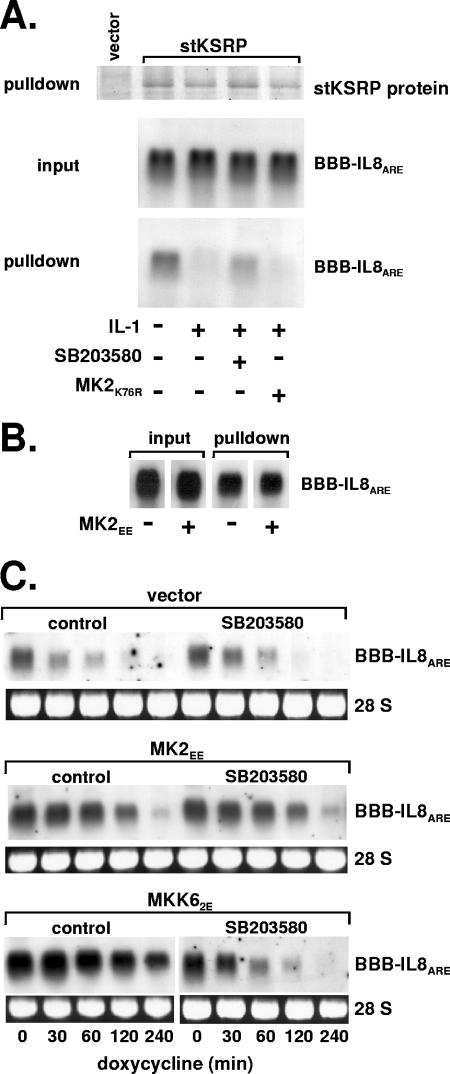
Rapid degradation of several ARE-containing mRNAs, including IL-8 mRNA, is impaired in cells exposed to IL-1 in a manner involving p38 MAP kinase and its substrate MK2 (46). In cells expressing stKSRP and β-globin mRNA containing the IL-8 ARE, significantly less of the mRNA was associated with stKSRP after treatment of the cells with IL-1 than in untreated cells (Fig. 5A). Inhibition of p38 MAP kinase using the specific inhibitor SB203580 largely reversed this effect of IL-1. Selective activation of the p38/MK2 pathway by expressing a constitutively active form of the p38-selective MAP kinase kinase MKK6 also impaired KSRP-RNA interaction (not shown). These results are in accordance with a decreased interaction of phosphorylated KSRP with ARE-containing RNA demonstrated in vitro (3). On the other hand, a kinase-inactive mutant of MK2 which interferes with IL-1-induced mRNA stabilization (46) could not reverse impairment of KSRP-mRNA interaction. Furthermore, expression of a constitutively active mutant of MK2 (MK2EE), which induced stabilization (46) (Fig. 5C), did not impair interaction (Fig. 5B). This and the observation by Briata et al. that MK2 does not phosphorylate KSRP indicates that the contribution of MK2 to stabilization of ARE-containing mRNAs is not reflected on this level.
FIG. 5.
Effect of the p38 MAP kinase cascade on the interaction of KSRP with the IL-8 ARE. (A) β-globin reporter mRNA containing the IL-8 ARE was coexpressed with stKSRP and a negative interfering mutant of MK2 (MK2K76R) where indicated. The cells were incubated without or with IL-1α (2 ng/ml) for 30 min in the absence or presence of SB203580 (2 μM) added 15 min earlier. After pulldown of stKSRP, the protein was detected by SDS-PAGE and Coomassie brilliant blue staining, and associated β-globin-IL-8 ARE mRNA was detected by Northern blot analysis. (B) Pulldown of stKSRP and mRNA detection were performed as described for panel A for cells cotransfected with an expression vector for MK2EE as indicated. (C) Degradation kinetics of β-globin-IL-8 ARE mRNA in cells transfected with empty vector or plasmids for expression of constitutively active mutants of MK2 (MK2EE) or MKK6 (MKK62E) in the absence or presence of SB203580 (2 μM).
We also tested whether MK2 has an indirect role in mRNA stabilization, e.g., facilitating export of active p38 MAP kinase from the nucleus to the cytoplasm (2, 14), where p38 MAP kinase might phosphorylate its target KSRP. This is unlikely, however: as shown in Fig. 5C, constitutively active MK2 induces mRNA stabilization (compare “control” in upper panel [vector] with “control” in middle panel [MK2EE], which is independent of p38 MAP kinase activity (middle, compare “control” and SB203580). Stabilization induced by the upstream kinase MKK6 was inhibited by SB203580 as expected.
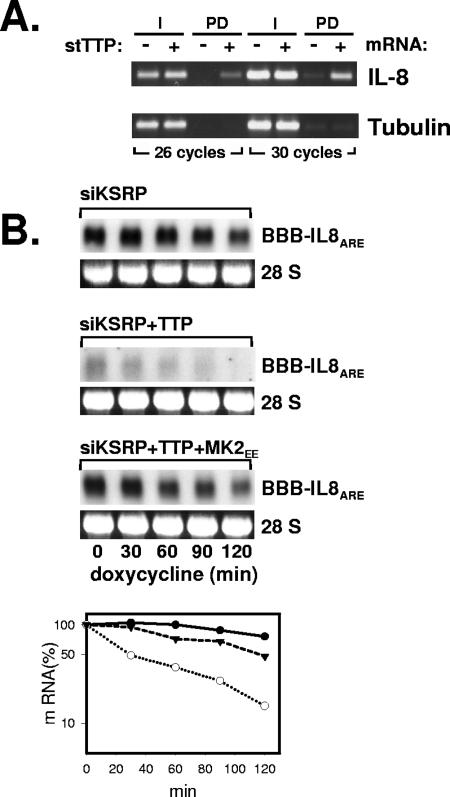
The IL-8 ARE is a target of TTP.
MK2 can phosphorylate the ARE-binding protein TTP, which can impair its destabilizing activity toward its target mRNAs (16). To test whether TTP can contribute to regulation of IL-8 mRNA stability, stTTP was expressed in HeLa cells and pulldown performed after induction of IL-8 mRNA by IL-1. IL-8 mRNA was enriched in the stTTP pulldown fraction (Fig. 6A). Furthermore, purified TTP interacted with the IL-8 ARE in vitro (not shown). Since detection of a destabilizing effect of TTP might be obscured by the predominant destabilization of KSRP, the effect of TTP on the degradation of the IL-8 ARE-containing reporter RNA in KSRP knockdown cells was determined. Coexpression of stTTP resulted in a marked decrease in the steady-state level and the stability of the reporter RNA (Fig. 6B). Coexpression of constitutively active MK2 (MK2EE) partly reverted this effect. These results indicate that TTP can participate in the control of IL-8 mRNA stability and may represent the MK2-sensitive component of rapid IL-8 mRNA degradation.
FIG. 6.
Interaction of TTP with the IL-8 ARE. (A) HeLa cells were transfected with a plasmid carrying stTTP or empty vector. Total RNA from cytoplasmic extract (I) or Strep-tactin pulldown (PD) was analyzed by RT-PCR, with primers specific for IL-8 mRNA or β-tubulin mRNA as a control. (B) HeLa cells were transfected with siRNA against KSRP, a plasmid carrying the β-globin-IL-8 ARE reporter RNA, and expression vectors for stTTP and MK2EE as indicated. The degradation kinetics of the reporter RNA was determined by Northern blot analysis and quantified as described for Fig. 1B. Closed circles, siKSRP; open circles, siKSRP plus TTP; triangles, siKSRP plus TTP plus MK2EE.
Microarray-based identification of mRNAs regulated by KSRP.
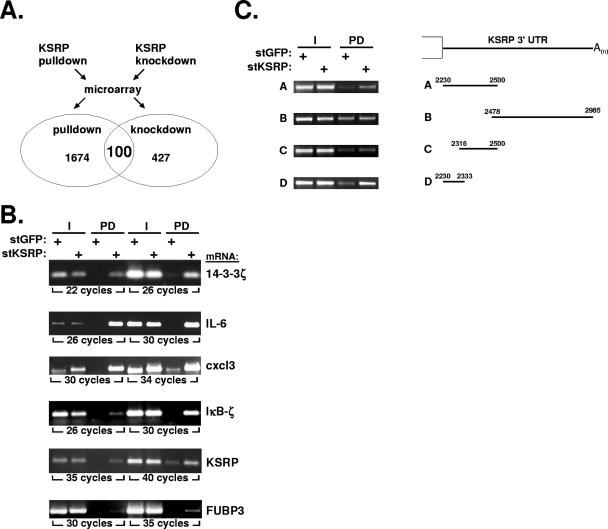
To obtain information on the population of mRNAs potentially regulated by KSRP, HeLa cells were stimulated with IL-1. A stimulation period of 2 hours was chosen because at this time IL-1-induced mRNA stabilization has ceased (20, 46) but expression of transiently induced mRNAs is still high enough for analysis. Two parameters were determined for RNAs from these cells, (i) enrichment in pulldown with KSRP and (ii) increase in amount upon knockdown of KSRP (Fig. 7A). RNA was analyzed on an oligonucleotide array covering 30,982 human genes. After pulldown of stKSRP, 1,734 mRNAs (i.e., 11% of all functionally annotated genes that were detected by the Agilent array) were enriched more than twofold in the stKSRP fraction (see Table S1 in the supplemental material). Determination of amounts in the cytoplasmic extract (input) excluded the possibility that this was due to different starting levels (not shown). Enrichment in the stKSRP fraction was verified by RT-PCR for several mRNAs (Fig. 7B).
FIG. 7.
Interaction of KSRP with endogenous mRNAs. (A) HeLa cells were transfected with stKSRP or stGFP as a control for pulldown assays and with siRNA against KSRP or against GFP as a control for knockdown assays. After stimulation for 2 h with IL-1α, total RNA was isolated and subjected to microarray analysis as described in Materials and Methods. mRNAs associated with stKSRP in pulldown and increased in KSRP knockdown cells were identified. Details on mRNAs positive for both parameters (with ratios of >2 for signal intensities of stKSRP/stGFP and siRNA against KSRP/GFP) are presented in Table 1. (B) Enrichment of the indicated mRNAs in the stKSRP pulldown was confirmed by RT-PCR. (C) Input (I) and stKSRP pulldown (PD) samples from cells expressing β-globin mRNA with the indicated insertions of the KSRP 3′ UTR were analyzed for the respective mRNA by RT-PCR with β-globin mRNA-specific primers.
mRNA from two independent KSRP knockdown experiments was analyzed to estimate the impact of KSRP on gene expression. The cells were stimulated with IL-1 for 2 hours, and RNA was isolated immediately or after additional incubation for 3 hours with actinomycin D to obtain information on changes in mRNA degradation. RNAs of 427 annotated genes showed a mean increase of twofold or higher in the KSRP knockdown cells compared to the level for GFP knockdown cells (see Table S2 in the supplemental material). For 1,674 of the 1,734 transcripts enriched in pulldown, complete data sets from both knockdown experiments were available. One hundred mRNAs were enriched in the stKSRP pulldown, and their levels also increased upon KSRP knockdown (Table 1). Forty-eight of them had short half-lives. Of these, 10 mRNAs, including that of IL-8, decayed more slowly in the KSRP knockdown cells (Table 1). This indicates that these mRNAs represent target transcripts for the destabilizing function of KSRP. Of note, 80% of them and 36% of all mRNAs in Table 1 were found in the ARED database (1), which lists mRNAs with AU-rich sequences in their 3′ UTRs as defined by a bioinformatic approach.
TABLE 1.
mRNAs enriched in pulldown and increased in knockdown of KSRPa
| RefSeq accession no. | Description | Gene name | mRNA status
|
ARE classd | |
|---|---|---|---|---|---|
| Unstableb | Stabilized in KSRP knockdownc | ||||
| NM_001200 | Bone morphogenetic protein 2 (BMP2), mRNA | BMP2 | + | + | |
| NM_000758 | Colony stimulating factor 2 (granulocyte-macrophage) (CSF2), mRNA | CSF2 | + | + | II |
| NM_002089 | Chemokine (C-X-C motif) ligand 2 (CXCL2), mRNA | CXCL2 | + | + | II |
| NM_002090 | Chemokine (C-X-C motif) ligand 3 (CXCL3), mRNA | CXCL3 | + | + | II |
| NM_018948 | ERBB receptor feedback inhibitor 1 (ERRFI1), mRNA | ERRFI1 | + | + | |
| NM_004864 | Growth differentiation factor 15 (GDF15), mRNA | GDF15 | + | + | II |
| NM_002166 | Inhibitor of DNA binding 2, dominant negative helix-loop-helix protein (ID2), mRNA | ID2 | + | + | I |
| NM_000600 | Interleukin 6 (beta 2 interferon, beta 2) (IL6), mRNA | IL6 | + | + | I |
| NM_000584 | Interleukin 8 (IL8), mRNA | IL8 | + | + | II |
| NM_000963 | Prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (PTGS2), mRNA | PTGS2 | + | + | II |
| NM_000024 | Adrenergic beta-2-, receptor, surface (ADRB2), mRNA | ADRB2 | + | ||
| NM_016201 | Angiomotin-like 2 (AMOTL2), mRNA | AMOTL2 | + | ||
| NM_025047 | ADP-ribosylation factor-like 14 (ARL14), mRNA | ARL14 | + | I | |
| NM_006577 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 (B3GNT2), mRNA | B3GNT2 | + | I | |
| NM_170735 | Brain-derived neurotrophic factor (BDNF), transcript variant 1, mRNA | BDNF | + | I | |
| NM_001901 | Connective tissue growth factor (CTGF), mRNA | CTGF | + | ||
| NM_001554 | Cysteine-rich angiogenic inducer 61 (CYR61), mRNA | CYR61 | + | ||
| NM_012242 | Dickkopf homolog 1 (Xenopus laevis) (DKK1), mRNA | DKK1 | + | ||
| NM_012266 | DnaJ (Hsp40) homolog, subfamily B, member 5 (DNAJB5), mRNA | DNAJB5 | + | ||
| NM_004419 | Dual specificity phosphatase 5 (DUSP5), mRNA | DUSP5 | + | ||
| NM_001946 | Dual specificity phosphatase 6 (DUSP6), transcript variant 1, mRNA | DUSP6 | + | ||
| NM_004951 | Epstein-Barr virus-induced gene 2 (lymphocyte-specific G protein-coupled receptor) (EBI2), mRNA | EBI2 | + | ||
| NM_001955 | Endothelin 1 (EDN1), mRNA | EDN1 | + | ||
| NM_001956 | Endothelin 2 (EDN2), mRNA | EDN2 | + | I | |
| NM_023037 | Furry homolog (Drosophila) (FRY), mRNA | FRY | + | ||
| NM_005261 | GTP binding protein overexpressed in skeletal muscle (GEM), transcript variant 1, mRNA | GEM | + | ||
| NM_005279 | G protein-coupled receptor 1 (GPR1), mRNA | GPR1 | + | ||
| NM_005328 | Hyaluronan synthase 2 (HAS2), mRNA | HAS2 | + | ||
| NM_001945 | Heparin-binding EGF-like growth factor (HBEGF), mRNA | HBEGF | + | II | |
| NM_182757 | IBR domain-containing 2 (IBRDC2), mRNA | IBRDC2 | + | ||
| NM_001547 | Interferon-induced protein with tetratricopeptide repeats 2 (IFIT2), mRNA | IFIT2 | + | ||
| NM_005544 | Insulin receptor substrate 1 (IRS1), mRNA | IRS1 | + | ||
| NM_000214 | Jagged 1 (Alagille syndrome) (JAG1), mRNA | JAG1 | + | II | |
| NM_031419 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor zeta (NFKBIZ), transcript variant 1, mRNA | NFKBIZ | + | ||
| NM_006186 | Nuclear receptor subfamily 4, group A, member 2 (NR4A2), transcript variant 1, mRNA | NR4A2 | + | I | |
| NM_021127 | Phorbol-12-myristate-13-acetate-induced protein 1 (PMAIP1), mRNA | PMAIP1 | + | I | |
| NM_005398 | Protein phosphatase 1, regulatory (inhibitor) subunit 3C (PPP1R3C), mRNA | PPP1R3C | + | ||
| NM_000958 | Prostaglandin E receptor 4 (subtype EP4) (PTGER4), mRNA | PTGER4 | + | I | |
| NM_198965 | Parathyroid hormone-like hormone (PTHLH), transcript variant 1, mRNA | PTHLH | + | I | |
| NM_144593 | Ras homolog enriched in brain-like 1 (RHEBL1), mRNA | RHEBL1 | + | ||
| NM_014575 | Schwannomin interacting protein 1 (SCHIP1), mRNA | SCHIP1 | + | ||
| NM_000450 | Selectin E (endothelial adhesion molecule 1) (SELE), mRNA | SELE | + | II | |
| NM_005842 | Sprouty homolog 2 (Drosophila) (SPRY2), mRNA | SPRY2 | + | I | |
| NM_003222 | Transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) (TFAP2C), mRNA | TFAP2C | + | ||
| NM_000594 | Tumor necrosis factor (TNF, superfamily, member 2) (TNF), mRNA | TNF | + | II | |
| NM_001003818 | Tripartite motif-containing 6 (TRIM6), transcript variant 1, mRNA | TRIM6 | + | ||
| NM_033035 | Thymic stromal lymphopoietin (TSLP), transcript variant 1, mRNA | TSLP | + | I | |
| NM_024640 | yrdC domain-containing (E. coli) (YRDC), mRNA | YRDC | + | ||
| NM_023039 | Ankyrin repeat, family A (RFXANK-like), 2 (ANKRA2), mRNA | ANKRA2 | |||
| NM_001233 | Caveolin 2 (CAV2), transcript variant 1, mRNA | CAV2 | I | ||
| NM_025214 | Coiled-coil domain-containing 68 (CCDC68), mRNA | CCDC68 | I | ||
| NM_004591 | Chemokine (C-C motif) ligand 20 (CCL20), mRNA | CCL20 | |||
| NM_012129 | Claudin 12 (CLDN12), mRNA | CLDN12 | |||
| NM_030627 | Cytoplasmic polyadenylation element binding protein 4 (CPEB4), mRNA | CPEB4 | II | ||
| NM_001912 | Cathepsin L (CTSL), transcript variant 1, mRNA | CTSL | |||
| NM_001565 | Chemokine (C-X-C motif) ligand 10 (CXCL10), mRNA | CXCL10 | |||
| NM_005409 | Chemokine (C-X-C motif) ligand 11 (CXCL11), mRNA | CXCL11 | |||
| NM_000782 | Cytochrome P450, family 24, subfamily A, polypeptide 1 (CYP24A1), nuclear gene encoding mitochondrial protein, mRNA | CYP24A1 | |||
| NM_019885 | Cytochrome P450, family 26, subfamily B, polypeptide 1 (CYP26B1), mRNA | CYP26B1 | |||
| AK026768 | cDNA: FLJ23115 fis, clone LNG07933 | DPY19L1P1 | |||
| NM_022726 | Elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4 (ELOVL4), mRNA | ELOVL4 | I | ||
| NM_001432 | Epiregulin (EREG), mRNA | EREG | I | ||
| NM_144503 | F11 receptor (F11R), transcript variant 4, mRNA | F11R | |||
| NM_001993 | Coagulation factor III (thromboplastin, tissue factor) (F3), mRNA | F3 | II | ||
| NM_020066 | Formin 2 (FMN2), mRNA | FMN2 | |||
| NM_013409 | Follistatin (FST), transcript variant FST344, mRNA | FST | |||
| NM_013372 | Gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) (GREM1), mRNA | GREM1 | |||
| NM_001548 | Interferon-induced protein with tetratricopeptide repeats 1 (IFIT1), transcript variant 2, mRNA | IFIT1 | |||
| NM_016584 | Interleukin 23, alpha subunit p19 (IL23A), mRNA | IL23A | |||
| NM_002192 | Inhibin, beta A (activin A, activin AB alpha polypeptide) (INHBA), mRNA | INHBA | I | ||
| NM_000210 | Integrin, alpha 6 (ITGA6), mRNA | ITGA6 | |||
| NM_005574 | LIM domain only 2 (rhombotin-like 1) (LMO2), mRNA | LMO2 | |||
| NM_033049 | Mucin 13, cell surface-associated (MUC13), mRNA | MUC13 | |||
| NM_021229 | Netrin 4 (NTN4), mRNA | NTN4 | |||
| NM_002581 | Pregnancy-associated plasma protein A, pappalysin 1 (PAPPA), mRNA | PAPPA | |||
| NM_002607 | Platelet-derived growth factor alpha polypeptide (PDGFA), transcript variant 1, mRNA | PDGFA | |||
| NM_016205 | Platelet-derived growth factor C (PDGFC), mRNA | PDGFC | |||
| NM_006823 | Protein kinase (cyclic AMP-dependent, catalytic) inhibitor alpha (PKIA), transcript variant 6, mRNA | PKIA | |||
| NM_001005377 | Plasminogen activator, urokinase receptor (PLAUR), transcript variant 3, mRNA | PLAUR | I | ||
| NM_000959 | Prostaglandin F receptor (FP) (PTGFR), transcript variant 1, mRNA | PTGFR | |||
| NM_003020 | Secretogranin V (7B2 protein) (SCG5), mRNA | SCG5 | |||
| NM_002998 | Syndecan 2 (heparan sulfate proteoglycan 1, cell surface-associated, fibroglycan) (SDC2), mRNA | SDC2 | |||
| NM_002999 | Syndecan 4 (amphiglycan, ryudocan) (SDC4), mRNA | SDC4 | |||
| NM_006216 | Serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 (SERPINE2), mRNA | SERPINE2 | |||
| NM_005025 | Serpin peptidase inhibitor, clade I (neuroserpin), member 1 (SERPINI1), mRNA | SERPINI1 | |||
| NM_203349 | SHC (Src homology 2 domain-containing) family, member 4 (SHC4), mRNA | SHC4 | |||
| NM_003759 | Solute carrier family 4, sodium bicarbonate cotransporter, member 4 (SLC4A4), mRNA | SLC4A4 | |||
| NM_032229 | SLIT and NTRK-like family, member 6 (SLITRK6), mRNA | SLITRK6 | I | ||
| NM_006714 | Sphingomyelin phosphodiesterase, acid-like 3A (SMPDL3A), mRNA | SMPDL3A | |||
| NM_003122 | Serine peptidase inhibitor, Kazal type 1 (SPINK1), mRNA | SPINK1 | |||
| NM_030964 | Sprouty homolog 4 (Drosophila) (SPRY4), mRNA | SPRY4 | I | ||
| NM_015000 | Serine/threonine kinase 38-like (STK38L), mRNA | STK38L | |||
| NM_018423 | Serine/threonine/tyrosine kinase 1 (STYK1), mRNA | STYK1 | I | ||
| NM_003236 | Transforming growth factor alpha (TGFA), mRNA | TGFA | I | ||
| NM_003243 | Transforming growth factor beta receptor III (betaglycan, 300 kDa) (TGFBR3), mRNA | TGFBR3 | |||
| NM_024847 | Transmembrane channel-like 7 (TMC7), mRNA | TMC7 | |||
| NM_003304 | Transient receptor potential cation channel, subfamily C, member 1 (TRPC1), mRNA | TRPC1 | I | ||
| NM_016061 | Yippee-like 5 (Drosophila) (YPEL5), mRNA | YPEL5 | |||
| NM_019006 | Zinc finger, AN1-type domain 6 (ZFAND6), mRNA | ZFAND6 | |||
| NM_003407 | Zinc finger protein 36, C3H type, homolog (mouse) (ZFP36), mRNA | ZFP36 | II | ||
Listed are mRNAs with >2-fold signal intensities in stKSRP pulldown compared to those in stGFP pulldown and >2-fold signal intensities in KSRP knockdown cells compared to those in GFP knockdown cells in two independent assays (for details, see Materials and Methods). Boldface indicates mRNAs destabilized by KSRP.
mRNAs labeled “unstable” (+) decreased to 50% or less at 3 h after actinomycin D addition in GFP knockdown cells in both assays.
For mRNAs labeled “stabilized” (+), the ratio of signal intensities for 3 h of actinomycin D treatment to those at 0 h is >1.33-fold higher for KSRP knockdown than for GFP knockdown cells.
ARE classification was done using the ARED3.0 database search tool (http://rc.kfshrc.edu.sa/ared). ARE-containing as well as unstable mRNAs are highly significantly overrepresented (P < 0.0001, Fisher's exact test).
Possible autoregulation of KSRP.
Interestingly, among the transcripts associated with stKSRP was that of its homolog FUBP3 and also the KSRP mRNA itself (see Table S1 in the supplemental material). To exclude the possibility that the latter result merely reflects a higher input level (due to the additional mRNAs derived from the stKSRP vector), RT-PCR was performed with primers that amplify 3′ UTRs not contained in the vector (Fig. 7B). The result clearly demonstrates an association of stKSRP with endogenous KSRP mRNA. The plasmid-derived KSRP mRNA contains part of the 3′ UTR. Since that mRNA was also enriched, while a KSRP mRNA without 3′ UTR sequences was not (not shown), a binding site for KSRP protein was further localized by expressing the β-globin mRNA with 3′ UTR sequences from KSRP mRNA. Association with stKSRP in pulldown assays was found to be mediated by a non-ARE region of 104 nt 3′ of the stop codon in the KSRP mRNA (Fig. 7C).
DISCUSSION
Rapid degradation of IL-8 mRNA and its inhibition by the p38/MK2 pathway are largely dependent on an ARE with a bipartite structure (44, 46). In this report, we define KSRP as a major factor involved in the function of this ARE, based on three lines of evidence obtained by (i) siRNA-mediated knockdown, (ii) in vitro protein-ARE interaction, and (iii) copulldown of RNA and protein. (i) siRNA-mediated knockdown of KSRP slowed down degradation of endogenous IL-8 mRNA and of a reporter mRNA containing the IL-8 ARE (Fig. 1). Consistent with prior reports that KSRP can recruit the deadenylating enzyme PARN to the mRNA (3, 9, 18), the IL-8 mRNA has long poly(A) tails in the KSRP knockdown cells. Evidence that, in addition to PARN, exosome components and, more recently, the decapping enzyme Dcp2 are recruited by KSRP has been presented (9, 18). Decapping and 5′-to-3′ degradation are involved in ARE-mediated decay according to other studies (33, 41). Whether KSRP is essential for deadenylation of all mRNAs that it destabilizes or only for some of them, as shown for IL-8 and CCL20 in this study, is not known. Perhaps recruiting the exosome or enzymes of 5′-to-3′ degradation is the major function for degradation of other mRNA targets. Of interest, it has been reported that KSRP interacts with the ARE-binding protein TTP (32), which may contribute to determining the subsequent events in degradation. (ii) KSRP directly interacts with the IL-8 ARE in vitro (Fig. 3A). One of the complexes formed with cytoplasmic extract from HeLa cells could be ascribed to KSRP interaction. We have reported a bipartite structure for the IL-8 ARE with a core domain containing the four AUUUA motifs that mediates moderate destabilization and an auxiliary domain which does not destabilize on its own but enhances the destabilizing effect of the core domain (44). The data in Fig. 4 indicate that this behavior reflects the weak interaction of KSRP with the individual domains compared to that with the complete ARE. (iii) Endogenous IL-8 mRNA as well as a reporter mRNA containing the IL-8 ARE is found associated with KSRP pulled down from cytoplasmic extracts. The extent of interaction assayed in this way for the complete ARE and its domains parallels that observed in gel shift assays and, most importantly, their destabilizing activity (Fig. 4). Clearly, the interaction of the IL-8 ARE with KSRP as assayed by gel shift as well as by copulldown from cell lysates shows that both the four AUUUA motifs and the auxiliary domain are as essential for maximal interaction with KSRP as they are for maximal mRNA destabilization in intact cells (44) (Fig. 4). This suggests that the degree of destabilization is directly dependent on the interaction with KSRP.
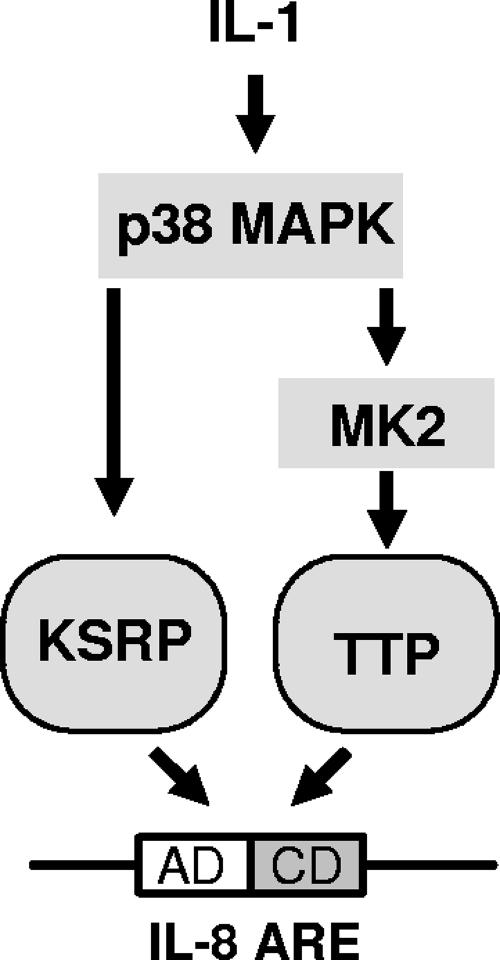
Rapid ARE-dependent degradation of IL-8 mRNA is impaired upon activation of the p38 MAP kinase/MK2 signaling pathway (46). Recently, p38 MAP kinase has been shown to phosphorylate KSRP at threonine 692, which inhibits its binding and destabilizing function for AREs of myogenin and p21 in a muscle cell differentiation model (3). In agreement with this report, IL-1α impairs association of KSRP with mRNA containing the IL-8 ARE in pulldown assays (Fig. 5A). This effect of IL-1, like its mRNA stabilization, is sensitive to inhibition of p38 MAP kinase. Interestingly, an inhibitory mutant of MK2 which interferes with mRNA stabilization (46) did not affect IL-1-induced impairment of KSRP-ARE interaction (Fig. 5A), and an active mutant of MK2 which induces stabilization did not mimic the effect of IL-1 (Fig. 5B). KSRP is not phosphorylated by MK2 (3). However, it cannot be ruled out that MK2 modifies KSRP function indirectly. The stabilization of the reporter-IL-8 ARE mRNA by an active form of MK2 in the presence of a p38 inhibitor (Fig. 5C) argues against a mere supportive role for MK2 for the effect of p38 MAP kinase, e.g., by facilitating its export from the nucleus (2, 14). MK2 may also affect other proteins involved in degradation. A known substrate of MK2 is the ARE-binding protein TTP, and activation of the p38/MK2 pathway has been associated with increased stability of TTP target mRNAs (6, 10, 24, 34, 42). We therefore investigated a possible role for TTP in degradation of IL-8 mRNA and its modulation by MK2. TTP, expressed in small amounts as suggested to detect its destabilizing activity (31) and to limit artifacts (4), accelerated degradation of an IL-8 ARE containing reporter RNA under conditions where basal degradation was slow due to KSRP knockdown. Acceleration of degradation was partly reversed by coexpressing active MK2 (Fig. 6). This, together with the copulldown of TTP and endogenous IL-8 mRNA, suggests that MK2 contributes to stabilization of IL-8 mRNA in part by impairing TTP function. It has to be noted, however, that TTP is not expressed strongly in the cells used here, and other targets of MK2 may be of relevance. Results from this study, together with the information cited above, are summarized in the scheme presented in Fig. 8. According to this model, IL-8 expression is increased by stabilization of its mRNA in response to a signaling pathway in which two consecutively activated kinases, p38 MAP kinase and MK2, impair the functions of distinct mRNA destabilizing proteins, KSRP and TTP, respectively.
FIG. 8.
Scheme of IL-8 ARE-dependent control of mRNA stability. The two destabilizing proteins KSRP and TTP can interact with the IL-8 ARE and promote degradation. Activators of the p38 MAP kinase pathway, like IL-1, can induce stabilization by impairing the function of KSRP via p38 MAP kinase and of TTP via MK2 (for details, see Discussion).
Il-8 is an important component of the inflammatory response (25). We used HeLa cells stimulated with the inflammatory cytokine IL-1 as an in vitro correlate of an inflammatory reaction for identifying other targets of KSRP besides IL-8.
The microarray data on KSRP-associated mRNAs show enrichment of relevant target mRNAs, such as those of IL-8 itself, IL-6, Cox 2 (PTGS2), CCL20, or CXCL3, which correspondingly accumulate in siRNA-mediated knockdown of KSRP (Table 1). However, though ARE-containing mRNAs are enriched in the KSRP-associated population, most of these mRNAs are not listed in the ARED database. This is likely due in part to limitations of the selection criteria which discard transcripts with AREs loosely related to the search pattern or lacking the AUUUA motif. On the other hand, relevant non-ARE targets may be enriched in the pulldown as well. Several studies provide evidence for interaction of KSRP with non-ARE sequences and functions distinct from mRNA destabilization (11, 23, 28, 35). It should be noted that mRNAs were enriched by pulldown of KSRP from cytoplasmic extracts, most likely causing underrepresentation of targets for nucleus-restricted functions of KSRP affecting, e.g., splicing (35) or transcription (11).
Interestingly, among the transcripts associated with KSRP were those of KSRP itself and of its homolog FUBP3. Recently, evidence has been presented for overlapping sets of mRNAs regulated by each of the three FUBPs (11). The association of two of their transcripts with KSRP suggests in addition auto- and cross-regulation of their own expression. RNA-targeted autoregulation of proteins involved in RNA metabolism has been described, e.g., for poly(A)-binding protein (27), for the ARE-binding protein TTP (5, 43), or for the yeast mRNA export factor Yra1p (13). Whether and how KSRP affects its own mRNA and that of FUBP3 will have to await further analysis.
siRNA-induced knockdown of KSRP resulted in >2-fold increased amounts of more than 400 transcripts. These could represent either direct targets of KSRP, like IL-8 mRNA, or indirect targets, upregulated as a consequence of increased expression of a direct target, like a transcription factor or receptor ligand. Further filtering in a rigorous way, including enrichment in KSRP pulldown, rapid degradation in control cells, and increased stability in KSRP knockdown cells as parameters, yielded 10 transcripts which are bona fide direct targets of KSRP-mediated destabilization. Most of them encode cytokines or other proteins connected with inflammatory and immune reactions. Structurally, they mostly are characterized by the presence of AREs. Among them are the mRNAs of IL-6 and CSF2 (also named GM-CSF), which we expected to be identified since IL-6 mRNA and a reporter with the GM-CSF ARE have been found stabilized by IL-1 stimulation or p38 MAP kinase activation in our initial studies (46). Of note, a set of seven KSRP targets was identified most recently by a similar strategy (37) in a different setting, namely, PI3K-AKT activation of pituitary α T3-1 cells. This may explain the lack of overlap with the transcripts identified here by investigating IL-1-stimulated HeLa cells. The true number of targets in the latter has to be considered much higher due to intrinsic limitations of the procedures applied. For example, significant stabilization of highly unstable RNAs might have been missed with the 3-h actinomycin D treatment; IL-1-induced RNAs with half-lives around or longer than 1.5 h would not be enriched twofold within the 2-h stimulation period with IL-1. Therefore, it is expected that more relevant targets are included in Table 1.
These data provide evidence for a role for KSRP in controlling the expression of inflammatory genes by limiting the half-lives of the respective mRNAs. This function of KSRP and its modulation by p38 MAP kinase may have important consequences for the quality, intensity, and duration of inflammatory reactions.
Supplementary Material
Acknowledgments
We thank H. Bujard, A. Stern, P. T. Lomedico, and A.-B. Shyu for previously supplied materials and C.-Y. Chen for monoclonal antibodies against KSRP and the His-KSRP expression vector. We are grateful to Roberto Gherzi for helpful discussion, to Axel Weber for help in the processing of microarray data, and to Heike Schneider for excellent technical assistance.
This work was supported by grants SFB566/A10, SFB566/Z2, and Ho1116/3 from the Deutsche Forschungsgemeinschaft. B.T.K. holds a scholarship of the Hannover Biomedical Research School.
Footnotes
Published ahead of print on 1 October 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bakheet, T., B. R. Williams, and K. S. Khabar. 2006. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res. 34:D111-D114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben-Levy, R., S. Hooper, R. Wilson, H. F. Paterson, and C. J. Marshall. 1998. Nuclear export of the stress-activated protein kinase p38 mediated by its substrate MAPKAP kinase-2. Curr. Biol. 8:1049-1057. [DOI] [PubMed] [Google Scholar]
- 3.Briata, P., S. V. Forcales, M. Ponassi, G. Corte, C. Y. Chen, M. Karin, P. L. Puri, and R. Gherzi. 2005. p38-dependent phosphorylation of the mRNA decay-promoting factor KSRP controls the stability of select myogenic transcripts. Mol. Cell 20:891-903. [DOI] [PubMed] [Google Scholar]
- 4.Brook, M., C. R. Tchen, T. Santalucia, J. McIlrath, J. S. Arthur, J. Saklatvala, and A. R. Clark. 2006. Posttranslational regulation of tristetraprolin subcellular localization and protein stability by p38 mitogen-activated protein kinase and extracellular signal-regulated kinase pathways. Mol. Cell. Biol. 26:2408-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brooks, S. A., J. E. Connolly, and W. F. Rigby. 2004. The role of mRNA turnover in the regulation of tristetraprolin expression: evidence for an extracellular signal-regulated kinase-specific, AU-rich element-dependent, autoregulatory pathway. J. Immunol. 172:7263-7271. [DOI] [PubMed] [Google Scholar]
- 6.Carballo, E., H. Cao, W. S. Lai, E. A. Kennington, D. Campbell, and P. J. Blackshear. 2001. Decreased sensitivity of tristetraprolin-deficient cells to p38 inhibitors suggests the involvement of tristetraprolin in the p38 signaling pathway. J. Biol. Chem. 276:42580-42587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, C. Y., R. Gherzi, S. E. Ong, E. L. Chan, R. Raijmakers, G. J. Pruijn, G. Stoecklin, C. Moroni, M. Mann, and M. Karin. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107:451-464. [DOI] [PubMed] [Google Scholar]
- 8.Chen, C. Y., and A. B. Shyu. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20:465-470. [DOI] [PubMed] [Google Scholar]
- 9.Chou, C. F., A. Mulky, S. Maitra, W. J. Lin, R. Gherzi, J. Kappes, and C. Y. Chen. 2006. Tethering KSRP, a decay-promoting AU-rich element-binding protein, to mRNAs elicits mRNA decay. Mol. Cell. Biol. 26:3695-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chrestensen, C. A., M. J. Schroeder, J. Shabanowitz, D. F. Hunt, J. W. Pelo, M. T. Worthington, and T. W. Sturgill. 2004. MAPKAP kinase 2 phosphorylates tristetraprolin on in vivo sites including Ser178, a site required for 14-3-3 binding. J. Biol. Chem. 279:10176-10184. [DOI] [PubMed] [Google Scholar]
- 11.Chung, H. J., J. Liu, M. Dundr, Z. Nie, S. Sanford, and D. Levens. 2006. FBPs are calibrated molecular tools to adjust gene expression. Mol. Cell. Biol. 26:6584-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dean, J. L., G. Sully, A. R. Clark, and J. Saklatvala. 2004. The involvement of AU-rich element-binding proteins in p38 mitogen-activated protein kinase pathway-mediated mRNA stabilisation. Cell. Signal. 16:1113-1121. [DOI] [PubMed] [Google Scholar]
- 13.Dong, S., C. Li, D. Zenklusen, R. H. Singer, A. Jacobson, and F. He. 2007. YRA1 autoregulation requires nuclear export and cytoplasmic Edc3p-mediated degradation of its pre-mRNA. Mol. Cell 25:559-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engel, K., A. Kotlyarov, and M. Gaestel. 1998. Leptomycin B-sensitive nuclear export of MAPKAP kinase 2 is regulated by phosphorylation. EMBO J. 17:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fechir, M., K. Linker, A. Pautz, T. Hubrich, U. Forstermann, F. Rodriguez-Pascual, and H. Kleinert. 2005. Tristetraprolin regulates the expression of the human inducible nitric-oxide synthase gene. Mol. Pharmacol. 67:2148-2161. [DOI] [PubMed] [Google Scholar]
- 16.Gaestel, M. 2006. MAPKAP kinases—MKs—two's company, three's a crowd. Nat. Rev. Mol. Cell Biol. 7:120-130. [DOI] [PubMed] [Google Scholar]
- 17.Garneau, N. L., J. Wilusz, and C. J. Wilusz. 2007. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 8:113-126. [DOI] [PubMed] [Google Scholar]
- 18.Gherzi, R., K. Y. Lee, P. Briata, D. Wegmuller, C. Moroni, M. Karin, and C. Y. Chen. 2004. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell 14:571-583. [DOI] [PubMed] [Google Scholar]
- 19.Gherzi, R., M. Trabucchi, M. Ponassi, T. Ruggiero, G. Corte, C. Moroni, C. Y. Chen, K. S. Khabar, J. S. Andersen, and P. Briata. 2006. The RNA-binding protein KSRP promotes decay of beta-catenin mRNA and is inactivated by PI3K-AKT signaling. PLoS Biol. 5:e5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Gowrishankar, G., R. Winzen, F. Bollig, B. Ghebremedhin, N. Redich, B. Ritter, K. Resch, M. Kracht, and H. Holtmann. 2005. Inhibition of mRNA deadenylation and degradation by ultraviolet light. Biol. Chem. 386:1287-1293. [DOI] [PubMed] [Google Scholar]
- 21.Gowrishankar, G., R. Winzen, O. Dittrich-Breiholz, N. Redich, M. Kracht, and H. Holtmann. 2006. Inhibition of mRNA deadenylation and degradation by different types of cell stress. Biol. Chem. 387:323-327. [DOI] [PubMed] [Google Scholar]
- 22.Green, J., K. S. Khabar, B. C. Koo, B. R. Williams, and S. J. Polyak. 2006. Stability of CXCL-8 and related AU-rich mRNAs in the context of hepatitis C virus replication in vitro. J. Infect. Dis. 193:802-811. [DOI] [PubMed] [Google Scholar]
- 23.Gu, W., F. Pan, H. Zhang, G. J. Bassell, and R. H. Singer. 2002. A predominantly nuclear protein affecting cytoplasmic localization of beta-actin mRNA in fibroblasts and neurons. J. Cell Biol. 156:41-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hitti, E., T. Iakovleva, M. Brook, S. Deppenmeier, A. D. Gruber, D. Radzioch, A. R. Clark, P. J. Blackshear, A. Kotlyarov, and M. Gaestel. 2006. Mitogen-activated protein kinase-activated protein kinase 2 regulates tumor necrosis factor mRNA stability and translation mainly by altering tristetraprolin expression, stability, and binding to adenine/uridine-rich element. Mol. Cell. Biol. 26:2399-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffmann, E., O. Dittrich-Breiholz, H. Holtmann, and M. Kracht. 2002. Multiple control of interleukin-8 gene expression. J. Leukoc. Biol. 72:847-855. [PubMed] [Google Scholar]
- 26.Holtmann, H., R. Winzen, P. Holland, S. Eickemeier, E. Hoffmann, D. Wallach, N. L. Malinin, J. A. Cooper, K. Resch, and M. Kracht. 1999. Induction of interleukin-8 synthesis integrates effects on transcription and mRNA degradation from at least three different cytokine- or stress-activated signal transduction pathways. Mol. Cell. Biol. 19:6742-6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hornstein, E., H. Harel, G. Levy, and O. Meyuhas. 1999. Overexpression of poly(A)-binding protein down-regulates the translation or the abundance of its own mRNA. FEBS Lett. 457:209-213. [DOI] [PubMed] [Google Scholar]
- 28.Huttelmaier, S., D. Zenklusen, M. Lederer, J. Dictenberg, M. Lorenz, X. Meng, G. J. Bassell, J. Condeelis, and R. H. Singer. 2005. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature 438:512-515. [DOI] [PubMed] [Google Scholar]
- 29.Junttila, M. R., S. Saarinen, T. Schmidt, J. Kast, and J. Westermarck. 2005. Single-step Strep-tag purification for the isolation and identification of protein complexes from mammalian cells. Proteomics 5:1199-1203. [DOI] [PubMed] [Google Scholar]
- 30.Kolev, N. G., and P. W. Huber. 2003. VgRBP71 stimulates cleavage at a polyadenylation signal in Vg1 mRNA, resulting in the removal of a cis-acting element that represses translation. Mol. Cell 11:745-755. [DOI] [PubMed] [Google Scholar]
- 31.Lai, W. S., E. Carballo, J. M. Thorn, E. A. Kennington, and P. J. Blackshear. 2000. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. J. Biol. Chem. 275:17827-17837. [DOI] [PubMed] [Google Scholar]
- 32.Linker, K., A. Pautz, M. Fechir, T. Hubrich, J. Greeve, and H. Kleinert. 2005. Involvement of KSRP in the post-transcriptional regulation of human iNOS expression-complex interplay of KSRP with TTP and HuR. Nucleic Acids Res. 33:4813-4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lykke-Andersen, J., and E. Wagner. 2005. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 19:351-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahtani, K. R., M. Brook, J. L. Dean, G. Sully, J. Saklatvala, and A. R. Clark. 2001. Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor alpha mRNA stability. Mol. Cell. Biol. 21:6461-6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Min, H., C. W. Turck, J. M. Nikolic, and D. L. Black. 1997. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 11:1023-1036. [DOI] [PubMed] [Google Scholar]
- 36.Rehbein, M., K. Wege, F. Buck, M. Schweizer, D. Richter, and S. Kindler. 2002. Molecular characterization of MARTA1, a protein interacting with the dendritic targeting element of MAP2 mRNAs. J. Neurochem. 82:1039-1046. [DOI] [PubMed] [Google Scholar]
- 37.Ruggiero, T., M. Trabucchi, M. Ponassi, G. Corte, C. Y. Chen, L. al-Haj, K. S. Khabar, P. Briata, and R. Gherzi. 2007. Identification of a set of KSRP target transcripts upregulated by PI3K-AKT signaling. BMC Mol. Biol. 8:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rydziel, S., A. M. Delany, and E. Canalis. 2004. AU-rich elements in the collagenase 3 mRNA mediate stabilization of the transcript by cortisol in osteoblasts. J. Biol. Chem. 279:5397-5404. [DOI] [PubMed] [Google Scholar]
- 39.Shaw, G., and R. Kamen. 1986. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 46:659-667. [DOI] [PubMed] [Google Scholar]
- 40.Snee, M., G. J. Kidd, T. P. Munro, and R. Smith. 2002. RNA trafficking and stabilization elements associate with multiple brain proteins. J. Cell Sci. 115:4661-4669. [DOI] [PubMed] [Google Scholar]
- 41.Stoecklin, G., T. Mayo, and P. Anderson. 2006. ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 7:72-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stoecklin, G., T. Stubbs, N. Kedersha, S. Wax, W. F. Rigby, T. K. Blackwell, and P. Anderson. 2004. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 23:1313-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tchen, C. R., M. Brook, J. Saklatvala, and A. R. Clark. 2004. The stability of tristetraprolin mRNA is regulated by mitogen-activated protein kinase p38 and by tristetraprolin itself. J. Biol. Chem. 279:32393-32400. [DOI] [PubMed] [Google Scholar]
- 44.Winzen, R., G. Gowrishankar, F. Bollig, N. Redich, K. Resch, and H. Holtmann. 2004. Distinct domains of AU-rich elements exert different functions in mRNA destabilization and stabilization by p38 mitogen-activated protein kinase or HuR. Mol. Cell. Biol. 24:4835-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winzen, R., S. Kafert, B. Preiss, H. A. Mylius-Spencker, K. Resch, and H. Holtmann. 1996. Interaction between the mRNA of the 55-kDa tumor necrosis factor receptor and cellular proteins. Possible involvement in post-transcriptional regulation of receptor expression. J. Biol. Chem. 271:13461-13467. [DOI] [PubMed] [Google Scholar]
- 46.Winzen, R., M. Kracht, B. Ritter, A. Wilhelm, C. Y. Chen, A. B. Shyu, M. Muller, M. Gaestel, K. Resch, and H. Holtmann. 1999. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 18:4969-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu, J. Y., S. L. DeRuiter, and D. L. Turner. 2002. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl. Acad. Sci. USA 99:6047-6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.