Abstract
During development, Sonic hedgehog (Shh) regulates the proliferation of cerebellar granule neuron precursors (GNPs) in part via expression of Nmyc. We present evidence supporting a novel role for the Mad family member Mad3 in the Shh pathway to regulate Nmyc expression and GNP proliferation. Mad3 mRNA is transiently expressed in GNPs during proliferation. Cultured GNPs express Mad3 in response to Shh stimulation in a cyclopamine-dependent manner. Mad3 is necessary for Shh-dependent GNP proliferation as measured by bromodeoxyuridine incorporation and Nmyc expression. Furthermore, Mad3 overexpression, but not that of other Mad proteins, is sufficient to induce GNP proliferation in the absence of Shh. Structure-function analysis revealed that Max dimerization and recruitment of the mSin3 corepressor are required for Mad3-mediated GNP proliferation. Surprisingly, basic-domain-dependent DNA binding of Mad3 is not required, suggesting that Mad3 interacts with other DNA binding proteins to repress transcription. Interestingly, cerebellar tumors and pretumor cells derived from patched heterozygous mice express high levels of Mad3 compared with adjacent normal cerebellar tissue. Our studies support a novel role for Mad3 in cerebellar GNP proliferation and possibly tumorigenesis, and they challenge the current paradigm that Mad3 should antagonize Nmyc by competition for direct DNA binding via Max dimerization.
Granule neuron precursors (GNPs) are generated in the rostral hindbrain during late embryogenesis. Expansion of the GNP pool takes place in the external granule layer (EGL) of the cerebellum, with peak proliferation of these cells occurring between postnatal day 5 (P5) and P8 in the mouse (17). GNP expansion is regulated by Sonic hedgehog (Shh), a secreted factor that plays a role in the patterning of many tissues. In the cerebellum, Shh is made by Purkinje neurons and regulates the division of GNPs during postnatal development (6, 54). Shh binds to the transmembrane receptor Patched (Ptc) and in turn relieves Ptc-mediated inhibition of Smoothened (Smo) activity (23). Smo, a G-protein-coupled receptor (51), activates an inhibitory G protein (7) that leads to activation of Gli transcription factors and the initiation of gene expression required for cell cycle progression. However, the Shh signaling intermediates that regulate GNP proliferation are just beginning to be understood.
The important role played by Shh in GNP proliferation has been linked directly to cell cycle regulation by the demonstration that Shh induces expression of D cyclins during development (4) via Nmyc (26, 38). Nmyc is a member of the Myc/Max/Mad family of basic helix-loop-helix leucine zipper (bHLHZ) DNA binding proteins that functions in most instances as a transcriptional activator. Both Myc and Mad proteins form heterodimers with the cofactor Max, thereby permitting binding to specific DNA motifs known as E-box sequences (15). These DNA-bound heterodimers recruit coactivator or corepressor complexes that generate alterations in chromatin structure and transcriptional activity. For example, Mad3 interacts with Max and the mSin3 corepressor to repress transcription from a reporter promoter containing an E-box CACGTG sequence in cultured fibroblasts (22). In the cerebellum, Nmyc is expressed in proliferating GNPs during the clonal expansion phase in vivo and is upregulated in GNPs in response to Shh treatment in vitro. Furthermore, overexpression of Nmyc in cultured GNPs leads to an increase in proliferation and the expression of D cyclins (26, 38). Conversely, inactivation of Nmyc in neural progenitor cells in vivo leads to a smaller and disorganized cerebellum with a reduced cell density in the internal granule layer (28).
Using a microarray-based approach, we identified genes that are transiently upregulated during GNP proliferation with profiles similar to those of known Shh target genes such as Cyclin D2 (8). One of these genes, Mad3, is a member of the Mad family of bHLHZ transcriptional regulators comprised of Mad1, Mxi1, Mad3, and Mad4 (22). Mad proteins are part of the larger family of Max binding proteins to which Nmyc also belongs (15). Mad family members have been shown to inhibit cell cycle progression and promote differentiation (2). However, the role of Mad3 in cerebellum development is unknown. The expression profile of Mad3 correlated highly with that of Cyclin D2 (Pearson correlation coefficient, 0.977 [8]), and Mad3 failed to be downregulated in weaver mice compared with wild-type littermates (8). Cerebellar granule cells in weaver mice fail to switch off the cell cycle and differentiate (36), suggesting that Mad3 may play a role in cell cycle progression of GNPs. Indeed, the expression profile of Cyclin D2 displayed a pattern in wild-type and weaver mice that was similar to that of Mad3 (Pearson correlation coefficient, 0.981). Because the products of genes with similar expression profiles have been shown to function in the same pathway (10), these data suggested that Mad3 might be a component of the Shh pathway in the cerebellum. Here we present evidence to support a novel role for Mad3 in the Shh pathway to promote proliferation of cerebellar GNPs. Using highly purified cultures of GNPs, we demonstrate that Mad3 is necessary for Shh-mediated proliferation. Furthermore, overexpression of Mad3, but not other family members such as Mad1, is sufficient to induce GNP proliferation in the absence of Shh. Structure-function analysis revealed that dimerization with Max and recruitment of the Sin3 corepressor are required for Mad3-mediated GNP proliferation. Surprisingly, DNA binding via the basic domain of Mad3 is not required, suggesting that Mad3 interacts with other DNA binding proteins to repress transcription. Our studies support a novel role for Mad3 in the Shh pathway to promote cerebellar GNP proliferation and challenge the current paradigm that Mad3 should antagonize Nmyc by competition for direct DNA binding via Max dimerization.
MATERIALS AND METHODS
Animals.
CD-1 and ptc+/− mice were bred and maintained in the animal facility at UC Davis. To examine actively proliferating GNPs, CD-1 mice were injected (100 μg per gram of body weight, intraperitoneally) with bromodeoxyuridine (BrdU) or phosphate-buffered saline (PBS) as a control. Animals were anesthetized at 2 h postinjection and immediately perfused transcardially with 4% paraformaldehyde (PFA), and fixed tissue was processed for immunohistochemistry.
Antibodies.
We used horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (IgG) or rabbit anti-mouse IgG (MP Biomedicals), mouse anti-HNK-1 (Chemicon), rabbit anti-Nmyc (Santa Cruz Biotechnology), mouse antihemagglutinin (anti-HA) (a gift of J. Trimmer), rat anti-HA (Roche), mouse anti-green fluorescent protein (anti-GFP) (BD Bioscience Clontech), Alexa 488-conjugated donkey anti-rat IgG and donkey anti-mouse IgG and Alexa 594-conjugated donkey anti-rabbit IgG (Invitrogen), biotinylated sheep anti-BrdU (Abcam), and the following anti-Mad3 antibodies (Santa Cruz Biotechnology): for immunohistochemistry, rabbit anti-Mad3 (sc-933) and Mad3 competing peptide (sc-933p) and for immunocytochemistry, rabbit anti-Mad3 (sc-sc-770). Both anti-Mad3 antibodies were verified by immunoblotting HEK293 extracts from cells transfected with HA-Mad3 as described for Fig. 2A.
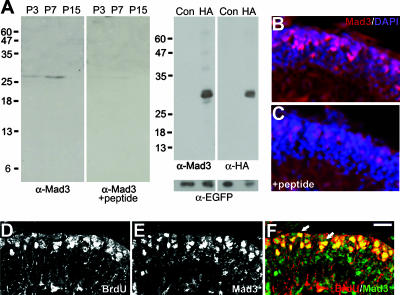
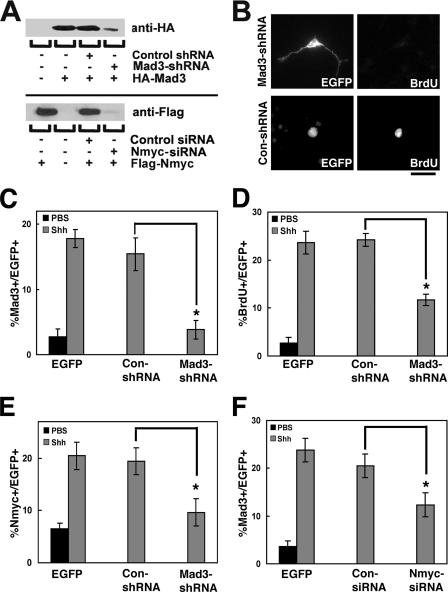
FIG. 2.
Mad3 is expressed in proliferating GNPs. (A) Left panels depict an immunoblot of nuclear extracts prepared from P3, P7, or P15 cerebellum probed with anti-Mad3 antibodies in the absence (left) or presence (right) of Mad3 competing peptide. Right panels depict an immunoblot of HEK293 cell extracts of HA-Mad3- and EGFP-cotransfected (HA) or control EGFP alone-transfected (Con) cells probed with anti-Mad3 (left), anti-HA (right), or anti-EGFP (bottom) antibodies. Molecular mass markers (kDa) are indicated. As expected, HA-Mad3 electrophoreses at a higher molecular mass (29 kDa) than endogenous Mad3 (27 kDa). (B and C) Immunohistochemistry of sagittal sections from P6 cerebellum stained with DAPI (blue) and anti-Mad3 antibodies (red) in the absence (B) or presence (C) of Mad3 competing peptide. Note that the image shown in panel C was exposed 25% longer than the image shown in panel B. (D to F) Immunohistochemistry of sagittal sections of cerebellum from P7 mice injected with BrdU for 2 h and stained with anti-BrdU (D) and anti-Mad3 (E) antibodies. The majority of BrdU+ cells (red) in the EGLa express Mad3 (green) as indicated by arrows in the overlay image (F). Bars, 40 μm (B and C) and 20 μm (D to F).
Constructs.
The Mad3-coding sequence was amplified from the RIKEN AV016640 cDNA clone and inserted into pHM6 (Roche) to create an in-frame HA tag at the N terminus. Mad4- and Nmyc-coding sequences were amplified from RIKEN AV057975 and IMAGE 3495446 cDNA clones, respectively, and inserted into pCMV-tag (Stratagene) to create an in-frame Flag tag at the N termini. Mad1- and Mxi1-coding sequences were amplified from IMAGE 1448816 and RIKEN AV031616 cDNA clones, respectively, and inserted into pCMV tag to create an in-frame Myc tag at the N termini. A two-step PCR protocol was used to make the Mad3 deletion constructs HA-Mad3-ΔSID (bp 16 to 78), HA-Mad3-Δbasic (bp 175 to 210), and HA-Mad3-ΔHLH (bp 211 to 344). For Mad3 knockdown experiments, the pSuper vector (Oligoengine) was used to generate a 19-nucleotide target sequence (CAT CCA CCT CCA GAT CCT C) or control sequence (TGT TCA GCG CGC CAG GAG C) for the mouse Mad3 gene. The sequences were incorporated into 64-nucleotide forward and reverse oligonucleotides, and the annealed oligonucleotides were subsequently cloned into the pSuper vector. To coexpress the short hairpin RNAs (shRNAs) and enhanced GFP (EGFP) from the same plasmid, the shRNA sequence and H1 promoter were cloned into a modified pBud vector (Invitrogen) expressing EGFP under the control of the cytomegalovirus promoter. The efficacy of the knockdown construct was assessed by cotransfection of the shRNA constructs and pHM6-HA-Mad3 into HEK293 cells and immunoblotting with anti-HA antibodies. Short interfering RNAs (siRNAs) directed against Nmyc and control siRNAs were purchased from Ambion (Nmyc siRNA, UUU UUC GUU CAC UGC GCG C; Nmyc control siRNA, UAU UCU UAC AGU ACU UAG G). All shRNA and siRNA sequences were verified by BLAST searches to not have potential overlap with other target sequences.
In situ hybridization.
In situ hybridization on frozen sections using digoxigenin (DIG)-labeled riboprobes was performed as described previously (8). Briefly, animals were perfused, and brains were dissected and fixed in 4% PFA-1× PBS for 2 h at 4°C, followed by incubation in 30% sucrose-1× PBS. Tissues were embedded in freezing medium and cut into 12- to 20-μm sagittal sections. Mad3 or Nmyc inserts were amplified with T7 and T3 primers and used as templates for in vitro transcription with T7 or T3 RNA polymerase (Ambion) to produce antisense or sense DIG-labeled riboprobes. Hybridizations were performed at 65°C for 16 h under standard washing conditions. Alkaline phosphatase-conjugated anti-DIG antibodies (Roche) bound to hybridized probe were visualized with nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate substrate (Roche).
Nuclear extracts and immunoblotting.
Dissected cerebella were washed with ice-cold PBS, disrupted by Dounce homogenization in cell lysis buffer [5 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) (pH 8), 85 mM KCl, 0.5% NP-40, and protease inhibitors], and the nuclei were pelleted by low-speed centrifugation. Nuclei were lysed in nuclei lysis buffer (50 mM Tris [pH 8], 10 mM EDTA, 1% sodium dodecyl sulfate, and protease inhibitors) and centrifuged, and the protein concentration in the supernatant was determined with bicinchoninic acid (Pierce). Sixty micrograms of protein was separated by 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose. Membranes were probed with primary antibodies in the presence or absence of a 10-fold molar excess of competing peptide. Bound primary antibodies were visualized with horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence substrate (Perkin-Elmer).
Cell culture.
Primary cultures of GNPs from CD-1 P5 to -7 mice were established as described previously (8). Briefly, isolated cerebella were treated with trypsin (Sigma) and DNase I (Roche). Dispersed cells after trituration were pelleted through a 4% bovine serum albumin cushion. GNPs were resuspended and centrifuged through a 40% Percoll step gradient (Amersham). Cells were resuspended in Neurobasal medium containing B27 supplement and Glutamax (Invitrogen) and plated on Lab-Tek 8-chamber slides coated with laminin (Roche) and polyornithine (Sigma) at a density of 2.5 × 105 cells/cm2. For some experiments, Shh-N (3 μg/ml) and/or cyclopamine (1 μg/ml; Calbiochem) was added at the beginning of culture. Shh-N was purchased from R&D Research or purified from bacteria harboring an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible Shh-N-expressing plasmid (a gift of D. Schaffer, UC Berkeley).
Immunofluorescence.
For immunohistochemistry, mice were perfused with 4% PFA, the cerebella were dissected, and 14-μm sagittal sections were cut and transferred to microscope slides. Sections were incubated with blocking solution (1× PBS, 4% milk, 1% Triton X-100), stained with primary antibodies for 16 h at 4°C, washed, and stained with secondary antibodies for 1 h at room temperature. For immunocytochemistry, cells were fixed with 4% PFA-10% sucrose and incubated in blocking buffer (1× PBS, 10% horse serum, 0.1% Triton X-100) for 30 min. Cells were stained with primary antibodies for 16 h at 4°C, washed, and incubated with secondary antibodies for 1 h at room temperature. Samples were mounted with Gel mount (Biomedia) and visualized with an epifluorescence microscope (Zeiss Axioplan2) with the appropriate filters. For BrdU incorporation, cells were pulsed with 10 μM BrdU (Sigma) for the final 12 h of culture according to published procedures (54). After labeling with primary and secondary antibodies, cells were postfixed, incubated with 2 N hydrochloric acid, and neutralized with 0.1 M sodium borate. Cells were stained with biotinylated anti-BrdU antibodies and tetramethyl rhodamine isocyanate-conjugated streptavidin (Jackson Immunoresearch). For terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assays (Roche), cells were stained according to the manufacturer's instructions.
Transfections.
GNPs were transfected using Lipofectamine 2000 (Invitrogen) for 30 min at 37°C prior to plating. For transfection experiments using Mad3 overexpression or Mad3 shRNA constructs, we used the following criteria to distinguish positive or negative stained cells. First, each transected cell (identified by EGFP or HA labeling) must be stained with DAPI (4′,6′-diamidino-2-phenylindole). Second, the total transfected cells were separated into two populations, negative and positive cells, according to the fluorescence intensity of Mad3, Nmyc, HNK-1, and/or BrdU expression, depending on the experiment and antibodies used. Negative cells did not have any signal or had a very weak signal similar to those of nontransfected cells in the same well, while positive cells showed a relatively strong signal in the cytoplasm or nucleus. Shh-N treatment quantification was performed using the AxioVision4.2 software (Zeiss). Statistical significance was determined using the Student t test in Excel.
RESULTS
Mad3 is expressed in cerebellar GNPs.
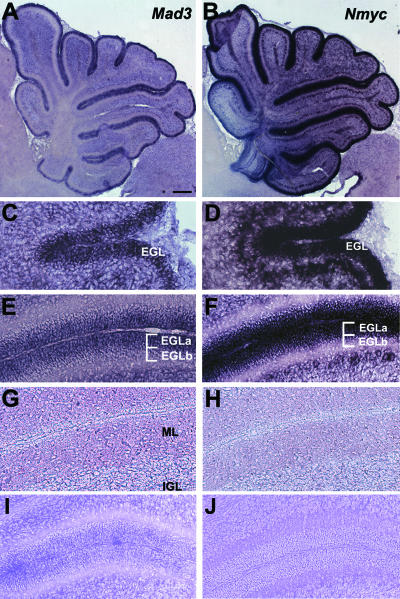
The expression profile for Mad3 (8) suggested that Mad3, like Cyclin D2 (45), should be expressed in the progenitor cells of the EGL. Therefore, the distribution of Mad3 mRNA in sections of mouse cerebellum was analyzed by in situ hybridization (Fig. 1). At P1, the mRNA encoding Mad3 is abundant in the EGL (Fig. 1C). At P7, Mad3 distribution is localized to the outer EGL (EGLa), the location of proliferating GNPs (Fig. 1A and E). At P15, expression of Mad3 is significantly reduced to nearly background levels (Fig. 1G). Nmyc mRNA displays a similar expression pattern (Fig. 1B, D, F, and H). For example, Nmyc is expressed in proliferating cells in the EGLa at P7 (Fig. 1F), consistent with previous studies (26). Control sense riboprobes for Mad3 or Nmyc yielded no significant signal (Fig. 1I and J). These observations validate the Mad3 expression profile defined using DNA microarrays (8) and are consistent with the expression pattern of other Shh signaling molecules, including Gli1, Ptc, and Nmyc (5, 26, 38), involved in the regulation of GNP proliferation during postnatal development.
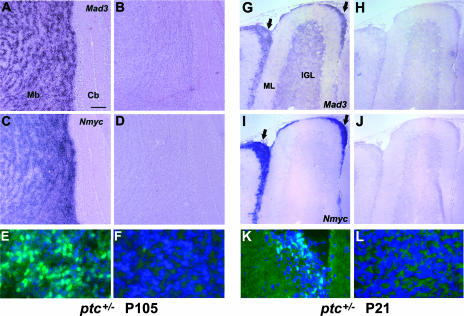
FIG. 1.
Mad3 and Nmyc distribution in postnatal cerebellum. Sagittal sections of mouse cerebellum at P1 (C and D), P7 (A, B, E, F, I, and J), or P15 (G and H) were hybridized in situ with DIG-labeled antisense probes for Mad3 (A, C, E, and G) or Nmyc (B, D, F, and H) or sense probes for Mad3 (I) or Nmyc (J). Abbreviations: EGLa, outer EGL; EGLb, inner EGL; IGL, internal granule layer; ML, molecular layer. Bars, 400 μm (A and B) and 100 μm (C to J).
To determine Mad3 protein distribution in the cerebellum, we prepared nuclear extracts from P3, P7, and P15 cerebellum for immunoblot analysis. Anti-Mad3 antibodies recognize a single band of approximately 27 kDa in P3 and P7 cerebellar nuclear extracts (Fig. 2A, far left), consistent with the peak of Mad3 mRNA expression (Fig. 1A) and the calculated molecular mass of Mad3 (23.6 kDa). Importantly, inclusion of a 10-fold molar excess of competing peptide to which the antibody was raised specifically abolished the presence of this band (Fig. 2A, left). To demonstrate further the specificity of the anti-Mad3 antibody, we performed an immunoblot analysis of extracts from HEK293 cells cotransfected with HA-tagged Mad3 and EGFP compared with control (EGFP only) transfected cells. Indeed, anti-Mad3 antibodies recognize a single band of the appropriate molecular mass (Fig. 2A, right), and the same band is recognized with anti-HA antibodies (Fig. 2A, far right). Immunoblotting with anti-EGFP antibodies demonstrated equivalent transfection and loading efficiencies. Thus, the commercial anti-Mad3 antibodies are specific for Mad3.
We next attempted to visualize Mad3 protein in cerebellar sections by immunohistochemistry. Anti-Mad3 antibodies revealed staining in GNPs in the EGLa at P7 (Fig. 2B). Importantly, the Mad3 staining pattern was dramatically decreased upon addition of the competing peptide (Fig. 2C), suggesting that the staining pattern that we observed is specific for Mad3. Interestingly, Mad3 has been reported to be specifically expressed in the S phase of the cell cycle in embryonic neuronal precursor cells and cultured fibroblasts (13, 43). To determine if Mad3 expression is specific to S phase in cerebellar GNPs, we analyzed the cerebella of P7 mice injected with BrdU for 2 hours by immunohistochemistry with anti-BrdU (Fig. 2D) and anti-Mad3 (Fig. 2E) antibodies. We observed significant overlap of BrdU-positive (BrdU+) cells compared with Mad3+ cells during the 2-h BrdU pulse-labeling (Fig. 2F). Quantification of confocal images indicated that 83.0% ± 7.9% of Mad3+ cells are colabeled with BrdU. Thus, Mad3 protein expression appears to be enriched selectively in the S phase of proliferating cerebellar GNPs in vivo, consistent with previous studies (13, 43).
Mad3 is a target of the Shh pathway in GNPs.
The transient expression of Mad3 in GNPs suggests that, like Nmyc, Mad3 could be regulated by Shh during granule cell proliferation. We tested this possibility using highly enriched cultures of granule cell precursors. Importantly, the in vitro culture system accurately reflects the in vivo situation for granule neuron development (18). In the absence of extracellular factors such as Shh, purified GNPs in culture undergo spontaneous differentiation as measured by expression of specific markers and cellular morphology. Furthermore, we have shown that genes known to be expressed in mature granule neurons or GNPs in vivo are similarly upregulated or downregulated, respectively, in cultured granule cells compared with in vivo development (8).
Purified GNPs treated with the biologically active portion of Shh (Shh-N) proliferate in vitro as measured by incorporation of BrdU and expression of a marker for GNPs (54). We hypothesized that GNPs treated with Shh-N will upregulate expression of Mad3, as has been demonstrated previously for Nmyc (26, 38). To this end, we analyzed GNPs treated with Shh-N or PBS as a control at the time of plating. Cells were cultured in the presence or absence of Shh-N for 2 days in vitro (DIV) and stained with anti-Mad3, anti-Nmyc, or anti-BrdU antibodies. Consistent with previous studies (54), GNPs cultured with Shh-N showed a significant increase in the number of BrdU+ cells compared to the PBS control group (1.0% ± 0.2% versus 20.0% ± 2.4% BrdU+/DAPI+ cells; P = 5.7 × 10−7).
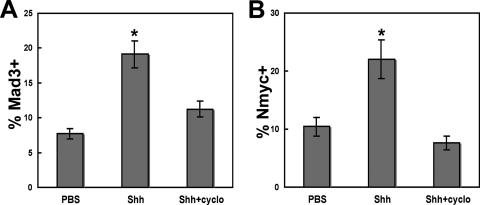
Similarly, Shh-N treatment of cerebellar GNPs resulted in a significant increase in the percentage of cells expressing Mad3 compared with PBS (7.7% ± 1.8% versus 19.1% ± 2.1% Mad3+/DAPI+ cells; P = 0.021) (Fig. 3A). To confirm that Mad3 is an Shh target, we used cyclopamine, a potent antagonist of Smo (50) that inhibits Shh-mediated GNP proliferation (26). We observed no significant increase in the percentage of Mad3+ cells upon Shh treatment in the presence of cyclopamine (7.7% ± 1.8% versus 11.2% ± 1.3% Mad3+/DAPI+ cells; P = 0.34) (Fig. 3A), suggesting that Mad3 acts downstream of Smo during Shh signaling. As expected, Shh-N treatment also increased the percentage of cells expressing Nmyc (10.4% ± 1.4% versus 22.0% ± 6.3% Nmyc+/DAPI+ cells; P = 0.045) (Fig. 3B) but not in the presence of cyclopamine (10.4% ± 1.4% versus 7.6% ± 1.2% Nmyc+/DAPI+ cells; P = 0.09) (Fig. 3B). Importantly, the majority of BrdU+ cells also express Mad3 (83.9% ± 7.5%), suggesting that cells expressing Mad3 are actively undergoing DNA replication upon Shh-N treatment. Because the majority of GNPs expressing Nmyc have previously been shown to incorporate BrdU (26, 38), it seems likely that GNPs express both Mad3 and Nmyc upon Shh-N treatment.
FIG. 3.
Mad3 is a target of the Shh pathway. Purified granule cells were treated with 3 μg/ml Shh-N or PBS at the time of plating, cultured for 2 days, and stained with DAPI and anti-Mad3 or anti-Nmyc antibodies. Graphs depict the percentage of DAPI+ cells that expressed Mad3 (A) or Nmyc (B) in the presence or absence of Shh and/or cyclopamine (cyclo). Error bars represent standard errors of the means for four to seven fields; each field contained 300 to 500 cells. Similar results were obtained in three independent experiments. Asterisks indicate P values of <0.05 compared with PBS-treated GNPs.
The percentage of Mad3+ and Nmyc+ cells appears to be higher than that of BrdU+ cells in the absence of Shh, suggesting the Mad3 and Nmyc may be expressed in nonproliferating cells. It is important to note that the GNP culture system represents an immature population of EGL progenitors with a mixture of proliferating and recently postmitotic but undifferentiated cells (32). Furthermore, in the absence of exogenous factors, the proliferating cells convert to immature postmitotic cells during the first two days in culture. Thus, we interpret these findings to suggest that the cells expressing Mad3 or Nmyc observed at 48 h in culture in the absence of Shh are EGL progenitors that were proliferating during the first 36 h of culture but have recently become postmitotic and thus are not labeled with BrdU during the final 12 h of culture. Taken together, these studies support the idea that Mad3 (like Nmyc) is an Shh target in cultured GNPs and indicate that Shh-N simultaneously induces proliferation of GNPs and expression of Mad3 and Nmyc.
Overexpression of Mad3 is sufficient to induce GNP proliferation.
Mad3 expression in Shh-N-treated GNPs suggested that Mad3 might play an active role during proliferation. To test this possibility, HA-Mad3 was transiently overexpressed in GNPs and the cells were cultured for 2 days in the absence of Shh and pulsed with BrdU for the final 12 h of culture. We counted the number of transfected cells (identified by labeling with anti-HA antibodies) that were colabeled with antibodies against BrdU or HNK-1, a cell surface antigen present on GNPs (55). As a control, we counted the number of transfected cells expressing EGFP (identified by labeling with anti-GFP antibodies) that incorporated BrdU or expressed HNK-1.
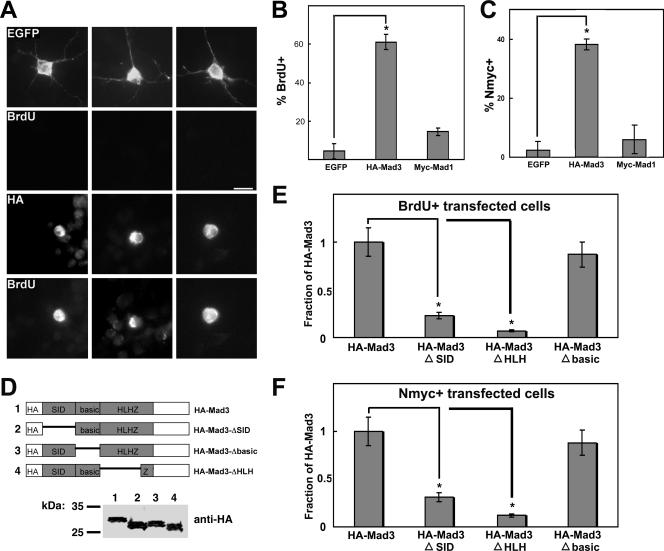
GNPs transfected with HA-Mad3 showed a sixfold increase in the percentage of cells that expressed HNK-1 compared with EGFP-transfected cells (10.0% ± 2.5% versus 60.7% ± 3.7%; P = 2.9 × 10−7), suggesting that overexpression of HA-Mad3 maintains the GNPs in a precursor state. Furthermore, GNPs transfected with HA-Mad3 were found to be frequently colabeled with anti-BrdU antibodies (Fig. 4A, bottom panels). Such colabeling was rarely observed with EGFP-transfected cells (Fig. 4A, top panels). Quantification of this experiment demonstrated a 14-fold increase in the percentage of HA-Mad3+-transfected cells compared with EGFP+-transfected cells that incorporated BrdU (4.34 ± 4.2% versus 61.1 ± 5.4%; P = 0.0006) (Fig. 4B), suggesting that transient overexpression of HA-Mad3 in the absence of Shh is sufficient to promote proliferation of GNPs in vitro. Importantly, overexpression of a different Mad family member, Myc-tagged Mad1, exhibited only a modest effect on BrdU incorporation in our culture system compared with EGFP+-transfected cells (4.34% ± 4.2% versus 14.5% ± 1.4%; P = 0.03) (Fig. 4B). Similar results were obtained upon overexpression of Flag-tagged Mad4 compared with EGFP-transfected cells (3.85% ± 0.96% versus 14.7% ± 3.9%; P = 0.017). Overexpression of Myc-tagged Mxi1 did not increase the percentage of BrdU+ cells compared with EGFP-transfected cells (3.85% ± 0.96% versus 5.88% ± 1.47%; P = 0.39). As expected, overexpression of Flag-tagged Nmyc increased the percentage of BrdU+ cells compared with EGFP-transfected cells (3.18% ± 1.6% versus 45.7% ± 9.1%; P = 0.02) to a similar extent as with HA-Mad3. Taken together, these data suggest that overexpression of Mad3 (like that of Nmyc) is sufficient to induce proliferation of cultured GNPs in the absence of Shh for 2 DIV and underscore the functional differences between Mad family members.
FIG. 4.
Mad3 is sufficient to induce GNP proliferation in the absence of Shh. (A) Granule cells were transfected with EGFP or HA-Mad3 and cultured for 2 days in the absence of Shh. During the final 12 h of culture, cells were pulsed with BrdU. Cells were fixed and stained with anti-EGFP antibodies or anti-HA antibodies and anti-BrdU. Bar, 10 μm. (B and C) Graphs depict the percentage of EGFP, HA-Mad3, or Myc-Mad1-transfected cells that incorporated BrdU (B) or expressed Nmyc (C). Error bars represent standard errors of the means for three to five wells; each well contained 25 to 50 transfected cells. Similar results were obtained in three independent experiments. Asterisks indicate P values of <0.01 compared with control EGFP-transfected cells. (D) Schematic of HA-Mad3 deletion constructs (top) and immunoblot of protein extracts from HEK293 cells expressing HA-Mad3 deletion constructs probed with anti-HA antibodies (bottom). Molecular mass markers are indicated. (E and F) Graphs depict the percentage of cells transfected with HA-Mad3 deletion constructs that incorporated BrdU (E) or expressed Nmyc (F) as a fraction of the value for the HA-Mad3 full-length construct. Asterisks indicate P values of <0.05 compared with HA-Mad3-transfected cells.
We next attempted to measure BrdU incorporation in GNPs overexpressing HA-Mad3 for 4 DIV and 6 DIV to determine how long HA-Mad3-overexpressing cells proliferate in culture. At 4 DIV we also observed an increase in the percentage of transfected cells incorporating BrdU among HA-Mad3-overexpressing cells compared with EGFP-transfected cells (1.04% ± 1.0% versus 30.3% ± 3.02%; P = 0.03). However, there was a 75% decrease in the number of HA-Mad3-transfected cells per well at 4 DIV compared with 2 DIV (34.2 ± 3.2 versus 8.7 ± 1.5; P = 0.003) while the number of EGFP-expressing cells per well remained constant (26.0 ± 3.1 versus 27.0 ± 1.2; P = 0.39). By 6 DIV the number of HA-Mad3-transfected cells per well was severely reduced compared with that at 2 DIV (34.2 ± 3.2 versus 2.5 ± 0.5; P = 0.004), while the number of EGFP-transfected cells per well at 6 DIV was not significantly different from that at 2 DIV (26.0 ± 3.1 versus 19.0 ± 1.0; P = 0.07). Nevertheless, the majority of the remaining HA-Mad3-transfected cells at 6 DIV were BrdU+ (83.3% ± 16.4%), suggesting that GNPs overexpressing HA-Mad3 at 4 DIV and 6 DIV were proliferating within the 12-h BrdU pulse.
Why does the total number of HA-Mad3 transfected GNPs decrease at 4 and 6 DIV? It is now well established that in addition to Myc's role in proliferation, ectopic expression of Myc can drive cells to undergo apoptosis (for a recent review, see reference 35). Thus, a likely possibility is that overexpression of HA-Mad3 for more than 2 DIV leads to programmed cell death, thereby decreasing the total number of HA-Mad3-transfected cells. Indeed, the majority of HA-Mad3+ cells at 4 and 6 DIV were undergoing the final stages of apoptosis as revealed by TUNEL staining (4 DIV, 85.9% ± 9.1%; 6 D IV, 92.9% ± 7.1%). In contrast, few cells overexpressing EGFP at 4 and 6 DIV were TUNEL+ (4 DIV, 13.2% ± 4.7%; 6 DIV, 21.8% ± 1.2%). Importantly, at 2 DIV the percentage of HA-Mad3+ cells that were stained by TUNEL was not statistically different from the percentage of EGFP+ cells that were stained by TUNEL (15.5% ± 2.4% versus 20.6% ± 1.7%; P = 0.086), suggesting that overexpression of HA-Mad3 for up to 2 DIV leads to increased proliferation, while overexpression of HA-Mad3 for more than 2 DIV ultimately leads to programmed cell death. Similar results were obtained with Flag-Nmyc overexpression, suggesting a common mechanism.
Nmyc is a key factor of Shh signaling in GNPs (26, 38); thus, one possibility is that overexpression of Mad3 for 2 DIV may increase proliferation in part by activating Nmyc. To test this possibility, EGFP- or HA-Mad3-transfected cells were costained with anti-Nmyc antibodies. Compared with the percentage of Nmyc+ cells upon EGFP transfection, overexpression of HA-Mad3 in GNPs resulted in a 16-fold increase in Nmyc-expressing cells (2.38% ± 2.3% versus 38.2% ± 1.6%; P = 0.025) (Fig. 4C). Again, this effect was specific for Mad3 because overexpression of Mad1 did not significantly increase the percentage of Nmyc-expressing transfected cells (2.38% ± 2.3% versus 6.0% ± 5.8%; P = 0.089) (Fig. 4C). A modest increase in the percentage of Nmyc+-transfected cells was observed upon overexpression of Flag-Mad4 compared with EGFP-transfected cells (4.78% ± 2.2% versus 13.5% ± 2.13%; P = 0.015), while overexpression of Myc-Mxi1 did not increase the percentage of Nmyc+-transfected cells compared with EGFP-transfected cells (4.78% ± 2.2% versus 5.30% ± 2.0%; P = 0.43). These data suggest that overexpression of Mad3 for 2 DIV is sufficient to induce GNP proliferation and expression of Nmyc in the absence of Shh. Intriguingly, overexpression of Flag-Nmyc increased the percentage of transfected cells that express Mad3 compared with EGFP-transfected cells (4.8% ± 4.6% versus 31.2% ± 1.9%; P = 0.039), suggesting that Mad3 and Nmyc may positively regulate each other, perhaps as a positive feedback mechanism for Shh signal amplification.
All Mad proteins contain two important domains: an N-terminal mSin3-interacting domain (SID) and a C-terminal bHLHZ segment that specifies dimerization with Max through the HLHZ region and DNA binding via the basic region. To determine the domains within Mad3 required for BrdU incorporation and Nmyc expression, we generated three deletion constructs: (i) HA-Mad3-ΔSID removed the SID, (ii) HA-Mad3-ΔHLH removed the Max dimerization domain, and (iii) HA-Mad3-Δbasic removed only the DNA binding domain (Fig. 4D). Deletion of the SID or the HLH domain significantly decreased the proliferation activity of Mad3 (Fig. 4E), suggesting that Max dimerization and recruitment of the mSin3 corepressor are required for its activity. Surprisingly, deletion of the basic region alone (HA-Mad3-Δbasic) did not abolish proliferation activity (Fig. 4E), suggesting that direct binding to DNA is dispensable for Mad3 activity. One possibility is that Mad3/Max heterodimers may bind DNA indirectly by interacting with other DNA binding proteins to exert their effect on transcription and GNP proliferation. We observed similar effects on the percentage of cells that express Nmyc upon overexpression of these Mad3 deletion mutants (Fig. 4F), suggesting that Mad3 direct binding to DNA is not required to increase the percentage of Nmyc+-transfected cells.
Mad3 is required for Shh-dependent GNP proliferation.
To determine if Mad3 is necessary for Shh-mediated GNP proliferation, we constructed shRNA constructs against the mouse Mad3 gene to silence Mad3 expression in GNPs. We modified an existing vector (see Materials and Methods) to allow simultaneous expression of the Mad3-shRNA construct and EGFP to identify transfected cells. First, we tested if this construct (pEGFP-Mad3-shRNA) could inhibit expression of HA-Mad3 in HEK293 cells. Cells were transiently cotransfected with HA-Mad3 and either pEGFP-Mad3-shRNA or a control shRNA construct (pEGFP-Con-shRNA), and protein extracts were analyzed by immunoblot analysis with anti-HA antibodies. The pEGFP-Mad3-shRNA construct could efficiently silence expression of HA-Mad3 in HEK293 cells compared with pEGFP-Con-shRNA (Fig. 5A, upper panel).
FIG. 5.
Mad3 is required for Shh-mediated GNP proliferation. (A) Anti-HA (top) or anti-Flag (bottom) immunoblot of protein extracts from HEK293 cells expressing HA-Mad3 or Flag-Nmyc with pEGFP-Con-shRNA or pEGFP-Mad3-shRNA or with Nmyc-siRNA or control-siRNA, respectively. (B) Granule cells were transfected with pEGFP-Con-shRNA or pEGFP-Mad3-shRNA and cultured for 2 days in the presence or absence of Shh. During the final 12 h of culture, cells were pulsed with BrdU. Cells were fixed and stained with anti-EGFP and anti-BrdU antibodies. (C to E) Graphs depict the percentage of transfected cells (EGFP+) that expressed Mad3 (C), incorporated BrdU (D), or expressed Nmyc (E) in PBS (black)- or Shh (gray)-treated cells upon Mad3 knockdown compared with control shRNA. (F) The graph depicts the percentage of transfected cells (EGFP+) that expressed Mad3 in PBS (black)- or Shh (gray)-treated cells upon Nmyc knockdown compared with a control siRNA. Error bars represent standard errors of the means for three or four wells; each well contained 25 to 50 transfected cells. Similar results were obtained in three independent experiments. Asterisks indicate P values of <0.05 for comparisons indicated with black lines. Bar, 20 μm.
We then proceeded to test the effect of Mad3 knockdown on GNP proliferation in our culture system. At the time of plating, GNPs were transfected with pEGFP-Con-shRNA or pEGFP-Mad3-shRNA in the absence or presence of Shh-N and assayed for Mad3 expression and BrdU incorporation. We confirmed the selective knockdown of Mad3 in GNPs by measuring the percentage of pEGFP-Mad3-shRNA-transfected cells expressing Mad3 compared with pEGFP-Con-shRNA-transfected cells (15.4% ± 3.5% versus 3.83% ± 2.1%; P = 0.03) (Fig. 5C). As expected, GNPs transfected with pEGFP-Con-shRNA were found to be frequently colabeled with anti-BrdU antibodies (Fig. 5B, bottom panels). In contrast, BrdU colabeling was rarely observed in granule cells transfected with pEGFP-Mad3-shRNA (Fig. 5B, top panels). Quantification of this experiment demonstrated that in the presence of Shh-N, transfection of pEGFP-Mad3-shRNA significantly decreased the number of BrdU+ cells compared with pEGFP-Con-shRNA-transfected cells (24.3% ± 1.54% versus 11.7% ± 2.72%; P = 0.012) (Fig. 5D), indicating that Shh-mediated proliferation of GNPs requires Mad3 expression. Interestingly, pEGFP-Con-shRNA-transfected cells displayed morphological properties consistent with proliferating cells (round with no processes), while pEGFP-Mad3-shRNA-transfected cells displayed morphological properties consistent with differentiated neurons (two or three elaborate processes) (Fig. 5B, compare EGFP staining in left panels), suggesting that knockdown of Mad3 not only inhibits GNP proliferation but also allows differentiation in the presence of Shh.
As discussed above, overexpression of HA-Mad3 leads to an increase in the percentage of Nmyc+ cells. Thus, Mad3 knockdown may alter the percentage of transfected cells expressing Nmyc. To test this possibility, similar experiments were carried out to determine the number of transfected cells expressing Nmyc in the presence or absence of Mad3. In the presence of Shh-N, knockdown of Mad3 led to a significant decrease in the percentage of transfected cells expressing Nmyc compared to pEGFP-Con-shRNA-transfected cells (19.4% ± 2.2% versus 9.6% ± 2.7%; P = 0.009) (Fig. 5E). Interestingly, previous studies have demonstrated that Nmyc is a direct target of the Shh signaling pathway (26, 38); thus, these data suggest that Nmyc is not only directly regulated by Shh signaling but perhaps also regulated indirectly by Mad3 and/or other factors regulated by Mad3. Does loss of Nmyc alter the levels of Mad3 upon Shh stimulation? In the presence of Shh-N, knockdown of Nmyc expression using commercially available siRNAs led to a significant decrease in the percentage of Mad3+-transfected cells compared to control siRNA-transfected cells (20.5% ± 2.7% versus 12.3% ± 2.3%; P = 0.044) (Fig. 5F). We confirmed the selective knockdown of Nmyc in GNPs by measuring the percentage of Nmyc-siRNA-transfected cells expressing Nmyc compared with control siRNA-transfected cells (24.4% ± 0.6% versus 8.1% ± 2.9%; P = 0.013). Furthermore, Nmyc-siRNA selectively knocked down Flag-Nmyc expressed in HEK293 cells compared with control siRNA (Fig. 5A, lower panel). Taken together, these results demonstrate that Mad3 (like Nmyc) is necessary for Shh-mediated GNP proliferation in vitro and suggest that Nmyc may regulate expression of Mad3.
Mad3 is upregulated in medulloblastoma and pretumor cells.
Aberrant Shh signaling contributes to medulloblastoma in both mice and humans (53), and Nmyc is upregulated in medulloblastomas derived from ptc+/− mice (26, 38). Thus, we hypothesized that Mad3 may also be upregulated in these tumors. To test this possibility, we analyzed Mad3 mRNA expression in the cerebella of 15-week-old ptc+/− mice (P105) that developed tumors with in situ hybridization. We observed a substantial induction of Mad3 message in tumors, in contrast to adjacent normal cerebellum (Fig. 6A). Consistent with previous studies, Nmyc expression is also specifically upregulated in tumors compared with normal cerebellum (Fig. 6C). No signal was observed in adjacent sections hybridized with sense riboprobes for Mad3 (Fig. 6B) or Nmyc (Fig. 6D). Immunohistochemistry of cerebellar sections from ptc+/− mice that developed tumors confirmed expression of Mad3 protein in cerebellar tumor tissue (Fig. 6E) but not in normal cerebellar tissue from the same animal (Fig. 6F). Interestingly, younger ptc+/− mice contain ectopic undifferentiated granule cells in the persistent EGL thought to represent a preneoplastic stage of cerebellar tumors (39). Thus, we analyzed the expression of Mad3 in ptc+/− mice at 3 weeks of age (P21), before tumors form. Indeed, Mad3 transcripts (Fig. 6G) and protein (Fig. 6K) are expressed in these ectopic EGL cells, and the expression pattern is similar to the Nmyc expression pattern (Fig. 6I), suggesting that Mad3 may be involved in tumorigenesis. These findings further support a novel role for Mad3 in the Shh signaling pathway and suggest that upregulation of Mad3 due to aberrant Shh activation may contribute to tumor formation.
FIG. 6.
Mad3 is upregulated in medulloblastoma and pretumor cells. (A to D and G to J) Sagittal sections of cerebella from 15-week-old ptc+/− mice (P105) that developed tumors (A to D) or from ptc+/− mice at P21 before tumors formed (G to J) were hybridized in situ with DIG-labeled Mad3 (A and G) or Nmyc (C and I) antisense riboprobes. Note that Mad3 and Nmyc transcripts are abundant in ectopic preneoplastic cells (arrows in panels G and I). Sense controls of adjacent sections are shown at right (B, D, H, and J). (E, F, K, and L) Immunohistochemistry of sagittal sections from 15-week-old ptc+/− mice (P105) that developed tumors (E and F) or from ptc+/− mice at P21 before tumors formed (K and L) stained with DAPI (blue) and anti-Mad3 antibodies (green). Mad3 protein is abundant in the tumor tissue (E) and ectopic preneoplastic cells (K); however, staining is absent in normal cerebellar tissue from the same animal (F and L). The area shown in panels F and L is the internal granule layer. Abbreviations: Cb, cerebellum; Mb, medulloblastoma; IGL, internal granule layer; ML, molecular layer. Bars, 200 μm (A to D), 115 μm (G to J), and 30 μm (E, F, K, and L).
DISCUSSION
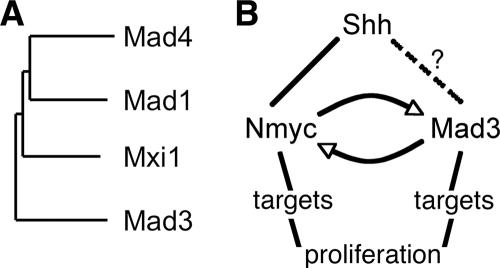
In this study, we have demonstrated a novel role for the Mad family member Mad3 in Shh-mediated GNP proliferation and Nmyc expression. Our study represents the first report, to our knowledge, demonstrating an active role in cellular proliferation for any Mad family member. Mad3 was initially implicated in cerebellar GNP proliferation based on its mRNA expression profile during postnatal cerebellum development (8), and the experiments described here represent important functional validation for one of the candidate genes identified in our previous microarray-based approach. Based on our studies using highly purified cultures of GNPs, we conclude the following: (i) Mad3 is necessary for Shh-mediated GNP proliferation, (ii) overexpression of Mad3 but not any other Mad family member is sufficient to promote GNP proliferation and Nmyc expression in the absence of Shh, (iii) the proliferation activity of Mad3 requires dimerization and repressor function but not direct DNA binding by Mad3; and (iv) overexpression of Mad3 due to aberrant Shh signaling may contribute to cerebellar tumor formation. Interestingly, Mad3 is the most diverse of the mouse Mad family members (Fig. 7A), and the selective role of Mad3 in GNP proliferation underscores the functional differences between Mad family members.
FIG. 7.
Mad3 is an unusual Mad family member that functions in cerebellar GNP proliferation. (A) Dendrogram of Mad family members. Amino acid sequences of the four mouse Mad family members were compared with ClustalW. (B) Model for relationship of Mad3 and Nmyc in Shh-mediated GNP proliferation. Shh signaling leads to upregulation of Nmyc and Mad3. Nmyc is a direct target gene, while it is unclear at present if Mad3 upregulation by Shh is direct or requires a newly synthesized intermediate (indicated by dashed lines). Nmyc and Mad3 can positively upregulate each other; however, we expect that Mad3 and Nmyc bind and regulate distinct target genes that are required for GNP proliferation.
Taken together, our results are surprising in the context of current models for Mad function for several reasons. First, nearly all studies to date have shown that Mad proteins inhibit proliferation, presumably by antagonizing the function of Myc proteins. For example, previous studies have demonstrated that overexpression or targeted disruption of Mad1 leads to suppression or induction, respectively, of Myc-dependent cellular proliferation and growth (12, 24, 47). Furthermore, an established method of blocking Myc-dependent proliferation is by overexpression of Mad family members (2). For instance, Mxi1-SR overexpression antagonized the function of Nmyc in in vitro assays of GNP proliferation (26) similar to those used in this study. Indeed, the original Mad3 cloning and characterization study demonstrated that overexpression of Mad3 (like that of Mad1) is capable of inhibiting Myc-dependent transformation of rat embryo fibroblasts (22), supporting the model that Mad proteins antagonize Myc function in cell growth and proliferation. However, the studies described here support the conclusion that Mad3 promotes proliferation in cultured GNPs and appears to have overlapping activity with Nmyc. The only previous report indicating a functional role of a Mad family member in processes related to proliferation is a recent study suggesting that Xenopus Mxi1 (Xmxi1) is essential for neurogenesis during embryogenesis (27). However, this study demonstrated that Xmxi1 plays an important role in the formation of the mature neural fate, which is necessary for activation by factors that induce neuronal differentiation, and this activity is not directly related to an active role in cell cycle progression.
Second, Mad proteins are thought to antagonize Myc proteins by competition for direct DNA binding to the same E-box sequences (1, 30). Like Myc family members, Mad family members utilize Max as a necessary cofactor for binding to E-box sequences, and the demonstration that Mad/Max heterodimers recruit corepressors to repress transcription at similar Myc DNA binding sites lead to the attractive model that Mad proteins function to antagonize Myc activity via opposing effects on chromatin modifications at shared target genes (19, 46, 57). The opposing functions of Myc and Mad proteins are thought to be mediated by competition for dimerization with Max, because Max dimerization is required for DNA binding and transcriptional regulation (2). Dimerization is controlled by the relative levels of protein expression and by the relative efficiencies of dimerization between different family members. Interestingly, Mad3 appears to form complexes with Max less efficiently than the bHLHZ domain of Myc (bMyc), whereas Mad4 competed more efficiently for dimerization with Max than bMyc (16), suggesting that if Mad3 is less abundant than Nmyc, Mad3 may not compete efficiently with Nmyc for Max dimerization. Our structure-function studies indicate that deletion of SID or the HLH domain significantly decreased the proliferation activity of Mad3, suggesting that Max dimerization and recruitment of the mSin3 corepressor are required for its activity. Thus, one might expect that a relatively high level of Mad3 protein, such as during the transient upregulation of Mad3 in S phase, will be required to compete with Nmyc to form Max heterodimers for DNA binding to promote proliferation. However, and quite unexpectedly, overexpression of Mad3 with a deletion of the basic region did not abolish proliferation activity, suggesting that Mad3 direct binding to DNA is not required for activity. One possibility is that Mad3Δbasic/Max heterodimers are still able to bind DNA, albeit presumably with reduced affinity, and thus are sufficient to support activity. However, previous studies reported that Mad3 lacking the basic region as well as the SID dimerizes efficiently with Max, while deletion of the Max basic region abolished Mad3/Max-mediated repression via a canonical E-box sequence (22). Thus, an intriguing alternative explanation is that Mad3 may dimerize with cofactors other than Max to exert its effect on transcription. For example, Mad1 and Mad4 can dimerize with the bHLHZ protein Mlx, although Mad/Mlt heterodimers appear to function similarly to Mad/Max heterodimers in terms of DNA binding specificity (3, 34). Another example is that Myc/Max repression of p21Cip1 and p15Ink5b promoters is thought to be mediated via binding to the zinc finger DNA binding protein Miz1 (49, 56). Thus, it is possible that Mad3 binds DNA indirectly via a Miz1-like protein. Identification of Mad3-specific interacting proteins should begin to address this possibility.
Third, the relationship between Mad3 and Nmyc suggests that the two molecules may function by a positive feedback mechanism, an unprecedented mechanism for the Myc/Max/Mad family. Overexpression of Mad3, but not any other Mad family member, is sufficient to increase significantly the percentage of cells expressing Nmyc in the absence of Shh, while knockdown of Mad3 leads to a significant decrease in the percentage of cells expressing Nmyc in the presence of Shh. These data suggest that Mad3 may positively regulate Nmyc expression. Inspection of the Nmyc promoter revealed two E-box sequences at −1660 and −3475 from the start of transcription. However, our structure-function analysis suggests that direct DNA binding of Mad3 is not required for Mad3-dependent Nmyc expression; thus, it is quite possible that Mad3 binds DNA indirectly at sites distinct from E-box sequences. Furthermore, the Sin3-interacting domain is required for Mad3-dependent Nmyc expression, supporting the expectation that Mad3 functions to repress transcription like other Mad family members and therefore would not be expected to be associated with the Nmyc promoter. One possibility is that Mad3 represses transcription of an as-yet-unidentified Nmyc repressor to exert its upregulation of Nmyc. Conversely, overexpression of Nmyc increases the percentage of cells expressing Mad3 in the absence of Shh, and loss of Nmyc decreases significantly the percentage of cells expressing Mad3 in the presence of Shh. Inspection of the Mad3 promoter revealed two consensus E-box sequences at positions −277 and −947 from the start of transcription, suggesting that Nmyc may bind and regulate transcription from the Mad3 promoter.
Fourth, the Myc antagonist model for Mad function is attractive because most Mad family members are expressed preferentially, although not exclusively, in differentiating cells in vitro and in vivo (2, 21, 22, 42, 52). In contrast, Myc expression is associated with proliferating cells and is rapidly upregulated upon mitogenic stimulation of quiescent cells (25, 44). However, unlike other Mad family members, Mad3 transcripts and proteins were detected in proliferating cells prior to differentiation (42). Furthermore, Mad3 expression is enriched selectively in S phase of proliferating cells (13, 41), suggesting that Mad3 regulates transcription in a cell cycle-dependent manner. The overlap in expression of Mad3 and Nmyc during the cell cycle of cerebellar GNPs suggests that Mad3 may impart S-phase-specific regulation of Nmyc target genes. However, this prediction is based on the assumption that Mad3 antagonizes Nmyc by competition for DNA binding to similar sites, and our data do not support such a model. Indeed, both Mad3 and Nmyc are required for GNP proliferation, as knockdown of Mad3 (this study) or interfering with Nmyc function (26, 38) inhibits BrdU incorporation in cultured GNPs. Thus, it appears that in cultured GNPs both Mad3 and Nmyc are required to promote GNP proliferation.
One might predict that Mad3 and Nmyc have the same function; however, the available in vivo evidence does not support this possibility. While mice with a targeted deletion of Nmyc in neural progenitor cells in vivo have a smaller and disorganized cerebellum with a reduced cell density in the internal granule layer (28), the role of Mad3 in postnatal cerebellum development in vivo is unknown. Mice with a targeted deletion of mad3 have been generated and are viable (43), similar to targeted deletions of either mad1 or mxi1 (12, 48). However, contrary to the expectation of other Mad family members, the effect of mad3 deletion was not associated with defects in cell cycle exit during embryogenesis like targeted deletions of mad1 or mxi1 (11, 12, 48). Our studies predict that GNP proliferation, and therefore the total numbers of GNPs, in the mad3−/− postnatal cerebellum will be reduced. As the effect of mad3 loss upon cerebellar granule cell development was not reported (43), future studies analyzing these mice will be necessary to test our prediction. The lack of an obvious phenotype in mad3-null mice (43) could be due to functional redundancy among Myc/Max/Mad family members or other compensatory mechanisms due to the chronic loss of Mad3. Because of the fact that no changes in the expression level of Mad family members were reported, coupled with our observation that other Mad family members besides Mad3 do not display proliferation activity in our culture system, we favor the interesting possibility that Mad3 function is redundant with or compensated by proteins other than those of the Mad family. For instance, synthetic effects are observed between mad1-null and p27Kip1-null mice, suggesting that the proteins encoded by these two genes may function in parallel to influence differentiation (33). In light of our studies supporting a role for Mad3 in proliferation, it is conceivable that Mad3 function is redundant with that of proteins involved in cell cycle checkpoint regulation such as Cyclin D2. Indeed, the S-phase-restricted expression of Mad3 in cultured fibroblast cells appears to be regulated in part by E2F1 (14), and E2F transcription factors are key regulators of S-phase entry (9). Interestingly, cyclin D2−/− mice are viable and exhibit only a modest reduction in proliferating GNPs in the cerebellum (20). Furthermore, the reduced cell number in cyclin D2−/− mice is associated with an increase in apoptosis in the EGL (20). Intriguingly, mad3-null mice exhibit increased sensitivity to radiation-induced apoptosis (43), and Myc sensitizes cells to apoptosis (37, 40). Thus, an interesting possibility that was recently put forth is that cells in S phase are particularly vulnerable to Myc-induced apoptosis, and expression of Mad3 during this period might orchestrate the selective downregulation of apoptosis-related target genes and render the cells less sensitive to Myc-dependent apoptosis (19).
Our model for the role of Mad3 in the Shh pathway and its relationship to Nmyc is shown in Fig. 7B. During postnatal cerebellum development, Shh signaling leads to expression of Mad3 and Nmyc. Nmyc is known to be directly upregulated by Shh, presumably via the Gli family of transcription factors; however, at present, it is unclear if the same is true for Mad3. One possibility is that Mad3 is a downstream target gene of Nmyc. Indeed, our studies demonstrating that knockdown of Nmyc in the presence of Shh decreases the percentage of cells expressing Mad3 support this model; however, the numbers of Shh-induced Mad3+ cells in the absence of Nmyc are still significantly higher than those of EGFP-transfected cells. We expect that Mad3 regulates expression of specific genes during S-phase progression to promote GNP proliferation. However, to date no transcriptional targets of Mad3 are known, and a small number of verified targets for Nmyc have thus far been identified. We define verification of Nmyc target genes as demonstration of gene promoters bound directly by Nmyc and transcriptional level alterations due to overexpression or loss of Nmyc. However, there are many studies identifying cMyc target genes (for a review, see reference 31), and most likely Nmyc and cMyc will overlap substantially in their target gene regulation. While regulation of specific genes is the general accepted mechanism for the Myc/Max/Mad family, a recent study has shown that Nmyc is required for the widespread maintenance of active chromatin in cerebellar GNPs (29). Because our studies suggest that Mad3 and Nmyc function similarly, it is possible that Mad3 may also function to regulate global chromatin structure. Future studies incorporating genomic approaches such as chromatin immunoprecipitation coupled with DNA microarrays (“ChIP-on-chip”) will allow us to directly address these questions.
Acknowledgments
We thank J. Trimmer for helpful comments on the manuscript and mouse anti-HA antibodies, P. Farnham and P. Knoepfler for suggestions and advice throughout the project, E. Kalashnikova for generation of the modified pBud vector, D. Schaffer for the Shh-N-expressing plasmid, and L. Vasquez for purification of Shh-N protein. We thank members of the Diaz lab, particularly G. Barisone and B. Hayes, for suggestions and assistance throughout the project. We thank the Chen, Farnham, Gelli, Trimmer, and Wulff labs for sharing equipment and reagents.
This work was supported by grants to E.D. from the Alfred P. Sloan Research Foundation, the James S. McDonnell Foundation 21st Century Award Program, and the UC Davis Health System Research Award Program and by an individual allocation of the UC Davis Institutional Research Grant from the American Cancer Society.
Footnotes
Published ahead of print on 24 September 2007.
REFERENCES
- 1.Amati, B., S. Dalton, M. W. Brooks, T. D. Littlewood, G. I. Evan, and H. Land. 1992. Transcriptional activation by the human c-Myc oncoprotein in yeast requires interaction with Max. Nature 359:423-426. [DOI] [PubMed] [Google Scholar]
- 2.Ayer, D. E., L. Kretzner, and R. N. Eisenman. 1993. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell 72:211-222. [DOI] [PubMed] [Google Scholar]
- 3.Billin, A. N., A. L. Eilers, C. Queva, and D. E. Ayer. 1999. Mlx, a novel Max-like BHLHZip protein that interacts with the Max network of transcription factors. J. Biol. Chem. 274:36344-36350. [DOI] [PubMed] [Google Scholar]
- 4.Ciemerych, M. A., A. M. Kenney, E. Sicinska, I. Kalaszczynska, R. T. Bronson, D. H. Rowitch, H. Gardner, and P. Sicinski. 2002. Development of mice expressing a single D-type cyclin. Genes Dev. 16:3277-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corrales, J. D., G. L. Rocco, S. Blaess, Q. Guo, and A. L. Joyner. 2004. Spatial pattern of sonic hedgehog signaling through Gli genes during cerebellum development. Development 131:5581-5590. [DOI] [PubMed] [Google Scholar]
- 6.Dahmane, N., and A. Ruiz-i-Altaba. 1999. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development 126:3089-3100. [DOI] [PubMed] [Google Scholar]
- 7.DeCamp, D. L., T. M. Thompson, F. J. de Sauvage, and M. R. Lerner. 2000. Smoothened activates Galphai-mediated signaling in frog melanophores. J. Biol. Chem. 275:26322-26327. [DOI] [PubMed] [Google Scholar]
- 8.Diaz, E., Y. Ge, Y. H. Yang, K. C. Loh, T. A. Serafini, Y. Okazaki, Y. Hayashizaki, T. P. Speed, J. Ngai, and P. Scheiffele. 2002. Molecular analysis of gene expression in the developing pontocerebellar projection system. Neuron 36:417-434. [DOI] [PubMed] [Google Scholar]
- 9.Dimova, D. K., and N. J. Dyson. 2005. The E2F transcriptional network: old acquaintances with new faces. Oncogene 24:2810-2826. [DOI] [PubMed] [Google Scholar]
- 10.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foley, K. P., and R. N. Eisenman. 1999. Two MAD tails: what the recent knockouts of Mad1 and Mxi1 tell us about the MYC/MAX/MAD network. Biochim. Biophys. Acta 1423:M37-47. [DOI] [PubMed] [Google Scholar]
- 12.Foley, K. P., G. A. McArthur, C. Queva, P. J. Hurlin, P. Soriano, and R. N. Eisenman. 1998. Targeted disruption of the MYC antagonist MAD1 inhibits cell cycle exit during granulocyte differentiation. EMBO J. 17:774-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox, E. J., and S. C. Wright. 2001. S-phase-specific expression of the Mad3 gene in proliferating and differentiating cells. Biochem. J. 359:361-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fox, E. J., and S. C. Wright. 2003. The transcriptional repressor gene Mad3 is a novel target for regulation by E2F1. Biochem. J. 370:307-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grandori, C., S. M. Cowley, L. P. James, and R. N. Eisenman. 2000. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 16:653-699. [DOI] [PubMed] [Google Scholar]
- 16.Grinberg, A., C.-D. Hu, and T. K. Kerppola. 2004. Visualization of Myc/Max/Mad family dimers and the competition for dimerization in living cells. Mol. Cell. Biol. 24:4294-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hatten, M. E., J. Alder, K. Zimmerman, and N. Heintz. 1997. Genes involved in cerebellar cell specification and differentiation. Curr. Opin. Neurobiol. 7:40-47. [DOI] [PubMed] [Google Scholar]
- 18.Hatten, M. E., W. O. Gao, M. E. Morrison, and C. A. Mason. 1998. The cerebellum, p. 419-459. In G. Banker and K. Goslin (ed.), Culturing nerve cells, 2nd ed. MIT Press, Cambridge, MA.
- 19.Hooker, C. W., and P. J. Hurlin. 2006. Of Myc and Mnt. J. Cell Sci. 119:208-216. [DOI] [PubMed] [Google Scholar]
- 20.Huard, J. M., C. C. Forster, M. L. Carter, P. Sicinski, and M. E. Ross. 1999. Cerebellar histogenesis is disturbed in mice lacking cyclin D2. Development 126:1927-1935. [DOI] [PubMed] [Google Scholar]
- 21.Hurlin, P. J., K. P. Foley, D. E. Ayer, R. N. Eisenman, D. Hanahan, and J. M. Arbeit. 1995. Regulation of Myc and Mad during epidermal differentiation and HPV-associated tumorigenesis. Oncogene 11:2487-2501. [PubMed] [Google Scholar]
- 22.Hurlin, P. J., C. Queva, P. J. Koskinen, E. Steingrimsson, D. E. Ayer, N. G. Copeland, N. A. Jenkins, and R. N. Eisenman. 1995. Mad3 and Mad4: novel Max-interacting transcriptional repressors that suppress c-myc dependent transformation and are expressed during neural and epidermal differentiation. EMBO J. 14:5646-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ingham, P. W., and A. P. McMahon. 2001. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 15:3059-3087. [DOI] [PubMed] [Google Scholar]
- 24.Iritani, B. M., J. Delrow, C. Grandori, I. Gomez, M. Klacking, L. S. Carlos, and R. N. Eisenman. 2002. Modulation of T-lymphocyte development, growth and cell size by the Myc antagonist and transcriptional repressor Mad1. EMBO J. 21:4820-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelly, K., B. H. Cochran, C. D. Stiles, and P. Leder. 1983. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell 35:603-610. [DOI] [PubMed] [Google Scholar]
- 26.Kenney, A. M., M. D. Cole, and D. H. Rowitch. 2003. Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development 130:15-28. [DOI] [PubMed] [Google Scholar]
- 27.Klisch, T. J., J. Souopgui, K. Juergens, B. Rust, T. Pieler, and K. A. Henningfeld. 2006. Mxi1 is essential for neurogenesis in Xenopus and acts by bridging the pan-neural and proneural genes. Dev. Biol. 292:470-485. [DOI] [PubMed] [Google Scholar]
- 28.Knoepfler, P. S., P. F. Cheng, and R. N. Eisenman. 2002. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 16:2699-2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knoepfler, P. S., X. Y. Zhang, P. F. Cheng, P. R. Gafken, S. B. McMahon, and R. N. Eisenman. 2006. Myc influences global chromatin structure. EMBO J. 25:2723-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kretzner, L., E. M. Blackwood, and R. N. Eisenman. 1992. Myc and Max proteins possess distinct transcriptional activities. Nature 359:426-429. [DOI] [PubMed] [Google Scholar]
- 31.Lee, L. A., and C. V. Dang. 2006. Myc target transcriptomes. Curr. Top. Microbiol. Immunol. 302:145-167. [DOI] [PubMed] [Google Scholar]
- 32.Manzini, M. C., M. S. Ward, Q. Zhang, M. D. Lieberman, and C. A. Mason. 2006. The stop signal revised: immature cerebellar granule neurons in the external germinal layer arrest pontine mossy fiber growth. J. Neurosci. 26:6040-6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McArthur, G. A., C. D. Laherty, C. Queva, P. J. Hurlin, L. Loo, L. James, C. Grandori, P. Gallant, Y. Shiio, W. C. Hokanson, A. C. Bush, P. F. Cheng, Q. A. Lawrence, B. Pulverer, P. J. Koskinen, K. P. Foley, D. E. Ayer, and R. N. Eisenman. 1998. The Mad protein family links transcriptional repression to cell differentiation. Cold Spring Harb. Symp. Quant. Biol. 63:423-433. [DOI] [PubMed] [Google Scholar]
- 34.Meroni, G., S. Cairo, G. Merla, S. Messali, R. Brent, A. Ballabio, and A. Reymond. 2000. Mlx, a new Max-like bHLHZip family member: the center stage of a novel transcription factors regulatory pathway? Oncogene 19:3266-3277. [DOI] [PubMed] [Google Scholar]
- 35.Meyer, N., S. S. Kim, and L. Z. Penn. 2006. The Oscar-worthy role of Myc in apoptosis. Semin. Cancer Biol. 16:275-287. [DOI] [PubMed] [Google Scholar]
- 36.Migheli, A., R. Piva, S. Casolino, C. Atzori, S. R. Dlouhy, and B. Ghetti. 1999. A cell cycle alteration precedes apoptosis of granule cell precursors in the weaver mouse cerebellum. Am. J. Pathol. 155:365-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nilsson, J. A., and J. L. Cleveland. 2003. Myc pathways provoking cell suicide and cancer. Oncogene 22:9007-9021. [DOI] [PubMed] [Google Scholar]
- 38.Oliver, T. G., L. L. Grasfeder, A. L. Carroll, C. Kaiser, C. L. Gillingham, S. M. Lin, R. Wickramasinghe, M. P. Scott, and R. J. Wechsler-Reya. 2003. Transcriptional profiling of the Sonic hedgehog response: a critical role for N-myc in proliferation of neuronal precursors. Proc. Natl. Acad. Sci. USA 100:7331-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oliver, T. G., T. A. Read, J. D. Kessler, A. Mehmeti, J. F. Wells, T. T. Huynh, S. M. Lin, and R. J. Wechsler-Reya. 2005. Loss of patched and disruption of granule cell development in a pre-neoplastic stage of medulloblastoma. Development 132:2425-2439. [DOI] [PubMed] [Google Scholar]
- 40.Pelengaris, S., M. Khan, and G. Evan. 2002. c-MYC: more than just a matter of life and death. Nat. Rev. Cancer 2:764-776. [DOI] [PubMed] [Google Scholar]
- 41.Pulverer, B., A. Sommer, G. A. McArthur, R. N. Eisenman, and B. Luscher. 2000. Analysis of Myc/Max/Mad network members in adipogenesis: inhibition of the proliferative burst and differentiation by ectopically expressed Mad1. J. Cell Physiol. 183:399-410. [DOI] [PubMed] [Google Scholar]
- 42.Queva, C., P. J. Hurlin, K. P. Foley, and R. N. Eisenman. 1998. Sequential expression of the MAD family of transcriptional repressors during differentiation and development. Oncogene 16:967-977. [DOI] [PubMed] [Google Scholar]
- 43.Queva, C., G. A. McArthur, B. M. Iritani, and R. N. Eisenman. 2001. Targeted deletion of the S-phase-specific Myc antagonist Mad3 sensitizes neuronal and lymphoid cells to radiation-induced apoptosis. Mol. Cell. Biol. 21:703-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rabbitts, P. H., J. V. Watson, A. Lamond, A. Forster, M. A. Stinson, G. Evan, W. Fischer, E. Atherton, R. Sheppard, and T. H. Rabbitts. 1985. Metabolism of c-myc gene products: c-myc mRNA and protein expression in the cell cycle. EMBO J. 4:2009-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ross, M. E., M. L. Carter, and J. H. Lee. 1996. MN20, a D2 cyclin, is transiently expressed in selected neural populations during embryogenesis. J. Neurosci. 16:210-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rottmann, S., and B. Luscher. 2006. The Mad side of the Max network: antagonizing the function of Myc and more. Curr. Top. Microbiol. Immunol. 302:63-122. [DOI] [PubMed] [Google Scholar]
- 47.Rudolph, B., A. O. Hueber, and G. I. Evan. 2001. Expression of Mad1 in T cells leads to reduced thymic cellularity and impaired mitogen-induced proliferation. Oncogene 20:1164-1175. [DOI] [PubMed] [Google Scholar]
- 48.Schreiber-Agus, N., Y. Meng, T. Hoang, H. Hou, Jr., K. Chen, R. Greenberg, C. Cordon-Cardo, H. W. Lee, and R. A. DePinho. 1998. Role of Mxi1 in ageing organ systems and the regulation of normal and neoplastic growth. Nature 393:483-487. [DOI] [PubMed] [Google Scholar]
- 49.Seoane, J., C. Pouponnot, P. Staller, M. Schader, M. Eilers, and J. Massague. 2001. TGFbeta influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat. Cell Biol. 3:400-408. [DOI] [PubMed] [Google Scholar]
- 50.Taipale, J., J. K. Chen, M. K. Cooper, B. Wang, R. K. Mann, L. Milenkovic, M. P. Scott, and P. A. Beachy. 2000. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature 406:1005-1009. [DOI] [PubMed] [Google Scholar]
- 51.van den Heuvel, M., and P. W. Ingham. 1996. smoothened encodes a receptor-like serpentine protein required for hedgehog signalling. Nature 382:547-551. [DOI] [PubMed] [Google Scholar]
- 52.Vastrik, I., A. Kaipainen, T. L. Penttila, A. Lymboussakis, R. Alitalo, M. Parvinen, and K. Alitalo. 1995. Expression of the mad gene during cell differentiation in vivo and its inhibition of cell growth in vitro. J. Cell Biol. 128:1197-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wechsler-Reya, R., and M. P. Scott. 2001. The developmental biology of brain tumors. Annu. Rev. Neurosci. 24:385-428. [DOI] [PubMed] [Google Scholar]
- 54.Wechsler-Reya, R. J., and M. P. Scott. 1999. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron 22:103-114. [DOI] [PubMed] [Google Scholar]
- 55.Wernecke, H., J. Lindner, and M. Schachner. 1985. Cell type specificity and developmental expression of the L2/HNK-1 epitopes in mouse cerebellum. J. Neuroimmunol. 9:115-130. [DOI] [PubMed] [Google Scholar]
- 56.Wu, S., C. Cetinkaya, M. J. Munoz-Alonso, N. von der Lehr, F. Bahram, V. Beuger, M. Eilers, J. Leon, and L. G. Larsson. 2003. Myc represses differentiation-induced p21CIP1 expression via Miz-1-dependent interaction with the p21 core promoter. Oncogene 22:351-360. [DOI] [PubMed] [Google Scholar]
- 57.Zhou, Z. Q., and P. J. Hurlin. 2001. The interplay between Mad and Myc in proliferation and differentiation. Trends Cell Biol. 11:S10-S14. [DOI] [PubMed] [Google Scholar]