Abstract
Bile acids (BAs) are water-soluble end products from cholesterol metabolism and are essential for efficient absorption of dietary lipids. By using targeted somatic mutagenesis of the nuclear receptor liver receptor homolog 1 (LRH-1) in mouse hepatocytes, we demonstrate here that LRH-1 critically regulates the physicochemical properties of BAs. The absence of LRH-1 and subsequent deficiency of Cyp8b1 eliminate the production of cholic acid and its amino acid conjugate taurocholic acid and increase the relative amounts of less amphipathic BA species. Intriguingly, while the expression of Cyp8b1 is almost extinguished in the livers of mice that lack LRH-1, the expression of the rate-limiting enzyme of BA synthesis, i.e., Cyp7a1, remains unchanged. The profound remodeling of the BA composition significantly reduces the efficacy of intestinal absorption of lipids and reuptake of BAs and facilitates the removal of lipids from the body. Our studies unequivocally demonstrate a pivotal role for LRH-1 in determining the composition of BAs, which, in turn has major consequences on whole-body lipid homeostasis.
Liver receptor homolog 1 (LRH-1 [NR5A2]) constitutes, together with steroidogenic factor 1 ([NR5A1]), the NR5A or Ftz-F1 subfamily of nuclear receptors. All members of this subfamily bind DNA as monomers, and to a large extent, the specificity of DNA recognition is dictated by the Ftz-F1 box, a unique domain at the C terminus of the DNA binding domain specific for the NR5A family (11). For a long time, LRH-1 was considered to be an orphan nuclear receptor, but recently, phospholipids were shown to bind human (23, 32, 44), but not mouse LRH-1 (23, 39). LRH-1 maintains embryonic stem cell pluripotency (17) and later in development contributes to the specification of the enterohepatic axis (1, 34, 36). In adult mammals, LRH-1 is expressed predominantly in ovaries (2), the liver (15), the exocrine pancreas (12), and the basal compartment of the gastrointestinal tract (4). In the intestine, LRH-1 facilitates cell renewal in synergy with β-catenin/Tcf4 signaling (4), a phenomenon which, under pathophysiological conditions, contributes to intestinal tumorigenesis (41). Recently, LRH-1 has been identified as a critical factor in the regulation of local glucocorticoid production in the gut, suggesting a role for LRH-1 in intestinal immune homeostasis (8, 30).
In addition to these functions, LRH-1 has been proposed to control complex metabolic pathways that govern hepatic and intestinal sterol homeostasis (11). Several genes involved in high-density lipoprotein metabolism and reverse cholesterol transport, including those encoding cholesterol ester transfer protein (28), apolipoprotein AI (10), and scavenger receptor BI, have been identified as targets for LRH-1 (40). In addition, LRH-1 has been proposed as a master regulator of genes implicated in the synthesis and transport of bile acids (BAs). In transient transfection assays, LRH-1 was shown to drive the expression of both cholesterol 7α-hydroxylase (CYP7A1), the rate-limiting enzyme of the BA biosynthesis pathway (31), and sterol 12α-hydroxylase (CYP8B1), which determines the synthesis of cholic acid (CA) (9). BA homeostasis is tightly regulated by a feedback mechanism involving the consecutive action of a number of nuclear receptors. LRH-1-mediated induction of BA synthesis provides ligands for the BA-activated farnesoid X receptor (FXR), which upon activation induces the expression of the short heterodimer partner (SHP) (16, 27). Apart from being a target gene of LRH-1 (25), SHP is a potent repressor of LRH-1 activity (24) and the induction of SHP by BAs represses LRH-1-mediated CYP7A1 gene expression (16, 27). Finally, LRH-1 has been shown to coordinately activate the gene expression of the ATP-binding cassette G5 and G8 proteins (ABCG5/8) (14), two half transporters involved in the biliary secretion of sterols, and to promote the ileal transport of BAs via the regulation of the apical sodium-dependent BA transporter (ASBT) (5) and basolateral multidrug resistance protein 3 (MRP3) (20).
Mice with homozygous germ line mutations in the LRH-1 gene are not viable (4, 17, 33). We report here for the first time the generation of a mouse model in which the LRH-1 gene is deleted in hepatocytes. By using these hepatocyte-specific LRH-1-deficient mice, we unequivocally demonstrate that Cyp7a1 gene expression, unlike what was previously thought, is not dependent on the presence of hepatic LRH-1. In contrast, the expression of Cyp8b1, which determines the physicochemical properties of BAs, is critically dependent on the presence of hepatic LRH-1, hence eliminating the production of CA and its amino acid conjugate taurocholic acid (TCA) and increasing the relative amounts of less amphipathic BA species. As a consequence, hepatocyte-specific LRH-1-deficient mice show severely altered BA composition, resulting in chronically reduced intestinal absorption of cholesterol and free fatty acids.
MATERIALS AND METHODS
Generation of LRH-1 mutant mice.
For the generation of LRH-1 floxed (LRH-1L2/L2) mice, genomic DNA covering the LRH-1 locus was amplified from the 129Sv strain by using high-fidelity PCR. The resulting DNA fragments were assembled into the targeting vector that, after linearization by NotI, was electroporated into 129Sv embryonic stem (ES) cells. G418-resistant colonies were selected and analyzed for homologous recombination by PCR and Southern blot hybridization. For the PCR screening strategy, primers ACE225 (5′GTCATAGGGAGTCAGGATACCATGG3′), ACE228 (5′GTTCTGACCACTTTCATCTCCTCACG3′), ACE229 (5′CTCAACTGCCGAAGAATGCTGCGG3′), and ACE231 (5′GTTAGCAATTTGGCAGATTTACGC3′) were used. Positive clones were verified by Southern blot hybridization. Therefore, genomic DNA was prepared from ES cells (or mouse tails), digested with XbaI or SacI, subjected to electrophoresis on a 0.8% agarose gel, and transferred to a positively charged nylon transfer membrane (Amersham Biosciences, Saclay, France). A 0.5-kb DNA fragment (NotI-NheI) located between exons 6 and 7 (3′ probe) and a 0.5-kb DNA fragment (NotI-SacII) placed between exons 2 and 3 (5′ probe) were used as probes. The karyotype was verified, and several correctly targeted ES cell clones were injected into blastocysts from C57BL/6J mice. These blastocysts were transferred into pseudopregnant females, resulting in chimeric offspring that were mated to female C57BL/6J mice that express the Flp recombinase under the control of the ubiquitous cytomegalovirus promoter (37). Offspring that transmitted the mutated allele, in which the selection marker was excised, and that lost the Flp transgene (LRH-1+/L2 mice) were selected, mated with serum albumin (SA)-Cre-ERT2 mice (42), and then further intercrossed to generate premutant SA-Cre-ERT2/LRH-1L2/L2 mice. The recombination of floxed alleles was induced by four consecutive intraperitoneal injections of 4-hydroxy-tamoxifen (1 mg; Sigma). pdLucFXR-LRH-1hep−/− reporter mice were generated by crossing LRH-1L2/L2 mice with previously described pdLucFXR reporter mice (19). The excision of LRH-1 in the liver was achieved by subsequently crossing pdLucFXR-LRH-1L2/L2 transgenic mice with Alb-Cre transgenic mice (35).
Animal procedures and biochemical measurements.
Mice were maintained on a 12-h light/12-h dark cycle. Animals had access to water and were fed a standard chow (D03; SAFE, Augy, France). For certain studies, mice were placed individually in metabolic cages so we could measure food intake and collect urine and feces. To assess the involvement of LRH-1 in mediating the transcriptional activity of liver X receptor (LXR), mice were treated with tamoxifen and 2 weeks later were either fed with a high-cholesterol (2%) diet or treated with T0901317 (Cayman; 50 mg/kg of body weight in 1% methylcellulose) or vehicle by oral gavage for 7 days. For the BA supplementation study, CA (Sigma; 0.2% wt/wt) was added to the chow diet for 2 days and was initiated 2 weeks after tamoxifen-induced recombination. All mice were killed approximately 3 weeks after the first tamoxifen injection, following a 3-h fast initiated at 6 a.m. Plasma clinical chemistry analysis was performed using an AU-400 automated laboratory workstation and commercial reagents (Olympus France SA, Rungis, France). Plasma insulin was determined by enzyme-linked immunosorbent assay (Mercodia AB, Uppsala, Sweden).
Imaging.
In vivo visualization of luciferase expression was performed during the dark phase (at 2:00 a.m.) as described previously (19). Just before imaging, mice were anesthetized by using 8 mg/kg Xylazine and 50 mg/kg ketamine.
Quantitative real-time PCR.
RNA from liver or ileum was isolated using the TRIzol method (Invitrogen, Carlsbad, CA). cDNAs were synthesized from total RNA with SuperScript II reverse transcriptase (Invitrogen) and random hexamer primers (Roche, Basel, Switzerland). The real-time PCR measurement of individual cDNAs was performed using SYBR green dye (QIAGEN, Courtaboeuf, France) to measure duplex DNA formation with the Roche LightCycler. Sequences of primers used for amplification are available at www-igbmc.u-strasbg.fr/recherche/Dep_GPSN/Eq_JAuwe/Publi/Paper.html.
Antibodies and Western blot analysis.
The rabbit polyclonal LRH-1 and mouse polyclonal SHP antibodies used in this study have been described previously (8). The anti-CYP8B1 antibody was purchased from Santa Cruz Biotechnology, Inc. (catalog no. sc-23515; Santa Cruz, CA). For immunoblotting, liver nuclear (50 μg) or whole-cell (80 μg) lysates were transferred to a nitrocellulose membrane. The membrane was incubated overnight at 4°C with the antibodies described above and then for 1 h at room temperature with a peroxidase-conjugated secondary antibody (Jackson ImmunoResearch, West Grove, PA). Proteins were visualized with SuperSignal West Pico chemiluminescence substrate (Pierce).
BA composition, BA pool size, and lipid measurements.
Measurement of BA pool size and composition was performed as follows. Livers/gallbladders and small intestines were cut in small pieces and boiled for 5 min in ethanol. Feces were freeze-dried, ground with a mortar and pestle, weighed, and boiled for 5 min in ethanol. Norcholic acid (Steraloids, Inc., Newport, RI; 1 mg/ml stock solution in 50% ethanol) was added to the ethanol as a recovery standard. The extract was filtered and adjusted to a specific volume by using volumetric flasks. Part of the extract was centrifuged at 12,000 × g for 5 min and subsequently diluted 10 times in water. This dilution was used for quantitative analyses by high-pressure liquid chromatography-tandem mass spectrometry using a method described before with minor modifications (50- by 1-mm C8 column, buffered mobile phase) (3). Plasma BAs were analyzed in deproteinized samples. Qualitative measurement of the BAs in livers/gallbladders, intestines, and feces extracts was performed by high-pressure liquid chromatography-mass spectrometry: the sample was injected onto a C18-guard column (20 by 2 mm) and washed with water, and subsequently, the BAs and BA conjugates were eluted with acetonitrile in a single peak. During the elution of this peak, spectra were taken.
Hepatic and fecal lipid content.
Fecal and liver lipids were extracted essentially by using the method of Folch et al. (13) and were quantified by using enzymatic assays. Briefly, for the analysis of fecal lipids, feces were collected from mice housed individually in metabolic cages over a 24-h period. One-hundred-milligram aliquots of feces were cleaned and dried for 1 h at 70°C, incubated with 2 ml of chloroform-methanol (2:1) for 30 min at 60°C with constant agitation, and then centrifuged. Water (1 ml) was added to the supernatant, and following vortexing, phase separation was induced by low-speed centrifugation (2,000 rpm for 10 min). The lower chloroform phase was then removed and transferred to a new tube, and the sample was evaporated to dryness. Samples were then resuspended in 500 μl chloroform-1% Triton X-100, evaporated to dryness, and finally resuspended in 500 μl of water, so that the final solvent was 1% Triton X-100 in water. For analysis of liver lipids, 100-mg aliquots of tissue were homogenized with 2 ml chloroform-methanol and then agitated overnight on an orbital shaker at 4°C. The homogenate was then centrifuged (5 min at 5,000 rpm), 0.9% NaCl solution was added to the liquid phase, and the samples were vortexed. Phase separation was induced by centrifugation (2,000 rpm for 10 min), and the bottom phase was removed to a new tube and then processed as above, so that the final solvent was 1% Triton X-100 in water. The quantities of total and free cholesterol (Wako, Neuss, Germany), triglycerides (BioMerieux, Marcy l'Etoile, France), free fatty acids (Wako), and phospholipids (Wako) in the fecal and liver lipid extracts were then assayed by using enzymatic kits according to the manufacturers' protocols, with the exception that the corresponding standard solutions or calibrators were diluted with 1% Triton X-100 in water. Cholesterol ester values were determined by subtracting the free cholesterol measurement from the total cholesterol value.
Histological analysis.
Hematoxylin-eosin staining was performed on 5-μm sections from paraffin-embedded mouse ilea. Oil Red O staining for lipid detection was performed on 10-μm frozen sections.
Statistical analysis.
Data represent the means ± standard errors of the means. Statistical differences between groups were determined by Student's t test and indicated when statistical significance was reached.
RESULTS
Liver function is preserved in mice with a deficiency of LRH-1 in the hepatocytes.
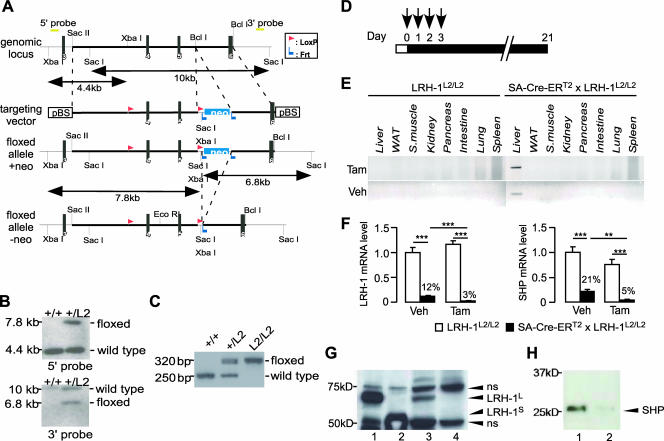
To enable the generation of temporally and spatially controlled mouse mutants for the LRH-1 gene in hepatocytes, mice carrying the floxed LRH-1 L2 alleles (LRH-1L2/L2 mice) were generated (Fig. 1A to C) and crossed with transgenic mice that express the Cre-ERT2 recombinase under the control of the mouse SA promoter (42). SA-Cre-ERT2 × LRH-1L2/L2 “bigenic” LRH-1 mice were then injected with tamoxifen or vehicle to induce Cre-mediated recombination of the floxed LRH-1 alleles in hepatocytes (Fig. 1D). Three weeks after the first tamoxifen injection, effective and selective recombination of the LRH-1 locus in liver could be detected as illustrated by PCR analysis of genomic DNA (Fig. 1E). The genomic PCR demonstrated that even with vehicle, the presence of SA-Cre-ERT2 resulted in partial excision of the LRH-1 locus (Fig. 1E, lower panel). This was further illustrated by quantitative real-time PCR analysis of hepatic LRH-1 expression, which was already reduced significantly after vehicle injection (Fig. 1F, left panel). Tamoxifen injection, however, further significantly reduced LRH-1 mRNA expression to almost undetectable levels (Fig. 1F, left panel). The expression of SHP, a well-established LRH-1 target gene (27), was reduced robustly (Fig. 1F, right panel). Consistent with the gene expression data, LRH-1 protein levels were undetectable in the livers of LRH-1hep−/− mice (Fig. 1G). Furthermore, very low levels of SHP protein were detected in the livers of the mutant animals (Fig. 1H), confirming in vivo the importance of LRH-1 in the regulation of basal SHP expression in the liver.
FIG. 1.
Generation of mice with a targeted mutation of LRH-1 in the hepatocytes. (A to C) Gene targeting and conditional deletion of exons 4 and 5 of the LRH-1 gene. (A) Restriction maps of the LRH-1 genomic locus, the targeting vector, the floxed allele with a neomycin cassette (+neo) and the floxed allele without a neomycin cassette (−neo). The indicated probes (5′ probe and 3′ probe) were used to assess recombination events. Black vertical bars represent the respective exons. (B) Southern blot analysis of homologous recombination in ES cells electroporated with the targeting vector. DNA derived from ES cells was digested with XbaI or SacI. Hybridizing fragments of wild-type (+/+) and floxed (+/L2) alleles to the 5′ probe or the 3′ probe and their respective sizes are indicated. (C) Recombination of loxP sites as demonstrated with PCR analysis of genomic DNA from ES cells. The sizes of the PCR products amplifying the floxed and wild-type LRH-1 loci are indicated. (D) Scheme depicting procedure of tamoxifen-induced recombination. Control LRH-1L2/L2 or bigenic SA-Cre-ERT2 × LRH-1L2/L2 male mice (10 weeks old) were intraperitoneally injected with 100 μl of olive oil-ethanol (10:1) (vehicle [Veh]) or the same volume of tamoxifen (Tam; 10 mg/ml) for four constitutive days (depicted by arrows). Twenty-one days after the first injection, mice were sacrificed and analyzed for recombination efficacy and specificity and general parameters for liver function were evaluated. (E) Recombination of the Lrh-1 locus is confined to the liver. Tissue specificity of recombination was assessed by PCR analysis of genomic DNA isolated from various tissues of LRH-1L2/L2 and bigenic SA-CreERT2 × LRH-1L2/L2 mice treated with vehicle or tamoxifen. WAT, white adipose tissue; S. muscle, skeletal muscle. (F) Percentage of remaining Lrh-1 (left panel) and Shp (right panel) mRNA in the livers of LRH-1L2/L2 and bigenic SA-CreERT2 × LRH-1L2/L2 mice after vehicle or tamoxifen treatment. mRNA levels were determined by quantitative real-time PCR analysis. Total RNA was extracted from the livers of the above-described mice. Results are the means ± standard errors of the means (error bars) of nine mice per group. Values were normalized to cyclophilin. P values were calculated with the Student t test and are indicated. **, P < 0.01; ***, P < 0.001. (G) LRH-1 immunoblotting performed on reticulocyte lysates programmed with cDNA encoding long (LRH-1L [lane 1]) and short (LRH-1S [lane 2]) isoforms of mouse LRH-1 and liver protein extracts of LRH-1L2/L2 (lane 3) and LRH-1hep−/− (lane 4) mice. Specific bands corresponding to LRH-1L and LRH-1S isoforms of mouse LRH-1 are indicated by arrowheads. ns, nonspecific band. (H) SHP immunoblotting performed on liver nuclear extracts of LRH-1L2/L2 (lane 1) and LRH-1hep−/− (lane 2) mice.
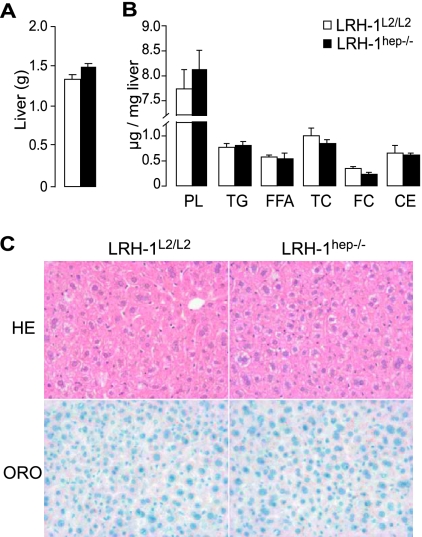
Given the leakiness of the SA-Cre-ERT2 activity in the absence of tamoxifen (Fig. 1E and F), we decided that in all subsequent analyses we would omit the vehicle-treated groups and compare only tamoxifen-treated groups. Cohorts of male LRH-1L2/L2 and SA-Cre-ERT2 × LRH-1L2/L2 mice were therefore injected with tamoxifen to generate respective control LRH-1L2/L2 and mutant LRH-1hep−/− mice, and these cohorts were phenotyped 3 weeks after injection. No changes in body weight were observed before and after tamoxifen injection between the two groups, suggesting that tamoxifen treatment was well tolerated (data not shown). The ablation of LRH-1 in the hepatocytes did not cause any specific biochemical abnormality indicative of liver dysfunction (Table 1). Compared to their LRH-1L2/L2 littermates, no significant difference in liver weight, architecture, or lipid content could be demonstrated for the LRH-1hep−/− mice (Fig. 2). Hence, short-term deletion of the LRH-1 gene in hepatocytes does not interfere with normal hepatobiliary function, has no major consequences on glucose and lipoprotein metabolism, and is rather well tolerated.
TABLE 1.
Liver function tests and metabolic parameters in plasma of LRH-1hepL2/L2 and LRH-1hep−/− micea
| Test or parameter | Result for:
|
|
|---|---|---|
| L2/L2 (n = 9) | hep−/− (n = 9) | |
| Liver function tests | ||
| Total protein (g/liter) | 61.9 ± 0.6 | 64.3 ± 0.9* |
| Albumin (g/liter) | 31.4 ± 0.9 | 30.6 ± 0.8 |
| Bilirubin (μmol/liter) | 1.9 ± 0.4 | 2.2 ± 0.6 |
| Aspartate aminotransferase (U/liter) | 51.2 ± 2.2 | 49.4 ± 3.5 |
| Alanine aminotransferase (U/liter) | 25.0 ± 2.6 | 27.9 ± 7.0 |
| Alkaline phosphatase (U/liter) | 84.2 ± 5.3 | 78.6 ± 4.8 |
| Metabolic parameters | ||
| Total cholesterol (mmol/liter) | 3.5 ± 0.2 | 3.8 ± 0.3 |
| HDL-cholesterol (mmol/liter) | 2.1 ± 0.1 | 2.2 ± 0.2 |
| LDL-cholesterol (mmol/liter) | 0.5 ± 0.0 | 0.7 ± 0.1 |
| Triglycerides (mmol/liter) | 1.5 ± 0.1 | 1.7 ± 0.1 |
| Free fatty acids (mEq/liter) | 2.4 ± 0.2 | 2.7 ± 0.2 |
| Glucose (mmol/liter) | 15.7 ± 1.0 | 14.6 ± 0.7 |
| Insulin (μg/liter) | 0.7 ± 0.2 | 0.5 ± 0.2 |
Mice were fasted for 3 h and parameters were determined 3 weeks after tamoxifen injection (*, P < 0.05). HDL, high-density lipoprotein; LDL, low-density lipoprotein.
FIG. 2.
Liver structure and function is conserved in LRH-1hep−/− mice. (A) Liver weight of tamoxifen-treated LRH-1L2/L2 (n = 9) and LRH-1hep−/− (n = 9) mice. (B) Liver lipid content of LRH-1L2/L2 (n = 7) and LRH-1hep−/− (n = 5) mice expressed in micrograms per milligram of liver weight. Phospholipids (PL), triglycerides (TG), free fatty acids (FFA), total cholesterol (TC), and free cholesterol (FC) were measured enzymatically as described in Materials and Methods. Cholesterol ester (CE) values were calculated by subtracting FC values from TC values. (C) Representative hematoxylin-eosin (HE [magnification, ×200]) and Oil Red O (ORO [magnification, ×400]) staining of liver sections of LRH-1L2/L2 and LRH-1hep−/− mice.
LRH-1 determines BA composition.
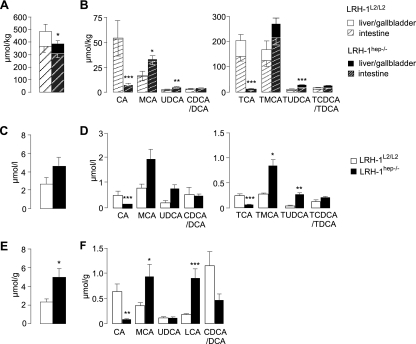
We next compared BA parameters between the LRH-1hep−/− and LRH-1L2/L2 mice. To measure BA pool size and composition in individual mouse samples, livers/gallbladders and intestines were collected separately, enabling us to determine how much each compartment contributed to the total pool size. The addition of data from the livers/gallbladders and intestines then yielded the total pool size and composition (Fig. 3A and B). BA pool size was moderately, but significantly, reduced in LRH-1hep−/− mice, subsequent to an equivalent reduction of BAs present in the livers/gallbladders and intestines (Fig. 3A). The reduced pool size was attributed mainly to the almost complete disappearance of CA and its tauroconjugated form, TCA (Fig. 3B). In contrast, levels of muricholic acid (MCA) and ursodeoxycholic acid (UDCA) and their taurine conjugates tauro-MCA (TMCA) and tauro-UDCA (TUDCA), respectively, were increased (Fig. 3B). Given the decreased BA pool size, it is of note that plasma BA levels had a tendency to increase (Fig. 3C). The composition of plasma BAs, however, mirrored the changes in BA pool composition, with a decrease in CA and TCA, and an increase in MCA, TMCA, UDCA, and TUDCA (Fig. 3D). Interestingly, the amount of total BAs in the feces was more than doubled in LRH-1hep−/− mice (Fig. 3E). The increase in total fecal BAs seemed accounted for mainly by the accumulation of fecal lithocholic acid (LCA), a hydrophobic secondary BA produced in the intestine by bacterial 7α-dehydroxylation of chenodeoxycholic acid (CDCA) and known to be poorly reabsorbed into the enterohepatic circulation. The increase of both MCA and LCA and the concomitant decrease of CA and CDCA/DCA peak (Fig. 3F), which in the mouse is predominantly DCA, were again consistent with our observation that LRH-1hep−/− mice fail to produce CA.
FIG. 3.
Loss of LRH-1 in hepatocytes leads to cholic acid deficiency and alters BA pool size, composition, and excretion. Total BA pool (A, C, and E) and BA composition (B, D, and F) in livers/gallbladders, intestinal extracts (A and B), plasma (C and D), and feces (E and F) of LRH-1L2/L2 (n = 4) and LRH-1hep−/− (n = 14) mice. Unconjugated CA and TCA, MCA and TMCA, UDCA and TUDCA, and CDCA/DCA and TCDCA/TDCA were measured in the enterohepatic compartment and in plasma. LCA and other unconjugated BA species were measured for feces samples. Statistical differences between both genotypes were calculated from the sum of liver/gallbladder and intestinal extracts. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
LRH-1 is essential for Cyp8b1, but not Cyp7a1, gene expression in vivo.
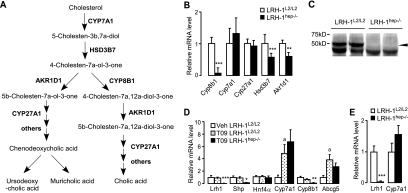
Synthesis of primary BAs occurs predominantly via the neutral pathway and involves numerous enzymes (reviewed in reference 38) (Fig. 4A), but only a few of the genes that encode these enzymes are controlled at the level of transcription. Since the observed abnormalities in BA composition were highly suggestive of a decrease in the activity of CYP8B1, which is responsible for 12α-hydroxylation of BAs, we first investigated the mRNA levels of this gene. Interestingly, the expression of Cyp8b1 seemed to be critically dependent on the presence of LRH-1, as its expression, both at the levels of mRNA (Fig. 4B) and protein (Fig. 4C), was almost completely abrogated in LRH-1hep−/− mice. These results corroborate the previous in vitro findings identifying Cyp8b1 as a target of LRH-1 (9) and provide unequivocal in vivo evidence for the critical role of LRH-1 in driving the expression of the gene whose product is required for CA production. In addition to Cyp8b1, but less striking, two other genes encoding 3β-hydroxy-Δ5-C27 steroid oxidoreductase (HSD3B7) and aldo-keto reductase family 1 member D1 (AKR1D1) were also reduced in LRH-1hep−/− mice (Fig. 4B). However, whether the decreased expression of those genes contributes to the decreased BA pool in LRH-1hep−/− mice is questionable, since the control of BA synthesis is exerted mainly by CYP7A1 activity.
FIG. 4.
Cyp8b1, but not Cyp7A1, is critically dependent on LRH-1. (A) Scheme depicting neutral BA biosynthetic enzymatic pathway. (B) Expression profile of key genes involved in BA biosynthesis with Cyp8b1, but not Cyp7a1, and impaired in LRH-1hep−/− mice. Relative mRNA expression levels of the indicated genes of the BA synthesis pathway in the livers of control LRH-1L2/L2 (n = 9) and mutant LRH-1hep−/− (n = 9) littermates are shown. Values were normalized to cyclophilin and expressed as percent change to control LRH-1L2/L2 mice. (C) CYP8B1 immunoblotting performed on liver extracts of LRH-1L2/L2 and LRH-1hep−/− mice. (D and E) LXR-mediated activation of Cyp7a1 is not dependent on the presence of LRH-1. (D) Relative mRNA expression levels of the indicated genes in the livers of either control LRH-1L2/L2 (n = 4) mice treated with vehicle (Veh) or control LRH-1L2/L2 (n = 5) and mutant LRH-1hep−/− (n = 4) mice treated with the LXR agonist T0901317 (T09; 50 mg/kg/day) for 7 days. Significant differences between vehicle and T0901317-treated cohorts are indicated with lowercase a's (P < 0.05), while significant differences between T0901317-treated control and mutant cohorts are indicated with asterisks. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (E) Relative mRNA expression levels of Lrh-1 and Cyp7a1 in the livers of control LRH-1L2/L2 (n = 4) and mutant LRH-1hep−/− (n = 10) mice fed a high-cholesterol (2%) diet for 1 week. Significant differences between groups are indicated. ***, P < 0.001.
We then analyzed whether the mRNA levels of CYP7A1, the rate-limiting enzyme of the neutral BA biosynthesis pathway, were altered. Contrary to what we expected, the expression of Cyp7a1 was not significantly different between livers of LRH-1L2/L2 and LRH-1hep−/− mice and even tended to increase in the mutant animals (Fig. 4B). The absence of a clear impact of the hepatic LRH-1 deficiency on Cyp7a1 gene expression incited us to explore whether in vivo LRH-1 could act as a competence factor for LXR activity as has been proposed previously (27). Surprisingly, we observed that upon treatment with the synthetic LXR agonist T0901317, mRNA levels of the two LXR targets Cyp7a1 and Abcg5 were induced to similar levels in control and LRH-1-null livers, indicating that the transcriptional activity of LXR is not dependent on LRH-1 (Fig. 4D). The apparent uncoupling between LRH-1 and LXR signaling was furthermore confirmed in mice fed a high-cholesterol diet for 1 week, where again, no differences in Cyp7a1 induction could be observed between LRH-1L2/L2 and LRH-1hep−/− mice (Fig. 4E). In combination, these data suggest that LRH-1 is not essential for the LXR-mediated induction of Cyp7a1 expression.
Lack of LRH-1 in liver alters nuclear receptor signaling.
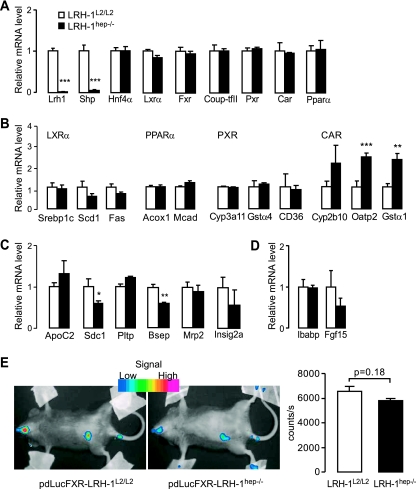
Several nuclear receptors have been shown to regulate, directly or indirectly, the biosynthesis of BAs (6, 21). To assess potential perturbances in these nuclear receptor signaling pathways in response to LRH-1 disruption, the expression and activity of these receptors were measured. Except for the robust decrease in Shp mRNA levels, no major changes in the expression of other nuclear receptors known to affect Cyp7a1 transcription, including Lxrα, Fxr, peroxisome proliferator-activated receptor α (Pparα), pregnane X receptor (Pxr), constitutive androstane receptor (Car) or hepatocyte nuclear factor 4α (HnF4α), were detected in the LRH-1hep−/− mice (Fig. 5A). The activity of LXRα, PXR, and PPARα was furthermore unchanged in control and mutant animals, as evidenced by the absence of changes in the expression of several established hepatic targets of these respective nuclear receptors (Fig. 5B). CAR, however, seemed activated since its target genes were induced in the mutant LRH-1hep−/− livers (Fig. 5B). As CAR has been previously reported to inhibit Cyp7a1 expression (29), the induction of its activity could counteract the derepression that is expected when the corepressor SHP is absent.
FIG. 5.
Effect of liver-specific LRH-1 deficiency on nuclear receptor signaling. (A to C) Gene expression profile in the livers of control LRH-1L2/L2 (n = 9) and mutant LRH-1hep−/− (n = 9) mice of (A) key nuclear receptors involved, directly or indirectly, in the regulation of Cyp7a1 gene transcription; (B) target genes of LXRα, FXR, PPARα, PXR, and CAR; and (C) target genes of FXR. Abbreviations: Srebp-1c, sterol regulatory element binding protein 1c; Scd1, stearoyl-coA desaturase 1; Fas, fatty acid synthase; Acox1, acyl-CoA oxidase 1; Mcad, acyl-CoA dehydrogenase, medium chain; Gstα, glutathione S-transferase alpha; Oatp2, organic anion transporting polypeptide 2; Bsep, bile salt export pump; Sdc1, syndecan 1; Pltp, phospholipid transfer protein; Mrp2, multidrug resistance-associated protein 2; Insig2a, insulin-induced gene 2a. Significant differences between groups are indicated. **, P < 0.01; ***, P < 0.001. (D) Gene expression of FXR target genes in the intestine of control and mutant mice described above. Abbreviations: Ibabp, ileal BA binding protein; Fgf15, fibroblast growth factor 15. (E) In vivo imaging (left panel) and quantification (right panel) of luciferase activity in pdLucFXR-LRH-1L2/L2 and pdLucFXR-LRH-1hep−/− mice.
Because of the well-known repressive function of FXR on Cyp7a1 expression and the marked remodeling of the BA pool observed in the LRH-1hep−/− mice, we also studied FXR activity in these mice. Intriguingly, while the expression of some liver FXR target genes, such as the bile salt export pump (Bsep [Abcb11]), syndecan 1, and Shp genes were decreased, the expression of other FXR targets, including apolipoprotein C2, phospholipid transfer protein, and multidrug resistance-associated protein 2, did not change significantly (Fig. 5A and C). In the intestines, no major changes in the expression of the intestinal BA binding protein and fibroblast growth factor 15 could be observed (Fig. 5D). To gain further insight in vivo on the impact of the observed BA remodeling in the mutant mice on FXR activity, FXR luciferase reporter mice (19) were bred with LRH-1hep−/− mice and basal endogenous FXR activity was quantified during the active dark period by in vivo imaging of luciferase activity (Fig. 5E). Consistent with previous reports, no luciferase activity was present in the liver in basal conditions (19). In the abdominal region, however, FXR activity tended to be lower in the pdLucFXR-LRH-1hep−/− mice compared to that in their control littermates (Fig. 5E). This tendency, which could reflect the existence of BA components with altered potency to activate FXR, could perhaps explain why the expression of only a subset of FXR target genes was modulated. The changes in only some of the FXR target genes may be FXR independent and result from the interference of other altered signaling pathways.
LRH-1 alters the expression of several liver lipid transporters.
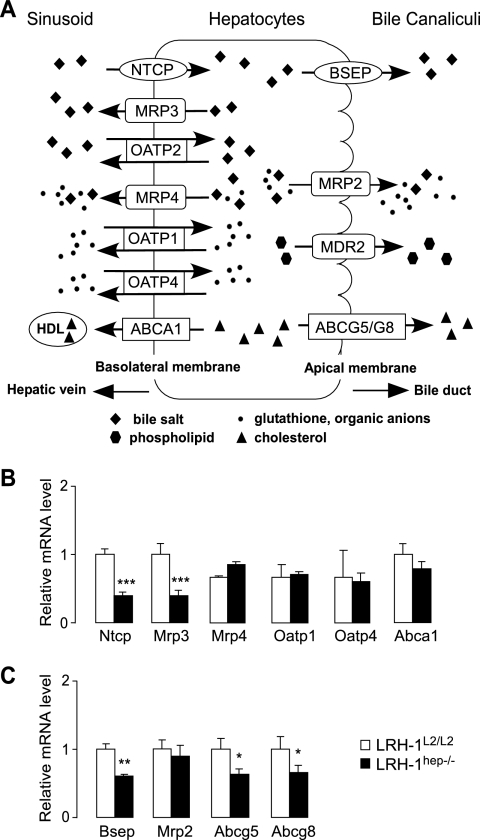
The absence of LRH-1 also affected the expression levels of several BA and sterol transporters that are present on the basolateral or apical/canalicular side of the hepatocytes (Fig. 6A). The most interesting change in expression among the basolateral transporters was the reduction of the sodium taurocholate cotransporting polypeptide (NTCP) (Fig. 6B), which is responsible for the selective uptake of BAs from the portal blood and which could contribute to the relative buildup of BAs in the plasma of the mutant mice (Fig. 3C). Consistent with earlier studies proposing LRH-1 as a transcriptional regulator of Mrp3 under cholestatic conditions, Mrp3 was also reduced in the livers of LRH-1hep−/− mice (Fig. 6B). The physiological impact of this regulation under basal conditions, however, is unclear since this transporter does not seem to play a crucial role in BA transport (45). Among the apical/canalicular transporters, the expression of Abcg8 and Abcg5, both involved in the biliary excretion of sterols, was decreased in the livers of LRH-1hep−/− mice (Fig. 6C). These data are in agreement with previous findings identifying LRH-1 as a transcriptional regulator of the Abcg5/Abcg8 intergenic region (14). The expression of Bsep, which is responsible for the export of the majority of BA into the bile canaliculi, was also reduced (Fig. 6C). In combination, these findings indicate that LRH-1 regulates not only the composition of BAs but also the genes that control the hepatic flux of the major components of bile.
FIG. 6.
LRH-1 regulates hepatic transport of BAs and sterols. (A) Schematic representation of basolateral and canalicular transporters. (B) Expression of indicated basolateral transporter genes in the livers of control LRH-1L2/L2 and mutant LRH-1hep−/− mice. Values were normalized to cyclophilin and expressed as percent change to control LRH-1L2/L2 mice in the above-described experiments. Significant differences between groups are indicated. ***, P < 0.001. (C) Expression profile of indicated apical transporter genes in the livers of control LRH-1L2/L2 and mutant LRH-1hep−/− mice shown in panel B. Significant differences between both groups are indicated. *, P < 0.05; **, P < 0.01.
Fecal lipid loss in LRH-1hep−/− mice as a result of BA-dependent intestinal lipid malabsorption.
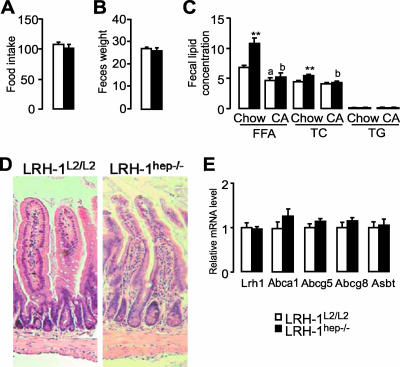
In view of the substantial differences observed in BA pool size and composition, we next investigated whether the deletion of hepatic LRH-1 could have an impact on intestinal lipid absorption. Individual animals were accustomed to metabolic cages and monitored for food intake, and stools were collected over a period of 24 h, 3 weeks after tamoxifen treatment. Food consumption (Fig. 7A) and total fecal mass (Fig. 7B) were essentially unchanged for LRH-1L2/L2 and LRH-1hep−/− mice, although there was a tendency toward increased fecal mass after a longer period of LRH-1 deletion (data not shown). Feces of LRH-1hep−/− mice, however, contained more fatty acids and cholesterol than did feces of LRH-1L2/L2 mice (Fig. 7C), indicating that the absorption of lipids was perturbed. Supplementing the diet with CA completely reversed the abnormalities in fecal lipid excretion (Fig. 7C), suggesting that the diminished lipid absorption is the direct consequence of the altered composition of the BA pool. Further arguments that the altered lipid absorption was not due to a direct effect on the intestine were provided by the absence of primary mucosal absorptive defects, such as that seen in inflammatory or infiltrative bowel diseases, as the histologies of the ilea of LRH-1hep−/− mice revealed no signs of inflammation or other pathologies (Fig. 7D and data not shown). Furthermore, the increase in cholesterol (Fig. 7C) and BAs (Fig. 3E and F) in the feces could not be explained by alterations of Lrh-1 expression in the small intestine or in the active transport of these molecules across the ileal intestinal epithelium, as the expression of the sterol transporters Abcg5/Abcg8, Abca1, and Asbt was unchanged (Fig. 7E). In combination, these data indicate that the disruption of LRH-1 gene function in the hepatocytes impacts the intestinal absorption of nutrients or metabolites of lipophilic origin and that this effect is mediated by the critical role that LRH-1 has in determining the physicochemical properties of the BA pool.
FIG. 7.
Absence of LRH-1 in hepatocytes induces fecal lipid loss. Control LRH-1L2/L2 (n = 7) and mutant LRH-1hep−/− (n = 5) mice were monitored in individual metabolic cages and feces were collected from each animal during a period of 24 h. (A) Food consumption normalized to body weight (mg/g) during a 24-h period. (B) Fecal mass normalized to body weight (mg/g). (C) Fecal lipid concentration expressed as micrograms of lipids per milligram of feces weight of chow-fed mice (Chow) or mice treated for 2 days with 0.2% wt/wt CA. Free fatty acids (FFA), total cholesterol (TC), and triglycerides (TG) were measured enzymatically. Significant differences between groups are indicated with asterisks. **, P < 0.01. Significant differences between chow versus CA are depicted in lowercase letters: a, P < 0.01; b, P < 0.001. (D) Ileal hematoxylin-eosin sections (magnification, ×100) from control LRH-1L2/L2 or mutant LRH-1hep−/− mice. (E) Gene expression of sterol and BA transporters in the ilea of control LRH-1L2/L2 or mutant LRH-1hep−/− mice.
DISCUSSION
BAs are water-soluble end products from cholesterol metabolism and are essential for efficient absorption of dietary lipids. Although many different BAs exist as a result of postsynthesis bacterial modification in the intestine, only two immediate end products are formed in the liver. The nature of these BAs, termed primary BAs, differs according to the species. In mice, the primary BAs formed in the liver are CA, a potent amphipathic molecule which requires CYP8B1 for its biosynthesis, and MCA, which is derived from CDCA and which exerts rather poor detergent properties compared with those of CA (18).
LRH-1 is a nuclear receptor that has been reported to control the transcription of both Cyp7a1 and Cyp8b1 (9, 31). As mice with homozygous germ line mutations in the LRH-1 gene are not viable (4, 17, 33), the biological role of LRH-1 is to a large extent derived from the study of in vitro systems and still requires confirmation in adequate in vivo models. The characterization of hepatocyte-specific LRH-1-deficient mice in this study unequivocally demonstrates that unlike previous reports, Cyp7a1 gene expression, which determines the BA production rate, is not dependent on the presence of hepatic LRH-1. In contrast, the expression of Cyp8b1, which determines the physicochemical properties of BA, is critically dependent on the presence of hepatic LRH-1, as evidenced by the almost complete absence of Cyp8b1 in hepatocyte-specific, LRH-1 mutant mice. Besides Cyp8b1, Shp mRNA levels were also abrogated, indicating that the livers of LRH-1hep−/− mice also behave as Cyp8b1/Shp hepatocyte-specific hypomorph mice. At first glance, these findings are difficult to reconcile with the modest but significant decrease in BA pool size that we observed for the LRH-1hep−/− mice. In fact, both CYP8B1- and SHP-deficient mice have increased BA pool sizes as a consequence of a compromised repression of Cyp7a1 gene transcription (22, 26). In this context, a possible explanation could be that effective derepression of BA synthesis induced by the absence of SHP or CYP8B1 can take place only if LRH-1 is present. Most likely, however, a combination of events that include the impaired expression of Cyp8b1, likely at the origin of the increased LCA excretion, together with the inability of the LRH-1-deficient mice to compensate their fecal loss of BAs by inducing Cyp7a1 gene expression, explains why these mice have a contracted BA pool size. This explanation would be in line with the fact that Cyp8b1-null mice also show increased fecal BA excretion (26).
Besides the demonstration that LRH-1 is not essential to drive liver-specific Cyp7a1 gene expression, our studies also demonstrate unequivocally that LRH-1 is not acting as a competence factor for LXR in vivo, as can be concluded from the fact that LXR-mediated induction of CYP7A1 and ABCG5 gene expression is intact despite the absence of LRH-1. Our data hence underscore the commanding role of hepatic LRH-1 in the determination of BA composition and amphipathicity. Interestingly, the role of LRH-1 in the control of BA homeostasis in the intact animal is uncoupled from the activity of the LXRs, suggesting that LXR-dependent signaling and LRH-1-dependent signaling represent two independent pathways to modulate BA synthesis.
As end products of cholesterol metabolism, BAs contribute to the removal of cholesterol and are crucial for intestinal absorption of lipids. With respect to the latter function, the composition and physicochemical properties of each constituent of the BA pool have been shown to be critical. While dietary supplementation of CA (the amphipathic BA), whose hydroxyl groups are all positioned on one side of the steroid nucleus, increases cholesterol absorption (7), dietary supplementation of MCA and UDCA (the more hydrophilic and less amphipathic BAs) decreases cholesterol absorption (43). Our results show that the absence of LRH-1 and subsequent deficiency of CYP8B1 in the hepatocytes result in the elimination of CA and its amino acid conjugate TCA and that the resulting shift in BA composition in animals with hepatocyte-specific LRH-1 deficiency selectively alters not only intestinal absorption of cholesterol and fatty acids but also the intestinal reuptake of BAs. In humans, who consume more fat than mice, intestinal lipid absorption contributes in a major fashion to whole-body energy homeostasis, and conditions in which lipid absorption is reduced induce a negative energy balance. Decreasing intestinal lipid absorption through inhibiting hepatic LRH-1 activity and altering BA composition seems well tolerated in mice and, hence, may be a route that merits exploration in an effort to combat obesity.
Acknowledgments
We thank J. Feige, E. Garo, A. Jauffre, T. Meyer and M. Selloum for technical and scientific input and I. Talianidis for the kind gift of SHP antibody.
We acknowledge research grants from CNRS, INSERM, ULP, Hôpital Universitaire de Strasbourg, ACI (no. 03-2488), ARC, and AFM and fellowship support from the Canon foundation to C.M. and the ARC to B.M.
Footnotes
Published ahead of print on 1 October 2007.
REFERENCES
- 1.Annicotte, J. S., E. Fayard, G. H. Swift, L. Selander, H. Edlund, T. Tanaka, T. Kodama, K. Schoonjans, and J. Auwerx. 2003. Pancreatic-duodenal homeobox 1 regulates expression of liver receptor homolog 1 during pancreas development. Mol. Cell. Biol. 23:6713-6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boerboom, D., N. Pilon, R. Behdjani, D. W. Silversides, and J. Sirois. 2000. Expression and regulation of transcripts encoding two members of the NR5A nuclear receptor subfamily of orphan nuclear receptors, steroidogenic factor-1 and NR5A2, in equine ovarian cells during the ovulatory process. Endocrinology 141:4647-4656. [DOI] [PubMed] [Google Scholar]
- 3.Bootsma, A. H., H. Overmars, A. van Rooij, A. E. van Lint, R. J. Wanders, A. H. van Gennip, and P. Vreken. 1999. Rapid analysis of conjugated bile acids in plasma using electrospray tandem mass spectrometry: application for selective screening of peroxisomal disorders. J. Inherit. Metab. Dis. 22:307-310. [DOI] [PubMed] [Google Scholar]
- 4.Botrugno, O. A., E. Fayard, J. S. Annicotte, C. Haby, T. Brennan, O. Wendling, T. Tanaka, T. Kodama, W. Thomas, J. Auwerx, and K. Schoonjans. 2004. Synergy between LRH-1 and beta-catenin induces G(1) cyclin-mediated cell proliferation. Mol. Cell 15:499-509. [DOI] [PubMed] [Google Scholar]
- 5.Chen, F., L. Ma, P. A. Dawson, C. J. Sinal, E. Sehayek, F. J. Gonzalez, J. L. Breslow, M. Ananthanarayanan, and B. L. Shneider. 2003. Liver receptor homologue-1 mediates species- and line-specific bile acid dependent negative feedback regulation of the apical sodium-dependent bile acid transporter. J. Biol. Chem. 278:19909-19916. [DOI] [PubMed] [Google Scholar]
- 6.Chiang, J. Y. 2002. Bile acid regulation of gene expression: roles of nuclear hormone receptors. Endocr. Rev. 23:443-463. [DOI] [PubMed] [Google Scholar]
- 7.Cohen, B. I., R. F. Raicht, and E. H. Mosbach. 1977. Sterol metabolism studies in the rat. Effects of primary bile acids (sodium taurochenodeoxycholate and sodium taurocholate) on sterol metabolism. J. Lipid Res. 18:223-231. [PubMed] [Google Scholar]
- 8.Coste, A., L. Dubuquoy, R. Barnouin, J. S. Annicotte, B. Magnier, M. Notti, N. Corazza, M. C. Antal, D. Metzger, P. Desreumaux, T. Brunner, J. Auwerx, and K. Schoonjans. 2007. LRH-1-mediated glucocorticoid synthesis in enterocytes protects against inflammatory bowel disease. Proc. Natl. Acad. Sci. USA 104:13098-13103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.del Castillo-Olivares, A., and G. Gil. 2000. α1-Fetoprotein transcription factor is required for the expression of sterol 12α-hydroxylase, the specific enzyme for cholic acid synthesis. Potential role in the bile acid-mediated regulation of gene transcription. J. Biol. Chem. 275:17793-17799. [DOI] [PubMed] [Google Scholar]
- 10.Delerive, P., C. M. Galardi, J. E. Bisi, E. Nicodeme, and B. Goodwin. 2004. Identification of liver receptor homolog-1 as a novel regulator of apolipoprotein AI gene transcription. Mol. Endocrinol. 18:2378-2387. [DOI] [PubMed] [Google Scholar]
- 11.Fayard, E., J. Auwerx, and K. Schoonjans. 2004. LRH-1: an orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol. 14:250-260. [DOI] [PubMed] [Google Scholar]
- 12.Fayard, E., K. Schoonjans, J. S. Annicotte, and J. Auwerx. 2003. Liver receptor homolog 1 controls the expression of carboxyl ester lipase. J. Biol. Chem. 278:35725-35731. [DOI] [PubMed] [Google Scholar]
- 13.Folch, J., M. Lees, and G. H. S. Stanley. 1957. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226:497-509. [PubMed] [Google Scholar]
- 14.Freeman, L. A., A. Kennedy, J. Wu, S. Bark, A. T. Remaley, S. Santamarina-Fojo, and H. B. Brewer, Jr. 2004. The orphan nuclear receptor LRH-1 activates the ABCG5/ABCG8 intergenic promoter. J. Lipid Res. 45:1197-1206. [DOI] [PubMed] [Google Scholar]
- 15.Galarneau, L., J.-F. Paré, D. Allard, D. Hamel, L. Lévesque, J. D. Tugwood, S. Green, and L. Bélanger. 1996. The α1-fetoprotein locus is activated by a nuclear receptor of the Drosophila FTZ-F1 family. Mol. Cell. Biol. 16:3853-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodwin, B., S. A. Jones, R. R. Price, M. A. Watson, D. D. McKee, L. B. Moore, C. Galardi, J. G. Wilson, M. C. Lewis, M. E. Roth, P. R. Maloney, T. M. Willson, and S. A. Kliewer. 2000. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell 6:517-526. [DOI] [PubMed] [Google Scholar]
- 17.Gu, P., B. Goodwin, A. C. Chung, X. Xu, D. A. Wheeler, R. R. Price, C. Galardi, L. Peng, A. M. Latour, B. H. Koller, J. Gossen, S. A. Kliewer, and A. J. Cooney. 2005. Orphan nuclear receptor LRH-1 is required to maintain Oct4 expression at the epiblast stage of embryonic development. Mol. Cell. Biol. 25:3492-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofmann, A. F., and A. Roda. 1984. Physicochemical properties of bile acids and their relationship to biological properties: an overview of the problem. J. Lipid Res. 25:1477-1489. [PubMed] [Google Scholar]
- 19.Houten, S. M., D. H. Volle, C. L. Cummins, D. J. Mangelsdorf, and J. Auwerx. 2007. In vivo imaging of farnesoid X receptor activity reveals the ileum as the primary bile acid signaling tissue. Mol. Endocrinol. 21:1312-1323. [DOI] [PubMed] [Google Scholar]
- 20.Inokuchi, A., E. Hinoshita, Y. Iwamoto, K. Kohno, M. Kuwano, and T. Uchiumi. 2001. Enhanced expression of the human multidrug resistance protein 3 by bile salt in human enterocytes. A transcriptional control of a plausible bile acid transporter. J. Biol. Chem. 276:46822-46829. [DOI] [PubMed] [Google Scholar]
- 21.Inoue, Y., L. L. Peters, S. H. Yim, J. Inoue, and F. J. Gonzalez. 2006. Role of hepatocyte nuclear factor 4α in control of blood coagulation factor gene expression. J. Mol. Med. 84:334-344. [DOI] [PubMed] [Google Scholar]
- 22.Kerr, T. A., S. Saeki, M. Schneider, K. Schaefer, S. Berdy, T. Redder, B. Shan, D. W. Russell, and M. Schwarz. 2002. Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev. Cell 2:713-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krylova, I. N., E. P. Sablin, J. Moore, R. X. Xu, G. M. Waitt, J. A. MacKay, D. Juzumiene, J. M. Bynum, K. Madauss, V. Montana, L. Lebedeva, M. Suzawa, J. D. Williams, S. P. Williams, R. K. Guy, J. W. Thornton, R. J. Fletterick, T. M. Willson, and H. A. Ingraham. 2005. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell 120:343-355. [DOI] [PubMed] [Google Scholar]
- 24.Lee, Y. K., and D. D. Moore. 2002. Dual mechanisms for repression of the monomeric orphan receptor liver receptor homologous protein-1 by the orphan small heterodimer partner. J. Biol. Chem. 277:2463-2467. [DOI] [PubMed] [Google Scholar]
- 25.Lee, Y. K., K. L. Parker, H. S. Choi, and D. D. Moore. 1999. Activation of the promoter of the orphan receptor SHP by orphan receptors that bind DNA as monomers. J. Biol. Chem. 274:20869-20873. [DOI] [PubMed] [Google Scholar]
- 26.Li-Hawkins, J., M. Gafvels, M. Olin, E. G. Lund, U. Andersson, G. Schuster, I. Bjorkhem, D. W. Russell, and G. Eggertsen. 2002. Cholic acid mediates negative feedback regulation of bile acid synthesis in mice. J. Clin. Investig. 110:1191-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu, T. T., M. Makishima, J. J. Repa, K. Schoonjans, T. A. Kerr, J. Auwerx, and D. J. Mangelsdorf. 2000. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell 6:507-515. [DOI] [PubMed] [Google Scholar]
- 28.Luo, Y., C. P. Liang, and A. R. Tall. 2001. The orphan nuclear receptor LRH-1 potentiates the sterol-mediated induction of the human CETP gene by liver X receptor. J. Biol. Chem. 276:24767-24773. [DOI] [PubMed] [Google Scholar]
- 29.Miao, J., S. Fang, Y. Bae, and J. K. Kemper. 2006. Functional inhibitory cross-talk between constitutive androstane receptor and hepatic nuclear factor-4 in hepatic lipid/glucose metabolism is mediated by competition for binding to the DR1 motif and to the common coactivators, GRIP-1 and PGC-1α. J. Biol. Chem. 281:14537-14546. [DOI] [PubMed] [Google Scholar]
- 30.Mueller, M., I. Cima, M. Noti, A. Fuhrer, S. Jakob, L. Dubuquoy, K. Schoonjans, and T. Brunner. 2006. The nuclear receptor LRH-1 critically regulates extra-adrenal glucocorticoid synthesis in the intestine. J. Exp. Med. 203:2057-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nitta, M., S. Ku, C. Brown, A. Y. Okamoto, and B. Shan. 1999. CPF: an orphan nuclear receptor that regulates liver-specific expression of the human cholesterol 7α-hydroxylase gene. Proc. Natl. Acad. Sci. USA 96:6660-6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ortlund, E. A., Y. Lee, I. H. Solomon, J. M. Hager, R. Safi, Y. Choi, Z. Guan, A. Tripathy, C. R. Raetz, D. P. McDonnell, D. D. Moore, and M. R. Redinbo. 2005. Modulation of human nuclear receptor LRH-1 activity by phospholipids and SHP. Nat. Struct. Mol. Biol. 12:357-363. [DOI] [PubMed] [Google Scholar]
- 33.Paré, J. F., D. Malenfant, C. Courtemanche, M. Jacob-Wagner, S. Roy, D. Allard, and L. Belanger. 2004. The fetoprotein transcription factor (FTF) gene is essential to embryogenesis and cholesterol homeostasis and is regulated by a DR4 element. J. Biol. Chem. 279:21206-21216. [DOI] [PubMed] [Google Scholar]
- 34.Pare, J. F., S. Roy, L. Galarneau, and L. Bélanger. 2001. The mouse fetoprotein transcription factor (FTF) gene promoter is regulated by three GATA elements with tandem E box and Nkx motifs, and FTF in turn activates the hnf3β, hnf4α, and hnf1α gene promoters. J. Biol. Chem. 276:13136-13144. [DOI] [PubMed] [Google Scholar]
- 35.Postic, C., M. Shiota, K. D. Niswender, T. L. Jetton, Y. Chen, J. M. Moates, K. D. Shelton, J. Lindner, A. D. Cherrington, and M. A. Magnuson. 1999. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J. Biol. Chem. 274:305-315. [DOI] [PubMed] [Google Scholar]
- 36.Rausa, F. M., L. Galarneau, L. Bélanger, and R. H. Costa. 1999. The nuclear receptor fetoprotein transcription factor is coexpressed with its target gene HNF-3b in the developing murine liver, intestine and pancreas. Mech. Dev. 89:185-188. [DOI] [PubMed] [Google Scholar]
- 37.Rodríguez, C. I., F. Buchholz, J. Galloway, R. Sequerra, J. Kasper, R. Ayala, A. F. Stewart, and S. M. Dymecki. 2000. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 25:139-140. [DOI] [PubMed] [Google Scholar]
- 38.Russell, D. W. 2003. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 72:137-174. [DOI] [PubMed] [Google Scholar]
- 39.Sablin, E. P., I. N. Krylova, R. J. Fletterick, and H. A. Ingraham. 2003. Structural basis for ligand-independent activation of the orphan nuclear receptor LRH-1. Mol. Cell 11:1575-1585. [DOI] [PubMed] [Google Scholar]
- 40.Schoonjans, K., J. S. Annicotte, T. Huby, O. A. Botrugno, E. Fayard, Y. Ueda, J. Chapman, and J. Auwerx. 2002. Liver receptor homolog-1 controls the expression of the scavenger receptor class B type I. EMBO Rep. 3:1181-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schoonjans, K., L. Dubuquoy, J. Mebis, E. Fayard, O. Wendling, C. Haby, K. Geboes, and J. Auwerx. 2005. Liver receptor homolog 1 contributes to intestinal tumor formation through effects on cell cycle and inflammation. Proc. Natl. Acad. Sci. USA 102:2058-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuler, M., A. Dierich, P. Chambon, and D. Metzger. 2004. Efficient temporally controlled targeted somatic mutagenesis in hepatocytes of the mouse. Genesis 39:167-172. [DOI] [PubMed] [Google Scholar]
- 43.Wang, D. Q., and S. Tazuma. 2002. Effect of beta-muricholic acid on the prevention and dissolution of cholesterol gallstones in C57L/J. mice. J. Lipid Res. 43:1960-1968. [DOI] [PubMed] [Google Scholar]
- 44.Wang, W., C. Zhang, A. Marimuthu, H. I. Krupka, M. Tabrizizad, R. Shelloe, U. Mehra, K. Eng, H. Nguyen, C. Settachatgul, B. Powell, M. V. Milburn, and B. L. West. 2005. The crystal structures of human steroidogenic factor-1 and liver receptor homologue-1. Proc. Natl. Acad. Sci. USA 102:7505-7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zelcer, N., K. van de Wetering, R. de Waart, G. L. Scheffer, H. U. Marschall, P. R. Wielinga, A. Kuil, C. Kunne, A. Smith, M. van der Valk, J. Wijnholds, R. O. Elferink, and P. Borst. 2006. Mice lacking Mrp3 (Abcc3) have normal bile salt transport, but altered hepatic transport of endogenous glucuronides. J. Hepatol. 44:768-775. [DOI] [PubMed] [Google Scholar]