Abstract
The Saccharomyces cerevisiae splicing factors Ntr1 (also known as Spp382) and Ntr2 form a stable complex and can further associate with DExD/H-box RNA helicase Prp43 to form a functional complex, termed the NTR complex, which catalyzes spliceosome disassembly. We show that Prp43 interacts with Ntr1-Ntr2 in a dynamic manner. The Ntr1-Ntr2 complex can also bind to the spliceosome first, before recruiting Prp43 to catalyze disassembly. Binding of Ntr1-Ntr2 or Prp43 does not require ATP, but disassembly of the spliceosome requires hydrolysis of ATP. The NTR complex also dynamically interacts with U5 snRNP. Ntr2 interacts with U5 component Brr2 and is essential for both interactions of NTR with U5 and with the spliceosome. Ntr2 alone can also bind to U5 and to the spliceosome, suggesting a role of Ntr2 in mediating the binding of NTR to the spliceosome through its interaction with U5. Our results demonstrate that dynamic interactions of NTR with U5, through the interaction of Ntr2 with Brr2, and interactions of Ntr1 and Prp43 govern the recruitment of Prp43 to the spliceosome to mediate spliceosome disassembly.
Splicing of nuclear pre-mRNA requires five small nuclear RNAs (snRNAs) and numerous protein factors (for reviews, see references 3 and 4). These factors bind to the pre-mRNA in a sequential manner, in the order of U1, U2, and then U4/U6.U5 as a tri-snRNP particle, to assemble the spliceosome. Subsequent to the binding of tri-snRNP, the spliceosome undergoes a major structural rearrangement, including the release of U1 and U4, and the addition of a large protein complex, the Prp19-associated complex (or NTC), and becomes catalytically competent. After the splicing reaction is complete, the postcatalytic spliceosome first releases the mature mRNA and then undergoes disassembly to dissociate all components for a new round of splicing. Extensive structural rearrangement of the pre-mRNA, including the formation and/or disruption of RNA base pairing, is associated with each step of the assembly and disassembly process. Mechanical devices that promote base pairing or facilitate unwinding assist in mediating such RNA rearrangements (for a review, see reference 32), and it is proposed that the U5 component Snu114 serves as a signal-dependent switch to control the spliceosome dynamics (31).
The DEXD/H-box RNA helicases belong to a large superfamily of proteins conserved from bacteria and viruses to humans (6, 35). They share a highly conserved helicase domain that includes the motif DEXD/H and play roles in all biological processes of RNA molecules, including transcription, editing, splicing, ribosome biogenesis, RNA export, translation, and RNA turnover. All of these proteins have ATPase or NTPase activities stimulated by RNA. Eight DEXD/H-box proteins are involved in various steps of the splicing reaction (29, 32). Each of them was thought to facilitate a structural transition at distinct steps, coupling the energy from ATP hydrolysis to remodeling of RNA-RNA or RNA-protein interactions. Among these proteins, Prp5, Prp28, Sub2, and Brr2 are required for early steps of spliceosome assembly. Prp2 is required for the first catalytic reaction, and Prp16 and Prp22 participate in the second catalytic reaction. Prp22 is also required for the release of mature message after the splicing reaction is complete, whereas Prp43 mediates final disassembly of the spliceosome (1, 22, 37). Prp16 and Prp22 have also been shown to play roles in modulating splicing fidelity by coupling ATP hydrolysis with a discard pathway (5, 23).
Although several of these DEXD/H-box RNA helicases have demonstrated activities of unwinding RNA-RNA or RNA-DNA duplexes (25, 33, 34, 39), they and many others do not show substrate specificity for unwinding of RNA duplexes in vitro. It is conceivable that they might require extrinsic factors to promote target recognition of correct substrates to execute their normal functions. Factors that interact with RNA helicases and possibly play roles in regulating their functions have been previously documented (29). The requirement of Spp2 for the function of Prp2 in promoting the step one reaction represents one such example (26, 30). Spp2 was initially identified as a high-copy-number suppressor of prp2-1 temperature sensitivity mutant (14) and has been shown to interact with Prp2 by both two-hybrid and glutathione S-transferase pull-down assays (26, 30). Spp2 is required for the interaction of Prp2 with the spliceosome to execute its function in the step one reaction (26). Another example is the requirement of Slu7 for recruiting Prp22, which is shown to interact with Slu7 by two-hybrid assays (38), to the spliceosome after Prp16-mediated conformational rearrangement to promote the second catalytic reaction (12). Prp2, Prp16, Prp22, and their cofactors do not remain stably associated with the spliceosome after execution of their functions.
Prp43 is required for spliceosome disassembly after mature mRNA is released (1, 22). We and others have recently shown that two splicing factors, Ntr1 and Ntr2, form a stable complex and then associate with Prp43 via the interaction of Prp43 and Ntr1 to form the NTR complex (2, 37). Ntr1 contains a G-patch domain at its N-terminal region, which is responsible for its interaction with Prp43. The NTR complex is functional in catalyzing disassembly of the spliceosome (37). Since Prp43 cannot function in the absence of Ntr1 or Ntr2, Ntr1-Ntr2 might act to target Prp43 to the spliceosome. In this report, we show that Prp43 interacts with the Ntr1-Ntr2 complex in a dynamic manner. Prp43 can associate with the spliceosome as the NTR complex or can be recruited by the Ntr1-Ntr2 complex, which can bind to the spliceosome independently of Prp43. We further show that Ntr1-Ntr2 also dynamically interacts with U5, which might serve to recruit the Ntr1-Ntr2 complex or the NTR complex to the spliceosome for disassembly. The interaction between NTR and U5 snRNP is mediated primarily through the interaction of Ntr2 and Brr2.
MATERIALS AND METHODS
Yeast strains.
Saccharomyces cerevisiae (yeast) strains used were BJ2168 (MATa prc1 prb1 pep4 leu2 trp1 ura3), YSCC1 (MATa prc1 prb1 pep4 leu2 trp1 ura3 PRP19-HA), YSCC131 (MATa prc1 prb1 pep4 leu2 trp1 ura3 NTR1HA), YSCC133 (MATa prc1 prb1 pep4 leu2 trp1 ura3 PRP43V5), YSCC134 (MATa prc1 prb1 pep4 leu2 trp1 SNU114-3HA), YSCC141 (MATa prc1 prb1 pep4 leu2 trp1 ura3 NTR1HA PRP43V5), and YSCC152 (MATa prc1 prb1 pep4 leu2 trp1 URA3::GAL-NTR2).
YSCC134 was constructed by the PCR epitope tagging method (27), using primers SN1 and SN2. Plasmid DNA pMPY-3XHA was used as the template in the PCR, and the PCR product was purified from agarose gel and used for transformation into S. cerevisiae strain BJ2168 selecting for URA3. Transformants were checked for correct integration by PCR across the junctions of integration using two pairs of primers, pair SN3 and UR1 and pair SN4 and UR2. Transformants with correct integration were grown in yeast extract-peptone-dextrose overnight and then selected on plates containing 5-fluoroorotic acid to select for uracil auxotrophy. All other strains were as previously described (37).
Oligonucleotides.
The oligonucleotide sequences were as follows: for SN1, ATATAAGCGCTGAATTATACGCTCAATTAAGAGAAAATGGCTTAGT ACCGAGGGAACAAAAGCTGGAGCT; for SN2, ATAAAAATATTGTGGACATATTGCTTAATTCTTATGCGCCAAGATTTTCACTATAGGGCGAATTGGGTAC; for SN3, GAAGGTTCCTGGTGATG; for SN4, GCGAAAGCAACTCCTTC; for UR1, GGTACGAACATCCAATG; and for UR2, TCTCTACAGGATCTGAC.
Antibodies and reagents.
Anti-V5 antibody was purchased from Invitrogen Inc., and 12CA5 was purchased from Berkeley Antibody Co. Antihemagglutinin (anti-HA) monoclonal antibody 8G5F, anti-Ntr1, anti-Ntr2, and anti-Prp43 polyclonal antibodies were as described previously (37). Anti-Snu114 antibody was produced by immunizing rabbits with the Escherichia coli-expressed glutathione S-transferase fusion of Snu114 N-terminal fragment of amino acid residues 1 to 130. Protein A-Sepharose was from Amersham Inc., and streptavidin-Sepharose was from Sigma Inc.
Preparation of Ntr2-depleted extracts.
S. cerevisiae strain YSCC152 was grown in uracil-dropout synthetic complete medium supplemented with 2% galactose for overnight culture and then inoculated to yeast extract-peptone-dextrose and grown for 18 h before harvesting. Splicing extracts were prepared according to the method of Cheng et al. (9).
Splicing assays, disassembly assays, immunoprecipitation, immunodepletion, and precipitation of the spliceosome with streptavidin-Sepharose.
Splicing assays were carried out according to the method of Cheng and Abelson (8). Immunoprecipitation was performed as described in the study of Tarn et al. (36) with anti-Ntc20, anti-Ntr1, anti-Ntr2, and anti-V5 antibodies. Each 20 μl of the splicing reaction mixture was precipitated with 1 μl of anti-Ntc20, 3 μl of anti-Ntr1, 3 μl of anti-Ntr2, or 1 μl of anti-V5 antibody conjugated to 10 μl of protein A-Sepharose. RNA was extracted from pellet fractions and analyzed by electrophoresis on 8% polyacrylamide-6 M urea gels. Depletion of Ntr1 was carried out by incubation of 100 μl of splicing extracts with 30 μl anti-Ntr1 antibody conjugated to 50 μl of protein A-Sepharose at 4°C for 1 h followed by removal of beads. To assay for spliceosome disassembly, precipitates were incubated at 25°C for 20 min with 30 μl of buffer DK (20 mM HEPES, pH 7.9, 60 mM KPO4, pH 7.0, 0.2 mM EDTA, 50 mM NaCl, 20% glycerol) with glycerol replaced with 3 mM MgCl2, 1 unit/μl of RNasin, 50 μg/ml tRNA, with/without 2 mM ATP, and with/without 10 μg of recombinant Prp43 or the D215A mutant of Prp43. After the separation of pellets and supernatants, RNA was extracted from both fractions and analyzed by electrophoresis on 8% polyacrylamide-6 M urea gels. Precipitation of the spliceosome with streptavidin agarose was carried out according to the method of Chan et al. (7).
Purification of NTR and Ntr1-Ntr2 complexes.
Purification of the NTR complex was according to the method of Tsai et al. (37). Approximately 300 mg of 40P (9) was mixed with KPO4, pH 7.0, to a final concentration of 60 mM and applied to a protein A-Sepharose column conjugated with 0.1 ml of the anti-HA antibody and preequilibrated with buffer DK. After recycling three times, the column was washed with 2 ml of buffer DK containing 0.05% Nonidet P-40 (NP-40) and then 1 ml of buffer DK without NP-40, all at 4°C. The resin was resuspended in 1 ml of buffer DK, equally divided into two Eppendorf tubes. After the removal of supernatant, one tube was incubated with 1 ml of 2 mM ATP in buffer DK at 25°C for 20 min, repeated once, to remove Prp43. Both tubes were then incubated at room temperature for 5 min with 0.1 ml of 0.2 mM HA-peptide diluted in buffer DK to elute bound materials. The elution step was repeated five times, and individual fractions were collected.
Gradient sedimentation.
Splicing reactions or released intron fractions were subjected to sedimentation analysis on 10 to 30% glycerol gradients in a buffer containing 20 mM HEPES, pH 7.9, 100 mM NaCl, and 0.2 mM EDTA. Gradients were centrifuged in an SW60 rotor at 50,000 rpm for 3 h at 4°C, and collected in 0.25-ml fractions.
Yeast two-hybrid assays.
The NTR1, NTR2, and BRR2 genes were fused to the LexA-DNA binding domain in plasmid pEG202 and the GAL4 activation domain in plasmid pACT2, and each pair of plasmids were transformed into S. cerevisiae strain EGY48 together with the β-galactosidase reporter plasmid pSH18-34. Interactions were examined for expression of β-galactosidase on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates.
RESULTS
ATP-independent binding of NTR complex to the spliceosome.
We have previously demonstrated that disassembly of the postcatalytic spliceosome mediated by NTR complex requires hydrolysis of ATP (37). It was conceivable that the disassembly requires energy to disassociate U2, U5, U6, and NTC from lariat intron and from each other, but whether binding of NTR complex to the spliceosome also requires ATP remained unknown. An assay to examine whether ATP was required for binding of NTR was set up as outlined in Fig. 1A. Splicing in an NTR complex-depleted extract was carried out by using a polyclonal antibody against Ntr1. Glucose was then added to a final concentration of 2%, and the reaction mixture was incubated for a further 10 min, followed by the addition of the affinity-purified NTR complex, on which Ntr1 was tagged with HA and Prp43 was tagged with V5 (37). In the control experiment, 2 mM of ATP was added instead of glucose. After incubation for 10 min, reaction mixtures were precipitated with antibodies against Ntc20, Ntr1, Ntr2, and V5. As shown in Fig. 1B, prior depletion of NTR complex resulted in the accumulation of greater amounts of lariat intron (compare lanes 6 and 11 with lane 1). Nearly twice as much spliceosome accumulated, and was precipitated by the anti-Ntc20 antibody, when ATP was depleted prior to the addition of NTR complex (compare lane 7 and lane 12). Precipitation with antibody against Ntr1, Ntr2, or V5 revealed that greater amounts of lariat intron were associated with NTR in extracts in which NTR was first depleted then readded after splicing (lanes 8 to 10 and 13 to 15), particularly when ATP was exhausted after the splicing reaction (lanes 13 to 15). These results suggest that binding of NTR to the spliceosome is ATP independent and that ATP facilitates its dissociation, preventing stable association of NTR with lariat intron.
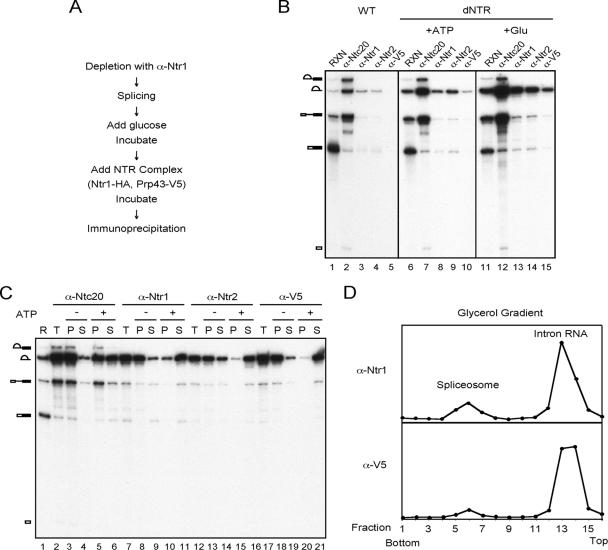
FIG. 1.
ATP-independent binding of NTR complex to the spliceosome. α-Ntr1, anti-Ntr1 antibody. (A) Scheme of assaying binding of NTR complex to the spliceosome. (B) Splicing reactions were carried out in wild-type (lanes 1 to 5) or Ntr1-depleted extracts (lanes 6 to 15) at 25°C for 30 min, then ATP (lanes 6 to 10) or glucose (lanes 11 to 15) was added to a final concentration of 2 mM or 2%, respectively, and the mixtures were further incubated for 10 min. Affinity-purified NTR complex was then added to the reaction mixtures and incubated for a further 10 minutes. The reaction mixtures were then precipitated with anti-Ntc20, anti-Ntr1, anti-Ntr2, or anti-V5 antibody, and RNA was isolated and analyzed by electrophoresis on 8% polyacrylamide-8 M urea gels. dNTR, Ntr1-depleted extracts; RXN, reaction. (C) Precipitates described for panel B (lanes 2, 7, 12, and 17) were incubated at 25°C for 5 min in the presence (lanes 5, 6, 10, 11, 15, 16, 20, and 21) or absence (lanes 3, 4, 8, 9, 13, 14, 18, and 19) of ATP, and supernatant and pellet fractions were collected. RNA was isolated and analyzed by electrophoresis on 8% polyacrylamide-8 M urea gels. R, reaction; T, total precipitate; P, pellet; S, supernatant. (D) Supernatant fractions shown in lanes 11 and 21 in panel C were sedimented on 10 to 30% glycerol gradients.
To confirm that ATP is required for spliceosome disassembly after binding of the NTR complex, ATP was added to the spliceosome precipitated with the antibodies described above (Fig. 1B, lanes 12 to 15), and the mixture was reincubated at 25°C for 10 min to see whether lariat intron could dissociate. As shown in Fig. 1C, only small amounts of lariat intron were released from beads when incubated in the absence of ATP (lanes 4, 9, 14, and 19). For lane 14, slightly more lariat intron was released when it was coprecipitated with Ntr2, possibly due to destabilization of Ntr2 from the spliceosome upon antibody binding (see below). In the presence of ATP, while approximately 50% of lariat intron that coprecipitated with Ntc20 was dissociated (Fig. 1C, lanes 5 and 6), the majority of the lariat intron coprecipitated with Ntr1 (lanes 11 and 12), Ntr2 (lanes 15 and 16), and Prp43 (lanes 20 and 21) was dissociated, indicating that ATP is required for the dissociation of lariat intron from these proteins. Sedimentation analysis of dissociated fractions from the spliceosome coprecipitated with Ntr1 or Prp43 on 10 to 30% glycerol gradients revealed that lariat intron was released as naked RNA (Fig. 1D) (37), suggesting complete disassembly of the spliceosome. Together, these results indicate that NTR does not require ATP for its binding to the spliceosome, and once bound, it could readily disassemble the spliceosome upon hydrolysis of ATP.
Prp43 could be recruited to the spliceosome by the Ntr1-Ntr2 complex.
In splicing extracts, a fraction of Ntr1 and Ntr2 is associated in a complex free of Prp43 (37). This raised the question of whether Ntr1-Ntr2 could also bind to the spliceosome without Prp43. To answer this, Prp43 was depleted from extracts prepared from Prp43-V5-containing cells by using anti-V5 antibody. Splicing reactions were carried out in Prp43-depleted extracts, followed by precipitation with antibodies against Ntc20, Ntr1, Ntr2, and V5 (Fig. 2A). While lariat intron was present only in small amounts in mock-depleted extracts (lane 1), a large amount of lariat intron accumulated in Prp43-depleted extracts (lane 6) and was coprecipitated with Ntr1 and Ntr2 (lanes 8 and 9) but not significantly with Prp43 (lane 10). This indicates that Ntr1 and Ntr2 can bind to the spliceosome in the absence of Prp43.
FIG. 2.
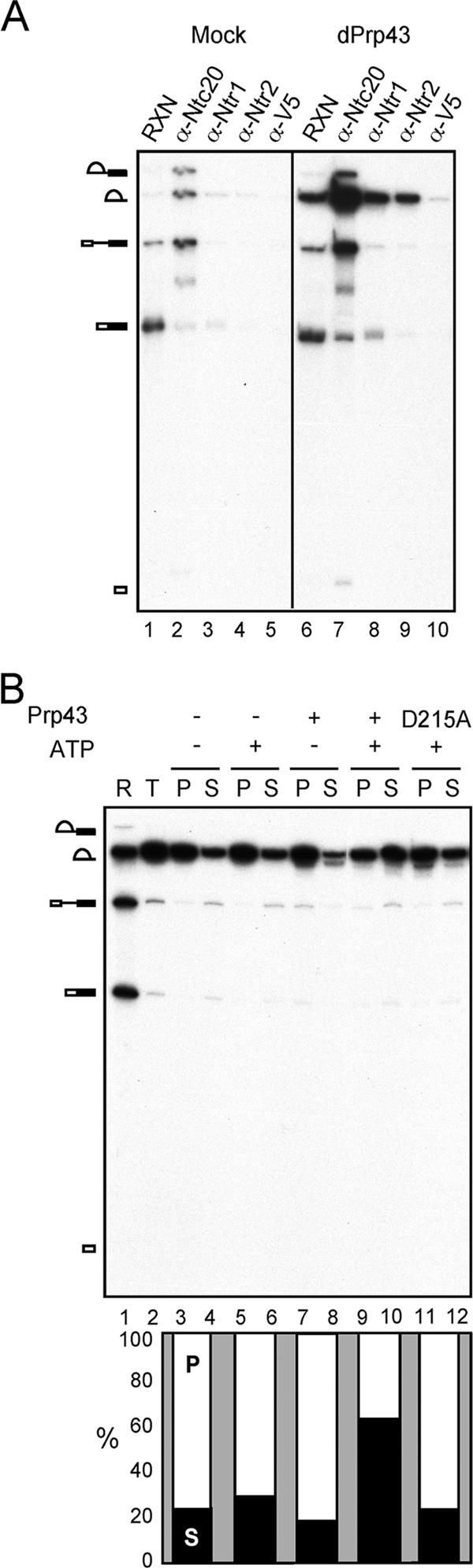
Ntr1 and Ntr2 could bind to the spliceosome independently of Prp43. (A) Splicing reactions were carried out in mock-depleted (lane 1 to 5) or Prp43-depleted Prp43-V5 extracts (lanes 6 to 10) and then precipitated with anti-Ntc20 (α-Ntc20), anti-Ntr1 (α-Ntr1), anti-Ntr2 (α-Ntr2), or anti-V5 (α-V5) antibody. RNA was isolated and analyzed by electrophoresis on 8% polyacrylaminde-8 M urea gels. dPrp43, Prp43-depleted extracts; RXN, reaction. (B) Precipitates with anti-Ntr1 antibody shown in lane 8 of panel A were incubated with (lanes 7, 8, 9, and 10) or without (lanes 3 to 6) Prp43 or with the D215A mutant of Prp43 (lanes 11 and 12) in the presence (lanes 5, 6, and 9 to 12) or absence (lanes 3, 4, 7, and 8) of ATP. Supernatant and pellet fractions were collected, and RNA was isolated and analyzed by electrophoresis on 8% polyacrylamide-8 M urea gels. Percentages of supernatant and pellet fractions are also presented in a bar graph with the sum of supernatant and pellet as 100%. R, reaction; T, total precipitate; P, pellet; S, supernatant; D215A, D215A mutant Prp43 protein.
We have previously demonstrated that in splicing extracts, there is much more Prp43 present than Ntr1 and Ntr2, and the majority of Prp43 is not in complex with Ntr1-Ntr2 (37). Since the Ntr1-Ntr2 complex could bind to the spliceosome, it is possible that following binding, Ntr1-Ntr2 might be able to recruit the free form of Prp43 to catalyze disassembly. This was examined by adding recombinant Prp43 to the spliceosome precipitated by the anti-Ntr1 antibody and seeing whether the spliceosome would undergo disassembly. As shown in Fig. 2B, upon addition of Prp43, lariat intron was released from beads in an ATP-dependent manner (lanes 9 and 10), whereas the ATPase mutant of Prp43 (D215A) was not functional in releasing lariat intron (lanes 11 and 12), as can be more clearly demonstrated with a bar graph. These results suggest that Ntr1-Ntr2 bound to the spliceosome is capable of recruiting Prp43 to catalyze spliceosome disassembly through hydrolysis of ATP. Thus, Ntr1-Ntr2 could associate with Prp43 to form NTR complex to bind to the spliceosome or bind to the spliceosome first and then recruit Prp43, which is the catalytic component for spliceosome disassembly.
Prp43 could be recruited to the spliceosome by the Ntr1-Ntr2 complex.
The fact that NTR could bind to the spliceosome in two modes, as the complete NTR or as Ntr1-Ntr2 complex, followed by recruitment of Prp43, suggests that the interaction between Ntr1-Ntr2 and Prp43 might be dynamic. As shown in Fig. 3A, when NTR complex was precipitated with anti-HA antibody by using Ntr1-HA extracts and then reincubated in buffer DK (see Materials and Methods) at 25°C for 20 min, Ntr1 and Ntr2 remained stably associated, whereas about 40% of Prp43 was dissociated when incubated in the absence of ATP (lanes 2 and 3), and approximately 65% was dissociated when incubated in the presence of 2 mM ATP (lanes 4 and 5), suggesting that the association of Prp43 with Ntr1 is not as stable as that of Ntr2 with Ntr1. This provides a means for the isolation of Ntr1-Ntr2 complex free of Prp43. The Ntr1-HA extract was fractionated by chromatography on anti-HA antibody-conjugated protein A-Sepharose column in the same way as for isolating NTR complex, except that after washing the column, the resin was transferred to an Eppendorf tube and incubated at 25°C for 20 min in 200 times the volume of buffer DK containing 2 mM ATP and repeated twice before elution. As shown in Fig. 3B, fractions eluted after preincubation contained no Prp43 (lanes 3 to 7). The purified Ntr1-Ntr2 complex, when added to NTR-depleted extracts, could reassociate with Prp43, as demonstrated by precipitation of the mixture with anti-Ntr1 antibody, in the presence or absence of ATP as shown in Fig. 3C, further confirming dynamic interactions between Ntr1-Ntr2 and Prp43.
FIG. 3.
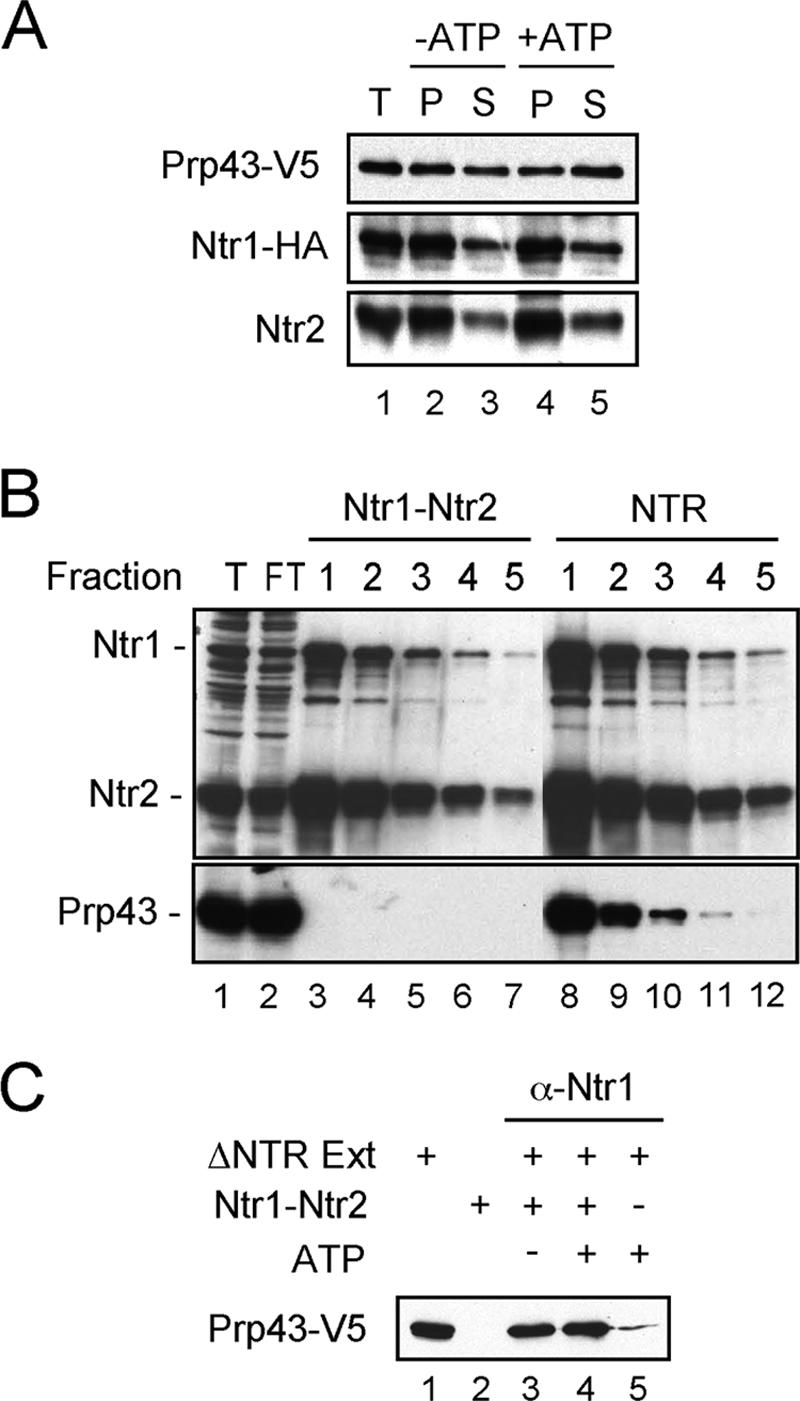
Dynamic interactions of Prp43 with Ntr1-Ntr2. (A) Extracts prepared from Ntr1-HA- and Prp43-V5-tagged strains were precipitated with anti-HA antibody (lane 1). The precipitates were then incubated in buffer DK alone (lanes 2 and 3) or with 2 mM ATP (lanes 4 and 5) at 25°C for 20 min, and supernatant and pellet fractions were collected for Western blotting, probing with antibodies against V5, Ntr1, and Ntr2. T, total precipitate; P, pellet; S, supernatant. (B) Extracts prepared from Ntr1-HA-tagged strain were fractionated on an anti-HA antibody-conjugated protein A-Sepharose column. After being washed, the beads were divided into two aliquots and transferred to Eppendorf tubes. One tube was incubated with buffer DK containing 2 mM ATP at room temperature for 20 min, and this was repeated once. Both tubes were then eluted with the HA-tagged peptide, and elution was repeated four times. Individual fractions were then Western blotted with antibodies against Ntr1, Ntr2, and Prp43. T, total precipitate; FT, flowthrough. (C) Ntr1-depleted extracts (ΔNTR Ext), prepared from Ntr1-HA- and Prp43-V5-tagged strains, were supplemented with affinity-purified Ntr1-Ntr2 complex and immunoprecipitated with anti-Ntr1 antibody (α-Ntr1) following incubation in the presence or absence of ATP. The precipitate was analyzed by Western blotting, probing with anti-V5 antibody.
Association of NTR complex with U5 snRNP.
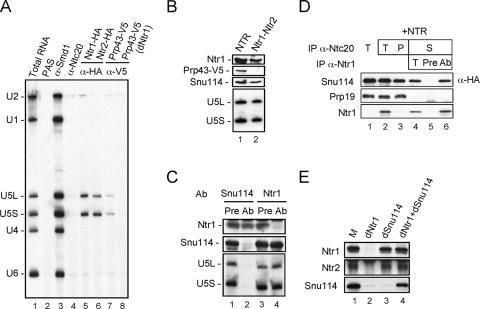
Prp43 was previously demonstrated to associate with small amounts of U2 and U6 but was demonstrated to have a much greater association with U5 (10, 15, 16). We examined whether Ntr1 and Ntr2 are also associated with U5 by immunoprecipitation of Ntr1-HA or Ntr2-HA extracts with anti-HA antibody. As shown in Fig. 4A, a small but significant amount of U5 snRNA was coprecipitated (lanes 5 and 6), suggesting that the Ntr1-Ntr2 complex is associated with U5. Precipitation of Prp43-V5 extracts with anti-V5 antibody also coprecipitated a small amount of U5 (lane 7). However, if the extract was previously depleted of NTR with anti-Ntr1 antibody, the anti-V5 antibody no longer coprecipitated U5 (Fig. 4A, lane 8) despite the fact that the majority of Prp43 was still present in the extract (37). This implies that the association of U5 with Prp43 is mediated through the association of U5 with the Ntr1-Ntr2 complex. Moreover, the association of U5 with Ntr1-Ntr2 was independent of Prp43, since U5 and its associated component Snu114 could be detected in the purified Ntr1-Ntr2 complex, as shown in Fig. 4B. Depletion analysis further revealed that only small amounts of U5 and Ntr1-Ntr2 are associated with each other, as depletion of Ntr1 did not significantly reduce the amount of U5 or Snu114 (Fig. 4C, lane 4), and depletion of Snu114 did not codeplete much Ntr1 (lane 2).
FIG. 4.
Association of NTR complex with U5 snRNP. (A) Extracts prepared from Ntr1-HA-, Ntr2-HA-, or Prp43-V5-tagged strains were precipitated with the anti-HA (α-HA) (lanes 5 and 6) or anti-V5 (α-V5) (lanes 7 and 8) antibody, and precipitates were analyzed by Northern blotting. Prp43-V5 extracts from which Ntr1 was depleted were also precipitated with the anti-V5 antibody (lane 8). Ntr1-HA extracts were precipitated with the anti-Smd1 (lane 3) or anti-Ntc20 (lane 4) antibody as controls. PAS, protein A-Sepharose. (B) Western and Northern blotting of affinity-purified NTR (lane 1) and Ntr1-Ntr2 complex (lane 2), probing with antibodies against Ntr1, V5, and Snu114 and with U5, respectively. (C) Western and Northern blotting of total extracts depleted with pre-Snu114 (lane 1), anti-Snu114 (lane 2), pre-Ntr1 (lane 3), or anti-Ntr1 (lane 4) serum, probing for Ntr1, Snu114, and U5. Pre, preimmune serum; Ab, antibody. (D) Splicing was carried out in Ntr1-depleted Snu114-3×HA extracts, and the reaction mixture was precipitated with the anti-Ntc20 antibody. The precipitate (lane 1) was then added to the affinity-purified NTR complex (lane 2), which contained untagged Snu114, and the supernatant (lane 4) was further precipitated with the pre-Ntr1 (lane 5) or anti-Ntr1 (lane 6) serum. (E) Snu114-3×HA mock-depleted extracts (M) (lane 1), Ntr1-depleted extracts (dNtr1) (lane 2), Snu114-depleted extracts (dSnu114) (lane 3), and the mixture of Ntr1- and Snu114-depleted extracts (dNtr1+dSnu114) were immunoprecipitated with the anti-Ntr1 antibody, followed by Western blotting, probing with anti-Ntr1 and anti-HA antibodies.
Since U4 or U6 was not efficiently coprecipitated with NTR, it can be presumed that NTR is associated with U5 but not with the tri-snRNP. A possible explanation for NTR-U5 association is that the NTR-U5 complex represents a disassembly intermediate of the spliceosome. We then examined whether spliceosome-bound U5 became associated with exogenously added NTR after spliceosome disassembly. Splicing was carried out in NTR-depleted extracts prepared from a strain in which Snu114 was tagged with three copies of the HA epitope, to accumulate large amounts of lariat intron, and the reaction mixture was precipitated with anti-Ntc20 antibody (Fig. 4D, lane 1). The NTR complex, affinity purified from untagged-Snu114 extracts, was then added to trigger spliceosome disassembly (Fig. 4D, lane 2). The association of U5 with NTR was examined by immunoprecipitation of the disassembled spliceosome in the supernatant fraction with anti-Ntr1 antibody (Fig. 4D, lane 4). There was a nearly quantitative coprecipitation of Snu114 with NTR (Fig. 4D, lane 6), strongly suggesting that U5 is associated with NTR either during spliceosome disassembly or after the spliceosome is disassembled, due to dynamic interaction between U5 and NTR.
Dynamic interaction between U5 and NTR was then examined by mixing extracts from which NTR was depleted and extracts from which U5 was depleted, followed by immunoprecipitation with anti-Ntr1 antibody, to see whether NTR and U5 could reassociate. U5 was depleted from the extract with anti-Snu114 antibody. Figure 4E shows that Snu114 was coprecipitated with Ntr1 in mock-depleted extracts (lane 1). There was no significant precipitation of Snu114 by anti-Ntr1 antibody when Ntr1 (Fig. 4E, lane 2) or Snu114 (Fig. 4E, lane 3) was depleted. However, when the two extracts were mixed together, Snu114 was coprecipitated with Ntr1 (Fig. 4E, lane 4), suggesting that the NTR-U5 interaction is dynamic and is independent of the splicing reaction.
Recruitment of NTR complex by U5 snRNP for spliceosome disassembly.
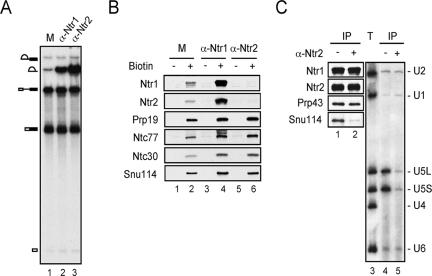
We speculated that dynamic interactions of NTR and U5 might play a role in the recruitment of NTR to the spliceosome and reasoned that in such a case, blocking the interaction between NTR and U5 would block binding of NTR to the spliceosome. Since Prp43 is not involved in binding of NTR to the spliceosome, we examined the requirement of Ntr1 and Ntr2 for the binding of NTR to the spliceosome by inhibition with antibodies against Ntr1 and Ntr2. As shown in Fig. 5A, preincubation of splicing extracts with either antibody resulted in the accumulation of lariat intron without affecting the splicing reaction, suggesting that both antibodies were effective in preventing NTR from functioning. To see whether the antibody blocks binding of NTR to the spliceosome, splicing reactions were carried out with biotinylated pre-mRNA so that the spliceosome could be isolated by precipitation with streptavidin agarose. As shown in Fig. 5B, anti-Ntr1 antibody did not inhibit the binding of Ntr1 or Ntr2 to the spliceosome (lane 4) but instead slightly increased the amount of the spliceosome accumulated (as judged by increasing amounts of Snu114 and NTC components associated with the spliceosome). Since the antibody was raised against an N-terminal fragment of the Ntr1 protein containing the G-patch domain required for interaction with Prp43 (37), it is speculated that the antibody precluded the Prp43 association, although this remains unproven, due to high background nonspecific binding of Prp43 to streptavidin-Sepharose. In contrast, preincubation with anti-Ntr2 antibody, raised against full-length protein, prevented binding of Ntr1 and Ntr2 to the spliceosome (Fig. 5B, lane 6), suggesting that Ntr2 might be involved in the binding of NTR to the spliceosome.
FIG. 5.
Antibody inhibition of splicing and the association of NTR with U5. (A) A 5-μl aliquot of extract was preincubated with 1.4 μg of anti-Ntr1 (lane 2) or 0.14 μg of anti-Ntr2 antibody (α-Ntr2) (lane 3) and used for splicing in a 10-μl reaction mixture. (B) Methods were as described for panel A, except splicing was carried out with five times the amounts of nonbiotinylated (lanes 1, 3, and 5) or biotinylated (lanes 2, 4, and 6) pre-mRNA, which was then precipitated with streptavidin-Sepharose followed by Western blotting. (C) Extracts were preincubated with (lanes 2 and 5) or without (lanes 1 and 4) anti-Ntr2 antibody and then precipitated with anti-Ntr1 antibody followed by Western (lanes 1 and 2) and Northern (lanes 3 to 5) blot analyses. M, mock-treated extracts; T, total RNA; IP, immunoprecipitation.
We then tested whether anti-Ntr2 antibody also inhibited interactions between NTR and U5. Extracts preincubated with anti-Ntr2 antibody were precipitated with purified anti-Ntr1 antibody conjugated to Sepharose, and the precipitate was analyzed for Snu114 and U5. As shown in Fig. 5C, preincubation with anti-Ntr2 antibody did not affect the association of Ntr2 or Prp43 with Ntr1 but greatly reduced the amount of Snu114 or U5 associated with Ntr1 (lanes 2 and 5), indicating that the anti-Ntr2 antibody interferes with the interaction between Ntr2 and U5 but not that between Ntr2 and Ntr1. This shows a strong correlation between the association of NTR and U5 snRNP and the binding of NTR to the spliceosome, suggesting that U5 might play a role in recruiting NTR to the spliceosome via an interaction with Ntr2.
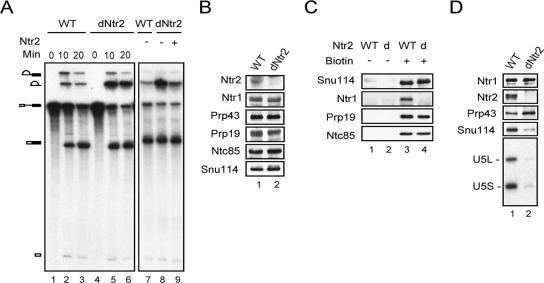
The role of Ntr2 in mediating the binding of NTR to the spliceosome was further confirmed by using an extract from which Ntr2 was depleted. We have previously shown that metabolic depletion of Ntr2 resulted in accumulation of pre-mRNA and lariat intron (37). Figure 6A shows that extracts prepared from such a strain grown in glucose-containing medium for 18 h also accumulated lariat intron (lanes 4 to 6) in the in vitro splicing reaction, in comparison with splicing in wild-type extracts (lanes 1 to 3). The addition of recombinant Ntr2 abolished the intron accumulation phenotype (Fig. 6A, lane 9), suggesting that the function of other NTR components was not affected in Ntr2-depleted extracts. Western blotting further revealed that depletion of Ntr2 did not greatly affect the amounts of Ntr1, Prp43, and NTC components Prp19 and Ntc85 in splicing extracts as shown in Fig. 6B. Using such an extract, we examined whether Ntr1 could bind to the spliceosome in the absence of Ntr2. Figure 6C shows that, while the binding of Snu114 or NTC components to the spliceosome was not affected, Ntr1 was not seen in the streptavidin-Sepharose pulled-down spliceosome (lane 4), suggesting a requirement of Ntr2 for binding of NTR to the spliceosome. Consistently, in the Ntr2-depleted extract, the amounts of Snu114 and U5 associated with Ntr1 (determined by immunoprecipitation with the anti-Ntr1 antibody) (Fig. 6D) were greatly reduced. Altogether, these findings strongly suggest that Ntr2 is required for the interaction of NTR with U5 for recruiting NTR to the spliceosome.
FIG. 6.
The association of NTR with U5 and with the spliceosomes formed in in vivo Ntr2-depleted extracts. (A) Splicing in wild-type (lanes 1 to 3 and 7) and in vivo Ntr2-depleted extracts alone (lanes 4 to 6 and 8) or with recombinant Ntr2 added (lane 9). (B) Western blotting of total extracts from wild-type (lane 1) and Ntr2-depleted (lane 2) strains. (C) Splicing in wild-type (lanes 1 and 3) or Ntr2-depleted (lanes 2 and 4) extracts by using nonbiotinylated (lanes 1 and 2) or biotinylated (lanes 3 and 4) pre-mRNA. Reaction mixtures were precipitated with streptavidin-Sepharose, followed by Western blot analysis of precipitates. (D) Wild-type (lane 1) or Ntr2-depleted (lane 2) extracts were immunoprecipitated with anti-Ntr1 antibody, and the precipitates were analyzed by Western blotting, probing for Ntr1, Ntr2, and Snu114, and by Northern blotting, probing for U5. WT, wild type; dNtr2 or d, in vivo Ntr2-depleted extract.
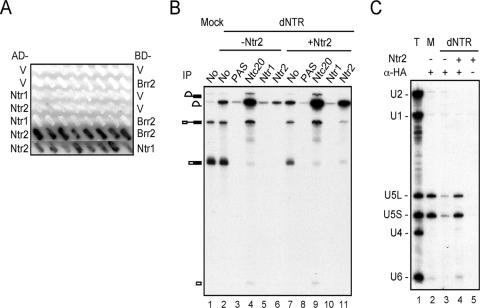
Ntr2 was previously shown to interact with Ntr1 but not with Prp43 in the NTR complex (37). No other known splicing factors are reported to interact with Ntr2. Among the U5 protein components, both Prp8 and Brr2 have been shown to interact with a wide range of splicing factors (18, 38), but their interactions with Ntr2 have not been reported. To identify protein components of U5 snRNP that interact with Ntr2, we examined the interactions of Ntr2 with the U5 proteins Prp8, Brr2, and Snu114. Two-hybrid assays shown in Fig. 7A reveal an interaction of Ntr2 with Brr2. Ntr2 does not interact with full-length Prp8 in such an assay (data not shown). The interaction with Snu114 could not be assessed, since both Snu114 and Ntr2 alone showed strong activation when fused to the DNA binding domain (data not shown). Further attempts by far-Western blotting of anti-Snu114 precipitated U5 snRNP, probed with 35S-labeled Ntr2 revealed only a very weak interaction of Ntr2 with Snu114 (data not shown). We conclude that the interaction of NTR with U5 is primarily mediated through the interaction of Ntr2 with Brr2.
FIG. 7.
Interaction of Ntr2 with U5 and with the spliceosome. (A) Two-hybrid interactions of Ntr2 and Brr2. Ntr1 or Ntr2 was fused to the LexA DNA binding domain (LexA-BD) and Brr2 was fused to GAL4 activation domain (GAL4-AD) for two-hybrid assays using β-galactosidase as a reporter. (B) Immunoprecipitation (IP) of the spliceosome formed in Ntr1-depleted extracts without (lanes 2 to 6) or with the supplement of 0.1 μg of recombinant Ntr2 protein (lanes 7 to 11) with no antibody (lanes 3 and 8) or anti-Ntc20 (lanes 4 and 9), anti-Ntr1 (lanes 5 and 10), or anti-Ntr2 antibody (lanes 6 and 11). Lanes 1, 2, and 7 had 2 μl of splicing reaction mixtures; lanes 4 to 6 and 8 to 11 had immunoprecipitated materials from 20 μl of splicing reaction mixtures. PAS, protein A-Sepharose. (C) The Ntr1-depleted extract was incubated with recombinant Ntr2-HA prebound to the anti-HA antibody (α-HA) coupled to protein A-Sepharose (lane 4). Bound materials were analyzed by Northern blotting, probing with U1, U2, U4, U5, and U6. Lane 1, total RNA from 1 μl extract; lane 2, mock-depleted extract; lane 3, Ntr2 not added; lane 5, anti-HA antibody not included.
To see whether Ntr2 is able to bind to the spliceosome independently of Ntr1, recombinant Ntr2 was added to extracts from which NTR was depleted by anti-Ntr1 antibody and splicing was performed. The association of Ntr2 with the spliceosome was examined by immunoprecipitation with anti-Ntr2 antibody. Depletion of NTR, with or without the addition of Ntr2, resulted in the accumulation of lariat intron (Fig. 7B, lanes 2 and 7), suggesting the absence of functional NTR. Failure to precipitate the spliceosome with anti-Ntr1 antibody further confirmed the absence of Ntr1 in the spliceosome (Fig. 7B, lanes 5 and 10). While anti-Ntc20 antibody precipitated pre-mRNA, spliced intermediates, and lariat intron regardless of the addition of Ntr2 (Fig. 7B, lanes 4 and 9), anti-Ntr2 antibody preferentially precipitated lariat intron only when Ntr2 was added (lanes 6 and 11), indicating that Ntr2 can bind to the spliceosome in the absence of Ntr1. To see whether binding of free Ntr2 to the spliceosome is mediated through U5, NTR-depleted extract was immunoprecipitated with anti-HA antibody after the addition of recombinant HA-tagged Ntr2, and the precipitate was analyzed by Northern blotting, probing with snRNAs. Figure 7C shows that recombinant Ntr2 coprecipitated U5 in the absence of Ntr1, suggesting that Ntr2 alone is sufficient to interact with U5. Taken together, these results demonstrate that Ntr2 is the key player mediating the interaction of NTR and U5 for the recruitment of NTR to the spliceosome.
DISCUSSION
We have previously shown that Ntr1 and Ntr2 form a stable complex, which further interacts with Prp43 to form the NTR complex through the interaction of Ntr1 and Prp43. The NTR complex is functional in mediating the disassembly of the lariat intron-containing spliceosome (37). We demonstrate here that the interaction between Prp43 and the Ntr1-Ntr2 complex is dynamic, and Prp43 could readily dissociate from Ntr1-Ntr2 upon dilution of the complex. This provides an explanation for the presence of a fraction of Ntr1-Ntr2 complex in the splicing extract, considering Prp43 is in great excess.
In the absence of Prp43, the Ntr1-Ntr2 complex could also bind to the spliceosome and recruit Prp43 to the spliceosome upon its addition. Neither the binding of Ntr1-Ntr2 nor the binding of Prp43 to the spliceosome requires ATP. ATP is required only for the disassembly reaction. Specifically, hydrolysis of ATP by Prp43 is required, because the D215A ATPase mutant of Prp43 is not functional in catalyzing disassembly of the Ntr1-associated spliceosome. Similar scenarios have been shown for Prp2 and Prp16, for which ATP is required only for their function in mediating conformational rearrangements of the spliceosome but not for their binding to the spliceosome (13, 28). Prp2 has been shown to interact with the G-patch domain-containing protein Spp2 to promote the step one reaction (26, 30). Spp2 is required for the function of Prp2 and presumably plays a role in the recruitment of Prp2 to the spliceosome similar to the function of Ntr1-Ntr2 in recruiting Prp43.
In addition to dynamic interactions with Prp43, we have shown here that the Ntr1-Ntr2 complex also interacts with U5 snRNP in a dynamic manner. A small fraction of U5 coprecipitated with Ntr1, Ntr2, and Prp43 from splicing extracts. Mixing of U5-depleted extracts and NTR-depleted extracts resulted in reassociation of U5 and NTR. Coprecipitation of U5 with Prp43 has previously been demonstrated (10, 16). We show that Prp43 does not directly interact with U5 and is associated with U5 through its association with Ntr1-Ntr2. It has also been reported recently that both Ntr1 and Ntr2 coprecipitated small amounts of U2, U5, and U6 by using TAP-tagged Ntr1 and Ntr2 extracts (2). Nevertheless, the fraction of U5 coprecipitated from the extract was greater than that of U2 or U6. Coprecipitation of amounts of U2 and U6 in the reported experiment larger than those in ours might be due to higher levels of the endogenous spliceosome in their extracts.
Proteomic analyses have revealed that protein complexes are widely present in the cell. However, the stoichiometry of the components in individual complex could not be assessed. A recent analysis of Yju2 protein has revealed its function in promoting the first catalytic step of splicing, despite its association with NTC, which is involved in spliceosome activation (19). Yju2 also interacts with NTC in a dynamic manner and may be recruited to the spliceosome by NTC through such dynamic interactions. In view of this, dynamic interactions might represent a significant portion of interactions in the proteomic database. Furthermore, dynamic interactions between protein components might be a general property for the recruitment of components to macromolecular complexes in various cellular pathways.
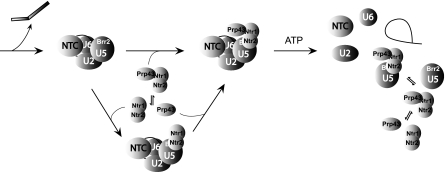
Several lines of evidence suggest that the association of Ntr1-Ntr2 with U5 is mediated through the interaction of Ntr2 with U5 snRNP. First, polyclonal antibodies against Ntr2 blocked the association of U5 with NTR complex as well as binding of NTR to the spliceosome. Second, genetic depletion of Ntr2 uncoupled the association of Ntr1 with U5 and with the spliceosome. Third, Ntr2 could bind to the spliceosome and also associate with U5 independently of Ntr1. Finally, Ntr2 interacts with U5 component Brr2 in two-hybrid assays. Thus, Ntr2 is essential and sufficient for the interaction of NTR with U5 and binding of NTR to the spliceosome, and such interactions are likely mediated through its interaction with Brr2. A diagram illustrating how dynamic interactions of Ntr1-Ntr2 with Prp43 and with U5 might mediate spliceosome disassembly is shown in Fig. 8.
FIG. 8.
A diagram illustrating the interactions of Ntr1-Ntr2 with Prp43 and with U5 to mediate spliceosome disassembly. Double arrows indicate equilibrium between association and dissociation forms.
Association of NTR with U5 but not with U4/U6.U5 tri-snRNP indicates that the interaction of U4/U6 prevents U5 from interacting with NTR. Protein-protein interactions within the human tri-snRNP have been analyzed in detail and demonstrate a key role of human Snu66 (hSnu66) and hPrp6 in bridging U4/U6 and U5 in the formation of tri-snRNP (17, 18). hPrp6 interacts with tri-snRNP-specific hSnu66 and U4/U6-specific hPrp3 and hPrp31 besides interacting with other U5 proteins and is required for tri-snRNP stability. Although hSnu66 also interacts with both U4/U6 and U5 proteins, it is not required for tri-snRNP stability (21). In view of this interaction network, Prp6 might play a role in modulating the structure of U5 snRNP for interactions with U4/U6 and with NTR.
NTR1 was also identified as SPP382 in a genetic screen for suppressors of prp38-1 mutation (24), which causes a temperature-sensitive growth defect and, in vitro, results in slow release of U1 and U4 during spliceosome activation (40). Mutant alleles of SPP382 show suppression of both prp38-1 and prp8-1, suggesting a link between NTR1 and U5. A mutant of AAR2 that encodes a U5 protein also acts as a prp38-1 suppressor, further supporting the U5-NTR linkage. Aar2 is specifically associated with U5 but not with the U4/U6.U5 tri-snRNP and has been shown to function as a recycling factor, possibly required for regeneration of the tri-snRNP during spliceosome cycling (11). Interestingly, several mutations in PRP43 affecting the ATPase activity also suppress the prp38-1 growth defect (24). These results suggest that reducing the rate of spliceosome cycling might partially compensate for impaired spliceosome assembly. The DEAH-box ATPases Prp16 and Prp22 have been demonstrated to play roles in modulating splicing fidelity by coupling ATP hydrolysis with a discard pathway (5, 23), but whether they are directly involved in disposing of the defective spliceosome is not known. Since NTR is recruited to the spliceosome by U5, it is conceivable that NTR can also bind to the spliceosome at earlier steps if spliceosome assembly is retarded. It will be of interest to see whether Prp43 can function to scavenge the impaired spliceosome in the discard pathway.
Proteomic analysis of mammalian splicing complexes has revealed the presence of a 35S, U5-containing particle in nuclear extracts. The complex also contains components of the Prp19-associated complex and other uncharacterized factors, many of which are present in the mature 45S spliceosome. It has been proposed that the 35S complex represents a disassembly intermediate of the spliceosome (20). Unlike in mammalian extracts, the U5-NTC complex has not been detected in yeast. Immunoprecipitation of NTC coprecipitated minute amounts of U2, U5, and U6 (Fig. 4A, lane 4), presumably representing the endogenous spliceosome, but has never coprecipitated a significantly greater amount of U5. In contrast, U5 was found to associate with NTR almost quantitatively in spliceosome disassembly assays (Fig. 5A). Although this suggests that U5 might be dissociated from the spliceosome in association with NTR, we cannot exclude the possibility that the association occurred after the disassembly due to dynamic interactions of U5 and NTR. Nevertheless, mammalian and yeast spliceosomes might undergo disassembly via distinct mechanisms.
Acknowledgments
We thank P. Lin for reading the manuscript and Harry Wilson for English editing.
This work was supported by Academia Sinica and National Science Council (Taiwan) grant NSC94-2321-B-001-021.
Footnotes
Published ahead of print on 24 September 2007.
REFERENCES
- 1.Arenas, J. E., and J. N. Abelson. 1997. Prp43: an RNA helicase-like factor involved in spliceosome disassembly. Proc. Natl. Acad. Sci. USA 94:11798-11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boon, K., T. Auchynnikava, G. Edwalds-Gilbert, J. D. Barrass, A. P. Droop, C. Dez, and J. D. Beggs. 2006. Yeast Ntr1/Spp382 mediates Prp43 function in postspliceosomes. Mol. Cell. Biol. 26:6016-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brow, D. A. 2002. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36:333-360. [DOI] [PubMed] [Google Scholar]
- 4.Burge, C. B., T. H. Tuschl, and P. A. Sharp. 1999. Splicing of precursors to mRNAs by the spliceosome, p. 525-560. In R. F. Gesteland, T. R. Cech, and J. F. Atkins (ed.), The RNA world, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 5.Burgess, S. M., and C. Guthrie. 1993. A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell 73:1377-1392. [DOI] [PubMed] [Google Scholar]
- 6.Caruthers, J. M., and D. B. Mckay. 2002. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 12:123-133. [DOI] [PubMed] [Google Scholar]
- 7.Chan, S.-P., D.-I. Kao, W.-Y. Tsai, and S.-C. Cheng. 2003. The Prp19p-associated complex in spliceosome activation. Science 302:279-282. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, S.-C., and J. Abelson. 1987. Spliceosome assembly in yeast. Genes Dev. 1:1014-1027. [DOI] [PubMed] [Google Scholar]
- 9.Cheng, S.-C., A. Newman, R.-J. Lin, G. D. McFarland, and J. N. Abelson. 1990. Preparation and fractionation of yeast splicing extract. Methods Enzymol. 181:89-96. [DOI] [PubMed] [Google Scholar]
- 10.Combs, D. J., R. J. Nagel, J. Ares, M., and S. W. Stevens. 2006. Prp43p is a DEAH-Box spliceosome disassembly factor essential for ribosome biogenesis. Mol. Cell. Biol. 26:523-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottschalk, A., B. Kastner, R. Lührmann, and P. Fabrizio. 2001. The yeast U5 snRNP coisolated with the U1 snRNP has an unexpected protein composition and includes the splicing factor Aar2p. RNA 7:1554-1565. [PMC free article] [PubMed] [Google Scholar]
- 12.James, S., W. Tirmer, and B. Schwer. 2002. How Slu7 and Prp18 cooperate in the second step of yeast pre-mRNA splicing. RNA 8:1068-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, S.-H., and R.-J. Lin. 1996. Spliceosome activation by PRP2 ATPase prior to the first transesterification reaction of pre-mRNA splicing. Mol. Cell. Biol. 16:6810-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Last, R. L., J. R. Maddock, and J. L. J. Woolford. 1987. Evidence for related functions of the RNA genes of Saccharomyces cerevisiae. Genetics 117:619-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lebaron, S., C. Froment, M. Fromont-Racine, J.-C. Rain, B. Monsarrat, M. Caizergues-Ferrer, and Y. Henry. 2005. The splicing ATPase Prp43p is a component of multiple preribosomal particles. Mol. Cell. Biol. 25:9269-9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leeds, N. B., E. C. Small, S. L. Hiley, T. R. Hughes, and J. P. Staley. 2006. The splicing factor Prp43p, a DEAH box ATPase, functions in ribosome biogenesis. Mol. Cell. Biol. 26:513-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, S., P. Li, O. Dybkow, S. Nottrott, K. Hartmuth, R. Lührmann, T. Carlomagno, and M. C. Wahl. 2007. Binding of the human Prp31 Nop domain to a composite RNA-protein platform in U4 snRNP. Science 316:115-120. [DOI] [PubMed] [Google Scholar]
- 18.Liu, S., R. Rauhut, H. Vornlocher, and R. Lührmann. 2006. The network of protein-protein interactions within the human U4/U6.U5 tri-snRNP. RNA 12:1418-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, Y.-C., H.-C. Chen, N.-Y. Wu, and S.-C. Cheng. 2007. A novel splicing factor Yju2 is associated with NTC and acts after Prp2 in promoting the first catalytic reaction of pre-mRNA splicing. Mol. Cell. Biol. 27:5403-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makarov, E. M., O. V. Makarova, H. Urlaub, M. Gentzel, C. L. Will, M. Wilm, and R. Lührmann. 2002. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298:2205-2208. [DOI] [PubMed] [Google Scholar]
- 21.Makarova, O. V., E. M. Makarov, and R. Lührmann. 2001. The 65 and 110 kDa SR-related proteins of the U4/U6.U5 tri-snRNP are essential for the assembly of mature spliceosome. EMBO J. 20:2553-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin, A., S. Schneider, and B. Schwer. 2002. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J. Biol. Chem. 277:17743-17750. [DOI] [PubMed] [Google Scholar]
- 23.Mayas, R. M., H. Maita, and J. P. Staley. 2006. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat. Struct. Mol. Biol. 13:467-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pandit, S., B. Lynn, and B. C. Rymond. 2006. Inhibition of a spliceosome turnover pathway suppresses splicing defects. Proc. Natl. Acad. Sci. USA 103:13700-13705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raghunathan, P. L., and C. Guthrie. 1998. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr. Biol. 8:847-855. [DOI] [PubMed] [Google Scholar]
- 26.Roy, J., K. Kim, J. R. Maddock, J. G. Anthony, and J. L. J. Woolford. 1995. The final stages of spliceosome maturation require Spp2p that can interact with the DEAH box protein Prp2p and promote step 1 of splicing. RNA 1:375-390. [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider, B. L., W. Seufert, B. Steiner, Q. H. Yang, and A. B. Futcher. 1995. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast 11:1265-1274. [DOI] [PubMed] [Google Scholar]
- 28.Schwer, B., and C. Guthrie. 1992. A dominant negative mutation in a spliceosomal ATPase affects ATP hydrolysis but not binding to the spliceosome. Mol. Cell. Biol. 12:3540-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silverman, E., G. Edwalds-Gilbert, and R.-J. Lin. 2003. DExD/H-box proteins and their partners: helping RNA helicases unwind. Gene 312:1-16. [DOI] [PubMed] [Google Scholar]
- 30.Silverman, E. J., A. Maeda, J. Wei, P. Smith, J. D. Beggs, and R.-J. Lin. 2004. Interaction between a G-patch protein and a spliceosome DEXD/H-box ATPase that is critical for splicing. Mol. Cell. Biol. 24:10101-10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Small, E. C., S. R. Leggett, A. A. Winans, and J. P. Staley. 2006. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H Box ATPase. Mol. Cell 23:389-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Staley, J. P., and C. Guthrie. 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92:315-326. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka, N., and B. Schwer. 2005. Characterization of the NTPase, RNA-binding, and RNA helicase activities of the DEAH-box splicing factor Prp22. Biochemistry 44:9795-9803. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka, N., and B. Schwer. 2006. Mutations in PRP43 that uncouple RNA-dependent NTPase activity and pre-mRNA splicing function. Biochemistry 45:6510-6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanner, N. K., and P. Linder. 2001. DexD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell 8:251-262. [DOI] [PubMed] [Google Scholar]
- 36.Tarn, W.-Y., K.-R. Lee, and S.-C. Cheng. 1993. The yeast PRP19 protein is not tightly associated with small nuclear RNAs, but appears to associate with the spliceosome after binding of U2 to the pre-mRNA and prior to formation of the functional spliceosome. Mol. Cell. Biol. 13:1883-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai, R.-T., R.-H. Fu, F.-L. Yeh, C.-K. Tseng, Y.-C. Lin, Y.-H. Huang, and S.-C. Cheng. 2005. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev. 19:2991-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Nues, R. W., and J. D. Beggs. 2001. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics 157:1451-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, Y., and C. Guthrie. 1998. PRP16, a DEAH-box RNA helicase, is recruited to the spliceosome primarily via its nonconserved n-terminal domain. RNA 4:1216-1229. [PMC free article] [PubMed] [Google Scholar]
- 40.Xie, J., K. Beickman, E. Otte, and B. C. Rymond. 1998. Progression through the spliceosome cycle requires Prp38p function for U4/U6 snRNA dissociation. EMBO J. 17:2938-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]