Abstract
HLAMatchmaker is a structurally based matching program. Each HLA antigen is viewed as a string of epitopes represented by short sequences (triplets) involving polymorphic amino acid residues in antibody-accessible positions. HLAMatchmaker determines which triplets are different between donor and recipient and this algorithm is clinically useful in determining HLA mismatch acceptability. Triplets provide however an incomplete description of the HLA epitope repertoire and expanded criteria must be used including longer sequences and polymorphic residues in discontinuous positions. Such criteria should consider the structural basis of antibody-antigen interactions including contact areas and binding energy, the essence of antigenicity.
This report describes the development of a structurally defined HLA epitope repertoire based on stereochemical modeling of crystallized complexes of antibodies and different protein antigens. This analysis considered also data in the literature about contributions of amino acid residues to antigen-antibody binding energy. The results have led to the concept that HLA antigens like other antigenic proteins have structural epitopes consisting of 15–22 residues that constitute the binding face with alloantibody. Each structural epitope has a functional epitope of about 2–5 residues that dominate the strength and specificity of binding with antibody. The remaining residues of a structural epitope provide supplementary interactions that increase the stability of the antigen-antibody complex. Each functional epitope has one or more non-self residues and the term “eplet” is used to describe polymorphic HLA residues within 3.0–3.5 Ångstroms of a given sequence position on the molecular surface. Many eplets represent short linear sequences identical to those referred to as triplets but others have residues in discontinuous sequence positions that cluster together on the molecular surface. Serologically defined HLA determinants correspond well to eplets. The eplet version of HLAMatchmaker represents therefore a more complete repertoire of structurally defined HLA epitopes and provides a more detailed assessment of HLA compatibility.
Keywords: HLAMatchmaker, HLA, epitope structure, histocompatibility, eplet
INTRODUCTION
Humoral sensitization to human leukocyte antigens (HLA) represents a considerable barrier in organ transplantation. Increasing proportions of kidney transplant candidates have preformed HLA-specific antibodies that decrease the probability of finding a suitably matched donor and it is widely accepted that anti-HLA antibodies play an important role in acute and chronic rejection leading to graft failure.
A better understanding of the epitope structure of HLA antigens is important not only for the identification of HLA-specific antibodies but also will permit a more efficient, structurally based strategy to determine HLA compatibility. HLAMatchmaker is a matching program that considers the structural basis of epitopes on class I HLA antigens [1]. Each HLA antigen is viewed as a string of short sequences (triplets) involving polymorphic amino acid residues in antibody-accessible positions; they are considered key elements of epitopes that can induce the formation of specific antibodies. The patient’s HLA phenotype represents the repertoire of self-triplets and HLAMatchmaker determines for each mismatched HLA antigen, which triplets in corresponding sequence positions are different. HLAMatchmaker-based matching improves transplant outcome [2–6], and is useful in serum analysis and the identification of acceptable mismatches for alloimmunized kidney transplant candidates [7–16] and refractory thrombocytopenic patients requiring matched platelet transfusions [17, 18] The original version of HLAMatchmaker considers triplets, i.e. linear sequences of three residues at least one of which would be polymorphic [1]. This algorithm has been verified by observations that many serologically defined private and public epitopes correspond to triplets and that an HLAMatchmaker-based analysis of serum reactivity is useful in predicting of cross-match results with potential donors [8, 10, 12, 13, 16]. Recent studies on human anti-HLA monoclonal antibodies have however, indicated that HLA epitopes include additional polymorphic residues located nearby triplets on the molecular surface [19]. Moreover, certain serologically defined antigenic determinants do not have corresponding triplets. This experience suggests that the structural definition of epitopes should use expanded criteria including longer sequences and residues in discontinuous sequence positions. Such criteria should consider the structural basis of antibody-antigen interactions including contact areas and binding energy, the essence of antigenicity [20–23]. This report describes how these concepts can be applied to the HLAMatchmaker algorithm to define structural histocompatibility at the humoral immune level.
METHODS AND RESULTS
Structural Analysis Tools
Studies on complexes of protein antigens and antibody domains (Fab and Fv) have provided detailed stereochemical descriptions of antigen-antibody recognition, interactions and shape complementarity. The Entrez Molecular Modeling Database (MMDB) of the National Center for Biotechnology Information (NCBI) stores on its website (http://www.ncbi.nlm.nih.gov/Structure) an extensive collection of crystallographic structures of antibody-antigen complexes that can be viewed with the Cn3D structure and sequence alignment software program [24]. The atomic coordinates of these molecular complexes are stored as specific PDB codes in the Protein Data Bank. The Cn3D molecular viewer identifies the locations of selected residues and their exposure on the molecular surface. This determines the shapes of epitopes defined by clusters of residues in linear and discontinuous sequences. The Cn3D program has also a “select by distance” (in Ångstroms) command that permits an assessment of the sizes of epitopes and paratopes and the intermolecular distances between them.
Sequence differences between antigenic proteins and corresponding self-proteins of the antibody producer were determined on the website (http://www.ncbi.nlm.nih.gov/BLAST) with the Basic Local Alignment Search Tool (BLAST) [25]. The “space fill” command of the Cn3D molecular viewer was used to identify on antigenic proteins, surface-exposed residues as potential contact sites for antibody.
Determinations of epitope structures were based on experimental findings reported in the literature about binding energy in antigen-antibody complexes selected for this analysis. This information was then applied to develop structural models of functional epitopes with the Cn3D molecular viewer.
Structural Aspects of the Antigen-Antibody Interface
The specific reactivity of antibody is determined by about 50 hypervariable amino acid residues in two variable domains (VH and VL) of heavy and light chains. Both VH and VL domains display high sequence diversity in three complementarity-determining regions (CDRs) separated by relatively conserved intervals termed framework regions [26, 27]. These CDRs form loops that constitute the antigen-binding site: CDR-H3 and CDR-L3 lie generally in the center of the traditional antigen binding site and CDR-H1, CDR-H2, CDR-L1, and CDR-L2 form the outside border [28–34]. CDR-H3 exhibits by far the greatest sequence diversity and plays a dominant role in determining the specificity and affinity of antibody. The other CDRs adopt limited sets of main-chain conformations referred to as canonical structures [29, 35].
Crystallographic models of antigen-antibody complexes have shown that small numbers of residues in the CDR loops make contact with a protein antigen [33, 36, 37]. Conversely, protein antigens have small clusters of amino acid residues that constitute the contact sites for the CDR loops. The resulting interface on average involves about 15–22 amino acid residues of the protein antigen and a similar number of antibody residues. The overall contact area on antigen ranges from 650 to 900 Å2. In comparison, an HLA molecule seen from the top (i.e. binding groove) has a surface area of about 750 Å2.
Structural and Functional Definition of an Epitope
Considering this information about the structure of the antigen-antibody interface we must address the question what constitutes an epitope. In 1960, Niels Jerne coined the term epitope when he proposed that an antigen particle carries several epitopes [38]. Many epitopes are antigenic determinants expressed on the molecular surface of antigen [20–23]. Others are hidden epitopes (or cryptotopes) that become immunologically available after fragmentation or denaturation of antigen. Processed antigenic peptides presented by major histocompatibility complex molecules to T-cells belong to this group and should be considered cryptotopes rather than T-cell epitopes [39]. Two groups of protein epitopes have been proposed: continuous (or linear) epitopes involving a single continuous amino acid sequence and, discontinuous epitopes that comprise amino acids separated in the primary sequence but clustered together on the molecular surface by folding of the native protein [40, 41]. Mapping studies of antibody reactivity patterns with natural variants and mutated protein antigens have generated information about the location of epitopes and have also suggested that epitopes can generally be defined by small numbers of amino acid residues.
Stereochemical analyses of crystals of antigen-antibody complexes have led to a structural definition of an epitope as that part of the antigen that is contacted by the CDR loops of antibody [36]. This means that with about 15–22 contact residues, a structural epitope comprises a rather large area on the antigen surface and involves many amino acid residues that make contact with a large group of residues on CDRs collectively referred to as the paratope of antibody. Direct contact between epitope and paratope residues is established through electrostatic forces such as hydrogen bonds, salt bridges, van der Waals forces of hydrophobic surfaces and shape complementarity [20–23]. The interface has also bound water molecules or other co-factors that contribute to the specificity and affinity of antigen-antibody interactions [42].
The binding energy of an antigen-antibody complex is primarily mediated by a small subset of contact residues in the epitope-paratope interface [22, 43, 44]. Substitutions of such “energetic” residues [45] as seen in naturally occurring antigenic variants or induced by site-directed or alanine scanning mutagenesis lead often to dramatic decreases in the binding of antigen to antibody [46–48]. Mapping studies have located energetic residues in “hot spots” of epitopes and paratopes, i.e. regions made up of small numbers of residues that contribute most of the binding energy [49]. Energetic residues are often located in the center of the epitope-paratope interface [50]. Contact residues in periphery of the interface make generally minor contributions to the binding energy; their replacements have frequently little effect on the binding with antigen. The considerable flexibility of CDR3 loops allows a mutual adaptation of both epitopes and paratopes, making it possible for a single antibody molecule to interact with a large number of related antigens [51]. This concept helps to understand the structural basis of serological cross-reactivity.
Thus, the binding or functional activity of an epitope involves a small subset of energetic residues centrally located in the structural epitope and contacted by the specificity-determining CDRs [52]. The assignment of a functional epitope on an antigenic protein should consider two criteria. In order to be immunogenic, a functional epitope must have at least one non-self residue, i.e. the antibody producer’s homologous proteins must have a different residue in the corresponding sequence position. Such residue must be on the molecular surface so it can make contact with the specificity-determining CDR.
In the epitope-paratope interface, the energetic residues of a functional epitope are often in close contact with the energetic residues of the specificity-determining CDRs. The latter can be identified by site directed mutagenesis of CDR loops [50, 53, 67] and constitute what might be called the functional paratope. Although CDR-H3 plays often a dominant role, other CDRs with energetic residues may provide important contacts with epitope [33].
The question must be raised how many and which residues might define a functional epitope. Such information would be useful in the design of a model for structurally based HLA compatibility.
Topography of Functional Epitopes on Protein Antigens
There is an extensive literature on the structural analysis of crystallized antigen-antibody complexes. This report addresses only anti-protein antibodies that have well-characterized reactivity patterns with natural antigenic variants and mutated antigens. In addition, sequence information about the antibody producer’s own structurally similar proteins will permit an assessment addressing which residues of the antigenic protein are non-self.
The murine antibody response to HEL represents an excellent model to address the assignment of functional epitopes. A great variety of HEL-specific monoclonal antibodies (mAbs) have been tested with structurally defined, naturally occurring antigenic variants (from different avian species) and HEL variants produced by site-directed mutagenesis. These findings have generated information about serological cross-reactivity patterns between structurally related lysozymes and residues associated with serological determinants. Extensive studies of a few dozen crystallized HEL-antibody complexes have provided detailed information about the epitope-paratope interface and how residue substitutions can affect the interactions between antibody and antigen.
HEL is a 129-residue, antiparallel α-helical protein internally cross-linked by four disulfide bonds [53]. A BLAST analysis of HEL and two mouse lysozymes, type P intestinal [54] and type M milk [55], identifies 49 non-self residues on HEL (they are indicated with the superscript NS), 30 of them are visible on the molecular surface and would therefore be antibody-accessible. This rather large number of exposed non-self residues is consistent with the experimental evidence for multiple epitopes on HEL antigen [56–59].
Reactivity patterns with various avian lysozymes and mutated HEL antigens have shown that anti-HEL mAbs react with serological determinants in one of four distinct antigenic regions that cover most of the lysozyme surface [56, 59]. Detailed structural information has become available on antigen-antibody complexes with mAbs specific for epitopes representative of three regions. This study addressed the reactivity of five HEL-specific mAbs but this report describes only one example in some detail.
Reactivity patterns of HEL-D1.3 with natural lysozyme variants and mutated HEL have indicated that this mAb reacts with an epitope that includes the 117–121 sequence in one of the loops of HEL [59]. In the crystalline antigen-antibody complex, the CDRs of D1.3 make contact with 16 residues in the 18–27 and 117–125 sequence segments of HEL [60, 61]. The paratope of D1.3 has 9 VH and 8 VL contact residues. Alanine scanning has indicated that HEL residues 121Q NS, 125R, 124I and 119A dominate the energetics of binding with D1.3 [50, 62, 63]. Turkey egg lysozyme differs from HEL by one amino acid in the interface (121H versus 121QNS) and has a 400-fold lower affinity with D1.3 than HEL [58, 64]. Other lysozymes that differ at position 121 (Glutamine to predominantly histidine) are non-reactive suggesting that the non-self 121QNS residue plays a critical role in the binding with D1.3.
Thus, the functional D1.3 epitope can be defined by the sequence 119A, 121QNS, 124I and 125R. The functional paratope of D1.3 has five energetic residues; three of them, H100D, H101Y and H102R are on CDR-H3.
Cn3D modeling has shown that the four residues of the functional HEL-D1.3 epitope form a single patch that is well exposed on the HEL surface (Figure 1a). The centrally located 121QNS protrudes from the HEL surface. The other residues 119D, 124I and 125R are within 3.0 Å of 121QNS. The molecular configuration of the functional paratope has three residues H101Y, L92W and L32Y that are less than 3.5 Å away from the functional HEL-D1.3 epitope (Figure 1a).
Figure 1.
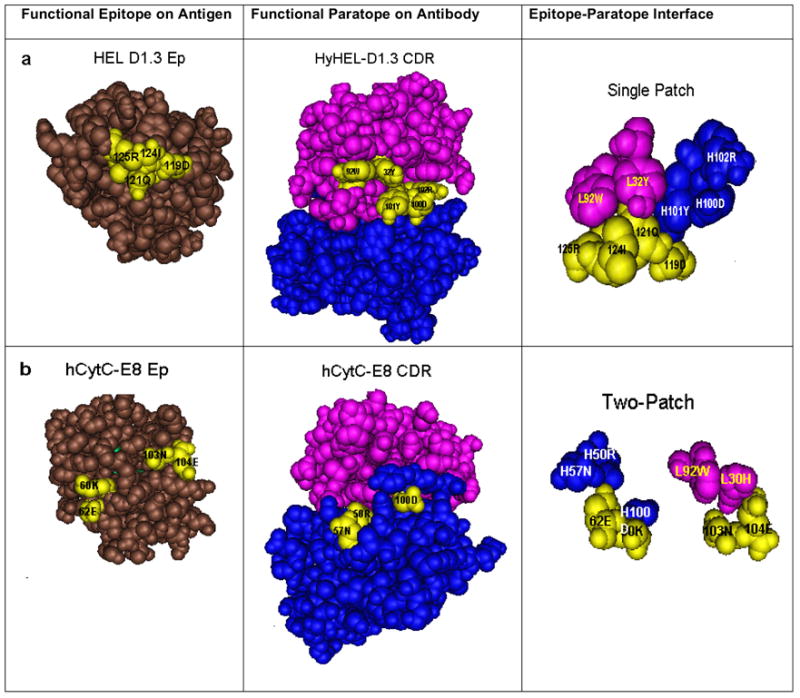
Single patch and two-patch shapes of energetic residues on antigen surface (functional epitope), antibody surface (functional paratope) and in the epitope-paratope interface
A similar analysis has been done of the reactivity of mAb E8 against horse cytochrome C (hCytC) [65]. Sequence comparison with mouse cytochrome C shows that this 105-residue antigenic protein has six non-self residues; all of them are exposed on the molecular surface and can elicit specific antibodies [20] [66]. Crystallographic analysis of the hCytC-FabE8 complex has shown that the structural E8 epitope constitutes a patch of ten residues located in three sequence segments of hCytC, namely 36F+37G, 60KNS+61E+62ENS and 96A+99K+100N+103D [67]. The two non-self residues 60KNS and 62ENS and are located in the center of the interface. Rat cytochrome C has an identical sequence as mouse cytochrome C and does not bind E8 [68]. Highly homologous cytochrome C from cow, rabbit, guanaco and dog, which have different residues in positions 60, 62 and/or 103, react poorly with E8 [68, 69]. A fourth and evolutionarily conserved residue 104E makes also a significant contribution to the binding with E8 [67]. Thus, the functional E8 epitope consists of 60KNS and 62ENS and two self-residues 103N and 104E. The E8 paratope has six VL and five VH residues in the CDR loops, five of which make contact with the functional epitope.[67].
Cn3D modeling has revealed that this functional epitope has two distinct patches (Figure 1b). One patch comprises 60KNS and 62ENS which are 3.3 Å apart and which make contact with VH residues H50N, H57N and H100D. The second pair consists of two self-residues 103N and 104E that interact with VL residues L30H and L92W. These two patches are well separated as shown by the 11.5 Å distance between 60KNS and 103N. The closest distance between the corresponding VH and VL contact residues is about 9.5 Å.
This type of analysis of functional epitope topography has been done for five additional mAbs. The results are summarized in Table 1 and more details are shown in a report on the HLAMatchmaker website [70]. In these seven crystal models, the structural epitopes consist of 10 to 21 amino acid residues in contiguous patterns on the protein surface. Small subsets ranging from 2 to 5 highly energetic contact residues in mostly central locations are considered to constitute the functional epitopes. These residues are largely in discontinuous sequences and at least one of them is non-self. This analysis has also shown that about 4–5 highly energetic antibody residues might define the functional paratopes (data not shown). As expected, the CDR-H3 loops play frequently but not always a major role and other CDRs are involved in the binding with functional epitopes.
Table 1.
Residue descriptions of interfaces of seven crystallized protein antigen-antibody complexes
| PDB | Antigen | Monoclonal Antibody | Number of Residues Structural Epitope | Residues in Functional Epitope 1 | Distances between Residues | Distance between Patches | References |
|---|---|---|---|---|---|---|---|
| Single | |||||||
| 1VFB | Hen Egg Lysozyme | D1.3 | 16 | 119A,121QNS, 1241, 125R | <3. 0–3.4 Å | - | [50,57–59,61,62, 64,95–97] |
| 3HFL | Hen Egg Lysozyme | HyHEL-5 | 13 | 45RNS,68RMS | 3.5 Å | [56,96,98–101] | |
| 1DQJ | Hen Egg Lysozyme | HyHEL-63 | 21 | 20Y,96Krls,97KNS | 0.0–3.5 Å | - | [102–104] |
| 1JRH | Interferon Receptor | A6 | 14 | 49YNS,40G,51VNS,52KNS,53NN | <3. 0–3.4 Å | - | [105–107] |
| Two- Patch | |||||||
| 1WEJ | Horse Cytochrome C | E8 | 10 | 60KNS,62ENS
103N, 104E |
3. 3 Å
<3.0 Å |
9.5 Å | [65–67,69,108, 109] |
| 1FBI | Hen Egg Lysozyme | F9.13.7 | 15 | 73RNS
20Y,96KHS |
- 3.1 Å | 13 Å | [110–112] |
| 1HFM | Hen Egg Lysozyme | HyHELIO | 20 | 93NNS,97KNS
20Y.21R |
3.3 Å
<3.0 Å |
7.5 Å | [100, 110,113–116] |
Functional epitope residues seem to cluster in two distinct shapes (Table 1). One reflects a single patch and there are four examples: HEL-D1.3, HyHEL-5, HyHEL-63 and IFNγR-A6. They are centrally located within the structural epitope (not shown) and their residues are 3–3.5 A apart from each other. Although a single patch consists of a short linear or discontinuous sequence of residues clustered closely together, it can make contact with several different CDR residues. These CDR residues might constitute the “functional” paratope that plays a determining role in the specific interaction with the functional epitope represented by a patch.
A functional epitope may also have a two-patch shape and there are two configurations. First, both patches have at least one non-self residue; hCytC-E8 and HEL-F9.13.7 are examples (Table 1). Second and illustrated by HyHEL-10, one patch has one (or more) non-self residues and the other patch consists of only self residues. The distance between patches ranges from 7.5 Å to 13 Å and within each patch the energetic residues are 3–3.5 Å apart from each other (Table 1).
Two-patch shape of HLA epitopes
Although most literature reports indicate that HLA epitopes correspond to single residues or clusters of few residues, the two-patch shape of a functional epitope may apply to some HLA epitopes. Our recent study on triplet-specific human monoclonal antibodies has shown that antibody binding depends on the presence of one or more additional residues shared between the immunizing antigen and the triplet-carrying reactive alleles [19]. For instance, the reactivity of an anti-62QE monoclonal antibody (mAb) requires the presence of a glycine residue in position 56. 62QE-carrying alleles are non-reactive if they have 56R. Similarly, the reactivity of two 142MI-specific mAbs require the presence of the GTLRG sequence in positions 79-83. These residues are located about 10–15 Å from these triplets and they appear to serve as critical contact sites for another CDR of antibody rather than the specificity-determining CDR. Interestingly, 56G and 79GTLRG are self-residues present in the HLA antigens of the antibody producer.
Absorption-elution analyses of allosera [71] and site-directed mutagenesis studies on class I HLA antigens [72–74] have identified additional residues that are critical for antibody-binding to epitope-defining residues. For instance the Bw6-specific antibody SFR8-B6 recognizes an epitope defined by 75R, 79R and 82R but its reactivity requires also the presence of the 90A residue which is about 10 Å away [74].
The two-patch shape may also explain the technique-dependency of certain alloantibodies that show negative reactions in complement-dependent cytotoxicity (CDC) but bind to HLA antigens, this refers to the cytotoxicity-negative, absorption positive (CYNAP) phenomenon. As an example, a multiparous woman was sensitized to HLA-A25 during pregnancy and her antibody had CDC reactivity with HLA-A25 but CYNAP activity with HLA-A26 [75]. These antigens are subtypes of HLA-A10 that carries a unique 150TAH triplet apparently recognized by this antibody. It seems likely that the binding energy of the specificity-determining CDR with 150TAH was not strong enough for the antibody to undergo the conformational change necessary for C1q binding, the first step of complement pathway leading to CDC. Apparently, another part of antigen must bind a second CDR so that sufficient energy is released for C1q activation. The only significant difference between HLA-A25 and HLA-A26 is in the 79-83 sequences, namely RIALR vs GTLRG. At a distance of about 14 Å from 150TAH, 79RIALR appears to be the critical contact site for the CDC activity of this antibody. This concept explains also the reactivity of antibodies with CDC reactivity against HLA-A26 but CYNAP activity with A25 [75].
Structural Aspects of Immune Complexes Involving HLA Molecules
No structural information is available about complexes of HLA antigens with alloantibodies, but Ziegler’s group has described the first crystallized antigen-antibody complex involving HLA (PDB code 1W72) [76]. They used a human monoclonal antibody specific for a melanoma-associated nona-peptide (MAGE-A1) bound to HLA-A1 and a high affinity Fab fragment (Hyb3) was used to generate the complex. Its binding interface comprises contact residues in eleven VH positions and four VL positions. The structural epitope involves four contact residues of the MAGE-A1 antigenic peptide and ten contact residues in the α-helices of HLA-A1. Hyb3 makes direct contact with MAGE-A1 via its CDR-H2 and CDR-H3 loops and with HLA-A1 via all three H chain CDR loops and CDR-L3. The contributions of CDR-L1 and CDR-L2 are only marginal.
The interactions between Hyb3 and MAGE-A1-HLA-A1 resemble those of T-cell receptor (TCR) with peptide-HLA [76]. This is not surprising because Hyb3 is specific for an HLA-restricted peptide. Similar to the CDRs of TCR α and β chains [20–23], Hyb3 uses its H and L chain CDRs to form an interface with the antigenic peptide and the α helices of HLA. The interface has however a different architecture in that Hyb3 has an angled orientation and contact the C-terminus rather than the center of peptide [76].
Although MAGE-A1 peptide residues 5T, 7H and 8S together with HLA-A1 α-helix residues 65R and 72Q appear to be important contact sites [76], there is insufficient information is about the functional epitope recognized by Hyb3.
Construction of a Structural Model for Functional Epitopes on HLA Class I Molecules
The above models of antigen-antibody complexes illustrate how structural epitopes contain patches of surface residues that play a dominant role in determining recognition by specific antibody. These residues cluster about 3–3.5 Å from each other. Non-self residues in these patches could be considered the driving force of functional epitopes in terms of immunogenicity (i.e. the ability to elicit antibody formation) and antigenicity (i.e. the ability to react with antibodies). The redesign of HLAMatchmaker incorporates these concepts. Instead of using triplets (i.e. linear sequences of three amino acid residues), the new algorithm considers patches of residues in linear and discontinuous sequences. These patches comprise residues clustered around polymorphic residues on the molecular surface.
Cn3D viewing of crystallized HLA molecular models stored on the NCBI website shows different patterns of surface polymorphisms for antigens encoded by HLA-A, HLA-B and HLA-C loci (Figure 2). The molecular surface around the bound peptide (see top view) has similar numbers of exposed polymorphic positions on the α1 helices of HLA-A and HLA-B antigens but more polymorphic positions are visible on the α2 helices of HLA-A antigens. The α helices of HLA-C antigens have much fewer polymorphic positions.
Figure 2.
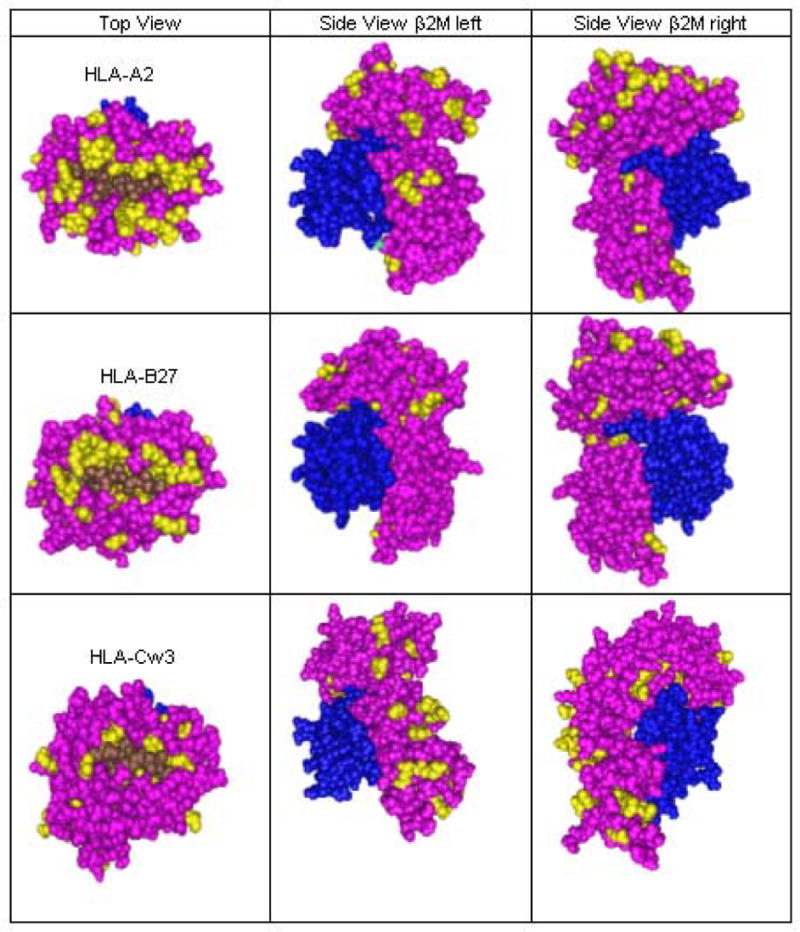
Polymorphic residues on class I molecules controlled by HLA-A, B and C loci
In contrast, HLA-C antigens have the most polymorphic positions in the membrane-proximal region, which becomes visible upon side viewing (Figure 2). HLA-A antigens have also more surface-exposed polymorphic positions in that region than HLA-B antigens. It should be noted that the sequence positions 200 to 275 of HLA-B antigens are all monomorphic.
Each surface-exposed polymorphic residue might be considered an essential element of a functional epitope that makes contact with the specificity-determining CDR of antibody. With reference to the findings with other protein antigens, one can expect that such residue might form a patch with other nearby residues. The “select by distance” command in Cn3D has been applied to identify residues around each exposed polymorphic position on representative class I molecules. Two distance parameters, 3.0 and 3.5 Å were chosen because as shown above, they provide the best estimate of the size of a functional epitope.
Sequence comparisons of the most common four-digit HLA class I alleles including all WHO-listed serological antigens have identified 75 polymorphic positions on the molecular surface, 47 of them involve one locus, 14 include two loci and 14 apply to all HLA-A, -B, -C loci (Table 2). Polymorphic positions are mostly on the top (N=25) or the side (N=36) of the molecule. Eight positions have “underside” locations (i.e. underneath the groove) and six are at the “bottom”, they become more readily visible if the molecule has been turned upside down. These positions seem less antibody-accessible if the HLA antigen is anchored in the cell membrane like in the lymphocytotoxicity test but they might react with antibody if the HLA molecule is fixed to a different surface like in a solubilized antigen-binding assay. Molecular surface expression of polymorphic residues has been graded as prominent (++), readily visible (+) and somewhat visible (−/+).
Table 2.
Polymorphic and monomorphic residue positions in 3.0 and 3.5 Angstrom patches of HLA class I antigens
| Sequence Position | Class 1 Locus | Molecular Location | Surface Exposure | Positions in 3.0 Angstrom Patches* | Positions in 3.5 Angstrom Patches* | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | C | side | ++ | 1 | 2 | 1 | 2 | 3 | ||||||||||
| 6 | C | side | + | 5 | 6 | 27 | 5 | 6 | 27 | |||||||||
| 9 | ABC | side | ± | 8 | 9 | 10 | 8 | 9 | 10 | 23 | ||||||||
| 12 | BC | underside | + | 11 | 12 | 13 | 21 | 11 | 12 | 13 | 21 | 92 | 94 | |||||
| 14 | C | side | + | 14 | 15 | 16 | 19 | 39 | 14 | 15 | 16 | 17 | 18 | 19 | 39 | |||
| 17 | A | side | ++ | 14 | 16 | 17 | 18 | 14 | 15 | 16 | 17 | 18 | ||||||
| 21 | C | underside | ± | 20 | 21 | 22 | 39 | 14 | 19 | 20 | 21 | 22 | 23 | 39 | ||||
| 30 | B | underside | + | 29 | 30 | 31 | 211 | 29 | 30 | 31 | 210 | 211 | ||||||
| 32 | B | underside | ± | 31 | 32 | 33 | 27 | 31 | 32 | 33 | 48 | |||||||
| 35 | C | side | ± | 35 | 36 | 48 | B53 | 35 | 36 | 46 | 48 | B53 | ||||||
| 41 | B | side | ++ | 40 | 41 | 42 | 40 | 41 | 42 | 43 | ||||||||
| 44 | A | side | ++ | 43 | 44 | 45 | 36 | 42 | 43 | 44 | 45 | 61 | ||||||
| 45 | B | side | + | 44 | 45 | 46 | 35 | 44 | 45 | 46 | 64 | |||||||
| 46 | B | side | + | 35 | 45 | 46 | 47 | 35 | 44 | 45 | 46 | 47 | 48 | |||||
| 56 | A | side | + | 55 | 56 | 57 | 59 | 54 | 55 | 56 | 57 | 58 | 59 | |||||
| 62 | A | top | ++ | 61 | 62 | 63 | 59 | 61 | 62 | 63 | 64 | 65 | 66 | |||||
| 63 | ABC | top | ± | 59 | 62 | 63 | 64 | P2 | 59 | 61 | 62 | 63 | 64 | 65 | 66 | P2 | ||
| 65 | ABC | top | ++ | 61 | 64 | 65 | 66 | 69 | 61 | 62 | 64 | 65 | 66 | 68 | 69 | |||
| 66 | ABC | top | ++ | 65 | 66 | 70 | P2 | 62 | 64 | 65 | 66 | 68 | 69 | 70 | P1 | P2 | ||
| 69 | B | top | ++ | 65 | 68 | 69 | 70 | 73 | 65 | 66 | 68 | 69 | 70 | 72 | 73 | |||
| 70 | ABC | top | + | 66 | 69 | 70 | 66 | 68 | 69 | 70 | 72 | 73 | ||||||
| 71 | B | side | + | 70 | 71 | 72 | 68 | 69 | 70 | 71 | 72 | 73 | 75 | |||||
| 73 | AC | top | + | 72 | 73 | 77 | 69 | 70 | 72 | 73 | 77 | |||||||
| 76 | A | top | ++ | 75 | 76 | 77 | 80 | 72 | 73 | 75 | 76 | 77 | 79 | 80 | P9 | |||
| 77 | ABC | top | ± | 73 | 76 | 77 | 78 | P8 | P9 | 73 | 75 | 76 | 77 | 78 | 79 | 80 | P8 | P9 |
| 79 | ABC | top | ++ | 78 | 79 | 80 | 76 | 78 | 79 | 80 | 82 | |||||||
| 80 | ABC | top | ++ | 76 | 79 | 80 | 84 | 76 | 78 | 79 | 80 | 82 | 83 | 84 | ||||
| 82 | AB | side | + | 80 | 82 | 86 | 87 | 79 | 80 | 82 | 83 | 86 | 87 | 88 | 89 | |||
| 83 | AB | top | ++ | 82 | 83 | 84 | 86 | 79 | 80 | 82 | 83 | 84 | 85 | 86 | ||||
| 90 | ABC | side | ++ | 89 | 90 | 91 | 88 | 89 | 90 | 91 | ||||||||
| 94 | BC | underside | ± | 93 | 94 | 95 | 119 | 93 | 94 | 95 | 118 | 119 | ||||||
| 103 | BC | side | ± | 2 | 103 | 104 | 2 | 103 | 104 | 108 | 110 | |||||||
| 105 | A | side | ++ | 104 | 105 | 106 | 1 | 104 | 105 | 106 | 107 | |||||||
| 107 | A | side | ++ | 106 | 107 | 106 | 169 | 105 | 106 | 107 | 108 | 169 | 173 | 180 | ||||
| 109 | A | side | + | 108 | 109 | 110 | 108 | 109 | 110 | 111 | 112 | |||||||
| 113 | BC | underside | + | 102 | 112 | 113 | 114 | 98 | 102 | 112 | 113 | 114 | ||||||
| 114 | ABC | underside | ± | 113 | 114 | 115 | 126 | 98 | 113 | 114 | 115 | 125 | 126 | |||||
| 116 | ABC | underside | ± | 115 | 116 | 123 | 124 | 115 | 116 | 123 | 124 | |||||||
| 127 | A | side | ++ | 127 | 128 | 132 | 127 | 128 | 129 | 132 | 133 | 134 | ||||||
| 131 | B | side | ++ | 129 | 130 | 131 | 132 | 129 | 130 | 131 | 132 | |||||||
| 138 | C | side | ++ | 137 | 138 | 139 | 141 | 137 | 138 | 139 | 140 | 141 | ||||||
| 142 | A | top | ++ | 138 | 141 | 142 | 138 | 139 | 141 | 142 | 144 | 145 | 146 | |||||
| 143 | B | top | ± | 142 | 143 | 144 | P9 | 141 | 142 | 143 | 144 | 145 | 146 | P9 | ||||
| 144 | A | side | + | 144 | 145 | 148 | 133 | 141 | 142 | 144 | 145 | 146 | 148 | |||||
| 145 | AB | top | ++ | 141 | 144 | 145 | 146 | 149 | 141 | 142 | 144 | 145 | 146 | 148 | 149 | |||
| 147 | BC | top | ± | 146 | 147 | 148 | 144 | 145 | 146 | 147 | 148 | 149 | 150 | 151 | P8 | |||
| 149 | A | top | ++ | 145 | 148 | 149 | 150 | 145 | 146 | 148 | 149 | 150 | 151 | |||||
| 150 | A | top | + | 149 | 150 | 151 | 146 | 148 | 149 | 150 | 151 | |||||||
| 151 | A | top | ++ | 150 | 151 | 152 | 148 | 149 | 150 | 151 | 154 | 155 | ||||||
| 152 | ABC | top | ± | 151 | 152 | 155 | 150 | 151 | 152 | 154 | 155 | P7 | ||||||
| 158 | AB | top | ++ | 157 | 158 | 159 | 154 | 155 | 157 | 158 | 159 | |||||||
| 161 | A | top | ++ | 157 | 161 | 162 | 157 | 159 | 161 | 162 | ||||||||
| 163 | A | top | + | 162 | 163 | 167 | 159 | 162 | 163 | 166 | 167 | P1 | ||||||
| 166 | A | top | ++ | 162 | 165 | 166 | 167 | 162 | 163 | 165 | 166 | 167 | 169 | 170 | ||||
| 167 | AB | top | ++ | 163 | 166 | 167 | 163 | 165 | 166 | 167 | 169 | 170 | ||||||
| 173 | C | side | ++ | 169 | 172 | 173 | 174 | 169 | 170 | 172 | 173 | 174 | 176 | |||||
| 177 | ABC | side | ++ | 176 | 177 | 178 | 176 | 177 | 178 | |||||||||
| 178 | B | side | + | 177 | 178 | 181 | 176 | 177 | 178 | 180 | 181 | |||||||
| 180 | B | side | ++ | 172 | 176 | 180 | 181 | 172 | 176 | 178 | 180 | 181 | ||||||
| 184 | AC | side | + | 183 | 184 | 185 | 183 | 184 | 185 | 266 | ||||||||
| 193 | AC | bottom | ++ | 192 | 193 | 194 | 192 | 193 | 194 | 198 | ||||||||
| 194 | ABC | side | + | 193 | 194 | 195 | 198 | 193 | 194 | 195 | 198 | 200 | ||||||
| 199 | B | bottom | + | 198 | 199 | 249 | 192 | 198 | 199 | 249 | ||||||||
| 207 | A | side | + | 206 | 207 | 208 | 206 | 207 | 208 | 240 | ||||||||
| 211 | C | side | ± | 210 | 211 | 212 | 30 | 208 | 210 | 211 | 212 | 213 | ||||||
| 219 | C | bottom | ++ | 219 | 220 | 222 | 219 | 220 | 221 | 222 | 224 | 256 | ||||||
| 246 | A | side | + | 229 | 245 | 246 | 247 | 228 | 229 | 245 | 246 | 247 | ||||||
| 248 | C | side | ++ | 247 | 248 | 249 | 247 | 248 | 249 | |||||||||
| 253 | AC | bottom | ++ | 251 | 252 | 253 | 256 | 250 | 251 | 252 | 253 | 256 | 257 | |||||
| 261 | C | side | + | 260 | 261 | 262 | 270 | 214 | 260 | 261 | 262 | 269 | 270 | |||||
| 267 | C | side | ++ | 266 | 267 | 268 | 266 | 267 | 268 | |||||||||
| 270 | C | side | + | 269 | 270 | 271 | 260 | 261 | 269 | 270 | 271 | |||||||
| 273 | C | side | ++ | 272 | 273 | 274 | 255 | 258 | 272 | 273 | 274 | |||||||
| 275 | C | bottom | ++ | 274 | 275 | 276 | 274 | 275 | 276 | |||||||||
| 276 | A | bottom | ++ | 275 | 276 | 278 | 274 | 275 | 276 | 278 | ||||||||
Polymorphic positions are marked in bold underlined font
Table 2 lists the sequence positions clustered within a 3.0 Å and 3.5 Å radius of each exposed polymorphic position. These patches are combinations of monomorphic and polymorphic positions; the latter are marked in bold, underlined font. The 3.0 Å patches have an average of 3.4 residues. Many of them correspond to triplets whereas others are slightly longer linear and discontinuous sequences. As expected, the 3.5 Å patches have more residues (average: 5.2), many of them have longer linear or discontinuous sequences. The vast majority of patches with multiple positions are in the 62–83 and 142–167 sequences of the α1 and α2 helices. This is not surprising because as Figure 4 illustrates, residue polymorphisms concentrate in the α helices. Moreover, α–helical structures have more residues in close proximity than β-sheets and the membrane-proximal domain. Many patches in α–helical regions share the same residue positions and this increases the likelihood of overlapping epitopes recognized by different antibodies. A few 3.0 Å and 3.5 Å patches in side locations comprise the same residue positions.
Several patches in the α helices include residues of peptides bound to the groove; their positions have the prefix P. For example, the 3.0 Å patch of position 66 has a partially exposed residue in peptide position 2 (P2) and the 3.0 Å patch of position 77 has two exposed peptide positions P8 and P9. Exposed peptide residues might contribute to the functional epitope recognized by alloantibody. Several studies have shown the influence of HLA-bound peptides on the reactivity of class I and class II specific antibodies [77–80]. The patterns listed in Table 2 might predict which HLA-specific antibodies might be peptide-dependent. Only the patches in position 32 include a β2-microglobulin position B53.
These findings on HLA patches are comparable to the functional epitopes reported for other protein antigens such the ones described in a previous section of this report. Especially, the 3.0 Å patches provide informative descriptions of HLA epitope structure. The polymorphic positions determine residue variability within each patch. The 3.0 Å patches have an average of 1.8 polymorphic positions (range 1–4) and 3.5 Å patches have between 1 and 6 polymorphic positions. Several patches have the same polymorphic positions although there differences between the monomorphic positions (e.g. the 3.0 Å patch in position 14 and the 3.0 Å and 3.5 Å patches in position 17 have same combination of polymorphic positions, namely 14 and 17). Such patches are considered equivalent.
The patch information in table 2 yielded 94 unique combinations of polymorphic positions and they appear to represent the basis of the complete repertoire of functional class I HLA epitopes. The residue composition of these polymorphic patches was determined with a Microsoft Excel macro developed by Grzegorz Dudek (Czestchowa University of Technology, Poland). This program is called HLA Patch Generator and can be downloaded from the HLAMatchmaker website http://tpis.upmc.edu
Eplet Version of HLAMatchmaker
An analysis of serologically defined HLA-A, B, C antigens and the more common 4-digit class I alleles has shown a total of 530 patches with different combinations of polymorphic residues. Most of them are in the α helix sequences 62–73 (N=192), 76–83 (N=91) and 142–152 (N=122) on the top of the HLA molecule and involve overlapping residues. The remaining 124 polymorphic patches are largely on the side of the molecule and include 39 at underside or bottom locations.
This rather extreme diversity and residue overlap among polymorphic patches especially in the α helices, might suggest that a structural definition of many functional epitopes might be too problematic. Further comparisons have shown however, that many overlapping polymorphic patches can be grouped together because they belong to a single antigen or a distinct group of antigens including those that are members of so-called cross-reacting groups (CREGs).
For example, the 62–70 sequence has seven overlapping patches shared by all six common HLA-A2 subtypes included in this analysis: A*0201, A*0202, A*0203, A*0205, A*0206 and A*0211 (Table 3). Two 3.0 Å patches (63GEK and 66ERKH) and three 3.5 Å patches (66GERK, 65GERKA and 66GERKAH) are unique to A2 and antibodies against any of them would be considered monospecific for A2. The 65RK patch is shared with A*3401 and the 62GE patch with B17 and antibodies against these patches would be considered specific for A2+A*3401 and A2+B17, respectively. 62GE readily explains the well-known serological cross-reactivity between A2 and B17 [81]. An immune response to an HLA-A2 mismatch might involve antibodies against all seven patches, except 65RK if the antibody producer types for A*3401 and 62GE if the antibody producer types for B17. Conversely, antibodies induced by a B17 mismatch might react with 62GE provided the antibody producer does not type for A2.
Table 3.
Examples of Antigen Groups with Multiple Patches in the 62–73 Sequence
| Alleles | 3.0 A Patches* | 3.5 A Patches* | Eplet |
|---|---|---|---|
| A*0201 A*0202 A*0203 A*0205 A*0206 A*0211 | 63GEK 66ERKH | 62GERK 65GERKA 66GERKAH | 63GEK |
| A*0201 A*0202 A*0203 A*0205 A*0206 A*0211 A*3401 | 65RK | 65RK | |
| A*0201 A*0202 A*0203 A*0205 A*0206 A*0211 B*5701 B*5703 B*5801 B*5802 | 62GE | 62GE | |
| B*5701 B*5703 B*5801 B*5802 | 63GEN | 62GERN 65GERNA 66GERNAS | 63GEN |
| B*1516 B*1517 B*5701 B*5703 B*5801 B*5802 | 66ERNS 69AS 70NASA 71SA | 69RNAST 70NASAT 71SAT 73ASATEN | 69AS |
| B*1516 B*1517 | 62RERN 65RERNA 66RERNAS | 65RN | |
| A*2301 A*2402 A*2403 A*2407 | 63EEK 65GK | 62EEGK 65EEGKA | 62EEK |
| A*2301 A*2402 A*2403 | 66EGKH | 66EEGKAH 69GKAHT | none |
| A*2407 | 66EGKQ | 66EEGKAQ 69GKAQT 73AQSTEN | none |
| A1,3,11,30,31,32,36,74 | 62QE 63QEN | 62QERN 65QERNA | 62QE |
| B7,22,42,67,81,82,83 | 66NQIQ 70IAQA | 66RNQIAQ 69QIAQT 70IAQAT 73AQATES | 70QA |
The number indicates the sequence position of the exposed polymorphic residue and the letters represent polymorphic residues in the corresponding patch as listed in Table 2
The common B17 subtypes B*5701, B*5703, B*5801 and B*5802 have four unique overlapping patches: 63GEN, 62GERN, 65GERNA and 66GERNAS (Table 3). An antibody against any of them would be considered monospecific for B17. Interestingly, an antibody against 62GE will react with A2+B17 whereas an antibody against 63GEN will react with only B17. When compared with 62GE, the overlapping 63GEN patch has one extra polymorphic residue, namely asparagine in position 66. This difference seems enough to distinguish antibody specificity against B17 vs A2+B17.
B17 shares eight overlapping patches with the B63-related alleles B*1516 and B*1517 (Table 3). An antibody against any of these patches will react with B17+B63, a known cross-reacting antigen combination [82]. B*1516 and B*1517 share three unique 3.5 Å patches and this may explain the specific recognition of previously reported monospecific anti-B63 antibodies [82].
As another example, four overlapping patches in the 62–73 sequence are unique to A9 subtypes A*2301, A*2402, A*2403 and A*2407 (Table 3). There are three additional patches shared by all A9 subtypes except A*2407. The latter allele has four unique patches all of them have a glutamine in position 70 whereas A*2301, A*2402 and A*2403 have a histidine in position 70. Since no monospecific antibodies against A*2407 have been reported, it is possible that H70Q substitutions contribute little to the functional epitopes defined by these patches. Permissible residue substitutions have also been observed for protein epitopes [22].
Table 3 shows also two examples of antigen groups with overlapping patches in the 62–73 sequence. The first is a group of antigens in the A1 CREG that share four patches. We have recently described a human monoclonal antibody reacting with this antigen group [19]. The second is a well-known group of B7-CREG antigens that has the same six overlapping patches.
The term “eplet” is used to represent one or an overlapping group of polymorphic patches shared by the same antigen(s). Table 3 shows the eplet assignments. The five 62–73 patches unique for A2 (63GEK, 66ERKH, 62GERK, 65GERKA and 66GERKAH) are collectively referred to as one eplet assigned as 63GEK. Although at this time, it is not known whether the 65RK patch shared between A2 and A*3401 represents a real epitope, this patch has also been converted to an eplet.
Table 3 demonstrates that serologically defined antigens such as B17 and B63 and, public epitopes represented by the A1 and B7 CREGs have corresponding eplets. The A9 alleles A*2301, A*2402, A*2403 and A*2407 have the 63EEK eplet that represents 63EEK, 65GK, 62EEGK and 65EEGKA. Since no monospecific antibodies against A*2407 have been identified, it seems reasonable that the 65GK eplet represents also the patches unique to A*2407 and the A*2301, A*2402 and A*2403 group.
Eplet conversions of overlapping patches have permitted an assessment of the repertoire of structurally defined functional epitopes. Interestingly, many eplets representing multiple and often overlapping patches seem to correspond with well-known serologically defined private and public determinants. The presence of such multiple patches might add to the immunogenicity of these antigenic determinants.
This analysis yielded a total of 199 distinct eplets, 110 of them are on the α helices on the top of the molecule. There are 60 eplets on the side surface, including 29 in less accessible positions at the bottom and under the peptide-binding groove. The HLAMatchmaker website: http://www.tpis.upmc.edu has a detailed description of eplets and downloadable Excel programs on eplet-based HLA compatibility.
DISCUSSION
This study addresses the concept that HLA antigens like other antigenic proteins have structural epitopes consisting of 15–20 residues that constitute the binding face with alloantibody. Each structural epitope has a functional epitope of about 2–5 residues that dominate the strength and specificity of binding with antibody. The remaining residues of a structural epitope provide supplementary interactions that increase the stability of the antigen-antibody complex. Each functional epitope has one or more non-self residues and the term eplet is used to describe polymorphic HLA residues within 3.0–3.5 Ångstroms of a given sequence position on the molecular surface. Many eplets represent short linear sequences identical to those referred to as triplets [1] but others have residues in discontinuous sequence positions that cluster together on the molecular surface. The eplet version of HLAMatchmaker considers therefore a more complete repertoire of structurally defined epitopes.
An epitope has two characteristics namely antigenicity, i.e. the reactivity with antibody, and immunogenicity, i.e. the ability of inducing an antibody response. Immunogenicity depends on the structural difference between an immunizing protein and the antibody responder’s homologous proteins [20]. Certain structural differences lead to immunodominant epitopes whereas others are associated with low immunogenicity. Antigenicity reflects the structural requirements for an epitope to react with a specific antibody. Sequence variability, secondary structure and conformational influences of nearby residues may affect epitope antigenicity.
The distinction between immunogenicity and antigenicity is important for the determination of HLA compatibility. HLAMatchmaker is based on the concept that a given eplet cannot induce specific alloantibodies if such eplet is present on any antigen of the antibody producer. Suppose three HLA antigens X, Y and Z have the eplet PQR in a given sequence location. Exposure to mismatched X will not lead to PQR-specific antibodies in recipients who type for Y or Z. This concept has been verified by observations that highly sensitized patients do not have antibodies against intralocus and interlocus triplet matches [7, 10, 11, 15]. On the other hand, PQR can be immunogenic for a recipient who does not type for X, Y or Z. Anti-PQR antibodies will react with the immunizing X and also with Y and Z but there might be exceptions for two reasons.
First, non-reactivity of the PQR eplet of antigens Y and/or Z might be due to a significantly altered conformation caused by nearby polymorphic residues different from those on the immunizing antigen X. Such residues are below the molecular surface and cannot make direct contact with antibody. Several reports have described a conformational effect of hidden residues on HLA epitope reactivity with antibody [78, 83, 84, 85, 86]. This scenario would only apply to epitope antigenicity but not immunogenicity because highly sensitized patients do not produce antibodies to self-epitopes expressed on mismatched HLA antigens [7, 10, 11, 15].
The second explanation reflects the two-patch shape of certain functional epitopes. In this scenario the reactivity of an anti-eplet antibody also requires contact with certain critical residues in another location on the immunizing antigen. This critical contact site must be within a sufficient distance from the eplet (<15 Å) so it can bind to other CDR loops of antibody. Suppose the PQR-carrying immunizing antigen X has the LMN sequence as a critical contact site and that Y but not Z has the same LMN sequence. In this scenario, anti-PQR antibodies will react only with X and Y because the PQR-carrying Z lacks the critical contact site necessary for binding with antibody. Recent studies have verified the role of critical contact sites in reactions with HLA antibodies [19] and other reported data are consistent with this notion [87–89]
Depending on the HLA type of the antibody producer, the polymorphic residues of a critical contact site on the immunizing antigen can be self or non-self. In the latter case, this might lead to antibodies against epitopes defined by combinations of non-self eplets and non-self critical contact sites. Such combinations may explain the Bw4/6 defined serological splits of HLA-B antigens. For instance, the Bw4-associated B38 and the Bw6-associated B39 splits of B16 share a distinct 158T eplet. These splits have identical sequences except in the 79–83 region that has the Bw4/6 epitopes [90, 91]. Immunization with 158T may lead to antibodies recognizing only B38 or B39; such antibodies require also reactivity with the Bw4 or Bw6 sequence. Other 158T-specific antibodies react with B16 (B38+B39) because they do not require interaction with a Bw4/6 defined critical contact site.
The critical contact site concept may also apply to antibodies against eplets shared by HLA antigens controlled by different loci, i.e. the interlocus matches. Some antibodies against these eplets may require a locus-specific sequence present on the immunizing antigen. For instance, the 82ALR eplet is shared by all Bw4-associated HLA-B antigens and the HLA-A antigens A23, A24, A25 and A32. Patients sensitized to 82ALR on a mismatched HLA-B antigen may have antibodies that react with both 82ALR -carrying HLA-A and HLA-B antigens [92] and/or they may anti-82ALR antibodies that react with only HLA-B antigens because they must react with a critical contact site unique to HLA-B antigens.
The different features of HLA immunogenicity and antigenicity are relevant to the application of structurally based algorithms for histocompatibility testing. HLAMatchmaker can be used as a quantitative tool to determine the degree of a mismatch. Recent studies have shown that the magnitude of kidney transplant-induced humoral sensitization correlates with the number of mismatched triplets on donor antigens [9]. Moreover, certain HLA antigen mismatches are compatible at the structural level and they are associated with similar kidney transplant survival rates as zero-antigen mismatches [2].
Eplet immunogenicity as determined by the frequency of a specific antibody response is important for histocompatibility [7]. High immunogenicity eplet mismatches should be avoided whereas low-immunogenicity eplets might be considered permissible mismatches. This information permits an expanded donor selection in platelet transfusions [17]. There is no structurally based prediction model for determining epitope immunogenicity. Possible factors include location and exposure of an epitope on the molecular surface, the relative difference in amino acid residue composition and the (HLA) genetic make up of the antibody responder. For the latter, the HLA-DR phenotype of the responder has been reported to influence antibody formation to class I mismatches [93]. At present, a practical approach is to collect information about the frequencies of epitope-specific antibody responses in context with the exposure rate to epitope mismatches [94].
HLAMatchmaker can be used to analyze serum screenings for sensitized patients. HLA typing differences between antibody producer and immunizer(s) will define the mismatched eplet repertoire the patient has been exposed to and this information facilitates the interpretation of serum screening results. Analysis of the antibody reactivity patterns with HLA panels may distinguish reactive and non-reactive eplets so that HLA mismatch acceptability can be established for sensitized patients.
The eplet versions of HLAMatchmaker can be downloaded from the website http://tpis.upmc.ed
Acknowledgments
This study is supported by grant AI-55933 from the National Institutes of Health
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Duquesnoy RJ. HLAMatchmaker: A Molecularly Based Algorithm For Histocompatibility Determination. I. Description of the Algorithm. Human Immunology. 2002;63:339–352. doi: 10.1016/s0198-8859(02)00382-8. [DOI] [PubMed] [Google Scholar]
- 2.Duquesnoy RJ, Takemoto S, De Lange P, Doxiadis IIN, Schreuder GMT, Claas FHJ. HLAMatchmaker: A Molecularly Based Algorithm For Histocompatibility Determination III. Effect of matching at the HLA-A,B amino acid triplet level on kidney transplant survival. Transplantation. 2003;75:884–889. doi: 10.1097/01.TP.0000055101.20821.AC. [DOI] [PubMed] [Google Scholar]
- 3.Boehringer D, Reinhard T, Duquesnoy R, Boehringer S, Enczmann J, de Lange P, Claas F, Sundmacher R. Beneficial Effect of Matching at the HLA-A and B Amino-Acid Triplet Level on Rejection Free Survival in Panetrating Keratoplasty. Transplantation. 2004;(77):417–421. doi: 10.1097/01.TP.0000110415.10401.94. [DOI] [PubMed] [Google Scholar]
- 4.Laux G, Mytilineos J, Opelz G. Critical evaluation of the amino acid triplet-epitope matching concept in cadaver kidney transplanation. Transplantation. 2004;77:902–907. doi: 10.1097/01.tp.0000114595.59168.3b. [DOI] [PubMed] [Google Scholar]
- 5.Duquesnoy R, Claas F. Is the Application of HLAMatchmaker Relevant in Kidney Transplantation? (Letter to the Editor) Transplantation. 2005;79:250–251. doi: 10.1097/01.tp.0000144327.92898.a6. [DOI] [PubMed] [Google Scholar]
- 6.Haririan A, Fagoaga O, Daneshvar H, Morawski K, Sillix D, El-Amm J, West M, Garnick J, Migdal S, Gruber S, Nehlsen-Cannarella S. Predictive value of HLA epitope matching using HLAMatchmaker for graft outcomes in a predominantly African-American renal transplant cohort. Clinical Transplantation. 2006 doi: 10.1111/j.1399-0012.2005.00473.x. In press. [DOI] [PubMed] [Google Scholar]
- 7.Duquesnoy RJ, Marrari M. HLAMatchmaker: A Molecularly Based Algorithm For Histocompatibility Determination. II. Verification of the Algorithm and Determination of the Relative Immunogenicity of Amino Acid Triplet-Defined Epitopes. Human Immunology. 2002;63:353–363. doi: 10.1016/s0198-8859(02)00381-6. [DOI] [PubMed] [Google Scholar]
- 8.Lobashevsky AL, Senkbeil RW, Shoaf JL, Stephenson AK, Skelton SB, Burke RM, Deierhoi MH, Thomas JM. The number of amino acid residues mismatches correlates with flow cytometry crossmatching results in high PRA renal patients. Human Immunology. 2002;63(5):364–74. doi: 10.1016/s0198-8859(02)00371-3. [DOI] [PubMed] [Google Scholar]
- 9.Dankers MKA, Witvliet MD, Roelen DL, De Lange P, Korfage N, Persijn GG, Duquesnoy RJ, Doxiadis IIN, Claas FHJ. The Number of Amino Acid Triplet Differences between Patient aand Donor is Predicitve for the Antibody Reactivity Against Mismatched HLA Antigens. Transplantation. 2004;I28:1236–1239. doi: 10.1097/01.tp.0000120385.03278.28. [DOI] [PubMed] [Google Scholar]
- 10.Duquesnoy RJ, Witvliet MJ, Doxiadis IIN, de Fijter H, Claas FHJ. HLAMatchmaker-Based Strategy To Identify Acceptable HLA Class I Mismatches For Highly Sensitized Kidney Transplant Candidates. Transplant International. 2004;7:31–38. doi: 10.1007/s00147-003-0641-z. [DOI] [PubMed] [Google Scholar]
- 11.Claas FHJ, Witvliet M, Duquesnoy RJ, Persijn G, Doxiadis IIN. The Acceptable Mismatch Program as a Fast Tool to Transplant Highly Sensitized Patients Awaiting a Post-Mortal Kidney: Short Waiting Time and Excellent Graft Outcome. Transplantation. 2004;78:190–193. doi: 10.1097/01.tp.0000129260.86766.67. [DOI] [PubMed] [Google Scholar]
- 12.Iniotaki-Theodoraki A, Kalogeropoulou E, Apostolaki M, Doxiadis I, Stavropoulos-Giokas C. Humoral sensitization against rejected grafts: Specific antibodies to graft immunogenic amino acid triplets. Transplant Proc. 2004;36:1728–1731. doi: 10.1016/j.transproceed.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Varnavidou-Nicolaidou A, Doxiadis I, Iniotaki-Theodoraki A, Patargias T, Stavropoulos-Giokas C, Kyriakides G. HLA class I donor-specific triplet antibodies detected after renal transplantation. Transplantation Proceedings. 2004;36:1732–1734. doi: 10.1016/j.transproceed.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 14.Adeyi O, Girnita A, Awadalla Y, Askar M, Shapiro R, Howe J, Martell J, Zeevi A, Nalesnik M, Rhandawa P, Demetris AJ, RJ D. Serum Analysis After Kidney Transplant Nephrectomy Reveals Restricted Antibody Specificity Patterns Against Donor HLA Class I Antigens. Transpl Immunol. 2005;14:53–62. doi: 10.1016/j.trim.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 15.Doxiadis IIN, Duquesnoy RJ, Claas HJ. Extending options for highly sensitized patients to receive a suitable kidney graft. Curr Opin Immunol. 2005;17:536–540. doi: 10.1016/j.coi.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 16.Goodman R, Taylor C, O’Rourke C, Lynch A, Bradley A, Key K. Utility of HLAMatchmaker and single-antigen HLA-antibody detection beads for identification of acceptable mismatches in highly sensitised patients awaiting kidney transplantation. Transplantation. 2006 doi: 10.1097/01.tp.0000205202.56915.f5. In Press. [DOI] [PubMed] [Google Scholar]
- 17.Nambiar A, Duquesnoy RJ, Adams S, Oblitas J, Leitman S, Stroncek DMF. HLAMatchmaker-Driven Analysis Of Response To HLA Matched Platelet Transfusions In Alloimmunized Patients. Blood. 2006;107:1680–1687. doi: 10.1182/blood-2004-10-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papassavas AC, Brown C, Brough S, Ouwenhand WH, Duquesnoy RJ, Navarrete C. The Use Of The Non-Immunogenic HLA Epitope Mismatched Strategy Improves The Outcome Of Platelet Transfusions In Refractory Patients. 2006. Submitted. [Google Scholar]
- 19.Duquesnoy R, Mulder A, Fernandez-Vina AMM, Claas F. HLAMatchmaker-based analysis of human monoclonal antibody reactivity demonstrates the importance of an additional contact site for specific recognition of triplet-defined epitopes. Hum Immunol. 2005;66:749–761. doi: 10.1016/j.humimm.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Benjamin DC, Berzofsky JA, East IJ, Gurd FRN, Hannum C, Leach SJ, Margoliash E, Michael JG, Miller A, Prager EM, Reichlin M, Sercarz EE, Smith-Gill SJ, Todd PE, Wilson AC. The antigenic structure of proteins: A reappraisal. Ann Rev lmmunol. 1984;2:67–101. doi: 10.1146/annurev.iy.02.040184.000435. [DOI] [PubMed] [Google Scholar]
- 21.Colman PM. Structure of antibody-antigen complexes: implications for immune recognition. Adv Immunol. 1988;43:99–132. doi: 10.1016/s0065-2776(08)60364-8. [DOI] [PubMed] [Google Scholar]
- 22.Getzoff ED, Tainer JA, Lerner RA, Geysen HM. The chemistry and mechanisnm of antibody binding to protein antigens. Adv Immunol. 1988;43:1–98. doi: 10.1016/s0065-2776(08)60363-6. [DOI] [PubMed] [Google Scholar]
- 23.Van Regenmortel MHV. Structural and functional approaches to the study of protein antigenicity. Immunol Today. 1989;10:266–272. doi: 10.1016/0167-5699(89)90140-0. [DOI] [PubMed] [Google Scholar]
- 24.Hogue C. Cn3D: a new generation of three-dimensional molecular structure viewer. Trends Biochem Sci. 1997;22:314–316. doi: 10.1016/s0968-0004(97)01093-1. [DOI] [PubMed] [Google Scholar]
- 25.Altschul S, Madden T, Schäffer A, Zhang J, Zhang Z, Miller W, Lipman D. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kabat EA, Wu T, Bilofsky H. Unusual distribution of amino acids in complementarity-determining (hypervarible) segments of heavy and light chains of immunoglobulins and their possible roles in specificity of antibody-combining sites. J Biol Chem. 1977;252:6609–6617. [PubMed] [Google Scholar]
- 27.Kabat EA. The structural basis of antibody complementarity. Adv Protein Chem. 1978;32:1–75. [PubMed] [Google Scholar]
- 28.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 29.Chothia C, Lesk A, Tramontano A, Smith-Gill S, Air G, Sheriff S, Padlan E, Davies D, Tulip W, Colman P, Spinelli S, Alzari P, Poljak R. Conformation of immunoglobulin hypervariable regions. Nature. 1989;342:877–883. doi: 10.1038/342877a0. [DOI] [PubMed] [Google Scholar]
- 30.Wilson IA, Stanfield RL. Antibody–antigen interactions: new structures and new conformational changes. Curr Opin Struct Biol. 1994;4:857–867. doi: 10.1016/0959-440x(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 31.Padlan EA. Anatomy of the antibody molecule. Mol Immunol. 1994;31:169–217. doi: 10.1016/0161-5890(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 32.Kirkham P, Schroeder H., Jr Antibody structure and the evolution of immunoglobulin V gene segments. Seminars in Immunology. 1994;6:347–360. doi: 10.1006/smim.1994.1045. [DOI] [PubMed] [Google Scholar]
- 33.MacCallum RM, Martin ACR, Thornton JM. Antibody-Antigen Interactions: Contact Analysis and Binding Site Topography. J Mol Biol. 1996;262:732–745. doi: 10.1006/jmbi.1996.0548. [DOI] [PubMed] [Google Scholar]
- 34.Zemlin M, Klinger M, Link J, Zemlin C, Bauer K, Englern J, Schroeder H, Jr, Kirkham P. Expressed Murine and Human CDR-H3 Intervals of Equal Length Exhibit Distinct Repertoires that Differ in their Amino Acid Composition and Predicted Range of Structures. J Mol Biol. 2003;334:733–749. doi: 10.1016/j.jmb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 35.Al-Lazikani B, Lesk A, Chothia C. Standard conformations for the canonical structures of immunoglobulins. J Mol Biol. 1997;273:927–948. doi: 10.1006/jmbi.1997.1354. [DOI] [PubMed] [Google Scholar]
- 36.Davies D, Padlan E, Sheriff S. Antibidy-antigen complexes. Annu Rev Biochem. 1990;59:439–473. doi: 10.1146/annurev.bi.59.070190.002255. [DOI] [PubMed] [Google Scholar]
- 37.Almagro J. Identification of differences in the specificity-determining residues of antibodies that recognize antigens of different size: implications for the rational design of antibody repertoires. J Mol Recognit. 2004;17:132–143. doi: 10.1002/jmr.659. [DOI] [PubMed] [Google Scholar]
- 38.Jerne N. Ann Rev Microbiol. 1960;14:341–358. doi: 10.1146/annurev.mi.14.100160.002013. [DOI] [PubMed] [Google Scholar]
- 39.Laver WG, Air GM, Webster RG, Smith-Gill SJ. Epitopes on protein antigens: misconception and realities. Cell. 1990;61:553–559. doi: 10.1016/0092-8674(90)90464-p. [DOI] [PubMed] [Google Scholar]
- 40.Sela M, Schechter B, Schechter I, Borek F. Antibodies to sequential and conformational determinants. Cold Spring Harbor Symp Quant Biol. 1967;32:537–545. [Google Scholar]
- 41.Atassi M, Smith J. A proposal for the nomenclature of antigenic sites in peptides and proteins. Immunochemistry. 1978;15:609–610. doi: 10.1016/0161-5890(78)90016-0. [DOI] [PubMed] [Google Scholar]
- 42.Braden B, Goldman E, Mariuzza R, Poljak R. Anatomy of an antibody molecule: structure, kinetics, thermodynamics and mutational studies of the antilysozyme antibody D. Immunol Rev. 1998;163:45–57. doi: 10.1111/j.1600-065x.1998.tb01187.x. [DOI] [PubMed] [Google Scholar]
- 43.Laune D, Molina K, Ferrieres G, Mani JC, Cohen P, Simon D, Bernardi T, Piechaczyk M, Pau B, Granier C. Systematic Exploration of the Antigen Binding Activity of Synthetic Peptides Isolated from the Variable Regions of Immunoglobulins. J Biol Chem. 1997;272:30937–30944. doi: 10.1074/jbc.272.49.30937. [DOI] [PubMed] [Google Scholar]
- 44.Van Regenmortel M. Reductionism and the search for structure-function relationships in antibody molecules. J Mol Recognit. 2002;15:240–247. doi: 10.1002/jmr.584. [DOI] [PubMed] [Google Scholar]
- 45.Novotny J. Protein antigenicity: a thermodynamic approach. Mol Immunol. 1991;28:201–208. doi: 10.1016/0161-5890(91)90062-o. [DOI] [PubMed] [Google Scholar]
- 46.Cunningham B, Wells J. Comparison of a structural and a functional epitope. J Mol Biol. 1993;234:554–563. doi: 10.1006/jmbi.1993.1611. [DOI] [PubMed] [Google Scholar]
- 47.Wells J. Binding in the growth hormone receptor complex. Proc Nat Acad Sci, USA. 1996;93:1–6. doi: 10.1073/pnas.93.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davies D, Cohen G. Interactions of protein antigens with antibodies. Proc Natl Acad Sci USA. 1996;93:7–12. doi: 10.1073/pnas.93.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeLano W. Unraveling hot spots in binding interfaces: progress and challenges. Curr Opin Struct Biol. 2002;12:14–20. doi: 10.1016/s0959-440x(02)00283-x. [DOI] [PubMed] [Google Scholar]
- 50.Hawkins R, Russell S, Baier M, Winter G. The contribution of contact and non-contact residues of antibody in the affinity of binding to antigen. The interaction of mutant D1.3 antibodies with lysozyme. J Mol Biol. 1993;234:958–964. doi: 10.1006/jmbi.1993.1650. [DOI] [PubMed] [Google Scholar]
- 51.Roberts V, Getzoff E, Tainer J. In: Structural basis of antigenic cross-reactivity, in Structure of Antignes. Van Regenmortel M, editor. CRC press; Boca Raon, FL: 1993. pp. 31–53. [Google Scholar]
- 52.Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 53.Tainer JT, Getzoff ED, Paterson Y, Olson AJ, Lerner RJ. The Atomic Mobility Component of Protein Antigenicity. Ann Rev lmmunol. 1985;3:501–35. doi: 10.1146/annurev.iy.03.040185.002441. [DOI] [PubMed] [Google Scholar]
- 54.Cortopassi GA, Wilson AC. Recent origin of the P lysozyme gene in mice. Nucleic Acids Res. 1990;18:1911–1920. doi: 10.1093/nar/18.7.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Obita T, Ueda T, Imoto T. Solution structure and activity of mouse lysozyme M. Cell Mol Life Sci. 2003;60:176–184. doi: 10.1007/s000180300012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith-Gill SJ, Lavoie TB, Mainhart C. Antigenic regions defined by monoclonal antibodies correspond to structural domains of avian lysozymes. J Immunol. 1984;133:384–393. [PubMed] [Google Scholar]
- 57.Metzger DW, Ch’ng LK, Miller A, Sercarz EE. The expressed lysozyme-specific B cell repertoire: 1. Heterogeneity in the monoclonal anti-hen egg white lysozyme specificity repertoire. and its difference from the in situ repertoire. Eur J Immunol. 1984;14:87–96. doi: 10.1002/eji.1830140116. [DOI] [PubMed] [Google Scholar]
- 58.Harper M, Lema F, Boulot G, Poljak RJ. Antigen specificity and cross-reactivity of monoclonal anti-lysozyme antibodies. Mol Immunol. 1987;2:97–108. doi: 10.1016/0161-5890(87)90081-2. [DOI] [PubMed] [Google Scholar]
- 59.Newman M, Mainhart C, Mallett C, Lavoie TB, Smith-Gill S. Patterns of antibody specificity during the balb/c immune response to hen eggwhite lysozyme’. J Immunol. 1992;149:3260–3272. [PubMed] [Google Scholar]
- 60.Amit A, Mariuzza R, Phillips S, Poljak R. Three-dimensional structure of an antigen-antibody complex at 2.8 Angstrom resolution. Science. 1986;233:747–752. doi: 10.1126/science.2426778. [DOI] [PubMed] [Google Scholar]
- 61.Fischmann TO, Bentley GA, Bhat TN, Boulot G, Mariuzza RA, Phillips SEV, Tello D, Poljak RJ. Crystallographic refinement of the three-dimensional structure of the FabD1.3-lysozyme complex at 2.5-A° resolution. J Biol Chem. 1991;266:12915–12920. [PubMed] [Google Scholar]
- 62.Dall’Acqua W, Goldman ER, Lin W, Teng C, Tsuchiya D, Li H, Ysern X, Braden BC, Li Y, Smith-Gill SJ, Mariuzza RA. A Mutational Analysis of Binding Interactions in an Antigen-Antibody Protein-Protein Complex. Biochemistry. 1998;37:7981–7991. doi: 10.1021/bi980148j. [DOI] [PubMed] [Google Scholar]
- 63.Bhat TN, Bentley GA, Boulot G, Greene MI, Tello D, Dall’Acqua W, Souchon H, Schwarz FP, Mariuzza RA, Poljak RJ. Bound water molecules and conformational stabilization help mediate an antigen-antibody association. Proc Natl Acad Sci USA. 1994;91:1089–1093. doi: 10.1073/pnas.91.3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fields B, Goldbaum AFA, Dall’Acqua W, Malchiodi EL, Cauerhff A, Schwarz FP, Ysern X, Poljak RJ, Mariuzza RA. Hydrogen Bonding and Solvent Structure in an Antigen-Antibody Interface. Crystal Structures and Thermodynamic Characterization of Three Fv Mutants Complexed with Lysozyme. Biochemistry. 1996;35:15494–15503. doi: 10.1021/bi961709e. [DOI] [PubMed] [Google Scholar]
- 65.Carbone FR, Paterson Y. Monoclonal antibodies to horse cytochrome c expressing four distinct idiotypes distribute among two sites on the native protein. J Immunol. 1985;135:2609–2616. [PubMed] [Google Scholar]
- 66.Bushnell GW, Louie GV, Brayer GD. High-resolution three-dimensional structure of horse heart Cytochrome c. J Mol Biol. 1990;214:585–595. doi: 10.1016/0022-2836(90)90200-6. [DOI] [PubMed] [Google Scholar]
- 67.Mylvaganam S, Paterson Y, Getzoff E. Structural Basis for the Binding of an Anti-cytochrome c Antibody to its Antigen: Crystal Structures of FabE8-Cytochrome c Complex to 1.8 AÊ Resolution and FabE8 to 2.26 AÊ Resolution. J Mol Biol. 1998;281:301–322. doi: 10.1006/jmbi.1998.1942. [DOI] [PubMed] [Google Scholar]
- 68.Jemmerson R, Paterson Y. Mapping antigenic sites on proteins: implications for the design of synthetic vaccines. BioTechniques. 1986;4:18–31. [Google Scholar]
- 69.Cooper HM, Jemmerson R, Hunt DF, Griffin PR, Yates JR, Shabanowitz J, Zhu N, Paterson Y. Site-directed chemical modification of horse cytochrome c results in changes in antigenicity due to local and long-range conformational perturbations. J Biol Chem. 1987;262:11591–11597. [PubMed] [Google Scholar]
- 70.Duquesnoy R. Structural and functional definitions of epitopes reacting with mouse monoclonal antibodies. 2006 HLAMatchmaker Website: http://tpis.upmc.edu.
- 71.Akkoc N, Scornik J. Amino acid residues on HLA molecules critical for alloantibody binding. Transplant Proc. 1991;23:389. [PubMed] [Google Scholar]
- 72.McCutcheon JA, Smith KD, Valenzuela A, Aalbers K, Lutz CT. HLA-B*0702 antibody epitopes are affected indirectly by distant antigen residues. Human Immunology. 1993;36(2):69–75. doi: 10.1016/0198-8859(93)90108-d. [DOI] [PubMed] [Google Scholar]
- 73.Lutz CT, Smith KD, Greazel NS, Mace BE, Jensen DA, McCutcheon JA, Goeken NE. Bw4-reactive and Bw6-reactive antibodies recognize multiple distinct HLA structures that partially overlap in the alpha-1 helix. Journal of Immunology. 1994;153(9):4099–110. [PubMed] [Google Scholar]
- 74.Smith KD, Kurago ZB, Lutz CT. Conformational changes in MHC class I molecules. Antibody, T-cell receptor, and NK cell recognition in an HLA-B7 model system. Immunologic Research. 1997;16(3):243–59. doi: 10.1007/BF02786393. [DOI] [PubMed] [Google Scholar]
- 75.Duquesnoy RJ. HLAMatchmaker: A Molecularly Based Algorithm For Histocompatibility Determination. ASHI Quarterly. 2002:60–62. doi: 10.1016/s0198-8859(02)00382-8. [DOI] [PubMed] [Google Scholar]
- 76.Hulsmeyer M, Chames P, Hillig R, Stanfield R, Held G, Coulie P, Alings C, Wille G, Saenger W, Uchanska-Ziegler B, Hoogenboom H, Ziegler A. A Major Histocompatibility Complex-Peptide-restricted antibody and T Cell Receptor molecules recognize their Target by distinct binding modes. Crystal structure of Human Leukocyte Antigen (HLA)-A1 -MAGE-A1 complex with Fab-Hyb3. J Biol Chem. 2005;280:2972–2980. doi: 10.1074/jbc.M411323200. [DOI] [PubMed] [Google Scholar]
- 77.Viken HD, Paulsen G, Drover S, Marshall WH, Sollid LM, Gaudernack G, Thorsby E. Influence on antibody recognition of amino acid substitutions in the cleft of HLA-DQ2 molecules. Suggestive evidence of peptide-dependent epitopes. Human Immunology. 1995;44(2):63–9. doi: 10.1016/0198-8859(95)00047-8. [DOI] [PubMed] [Google Scholar]
- 78.Smith KD, Mace BE, Valenzuela A, Vigna JL, McCutcheon JA, Barbosa JA, Huczko E, Engelhard VH, Lutz CT. Probing HLA-B7 conformational shifts induced by peptide-binding groove mutations and bound peptide with anti-HLA monoclonal antibodies. Journal of Immunology. 1996;157(6):2470–8. [PubMed] [Google Scholar]
- 79.Takamiya Y, Sakaguchi T, Miwa K, Takiguchi M. Role of HLA-B*5101 binding nonamer peptides in formation of the HLA-Bw4 public epitope. International Immunology. 1996;8(7):1027–34. doi: 10.1093/intimm/8.7.1027. [DOI] [PubMed] [Google Scholar]
- 80.Mulder M, Eijsink C, Kester MGD, Franke MEI, Kardol MJ, Heemskerk MHM, van Kooten C, Verreck FA, Drijfhout JW, Koning F, Doxiadis IN, Claas FJH. Impact of Peptides on the Recognition of HLA Class I Molecules by Human HLA Antibodies. J Immunol. 2005;175:5950–5957. doi: 10.4049/jimmunol.175.9.5950. [DOI] [PubMed] [Google Scholar]
- 81.Ways JP, Rothbard JB, Parham P. Amino acid residues 56 to 69 of HLA-A2 specify an antigenic determinant shared by HLA-A2 and HLA-B17. Journal of Immunology. 1986;137(1):217–22. [PubMed] [Google Scholar]
- 82.Hildebrand WH, Domena JD, Shen SY, Lau M, Terasaki PI, Bunce M, Marsh SG, Guttridge MG, Bias WB, Parham P. HLA-B15: a widespread and diverse family of HLA-B alleles. Tissue Antigens. 1994;43(4):209–18. doi: 10.1111/j.1399-0039.1994.tb02327.x. [DOI] [PubMed] [Google Scholar]
- 83.Marsh SG, Bodmer JG. HLA-DR and -DQ epitopes and monoclonal antibody specificity. Immunology Today. 1989;10:305–312. doi: 10.1016/0167-5699(89)90086-8. [DOI] [PubMed] [Google Scholar]
- 84.Klohe E, Fu XT, Ballas M, Karr RW. HLA-DR beta chain residues that are predicted to be located in the floor of the peptide-binding groove contribute to antibody-binding epitopes. Human Immunology. 1993;37(1):51–8. doi: 10.1016/0198-8859(93)90142-n. [DOI] [PubMed] [Google Scholar]
- 85.Drover S, Marshall WH, Kwok WW, Nepom GT, Karr RW. Amino acids in the peptide-binding groove influence an antibody-defined, disease-associated HLA-DR epitope. Scandinavian Journal of Immunology. 1994;39(6):539–50. doi: 10.1111/j.1365-3083.1994.tb03411.x. [DOI] [PubMed] [Google Scholar]
- 86.Fu XT, Drover S, Marshall WH, Karr RW. HLA-DR residues accessible under the peptide-binding groove contribute to polymorphic antibody epitopes. Hum Immunol. 1995;43:243–250. doi: 10.1016/0198-8859(95)00036-4. [DOI] [PubMed] [Google Scholar]
- 87.Akkoc N, Scornik JC. HLA epitope matching. Contribution of matched residues to epitopes recognized by alloantibodies. Transplantation. 1991;52(5):903–7. [PubMed] [Google Scholar]
- 88.McCutcheon JA, Lutz CT. Mutagenesis around residue 176 on HLA-B*0702 characterizes multiple distinct epitopes for anti-HLA antibodies. Human Immunology. 1992;35(2):125–31. doi: 10.1016/0198-8859(92)90020-n. [DOI] [PubMed] [Google Scholar]
- 89.Smith KD, Lutz CT. Alloreactive T Cell Recognition of MHC Class I Molecules. The T Cell Receptor Interacts with Limited Regions of the MHC Class I Long Alpha Helices. J Immunol. 1997;158:2805–2812. [PubMed] [Google Scholar]
- 90.Muller CA, Engler-Blum G, Gekeler V, Steiert I, Weiss E, Schmidt H. Genetic and serological heterogeneity of the supertypic HLA-B locus specificities Bw4 and Bw6. Immunogenetics. 1989;30(3):200–7. doi: 10.1007/BF02421207. [DOI] [PubMed] [Google Scholar]
- 91.Adams EJ, Martinez-Naves E, Arnett KL, Little AM, Tyan DB, Parham P. HLA-B16 antigens: sequence of the ST-16 antigen, further definition of two B38 subtypes and evidence for convergent evolution of B*3902. Tissue Antigens. 1995;45(1):18–26. doi: 10.1111/j.1399-0039.1995.tb02410.x. [DOI] [PubMed] [Google Scholar]
- 92.Nosek J, Hinzova E, Vorlicek J. Quantitative estimation of HLA-A and HLA-B antigens carrying the Bw4 supertypic specificity in human peripheral blood lymphocytes. Immunogenetics. 1987;26(4–5):273–81. doi: 10.1007/BF00346522. [DOI] [PubMed] [Google Scholar]
- 93.Fuller TC, Fuller A. The humoral immune response against an HLA class I allodeterminant correlates with the HLA-DR phenotype of the responder. Transplantation. 1999;68(2):173–82. doi: 10.1097/00007890-199907270-00002. [DOI] [PubMed] [Google Scholar]
- 94.Duquesnoy RJ, Claas FHJ. Progress Report of 14th International Histocompatibility Workshop Project on the Structural Basis of HLA Compatibility. Tissue Antigens. 2006 doi: 10.1111/j.1399-0039.2006.00766.x. In Press. [DOI] [PubMed] [Google Scholar]
- 95.Bhat TN, Bentley GA, Fischmann TO, Boulot G, Poljak RJ. Small rearrangements in structures of Fv and Fab fragments of antibody D1.3 on antigen binding. Nature. 1990;347:483–485. doi: 10.1038/347483a0. [DOI] [PubMed] [Google Scholar]
- 96.Novotny J, Bruccolery RE, Saul FE. On the attribution of binding energy in antigen-antibody complexes McPC603, D1.3, and HyHEL-5. Biochemistry. 1989;28:4735–4749. doi: 10.1021/bi00437a034. [DOI] [PubMed] [Google Scholar]
- 97.England P, Bregegere F, Bedouelle H. Energetic and Kinetic Contributions of Contact Residues of Antibody D1.3 in the Interaction with Lysozyme. Biochemistry. 1997;36:164–172. doi: 10.1021/bi961419y. [DOI] [PubMed] [Google Scholar]
- 98.Shick K, Xavier K, Rajpal A, Smith-Gill SJ, Willson R. Association of the anti-hen egg lysozyme antibody HyHEL-5 with avian species variant and mutant lysozymes. Biocm Biophys Acta. 1997;1340:205–214. doi: 10.1016/s0167-4838(97)00035-6. [DOI] [PubMed] [Google Scholar]
- 99.Sheriff S, Silverton EW, Padlan EA, Cohen GH, Smith-Gill SJ, Finzel BC, Davies DR. Three-dimensional structure of an antibody-antigen complex. Proc Natl Acad Sci USA. 1987;84:8075–8079. doi: 10.1073/pnas.84.22.8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lavoie TB, Drohan WN, Smith-Gill SJ. Experimental analysis by site-directed mutagenesis of somatic mutation effects on affinity and fine specificity in antibodies specific for lysozyme. J Immunol. 1992;148:503–513. [PubMed] [Google Scholar]
- 101.Chacko S, Silverton E, Kam-Morgan L, Smith-Gill S, Cohen GH, Davies D. Structure of an Antibody-Lysozyme Complex Unexpected Effect of a Conservative Mutation. JMol Biol. 1995;245:261–274. doi: 10.1006/jmbi.1994.0022. [DOI] [PubMed] [Google Scholar]
- 102.Li Y, Li H, Smith-Gill S, Mariuzza R. Three-Dimensional Structures of the Free and Antigen-Bound Fab from Monoclonal Antilysozyme Antibody HyHEL-63. Biochemistry. 2000;39:6296–6309. doi: 10.1021/bi000054l. [DOI] [PubMed] [Google Scholar]
- 103.Li Y, Huang YM, Swaminathan CP, Smith-Gill SJ, Mariuzza RA. Magnitude of the Hydrophobic Effect at Central versus Peripheral Sites in Protein-Protein Interfaces. Structure. 2005;13:297–307. doi: 10.1016/j.str.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 104.Li Y, Urrutia M, Smith-Gill SJ, Mariuzza RA. Dissection of binding interactions in the complex between the anti-lysozyme antibody HyHEL-63 and its antigen. Biochemistry. 2003;42:11–22. doi: 10.1021/bi020589+. [DOI] [PubMed] [Google Scholar]
- 105.Aguet M, Merlin G. Purification of human γ interferon receptors by sequential affinity chromatography on immobilized monoclonal antireceptor antibodies and human g interferon. J Expt Med. 1987;165:988–999. doi: 10.1084/jem.165.4.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sogabe S, Stuart F, Henke C, Bridges A, Williams G, Birch A, Winkler FK, Robinson JA. Neutralizing Epitopes on the Extracellular Interferon γ Receptor (IFNγR) α-Chain Characterized Homolog Scanning Mutagenesis and X-ray Crystal Structure of the A6 Fab-IFNγR1-108 Complex. J Mol Biol. 1997;273:882–897. doi: 10.1006/jmbi.1997.1336. [DOI] [PubMed] [Google Scholar]
- 107.Gray PW, Leong S, Fennie EH, Farrar MA, Pingel JT, Fernandez-Luna J, Schreiber RD. Cloning and expression of the cDNA for the murine interferon gamma receptor. Proc Natl Acad Sci USA. 1989;86:8497–8501. doi: 10.1073/pnas.86.21.8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mylvaganam SE, Paterson Y, Kaiser K, Bowdish K, Getzoff ED. Biochemical implications from the variable gene sequences of an anti-cytochrome c antibody and crystallographic characterization of its antigen-binding fragment in free and antigen-complexed forms. J Mol Biol. 1991;221:455–462. doi: 10.1016/0022-2836(91)80066-4. [DOI] [PubMed] [Google Scholar]
- 109.Collawn JF, Wallace CJA, Proudfoot AEI, Paterson Y. Monoclonal antibodies as probes of conformational changes in protein-engineered cytochrome c. J Biol Chem. 1988;263:8625–8634. [PubMed] [Google Scholar]
- 110.Pons J, Stratton J, Kirsch J. How do two unrelated antibodies, HyHEL-10 and F9.13.7, recognize the same epitope of hen egg-white lysozyme? Protein Science. 2002;11:2308–2315. doi: 10.1110/ps.0209102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lescar J, Pellegrini M, Souchon H, Tello D, Poljak RJ, Peterson N, Greene M, Alzari PM. Crystal structure of a cross-reaction complex between Fab F9.13.7 and guinea fowl lysozyme. J Biol Chem. 1995;270:18067–18076. doi: 10.1074/jbc.270.30.18067. [DOI] [PubMed] [Google Scholar]
- 112.Sinha N, Mohan S, Lipschultz C, Smith-Gill S. Differences in Electrostatic Properties at Antibody–Antigen Binding Sites: Implications for Specificity and Cross-Reactivity. Biophys J. 2002;83:2946–2968. doi: 10.1016/S0006-3495(02)75302-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kam-Morgan LNW, Smith-Gill SJT, Zhang MGL, Wilson AC, Kirsch JF. High resolution mapping of the HyHEL-10 epitope of chicken lysozyme by site-directed mutagenesis. Proc Nat Acad Sci, USA. 1993;90:3958–3962. doi: 10.1073/pnas.90.9.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Padlan EA, Silverton EW, Sheriff S, Cohen GH, Smith-Gill SJ, Davies DR. Structure of an antibody-antigen complex: crystal structure of the HyHEL-10 Fab-lysozyme complex. Proc Nat Acad Sci, USA. 1989;86:5938–5942. doi: 10.1073/pnas.86.15.5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kondo H, Shiroishi M, Matsushimai M, Tsumoto K, Kumagai I. Crystal Structure of Anti-Hen Egg White Lysozyme Antibody (HyHEL-10) Fv-Antigen Complex. J Biol Chem. 1999;274:27623–2731. doi: 10.1074/jbc.274.39.27623. [DOI] [PubMed] [Google Scholar]
- 116.Pons J, Rajpal A, Kirsch JF. Energetic analysis of an antigen/antibody interface: alanine scanning mutagenesis and double mutant cycles on the HyHEL-10/lysozyme interaction. Protein Sci. 1999;8:958–968. doi: 10.1110/ps.8.5.958. [DOI] [PMC free article] [PubMed] [Google Scholar]


