Abstract
Escherichia coli RNase E, an essential single-stranded specific endoribonuclease, is required for both ribosomal RNA processing and the rapid degradation of mRNA. The availability of the complete sequences of a number of bacterial genomes prompted us to assess the evolutionarily conservation of bacterial RNase E. We show here that the sequence of the N-terminal endoribonucleolytic domain of RNase E is evolutionarily conserved in Synechocystis sp. and other bacteria. Furthermore, we demonstrate that the Synechocystis sp. homologue binds RNase E substrates and cleaves them at the same position as the E. coli enzyme. Taken together these results suggest that RNase E-mediated mechanisms of RNA decay are not confined to E. coli and its close relatives. We also show that the C-terminal half of E. coli RNase E is both sufficient and necessary for its physical interaction with the 3′–5′ exoribonuclease polynucleotide phosphorylase, the RhlB helicase, and the glycolytic enzyme enolase, which are components of a “degradosome” complex. Interestingly, however, the sequence of the C-terminal half of E. coli RNase E is not highly conserved evolutionarily, suggesting diversity of RNase E interactions with other RNA decay components in different organisms. This notion is supported by our finding that the Synechocystis sp. RNase E homologue does not function as a platform for assembly of E. coli degradosome components.
Keywords: RNA degradosome/mRNA degradation
E. coli RNase E is a site-specific endoribonuclease (for review, see ref. 1) that was originally identified as an activity essential for cell viability and the generation of 5S rRNA from 9S RNA, a larger precursor (2). The endoribonucleolytic activity of this enzyme is now known also to have an important role in the degradation of a number of mRNAs (for reviews, see refs. 3 and 4) and antisense RNAs that control the replication of ColE1-type and IncFII plasmids (5–7). Very recently, RNase E has been reported to shorten 3′ poly(A) tails (8), which also are involved in determining E. coli RNA stability (for review, see ref. 9), suggesting another mechanism by which this enzyme can exercise control over RNA decay.
In E. coli, RNase E is a component of the degradosome, a multiprotein complex whose other major components are polynucleotide phosphorylase (PNPase), a 3′–5′ exoribonuclease, the RhlB RNA helicase, and the glycolytic enzyme enolase (10–13). Three minor components of the degradosome, DnaK, GroEL, and polyphosphate kinase also have been identified (12, 14). Although RhlB has been shown to assist PNPase-mediated attack of structured RNAs (13), and there are data suggesting that polyphosphate kinase maintains an appropriate microenvironment for the degradosome by removing polyphosphate and NDPs that inhibit PNPase activity (14), the role of DnaK, GroEL, and enolase as they related to degradosome function is not known.
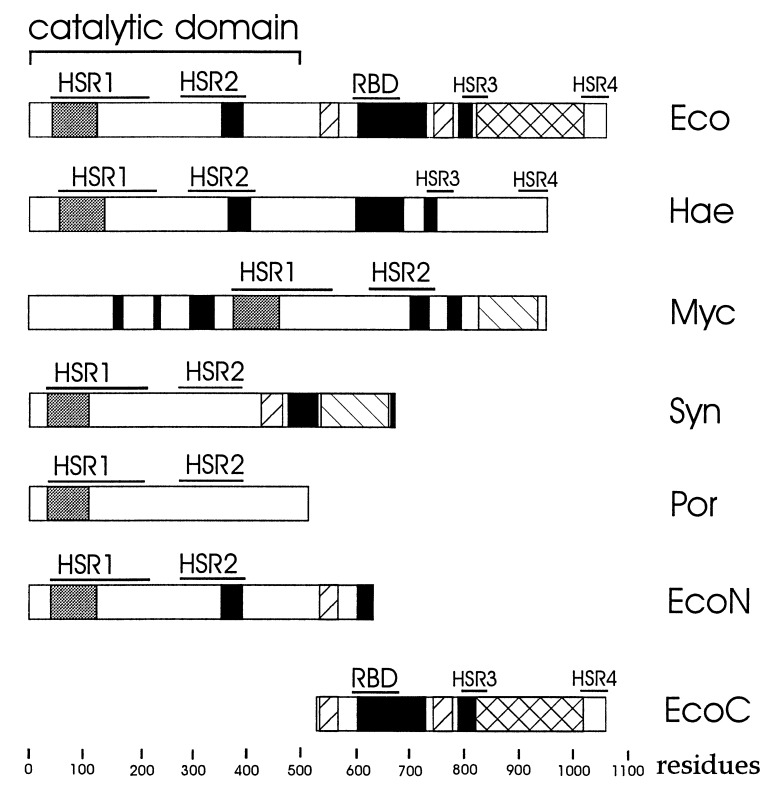
E. coli RNase E is a relatively large protein of 1,061 amino acids (15); however, its N-terminal half (Fig. 1) is sufficient in vitro for endoribonucleolytic cutting (16) and the removal of poly(A) tails (8). Interestingly, this domain of RNase E has high sequence similarity to the E. coli CafA protein (17), implicated in cell division and chromosomal segregation (18). Although, the precise role of CafA has yet to be determined, overexpression of the CafA protein partially complements temperature-sensitive mutations in RNase E (19), suggesting that CafA and the N-terminal half of RNase E may be related functionally. Recently, near the N terminus of RNase E an S1-like RNA-binding domain has been identified (ref. 20, Fig. 1), which is presumably required for the endoribonucleolytic activity of this enzyme.
Figure 1.
Structure of E. coli Rne (Eco) and proteins having similar sequences in Haemophilus influenzae Rd (Hae), Mycobacterium tuberculosis (Myc), Synechocystis sp. (Syn), Porphyra purpurea (Por). The N- and C-terminal segments of E. coli RNase E (EcoN and EcoC, respectively) also are shown. Horizontal bars indicate the location of two regions (designated HSR1 and HSR2; see Table 1) within the N-terminal endoribonucleolytic domain of E. coli RNase E, which are highly conserved in other bacteria, two regions (designated HSR3 and HSR4) within the C-terminal half of E. coli RNase E having significant sequence similarity to the corresponding region of the H. influenzae Rd Rne homologue, and an RNA-binding domain of E. coli RNase E (16). The putative S1-like RNA-binding domain predicted by Bycroft et al. (20) and regions enriched in arginine, proline, and acidic residues (see Table 1) are indicated by gray, black, and back-hatched and forward-hatched boxes, respectively.
Analyses of the sequence of the C-terminal half of RNase E have revealed segments rich in arginines, prolines, or acidic residues (Fig. 1; refs. 21 and 22 and this work), and a segment overlapping the centrally located arginine-rich region has been shown to bind RNase E substrates (16, 23) altering overall RNA conformation (23); however, the precise role of this arginine-rich RNA-binding domain in RNA decay has yet to be determined. Kido and associates (24) have isolated mutants containing RNase E truncated at its C terminus and shown that the protein lacking its C-terminal half is not able to interact with PNPase; therefore, although the C-terminal half contains region(s) required for degradosome assembly in addition to an RNA-binding site, it is dispensable for cell viability (24), only the N-terminal catalytic domain appears to be essential in E. coli.
Given the central importance of RNase E in E. coli RNA processing and decay there is considerable interest in identifying sequence and functional homologues of this enzyme in other organisms (for review, see ref. 1). Sequences with similarity to E. coli RNase E have been reported in the genomes of Haemophilus influenzae Rd (25), Rickettsia prowazekii (S. Andersson, personal communication), and cyanobacterium Synechocystis sp. (26) as well as in the plastid genome of the red algae Porphyra purpurea (27); however, to our knowledge none of these sequence homologues have been reported to have RNase E activity, bind RNA, or to be associated with a degradosome-like complex. Interestingly, although there is no sequence homologue in the genome of Bacillus subtilis (28), there is evidence for an RNase E-like activity in this organism (29). Therefore, not all of the RNase E-like activities discovered in other eubacteria, including the photosynthetic bacterium Rhodobacter capsulatus (30) and Streptomyces species S. lividans and S. coelicolor (31), and in Haloarcula marismortui (32) a member of the archaebacteria are necessarily evolutionarily related to E. coli RNase E. A functional RNase E homologue also has been identified in human cells (33). Furthermore, a cDNA clone derived from human cells with, however, no similarity to the rne gene has been reported to encode a gene product with an RNase E-like activity (34, 35). To gain insight into the extent to which the multifaceted functions of E. coli RNase E protein are evolutionarily conserved, we have taken both phylogenetic and experimental approaches. We report here that sequences within the N-terminal catalytic domain of E. coli RNase E are highly conserved in eubacteria and that the RNase E sequence homologue in a Synechocystis sp. binds RNA and cleaves RNase E substrates at the same sites as the E. coli enzyme, suggesting that RNase E-mediated mechanisms of RNA decay are not a unique feature of E. coli and it closest relatives. However, sequences within the RNase E C-terminal half, which we show here is sufficient for degradosome assembly in E. coli, are not highly conserved in the Synechocystis sp. and other bacteria, suggesting that in other RNase E-containing organisms this enzyme may differ in its interactions with other components of the RNA decay machinery. Support for this notion is provided by our finding that the Synechocystis sp. homologue when expressed in E. coli is unable to associate with any components of the degradosome.
MATERIALS AND METHODS
Construction of Expression Plasmids.
Construction of pVK–SSP010: a 2047-bp fragment containing the entire sequence of the Synechocystis sp. rne gene was generated by PCR amplification of the corresponding region of cosmid slr1129 (kindly provided by S. Tabata; see ref. 26) by using oligonucleotide primers 5′-GCATCTGTGCATATGCCAAAACAAATTGT-3′ and 5′-CGCAGGATCCTACTCCGCTGAAG-3′. The PCR product was cut with NdeI and BamHI, gel purified, and ligated into NdeI- and BamHI-cut pRE196 (12). Construction of pRE220 encoding FLAG-tagged C terminus of E. coli RNase E: a DNA fragment of pRE196 spanning the E. coli rne-coding region 2,140–3,648 bp (numbering according to ref. 22) was amplified by using primers: 5′-ATATCATATGCCGGATGTGCCG-3′ and 5′-GCCGAATTCGAAGGCAAAAGTAG-3′. The resulting PCR product was cut with NdeI and EcoRI and inserted into pRE196 cut with the same enzymes. The missing T7 terminator region was reconstituted by replacing the 573 bp BamHI/EcoRI-fragment with the 1,062-bp BamHI/EcoRI fragment from the original pRE196 (12). Construction of pRE197 encoding FLAG-tagged N terminus of E. coli RNase E: pRE196 was digested with BamHI and NruI, and the resulting ends of the plasmid were filled and religated.
Expression and Purification of Proteins.
FLAG-tagged full-length E. coli Rne, its N- and C-terminal segments, and Synechocystis sp. Rne were overexpressed in E. coli BL21(DE3) cells (Novagen) containing pRE196 (12), pRE197, pRE220, and pVK-SSP010, respectively. Cells containing the appropriate plasmid were grown exponentially in Luria–Bertani medium containing carbenicillin (100 μg/ml) at 30°C with shaking (150 rpm). At an OD600 of 0.5–0.6, isopropyl β-d-thiogalactoside was added to a final concentration of 1 mM, and the cultures were incubated for an additional period of 2 h. The cells were collected by centrifugation at 4°C, and the FLAG-tagged polypeptide was purified as described previously (12), including the micrococcal nuclease treatment, which prevents the copurification of proteins dependent on the presence of RNA (12).
RNA Synthesis and Labeling.
Internally labeled E. coli 9S RNA and RNAI were synthesized by using HaeIII-linearized plasmid pTH90 (36) and a PCR-generated template (23), respectively, and an in vitro transcription kit from Stratagene.
In Vitro RNase E Assay.
The RNAI or 9S RNA substrates were incubated separately with equimolar amounts of the E. coli degradosome or Synechocystis sp. Rne protein in RNase E reaction buffer (23) at 30°C. The reaction mixture was supplemented with the EGTA (to a final concentration of 5 mM), which specifically chelates calcium and thereby inhibits the activity of any traces of micrococcal nuclease. Aliquots were removed from the reactions at different time points, extracted with phenol, ethanol precipitated, and mixed with an equal volume of sequencing dye (37). The samples were denatured for 3 min at 85°C before running in a 7% polyacrylamide/urea sequencing gel.
Northwestern Analysis.
The preparations of purified E. coli degradosome and Synechocystis sp. Rne protein were mixed separately with 2× SDS/polyacrylamide gel-loading buffer (37) and incubated at 100°C for 5 min, and aliquots of each sample were run on an 8% polyacrylamide/SDS gel (38). The proteins were transferred to poly(vinylidene difluoride) membrane (Millipore) by electroblotting for 2 h at 0.5 A/cm2 and probed with internally labeled RNAI and 9S RNA as described (39).
Western Blot Analysis.
Protein preparations were probed with anti-FLAG (Eastman Kodak), anti-Rne, anti-Pnp, anti-RhlB, and anti-enolase (generous gifts from A. J. Carpousis, Toulouse, France) antibodies by using the ECL Western blot detection kit (Amersham).
Protein Sequencing.
Proteins were separated by SDS/PAGE, transferred to poly(vinylidene difluoride) membrane (Immobilon, Millipore), stained with Coomassie brilliant blue R250, and submitted for N-terminal sequencing (Biocenter Core Facility, Vienna, Austria).
Sequence Comparisons.
The Wisconsin Package, version 8, Genetic Computer Group (Madison, WI) was used to identify and align RNase E-like protein sequences in the GenBank and SwissProt databases.
RESULTS AND DISCUSSION
Comparative Analysis of RNase E-Like Proteins of Bacterial and Organellic Origin.
Sequences reported to have similarity to E. coli RNase E (see above) and a sequence of a Mycobacterium tuberculosis homologue found in the European Molecular Biology Laboratory (EMBL) database were aligned by using the pileup program (see Materials and Methods) to identify conserved sequences and were analyzed for unusually arginine-, proline- and acidic-rich segments, which have been noted in the C-terminal half of E. coli RNase E (21, 22). As shown in Fig. 1, sequences within the N-terminal endoribonucleolytic half of RNase E (residues 1–498) are highly conserved. For example, the Porphyra purpurea homologue, which in this study is the one most distantly related to E. coli (27), has 54% similarity (25% identity) to the N-terminal half of E. coli RNase E. For all of the RNase E homologues in this study, the sequence similarity is highest in two regions of the E. coli enzyme between residues 41 and 222 and residues 278 and 398, respectively (Fig. 1 and Table 1). The first of these regions in E. coli RNase E contains the S1-like RNA-binding domain (residues 41–122; Fig. 1; see ref. 20), whereas the second may be required for endoribonucleolytic activity (16). The high degree of sequence similarity to these regions in the N-terminal half of E. coli RNase E suggests that the corresponding regions in the homologues may have similar functions.
Table 1.
General characteristics of RNase E-like proteins showing sequence similarity to the E. coli Rne
| Organism, ref. no. | Acession no. in nucleic acid (and/or protein) database | Length, aa | % identity to the N terminus (residues 1–498) of E. coli RNase E | Coordinates of the regions
|
|
|---|---|---|---|---|---|
| High sequence similarity (HSR1 and HSR2) | Enriched in arginine, prolin, and acidic residues | ||||
| E. coli (22) | X67470 (GenBank) P21513 (SwissProt) | 1,061 | 100 | 41–222 (HSR1), 278–398 (HSR2) | 354–391, 604–730, 789–819; 534–568, 743–778; 821–1023 |
| Haemophilus influenzae Rd (25) | U3272 (GenBank) P4443 (SwissProt) | 951 | 82 | 57–238 (HSR1), 293–413 (HSR2) | 369–406, 598–788, 727–751 |
| Synechocystis sp. PCC6803 (26) | D90899 (GenBank) | 674 | 34.9 | 37–213 (HSR1), 271–391 (HSR2) | 473–527, 663–670; 428–465, 504–524; 536–660 |
| Porphyra purpurea (27) | P51211 (SwissProt) | 511 | 25 | 37–213 (HSR1), 271–391 (HSR2) | No extensive arginine-rich, proline-rich, or acidic stretches |
| Mycobacterium tuberculosis (50) | Z81451 (GenBank) | 953 | 35.5 | 378–559 (HSR1), 622–743 (HSR2) | 154–171, 230–238, 295–340, 701–736, 771–795; 827–934 |
In contrast to the N-terminal half (1–498 aa) of E. coli RNase E, the C-terminal half of this enzyme (499–1061 aa) is not highly conserved. All of the C-terminal portions of the homologues in this study are shorter than that of E. coli RNase E (Fig. 1); furthermore, by using the fasta program, we were unable to identify any segments in the C-terminal half of the E. coli RNase E where the order of amino acid residues is conserved in all of the homologues. The only C-terminal sequence similarity we identified was between two regions [Fig. 1, high sequence similarity region (HSR) 3 and 4] in E. coli RNase E (residues 797–846 and 1,005–1,056) and the H. influenzae Rd homologue (residues 733–779 and 896–927, respectively), which in this study is the closest evolutionarily to the E. coli enzyme. As H. influenzae Rd also contains homologues of PNPase, RhlB, and enolase (25) that are highly similar in sequence to the corresponding enzymes in E. coli, the sequence similarity between two regions in the C-terminal half of E. coli RNase E and the H. influenzae Rd homologue may reflect conservation of function such as the interaction with other degradosome components.
All of the homologues with the exception of Porphyra purpurea contain at least one arginine-rich segment; the M. tuberculosis and Synechocystis sp. homologues also contained an acidic region, and a proline-rich region was identified in Synechocystis sp. (Fig. 1). Therefore, the C-terminal end of RNase E may share with some of its homologues function(s) requiring segments biased in amino acid composition but not identifiable by a particular order of amino acid residues. Interestingly, in contrast to E. coli RNase E in which the two arginine-rich regions are in the C-terminal half, the largest of the two arginine-rich domains in M. tuberculosis precedes the segment having high similarity to the N-terminal catalytic domain of RNase E raising the possibility that there is flexibility in the ordering of functional modules within RNase E and its homologues.
Synechocystis sp. and E. coli RNase E have Similar Catalytic and RNA-Binding Properties.
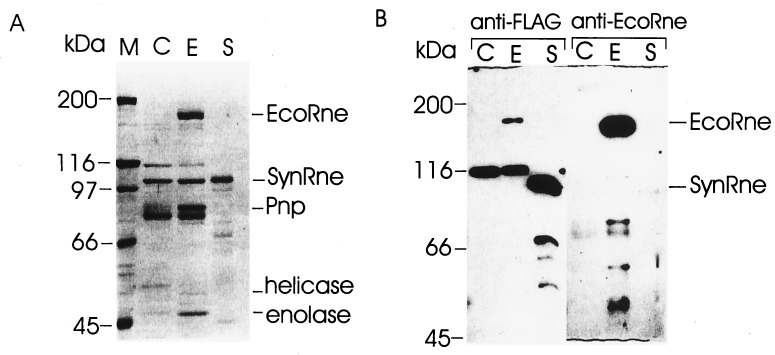
To determine whether sequence similarity to E. coli RNase E reflects evolutionary conservation at the level of function, the Synechocystis sp. homologue, which we will refer to as SynRne, was purified as a fusion to the FLAG tag (40) and assayed for the ability to cleave and bind RNase E substrates. As shown in Fig. 2A, the SynRne preparation contained a 105-kDa protein (as judged by its mobility in the SDS/polyacrylamide gel) that was >95% pure (lane S) and did not contain PNPase, RhlB helicase, and enolase usually copurifying with E. coli RNase E (lane E vs. lane S). The polypeptides of 80, 100, and 116 kDa (lane C) are retained at the column even if the extracts contain low or no FLAG-tagged RNase E and seem to copurify with the degradosome (12). Thus, they are not degradosomal components because this copurification is not dependent on the interaction with RNase E. Probing of the SynRne preparation with anti-FLAG antibodies (Fig. 2B) identified species smaller than 105 kDa, suggesting that minor contaminating species in the preparation are proteolytic products of full-length SynRne. Furthermore, as a control, we showed by using polyclonal antibodies raised against E. coli RNase E that the SynRne preparation does not contain detectable amounts of the E. coli enzyme (Fig. 2B).
Figure 2.
Purification of the Synechocystis sp. RNase E. E. coli cell extracts lacking overexpressed protein (C) or containing overexpressed E. coli (E) or Synechocystis sp. (S) RNase E were treated with micrococcal nuclease and purified on the anti-FLAG gel column as described (12). The preparations were run in triplicate in a 8% polyacrylamide/SDS gel. One-third of the gel was stained with Coomassie brilliant blue (A), whereas the remainder of the gel was used for blotting the proteins onto poly(vinylidene difluoride) membrane, and the remaining two-thirds probed either with anti-FLAG or anti-EcoRne antibodies (B). Indicated are protein size markers (kDa), Synechocystis sp. Rne (SynRne), and the major components of the E. coli degradosome; RNase E (EcoRne), polynucleotide phosphorylase (Pnp), the RhlB RNA helicase, and enolase. Although SynRne migrates in SDS/polyacrylamide gels with an apparent molecular mass of 105 kDa, the predicted molecular mass of SynRne is 78 kDa. A possible explanation for this observation is that SynRne contains a region rich in prolines, which have been shown to affect the migration of other proteins, including E. coli RNase E (22).
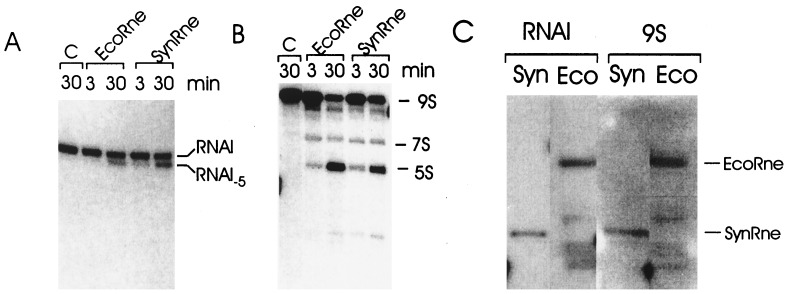
To determine whether SynRne has endoribonucleolytic activity, it was incubated with 9S RNA and RNAI, both of which are well characterized substrates of E. coli RNase E. We found that both these RNAs were cut by SynRne (Fig. 3 A and B); moreover, the pattern of cleavage products generated by the Synechocystis sp. enzyme was the same as that produced by E. coli RNase E. We therefore conclude that SynRne is an endoribonuclease whose cleavage specificity is indistinguishable from that of E. coli RNase E and that some and possibly all of the other sequence homologues in this study also may have RNase E-like RNA cleavage activity. In addition, by using a Northwestern assay, we detected RNA binding by SynRne (Fig. 3C). Interestingly, however, in the case of E. coli RNase E, this approach successfully detected RNA binding to a centrally located arginine-rich region (16) but not to the N-terminal endoribonucleolytic domain. It should therefore not be assumed for SynRne that the RNA binding we have detected is due to sequences having similarity to the N-terminal half of E. coli RNase E, it could well be due to one or both of its arginine-rich regions.
Figure 3.
In vitro cleavage of RNAI (A) and 9S RNA (B) by the E. coli and Synechocystis sp. Rne proteins. Internally labeled RNAI or 9S RNA was incubated in RNase E reaction buffer for 3 and 30 min with either E. coli RNase E (EcoRne) or Synechocystis sp. Rne (SynRne), or without protein for 30 min (C), as a negative control. The positions of the original substrates (RNAI and 9S RNA) and their cleavage products (RNA I-5 and 5S rRNA (5S), respectively) are indicated. (C) Protein blots probed with RNAI and 9S RNA. Samples of the Synechocystis sp. Rne protein (Syn) and the E. coli RNA degradosome (Eco) were resolved by SDS/PAGE, electroblotted onto poly(vinylidene difluoride) membrane, and probed with 32P-labeled RNAI or 9S RNA as described in Materials and Methods. The Synechocystis sp. Rne (SynRne) and E. coli Rne (EcoRne) proteins complexed with RNA are indicated.
C-Terminal Half of E. coli RNase E Is Sufficient to Recruit the Major Degradosome Proteins.
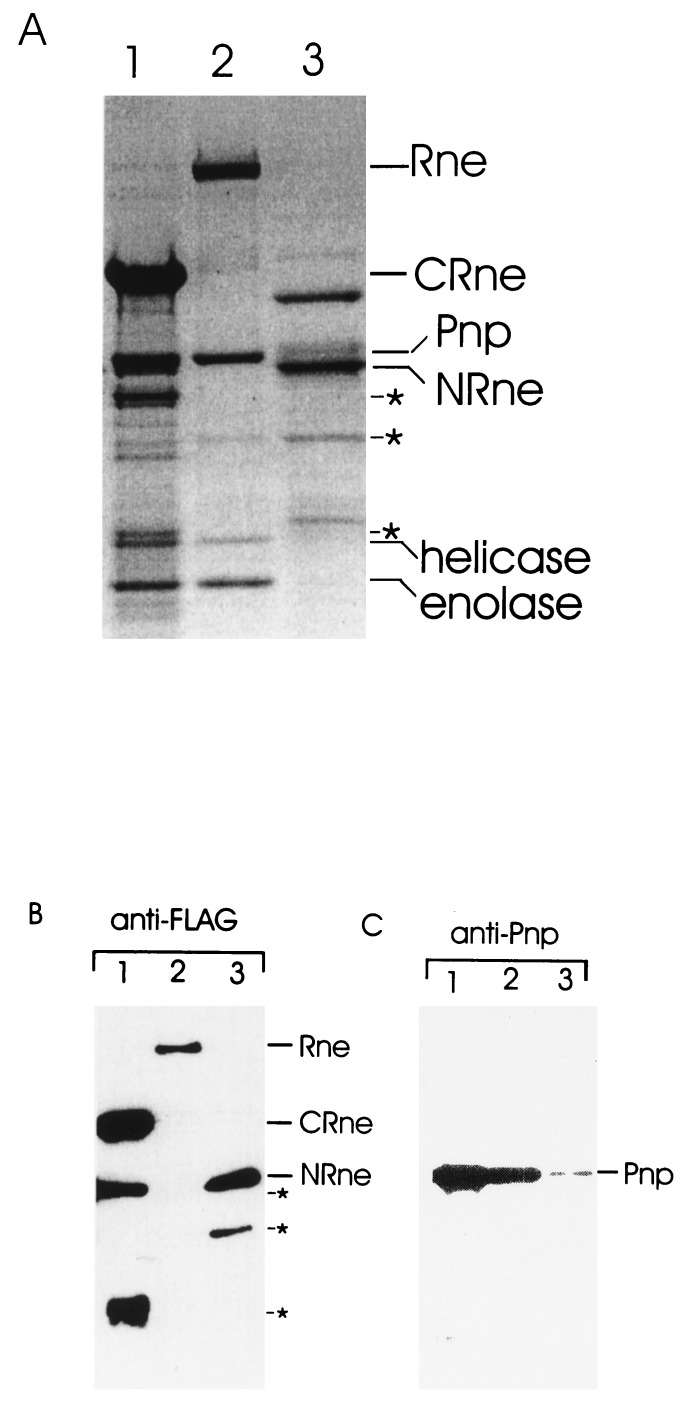
Our finding that SynRne is unable to recruit any components of the E. coli degradosome even though it has significant sequence similarity to the N-terminal half of RNase E prompted us to investigate whether this was because all of the sites required for degradosome assembly are contained within the C-terminal end of E. coli RNase E. We expressed and purified a FLAG-tagged RNase E polypeptide containing residues 530–1,061 (CRne) and analyzed the preparation for the other major degradosome components. FLAG-tagged full-length RNase E (Rne) and an Rne polypeptide containing residues 1–634 (NRne) also were purified as controls. As shown in Fig. 4A, we found copurifying with the C-terminal RNase E polypeptide (lane 1) polypeptides having the expected molecular masses of PNPase (85 kDa), RhlB (50 kDa), and enolase (48 kDa). Moreover, the stoichiometry of these polypeptides in the CRne preparation was indistinguishable from that of the full-length RNase E preparation (lane 2). In contrast, we did not detect the stoichiometric association of polypeptides of 85, 50, and 48 kDa with NRne (lane 3). Probing of the preparations with anti-FLAG antibodies (Fig. 4B) confirmed the identity of the CRne (120 kDa), Rne (180 kDa), and NRne (80 kDa) polypeptides and revealed minor proteolytic products of CRne and NRne (lanes 1 and 3, respectively). The identity of PNPase and RhlB helicase and enolase in the CRne and full-length Rne preparations (lanes 1 and 2, respectively) was confirmed by probing the preparation with anti-PNPase antibodies (Fig. 4C), with anti-enolase and anti-RhlB antibodies (data not shown) and/or N-terminal sequencing of the candidate proteins. Taken together the above results provide good evidence that the C-terminal half of RNase E is sufficient as a platform for degradosome assembly. Interestingly, probing of the NRne preparation with antibodies against PNPase revealed the presence of this 3′ exonuclease; however, the actual amount was at least an order of magnitude lower than that found in the full-length or C-terminal preparations suggesting that in the region of overlap between NRne and CRne (residues 530–634) there is a segment(s) that binds PNPase but is not sufficient for the assembly of stable degradosomes.
Figure 4.
Identification of the proteins associated with the C-terminal half of E. coli Rne. Cell extracts containing overexpressed full-length E. coli Rne or N- or C-terminal portions of this polypeptide (see Fig. 1) were treated with micrococcal nuclease and purified by affinity chromatography as described in Materials and Methods. Samples of each preparation were run in triplicate on an 8% polyacrylamide/SDS gel (lanes 2, 3, and 1, respectively). As described for Fig. 2B, one-third of the gel was stained with Coomassie brilliant blue (A), whereas the remaining two-thirds were either probed with anti-FLAG (B) or anti-Pnp (C) antibodies after transfer to a poly(vinylidene difluoride) membrane. The position of the major components of the E. coli RNA degradosome, RNase E (Rne), polynucleotide phosphorylase (Pnp), enolase, RhlB helicase as well as N- and C-terminal fragments (NRne and CRne, respectively) of E. coli Rne are indicated. The proteins marked by asterisks and detected by Western blot with anti-FLAG antibodies are proteolytic products of NRne and CRne.
CONCLUSIONS
Over the last few years, the explosion in genome sequencing has facilitated the extent to which genes having important roles in one organism are conserved in others to be determined, thereby, providing important insight into the evolution of cellular processes (cf. 41). The results of the computer-assisted sequence comparisons reported here (Fig. 1 and Table 1) showed that the N-terminal half of the multifaceted E. coli RNase E protein is evolutionarily conserved in other bacteria and in a plastid and led us to test whether the RNase E sequence homologue in a cyanobacterium Synechocystis sp., which is thought to have diverged from E. coli over 3 billion years ago (42), has endoribonucleolytic activity. Our finding that the Synechocystis sp. homologue not only cuts RNA but cuts at the same sites as E. coli RNase E (Fig. 2) provides good experimental support for the notion that the factors controlling mRNA processing and decay are ancient and highly conserved evolutionarily (43). However, although homologues of the N-terminal half of RNase E have been identified in the genomes of all the Gram-negative bacteria that have been completely sequenced, there appears to be a sporadic distribution of RNase E sequence homologues in the Gram-positive bacteria. In addition to the M. tuberculosis homologue, we have identified sequence similarity to the N-terminal half of RNase E in the partially sequenced genome of Streptomyces coelicolor (data not shown), but no sequence homologues of RNase E have been identified in the completely sequenced genomes of Bacillus subtilis (28), Mycoplasma genitalium and pneumoniae (44, 45), Helicobacter pylori (46), and Borrelia burgdorferi (47). However, although there is no RNase E sequence homologue in B. subtilis, there is good evidence that this bacterium does contain an RNase E-like endoribonucleolytic activity (29), suggesting that in some eubacteria RNase E sequence homologues became redundant through the acquisition of another gene (for review, see ref. 48) encoding RNase E-like activity.
As previous Northwestern analyses of E. coli RNase E detected RNA binding to a segment rich in arginine residues, but not to its N-terminal endoribonucleolytic domain (16, 49), the RNA-binding activity we have found for the Synechocystis sp. homologue (Fig. 3C) may be conferred by its arginine-rich region rather than the N-terminal region having sequence similarity to the endoribonucleolytic domain of E. coli RNase E. Whether or not this proves to be correct, our results show that RNA binding is another facet shared by E. coli RNase E and the Synechocystis sp. homologue. Our finding that the Synechocystis sp. homologue does not interact physically with any of the major components of the E. coli degradosome (Fig. 2A) reflects the overall lack of evolutionary conservation of the E. coli RNase E C-terminal half (Fig. 1), which we have shown here is sufficient as a platform for degradosome assembly in E. coli (Fig. 4). This result together with the recent finding that the C-terminal half of RNase E is not essential for cell viability (24), only for the normal rapid decay of RNA in E. coli, suggest that during evolution there has been scope for divergence in the interactions between RNase E and other components of the decay machinery.
Acknowledgments
We thank Dr. S. Tabata for the generous gift of the cosmid slr1129 and Dr. A. J. Carpousis for providing anti-Rne, anti-Pnp, anti-RhlB, and anti-enolase antibodies. We also thank Dr. R.-P. Nilsson and P. Camaj for valuable advice on computer graphics. This work was supported by grants from Fonds zur Förderung der Wissenschaftlichen Forschung to A.v.G., from the Austrian government to A.M, and from the National Science Council of the Republic of China to S.L.-C. and A.M.
ABBREVIATIONS
- PNPase
polynucleotide phosphorylase
- RhlB
RhlB RNA helicase
- FLAG
oligopeptide Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys
- HSR
high sequence similarity region
Note Added in Proof
A detailed analysis of the protein–protein interactions with the E. coli RNA degradosome is reported in ref. 51.
References
- 1. Cohen S N, McDowall K J. Mol Microbiol. 1997;23:1099–1106. doi: 10.1111/j.1365-2958.1997.tb02593.x. [DOI] [PubMed] [Google Scholar]
- 2.Apirion D, Lassar A B. J Biol Chem. 1978;253:1738–1742. [PubMed] [Google Scholar]
- 3.Ehretsmann C P, Carpousis A J, Krisch H M. FASEB J. 1992;6:3186–3192. doi: 10.1096/fasebj.6.13.1397840. [DOI] [PubMed] [Google Scholar]
- 4.Melefors Ö, Lundberg U, von Gabain A. In: Control of Messenger RNA Stability. Belasco J, Brawerman G, editors. San Diego: Academic; 1993. pp. 53–70. [Google Scholar]
- 5.Lin-Chao S, Cohen S N. Cell. 1991;65:1233–1242. doi: 10.1016/0092-8674(91)90018-t. [DOI] [PubMed] [Google Scholar]
- 6.Söderbom F, Binnie U, Masters M, Wagner G. Mol Microbiol. 1997;26:493–504. doi: 10.1046/j.1365-2958.1997.5871953.x. [DOI] [PubMed] [Google Scholar]
- 7.Mikkelsen N D, Gerdes K. Mol Microbiol. 1997;26:311–320. doi: 10.1046/j.1365-2958.1997.5751936.x. [DOI] [PubMed] [Google Scholar]
- 8.Huang H, Liao J, Cohen S N. Nature (London) 1998;391:99–102. doi: 10.1038/34219. [DOI] [PubMed] [Google Scholar]
- 9.Cohen S N. Cell. 1995;80:829–832. doi: 10.1016/0092-8674(95)90284-8. [DOI] [PubMed] [Google Scholar]
- 10.Carpousis A J, Van Houwe G, Ehretsmann C, Krisch H M. Cell. 1994;76:889–900. doi: 10.1016/0092-8674(94)90363-8. [DOI] [PubMed] [Google Scholar]
- 11.Py B, Causton H, Mudd E A, Higgins C F. Mol Microbiol. 1994;14:717–729. doi: 10.1111/j.1365-2958.1994.tb01309.x. [DOI] [PubMed] [Google Scholar]
- 12.Miczak A, Kaberdin V R, Wei C-L, Lin-Chao S. Proc Natl Acad Sci USA. 1996;93:3865–3869. doi: 10.1073/pnas.93.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Py B, Higgins C F, Krisch H M, Carpousis A J. Nature (London) 1996;381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 14.Blum E, Py B, Carpousis A J, Higgins C F. Mol Microbiol. 1997;26:387–398. doi: 10.1046/j.1365-2958.1997.5901947.x. [DOI] [PubMed] [Google Scholar]
- 15.Casaregola S, Jacq A, Laoudj D, McGurk G, Margarson S, Tempete M, Norris V, Holland I B. J Mol Biol. 1994;238:867. doi: 10.1006/jmbi.1994.1344. [DOI] [PubMed] [Google Scholar]
- 16.McDowall K J, Cohen S N. J Mol Biol. 1996;255:349–355. doi: 10.1006/jmbi.1996.0027. [DOI] [PubMed] [Google Scholar]
- 17.McDowall K J, Hernandez R G, Lin-Chao S, Cohen S N. J Bacteriol. 1993;175:4245–4249. doi: 10.1128/jb.175.13.4245-4249.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okada Y, Wachi M, Hirata A, Suzuki K, Nagai K, Matsuhashi M. J Bacteriol. 1994;176:917–922. doi: 10.1128/jb.176.3.917-922.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wachi M, Umitsuki G, Nagai K. Mol Gen Genet. 1997;253:515–519. doi: 10.1007/s004380050352. [DOI] [PubMed] [Google Scholar]
- 20.Bycroft M, Hubbard T J P, Proctor M, Freund S M V, Murzin A G. Cell. 1997;88:235–242. doi: 10.1016/s0092-8674(00)81844-9. [DOI] [PubMed] [Google Scholar]
- 21.Claverie-Martin F, Diaz-Torres M, Yancey S D, Kushner S R. J Biol Chem. 1991;266:2843–2851. [PubMed] [Google Scholar]
- 22.Casaregola S, Jacq A, Laoudj D, McGurk G, Margarson S, Tempete M, Norris V, Holland I B. J Mol Biol. 1992;228:30–40. doi: 10.1016/0022-2836(92)90489-7. [DOI] [PubMed] [Google Scholar]
- 23.Kaberdin V R, Chao Y-H, Lin-Chao S. J Biol Chem. 1996;271:13103–13109. doi: 10.1074/jbc.271.22.13103. [DOI] [PubMed] [Google Scholar]
- 24.Kido M, Yamanaka K, Mitani T, Niki H, Ogura T, Hiraga S. J Bacteriol. 1996;178:3917–3925. doi: 10.1128/jb.178.13.3917-3925.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, et al. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 26.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosava M, Sugiura M, Sasamoto S, et al. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 27.Reith M, Munholland J. Plant Mol Biol Rep. 1995;13:333–335. [Google Scholar]
- 28.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, et al. Nature (London) 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 29.Condon C, Putzer H, Luo D, Grunberg-Manago M. J Mol Biol. 1997;268:235–242. doi: 10.1006/jmbi.1997.0971. [DOI] [PubMed] [Google Scholar]
- 30.Fritsch J, Rothfuchs R, Rauhut R, Klug G. Mol Microbiol. 1995;15:1017–1029. doi: 10.1111/j.1365-2958.1995.tb02277.x. [DOI] [PubMed] [Google Scholar]
- 31.Hagege J M, Cohen S N. Mol Microbiol. 1997;25:1077–1090. doi: 10.1046/j.1365-2958.1997.5311904.x. [DOI] [PubMed] [Google Scholar]
- 32.Franzetti B, Sohlberg B, Zaccai G, von Gabain A. J Bacteriol. 1997;179:1180–1185. doi: 10.1128/jb.179.4.1180-1185.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wennborg A, Sohlberg B, Angerer D, Klein G, von Gabain A. Proc Natl Acad Sci USA. 1995;92:7322–7326. doi: 10.1073/pnas.92.16.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang M, Cohen S N. Proc Natl Acad Sci USA. 1994;91:10591–10595. doi: 10.1073/pnas.91.22.10591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Claverie-Martin F, Wang M, Cohen S N. J Biol Chem. 1997;272:13823–13828. doi: 10.1074/jbc.272.21.13823. [DOI] [PubMed] [Google Scholar]
- 36.Sohlberg B, Lundberg U, Hartl F-U, von Gabain A. Proc Natl Acad Sci USA. 1993;90:277–281. doi: 10.1073/pnas.90.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 38.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 39.Cormack R S, Genereaux J L, Mackie G A. Proc Natl Acad Sci USA. 1993;90:9006–9010. doi: 10.1073/pnas.90.19.9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiang C M, Roeder R G. Peptide Res. 1993;6:62–64. [PubMed] [Google Scholar]
- 41.Quail P H. BioEssays. 1997;19:571–579. doi: 10.1002/bies.950190708. [DOI] [PubMed] [Google Scholar]
- 42.Dickerson R E. Sci Am. 1980;242:136–153. [PubMed] [Google Scholar]
- 43.Darr S C, Brown J W, Pace N R. Trends Biochem Sci. 1992;17:178–182. doi: 10.1016/0968-0004(92)90262-8. [DOI] [PubMed] [Google Scholar]
- 44.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, et al. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 45.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li B C, Herrmann R. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, et al. Nature (London) 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 47.Fraser C M, Casjens S, Huang W-M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E, et al. Nature (London) 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 48.Doolittle R F. Nature (London) 1997;392:339–342. doi: 10.1038/32789. [DOI] [PubMed] [Google Scholar]
- 49.Taraseviciene L, Björk G, Uhlin B E. J Biol Chem. 1995;270:26391–26398. doi: 10.1074/jbc.270.44.26391. [DOI] [PubMed] [Google Scholar]
- 50.Cole S T, Brosch R, Parkhill J, Garnier J, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, et al. Nature (London) 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 51.Vanzo, N. F., Li, Y. S., Py, B., Blum, E., Higgins, C. F., Raynal, L. C., Krisch, H. M. & Carpousis, A. J. (1998) Genes Dev., in press. [DOI] [PMC free article] [PubMed]