Abstract
Translocation of nuclear-encoded preproteins across the outer membrane of mitochondria is mediated by the multicomponent transmembrane TOM complex. We have isolated the TOM core complex of Neurospora crassa by removing the receptors Tom70 and Tom20 from the isolated TOM holo complex by treatment with the detergent dodecyl maltoside. It consists of Tom40, Tom22, and the small Tom components, Tom6 and Tom7. This core complex was also purified directly from mitochondria after solubilization with dodecyl maltoside. The TOM core complex has the characteristics of the general insertion pore; it contains high-conductance channels and binds preprotein in a targeting sequence-dependent manner. It forms a double ring structure that, in contrast to the holo complex, lacks the third density seen in the latter particles. Three-dimensional reconstruction by electron tomography exhibits two open pores traversing the complex with a diameter of ∼2.1 nm and a height of ∼7 nm. Tom40 is the key structural element of the TOM core complex.
Keywords: TOM complex, mitochondria, protein translocation channel, electron tomography, protein targeting
Transport of nuclear-encoded proteins into mitochondria is mediated by distinct translocation machineries located in the outer and inner membranes of mitochondria. Components in the outer membrane, which facilitate the recognition of preproteins, their transfer through the outer membrane, and the insertion of resident outer membrane proteins, are organized in the TOM complex (for review see Haucke and Schatz 1997; Neupert 1997; Ryan and Pfanner 1998; Bains and Lithgow 1999). Two translocation machineries in the inner membrane (TIM complexes), which are specific for different subsets of preproteins, mediate the transfer of preproteins across or into the inner membrane (Sirrenberg et al. 1996, Sirrenberg et al. 1998; Neupert 1997; Pfanner and Meijer 1997; Köhler et al. 1998). Studies with Neurospora crassa and Saccharomyces cerevisiae have shown that preproteins are first recognized by specific receptor components of the TOM complex exposing hydrophilic domains into the cytosol. Preproteins are bound and then transferred to a specific protein conducting channel, also known as the general import/insertion pore (GIP), that translocates proteins through the outer membrane (Pfaller et al. 1988; Kiebler et al. 1990).
Three import receptors, Tom20 (Söllner et al. 1989; Ramage et al. 1993), Tom22 (Kiebler et al. 1993; Lithgow et al. 1994; Hönlinger et al. 1995; Nakai and Endo 1995), and Tom70 (Hines et al. 1990; Söllner et al. 1990) were identified in both Neurospora and S. cerevisiae. Two other components with predicted receptor function, Tom71, a Tom70 homologue (Schlossmann et al. 1996), and Tom37 (Gratzer et al. 1995), have been identified only in S. cerevisiae, so far. Tom22 appears to be involved in the recognition of preproteins and is associated with the insertion pore (Dekker et al. 1998; Künkele et al. 1998). Tom20, together with Tom22, mainly binds preproteins with NH2-terminal matrix targeting sequences and proteins destined for insertion into the outer membrane (Mayer et al. 1995; Brix et al. 1997). Tom70 and Tom71 were found to preferentially bind preproteins with internal targeting information (Schlossmann et al. 1994; Brix et al. 1997). Those components, which appear to form the general protein import pore, include Tom40 (Vestweber et al. 1989; Kiebler et al. 1990) and three smaller proteins, Tom7 (Hönlinger et al. 1996), Tom6 (Kassenbrock et al. 1993; Alconada et al. 1995; Cao and Douglas 1995), and Tom5 (Dietmeier et al. 1997). Tom22 has hydrophilic domains in the cytosol and the intermembrane space; a function beyond initial precursor binding has been proposed (Nakai et al. 1995; Court et al. 1996; Moczko et al. 1997; Komiya et al. 1998).
The recent isolation and purification of the TOM holo complex of Neurospora has provided information about its composition, structure, and channel function (Künkele et al. 1998). The isolated complex contained all established import receptors, as well as the GIP components. A Tom5 equivalent has not been identified in Neurospora so far. No other components were present.
A deep understanding of protein translocation across the outer membrane of mitochondria requires structural information of the TOM protein-conducting channel itself. Here, we report on the isolation and the structure of the TOM core complex comprising the general import pore. Its constituents are Tom40, the major pore forming protein, Tom22, and the small Tom components, Tom7 and Tom6. An as yet unidentified band of the size of yeast Tom5 is also present in the core complex. The isolated core complex forms a high conductance channel in lipid membranes with properties similar to those of the holo complex. Analysis of the binding activity of the core complex to a translocation substrate in detergent solution indicates that it fulfills protein import functions. Single particle EM reveals particles, the majority of which contain two centers of stain accumulation and a less abundant complex with one center. These likely represent protein-conducting channels. Electron tomography and three-dimensional (3D) image reconstruction yielded a map of the TOM core complex with two channels crossing the outer membrane.
Materials and Methods
Isolation and Purification of the TOM Core Complex
TOM holo complex and TOM core complex were isolated and purified from mitochondrial membranes of a Neurospora strain (GR-107) in which the wild-type Tom22 is replaced with a version of Tom22 encoding a protein with a hexahistidinyl tag at its COOH terminus. Growth of the Neurospora cells and preparation of mitochondria were performed as described previously (Sebald et al. 1979; Künkele et al. 1998). Mitochondria were solubilized for 30 min at 4°C in 50 mM potassium acetate, 10 mM MOPS, pH 7.0, 20% glycerol, and 1% (wt/vol) n-dodecyl β-D-maltoside (DDM; Anatrace Inc.) in the presence of 1 mM PMSF at a protein concentration of 10 mg ml−1. Insoluble material was removed by centrifugation at 257,320 g for 30 min. The clarified extract was loaded onto a nickel-nitrilotriacetic acid-agarose (Ni-NTA; Quiagen) column using 4 ml resin per 1 g of total mitochondrial protein. The column was washed with 20 vol 50 mM potassium acetate, 10 mM MOPS, pH 7.0, 20% glycerol, 0.1% DDM, and 40 mM imidazole. Specifically bound material was eluted with 300 mM imidazole in the same buffer. For further purification, the TOM complex containing fractions were pooled and loaded onto a MonoQ (Pharmacia Biotech) anion-exchange column equilibrated with 50 mM potassium acetate, 10 mM MOPS, pH 7.0, 20% glycerol, and 0.1% DDM. The TOM complex was then eluted by a linear 0–500 mM KCl gradient in the same buffer. Stock solutions of purified TOM complex were stored at a protein concentration of ∼5–10 mg ml−1 at 4°C. The identity of the individual Tom proteins was verified by immunodetection with antibodies specific for the Tom components. An average preparation of the TOM core complex started with ∼1.5 kg of Neurospora cells (wet wt) which yielded ∼7 g of mitochondrial protein. The final preparation contained ∼10–15 mg pure TOM core complex.
For determination of the stoichiometry of Tom components, core complex was isolated from strain GR-107 grown in the presence of 35S-sulfate. The purified radio-labeled TOM core complex was subjected to SDS-PAGE. For the detection and quantification of radio-labeled proteins, dried gels were analyzed by phosphorimaging analysis.
Holo complex containing all established Tom components was purified from isolated Neurospora mitochondrial outer membrane vesicles in 0.5% (wt/vol) digitonin as previously described (Künkele et al. 1998).
For preparing of TOM core complex that lacks the hydrophilic receptor domains, core complex (900 μg) was incubated with 100 μg ml−1 trypsin in 100 μl 50 mM potassium acetate, 10 mM MOPS, pH 7.0, 20% glycerol, and 0.1% DDM at 0°C for 30 min. Proteolysis was stopped with trypsin inhibitor (0.5 mg ml−1) and proteolytic cleavage of TOM complex was assessed by SDS-PAGE.
Preparation of Chemical Amounts of pSu9-DHFR
A COOH terminally His-tagged fusion protein consisting of the presequence of subunit 9 of the F0-ATPase (residues 1–69) and dihydrofolate reductase (pSu9-DHFR) was expressed in Escherichia coli and purified by Ni-NTA chromatography. In brief, bacteria were grown overnight in 250 ml LB medium at 37°C. The overnight culture was diluted and grown to an OD600 of 0.8–0.9. Expression of pSu9-DHFR was induced by adding isopropyl thiogalactoside to a final concentration of 2 mM. Cells were grown for 1 h and harvested by centrifugation. The bacterial pellet was resuspended in buffer containing 50 mM NaHPO4, pH 8.0, 300 mM NaCl, 10 mM imidazole, 10% glycerol, 1 mM PMSF, 10 μg ml−1 α-macroglobulin, 10 μg ml−1 leupeptin, and 10 μg ml−1 pepstatin, and then sonicated using a Branson 250 sonifier. Unbroken cells were removed by centrifugation and supernatants were loaded onto a Ni-NTA affinity column. The column was washed with 10 column vol phosphate buffer, and bound material was eluted by a linear 10–300 mM imidazole gradient in the same buffer. The peak fractions containing 3–4 mg ml−1 of purified pSu9-DHFR were stored at −80°C.
Size Exclusion Chromatography
Purified TOM complex (100 μg) was loaded onto a TosoHaas TSK G4000 PWXL size-exclusion column equilibrated with 50 mM potassium acetate, 10 mM MOPS, pH 7.0, 10% glycerol, and 0.5% digitonin at room temperature using an Äkta chromatography system (Pharmacia Biotech, Inc.). Protein was eluted at a flow rate of 0.45 ml min−1. The absorbance of the eluant was monitored at 280 nm. The molecular weights of TOM complexes were calculated using thyroglobulin (669 kD), apoferritin (443 kD), alcohol dehydrogenase (155 kD), and carboanhydrase (29 kD) as protein standards.
TOM core complex used for EM was passed over a Superose 6 gel filtration column (Pharmacia Biotech), equilibrated with 50 mM potassium acetate, 10 mM MOPS, pH 7.0, 20% glycerol, and 0.1% DDM.
Gel Electrophoresis
Native PAGE was performed using a 4–15% acrylamide gradient (PhastGel; Pharmacia Biotech, Inc.). For blue native polyacrylamide electrophoresis (Schägger and von Jagow 1991), purified TOM complex (50 μg) was dissolved in 25 μl sample buffer, 0.5% (wt/vol) Coomassie brilliant blue G-250, 10 mM bis-Tris, pH 7.0, and 50 mM aminocaproic acid, and electrophoresed through 6–16.5% polyacrylamide gradient gels. Immunodecoration was performed by standard procedures and detection was achieved by the enhanced chemiluminescence method (Nycomed Amersham).
SDS-PAGE was performed according to the procedure described by Laemmli 1970, using 16% acrylamide and 0.6% bisacrylamide. To achieve a higher resolution of the smaller TOM components, high Tris/urea gels were used (Künkele et al. 1998).
Conductance Measurements
Conductance measurements of TOM complex in planar black lipid membranes were carried out as previously described (Benz et al. 1978; Künkele et al. 1998). Membranes were formed from a 1% (wt/vol) solution of dipytanoyl phosphatidyl choline (DPPC; Avanti Polar Lipids) in n-decane/butanol (9:1 vol/vol) across circular holes (surface area ∼0.1 mm2) in the wall of a Teflon cell separating two aqueous compartments of 5 ml each. The aqueous solutions contained 1 M KCl, 5 mM Hepes-KOH, pH 7.0 (σ0 = 96.7 mS cm−1).
The channel size of native TOM complex and TOM subcomplexes was determined by analyzing the partitioning of differently sized PEGs into the TOM complex channel. The electrolyte contained 1 M KCl, 5 mM Hepes-KOH, pH 7.0, and 20% (wt/vol) polyethylene glycol (PEG) of various molecular weight (Fluka; Sigma Chemical Co.). The bulk electrolyte conductances, σPEG, were the same for all PEG solutions (σPEG = 58.1 ± 0.4 mS cm−1, mean ± SEM). Membrane currents were measured at a membrane potential of +20 mV with a pair of Ag/AgCl electrodes (Metrohm) using a Keithley 428 current amplifier. Amplified signals were monitored with an analogue/digital storage oscilloscope (Hameg HM 407) and recorded with a strip chart recorder (Philips PM8100). The conductance of each buffer solution, σ0and σPEG, was measured using a Greisinger GLM 200A conductance meter.
Electron Microscopy Analysis
Purified TOM complex preparations (0.1 mg protein ml−1) were adsorbed to glow-discharged carbon-coated specimen support grids (Cu, 600 mesh or 400 × 100 mesh) for 30 s. The grids were washed twice with deionized water, blotted with filter paper, and stained with 2% (wt/vol) aqueous uranyl acetate for 60 s.
Projection images of isolated TOM complex were recorded at 0° tilt in a Philips CM12 transmission electron microscope operating at 120 kV at 43,800× and an underfocus of between 1 and 2.5 μm. Single particle images were processed as described (Künkele et al. 1998). In brief, micrographs were digitized into 2048 × 2048 × 16 bit arrays using an Eikonix densitometer at a step size of 0.34 nm/pixel. Further image processing was carried out on Silicon graphics workstations using the EM software described by Hegerl 1996. Images were low-pass filtered to the first order of the electron microscope contrast function, corresponding to cutoff frequencies between (0.18 nm)−1 and (0.3 nm)−1. 1,598 particles were manually marked in the digitized images. After extraction of frames with 64 × 64 pixels, the images were iteratively aligned with respect to translation and rotation via cross-correlation (Frank et al. 1981) to a synthetic double ring model with smooth borders as a first reference. Using multivariate statistical analysis (MSA; Frank and van Heel 1982), 30 eigenimages were calculated. Using the 10 most significant eigenimages, the data set was divided into 20 classes. Two sets of images were generated from classes representing particles with one and two pores. For the images showing two pores, two independent averages were calculated from images with odd and even numbers. From the Fourier-ring correlation (Saxton and Baumeister 1982) between those averages, a resolution of (2.5 nm)−1 was determined.
Electron tomography of TOM core complex stained with 2% (wt/vol) uranyl acetate was carried out using a Philips 200 FEG electron transmission microscope equipped with a VIPS-1000 computer (TVIPS) and a large-area CCD camera (Photometrics; Dierksen et al. 1992). The microscope was operated at 120 kV at an underfocus of 1.5 μm, an EM of 26,950× and a postmagnification factor of 2.05 on the CCD camera. This corresponded to a pixel size of 0.344 nm at the specimen. Data were collected within a tilt range of ±60°, with 6° angular increments. The mean dose per image was ∼1,600 e− nm−2. The direction of the tilt axis was determined from an independent tilt series of a specimen with 10-nm gold clusters.
Image processing of the tomographic data included the alignment of the projections of each tilt series to a common origin, the selection of single particles at 0° tilt, and the 3D reconstruction of individual particles by means of weighted backprojection.
From the 3D maps of 321 particles, projection images were calculated, aligned, and subjected to MSA. Particles that did not show two pores were excluded from the data set. For calculation of the final 3D model, the largest homogenous class of particles (n = 116) was subjected to refined 3D alignment with respect to all six alignment parameters (i.e., three Cartesian coordinates and three angles) using the 3D map of the previous cycle as a reference. To avoid biased 3D alignment and merging molecules with different up and down orientation, each individual particle was allowed to rotate by all three Euler angles in each refinement cycle.
For the visualization of the 3D model of the TOM core complex, the AVS/Express 4.0 software package (Advanced Visual Systems Inc.) was used. The surface models were, based on a protein density of 1.3 g cm−3, thresholded to a volume with an expected mass of 410 kD.
Results
Isolation of TOM Core Complex
The TOM core complex was prepared in two different ways: either by treatment of purified TOM holo complex with high concentrations of nonionic detergent and size-exclusion chromatography or by direct isolation from detergent-solubilized mitochondria in DDM.
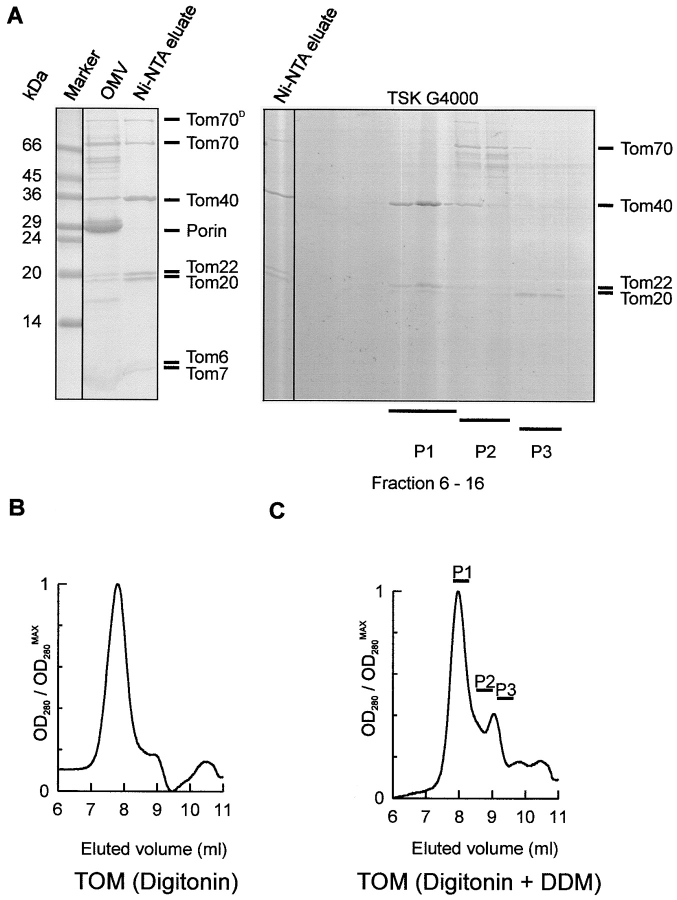
The TOM holo complex containing all the established Tom components was purified from isolated mitochondrial outer membrane vesicles prepared from a Neurospora strain bearing a hexahistidinyl-tagged form of Tom22 (Fig. 1 A, left; Künkele et al. 1998). Fig. 1 B shows the elution profile of TOM holo complex fractionated by size exclusion chromatography. The fractions were analyzed for polypeptide composition by PAGE. The peak fractions contained the established components, Tom70, Tom40, Tom22, Tom20, Tom7, and Tom6. According to the elution profile, the apparent molecular mass of the holo complex was estimated to be ∼490 kD, in agreement with earlier studies (Künkele et al. 1998).
Figure 1.
Dissociation of the purified TOM holo complex by detergent treatment into a TOM subcomplex and the import receptors Tom70 and Tom20. A, Left, SDS-PAGE of TOM holo complex. TOM holo complex was isolated from mitochondrial outer membrane vesicles of a Neurospora strain that carried a Tom22 with a hexahistidinyl tag. Isolated outer membrane vesicles were solubilized in digitonin and subjected to Ni-NTA chromatography. The lanes marked with OMV and Ni-NTA eluate represent solubilized mitochondrial outer membrane proteins, and TOM complex purified by Ni-NTA chromatography, respectively. Tom70D denotes the dimeric form of Tom70. The gel was stained with Coomassie brilliant blue. Right, Coomassie-stained SDS polyacrylamide gel of fractions from the gel filtration shown in C. B, Size-exclusion chromatography of TOM holo complex on a TSK G4000 PWXL column. The peak fraction corresponds to ∼490 kD and contained all the TOM complex proteins. C, Gel chromatography of purified Tom holo complex as in B, after incubation with 0.33% DDM. Elution was followed by monitoring absorption at 280 nm. P1, P2, and P3 represent fractions analyzed by gel electrophoresis. The peak fraction corresponds to ∼410 kD.
Incubation of the holo complex with DDM at a concentration of 0.33% (wt/vol) at 37°C for 1 h led to dissociation of the import receptors, Tom70 and Tom20, and formation of a defined subcomplex. Size-exclusion chromatography of this material gave a profile similar to that of the holo complex, which was slightly shifted towards the low molecular weight range (Fig. 1 C). The main fraction contained nearly all of Tom40, Tom22, and the smaller Tom components, Tom7 and Tom6. Tom40 and Tom22 were present at roughly the same proportion as in the holo complex, as indicated by quantification of Coomassie staining (Fig. 1 A, right). Most of Tom70 and Tom20 eluted in fractions corresponding to lower molecular weight. Similar results were obtained when intact TOM complex was treated with the nonionic detergent Triton X-100 at concentrations above 0.33% (data not shown). Treatment of the TOM holo complex with SDS, in contrast, completely dissociated the complex into its individual components (data not shown). Thus, Tom40, Tom22, Tom7, and Tom6 form a defined, and rather stable subcomplex that we designated as the TOM core complex. According to the elution from the sizing column, the apparent molecular mass of this complex was estimated to be ∼410 kD.
To isolate the TOM core complex directly from mitochondria in high amounts, mitochondria from a strain with a hexahistidinyl-tag on Tom22 were solubilized in DDM at a concentration of 1% (wt/vol). The extract was loaded onto a Ni-NTA affinity column and, after extensive washing, bound material was eluted with an imidazole gradient. Tom40, Tom22, and the smaller Tom components all coeluted within five major fractions that accounted for ∼0.2% of protein loaded onto the column (Fig. 2 A, lane 2). Further anion-exchange chromatography resulted in a virtually pure TOM core complex (Fig. 2 A, lane 3). The yield of purified core complex was ∼2 mg complex per 1 g isolated mitochondrial protein.
Figure 2.
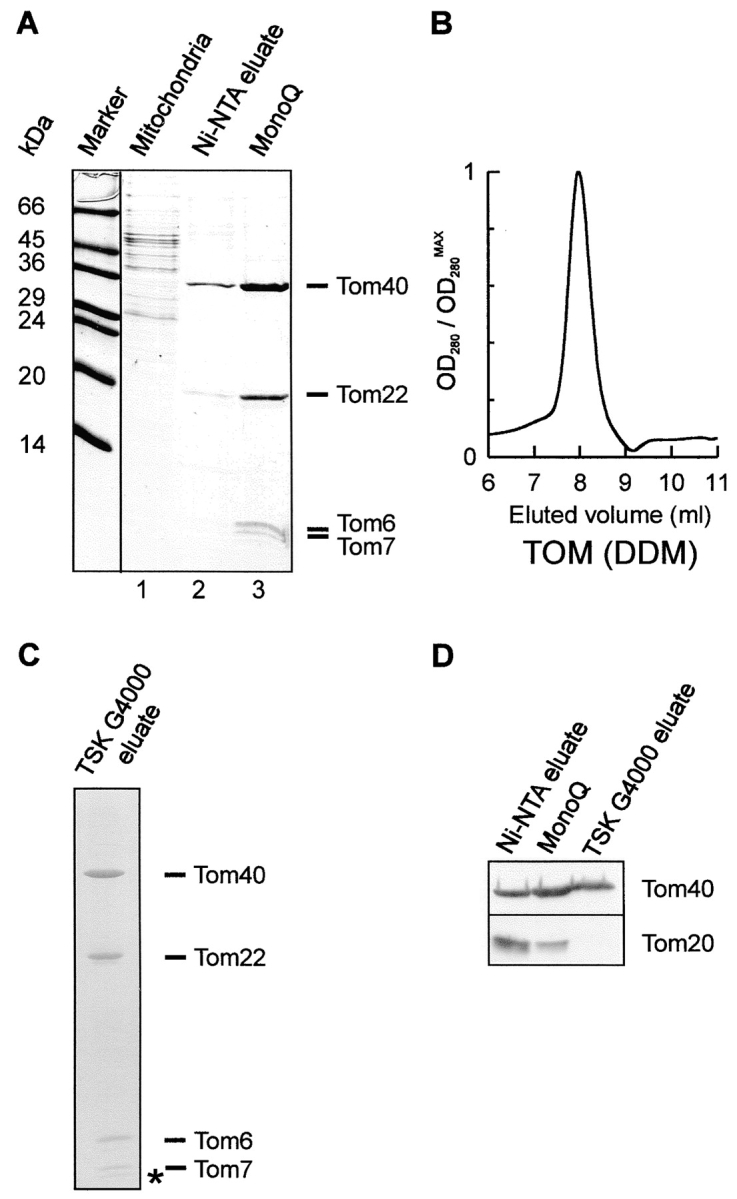
Purification of the TOM core complex. A, Isolation of the TOM core complex from mitochondria. Mitochondria from a Neurospora strain containing a Tom22 with a 6×His tag were solubilized in DDM and bound to Ni-NTA. Bound complex was eluted with imidazole. Fractions containing Tom40 were pooled and further purified by anion-exchange chromatography on a MonoQ column. Fractions were analyzed by SDS-PAGE and proteins were stained with Coomassie brilliant blue. Lane 1, Total mitochondrial protein solubilized in DDM; lane 2, eluant from Ni-NTA; lane 3, peak fraction of MonoQ chromatography. B, Size-exclusion chromatography on a TSK G4000 PWXL column of purified TOM core complex. The elution profile was revealed by monitoring absorption at 280 nm. The peak fraction corresponds to ∼410 kD. C, The peak fraction of the TSK sizing column analyzed by high Tris/urea SDS-PAGE and Coomassie staining. The asterisk denotes the band that possibly represents Tom5. D, Analysis of peak column fractions by SDS-PAGE and immunoblotting using antibodies against Tom40 and Tom20. Tom20 was completely removed from the core complex after passage over a Ni-NTA affinity, MonoQ, and TSK G4000 sizing column.
Upon size exclusion chromatography, the isolated TOM core complex was recovered in a single peak (Fig. 2 B) that contained only Tom40, Tom22, and the smaller Tom components (Fig. 2 C). Tom70 and Tom20 were not detected in this complex. Low amounts of Tom20 were present in the preparation eluted from the Ni-NTA column but, as determined by immunoblotting, these were completely removed after the gel filtration step (Fig. 2 D).
TOM core complex was purified from Neurospora cells that were grown in the presence of 35S-sulfate. In the purified complex, Tom40, Tom22, Tom7, and Tom6 were present in molar ratios of ∼8:4:2:2 (n = 2).
Characterization of the Isolated Core Complex
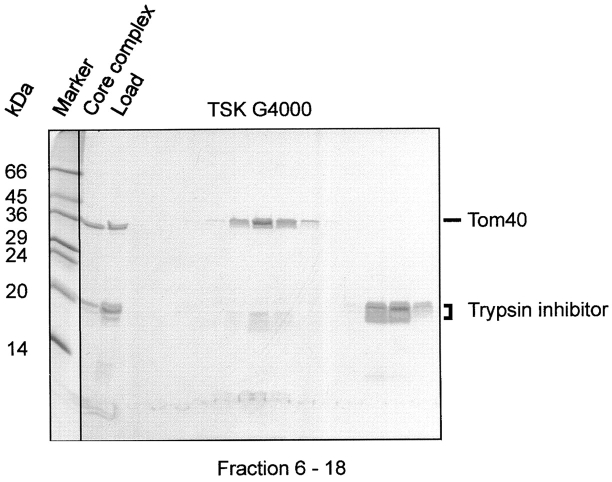
Isolated core complex was incubated with low amounts of trypsin (Fig. 3) and analyzed by size-exclusion chromatography. Proteolytic cleavage left Tom40 intact and removed the hydrophilic domains of Tom22 and the small Tom components, yielding fragments <3–5 kD. Immunoblotting using specific antisera against the COOH and NH2 terminus of Tom22 did not recognize a fragment of the protein. With antiserum against Tom6, no protein could be detected (data not shown).
Figure 3.
Limited proteolysis of purified TOM core complex. Purified TOM core complex was treated with 100 μg ml−1 trypsin. After addition of trypsin inhibitor, the complex was subjected to size-exclusion chromatography and SDS-PAGE. No intact Tom22 protein was detected after trypsin treatment. Proteins were stained with Coomassie brilliant blue.
The trypsinized complex eluted in a defined peak corresponding to a high molecular mass complex of ∼410 kD. A similar observation was made when the TOM holo complex, isolated in digitonin, was treated with protease (data not shown). This result indicated that the hydrophilic domains of Tom22 and of the small Tom components are not important for the structural integrity of the core complex.
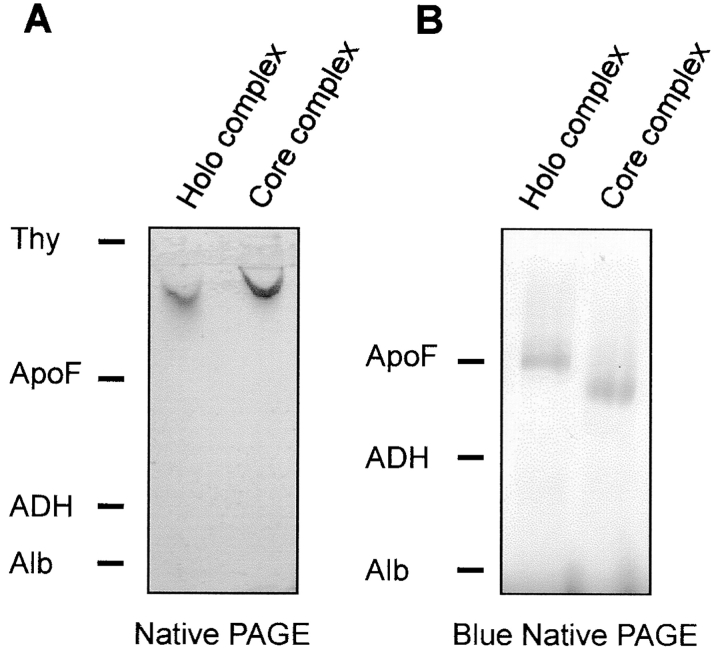
To further confirm the tight association of Tom40, Tom22, and the smaller Tom proteins in a defined subcomplex, purified TOM holo complex and TOM core complex were examined by native PAGE. Single high molecular weight bands were observed upon staining with Coomassie brilliant blue (Fig. 4 A). The different migration behavior of the complexes is due to the different detergents. The holo complex was solubilized in digitonin whereas the core complex has been purified in DDM.
Figure 4.
Native gel electrophoresis of the TOM complexes. A, Native PAGE (Phast) of purified TOM holo complex and isolated core complex. B, Blue native gel electrophoresis of TOM holo complex and core complex. Gels were stained with Coomassie. Marker proteins: Thy, thyroglobulin (669 kD); ApoF, apoferritin (443 kD); ADH, alcohol dehydrogenase (155 kD); Alb, albumin (66 kD).
Immunoblotting of the holo complex with monospecific antisera confirmed the presence of Tom70, Tom40, Tom22, Tom20, Tom7, and Tom6 (data not shown). The band representing the core complex yielded a positive signal using antibodies against Tom40, Tom22, and Tom6.
When the holo complex was treated with DDM (0.33%) or Triton X-100 (0.33%), the resulting core complex had the same electrophoretic mobility as the core complex isolated from mitochondria, and Tom70 migrated close to the running front (data not shown). Only Tom40, Tom22, and the smaller Tom components were detected in the band corresponding to the core complex.
Examination of the holo and core complexes by blue native gel electrophoresis, a method in which the binding of Coomassie brilliant blue adds negative charges to the protein complexes, gave results similar to those obtained without inclusion of a dye (Fig. 4 B). However, these conditions resulted in partial dissociation of the TOM holo complex since Western blotting and decoration of proteins with specific antibodies revealed that Tom70, and most of Tom20, no longer comigrated with Tom40 (data not shown). This partial disintegration of the TOM holo complex can be attributed to a destabilizing effect of the negatively charged dye used on the complex (Dekker et al. 1998).
Channel Activity of the Isolated Core Complex
To test whether the isolated core complex contains pores, we analyzed its channel forming activity after reconstitution into lipid membranes. Purified core complex was added to both sides of a black lipid membrane bilayer. Current recordings showed characteristic steps of conductance increase that reflect insertion of the core complex into the lipid bilayer. An average conductance of ∼2.3 nS in the presence of 1 M KCl was observed (Fig. 5 A). The trypsin-treated TOM holo complex had an average conductance of 2.7 nS (data not shown). These average conductances were similar to that of the holo complex (2.3 nS in 1 M KCl; Künkele et al. 1998). The hydrophilic import receptor domains of Tom70, Tom22, and Tom20 apparently play only a minor role in the channel properties of the TOM complex.
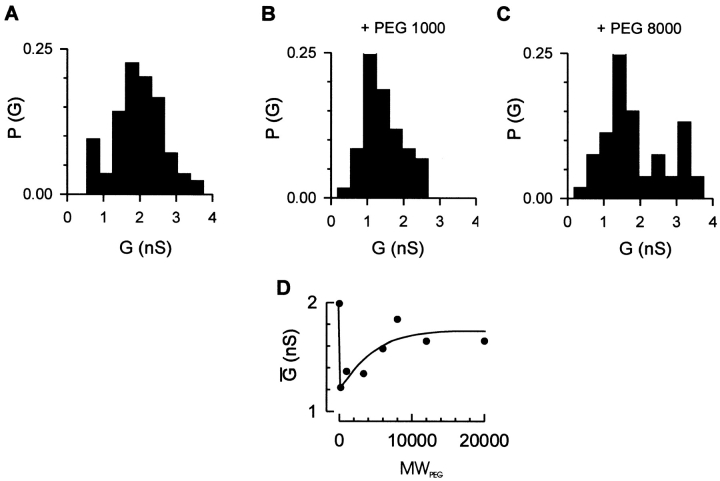
Figure 5.
Channel conductance of the TOM core complex in the presence of differently sized nonelectrolyte polymers. Purified TOM core complex (2 μg ml−1 final concentration) was added to both sides of a black lipid bilayer formed of diphytanoyl phosphatidyl choline/n-decane. Single channel conductances were measured at a membrane potential of ΔV = +20 mV. Histograms of channel conductances in polymer-free solution (A), in the presence of PEG1000 (B), and in the presence of PEG8000 (C). P(G) is the probability that a given conductance increment G is observed. D, Dependence of PEG-induced channel conductance change on the polymer weight. The electrolytes contained 1 M KCl, 5 mM Hepes, pH 7.0; the PEG concentrations were 20% (wt/vol), respectively. The data represent the mean conductances of n = 84, 56, 59, 22, 97, 53, 54, and 31 measurements in the absence and the presence of PEG 200, PEG 1000, PEG 3,350, PEG 6000, PEG 8,000, PEG 12,000, and PEG 20,000, respectively. Note that addition of PEG to a polymer-free solution decreases the single channel conductance due to the reduced bulk electrolyte conductance.
To probe the channel size of the isolated core complex, we studied its conductance in the presence of differently sized nonelectrolyte polymers. Low molecular weight polyethylene glycol, PEG1000, led to decreased channel conductances (Fig. 5 B). High molecular weight polyethylene glycols, such as PEG8000, affected the channel conductance to a lesser degree (Fig. 5 C). The results indicate that the TOM core complex channel can be blocked by molecules of up to ∼6 kD (Fig. 5 D).
Binding of Preprotein by the TOM Core Complex
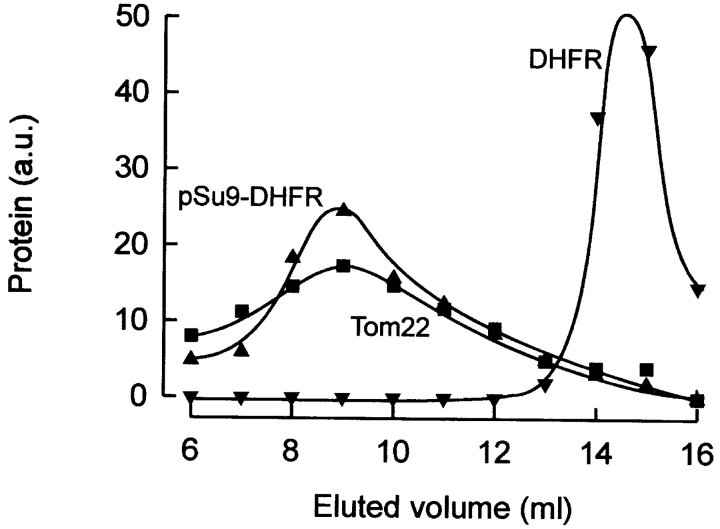
Does the TOM core complex retain its ability to bind a chemically pure preprotein in the presence of detergent? To address this question, we incubated chemical amounts of pure preprotein (pSu9-DHFR) with mitochondrial outer membrane vesicles. Membranes were solubilized with DDM at a concentration identical to that used for the isolation of the core complex directly from mitochondria, and the lysate was subjected to size exclusion chromatography. All column fractions were analyzed by SDS-PAGE and immunoblotting. A large fraction of pSu9-DHFR coeluted with Tom22 (Fig. 6) and Tom40 (not shown) in a high molecular weight complex. Only background binding was observed when DHFR lacking a mitochondrial presequence was analyzed, excluding the possibility that formation of the TOM-pSu9-DHFR complex was the result of unspecific binding. Thus, pSu9-DHFR remained firmly bound to the TOM core complex in a signal-sequence dependent manner, even at high levels of nonionic detergent.
Figure 6.
Preprotein binding to TOM core complex. Recombinant pSu9-DHFR (100 μg) was incubated with purified mitochondrial outer membrane vesicles (200 μg). They were incubated with DDM and subjected to gel filtration on TSK G-4000 PWXL. TOM complex and pSu9-DHFR in the eluate was analyzed by SDS-PAGE and immunoblotting with antibodies recognizing Tom22 and DHFR. As a control, the same protocol was performed with DHFR instead of pSu9-DHFR.
Structure of the Isolated TOM Core Complex
Electron micrographs of negatively stained TOM core complex particles displayed predominantly two stain filled openings or pores, but particles representing a single ring were also present (Fig. 7 A). The length of the two pore particles was ∼12 nm, with a width of ∼7 nm. For further image processing, a total of 1,598 particle images were extracted and aligned with respect to translation and rotation via cross-correlation (Frank et al. 1981). Using MSA (Frank and van Heel 1982), 30 eigenimages were calculated and the data set was broken up into 20 classes using the 10 most significant eigenimages. The class averages contained predominantly two or one pores (Fig. 7 B). Preparations of the TOM holo complex contained roughly equal amounts of particles with either two or three rings (Künkele et al. 1998). None of the three ring structures were observed in the core complex preparations. Group averages that showed one and two pores, respectively, were merged, yielding two main groups. Classification analysis was then used to eliminate remaining core complexes with poorly defined structures. This analysis resulted in projection maps of two core complex classes (Fig. 7C and Fig. D).
Figure 7.
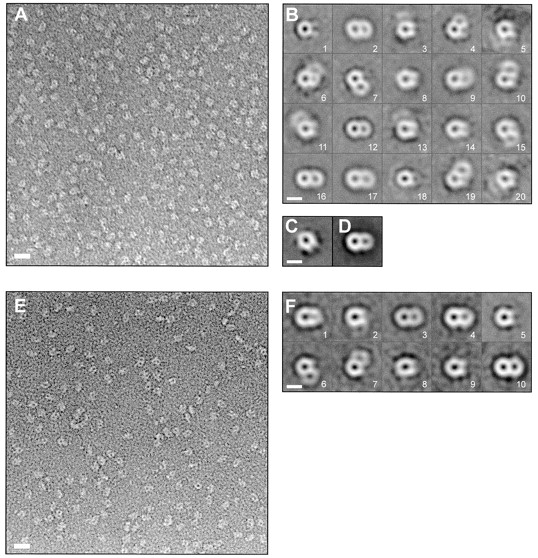
EM and projection map of the TOM core complex. A, Survey view of negatively stained TOM core complex. The image was filtered to the first zero of the electron microscope contrast transfer function. Bar, 11 nm. B, Classification analysis of 1,598 TOM core complex particles. Using MSA, the data set was split into 20 classes. Classes 1–20, represent the averages of 77, 175, 36, 59, 32, 39, 50, 102, 121, 40, 59, 163, 26, 59, 79, 159, 211, 50, 46, and 15 particle images, respectively. Bar, 7 nm. C and D, Group averages of the core complex that showed one and two pores, respectively, were merged, yielding two main groups that were subjected to further alignment, classification, and averaging. The maps shown in C and D were calculated from 306 and 866 particles, respectively. Bar, 7 nm. E, Survey view of trypsin-treated core complex. Bar, 11 nm. F, Classification analysis of 777 trypsin-treated TOM core complex particles. The data set was split into 20 classes, as in B. Classes 1–10 represent the averages of 51, 59, 131, 93, 120, 88, 34, 40, 35 and 102 particle images, respectively. Class averages of <10 particle images are not shown. Bar, 7 nm.
Projection maps of the TOM core complex treated with trypsin (Fig. 7 E) were calculated. The class averages of the trypsinized complex predominantly showed particles with two pores (n = 326) and one pore (n = 254; Fig. 7 F). Frequently, one of the channels appeared less distinct. This may be due to stain fluctuations, as can be seen in the original micrograph (Fig. 7A and Fig. E). The overall structure of the trypsinized core complex was similar to that of the intact core complex.
Do the stain filled openings of the TOM core complex, as seen in the 2D projections, span the entire complex? A 3D map of the TOM core complex was constructed by means of electron tomography. A total of 321 core complex particles were individually reconstructed in three dimensions from 6,741 projections, and subjected to 3D alignment and classification. As mentioned in Materials and Methods, the orientation of each particle was checked before averaging. As a result, 19% of the particles had to be flipped from up to down orientation. We cannot exclude, however, that the final 3D average is slightly distorted, due to possible flattening of the molecules that would mimic a common orientation of actually differently oriented particles.
An average of 116 reconstructions corresponding to the most prominent class of particles is shown in Fig. 8. The resolution of this average was 1/2.4 nm−1, based on the Fourier shell correlation function and 1/3.4 nm−1 following the stronger phase residual criterion (Saxton and Baumeister 1982).
Figure 8.
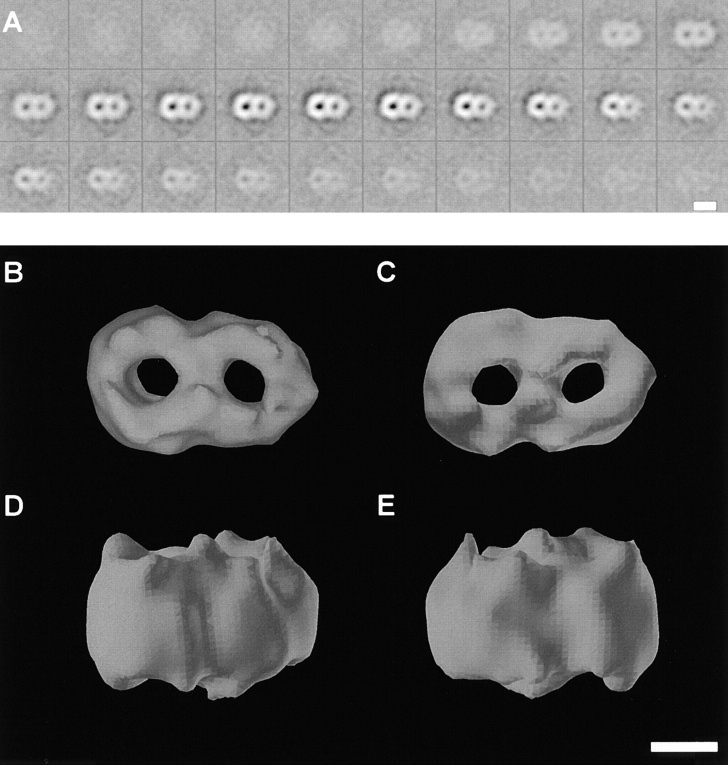
3D map of the TOM core complex obtained by electron tomography. Negatively stained TOM core complex particles were reconstructed individually, before the data set was subjected to 3D alignment, classification, and averaging. A, Gray level representation of horizontal slices through the average volume at a distance of 0.344 nm. Bar, 7 nm. B, Top view. C, Bottom view. D and E, Two different side views. The threshold for the isosurface representation was set to 64% of the molecular mass of 410 kD to achieve noise free representation. Bar, 3.5 nm.
The top view of the 3D model shows a two-ring structure. The density of the contacts between the two rings is as strong as the density of the walls of the rings, thus excluding the possibility that the two-ring structure was due to association of two independent translocation pores. The diameters of the two channels measure ∼2.1 nm. Based on the final reconstruction, and taking into account possible flattening and incomplete staining effects occurring during specimen preparation, the height of the TOM complex is ∼7 nm.
Discussion
We have isolated and analyzed the TOM core complex, which lacks the receptors Tom70 and Tom20, but retains several of the essential properties of the TOM holo complex. Tom40 is present in the holo and the core complex in the same number and constitutes the main component of the protein conducting channel. Tom22, which appears to have both a receptor function and a role in translocation, is firmly associated with Tom40. Likewise, the small Tom components, Tom6 and Tom7, which are believed to be required for the stability of the TOM complex (Hönlinger et al. 1996), are present in the core complex. An equivalent of the yeast Tom5 has not been identified in the Neurospora TOM complex. However, a band in the size range of yeast Tom5 was resolved upon SDS gel electrophoresis of the Neurospora complex. Both the core and the holo complex contain the same two pores likely representing protein conducting channels. On the other hand, the TOM core complex lacks the third density seen in the holo complex (Künkele et al. 1998) that could represent another protein mass with or without pore character. It is conceivable that the third density is due to the presence of the receptors Tom70 and Tom20. The Tom70 and Tom20 molecules seem to be attached to the periphery of the holo complex, as they are easily released in the presence of very mild detergents. The ion conductance properties, as defined by electrophysiology, are essentially the same with both types of complexes (Künkele et al. 1998). The isolated TOM core complex, after binding preprotein to intact mitochondrial outer membrane vesicles, retains the preprotein. Furthermore, the purified TOM core complex is able to bind preproteins in a specific manner (our unpublished results). The hydrophilic domains of Tom70 and Tom20 may increase the rate and specificity of preprotein binding. We conclude that the TOM core complex encompasses the function of the protein conducting channel.
Our data also allows us to define the minimal structural requirement of the translocation pore to Tom40, the transmembrane segments of Tom22, and the small Tom components. Whether the transmembrane segments are essential for the formation of the double pore structure remains an open question.
Although the sequence homology between Tom40, mitochondrial and bacterial porins is limited, circular dichroism data suggest a common β-barrel-like structure (Hill et al. 1998). Renaturation and reconstitution of E. coli-expressed Tom40 into lipid vesicles yielded channels similar in conductance to that of the core complex (Hill et al. 1998). It is presently unclear whether single Tom40 molecules formed these pores, or whether renatured Tom40 assembled into a multisubunit structure to create pores.
EM and image analysis of the isolated TOM core complex revealed mainly double ring structures, but a significant fraction of single ring particles were also observed (∼19%). This percentage of single ring particles is higher than observed with the holo complex (∼2%; Künkele et al. 1998). Analysis of the TOM core complex by both native gel electrophoresis and size exclusion chromatography yielded a homogenous population of molecules of ∼410 kD, which correspond to the double ring structures. Therefore, the TOM core complex may display a somewhat increased instability, or the placement on grids and negative staining of samples may promote disassembly.
The height of the TOM core complex of ∼7 nm is ∼2 nm larger than the thickness of a lipid bilayer. In fact, the extra membrane loops of Tom40 and receptors, or intermembrane space domains of Tom22, and the small Tom components would not be able to form large masses on either side of the complex. When edge-on views of the core complex become available, it should be possible to resolve the cytosolic and intermembrane space domains of the TOM core complex in more detail.
The 3D reconstruction of the TOM core complex shows several globular elements. Given that the TOM core complex is composed of about eight Tom40 molecules, these elements could represent dimers of Tom40.
At present, the cytosolic and intermembrane space sides of the TOM core complex cannot be distinguished. Probing both surfaces of the molecule with tags or antibodies should help resolve this issue. As higher resolution images become available, it will also be possible to better resolve the surface boundaries and internal surfaces of the putative translocation channels.
The size of the pores of the TOM core complex, as derived from single particle analysis, can be compared with that calculated from conductance measurements of the TOM core complex in the presence of differently sized nonelectrolyte polymers. Low molecular weight polyethylene glycols that were able to partition into the pores of the core complex reduced the mean conductance of the complex. Intermediate and large size polyethylene glycols with molecular weights of >6,000 reduced the currents mediated by the TOM complex to a lesser extent. Apparently, molecules with hydrodynamic radii larger than that of PEG6000 were not able to penetrate the core complex channel. Given a radius of ∼2.5 nm of PEG6000 (Carneiro et al. 1997), the size of the core complex channel should not exceed 5 nm. This value is roughly two times larger than that indicated by EM (∼2.1 nm). High molecular weight PEG molecules might block the entrances of the import channels of the TOM complex.
Conceivably, the two-ring structure is a dynamic assembly. A structural flexibility of the TOM complex in terms of alterations of subunit interactions during import of matrix-targeted preproteins has indeed been observed (Rapaport et al. 1998). Furthermore, it seems possible that the two rings undergo a rearrangement to form a structure with a single large pore, when preproteins are imported that appear to cross the TOM complex not in an extended state, but rather in a folded state, as suggested for the precursors of Tom40 and the ADP/ATP carrier (Endres et al. 1999; Rapaport and Neupert 1999). In the case of import of integral proteins of the outer membrane, the rings of the TOM complex may have to open to release the preprotein into the lipid phase. Furthermore, it remains to be determined whether the assembled TOM complex is in an equilibrium with its individual subunit constituents, e.g., with monomers or dimers of Tom40 (Rapaport and Neupert 1999).
The TOM core complex shares a number of interesting characteristics with the Sec61p complex, which facilitates protein translocation in the ER (Hanein et al. 1996; Matlack et al. 1998). Although there are no structural relationships between the proteins constituting the two complexes, both complexes appear to form a functionally similar passive conduit for polypeptides. Both complexes are organized as oligomers, with a major component that spans the membrane several times, Tom40 and Sec61α, respectively, and contain additional small components that span the membrane only once. Further, both complexes form ring-like structures with the characteristics of hydrophilic pores (Simon and Blobel 1991). Even the size of the pore appears similar, and the putative protein conducting channels appear to be dynamic in terms of their sizes (Crowley et al. 1994; Hamman et al. 1997; Liao et al. 1997; Hamman et al. 1998). Finally, the TOM core complex and the Sec61p complex are basic elements of larger, more complex assemblies, and they associate with auxiliary proteins, such as receptors for targeting signals, or, as shown in the case of the ER translocon, enzymes modifying the translocating chains. So, it appears that during evolution, protein conducting channels have been generated that have certain functional properties in common, but differ entirely in the origin of their constituents (Kellaris et al. 1991; Schatz and Dobberstein 1996; Powers and Walter 1997; Lyman and Schekman 1997; Hamman et al. 1998; Matlack et al. 1998).
Acknowledgments
We thank W. Baumeister for continuous support and R. Benz for his support in the electrophysiology measurements. We also thank D. Rapaport for critically reading the manuscript, and M. Braun and U. Staudinger for technical assistance.
This research was supported by grants of the Deutsche Forschungsgemeinschaft (S. Nussberger and W. Neupert), the Münchener Medizinische Wochenschrift (S. Nussberger), and the Medical Research Council of Canada (F.E. Nargang).
Footnotes
Abbreviations used in this paper: 3D, three-dimensional; DHFR, dihydrofolate reductase; DDM, n-dodecyl β-D-maltoside; MSA, multivariate statistical analysis; Ni-NTA, nickel-nitrilotriacetic acid; pSu9, presequence of subunit 9 of the F0-ATPase.
References
- Alconada A., Kubrich M., Moczko M., Hönlinger A., Pfanner N. The mitochondrial receptor complexthe small subunit Mom8b/Isp6 supports association of receptors with the general insertion pore and transfer of preproteins. Mol. Cell. Biol. 1995;15:6196–6205. doi: 10.1128/mcb.15.11.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bains G., Lithgow T. The Tom channel in the outer membranealive and kicking. BioEssays. 1999;21:1–4. doi: 10.1002/(SICI)1521-1878(199901)21:1<1::AID-BIES1>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Benz R., Janko K., Boos W., Läuger P. Formation of large, ion-permeable membrane channels by the matrix protein (porin) of Escherichia coli . Biochim. Biophys. Acta. 1978;511:305–319. doi: 10.1016/0005-2736(78)90269-9. [DOI] [PubMed] [Google Scholar]
- Brix J., Dietmeier K., Pfanner N. Differential recognition of preproteins by the purified cytosolic domains of the mitochondrial import receptors Tom20, Tom22, and Tom70. J. Biol. Chem. 1997;272:20730–20735. doi: 10.1074/jbc.272.33.20730. [DOI] [PubMed] [Google Scholar]
- Cao W., Douglas M.G. Biogenesis of ISP6, a small carboxyl-terminal anchored protein of the receptor complex of the mitochondrial outer membrane. J. Biol. Chem. 1995;270:5674–5679. doi: 10.1074/jbc.270.10.5674. [DOI] [PubMed] [Google Scholar]
- Carneiro C.M.M., Krasilnikov O.V., Yuldasheva L.N., Campos de Carvalho A.C., Nogueira R.A. Is the mammalian porin channel, VDAC, a perfect cylinder in the high conductance state? FEBS Lett. 1997;416:187–189. doi: 10.1016/s0014-5793(97)01198-8. [DOI] [PubMed] [Google Scholar]
- Court D.A., Nargang F.E., Steiner H., Hodges R.S., Neupert W., Lill R. Role of the intermembrane space domain of the preprotein receptor Tom22 in protein import into mitochondria. Mol. Cell. Biol. 1996;16:4035–4042. doi: 10.1128/mcb.16.8.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley K.S., Liao S., Worrell V.E., Reinhart G.D., Johnson A.E., Crowley K.S., Reinhart G.D., Johnson A.E. Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore. Cell. 1994;78:461–471. doi: 10.1016/0092-8674(94)90424-3. [DOI] [PubMed] [Google Scholar]
- Dekker P.J.T., Ryan M.T., Brix J., Müller H., Hönlinger A., Pfanner N. Preprotein translocase of the outer mitochondrial membranemolecular dissection and assembly of the general import pore complex. Mol. Cell. Biol. 1998;18:6515–6524. doi: 10.1128/mcb.18.11.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierksen K., Typke D., Hegerl R., Koster A.J., Baumeister W. Towards automatic electron tomography. Ultramicroscopy. 1992;40:71–87. [Google Scholar]
- Dietmeier K., Hönlinger A., Bömer U., Dekker P.J.T., Eckerskorn C., Lottspeich F., Kübrich M., Pfanner N. Tom5 functionally links mitochondrial preprotein receptors to the general import pore. Nature. 1997;388:195–200. doi: 10.1038/40663. [DOI] [PubMed] [Google Scholar]
- Endres M., Neupert W., Brunner M. Transport of the ADP/ATP carrier of mitochondria from the TOM complex to the TIM22-54 complex. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:3214–3221. doi: 10.1093/emboj/18.12.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J., van Heel M. Correspondence analysis of aligned images of biological particles. J. Mol Biol. 1982;161:134–137. doi: 10.1016/0022-2836(82)90282-0. [DOI] [PubMed] [Google Scholar]
- Frank J., Verschoor A., Boublik M. Computer averaging of electron micrographs of 40S ribosomal subunits. Science. 1981;214:1353–1355. doi: 10.1126/science.7313694. [DOI] [PubMed] [Google Scholar]
- Gratzer S., Lithgow T., Bauer R.E., Lamping E., Paltauf F., Kohlwein S.D., Haucke V., Junne T., Schatz G., Horst M. Mas37p, a novel receptor subunit for protein import into mitochondria. J. Cell Biol. 1995;129:25–34. doi: 10.1083/jcb.129.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamman B.D., Chen J.C., Johnson E.E., Johnson A.E. The aqueous pore through the translocon has a diameter of 40–60 A during cotranslational protein translocation at the ER membrane. Cell. 1997;89:535–544. doi: 10.1016/s0092-8674(00)80235-4. [DOI] [PubMed] [Google Scholar]
- Hamman B.D., Hendershot L.M., Johnson A.E. BiP maintains the permeability barrier of the ER membrane by sealing the lumenal end of the translocon pore before and early in translocation. Cell. 1998;20:747–758. doi: 10.1016/s0092-8674(00)81403-8. [DOI] [PubMed] [Google Scholar]
- Hanein D., Matlack K.E., Jungnickel B., Plath K., Kalies K.U., Miller K.R., Rapoport T.A., Akey C.W. Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell. 1996;87:721–732. doi: 10.1016/s0092-8674(00)81391-4. [DOI] [PubMed] [Google Scholar]
- Haucke V., Schatz G. Import of proteins into mitochondria and chloroplasts. Trends Cell Biol. 1997;7:103–106. doi: 10.1016/S0962-8924(96)10052-0. [DOI] [PubMed] [Google Scholar]
- Hegerl R. The EM program packagea platform for image processing in biological electron microscopy. J. Struct. Biol. 1996;116:30–34. doi: 10.1006/jsbi.1996.0006. [DOI] [PubMed] [Google Scholar]
- Hill K., Model K., Ryan M.T., Dietmeier K., Martin F., Wagner R., Pfanner N. Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature. 1998;395:516–521. doi: 10.1038/26780. [DOI] [PubMed] [Google Scholar]
- Hines V., Brandt A., Griffiths G., Horstmann H., Brutsch H., Schatz G. Protein import into yeast mitochondria is accelerated by the outer membrane protein MAS70. EMBO (Eur. Mol. Biol. Organ.) J. 1990;9:3191–3200. doi: 10.1002/j.1460-2075.1990.tb07517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hönlinger A., Kübrich M., Moczko M., Gärtner F., Mallet L., Bussereau F., Eckerskorn C., Lottspeich F., Dietmeier K., Jacquet M. The mitochondrial receptor complexMom22 is essential for cell viability and directly interacts with preproteins. Mol. Cell. Biol. 1995;15:3382–3389. doi: 10.1128/mcb.15.6.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hönlinger A., Bömer U., Alconada A., Eckerskorn C., Lottspeich F., Dietmeier K., Pfanner N. Tom7 modulates the dynamics of the mitochondrial outer membrane translocase and plays a pathway-related role in protein import. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:2125–2137. [PMC free article] [PubMed] [Google Scholar]
- Kassenbrock C.K., Cao W., Douglas M.G. Genetic and biochemical characterization of ISP6, a small mitochondrial outer membrane protein associated with the protein translocation complex. EMBO (Eur. Mol. Biol. Organ.) J. 1993;12:3023–3034. doi: 10.1002/j.1460-2075.1993.tb05971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellaris K.V., Bowen S., Gilmore R. ER translocation intermediates are adjacent to a nonglycosylated 34-kD integral membrane protein. J. Cell Biol. 1991;114:21–33. doi: 10.1083/jcb.114.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler M., Pfaller R., Söllner T., Griffiths G., Horstmann H., Pfanner N., Neupert W. Identification of a mitochondrial receptor complex required for recognition and membrane insertion of precursor proteins. Nature. 1990;348:610–616. doi: 10.1038/348610a0. [DOI] [PubMed] [Google Scholar]
- Kiebler M., Keil P., Schneider H., van der Klei I.J., Pfanner N., Neupert W. The mitochondrial receptor complexa central role of MOM22 in mediating preprotein transfer from receptors to the general insertion pore. Cell. 1993;74:483–492. doi: 10.1016/0092-8674(93)80050-o. [DOI] [PubMed] [Google Scholar]
- Köhler C.M., Jarosch E., Tokatlidis K., Schmid K., Schweyen R.J., Schatz G. Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science. 1998;279:369–373. doi: 10.1126/science.279.5349.369. [DOI] [PubMed] [Google Scholar]
- Komiya T., Rospert S., Köehler C., Looser R., Schatz G., Mihara K. Interaction of mitochondrial targeting signals with acidic receptor domains along the protein import pathwayevidence for the “acid chain” hypothesis. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:3886–3898. doi: 10.1093/emboj/17.14.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Künkele K.P., Heins S., Dembowski M., Nargang F.E., Benz R., Thieffry M., Walz J., Lill R., Nussberger S., Neupert W. The preprotein translocation channel of the outer membrane of mitochondria. Cell. 1998;93:1009–1019. doi: 10.1016/s0092-8674(00)81206-4. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liao S., Lin J., Do H., Johnson A.E. Both lumenal and cytosolic gating of the aqueous ER translocon pore are regulated from inside the ribosome during membrane protein integration. Cell. 1997;90:31–41. doi: 10.1016/s0092-8674(00)80311-6. [DOI] [PubMed] [Google Scholar]
- Lithgow T., Junne T., Suda K., Gratzer S., Schatz G. The mitochondrial outer membrane protein Mas22p is essential for protein import and viability of yeast. Proc. Natl. Acad. Sci. USA. 1994;91:11973–11977. doi: 10.1073/pnas.91.25.11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman S.K., Schekman R. Binding of secretory precursor polypeptides to a translocon subcomplex is regulated by BiP. Cell. 1997;88:85–96. doi: 10.1016/s0092-8674(00)81861-9. [DOI] [PubMed] [Google Scholar]
- Matlack K.E.S., Mothes W., Rapoport T.A. Protein translocationtunnel vision. Cell. 1998;92:381–390. doi: 10.1016/s0092-8674(00)80930-7. [DOI] [PubMed] [Google Scholar]
- Mayer A., Nargang F.E., Neupert W., Lill R. MOM22 is a receptor for mitochondrial targeting sequences and cooperates with MOM19. EMBO (Eur. Mol. Biol. Organ.) J. 1995;14:4204–4211. doi: 10.1002/j.1460-2075.1995.tb00094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moczko M., Bömer U., Kübrich M., Zufall N., Hönlinger A., Pfanner N. The intermembrane space domain of mitochondrial Tom22 functions as a trans binding site for preproteins with N-terminal targeting sequences. Mol. Cell. Biol. 1997;17:6574–6584. doi: 10.1128/mcb.17.11.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai M., Endo T. Identification of yeast MAS17 encoding the functional counterpart of the mitochondrial receptor complex protein MOM22 of Neurospora crassa . FEBS Lett. 1995;357:202–206. doi: 10.1016/0014-5793(94)01362-5. [DOI] [PubMed] [Google Scholar]
- Nakai M., Kinoshita K., Endo T. Mitochondrial receptor complex protein. The intermembrane space domain of yeast MAS17 is not essential for its targeting or function. J. Biol. Chem. 1995;270:30571–30575. doi: 10.1074/jbc.270.51.30571. [DOI] [PubMed] [Google Scholar]
- Neupert W. Protein import into mitochondria. Annu. Rev. Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- Pfaller R., Steger H.F., Rassow J., Pfanner N., Neupert W. Import pathways of precursor proteins into mitochondriamultiple receptor sites are followed by a common membrane insertion site. J. Cell Biol. 1988;107:2483–2490. doi: 10.1083/jcb.107.6.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N., Meijer M. The Tom and Tim machine. Curr. Biol. 1997;7:R100–R103. doi: 10.1016/s0960-9822(06)00048-0. [DOI] [PubMed] [Google Scholar]
- Powers T., Walter P. A ribosome at the end of the tunnel. Science. 1997;278:2072–2073. doi: 10.1126/science.278.5346.2072. [DOI] [PubMed] [Google Scholar]
- Ramage L., Junne T., Hahne K., Lithgow T., Schatz G. Functional cooperation of mitochondrial protein import receptors in yeast. EMBO (Eur. Mol. Biol. Organ.) J. 1993;12:4115–4123. doi: 10.1002/j.1460-2075.1993.tb06095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D., Neupert W. Biogenesis of Tom40, core component of the TOM complex of mitochondria. J. Cell Biol. 1999;146:321–331. doi: 10.1083/jcb.146.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D., Künkele K.P., Dembowski M., Ahting U., Nargang F.E., Neupert W., Lill R. Dynamics of the TOM complex of mitochondria during binding and translocation of preproteins. Mol Cell Biol. 1998;18:5256–5262. doi: 10.1128/mcb.18.9.5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan M.T., Pfanner N. The preprotein translocase of the mitochondrial outer membrane. Biol. Chem. 1998;379:289–294. [PubMed] [Google Scholar]
- Saxton W.O., Baumeister W. The correlation averaging of a regularly arranged bacterial cell envelope protein. J. Microsc. 1982;127:127–138. doi: 10.1111/j.1365-2818.1982.tb00405.x. [DOI] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- Schatz G., Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- Schlossmann J., Dietmeier K., Pfanner N., Neupert W. Specific recognition of mitochondrial preproteins by the cytosolic domain of the import receptor MOM72. J. Biol Chem. 1994;269:11893–11901. [PubMed] [Google Scholar]
- Schlossmann J., Lill R., Neupert W., Court D.A. Tom71, a novel homologue of the mitochondrial preprotein receptor Tom70. J. Biol. Chem. 1996;271:17890–17895. doi: 10.1074/jbc.271.30.17890. [DOI] [PubMed] [Google Scholar]
- Sebald W., Neupert W., Weiss H. Preparation of Neurospora crassa mitochondria. Methods Enzymol. 1979;55:144–148. doi: 10.1016/0076-6879(79)55020-4. [DOI] [PubMed] [Google Scholar]
- Simon S.M., Blobel G. A protein-conducting channel in the endoplasmic reticulum. Cell. 1991;65:371–380. doi: 10.1016/0092-8674(91)90455-8. [DOI] [PubMed] [Google Scholar]
- Sirrenberg C., Bauer M.F., Guiard B., Neupert W., Brunner M. Import of carrier proteins into the mitochondrial inner membrane mediated by Tim22. Nature. 1996;384:582–585. doi: 10.1038/384582a0. [DOI] [PubMed] [Google Scholar]
- Sirrenberg C., Endres M., Folsch H., Stuart R.A., Neupert W., Brunner M. Carrier protein import into mitochondria mediated by the intermembrane proteins Tim10/Mrs11 and Tim12/Mrs5. Nature. 1998;391:912–915. doi: 10.1038/36136. [DOI] [PubMed] [Google Scholar]
- Söllner T., Griffiths G., Pfaller R., Pfanner N., Neupert W. MOM19, an import receptor for mitochondrial precursor proteins. Cell. 1989;59:1061–1070. doi: 10.1016/0092-8674(89)90762-9. [DOI] [PubMed] [Google Scholar]
- Söllner T., Pfaller R., Griffiths G., Pfanner N., Neupert W. A mitochondrial import receptor for the ADP/ATP carrier. Cell. 1990;62:107–115. doi: 10.1016/0092-8674(90)90244-9. [DOI] [PubMed] [Google Scholar]
- Vestweber D., Brunner J., Baker A., Schatz G. A 42K outer-membrane protein is a component of the yeast mitochondrial protein import site. Nature. 1989;341:205–209. doi: 10.1038/341205a0. [DOI] [PubMed] [Google Scholar]