Three-dimensional (3D) image reconstruction from electron micrographs is providing a wealth of new structural information on complexes formerly intractable to solution. Nowhere will this technique be more important than in the analysis of multisubunit membrane-embedded complexes, where the nature of the membrane interactions makes crystallization of the intact complex exceedingly difficult. This is especially true in the case of highly dynamic membrane-embedded complexes, for which formation of perfect crystals is highly improbable.
The study by Ahting et al. 1999 represents a case in point. The TOM complex in the mitochondrial outer membrane consists of at least six distinct protein subunits, and functions as the gateway through which nuclear-encoded polypeptides can be imported into a mitochondrion. The TOM complex must recognize mitochondrial precursor proteins from amongst all other nascent polypeptides synthesized in the cytosol, bind the mitochondrial precursor proteins productively, and translocate them across the outer membrane. It does all this without a requirement for ATP hydrolysis or a transmembrane potential (Schatz 1997), with the energy to complete translocation coming from subsequent components of the import machinery (Neupert 1997).
Previous work from the same group reported the purification and low-resolution two-dimensional structure of the TOM holo complex containing all six protein subunits, using negative staining and electron microscopy (Kunkele et al. 1998a). The surprising projection structure revealed from one to three centers of stain accumulation, which appeared to represent channels in the complex through which a translocating polypeptide would be threaded in situ. Electrophysiological measurements confirmed that such channels did exist in the complexes, and remained operational, even after detergent treatment, to solubilize the TOM complex (Kunkele et al. 1998a,Kunkele et al. 1998b).
By differential solubilization with detergent, a core complex lacking the receptor subunits Tom20 and Tom70 has now been purified. The core complex usually appears as two (less often as one or three) annular features that accumulate stain, and reconstruction of the two-feature type of image in three dimensions strongly suggests that these annuli represent channels traversing the complex. The reconstruction also reveals some surface topology of the complex, or rather, reveals that there is very little in the way of topologically distinct detail in the structure. Presumably, the extramembrane domains of the complex are small (which is predicted for several of the component subunits), closely apposed against the complex, or have been denatured by drying or heavy metal contrasting during sample preparation. In the tomographic type of reconstruction that was used, resolution is anisotropic, and the geometry is such that the region of lowest resolution corresponds to only those regions of the complex that might protrude from the bilayer, towards either the cytosol or the intermembrane space. Nonetheless, this first glimpse at the TOM complex in three dimensions provides a foundation for further studies aimed towards a full understanding of this fascinating molecular machine.
The Holo Complex and the Core Complex
What happened to the third region accumulating stain in the holo complex, which is seen only rarely in the core complex? One explanation is that stain was accumulating in the tetratricopeptide repeat (TPR) proteins, Tom70 and Tom20. The single TPR unit of Tom20 is required for interaction with Tom70, which has seven TPR units (Haucke et al. 1996). Based on the 3D structure of the TPR domains of protein phosphatase 5, Das et al. 1998 demonstrated that eight TPR units would form a tall helix surrounding a central channel. Intriguingly, the radial dimensions of the TPR twist are similar to the size of the third region accumulating stain in the TOM holo complex. Another possibility is that the presence of the Tom20 and Tom70 subunits in the holo complex induces a docking or rearrangement of the Tom40 subunits to form three channels in the plane of what would be the mitochondrial membrane.
Pushing the Boundaries of Low-resolution Structures
Beyond this study of a mitochondrial outer membrane complex, EM has been applied to structural analysis of a host of biologically important complexes. Structures of ribosomes, nuclear pore complexes, kinesin–microtubule complexes, and clathrin coats are being solved to ever higher resolution. This limited list of some of the beautiful structures becoming available already highlights the impact that EM is having for molecular cell biologists.
In addition to revealing the structure of the eukaryotic ribosome, images have been reconstructed that depict how the ribosome docks with the Sec61 complex (Beckmann et al. 1997; Fig. 1 A). The 3D structure of the complex provides a framework in which to understand the multiple and sequential interactions made by a nascent polypeptide as it leaves a cytoplasmic ribosome to be translocated through an intracellular membrane. Like the structure of the TOM complex, the ribosome–Sec61 structure prompts new experiments and new ideas on the function and mechanics of the system.
Figure 1.
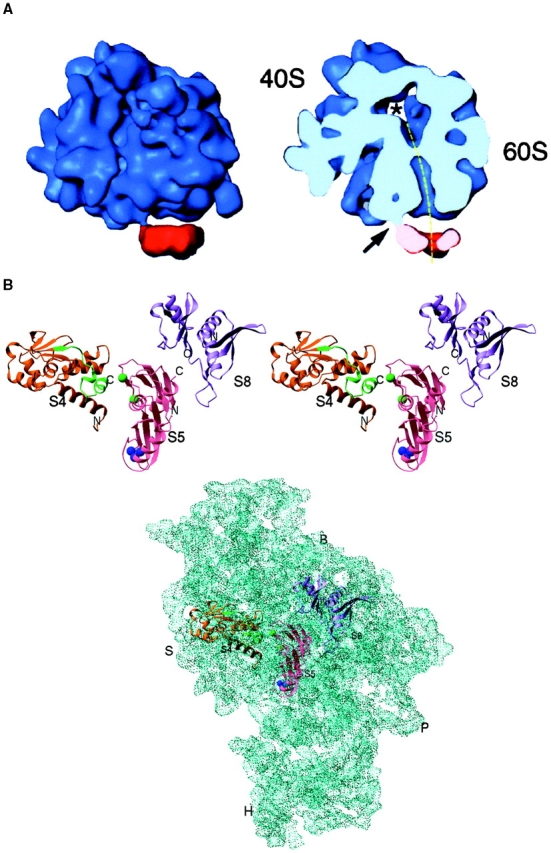
A, Alignment of the nascent polypeptide tunnel and translocation pore of the Sec61 complex. Cryo-EM was used to visualize the interaction of a solubilized membrane protein, intractable to crystallization, with the ribosome. The complexes were reconstituted in vitro using purified yeast ribosomes and Sec61 complex solubilized from ER with detergent. The path followed by the nascent polypeptide as it exits the ribosome and crosses the ER can be traced in the transverse section (dashed line). The 40S and 60S subunits of the ribosome are labeled. Modified after Beckmann et al. 1997. B, Atomic resolution structures for components of a macromolecular complex, such as the ribosome, can be fitted into a lower resolution map of the entire complex (derived from either X-ray or cryo-EM), and their detailed interactions can be interpreted. The ribosomal proteins S4, S5, and S8 have been crystallized, and their structures resolved at atomic resolution. The crystal structures are shown fitted to the 5.5 Å X-ray map of the 30S ribosomal subunit from Thermus thermophilus. B, Ribosomal body landmark; S, ribosomal shoulder landmark; H, ribosomal head landmark; and P, ribosomal platform landmark. Modified after Clemons et al. 1999.
The key to solving large macromolecular structures that are not amenable to 3D crystallization has been the development of cryo-EM and image processing techniques. Methodologies exist for analysis of two-dimensional crystals or sheets (e.g., many membrane proteins); helical structures (e.g., microtubules and flagella); icosahedral structures (e.g., viruses); and other objects with inherent symmetries. Single-particle image processing techniques applicable to objects lacking symmetries were pioneered by Frank and coworkers (e.g., Frank et al. 1978), and recently, such methods have begun to reveal features of secondary structure at resolutions approaching 10 Å. Images recorded from specimens that assume either random or preferred orientations within a layer of vitreous ice on a specimen grid (Dubochet et al. 1988) can be aligned, classified, and combined mathematically to give a 3D structure of the macromolecule in its native conformation, without the distortions induced by air-drying or heavy metal-contrasting. For images obtained by cryo-EM, optical densities within the reconstructed volume are proportional to actual mass density in the original object. Indeed, the next logical step for the solubilized TOM complex will be cryoimaging and 3D reconstruction by single-particle methods.
Adding in Atomic Resolution to Moderate Resolution Complex Structures
Even where the structural detail obtained from EM can be pushed to resolutions near 10 Å, individual protein subunits usually cannot be visualized within a complex. One approach towards pinpointing individual subunits is to label the isolated complexes with mAbs or with large peptide tags, or to use natural ligands to lock the complex in two discrete conformations, and use difference mapping of the two forms of the complex. An elegant example of what can be done is seen in recent work with gold cluster labeling of precise segments of kinesin motors docked onto microtubule lattices, providing much needed detail in the conformational changes that occur during the ATP-driven conformational changes in the motor protein, kinesin (R. Milligan, personal communication).
Ultimately, the atomic-resolution structures obtained with X-ray diffraction from crystallized protein and ribonucleoprotein domains can be mapped onto either a moderate resolution EM map or a high, but not quite atomic, resolution map from X-ray crystallography. The lower resolution map provides a framework or context in which the fine structure of the domain can be interpreted. This divide and conquer strategy is being applied to solve the structure of the prokaryotic ribosome, where individual subunits and subcomplexes have been crystallized, and their structures mathematically fitted or docked into 11.5 Å electron microscopic maps (Gabashvili, I.S., R.K. Agrawal, C.M.T. Spahn, R.A. Grassucci, J. Frank, and P. Penczek, manuscript in preparation) or 5.0–7.8 Å resolution crystallographic maps of the intact ribosome or ribosomal subunits (Cate et al. 1999; Clemons et al. 1999; Ban et al. 1999; Fig. 1 B). Now that several laboratories are progressing towards atomic resolution domain structures for components of the TOM complex, the same strategy could well provide a means to visualize the intact protein translocation machinery in three dimensions, with the phospholipid bilayer stripped away.
Acknowledgments
We thank Joachim Frank and Venki Ramakrishnan and their coworkers for permission to reproduce their data, Ron Milligan for communication of results before publication, and Amy Heagle for assistance with the artwork.
Funding from the Human Frontiers Science Program is gratefully acknowledged.
Footnotes
Abbreviations used in this paper: 3D, three-dimensional; TPR, tetratricopeptide repeat.
References
- Ahting U., Thun C., Hegerl R., Typke D., Nargang F.E., Neupert W., Nussberger S. The TOM core complexthe general protein import pore of the outer membrane of mitochondria. J. Cell Biol. 1999;147:959–968. doi: 10.1083/jcb.147.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N., Nissen P., Hansen J., Capel M., Moore P.B., Steitz T.A. Placement of protein and RNA structures into a 5 Å-resolution map of the 50S ribosomal subunit. Nature. 1999;400:841–847. doi: 10.1038/23641. [DOI] [PubMed] [Google Scholar]
- Beckmann R., Bubeck D., Grassucci R., Penczek P., Verschoor A., Blobel G., Frank J. Alignment of conduits for the nascent polypeptide chain in the ribosome–Sec61 complex. Science. 1997;278:2123–2126. doi: 10.1126/science.278.5346.2123. [DOI] [PubMed] [Google Scholar]
- Cate J.H., Yusupov M.M., Yuspova G., Earnest T.N., Noller H.F. X-ray crystal structures of 70S ribosome functional complexes. Science. 1999;285:2095–2104. doi: 10.1126/science.285.5436.2095. [DOI] [PubMed] [Google Scholar]
- Clemons W.M., Jr., May J.L.C., Wimberly B.T., McCutcheon J.P., Capel M.S., Ramakrishnan V. Structure of a bacterial 30S ribosomal subunit at 5.5 Å resolution. Nature. 1999;400:833–840. doi: 10.1038/23631. [DOI] [PubMed] [Google Scholar]
- Das A.K., Cohen P.T.W., Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5implications for TPR-mediated protein–protein interactions. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubochet J., Adrian M., Chang J.J., Homo J.-C., Lepault J., McDowall A.W., Schultz P. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- Frank J., Goldfarb W., Eisenberg D., Baker T.S. Reconstruction of glutamine synthetase using computer averaging. Ultramicroscopy. 1978;3:283–290. doi: 10.1016/s0304-3991(78)80038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haucke V., Horst M., Schatz G., Lithgow T. The Mas20p and Mas70p subunits of the protein import receptor of yeast mitochondria interact via the tetratricopeptide repeat motif in Mas20pevidence for a single hetero-oligomeric receptor. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:1231–1237. [PMC free article] [PubMed] [Google Scholar]
- Kunkele K.P., Heins S., Dembowski M., Nargang F.E., Benz R., Thieffrey M., Walz J., Lill R., Nussberger S., Neupert W. The preprotein translocation channel of the outer membrane of mitochondria Cell. 93 1998. 1009 1019a [DOI] [PubMed] [Google Scholar]
- Kunkele K.P., Juin P., Pompa C., Nargang F.E., Henry J.P., Neupert W., Lill R., Thieffrey M. The isolated TOM complex of the mitochondrial outer membrane. Characterisation of the cation-selective and voltage-gated preprotein-conducting pore J. Biol. Chem. 273 1998. 31032 31039b [DOI] [PubMed] [Google Scholar]
- Neupert W. Protein import into mitochondria. Annu. Rev. Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- Schatz G. Just follow the acid chain. Nature. 1997;388:121–122. doi: 10.1038/40510. [DOI] [PubMed] [Google Scholar]


