Abstract
Little is known about the mechanisms of intracellular targeting of viral nucleic acids within infected cells. We used in situ hybridization to visualize the distribution of tobacco mosaic virus (TMV) viral RNA (vRNA) in infected tobacco protoplasts. Immunostaining of the ER lumenal binding protein (BiP) concurrent with in situ hybridization revealed that vRNA colocalized with the ER, including perinuclear ER. At midstages of infection, vRNA accumulated in large irregular bodies associated with cytoplasmic filaments while at late stages, vRNA was dispersed throughout the cytoplasm and was associated with hair-like protrusions from the plasma membrane containing ER. TMV movement protein (MP) and replicase colocalized with vRNA, suggesting that viral replication and translation occur in the same subcellular sites. Immunostaining with tubulin provided evidence of colocalization of vRNA with microtubules, while disruption of the cytoskeleton with pharmacological agents produced severe changes in vRNA localization. Mutants of TMV lacking functional MP accumulated vRNA, but the distribution of vRNA was different from that observed in wild-type infection. MP was not required for association of vRNA with perinuclear ER, but was required for the formation of the large irregular bodies and association of vRNA with the hair-like protrusions.
Keywords: in situ hybridization, tobacco mosaic virus, virus replication, endoplasmic reticulum, cytoskeleton
Specific virus–host interactions determine the capability of viruses to successfully replicate in a cell and spread local and systemically in the host. Plant viruses, like those of animals, compartmentalize viral RNA synthesis by association with the cellular endomembrane system (e.g., Restrepo-Hartwig and Carrington 1994; Restrepo-Hartwig and Ahlquist 1996; Schaad et al. 1997; Ward et al. 1997). However, despite their central role in viral pathology, little is known about how viral RNA–protein complexes are transported through the cytoplasm, anchored and replicated in cellular membranes, and targeted to plasmodesmata to initiate spread to adjacent plant cells. Defining those mechanisms will give new insights about the specific virus–host interactions that ultimately determine a successful viral infection.
Tobacco mosaic virus (TMV)1 is the type member of the tobamovirus group. The genome of TMV consists of one single-stranded 6.4-kb RNA containing four open reading frames. The genomic RNA serves as messenger RNA (mRNA) for the production of a 126-kD protein and, by readthrough of an amber termination codon, a 183-kD protein (Beier et al. 1984). Both proteins are involved in replication of viral RNA (Ishikawa et al. 1986). The 30-kD movement protein (MP), which is required for cell-to-cell movement (Deom et al. 1987), and the 17.5-kD coat protein (CP) are dispensable for replication (Meshi et al. 1987; Takamatsu et al. 1987).
Cytological (Bald 1964) and radioautographic studies (Smith and Schlegel 1965) suggested that replication of TMV RNA was associated with the nucleus. However, other approaches later showed that double-stranded replicative intermediates of TMV RNA (Ralph et al. 1971; Nilsson-Tillgren et al. 1974; Beachy and Zaitlin 1975) and virus-specific RNA-dependent RNA polymerase (Zaitlin et al. 1973; Osman and Buck 1996) were localized and/or associated with cellular cytoplasmic membranes. Biochemical fractionation of TMV-infected protoplasts revealed that TMV minus-strand RNA accumulated primarily in a cytoplasmic, extranuclear fraction (Okamoto et al. 1988).
Cytological analyses of TMV-infected cells showed virus replication complexes associated with cytoplasmic inclusions, called viroplasms, which expanded throughout the infection. These inclusions contained ribosomes, tubules, and 126/183-kD replication proteins (Esau and Cronshaw 1967; Martelli and Russo 1977; Hills et al. 1987; Saito et al. 1987). Recent studies with TMV that encodes MP:green fluorescent protein (GFP) fusion protein have suggested that the fluorescent irregularly shaped structures in cells that contain MP and replicase were derived from the ER membranes (Heinlein et al. 1998; Reichel and Beachy 1998).
Since the MP binds single-stranded nucleic acids in vitro (Citovsky et al. 1990) and accumulates in plasmodesmata (Tomenius et al. 1987; Deom et al. 1990), apparently increasing the plasmodesmata size exclusion limit (Wolf et al. 1989; Oparka et al. 1997), it was suggested that the MP and viral TMV RNA form ribonucleoprotein complexes that are directed to and transported through plasmodesmata (Citovsky and Zambryski 1991). Colocalization of the MP with microtubules (Heinlein et al. 1995) and microfilaments (McLean et al. 1995) prompted the hypothesis that elements of the cytoskeleton play a role in the intracellular transport of the MP-viral RNA complexes (Zambryski 1995; Carrington et al. 1996). However, the specific sites of viral RNA accumulation are not known; likewise, it is not known if cytoskeletal components are associated with viral RNA.
In many different biological systems, the coordinated activities of components of the cytoskeleton are responsible for the specific transport of RNAs, as well as the anchoring of RNAs at their final locations (e.g., Wilhelm and Vale 1993; Hovland et al. 1996; Bassell and Singer 1997; Gavis 1997; Arn and Macdonald 1998; Hazelrigg 1998). In some cases, mRNAs are transported as RNA–protein complexes or “granular particles” (e.g., Ainger et al. 1993; Kloc and Etkin 1993; Forristal et al. 1995; Broadus et al. 1998; Schuldt et al. 1998) that are formed by RNA–RNA and RNA–protein interactions (Ferrandon et al. 1998). Recent observations suggest that in different cell types, the RNA-binding protein involved in the formation of an RNA–protein complex may mediate RNA localization by using different cytoskeletal components (Oleynikov and Singer 1998).
In this work, nonradioactive methods of in situ hybridization were used to localize the intracellular distribution of TMV RNA during virus infection. The combined use of immunostaining and in situ hybridization techniques allowed examination of the relationship between the sites of vRNA accumulation and the distribution of selected host (microtubules, ER) and viral (MP, replicase) components. The pattern of vRNA accumulation after treatment with oryzalin and cytochalasin D gave new insights about the role of actin filaments and microtubules in vRNA transport and replication. Furthermore, the use of TMV derivatives lacking a functional MP allowed us to determine the role of the MP in establishing the sites of vRNA infection and association with host components.
Materials and Methods
Plasmids
Diagrams of the infectious clones used in this study are shown in Fig. 1 A (below). The plasmids pU3/12 (Holt and Beachy 1991) and pU3/12 ΔM-RV (Nejidat et al. 1991) were used to generate infectious transcripts of wild-type (wt) TMV and TMV that does not express a functional MP (ΔM), respectively. The plasmids pT-MfCP (MP:GFP-CP) and pTMV-M:GfusBr (MP:GFP-ΔC) (Oparka et al. 1997) contain the MP fused to GFP in which serine at amino acid position 65 is replaced by threonine (Heim et al. 1995). vRNA-MP:GFP-ΔC does not contain the CP open reading frame (see Fig. 1 A).
Figure 1.
(A) Diagrammatic representation of the genomic structures of wt TMV RNA and derivatives used in this study (indicated in the left margin). Coding regions for the replicase, MP, GFP, and CP are shown by open boxes. Broken lines represent fusion proteins (MP fused to GFP), while solid lines represent expression of free proteins. (B) Accumulation of genome-length TMV plus-strand RNA in tobacco protoplasts inoculated with wt TMV RNA (w.t.) and with different TMV derivatives (indicated above each lane). “Mock” corresponds to protoplasts treated in absence of inoculum. Total RNA was extracted at 4, 14, and 30 hpi, glyoxylated, and subjected by electrophoresis on a 1% denaturing agarose gel. Northern blots were probed with a digoxigenin-labeled RNA probe that recognized the plus strand of TMV RNA. Each construct was tested in at least three independent experiments; results consistent with those shown were obtained in each experiment.
The plasmid pTR447 was used as template to produce the nonradioactive (digoxigenin or fluorescein) labeled RNA probes used in Northern blot and in situ hybridization experiments. To create pTR447, the XbaI–HindIII fragment of the TMV replicase coding sequence (comprising nucleotides 999 to 1446) was removed from pU3/12 ΔM-RV and subcloned into the plasmid vector Bluescript SK+ (Stratagene Inc.) previously digested with the same restriction endonucleases.
Protoplast Infection, RNA Extraction, and Northern Hybridization Analysis
Infectious RNAs were obtained by in vitro transcription of TMV clones using the MEGAscript T7 kit (Ambion Inc.). Protoplasts were transfected by electroporation as described by Watanabe et al. 1987 and collected by centrifugation at 4, 8, 14, 20, and 30 h post-infection (hpi). Total RNA was isolated from 5 × 104 protoplasts using TRIzol reagent following the recommendations of the manufacturer (GIBCO-BRL). RNA was recovered from the aqueous phase after centrifugation at 12,000 g for 15 min at 4°C, precipitated with isopropanol, and resuspended in an adequate volume of sterile water.
To monitor plus- and minus-strand accumulation, the RNAs were denatured with glyoxal following the procedure described by McMaster and Carmichel 1977. RNAs extracted from equal numbers of protoplasts were loaded onto the gel and electrophoresed in 20 mM Hepes, pH 7.00, 1 mM EDTA. After electrophoresis, the gels were soaked in 50 mM NaOH for 15 min and 50 mM sodium acetate/acetic acid for an additional 15 min. The nucleic acids were then transferred to nitrocellulose membranes in 10× SSC (1.5 M NaCl, 0.15 M sodium citrate, pH 7.0) according to standard procedures (Sambrook et al. 1989). Before hybridization, the membranes were stained with methylene blue (Sambrook et al. 1989) to verify that the levels of ribosomal RNA in each sample were similar.
Prehybridization, hybridization, and colorimetric detection were performed as previously described by Más and Pallás 1996. Single-stranded digoxigenin-RNA probes that recognized plus or minus strands of the genomic TMV RNA were obtained by in vitro transcription of the plasmid pTR447 clone previously linearized with XbaI or HindIII and using T7 or T3 RNA polymerases, respectively (Boehringer Mannheim Biochemicals).
Immunofluorescent Labeling
Protoplasts were fixed in paraformaldehyde and spun onto polylysine-coated slides (Sigma Chemical Co.) as described by Kahn et al. 1998. For immunostaining, the samples were washed two times in PBS-TE buffer (140 mM NaCl, 2.5 mM KCl, 10 mM Na2HPO4, 1.5 mM KH2PO4, 0.5% Tween 20, and 5 mM EGTA). The slides were then immersed in cold methanol for 10 min and washed again in PBS-TE before incubation for 2 h at room temperature with the primary antibody. Monoclonal mouse anti–α-tubulin and anti–actin antibodies (Amersham Corp.) were diluted 1:100 in PBS-TE. The antibody against the rabbit anti–ER lumenal binding protein (BiP), kindly provided by R.S. Boston (North Caroline State University, Raleigh, NC), was used at 1:100 dilution in PBS-TE. To immunolocalize TMV replicase, a polyclonal antibody against the 126/183-kD replicase protein (Nelson et al. 1993) was diluted 1:50 in PBS-TE. After incubation with the primary antibody, the samples were washed two times in PBS-TE and incubated for 2 h at room temperature with 1:100 dilution of the secondary antibody:TRITC-conjugated goat anti–mouse IgG (Jackson ImmunoResearch Laboratories, Inc.) or TRITC-conjugated goat anti–rabbit IgG (Pierce Chemical Co.). The samples were then washed again and either mounted in Mowiol (Calbiochem Corp.) or processed by in situ hybridization to detect vRNA.
In Situ Hybridization
Before hybridization, the samples were treated with proteinase K (1 μg/ml) for 5 min, washed, and refixed with paraformaldehyde at room temperature for 30 min. After fixation, the samples were washed again and immersed in 0.25% acetic anhydride, 100 mM triethanolamine-HCl, pH 8.0, for 10 min to prevent nonspecific binding of the probe to positively charged amino groups. After acetylation, the samples were dehydrated by 10 min washes in 70, 80, 90, 95, and 100% ethanol solutions. The samples were then hybridized overnight at 55°C in a moist chamber with the hybridization solution containing 50% (vol/vol) of deionized formamide, 4× SSC, 0.1% (wt/vol) SDS, 8% (wt/vol) dextran sulphate, and 10 ng/μl of fluorescein-RNA (fluor-RNA) probe. The fluor-RNA probes that recognized plus or minus strand of the genomic TMV RNA were obtained by in vitro transcription of pTR447 as described above. Probes were labeled with fluorescein-12-UTP (Boehringer Mannheim Biochemicals) following the recommendations of the manufacturer. After hybridization, the samples were washed two times in 2× SSC for 10 min at room temperature and two times in 0.1× SSC for 10 min at 50°C, air-dried, and mounted as described above.
Treatment with Cytoskeleton Depolymeration Agents
To disrupt microtubules, infected protoplasts were treated with 10 μM oryzalin (ChemService) for 2 h. To disrupt microfilaments, infected protoplasts were treated with 100 μM cytochalasin D (Calbiochem Corp.) for 2 h. Stock solutions of oryzalin and cytochalasin D were prepared in DMSO so that the final concentration of DMSO added to the protoplasts did not exceed 0.1% (vol/vol). After these treatments, the protoplasts were fixed and processed for in situ hybridization following the procedure described above.
Confocal Microscopy
Confocal imaging was performed as previously described (Más and Beachy 1998) using a confocal laser scanning microscope (IX70; Olympus Corp.). Optical sections were made at 0.8-μm intervals and projections of serial optical sections were obtained using the software provided by the manufacturer. To examine fluorescein and GFP fluorescence, optical planes were scanned with the 488-nm argon laser using a 550-nm (FVX-BA550RIF) barrier filter and 60× 1.4 NA PlanApo or 100× 1.35 NA UPlanApo oil-immersion objectives. The TRITC signal was detected with the 568-nm krypton laser (FVX-BA585IF). In experiments of dual localization (with fluorescein and TRITC), both fluorophores were examined independently, or by attenuating the 488-nm line to 25% intensity to reduce the possibility of crossover between the channels. Furthermore, single immunodetection controls verified the absence of fluorescence crossover.
Results
Northern Hybridization of Protoplasts Infected with TMV
Before in situ hybridization analysis, the specificity of the RNA probe and the accumulation of viral RNA of the different TMV constructs used in this study (Fig. 1 A) were analyzed by Northern blot hybridization (Fig. 1 B). Total RNAs from equal numbers of protoplasts were extracted at different times after infection and subjected to Northern hybridization with a probe labeled with digoxigenin (dig-RNA). The RNA probe contains a fragment of the replicase sequence (see details in Materials and Methods). The probe hybridized with an RNA of ∼6.4 kb, representing the full-length, plus-strand genome of TMV (Fig. 1 B, w.t.). As anticipated, vRNA accumulated to approximately the same level in cells infected with wt TMV RNA and in cells infected with TMV RNA that lacked the MP (ΔM), confirming previous studies showing that the MP is not required for replication (Meshi et al. 1987). RNAs from mock-inoculated protoplasts did not react with the probe (Fig. 1 B, Mock).
Time-course analysis showed that vRNA accumulated to detectable levels at 4 hpi, increased until 14 hpi, and then gradually decreased to a lower level through 30 hpi (Fig. 1 B). The levels of accumulation of vRNA in mutants that lack the CP sequence and express the MP:GFP fusion protein (MP:GFP-ΔC in Fig. 1) were somewhat lower than in wt infection, especially at 30 hpi (Fig. 1 B). Since MP:GFP-ΔC produced no CP, the lower levels of vRNA in these samples are presumed to reflect the lack of encapsidated vRNA. Protoplasts infected with RNAs derived from constructs containing the CP open reading frame and MP:GFP fusion protein (MP:GFP-CP) showed lower levels of vRNA than those observed in wt infection (compare wt and MP:GFP-CP in Fig. 1 B).
The levels of ribosomal RNAs in each sample were similar, as visualized by staining with methylene blue before hybridization (not shown).
When samples were hybridized with a probe to detect minus-strand RNA, the signal was observed only at early stages of infection (4–8 hpi) and in much lower amounts than plus strands (not shown). Similar results were described by Ishikawa et al. 1991 in TMV (L strain)-infected protoplasts.
These studies show that the nonradioactive RNA probes were sufficiently sensitive and specific for use in situ hybridization procedures. They also indicate that vRNA produced by wt, ΔM, and MP:GFP virus infections accumulate to approximately the same degree throughout infection.
Subcellular Distribution of TMV Genomic RNA in Infected Protoplasts
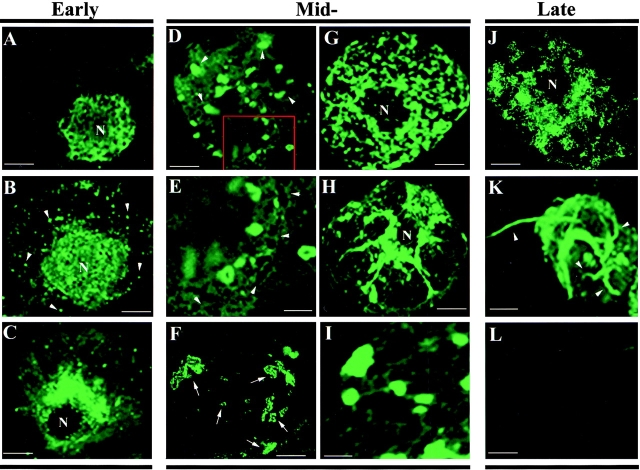
The intracellular accumulation of TMV RNA during infection was visualized by in situ hybridization. Protoplasts isolated from BY-2 tobacco cells were transfected with wt RNA, collected at different times after infection, and hybridized with fluor-RNA probes. The fluorescent signals were visualized by confocal microscopy after collecting optical sections with a focal depth of 0.8 μm. Since infection of BY-2 protoplasts is apparently not synchronous (Heinlein et al. 1998), the accumulation of vRNA was considered in three stages: early, 6–12 hpi; mid, 12–22 hpi; and late, 22–36 hpi. Representative examples are shown in Fig. 2. At early stages of infection (Fig. 2, A–C), vRNA was primarily localized in faint fluorescent vesicle-like structures surrounding the nucleus (Fig. 2 A). The fluorescent signal was also observed in small, discrete cytoplasmic patches of uniform size, dispersed randomly throughout the cell (Fig. 2 B, arrowheads). In some cells, vRNA tended to accumulate in large amounts around the nucleus as shown in an optical section in Fig. 2 C.
Figure 2.
Intracellular distribution of vRNA in protoplasts infected with wt vRNA. Protoplasts were collected at different times after infection, fixed, and hybridized with a fluorescein-RNA probe that recognized TMV RNA. Fluorescent signals were visualized by confocal microscopy, collecting optical sections with a focal depth of 0.8 μm. (A–C) At early stages of infection (6–12 hpi), vRNA is associated with vesicle-like structures surrounding the nucleus (A) and in small patches or particles in the cytoplasm (B and C). Note the large amount of vRNA in perinuclear regions (C). (D–I) vRNA at midstages of infection (12–22 hpi) localized in large and small bodies (D and E), a weakly fluorescent reticulated network (E), large vesicles (F), large bodies (G) that appear to be associated with filaments (H and I). (J and K) vRNA at late stages of infection (22–36 hpi). vRNA is associated with small, intensely labeled spots around the nucleus, dispersed throughout the cytoplasm, and at the periphery of the cell (J) and localized in filamentous structures, some of which protrude from the surface of the cells (K). (L) Mock-inoculated protoplasts identically processed and imaged. The images shown in A, B, K, and L correspond to projections of serial optical sections, while C, D, F–H, and J represent single optical sections. Bars: (A–D and F–G) 10 μm; (E–I) 5 μm.
At midstages of infection (Fig. 2, D–I), vRNA was associated with large irregularly shaped bodies (Fig. 2 D, arrowheads). High magnification of the image revealed that these bodies were, in many cases, associated with a weakly fluorescent reticulated network (Fig. 2 E) that resembled the cortical ER. Short fluorescent strands of tubules, small fluorescent circles, and vesicles of variable size and shape were also observed (Fig. 2 F, arrows). During the middle stage of infection, most of the large bodies were localized around the nucleus, although some were dispersed throughout the cytoplasm (Fig. 2 D) and in some cells occupied much of the cytoplasm (Fig. 2 G). Clear localization of vRNA in filamentous structures was visible during this period (Fig. 2H and Fig. I); most of the filaments appeared to be associated with the fluorescent bodies (Fig. 2H and Fig. I). At late stages of infection (Fig. 2J and Fig. K), vRNA was localized to small, intensely labeled spots around the nucleus, throughout the cytoplasm, and at the periphery of the cell (Fig. 2 J). The number and intensity of these spots decreased over time. By 40 hpi, 15% of infected protoplasts showed vRNA localized to elongated structures that appeared to protrude through the plasma membrane (Fig. 2 K, arrowheads). The protrusions resembled those previously described as binding the MP:GFP fusion protein (Heinlein et al. 1998; Más and Beachy 1998).
We conclude from these studies that vRNA accumulates in different subcellular compartments throughout infection. This compartmentalization most likely reflects the roles of vRNA in replication, translation, and assembly in viral particles.
Mock-inoculated protoplasts identically processed and imaged did not show fluorescent signals (Fig. 2 L). In addition, no signal was observed when either the fluor-RNA probe was omitted from the hybridization reaction or when the samples were treated with RNAse A before the hybridization reaction (not shown). Furthermore, 90–95% of the inoculated protoplasts showed hybridization signal, coinciding with the percentage of protoplasts that are infected after inoculation with wt vRNA.
As anticipated from the Northern hybridization studies described above and those described by Ishikawa et al. 1991, in situ hybridization experiments using a probe that bound minus-strand RNA revealed fluorescent signals only at early stages of infection (4–8 hpi). As illustrated in Fig. 3, fluorescence accumulated primarily in perinuclear vesicles and in small cytoplasmic patches, similar to the accumulation of plus-strand vRNA (compare with Fig. 2 B). Interestingly, the surface of the nucleus was not uniformly covered by fluorescence in these reactions. In protoplasts visualized at 4–6 hpi, the fluorescent signal was clearly absent from a discrete region of most nuclei (Fig. 3 B). Visualization at high magnification revealed circular regions free of RNA in protoplasts with weak (Fig. 3 C) or strong (Fig. 3 D) fluorescence around the nucleus.
Figure 3.
Intracellular distribution of TMV minus-strand RNA in protoplasts infected with wt RNA. Protoplasts collected at early stages of infection (4–6 hpi) were fixed and hybridized with a fluorescein-RNA probe that recognized the complementary strand of vRNA. (A) Minus-strand RNA accumulation in a perinuclear pattern and in small cytoplasmic patches. (B) RNA surrounding the nucleus with a discrete region that lacked hybridization. (C and D) Nuclei of protoplasts at 6 hpi showing vRNA-free zone. A, C, and D correspond to single optical sections, while B corresponds to a projection of serial optical sections. N, nucleus. Bars: (A and B) 10 μm; (C and D) 5 μm.
Taken together, these results indicate that the fluorescent signal obtained after in situ hybridization of U1-infected protoplasts reflects the intracellular distribution of TMV RNA.
Spatial Relationships between vRNA and Viral Proteins
Viral RNA Colocalizes with the Replicase.
To examine the relationship between the sites of vRNA accumulation and the location of the viral replicase, protoplasts infected with wt TMV RNA were collected at early stages of infection, first immunostained with antibody raised against the TMV replicase protein (Nelson et al. 1993), and subsequently processed for in situ hybridization with the fluor-RNA probe. The distribution of the replicase, visualized with TRITC-conjugated secondary antibody (Fig. 4, red) was similar to that of vRNA (green). Superimposition of coincident green and red signals are presented in yellow (merged image) and are most apparent in bodies and filaments in the cytoplasm. The antireplicase antibody also labeled structures that lacked vRNA (red in merged image). It is not known whether these differences reflect differences in distribution of replicase protein and vRNA or differences in intensity of each fluorophore.
Figure 4.
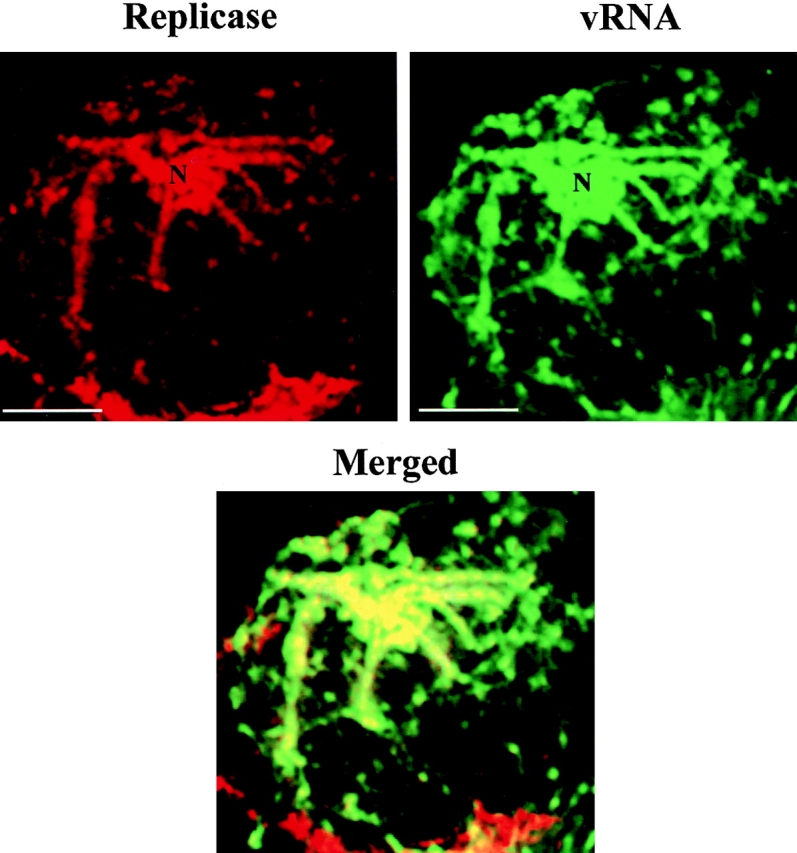
vRNA colocalizes with the replicase. Protoplasts infected with wt vRNA were fixed at early stages of infection and processed for immunofluorescence using antireplicase antibody and in situ hybridization for detecting vRNA. Most of the replicase (red signal) was localized in sites were vRNA (green signal) accumulated. Merging both images demonstrates colocalization of both signals (yellow in merged image). N, nucleus. Bars, 10 μm.
In double labeling experiments, the fluorophores were scanned independently to minimize crossover between the two channels. Similar patterns of distribution of replicase were observed when the samples were processed only by immunostaining with the replicase antibody, indicating that the results were not due to artifacts produced as a result of the in situ hybridization procedure.
Viral RNA Colocalizes with the MP.
The patterns of vRNA accumulation over time resembled the pattern of synthesis, accumulation, and degradation of the MP during TMV infection (Heinlein et al. 1998; Más and Beachy 1998). To investigate the spatial relationship between vRNA and MP, protoplasts infected with vRNA-MP:GFP-CP were hybridized with the digoxigenin-RNA probe and hybridization was visualized by immunostaining with a rhodamine antidigoxigenin antibody. The distribution of vRNA-MP:GFP-CP was similar to that of vRNA in wt infection, indicating that the presence of the MP:GFP fusion protein did not significantly alter the pattern of vRNA distribution. Similar patterns of fluorescence were also observed in protoplasts infected with vRNA-MP:GFP-ΔC, a mutant virus that expresses the MP:GFP fusion and lacks the CP sequence (not shown).
The hybridization procedure was incompatible with simultaneous detection of MP:GFP due to bleaching of GFP fluorescence during the treatment of the samples. Therefore, to compare the accumulation of both the MP and vRNA in the same cell, protoplasts infected with vRNA-MP:GFP-ΔC were harvested at midstages of infection, fixed, and spun onto poly-lysine–coated slides (see Materials and Methods). Cells showing the characteristic fluorescence pattern of MP:GFP accumulation as previously described (Heinlein et al. 1998; Más and Beachy 1998) were scanned using the 488-nm argon laser of the confocal microscope and the samples were then processed for in situ hybridization. Before the hybridization reaction, the cells were imaged to verify the absence of fluorescence due to GFP.
After hybridization with the fluor-RNA probe, the samples were examined to detect the products of hybridization (vRNA). As shown in Fig. 5, there was a striking coincidence between the distribution of MP and the sites of vRNA accumulation. vRNA was also found in small cytoplasmic patches that lacked detectable MP (arrows). Similar patterns of distribution of replicase, MP, and vRNA distribution confirmed the hypothesis that the replication of vRNA and the accumulation of the MP occur at the same subcellular sites.
Figure 5.
vRNA colocalizes with the MP. Protoplasts infected with vRNA-MP:GFP-ΔC (see Fig. 1) were fixed at midstages of infection, and the pattern of MP accumulation was observed as fluorescence emitted by the MP:GFP. The samples were then treated to eliminate fluorescence by GFP and hybridized with the fluor-RNA probe to detect vRNA. A cell showing the MP distribution was identified by its position and shape and visualized to detect the products of hybridization (vRNA). Comparison of both images reflects colocalization of MP:GFP and vRNA. N, nucleus. Bars, 10 μm.
Role of the MP in Establishing the Sites of RNA Accumulation in Infected Cells.
As shown above, MP colocalizes with the main sites of vRNA accumulation during TMV infection. To clarify the role of the MP in establishing the sites of virus replication, protoplasts infected with vRNA-ΔM were processed for in situ hybridization. At early stages of infection, a similar pattern of fluorescence was observed in protoplasts infected with wt vRNA or vRNA-ΔM. The fluorescent signal was localized in vesicle-like structures surrounding the nucleus and in small cytoplasmic patches (Fig. 6 A). In contrast to infection by wt TMV, vRNA-ΔM also accumulated in stacks of tubular structures near the nucleus (Fig. 6 B, arrowheads). Higher magnification revealed vesicles of different sizes (2–5 μm diameter) around the nucleus as well as throughout the cytoplasm (Fig. 6 C, arrowheads). At midstages of infection, the most striking feature that distinguished this pattern of vRNA-ΔM distribution from that of wt vRNA was the absence of large fluorescent bodies. Instead, smaller fluorescent spots were distributed throughout the cell, interconnected by fewer fluorescent reticulated strands than in wt infection (Fig. 6 D, arrowheads). We often observed stacks of large lamellar cisternae that were up to 10 μm in length and 4–5 μm in depth (Fig. 6 E, arrows). At late stages of infection, the fluorescent signal was observed only as small, intensely labeled spots around the nucleus and scattered throughout the cytoplasm (Fig. 6 F). Cells infected with vRNA-ΔM did not exhibit fluorescent protrusions from the surface of the cells as in cells infected with wt vRNA (e.g., Fig. 2 K).
Figure 6.

Intracellular distribution of vRNA-ΔM. Infected protoplasts were fixed and hybridized with a fluorescein-RNA probe to localize vRNA. Fluorescence was visualized by confocal microscopy of optical sections with a focal depth of 0.8 μm. (A–C) Accumulation of vRNA-ΔM at early stages of infection in vesicle-like structures surrounding the nucleus and in small cytoplasmic patches (A) and in stacks of tubular strands (B). vRNA-ΔM was also associated with small vesicles surrounding the nucleus and throughout the cytoplasm (C). At midstages of infection, vRNA-ΔM was observed in bright fluorescent spots interconnected by short filaments (D) and in stacks of lamellar structures (E). At late stages of infection (F), fluorescent signal was observed only in small, intensely labeled spots around the nucleus and scattered throughout the cytoplasm. Note the absence of vRNA-ΔM in association with large fluorescent bodies or with clearly defined filaments. A corresponds to the projection of 12 serial optical sections, while B–F correspond to single optical sections. N, nucleus. Bars: (A, B, E, and F) 10 μm; (C and D) 5 μm.
Similar distribution of vRNA was observed when protoplasts were infected with vRNA-ΔM-GFP-ΔC, a virus construct that lacks CP and MP and produces free GFP (not shown).
Spatial Relationship between Viral RNA and Host Components
Viral RNA Colocalizes with the Endoplasmic Reticulum.
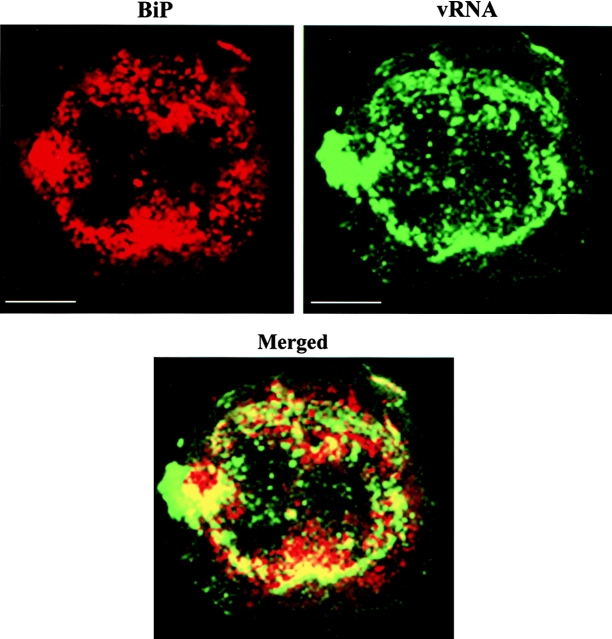
As noted above, the reticulated pattern of fluorescence that represents the location of wt vRNA resembled the distribution of the ER. Furthermore, previous studies demonstrated colocalization of TMV MP and the replicase with ER on the fluorescent irregularly shaped bodies (Heinlein et al. 1998; Reichel and Beachy 1998). To determine if vRNA is associated with the ER, in situ hybridization was performed on protoplasts collected at early stages of infection. The samples were also stained using an antibody to the ER BiP (Boston et al. 1996) and a TRITC-conjugated secondary antibody. As shown in Fig. 7, BiP was localized to perinuclear areas and small aggregates dispersed in the cytoplasm. vRNA was localized in structures surrounding the nucleus and scattered in the cytoplasm and in fluorescent bodies. After superimposing the images, we concluded that a portion of the vRNA detected is associated with the ER (yellow in merged image). BiP and vRNA were not fully coincident, indicating that a fraction of vRNA was independent of the ER.
Figure 7.
vRNA colocalizes with BiP (ER marker). Protoplasts infected with wt vRNA were fixed at early stages of infection and processed for immunofluorescence using anti–BiP antibody and in situ hybridization. Most of the BiP (red) was associated with sites that contain vRNA (green). Merging the two images demonstrates colocalization of the signals (yellow in merged image). Bars, 10 μm.
In protoplasts treated at early stages of infection with 50 μg/ml of Brefeldin A (BFA), a fungal metabolite that disrupts the endomembrane system in plant cells (Henderson et al. 1994; Satiat-Jeunemaitre et al. 1996), the vRNA was dispersed throughout the cytoplasm in small fluorescent spots. No fluorescent vesicle-like structures were observed surrounding the nucleus in treated samples (Fig. 8). These results corroborate the hypothesis that at early stages of infection vRNA is associated with ER.
Figure 8.

Disruption of the ER induces changes in the localization of wt vRNA. In protoplasts treated at early stages of infection with 50 μg/ml of BFA, the vRNA was accumulated in small fluorescent spots dispersed throughout the cytoplasm (vRNA/BFA). Note the absence of the perinuclear fluorescent structures that accumulate in nontreated protoplasts (see Fig. 2, A–C, for comparisons). N, nucleus. Bar, 10 μm.
Viral RNA Colocalizes with Microtubules.
At early and midstages of infection, vRNA was associated with fluorescent cytoplasmic filaments (Fig. 2H and Fig. I). To determine if vRNA is associated with elements of the cytoskeleton, infected protoplasts that were immunostained with a monoclonal antibody to α-tubulin were processed for in situ hybridization. A variety of different experimental conditions was evaluated before selecting conditions that permitted visualization of both signals. The procedure that was used involved treating fixed protoplasts with 1 μg/ml of proteinase K for 5 min to partially digest cross-linked proteins and subsequent refixation to avoid disintegration of the cells. These procedures improved the accessibility of the probe while preserving the integrity of the microtubules. Fig. 9 shows confocal micrographs of tubulin (in red) and vRNA (in green) in an infected protoplast processed at midstage of infection. Merging the images clearly showed the coalignment of both signals (merged image). Most of the green fluorescent filaments corresponding to sites of vRNA accumulation coaligned with the cytoplasmic strands of microtubules (Fig. 9 A, yellow in merged image). Furthermore, some of the bright fluorescent spots that contained vRNA accumulated in tracks that were aligned with microtubule filaments (Fig. 9 B).
Figure 9.
vRNA colocalizes with microtubules. Protoplasts infected with wt vRNA were fixed and processed for immunofluorescence using antitubulin antibody and in situ hybridization. (A) Most of the tubulin (red) was associated with filaments that coaligned with the filaments that contain vRNA (green). Merging the images demonstrates colocalization of the signals (yellow in merged image). (B) Some of the bright fluorescent patches containing vRNA (green) appear to be aligned with microtubule filaments (red). Bars, (A) 10 μm; (B) 2 μm.
Protoplasts infected with vRNA-ΔM that were identically processed and imaged did not show fluorescent signals associated with cytoskeleton (except in cytochalasin D–treated protoplasts, see below). The results indicate that colocalization of vRNA with microtubules was not due to artifactual aggregation that occurred during fixation procedures. These conclusions are based on replicated data, either in the number of experiments or in the number of protoplasts analyzed per experiment.
In companion studies, we attempted to stain with antiactin antibodies and with the probe for vRNA. However, we did not resolve a typical pattern of filamentous actin distribution (not shown). Most of the cells showed very short fluorescent strands of actin dispersed throughout the cytoplasm. In some cells, we observed a coincidence between the short strands of TRITC-fluorescent signal and vRNA (not shown). It is possible that under the conditions of these experiments, the integrity of the microfilaments was not maintained. A relationship between actin filaments and vRNA can not be ruled out since disruption of microfilaments by treating infected protoplasts with cytochalasin D altered the pattern of vRNA distribution (see below).
Effects of Cytoskeletal Inhibitors on Localization of vRNA.
The results provide strong evidence for colocalization of vRNA with microtubules and suggest that cytoskeletal elements may be involved in distribution of vRNA in protoplasts. To clarify the role of the cytoskeleton in vRNA distribution, protoplasts infected with wt vRNA were treated with specific cytoskeletal inhibitors and vRNA accumulation was examined by in situ hybridization. Representative examples are shown in Fig. 10. Treatment of protoplasts at early stages of infection with oryzalin, a plant microtubule depolymerizing agent (Hugdahl and Morejohn 1993), abolished the accumulation of vRNA around the nucleus and most vRNA was dispersed throughout the cytoplasm (not shown). When oryzalin was added at midstages of infection, nearly all of the fluorescence was associated with enlarged fluorescent bodies on or near the nuclear envelope (Fig. 10, vRNA/Oryzalin). In nontreated protoplasts, most of the fluorescent bodies were dispersed throughout the cytoplasm at this stage of infection (e.g., Fig. 2 G). When oryzalin was added late in infection, we did not observe changes in distribution of vRNA. In treated as well as nontreated protoplasts, vRNA was associated with the filament-like structures protruding from the surface of the cells (see Fig. 2 K).
Figure 10.

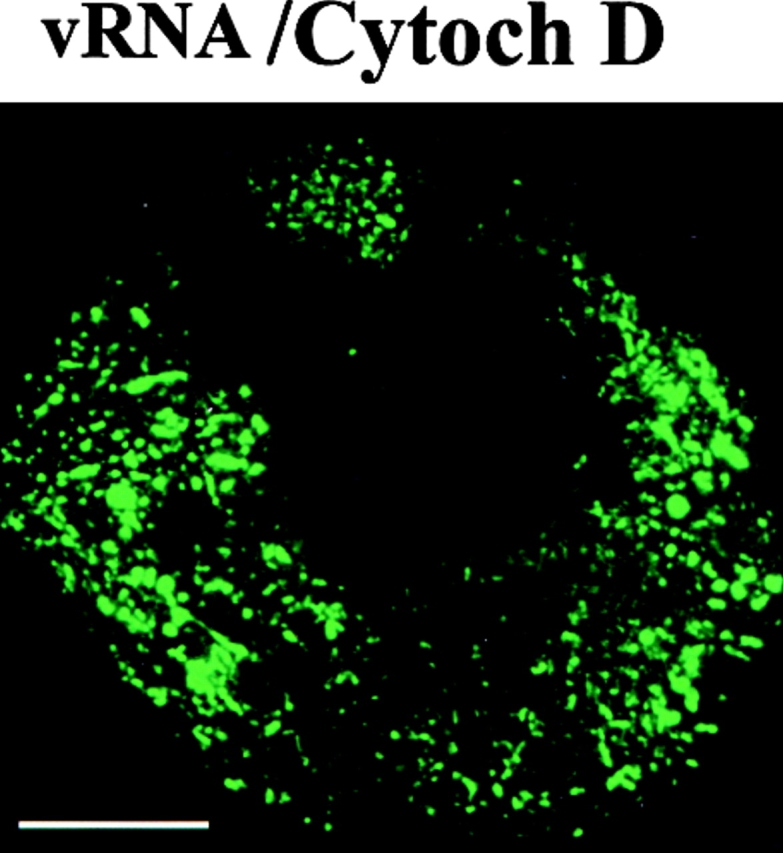

Disruption of cytoskeleton induces changes in the localization of wt vRNA and vRNA-ΔM. Protoplasts treated at midstages of infection with 10 μM oryzalin for 2 h induces the accumulation of vRNA in enlarged fluorescent bodies surrounding the nucleus (vRNA/Oryzalin). Treatment at early stages of infection with cytochalasin D delays the formation of large bodies, and the pattern of vRNA accumulation was similar to that observed in infection by vRNA-ΔM (vRNA/Cytoch D). In protoplasts infected with vRNA-ΔM and treated at early stages of infection with cytochalasin D, vRNA-ΔM accumulated in filaments and narrow structures at the periphery of the cell (vRNA-ΔM/Cytoch D). Bars, 10 μm.
In protoplasts treated during early stages of infection with cytochalasin D, there was a clear delay in the formation of the cytoplasmic bodies compared with nontreated protoplasts. Interestingly, the pattern of vRNA accumulation in cytochalasin D–treated cells was similar to that observed in cells infected with vRNA-ΔM (compare Fig. 10, vRNA/Cytoch D, with Fig. 6). At midstages of infection, at a time when the large cytoplasmic bodies were formed, the vRNA in cytochalasin D–treated protoplasts (not shown) was detected in enlarged bodies that appeared as more elongated structures than those observed in nontreated cells.
In protoplasts infected with vRNA-ΔM and treated with cytochalasin D at early stages of infection, vRNA-ΔM was associated with filaments resembling microtubules (Fig. 10, ΔM/Cytoch D). In these protoplasts, vRNA-ΔM also accumulated at the periphery of the cell, but no fluorescence was observed around the nucleus or in small bodies throughout the cytoplasm, as in the case of nontreated, vRNA-ΔM–infected protoplasts (see Fig. 6 for comparisons). The effects of cytochalasin D were more dramatic in infections with vRNA-ΔM than with wt vRNA, suggesting an influence of the MP in the localization of vRNA in cytochalasin D–treated protoplasts.
The results of these studies demonstrate that disruption of the cytoskeleton produces changes in the pattern of vRNA localization and suggest an important role of microtubules and microfilaments in the distribution of vRNA during virus infection.
Discussion
Early Stages of TMV Infection
The replication of many positive-strand RNA viruses occurs in close association with membranes (e.g., Wu et al. 1992; Molla et al. 1993; Strauss and Strauss 1994; Osman and Buck 1996). In this study, TMV vRNA was specifically localized with several types of cellular membranes. Early in infection, vRNA was associated with a perinuclear reticulated network and with strands of tubules and small vesicles. These structures closely resembled the cortical polygonal ER network of tubules and sheets with intervening lamellar segments connected to the nuclear envelope, previously described in plant cells (Allen and Brown 1988; Hepler et al. 1990; Staehelin 1997). Colocalization of vRNA with BiP, a resident protein of the ER (Denecke et al. 1995; Boston et al. 1996) and the disruption of the fluorescent structures by BFA (Fig. 8), support the hypothesis that vRNA is associated with ER. Our observations that viral replicase (Ishikawa et al. 1986) and the complementary minus-strand RNA (used as template for vRNA replication) were localized in these membranous structures strongly suggest that replication of TMV vRNA takes place in close association with the ER. Earlier reports described a relationship between TMV replication complexes and membranous extracts from infected cells (Watanabe and Okada 1986; Osman and Buck 1996).
Minus-strand RNA was also localized to structures (presumed to be ER) that surround the nucleus, with the exception of a discrete region. The absence of minus-strand RNA from this region could reflect compartmentalization of the perinuclear ER, or it may indicate that the ER is divided into subdomains with specific morphological or functional properties (Staehelin 1997). Ishikawa et al. 1986 suggested that the synthesis of plus and minus strands of vRNA requires different types of factors or molecular interactions, an observation that may have relevance to our studies.
vRNA and viral replicase were colocalized in small patches that are distributed throughout the cytoplasm. As previously indicated in poliovirus infection (Bienz et al. 1983), we suggest that patches that contain vRNA and replicase correspond to replication complexes in association with ER that compartmentalize the required components of replication to enhance virus production.
At early stages of infection, vRNA was associated with fluorescent filaments that resembled elements of the cytoskeleton (not shown). Based on the effects of oryzalin and cytochalasin D on the distribution of vRNA in the cytoplasm, we suggest that there is association of vRNA with the cytoskeleton at a very early stage of infection. In this scenario, vRNA exploits the cytoskeleton for transport from the cytoplasm to perinuclear positions. This hypothesis is based in part on the observation that treating protoplasts with oryzalin at the time of inoculation prevented the localization of vRNA to the perinuclear region. A recent report described a mechanism in newt lung epithelial cells by which the ER membranes attached to microtubules are transported toward the cell center through actomyosin-based retrograde flow of microtubules with ER attached as cargo (Waterman-Storer and Salmon 1998). Following this model, it is possible that vRNA-replicase complexes associated with ER are transported via microtubules to perinuclear positions using a similar mechanism.
Throughout the infection, vRNA was localized in different subcellular compartments. We suggest that this reflects movement of vRNA to different compartments, but cannot eliminate the possibility that compartmentalization represents the synthesis and subsequent degradation of vRNA rather than movement of vRNA from one compartment to another.
When protoplasts were infected with vRNA-ΔM, a mutant of TMV that does not produce MP, vRNA-ΔM was localized to vesicle-like structures around the nucleus and in small patches in the cytoplasm. These results indicate that at an early stage of infection, association with the ER is an intrinsic property of vRNA and/or the replicase and does not require MP. It is possible that vRNA contains sequences that target to the ER. Some cellular mRNAs are known to contain specific signals that direct them to the rough ER for translation (St. Johnston 1995).
Midstages of Infection: Virus Accumulation and Intracellular Transport
At midstages of infection, vRNA was localized in fluorescent, irregularly shaped bodies, some of which were vesicle-like in appearance. Furthermore, the replicase (Fig. 4) and MP (Fig. 5) colocalized with vRNA on these bodies. Since such structures were not observed in protoplasts infected with vRNA-ΔM, we conclude that MP is required for the formation and/or stabilization of the bodies. These structures may correspond to the previously described “viroplasms or amorphous inclusions” induced by TMV infection (Martelli and Russo 1977). Our observations support an earlier suggestion that replication complexes associated with rough ER function as mRNAs (Beachy and Zaitlin 1975). During its synthesis, MP may remain associated with vRNA, acting as an anchoring protein and trapping vRNA on ER-derived structures. Accordingly, the large accumulation of MP is coincident with dramatic morphological changes that take place on the ER (Reichel and Beachy 1998).
Over time, the cytoplasmic bodies become elongated structures, often associated with fluorescent filaments, directed toward the periphery of the cell. Immunostaining with antitubulin antibody and in situ hybridization reactions provide clear evidence of colocalization of vRNA with microtubules. Treatment of protoplasts at midstage of infection with oryzalin prevented the dispersion of the bodies to the periphery of the cell, suggesting that microtubules play a role in intracellular distribution of vRNA. Two different microtubule-based mechanisms, membrane sliding and tip attachment complexes, participate in the movement of ER from the cell center to the periphery of newt lung epithelial cells (Waterman-Storer and Salmon 1998). In TMV infection, the complexes that contain MP, vRNA, and replicase that are associated with ER may be transported towards the periphery of the cells using such mechanisms.
Treatment of infected protoplasts with cytochalasin D clearly altered the pattern of vRNA distribution and caused a delay in the appearance of the large bodies, suggesting a role of microfilaments in their formation and/or stabilization. However, depolymerization of microtubules can also lead to disruption of microfilaments (Panteris et al. 1992); it is therefore difficult to define the specific role of each cytoskeletal component in the intracellular spread of vRNA.
vRNA-ΔM was not associated with elements of the cytoskeleton unless the cells were treated at early stages of infection with cytochalasin D. Such treatment resulted in accumulation of vRNA-ΔM in fluorescent spots at the periphery of the cell, as well as on filaments that were similar in appearance to microtubules. These results suggest that vRNA was associated with microtubules when both MP and microfilaments were absent. The transport of mRNAs along cytoskeletal components has been described in a variety of biological systems, especially in relation to cell differentiation and development (Ferrandon et al. 1994; Kloc and Etkin 1995; Ainger et al. 1997; Broadus et al. 1998). In those cases, RNA transport mechanisms involved specific sequences in the 3′ untranslated region that are recognized by proteins that bind these sequences and mediate the interaction with the cytoskeleton (Oleynikov and Singer 1998). In our study, the change in the distribution pattern of vRNA that is induced by cytochalasin D was more evident in protoplasts infected with vRNA-ΔM vs. wt vRNA. These data suggest some influence of the MP in localization of vRNA in cytochalasin D–treated protoplasts. The implications of MP and microfilaments in the formation and anchoring of the cytoplasmic bodies are discussed below.
Late Stages of Infection: Virus Spread and/or Degradation?
Throughout infection, vRNA was localized around the nucleus, although at mid and late stages, vRNA was also dispersed throughout the cytoplasm and at the periphery of the cell. Accumulation of vRNA around the nucleus may facilitate association with host components that are required for virus infection. However, the accumulation of the much larger bodies that surround the nucleus may imply a different biological role in replication. Recently, a model was proposed by which misfolded proteins that escape the proteosome-mediated pathway of degradation can aggregate to form large structures, referred to as “aggresomes.” Aggresomes are transported on microtubules from peripheral sites to a ubiquitin-rich nuclear location at the microtubule-organizing center, where they are entangled with collapsed intermediate filaments (Johnston et al. 1998). Following this model, during TMV infection, the bodies that contain MP, vRNA, and replicase may be transported on microtubules in order to be degraded. In other recent studies, we described the ubiquitination of MP and the role of proteosomes in degradation of MP (C. Reichel and R.N. Beachy, manuscript submitted for publication).
At late stages of infection, vRNA was localized in structures that protrude from the surface of the cell. These protrusions were not disrupted by oryzalin or cytochalasin D. Furthermore, in other studies, we observed that the protrusions were stained with DiOC6(3) (3, 3′-dihexyloxacarbocyanine iodide), a vital fluorescent stain of ER (not shown). Other studies indicate that these ER-containing structures are not induced by virus infection per se, but may be stimulated by infection (P. Más and R.N. Beachy, manuscript in preparation). We propose that the protruding structures are related to desmotubules, the appressed ER that comprises the central component of plasmodesmata (Lucas et al. 1993). Alternatively, they could be a consequence of cell damage at late stages of infection, which results in the extrusion of ER through the plasma membrane. Interestingly, vRNA-ΔM was not associated with the protrusions, indicating a role of the MP in localization of vRNA with these structures. Together, these results may imply that at least two different types of ER are involved in TMV infection. One type of ER is involved in vRNA replication and does not require the presence of the MP. A second type corresponds to the filamentous protrusions that may be involved in the intercellular spread of the virus and requires a functional MP.
Model of TMV Infection
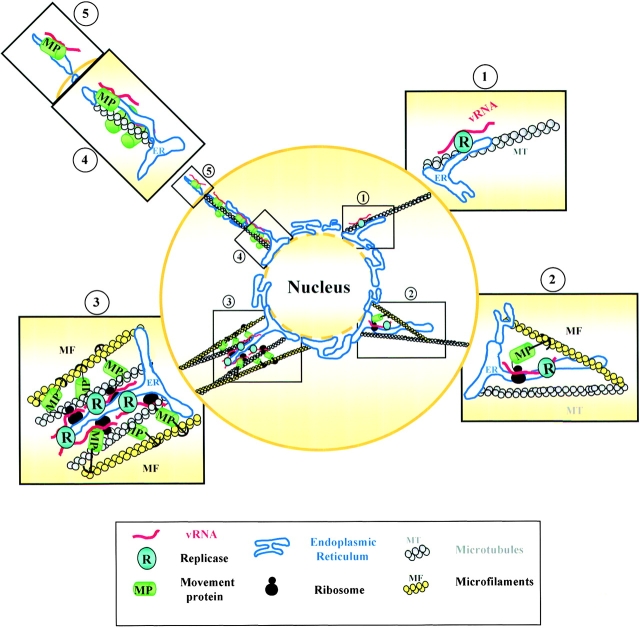
The data presented here and in previous publications are consistent with a model of TMV infection (Fig. 11) in which the replication of vRNA takes place in close association with membranes of the ER (Fig. 11, Fig. 1). In this model, cytoskeletal elements are involved in targeting vRNA/replicase complexes to the perinuclear ER, perhaps via a retrograde flow of microtubules with ER attached as cargo. The ER-associated nascent vRNAs in replication complexes function as mRNAs for the synthesis of MP (Fig. 11, Fig. 2). The MP remains associated with vRNA in the complex, resulting in the formation of large ER-derived structures (Fig. 11, Fig. 3). At this point, the distribution of vRNA would be determined by a balance between the formation and anchoring of the large structures and their spread towards the periphery of the cell. Our results are consistent with a model in which MP and microfilaments participate in the formation and anchoring of the ER-derived structures (Fig. 11, Fig. 3), while microtubules are involved in the transport to their final destinations; i.e., to the periphery for intercellular spread, or toward the nucleus for degradation (Fig. 11, Fig. 4).
Figure 11.
Model of TMV infection in BY2 protoplasts. (1) Early in infection, TMV vRNA comes in close association with membranes of the ER, and vRNA-replicase complexes associated with ER are transported via microtubules to perinuclear positions. (2) Nascent vRNAs synthesize MP that remains associated with vRNA. (3) Formation and anchoring of large ER-derived structures containing MP, replicase, and vRNA that are stabilized by MP and microfilament interactions. (4) Microtubule-based transport system of vRNA-MP complexes towards the periphery of the cell to initiate cell-to-cell spread. (5) Protrusion of ER containing vRNA and MP through the plasma membrane. Note that the model is not drawn in scale.
Several types of experimental evidence support this model. First, there is a dramatic effect on distribution of vRNA-ΔM in cytochalasin D–treated protoplasts. Not only were bodies not found in these protoplasts, in contrast to wt vRNA, but vRNA-ΔM was located on or near the cell periphery but not in the protrusions from the plasma membrane. Since in nontreated protoplasts vRNA-ΔM was not localized at the periphery in early stages of infection, the intracellular spread that normally occurs later in infection was apparently accelerated in the absence of microfilaments. Second, the clear association of vRNA-ΔM with microtubules would explain the role of microtubules in the intracellular spread of the virus towards the periphery of the cell. Third, at late stages of infection, the close relationship between the ER and microtubules explains the association of vRNA in the presence of the MP to a precursor of the plasmodesmata (Fig. 11, Fig. 5) that would result in the cell-to-cell spread of the virus in leaf tissues.
Acknowledgments
We thank Drs. S. Halpain, C. Reichel, and B. Cooper for critical comments of this manuscript. We are also grateful to the Olympus Co. and Drs. S. Kay and S. Halpain (Department of Cell Biology, The Scripps Research Institute, La Jolla, CA) for use of the confocal laser scanning microscope.
This research was supported by National Science Foundation grant MCB 9631124 and by The Scripps Family Chair. P. Más was supported by fellowship from Ministerio de Educación y Cultura, Spain.
Footnotes
BFA, Brefeldin ABiP, lumenal binding proteinCP, coat proteinfluor-RNA, fluorescein-RNAGFP, green fluorescent proteinhpi, hours post-infectionMP, movement proteinTMV, tobacco mosaic viruswt, wild-type
Roger N. Beachy's current address is Donald Danforth Plant Science Center, Clayton, MO 63130.
References
- Ainger K., Avossa D., Diana A.S., Barry C., Barbarese E., Carson J.H. Transport and localization elements in myelin basic protein mRNA. J. Cell Biol. 1997;138:1077–1087. doi: 10.1083/jcb.138.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainger K., Avossa D., Morgan F., Hill S.J., Barry C., Barbarese E., Carson J.H. Transport and localization of exogenous MBP mRNA microinjected into oligodendrocytes. J. Cell Biol. 1993;123:431–441. doi: 10.1083/jcb.123.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen N.S., Brown D.T. Dynamics of the endoplasmic reticulum in living onion epidermal cells in relation to microtubules, microfilaments, and intracellular particle movement. Cell. Motil. Cytoskelet. 1988;10:153–163. [Google Scholar]
- Arn E.A., Macdonald P.M. Motors driving mRNA localizationnew insights from in vivo imaging. Cell. 1998;95:151–154. doi: 10.1016/s0092-8674(00)81745-6. [DOI] [PubMed] [Google Scholar]
- Bald J.G. Cytological evidence for the replication of plant virus ribonucleic acid in the nucleus. Virology. 1964;22:377–387. doi: 10.1016/0042-6822(64)90028-5. [DOI] [PubMed] [Google Scholar]
- Bassell G., Singer R.H. mRNA and cytoskeletal filaments. Curr. Opin. Cell Biol. 1997;9:109–115. doi: 10.1016/s0955-0674(97)80159-7. [DOI] [PubMed] [Google Scholar]
- Beachy R.N., Zaitlin M. Replication of tobacco mosaic virus. Replicative intermediate and other viral related RNAs associated with polyribosomes. Virology. 1975;63:84–97. doi: 10.1016/0042-6822(75)90373-6. [DOI] [PubMed] [Google Scholar]
- Beier H., Barciszewska M., Krupp G., Mitnacht R., Gross H.J. UAG readthrough during TMV RNA translationisolation and sequence of two tRNAs Tyr with supressor activity from tobacco plants. EMBO (Eur. Mol. Biol. Organ.) J. 1984;3:351–356. doi: 10.1002/j.1460-2075.1984.tb01810.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz K., Egger D., Rasser Y., Bossart W. Intracellular distribution of poliovirus proteins and the induction of virus-specific cytoplasmic structures. Virology. 1983;131:39–48. doi: 10.1016/0042-6822(83)90531-7. [DOI] [PubMed] [Google Scholar]
- Boston R., Viitanen P.V., Vierling E. Molecular chaperones and protein folding in plants. Plant Mol. Biol. 1996;32:191–222. doi: 10.1007/BF00039383. [DOI] [PubMed] [Google Scholar]
- Broadus J., Fuerstenberg S., Doe C.Q. Staufen-dependent localization of prospero mRNA contributes to neuroblats daughter-cell fate. Nature. 1998;391:792–795. doi: 10.1038/35861. [DOI] [PubMed] [Google Scholar]
- Carrington J.C., Kasschau K.D., Mahajan S.K., Schaad M.C. Cell-to-cell and long-distance transport of viruses in plants. Plant Cell. 1996;8:1669–1681. doi: 10.1105/tpc.8.10.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V., Knorr D., Schuster G., Zambryski P. The P30 movement protein of tobacco mosaic virus is a single-stranded nucleic acid binding protein. Cell. 1990;60:637–647. doi: 10.1016/0092-8674(90)90667-4. [DOI] [PubMed] [Google Scholar]
- Citovsky V., Zambryski P. How do plant virus nucleic acids move through intercellular connections? Bioessays. 1991;13:373–379. doi: 10.1002/bies.950130802. [DOI] [PubMed] [Google Scholar]
- Denecke J., Carlsson L.E., Vidal S., Hoglund A.S., Ek B., van Zeijil M.J., Sinjorjo K.M.C., Palva E.T. The tobacco homolog of mammalian calreticulin is present in protein complexes in vivo. Plant Cell. 1995;7:391–406. doi: 10.1105/tpc.7.4.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deom C.M., Oliver M.J., Beachy R.N. The 30-kilodalton gene product of tobacco mosaic virus potentiates virus movement. Science. 1987;237:389–394. doi: 10.1126/science.237.4813.389. [DOI] [PubMed] [Google Scholar]
- Deom C.M., Schubert K.R., Wolf S., Holt C.A., Lucas W.J., Beachy R.N. Molecular characterization and biological function of the movement protein of tobacco mosaic virus in transgenic plants. Proc. Natl. Acad. Sci. USA. 1990;87:3284–3288. doi: 10.1073/pnas.87.9.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau K., Cronshaw J. Relation of tobacco mosaic virus to the host cells. J. Cell Biol. 1967;33:665–678. doi: 10.1083/jcb.33.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandon D., Elphick L., Nusslein-Volhard C., St. Johnston D. Staufen associates with the 3′ UTR of bicoid RNA to form particles that move in a microtubule dependent manner. Cell. 1994;79:1221–1232. doi: 10.1016/0092-8674(94)90013-2. [DOI] [PubMed] [Google Scholar]
- Ferrandon D., Koch I., Westhof E., Nusslein-Volhard C. RNA–RNA interaction is required for the formation of specific bicoid mRNA 3′ UTR-STAUFEN ribonucleoprotein particles. EMBO (Eur. Mol. Biol. Organ.) J. 1998;16:1751–1758. doi: 10.1093/emboj/16.7.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forristal C., Pondel M., Chen L., King M.L. Patterns of localization and cytoskeletal association of two vegetally localized RNAs, Vg1 and Xcat-2. Development (Camb.) 1995;121:201–208. doi: 10.1242/dev.121.1.201. [DOI] [PubMed] [Google Scholar]
- Gavis E.R. Expeditions to the poleRNA localization in Xenopus and Drosophila . Trends Cell Biol. 1997;7:485–492. doi: 10.1016/S0962-8924(97)01162-8. [DOI] [PubMed] [Google Scholar]
- Hazelrigg T. The destinies and destinations of RNAs. Cell. 1998;95:451–460. doi: 10.1016/s0092-8674(00)81613-x. [DOI] [PubMed] [Google Scholar]
- Heim R., Cubitt A.B., Tsien R.Y. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- Heinlein M., Epel B.L., Padgett H.S., Beachy R.N. Interaction of tobamovirus movement proteins with the plant cytoskeleton. Science. 1995;270:1983–1985. doi: 10.1126/science.270.5244.1983. [DOI] [PubMed] [Google Scholar]
- Heinlein M., Padgett H.S., Gens J.S., Pickard B.G., Casper S.J., Epel B.L., Beachy R.N. Changing patterns of localization of TMV movement protein and replicase to endoplasmic reticulum and microtubules during infection. Plant Cell. 1998;10:1107–1120. doi: 10.1105/tpc.10.7.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson J., Satiat-Jeunemaitre B., Napier R., Hawes C. Brefeldin A–induced disassembly of the Golgi apparatus is followed by disruption of the endoplasmic reticulum in plant cells. J. Exp. Bot. 1994;45:1347–1351. [Google Scholar]
- Hepler P.K., Palevitz B.A., Lancelle S.A., McCauley M.M., Lichtscheidl I. Cortical endoplasmic reticulum in plants. J. Cell Sci. 1990;96:355–373. [Google Scholar]
- Hills G.J., Plaskitt K.A., Young N.D., Dunigan D.D., Watts J.W., Wilson T.M.A., Zaitlin M. Immunogold localization of the intracellular sites of structural and nonstructural tobacco mosaic virus proteins. Virology. 1987;161:488–496. doi: 10.1016/0042-6822(87)90143-7. [DOI] [PubMed] [Google Scholar]
- Holt C.A., Beachy R.N. In vivo complementation of infectious transcripts from mutant tobacco mosaic virus cDNAs in transgenic plants. Virology. 1991;181:109–117. doi: 10.1016/0042-6822(91)90475-q. [DOI] [PubMed] [Google Scholar]
- Hovland R., Hesketh J.E., Pryme I.F. The compartmentalization of protein synthesisimportance of cytoskeleton and role in mRNA targeting. Int. J. Biochem. Cell Biol. 1996;28:1089–1105. doi: 10.1016/1357-2725(96)00059-3. [DOI] [PubMed] [Google Scholar]
- Hugdahl J.D., Morejohn L.C. Rapid and reversible high-affinity binding of the dinitroaniline herbicide oryzalin to tubulin of Zea mays L. Plant Physiol. 1993;102:725–740. doi: 10.1104/pp.102.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa M., Meshi T., Motoyoshi F., Takamatsu N., Okada Y. In vitro mutagenesis of the putative replicase genes of tobacco mosaic virus. Nucleic Acids Res. 1986;14:8291–8305. doi: 10.1093/nar/14.21.8291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa M., Meshi T., Ohno T., Okada Y. Specific cessation of minus-strand RNA accumulation at an early stage of tobacco mosaic virus infection. J. Virol. 1991;65:861–868. doi: 10.1128/jvi.65.2.861-868.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston J.A., Ward C.L., Kopito R.R. Aggresomesa cellular response to misfolded proteins. J. Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn T.W., Lapidot M., Heinlein M., Reichel C., Cooper B., Gafny R., Beachy R.N. Domains of the TMV movement protein involved in subcellular localization. Plant J. 1998;15:15–25. doi: 10.1046/j.1365-313x.1998.00172.x. [DOI] [PubMed] [Google Scholar]
- Kloc M., Etkin L.D. Translocation of repetitive RNA sequences with the germ plasm in Xenopus oocytes. Science. 1993;262:1712–1714. doi: 10.1126/science.7505061. [DOI] [PubMed] [Google Scholar]
- Kloc M., Etkin L.D. Two distinct pathways for the localization of RNAs at the vegetal cortex in Xenopus oocytes. Development (Camb.) 1995;121:287–297. doi: 10.1242/dev.121.2.287. [DOI] [PubMed] [Google Scholar]
- Lucas W.J., Ding B., van der Schoot C. Plasmodesmata and the supracellular nature of plants. New Phytol. 1993;125:435–476. doi: 10.1111/j.1469-8137.1993.tb03897.x. [DOI] [PubMed] [Google Scholar]
- Martelli G.P., Russo M. Plant virus inclusion bodies. Adv. Virus Res. 1977;21:175–266. doi: 10.1016/s0065-3527(08)60763-0. [DOI] [PubMed] [Google Scholar]
- Más P., Beachy R.N. Distribution of TMV movement protein in single living protoplasts immobilized in agarose. Plant J. 1998;15:835–842. doi: 10.1046/j.1365-313X.1998.00253.x. [DOI] [PubMed] [Google Scholar]
- Más P., Pallás V. Long-distance movement of cherry leaf roll virus in infected tobacco plants. J. Gen. Virol. 1996;77:531–540. doi: 10.1099/0022-1317-77-3-531. [DOI] [PubMed] [Google Scholar]
- McLean B.G., Zupan J., Zambryski P. Tobacco mosaic virus movement protein associates with the cytoskeleton in tobacco cells. Plant Cell. 1995;7:2101–2114. doi: 10.1105/tpc.7.12.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMaster G.K., Carmichel G.G. Analysis of single and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc. Natl. Acad. Sci. USA. 1977;74:4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshi T., Watanabe Y., Saito T., Sugimoto A., Maeda T., Okada Y. Function of the 30 kd protein of tobacco mosaic virusinvolvement in cell-to-cell movement and dispensability for replication. EMBO (Eur. Mol. Biol. Organ.) J. 1987;6:2557–2563. doi: 10.1002/j.1460-2075.1987.tb02544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molla A., Paul A.V., Wimmer E. Effects of temperature and lipophilic agents on poliovirus formation and RNA synthesis in a cell-free system. J. Virol. 1993;67:5932–5938. doi: 10.1128/jvi.67.10.5932-5938.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejidat A., Cellier F., Holt C.A., Gafny R., Eggenberger A.L., Beachy R.N. Transfer of the movement protein gene between two tobamovirusesinfluence on local lesion development. Virology. 1991;180:318–326. doi: 10.1016/0042-6822(91)90036-b. [DOI] [PubMed] [Google Scholar]
- Nelson R.S., Hodgson R.A.J., Beachy R.N., Shintaku M.H. Impeded systemic accumulation of the masked starin of tobacco mosaic virus. Mol. Plant-Microbe Interact. 1993;6:45–54. [Google Scholar]
- Nilsson-Tillgren T., Kielland-Brandt M.C., Bekke B. Studies on the biosynthesis of tobacco mosaic virus VI. On the subcellular localization of double-stranded viral RNA. Mol. Gen. Genet. 1974;128:157–169. doi: 10.1007/BF02654488. [DOI] [PubMed] [Google Scholar]
- Okamoto S., Machida Y., Takebe I. Subcellular localization of tobacco mosaic virus minus strand RNA in infected protoplasts. Virology. 1988;167:194–200. doi: 10.1016/0042-6822(88)90069-4. [DOI] [PubMed] [Google Scholar]
- Oleynikov Y., Singer R.H. RNA localizationdifferent zipcodes, same postman? Trends Cell Biol. 1998;8:381–383. doi: 10.1016/s0962-8924(98)01348-8. [DOI] [PubMed] [Google Scholar]
- Oparka K.J., Prior D.A.M., Santa Cruz S., Padgett H.S., Beachy R.N. Gating of epidermal plasmodesmata is restricted to the leading edge of expanding infection sites of tobacco mosaic virus (TMV) Plant J. 1997;12:781–789. doi: 10.1046/j.1365-313x.1997.12040781.x. [DOI] [PubMed] [Google Scholar]
- Osman T.A.M., Buck K.W. Complete replication in vitro of tobacco mosaic virus RNA by a template-dependent, membrane-bound RNA polymerase. J. Virol. 1996;70:6227–6234. doi: 10.1128/jvi.70.9.6227-6234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panteris E., Apostolakos P., Galatis B. The organization of F-actin in root-tip cells of Adiantum-capillus-veneris throughout the cell-cycle. A double label fluorescence microscopy study. Protoplasma. 1992;170:128–137. [Google Scholar]
- Ralph R.K., Bullivant S., Wojcik S.J. Cytoplasmic membranes as a possible site of tobacco mosaic virus RNA replication. Virology. 1971;43:713–716. doi: 10.1016/0042-6822(71)90295-9. [DOI] [PubMed] [Google Scholar]
- Reichel C., Beachy R.N. Tobacco mosaic virus infection induces severe morphological changes of the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 1998;95:11169–11174. doi: 10.1073/pnas.95.19.11169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo-Hartwig M.A., Ahlquist P. Brome Mosaic virus Helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J. Virol. 1996;70:8908–8916. doi: 10.1128/jvi.70.12.8908-8916.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo-Hartwig M.A., Carrington J.C. The tobacco etch potyvirus 6-kilodalton protein is membrane-associated and involved in viral replication. J. Virol. 1994;68:2388–2397. doi: 10.1128/jvi.68.4.2388-2397.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T., Hosokawa D., Meshi T., Okada Y. Immunocytochemical localization of the 130K and 180K proteins (putative replicase components) of tobacco mosaic virus. Virology. 1987;160:477–481. doi: 10.1016/0042-6822(87)90020-1. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T.A. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. C. Wolan, editor. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 7.37–7.51.
- Satiat-Jeunemaitre B., Cole L., Bouret T., Howard R., Hawes C. Brefeldin A effects in plant and fungal cellssomething new about vesicle trafficking? J. Microsc. 1996;181:162–177. doi: 10.1046/j.1365-2818.1996.112393.x. [DOI] [PubMed] [Google Scholar]
- Schaad M.C., Jensen P.E., Carrington J.C. Formation of plant RNA virus replication complexes on membranesrole of an endoplasmic reticulum-targeted viral protein. EMBO (Eur. Mol. Biol. Organ) J. 1997;16:4049–4059. doi: 10.1093/emboj/16.13.4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldt A.J., Adams J.H., Davidson C.M., Micklem D.R., Haseloff J., Johnston D.S., Brand A.H. Miranda mediates asymmetric protein and RNA localization in developing nervous sytem. Genes Dev. 1998;12:1847–1857. doi: 10.1101/gad.12.12.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.H., Schlegel D.E. The incorporation of nucleic acid precursors in healthy and virus-infected plant. Virology. 1965;26:180–189. doi: 10.1016/0042-6822(65)90045-0. [DOI] [PubMed] [Google Scholar]
- St. Johnston D. The intracellular localization of messenger RNAs. Cell. 1995;81:161–170. doi: 10.1016/0092-8674(95)90324-0. [DOI] [PubMed] [Google Scholar]
- Staehelin L.A. The plant ERa dynamic organelle composed of a large number of discrete functional domains. Plant J. 1997;11:1151–1165. doi: 10.1046/j.1365-313x.1997.11061151.x. [DOI] [PubMed] [Google Scholar]
- Strauss J.H., Strauss E.G. The aphavirusesgene expression, replication and evolution. Microbiol. Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamatsu N., Ishikawa M., Meshi T., Okada Y. Expression of bacterial cloramphenicolacetyltransferase gene in tobacco plants mediated by TMV-RNA. EMBO (Eur. Mol. Biol. Organ.) J. 1987;6:307–311. doi: 10.1002/j.1460-2075.1987.tb04755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomenius K., Clapham D., Meshi T. Localization by immunogold cytochemistry of the virus-coded 30K protein in plasmodesmata of leaves infected with tobacco mosaic virus. Virology. 1987;160:363–371. doi: 10.1016/0042-6822(87)90007-9. [DOI] [PubMed] [Google Scholar]
- Ward B.M., Medville R., Lazarowitz S.G., Turgeon R. The geminivirus BL1 movement protein is associated with endoplasmic reticulum-derived tubules in developing phloem cells. J. Virol. 1997;71:3726–3733. doi: 10.1128/jvi.71.5.3726-3733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Meshi T., Okada Y. Infection of tobacco protoplasts with in vitro transcribed tobacco mosaic virus RNA using an improved electroporation method. FEBS Lett. 1987;219:65–69. [Google Scholar]
- Watanabe Y., Okada Y. In vitro viral RNA synthesis by a subcellular fraction of TMV-inoculated tobacco protoplasts. Virology. 1986;149:74–83. doi: 10.1016/0042-6822(86)90087-5. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer C., Salmon E.D. Endoplasmic reticulum membrane tubules are distributed by microtubules in living cells using three distinct mechanisms. Curr. Biol. 1998;8:798–806. doi: 10.1016/s0960-9822(98)70321-5. [DOI] [PubMed] [Google Scholar]
- Wilhelm J., Vale R.D. RNA on the movethe mRNA localization pathway. J. Cell Biol. 1993;123:269–274. doi: 10.1083/jcb.123.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S., Deom C.M., Beachy R.N., Lucas W.J. Movement protein of tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science. 1989;246:377–379. doi: 10.1126/science.246.4928.377. [DOI] [PubMed] [Google Scholar]
- Wu S.X., Ahlquist P., Kaesberg P. Active complete in vitro replication of nodavirus RNA requires glycerophospholipid. Proc. Natl. Acad. Sci. USA. 1992;89:11136–11140. doi: 10.1073/pnas.89.23.11136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitlin M., Duda C.T., Petti M.A. Replication of tobacco mosaic virus. V. Properties of the bound and solubilized replicase. Virology. 1973;53:300–311. doi: 10.1016/0042-6822(73)90207-9. [DOI] [PubMed] [Google Scholar]
- Zambryski P. Plasmodesmataplant channels for molecules on the move. Science. 1995;270:1943–1944. doi: 10.1126/science.270.5244.1943. [DOI] [PubMed] [Google Scholar]