Abstract
An understanding of how the nuclear pore complex (NPC) mediates nucleocytoplasmic exchange requires a comprehensive inventory of the molecular components of the NPC and a knowledge of how each component contributes to the overall structure of this large molecular translocation machine. Therefore, we have taken a comprehensive approach to classify all components of the yeast NPC (nucleoporins). This involved identifying all the proteins present in a highly enriched NPC fraction, determining which of these proteins were nucleoporins, and localizing each nucleoporin within the NPC. Using these data, we present a map of the molecular architecture of the yeast NPC and provide evidence for a Brownian affinity gating mechanism for nucleocytoplasmic transport.
Keywords: nuclear pore complex, nucleoporins, nucleocytoplasmic transport, mass spectrometry, proteomics
Introduction
The presence of a nucleus requires that a diverse set of macromolecules must be efficiently transported across the double-membraned nuclear envelope (NE). The mediators of this exchange are large assemblies termed nuclear pore complexes (NPCs) which span pores in the NE in order to connect the nuclear and cytoplasmic compartments (Davis 1995). The NPC is octagonally symmetric around its cylindrical axis. It is composed of a cylindrical core, with a plane of pseudo-mirror symmetry running parallel to the NE, and contains eight interconnected spokes which surround a central channel (or central transporter). Peripheral filaments emanate from the core into the nucleoplasm and cytoplasm; while the cytoplasmic filaments spread outwards, the nuclear filaments conjoin distally to form a basket-like structure. This general architecture is shared by NPCs in all eukaryotes, and many of the NPC proteins (collectively termed nucleoporins, or nups) are conserved across phyla (Davis 1995; Fabre and Hurt 1997).
Nucleocytoplasmic transport depends on the interplay between soluble transport factors, their cargoes, and the NPC. Karyopherins (kaps, also known as importins/exportins) bind to specific import or export signals and mediate substrate docking to the NPC. Transport also requires energy, the small GTPase Ran and factors that regulate its nucleotide-bound state (Mattaj and Englmeier 1998; Talcott and Moore 1999). Several lines of evidence indicate that the docking sites on NPCs for various transport factors are composed of a specific subset of nups that contains degenerate Phe-Gly repeats (FG nups) and appear to constitute most of the filamentous structures emanating from the NPC (Buss et al. 1994; Radu et al. 1995; Katahira et al. 1999; Kehlenbach et al. 1999). How these and other nups cooperate within the context of the NPC to mediate transport remains a mystery (Mattaj and Englmeier 1998; Talcott and Moore 1999). The functions of the NPC arise from the complex overlapping contributions of individual nups; hence, a comprehensive approach is essential to understanding the mechanism of nucleocytoplasmic transport.
To this end, we have identified all the proteins present in a subcellular fraction that is highly enriched in NPCs. Furthermore, we have determined which of these proteins are nups and localized each nup within the NPC. Using these data, we present a map of the molecular architecture of the yeast NPC and propose a mechanism for nucleocytoplasmic transport.
Materials and Methods
Separation and Identification of Yeast NPC Proteins
For HPLC, highly enriched yeast NPC proteins (Rout and Blobel 1993) were precipitated by the addition of 4 vol of filtered methanol, resuspended by incubation in 10 mM Tris base, 10 mM DTT, 2% SDS for 10 min at 65°C, and then diluted with 9 vol of 10 mM sodium phosphate and 0.1 mM CaCl2, pH 6.8. Ceramic hydroxyapatite HPLC gave very efficient recovery of the loaded proteins (∼70% for most proteins), regardless of their size or charge, with moderate resolution. A volume of the NPC preparation containing ∼5 mg protein was loaded at 0.5 ml/min onto a Pentax SH-0710M hydroxyapatite column (American International Chemical, Inc.) preequilibrated in buffer A (10 mM sodium phosphate, 1 mM DTT, 0.1% SDS, and 0.1 mM CaCl2, pH 6.8). Proteins were differentially eluted at 0.5 ml/min with a gradient of 100% buffer A to 100% buffer B (1 M sodium phosphate, 1 mM DTT, and 0.1% SDS, pH 6.8). 80 2-ml fractions were collected and polypeptides precipitated with TCA. Reverse phase TFA-HPLC had excellent resolution in the low molecular mass protein range (up to ∼50 kD). A similar quantity of the NPC preparation was thus loaded at 0.15 ml/min onto a Vydac narrow bore 2.1 × 250-mm C4 reverse-phase column preequilibrated in 88% solvent A2 (0.1% TFA in H2O), 12% solvent B2 (0.08% TFA in acetonitrile). A linear gradient to 100% B2 differentially eluted the bound proteins (180 fractions were collected). Controls ensured that all the proteins bound to both columns before fractionation. Fractions from both chromatographic preparations were separated by SDS-PAGE on 4–20% Novex gels and the protein bands visualized by copper staining (Bio Rad). The bands of interest were excised, subjected to in-gel digestion with trypsin, and the resulting peptide mixtures extracted as described (Qin et al. 1997). Peptide mixtures were analyzed with matrix-assisted laser desorption/ionization mass spectrometry (MALDI-TOF MS) using a delayed ion extraction and ion mirror reflector mass spectrometer (Voyager-DE STR; Perseptive Biosystems), a delayed ion extraction linear system (Voyager-DE; Perseptive Biosystems), or a MALDI ion trap mass spectrometer that was constructed in-house (Qin et al. 1996). The accurately measured masses of the tryptic peptides were used to search for protein candidates in the nonredundant protein sequence database with the program ProFound (http://prowl.rockefeller.edu or http://ProteoMetrics.com). The tandem MS fragmentation analyses were carried out as described (Qin et al. 1997) using the protein search algorithm PepFrag (Fenyo et al. 1998; http://prowl.rockefeller.edu or http://ProteoMetrics.com). Formic acid HPLC separation was performed as described; while the resolution and recovery using this method was superior to TFA-HPLC, formylation of residues prevented mass spectrometric identification of the separated proteins and instead they were analyzed by peptide microsequencing (Rout and Blobel 1993; Fernandez et al. 1994; Wozniak et al. 1994).
Tagged Strains
The genomic copy of each gene was tagged by a COOH-terminal, in-frame integration of a PCR-derived DNA fragment encoding the IgG binding domains of protein A amplified from the plasmid pProtA/HU (Aitchison et al. 1995a) or from pProtA/HIS5 which instead carries the S. pombe HIS5 gene as its selectable marker (Wach et al. 1997). Genes were tagged similarly with the 3HA (FLU) epitope (Longtine et al. 1998). This fast, well-proven, and reliable genomic tagging method ensures that the chimeras are expressed from their own promoters, and, by virtue of the COOH-terminal placement, minimizes the possibility of the tag affecting normal polypeptide chain folding during translation. Correct integration of the tag was confirmed by immunoblotting and, where the molecular mass of the chimera was close to those of other tagged strains, by PCR analysis (Aitchison et al., 1995). Diploid cells were sporulated and haploids containing the tagged gene of interest were isolated by tetrad dissection. In only one case (Cdc31p) did the tag affect the viability of the haploid. Therefore, the untagged protein was monitored with monospecific polyclonal antibodies. To reveal NPC associations, tagged strains were crossed with nup120Δ cells (NP120-25-3 or its sister spore NP120-25-4 (Aitchison et al. 1995b). Diploid cells were sporulated and nup120Δ haploids containing the tagged gene of interest were selected.
Immunoblotting Assays
Cells were fractionated using published protocols to produce NEs, NPCs (Rout and Blobel 1993; Strambio-de-Castillia et al. 1995), or carbonate-extracted nuclear membranes (Wozniak et al. 1994). The relative coenrichment of the tagged proteins was detected as described (Rout et al. 1997), with the FLU tag being visualized by mAb12CA5 (BAbCo). For quantitative immunoblotting, 3-, 5-, 8-, and 12-μg aliquots of tagged NEs were separated by SDS-PAGE, transferred to nitrocellulose and stained sequentially with mAb118C3 (to detect the internal standard Pom152p; Strambio-de-Castillia et al. 1995) and Cy5-conjugated donkey anti–mouse (to detect both the Mab118C3 and the protein A tag; Jackson ImmunoResearch Laboratories, Inc.). Band intensities were quantified (using ImageQuant v.1.1 software in the PhosphorImager; Molecular Dynamics) and the signals of the tag were plotted as a function of the internal standard signals. The resulting slopes (calculated by regression analysis in Microsoft Excel, which also confirmed that the plot was linear, and hence not saturated) yielded a value that represented the relative abundance of the tagged protein to the internal standard in the NPC. In some cases the Pom152p band overlapped the tag signal, and so Nup57p (detected with mAb350) was used as the internal standard; parallel incubations with both mAb350 and mAb118C3 on nonoverlapping tagged proteins allowed the calculation of a conversion factor. We conducted numerous controls to limit measurement errors. Thus, semiquantitative analyses confirmed that transfer of proteins to the immunoblot membrane was efficient, such that there was no signal bias due to the molecular mass of the tagged protein. We ensured that both the first and second antibodies were saturated, so the signal did not become antibody-limited. Mock quantitation with untagged NEs indicated the contribution of signal from the internal standard's breakdown products was negligible. By quantitating major PrA-tagged non-NPC proteins we could show that we had not saturated the maximum limit of the ratios that could be measured, nor had we saturated the detection limit of the PhosphorImager (data not shown).
Immunolocalization
Images of cells fixed and stained for immunofluorescence microscopy (Rout et al. 1997) were recorded with a Hamamatsu Orca digital camera on a Zeiss Axioplan 2 microscope using Openlab software (Improvision) or a Zeiss Axioskop 2 and Spot camera (Diagnostic Instruments Inc.). Immunoelectron microscopy was performed on NEs isolated from tagged haploid strains (Strambio-de-Castillia et al. 1995), except for the NH2-terminal fragment of Nup145p, which from necessity used NEs made from a heterozygous tagged diploid strain (Emtage et al. 1997). An optimum heparin extraction range was first tested by diluting NEs with 4 vol of 10 mM bisTris-Cl, pH 6.50, 0.1 mM MgCl2, and 20% DMSO containing various concentrations of heparin. After incubation on ice for 1 h, the NEs were centrifuged at 100,000 g for 20 min at 4°C, and the supernatant and pellet fractions checked by immunoblotting for retention of the tagged protein in the NE pellet, to ensure no significant loss of the tagged protein had occurred. NEs similarly extracted with the correct concentrations of heparin were processed for immunoelectron microscopy (Kraemer et al. 1995), using affinity-purified rabbit IgG (ICN) as primary and 5-nm gold-labeled anti-rabbit IgG (Amersham Life Sciences) as secondary antibodies, always using the same labeling conditions because the tags were identical. We found no extraction-specific effects other than the expected increase in signal. We controlled for possible experimenter bias by performing the immunoelectron microscopy double blind. The absence of any signal without the first antibody indicated we were detecting specific labeling. We also excluded nucleocytoplasmic misorientation as a possible explanation of apparent bilateral localization of a nup, as we always found examples of the signal on both sides of the same NPC (data not shown). Montages were created in Adobe Photoshop v.4.0.1 and the gold particle positions measured with NIH Image v.1.61.
To estimate the position of each nup within the NPC from the immunoelectron microscopy labeling distributions, we first measured the distances of each gold particle (2,496 particles for the montages presented in Fig. 7) from both the cylindrical axis of the NPC and from its mirror plane (Kraemer et al. 1995). The resulting distributions of particle positions represent raw data uncorrected for cylindrical averaging, steric hindrance from the dense NPC core, and blurring effects arising from the distribution of antibody orientations. We developed a modeling procedure that corrected for each of these effects. Using this model, we calculated the expected cylindrically averaged and projected distributions for distance intervals of 2.5 nm, in both the R (position from the cylindrical axis) and Z (position from the mirror plane) directions, and determined the values of R and Z that gave the best fit to the raw experimental data for each nup. Rare examples of sagittally sectioned labeled NPCs allowed us to compare the accuracy of our R estimates with a directly measured R average figure; the two values were within 4 nm of each other, well within our estimated experimental error of ∼7 nm (data not shown). For symmetrically localized nups we pooled the distributions on both sides to increase the accuracy of our estimates.
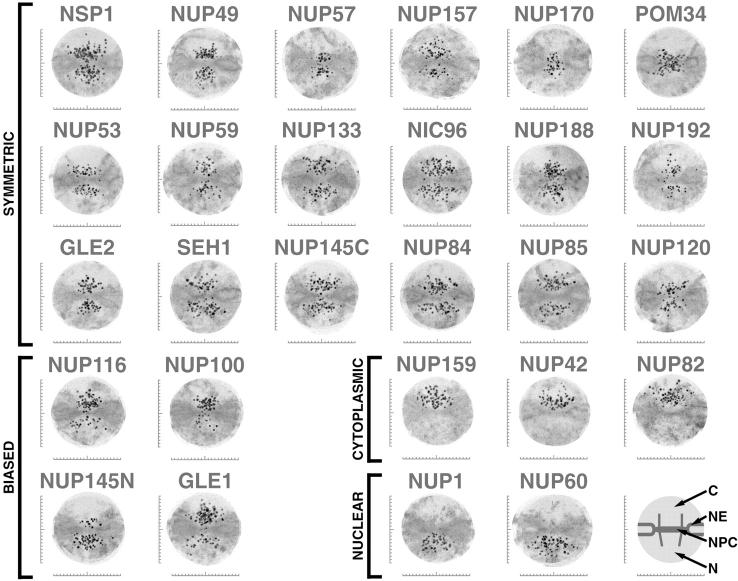
Figure 7.
Localization of the tagged nucleoporins by immunoelectron microscopy. To estimate the position of the tagged nups within the NPC, we immunolabeled NEs from each protein A–tagged strain using a gold-conjugated antibody to visualize the tag. We selected labeled NPC images that had no obvious signs of damage or occlusion, and whose nucleocytoplasmic orientation could be unambiguously determined (Nehrbass et al. 1996). We chose only those NPCs sectioned perpendicular to the NE plane with a clearly visible double membrane. We selected a radius of 100 nm around the estimated center of each NPC as an excision limit and then created an aligned superimposed montage using 20 of the resulting excised NPC images (Strambio-de-Castillia et al. 1999). Scale bars are graduated in 10-nm intervals, with the horizontal bar centered on the cylindrical axis of the NPC and the vertical bar centered on the plane of NPC pseudo-mirror symmetry. The major features in each montage are diagrammed schematically on the lower right (C, cytoplasm; N, nucleoplasm). Montages from extraction conditions with the highest degree of specific labeling (averages ranging between 3–10 gold particles/NPC) are shown. They are arranged into those showing approximately symmetric labeling on both sides of the NPC, those with labeling biased towards one face, and those localized exclusively either to the cytoplasmic face or the nucleoplasmic face. NUP145N and NUP145C, NH2-terminal and COOH-terminal tagged fragments respectively of Nup145p (in all other figures and tables NUP145 refers only to the COOH-terminal tagged fragment).
Results
Identification of Proteins Associated with the Yeast Nuclear Pore Complex
A systematic compositional analysis of the NPC has the potential to identify virtually all of the NPC components. However, an exhaustive analysis requires the isolation and identification of every detectable protein in a highly enriched preparation of intact NPCs. This has become feasible with the development of a method that produces large quantities of highly enriched yeast (Saccharomyces) NPCs (Rout and Blobel 1993), together with the completion of the yeast genome sequence and advances in protein identification techniques. This NPC fraction is the most highly enriched preparation available. The NPCs isolated by this mild procedure appear morphologically intact (Rout and Blobel 1993; Yang et al. 1998) and no significant loss has been found for any confirmed nup, including the most peripheral nups and all of the known pore membrane proteins (see Fig. 5, Table ). Indeed, many peripherally associated nucleocytoplasmic transport factors partially cofractionate with the NPCs during this procedure.
Figure 5.
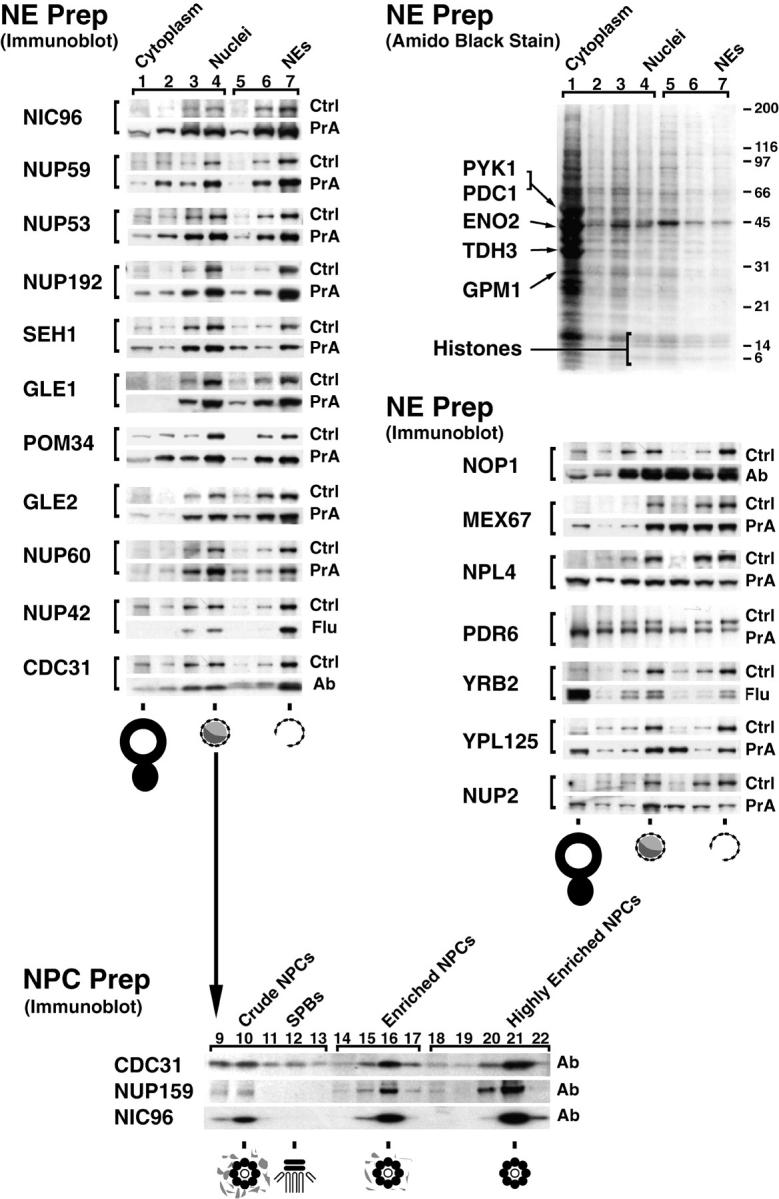
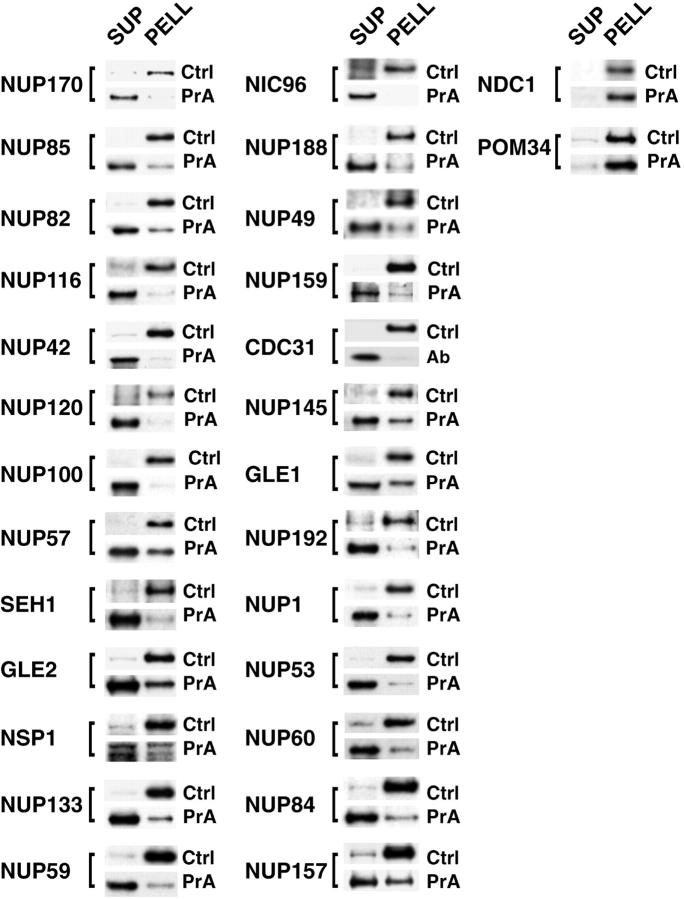
Relative distribution of tagged proteins in subcellular fractions. (Top right) Cells from the Nup42p Flu-tagged strain were fractionated and the proteins from each fraction were separated by SDS-PAGE and visualized by amido black. Lanes 1–4 were loaded at one cell equivalent and lanes 5–7 were loaded at three cell equivalents. Fraction 1 contains cytoplasmic material, 2 and 3 contain membranes, and 4 contains mainly nuclei. The nuclei were subjected to a second round of fractionation to obtain the two chromatin fractions 5 and 6, and fraction 7 enriched in NEs. The majority of proteins, including the indicated abundant cytoplasmic markers (Pyk1p, Pdc1p, Eno2p, Tdh3p, and Gpm1p) identified by mass spectrometry did not coenrich with the nuclei or the NEs. The histone triplet at ∼14 kD coenriched with nuclei but not NEs. (Top left) Proteins that cofractionate with NEs. Fractions from the same enrichment procedure for various tagged strains were probed for the internal standard Pom152p (Ctrl) and the protein of interest, mostly through a protein A tag (PrA). In a few cases, a FLU tag (Flu) or a monospecific antibody (Ab) was used. As expected, known nups coenriched with the NE-containing fractions, as did the newly identified Nup60p, Nup192p, and Pom34p. Seh1p, Gle1p, Gle2p, and Nup42p/Rip1p also coenriched with NEs, securing the classification of these proteins as nups. (Middle right) Proteins that do not cofractionate with NEs. Mex67p coenriched mainly with nuclei and partially with NEs, but a significant amount remained in the nucleoplasm in agreement with its recent classification as an important new mRNA export factor (Katahira et al. 1999). Similarly, both Pdr6p/Kap122p and Ypl125p/Kap120p showed a partial association with the highly enriched NEs during fractionation which, taken together with the immunofluorescence microscopy data, confirmed them as karyopherins. Neither Yrb2p nor Nup2p, which are closely related to each other at the primary sequence level, cofractionated with the NPC-containing fractions. Yrb2p is now known not to be a nup (see Fig. 4), but the significant amount of Nup2p remaining with the NE fraction points to a strong association with the NPCs. As Npl4p did not cofractionate with NPCs (and showed no NPC association by immunofluorescence microscopy; Fig. 4), we concluded that it is not a strongly bound component of the NPC. (Bottom) Nuclei were separated into fractions containing spindle pole bodies (SPBs), crude NPCs, enriched NPCs, and highly enriched NPCs; fraction numbers are as previously described (Rout and Kilmartin 1990). Cdc31p, which coenriched with the Pom152p control in the NE preparation, also cofractionated with Nic96p and Nup159p, two previously known nucleoporins, in the highly enriched NPC preparation, which allowed us to characterize Cdc31p as a component of the NPC. However, unlike the controls, significant amounts of Cdc31p were found in the SPB fraction. Cdc31p had given SPB staining plus some peripheral nuclear signal by immunofluorescence microscopy (Biggins and Rose 1994). Thus, Cdc31p appears to be present in both NPCs and SPBs, behaving in a similar fashion to Ndc1p (Chial et al. 1998).
Table 1.
Characterization of Proteins in the NPC Fraction
| Protein | Tag | Immunofluorescence localization | Coenrich with: | References | |||||
|---|---|---|---|---|---|---|---|---|---|
| YPD code | Gene name | Mol wt | PrA | FLU | Nuclei? | NE? | NPC? | ||
| kD | |||||||||
| YAR002W | NUP60 | 59 | Y | ND | NPC | Y | Y | ND | This study |
| YBL079W | NUP170 | 169 | Y | ND | NPC | Y | Y | ND | This study (not shown) |
| YDL088C | NUP59 | 59 | Y | ND | NPC | Y | Y | ND | This study |
| YDL116W | NUP84 | 84 | Y | ND | NPC | Y | Y | ND | This study (not shown) |
| YDL207W | GLE1 | 62 | Y | ND | NPC | Y | Y | ND | This study |
| YDR192C | NUP42 | 43 | Y | Y | NPC | Y | Y | ND | This study |
| YER105C | NUP157 | 157 | Y | ND | NPC | Y | Y | ND | This study (not shown) |
| YER107C | GLE2 | 41 | Y | ND | NPC | Y | Y | ND | This study |
| YFR002W | NIC96 | 96 | Y | ND | NPC | Y | Y | Y | This study |
| YGL092W | NUP145 | 146 | Y | ND | NPC | Y | Y | Y | (3), (4) |
| YGL100W | SEH1 | 39 | Y | ND | NPC | Y | Y | ND | This study |
| YGL172W | NUP49 | 49 | Y | ND | NPC | Y | Y | Y | (3), (4) |
| YGR119C | NUP57 | 58 | Y | ND | NPC | Y | Y | Y | (3), (4) |
| YIL115C | NUP159 | 159 | Y | ND | NPC | Y | Y | Y | This study, (6) |
| YJL039C | NUP192 | 191 | Y | ND | NPC | Y | Y | ND | This study |
| YJL041W | NSP1 | 87 | Y | ND | NPC | Y | Y | Y | (3), (4) |
| YJL061W | NUP82 | 82 | Y | ND | NPC | Y | Y | ND | This study (not shown) |
| YJR042W | NUP85 | 85 | Y | ND | NPC | Y | Y | ND | This study (not shown) |
| YKL057C | NUP120 | 120 | Y | ND | NPC | Y | Y | ND | This study (not shown) |
| YKL068W | NUP100 | 100 | Y | ND | NPC | Y | Y | Y | (3), (4) |
| YKR082W | NUP133 | 133 | Y | ND | NPC | Y | Y | Y | (7) |
| YLR018C | POM34 | 34 | Y | Y | NPC | Y | Y | ND | This study |
| YML031W | NDC1 | 74 | Y | ND | NPC, SPB | Y | Y | Y | (8) |
| YML103C | NUP188 | 189 | Y | ND | NPC | Y | Y | Y | (9) |
| YMR047C | NUP116 | 116 | Y | ND | NPC | Y | Y | Y | (3), (4) |
| YMR129W | POM152 | 152 | Y | ND | NPC | Y | Y | ND | This study, (4), (5) |
| YMR153W | NUP53 | 53 | Y | ND | NPC | Y | Y | ND | This study (not shown) |
| YOR098C | NUP1 | 114 | Y | ND | NPC | Y | Y | Y | (3), (4) |
| YOR257W | CDC31 | 19 | Not functional | NPC | Y | Y | Y | This study | |
| YER110C | KAP123 | 123 | Y | ND | Cytoplasmic/NPC | N | N | ND | (1) |
| YGL016W | PDR6 | 123 | Y | ND | Cytoplasmic/NPC | N | N | ND | This study |
| YIL063C | YRB2 | 36 | Y | Y | Nuclear, NPC | N | N | ND | This study |
| YIL149C | MLP2 | 195 | Y | ND | Nuclear envelope | Y | Y | ND | (2) |
| YKR095W | MLP1 | 218 | Y | ND | Nuclear envelope | Y | Y | N | (2) |
| YLR335W | NUP2 | 78 | Y | ND | Nuclear, NPC | N | N | ND | This study |
| YLR347C | KAP95 | 95 | Y | ND | Cytoplasmic, NPC | N | N | ND | (1) |
| YNL189W | KAP60 | 60 | Y | ND | Cytoplasmic, NPC | N | N | ND | This study |
| YPL125W | KAP120 | 120 | Y | ND | Cytoplasmic, NPC | N | N | ND | This study |
| YPL169C | MEX67 | 67 | Y | ND | Cytoplasmic, nuclear, NPC | Y | N | ND | This study |
| YBR025C | 44 | ND | Y | Cytoplasmic | |||||
| YBR194W | 14 | ND | Y | Cytoplasmic | |||||
| YCL059C | KRR1 | 37 | ND | Y | Nuclear rim | ||||
| YDL121C | 17 | ND | Y | Endoplasmic reticulum | |||||
| YDL148C | 94 | Y | ND | Nuclear/portion of nucleolar | |||||
| YDR190C | 50 | Y | ND | Nuclear | |||||
| YDR412W | 28 | Y | Y | Granular cytoplasmic | |||||
| YEL026W | SNU13 | 14 | ND | Y | Nucleolar | ||||
| YER002W | 27 | ND | Y | Nuclear/nucleolar | |||||
| YER006W | 58 | Y | ND | Nuclear/nucleolar | |||||
| YER049W | 74 | N | N | High CAI (0.255) | |||||
| YER126C | 30 | ND | Y | Nuclear | |||||
| YGR103W | 70 | Y | ND | Nuclear/nuclear rim/nucleolar | |||||
| YHR009C | 57 | Y | ND | Cytoplasmic | |||||
| YLR074C | 19 | ND | ND | DNA-binding protein homolog | |||||
| YHR127W | HSN1 | 27 | ND | Y | Nuclear | ||||
| YHR052W | 43 | ND | ND | High CAI (0.240) | |||||
| YMR131C | 57 | Y | ND | Portion of nucleolus | |||||
| YMR290C | HAS1 | 57 | Y | ND | Peripheral nuclear, nucleolar | ||||
| YNR053C | 56 | Y | ND | Nuclear | |||||
| YOL145C | CTR9 | 125 | Y | ND | Strong nuclear, some cytoplasmic | ||||
| YOR051C | 47 | Y | ND | Nuclear | |||||
| YOR091W | 46 | Y | ND | Cytoplasmic | |||||
| YOR145C | 30 | Y | Y | Cytoplasmic | |||||
| YOR206W | 82 | Y | ND | Crescent, peripheral nuclear | |||||
| YPL093W | 74 | Y | ND | Nucleolar/nuclear | |||||
| YPL146C | 53 | Y | ND | Nucleolar, portion of nucleus | |||||
| YPL217C | 136 | Y | ND | Nuclear, nucleolar, some cytoplasmic | |||||
| YPR144C | 64 | Y | ND | Nuclear speckles | |||||
(1) Rout et al., 1997; (2) Strambio-de-Castillia et al., 1999; (3) Rout and Blobel, 1993; (4) Strambio-de-Castillia et al., 1995; (5) Wozniak et al., 1994; (6) Kraemer et al., 1995; (7) Pemberton et al. 1995; (8) Chial et al., 1997; (9) Nehrbass et al., 1996.
To identify individual proteins associated with the NPC, we subjected the highly enriched NPC fractions to two-dimensional separations involving HPLC followed by SDS-PAGE. To obtain the highest resolution and yields, three different HPLC separation techniques were used. Fractions collected from each separation were then resolved by SDS-PAGE to generate a two-dimensional pattern of proteins. The pattern produced by hydroxyapatite HPLC/SDS-PAGE is shown in Fig. 1. Individual proteins in each band were identified by tryptic digestion followed by MALDI-TOF mass spectrometry. A sufficient number of tryptic peptide masses were determined from each band to unambiguously distinguish its corresponding open reading frame (ORF) in a genome database search (Kuster and Mann 1998). As an example, Fig. 2 shows the MALDI-TOF mass spectrum of the tryptic peptides obtained from a relatively weak band (no. 217). Three proteins were identified in the band: Nup120p is a known nup (Aitchison et al. 1995b; Heath et al. 1995), while Pdr6p/Kap120p and Ypl125p/Kap122p are both transport factors (see below). These three proteins were also detected in surrounding bands, providing independent verification of their presence in the NPC fraction. The MALDI-TOF peptide mapping procedure was used to analyze 465 bands from three independent hydroxyapatite HPLC/SDS-PAGE separations. In addition, 177 bands from the TFA-HPLC/SDS-PAGE separations were analyzed by MALDI-ion trap tandem mass spectrometry of the tryptic peptides (Qin et al. 1997). Finally, these data were combined with those obtained by the peptide microsequencing analysis of 49 prominent bands in the formic acid-HPLC/SDS-PAGE separation (Fernandez et al. 1994; Aitchison et al. 1995a,Aitchison et al. 1995b).
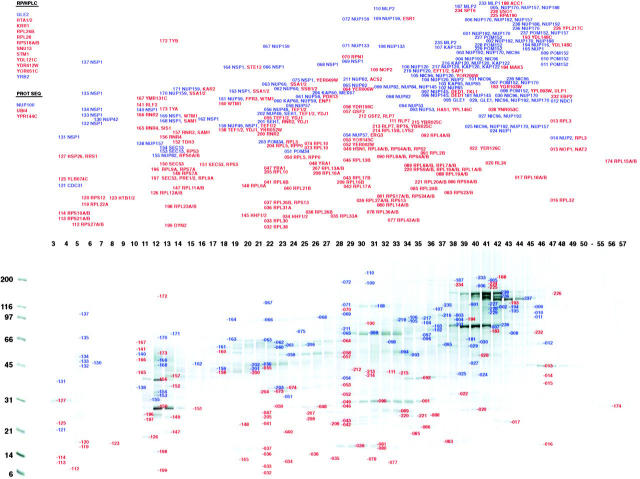
Figure 1.
Identification of proteins in the highly enriched NPC fraction. Proteins in the highly enriched NPC fraction were separated by hydroxyapatite HPLC and SDS-PAGE. The number above each lane indicates the corresponding fraction number. Proteins were visualized with Coomassie blue; the approximate molecular mass of proteins as estimated from standards shown on the left side. Bands analyzed by MALDI-TOF mass spectrometry are indicated by the adjacent numbers. The proteins identified in each band are shown in the top panel. Proteins known to directly associate with the NPC are colored blue, while proteins believed not to be NPC-associated are colored red. On the top left are listed proteins not found in this separation but identified by MS/MS of reverse phase HPLC-separated peptides (RP/HPLC) or peptide microsequencing (PROT SEQ).
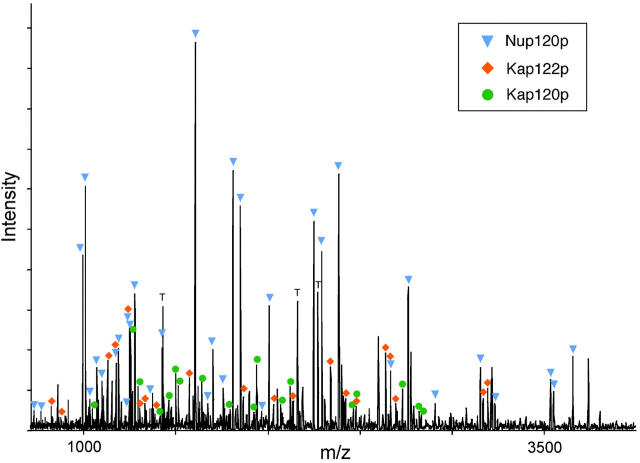
Figure 2.
Identification of proteins in a gel band by mass spectrometry. MALDI-TOF mass spectrum of the peptides produced by in-gel trypsin digestion of band no. 217 (molecular mass of ∼100 kD, fraction 35). The peak masses were used to identify proteins present in the band with the protein identification algorithm ProFound. The dominant protein component was Nup120p, which was identified with a probability 5 × 1022 higher than the next most probable protein. The peaks arising from Nup120p were subtracted from the spectrum and a new search was initiated, identifying Kap120p with a probability 3 × 109 higher than the next most probable protein. Finally, the peaks arising from Kap120p were subtracted from the spectrum and a new search was initiated identifying Pdr6p/Kap122p with a probability 1 × 108 higher than the next most probable protein. Colored markers indicate which peaks arise from each of the three identified proteins. The peaks labeled T arise from trypsin self-digestion. Unlabeled peaks likely originate from modified or incompletely digested peptides in Nup120p, Kap120p, and Kap122p, or from additional proteins.
Our results are summarized in Fig. 1. We identified a total of 174 proteins, of which 40 were ultimately found to be associated with the NPC (see below and Table ). Of the identified proteins, 34 corresponded to uncharacterized ORFs. Each of these proteins was characterized to assess whether it was likely to be a nup (see below). The mass spectrometric analysis was estimated to be 10–100-fold more sensitive than was necessary to detect the known coenriching nups. Indeed, we detected proteins that are known to dissociate readily from the NPC (e.g., transport factors) or are located at considerable distances from the NPC (e.g., Mlp1p and Mlp2p). Thus, even if some genuine nups were to fall off during the enrichment process, we would still expect to detect them. This high level of sensitivity is reflected in our observation of 134 contaminating proteins, in addition to 29 nups and 11 transport factors and NPC-associated proteins (Fig. 1, Table ). Indeed, most nups were observed in three or more independent measurements (e.g., Pom34p was identified four times, and Nsp1p 29 times). Because the investigation involved large amounts of material and a high level of oversampling, this approach likely provides a relatively complete inventory of NPC components.
Protein Characterization and the Identification of Nups and Transport Factors
Database searches first sorted all the proteins identified in the highly enriched NPC fraction into three general classes: known NPC-associated proteins (nups and transport factors), previously uncharacterized proteins, which may or may not be nups or transport factors, and proteins with well-characterized functions, apparently unrelated to transport. Most proteins in this latter class were not analyzed further, but may prove informative in future studies. These include numerous nuclear and nucleolar components, which may have contaminated the fraction, or were perhaps caught in transit across the NPC. The histones (bands 034, 123, and 145 in Fig. 1) and Nop1p (band 015) fall into this category. Some highly abundant cytoplasmic or ER proteins were also found, such as GAPDH (Tdh3p, band 152). Certain proteins could be contaminants but may have specific associations with the NPC. For instance, roles in nucleocytoplasmic transport have been ascribed to heat shock proteins (bands 162, 062, 063, 163, and 170; Shulga et al. 1996). We also identified a large number of ribosomal proteins, which could be contaminants, abundant transport substrates, or may be accounted for by specific associations between polysomes, NPCs and mRNPs exiting the NPC (Visa et al. 1996).
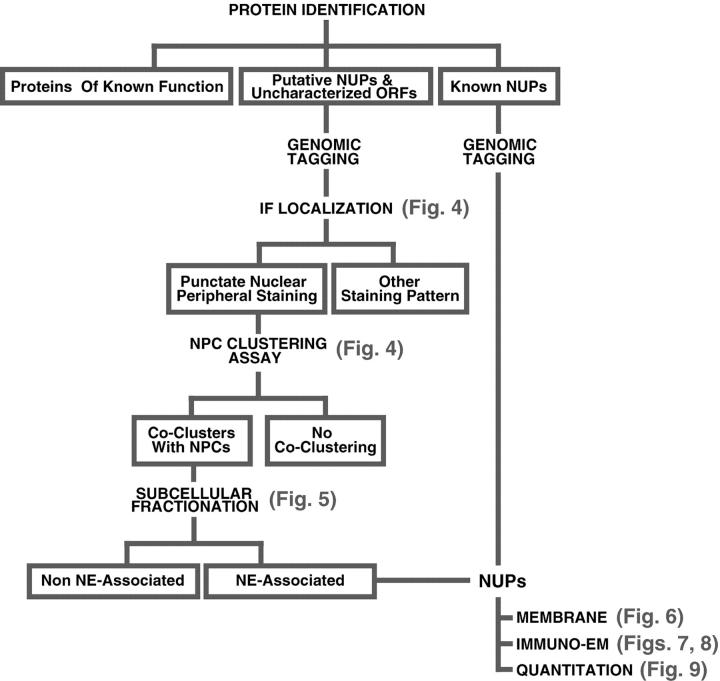
Fig. 3 shows the design of a series of classification assays to determine which of the uncharacterized proteins were also nups. As discussed above, many proteins transiently associate with the NPC. We thus created an operational definition of a nup as a protein, the majority of which spends most of its time as a constituent of the octagonally symmetric NPC. For this study, proteins whose predominant localization is elsewhere, such as Sec13p (Siniossoglou et al. 1996) or nucleocytoplasmic transport factors (Rout et al. 1997), are not considered nups. Although such proteins may indeed behave as functional nups when associated with the NPC, the scope of our study precludes a detailed characterization of their behavior. According to our definition, a nup should colocalize with NPCs in the cell, and cofractionate with NPCs upon their isolation. Therefore, we tested each uncharacterized protein and (where necessary) proteins previously described as nups for these attributes. These proteins were genomically tagged at their COOH termini (ensuring normal expression levels) with a protein A (PrA) epitope in order to follow them during their characterization (Aitchison et al. 1995a). We also genomically tagged numerous proteins with the much smaller FLU moiety (Longtine et al. 1998) to check for tag-specific effects on protein localization. Our first screen involved localization of the epitope-tagged proteins in cells by immunofluorescence microscopy, because nups give a characteristic punctate nuclear rim staining pattern. However, as some nucleocytoplasmic transport factors and NE proteins also give a similar staining pattern, a second screen tested for colocalization with NPCs in a strain in which the NPCs themselves cluster into patches on the NE. Upon deletion of the gene encoding Nup120p the NPCs cluster to one side of the nuclear envelope (Fabre and Hurt 1997), which allows NE components to be distinguished from the coclustering NPC-associated nups and transport factors (Aitchison et al. 1996; Siniossoglou et al. 1996; Strambio-de-Castillia et al. 1999). Although most of the unknown ORFs gave immunostaining patterns typical for proteins of the ER, nucleus or nucleolus, 39 proteins showed associations with the NPCs (Fig. 4, Table ). In our third screen, the tagged proteins were followed by subcellular fractionation (Strambio-de-Castillia et al. 1995). We mainly used a preparation of NEs for this assay (Strambio-de-Castillia et al. 1999). While NE components and nups coenrich with the NE-containing fractions, nucleocytoplasmic transport factors and other proteins only partially associated with the NPC will be found mainly in the nucleoplasmic or cytoplasmic fractions (Fig. 5). Thus, the nups emerge as proteins that both cocluster with NPCs and preferentially cofractionate with the NEs.
Figure 3.
Flow chart outlining our experimental strategy for protein classification.
Figure 4.
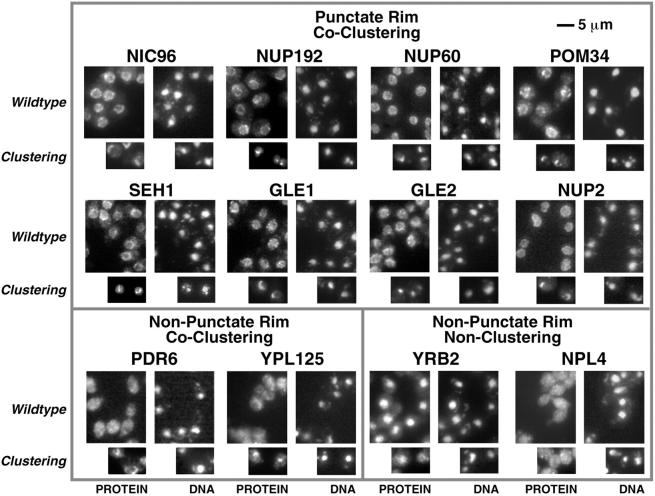
Localization of PrA-tagged chimeras by immunofluorescence microscopy. Four images are shown for each protein. Scale bar is shown top right. The left pair shows the tagged protein localization (PROTEIN), while the right pair shows the corresponding positions of the nuclei in the same cells (DNA). The larger images illustrate the tag localization in otherwise wildtype cells, while the smaller images show localization in a nup120Δ strain in which the NPCs cluster to large discrete patches on the NE. NPC-associated proteins exhibit punctate staining around the nucleus in the Wildtype strain and cocluster with the NPCs in the Clustering strain. A sample of these is shown in the top box. This includes Gle1p and Gle2p, which had not been defined as nups. We also identified a typical NPC staining pattern for three previously uncharacterized ORFs: yar002w, yjl039c, and ylr018c. Further characterization of these three proteins demonstrated that they are indeed nups and that Ylr018p is an integral membrane protein (Fig. 5 and Fig. 6). In keeping with standard nomenclature, we term these proteins, Nup60p, Nup192p and Pom34p, respectively. Nup192p is the previously described 170-kD NPC-associated protein identified by peptide sequencing (Aitchison et al. 1995a). It has also recently been described as a potential nup by others (Kosova et al. 1999). Examples of proteins that do not display the punctate staining pattern, but still colocalize with the NPCs (Non-Punctate Rim, Co-Clustering) are shown in the lower left box, and include putative transport factors that partially associate with the NPC such as the karyopherin homologues Pdr6p and Ypl125p. Proteins that do not localize to the nuclear periphery and do not co-cluster with the NPCs (Non-Punctate Rim, Non-Clustering) are shown in the lower right box. The FG motif protein Yrb2p was found primarily in the nucleoplasm in agreement with recent studies (Noguchi et al. 1997; Taura et al. 1997). Although we did not find Npl4p in the highly enriched NPC fraction, it has been reported to be a possible nup (DeHoratius and Silver 1996). We detected this protein throughout the cytoplasm and observed no association with the NPC. The Npl4p-PrA chimera was functional, because although deletion of the NPL4 gene is lethal, we saw no growth defects in the haploid-tagged strain.
Defining NPC Architecture: Characterization of the Nucleoporins
The assays described above found some 30 proteins that conform to our definition of nups (Table ). Some proteins previously described as nups were eliminated on the basis of our definition. Also some NPC components with a major function elsewhere (such as Sec13p) fall outside this categorization. Despite our best efforts, other bona fide nups may have been missed or miscategorized during our analysis. However, we believe we have found virtually all the nups and a host of auxiliary transport factors (some previously uncharacterized). We then determined the position and stoichiometry of each nup within the NPC, to provide a comprehensive overview of the molecular architecture of this assembly.
Transmembrane Nucleoporins
A straightforward way of delimiting the localization of proteins within the NPC is to determine which nups contain transmembrane domains. Such proteins could only be found in a region immediately surrounding the pore membrane that transits the outer spoke domain of the NPC (Yang et al. 1998). We extracted NEs made from each of the PrA-tagged nup strains, using a standard high pH method (Fujiki et al. 1982). We found only three nups that cosedimented with the resulting membranes and are therefore termed pore membrane proteins (pom). Ndc1p and Pom152p have been previously described (Wozniak et al. 1994; Chial et al. 1998). The previously uncharacterized protein Ylr018p also conforms to the definition of a POM and is correspondingly named Pom34p (Fig. 6). All other nups were extracted under these conditions and were therefore not considered to be integral membrane proteins.
Figure 6.
Ndc1p, Pom152p, and Pom34p are integral membrane proteins in the NPC. The NE fraction for each strain was extracted to obtain a supernatant fraction (SUP) and a pellet containing the integral membrane proteins (PELL). For each extraction, these fractions were probed for the protein of interest (PrA, Ab), the control transmembrane nup Pom152p (Ctrl), and known peripheral nups (not shown).
Immunolocalization of the Nucleoporins within the NPC
We used preembedding immunoelectron microscopy on the isolated NEs from the tagged strains to sublocalize nups within the NPC. Occluding material can interfere with antibody accessibility leading to mislocalization of a protein (Rout and Kilmartin 1990). Therefore, we used isolated NEs, because this provides an opportunity to biochemically remove such occluding material in a highly controlled manner, while leaving the NEs themselves biochemically and structurally well preserved (Kilmartin and Fogg 1982; Strambio-de-Castillia et al. 1995; Strambio-de-Castillia et al. 1999). However, a three-dimensional model obtained from electron cryomicroscopy studies reveals structures within the NPC that would be inaccessible to an antibody molecule (Yang et al. 1998). A similar accessibility problem for SPB proteins was solved by using limited, controlled extractions to expose the epitopes without altering the gross SPB architecture (Rout and Kilmartin 1990). We used a comparable approach, involving mild heparin treatments with a variety of controls to ensure specificity and accuracy of labeling.
We generated aligned montages to show the distribution of labeling around the NPC for 27 nups (Fig. 7). The montages preserved the morphology and dimensions of individual NPCs, indicating that our alignments were accurate. Our results disagree with certain localizations performed using one particular technique (Fahrenkrog et al. 1998; Kosova et al. 1999; Strahm et al. 1999). That technique yields localizations that include a distribution of Gle1p throughout the cytoplasm, and Nup42p throughout the nucleoplasm; these seem highly unlikely, as both of these proteins cofractionate absolutely with the NPC-containing fractions (Fig. 5). Furthermore, the authors acknowledge potential problems with their methodology, including overexpression of the tagged protein and epitope inaccessibility, which we avoided. By contrast, considerable credence is given to our data by their consistency with results obtained by electron microscopy (Rout and Blobel 1993; Yang et al. 1998), with all other yeast nup localizations (Kraemer et al. 1995; Nehrbass et al. 1996; Hurwitz et al. 1998; Marelli et al. 1998; Wente, S.R., personal communication), with the proximity of nups deduced from the isolation of nup subcomplexes (Grandi et al. 1993, Grandi et al. 1995; Siniossoglou et al. 1996), and with our immunofluorescence localization, cofractionation, and quantitation data (see Fig. 4, Fig. 5, and Fig. 9).
Figure 9.
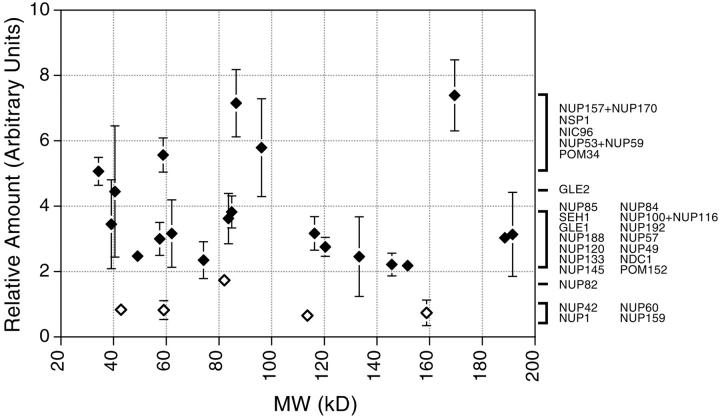
Relative abundance of nucleoporins in the NPC. Each data point represents the ratio of the signal from the tagged nucleoporin to the signal for the internal standard (Table ), which are proportional to the relative quantity of each protein in the NPC, averaged from at least two independent measurements for each nup (each generated from four different ratio measurements). The brackets indicate clusters of relative abundance containing the nups indicated at the right. Nups found exclusively on one side of the NPC are shown as open diamonds.
The signal for some nups concentrated around the central axis (Nup57p), while others (Nup60p) spread up to 80 nm from it. The large amount of gold particle scatter is due in part to alignment inaccuracies, distortions, and random rotations of the tags themselves, the antibodies around the tags, and of the NPCs around their axes (cylindrical averaging). However, the spread likely also reflects a genuine flexibility of structure within the NPC, particularly in its filamentous structures (Talcott and Moore 1999).
We devised an analytical method to extract the position of each nup within the NPC from the labeling distributions shown in the montages. The most obvious feature of the resulting cylindrical map (summarized in Fig. 8 and Table ) is how the nups followed the contours of the NPC, even out into the cytoplasmic filaments and nuclear basket. This indicated that we had generated a surface map for most nups, under conditions of minimal damage to the NPC structure. There appears to be room for only one large central channel for the transit of macromolecules, which agrees with recent biophysical measurements (Keminer et al. 1999).
Figure 8.
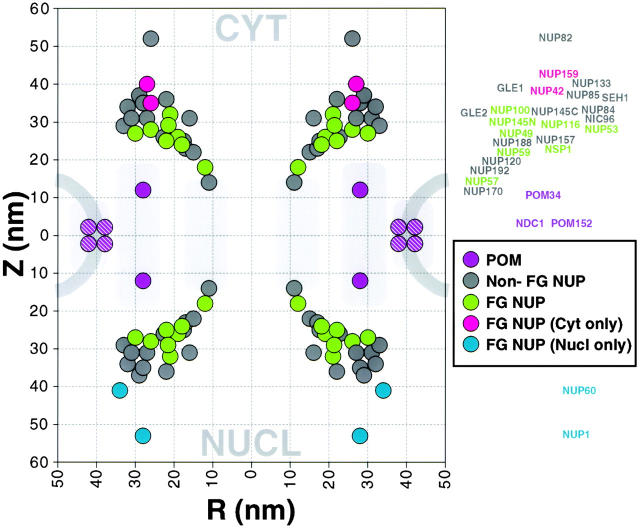
Plot of the position of nucleoporins in the NPC. Statistical analysis of the distribution of the gold particles in each montage allows determination of the position of the proteins relative to the NPC cylindrical axis (R) and mirror plane of pseudosymmetry (Z). The plotted circle size was arbitrarily chosen for the sake of clarity. A mask for a cross-section of the yeast NPC and pore membrane is shown schematically to scale in light gray. We have highlighted the FG nups, the majority of which were found on both sides of the NPC (green), and a few that were found towards the periphery and exclusively on the nuclear side (blue) or the cytoplasmic side of the NPC (red). Most of the non-FG nups (dark gray) were found on both sides. The integral membrane protein Pom34p (purple) was close to the membrane, as were the inferred positions of Pom152p and Ndc1p (purple stripes).
Table 2.
Localizations of Stoichiometries of Nucleoporins
| Gene name | Molecular mass | FG repeats? | Quantitation | Localizations | ||
|---|---|---|---|---|---|---|
| Relative amount | R | Z | Number of particles | |||
| kD | nm | nm | ||||
| CDC31 | 19 | N | ND | ND | ND | ND |
| GLE1 | 62 | N | 3.2 ± 1.0 | 22 | 36 | 140 |
| GLE2 | 41 | N | 4.4 ± 2.0 | 16 | 31 | 80 |
| NDC1 | 74 | N | 2.3 ± 0.6 | ND | ND | ND |
| NIC96 | 96 | N | 5.8 ± 1.5 | 33 | 29 | 158 |
| NSP1 | 87 | Y | 7.2 ± 1.0 | 22 | 25 | 193 |
| NUP1 | 114 | Y | 0.7 ± 0.1 | 28 | 53 | 58 |
| NUP100 | 100 | Y | 1.3 ± 0.4 | 21 | 32 | 73 |
| NUP116 | 116 | Y | 1.8 ± 0.2 | 26 | 28 | 93 |
| NUP120 | 120 | N | 2.8 ± 0.3 | 17 | 23 | 60 |
| NUP133 | 133 | N | 2.5 ± 1.2 | 29 | 37 | 95 |
| NUP145C | 81 | N | 2.2 ± 0.3 | 27 | 31 | 113 |
| NUP145N | 65 | Y | ND | 22 | 28 | 76 |
| NUP157 | 157 | N | 3.0 ± 0.9 | 22 | 26 | 100 |
| NUP159 | 159 | Y | 0.7 ± 0.4 | 27 | 40 | 66 |
| NUP170 | 169 | N | 4.4 ± 0.5 | 11 | 14 | 48 |
| NUP188 | 189 | N | 3.0 ± 0.2 | 18 | 25 | 154 |
| NUP192 | 191 | N | 3.1 ± 1.3 | 15 | 22 | 58 |
| NUP42 | 43 | Y | 0.8 ± 0.1 | 26 | 35 | 72 |
| NUP49 | 49 | Y | 2.5 ± 0.1 | 19 | 26 | 122 |
| NUP53 | 53 | Y | 3.6 ± 0.5 | 30 | 27 | 89 |
| NUP57 | 58 | Y | 3.0 ± 0.5 | 12 | 18 | 65 |
| NUP59 | 59 | Y | 2.0 ± 0.2 | 18 | 24 | 73 |
| NUP60 | 59 | Y | 0.8 ± 0.3 | 34 | 41 | 83 |
| NUP82 | 82 | N | 1.7 ± 0.1 | 26 | 52 | 85 |
| NUP84 | 84 | N | 3.6 ± 0.8 | 31 | 31 | 98 |
| NUP85 | 85 | N | 3.8 ± 0.5 | 28 | 35 | 115 |
| POM152 | 152 | N | 2.2 ± 0.1 | ND | ND | ND |
| POM34 | 34 | N | 5.1 ± 0.4 | 28 | 12 | 104 |
| SEH1 | 39 | N | 3.4 ± 1.4 | 32 | 34 | 101 |
We did not attempt to localize Sec13p or Cdc31p, the former because of the likelihood of false signal from ER contamination and the latter because no functional PrA tagged strain was available. We also did not analyze Pom152p or Ndc1p, as their transmembranous nature already localized them to the region surrounding the pore membrane domain. Indeed, we found the membrane protein Pom34p adjacent to the nuclear membrane, and we have surmised that the other two pore membrane proteins are similarly located (Fig. 8); Pom152p is already known to form part of a peripheral membrane ring (Wozniak et al. 1994; Strambio-de-Castillia et al. 1995).
Analysis of both the montages and the cylindrical map showed that the nups fell into three localization categories. Remarkably, most of the nups were symmetrically localized, being found equally on both the nucleoplasmic and cytoplasmic faces of the NPC and roughly equidistant from its central mirror plane. Though still localized on both sides, four nups showed a marked biased localization. This observation did not appear to be a result of antigen inaccessibility, because this bias was seen in a variety of extraction conditions; the structural basis for this bias is not yet clear. Of three nups showing a cytoplasmic bias, two were the FG-containing gene duplicates, Nup100p and Nup116p, and the third was Gle1p. Similarly, the FG-containing NH2-terminal fragment of Nup145p (Emtage et al. 1997) showed a marked bias towards a localization on the nucleoplasmic face, though the COOH-terminal fragment lacking FG motifs was symmetrically localized. This may explain the reason for autoproteolysis of Nup145p, which would allow the COOH-terminal portion to remain associated with the symmetrical Nup84p complex while the NH2-terminal fragment could redistribute to favor the nuclear face. Only a minority of nups displayed one-sided localizations. Two FG nups (Nup1p and Nup60p) were exclusively on the nucleoplasmic face of the NPC, and significantly further from the NPC midplane than the majority of nups. It is likely that these two proteins constitute the nuclear basket since their positions coincide with that determined for the basket (Rout and Blobel 1993; Fahrenkrog et al. 1998). Nup159p, Nup42p (FG nups sharing significant sequence similarities), and Nup82p (a non-FG nup) were similarly distal to the NPC midplane but exclusively cytoplasmic. All three proteins thus appear to be part of the cytoplasmic filaments (Kraemer et al. 1995; Hurwitz et al. 1998).
Stoichiometry of the Nucleoporins
We modified a previously described method of quantitative immunoblotting for SDS-PAGE-separated samples (Strambio-de-Castillia et al. 1999) to estimate the relative stoichiometry of the nups. Our method relied on the fact that all the nups were tagged with the same PrA moiety and then detected with the same antibody. Thus, for NE samples prepared from different tagged strains, the immunoblot signal of each nup (relative to an internal standard nup) should depend only upon the copy number of that nup in the NPC. The nups appeared to fall into three clusters of abundance (Fig. 9, Table ). Four of the five nups that were localized to only one face of the NPC were found in the lowest relative abundance cluster (between 0.7–0.8). The middle cluster (∼2.0–4.0) contained most of the nups, while the highest (>5.0) contained only a handful. We combined the values of the abundance measurements for the gene duplicate nups, as it is not clear that such duplicates exist in the nups of other eukaryotes and because many such duplicates are functionally and structurally analogous (though not necessarily equivalent; Aitchison et al. 1995a; Bailer et al. 1998). Thus, although we cannot exclude the possibility that the stoichiometry of some nups varies between individual NPCs, these three levels of abundance suggest that most nups use a simple stoichiometry in constructing the NPC. If the lowest common stoichiometric denominator is one nup copy per spoke, then Nup1p, Nup60p, Nup42p, and Nup159p would fall into this category, agreeing well with their unilateral localization on the NPC. The exception may be Nup82p; perhaps two copies are found on the cytoplasmic face of each NPC spoke. Most of the bilaterally localized proteins would then be present in two copies per spoke, while the abundant group of nups would be present in four (or more) copies per spoke. The overall correlation between the localization of each nup and its abundance was striking and provided an independent confirmation of our results. Based on these stoichiometry estimates and the molecular mass of each protein (and assuming that Cdc31p and Sec13p are present at two copies per spoke), we calculated an expected mass of ∼44 MD for the isolated NPC. This value is somewhat lower than the 56–72 MD estimated by hydrodynamic methods but falls well within the range calculated by STEM measurements (Rout and Blobel 1993; Yang et al. 1998). Quantitation of the numerous transport factors associated with isolated NPCs (Fig. 1 and Fig. 5) indicated that they may easily account for these differences (data not shown). Thus, it appears that we have not overlooked a significant fraction of nups in our analysis, and that our inventory and structural overview of NPC components is essentially complete.
Discussion
The NPC is Largely Symmetrical and Constructed of Surprisingly Few Distinct Proteins
We present the results of an oversampled analysis that has identified the complement of yeast nups, and set them within the structural context of the NPC. In addition to previously characterized nups, our work has allowed the classification of Nup60p/Yar002p, Pom34p/Ylr018p, Cdc31p, Rip1p/Nup42p, Nup192p/Yjl039p, Gle1p, and Gle2p as nups. It is possible that our nup definition and subsequent classification system may exclude some bona fide constituents; nonetheless, the number so excluded is likely small, and we can, therefore, set an upper limit of ∼30 distinct NPC components. This indicates a surprisingly simple composition for such a massive structure (e.g., given ∼75 different proteins in a ribosome), and is significantly lower than previous estimates (Rout and Wente 1994). We found three factors that explain this.
First, most nups were found on both the nucleoplasmic and cytoplasmic sides of the NPC, equidistant from the midplane of the NPC running parallel to the NE, and were estimated to be present in two or four copies per spoke (16 or 32 copies/NPC). Thus, the NPC appears to be mainly composed of 16 copies of a nup subcomplex, 8 copies on each side of the NPC midplane. This agrees with the high degree of twofold symmetry shown by the three-dimensional map of the isolated yeast NPC (Yang et al. 1998), and indeed our positional plot for the nups superimposes well around a mask derived from a central vertical section from this map (Fig. 8). Complications from the biased and gene duplicate nups do not change this overall picture. A relatively minor degree of asymmetry is introduced by placing proteins unequally on either the nucleoplasmic or cytoplasmic face of this symmetrical superstructure, which could contribute to the observed minor morphological asymmetry of the yeast NPC (Rout and Blobel 1993; Yang et al. 1998). Second, economy of composition is also achieved by the NPC sharing proteins with other cellular components, such as the COPII component Sec13p, and the spindle pole components Ndc1p and (as shown here) Cdc31p. Third, the high NPC mass is also a result of the large average molecular weight (∼100 kD) of yeast nups. Thus only 30 such proteins, each present in 16 copies, would produce a structure of ∼50 MD. This combination of a limited number of large nups, present in multiple copies in a highly symmetric structure, can thus completely account for the mass of the NPC.
The Composition and Architecture of the NPC Indicate a Possible Mechanism for Nucleocytoplasmic Transport
Four major observations concerning the mechanisms of NPC-mediated transport result from our study.
The NPC Lacks Proteins Normally Associated With Mechanochemical Transport.
Nucleocytoplasmic transport requires a source of energy, as it accumulates material against a concentration gradient (Shulga et al. 1996). Earlier models proposed that myosin (Berrios et al. 1991) or other mechanoenzymes may be involved in transport (Akey 1990). However, we found no NPC-associated motor proteins, nor any other nups that are either ATPases or GTPases, based on sequence comparisons. At least in part, the required energy likely comes from the maintenance of two distinct pools of Ran in the cell (Mattaj and Englmeier 1998; Talcott and Moore 1999). In the nucleus, Ran is maintained in its GTP-bound state, while the cytoplasmic pool is in its GDP-bound form. This distribution can be used by transport factors to determine which side of the NE they are on. Thus, the formation of an import complex between a kap and its cargo is stable in the presence of cytoplasmic Ran-GDP, but, in the nucleoplasm, Ran-GTP triggers its disassembly. In contrast, the formation of an export complex is stabilized in the nucleus by Ran-GTP, but as this complex reaches the cytoplasm, GTP on Ran is hydrolyzed and the complex disassembles (Gorlich et al. 1996; Koepp and Silver 1996; Fornerod et al. 1997; Kutay et al. 1997; Mattaj and Englmeier 1998).
It is, therefore, interesting that we found no yeast nups with obvious Ran binding sites (with the possible exception of the nonessential Nup2p; Fig. 4 and Fig. 5), suggesting that Ran does not need to be bound at the NPC. Another proposal involved the movement of transport factors along FG nups, propelled by cycles of Ran GTP hydrolysis (Rexach and Blobel 1995; Koepp and Silver 1996). However, it has recently been shown that GTP hydrolysis is not required for the translocation step across the NPC (Schwoebel et al. 1998; Englmeier et al. 1999). Taken together, it seems that neither mechanoenzymes nor other nucleotide-driven processes are involved in the actual translocation step of transport. Binding alone seems sufficient to mediate exchange across the NPC (Kose et al. 1997). In the absence of motors, translocation across the NPC is likely propelled by Brownian motion (Simon et al. 1992; Magnasco 1993). However, such facilitated diffusion models still invoke a conceptual gate somewhere in the NPC (Koepp and Silver 1996), as nucleocytoplasmic transport involves a restriction of passive diffusion, apparently within the central channel (Feldherr and Akin 1997; Keminer et al. 1999).
The Transport Path Is Surrounded by a Large Number of Closely Packed Docking Site Proteins.
Each NPC possesses an abundance of binding sites for transport factors. It seems the FG nups form a phalanx of docking filaments bristling out from the NPC towards the nucleoplasm and cytoplasm and clustered around both ends of the mouth of the central channel. Based on our data, every NPC can, in principle, accommodate at least 160 transport factor molecules, even if each FG nup molecule binds only one transport factor (Table ). These FG nups are found throughout the NPC from its cytoplasmic to nuclear extremities, such that they may be available to a transport factor at almost all stages of its journey across the NPC (Fig. 7 and Fig. 8).
Most Docking Site Proteins Are Found on both the Nuclear and Cytoplasmic Sides of the NPC.
8 of the 12 FG nups are found on both faces of the NPC at similar distances from the NPC mirror plane. This high degree of symmetry appears inconsistent with the level of NPC asymmetry necessary to support models in which a continuous affinity gradient drives the directionality of transport (Floer et al. 1997).
A Minority of Docking Sites Displays a Limited and Organized Asymmetry.
The idea that Ran may be the only essential vectorial cue (Nachury and Weis 1999) is countered by the continued directional transport of transport factors unable to interact with Ran (Gorlich et al. 1996; Kose et al. 1997), and the observation that transport is irreversible across single NPCs (Keminer et al. 1999). The fact that we observed a marked asymmetry in a subset of the FG nups also indicates that differences between the nuclear and cytoplasmic NPC docking sites are important for transport (Blobel 1995). Indeed, in vertebrates one-sided FG nups have been implicated in the termination of translocation, leading to the suggestion that these play a crucial role in targeting transport factors to the correct side of the NPC (Delphin et al. 1997; Kehlenbach et al. 1999). We note that certain homologous yeast one-sided FG nups, apparently involved in similar termination reactions (Rexach and Blobel 1995; Floer et al. 1997; Floer and Blobel 1999), are placed further away from the NE plane than all other FG nups, exposing them and their bound transport factors to the Ran environment particular to the compartment they face.
A Brownian Affinity Gating Model for Nucleocytoplasmic Transport
Constrained by the above observations, a coherent model of nucleocytoplasmic transport must explain the following: (a) NLS or NES signal–mediated gating at the NPC, (b) unidirectional (vectorial) transport across the NPC, (c) recycling to allow unlimited rounds of transport, and (d) the energy requirements of these processes. We propose that Brownian motion (diffusion) accounts for translocation while vectoriality is a combined effort of both the asymmetric nups and the asymmetric distribution of soluble transport factors. Two gating mechanisms remain possible. One involves a dilatory gate. However, given the number and disposition of FG nups, we propose another mechanism that does not require conventional mechanical gating.
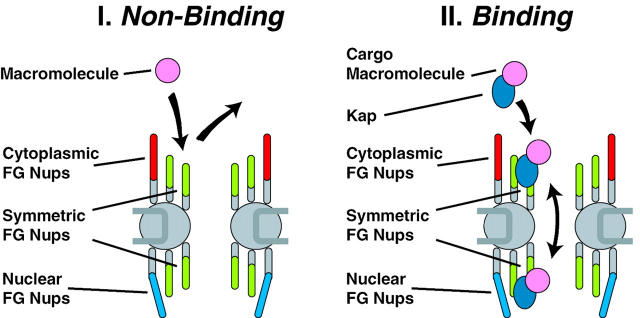
Two characteristics of the NPC could account for gating (Fig. 10). On one hand, the NPC contains a narrow central tube, ringed at each end by a dense array of filamentous FG nups (Fig. 8). This confined channel (which may be further occluded by the filamentous FG nups) presents a significant barrier to the passive diffusion of macromolecules (Paine et al. 1975; Feldherr and Akin 1997). Thus, macromolecules that do not bind to nups are less likely to diffuse across the NPC and are largely barred from passage across the NE (Fig. 10 I). On the other hand, macromolecules that bind to nups (specifically cargo-carrying transport factors binding to FG nups) increase their residence time at the entrance of the central tube, and so their diffusion across the NPC is greatly facilitated (Fig. 10 II). In a sense, the FG nups provide an attraction force for transport factors that could counter the repulsion force from diffusive exclusion. Given a sufficiently high release rate, rapid and reversible binding would promote a fast diffusional exchange of transport factors among the symmetrically disposed docking sites and so between the two faces of the NPC. By restricting the number of diffusional degrees of freedom (e.g., along the filaments), the FG nups may further facilitate the passage of transport factors across the NPC. In effect, the apparent size of the diffusion channel would become very much larger for proteins that can bind the FG nups than for those proteins unable to bind. This difference provides a virtual gate, in which the apparent size of the NPC increases for a cargo only while its signal-mediated translocation is active, but which does not necessarily involve a physical dilation of the NPC (Blobel 1995).
Figure 10.
The Brownian affinity gate model. (I) The narrow bore of the central tube ensures that macromolecules that do not bind to nups find in hard to diffuse across the NPC, and are thus largely excluded; the diffusive movement of the filamentous nups may contribute to this exclusion. (II) Macromolecules that bind to nups increase their residence time at the entrance of the central tube, and so their diffusion across the NPC is greatly facilitated.
Our model is clearly a simplified one. In particular, some aspects of gating still seem to require the existence of a mechanical gate at the central tube. However, we suggest an alternative that does not require such large-scale concerted motions of the central tube. Thus, we propose that diffusive movement of the tethered filamentous FG nups may contribute to the discrimination between nonbinding and binding macromolecules. Brownian motion would drive the rapid motion of these filaments; small molecules could readily slip past the flailing filaments, while large molecules would tend to be deflected away from the channel by frequent impacts. Such entropic exclusion has been demonstrated (Brown and Hoh 1997). The dilation of the central channel and nuclear basket observed for the largest transport substrates (Akey 1990; Kiseleva et al. 1996) would then be a consequence, rather than cause, of substrate translocation.
For unidirectional transport across the NPC, translocating transport factors need cues to determine which side of the NPC they are on. Vectorial cues arise from two potential sources, the asymmetry of the FG nups and the asymmetric distribution of Ran-GTP (Mattaj and Englmeier 1998). Transport is then concluded by a vectorial step.
Consider a cargo imported by its kap into the nucleoplasm (Fig. 11). The cargo alone cannot easily pass through the diffusion barrier of the NPC. In the cytoplasm, the kap is exposed only to Ran-GDP, allowing it to acquire the cargo. This cargo–kap complex can now bind the NPC (Fig. 11 I), passing through the now effectively open Brownian gate and diffusing across the NPC by binding symmetric FG nups. This allows the cargo–kap complex access to both faces of the NPC (Fig. 11 II). We propose that in this state, the cargo–kap complex has its highest affinity for the one-sided FG nups on the nuclear face. At the nuclear face, the kap moves preferentially away from the symmetric region of the nuclear pore to these higher affinity binding sites, preventing its exchange back to the cytoplasmic face. In the absence of any other influence this high affinity interaction would be stable, and the cargo–kap complex would arrest here, bound to the FG nups at the nuclear extremity of the NPC (Fig. 11 III). This situation is seen in the case of kap mutants that cannot bind Ran (Gorlich et al. 1996). However, the peripheral localization of these nups exposes their bound kap complex to the nucleoplasmic milieu and in particular, to Ran-GTP. The binding of Ran-GTP to the kap induces cargo release and undocking from the NPC, which causes the cargo and kap to be liberated into the nucleoplasm (Fig. 11 IV). Once nucleoplasmic, the cargo can no longer bind the NPC, and the Brownian gate, in effect, shuts.
Figure 11.
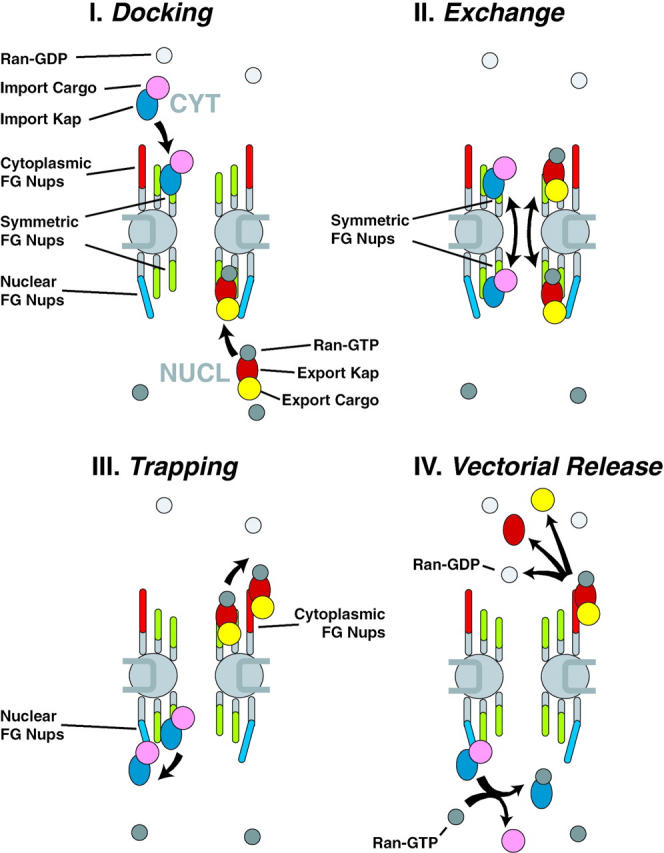
Diagram of NPC illustrating a model for nucleocytoplasmic transport. See Discussion for details.
Export would be similar (Fig. 11 I–IV). An exporting kap binds both Ran-GTP and its export cargo in the nucleoplasm. After passing through the symmetric FG nups, the cargo-Ran-kap complex is preferentially drawn to the high affinity binding sites, this time on the NPC cytoplasmic face. In isolation, this complex would be stable, as has been observed (Floer and Blobel 1999; Kehlenbach et al. 1999). However, the kap-bound Ran-GTP is now at the cytoplasmic periphery of the NPC, exposed to the cytoplasmic RanGAP, which induces the hydrolysis of the GTP on Ran. This in turn, causes undocking of the kap complex from the cytoplasmic binding sites and the release of both Ran and the cargo from the kap. It can readily be seen how recycling of the export kap back to the nucleoplasm would be akin to cargoless import, and likewise how recycling of an importing kap akin to cargoless export. In this fashion, the transport cycles are completed and the system restored, powered by the hydrolysis of GTP.
Numerous mechanistic refinements can be added. For instance, as exporting and importing kaps differ only in which Ran-bound state they load and dispose of cargo, it is possible that some kaps may carry material in both directions. The various transport factors apparently favor different FG nups (Aitchison et al. 1996; Rout et al. 1997; Marelli et al. 1998), and may also differ as to which vectorial cues (asymmetric FG nups versus soluble factors) they emphasize; some may only require symmetric FG nups and Ran. Such techniques for segregating transport factors into alternative pathways would serve to decrease congestion at the NPC, and furthermore provide opportunities for differential regulation by altering the affinity for a particular docking site to its particular preferred class of transport factor (Saavedra et al. 1997; Marelli et al. 1998). Furthermore, some transport factors may not require Ran at all, gaining directional cues and energy from alternative sources. For example, NPC-associated, ATP-driven RNA helicases (Snay-Hodge et al. 1998; Tseng et al. 1998), plus the exchange of RNA-binding proteins at the NPC, might reel mRNPs across the NPC in a process that is largely independent of Ran-mediated GTP hydrolysis. We also expect that in higher eukaryotes other mechanisms, like incorporating binding sites for Ran and its cofactors within the NPC, have been called into play to improve the efficiency and regulation of transport.
Perspectives
The yeast NPC provides a simple and accessible model system for studying nucleocytoplasmic transport, in which for the first time we have defined and mapped virtually all the components. This information can now be used to explore the fine structure of the NPC, determine the assembly pathway of the NPC, and further elucidate the mechanisms of nucleocytoplasmic transport. The tagged strains also constitute a series of powerful new tools that can be used to probe the function of nups in the context of the whole machine.
Acknowledgments
We would like to apologize to those authors whose primary work we could not cite due to space limitations. We are particularly grateful to Helen Shio for her superb electron microscopy work. We thank Julia Kipper, Rosemary Williams, Yongmin Oh, Dwayne Weber, and other members of the Rout, Chait, and Aitchison laboratories, as well as Rick Wozniak and Christopher Akey for many useful suggestions, much practical help, and materials; Wenzhu Zhang for ProFound; Mark Rose for the anti-Cdc31p antibodies; Ed Hurt for the anti-Nic96p antibodies; and Joe Fernandez for his excellent protein sequence data. We also thank Marcelo Magnasco, Jan Hoh, Kathy Wilson, and Rod MacKinnon for insightful suggestions regarding the model, and John Kuriyan and Stephen Burley for their useful critiques of the manuscript. Particular thanks go to Günter Blobel for much support, useful discussion, and encouragement.
We acknowledge funding from the National Institutes of Health (grant RR00862 to B.T. Chait), the Rockefeller University (to M.P. Rout), the Howard Hughes Medical Institute (A. Suprapto), and the Medical Research Council of Canada and Alberta Heritage Foundation for Medical Research (J.D. Aitchison).
Footnotes
Abbreviations used in this paper: MALDI-TOF, matrix-assisted laser desorption/ionization time of flight; NE, nuclear envelope; NPCs, nuclear pore complexes; nup, nucleoporin; ORF, open reading frame; pom, pore membrane proteins; PrA, protein A.
References
- Aitchison J.D., Rout M.P., Marelli M., Blobel G., Wozniak R.W. Two novel related yeast nucleoporins Nup170p and Nup157pcomplementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p J. Cell Biol. 131 1995. 1133 1148a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison J.D., Blobel G., Rout M.P. Nup120pa yeast nucleoporin required for NPC distribution and mRNA transport J. Cell Biol. 131 1995. 1659 1675b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison J.D., Blobel G., Rout M.P. Kap104pa karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- Akey C.W. Visualization of transport-related configurations of the nuclear pore transporter. Biophys. J. 1990;58:341–355. doi: 10.1016/S0006-3495(90)82381-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailer S.M., Siniossoglou S., Podtelejnikov A., Hellwig A., Mann M., Hurt E. Nup116p and Nup100p are interchangeable through a conserved motif which constitutes a docking site for the mRNA transport factor gle2p. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:1107–1119. doi: 10.1093/emboj/17.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrios M., Fisher P.A., Matz E.C. Localization of a myosin heavy chain-like polypeptide to Drosophila nuclear pore complexes. Proc. Natl. Acad. Sci. USA. 1991;88:219–223. doi: 10.1073/pnas.88.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S., Rose M.D. Direct interaction between yeast spindle pole body componentsKar1p is required for Cdc31p localization to the spindle pole body. J. Cell Biol. 1994;125:843–852. doi: 10.1083/jcb.125.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G. Unidirectional and bidirectional protein traffic across membranes. Cold Spring Harb. Symp. Quant. Biol. 1995;60:1–10. doi: 10.1101/sqb.1995.060.01.003. [DOI] [PubMed] [Google Scholar]
- Brown H.G., Hoh J.H. Entropic exclusion by neurofilament sidearmsa mechanism for maintaining interfilament spacing. Biochemistry. 1997;36:15035–15040. doi: 10.1021/bi9721748. [DOI] [PubMed] [Google Scholar]
- Buss F., Kent H., Stewart M., Bailer S.M., Hanover J.A. Role of different domains in the self-association of rat nucleoporin p62. J. Cell Sci. 1994;107:631–638. doi: 10.1242/jcs.107.2.631. [DOI] [PubMed] [Google Scholar]
- Chial H.J., Rout M.P., Giddings T.H., Winey M. Saccharomyces cerevisiae Ndc1p is a shared component of nuclear pore complexes and spindle pole bodies. J. Cell Biol. 1998;143:1789–1800. doi: 10.1083/jcb.143.7.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L.I. The nuclear pore complex. Annu. Rev. Biochem. 1995;64:865–896. doi: 10.1146/annurev.bi.64.070195.004245. [DOI] [PubMed] [Google Scholar]
- DeHoratius C., Silver P.A. Nuclear transport defects and nuclear envelope alterations are associated with mutation of the Saccharomyces cerevisiae NPL4 gene. Mol. Biol. Cell. 1996;7:1835–1855. doi: 10.1091/mbc.7.11.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delphin C., Guan T., Melchior F., Gerace L. RanGTP targets p97 to RanBP2, a filamentous protein localized at the cytoplasmic periphery of the nuclear pore complex. Mol. Biol. Cell. 1997;8:2379–2390. doi: 10.1091/mbc.8.12.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emtage J.L., Bucci M., Watkins J.L., Wente S.R. Defining the essential functional regions of the nucleoporin Nup145p. J. Cell Sci. 1997;110:911–925. doi: 10.1242/jcs.110.7.911. [DOI] [PubMed] [Google Scholar]
- Englmeier L., Olivo J.C., Mattaj I.W. Receptor-mediated substrate translocation through the nuclear pore complex without nucleotide triphosphate hydrolysis. Curr. Biol. 1999;9:30–41. doi: 10.1016/s0960-9822(99)80044-x. [DOI] [PubMed] [Google Scholar]
- Fabre E., Hurt E. Yeast genetics to dissect the nuclear pore complex and nucleocytoplasmic trafficking. Annu. Rev. Genet. 1997;31:277–313. doi: 10.1146/annurev.genet.31.1.277. [DOI] [PubMed] [Google Scholar]
- Fahrenkrog B., Hurt E.C., Aebi U., Pante N. Molecular architecture of the yeast nuclear pore complexlocalization of Nsp1p subcomplexes. J. Cell Biol. 1998;143:577–588. doi: 10.1083/jcb.143.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldherr C.M., Akin D. The location of the transport gate in the nuclear pore complex. J. Cell Sci. 1997;110:3065–3070. doi: 10.1242/jcs.110.24.3065. [DOI] [PubMed] [Google Scholar]
- Fenyo D., Qin J., Chait B.T. Protein identification using mass spectrometric information. Electrophoresis. 1998;19:998–1005. doi: 10.1002/elps.1150190615. [DOI] [PubMed] [Google Scholar]
- Fernandez J., Andrews L., Mische S.M. An improved procedure for enzymatic digestion of polyvinylidene difluoride-bound proteins for internal sequence analysis. Anal. Biochem. 1994;218:112–117. doi: 10.1006/abio.1994.1148. [DOI] [PubMed] [Google Scholar]
- Floer M., Blobel G. Putative reaction intermediates in Crm1-mediated nuclear protein export. J. Biol. Chem. 1999;274:16279–16286. doi: 10.1074/jbc.274.23.16279. [DOI] [PubMed] [Google Scholar]
- Floer M., Blobel G., Rexach M. Disassembly of RanGTP-karyopherin beta complex, an intermediate in nuclear protein import. J. Biol. Chem. 1997;272:19538–19546. doi: 10.1074/jbc.272.31.19538. [DOI] [PubMed] [Google Scholar]
- Fornerod M., Ohno M., Yoshida M., Mattaj I.W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Fujiki Y., Hubbard A.L., Fowler S., Lazarow P.B. Isolation of intracellular membranes by means of sodium carbonate treatmentapplication to endoplasmic reticulum. J. Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D., Pante N., Kutay U., Aebi U., Bischoff F.R. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Grandi P., Doye V., Hurt E.C. Purification of NSP1 reveals complex formation with ‘GLFG’ nucleoporins and a novel nuclear pore protein NIC96. EMBO (Eur. Mol. Biol. Organ.) J. 1993;12:3061–3071. doi: 10.1002/j.1460-2075.1993.tb05975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P., Emig S., Weise C., Hucho F., Pohl T., Hurt E.C. A novel nuclear pore protein Nup82p which specifically binds to a fraction of Nsp1p. J. Cell Biol. 1995;130:1263–1273. doi: 10.1083/jcb.130.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath C.V., Copeland C.S., Amberg D.C., Del Priore V., Snyder M., Cole C.N. Nuclear pore complex clustering and nuclear accumulation of poly(A)+ RNA associated with mutation of the Saccharomyces cerevisiae RAT2/NUP120 gene. J. Cell Biol. 1995;131:1677–1697. doi: 10.1083/jcb.131.6.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz M.E., Strambio-de-Castillia C., Blobel G. Two yeast nuclear pore complex proteins involved in mRNA export form a cytoplasmically oriented subcomplex. Proc. Natl. Acad. Sci. USA. 1998;95:11241–11245. doi: 10.1073/pnas.95.19.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J., Strasser K., Podtelejnikov A., Mann M., Jung J.U., Hurt E. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:2593–2609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehlenbach R.H., Dickmanns A., Kehlenbach A., Guan T., Gerace L. A role for RanBP1 in the release of CRM1 from the nuclear pore complex in a terminal step of nuclear export. J. Cell Biol. 1999;145:645–657. doi: 10.1083/jcb.145.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keminer O., Siebrasse J.P., Zerf K., Peters R. Optical recording of signal-mediated protein transport through single nuclear pore complexes. Proc. Natl. Acad. Sci. USA. 1999;96:11842–11847. doi: 10.1073/pnas.96.21.11842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin J.V., Fogg J. Partial purification of yeast spindle pole bodies. In: Cappuccinelli P., Morris N.R., editors. Microtubules in Microorganisms. Marcel Dekker, Inc.; New York: 1982. pp. 157–169. [Google Scholar]
- Kiseleva E., Goldberg M.W., Daneholt B., Allen T.D. RNP export is mediated by structural reorganization of the nuclear pore basket. J. Mol. Biol. 1996;260:304–311. doi: 10.1006/jmbi.1996.0401. [DOI] [PubMed] [Google Scholar]
- Koepp D.M., Silver P.A. A GTPase controlling nuclear traffickingrunning the right way or walking RANdomly? Cell. 1996;87:1–4. doi: 10.1016/s0092-8674(00)81315-x. [DOI] [PubMed] [Google Scholar]
- Kose S., Imamoto N., Tachibana T., Shimamoto T., Yoneda Y. Ran-unassisted nuclear migration of a 97-kD component of nuclear pore-targeting complex. J. Cell Biol. 1997;139:841–849. doi: 10.1083/jcb.139.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosova B., Pante N., Rollenhagen C., Hurt E. Nup192p is a conserved nucleoporin with a preferential location at the inner site of the nuclear membrane. J. Biol. Chem. 1999;274:22646–22651. doi: 10.1074/jbc.274.32.22646. [DOI] [PubMed] [Google Scholar]
- Kraemer D.M., Strambio-de-Castillia C., Blobel G., Rout M.P. The essential yeast nuceloporin NUP159 is located on the cytoplasmic side of the nuclear pore complex and serves in karyopherin-mediated binding of transport substrate. J. Biol. Chem. 1995;270:19017–19021. doi: 10.1074/jbc.270.32.19017. [DOI] [PubMed] [Google Scholar]
- Kuster B., Mann M. Identifying proteins and post-translational modifications by mass spectrometry. Curr. Opin. Struct. Biol. 1998;8:393–400. doi: 10.1016/s0959-440x(98)80075-4. [DOI] [PubMed] [Google Scholar]
- Kutay U., Bischoff F.R., Kostka S., Kraft R., Gorlich D. Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae . Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Magnasco M. Forced thermal ratchets. Phys. Rev. Lett. 1993;71:1477–1481. doi: 10.1103/PhysRevLett.71.1477. [DOI] [PubMed] [Google Scholar]
- Marelli M., Aitchison J.D., Wozniak R.W. Specific binding of the karyopherin Kap121p to a subunit of the nuclear pore complex containing Nup53p, Nup59p, and Nup170p. J. Cell Biol. 1998;143:1813–1830. doi: 10.1083/jcb.143.7.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I.W., Englmeier L. Nucleocytoplasmic transportthe soluble phase. Annu. Rev. Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- Nachury M.V., Weis K. The direction of transport through the nuclear pore can be inverted. Proc. Natl. Acad. Sci. USA. 1999;96:9622–9627. doi: 10.1073/pnas.96.17.9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrbass U., Rout M.P., Maguire S., Blobel G., Wozniak R.W. The yeast nucleoporin Nup188p interacts genetically and physically with the core structures of the nuclear pore complex. J. Cell Biol. 1996;133:1153–1162. doi: 10.1083/jcb.133.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi E., Hayashi N., Nakashima N., Nishimoto T. Yrb2p, a Nup2p-related yeast protein, has a functional overlap with Rna1p, a yeast Ran-GTPase-activating protein. Mol. Cell Biol. 1997;17:2235–2246. doi: 10.1128/mcb.17.4.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paine P.L., Moore L.C., Horowitz S.B. Nuclear envelope permeability. Nature. 1975;254:109–114. doi: 10.1038/254109a0. [DOI] [PubMed] [Google Scholar]
- Pemberton L.F., Rout M.P., Blobel G. Disruption of the nucleoporin gene NUP133 results in clustering of nuclear pore complexes. Proc. Natl. Acad. Sci. USA. 1995;92:1187–1191. doi: 10.1073/pnas.92.4.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Ruud J., Chait B.T. A practical ion trap mass spectrometer for the analysis of peptides by matrix-assisted laser desorption/ionization. Anal. Chem. 1996;68:1784–1791. doi: 10.1021/ac9511612. [DOI] [PubMed] [Google Scholar]
- Qin J., Fenyo D., Zhao Y., Hall W.W., Chao D.M., Wilson C.J., Young R.A., Chait B.T. A strategy for rapid, high-confidence protein identification. Anal. Chem. 1997;69:3995–4001. doi: 10.1021/ac970488v. [DOI] [PubMed] [Google Scholar]
- Radu A., Moore M.S., Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- Rexach M., Blobel G. Protein import into nucleiassociation and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- Rout M.P., Kilmartin J.V. Components of the yeast spindle and spindle pole body. J. Cell Biol. 1990;111:1913–1927. doi: 10.1083/jcb.111.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout M.P., Blobel G. Isolation of the yeast nuclear pore complex. J. Cell Biol. 1993;123:771–783. doi: 10.1083/jcb.123.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout M.P., Wente S.R. Pores for thoughtnuclear pore proteins. Trends Cell Biol. 1994;4:357–365. doi: 10.1016/0962-8924(94)90085-x. [DOI] [PubMed] [Google Scholar]
- Rout M.P., Blobel G., Aitchison J.D. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- Saavedra C.A., Hammell C.M., Heath C.V., Cole C.N. Yeast heat shock mRNAs are exported through a distinct pathway defined by Rip1p. Genes Dev. 1997;11:2845–2856. doi: 10.1101/gad.11.21.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwoebel E.D., Talcott B., Cushman I., Moore M.S. Ran-dependent signal-mediated nuclear import does not require GTP hydrolysis by Ran. J. Biol. Chem. 1998;273:35170–35175. doi: 10.1074/jbc.273.52.35170. [DOI] [PubMed] [Google Scholar]
- Shulga N., Roberts P., Gu Z., Spitz L., Tabb M.M., Nomura M., Goldfarb D.S. In vivo nuclear transport kinetics in Saccharomyces cerevisiaea role for heat shock protein 70 during targeting and translocation. J. Cell Biol. 1996;135:329–339. doi: 10.1083/jcb.135.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S.M., Peskin C.S., Oster G.F. What drives the translocation of proteins? Proc. Natl. Acad. Sci. USA. 1992;89:3770–3774. doi: 10.1073/pnas.89.9.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S., Wimmer C., Rieger M., Doye V., Tekotte H., Weise C., Emig S., Segref A., Hurt E.C. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84:265–275. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- Snay-Hodge C.A., Colot H.V., Goldstein A.L., Cole C.N. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:2663–2676. doi: 10.1093/emboj/17.9.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahm Y., Fahrenkrog B., Zenklusen D., Rychner E., Kantor J., Rosbash M., Stutz F. The RNA export factor Gle1p is located on the cytoplasmic fibrils of the NPC and physically interacts with the FG-nucleoporin Rip1p, the DEAD-box protein Rat8p/Dbp5p and a new protein Ymr255p. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:5761–5777. doi: 10.1093/emboj/18.20.5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-de-Castillia C., Blobel G., Rout M.P. Isolation and characterization of nuclear envelopes from the yeast Saccharomyces . J. Cell Biol. 1995;131:19–31. doi: 10.1083/jcb.131.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-de-Castillia C., Blobel G., Rout M.P. Proteins connecting the nuclear pore complex with the nuclear interior. J. Cell Biol. 1999;144:839–855. doi: 10.1083/jcb.144.5.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talcott B., Moore M.S. Getting across the nuclear pore complex. Trends Cell Biol. 1999;9:312–318. doi: 10.1016/s0962-8924(99)01608-6. [DOI] [PubMed] [Google Scholar]
- Taura T., Schlenstedt G., Silver P.A. Yrb2p is a nuclear protein that interacts with Prp20p, a yeast Rcc1 homologue. J. Biol. Chem. 1997;272:31877–31884. doi: 10.1074/jbc.272.50.31877. [DOI] [PubMed] [Google Scholar]
- Tseng S.S., Weaver P.L., Liu Y., Hitomi M., Tartakoff A.M., Chang T.H. Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:2651–2662. doi: 10.1093/emboj/17.9.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa N., Alzhanova-Ericsson A.T., Sun X., Kiseleva E., Bjorkroth B., Wurtz T., Daneholt B. A pre-mRNA-binding protein accompanies the RNA from the gene through the nuclear pores and into polysomes. Cell. 1996;84:253–264. doi: 10.1016/s0092-8674(00)80980-0. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat A., Alberti-Segui C., Rebischung C., Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae . Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Wozniak R.W., Blobel G., Rout M.P. POM152 is an integral protein of the pore membrane domain of the yeast nuclear envelope. J. Cell Biol. 1994;125:31–42. doi: 10.1083/jcb.125.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Rout M.P., Akey C.W. Three-dimensional architecture of the isolated yeast nuclear pore complexfunctional and evolutionary implications. Mol. Cell. 1998;1:223–234. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]