Abstract
Regulatory mechanisms controlling the timing of developmental events are crucial for proper development to occur. ftz-f1 is expressed in a temporally regulated manner following pulses of ecdysteroid and this precise expression is necessary for the development of Drosophila melanogaster. To understand how insect hormone ecdysteroids regulate the timing of FTZ-F1 expression, we purified a DNA binding regulator of ftz-f1. Mass spectroscopy analysis revealed this protein to be a fly homolog of mammalian B lymphocyte-induced maturation protein 1 (Blimp-1). Drosophila Blimp-1 (dBlimp-1) is induced directly by 20-hydroxyecdysone, and its product exists during high-ecdysteroid periods and turns over rapidly. Forced expression of dBlimp-1 and RNA interference analysis indicate that dBlimp-1 acts as a repressor and controls the timing of FTZ-F1 expression. Furthermore, its prolonged expression results in delay of pupation timing. These results suggest that the transient transcriptional repressor dBlimp-1 is important for determining developmental timing in the ecdysone-induced pathway.
The steroid hormone ecdysone and its active metabolite 20-hydroxyecdysone (20E) (hereafter referred to collectively as ecdysone) are responsible for many essential developmental processes, including insect molting, metamorphosis, oogenesis, and embryogenesis (25, 40). The insect ecdysone response provides an excellent model for studying hormone function, in which temporally regulated induction of multiple genes is required to control complex developmental events. For instance, at the onset of metamorphosis in Drosophila melanogaster, a large pulse of ecdysone causes the third-instar larval-to-prepupal transition. Based on the observation of puffs on polytene chromosomes in cultured salivary glands more than 30 years ago, it has long been known that there are at least four categories of ecdysone-inducible genes (1-4, 38). The early genes are induced directly by the ecdysone-receptor complex and are repressed by their products. The early-late genes are also induced directly by ecdysone but require an ecdysone-induced gene product(s) for maximal induction. The late genes are induced by the early gene products, and the mid-prepupal genes are induced only after ecdysone levels have declined. In the last two decades, many of the genes belonging to these four groups have been cloned, and their regulated expression profile has been confirmed. These include multiple transcription factors, which constitute an ecdysone-induced gene cascade.
ftz-f1 is a mid-prepupal gene (29) that encodes a nuclear receptor-type transcription factor (30). The beta isoform of the ftz-f1 gene product is expressed not only during the mid-prepupal period at the onset of metamorphosis but also during late embryogenesis, just before larval ecdysis and eclosion (45, 51, 54, 55). All of these periods closely follow declines in ecdysone levels. The importance of timing of ftz-f1 expression has been shown by rescue of ftz-f1 mutants by temporally specific expression of βFTZ-F1 as well as developmental arrest by premature expression of βFTZ-F1 (55). Expression and/or induction of ftz-f1 after a decline in ecdysone levels has been reported to occur in several insects besides Drosophila (17, 31, 46), suggesting that the temporally regulated expression of ftz-f1 is crucial for insect development.
However, the mechanism by which ftz-f1 is temporally regulated is still unclear; only two transcriptional regulators have been identified to date (20, 28, 52). One is the early-late gene product DHR3, a nuclear receptor-type transcription factor that is expressed from just before puparium formation to the mid-prepupal period. Several lines of evidence indicate it to be a transcriptional activator for the ftz-f1 gene: (i) premature expression of DHR3 under the control of a heat shock promoter induces βFTZ-F1 (28, 52), (ii) βFTZ-F1 expression is reduced in a DHR3 mutant (27), and (iii) DHR3 binds to three sites downstream of the transcription initiation site of the ftz-f1 locus (+150, +240, and + 300), and mutations in these sites reduce expression of the ftz-f1 promoter-lacZ fusion gene in transgenic flies (20).
The other transcription factor known to regulate ftz-f1 is the early gene product E75B, which is also a member of the nuclear receptor superfamily but lacks one zinc finger and thus cannot bind to DNA by itself. E75B is expressed around puparium formation, when ecdysone levels are high, and disappears after ecdysone levels decline during the mid-prepupal period. Because E75B binds directly to DHR3 and inhibits its activator function, DHR3 can activate ftz-f1 only after ecdysone levels have declined (52). However, the temporal pattern of ftz-f1 expression is preserved in DHR3 and E75B mutants (8, 27), and mutations in the DHR3 binding sites of the ftz-f1 promoter-lacZ fusion gene have no effect on the timing of β-galactosidase expression in transgenic flies (20). Thus, all these results suggest that temporal regulation of ftz-f1 expression is achieved by other factors.
To understand how ftz-f1 expression is temporally regulated, we have analyzed the cis-regulatory region of the ftz-f1 locus to identify developmentally regulated factors that bind to these regions (20). One factor, designated factor I-4, binds to the region upstream of the transcriptional start site of ftz-f1 and is expressed during mid-embryogenesis and the early prepupal period. Here, we determined the binding site of factor I-4, identified its gene based on the information on the purified protein, and analyzed its biological function during development, including the regulation of the ftz-f1 gene.
MATERIALS AND METHODS
Gel mobility shift assay.
Binding was performed at 25°C for 1 h in 10 μl reaction buffer (15 mM Tris-HCl at pH 7.8, 150 mM NaCl, 0.1 mM EGTA, 1 mM EDTA, 1 mM dithiothreitol, 0.2% Igepal CA-630 [Sigma], 1 mM NaPO4 at pH 7.8, 10% glycerol), containing 20 fmol 32P-labeled DNA probe, 2 μg poly(dI-dC)-poly(dI-dC) (Pharmacia), 100 ng salmon sperm DNA, 10 μg yeast tRNA, 50 μg bovine serum albumin fraction V (Sigma), and 1 μl nuclear extract or fraction. Complex and free probe were separated by agarose gel electrophoresis and detected as previously described (50). Site I-4 DNA was obtained by hybridization of synthesized oligonucleotides carrying 5′-GTTTCACTTTGGCTTTCCGTTTTGG and the complementary sequence. Site I-4m DNA was similarly obtained using synthesized oligonucleotides carrying 5′-GTTTCACTTTAGATCTCCGTTTTGG and the complementary sequence. The mutant site I-4m2 DNA was generated similarly, using the sequence 5′-AAAAGTCTGACTCTGGCTCTGCGTTTGGG. For the supershift assay, 1 μl of anti-Drosophila Blimp-1 (anti-dBlimp-1) or anti-βFTZ-F1 serum was added to the incubation mixture, further incubated for 30 min, and loaded on the gel. An oligonucleotide carrying the FTZ-F1 binding site in the EDG84A promoter, EDG84F1, was used to detect βFTZ-F1 (33).
Methylation interference.
Methylation interference was performed as previously described by Kageyama et al. (20) using an Eco52I-HincII fragment carrying base pairs −70 to −470 and labeled at the Eco52I site by T4 polynucleotide kinase and [α-32P]ATP.
Preparation of nuclear extracts and purification of factor I-4.
Staged nuclear extracts were prepared as previously described by Ueda et al. and Kageyama et al. (20, 51). Nuclear extracts from 8- to 16-h embryos were prepared as previously described by Ueda et al. (51), except that 0.2% Igepal CA-630 was added to solutions II and III. All purification steps were performed on ice or in the cold room. Twenty-five milliliters of nuclear extract from 130-g mid-stage embryos was loaded on S Sepharose columns. After washing with buffer L (10 mM HEPES-NaOH at pH 7.9, 1 mM EDTA, 0.1% Igepal CA-630, 20% glycerol, 1 mM dithiothreitol) containing 150 mM NaCl, factor I-4 was eluted with buffer L containing 250 mM NaCl. The S Sepharose fraction containing high factor I-4 activity (2.4 ml) was adjusted to 5.6 ml with L buffer after addition of 7 mg site I DNA-latex resin, 112 μg sheared salmon sperm DNA, 11.2 μg poly(dI-dC)-poly(dI-dC), and 112 μg yeast tRNA. The mixture was incubated for 30 min on ice, and then the supernatant was removed after centrifugation at 15,000 rpm for 15 min. After the resin was washed three times with 1 ml L buffer containing 200 mM NaCl, factor I-4 was recovered as supernatant by incubating with 230 μl L buffer containing 500 mM NaCl for 10 min. Two hundred microliters of the supernatant was diluted with 470 μl L buffer containing 40 μg sheared salmon sperm DNA, 4 μg poly(dI-dC)-poly(dI-dC), and 40 μg yeast tRNA and was mixed with 5 mg latex resin carrying mutated site I-4 DNA. After incubation for 30 min and centrifugation, 660 μl supernatant was recovered and mixed with 3.5 mg wild-type site I-4 DNA-latex resin, and then the mixture was incubated for 30 min. After the supernatant was removed, the resin was washed three times with 1 ml L Buffer containing 200 mM NaCl, and then the factor was eluted twice with 200 μl L Buffer containing 500 mM NaCl. Site I-4 DNA-affinity resin and mutated site I-4 DNA-affinity resin were prepared as previously described (16) using latex beads as resin. Synthesized oligonucleotides 5′-TTTCACTTTCGCTTTCCGTTTGGGGG and 5′-AAACGGAAAGCGAAAGTGAAACCCCC were used for making wild-type and site I-4 DNA-latex resin, and synthesized oligonucleotides 5′-GATCCGTCTGACTCTGGCTCTGGCTCTGGCTCTGGCTCTGCGTTTGA and 5′-GATCTCAAACGCAGAGCCAGAGCCAGAGCCAGAGCCAGAGTCAGACG were used for making mutated site I-4 DNA-latex resin.
Plasmid construction for forced expression of dBlimp-1.
A cDNA clone (RE26660) containing the entire dBlimp-1 coding region was obtained from Research Genetics. Double-stranded oligonucleotides obtained by hybridization of two synthesized oligonucleotides, 5′-AATTCTAGTCGCCATGCA and 5′-TGGCGACTAG, and a 1.1-kb EcoT22I-SalI fragment of RE26660 were inserted between the EcoRI and SalI sites in pBluescript II. The established plasmid was digested with SalI and ApaI, and a 2-kb SalI-ApaI fragment from RE26660 was inserted. After a NotI linker was inserted at the blunt-ended KpnI site, an EcoRI and NotI digest of this plasmid was ligated into the EcoRI and NotI sites of pCaSpeR-HS plasmid to establish transgenic fly lines expressing dBlimp-1 under control of the heat shock promoter. To construct a P element expressing Flag-tagged dBlimp-1 protein, double-stranded oligonucleotides obtained by hybridization of two synthesized oligonucleotides, 5′-GATCATCGAATGCACGTAGATCTGGTAC and 5′-CAGATCTACGTGCATTCGAT, were inserted into the FbaI and KpnI sites of RE26660, and then the EcoRI-NotI fragment of the obtained plasmid was inserted into the pCaSpeR-HS plasmid as described above.
Plasmid construction for dBlimp-1 RNA interference (RNAi).
A 550-bp DNA fragment spanning the beginning of the second exon to the beginning of third exon was obtained by PCR on genomic DNA using primer 5′-ATCAGATCTTGCATGGACATCACAACCACAACCAT, which contains a BglII site, and primer 5′-TAGAATTCGCTGCTCCAAACTCCTTCAGTCTGCAAG, which contains an EcoRI site. A 450-bp DNA fragment from the beginning to the end of the second exon was obtained by PCR on genomic DNA using primer 5′-ATAGCGGCCGCTTGCATGGACATCACAACCACAACCATCT, which contains an Eco52I site, and primer 5′-AAGAATTCACATTTGGCGTTGAGTAGACCATGGA, which contains an EcoRI site. After ligation of the two fragments using their EcoRI sites, ligated DNA was digested with BglII and Eco52I and was inserted into the pUAST vector using the BglII and NotI sites.
Antibody preparation.
An EcoRI-SalI digest of dBlimp-1 cDNA in pCaSpeR-HS was inserted into the EcoRI and SalI sites of pET28b to express the N-terminal half (from amino acid 1 to 372) of dBlimp-1 in Escherichia coli. The established plasmid was transformed into E. coli BL21DE3(LysS), and the N-terminal half of Blimp-1 was expressed according to the manufacturer's protocol and subjected to immunization after purification.
RNA extraction and Northern blotting.
RNA was prepared using Sepasol-I super (Nakarai) according to the manufacturer's protocol. Northern blotting was performed as described previously (43).
RT-PCR.
For quantitative real-time reverse transcription-PCR (RT-PCR), cDNA was synthesized using random 9-mer oligonucleotides and ReverTra Ace (Toyobo), and RNA was treated with RNase-free DNase I (Takara) and used as a template for real-time PCR using a LightCycler system (Roche). The following synthetic oligonucleotides were used for detecting reverse transcripts: 5′-CGCACCTCCAGAAGCATCAT and 5′-GGGCAGAGATCACAGGCATA for dBlimp-1, 5′-AGCCGCAGCAGCAAATG and 5′-ACCCGAGTGGTGCAGAT for E75A, and 5′-CCACCAGTCGGATCGATATG and 5′-CACGTTGTGCACCAGGAACT for rp49 (23).
In vitro culture of salivary glands.
Thirty pairs of salivary glands from the middle stage of third-instar larvae were cultured in Schneider medium in the presence or absence of 5 mM 20E or 70 mM cycloheximide.
Fly work.
All flies used in the transformation study had a y1 Df(1)w67c1 background. Nuclear extracts were prepared from an Oregon-R strain. dBlimp-1P14751 was a kind gift from T. Aigaki, and Sgs-2 flies were from A. J. Andres. hs-Gal4 lines were obtained from the Genetic Stock Research Center, National Institute of Genetics. Flies were raised at 25°C on 10% glucose, 8% cornmeal, 4% ebios, and 0.7% agar medium containing propionic acid and butyl-p-hydroxybenzoate as antifungal agents. Staging of mid- to late third-instar larvae was determined by observation of green fluorescent protein signals in Sgs-2 larvae (9) or of signals in guts of larvae cultured in bromophenol blue-containing food (21). Staging after puparium formation was done by incubating newly transformed white prepupae at 25°C. Transgenic fly lines were established by germ line transformation using the established plasmid.
Western blotting.
Western blotting was performed as described previously (33). Amounts of protein loaded in each lane were checked by staining the membrane again using anti-α-tubulin antibody.
RESULTS
Determination of the factor I-4 binding site in the ftz-f1 promoter.
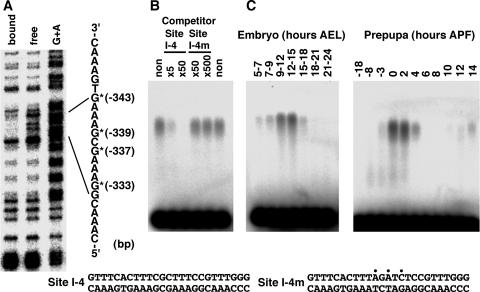
To determine the precise binding site of factor I-4 in the ftz-f1 promoter, a methylation interference assay was performed using an Eco52I-HincII fragment (bases −470 to −70) that includes the restriction fragment to which factor I-4 was previously shown to bind (20). As shown in Fig. 1A, four guanine residues located at positions −333, −337, −339, and −343 exhibited methylation interference upon factor I-4 binding. To confirm this result, a gel mobility shift assay was performed using a 25-bp double-stranded oligonucleotide encompassing these positions (site I-4) as a probe (Fig. 1B). As expected, a complex was clearly observed. This complex disappeared upon addition of cold site I-4 DNA but not site I-4m DNA carrying base substitutions, indicating that the factor binds with strong sequence specificity to the identified position. A developmental gel mobility shift assay revealed that the factor is present at high levels from 9 to 15 h after egg laying and then rapidly disappears during the embryonic stage (Fig. 1C, left). At the onset of metamorphosis, the factor is present from −3 h after puparium formation (APF) to 4 h APF, with a peak from 0 to 2 h APF. It then reappears from 10 to 14 h APF (Fig. 1C, right). These observations corroborate previous results obtained using the larger restriction fragment (20). These results indicate that factor I-4 binds to the DNA sequence around 340 bp upstream of the ftz-f1 transcriptional start site.
FIG. 1.
Determination of the binding site and developmental expression pattern of factor I-4. (A) Methylation interference using a 400-bp Eco52I-HincII fragment. Positions of nucleotides showing methylation interference are represented by asterisks with the distance from the transcription start site. The G+A Maxam-Gilbert sequencing reaction was used as a marker. (B) Confirmation of sequence-specific binding to the identified site by a gel mobility shift competition assay. 32P-labeled site I-4 DNA was used as a probe, and the indicated amounts of site I-4 or site I-4m competitor DNA compared with the probed site I-4 DNA were added to the binding reaction mixtures. The nucleotide sequences of site I-4 and site I-4m DNAs are indicated at the bottom. Positions of introduced mutations are indicated by dots. (C) Confirmation of factor I-4 binding by gel mobility shift assays using developmentally staged nuclear extracts at embryonic stages (left) and at the onset of metamorphosis (right). 32P-labeled site I-4 DNA was used as a probe. AEL, after egg laying.
Purification of factor I-4 from embryonic nuclear extract.
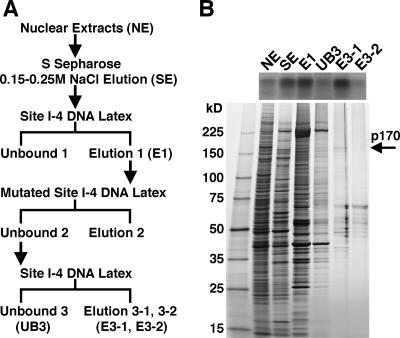
To identify factor I-4, we purified it from an embryonic nuclear extract, as outlined in Fig. 2A. Mid-embryonic-stage nuclear extract was fractionated by S Sepharose column chromatography, and then the active fraction was further fractionated through a latex resin conjugated with multimeric wild-type site I-4 DNA. After removing nonspecific binding using a latex resin conjugated with polymerized mutated site I-4 DNA, the factor was purified using the latex resin carrying wild-type site I-4 DNA. Table 1 shows a summary of purification of the factor, and Fig. 2B shows the results of a gel mobility assay using typical fractions of the purification steps and silver staining after sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis using the same fractions. Even after purification by the second wild-type site I-4 DNA affinity resin, several stained bands were observed in the eluted fractions (E3-1 and E3-2). However, the intensity of the 170-kDa band in the SDS-polyacrylamide gel correlated well with its binding to the site I-4 probe in the gel mobility shift assay, suggesting that the 170-kDa protein is indeed factor I-4.
FIG. 2.
Identification of factor I-4 as a Blimp-1/CG5249-encoded protein. (A) Purification of factor I-4. Nuclear extract (NE) derived from embryos at 8 to 16 h after egg laying was loaded onto an S Sepharose column. Fractions eluted between 0.15 and 0.25 M NaCl (SE) were subjected to affinity purification. The first affinity purification was performed using site I-4 DNA-conjugated latex beads. The eluate from the first affinity chromatography (E1) was then incubated with mutated site I-4 DNA-conjugated beads to perform subtraction. The supernatant from the subtraction was subjected to a second affinity purification. Eluate from the second affinity chromatography (E3-1 and E3-2) was obtained. (B) Detection of the binding activity and proteins in typical fractions during purification by a gel mobility shift assay (upper panel) and SDS-PAGE (lower panel). One microliter of the fractions in the purification step was used for the gel mobility shift assay, and 10 μl of the same fraction was reserved for SDS-PAGE, except for NE and SE, which were loaded at only 0.5 μl. Proteins were detected by silver staining.
TABLE 1.
Purification of factor I-4
| Step | Total protein (mg) | Total activity (U)a | Sp act (U/mg) | Recovery (%) | Purification (fold) |
|---|---|---|---|---|---|
| Nuclear extract | 260 | 12,000 | 45 | 100 | 1 |
| S Sepharose | 80 | 4,800 | 60 | 40 | 1.3 |
| Wild-type I-4E latex | |||||
| First | 0.165 | 800 | 4,850 | 6.6 | 107 |
| Second | 0.0012b | 300 | 250,000 | 2.5 | 5,500 |
One unit corresponds to the activity that can shift 1 fmol of the probe.
The amount of protein was estimated by the intensity of the band.
Factor I-4 is a homolog of mammalian transcriptional repressor Blimp-1/PRDI-BF1.
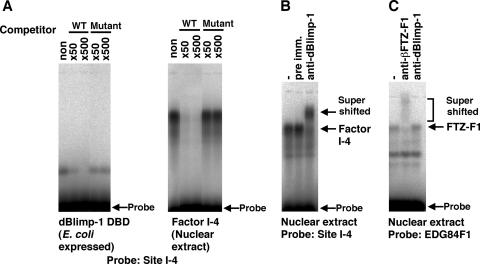
To identify the 170-kDa factor, protein in the final fraction was separated through an SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride membrane. The 170-kDa band was excised and subjected to time-of-flight mass spectroscopy analysis after lysyl endopeptidase treatment. These data show that factor I-4 is encoded by CG5249/Blimp-1 (data not shown). The deduced amino acid sequence contains zinc fingers sharing strong homology with those of the mammalian transcriptional repressor Blimp-1/PRDI-BF1 (22, 37, 49) (75% identity). Furthermore, it has been shown that Blimp-1/PRDI-BF1 binds to a similar sequence (22) as the identified binding site of factor I-4. To confirm this, the zinc finger region of dBlimp-1 was expressed in E. coli and examined by gel mobility shift assay, using site I-4 DNA as a probe. A complex produced by the recombinant protein was indeed observed. This complex disappeared upon addition of cold site I-4 DNA but not mutated site I DNA (Fig. 3A, left panel). Similar results were obtained when nuclear extract containing factor I-4 was used (Fig. 3A, right panel). In addition, antiserum against the N-terminal region of dBlimp-1 produced a supershifted complex in nuclear extracts, while preimmune serum did not (Fig. 3B). The supershift was not due to nonspecific binding of the antibody, as the antiserum did not react to a complex with βFTZ-F1 (Fig. 3C). From these results, we conclude that factor I-4 is encoded by CG5249/Blimp-1, and we refer to it as dBlimp-1 to distinguish it from homologs in other species.
FIG. 3.
Confirmation of factor I-4 as dBlimp-1. (A) Gel mobility shift competition assay using in vitro-expressed dBlimp-1 (left) and factor I-4 in the nuclear extract (right). The indicated amounts of site I-4 or site I-4m2 DNA compared with the probed site I-4 DNA were added to the reaction mixtures for the gel mobility shift assay. A 6.9-ng amount of purified recombinant proteins or 1 μl of nuclear extract was used in each binding reaction. WT, wild type. (B) Supershift of factor I-4 by anti-dBlimp-1. Anti-dBlimp-1 or preimmune serum was added to the reaction mixture for the gel mobility shift assay to detect dBlimp-1 in the complex. (C) Specificity of anti-dBlimp-1. βFTZ-F1 was detected by gel mobility shift assay. Anti-dBlimp-1 or anti-βFTZ-F1 serum was added to the reaction mixture for the gel mobility shift assay to examine specificity.
Besides the zinc finger motif, dBlimp-1 has another conserved motif at its N-terminal region, the PR domain (34, 48). The PR domain has strong sequence similarity to the SET domain, which is found in methyltransferase proteins. However, the PR domain in Blimp-1 is thought to lack methyltransferase activity, because it does not contain the NHSC(I) sequence, which is conserved in other SET domain proteins with methyltransferase activity (24, 32). In addition to these two conserved motifs, dBlimp-1 and mammalian Blimp-1 share a central, proline-rich region and a short, conserved N-terminal region that is not present in other SET domain proteins.
Expression pattern of dBlimp-1 mRNA.
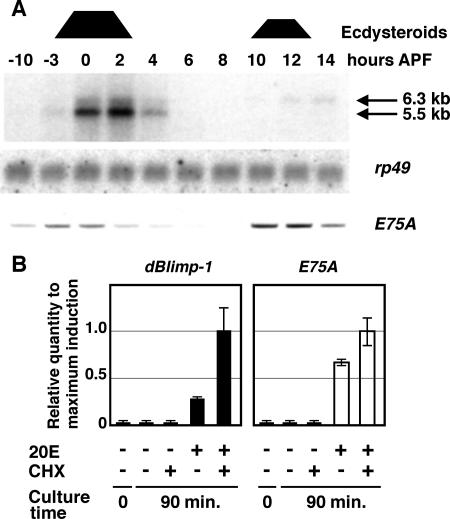
To determine the expression pattern of dBlimp-1, we performed Northern blot analysis using total RNA prepared from animals around the prepupal stage. As shown in Fig. 4A (upper panel), a strong 5.5-kb band was detected between 0 and 2 h APF. The bands were detected from 3 h before puparium formation and disappeared at 4 h APF. A 6.3-kb band was detected at 10 to 14 h APF, which corresponds to the increase of ecdysteroid that leads to head eversion and the completion of the prepupal stage. This result is consistent with developmental profiles of the dBlimp-1/factor I-4 binding activity detected by gel mobility shift assays (Fig. 1C), indicating that temporal regulation of dBlimp-1 occurs at the level of mRNA expression. The coincidence of dBlimp-1 mRNA expression with the ecdysone peaks raises the possibility that dBlimp-1 mRNA is induced by ecdysone. Interestingly, the expression profile of the transcript did not completely coincide with that of the E75A early gene transcript detected by RT-PCR method using the same staged RNA preparation; the appearance and disappearance of dBlimp-1 mRNA were delayed slightly (Fig. 4A, lower panel), suggesting that the regulation mechanisms are slightly different for these two genes.
FIG. 4.
Characterization of dBlimp-1 transcripts. (A) Detection of dBlimp-1 mRNA by Northern blotting using staged total RNA at the onset of metamorphosis. High-ecdysone periods are indicated at the top by trapezoids. Positions of dBlimp-1 mRNA are indicated by arrows. Middle panel, detection of rp49 mRNA using the same membrane. Bottom panel, level of E75A transcript detected by RT-PCR using the same staged RNA. (B) Induction of dBlimp-1 (left) and E75A (right) mRNAs by 20E in cultured salivary glands. Expression levels were measured by quantitative real-time RT-PCR using total RNA from salivary glands cultured for 90 min in the presence of 20E and/or cycloheximide (CHX). The value for each transcript was normalized to that of rp49 transcripts, with the level obtained with 20E and cycloheximide set as 1 for each transcript. The same template was used to measure the amounts of dBlimp-1 and E75A mRNA. Error bars indicate standard deviations.
Properties of the dBlimp-1 transcript.
To examine whether dBlimp-1 mRNA is induced by ecdysone like E75A mRNA, salivary glands from late third-instar larvae (more than 10 h before puparium formation) were cultured for 1.5 h in the presence or absence of 20E, and the expression level of the dBlimp-1 transcript was determined using quantitative RT-PCR (Fig. 4B). While no dBlimp-1 expression was detected when salivary glands were cultured in the absence of 20E, transcript was observed when 20E was added to the culture medium, even in the presence of cycloheximide. These results suggest that the dBlimp-1 transcript is directly induced by 20E. Interestingly, the level of induction in the presence of both 20E and cycloheximide was four times higher than that in the presence of 20E alone. This difference was less prominent for E75A mRNA. As cycloheximide is known to stabilize some mRNA species (15, 19, 39), these results suggest that the normal turnover rate of dBlimp-1 mRNA is much higher than that of E75A mRNA.
Knockdown of dBlimp-1 results in prepupal lethality and altered timing of βFTZ-F1 expression.
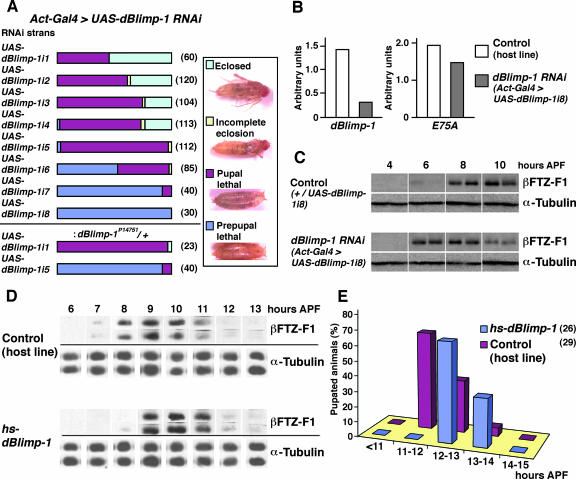
To elucidate the function of dBlimp-1 during Drosophila development, we knocked it down by RNAi. We established transgenic UAS-dBlimp-1i fly lines that express hairpin-type dBlimp-1 RNA under the control of GAL4 and observed the phenotype after mating with an Act5C-GAL4 strain expressing the GAL4 activator ubiquitously under the control of the Actin5C promoter. As shown in Fig. 5A, five out of eight established dBlimp-1i lines showed lethality at pupal stages in most of the observed animals. Many of them eclosed but died shortly thereafter (eclosed) or died during eclosion (incomplete eclosion). The rest of the lines showed prepupal lethality in more than half of the observed animals (Fig. 5A). The level of the dBlimp-1 transcript in Act5c-GAL4>UAS-dBlimp-1i7 line with the strongest phenotype was reduced about one-fourth compared to that in the control line, as revealed by quantitative RT-PCR (Fig. 5B) at 2 h APF. To test whether these phenotypes were caused by reduction of dBlimp-1 function, we observed the RNAi phenotype in the presence of a hypomorphic dBlimp-1P14751 mutation, which carries a P element insertion in the first intron of the gene. dBlimp-1P14751 homozygous mutants show disintegration of the tracheal network, closely resembling that of the deficiency strain (34), and die before hatching. Introduction of this mutation into ActGAL4>UAS-Blimp-1 flies clearly enhanced the RNAi phenotype (Fig. 5A, lower panel), strongly suggesting that the observed RNAi phenotype was caused by a specific effect on the dBlimp-1 gene and that dBlimp-1 is required for metamorphosis to be completed and may be involved in regulating the prepupal-to-pupal transition.
FIG. 5.
dBlimp-1 has repressor activity and controls the timing of βFTZ-F1 expression and pupation. (A) Phenotypes of dBlimp-1 RNAi lines. Eight independent UAS-dBlimp-1i lines were mated with the Act-Gal4 line, and prepupae of their progeny were collected and their lethal phases were scored from the prepupal stage to adult. The effect of mutation in the dBlimp-1 gene by P-element insertion (dBlimp-1P14751) was examined for the UAS-dBlimp-1i1 and -5 lines. Numbers in parentheses represent the number of scored animals. Typical examples of arrested animals are shown on the right. (B) Reduction of dBlimp-1 transcript levels in RNAi animals. Expression levels were measured by quantitative real-time RT-PCR using total RNA from prepupae at 2 h APF in the indicated lines. The same template was used to measure the amounts of dBlimp-1 and E75A mRNAs. (C) Premature expression of βFTZ-F1 by RNAi of dBlimp-1. The expression of βFTZ-F1 in dBlimp-1 RNAi (Act5c-Gal4>UAS-dBlimp-1i8) and control (+/UAS-dBlimp-1i8) animals from 4 to 10 h APF was estimated by Western blotting. Anti-α-tubulin antibody was used to confirm the amount of loaded protein in each lane. (D) Delay of βFTZ-F1 expression by induction of dBlimp-1. Prepupae at 5 h APF of the hs-dBlimp-1 line or the host strain were heat shocked at 34°C for 1 h and then reared at 25°C. The expression levels of βFTZ-F1 in two individual prepupae at the indicated times were estimated by Western blotting. Anti-α-tubulin antibody was used to confirm the amount of protein loaded in each lane. (E) Delay of pupation timing by forced induction of dBlimp-1. Prepupae at 5 h APF of the hs-dBlimp-1 line or the host strain were heat shocked at 34°C for 1 h and then reared at 25°C. Numbers of newly pupated animals were counted every hour, and the percentage of pupated animals in each period was plotted. Numbers in parentheses represent the number of scored animals. Four out of 26 animals of the hs-dBlimp-1 line failed to pupate.
Because dBlimp-1 is thought to bind to the cis-regulatory region of ftz-f1, we analyzed the effect of RNAi on βFTZ-F1 expression during the prepupal period by Western blotting (Fig. 5C). Two independent Act5c-GAL4>UAS-dBlimp-1i8 lines that showed prepupal lethality were collected every 2 h and subjected to Western blot analysis to obtain reliable results. In the animals from the control line, high-level expression of βFTZ-F1 was detected from 8 to 10 h APF, as previously reported (33), and very low-level expression was occasionally detected at 6 h APF. However, in dBlimp-1 knockdown animals, high-level expression was detected even at 6 h APF (Fig. 5C, lanes 3 and 4). Although the penetrance of this phenotype was not 100%, the higher-level expression at 6 h APF was observed in 60% of animals in duplicate experiments using two independently established RNAi lines. Furthermore, the high-level expression in RNAi animals persisted only until 8 h APF; by 10 h APF, the expression level was greatly reduced. These results suggest that dBlimp-1 prevents premature expression of the ftz-f1 gene by acting as a transcriptional repressor during the high-ecdysone periods.
Prolonged expression of dBlimp-1 results in reduced βFTZ-F1 expression and delayed pupation.
To test the possibility that dBlimp-1 functions as a transcriptional repressor of ftz-f1, we established transgenic hs-dBlimp-1 lines that express dBlimp-1 under the control of the heat shock promoter and analyzed the effect of forced dBlimp-1 expression on the expression of βFTZ-F1 during the prepupal period by Western blotting. When prepupae of the hs-dBlimp-1 line were treated at 34°C for 1 h at 5 h APF, the expression level of βFTZ-F1 was significantly reduced in prepupae at 8 h APF, although the same treatment did not cause any effect on βFTZ-F1 expression in the control animals (Fig. 5D). This result supports the idea that dBlimp-1 acts as a repressor for the ftz-f1 gene. To further explore the effect of prolonged dBlimp-1 expression, heat-treated animals were observed at later developmental stages. The hs-dBlimp-1 animals exhibited a delay in pupation (Fig. 5E), suggesting that dBlimp-1 has an important role not only in controlling the timing of βFTZ-F1 expression but also in pupation.
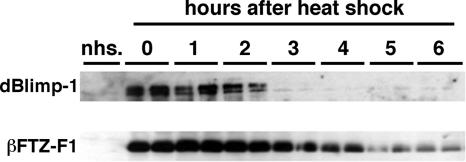
dBlimp-1 protein is unstable.
As the timing of dBlimp-1 expression is important for temporal regulation of the ecdysone-induced pathway and our results suggested that dBlimp-1 mRNA might be unstable, we examined the stability of dBlimp-1 directly. Transgenic fly lines expressing Flag-tagged dBlimp-1 under the control of the heat shock promoter were established, and prepupae were heat shocked at 0 h APF for 1 h at 37°C and examined by Western blotting using anti-Flag antibody (Fig. 6). Strong expression of dBlimp-1 was detectable soon after the heat shock, and the protein level rapidly decreased and became undetectable 3 h after heat induction. A similar turnover profile was observed using heat shock-induced dBlimp-1 without the Flag tag (data not shown). In contrast, such rapid turnover was not observed for heat shock-induced βFTZ-F1, which was detectable at least 6 h after heat shock. This prolonged βFTZ-F1 protein expression is not caused by the stability of its mRNA, because induced βFTZ-F1 mRNA disappeared within 3 h after heat shock (data not shown). These results indicate that dBlimp-1 protein is less stable than βFTZ-F1.
FIG. 6.
dBlimp-1 is a labile protein. dBlimp-1 or βFTZ-F1 was expressed under the control of the heat shock promoter at 0 h APF, and the level of induced protein was detected by Western blotting using either an anti-Flag antibody or anti-βFTZ-F1 serum. Samples from two independent animals of the hs-Blimp-1 or hs-βFTZ-F1 line were examined every hour. Non-heat-shock control animals were examined at 0 h APF.
DISCUSSION
Over 30 years ago, the existence of a factor that is directly induced by ecdysone and that represses early genes at the onset of metamorphosis was proposed based on observations of chromosomal puff patterns in cultured salivary glands (3). Here, we show that the expression profile of dBlimp-1 mRNA mirrors that of a typical early gene and that dBlimp-1 transcript is directly induced by 20E in cultured salivary glands. Similar findings from cultured organs in a recent independent report corroborate our results (7). Moreover, we demonstrate that dBlimp-1 acts as a repressor, making it a good candidate for the factor predicted by Ashburner and colleagues (3). Intriguingly, although there is no direct evidence showing that dBlimp-1 can repress early genes, the 5′ upstream region of the early gene br contains putative dBlimp-1 binding sites. On the other hand, the map position of dBlimp-1 was not identified as an early puff locus. This might be due to the low level of dBlimp-1 expression compared to that of other early genes. Further study is necessary to examine the effect of dBlimp-1 on early genes.
Although the dBlimp-1 transcript is directly induced by 20E, its expression profile is slightly delayed compared to that of the E75A transcript. The delay in its disappearance APF and rough coincidence between the time of its disappearance and the decline of the ecdysone level suggest that the gene may not be repressed by early gene product as are known early genes. Rather, its transcription may require the continual presence of 20E so that it is reduced when the ecdysteroid titer falls. Since the ecdysteroid titer is already quite high at 3 h before puparium formation, the reason for the delay in the appearance of dBlimp-1 mRNA is not clear. These questions require further study.
The discovery of an ecdysone-inducible repressor provides new insights into the regulatory mechanisms of ftz-f1, which is induced by pulses of ecdysone. Although the ecdysone-inducible transcription factors DHR3 and E75B were previously identified (8, 20, 27, 52), they cannot entirely account for the regulatory mechanism for βFTZ-F1 expression. Our results show that the timing of βFTZ-F1 expression is altered in prepupae in which dBlimp-1 is knocked down or expressed for a longer period of time, indicating that the timing of dBlimp-1 expression is crucial for temporal control of the ftz-f1 gene. In our RNAi experiment, however, we observed only 2-h-earlier expression. This might be due to the incomplete knockdown of dBlimp-1 (see below) or to other redundant repression mechanisms, such as DHR3 and E75B. In spite of these unresolved questions, our results provide clear evidence that dBlimp-1 plays a key role in determining the timing of ftz-f1 expression by acting as a repressor during the high-ecdysone period at the onset of metamorphosis.
We also obtained unexpected evidence suggesting that the turnover rate of dBlimp-1 mRNA is quite high. Whereas dBlimp-1 mRNA levels increased upon addition of cycloheximide in cultured organs, other ecdysone-induced early genes, including br (6), E74A (7, 47), and E75A and E75B (44), did not show significant increases in mRNA levels during the 2 hours of culture. The instability of dBlimp-1 mRNA may have affected our RNAi experiment, in which we were able to reduce the level of dBlimp-1 mRNA only to one-fourth of the normal level at 2 h APF.
Furthermore, we found that transgenic dBlimp-1 protein expressed under the control of the heat shock promoter disappeared rapidly. In contrast, BR-C proteins, which are early gene products, have been shown to persist as long as βFTZ-F1 when expressed under the control of the heat shock promoter (13, 26). Furthermore, the dBlimp-1 mRNA peak detected by Northern blotting and the protein activity peak detected by gel mobility shift assay coincided well. In contrast, the protein peaks for other ecdysone-induced transcription factors, such as E74A (10) and E75B (8, 52) were roughly 2 h later than their mRNA peaks. These observations support the idea that the rate of degradation of endogenous dBlimp-1 is also higher than those of many other ecdysone-inducible transcription factors. The degradation rate of each protein is controlled by signals within its own sequence. For example, PEST sequences are proline-, glutamic acid-, serine-, and threonine-rich sequences that target proteins for degradation (36, 41). Indeed, dBlimp-1 contains a proline-rich PEST sequence that may be responsible for its instability, since removal of this region stabilizes protein expressed under the control of the heat shock promoter (M. Sarhan and H. Ueda, unpublished data). Whatever the mechanism of the instability, our results indicate that instability of dBlimp-1 mRNA and protein plays a crucial role in determining the timing of βFTZ-F1 expression and pupation. In strongly affected dBlimp-1 RNAi lines, most animals arrested development at the prepupal stage and expressed βFTZ-F1 prematurely. We have previously shown that premature expression of βFTZ-F1 during the prepupal period causes developmental arrest at the prepupal stage (55). Thus, developmental arrest in the dBlimp-1 RNAi animals might be mediated through the premature expression of βFTZ-F1. On the other hand, we found that forced expression of dBlimp-1 caused delays in the timing of both βFTZ-F1 expression and pupation. Thus, the timing of pupation might be controlled by the timing of βFTZ-F1 expression.
Recently, it has been reported that dBlimp-1 expression in the tracheal system in Drosophila embryos is important for development of this tissue (34). In addition, dBlimp-1 is expressed in a spatially restricted manner in other regions during early embryogenesis, although the functions of these early-expression domains remain unknown. Blimp-1 is similarly expressed in many different tissues in vertebrates, where it is known to play important roles in embryogenesis, germ cell determination, specification in nerve and muscle cells, linage determination in epidermis, and B-cell maturation (5, 11, 12, 14, 18, 35, 42, 49, 53). Thus, dBlimp-1 may be involved in many other developmental events in the fly.
Acknowledgments
We thank all members of the Hirose laboratory for helping us and giving useful comments during our research, especially K. Nishioka for critical advice on antibody preparation and H. Furuhashi for help with quantitative PCR. We thank K. Kusano, A. Ishihama, H. Araki, S. Hayashi, and Y. Hiromi for helpful discussions about our research. We are grateful to A. J. Andres, T. Aigaki, and the Genetic Stock Research Center, National Institute of Genetics, for providing fly strains.
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology. Y. Agawa and Y. Kageyama were supported by the Center of Excellence Program of Japan and the Japan Society for the Promotion of Science, respectively. M. Sarhan was supported by the Ministry of Higher Education, Cultural Affairs and Missions Sector, Egyptian government.
Footnotes
Published ahead of print on 8 October 2007.
REFERENCES
- 1.Ashburner, M. 1972. Patterns of puffing activity in the salivary gland chromosomes of Drosophila. VI. Induction by ecdysone in salivary glands of D. melanogaster cultured in vitro. Chromosoma 38:255-281. [DOI] [PubMed] [Google Scholar]
- 2.Ashburner, M. 1974. Sequential gene activation by ecdysone in polytene chromosomes of Drosophila melanogaster. II. The effects of inhibitors of protein synthesis. Dev. Biol. 39:141-157. [DOI] [PubMed] [Google Scholar]
- 3.Ashburner, M., C. Chihara, P. Meltzer, and G. Richards. 1974. Temporal control of puffing activity in polyten chromosome. Cold Spring Harbor Symp. Quant. Biol. 38:655-662. [DOI] [PubMed] [Google Scholar]
- 4.Ashburner, M., and G. Richards. 1976. Sequential gene activation by ecdysone in polytene chromosomes of Drosophila melanogaster. III. Consequences of ecdysone withdrawal. Dev. Biol. 54:241-255. [DOI] [PubMed] [Google Scholar]
- 5.Baxendale, S., C. Davison, C. Muxworthy, C. Wolff, P. W. Ingham, and S. Roy. 2004. The B-cell maturation factor Blimp-1 specifies vertebrate slow-twitch muscle fiber identity in response to Hedgehog signaling. Nat. Genet. 36:88-93. [DOI] [PubMed] [Google Scholar]
- 6.Bayer, C. A., B. Holley, and J. W. Fristrom. 1996. A switch in broad-complex zinc-finger isoform expression is regulated posttranscriptionally during the metamorphosis of Drosophila imaginal discs. Dev. Biol. 177:1-14. [DOI] [PubMed] [Google Scholar]
- 7.Beckstead, R. B., G. Lam, and C. S. Thummel. 2005. The genomic response to 20-hydroxyecdysone at the onset of Drosophila metamorphosis. Genome Biol. 6:R99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bialecki, M., A. Shilton, C. Fichtenberg, W. A. Segraves, and C. S. Thummel. 2002. Loss of the ecdysteroid-inducible E75A orphan nuclear receptor uncouples molting from metamorphosis in Drosophila. Dev. Cell 3:209-220. [DOI] [PubMed] [Google Scholar]
- 9.Biyasheva, A., T. V. Do, Y. Lu, M. Vaskova, and A. J. Andres. 2001. Glue secretion in the Drosophila salivary gland: a model for steroid-regulated exocytosis. Dev. Biol. 231:234-251. [DOI] [PubMed] [Google Scholar]
- 10.Boyd, L., E. O'Toole, and C. S. Thummel. 1991. Patterns of E74A RNA and protein expression at the onset of metamorphosis in Drosophila. Development 112:981-995. [DOI] [PubMed] [Google Scholar]
- 11.Chang, D. H., C. Angelin-Duclos, and K. Calame. 2000. BLIMP-1: trigger for differentiation of myeloid lineage. Nat. Immunol. 1:169-176. [DOI] [PubMed] [Google Scholar]
- 12.Chang, D. H., G. Cattoretti, and K. L. Calame. 2002. The dynamic expression pattern of B lymphocyte induced maturation protein-1 (Blimp-1) during mouse embryonic development. Mech. Dev. 117:305-309. [DOI] [PubMed] [Google Scholar]
- 13.Crossgrove, K., C. A. Bayer, J. W. Fristrom, and G. M. Guild. 1996. The Drosophila Broad-Complex early gene directly regulates late gene transcription during the ecdysone-induced puffing cascade. Dev. Biol. 180:745-758. [DOI] [PubMed] [Google Scholar]
- 14.de Souza, F. S., V. Gawantka, A. P. Gomez, H. Delius, S. L. Ang, and C. Niehrs. 1999. The zinc finger gene Xblimp1 controls anterior endomesodermal cell fate in Spemann's organizer. EMBO J. 18:6062-6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edgar, B. A., M. P. Weir, G. Schubiger, and T. Kornberg. 1986. Repression and turnover pattern fushi tarazu RNA in the early Drosophila embryo. Cell 47:747-754. [DOI] [PubMed] [Google Scholar]
- 16.Handa, H., Y. Yamaguchi, and T. Wada. 1999. Purification of DNA binding poteins. Oxford University Press, Ocford, United Kingdom.
- 17.Hiruma, K., and L. M. Riddiford. 2001. Regulation of transcription factors MHR4 and βFTZ-F1 by 20-hydroxyecdysone during a larval molt in the tobacco hornworm, Manduca sexta. Dev. Biol. 232:265-274. [DOI] [PubMed] [Google Scholar]
- 18.Horsley, V., D. O'Carroll, R. Tooze, Y. Ohinata, M. Saitou, T. Obukhanych, M. Nussenzweig, A. Tarakhovsky, and E. Fuchs. 2006. Blimp1 defines a progenitor population that governs cellular input to the sebaceous gland. Cell 126:597-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hurban, P., and C. S. Thummel. 1993. Isolation and characterization of 15 ecdysone-inducible Drosophila genes reveal unexpected complexities in ecdysone regulation. Mol. Cell. Biol. 13:7101-7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kageyama, Y., S. Masuda, S. Hirose, and H. Ueda. 1997. Temporal regulation of the mid-prepupal gene FTZ-F1: DHR3 early late gene product is one of the plural positive regulators. Genes Cells 2:559-569. [DOI] [PubMed] [Google Scholar]
- 21.Karim, F. D., and C. S. Thummel. 1992. Temporal coordination of regulatory gene expression by the steroid hormone ecdysone. EMBO J. 11:4083-4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keller, A. D., and T. Maniatis. 1991. Identification and characterization of a novel repressor of β-interferon gene expression. Genes Dev. 5:868-879. [DOI] [PubMed] [Google Scholar]
- 23.Kongsuwan, K., Q. Yu, A. Vincent, M. C. Frisardi, M. Rosbash, J. A. Lengyel, and J. Merriam. 1985. A Drosophila Minute gene encodes a ribosomal protein. Nature 317:555-558. [DOI] [PubMed] [Google Scholar]
- 24.Kouzarides, T. 2002. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 12:198-209. [DOI] [PubMed] [Google Scholar]
- 25.Kozlova, T., and C. S. Thummel. 2003. Essential roles for ecdysone signaling during Drosophila mid-embryonic development. Science 301:1911-1914. [DOI] [PubMed] [Google Scholar]
- 26.Kucharova-Mahmood, S., I. Raska, B. M. Mechler, and R. Farkas. 2002. Temporal regulation of Drosophila salivary gland degeneration by the Broad-Complex transcription factors. J. Struct. Biol. 140:67-78. [DOI] [PubMed] [Google Scholar]
- 27.Lam, G., B. L. Hall, M. Bender, and C. S. Thummel. 1999. DHR3 is required for the prepupal-pupal transition and differentiation of adult structures during Drosophila metamorphosis. Dev. Biol. 212:204-216. [DOI] [PubMed] [Google Scholar]
- 28.Lam, G. T., C. Jiang, and C. S. Thummel. 1997. Coordination of larval and prepupal gene expression by the DHR3 orphan receptor during Drosophila metamorphosis. Development 124:1757-1769. [DOI] [PubMed] [Google Scholar]
- 29.Lavorgna, G., F. D. Karim, C. S. Thummel, and C. Wu. 1993. Potential role for a FTZ-F1 steroid receptor superfamily member in the control of Drosophila metamorphosis. Proc. Natl. Acad. Sci. USA 90:3004-3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lavorgna, G., H. Ueda, J. Clos, and C. Wu. 1991. FTZ-F1, a steroid hormone receptor-like protein implicated in the activation of fushi tarazu. Science 252:848-851. [DOI] [PubMed] [Google Scholar]
- 31.Li, C., M. Z. Kapitskaya, J. Zhu, K. Miura, W. Segraves, and A. S. Raikhel. 2000. Conserved molecular mechanism for the stage specificity of the mosquito vitellogenic response to ecdysone. Dev. Biol. 224:96-110. [DOI] [PubMed] [Google Scholar]
- 32.Marmorstein, R. 2003. Structure of SET domain proteins: a new twist on histone methylation. Trends Biochem. Sci. 28:59-62. [DOI] [PubMed] [Google Scholar]
- 33.Murata, T., Y. Kageyama, S. Hirose, and H. Ueda. 1996. Regulation of the EDG84A gene by FTZ-F1 during metamorphosis in Drosophila melanogaster. Mol. Cell. Biol. 16:6509-6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ng, T., F. Yu, and S. Roy. 2006. A homologue of the vertebrate SET domain and zinc finger protein Blimp-1 regulates terminal differentiation of the tracheal system in the Drosophila embryo. Dev. Genes Evol. 216:243-252. [DOI] [PubMed] [Google Scholar]
- 35.Ohinata, Y., B. Payer, D. O'Carroll, K. Ancelin, Y. Ono, M. Sano, S. C. Barton, T. Obukhanych, M. Nussenzweig, A. Tarakhovsky, M. Saitou, and M. A. Surani. 2005. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 436:207-213. [DOI] [PubMed] [Google Scholar]
- 36.Rechsteiner, M., and S. W. Rogers. 1996. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 21:267-271. [PubMed] [Google Scholar]
- 37.Ren, B., K. J. Chee, T. H. Kim, and T. Maniatis. 1999. PRDI-BF1/Blimp-1 repression is mediated by corepressors of the Groucho family of proteins. Genes Dev. 13:125-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richards, G. 1976. Sequential gene activation by ecdysone in polytene chromosomes of Drosophila melanogaster. IV. The mid prepupal piriod. Dev. Biol. 54:264-275. [DOI] [PubMed] [Google Scholar]
- 39.Richards, G., J. L. Da Lage, F. Huet, and C. Ruiz. 1999. The acquisition of competence to respond to ecdysone in Drosophila is transcript specific. Mech. Dev. 82:131-139. [DOI] [PubMed] [Google Scholar]
- 40.Riddiford, L. M. 1993. Hormones and Drosophila Development, p. 899-940. In M. Bate and A. Martinez-Arias (ed.), The development of Drosophila melanogaster, vol. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 41.Rogers, S., R. Wells, and M. Rechsteiner. 1986. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science 234:364-368. [DOI] [PubMed] [Google Scholar]
- 42.Roy, S., and T. Ng. 2004. Blimp-1 specifies neural crest and sensory neuron progenitors in the zebrafish embryo. Curr. Biol. 14:1772-1777. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 44.Segraves, W. A., and D. S. Hogness. 1990. The E75 ecdysone-inducible gene responsible for the 75B early puff in Drosophila encodes two new members of the steroid receptor superfamily. Genes Dev. 4:204-219. [DOI] [PubMed] [Google Scholar]
- 45.Sullivan, A. A., and C. S. Thummel. 2003. Temporal profiles of nuclear receptor gene expression reveal coordinate transcriptional responses during Drosophila development. Mol. Endocrinol. 17:2125-2137. [DOI] [PubMed] [Google Scholar]
- 46.Sun, G. C., S. Hirose, and H. Ueda. 1994. Intermittent expression of BmFTZ-F1, a member of the nuclear hormone receptor superfamily, during development of the silkworm Bombyx mori. Dev. Biol. 162:426-437. [DOI] [PubMed] [Google Scholar]
- 47.Thummel, C. S., K. C. Burtis, and D. S. Hogness. 1990. Spatial and temporal patterns of E74 transcription during Drosophila development. Cell 61:101-111. [DOI] [PubMed] [Google Scholar]
- 48.Tunyaplin, C., M. A. Shapiro, and K. L. Calame. 2000. Characterization of the B lymphocyte-induced maturation protein-1 (Blimp-1) gene, mRNA isoforms and basal promoter. Nucleic Acids Res. 28:4846-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turner, C. A., Jr., D. H. Mack, and M. M. Davis. 1994. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell 77:297-306. [DOI] [PubMed] [Google Scholar]
- 50.Ueda, H., and S. Hirose. 1990. Identification and purification of a Bombyx mori homologue of FTZ-F1. Nucleic Acids Res. 18:7229-7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ueda, H., S. Sonoda, J. L. Brown, M. P. Scott, and C. Wu. 1990. A sequence-specific DNA-binding protein that activates fushi tarazu segmentation gene expression. Genes Dev. 4:624-635. [DOI] [PubMed] [Google Scholar]
- 52.White, K. P., P. Hurban, T. Watanabe, and D. S. Hogness. 1997. Coordination of Drosophila metamorphosis by two ecdysone-induced nuclear receptors. Science 276:114-117. [DOI] [PubMed] [Google Scholar]
- 53.Wilm, T. P., and L. Solnica-Krezel. 2005. Essential roles of a zebrafish prdm1/blimp1 homolog in embryo patterning and organogenesis. Development 132:393-404. [DOI] [PubMed] [Google Scholar]
- 54.Woodard, C. T., E. H. Baehrecke, and C. S. Thummel. 1994. A molecular mechanism for the stage specificity of the Drosophila prepupal genetic response to ecdysone. Cell 79:607-615. [DOI] [PubMed] [Google Scholar]
- 55.Yamada, M., T. Murata, S. Hirose, G. Lavorgna, E. Suzuki, and H. Ueda. 2000. Temporally restricted expression of transcription factor βFTZ-F1: significance for embryogenesis, molting and metamorphosis in Drosophila melanogaster. Development 127:5083-5092. [DOI] [PubMed] [Google Scholar]