Abstract
The nucleolus has begun to emerge as a subnuclear organelle capable of modulating the activities of nuclear proteins in a dynamic and cell type-dependent manner. It remains unclear whether one can extrapolate a rule that predicts the nucleolar localization of multiple proteins based on protein sequence. Here, we address this issue by determining the shared and unique mechanisms that regulate the static and dynamic distributions of a family of nucleolar GTP-binding proteins, consisting of nucleostemin (NS), guanine nucleotide binding protein-like 3 (GNL3L), and Ngp1. The nucleolar residence of GNL3L is short and primarily controlled by its basic-coiled-coil domain, whereas the nucleolar residence of NS and Ngp1 is long and requires the basic and the GTP-binding domains, the latter of which functions as a retention signal. All three proteins contain a nucleoplasmic localization signal (NpLS) that prevents their nucleolar accumulation. Unlike that of the basic domain, the activity of NpLS is dynamically controlled by the GTP-binding domain. The nucleolar retention and the NpLS-regulating functions of the G domain involve specific residues that cannot be predicted by overall protein homology. This work reveals common and protein-specific mechanisms underlying the nucleolar movement of NS family proteins.
The nucleolus is a nonmembrane-bound subnuclear organelle where ribosome biogenesis takes place (7, 40). Increasing evidence has shown that the nucleolus is also involved in nonribosomal activities, the best-characterized example of which is the assembly of signal recognition particle (11, 15, 36). Proteins involved in cell cycle regulation, cell growth, telomere maintenance, and protein degradation have also been spotted in the nucleolus (26, 30). Many of these nonconventional nucleolar proteins are primarily localized in or transiently associated with other nuclear subdomains, suggesting that nucleolar localization may serve as a general mechanism to modulate the functions of nuclear and viral proteins that may not accumulate in the nucleolus in steady state. Recent work has begun to build a dynamic picture of nuclear proteins moving between different subnuclear compartments (1, 2, 4, 6, 9, 14, 18, 19, 22, 23, 25, 27, 33, 35, 38, 42, 43). The shuttling speed of proteins between the nucleolus and the nucleoplasm is several orders of magnitude faster than most transcriptional and translational processes, which allows cells to respond to a variety of environmental stimuli in a rapid and dynamic fashion (10). Therefore, understanding this molecular behavior will provide crucial information on the fast regulation of biological events.
Unlike nucleocytoplasmic translocation, proteins accumulate in the nucleolus via multiple mechanisms involving protein-protein, protein-RNA, and protein-DNA interactions. To date, no strong consensus sequence has been identified as the nucleolar localization signal (NoLS) for multiple proteins in a necessary and sufficient manner (3, 17, 31, 34, 41, 45), and very little is known about how proteins accumulate in the nucleolus and move between different subnuclear compartments at the molecular level. A common feature of most nucleolar localization regions is a stretch of positively charged residues that often overlap with the predicted nuclear localization signal. Yet, proteins with basic residue repeats do not always accumulate in the nucleolus. In addition, nucleolar localization can be modulated by signals beyond the primary protein sequence, such as GTP binding, H+ ion concentration, and phosphorylation (3, 18, 24, 38). In previous work, we used a stem cell-enriched factor, nucleostemin (NS), as a model molecule to address the mechanism controlling the dynamic distribution of nucleolar proteins. We have shown that the partitioning of NS between the nucleolus and the nucleoplasm involves multiple components and that GTP binding plays a key role in driving the dynamic cycling of NS (20, 21, 38, 39). The basic (B) domain of NS alone can accumulate in the nucleolus but is not sufficient to recapitulate its dynamic movement. Further modification of its nucleolar entry and retention is achieved by its GTP-binding (G) domain and the adjacent intermediate (I) domain. It is not entirely clear which of these mechanisms is shared by other proteins and which is unique for NS.
We reason that the conservation and divergence of nucleolar localization mechanisms may best be revealed by looking at multiple proteins that are structurally and evolutionarily related. The gene encoding NS is a member of a gene family that features an MMR_HSR1 domain. The MMR_HSR1 domain consists of five GTP-binding motifs arranged in a circularly permuted order, where a highly conserved G5 variant motif (hereafter referred to as a G5* motif) (DARXP) and the G4 (NKXDL) motif are positioned N terminally to the G1 (GXPNVGKSS), G2 (GXT), and G3 (DXPG) motifs (5, 16). Members of the MMR_HSR1 family are localized in different subcellular compartments of organisms from single-celled microorganisms to high vertebrates (32). Among them, NS, GNL3L (guanine nucleotide binding protein-like 3), and Ngp-1 (hereafter referred to as Ngp1) (28) form a subfamily of proteins found in the nucleolus. In this study, we dissect the shared and unique mechanisms controlling the nucleolar distribution of these three proteins. Based on our findings, we propose a model that depicts the nucleolar localization action as a combination of a positively charged region and a nucleoplasmic localization signal (NpLS), both of which can be further modulated in a protein-specific manner by a retention and an NpLS-regulating signal in the G domain.
MATERIALS AND METHODS
Phylogenetic analysis.
A rooted phylogenetic tree for NS family genes was drawn from ClustalW-aligned sequences using PHYLIP Drawgram version 3.2, available at http://workbench.sdsc.edu (8, 37).
Generation of deletion, point mutation, chimeric, and heterologous fusion constructs.
Deletions, point mutations, and chimeric mutants were created by using the stitching PCR strategy as described previously (38, 39). The final products were subcloned into various expression vectors at the SalI and NotI sites for the N-terminal fusion or the SalI and AgeI sites for the C-terminal fusion. To generate C-terminally green fluorescent protein (GFP)-fused constructs, PCR fragments were subcloned into the pEGFP-N1 vector (Clontech). The N-terminal Myc epitope (EQKLISEED; EcoRI-SalI fragment) and the C-terminal hemagglutinin (HA) epitope (YPYDVPDYA; AgeI-NotI fragment) were engineered into the pCIS expression vector. All constructs were confirmed by sequencing reactions.
GTP-binding assay.
GTP-binding assays were conducted as described previously (13, 38), using 1 μg of purified proteins and 80 μl of GTP-conjugated agarose (2.2 μmol/ml). After an extensive wash procedure, the amount of protein retained by the GTP-conjugated agarose was fractionated on a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel and detected by Western blotting.
Cell culture, transfection, indirect immunofluorescence, and image acquisition.
U2OS cells were used for all analyses in this study. Cells were maintained in Dulbecco's modified Eagle's medium supplemented with 5% fetal bovine serum (HyClone), penicillin (50 IU/ml), streptomycin (50 μg/ml), and glutamine (1%). Plasmid transfections were performed using Lipofectamine-plus reagent (Invitrogen) and analyzed 1 day after transfection. Immunofluorescence studies were performed as described previously (20, 38). The primary antibodies included monoclonal anti-HA antibody (1:2,000) (HA.11; Covance), monoclonal anti-Myc antibody (1:2,000) (9E10; Covance), monoclonal antifibrillarin antibody (1:1,000) (38F3; EnCor), and monoclonal anti-B23 antibody (1:4,000) (Zymed). All images were acquired by using a Zeiss LSM510 confocal microscope with a 63× plan-apochromat oil objective (numerical aperture, 1.4) and scanned with a 512 by 512 frame size, 3× zoom, and <1.4-μm optical thickness. The detector gain and amplifier offset were adjusted to ensure that all signals were appropriately displayed within the linear range of intensities.
FRAP and FLIP.
U2OS cells were grown on Nalgene Lab Tek II chamber slides and transfected with 0.6 μg plasmid DNA 1 day before the measurement. Bleaching experiments were performed on a Zeiss LSM510 confocal microscope with a 63× plan-apochromat oil objective. The GFP signal was excited with a 488-nm argon laser (21 mW nominal output), and emissions above 505 nm were monitored. The cells were maintained at 35°C with a heat blower throughout the entire course of the fluorescence recovery after photobleaching (FRAP) and fluorescence loss in photobleaching (FLIP) experiments. To minimize the evaporation of the medium, the chambers were covered with lids bearing a small opening to allow contact between the thermosensor probe and the medium. The FRAP paradigm, modified based on previous reports (6, 27), was designed such that a circle of 1 μm in diameter was bleached inside the nucleolus by using a short laser pulse administered at 70% of the power of the 488-nm argon laser (21 mW nominal output) for three iterations which lasted for 256 ms. For image acquisition, the laser power was attenuated to 0.6% of the bleaching intensity, and cells were scanned with 5× zoom at 0.5-s intervals for 45 s after photobleaching. For quantification, the fluorescence intensities of the bleached area, the entire nucleus, and the area outside of the nucleus were measured. The relative fluorescence intensity (RFI) in the bleached area was normalized to the total intensity in the nucleus after background subtraction using the following calculation: RFI = (It/I0) × (TN0/TNt), where It and I0 are the background-subtracted intensities of the bleached spot at time point t and before photobleaching, respectively, and TNt and TN0 are the background-subtracted intensities of the entire nucleus at time point t and before photobleaching, respectively. Cells with a signal loss of >10% during the imaging phase were discarded. FLIP experiments were performed in which a 2- by 4-μm rectangular region in the nucleoplasm was bleached with repetitive pulses at 70% of the power of the 488-nm argon laser, with a 150-ms duration per pulse and a 0.59-s interval between consecutive pulses. For image acquisition, the laser power was attenuated to 0.6% of the bleach intensity, and the cells were scanned with 3× zoom at 0.74-s intervals. The RFI in the nucleolus of bleached cells was normalized to the nucleolar intensity of neighboring nonbleached cells after background subtraction using the following calculation: RFI = (It/I0) × (C0/Ct), where It and I0 are the background-subtracted intensities of the nucleolus in the bleached cell at time point t and before photobleaching, respectively, and Ct and C0 are the background-subtracted intensities of the nucleolus in the neighboring control cell at time point t and before photobleaching, respectively. Both the FRAP and the FLIP data represent the averages of the results for over 20 cells from two to four independent experiments.
RESULTS
NS, GNL3L, and Ngp1 constitute a subfamily of GTP-binding proteins with distinct nucleolar localization properties.
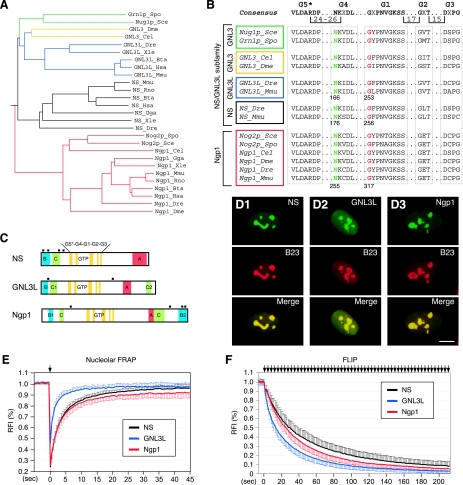
A genome-wide search revealed only two genes (whose products are GNL3L and Ngp1) that display high homology with the NS gene in humans. Phylogenetic analyses showed that NS and GNL3L share higher homology with each other than with Ngp1 (Fig. 1A). While Ngp1 is represented by a single gene from yeasts to humans, NS and GNL3L share the same orthologues in yeast (Saccharomyces cerevisiae), fruit flies, and pinworms. Only in vertebrates do they begin to emerge as separate genes. The yeast, fly, and worm orthologues of NS and GNL3L are more similar to the human GNL3L than to the human NS in their protein sequences. A key feature of the NS gene family is the MMR_HSR1 domain that contains five highly conserved GTP-binding motifs arranged in a circularly permutated order (G5*-G4-G1-G2-G3). The Asn176 and Gly256 residues in the G4 and G1 motifs of NS, which are important for mediating the GTP binding of NS (38), are also conserved in GNL3L and Ngp1 (Fig. 1B). In addition to the MMR_HSR1 domain, one or two B domains, one or two coiled-coil (C) structures, an acidic (A) domain, and multiple nuclear localization signals are found in all of the NS family proteins (Fig. 1C). Structurally, NS and GNL3L differ from Ngp1 in the number and position(s) of the B domain(s) and in the spacing between the G4 and G1 motifs (81 bp, 75 bp, and 57 bp for NS, GNL3L, and Ngp1, respectively).
FIG. 1.
NS, GNL3L, and Ngp1 represent a subfamily of GTP-binding proteins with distinct nucleolar localization properties. (A) A rooted phylogenetic tree of NS, GNL3L, and Ngp1 drawn with PHYLIP Drawgram version 3.2. The vertebrate NS genes (black lines) and the GNL3L genes (blue lines) share the same orthologues in the yeast (green lines), fruit fly, and pinworm (yellow lines), whereas the Ngp1 gene (red lines) is the same from yeast to humans. Species abbreviations: Bta, Bos taurus; Cel, Caenorhabditis elegans; Dme, Drosophila melanogaster; Dre, Danio rerio; Gga, Gallus gallus; Hsa, Homo sapiens; Mmu, Mus musculus; Rno, Rattus norvegicus; Sce, Saccharomyces cerevisiae; Spo, Schizosaccharomyces pombe; Xle, Xenopus laevis. (B) The protein sequences of the GTP-binding motifs of the NS family genes were aligned, and the positions of the highly conserved Asn residue (green) in the G4 motif and the Gly residue (red) in the G1 motif were numbered at the bottom. (C) Schematic diagrams of mouse NS, GNL3L, and Ngp1. All genes share an MMR_HSR1 structure, consisting of five circularly permuted GTP-binding motifs (G5*, G4, G1, G2, and G3) and some variations in the B, C, and A domains. Black square boxes indicate nuclear localization signals. (D) The subcellular distributions of NS, GNL3L, and Ngp1 in U2OS cells were revealed by a C-terminally fused GFP (green) and counterstained with anti-B23 immunofluorescence (red). Scale bar, 10 μm. (E) The FRAP recovery rates of NS, GNL3L, and Ngp1 were determined in the nucleoli of U2OS cells transfected with their respective GFP fusion constructs. (F) The FLIP rates of NS, GNL3L, and Ngp1 were measured in U2OS cells, where a small region in the nucleoplasm was repeatedly bleached and the loss of fluorescence signal in the nearest nucleolus was measured over time (see Materials and Methods and Fig. S1B in the supplemental material). The y axes represent the RFI (see Materials and Methods) in the bleached area (for FRAP) or in the nonbleached nucleolus (for FLIP). Error bars represent the standard deviations and are omitted on one side of the curves for clarity. Arrows indicate the bleaching events.
The subcellular distributions of the NS family proteins were shown by using a C-terminally fused GFP in U2OS cells (Fig. 1D), as well as by using an N-terminally fused GFP (data not shown). All three proteins are localized primarily in the nucleolus. GNL3L displays a higher nucleoplasmic intensity relative to its nucleolar intensity than NS or Ngp1 does. To determine whether the dynamic properties of GNL3L and Ngp1 resemble those of NS, FRAP experiments were conducted on C-terminal GFP fusion proteins of NS, GNL3L, and Ngp1. A circle of 1 μm in diameter within the nucleolus was bleached, and the fluorescence recovery in the bleached area was recorded for 45 s (Fig. 1E; also see Fig. S1A in the supplemental material). Our results showed that the fluorescence recovery of GNL3L is significantly faster than that of NS, and the GNL3L signal is harder to bleach than the NS and Ngp1 signals using the same bleaching paradigm. Although Ngp1 resembles NS less than GNL3L does in the protein sequence, its FRAP recovery rate is the same as that of NS initially but reaches a plateau that is slightly lower and faster than that of NS. To validate the use of C-terminally GFP-fused proteins to track the dynamic distribution of GNL3L and Ngp1, we measured the FRAP recovery rates of the N-terminally GFP-fused GNL3L and Ngp1 and determined that they are identical to the FRAP kinetics of the C-terminally GFP-fused GNL3L and Ngp1 (see Fig. S2 in the supplemental material). FLIP experiments, where the fluorescence loss in the nucleolus was recorded while a small region in the nucleoplasm was repeatedly bleached, demonstrated that GNL3L has the fastest protein shuttling speed between the nucleolus and the nucleoplasm, followed by Ngp1 and NS in decreasing order (Fig. 1F; also see Fig. S1B in the supplemental material).
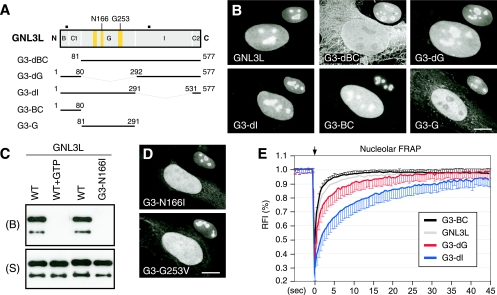
Nucleolar accumulation of GNL3L requires the B-C domain.
To determine the protein structure required for the nucleolar localization of GNL3L, C-terminally GFP-fused GNL3L mutants were examined for their static and dynamic distributions (Fig. 2A). Our data showed that the N-terminal B and C1 domains of GNL3L of 80 amino acids are both necessary and sufficient for mediating nucleolar localization (Fig. 2B, mutants G3-dBC and G3-BC), consistent with the findings of a recent study (29). Deleting the B-C domain also reduces the nuclear translocation of GNL3L, and the cytoplasmic signal displays a significant colocalization with the cytoskeleton. By contrast, deleting the G domain (mutant G3-dG) or the I domain (mutant G3-dI) does not affect the nucleolar accumulation of GNL3L, and the G domain alone does not possess the nucleolar localization capability. To determine whether the distribution of GNL3L is regulated by its GTP-binding property, we first established its ability to bind GTP by using a biochemical assay in which GTP-conjugated agarose was incubated with purified GNL3L protein and washed extensively to remove unbound proteins (13, 38). Western blot analyses showed that GNL3L is retained by the GTP-conjugated agarose (Fig. 2C, WT). The pull-down of GNL3L can be specifically blocked by preincubating the purified GNL3L protein with 10 mM free GTP (Fig. 2C, WT+GTP) or by mutating the Asn166 residue to Ile (mutant G3-N166I). The Asn166 residue in the G4 motif and the Gly253 residue in the G1 motif of GNL3L correspond to the conserved Asn176 and Gly256 residues in NS (Fig. 1B, 2A) that have been shown to be essential for the GTP binding of NS (38). The N166I mutation also mimics the constitutive negative Ras mutant N116I (12). Mutating the Asn166 residue to Ile (mutant G3-N166I) or the Gly253 residue to Val (mutant G3-G253V) disrupts the nucleolar accumulation of GNL3L (Fig. 2D), supporting the idea that GTP binding regulates its nucleolar localization. While the FRAP recovery rate of the B-C domain (mutant G3-BC) resembles that of the full-length GNL3L, a deletion of the G domain (mutant G3-dG) or the I domain (mutant G3-dI) increases the nucleolar residence time of GNL3L (Fig. 2E). A subtle increase in the FRAP recovery rate of mutant G3-BC compared to the recovery rate of the wild-type GNL3L was seen. Whether it is caused by the different molecular weights of these two proteins or not is unclear. These results show that the B-C domain of GNL3L mimics its nucleolar distribution both statically and dynamically. Although the G domain of GNL3L is not sufficient to mediate nucleolar localization and lacks nucleolar retention activity like that of NS, GTP binding plays a regulatory role in the nucleolar accumulation of the full-length GNL3L.
FIG. 2.
The B-C domain of GNL3L is both necessary and sufficient to mediate nucleolar localization. For nucleolar accumulation of the full-length GNL3L, GTP binding is required. (A) Truncated mutants with C-terminally fused GFP were designed to determine the structural requirement for the nucleolar localization of GNL3L. Numbers and black boxes indicate amino acid positions and nuclear localization signals, respectively. Yellow boxes represent, from left to right, the G5*, G1, and G4 motifs. (B) The B-C1 domain is both necessary and sufficient for mediating nucleolar localization. Anti-B23 immunofluorescence of the same cell at a 60% scale is shown in the upper right quadrant of each cell. (C) Wild-type GNL3L (WT) can be retained by GTP-conjugated agarose. The GTP binding of GNL3L is blocked by preincubating GNL3L with 10 mM free GTP (WT+GTP) or by mutating the Asn166 residue in the G4 domain to Ile (mutant G3-N166I). B, bound fraction; S, supernatant. (D) Mutating the Asn166 residue to Ile (mutant G3-N166I) or the Gly253 residue to Val (mutant G3-G253V) perturbs the nucleolar distribution of GNL3L. Anti-B23 staining is shown in the upper right quadrant of each panel. (E) The nucleolar FRAP curves depict the averages of the RFI results in the bleached area relative to the prebleach intensity (set at 1; n = 20) over a 45-s period following photobleaching. Error bars showing standard deviations are omitted on one side for clarity. The FRAP recovery rate of the wild-type protein (GNL3L) mostly resembles the recovery rate of the B-C domain (mutant G3-BC). The arrow indicates the bleach pulse. Scale bars in panels B and D show 10 μm.
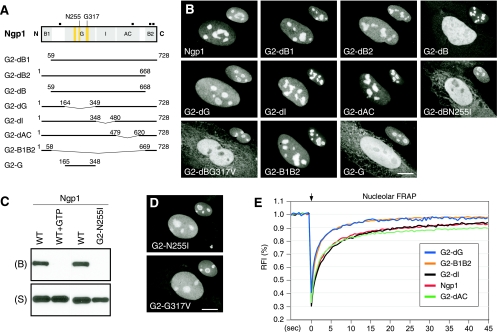
The static and dynamic distributions of Ngp1 are controlled by its B domains and G domain separately.
Deletion and point mutations were created on Ngp1 to identify the protein domains required for its static or dynamic distribution (Fig. 3A). Compared to the nucleolar signal of the full-length protein, the nucleolar signal of Ngp1 is partially dispersed by deletion of the B2 or the G domain (Fig. 3B, mutants G2-dB2 and G2-dG). While a deletion of the B1 domain (mutant G2-dB1), the I domain (mutant G2-dI), or the AC domain (mutant G2-dAC) has no visible effect on the static distribution of Ngp1, deleting both the B1 and B2 domains (mutant G2-dB) abolishes its nucleolar accumulation. When combined with a single-amino-acid mutation on the Asn255 residue (to Ile; mutant G2-dBN255I) or on the Gly317 residue (to Val; mutant G2-dBG317V), this B1-B2 deletion of Ngp1 completely abolishes its nucleolar distribution. The Asn255 and Gly317 residues of Ngp1 correspond to the Asn176 and Gly256 residues of NS, respectively (Fig. 1B and 3A). Notably, the B1-B2 domain alone can accumulate in the nucleolus (mutant G2-B1B2), but the G domain by itself cannot (mutant G2-G). We used GTP-binding assays to confirm that Ngp1 can bind the GTP-conjugated agarose specifically (Fig. 3C, WT versus WT+GTP), and its GTP-binding activity is abolished by the N255I mutation (mutant G2-N255I). While the G2-N255I and G2-G317V mutants still accumulate in the nucleolus, their nucleoplasmic-to-nucleolar intensity ratios are significantly higher than that of the wild-type Ngp1, supporting the idea that GTP binding is involved in the nucleolar localization of Ngp1 (Fig. 3D). Although the G domain alone does not accumulate in the nucleolus, it can increase the nucleolar retention time of Ngp1 (Fig. 3E). The FRAP recovery rate of the G-domain deletion mutant (G2-dG) is faster than the recovery rate of the full-length Ngp1 and identical to that of the B1-B2 domain (mutant G2-B1B2). Deleting the I domain (mutant G2-dI) or the AC domain (mutant G2-dAC) does not alter the FRAP kinetics of Ngp1. These data demonstrate that the nucleolar accumulation of Ngp1 is mediated mostly by the B2 domain and to a lesser extent by the B1 domain and that efficient nucleolar retention of Ngp1 requires the G domain, which is incapable of accumulating in the nucleolus by itself.
FIG. 3.
The B domains of Ngp1 mediate its nucleolar accumulation, and the G domain controls its nucleolar retention time. (A) C-terminally GFP-fused mutants of Ngp1 were constructed to map the protein domains involved in its static and dynamic distributions. Numbers and black boxes indicate amino acid positions and nuclear localization signals, respectively. Bent line segments indicate deleted protein regions. (B) Distribution analyses showed that deleting both the B1 and B2 domains (mutant G2-dB) disrupts the nucleolar accumulation of Ngp1. The B1-B2 deletion plus a mutation on the Asn255 residue in the G4 motif (mutant G2-dBN255I) or on the Gly317 residue in the G1 motif (mutant G2-dBG317V) completely exclude the protein from the nucleolus. The B1 plus B2 domain (mutant G2-B1B2), but not the G domain (mutant G2-G), is sufficient for accumulation in the nucleolus. Anti-B23 immunofluorescence of the same cells at a 60% scale is shown in the upper right quadrant of each panel. (C) Ngp1 can be retained by GTP-conjugated agarose. The GTP binding of Ngp1 is abolished by preincubating Ngp1 with 10 mM free GTP (WT+GTP) or by mutating the Asn255 residue to Ile (mutant G2-N255I). B, bound fraction; S, supernatant. (D) A single-residue mutation on Asn255 or Gly317 partially perturbs the nucleolar accumulation of Ngp1. (E) Nucleolar FRAP experiments showed that while the B1-B2 domain is necessary and sufficient for mediating nucleolar distribution, it is not enough to recapitulate the dynamic property of the full-length Ngp1 (mutant G2-B1B2). It is the non-nucleolus-targeting G domain that contributes to the retention property of Ngp1 (mutant G2-dG). Deleting the I or AC domain does not alter the dynamic property of Ngp1. Scale bars in panels B and D show 10 μm.
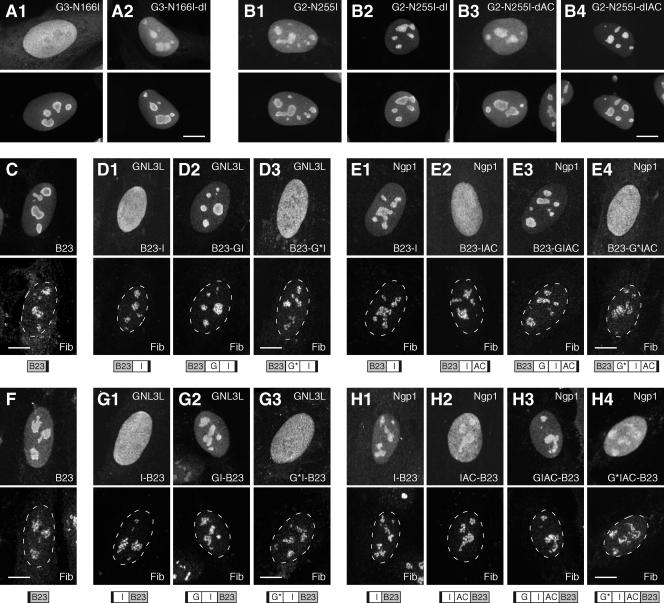
Nucleolar accumulation of GNL3L and Ngp1 is blocked by a GTP-regulated NpLS.
We have shown that deleting the I domain can restore the nucleolar localization of the G256V mutant of NS, indicating an antinucleolus-targeting activity in its I domain (38). To determine if the I domains of GNL3L and Ngp1 have the same function, a deletion of the I domain was created in the N166I mutant of GNL3L, and deletions of the I, AC, or IAC domains were created in the N255I mutant of Ngp1. Our data revealed that a deletion of the I domain of GNL3L is able to restore the nucleolar localization of the G3-N166I mutant in 30 to 40% of the cells (Fig. 4A). Similarly, the nucleolar localization of the N255I mutant of Ngp1 (Fig. 4B1) can be increased by a deletion of the I domain (Fig. 4B2) or the IAC domain (Fig. 4B4), but not by a deletion of the AC domain (Fig. 4B3). We further demonstrated that the G253V mutant of GNL3L (G3-G253V) and the G317V mutant of Ngp1 (G2-G317V) can regain their wild-type nucleolar distributions by deletion of the I domain of GNL3L or the I (or IAC) domain of Ngp1, respectively (see Fig. S3 in the supplemental material). These results indicate that the I domain of GNL3L and the I or IAC domain of Ngp1 each function as an NpLS that prevents the non-GTP-bound GNL3L and Ngp1 from accumulating in the nucleolus. To determine if the NpLS activity of the I or IAC domains of GNL3L and Ngp1 depends on their respective nucleolus-targeting B domains, heterologous chimeric proteins were designed in which the NpLS regions of GNL3L and Ngp1 were fused N or C terminally to another nucleolar protein, B23, and tested for their distribution by using a C-terminal HA tag (Fig. 4C to E) or an N-terminal Myc tag (Fig. 4F to H). Compared to the full-length B23 protein (Fig. 4C and F), the I domain of GNL3L (Fig. 4D1 and G1) and the IAC domain of Ngp1 (Fig. 4E2 and H2), but not the I domain of Ngp1 (Fig. 4E1 and H1), can each function as an NpLS to reduce the nucleolar accumulation of B23 fusion proteins. The NpLS-fused B23 proteins can recover their nucleolar localization capabilities by the addition of their respective G domains (Fig. 4D2, E3, G2, and H3), but not by the addition of the G domains containing the N166I mutation (for GNL3L) (Fig. 4D3 and G3) or the N255I mutation (for Ngp1) (Fig. 4E4 and H4). Because these phenotypes are observed independent of the fusion sites on B23, the tagged epitopes, or the lengths of the fusion proteins, one can eliminate the possibility that they are caused by fusion artifacts.
FIG. 4.
Nucleolar accumulation of GNL3L and Ngp1 are controlled by a G-domain-regulated NpLS in the I or IAC domain. (A and B) The nucleolar localization of the G3-N166I (A1) and G2-N255I (B1) mutants can be restored by a deletion of the I domain of GNL3L (A2) or the I or IAC domain of Ngp1 (B2 and B4), but not by a deletion of the AC domain of Ngp1 (B3). The distributions of the mutant proteins were detected by using a C-terminally fused GFP (top panels) and counterstained with anti-B23 immunofluorescence (bottom panels). (C to H) The NpLS activity of the I and IAC domains of GNL3L and Ngp1 remains functional when they are fused to another nucleolar protein, B23. The distribution patterns of B23 (C and F) and B23 fusion proteins (D, E, G, and H) were detected by using a C-terminal HA tag (C to E, upper panels) or an N-terminal Myc tag (F to H, upper panels) and double labeled with antifibrillarin immunofluorescence (Fib, lower panels). The I domain of GNL3L (D1 and G1) and the IAC domain of Ngp1 (E2 and H2), but not the I domain of Ngp1 (E1 and H1), can reduce the nucleolar accumulation of B23. The NpLS activities of the I domain of GNL3L and the IAC domain of Ngp1 can be neutralized by their respective wild-type G domains (D2, E3, G2, and H3), but not by the mutant G domain (G*) with the N166I mutation (for GNL3L; D3 and G3) or the N255I mutation (for Ngp1; E4 and H4). Dashed lines demarcate the nucleocytoplasmic boundaries. The designs of the fusion constructs are depicted below the panels. Black boxes indicate the HA or Myc epitope. Scale bars, 10 μm.
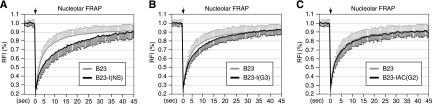
The NpLS activities of the I and IAC domains can be achieved by an NoLS-masking mechanism or by a nucleoplasmic retention mechanism. To differentiate these two possibilities, the protein flux rates between the nucleolus and the nucleoplasm of the I domains fused to the B23 proteins were measured by nucleolar FRAP experiments. Compared to the recovery rate of the wild-type B23 protein, the nucleolar FRAP recovery rate of the NS I domain fused to the B23 protein is significantly delayed (P < 0.001) (Fig. 5A), and the FRAP recovery rate of the GNL3L I domain fused to the B23 protein displays a mild but statistically significant delay at 5, 10, and 15 s after photobleaching (P < 0.005) (Fig. 5B). Although their static distribution patterns are distinctively different, the dynamic properties of the Ngp1 IAC domain fused to the B23 protein and of the wild-type B23 appear identical (Fig. 5C). Together, these data demonstrate that the I and IAC domains of the NS family proteins possess an intrinsic activity to antagonize nucleolar localization. This activity is dynamically regulated by their adjacent G domains in a GTP-dependent manner. In the case of NS and, to a lesser extent, GNL3L, this NpLS activity employs a nucleoplasmic retention mechanism.
FIG. 5.
The NpLS activity of the I domain of NS and, to a lesser extent, GNL3L, is mediated by a nucleoplasmic retention mechanism. The exchange rates between the nucleolus and the nucleoplasm of the B23 and B23-NpLS domain fusion proteins were measured by the nucleolar FRAP paradigm described in the legend to Fig. 1E. (A) The FRAP recovery rate of the NS I domain fused to the B23 protein [B23-I(NS)] is significantly delayed compared to the recovery rate of the control B23 protein. (B) The FRAP recovery rate of the GNL3L I domain fused to the B23 protein shows a slight but statistically significant decrease at the 5-, 10-, and 15-s time points compared to the recovery rate of the control B23 protein (P < 0.005; n = 50). (C) The FRAP recovery rates of the Ngp1 IAC domain fused to the B23 protein and the control B23 protein are identical. Error bars showing standard deviations are omitted on one side of the curves for clarity.
The nucleolar localization activities of the B-C regions of NS and GNL3L are functionally separable from the rest of the proteins.
Despite the close protein homology and evolutionary connection between NS and GNL3L, these two proteins exhibit distinct static and dynamic properties in their nucleolar localization, which allows us to determine systematically whether their nucleolar accumulation, retention, and NpLS activities can substitute for one another or whether they act in a protein-specific manner. We utilized two 7-amino-acid stretches in the G5* motif (VLDARDP) and the G1 motif (PNVGKSS) that were found to be identical in NS and GNL3L and created a “perfect” chimera composed of an N-terminal B-C domain, a middle G domain, and a C-terminal NpLS domain derived from either NS or GNL3L (see Fig. S4 in the supplemental material). Distribution studies showed that swapping the B-C domains between these two proteins does not change their nucleolus-predominant localization (Fig. 6A). By comparison, the nucleolar-to-nucleoplasmic intensity ratio of chimeric protein NS122 (Fig. 6A2) is the same as or slightly higher than that of chimeric protein NS211 (Fig. 6A1). Dynamically, the FRAP recovery rates of mutants NS122 and NS211 are faster than that of NS and slower than that of GNL3L (Fig. 6A3). These results demonstrate that the B-C domains of NS and GNL3L can be functionally separated from the rest of the protein and still maintain their relative activities in mediating nucleolar localization.
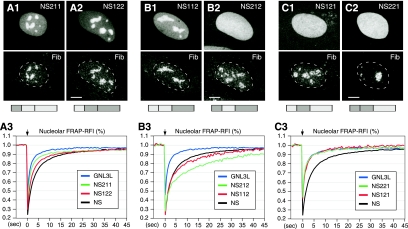
FIG. 6.
The B-C domains can be exchanged between NS and GNL3L and still maintain their relative activities, but the ability of the G domain to regulate the NpLS activity is protein specific. Chimeric proteins of NS and GNL3L were created that cross over at the highly conserved G5* and G1 motifs (see Fig. S4 in the supplemental material for details). Protein distribution was detected by using a C-terminally fused GFP and counterstained with antifibrillarin (Fib) antibody. When only the B-C domains are switched, the chimeric proteins NS211 (A1) and NS122 (A2) maintain their nucleolar distribution, and their FRAP recovery rates fall between those of NS and GNL3L (A3). When the G domain of NS is fused to the NpLS domain of GNL3L, the chimeric protein appears nucleolar (B1; NS112) or diffuse (B2; NS212), depending on which B-C domain it is coupled to. The FRAP recovery rates of these two mutants are as slow as or slower than that of NS (B3). When the G domain of GNL3L is fused to the NpLS domain of NS, the chimeric proteins are diffusely localized in the nucleus (C1 and C2), and their protein exchange rates are as fast as that of GNL3L (C3). The chimeric protein structures are depicted below the panels, with light and dark grey boxes representing protein origin from NS or GNL3L, respectively.
The nucleolar retention and the NpLS-regulating activities of the G domain involve specific residues that are nonpredictable based on overall protein homology.
When the G and the NpLS domains were mismatched, the nucleolar distributions of the chimeric proteins were significantly perturbed. When the G domain of NS is fused to the NpLS domain of GNL3L (Fig. 6B), the chimeric protein accumulates either in the nucleolus when conjugated to the B-C domain of NS (chimeric protein NS112; Fig. 6B1) or in the nucleoplasm when conjugated to the B-C domain of GNL3L (chimeric protein NS212; Fig. 6B2). Although the NS112 mutant appears nucleolar, its nucleoplasmic intensity is higher than that of NS. Compared to the recovery rate of NS, the nucleolar FRAP recovery rates of these two mutants are slow. Notably, the decrease in the FRAP recovery rate is more evident in NS212 than in NS112, showing that the nucleolar accumulation and protein exchange rate are controlled independently (Fig. 6B3). Conversely, a fusion between the G domain of GNL3L and the NpLS domain of NS abolishes its nucleolar accumulation regardless of which B-C domain it is coupled to (Fig. 6C1 and C2). Contrary to the activities of the NS112 and NS212 mutants, the NS121 and NS221 mutants shuttle between the nucleolus and the nucleoplasm at the same speed as the full-length GNL3L (Fig. 6C3). These results demonstrate that the G domain of GNL3L lacks the retention activity and is incapable of antagonizing the NpLS activity of NS. Based on the data for the chimeric proteins, we conclude that the NpLS-regulating activity is modulated by the G domain between the G5* and G1 motifs in a highly protein-specific manner and that the retention signal reduces the nucleolar-nucleoplasmic exchange rate of proteins no matter whether they accumulate in the nucleolus or in the nucleoplasm.
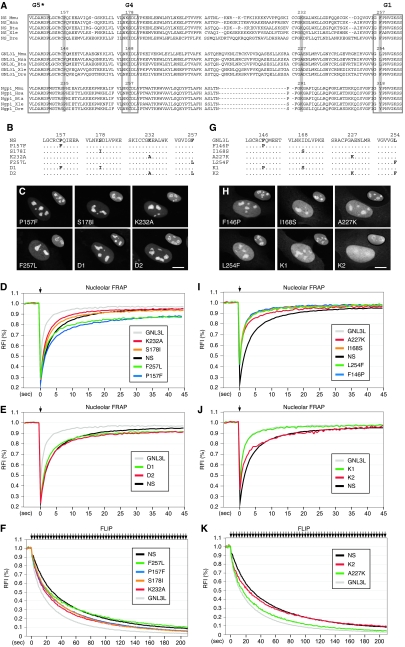
The fact that the G-domain activity cannot be predicted based on the overall protein homology suggests that a few amino acids may play a decisive role. To resolve the protein-specific activity of the G domain at the single-residue level, we targeted specific amino acids that are located between the G5* and G1 motifs and are highly conserved among the NS and Ngp1 genes of different species but are not shared by GNL3L. Using these three criteria, we identified a Lys residue in NS (Lys232) and Ngp1 (Lys291) that is highly conserved throughout evolution and corresponds to an Ala residue in GNL3L (A227). Another amino acid, Pro157 in NS or Pro235 in Ngp1, fulfills the same criteria, except for the frog and zebra fish GNL3L proteins (Fig. 7A). Each member of the NS family proteins also possesses a unique amino acid in the G4 motif (Ser178 in NS, Ile168 in GNL3L, and Cys257 in Ngp1) and in the G1 motif (Phe257 in NS, Leu254 in GNL3L, and Tyr318 in Ngp1) (Fig. 7A). In the first set of experiments, we replaced a single amino acid in NS at one of these four sites with the corresponding amino acid found in GNL3L (P157F, S178I, K232A, and F257L), as well as creating mutants with combined mutations of P157F and S178I (P157F-S178I; mutant D1) or K232A-F257L (mutant D2) (Fig. 7B). Statically, all six mutants are localized predominantly in the nucleolus (Fig. 7C). Dynamically, the K232A and S178I mutants exhibit a faster FRAP recovery rate than the wild-type NS, whereas the P157F and F257L mutants differ from the wild-type NS primarily in their recovery plateau level (Fig. 7D and Table 1). The FRAP recovery rates of the D1 and D2 mutants fall between those of the mutants with their respective single residues, indicating that the effects of these single-residue mutations on the dynamic property of NS are additive (Fig. 7E). The fast protein exchange rates of the K232A and S178I mutants were confirmed by FLIP experiments in which fluorescence loss in the nucleolus was recorded while a 2- by 4-μm area in the nucleoplasm was repeatedly bleached (Fig. 7F). Notably, the FLIP rate of the P157F mutant is faster than that of wild-type NS, and the loss of F257L signal in the nucleolus is faster than the loss of wild-type signal during the first 30 s of photobleaching.
FIG. 7.
A combined A227K-L254F mutation on GNL3L mimics the effect of swapping its G domain with that of NS. (A) To identify the amino acids located between the G5* and G1 motifs that play a key role in mediating the nucleolar retention and the NpLS-regulating activities of the G domain, the protein sequences of the G5*-to-G1 domain of the vertebrate NS family proteins were aligned by using the ClustalW program. The four residues that are either shared by NS and Ngp1 but not by GNL3L or uniquely conserved in each member of the NS family (numbered and highlighted) were chosen for the amino acid substitution experiments. (B) Single-amino-acid substitutions were made on NS at one or two of these four sites. (C) Static distributions of NS mutant proteins were detected by using C-terminally fused GFP and counterstained with anti-B23 immunofluorescence, shown on a 60% scale in the upper right quadrant of each panel. (D) FRAP analyses showed that single-residue mutants K232A and S178I display a faster recovery rate than the wild-type NS. The P157F and F257L mutants reach a lower plateau level than the wild-type NS. (E) The FRAP recovery rates of double-residue mutants D1 and D2 fall between those of their respective single-residue mutants. (F) FLIP experiments showed that the nucleolar retention times of the P157L, S178I, and K232A mutants are decreased compared to that of the wild-type NS. The FLIP rate of the F257L mutant is faster than that of the wild-type NS during the first 30 s of photobleaching. (G) Single-amino-acid substitutions were made on GNL3L at one or two of the four targeted sites. (H) The distributions of mutant GNL3L proteins were shown by a C-terminally fused GFP and counterstained with anti-B23 immunofluorescence, shown on a 60% scale in the upper right quadrant of each panel. Unlike the wild-type GNL3L, the double-residue GNL3L mutant K2 (A227K-L254F) is distributed diffusely in the nucleus. (I and J) FRAP analyses showed that the single-residue GNL3L mutant A227K (I) has a mild increase and the double-residue mutant K2 (J) has a significant increase in the nucleolar retention time. (K) FLIP experiments confirm the FRAP findings for A227K and K2 mutants. Scale bars in panels C and H show 10 μm.
TABLE 1.
Statistical analyses of FRAP recovery rates of single- and double-residue mutations on NS and GNL3L
| Protein or mutation(s) (mutant name) | % FRAP recovery (mean ± SD) at:a
|
||
|---|---|---|---|
| 5 s | 10 s | 40 s | |
| NS | 74.0 ± 3.3 | 84.7 ± 2.4 | 94.6 ± 2.9 |
| P157F | 68.4 ± 4.8 ** | 76.9 ± 4.4 ** | 87.6 ± 4.0 ** |
| S178I | 79.0 ± 3.0 ** | 86.7 ± 2.5 ** | 93.4 ± 3.8 |
| K232A | 82.2 ± 2.4 ** | 89.3 ± 2.0 ** | 94.8 ± 2.5 |
| F257L | 72.9 ± 6.0 | 79.7 ± 5.6 ** | 86.8 ± 5.4 ** |
| P157F-S178I (D1) | 77.0 ± 4.4 * | 84.6 ± 3.5 | 91.4 ± 4.2 ** |
| K232A-F257L (D2) | 73.8 ± 4.6 | 83.1 ± 4.0 | 91.1 ± 4.1 ** |
| GNL3L | 90.2 ± 3.2 | 94.4 ± 2.6 | 96.7 ± 3.2 |
| F146P | 90.5 ± 1.9 | 94.6 ± 2.2 | 97.6 ± 3.0 |
| I168S | 89.9 ± 3.8 | 93.6 ± 2.9 | 97.4 ± 2.6 |
| A227K | 85.5 ± 4.0 ** | 90.4 ± 2.8 ** | 96.5 ± 3.0 |
| L254F | 89.6 ± 4.1 | 93.0 ± 4.0 | 97.7 ± 4.5 |
| F146P-I168S (K1) | 90.0 ± 3.2 | 93.4 ± 2.8 | 96.9 ± 2.9 |
| A227K-L254F (K2) | 77.4 ± 8.5 ** | 84.3 ± 7.6 ** | 95.3 ± 9.3 |
*, P value is <0.01 compared to NS or GNL3L; **, P value is <0.001 compared to NS or GNL3L.
In the second set of experiments, reciprocal mutations were made on the F146, I168, A227, and/or L254 residue of GNL3L (Fig. 7G). The four single-residue mutants and double-residue mutant K1 (F146P-I168S) of GNL3L show the same distribution patterns as the wild-type GNL3L (Fig. 7H). Double-residue mutant K2 (A227K-L254F), on the other hand, is diffusely localized in the whole nucleus. While the F146P, I168S, L254F, and K1 mutants behave the same as the wild-type GNL3L in terms of their dynamic properties, the A227K mutation produces a mild but statistically significant delay in the FRAP recovery rate of GNL3L (Fig. 7I and J and Table 1). Notably, a combined mutation of A227K-L254F (mutant K2) significantly increases the nucleolar residence time of GNL3L (Fig. 7J and Table 1), mimicking the static and dynamic properties of the NS212 mutant. The effects of the A227K and A227K-L254F mutations (mutant K2) on the protein exchange rate of GNL3L were also confirmed by FLIP experiments (Fig. 7K) performed in the same way as described above and whose results are shown in Fig. 1F and 7F. These results show that the P157L, S178I, and K232A mutations on NS can increase its nucleolar-nucleoplasmic exchange rate. Reciprocally, the A227K and the A227K-L254F (mutant K2) mutations can decrease the nucleolar-nucleoplasmic exchange rate of GNL3L. Together, our data demonstrate that the nucleolar retention and NpLS-regulating activities of the G domain are determined by a few key residues.
DISCUSSION
The goal of this study is to determine the molecular mechanism and the protein code that govern the nucleolar accumulation and the nucleolar-nucleoplasmic exchange rate of a subfamily of GTP-binding proteins, NS, GNL3L, and Ngp1. These proteins are structurally and evolutionarily related but differ in their nucleolar dynamics. Like most nucleolar proteins, all of them contain one or two stretches of basic residues that are necessary and sufficient for mediating nucleolar distribution. These basic regions can be fused to a heterologous protein (i.e., GFP) or exchanged between NS and GNL3L and still maintain their relative nucleolus-targeting activities. However, the basic regions of NS and Ngp1 shuttle between the nucleolus and the nucleoplasm at a much faster speed than the full-length proteins, suggesting that additional modules exist in the rest of the protein that provide regulatory controls over their nucleolar-nucleoplasmic exchange rate. Further investigation identified a nucleolar retention activity associated with the G domain of NS and Ngp1 that can increase the nucleolar residence time of the full-length protein. Despite the close homology between GNL3L and NS, GNL3L does not contain such a retention signal. Given these results and that the protein sequence identity (or similarity) in the G domain is 53% (or 65%) between NS and GNL3L and 43% (or 50%) between NS and Ngp1, we reason that the retention signal is determined by a few key residues rather than by the overall protein homology. In addition to the retention signal, all three proteins contain an NpLS within their I (or IAC) domain that can reduce their nucleolar accumulation. The NpLS activities of the I domains are regulated by the G domains, depending on their GTP-binding state, and can function independently of the NoLS domains of NS family proteins.
Analyses of the NS-GNL3L chimeric proteins with mismatched G and NpLS domains reveal that the nucleolar retention signal of NS can slow down the protein exchange rate regardless of whether the protein accumulates in the nucleolus or in the nucleoplasm (Fig. 6B, mutants NS112 and NS212). Furthermore, the ability of the G domain to neutralize the activity of NpLS is distinct among the NS family proteins. Even though they share 53% protein sequence identity, the same domain structure, and the same yeast orthologue, neither the G domain of NS nor that of GNL3L can fully cross-regulate the NpLS activity of the other protein. Amino acid substitution experiments showed that a Lys-to-Ala switch can reciprocally affect the nucleolar retention times of NS and GNL3L. The combined mutation A227K-L254F on GNL3L disrupts its nucleolar accumulation and significantly increases its retention time in much the same way as swapping the G5*-to-G1 domain of GNL3L with that of NS (mutant NS212). Although the F146P and L254F mutations have no apparent effect on the dynamic distribution of GNL3L, the reverse mutations on NS (P157F and F257L) lead to a lower FRAP recovery plateau in a way that is similar to the recovery plateau of Ngp1. This finding suggests that such mutations render a portion of NS in the nucleolus unexchangeable within the time frame of our recording. Notably, the FLIP rate of P157F is faster than that of the wild-type NS, which resembles the dynamic property of Ngp1 as well. In this study, we specifically targeted the residues that are shared by NS and Ngp1, but not by GNL3L, based on the rationale that NS and Ngp1 reside in the nucleolus longer than GNL3L does. We also investigated the function of two residues in the G4 and G1 domains that are distinct among NS, GNL3L, and Ngp1 but highly conserved throughout evolution. Although our data do not exclude the possibility that other, nonconserved amino acids or the peptidyl length may contribute to the G-domain activity, they demonstrate that the protein-specific retention and NpLS-regulating activities of the G domain are mediated largely by a few key residues that are difficult to predict based on the overall protein homology.
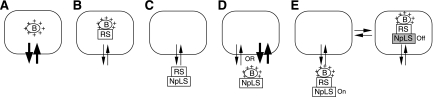
Based on our data, one can envision several molecular models to explain the accumulation and dynamic exchange of nuclear proteins between the nucleolar and nucleoplasmic compartments. A common mechanism shared by most, if not all, nucleolar proteins involves a stretch of positively charged residues that permits them to accumulate in the nucleolus (Fig. 8A). This region can be transferred to another protein and still maintain its nucleolar localization activity. In addition to this most commonly identified nucleolar localization sequence, a retention signal and an NpLS exist in some or all of the NS family proteins which modulate their static and dynamic distributions. The retention signal can increase the retention time of proteins localized in the nucleolus (Fig. 8B) or in the nucleoplasm (Fig. 8C). The NpLS possesses the ability to redistribute nucleolar proteins from the nucleolus to the nucleoplasm. This NpLS activity can be constitutively active, in which case the protein appears nucleoplasmic (Fig. 8D), or it can be turned on and off to allow for a regulated distribution between the nucleolus and the nucleoplasm (Fig. 8E). Finally, the nucleolar retention and the NpLS-regulating activities of the G domain involve specific residues that cannot be predicted based on the overall protein homology.
FIG. 8.
The mechanism underlying nucleolar localization may include a nucleolus-targeting B domain, a retention signal, and an NpLS coupled with a regulatory module. In addition to the commonly identified, positively charged (+) B domain, the static and dynamic distribution of nucleolar proteins between the nucleolus (rounded rectangles) and the nucleoplasm can be further modified by a retention signal, an NpLS, or a combination of both. By itself, the B domain can accumulate in the nucleolus with a short nucleolar retention time, represented by thick arrows (A). The addition of a retention signal (RS) can slow down the protein exchange rate, indicated by thin arrows, of both nucleolar (B) and nucleoplasmic proteins (C). The ability of the NpLS to detain proteins with nucleolar localization capability in the nucleoplasm can be constitutively active (D) or regulated by a regulatory module (E). For NS and Ngp1, the G domain functions as both the retention signal and the regulatory module for the NpLS.
This work also raises several issues that require further investigation. First, although we can demonstrate experimentally that the NpLS activity of the I domain of NS works through a retention mechanism, the abilities of the I domain of GNL3L and the IAC domains of Ngp1 to decrease the FRAP recovery rates of B23 are either subtle or undetectable. Given that the I and IAC domains of GNL3L and Ngp1 are equally capable of disrupting the nucleolar accumulation of B23 as NS is (Fig. 4) (our previous study [20]), the lack of effect of these regions on the dynamic distribution of B23 suggests that their NpLS activities may involve intramolecular masking of NoLS. However, it is important to note that the FRAP approach may not be sensitive enough to detect small changes in protein dynamics that can be amplified to a significant effect on the static distribution in steady state. Second, in addition to blocking nucleolar localization, the I domains of NS and GNL3L, but not that of Ngp1, may participate in protein dissociation from the nucleolus. This is demonstrated by a delay in the FRAP recovery rate in the I-domain deletion mutants of NS (38) and GNL3L (Fig. 2E). Third, a single-residue mutation on the conserved Asn166 or Gly253 of GNL3L has a more profound effect on its static distribution than deleting the G5*-to-G1 domain does. These findings suggest that the NpLS activity of GNL3L may involve part of the G domain or that deleting the G domain of GNL3L may perturb the function and protein conformation of its NpLS domain. Finally, different nucleolar proteins display distinct nucleolar retention times. The physiological significance of that remains incompletely understood. We have shown that the ability of GNL3L to inhibit the transcriptional activity of estrogen-related protein γ does not require its nucleolus-localizing B-C domain and can be increased only slightly by the NS122 mutation that increases its nucleolar retention (44). One inference of this finding is that it is less likely that the mechanism of nucleolar sequestration is used by proteins with a short nucleolar residence time, like GNL3L, than by proteins with a long nucleolar residence time, like NS or Ngp1. In conclusion, our data reveal two novel activities associated with the G domain that function as a retention signal and an NpLS regulator, as well as an NpLS activity in the I (or IAC) domain. We propose that nucleolar localization is determined by a balance between the positively charged domain, a protein-specific retention signal, and an NpLS, which operate in concert to achieve a controlled distribution of proteins inside the nucleus. This knowledge of dynamic protein trafficking to and from the nucleolus will cast new light on the regulation of fast biological events taking place in the nucleus.
Supplementary Material
Acknowledgments
We thank Seokwoon Kim for his technical help at the beginning of this work.
This work was supported by Public Health Service grant CA113750 awarded to R. Y. L. Tsai.
Footnotes
Published ahead of print on 8 October 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Andersen, J. S., Y. W. Lam, A. K. Leung, S. E. Ong, C. E. Lyon, A. I. Lamond, and M. Mann. 2005. Nucleolar proteome dynamics. Nature 433:77-83. [DOI] [PubMed] [Google Scholar]
- 2.Bernardi, R., P. P. Scaglioni, S. Bergmann, H. F. Horn, K. H. Vousden, and P. P. Pandolfi. 2004. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat. Cell Biol. 6:665-672. [DOI] [PubMed] [Google Scholar]
- 3.Catez, F., M. Erard, N. Schaerer-Uthurralt, K. Kindbeiter, J. J. Madjar, and J. J. Diaz. 2002. Unique motif for nucleolar retention and nuclear export regulated by phosphorylation. Mol. Cell. Biol. 22:1126-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, D., and S. Huang. 2001. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J. Cell Biol. 153:169-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daigle, D. M., L. Rossi, A. M. Berghuis, L. Aravind, E. V. Koonin, and E. D. Brown. 2002. YjeQ, an essential, conserved, uncharacterized protein from Escherichia coli, is an unusual GTPase with circularly permuted G-motifs and marked burst kinetics. Biochemistry 41:11109-11117. [DOI] [PubMed] [Google Scholar]
- 6.Dundr, M., T. Misteli, and M. O. Olson. 2000. The dynamics of postmitotic reassembly of the nucleolus. J. Cell Biol. 150:433-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fatica, A., and D. Tollervey. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14:313-318. [DOI] [PubMed] [Google Scholar]
- 8.Felsenstein, J. 1997. An alternating least squares approach to inferring phylogenies from pairwise distances. Syst. Biol. 46:101-111. [DOI] [PubMed] [Google Scholar]
- 9.Fox, A. H., Y. W. Lam, A. K. Leung, C. E. Lyon, J. Andersen, M. Mann, and A. I. Lamond. 2002. Paraspeckles: a novel nuclear domain. Curr. Biol. 12:13-25. [DOI] [PubMed] [Google Scholar]
- 10.Gorski, S. A., M. Dundr, and T. Misteli. 2006. The road much traveled: trafficking in the cell nucleus. Curr. Opin. Cell Biol. 18:284-290. [DOI] [PubMed] [Google Scholar]
- 11.Grosshans, H., K. Deinert, E. Hurt, and G. Simos. 2001. Biogenesis of the signal recognition particle (SRP) involves import of SRP proteins into the nucleolus, assembly with the SRP-RNA, and Xpo1p-mediated export. J. Cell Biol. 153:745-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang, C. F., and N. N. Chuang. 2000. Disrupting the geranylgeranylation at the C-termini of the shrimp Ras by depriving guanine nucleotide binding at the N-terminal. J. Exp. Zool. 286:441-449. [DOI] [PubMed] [Google Scholar]
- 13.Iismaa, S. E., L. Chung, M. J. Wu, D. C. Teller, V. C. Yee, and R. M. Graham. 1997. The core domain of the tissue transglutaminase Gh hydrolyzes GTP and ATP. Biochemistry 36:11655-11664. [DOI] [PubMed] [Google Scholar]
- 14.Isogai, Y., and R. Tjian. 2003. Targeting genes and transcription factors to segregated nuclear compartments. Curr. Opin. Cell Biol. 15:296-303. [DOI] [PubMed] [Google Scholar]
- 15.Jacobson, M. R., and T. Pederson. 1998. Localization of signal recognition particle RNA in the nucleolus of mammalian cells. Proc. Natl. Acad. Sci. USA 95:7981-7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leipe, D. D., Y. I. Wolf, E. V. Koonin, and L. Aravind. 2002. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317:41-72. [DOI] [PubMed] [Google Scholar]
- 17.Martel, C., P. Macchi, L. Furic, M. A. Kiebler, and L. Desgroseillers. 2006. Staufen1 is imported into the nucleolus via a bipartite nuclear localization signal and several modulatory determinants. Biochem. J. 393:245-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mekhail, K., L. Gunaratnam, M. E. Bonicalzi, and S. Lee. 2004. HIF activation by pH-dependent nucleolar sequestration of VHL. Nat. Cell Biol. 6:642-647. [DOI] [PubMed] [Google Scholar]
- 19.Mekhail, K., M. Khacho, A. Carrigan, R. R. Hache, L. Gunaratnam, and S. Lee. 2005. Regulation of ubiquitin ligase dynamics by the nucleolus. J. Cell Biol. 170:733-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng, L., H. Yasumoto, and R. Y. Tsai. 2006. Multiple controls regulate nucleostemin partitioning between nucleolus and nucleoplasm. J. Cell Sci. 119:5124-5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Misteli, T. 2005. Going in GTP cycles in the nucleolus. J. Cell Biol. 168:177-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Misteli, T. 2001. Protein dynamics: implications for nuclear architecture and gene expression. Science 291:843-847. [DOI] [PubMed] [Google Scholar]
- 23.Misteli, T. 2004. Spatial positioning: a new dimension in genome function. Cell 119:153-156. [DOI] [PubMed] [Google Scholar]
- 24.Negi, S. S., and M. O. Olson. 2006. Effects of interphase and mitotic phosphorylation on the mobility and location of nucleolar protein B23. J. Cell Sci. 119:3676-3685. [DOI] [PubMed] [Google Scholar]
- 25.Olson, M. O., and M. Dundr. 2005. The moving parts of the nucleolus. Histochem. Cell Biol. 123:203-216. [DOI] [PubMed] [Google Scholar]
- 26.Pederson, T. 1998. The plurifunctional nucleolus. Nucleic Acids Res. 26:3871-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phair, R. D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404:604-609. [DOI] [PubMed] [Google Scholar]
- 28.Racevskis, J., A. Dill, R. Stockert, and S. A. Fineberg. 1996. Cloning of a novel nucleolar guanosine 5′-triphosphate binding protein autoantigen from a breast tumor. Cell Growth Differ. 7:271-280. [PubMed] [Google Scholar]
- 29.Rao, M. R., G. Kumari, D. Balasundaram, R. Sankaranarayanan, and S. Mahalingam. 2006. A novel lysine-rich domain and GTP binding motifs regulate the nucleolar retention of human guanine nucleotide binding protein, GNL3L. J. Mol. Biol. 364:637-654. [DOI] [PubMed] [Google Scholar]
- 30.Raska, I., P. J. Shaw, and D. Cmarko. 2006. Structure and function of the nucleolus in the spotlight. Curr. Opin. Cell Biol. 18:325-334. [DOI] [PubMed] [Google Scholar]
- 31.Reed, M. L., B. K. Dove, R. M. Jackson, R. Collins, G. Brooks, and J. A. Hiscox. 2006. Delineation and modelling of a nucleolar retention signal in the coronavirus nucleocapsid protein. Traffic 7:833-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reynaud, E. G., M. A. Andrade, F. Bonneau, T. B. Ly, M. Knop, K. Scheffzek, and R. Pepperkok. 2005. Human Lsg1 defines a family of essential GTPases that correlates with the evolution of compartmentalization. BMC Biol. 3:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rubbi, C. P., and J. Milner. 2000. Non-activated p53 co-localizes with sites of transcription within both the nucleoplasm and the nucleolus. Oncogene 19:85-96. [DOI] [PubMed] [Google Scholar]
- 34.Sheng, Z., J. A. Lewis, and W. J. Chirico. 2004. Nuclear and nucleolar localization of 18-kDa fibroblast growth factor-2 is controlled by C-terminal signals. J. Biol. Chem. 279:40153-40160. [DOI] [PubMed] [Google Scholar]
- 35.Sleeman, J. E., L. Trinkle-Mulcahy, A. R. Prescott, S. C. Ogg, and A. I. Lamond. 2003. Cajal body proteins SMN and Coilin show differential dynamic behaviour in vivo. J. Cell Sci. 116:2039-2050. [DOI] [PubMed] [Google Scholar]
- 36.Sommerville, J., C. L. Brumwell, J. C. Politz, and T. Pederson. 2005. Signal recognition particle assembly in relation to the function of amplified nucleoli of Xenopus oocytes. J. Cell Sci. 118:1299-1307. [DOI] [PubMed] [Google Scholar]
- 37.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai, R. Y., and R. D. McKay. 2005. A multistep, GTP-driven mechanism controlling the dynamic cycling of nucleostemin. J. Cell Biol. 168:179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai, R. Y., and R. D. McKay. 2002. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 16:2991-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tschochner, H., and E. Hurt. 2003. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 13:255-263. [DOI] [PubMed] [Google Scholar]
- 41.Weber, J. D., M. L. Kuo, B. Bothner, E. L. DiGiammarino, R. W. Kriwacki, M. F. Roussel, and C. J. Sherr. 2000. Cooperative signals governing ARF-Mdm2 interaction and nucleolar localization of the complex. Mol. Cell. Biol. 20:2517-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weber, J. D., L. J. Taylor, M. F. Roussel, C. J. Sherr, and D. Bar-Sagi. 1999. Nucleolar Arf sequesters Mdm2 and activates p53. Nat. Cell Biol. 1:20-26. [DOI] [PubMed] [Google Scholar]
- 43.Wong, J. M., L. Kusdra, and K. Collins. 2002. Subnuclear shuttling of human telomerase induced by transformation and DNA damage. Nat. Cell Biol. 4:731-736. [DOI] [PubMed] [Google Scholar]
- 44.Yasumoto, H., L. Meng, T. Lin, Q. Zhu, and R. Y. Tsai. 2007. GNL3L inhibits activity of estrogen-related receptor γ by competing for coactivator binding. J. Cell Sci. 120:2532-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.You, J., B. K. Dove, L. Enjuanes, M. L. DeDiego, E. Alvarez, G. Howell, P. Heinen, M. Zambon, and J. A. Hiscox. 2005. Subcellular localization of the severe acute respiratory syndrome coronavirus nucleocapsid protein. J. Gen. Virol. 86:3303-3310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.