Abstract
Mitotic chromatin condensation is essential for cell division in eukaryotes. Posttranslational modification of the N-terminal tail of histone proteins, particularly by phosphorylation by mitotic histone kinases, may facilitate this process. In mammals, aurora B is believed to be the mitotic histone H3 Ser10 kinase; however, it is not sufficient to phosphorylate H3 Ser10 with aurora B alone. We show that histone H3 is phosphorylated by vaccinia-related kinase 1 (VRK1). Direct phosphorylation of Thr3 and Ser10 in H3 by VRK1 both in vitro and in vivo was observed. Loss of VRK1 activity was associated with a marked decrease in H3 phosphorylation during mitosis. Phosphorylation of Ser10 by VRK1 is similar to that by aurora B. Moreover, expression and chromatin localization of VRK1 depended on the cell cycle phase. Overexpression of VRK1 resulted in a dramatic condensation of nuclei. Our findings collectively support a role of VRK1 as a novel mitotic histone H3 kinase in mammals.
Chromatin congregates to chromosomes during mitosis to facilitate the even segregation of genetic information to two daughter cells. In nucleosomes, the combinational modification of histone tails, the so-called “histone code,” controls chromatin-templated processes from gene expression to cell fate decision (20, 30). Phosphorylation of the N-terminal tail of histone H3 may be responsible for chromatin condensation (21). During mitosis, the N-terminal tail of histone H3 is phosphorylated at several residues, including Thr3 (5, 36), Ser10 (3, 7, 17, 18), Thr11 (37), and Ser28 (12). A correlation between histone H3 Ser10 phosphorylation and chromatin condensation in Aspergillus nidulans (6) and Tetrahymena thermophila (47) is well established. However, in other species, condensation is not accomplished simply by Ser10 phosphorylation, and additional phosphorylation or modification of histone tails is required (21).
A number of studies have shown that members of the aurora kinase family are responsible for phosphorylation of histone H3 (3, 7, 17, 18). Mammals contain three isotypes of aurora kinase designated aurora A, B, and C (11). Among these, aurora B is a strong candidate phosphorylator of Ser10 in histone H3 as is evident from data obtained with hesperadin, the aurora B inhibitor (14), which suppressed Ser10 phosphorylation during mitosis (7, 17). However, residual Ser10 phosphorylation was detected, even upon depletion of aurora B in cells, suggesting the presence of an additional histone H3 kinase (29).
NIMA (never in mitosis), the histone H3 Ser10 kinase in Aspergillus nidulans (6, 34), triggers chromatin condensation in cells arrested at the interphase (28). In mammals, Nercc1, the functional ortholog of NIMA, was found to be phosphorylating histone H3 (39). Nucleosomal histone kinase 1 (NHK1) from Drosophila melanogaster is the kinase shown to phosphorylate histone protein in chromatin as a substrate. NHK1 phosphorylated H2A at Thr119 in chromatin but not with free histone as the substrate (1). Recent studies showed that NHK1 participates in mitotic progression (4) and maintenance of proper chromosomal architecture (19). These data strongly indicate that NHK1 is a bona fide mitotic histone kinase.
Vaccinia-related kinase 1 (VRK1) is the mammalian homolog of NHK1 (1). Sequence similarities between NHK1 and VRK1 are evident in the kinase domain (approximately 40% identity), and the carboxyl termini contain a characteristic basic-acidic-basic motif (1). VRK1, identified from the screening of novel genes involved in cell cycle regulation from fetal liver (31), is designated on the basis of 40% sequence identity with vaccinia virus B1 kinase, which plays a critical role in viral DNA replication (38). The kinase is highly expressed in proliferative tissues, including embryonic tissues, adult testis, and thymus, as well as in several cancer cell lines, implying a functional role in cell cycle regulation and tumorigenicity (31). VRK1 participates in cell cycle progression by means of phosphorylation of a barrier-to-autointegration factor (BAF) that plays structural roles in chromatin and the nuclear envelope and displays subcellular localization changes during the cell cycle (33) and by activating the transcription of proliferation-related proteins such as retinoblastoma, cyclin-dependent kinase 2, and survivin (40).
In this report, we demonstrate that VRK1 is a chromatin-associated protein displaying cell cycle-dependent expression and subcellular localization. VRK1 phosphorylates Thr3 and Ser10 in free and core histone H3 and in nucleosomes. Overexpression of the constitutively active form of VRK1 leads to hypercondensation of the nucleus, in similarity to data obtained in studies of NIMA, the fungal enzyme, in eukaryotic cells.
MATERIALS AND METHODS
Plasmids and antibodies.
To generate VRK1 expression constructs, mouse, rat, and human full-length VRK1 was amplified by PCR from a day-16 mouse embryo cDNA library (Clontech), from Rat-1 cell cDNA, and from HeLa cell cDNA, respectively. For mammalian expression constructs, VRK1 and its kinase-dead mutant generated by site-directed mutation (Lys179 to Glu) were subcloned into pcDNA3.1 (Invitrogen), pFlag-CMV2 (Sigma), and pEGFP-N1, pEGFP-C1, pDsRed1-N1, and pDsRed1-C1 (BD Biosciences). Human VRK3 and HP1γ cDNAs were subcloned into pDsRed1-C1 and pEGFP-N1, respectively. For expression in Escherichia coli, VRK1 and its mutants (VRK2, VRK3, and histone H3 mutants) were subcloned into pPosKJ, pProEX, or pGEX-4T-1 (Amersham) as described previously (25). VRK antisera were prepared as described previously (23). The following antibodies were purchased from commercial sources: anti-Flag epitope (M2) from Sigma; anti-green fluorescent protein (anti-GFP; C163) from Zymed; anti-glyceraldehyde 3-phosphate dehydrogenase (anti-GAPDH) and antibromodeoxyuridine (anti-BrdU) from Calbiochem; anti-phospho-cdc2 (Tyr15) and anti-cleaved caspase3 from Cell Signaling Technology; anti-RNA polymerase II from Abcam; anti-DsRed1 and anti-aurora B (AIM-1) from BD Biosciences; anti-lamin B, anti-α-tubulin, anti-cyclin D1, anti- GFP, and anti-γ-tubulin from Santa Cruz Biotechnology; and all antihistone antibodies used in this study as well as anti-HP1α from Upstate Biotechnology.
Extraction of subcellular fractions.
Cytoplasmic and nucleoplasmic extracts were prepared as described previously (23). The pellet obtained following the extraction of the nucleoplasm, which contained the subnuclear organelles and chromatins, was further extracted as follows. A euchromatin-enriched fraction was isolated as the soluble fraction following treatment of the pellet with DNase I for 1 h at room temperature. Next, the pellet remaining after the euchromatin extraction was vigorously sonicated to obtain the nuclear matrix. Finally, following the nuclear matrix extraction, the pellet was treated with 6 M urea and 2% sodium dodecyl sulfate to solubilize the heterochromatin-enriched fraction. To prepare crude soluble chromatin, the pellet from the nucleoplasmic extraction was treated with micrococcal nuclease (100 U) for 1 h at 30°C. Each fraction was evaluated by Ponceau S staining and immunoblotting.
Immunoprecipitation, immunoblotting and immunocytochemistry.
Immunoprecipitation, immunoblotting, and immunocytochemistry were performed as described previously (23).
TUNEL assay.
For the TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling) assay, HeLa cells transfected with DsRed1-C1-VRK1 for various time periods were grown on glass chips coated with poly-d-lysine (Sigma). Cells were then fixed with 4% paraformaldehyde, the glass chips were removed, and the cells were permeabilized and stained with a DeadEnd fluorometric TUNEL system kit (Promega) according to the manufacturer's instructions.
Protein kinase assay.
An in vitro kinase assay was performed with 100 ng (see Fig. 4E, G, and H) to 1 μg (see Fig. 4B to D and F) of either recombinant glutathione S-transferase-VRK1 (GST-VRK1) or VRK1 protein and 100 ng (see Fig. 4E, G, and H) or 1 μg (see Fig. 4B to D and F) of substrates, including core histones, nucleosomes, each histone protein, and BAF. The duration of the kinase assay was 30 min except for that of one assay (see Fig. 4H). The standard procedure for the in vitro kinase assay was carried out as described previously (23). The kinase activity of VRK1 in cell extracts was analyzed using immunoprecipitated VRK1 with anti-VRK1 antibody.
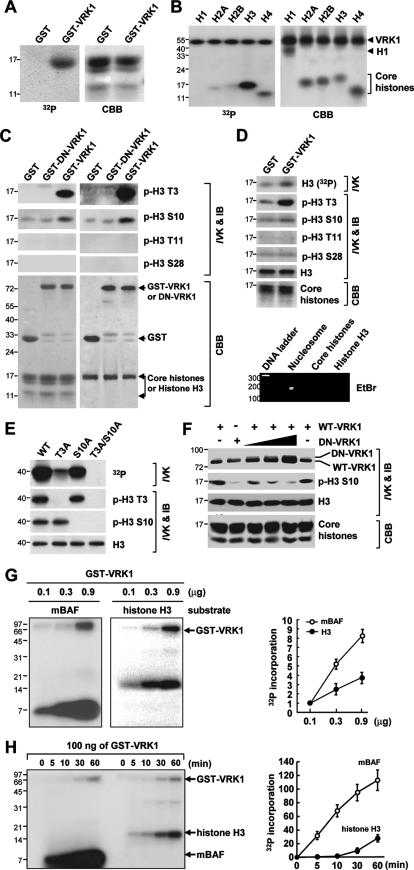
FIG. 4.
Phosphorylation of Thr3 and Ser10 in H3 by VRK1 in vitro. (A and B) Phosphorylation of H3 from purified free core histones (A) or individual histone proteins (B) by VRK1. “32P” indicates the autoradiogram results. (C and D) Identification of sites in H3 phosphorylated by VRK1. Following an in vitro kinase assay (IVK), purified core histone proteins (panel C, left half), purified H3 protein (C, right half), and nucleosomes (D) from HeLa cells were analyzed by immunoblotting (IB) with site-specific anti-phosphorylated histone antibodies. Agarose gel electrophoresis followed by ethidium bromide (EtBr) staining confirmed the presence of monomeric nucleosomes containing DNA (panel D, bottom). (E) Confirmation of phosphorylation sites of histone H3 by VRK1 by use of either wild-type (WT) or mutated histone H3. (F) Requirement of VRK1 activity for H3 phosphorylation. Addition of dominant-negative VRK1 (DN-VRK1) reduced the phosphorylation of histone H3 Ser10 by wild-type VRK in vitro. (G and H) Phosphorylation results for monomeric BAF (mBAF) and histone H3 as substrates were compared in different concentrations of VRK1 (G) and for various time periods of incubation (H). Bars and error bars represent means ± standard deviations (n = 3) (G and H). In all panels, the quantity of loaded sample was visualized by staining with Coomassie brilliant blue (CBB). VRK1 lacking a GST moiety was used in panels B, E, and F.
Cell culture and transfection.
HeLa and H1299 cells were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100 U/ml of both penicillin G and streptomycin. Transient transfection of HeLa and H1299 cells was carried out using Metafectene reagent (Biontex, Munich, Germany) according to the manufacturer's protocol. For immunocytochemical analysis, cells were grown on a glass chip coated with poly-d-lysine. Transfected cells were maintained or treated with drugs for appropriate times before microscopic observation. The small interfering RNA (siRNA) duplex targeting human VRK1 (siVRK1 combined with two siRNA duplexes, GCTAAGCTTAAGAATTCTG and CAAGGAACCTGGTGTTGAA) and targeting human aurora B (siAurB; catalog no. L-003326-00) and the control scrambled siRNA (siCont) were obtained from Dharmacon (Lafayette, CO).
Cell cycle synchronization and flow cytometry.
HeLa cells were synchronized as described previously (26). Briefly, cells were arrested in mitotic phase by treatment for 12 h with 10 nM taxol (Paclitaxel; Sigma) and 0.4 μg/ml nocodazole (Sigma), in G1 by treatment for 24 h with 2 mM hydroxyurea, and in G1/S by treatment for 20 h with 200 μM mimosine. A double thymidine blocking was performed as described previously (16). Briefly, cells were treated with 2 mM thymidine for 20 h, released for 6 h in fresh DMEM containing 10% FBS, and treated again with 2 mM thymidine for 12 h. Cells arrested in G1/S were released by switching the medium to fresh DMEM containing 10% FBS. Aliquots were analyzed by flow cytometry, and the remaining cells were used for the preparation of whole-cell extracts or subcellular fractions. For flow cytometric analysis, the cells were fixed with 70% ethanol for 20 min, stained with 20 μg/ml propidium iodide, and treated with 1 μg/ml RNase A for 10 min at room temperature. Samples containing 10,000 cells were then analyzed on a FACSCalibur system (Becton Dickinson).
Purification of recombinant proteins and GST pulldown assay.
Monomeric BAF protein was prepared from the insoluble fraction by tobacco etch virus protease treatment to cleave the hexameric His tag as described previously (33). The purification of GST fusion proteins and the GST pulldown assay were carried out as described previously (23, 25).
Semiquantitative RT-PCR.
Total RNA was isolated using TRI reagent (Molecular Research Center) according to the manufacturer's instructions. Reverse transcription-PCR (RT-PCR) was carried out as described previously (22). Briefly, following DNase I (Promega) treatment, RNA was reverse transcribed using oligo(dT) (Promega) and monkey murine leukemia virus reverse transcriptase (Roche) in the presence of RNasin (Promega). To obtain semiquantitative results, primers for both GAPDH and VRK1 were included in each reaction mixture.
RESULTS
Cell cycle-regulated expression of VRK1.
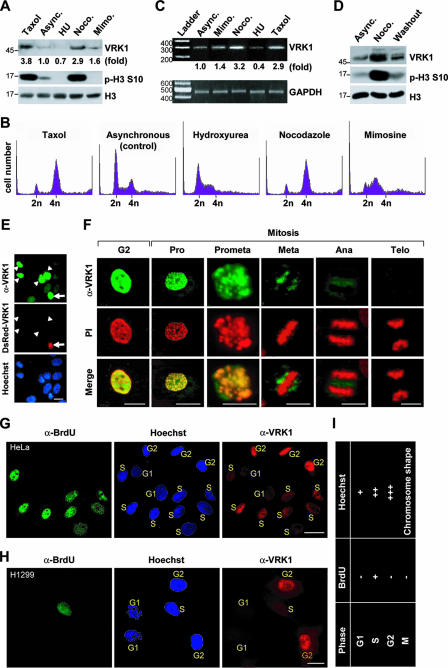
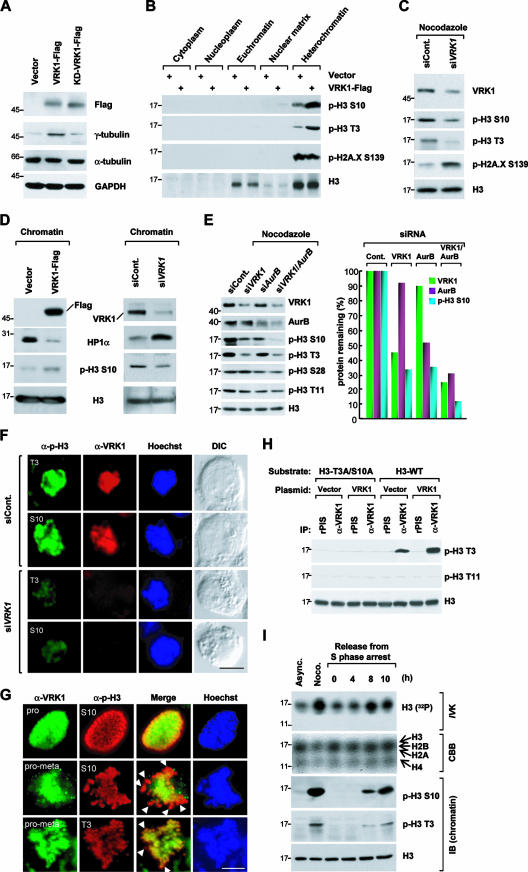
We monitored VRK1 expression during cell cycle progression by use of a VRK1-specific antibody which only recognized VRK1 in various experiments, including immunoblotting, immunoprecipitation, and immunocytochemistry (see Fig. S1 in the supplemental material). Mitotic arrest induced by treatment with nocodazole or taxol elevated the VRK1 protein (Fig. 1A) and mRNA (Fig. 1C) levels. In contrast, cells arrested at the G1 or S phase by treatment with hydroxyurea or mimosine, respectively, showed decreased or moderately elevated VRK1 expression compared to asynchronous control cells (Fig. 1A and C). The increase in VRK1 expression during mitosis was further confirmed using a washout study. Nocodazole washout triggered the transition of synchronized cells from mitotic phase to G1 phase accompanied by a decrease in VRK1 expression (Fig. 1D). Nuclear localization of VRK1 was confirmed by immunostaining with an anti-VRK1 antibody (Fig. 1E). Unexpectedly, the endogenous VRK1 content differed among cells at different phases of the cycle (Fig. 1E). Specifically, in mitosis, intense VRK1 staining at the prophase was observed which gradually decreased and was barely detectable from the anaphase to the telophase (Fig. 1F). To determine the cellular expression of VRK1 in asynchronously growing HeLa, cells in the S phase were labeled with BrdU for 1 h. We additionally applied Hoechst staining to identify cells at the G1 (relatively weak staining) or G2 (relatively intense staining) phase. Cells in the M phase were identified according to their unique chromosomal shape following Hoechst staining. Thus, the cell cycle stages for individual cells were determined using a combination of Hoechst staining and BrdU labeling (see Fig. 1I for a cell cycle chart based on staining patterns). This method disclosed clear differences in the cellular content of VRK1 according to the cell cycle stage. At G1, VRK1 was barely detectable, but it gradually accumulated from the S to G2 phase in both HeLa (Fig. 1G) and H1299 (Fig. 1H) cells. Based on these results, we conclude that VRK1 is differentially expressed during cell cycle progression and peaks in the G2/M phase.
FIG. 1.
Profile of VRK1 expression during cell cycle progression. (A to C) Differential expression of VRK1 during cell cycle progression. Total extracts were prepared from asynchronously growing HeLa cells (Async. [control]) or cells treated with taxol and nocodazole (Noco.) for mitotic arrest or hydroxyurea (HU) and mimosine (Mimo) for G1/S arrest. The extracts were analyzed by immunoblotting with the indicated antibodies (A). Cell cycle synchronization was validated by flow cytometric analyses (B). The level of VRK1 transcript during each phase of the cell cycle was measured by semiquantitative RT-PCR (C). (D) VRK1 protein levels from asynchronously growing HeLa cells following a 12-h treatment with nocodazole and then a 1-h washout with fresh medium. (E) Immunostaining for VRK1 (green) and DNA (Hoechst; blue) in HeLa cells transfected with DsRed1-C1-VRK1. The arrow indicates a transfected cell, and arrowheads indicate nontransfected cells with a high level of endogenous VRK1 expression. (F) Immunostaining for VRK1 (green) and DNA staining (propidium iodide [PI]; red) during interphase and mitosis. Pro, prophase; Prometa, prometaphase; Meta, metaphase; Ana, anaphase; Telo, telophase. (G and H) Immunostaining for S-phase cells (BrdU-positive; green), VRK1 (red), and DNA staining (Hoechst; blue) in HeLa (G) or H1299 (H) cells. The stages of the cell cycle are indicated in yellow and were evaluated according to the criteria presented in panel I. (I) Chart to evaluate the stage of the cell cycle according to BrdU and Hoechst staining results. The scale bars in panels E to H represent 10 μm.
Chromatin localization of VRK1.
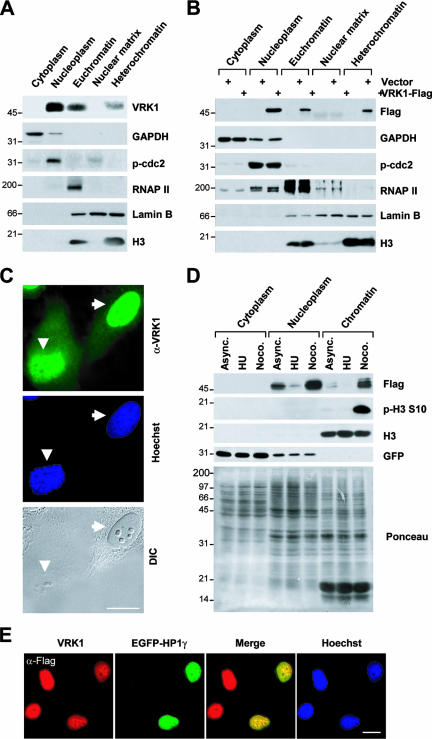
VRK1 is a nuclear protein (27, 32). To determine its subnuclear localization, nuclei were biochemically fractionated. Both endogenous VRK1 (Fig. 2A) and Flag-tagged VRK1 (Fig. 2B) were predominantly located in the nucleoplasm; a substantial amount of VRK1 was identified in the chromatin-enriched fractions (Fig. 2A and B). Chromatin localization of VRK1 was further confirmed by immunocytochemical analysis. A high VRK1 level was observed in the area of dense chromosomes, even in cells lacking a nuclear envelope (Fig. 2C). Cell cycle synchronization by drug treatment after transient expression of Flag-VRK1 further confirmed subcellular localization. The Flag-VRK1 level varied depending on the stage of cell cycle arrest (Fig. 2D). Compared to control asynchronous cell results, the VRK1 protein level was lower in G1/S and higher in mitotic phase (Fig. 2D). Interestingly, the chromatin-localized VRK1 level was higher in mitotic arrested cells than in asynchronously growing or G1/S-arrested cells (Fig. 2D). Moreover, VRK1 colocalized with gamma heterochromatin protein 1 (HP1γ) (Fig. 2E), a well-known chromatin-associated protein (15), which is consistent with results from a previous proteomic analysis showing VRK1 to be a chromatin-associated protein (45). Similarly, affinity chromatography with the DNA fragment of Cyp2b10, the phenobarbital-inducible p450 gene, was applied to identify VRK1 (48). The data strongly suggest that VRK1 is localized not only in nucleoplasm but also in chromatin.
FIG. 2.
Subnuclear localization of VRK1 protein. (A and B) Fractions from untransfected cells (A) or pFlag-VRK1-transfected cells (B) were analyzed by immunoblotting with the indicated antibodies. Antibodies against GAPDH, phospho-cdc2 (p-cdc2), RNA polymerase II (RNAP II), lamin B, and histone H3 (H3) were used as markers for the various fractions. (C) Immunostaining for VRK1 staining (green) and DNA staining (Hoechst; blue). Arrows indicate cells in interphase (nuclear envelope present), and arrowheads indicate mitotic cells (nuclear envelope absent). DIC, differential interference contrast. (D) Expression of Flag-VRK1 and its accumulation in chromatin during the cell cycle. HeLa cells were cotransfected with pFlag-VRK1 and pEGFP-N1 for 12 h, and then cell cycle was arrested at G1/S phase induced by hydroxyurea (HU) and mitotic phase induced by nocodazole (Noco.). Subcellular fractions were prepared as described in Materials and Methods. Ponceau S staining of the blot shows the quantity and quality of the fractions. It is notable that there is no nucleoplasm in mitotic cells (Noco. treated); the label nucleoplasm was used here for convenience to indicate the biochemical preparation for the asynchronous (Async.) control and G1/S-arrested cells. (E) Colocalization of VRK1 and HP1γ. Cells were cotransfected with pFlag-VRK1 (upper panel) and pEGFP-HP1γ and then stained for Flag (red; upper panel) and DNA (Hoechst; blue). Scale bars in panels C and E represent 10 μm.
Association of VRK1 and core histones.
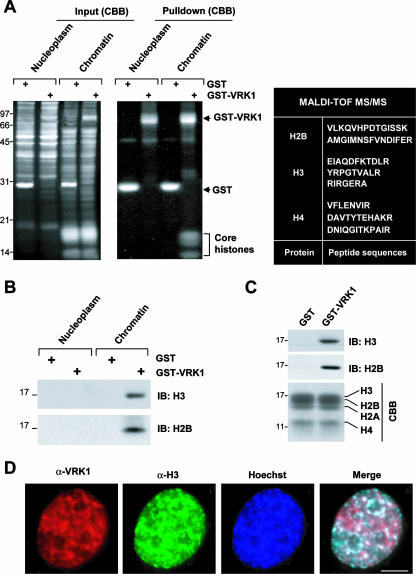
To investigate the significance of VRK1 association with chromatin, we isolated VRK1-binding proteins from soluble chromatin extracts. A GST pulldown assay led to the identification of four bands ranging from 14 to 17 kDa (Fig. 3A, middle panel). Matrix-assisted laser desorption ionization-time of flight (tandem mass spectrometry) (MALDI-TOF MS/MS) analyses revealed that three of these proteins were core histones H2B, H3, and H4 (Fig. 3A, right panel). Immunoblotting of the gel (shown in the middle panel in Fig. 3A) confirmed interactions of histone H3 and H2B with VRK1 (Fig. 3B). Pulldown assays using GST (control) or GST-VRK1, followed by immunoblotting for H3 and H2B, disclosed direct binding (Fig. 3C). Immunocytochemistry analyses further supported the colocalization of VRK1 and H3 (Fig. 3D). Since histones are basic proteins and VRK1 autophosphorylates at several sites as well (27), we examined whether autophosphorylation is required for histone binding. Dominant-negative and kinase-dead mutants of VRK1 generated by deletion of a partial ATP-binding site (amino acids 43 to 49) of wild-type VRK1 and by point mutation of Lys179 to Glu, respectively, still bound H3 (see Fig. S2 in the supplemental material), indicating that VRK1 autophosphorylation is not necessary for H3 binding or colocalization with core histones in chromatin.
FIG. 3.
Association and colocalization of VRK1 and core histones in chromatin. (A) Identification of core histones as binding partners of VRK1. VRK1 binding proteins were identified in the nucleoplasm and solubilized chromatin fractions by use of a GST pulldown assay. Left panel, Coomassie brilliant blue (CBB)-stained gel containing total extract from each fraction and GST or GST-VRK1; middle panel, CBB-stained gel after pulldown using GST or GST-VRK1. Core histones were identified by MALDI-TOF MS/MS, and the peptide sequences are presented in the right panel. (B) Immunoblotting (IB) results for H3 and H2B. The gel shown in the middle of panel A was transferred to a nitrocellulose membrane and analyzed by immunoblotting with antibodies against histones H3 and H2B. (C) Direct binding of VRK1 and core histone proteins. Purified free core histones were pulled down with GST or GST-VRK1. (D) Colocalization of VRK1 and H3. HeLa cells were stained for VRK1 (red), histone H3 (green), and nuclei (Hoechst; blue). Bar, 5 μm.
Phosphorylation of H3 by VRK1 in vitro.
NHK-1 phosphorylates histones in the context of chromatin (1). Our binding data showing interaction between VRK1 and core histones (Fig. 3), as well as sequence and structural similarities between VRK1 and NHK1 (1), suggest that VRK1 phosphorylates histone proteins. To examine this possibility, we performed an in vitro kinase assay using free core histones as substrates. The upper band observed with free histone H3 or H2B was a VRK1-phosphorylated form (Fig. 4A). To confirm the relevance of these results, we examined the phosphorylation of pure histone proteins. VRK1 phosphorylated H3 strongly and H2A, H2B, and H4 weakly and did not phosphorylate H1 (Fig. 4B). In contrast, NHK1 phosphorylated histone proteins only when present in the nucleosome (1). Thus, VRK1 displays histone kinase activity, but its specificity is somewhat different from that of NHK1. The N-terminal tail of H3 is phosphorylated at several sites, including Thr3 by haspin (5), Ser10 and Ser28 by aurora B (3, 12, 18), and Thr11 by Dlk/ZIP kinase (37). To identify the VRK1 phosphorylation sites on histone H3, we conducted an in vitro kinase assay and analyzed the phosphorylated protein by immunoblotting with site-specific anti-phospho-H3 antibodies. VRK1 phosphorylated purified histone H3 at two sites, specifically, Thr3 and Ser10 (Fig. 4C). Phosphorylation at these sites was additionally observed in purified free core histones (Fig. 4C) and monomeric nucleosomes (Fig. 4D). To exclude other phosphorylation sites, we generated site-directed mutants of H3 designated T3A, S10A, and T3A/S10A. VRK1 phosphorylated T3A or S10A at the nonmutated site but could not phosphorylate the T3A/S10A mutant, which contained substitutions at both sites (Fig. 4E). Our results indicate that VRK1 phosphorylates H3 at both Thr3 and Ser10.
Finally, a dominant-negative mutant of VRK1 eliminated Ser10 phosphorylation (Fig. 4F), confirming that this process is mediated by VRK1. It is known that VRK1 phosphorylates BAF (33). Thus, we compared VRK1-mediated protein phosphorylation results using BAF and histone H3. The phosphorylation patterns of BAF and histone H3 were similar, and the intensity was in proportion to VRK1 concentration (Fig. 4G) and incubation duration (Fig. 4H), although phosphorylation of greater intensity was found when monomeric BAF was used as a substrate (Fig. 4G and H). We thought that one of the reasons that BAF showed stronger phosphorylation than histone H3 during the in vitro kinase assay might be the difference in the number of residues phosphorylated by VRK1. VRK1 phosphorylates at three different residues of BAF with identical levels of potency (33), while VRK1 phosphorylates at Thr3 similarly strongly but phosphorylates Ser10 at the weaker level seen with histone H3 in vitro.
Phosphorylation of H3 by VRK1 in vivo.
To establish whether VRK1 kinase is required for histone H3 phosphorylation in vivo, we examined the effects of overexpressing Flag-VRK1 or knocking down VRK1 expression by use of siRNA. Overexpression of wild-type VRK1 increased the level of the mitotic index protein γ-tubulin approximately fivefold in comparison to the results seen with the vector control or the kinase-dead mutant of VRK1 (Fig. 5A) as well as the level of H3 phosphorylation at both Thr3 and Ser10 in the heterochromatin-enriched fraction (Fig. 5B). Consistently, knockdown of endogenous VRK1 by siRNA reduced phosphorylation of H3 at both Thr3 and Ser10 (Fig. 5C). Ser10 but not Thr3 phosphorylation can be confirmed indirectly, as previous studies showed that HP1 binding to chromatin and H3 phosphorylation at Ser10 are mutually exclusive (41). HP1 is loaded on chromatin via binding to triple-methylated Lys9 of H3 (8), which suppresses Ser10 phosphorylation. However, when Ser10 is phosphorylated first, HP1 binding to chromatin is inhibited (7, 17).
FIG. 5.
Phosphorylation of Thr3 and Ser10 in H3 by VRK1 in vivo. (A) Increase of the mitotic index (γ-tubulin) after transient expression of VRK1. Total extracts from HeLa cells transfected with vector (pFlag), Flag-VRK1, or Flag-tagged kinase-dead VRK1 (KD-VRK1) were analyzed by immunoblotting (IB) with the indicated antibodies. The mitotic index was determined from the level of γ-tubulin. (B) Increased phosphorylation of histone H3 at Thr3 and Ser10 in heterochromatin induced by overexpression of VRK1. Subcellular fractions from HeLa cells transfected with either Flag-VRK1 or vector alone were analyzed by immunoblotting. (C) Reduction of phosphorylation of Thr3 and Ser10 in H3 by knockdown of VRK1 by use of siRNA. HeLa cells transfected with siControl or siVRK1 were treated with nocodazole (Noco.) for 12 h to enhance the basal level of phospho-Thr3 and phospho-Ser10. (D) Regulation of the localization of HP1α in chromatin by VRK1. Overexpression of VRK1 decreases the level of HP1α in the chromatin fraction, whereas knockdown of VRK1 increases it. (E) VRK1 and aurora B independently phosphorylate Ser10 of H3. HeLa cells transfected with siControl, siVRK1, siAurB, and both siVRK1 and siAurB were treated with nocodazole for 12 h to enhance the basal level of phospho-Ser10. Cells on the mitotic phase were collected by shaking-off method, and then total extracts from each transfected cells were analyzed by immunoblotting with the indicated antibodies. Representative results from three independent experiments are presented. (F) HeLa cells transfected with siControl and siVRK1 for 24 h were further treated with nocodazole for 12 h. Cells on the mitotic phase were observed using fluorescence microscopy. Cells were stained for VRK1 (red), phospho-histone H3 (green), and nuclei (Hoechst; blue). DIC, differential interference contrast. Scale bar, 5 μm. (G) Colocalization of phospho-Thr3 and phospho-Ser10 with VRK1 in early mitotic cells. Immunostaining for VRK1 (green), phospho-H3 Thr3, or phospho-H3 Ser10 (red) and DNA (Hoechst; blue). Arrowheads indicate highly phosphorylated regions in the DNA. Scale bar, 5 μm. pro, prophase; pro-meta, prometaphase. (H) Control experiment for the validation of in vitro kinase assay using immunoprecipitated (IP) VRK1. Immunoprecipitated VRK1 prepared from control or pFlag-VRK1-transfected cells were used in the in vitro kinase assay with wild-type or mutated histones as substrates. The VRK1 kinase activity was assessed by immunoblotting with an anti-phospho-Thr3 antibodies. (I) Endogenous VRK1 kinase activity during the cell cycle. HeLa cells were manipulated in the cell cycle by treatment with nocodazole (Noco.; for mitotic arrest) or by double thymidine blocking (0 h for S arrest; 4 h for G2 arrest; 8 and 10 h for mitotic arrest). VRK1 was immunoprecipitated and used in an in vitro kinase (IVK) assay with free core histones as substrates (upper panel). Coomassie brilliant blue (CBB) staining shows the quantity of substrate in each reaction. The stage of the cell cycle was validated by immunoblotting using anti-phosphorylated histone antibodies. Async., asynchronous.
In accordance with the theory of mutually exclusive binding, overexpression or knockdown of VRK1 altered the association of HP1α in chromatin fractions (Fig. 5D). By examining chromatin-loaded HP1α, we could indirectly verify VRK1-mediated Ser10 phosphorylation of H3. Since aurora B is a known histone H3 kinase during mitosis (3, 18), we examined its relationship with VRK1 with regard to H3 phosphorylation at Ser10. HeLa cells transfected with siRNAs indicated in Fig. 5E for 24 h were further grown in the culture media for 12 h in the presence of nocodazole to be arrested at the mitotic prometaphase. Cells on the mitotic phase were collected by a shaking-off method. Expectedly, depletion of VRK1 resulted in the reduction of phosphorylation at both Thr3 and Ser10 in histone H3 but not at Thr11 and Ser28, whereas depletion of aurora B resulted in the reduction of phosphorylation at both Ser10 and Ser28 but not at Thr3 and Thr11 (Fig. 5E). Interestingly, both aurora B and VRK1 were required for phosphorylation of Ser10. Furthermore, a double knockdown of the two genes dramatically reduced Ser10 phosphorylation (Fig. 5E). When we treated siVRK1, we observed a dramatic decrease in histone H3 Thr3 and Ser10 phosphorylation even though the cells were synchronized at prometaphase in the presence of nocodazole (Fig. 5F). Phosphorylation of histone H3 on Thr3 is very prominent at the prometaphase in HeLa cells (5). The incomplete knockdown of histone H3 phosphorylation shown in Fig. 5F might have been due to the action of other mitotic histone kinases, such as haspin and aurora B, that can phosphorylate Thr3 and Ser10, respectively. VRK1 expression and H3 phosphorylation at Thr3 and Ser10 coincided in mitotic cells. Specifically, during the prophase, VRK1 and phosphorylated H3 colocalized throughout the nucleus, and during prometaphase, VRK1 was delocalized from a more restricted area containing a high level of phosphorylated H3 (Fig. 5G). To measure the intrinsic activity of VRK1 during the cell cycle, we used immunoprecipitated VRK1, which was validated to check the phosphorylation at Thr3 but not at Thr11 of histone H3 (Fig. 5H). VRK1 kinase activity and expression were increased at late G2 or early M phase (Fig. 5I). The results collectively suggest that VRK1 is a key mediator of phosphorylation of histone H3 at Thr3 and Ser10 in vivo.
Hyperphosphorylation of histone H3 by a hyperactive form of VRK1.
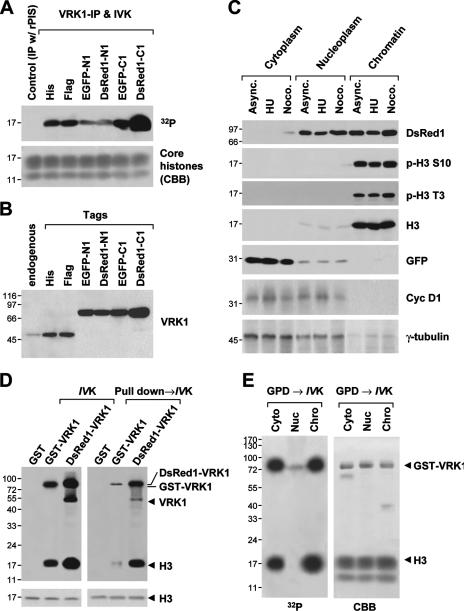
Among the tagged (including His, Flag, enhanced GFP, and DsRed1) VRK1 constructs, VRK1 fused to the C terminus of DsRed1 (DsRed1-C1-VRK1) displayed the strongest kinase activity (Fig. 6A) and highest protein level (Fig. 6B). While Flag-VRK1 was localized in chromatin of mitotic cells (Fig. 2D), DsRed1-C1-VRK1, which was highly expressed in chromatin, bypassed cell cycle stages determined by the levels of marker proteins such as cyclin D1 for G1/S phase and γ-tubulin for mitotic phase and led to the hyperphosphorylation of Thr3 and Ser10 in histone H3 (Fig. 6C) compared to Flag-VRK1 results (Fig. 2D). The mechanism underlying the strong kinase activity of DsRed1-C1-VRK1 is unclear at present and requires further structural analysis. During an in vitro kinase assay, both purified GST-VRK1 and immunoprecipitated-DsRed-VRK1 phosphorylated core histone as a substrate (Fig. 6D, left panel). However, upon the incubation with total cell extracts, while the activity of DsRed-VRK1 was reduced only 1.4-fold, that of GST-VRK1 was reduced more than 10-fold (Fig. 6D, right panel). Accordingly, we speculated that a strong inhibitory factor in total cell extracts may suppress VRK1 activity. To determine where this inhibition occurs in cells, we fractionated cell extracts into cytoplasmic, nucleoplasmic, and crude chromatin fractions followed by incubation with VRK1 to analyze its activity. Inhibition of VRK1 was evident upon incubation with the nucleoplasm fraction (Fig. 6E). However, we could not identify the inhibitory binding partner in nucleoplasm. The data collectively suggest that DsRed1-C1-VRK1 is a constitutively active form of VRK1 and is not affected by an unidentified nuclear inhibitor that strongly suppresses VRK1 activity and hence shows extraordinarily stronger kinase activity than any other tag-fused VRK1 proteins.
FIG. 6.
DsRed1-C1-VRK1 is a constitutively active form of VRK1. (A) In vitro kinase assay (IVK) of immunoprecipitated VRK1 (VRK1-IP) from cells transfected with various VRK1-expressing vectors. DsRed1-C1-tagged VRK1 displayed the strongest kinase activity. Purified core histones were used as the substrate. Coomassie brilliant blue (CBB) staining was used to estimate the substrate amounts. (B) Immunoblot of anti-VRK1 from total cell extracts of each VRK1-expressing sample. (C) Subcellular distribution and expression of DsRed1-C1-VRK1 in asynchronously growing, G1/S arrest induced by hydroxyurea (HU) and mitotic arrest induced by nocodazole (Noco.) cells. Cells asynchronously growing (Async.; control) cotransfected with DsRed1-C1-VRK1 and enhanced GFP-N1 were fractionated as described in Materials and Methods. The levels of cyclin D1 and γ-tubulin were used as cell cycle markers for the G1/S and mitotic phases, respectively. (D) Preincubation of VRK1 with total cell extracts reduced the kinase activity of VRK1. A conventional in vitro kinase assay (IVK) of GST-VRK1 and DsRed1-VRK1 prepared by immunoprecipitation using an anti-VRK1 antibody was performed with histone H3 as a substrate. GST-VRK1 employed in a pulldown assay (pull down→IVK) was prepared as follows: GST-VRK1 was initially preincubated with total cell extracts overnight at 4°C and collected by affinity pulldown (GPD) using glutathione beads. GST-VRK1 on beads was released by treatment with reduced glutathione. (E) Nucleoplasmic inhibition of VRK1. An in vitro kinase assay of GST-VRK1 was carried out as described for panel D except with preincubation with extracts from cytoplasm (cyto), nucleoplasm (nuc), and chromatin (chro).
Nuclear condensation by overexpressed VRK1.
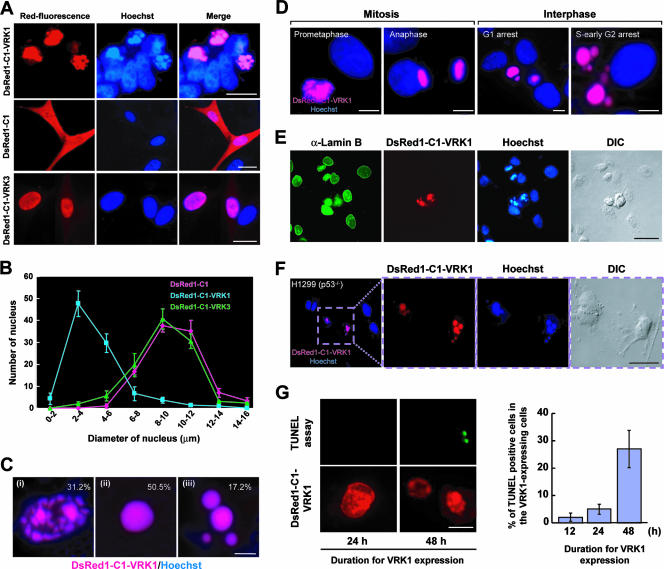
Never-in-mitosis in Aspergillus nidulans (NIMA) histone H3 Ser10 kinase is the single protein known to date that induces shrinkage of the nucleus when ectopically overexpressed in mammalian cells (28). In our experiments, transfection of cells with DsRed1-C1-VRK1 led to dramatic nuclear condensation (Fig. 7A) similar to that observed in transient expression of NIMA, the fungal enzyme, in eukaryotic cells (28). In addition, cells transfected with DsRed1-C1-VRK1 displayed significantly more condensed nuclear shrinkage than control (DsRed1-C1-transfected) cells and those transfected with DsRed1-C1-VRK3 (Fig. 7B). Condensed nuclei were classified into three types according to morphology, specifically, heterochromatinized foci (Fig. 7Ci), uniform comprehensive condensation (Fig. 7Cii), and partitioned comprehensive condensation (Fig. 7Ciii). A percentage of the three classification types are shown in Fig. 7C. The highest levels of uniform and partitioned comprehensive condensation occurred in G1 and G2/S cells, respectively (Fig. 7D). Partially condensed foci were detected from G1 to G2, which may represent an intermediate stage prior to comprehensive condensation. Endogenous VRK1 was dissociated from chromatin during mitosis progression, and its expression decreased from the prometaphase to the anaphase (Fig. 1F), whereas DsRed1-C1-VRK1 remained associated with chromatin even at the anaphase (Fig. 7D). Therefore, the phosphorylation of histone H3 by DsRed1-C1-VRK1 may overwhelm the endogenous histone phosphatase activity, resulting in hyperphosphorylation of histone H3 at Thr3 and Ser10 and abnormal nuclear morphology. Immunostaining for lamin B (Fig. 7E) revealed that nuclear hypercondensation caused by DsRed1-C1-VRK1 triggered crushing of the nuclear envelope. This finding is similar to previous data obtained from immunostaining for NIMA kinase (28). Since VRK1 phosphorylates p53 protein (27, 46), we tested whether this nuclear condensation occurs in H1299 cells that do not express p53. Our data show that nuclear condensation caused by DsRed1-C1-VRK1 is independent of the presence of p53 protein (Fig. 7F). In addition, we demonstrated that the DsRed1-C1-VRK1-expressing cells were alive by showing negative staining for the TUNEL assay (Fig. 7G); however, when expression continued for a longer duration, approximately 30% of VRK1-expressing cells gave positive results in the TUNEL assay (Fig. 7G). These results imply that this nuclear condensation is not associated with potential apoptosis events. To our knowledge, DsRed1-C1-VRK1 is the first mammalian mitotic protein kinase that induces nuclear condensation.
FIG. 7.
Induction of nuclear condensation by DsRed1-C1-VRK1. (A and B) HeLa cells were transfected with control vector (DsRed1-C1), DsRed1-C1-VRK1, or DsRed1-C1-VRK3 and then analyzed by microscopy after 24 h (A). The diameter of each nucleus measured as the long axis was analyzed using Axiovision software (Carl Zeiss) (B). (C) Representative nuclear morphologies of DsRed1-C1-VRK1-expressing cells. Condensed nuclei were composed of heterochromatinized foci (panel i; 31.2%), uniform comprehensive condensation (panel ii; 50.5%), and partitioned comprehensive condensation (panel iii; 17.2%). (D) Chromatin localization and nuclear condensation during cell cycle in the cells transfected with DsRed1-C1-VRK1. A combination of hydroxyurea and mimosine was used to arrest cells in G1 phase, and double thymidine blocking and subsequent release for 1 h was used to obtain cells in early S or G2 phase. Mitotic cells were identified on the basis of the DNA morphology. (E) Disorganization of the nuclear envelope induced by the presence of DsRed1-C1-VRK1. HeLa cells transfected with DsRed1-C1-VRK1 were immunostained for lamin B (green) and DNA (Hoechst; blue). (F) Nuclear condensation of VRK1 in p53 null H1299 cells. The three panels at the right show magnifications of the dotted square in the leftmost panel. (G) Apoptotic DNA fragmentation was determined by TUNEL assays in the cells transfected with DsRed1-C1-VRK1 for the indicated durations. Scale bars represent 10 μm in panel A, 5 μm in panels C, D, and H, and 20 μm in panels E and F. DIC, differential interference contrast.
DISCUSSION
Chromatin-localized VRK1 as a histone kinase.
VRK1 phosphorylates transcription factors, including p53 (27), c-Jun (43), and ATF2 (44). These phosphorylation reactions predominantly occur in the nucleus. The cytosol contains several structural and functional organelles, such as endoplasmic reticulum, Golgi body, and lysosome. Notably, the nucleus also contains subnuclear structures including nucleolus, promyelocytic oncogenic domain, Cajal body, and histone deacetylase-enriched foci as well as euchromatin and heterochromatin regions (13). These subnuclear structures lack a membranous barrier, which facilitates translocation among them. However, the intrinsic functions of a nuclear protein are restricted by the requirements of the exact subnuclear localization. The present report shows that endogenous VRK1 localizes in the chromatin and the nucleoplasm as well. First, biochemical analyses of subnuclear distribution revealed significant levels of both endogenous and transiently expressed ectopic VRK1 in chromatin. Second, core histones, the fundamental components of chromatin, were identified as binding partners of VRK1, while there was no evident binding protein in nucleoplasm that interacted with VRK1 during an in vitro binding assay. Third, HP1γ, a well-characterized chromatin-localized protein, colocalized with VRK1. Moreover, BAF, a newly identified substrate protein of VRK1, localizes in chromatin (33).
The chromatin localization of VRK1 indicates a role in the phosphorylation of core histones, which are binding proteins in the nucleus. In this report we show that VRK1 phosphorylates histone H3 at two sites, Thr3 and Ser10, both in vitro and in vivo. Earlier studies provide evidence that these sites are phosphorylated by haspin (Thr3) and aurora B (Ser10). The phosphorylation patterns of these two sites were similar with respect to elapsed time. Specifically, phosphorylation become apparent at early mitosis (from late G2 to prophase), peaked at the metaphase, and disappeared at late mitosis (telophase). Our results suggest that VRK1 plays an essential role in the mitotic phase by phosphorylating Thr3 and Ser10 of histone H3. Moreover, the contribution of VRK1 with respect to Ser10 phosphorylation was similar with aurora B. It is reported that lack of histone H3 phosphorylation at Thr3 causes accumulation of cells in prometaphase and leads to failure of metaphase chromosome alignment (5). Inhibition of Ser10 phosphorylation by inactivating aurora B with hesperadin, an inhibitor, also leads to improper attachment of kinetochore fibers to the chromosome (24), changes in binding properties of chromatin proteins such as HP1 (7, 17), and failure to recruit the condensin complex and mitotic spindle assembly (10). NHK-1, a Drosophila homolog of VRK1, phosphorylates T119 in histone H2A and plays an essential role in mitotic progression and in the formation of chromosome architecture in meiosis (1, 4, 19). Although the net level of phosphorylation of histone H2A by VRK1 was weaker than that seen with H3 during in vitro kinase assay (Fig. 4B), it is possible that VRK1 phosphorylates T119 in H2A in vivo. Hence, future study on the relationship between the distinct phosphorylation patterns of different histones will open the door to deciphering the histone code, especially with respect to maintenance and the dynamics of chromatin architecture. In accordance with these previous studies, our results suggest that VRK1-mediated phosphorylation of Thr3 and Ser10 in histone H3 is functionally linked to the formation of proper chromosome architecture.
Overexpression of DsRed1-C1-VRK1, the constitutively active form of the kinase, induced marked nuclear condensation in transfected cells regardless of the cell cycle stages, in similarity to data obtained following ectopic expression of NIMA in mammalian cells. The existence of a NIMA-like pathway in mammalian cells has been proposed previously (28). Our data provide evidence that the NIMA-like pathway in mammals is mediated by VRK1. Transient expression of DsRed1-C1-VRK1 caused hyperphosphorylation of Thr3 and Ser10 of histone H3, regardless of the cell cycle stage, and concomitantly induced dramatic nuclear condensation, supporting the theory that VRK1 is the mammalian functional counterpart of NIMA. Another possibility is that VRK1 phosphorylates alternative sites in histones or other target proteins that play critical roles in chromatin condensation.
Cell cycle-regulated expression of VRK1.
Recently, BAF was revealed as a bona fide substrate of VRK1 (33). VRK1 phosphorylates BAF following a short incubation period during an in vitro kinase assay. Subcellular localization of BAF is regulated during cell cycle progression (42). During interphase, BAF is present in the nucleus as a complex with chromatin formed via direct interactions with DNA and with the nuclear envelope via association with components of a nuclear lamin complex such as LAP2β, emerin, and MAN1. In contrast, BAF localization becomes dynamic during mitotic progression. From the metaphase to the anaphase, BAF is dissociated from chromatin and localizes to the cytoplasm. In the telophase, BAF relocalizes to chromatin. Such dynamic subcellular localization of BAF is possibly regulated by its phosphorylation status (2), which in turn is modulated by VRK1 (33). Our present findings demonstrate that VRK1 expression and activity are regulated in a cell cycle-dependent manner. A gradual increase in VRK1 expression was observed from the G1 to the mitotic phase. Moreover, VRK1 activity correlated with expression. Thus, the expression and activity profiles of VRK1 may affect phosphorylation status and trigger dynamic localization of BAF during the cell cycle.
Interestingly, expression of Flag-tagged VRK1 under control of the cytomegalovirus promoter was variable in transfected cells, suggesting the existence of another mechanism to regulate the amount of VRK1 protein. Protein degradation mediated by APC/C (anaphase promoting complex-cyclosome) is critical for modulating the amount of cell cycle regulators and mitotic progression (35). APC/C is a major ubiquitin E3 enzyme responsible for the degradation of mitotic kinases, including aurora A and B, Nek2A, and Plk1, via a proteasome-dependent pathway (9, 35). The VRK1 protein level was also altered during the cell cycle. Following nocodazole washout to induce cell cycle progression, MG132 treatment blocked the degradation of VRK1 and inhibited exit from the mitotic phase, as evaluated by assays of the phospho-H3 S10 level (see Fig. S3 in the supplemental material). Thus, while the nature of the precise molecular mechanism involved in VRK1 degradation awaits further investigation, regulation of VRK1 stability by the proteasome is one of the potential mechanisms for its expression profile during the cell cycle.
In summary, our results provide evidence for the function and expression of chromatin-associated VRK1 and establish the mitotic histone kinase activity profile during cell cycle progression. We propose that VRK1 phosphorylates core histones mainly at the G2/M transition to trigger chromosome condensation.
Supplementary Material
Acknowledgments
We thank C. W. Lee (Sungkyunkwan University, Seoul, Republic of Korea) for critical discussion and providing histone-related antibodies, J. B. Kwon (Ewha Womans University, Seoul, Republic of Korea) for providing purified free core histone proteins, S. H. Ryu (POSTECH, Pohang, Republic of Korea) for analyzing MALDI-TOF MS/MS, and D. J. Jun (POSTECH, Pohang, Republic of Korea) for assistance with the FACS analysis.
This study was supported by a grant from the National R&D Program for Cancer Control (0520250-2), by the Ministry of Health & Welfare, by a Biotech grant supported by POSCO, and the Brain Korea 21 program of the Ministry of Education, Republic of Korea.
Footnotes
Published ahead of print on 15 October 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aihara, H., T. Nakagawa, K. Yasui, T. Ohta, S. Hirose, N. Dhomae, K. Takio, M. Kaneko, Y. Takeshima, M. Muramatsu, and T. Ito. 2004. Nucleosomal histone kinase-1 phosphorylates H2A Thr 119 during mitosis in the early Drosophila embryo. Genes Dev. 18:877-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bengtsson, L., and K. L. Wilson. 2006. Barrier-to-autointegration factor phosphorylation on Ser-4 regulates emerin binding to lamin A in vitro and emerin localization in vivo. Mol. Biol. Cell 17:1154-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crosio, C., G. M. Fimia, R. Loury, M. Kimura, Y. Okano, H. Zhou, S. Sen, C. D. Allis, and P. Sassone-Corsi. 2002. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol. Cell. Biol. 22:874-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cullen, C. F., A. L. Brittle, T. Ito, and H. Ohkura. 2005. The conserved kinase NHK-1 is essential for mitotic progression and unifying acentrosomal meiotic spindles in Drosophila melanogaster. J. Cell Biol. 171:593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dai, J., S. Sultan, S. S. Taylor, and J. M. Higgins. 2005. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev. 19:472-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Souza, C. P., A. H. Osmani, L. P. Wu, J. L. Spotts, and S. A. Osmani. 2000. Mitotic histone H3 phosphorylation by the NIMA kinase in Aspergillus nidulans. Cell 102:293-302. [DOI] [PubMed] [Google Scholar]
- 7.Fischle, W., B. S. Tseng, H. L. Dormann, B. M. Ueberheide, B. A. Garcia, J. Shabanowitz, D. F. Hunt, H. Funabiki, and C. D. Allis. 2005. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438:1116-1122. [DOI] [PubMed] [Google Scholar]
- 8.Fischle, W., Y. Wang, S. A. Jacobs, Y. Kim, C. D. Allis, and S. Khorasanizadeh. 2003. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 17:1870-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fry, A. M., P. Meraldi, and E. A. Nigg. 1998. A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J. 17:470-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giet, R., and D. M. Glover. 2001. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J. Cell Biol. 152:669-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giet, R., C. Petretti, and C. Prigent. 2005. Aurora kinases, aneuploidy and cancer, a coincidence or a real link? Trends Cell Biol. 15:241-250. [DOI] [PubMed] [Google Scholar]
- 12.Goto, H., Y. Yasui, E. A. Nigg, and M. Inagaki. 2002. Aurora-B phosphorylates histone H3 at serine28 with regard to the mitotic chromosome condensation. Genes Cells 7:11-17. [DOI] [PubMed] [Google Scholar]
- 13.Hatta, M., and A. Fukamizu. 2001. PODs in the nuclear spot: enigmas in the magician's pot. Sci. STKE 2001:PE1. [DOI] [PubMed] [Google Scholar]
- 14.Hauf, S., R. W. Cole, S. LaTerra, C. Zimmer, G Schnapp, R. Walter, A. Heckel, J. van Meel, C. L. Rieder, and J. M. Peters. 2003. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161:281-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hediger, F., and S. M. Gasser. 2006. Heterochromatin protein 1: don't judge the book by its cover! Curr. Opin. Genet. Dev. 16:143-150. [DOI] [PubMed] [Google Scholar]
- 16.Hirota, T., N. Kunitoku, T. Sasayama, T. Marumoto, D. Zhang, M. Nitta, K. Hatakeyama, and H. Saya. 2003. Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell 114:585-598. [DOI] [PubMed] [Google Scholar]
- 17.Hirota, T., J. J. Lipp, B. H. Toh, and J. M. Peters. 2005. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 438:1176-1180. [DOI] [PubMed] [Google Scholar]
- 18.Hsu, J. Y., Z. W. Sun, X. Li, M. Reuben, K. Tatchell, D. K. Bishop, J. M. Grushcow, C. J. Brame, J. A. Caldwell, D. F. Hunt, R. Lin, M. M. Smith, and C. D. Allis. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102:279-291. [DOI] [PubMed] [Google Scholar]
- 19.Ivanovska, I., T. Khandan, T. Ito, and T. L. Orr-Weaver. 2005. A histone code in meiosis: the histone kinase, NHK-1, is required for proper chromosomal architecture in Drosophila oocytes. Genes Dev. 19:2571-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 21.Johansen, K. M., and J. Johansen. 2006. Regulation of chromatin structure by histone H3S10 phosphorylation. Chromosome Res. 14:393-404. [DOI] [PubMed] [Google Scholar]
- 22.Jun, D. J., J. H. Lee, B. H. Choi, T. K. Koh, D. C. Ha, M. W. Jeong, and K. T. Kim. 2006. Sphingosine-1-phosphate modulates both lipolysis and leptin production in differentiated rat white adipocytes. Endocrinology 147:5835-5844. [DOI] [PubMed] [Google Scholar]
- 23.Kang, T. H., and K. T. Kim. 2006. Negative regulation of ERK activity by VRK3-mediated activation of VHR phosphatase. Nat. Cell Biol. 8:863-869. [DOI] [PubMed] [Google Scholar]
- 24.Kapoor, T. M., M. A. Lampson, P. Hergert, L. Cameron, D. Cimini, E. D. Salmon, B. F. McEwen, and A. Khodjakov. 2006. Chromosomes can congress to the metaphase plate before biorientation. Science 311:388-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon, S. Y., Y. J. Choi, T. H. Kang, K. H. Lee, S. S. Cha, G. H. Kim, H. S. Lee, K. T. Kim, and K. J. Kim. 2005. Highly efficient protein expression and purification using bacterial hemoglobin fusion vector. Plasmid 53:274-282. [DOI] [PubMed] [Google Scholar]
- 26.Li, F., G. Ambrosini, E. Y. Chu, J. Plescia, S. Tognin, P. C. Marchisio, and D. C. Altieri. 1998. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature 396:580-584. [DOI] [PubMed] [Google Scholar]
- 27.Lopez-Borges, S., and P. A. Lazo. 2000. The human vaccinia-related kinase 1 (VRK1) phosphorylates threonine-18 within the mdm-2 binding site of the p53 tumour suppressor protein. Oncogene 19:3656-3664. [DOI] [PubMed] [Google Scholar]
- 28.Lu, K. P., and T. Hunter. 1995. Evidence for a NIMA-like mitotic pathway in vertebrate cells. Cell 81:413-424. [DOI] [PubMed] [Google Scholar]
- 29.MacCallum, D. E., A. Losada, R. Kobayashi, and T. Hirano. 2002. ISWI remodeling complexes in Xenopus egg extracts: identification as major chromosomal components that are regulated by INCENP-aurora B. Mol. Biol. Cell 13:25-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nasmyth, K. 2002. Segregating sister genomes: the molecular biology of chromosome separation. Science 297:559-565. [DOI] [PubMed] [Google Scholar]
- 31.Nezu, J., A. Oku, M. H. Jones, and M. Shimane. 1997. Identification of two novel human putative serine/threonine kinases, VRK1 and VRK2, with structural similarity to vaccinia virus B1R kinase. Genomics 45:327-331. [DOI] [PubMed] [Google Scholar]
- 32.Nichols, R. J., and P. Traktman. 2004. Characterization of three paralogous members of the Mammalian vaccinia related kinase family. J. Biol. Chem. 279:7934-7946. [DOI] [PubMed] [Google Scholar]
- 33.Nichols, R. J., M. S. Wiebe, and P. Traktman. 2006. The vaccinia-related kinases phosphorylate the N′ terminus of BAF, regulating its interaction with DNA and its retention in the nucleus. Mol. Biol. Cell 17:2451-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osmani, S. A., R. T. Pu, and N. R. Morris. 1988. Mitotic induction and maintenance by overexpression of a G2-specific gene that encodes a potential protein kinase. Cell 53:237-244. [DOI] [PubMed] [Google Scholar]
- 35.Peters, J. M. 2002. The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol. Cell 9:931-943. [DOI] [PubMed] [Google Scholar]
- 36.Polioudaki, H., Y. Markaki, N. Kourmouli, G. Dialynas, P. A. Theodoropoulos, P. B. Singh, and S. D. Georgatos. 2004. Mitotic phosphorylation of histone H3 at threonine 3. FEBS Lett. 560:39-44. [DOI] [PubMed] [Google Scholar]
- 37.Preuss, U., G. Landsberg, and K. H. Scheidtmann. 2003. Novel mitosis-specific phosphorylation of histone H3 at Thr11 mediated by Dlk/ZIP kinase. Nucleic Acids Res. 31:878-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rempel, R. E., M. K. Anderson, E. Evans, and P. Traktman. 1990. Temperature-sensitive vaccinia virus mutants identify a gene with an essential role in viral replication. J. Virol. 64:574-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roig, J., A. Mikhailov, C. Belham, and J. Avruch. 2002. Nercc1, a mammalian NIMA-family kinase, binds the Ran GTPase and regulates mitotic progression. Genes Dev. 16:1640-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santos, C. R., M. Rodriguez-Pinilla, F. M. Vega, J. L. Rodriguez-Peralto, S. Blanco, A. Sevilla, A. Valbuena, T. Hernandez, A. J. van Wijnen, F. Li, E. de Alava, M. Sanchez-Cespedes, and P. A. Lazo. 2006. VRK1 signaling pathway in the context of the proliferation phenotype in head and neck squamous cell carcinoma. Mol. Cancer Res. 4:177-185. [DOI] [PubMed] [Google Scholar]
- 41.Schreiber, S. L., and B. E. Bernstein. 2002. Signaling network model of chromatin. Cell 111:771-778. [DOI] [PubMed] [Google Scholar]
- 42.Segura-Totten, M., and K. L. Wilson. 2004. BAF: roles in chromatin, nuclear structure and retrovirus integration. Trends Cell Biol. 14:261-266. [DOI] [PubMed] [Google Scholar]
- 43.Sevilla, A., C. R. Santos, R. Barcia, F. M. Vega, and P. A. Lazo. 2004. c-Jun phosphorylation by the human vaccinia-related kinase 1 (VRK1) and its cooperation with the N-terminal kinase of c-Jun (JNK). Oncogene 23:8950-8958. [DOI] [PubMed] [Google Scholar]
- 44.Sevilla, A., C. R. Santos, F. M. Vega, and P. A. Lazo. 2004. Human vaccinia-related kinase 1 (VRK1) activates the ATF2 transcriptional activity by novel phosphorylation on Thr-73 and Ser-62 and cooperates with JNK. J. Biol. Chem. 279:27458-27465. [DOI] [PubMed] [Google Scholar]
- 45.Shiio, Y., R. N. Eisenman, E. C. Yi, S. Donohoe, D. R. Goodlett, and R. Aebersold. 2003. Quantitative proteomic analysis of chromatin-associated factors. J. Am. Soc. Mass Spectrom. 14:696-703. [DOI] [PubMed] [Google Scholar]
- 46.Vega, F. M., A. Sevilla, and P. A. Lazo. 2004. p53 stabilization and accumulation induced by human vaccinia-related kinase 1. Mol. Cell. Biol. 24:10366-10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei, Y., L. Yu, J. Bowen, M. A. Gorovsky, and C. D. Allis. 1999. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell 97:99-109. [DOI] [PubMed] [Google Scholar]
- 48.Zelko, I., R. Kobayashi, P. Honkakoski, and M. Negishi. 1998. Molecular cloning and characterization of a novel nuclear protein kinase in mice. Arch. Biochem. Biophys. 352:31-36. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.