Abstract
By assessing the contribution of deregulated E2F activity to erythroid defects in Rb null mice, we have identified E2f-2 as being upregulated in end-stage red cells, where we show it is the major pRb-associated E2f and the predominant E2f detected at key target gene promoters. Consistent with its expression pattern, E2f-2 loss restored terminal erythroid maturation to Rb null red cells, including the ability to undergo enucleation. Deletion of E2f-2 also extended the life span of Rb null mice despite persistent defects in placental development, indicating that deregulated E2f-2 activity in differentiating erythroblasts contributes to the premature lethality of Rb null mice. We show that the aberrant entry of Rb null erythroblasts into S phase at times in differentiation when wild-type erythroblasts are exiting the cell cycle is inhibited by E2f-2 deletion. E2f-2 loss induced cell cycle arrest in both wild-type and Rb null erythroblasts and was associated with increased DNA double-strand breaks. These results implicate deregulated E2f-2 in the cell cycle defects observed in Rb null erythroblasts and reveal a novel role for E2f-2 during terminal red blood cell differentiation. The identification of a tissue-restricted role for E2f-2 in erythropoiesis highlights the nonredundant nature of E2f transcription factor activities in cell growth and differentiation.
The cell cycle withdrawal that accompanies terminal differentiation is generally thought to be permanent and in many respects resembles cellular senescence since it is accompanied by chromatin condensation and global gene silencing (18). Terminal differentiation of red blood cells, lens fibers, and keratinocytes is unusual in that it is accompanied by the ultimate gene-silencing process, loss of the nucleus (45). Intriguingly, the Rb tumor suppressor is essential for the normal differentiation and enucleation of both mature red blood cells and lens fibers (5, 24, 37).
Most characterized functions of pRb are mediated through its interaction with members of the E2f family of transcription factors (41). Hypophosphorylated pRb binds to and inhibits the “activator” E2f-1, -2, and -3a in a cell cycle-dependent manner and in response to checkpoint activation. Active repression of cell cycle genes is key to the tumor suppressor properties of both pRb and E2f-1 (41). E2f-4 and -5 are “repressor” E2fs that interact preferentially with the pRb-related pocket proteins, p107 and p130, to repress the transcription of cell cycle target genes and also cooperate with transforming growth factor β-induced Smad activity to repress c-Myc (4).
E2f-1, -2, and -3a function as transcriptional activators of genes required for cell cycle progression and checkpoint control (7). Loss of all three activating E2fs resulted in fibroblasts that failed to cycle (44). E2f-regulated genes include those for enzymes involved in nucleotide biosynthesis (thymidine kinase, thymidylate synthase, and dihydrofolate reductase) and DNA replication (cdc6, orc, and DNA polymerase α), regulators of cell cycle phase transition such as cyclin E and cyclin A2, and more recently identified targets involved in mitosis such as Mad2 (11). E2fs also regulate key mediators of programmed cell death such as Apaf-1 (25), caspases (28), and p73 (15), and more recently, E2fs have been shown to directly regulate genes involved in terminal differentiation (8, 26).
The E2f-2 transcription factor is less characterized than either E2f-1 or E2f-3 and has unique properties that cannot be explained in terms of current knowledge. E2f-2 is specifically required for normal T-cell function in the adult mouse, and surprisingly, given its role as an activator E2F, its ablation by gene targeting resulted in hyperproliferation of effector T cells and autoimmune disease (27, 46). E2f-2 is expressed with delayed kinetics compared to E2f-1 following T-cell activation, suggesting a negative role for E2f-2 during the proliferative response to antigen (27). E2f-2 can also promote terminal differentiation and irreversible cell cycle exit of neurotrophin-dependent sympathetic neurons (29). These antiproliferative functions of E2f-2 in differentiating cells contrast with its proposed role in proliferating hematopoietic progenitors (21) or in cycling fibroblasts (44).
The phenotype of Rb null mice is complex, with both cell-intrinsic and -extrinsic stresses contributing to defects in cellular differentiation (5, 36, 37, 42). The extrinsic stresses are largely due to defective placental development since conditional deletion of Rb in the embryo, and not the placenta, allowed mice to develop to birth with relatively normal tissue differentiation (6, 42, 43). However, red blood cell maturation defects remained evident in conditionally targeted Rb null embryos during gestation despite a normal placenta (6, 37, 42). By examining how E2fs contribute to the role of pRb in stress erythropoiesis, we have identified E2f-2 as a key effector of pRb in end-stage erythroblasts and demonstrate a novel role for E2f-2 in promoting proper cell cycle checkpoint control and preventing DNA damage during terminal erythroid differentiation.
MATERIALS AND METHODS
Mice.
E2f-2 null mice were kindly provided by Michael Greenberg of Harvard University and Joseph Nevins of Duke University. E2f-1 null and Rb heterozygous mice were obtained from Jackson Laboratories (Bar Harbor, ME). All of the mice used in these studies were maintained on a mixed 129/B6 genetic background. Timed matings were set up as described previously (22). Mice were injected intraperitoneally with 3 mg/kg bromodeoxyuridine (BrdU) plus 0.3 mg/kg fluorodeoxyuridine prepared in sterile phosphate-buffered saline as described previously (22). Fetal livers dissected from embryos were disaggregated in DPBS (Invitrogen) with 2% fetal calf serum to a single-cell suspension by serial passage through a 23-gauge needle, followed by a 27-gauge needle.
Flow cytometry.
Flow cytometric analysis of live cells and sorting were carried out as described previously (37). Intracellular detection of phospho-histone H3 (PH3; Upstate catalog no. 06-570) and phospho-H2AX-fluorescein isothiocyanate (Upstate catalog no. 16-202A) was performed as described previously (13). BrdU-fluorescein isothiocyanate staining was done according to the manufacturer's specifications (BD Biosciences catalog no. 347583).
Western blotting.
Nuclear lysates and radioimmunoprecipitation assay lysates were prepared as described previously (22). Equal quantities (50 μg) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Membranes were blocked in 5% powdered milk in phosphate-buffered saline for 1 h at room temperature and then incubated with primary antibodies specific for E2f-1 (sc-2280; Santa Cruz), E2f-2 (sc-633; Santa Cruz), E2f-3 (sc-878; Santa Cruz), E2f-4 (sc-866; Santa Cruz), pRb (clone G3-245; BD Biosciences catalog no. 554136), p107 (sc-318; Santa Cruz), phospho-ATM Ser1981 (4526; Cell Signaling Technologies), ATM (NB100-270; Novus), and p53 (AF1355; R&D Systems). Secondary antibodies (DAKO) were used at 1:5,000 for 1 h at room temperature, and detection was carried out by enhanced chemiluminescence.
Immunoprecipitation (IP), gel shift, and kinase assays.
IPs were carried out as described previously (23). Antibodies used for IP were specific for E2f-1 (sc-251; Santa Cruz); E2f-2, -3, and -4 and pRb (same as above); cdc2 (554161; Pharmingen); and Raf-1 (sc-227; Santa Cruz). Gel shifts for E2fs were carried out as described previously (22) with nuclear extracts. IP-gel shift analyses were carried out as described previously (20). Supershifting antibodies for E2f-1, E2f-2, E2f-3, and E2f-4 were the same as above. pRb and p107 were supershifted with the 21C9 and SD15 antibodies described previously (22).
Primary erythroblast culture.
Erythroblasts were differentiated in StemSpan SFEM (Stem Cell Technologies) supplemented with 20% fetal calf serum, 1 μM mifepristone, and 2 U/ml erythropoietin. For cell cycle analysis, culture was carried out in the presence of 30 mg/ml BrdU plus 3 mg/ml fluorodeoxyuridine (Sigma) with or without 600 ng/ml nocodazole. Cells were harvested for flow cytometry or cytospun onto slides for staining with benzidine-Wright-Giemsa as described previously (37).
RNA analysis.
In situ hybridization for E2f-2 mRNA was carried out as described previously (22), with a probe to the 3′ untranslated region.
RNA was extracted with TRIzol, and cDNA was generated with SuperScript II reverse transcriptase (RT) (Invitrogen).
Primers used in RT-PCR for E2f-1, -2, -3, and -4 were as follows: E2f-1 FOR, TGA CAG AAG CCT AGG GAT TCA GGG TGT CTG; E2f-1 REV, TTG GTC TGC TCA ATG TGG CAG CAA CCA; E2f-2 FOR, AGC GTC CTG CAT CTG TCT ACC TCC TCA CAG; E2f-2 REV, ATG TGC GCT TAG GCA CTG GGC GTG GGT; E2f-3 FOR, TGA TTG GGC TTT GTA CAA ACT GTC CTT; E2f-3 REV, CAT TCC GTG GTA GCA GAC TCA CAA AGA; E2f-4 FOR, TAC AAC CTG GAC GAG AGT GAA GGT GTC TGT; E2f-4 REV, CAG CTA TGC TAG CAC AGT CAC AAG TGC; HPRT FOR, CAC AGG ACT AGA ACA CCT GC; HPRT REV, GCT GGT GAA AAG GAC CTC T.
Histological procedures and immunohistochemistry.
Embryos were fixed in 10% neutral buffered formalin, processed, embedded, and sectioned as described previously (22). BrdU (BD Biosciences catalog no. 347580), PH3 Ser10 (Upstate catalog no. 06-570), and phospho-H2A.X Ser139 (Upstate catalog no. 07-164) immunohistochemistry was carried out as described previously (22, 37).
Chromatin IP.
E15.5 FL was disaggregated to a single-cell suspension as described above. Cross-linking with formaldehyde and nuclear extract isolation were performed as previously described (1). Chromatin was sonicated to an average length of 600 bp and immunoprecipitated with 1 μg of antibody specific for E2f-1 (sc-251), E2f-2 (sc-633), E2f-3 (sc-878), E2f-4 (sc-1082) (all from Santa Cruz), or pRb (554136) (from BD Biosciences). Cross-linked protein-DNA complexes were washed, and immunoprecipitated DNA or input DNA was amplified with E2f-1 (5′-CCAAT GCTCGCCATCGGAGCCTCCGTC-3′ and 5′-ACGATCTGCGAGGAGTCGAGCAGC CGC-3′), E2f-3a (5′-GGCGACAGGGATCCAATGGACTGTGGGAAC-3′ and 5′-CAG CCGATGCAACGGATTGCGAGGCGGTGG-3′), Cdc6 (5′-GAGTGGGTGTGACTTCGTGGTGGTCGC-3′ and 5′-CGCTCAGTGCA CACTCCTCGCGTCTCG-3′), Cyclin E (5′-GGGAGGCCACGCCC CCACCAGAGCTCCTCG-3′ and 5′-CACCGCGCTGGCTGTCGAGCCGGCTGCTCC-3′), and Bcl-XL (5′-GGCAGGTTCCTAAGCTTCGCAATTCCT-3′ and 5′-CTTGTAGGAGAGAAAGTCGACCACCAG-3′).
PCR products were visually resolved on 2% agarose gels and stained with ethidium bromide. Quantification of immunoprecipitate-enriched DNA sequences was conducted as described previously (9), by real-time PCR with QuantiTect SYBR green PCR and the Applied Biosystems 7900 Fast Real-Time PCR System, and analysis was done with the associated SDS 2.3 software. Data are represented as percentages of the input, calculated by 2ΔCT × 20, where ΔCT is determined by CTInput − CTIP sample and 20 refers to the input being 20% of the chromatin amount exposed to IP.
RESULTS
E2f-2 expression is upregulated at end stages of erythroid differentiation.
To determine which members of the E2f family were likely to be deregulated by Rb loss in the erythroid system and therefore could contribute to the erythroid phenotype in Rb null mice, we examined the expression of E2f-1, -2, -3, and -4 (the four E2fs that are reported to interact with pRb) in hematopoietic and nonhematopoietic tissues of the mouse (Fig. 1).
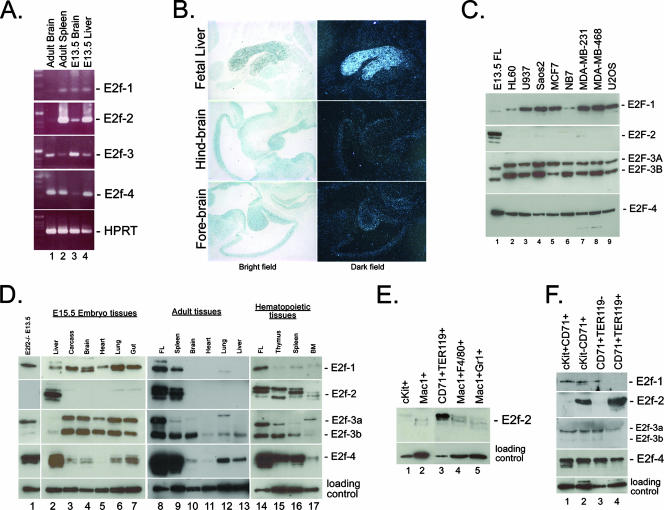
FIG. 1.
E2f-2 is upregulated during terminal erythroid differentiation. (A) Expression of E2f-1, -2, -3, and -4 was examined by semiquantitative RT-PCR with RNA derived from adult brain, adult spleen, E13.5 fetal liver, and E13.5 fetal brain. HPRT expression was used as a control. (B) E2f2-2 mRNA expression in the E13.5 embryo was examined by in situ hybridization and imaged with both bright-field (left) and dark-field (right) objectives. (C) Western blot analysis of E2F expression in exponentially growing human tumor cell lines. (D) Western blot analysis of E2f expression in nuclear lysates prepared from wild-type E15.5 mouse (lanes 2 to 7) or adult mouse (lanes 9 to 13 and 15 to 17) tissues. Control samples from E13.5 E2f2−/− fetal liver (FL; lane 1) and E13.5 wild-type fetal liver are indicated (lanes 8 and 14). BM, bone marrow. (E) Specific adult bone marrow populations were sorted to >95% purity by flow cytometry on the basis of the expression of c-Kit, Mac1, CD71, TER119, F4/80, and Gr1, and the level of E2f-2 in each population was determined by Western blotting. (F) E13.5 fetal liver erythroblasts were sorted to >95% purity by flow cytometry on the basis of the expression of c-Kit, CD71, and TER119, and levels of E2f-1, -2, -3, and -4 in each population were determined by Western blotting.
As shown in Fig. 1A, by RT-PCR, we observed increased expression of E2f-2 in adult spleen and fetal liver compared to either fetal or adult brain. In situ hybridization to detect E2f-2 message in embryonic day 13.5 (E13.5) mouse tissues confirmed that E2f-2 is expressed at much higher levels in the fetal liver than in all other tissues at this stage of development (Fig. 1B). E2f-4 was also expressed at high levels in adult spleen and fetal liver, but in contrast to E2f-2, it was detected in adult brain as well (Fig. 1A). Expression of E2f-1 or E2f-3a was significantly lower in hematopoietic tissues than either E2f-2 or E2f-4 (Fig. 1A).
When we analyzed E2f expression in commonly used tumor cell lines, we observed that in contrast to the expression of E2f-1, E2f-3, or E2f-4, expression of E2f-2 was largely undetectable (Fig. 1C), indicating that E2f-2 is subject to distinct mechanisms of gene regulation compared to other E2f family members.
During embryonic development, many tissues are actively proliferating and most tissues expressed the E2f-1, -3, and -4 proteins (Fig. 1D, lanes 2 to 7). E2f-4 was most highly expressed in the fetal liver (Fig. 1D, lane 2), but like E2f-1 and E2f-3, it was also expressed in other tissues. At E15.5 of mouse development, the E2f-2 protein was strongly expressed in the fetal liver (Fig. 1D, lane 2) but not in other tissues of the embryo that were examined (Fig. 1D, lanes 3 to 7), including the central nervous system (Fig. 1D, lane 4).
E2f-1 and E2f-3a expression was undetectable in the adult brain (Fig. 1D, lane 10), heart (lane 11), or liver (lane 13). E2f-3b was expressed at moderate levels in all of the adult tissues examined (Fig. 1D, lanes 8 to 13), consistent with its role as a repressor E2f (1). Similarly, E2f-4 was expressed in most of the adult tissues examined, including the brain, lung, and liver (lanes 9, 11, and 12). In the self-renewing hematopoietic system, E2f-1, -3b, and -4 were all expressed at high levels, as shown here for the spleen, thymus, and bone marrow (Fig. 1D, lanes 9, 15, 16, and 17). E2f-3a was expressed at very low levels compared to E2f-3b and was barely detectable in the mouse spleen (Fig. 1D, lanes 9 and 16). In contrast to the other activator E2fs and to E2f-4, E2f-2 was only detected in hematopoietic tissues in the adult mouse (Fig. 1D, lanes 8 to 13) and not in the adult brain, heart, lung, or liver (lanes 10 to 13). In summary, our results demonstrate strong upregulation of E2f-2 in the embryonic and adult mouse hematopoietic system compared to nonhematopoietic tissues or tumor cell lines.
To determine whether the robust expression of E2f-2 in the hematopoietic system was attributable to any particular cell population or stage of differentiation, we used flow cytometric sorting techniques to isolate specific hematopoietic cell types from both adult bone marrow (Fig. 1E) and fetal liver (Fig. 1F). Surprisingly, we observed that E2f-2 was most highly expressed in bone marrow erythroblasts (Fig. 1E, lane 3) that are distinguished by coexpression of CD71 (transferrin receptor) and TER119 (a marker of mature red cells that is upregulated as erythroblasts exit the cell cycle) (16). E2f-2 was expressed at significantly lower levels in c-Kit+ progenitors (Fig. 1E, lane 1), Mac1+ F4/80+ macrophages (Fig. 1E, lane 4), and Mac1+ Gr1+ granulocytes (Fig. 1E, lane 5).
In the developing fetal liver (Fig. 1F), which is predominantly erythroid, we observed high-level expression of E2f-2 in the more mature c-Kit− CD71+ (Fig. 1F, lane 2) and CD71+ TER119+ (Fig. 1F, lane 4) erythroblasts but minimal expression in immature c-Kit+ CD71+ (Fig. 1F, lane 1) or CD71+ TER119− erythroblasts (Fig. 1F, lane 3). In contrast to the expression of E2f-2, expression of the other two activator E2fs (E2f-1 and E2f-3a) was downregulated to barely detectable levels in mature TER119+ erythroblasts (Fig. 1F, lane 4) compared to immature TER119− erythroblasts (Fig. 1F, lane 3). E2f-4 was also downregulated in mature TER119+ erythroblasts (Fig. 1F, lane 4) compared to immature TER119− erythroblasts (Fig. 1F, lane 3), although it was still readily detectable. In addition to full-length E2f-2, we detected three closely migrating lower-molecular-weight forms of E2f-2 in unsorted hematopoietic tissues (Fig. 1D, lanes 2, 8, 9, and 14 to 17). However, the absence of these lower-molecular-weight forms of E2f-2 in sorted erythroblasts (Fig. 1E, lane 3, and F, lanes 2 and 4) suggested that the processing of E2f-2 that is observed in these tissues is taking place not in erythroblasts but in nonerythroid cell types.
In summary, our data demonstrate that E2f-2 is upregulated at end stages of erythroid maturation and is highly expressed in mature TER119+ erythroblasts within the fetal and adult hematopoietic systems.
E2f-2 is the major pRb-associated E2f in end-stage erythroblasts.
To determine whether high-level E2f-2 expression was reflected in increased association with pRb, we examined the interaction of pRb with different E2fs in wild-type fetal liver. We observed that E2f-2 was the major pRb-associated E2f (Fig. 2A, lane 3) at E15.5 of gestation, most likely reflecting the predominant representation of TER119+ erythroblasts in the fetal liver at this stage of development (36). Low levels of E2f-1 and E2f-3 were found in a complex with pRb at E15.5 of gestation (Fig. 2A, lanes 2 and 4), but we were unable to detect any interaction between pRb and E2f-4 in the fetal liver at midgestation (Fig. 2A, lane 5, top), although p107 was readily immunoprecipitated with E2f-4 in the same lysates (Fig. 2A, lane 5, bottom). Failure to immunoprecipitate higher levels of pRb from E15.5 fetal liver lysates with antibody to E2f-3 could not be attributed to poor affinity of the antibody since we were able to efficiently coimmunoprecipitate pRb with E2f-3 from fetal brain lysates with the same antibody (Fig. 2B, lane 3). Conversely, we were unable to coimmunoprecipitate pRb from fetal brain lysates with antibody to E2f-2 (Fig. 2B, lane 1), even though pRb was readily coimmunoprecipitated from fetal liver with this anti-E2f2 antibody (Fig. 2B, lane2). Thus, the co-IP of pRb with anti-E2f antibodies was more readily explained by the pattern of E2f expression (Fig. 1D, lanes 2 and 4) than by issues of antibody affinity for specific E2fs.
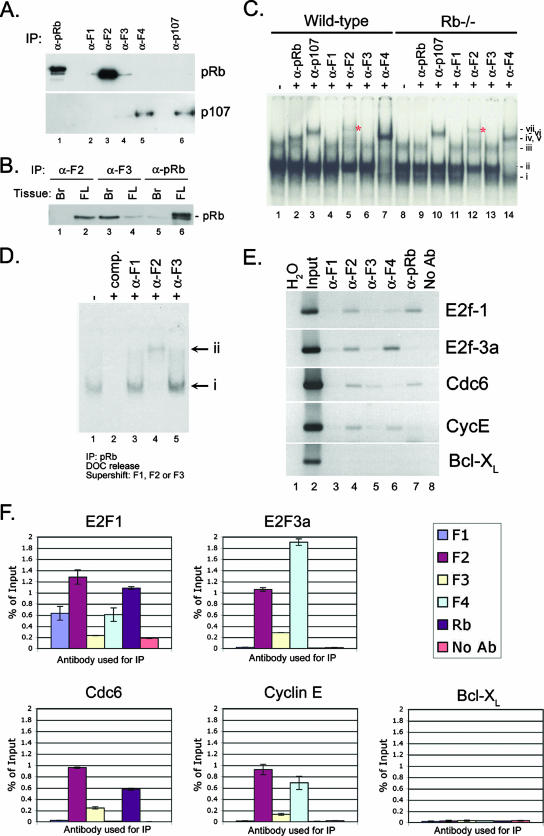
FIG. 2.
E2f-2 is the major pRb-associated E2f in end-stage erythroblasts. (A) IP of pRb, E2f-1, E2f-2, E2f-3, E2f-4, and p107 from E15.5 fetal liver was carried out, and immunoprecipitates were examined by Western blotting for pRb (top) or p107 (bottom). (B) IP of E2f-2 (lanes 1 and 2), E2f-3 (lanes 3 and 4), or pRb (lanes 5 and 6) from E15.5 fetal brain (Br) or fetal liver (FL) was carried out, and immunoprecipitates were examined by Western blotting for pRb. (C) Gel shift analysis of wild-type and Rb null fetal liver nuclear lysates for E2f DNA binding activity. Complexes were identified with supershifting antibodies as (i) free E2f-2, (ii) free E2f-4, (iii) pRb/E2f and p107/E2f complexes, (iv) supershifted E2f-4, (v) supershifted pRb/E2f complexes, (vi) supershifted p107/E2f complexes, and (vi) supershifted E2f-2. (D) The E2f DNA binding activity of pRb immunoprecipitates was examined following deoxycholate (DOC) release, with E2F-specific supershifting antibodies that identified complex i as free E2F and complex ii as supershifted E2f-2. (E) Ethidium bromide gel resolution of PCR products amplified from chromatin IP. Individual E2fs and pRb were immunoprecipitated and examined for in vivo binding at validated E2f target gene promoters (E2f1, E2f-3a, Cdc6, and cyclin E) and at a control promoter (Bcl-XL) with input DNA as a positive control and a no-antibody (No Ab) immunoprecipitate sample as a negative control. (F) Quantification of precipitated DNA retrieved from chromatin IP experiments was carried out by real-time PCR where the amount of immunoprecipitated promoter DNA was expressed as a percentage of the total input chromatin.
When we examined E2F DNA binding activity in nuclear lysates from E13.5 wild-type fetal livers, we observed the presence of a complex that supershifted with antibodies to E2f-2 (Fig. 2C, lane 5, vii, *). Furthermore, when we examined E2F DNA binding activity in fetal livers from Rb−/− embryos, we observed the presence of a novel, faster-migrating E2F complex (lane 8, i) that was supershifted with antibodies to E2f-2 (lane 12, vii, *) but not with antibodies to E2f-1, -3, or -4, indicating that the novel faster-migrating complex (i) contained E2f-2.
To confirm that pRb preferentially complexed with E2f-2, we carried out immunoprecipitation-gel shift analysis in which the E2F DNA binding activity of deoxycholate-released pRb immunocomplexes from E13.5 fetal liver was examined by supershifting with E2f-specific antibodies. We observed a complete supershift of all E2f activity that immunoprecipitated with pRb from E13.5 fetal liver nuclear extracts (Fig. 2D, lane 1) with an antibody to E2f-2 (Fig. 2C, lane 4) but not with an antibody to E2f-1 or -3 (Fig. 2D, lanes 3 and 5).
We next examined whether E2f-2 associated with pRb at the promoters of characterized E2f target genes in vivo by chromatin IP experiments with antibodies to E2f-1, -2, -3, and -4 and pRb. Quantitative analysis of immunoprecipitated DNA carried out by real-time PCR on duplicate samples (Fig. 2F) confirmed results obtained by gel electrophoresis (Fig. 2E). We observed that E2f-2 was found at the promoters of E2f-1, E2f-3a, Cdc6, and cyclin E in E15.5 fetal liver erythroblasts (Fig. 2E, lane 4, and F) but not at the promoter of Bcl-XL, a gene that is not an E2F target gene but is highly expressed in erythroblasts (10). E2f-4 was also detected at some of these promoters and was particularly prevalent at the E2f-3a promoter, although the significance of this remains to be determined. We detected pRb bound at the Cdc6 and E2f-1 promoters with E2f-2, but not at the E2f-3a or cyclin E promoters. Intriguingly, the amount of pRb bound at E2f-regulated promoters correlated inversely with the levels of E2f-4 (Fig. 2E, lanes 6 and 7, and F).
In summary, these results indicate that E2f-2 is the major pRb-associated E2f in the midgestational fetal liver and the predominant E2f bound to target gene promoters, although pRb only appears bound with E2f-2 at a subset of these promoters.
E2f-2 loss restores erythroid maturation capacity to Rb null red blood cells.
Given elevated E2f-2 expression during red cell maturation and the predominance of E2f-2 in pRb complexes in fetal liver extracts, we used mouse genetics to explore the role of deregulated E2f-2 in the erythroid phenotype of Rb null mice. As we reported previously (37), Rb loss resulted in reduced representation by CD71+ TER119+ erythroblasts (Fig. 3B and I; mean value, 38.2%; standard deviation [SD], 3.8%) in E12.5 fetal liver compared to the wild type (Fig. 3A and I; mean value, 82.4%; SD, 3.5%), consistent with reduced numbers of mature red blood cells in the Rb null mouse. The numbers of CD71+ TER119+ erythroblasts in E2f2−/− fetal liver were similar (Fig. 3C and I; mean value, 82.6%; SD, 3.3%) to those observed in the wild type (Fig. 3A and I; mean value, 82.4%; SD, 3.5%). Importantly, E2f-2 loss restored representation by CD71+ TER119+ erythroblasts (Fig. 3D; mean value 70.9%; SD, 2.6%) to the Rb null fetal liver, indicating that E2f-2 loss promoted red blood cell differentiation in Rb null mice.
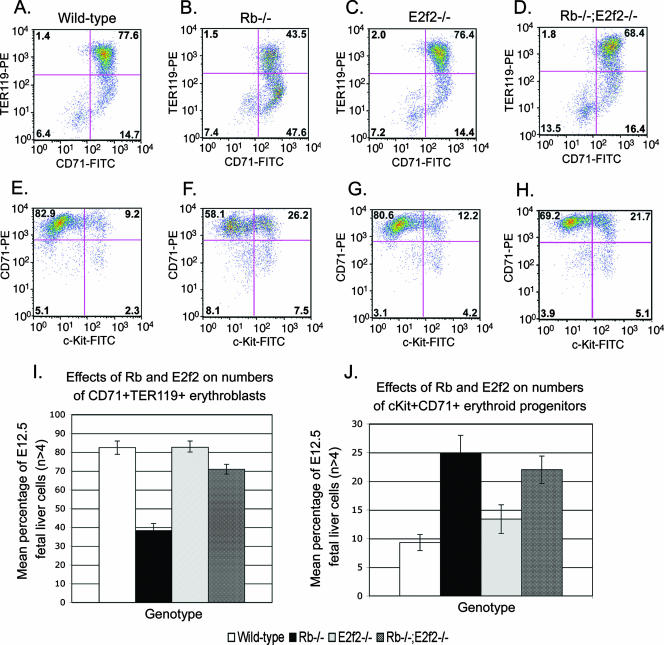
FIG. 3.
E2f-2 loss restores erythroid differentiation to Rb null erythroblasts. (A to D) Flow cytometric labeling of E12.5 cells to quantify the relative numbers of mature CD71+ TER119+ erythroblasts in wild-type (A), Rb−/− (B), E2f2−/− (C), and Rb−/−; E2f2−/− (D) fetal livers. (E to H) Flow cytometric labeling of E12.5 cells to quantify the relative numbers of c-Kit+ CD71+ erythroblast progenitors in wild-type (E), Rb−/− (F), E2f2−/− (G), and Rb−/−; E2f2−/− (H) fetal liver. (I to J) Quantitative analysis of the effect Rb loss and E2f-2 loss had on the representation of CD71+ TER119+ erythroblasts (I) or c-Kit+ CD71+ progenitors (J) in E12.5 fetal liver samples. Data are shown as the mean value of at least four independent samples per genotype, and error bars represent the SD from the mean. FITC, fluorescein isothiocyanate; PE, phycoerythrin.
Immature erythroblasts that were c-Kit+ CD71+ showed increased representation in Rb null fetal liver (Fig. 3F and J; mean value, 24.9%; SD, 1.4%) compared to the wild type (Fig. 3E and J; mean value, 9.3%; SD, 1.4%). E2f-2 loss did not significantly reduce the numbers of c-Kit+ CD71+ progenitors in the Rb null fetal liver (Fig. 3H and J; mean value, 22.0%; SD, 2.4%), consistent with the observed pattern of E2f-2 expression in CD71+ TER119+ erythroblasts (Fig. 1F, lane 4) but not in c-Kit+ CD71+ progenitors (Fig. 1F, lane 1). Increased representation by c-Kit+ CD71+ erythroid progenitors in Rb null fetal liver is likely mediated by other activator E2Fs expressed in this cell type (Fig. 1F, lane 1).
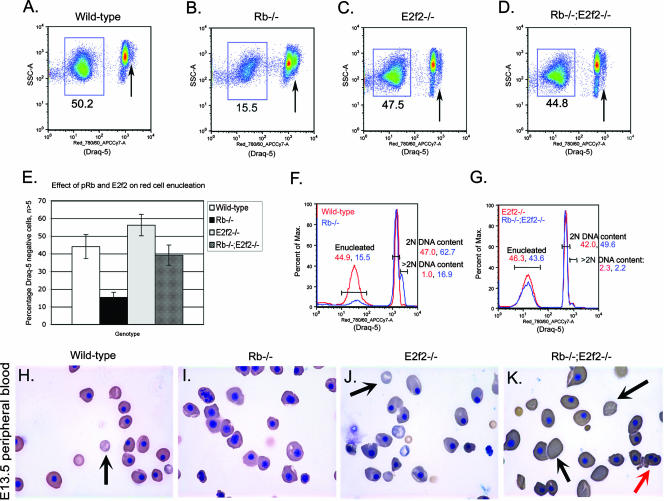
To determine whether E2f-2 loss affected the enucleation efficiency of Rb null erythroblasts, we examined the peripheral blood of E13.5 embryos by flow cytometry for uptake of the vital DNA dye Draq5. Consistent with defective red cell enucleation, the representation of Draq5-negative cells in the peripheral blood of Rb null embryos at E13.5 was markedly reduced (Fig. 4B and E; mean value, 15.3%; SD, 2.9%) compared to that in the wild type (Fig. 4A and E; mean value, 44.0%; SD, 6.8%). Furthermore, nucleated red cells in the peripheral blood of Rb null embryos showed an increased proportion of cells with greater-than-2N DNA content (Fig. 4B, arrow, and F). Enucleation in E13.5 E2f2−/− embryos (Fig. 4C and E; mean value, 56.2%; SD, 6.1%) was similar to wild-type levels (Fig. 4A and E). When we examined the uptake of Draq5 by cells in the peripheral blood of E13.5 Rb−/−; E2f2−/− embryos, we observed a significant increase in the percentage of Draq5neg enucleated cells (Fig. 4D and E; mean value, 39.2%; SD, 5.7%) compared to that in Rb−/− embryos (Fig. 4B and E), indicating that loss of E2f-2 restored enucleation capacity to Rb null erythroblasts. Cells with greater-than-2N DNA content were not elevated in the peripheral blood of Rb−/−; E2f2−/− embryos (Fig. 4G). Cytological analysis of peripheral blood at E13.5 confirmed an increased efficiency of red cell enucleation in Rb−/−; E2f2−/− embryos (Fig. 4K) compared to peripheral blood from Rb null embryos (Fig. 4I), further demonstrating that loss of E2f-2 promoted maturation of Rb null erythroblasts. However, enucleated peripheral blood erythrocytes from both Rb−/−; E2f2−/− and E2f2−/− mice were noticeably abnormal in size and shape, with abnormally large cells (macrocytes) detected at E13.5 (Fig. 4J and K). These results support a previous report of increased red blood cell volume in adult E2f-2 null mice (21).
FIG. 4.
E2f-2 loss promotes enucleation of Rb null erythroblasts. (A to D) Representative Draq5 staining of DNA content in peripheral blood cells to quantify percentages of enucleated cells in wild-type (A), Rb−/− (B), E2f2−/− (C), and Rb−/−; E2f2−/− (D) E13.5 mouse embryos. Enucleated cells stain negatively for Draq5 and are boxed in each plot. Nucleated cells stain positively for Draq5, and arrows indicate >2N DNA content. (E) Quantitative analysis of the effect Rb loss and E2f-2 loss had on enucleation efficiency. Data are shown as the mean value of at least five independent samples per genotype, and error bars represent the SD from the mean. (F to G) Flow cytometric analysis of Draq5 uptake by wild-type (F, red), Rb−/− (F, blue), E2f2−/− (G, red), and Rb−/−; E2f2−/− (G, blue) peripheral blood cells. DNA content in nucleated, Draq5-positive cells is indicated as either 2N or >2N to quantify arrested or cycling cells in the peripheral blood, respectively. Max., maximum. (H to K) Representative peripheral blood smears stained with benzidine-Wright-Giemsa from E13.5 wild-type (H), Rb−/− (I), E2f2−/− (J), and Rb−/−; E2f2−/− (K) embryos. Black arrows indicate enucleated red cells, and the red arrow indicates the increased presence of binucleate red cells in Rb−/−; E2f2−/− peripheral blood.
In summary, these results indicate that E2f-2 loss restored red cell maturation to Rb null erythroblasts, consistent with the expression pattern of E2f-2 in end-stage erythroblasts and implicating deregulated E2f-2 activity in the enucleation defect in Rb null mice.
Rb−/−; E2F2−/− mice have an extended life span compared to Rb−/− mice.
Considering the positive effect E2f-2 loss had on red cell maturation, we examined the effect of E2f-2 loss on the survival of Rb null embryos. We collected embryos at increasing developmental age from timed matings of Rb+/−; E2f2+/− with Rb+/−; E2f2−/− mice or Rb+/−; E2f2−/− with Rb+/−; E2f2−/− mice. As shown in Table 1, we failed to observe any viable Rb−/−; E2F2+/− embryos beyond E13.5, whereas we observed seven Rb−/−; E2f2−/− embryos alive at E15.5 and two Rb−/−; E2f2−/− embryos alive at E16.5. Our data showed that most Rb−/−; E2f2−/− embryos died between E15.5 and E17.5, whereas Rb−/−; E2f2+/− embryos died between E13.5 and E14.5. These data indicate that restoring red cell maturation to Rb null mice through targeted deletion of E2f-2 extended their life span by 2 to 3 gestational days.
TABLE 1.
E2f-2 loss extends the viability of Rb null micea
| Mouse genetic background and age | No. of mice
|
Total | ||
|---|---|---|---|---|
| Rb+/+ | Rb+/− | Rb−/− | ||
| E2f2+/− | ||||
| E12.5 | 6 | 15 | 4 | 25 |
| E13.5 | 10 | 18 | 7 | 35 |
| E15.5 | 7 | 13 | 0 | 20 |
| E16.5 | 2 | 7 | 0 | 9 |
| E2f2−/− | ||||
| E12.5 | 12 | 36 | 16 | 64 |
| E13.5 | 23 | 39 | 16 | 78 |
| E15.5 | 11 | 30 | 7 | 48 |
| E16.5 | 10 | 17 | 2 | 29 |
Mice were determined to be alive on the basis of the presence of a heartbeat. The numbers of live Rb null embryos recovered at different gestational ages were assessed as a function of the E2f-2 status of the mice. Rb null mice lacking E2f-2 were collected at E16.5, whereas Rb null mice on an E2f2+/− background were not collected alive after E13.5.
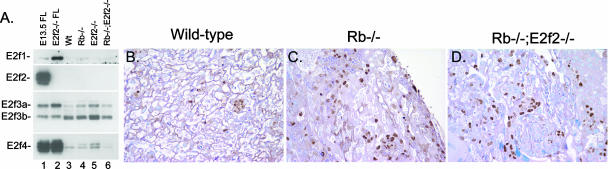
Although E2f-2 loss restored red cell maturation to Rb null erythroblasts, E2f-2 loss failed to prevent ectopic proliferation and developmental defects observed in the Rb null placenta (Fig. 5C). The persistence of ectopic proliferation in the Rb−/−; E2f2−/− placenta (Fig. 5D) was not surprising given that E2f-2 protein was not detected in the wild-type or Rb null placenta (Fig. 5A). Thus, while improved red cell maturation extended the life span of Rb null mice, it was insufficient to rescue to birth, most likely due to persistent placental defects.
FIG. 5.
Loss of E2f-2 does not improve the phenotype of the Rb null placenta. (A) E2f expression in E13.5 wild-type (Wt), Rb−/−, E2f2−/−, and Rb−/−; E2f2−/− placentas was determined by Western blotting (A, lanes 3 to 6) and compared to expression in wild-type and E2f2−/− fetal livers (A, lanes 1 to 2). (B to D) Histological sections of E13.5 wild-type (B), Rb−/− (C), and Rb−/−; E2f2−/− (D) placentas were immunostained for BrdU incorporation, and ectopic proliferation in the labyrinth layer is indicated by dark-brown-staining nuclei. Structural abnormalities in Rb−/− (C) and Rb−/−; E2f2−/− (D) placenta samples are also evidenced by deficits in maternal-fetal blood spaces.
These observations suggest that E2f-2 loss extends the life span of Rb null embryos by 2 to 3 gestational days through improvements in red cell development but that persistent defects in placental development likely explain the death of compound Rb−/−; E2f2−/− embryos between E15.5 and E17.5.
E2f-2 loss promotes cell cycle arrest of Rb null erythroblasts.
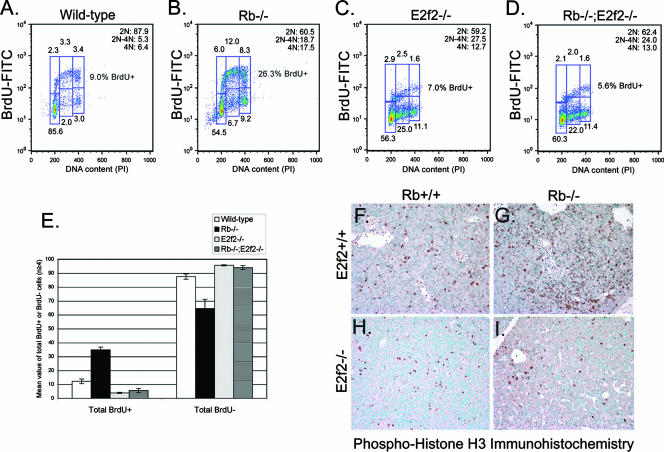
Given the increased presence of red blood cells with greater-than-2N DNA content in the peripheral blood of Rb null embryos compared to wild-type mice (Fig. 4F), we assessed the effect of Rb loss on cell cycle exit during terminal stages of erythroid maturation. We sorted CD71+ TER119+ erythroblasts from E13.5 wild-type and Rb null fetal livers and examined their ability to incorporate BrdU as a measure of entry into S phase of the cell cycle. We observed that while greater than 85% of wild-type erythroblasts failed to incorporate BrdU after 15 h of differentiation culture and predominated in G1 of the cell cycle (Fig. 6A), significantly fewer Rb null erythroblasts arrested in G1 (Fig. 6B) and there was an approximately threefold increase in the number of cells still incorporating BrdU (Fig. 6B and E; mean value, 34.8%; SD, 1.9%) compared to the wild type (Fig. 6A and E; mean value, 12.1%; SD, 1.7%), indicating that fewer Rb null cells had undergone the cell cycle exit that normally accompanies terminal erythroid differentiation.
FIG. 6.
E2f-2 loss promotes cell cycle arrest of Rb null erythroblasts. (A to D) Representative BrdU labeling and propidium iodide (PI) staining of E13.5 CD71+ TER119+ erythroblasts sorted to >95% purity from wild-type (A), Rb−/− (B), E2f2−/− (C), and Rb−/−; E2f2−/− (D) fetal livers. Cells were differentiated for 15 h in culture before BrdU was added for an additional 3 h of incubation to label cells progressing through the cell cycle. (E) Quantitative analysis of the effect Rb loss and E2f-2 loss had on the total number of cells that incorporated BrdU (BrdU positive) and were therefore determined to be in cycle, alongside the total numbers of cells that failed to incorporate BrdU (BrdU negative) and were therefore determined to have undergone cell cycle arrest. Data are shown as the mean value of at least four independent samples per genotype, and error bars represent the SD from the mean. (F to I) Immunohistochemical staining for PH3 on sections of E13.5 fetal livers from wild-type (F), Rb−/− (G), E2f2−/− (H), and Rb−/−; E2f2−/− (I) embryos. FITC, fluorescein isothiocyanate.
To determine whether E2f-2 loss promotes end-stage maturation of Rb null erythroblasts through effects on the cell cycle, we examined BrdU incorporation by CD71+ TER119+ erythroblasts from E13.5 E2f2−/− and Rb−/−; E2f2−/− fetal livers (Fig. 6C and D). We observed that both E2f2−/− and Rb−/−; E2f2−/− erythroblasts displayed a marked failure to incorporate BrdU after 15 h in differentiation culture, consistent with cell cycle arrest (Fig. 6C to E). Indeed, E2f2−/− CD71+ TER119+ erythroblasts were even less efficient than wild-type erythroblasts at incorporating BrdU (Fig. 6C and E; mean value, 3.9%; SD, 0.4%). Importantly, loss of E2f-2 prevented Rb null erythroblasts from incorporating BrdU (Fig. 6D and E; mean value, 5.6%; SD, 1.5%), indicating that deregulated E2f-2 was driving the aberrant cell cycle observed in end-stage Rb null erythroblasts (Fig. 6B and E; mean value, 34.8%; SD, 1.9%).
To further assess the effect of E2f-2 loss on the cell cycle of differentiating erythroblasts, we examined the expression of PH3, which is commonly used as a marker of mitosis (12). Consistent with a higher percentage of Rb null erythroblasts being in cycle (Fig. 6E), we noted increased staining for PH3 in sections of Rb null fetal liver (Fig. 6G) compared to the wild type (Fig. 6F). Furthermore, we also noted markedly reduced PH3 staining in fetal liver sections from E2f2−/− and Rb−/−; E2f2−/− embryos (Fig. 6H and I) compared to either wild-type or Rb null fetal livers (Fig. 6F and G), supporting the conclusion drawn from analysis of BrdU incorporation that loss of E2f-2 induced cell cycle arrest in differentiating erythroblasts.
Multiple cell cycle defects in E2f-2-deficient erythroblasts.
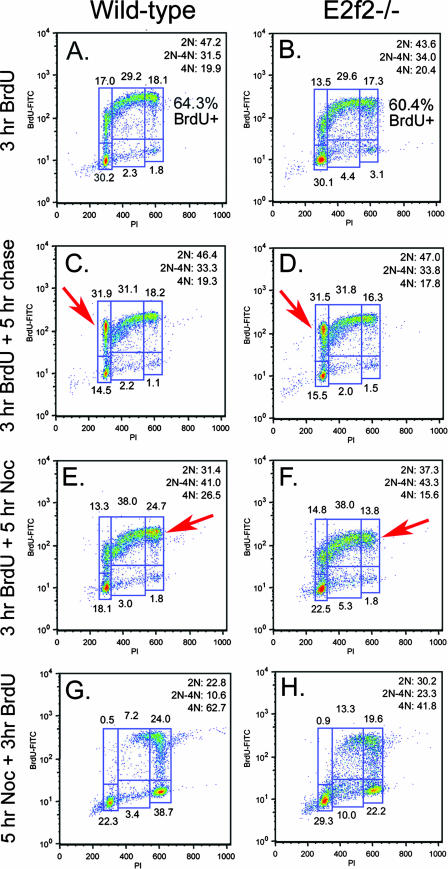
E2f-2−/− CD71+ TER119+ erythroblasts accumulated as BrdU-negative cells with increased 2N to 4N DNA content during their first 15 h in culture (Fig. 6C and D), suggesting that these cells may have arrested in S phase. To determine whether cell cycle arrest in S phase explained the increased number of E2f2−/− erythroblasts arrested with BrdU-negative 2N to 4N DNA content, we used BrdU pulse-chase experiments to assess the ability of E2f-2 null erythroblasts to enter into and progress through the cell cycle. E2f2−/− erythroblasts appeared to cycle as efficiently as wild-type erythroblasts during their first 3 h in culture, since the percentage of BrdU-positive cells in E2f2−/− cultures (Fig. 7B, 60.4%) was found to be as high as in the wild type (Fig. 7A, 64.3%). Furthermore, when we chased BrdU-labeled erythroblasts for a further 5 h in culture, we observed that E2f2−/− erythroblasts progressed through the cell cycle to a BrdU-positive G1 as efficiently (Fig. 7D, 31.5%, red arrow) as wild-type erythroblasts (Fig. 7C, 31.9%, red arrow), indicating that E2f-2 loss did not prevent cell cycle progression prior to their arrest.
FIG. 7.
E2f-2 loss is associated with both a delay in S-phase progression and a failure to maintain mitotic arrest. (A to H) Representative BrdU labeling and propidium iodide (PI) staining of E13.5 wild-type (A, C, E, and G) or E2f2−/− (B, D, F, and H) fetal liver erythroblasts cultured for 3 h in the presence of BrdU to assess the cell cycle ex vivo (A and B); cultured for 3 h in the presence of BrdU, followed by 5 h in the absence of BrdU, to assess cell cycle progression to BrdU-positive G1 (C and D); cultured for 3 h in the presence of BrdU, followed by 5 h in the presence of nocodazole (Noc), to assess cell cycle progression to BrdU-positive G2/M and maintenance of a mitotic arrest (E and F); and cultured in nocodazole for 5 h to arrest cells at mitosis, followed by the addition of BrdU for 3 h of incubation, to assess S-phase entry, S-phase clearance to G2/M, and maintenance of a mitotic arrest (G and H).
To separate effects in S phase from possible defects elsewhere in the cell cycle, we examined the effect of nocodazole (a spindle-depolymerizing agent) on the kinetics of cell cycle progression in wild-type and E2f-2 null erythroblasts. We noted that E2f2−/− erythroblasts failed to arrest effectively in mitosis when treated with nocodazole, as determined by reduced numbers of cells accumulating with 4N DNA content (Fig. 7F, red arrows) compared to wild-type erythroblasts (Fig. 7E). The failure to arrest with 4N DNA content in response to nocodazole could be explained by a premature arrest in S phase. However, the demonstrated ability of E2f2−/− erythroblasts to progress through to a BrdU-positive G1 (Fig. 7D, red arrow) as efficiently as wild-type erythroblasts (Fig. 7C, red arrow) argues that this cannot be the sole explanation for a failure to undergo nocodazole-induced arrest. We considered whether E2f2−/− erythroblasts manifested cell cycle defects that included both defective S-phase progression and an impaired mitotic checkpoint. Consistent with multiple cell cycle defects, E2f2−/− erythroblasts showed delayed clearance from S phase (Fig. 7H, 13.3% in S) compared to the wild type (Fig. 7G, 7.2% in S) when treated with nocodazole but also reduced mitotic arrest (Fig. 7H, 4N = 41.8%) compared to the wild type (Fig. 7G, 4N = 62.7%).
In summary, E2f-2 null erythroblasts appeared to cycle as efficiently as wild-type erythroblasts for the first several hours in culture but exhibited both delayed S-phase progression and defective mitotic checkpoint control and by 15 h in differentiation culture had undergone premature cell cycle arrest.
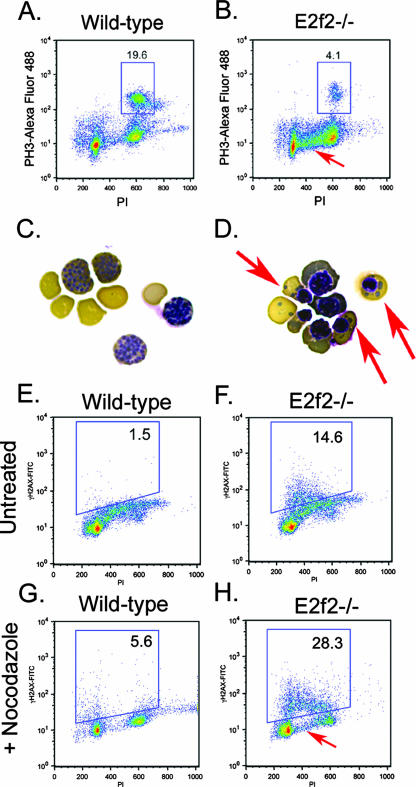
Failure to undergo nocodazole-induced arrest in mitosis was confirmed by flow cytometric staining for PH3 expression (Fig. 8B). Wild-type erythroblasts induced high levels of PH3 expression (Fig. 8A), in contrast to E2f2−/− erythroblasts, which expressed PH3 at basal levels (Fig. 8B, data not shown). Furthermore, nocodazole-treated E2f2−/− erythroblast cultures contained markedly increased numbers of cells with 2N to 4N DNA content (Fig. 8B, arrow) compared to the wild type (Fig. 8A). When we examined the cytological effect of nocodazole, we observed chromosome condensation characteristic of mitotic arrest in nocodazole-treated wild-type erythroblasts (Fig. 8C) but a failure of E2f2−/− erythroblasts to accumulate with condensed chromosomes (Fig. 8D). Rather, we observed erythroblast nuclei of various sizes and the appearance of chromatin fragments or micronuclei (Fig. 8D, red arrows). The appearance of micronuclei in E2f2−/− erythroblasts (Fig. 8D, red arrows) is consistent with a mitotic checkpoint defect, and loss of such chromatin fragments at cytokinesis may provide an additional explanation for why E2f2−/− erythroblasts accumulate with 2N to 4N DNA content.
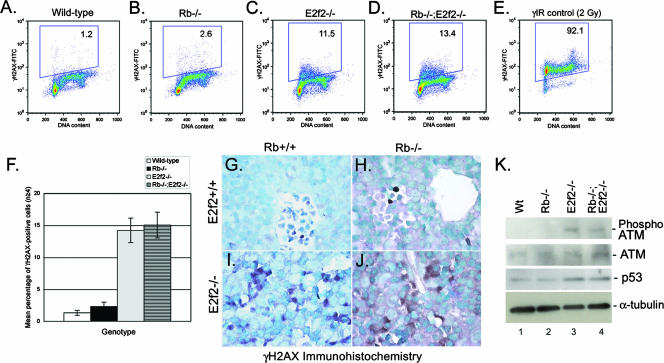
FIG. 8.
Mitotic defects are associated with DNA damage in E2f2−/− erythroblasts. (A and B) Flow cytometric analysis of intracellular staining for PH3 on nocodazole-treated wild-type (A) and E2f2−/− (B) E13.5 fetal liver erythroblasts. The red arrow indicates increased 2N to 4N DNA content in E2f2−/− cultures. (C and D) Benzidine-Wright-Giemsa staining of wild-type (C) and E2f2−/− (D) erythroblasts treated for 18 h with nocodazole to induce mitotic arrest and chromatin condensation at metaphase. Red arrows indicate the presence of cytoplasmic chromatin fragments, or micronuclei, in E2f2−/− cultures. (E to H) Flow cytometric analysis of intracellular staining for γH2AX on E13.5 erythroblasts derived from wild-type (E and G) or E2f2−/− (F and H) fetal liver differentiated in culture for 18 h in the presence or absence of nocodazole. FITC, fluorescein isothiocyanate; PI, propidium iodide.
E2f-2 loss causes increased DNA damage in maturing erythroblasts.
Given the presence of DNA fragments in nocodazole-treated E2f2−/− erythroblasts (Fig. 8D, red arrows), we considered whether E2f2−/− erythroblasts underwent premature cell cycle arrest (Fig. 6C) as a consequence of having sustained DNA damage. With intracellular staining for γH2AX as a measure of DNA double-strand breaks, we observed increased DNA damage in untreated E2f2−/− erythroblasts (Fig. 8F, 14.6%), independent of nocodazole treatment, compared to wild-type erythroblasts (Fig. 8E, 1.5%). When we treated erythroblasts with nocodazole, wild-type cells cleared S phase, as evidenced by the loss of cells with 2N to 4N DNA content (Fig. 8G), and only low levels of DNA damage were sustained (Fig. 8G; 5.6% γH2AX positive). Nocodazole treatment appeared to increase the amount of DNA damage sustained by E2f2−/− erythroblasts (Fig. 8H; 28.3% γH2AX positive), although this may be due to underestimation of basal levels of γH2AX expression in untreated cells as a result of profile overlap with cells in S phase. Cells with early 2N to 4N DNA content stained most strongly with antibody to γH2AX (Fig. 8H, arrow), indicating that most of the DNA damage in E2f2−/− erythroblast cultures occurred in cells entering S phase or, conversely, that the damage gave rise to cells with increased 2N to 4N DNA content due to loss of DNA fragments.
To determine whether the observed increase in DNA damage in E2f-2 null erythroblasts contributed to rescue of the red cell maturation defect in Rb null cells, we examined γH2AX staining in Rb−/− and Rb−/−; E2f2−/− fetal livers. While neither wild-type (Fig. 9A) nor Rb null (Fig. 9B) erythroblasts showed detectable staining for γH2AX after 18 h in differentiation culture, both E2f2−/− and Rb−/−; E2f2−/− erythroblasts showed increased levels of γH2AX staining (Fig. 9C, D, and F). Furthermore, when we examined levels of γH2AX staining in vivo on sections of E13.5 fetal liver, we observed increased γH2AX staining in both E2f2−/− (Fig. 9I) and Rb−/−; E2f2−/− (Fig. 9J) fetal livers compared to either wild-type (Fig. 9G) or Rb null (Fig. 9H) fetal liver. Additionally, levels of p53 and phospho-ATM were increased in E2f2−/− (Fig. 9K, lane 3) and Rb−/−; E2f2−/− (Fig. 9K, lane 4) fetal liver compared to either wild-type (Fig. 9K, lane 1) or Rb−/− (Fig. 9K, lane 2) fetal liver, supporting the conclusion that E2f2−/− and Rb−/−; E2f2−/− erythroblasts acquire increased levels of DNA double-strand breaks.
FIG. 9.
E2f-2 loss induces DNA double-strand breaks in wild-type and Rb null erythroblasts. (A to D) Flow cytometric analysis of intracellular staining for γH2AX on E13.5 erythroblasts derived from wild-type (A), Rb−/− (B), E2f2−/− (C), or Rb−/−; E2f2−/− (D) fetal liver and differentiated in culture for 18 h. (E) Control staining for γH2AX at 1 h after gamma irradiation of wild-type erythroblasts. (F) Quantitative analysis of the effect Rb loss and E2f-2 loss had on the incidence of DNA double-strand breaks. Data are shown as the mean percentage of γH2AX-positive cells for at least four different samples per genotype, and error bars represent the SD from the mean. (G to J) Immunohistochemical staining for γH2AX on sections of E13.5 wild-type (G), Rb−/− (H), E2f2−/− (I), and Rb−/−; E2f2−/− (J) fetal livers. (K) Western blotting analysis of phospho-ATM (ser1981), ATM, and p53 levels in wild-type (Wt), Rb−/−, E2f2−/−, and Rb−/−; E2f2−/− E13.5 fetal liver extracts. Protein loading was controlled for by α-tubulin. FITC, fluorescein isothiocyanate.
On the basis of these findings, we examined whether inducing DNA double-strand breaks in Rb null erythroblasts caused a cell cycle arrest that, in turn, would promote enucleation. We showed that 2 Gy of gamma irradiation induced high levels of DNA double-strand breaks at 1 h postirradiation in Rb null erythroblasts, as determined by intracellular staining for γH2AX (Fig. 9E). However, DNA damage had no impact on the extent of red cell enucleation in Rb null erythroblast cultures, despite inducing an effective cell cycle arrest, predominantly in the G2 phase of the cell cycle (data not shown). These results suggest that while DNA damage and damage-induced arrest may contribute to improved red cell maturation upon E2f-2 loss, it is not sufficient to induce enucleation of Rb null erythroblasts. This, in turn, suggests that the DNA damage and cell cycle arrest induced by loss of E2f-2 are distinct in the ability to promote enucleation of Rb null erythroblasts.
In summary, we have shown that E2f-2 is upregulated in end-stage red cells and is the principal E2f associated with pRb at this stage of erythroid differentiation. Targeted deletion of E2f-2 promotes erythroid maturation of Rb null red cells, including upregulation of end-stage differentiation markers and induction of enucleation. Loss of E2f-2 also prolonged the survival of Rb null embryos by several days, and this is likely due to the improved red cell phenotype since placental defects were still evident in Rb−/−; E2f2−/− mice. The rescue of the erythroid defects in Rb null red cells was associated with elevated DNA damage and cell cycle arrest, although additional features of this arrest are clearly important to explain the rescue, as inducing DNA damage and growth arrest per se was not sufficient to achieve the rescue. These results identify a unique tissue-restricted role for an E2f family member that highlights the nonredundant nature of the family's activities in cell growth and differentiation.
DISCUSSION
We show here that E2f-2 is a key target of pRb in end-stage erythroblasts and that E2f-2 loss rescues many aspects of the Rb null erythroid defect and extends the life span of Rb null mice. The ability of E2f-2 loss to rescue red cell maturation and extend the survival of Rb null mice is unlikely to be due to improved placental development in Rb−/−; E2f2−/− mice since E2f-2 is not expressed in wild-type or Rb null placenta, and the placental phenotype in compound mutant mice is similar to that observed in the Rb null mouse. We found that, in contrast to E2f-2, E2f-3b is highly expressed in the placenta, supporting a previous report implicating deregulated E2f-3 in placental development defects observed in Rb null mice (42). We also confirmed that loss of E2f-2 had a limited impact on ectopic proliferation or cell survival in the developing nervous system of the Rb null mouse (data not shown; 33). Therefore, we suggest that E2f-2 loss extends the survival of Rb null mice by promoting erythroid maturation. Given that systemic hypoxia and nutrient deprivation are the likely cause of the demise of Rb null embryos by E14.5 (36, 42, 43), improved red blood cell function brought about by E2f-2 deletion could delay the onset of death by improving oxygen delivery to peripheral tissues. This is a temporary effect, however, as Rb−/−; E2f2−/− embryos succumb to death between E15.5 and E17.5 with persistent placental defects.
Effects of DNA damage on cell cycle regulation during terminal differentiation.
We show that failure of differentiating Rb null erythroblasts to mature is associated with aberrant cell cycle entry and that this is overcome by targeted deletion of E2f-2. E2f-2 loss also promoted end-stage differentiation and enucleation in Rb null erythroblasts. Interestingly, cell cycle arrest of E2f-2 null erythroblasts was associated with increased DNA double-strand breaks; however, the source of the DNA damage in E2f-2 null erythroblasts is not clear. Our data demonstrate that loss of E2f-2 causes erythroblasts to be delayed for S-phase progression, consistent with a previous report (21), but also to exhibit defective mitotic checkpoint activation. Failure to enforce the mitotic checkpoint has been demonstrated to induce DNA double-strand breaks and micronuclei (39), as we observed in E2f-2 null erythroblasts, and future work will explore the molecular basis of this defect.
DNA damage has been previously implicated in a so-called “differentiation checkpoint” during myogenesis whereby genotoxic agents inhibited muscle differentiation if applied to myoblasts prior to the establishment of the terminal gene expression program (31). This reversible differentiation checkpoint was postulated to delay the onset of differentiation as a consequence of DNA repair mechanisms active at expressed genes. Conversely, it has been suggested that growth arrest induced by DNA damage is not reversible in differentiating cells but rather provokes inappropriate differentiation, leading to the formation of abnormal tissues (30). Alternatively, DNA damage may induce cell cycle exit and premature differentiation without inhibiting the tissue-specific gene expression program. Our data show that E2f-2 null erythroblasts undergo cell cycle exit earlier than wild-type erythroblasts and that end-stage red cells in E2f-2 null embryos are abnormal in size but that they express normal levels of end-stage differentiation markers, such as TER119. This suggests that DNA damage does not inhibit differentiation but, at least in red cells, may actually promote it.
Erythroblasts become smaller as they differentiate (19), and the appearance of enlarged red cells in the periphery of E2f-2 null mice is consistent with premature cell cycle exit leading to aberrantly sized red cells. Thus, loss of E2f-2 may restore enucleation capacity to Rb null erythroblasts by imposing a growth arrest at an earlier stage in differentiation, thereby pre-empting the differentiation defects induced by loss of pRb. The effect of premature cell cycle arrest induced by E2f-2 loss on promoting differentiation and, conversely, the ability of Rb loss and aberrant cell cycling to inhibit differentiation suggest that terminal erythroid differentiation is coupled to cell cycle exit. However, we were unable to induce terminal differentiation in Rb null erythroblasts by treatment with exogenous cytostatic agents (including Cdk inhibitors and transforming growth factor β) or DNA-damaging agents (ionizing radiation and doxorubicin [Adriamycin]) despite an effective arrest in G2 of the cell cycle (data not shown). This indicates that G2 arrest is not effective for induction of terminal erythroid maturation and also leads us to conclude that the arrest elicited by E2f-2 loss is unique thus far in its ability to induce both pRb-independent cell cycle arrest of erythroblasts and their terminal differentiation.
Mouse embryo fibroblasts (MEFs) lacking all three activator E2fs (E2f-1, -2, and -3a) fail to cycle and incorporate BrdU, reportedly because of a failure to activate E2f target genes critical for DNA replication and cell cycle progression (44). However, cell cycle arrest and repression of E2f targets was p53 dependent and inactivation of the p53 tumor suppressor promoted E2f target gene expression and overcame proliferation defects in triple-knockout (TKO) MEFs (40). Our data suggest that E2f2−/− TER119+ erythroblasts are functionally TKO erythroblasts because of the low levels of E2f-1 and E2f-3a in these cells. The increased numbers of TKO MEFs with 2N to 4N DNA content (44) and the p53-dependent cell cycle arrest in TKO MEFs (40) suggest that, as in E2f2−/− erythroblasts, cell cycle arrest in TKO MEFs may be due to increased DNA damage. These results, together with our results presented here, raise the intriguing possibility that the activator E2Fs play a more important role in preventing DNA damage, possibly by enforcing cell cycle checkpoints, than in actively promoting cell cycle progression.
E2f-2 in erythropoiesis.
We have shown that during normal erythropoiesis, E2f-2 expression is upregulated as cells exit the cell cycle and terminally differentiate. The mechanism by which E2f-2 is upregulated in end-stage red cells is not known, although the E2f-2 promoter contains functional c-Myc binding sites (34) and c-Myc is known to be downregulated during terminal differentiation of mouse erythroleukemia cells (2). It will be interesting to determine whether upregulation of E2f-2 in end-stage red cells is mediated through derepression brought about by c-Myc downregulation or whether erythroid-specific factors, such as EKLF, are involved in its upregulation.
The role of E2f activity in the erythroid phenotype of Rb null mice was previously investigated in studies involving a naturally occurring point mutant form of pRB isolated from a partially penetrant form of retinoblastoma (R661W). This mutant form of pRB was reported to retain the capacity to induce terminal differentiation (38), despite being defective for E2f binding and cell cycle arrest (35). The generation of mice expressing the mouse variant (R654W) of the human R661W allele indicated that while pRb functions to repress the cell cycle through interactions with E2fs, it also had E2f-independent functions in terminal differentiation (38). In particular, it was shown that this mutant form of pRb could promote erythroid maturation and extend the life span of Rb null mice (38). However, the R661W mutant retains the ability to bind E2f-2 (3), a fact that was not realized in earlier studies (35) because of the low expression of E2f-2 in tumor cell lines, as we show here (Fig. 1C). Thus, rescue of erythroid maturation in Rb null mice by expression of the R654W Rb mutant may be explained by its continued ability to repress through E2f-2 and not to secondary non-E2f effects. Interestingly, we show that deletion of E2f-2 in mice restored normal erythroid differentiation and extended the life span of Rb null mice similar to mice expressing the R654W mutant (38).
E2f-4 has also been shown to play an important role in erythropoiesis (14, 17, 32), but in contrast to E2f-2, it appears to be involved in regulating the proliferation of less mature erythroid progenitors (17). Counter to the conventional role attributed to E2f-4 as a repressor E2f, E2f-4 appeared to be required for expansion of proerythroblasts (17). Whether this was due to a direct role in promoting E2f target gene expression or to indirect effects on activator E2fs mediated by “pocket protein shuffling” (20) has not been determined, although the presence of E2f-4 at the promoters of these genes supports the former conclusion. Together with our work, these studies highlight the importance of E2fs and pocket proteins in erythroid differentiation.
Acknowledgments
Work on this project was funded by NIH grant RO1 HL080262 (K.M.) and by a Markey scholarship (A.D.).
We acknowledge the excellent technical support provided by the University of Chicago flow cytometry facility and Ryan Duggan in particular.
Footnotes
Published ahead of print on 8 October 2007.
REFERENCES
- 1.Aslanian, A., P. J. Iaqunita, R. Verona, and J. A. Lees. 2004. Repression of the Arf tumor suppressor by E2f-3 is required for normal cell cycle kinetics. Genes Dev. 18:1413-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben-David, Y., and A. Bernstein. 1991. Friend virus-induced erythroleukemia and the multistage nature of cancer. Cell 66:831-834. [DOI] [PubMed] [Google Scholar]
- 3.Benevolenskaya, E. V., H. L. Murray, P. Branton, R. A. Young, and W. G. Kaelin, Jr. 2005. Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol. Cell 18:623-635. [DOI] [PubMed] [Google Scholar]
- 4.Chen, C., Y. Kang, P. M. Siegel, and J. Massague. 2002. E2F4/5 and p107 as Smad cofactors linking the TGFβ receptor to c-myc repression. Cell 110:19-32. [DOI] [PubMed] [Google Scholar]
- 5.Clark, A. J., K. M. Doyle, and P. O. Humbert. 2004. Cell intrinsic functions of pRb in erythropoiesis. Blood 104:1324-1326. [DOI] [PubMed] [Google Scholar]
- 6.de Bruin, A., L. Wu, H. I. Saavedra, P. Wilson, Y. Yang, T. J. Rosol, M. Weinstein, M. L. Robinson, and G. Leone. 2003. Rb function in extraembryonic tissue lineages suppresses apoptosis in the CNS of Rb-deficient mice. Proc. Natl. Acad. Sci. USA 100:6546-6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dimova, D. K., and N. J. Dyson. 2005. The E2F transcriptional network: old acquaintances with new faces. Oncogene 24:2810-2826. [DOI] [PubMed] [Google Scholar]
- 8.Dimova, D. K., O. Stevaux, M. V. Frolov, and N. J. Dyson. 2003. Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev. 17:2308-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frank, S. R. 2001. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 15:2069-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gregory, T., C. Yu, S. H. Orkin, G. A. Blobel, and M. J. Weiss. 1999. GATA-1 and erythropoietin cooperate to promote erythroid cell survival by regulating Bcl-XL expression. Blood 94:87-96. [PubMed] [Google Scholar]
- 11.Hernando, E., Z. Nahle, G. Juan, E. Diaz-Rodriguez, M. Alaminos, M. M. Hemann, L. Michel, V. Mittal, W. Gerald, R. Benezra, S. W. Lowe, and C. Cordon-Cardo. 2004. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature 430:797-802. [DOI] [PubMed] [Google Scholar]
- 12.Hirota, T., J. J. Lipp, B. H. Toh, and J. M. Peters. 2005. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 438:1176-1180. [DOI] [PubMed] [Google Scholar]
- 13.Huang, X., and Z. Darzynkiewicz. 2006. Cytometric assessment of histone H2AX phosphorylation: a reporter of DNA damage. Methods Mol. Biol. 314:73-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humbert, P. O., C. Rogers, S. Ganiatsas, R. L. Landsberg, J. M. Trimarchi, S. Dandapani, C. Brugnara, S. Erdman, M. Schrenzel, R. T. Bronson, and J. A. Lees. 2000. E2f4 is essential for normal erythrocyte maturation and neonatal viability. Mol. Cell 6:281-291. [DOI] [PubMed] [Google Scholar]
- 15.Irwin, M., M. C. Marin, A. C. Phillips, R. S. Seelam, D. I. Smith, W. Liu, E. R. Flores, K. Y. Tsai, T. Jacks, K. H. Vousden, and W. G. Kaelin. 2000. Role for the p53 homologue p73 in E2F-1 induced apoptosis. Nature 407:645-648. [DOI] [PubMed] [Google Scholar]
- 16.Kina, T., K. Ikuta, E. Takayama, K. Wada, A. Majumdar, I. L. Weissman, and Y. Katsura. 2000. The monoclonal antibody TER-119 recognizes a molecule associated with glycophorin A and specifically marks the late stages of murine erythroid lineage. Br. J. Haematol. 109:280-287. [DOI] [PubMed] [Google Scholar]
- 17.Kinross, K. M., A. J. Clark, R. M. Iazzolino, and P. O. Humbert. 2006. E2f4 regulates fetal erythropoiesis through the promotion of cellular proliferation. Blood 108:886-895. [DOI] [PubMed] [Google Scholar]
- 18.Korenjak, M., and A. Brehm. 2005. E2F-Rb complexes regulating transcription of genes important for differentiation and development. Curr. Opin. Genet. Dev. 15:1-8. [DOI] [PubMed] [Google Scholar]
- 19.Koury, M. J., S. T. Sawyer, and S. J. Brandt. 2002. New insights into erythropoiesis. Curr. Opin. Hematol. 9:93-100. [DOI] [PubMed] [Google Scholar]
- 20.Lee, E. Y., H. Cam, U. Ziebold, J. B. Rayman, J. A. Lees, and B. D. Dynlacht. 2002. E2F4 loss suppresses tumorigenesis in Rb mutant mice. Cancer Cell 2:463-472. [DOI] [PubMed] [Google Scholar]
- 21.Li, F. X., J. W. Zhu, C. J. Hogan, and J. DeGregori. 2003. Defective gene expression, S phase progression, and maturation during hematopoiesis in E2F1/E2F2 mutant mice. Mol. Cell. Biol. 23:3607-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macleod, K., Y. Hu, and T. Jacks. 1996. Loss of Rb activates both p53-dependent and -independent cell death pathways in the developing mouse nervous system. EMBO J. 15:6178-6188. [PMC free article] [PubMed] [Google Scholar]
- 23.Macleod, K. F., N. Sherry, G. Hannon, D. Beach, T. Tokino, K. Kinzler, B. Vogelstein, and T. Jacks. 1995. p53-dependent and independent expression of p21 during cell growth, differentiation and DNA damage. Genes Dev. 9:935-944. [DOI] [PubMed] [Google Scholar]
- 24.Morgenbesser, S. D., B. O. Williams, T. Jacks, and R. A. DePinho. 1994. p53-dependent apoptosis produced by Rb-deficiency in the developing mouse lens. Nature 371:72-74. [DOI] [PubMed] [Google Scholar]
- 25.Moroni, M. C., E. S. Hickman, E. L. Denchi, G. Caprara, E. Colli, F. Cecconi, H. Muller, and K. Heliin. 2001. Apaf-1 is a transcriptional target for E2F and p53. Nat. Cell Biol. 3:552-558. [DOI] [PubMed] [Google Scholar]
- 26.Müller, H., A. P. Bracken, R. Vernell, M. C. Moroni, C. F., E. Grassilli, E. Prosperini, E. Vigo, J. D. Oliner, and K. Helin. 2001. E2Fs regulate the expression of genes involved in differentiation, development, proliferation and apoptosis. Genes Dev. 15:267-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murga, M., O. Fernandez-Capetillo, S. J. Field, B. Moreno, L. R. Borlado, Y. Fujiwara, D. Balomenos, A. Vicario, A. C. Carrera, S. H. Orkin, M. E. Greenberg, and A. M. Zubiaga. 2001. Mutation of E2F2 in mice causes enhanced T lymphocyte proliferation, leading to the development of autoimmunity. Immunity 15:959-970. [DOI] [PubMed] [Google Scholar]
- 28.Nahle, Z., J. Polakoff, R. V. Davuluri, M. E. McCurrach, M. D. Jacobson, M. Narita, M. Q. Zhang, Y. Lazebnik, D. Bar-Sagi, and S. W. Lowe. 2002. Direct coupling of the cell cycle and cell death machinery by E2F. Nat. Cell Biol. 4:859-864. [DOI] [PubMed] [Google Scholar]
- 29.Persengiev, S. P., J. Li, M. L. Poulin, and D. L. Kilpatrick. 2001. E2F2 converts reversibly differentiated PC12 cells to an irreversible neurotrophin-dependent state. Oncogene 20:5124-5131. [DOI] [PubMed] [Google Scholar]
- 30.Polesskaya, A., and M. A. Rudnicki. 2002. A MyoD dependent differentiation checkpoint: ensuring genome integrity. Dev. Cell 3:757-764. [DOI] [PubMed] [Google Scholar]
- 31.Puri, P. L., K. Bhakta, L. D. Wood, A. Costano, J. Zhu, and J. Y. Wang. 2002. A myogenic differentiation checkpoint activated by genotoxic stress. Nat. Genet. 32:585-593. [DOI] [PubMed] [Google Scholar]
- 32.Rempel, R. E., M. T. Saenz-Robles, R. Storms, S. Morham, S. Ishida, A. Engel, L. Jakoi, M. F. Melhem, J. M. Pipas, C. Smith, and J. R. Nevins. 2000. Loss of E2f4 activity leads to abnormal development of multiple cellular lineages. Mol. Cell 6:293-306. [DOI] [PubMed] [Google Scholar]
- 33.Saavedra, H. I., L. Wu, A. de Bruin, C. Timmers, T. J. Rosol, M. Weinstein, M. L. Robinson, and G. Leone. 2002. Specificity of E2F1, E2F2 and E2F3 in mediating phenotypes induced by loss of Rb. Cell Growth Differ. 13:215-225. [PubMed] [Google Scholar]
- 34.Sears, R., K. Ohtani, and J. R. Nevins. 1997. Identification of positively and negatively acting elements regulating expression of the E2F2 gene in response to cell growth signals. Mol. Cell. Biol. 17:5227-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sellers, W. R., B. G. Novitch, S. Miyake, A. Heith, G. A. Otterson, F. J. Kaye, A. B. Lassar, and W. G. Kaelin. 1998. Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation and suppress tumor cell growth. Genes Dev. 12:95-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spike, B. T., B. C. Dibling, and K. F. Macleod. 2007. Hypoxic stress underlies defects in erythroblast island formation in the Rb null mouse. Blood 110:2173-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spike, B. T., A. Dirlam, B. C. Dibling, J. Marvin, B. O. Williams, T. Jacks, and K. F. Macleod. 2004. The Rb tumor suppressor is required for stress erythropoiesis. EMBO J. 23:4319-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun, H., Y. Chang, B. Schweers, M. A. Dyer, X. Zhang, S. W. Hayward, and D. W. Goodrich. 2006. An E2F binding-deficient Rb1 protein partially rescues developmental defects associated with Rb1 nullizygosity. Mol. Cell. Biol. 26:1527-1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanaka, T., and N. Shimizu. 2000. Induced detachment of acentric chromatin from mitotic chromosomes leads to their cytoplasmic localization at G1 and the micronucleation by lamin reorganization at S phase. J. Cell Sci. 113:697-707. [DOI] [PubMed] [Google Scholar]
- 40.Timmers, C., N. Sharma, R. Opavsky, B. Maiti, L. Wu, J. Wu, D. Orringer, P. Trikha, H. I. Saavedra, and G. Leone. 2007. E2f1, E2f2 and E2f3 control E2F target expression and cellular proliferation via a p53-dependent negative feedback loop. Mol. Cell. Biol. 27:65-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trimarchi, J., and J. A. Lees. 2002. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 3:11-20. [DOI] [PubMed] [Google Scholar]
- 42.Wenzel, P. L., L. Wu, A. de Bruin, J. L. Chong, W. Y. Chen, G. Dureska, E. Sites, T. Pan, A. Sharma, K. Huang, R. Ridgway, K. Mosaliganti, R. Sharp, R. Machiraju, J. Salz, H. Yamamoto, J. C. Cross, M. L. Robinson, and G. Leone. 2007. Rb is critical in a mammalian tissue stem cell population. Genes Dev. 21:85-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu, L., A. de Bruin, H. I. Saavedra, M. Starovic, A. Trimboli, Y. Yang, J. Opavska, P. Wilson, J. C. Thompson, M. C. Ostrowski, T. J. Rosol, L. A. Woollett, M. Weinstein, J. C. Cross, M. L. Robinson, and G. Leone. 2003. Extra-embryonic function of Rb is essential for embryonic development and viability. Nature 421:942-947. [DOI] [PubMed] [Google Scholar]
- 44.Wu, L., C. Timmers, B. Maiti, H. I. Saavedra, L. Sang, G. T. Chong, F. Nuckolls, P. Giangrande, F. A. Wright, S. J. Field, M. E. Greenberg, S. H. Orkin, J. R. Nevins, M. L. Robinson, and G. Leone. 2001. The E2F1-3 transcription factors are essential for cellular proliferation. Nature 414:457-462. [DOI] [PubMed] [Google Scholar]
- 45.Yoshida, H., K. Kawane, M. Koike, Y. Mori, Y. Uchiyama, and S. Nagata. 2005. Phosphatidlyserine-dependent engulfment by macrophages of nuclei from erythroid precursor cells. Nature 437:754-758. [DOI] [PubMed] [Google Scholar]
- 46.Zhu, J. W., S. J. Field, L. Gore, M. Thompson, H. Yang, Y. Fujiwara, R. D. Cardiff, M. Greenberg, S. H. Orkin, and J. DeGregori. 2001. E2F1 and E2F2 determine thresholds for antigen-induced T-cell proliferation and suppress tumorigenesis. Mol. Cell. Biol. 21:8547-8564. [DOI] [PMC free article] [PubMed] [Google Scholar]