Abstract
Arsenite is a human carcinogen causing skin, bladder, and lung tumors, but the cellular mechanisms underlying these effects remain unclear. We investigated expression of the essential base excision DNA repair enzyme apurinic endonuclease 1 (Ape1) in response to sodium arsenite. In mouse 10T½ fibroblasts, Ape1 induction in response to arsenite occurred about equally at the mRNA, protein, and enzyme activity levels. Analysis of the APE1 promoter region revealed an AP-1/CREB binding site essential for arsenite-induced transcriptional activation in both mouse and human cells. Electrophoretic mobility shift assays indicated that an ATF4/c-Jun heterodimer was the responsible transcription factor. RNA interference targeting c-Jun or ATF4 eliminated arsenite-induced APE1 transcription. Suppression of Ape1 or ATF4 sensitized both mouse fibroblasts (10T½) and human lymphoblastoid cells (TK6) to arsenite cytotoxicity. Expression of Ape1 from a transgene did not efficiently restore arsenite resistance in ATF4-depleted cells but did offset initial accumulation of abasic DNA damage following arsenite treatment. Mutagenesis by arsenite (at the TK and HPRT loci in TK6 cells) was observed only for ATF4-depleted cells, which was strongly offset by Ape1 expression from a transgene. Therefore, the ATF4-mediated up-regulation of Ape1 and other genes plays a key role against arsenite-mediated toxicity and mutagenesis.
Epidemiological studies from Bangladesh, India (West Bengal), Taiwan, and South America indicate that arsenic ingestion, typically via contaminated drinking water, is associated with increased incidence of diverse human diseases, such as atherosclerosis, diabetes, and cancers of the skin, bladder, and lung (55). The arsenic found in drinking water is overwhelmingly one of the inorganic arsenic forms, mostly pentavalent arsenate and trivalent arsenite (55).
Arsenite treatment has recently been introduced as therapy for acute promyelocytic leukemia (61). The pathological and therapeutic mechanisms of arsenic are actively debated, and DNA may be an important target for arsenic-related damage. For example, comet assays demonstrate that exposure to as little as 1 nM of sodium arsenite could rapidly (within 30 min) induce DNA base damage in human HeLa, neutrophilic NB4, and HL-60 cells (59, 69). For comparison, the reported mean blood arsenic concentration in people consuming highly contaminated water was 560 nM (49), while the mean plasma arsenite concentration for cancer treatment was 1,000 to 6,000 nM (60). However, many cell-based assays have failed to detect a significant increase in point mutations due to arsenite (55). One conclusion from these observations is that a strong cellular antimutagenesis mechanism counteracts the mutational potential of arsenite-induced DNA damage.
Oxidized and other small base lesions are excised by DNA glycosylases, which channel damage into the base excision DNA repair (BER) pathway. In mammalian cells, glycosylase products are processed by the abasic (apurinic/apyrimidinic [AP]) endonuclease (apurinic endonuclease 1 [Ape1], human Ape1 [hApe1] and mouse Ape1 [mApe1]) which incises on the 5′ side of AP sites or removes the 3′-terminal unsaturated deoxyribose produced by the AP lyase activities associated with some DNA glycosylases (10). Although an Ape1-independent pathway is triggered by the NEIL1 and NEIL2 glycosylases (71), Ape1 is clearly the predominant activity for processing abasic sites in various human cell types (18) and in murine fibroblasts (30). In the main BER pathway, the 5′-terminal deoxyribose-5-phosphate generated by Ape1 incision is removed by the deoxyribose-phosphate lyase activity of DNA polymerase β (10), which also replaces the single missing nucleotide to allow DNA ligase to complete the repair (10). Longer repair patches are also generated at significant levels, and the switching mechanism that governs the short-patch/long-patch distribution remains a major unresolved issue (11).
To investigate the process of arsenite-induced cell damage and the role of Ape1 in arsenite-related mutagenesis and carcinogenesis, we examined the response of mApe1 in mouse fibroblasts in culture upon exposure to sodium arsenite. We show that mApe1 protein induction is due to increased transcription, and we identified an essential activator protein 1 (AP-1)/cyclic AMP response element-binding protein (CREB) binding site for the response. Further experiments demonstrated the roles of the transcription factors c-Jun and activating transcription factor 4 (ATF4) in this homeostatic defense response to arsenite-induced stress. Depleting ATF4 potentiates arsenite-induced mutation at the thymidine kinase (TK) and hypoxanthine phosphoribosyltransferase (HPRT) loci of human B lymphoblast TK6 cells. However, the mutagenic action in ATF4-deficient cells can be efficiently prevented by reexpression of mApe1. Thus, these data suggest an ATF4-mediated cell response against arsenite-induced mutagenesis, with Ape1 as a key component in this pathway.
MATERIALS AND METHODS
Cell culture and arsenite treatment.
The immortalized mouse embryonic fibroblast cell line 10T½ (C3H clone 8) and the immortalized TK6 human B lymphoblast cells were kindly provided by John B. Little of the Harvard School of Public Health. Both cell lines have been confirmed to exhibit normal p53 function (63, 76). The NIH 3T3 cell line was purchased from ATCC. The 10T½ and NIH 3T3 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% (vol/vol) bovine serum (HyClone), and TK6 cells were maintained in RPMI 1640 medium supplemented with 10% horse serum. All cultures were grown in 5% (vol/vol) CO2 at 37°C. Growing (50% confluence) or confluent 10T½ cultures were treated with sodium arsenite for 0.5 to 8 h, washed twice with phosphate-buffered saline (PBS), and shifted to fresh supplemented medium for 2 h for Northern blot samples, 4 h for immunoblotting samples, and 6 h for AP endonuclease assays and for promoter-reporter analysis. TK6 cells (106/ml) were treated as described above for 10T½ cells, but the removal of arsenite and washing steps were conducted by centrifugation at 500 × g for 5 min, and the pellets were resuspended in fresh medium. For ATF4-deficient cells, the culture medium was supplemented with 20 μM β-mercaptoethanol and 1× nonessential amino acid mix containing alanine, aspartate, asparagine, glutamate, glycine, proline, and serine (Invitrogen). For treatment with N-acetyl-l-cysteine (NAC), solutions were freshly prepared on the day of treatment and adjusted to pH 7.4 by the addition of NaOH.
Plasmid construction and site-directed mutagenesis.
For analyzing the mAPE1 promoter, plasmids in the pCAT-Basic vector (Promega) carrying the chloramphenicol acetyltransferase (CAT) reporter gene were constructed. The plasmids for the promoter studies of mAPE1 (pCBM1, pCBM2, pCBM6, pCBM7, pCBM10, pCBM14, pCBM16, pCBM18, and pCBM20) and hAPE1 (pCB9 and pCB10) have been described previously (17, 24, 25). The construction of the small interfering RNA (siRNA) retroviral vector pSUPER has been described previously (18). The retroviral vector hairpin siRNAs and their sequences were as follows: mouse ATF4, 5′-TCCCTCCATGTGTAAAGGA-3′; human ATF4, 5′-TCCCTCAGTGCATAAAGGA-3′; mouse c-Jun, 5′-GTCTCAGGAGCGGATCAAG-3′; hAPE1, 5′-ATGACAAAGAGGCAGCAGG-3′; mAPE1, 5′-CCGAGAAGGAGGCCGCGGG-3′; and siRNA control targeting luciferase (LUC), 5′-CTTACGCTGAGTACTTCGA-3′. Mutation of the wild-type mAPE1 promoter to generate a series of mutant mAPE1-CAT constructs was carried out on pCBM14 using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and following the manufacturer's instructions. The expression vectors pQCXIH-hAPE1 and pQCXIH-mAPE1 were generated by inserting their coding sequences into the retroviral vector pQCXIH (Clontech). Each construct was confirmed by DNA sequencing at the Dana Farber/Harvard Cancer Center facility. Viral vector infection and cell selection were performed as described previously (18).
Northern blot assay.
After a 2-h incubation following the exposure to sodium arsenite, the cells were harvested for Northern blot analysis. Total RNA extraction was carried out using a commercially available kit (RNeasy; Qiagen, Valencia, CA). Samples (15 μg) of total RNA were resolved by electrophoresis in a 1% denaturing agarose gel, transferred to nylon membranes, and hybridized with a 450-bp SmaI-SalI mAPE1 probe from pBSK-mAPE1 (24). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA was used as a control probe. The radioactive signal was measured by phosphorimaging (Bio-Rad GS-525 molecular image analyzer).
Immunoblotting.
Cells were incubated for 4 h in fresh medium following sodium arsenite exposure. Samples of 107 cells were resuspended in 100 μl of buffer I containing 10 mM Tris-HCl, pH 7.8, and 200 mM KCl and mixed with 100 μl of buffer II containing 10 mM Tris-HCl, pH 7.8, 200 mM KCl, 2 mM EDTA, 10% (vol/vol) glycerol, 0.2% (vol/vol) Nonidet P-40, 2 mM dithiothreitol (DTT), 0.5 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail (Sigma Chemical Co., St. Louis, MO). After the mixture was gently rocked at 4°C for 40 min, the lysate was clarified by centrifugation at 16,000 × g for 10 min. Protein concentrations were determined by the Bradford assay (7). After electrophoresis on sodium dodecyl sulfate-polyacrylamide (12%) gels and electroblotting onto nitrocellulose (Schleicher & Schuell, Wiltshire, United Kingdom), the blots were probed with primary antibodies: mouse anti-human Ape1 (sc-17774; Santa Cruz Biotechnology, Santa Cruz, CA), anti-Apn1 (52), anti-c-Jun or anti-ATF4 (both from Santa Cruz Biotechnology), or mouse anti-human β-actin (AC-15; Sigma). After the blots were washed and probed with the appropriate secondary (anti-mouse or anti-rabbit) antibodies, the proteins were visualized with a chemiluminescence or chemifluorescence detection system (Amersham, Piscataway, NJ).
AP endonuclease activity assay.
The AP-specific endonuclease activity of Ape1 was analyzed using a quantitative in vitro assay that measures the incision of a 35-mer duplex oligonucleotide substrate containing a synthetic tetrahydrofuran (F) AP site (5′-CGCAATGGACCGGFACCCCCGTATGGATCCAACCA-3′) as previously described with some modifications (72). Briefly, 0.1 pmol of 5′-labeled, purified duplex DNA substrate was incubated with 5 ng of crude whole-cell extract in a 10-μl reaction mixture containing 50 mM HEPES, pH 7.5, 50 mM KCl, 10 mM MgCl2, 2 mM DTT, 10 mM dAMP, 1 μg/ml bovine serum albumin, and 0.05% Triton X-100. The reaction mixtures were incubated for 15 min at 37°C and terminated by the addition of formamide loading buffer containing 90% formamide, 10 mM EDTA, bromphenol blue, and xylene cyanol and heating at 100°C for 2 min. The DNA products were then resolved by electrophoresis on 12% acrylamide gels containing 7 M urea. After drying, the gels were analyzed using a Molecular Imager System (model GS-525; Bio-Rad, Hercules, CA), and the results were quantified using Molecular Analyst software (Bio-Rad). The percent AP endonuclease activity of each reaction was calculated by using the relation product/(product + substrate) × 100% to indicate relative activity.
Reporter assay.
Transfection of 10T½ cells and LUC assay were performed as described previously (24). Reporter plasmids were introduced into cells by the calcium phosphate method (31). After a 6-h incubation without arsenite in fresh medium, the cells were harvested to determine CAT activity using the Fast CAT Green (deoxy) kit (Molecular Probes, Inc.). The β-galactosidase expression vector pCMV-β-gal was cotransfected as an internal control to monitor transfection efficiency, assayed using the β-galactosidase enzyme assay system (Promega, Madison, WI). Lysates for the CAT and β-galactosidase assays were generated according to the β-galactosidase kit protocol. Luciferase and CAT activities were normalized to the β-galactosidase activity in each transfection sample.
Electrophoretic mobility shift assay (EMSA).
Whole-cell extracts were prepared at various times after incubation with arsenite in fresh medium according to a previously published method with some modifications (65). Briefly, 5 × 106 cells were scraped into PBS, washed, and collected by centrifugation. Cell pellets were suspended in 100 μl extraction buffer containing 20 mM HEPES (pH 7.9), 100 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, 25% glycerol, 0.2% Nonidet P-40, 0.2 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail (Sigma; diluted according to the supplier) and lysed by gently rocking at 4°C for 40 min. The lysate was centrifuged at 100,000 × g for 5 min, and the supernatant was stored frozen at −80°C. DNA binding reactions were carried out in a volume of 15 μl by incubating 5 μg of whole-cell extracts and 1 μg of poly(dG-dC) in a buffer containing 20 mM HEPES-KOH, pH 7.9, 40 mM KCl, 1 mM MgCl2, 4% Ficoll, 0.1 mM EDTA, 0.1% Nonidet P-40, and 1 mM DTT. After incubation for 10 min on ice, the end-labeled duplex 35-bp oligonucleotide 5′-ACCCGTGCCCGGGGCCGTGACGTAAGTGCGCCGCG-3′ was added, and the incubation was continued at room temperature for 20 min. This was followed by electrophoresis on a 4% polyacrylamide nondenaturing gel at a constant voltage of 8 V/cm applied for 2 h at 4°C and then by autoradiography of the dried gel. For competition experiments, a 10- to 100-fold excess of unlabeled competitor duplex DNA oligonucleotide was incubated with the protein extract for 10 min before the labeled oligonucleotide probes were added. For supershift assays, monoclonal/polyclonal antibodies (1 μg in 0.5 μl) against FosB, c-Fos, c-Jun, JunB, JunD, Fra-1, Fra-2, CREB-1, ATF1, ATF2, ATF3, and ATF4 (all from Santa Cruz Biotechnology) were added to the binding buffer for 1 h at 0 to 4°C before the addition of the labeled DNA probes.
siRNA treatment and Ape1 rescue.
Gene “knockdown” via siRNA targeting ATF4, c-Jun, or APE1 were carried out using the retroviral vector pSUPER essentially as described previously (18). The retroviral vector hairpin siRNAs and their sequences were as follows: mouse ATF4, 5′-TCCCTCCATGTGTAAAGGA-3′; human ATF4, 5′-TCCCTCAGTGCATAAAGGA-3′; mouse c-Jun, 5′-GTCTCAGGAGCGGATCAAG-3′; hAPE1, 5′-AAATGACAAAGAGGCAGCAGG-3′; mAPE1, 5′-AACCGAGAAGGAGGCCGCGGG-3′; and LUC, 5′-CTTACGCTGAGTACTTCGA-3′. Following infection and a 48-h incubation in nonselective medium, puromycin selection was performed for 4 to 7 days. The expression vectors pQCXIH-hAPE1 (18) and pQCXIH-mAPE1 were generated by inserting their coding sequences into the retroviral vector pQCXIH (Clontech).
TK and HPRT mutation assays.
siRNA-expressing TK6 cells were first treated with CHAT medium (RPMI 1640 medium plus 10% horse serum plus 10 μM deoxycytidine, 200 μM hypoxanthine, 0.2 μM aminopterin, and 17.5 μM thymidine) for 48 h to eliminate preexisting HPRT and TK mutants (27). The cells were then treated with sodium arsenite as described above. After treatment, each culture was centrifuged, washed twice with 10 ml PBS, and resuspended in fresh medium. Cultures were grown in nonselective medium to allow phenotypic expression prior to plating for determination of the mutant fraction. Expression times were 3 days for TK and 6 days for HPRT. After the expression time, the cells were seeded into 96-well microtiter plates in the presence of the appropriate selective agent. TK6 cells (50,000 cells/well) were plated in 2.0 μg/ml trifluorothymidine to isolate TK mutants. For HPRT mutants, TK6 cells (50,000 cells/well) were plated in 0.5 μg/ml 6-thioguanine. Cells from each culture were also plated in 20 wells at 0.3 cell/well in the absence of the selective agent, and five wells with growing cells were used to determine the average plating efficiency. All plates were incubated at 37°C and 5% CO2 in a humidified incubator for 11 days and then scored for growth in the wells. Plates containing trifluorothymidine were then resupplemented with fresh selecting medium and incubated for an additional 10 days to score for the appearance of slow-growing TK mutants. Mutation frequencies were calculated as described previously (8).
Cell survival assay.
Cells were plated at a density of 5 × 105 cells per 60-mm dish. The following day, they were exposed to sodium arsenite as described above. Triplicate plates were used for each dose. The treated cells were washed with PBS, and fresh medium was added, followed by 5 or 6 days of incubation at 37°C in a 5% CO2 incubator. For TK6 cells, dishes were incubated for 6 days at 37°C in a 5% CO2 incubator. The cells were trypsinized, stained with trypan blue (Sigma Chemical Co., St. Louis, MO), and counted using a microscope. The results are expressed as the number of cells in the arsenite-treated plates relative to the number in control plates. For 10T½ cells, 5,000 cells following arsenite treatment were reseeded in 100-mm-diameter petri dishes for colony formation. Cultures were incubated for 12 days, at which time they were fixed with formaldehyde and stained with Giemsa and the number of colonies was counted. In addition, a parallel viability assay using the Cell Counting kit 8 (Dojindo Laboratories, Gaithersburg, MD) was performed for both 10T½ and TK6 cells, following the manufacturer's instructions.
Apoptosis assays.
Controls and sodium arsenite-treated cells (about 105 cells) were harvested and then immediately stained with annexin V-fluorescein isothiocyanate (FITC) and propidium iodide and assayed by flow cytometry following the protocol of the manufacturer (ApoAlert kits; BD Bioscience). Flow cytometric analysis was performed to monitor the green fluorescence of FITC-conjugated annexin V (530 nm) and the red fluorescence of DNA-bound propidium iodide (630 nm) by using a Beckman-Coulter Elite flow cytometer (Harvard-NIEHS Environmental Health Sciences Center, Harvard School of Public Health), and the data were analyzed with Cell Quest software (BD Bioscience).
Abasic sites and ARP assay.
Cellular genomic DNA was isolated from 2 × 106 cells using an anion-exchange resin-based kits (Qiagen Blood & Cell Culture DNA). Samples of 1 μg DNA were subjected to an AP site quantification assay using an aldehyde-reactive probe (ARP) kit (Kamiya Biomedical Company, Seattle, WA) according to the manufacturer's instructions. Data are expressed as the number of AP sites per 105 nucleotides, as determined against a standard supplied by the manufacturer. In some experiments, to control for the artificial formation of AP sites during the sample preparation of DNA extraction (10 min at 37°C), the cells were treated with methoxyamine (Sigma Chemical Co., St. Louis, MO). Then, the methoxyamine-treated cells were washed three times with PBS at 37°C, followed by DNA extraction and ARP assay. The number of cellular AP sites was then calculated based on the difference between methoxyamine-treated and untreated samples (1).
Statistical analysis.
Statistical significance between means was determined by using the standard t test. Data were graphed showing the standard deviations.
RESULTS
Arsenite stress induces APE1 expression.
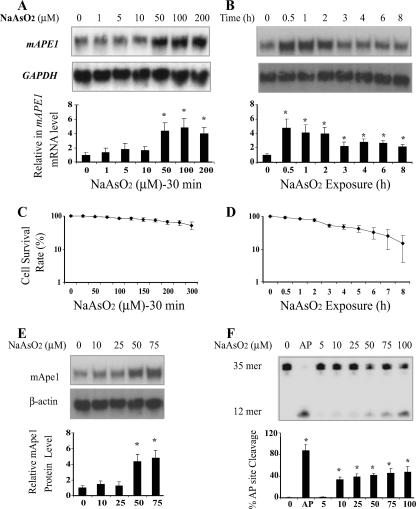
Previous studies indicated that APE1 is transcriptionally induced in response to the oxidants hydrogen peroxide (H2O2) and sodium hypochlorite (20, 51) or the chemical carcinogen asbestos (19). We tested whether such a response could be elicited by sodium arsenite, which has been connected to oxidative stress (6, 39). Different arsenite exposure conditions (varying the time and dose) have been tested. We initially used a low to moderate concentration of sodium arsenite (10 to 10,000 nM) and 1 to 3 days treatment time. The cells either failed to induce APE1 or exhibited some cytotoxic effects. A similar result was also reported by Hu et al. (29) using human fibroblasts, in which 100 to 5,000 nM sodium arsenite treatment for 24 h did not change the Ape1 protein level. Experiments involving shorter exposures were then conducted in growing and nongrowing (fully confluent) cells. Mouse fibroblast 10T½ cells were treated with increasing concentrations of sodium arsenite for 30 min, followed by incubation in fresh arsenite-free medium for 2 hours. Total RNA was then isolated for Northern blot analysis, with the intensity of the mAPE1 mRNA signals normalized to those for GAPDH mRNA in the same samples. While actively proliferating cells did not exhibit induction, in confluent 10T½ cells, the steady-state level of mAPE1 mRNA increased in a dose-dependent manner with exposures from 5 to 100 μM arsenite (Fig. 1A). The 30-min treatments showed minimal cytotoxicity below ∼100 μM arsenite (Fig. 1C), but longer exposures (1 to 8 h) increased cell killing (Fig. 1D). Using 50 μM arsenite, which gives maximal mAPE1 induction with only moderate toxic effects, a kinetic study showed that mAPE1 induction could be sustained for up to 8 h, after a maximum was reached at 0.5 h (Fig. 1B). The decreased mAPE1 expression over time coincided with increasing cytotoxicity (Fig. 1B and D). A similar but smaller-scale mAPE1 induction was also observed in arsenite-treated mouse NIH 3T3 fibroblasts (data not shown).
FIG. 1.
Expression of mAPE1 in response to sodium arsenite in 10T½ cells. (A) Dose-response experiments. Confluent cells were treated for 30 min in medium containing the indicated concentration of arsenite and then washed and incubated in fresh medium for 2 h before harvesting for Northern blotting analysis. (B) Kinetics. Confluent cells were treated with 50 μM arsenite for the indicated times, then washed, and incubated in fresh medium for 2 h before harvesting for Northern blotting analysis. (C and D) Cellular viability under conditions corresponding to the experiments shown in panels A and B, respectively. (E) Four hours after arsenite treatment, Ape1 protein levels were detected by immunoblotting. (F) Six hours after arsenite treatment, the AP endonuclease assay of Ape1 activity was performed. The 35-mer band is the substrate, and the 12-mer band is the AP endonuclease cleavage product. AP, purified hApe1 as a positive control. Three independent sets of assays were performed and normalized to GAPDH mRNA for Northern blotting or to β-actin for immunoblotting. Standard deviations are indicated by error bars. Values that were significantly different (P < 0.05) from the value for the untreated control are indicated by an asterisk.
Ape1 protein expression in response to arsenite was examined by immunoblotting in 10T½ cells. As shown in Fig. 1E, after normalizing against the β-actin protein signal, the mApe1 protein level exhibited a degree of induction and a dose response similar to that for the mAPE1 mRNA. Maximal induction of the protein was reached 4 to 6 h after a 50 μM arsenite treatment (see Fig. S1 in the supplemental material). The induced protein was active as confirmed by an AP site cleavage assay that demonstrated a peak of 5.6-fold increase in the activity 6 h after treatment with 100 μM sodium arsenite (Fig. 1F).
Arsenite activation of the APE1 promoter.
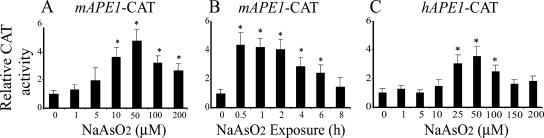
The preceding results suggested that mAPE1 induction in response to arsenite in this system is primarily at the mRNA level. To analyze whether mAPE1 mRNA induction by arsenite is regulated transcriptionally, we transiently transfected 10T½ cells with a CAT reporter gene driven by the mAPE1 promoter (spanning sequences from −1534 to +295 bp) (24). After transient transfection for 24 h, the cells were treated with various concentrations of sodium arsenite for 30 min and washed, and 6 h later, the cells were assayed for CAT activity. The arsenite concentration-dependent profile showed a significant increase starting at 10 μM sodium arsenite, with maximum expression at 50 μM (Fig. 2A). A kinetic analysis (Fig. 2B) demonstrated a pattern similar to that in the Northern blotting data (Fig. 1B), with a maximum at 0.5 h of exposure (Fig. 2B). A nearly identical result was also found for a human APE1 promoter-CAT vector transfected into 10T½ cells (Fig. 2C) or the human p53+ osteosarcoma cell line U2OS (see Fig. S2 in the supplemental material) (16). On the other hand, no change was observed in the mAPE1 mRNA decay rate over 24 h following an inducing treatment with sodium arsenite (data not shown). Thus, arsenite-induced mAPE1 expression results from promoter activation with little or no posttranscriptional regulation.
FIG. 2.
mAPE1 promoter activity in response to arsenite exposure. 10T½ cells were plated in 60-mm dishes, transfected with a DNA mixture consisting of (per plate) 2 μg of mAPE1 promoter-CAT reporter (1.8 kb of the promoter region of the mouse mAPE1 gene fused to the CAT reporter gene), and 2 μg of pCMV-β-gal as a control. After 36 h, the confluent cells were treated either with the indicated concentrations of sodium arsenite for 30 min (A) or with 50 μM sodium arsenite for the indicated times (B). Following the arsenite treatment, the medium was replaced and the cells were incubated a further 6 h before harvesting for assay of CAT and β-galactosidase activities. (C) Arsenite activation of the hAPE1 promoter. Following the procedures described above for panels A and B, 10T½ cells were transfected with pCB9 containing a 1.9-kb fragment of the hAPE1 promoter (17, 24, 25) and treated with sodium arsenite. Background CAT activity (from mock-transfected cells) was subtracted from each experimental measurement, and the resulting value was corrected for variation in transfection efficiency by normalization with background-subtracted β-galactosidase activity. Normalized CAT activity in the absence of arsenite of pCAT-Basic was arbitrarily assigned a value of 1, and each data point represents the mean plus standard deviation (error bar) from three independent experiments. Values that were significantly different (P < 0.05) from the values for the untreated control are indicated by an asterisk. The CAT reporter used is indicated at the top of each panel.
Identification of arsenite response elements in the mAPE1 promoter.
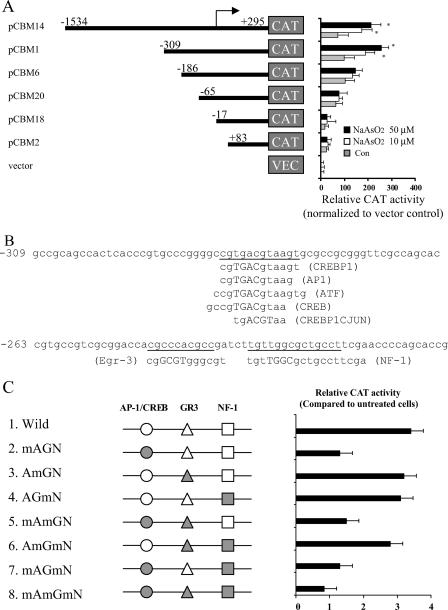
To identify the transcription factors involved in regulating the mAPE1 gene in response to arsenite, a detailed promoter-reporter analysis was performed. A set of progressive 5′ deletions within the mAPE1 promoter fused to the CAT reporter gene (24) was used to follow the arsenite response in transient-transfection assays. The transfected cells were treated with sodium arsenite (10 or 50 μM) for 30 min, followed by a change to fresh medium for 6 h before harvesting for assays. As shown in Fig. 3A, the full-length (1.8-kb) mAPE1 promoter and a 604-bp fragment (bp −309 to bp +295) conferred two- to threefold-increased reporter expression in response to arsenite. However, fragments retaining only 186 bp or less of upstream sequence lost most or all of the promoter response to arsenite. Thus, there seems to be an essential element(s) in the region from bp −309 to −186 of the mAPE1 promoter. As analyzed using the transcription factor binding site search software TFSEARCH (http://molsun1.cbrc.aist.go.jp/research/db/TFSEARCH.html), the 124-bp mAPE1 promoter region encompassed by bp −309 to −186 contains three potential binding sequences for major transcription factors: AP-1/CREB, early growth response 3, and nuclear factor 1 (Fig. 3B). The sequence from bp −280 to −269, CGTGACGTAAGT, actually overlaps with possible AP-1, CREB, and ATF binding sites. To identify possible transcription factors involved in regulating the mAPE1 response to arsenite, site-directed mutagenesis targeting three binding sites in the pCMB14 vector (Fig. 3A) was carried out (the mutated sequences are shown in Table 1). Transient-transfection assays showed that mutation of the AP-1/CREB site nearly eliminated mAPE1 transcriptional induction by arsenite, while mutations in the other transcription factor sequences showed little effect (Fig. 3C). These results indicate that the interaction of the AP-1/CREB sequence in mAPE1 and its binding proteins could play key roles in mAPE1 regulation under arsenite stress. The AP-1 transcription factor is a dimeric complex comprised of members of the Jun, Fos, ATF, and musculoaponeurotic fibrosarcoma protein families (12). The AP-1 complex can therefore include many different combinations of heterodimers and homodimers involving c-Jun, JunB, JunD, c-Fos, FosB, Fra-1, Fra-2, ATF1, ATF2, ATF3, and ATF4.
FIG. 3.
Identification of the mAPE1 promoter region responsible for arsenite-mediated gene induction. (A) mAPE1 promoter-CAT constructs. Plasmid pCBM contains approximately 1.8 kb of the promoter region of the mouse mAPE1 gene fused to the CAT reporter gene. Derivatives of pCBM14, generated by successive 5′ deletions of the promoter start site (pCBM1, pCBM6, pCBM20, pCBM18, and pCBM2), are diagrammed. The cells were treated with 0, 10, and 50 μM sodium arsenite for 30 min. The promoter-reporter assay was carried out as described in the legend to Fig. 2. The change (n-fold) in normalized CAT activity (relative to that in empty vector-treated cells) is presented as the mean, and the error bars correspond to standard deviations. Values that are significantly different (P < 0.05) from the value for untreated cells are indicated by an asterisk. Con, control. (B) Consensus transcription factor binding sites in the mAPE1 promoter region (−309 to −186 bp). CREBP1, cyclic AMP response element-binding protein 1. (C) Schematic diagram of the mapped 123-bp fragment. Experimentally identified or putative transcription factor binding sites are indicated on the line: AP-1/CREB, activator protein 1/cAMP response element-binding protein (circles); GR-3, early growth response 3 (triangles); NF-1, nuclear factor 1 (squares). The DNA sequence was altered to generate the eight mutated promoters shown in gray. The mutated promoters were located in an mAPE1-CAT reporter vector (Table 1). The mutated binding sites are indicated by mA (mutated AP-1/CREB site), mG (mutated GR3 site), and mN (mutated NF-1 site), while wild-type binding sites are indicated by A (AP-1/CREB site), G (GR3 site), and N (NF-1 site).
TABLE 1.
mAPE1 oligonucleotides used for mapping the 123-bp promoter fragment
| Oligonucleotidea | DNA sequenceb |
|---|---|
| AP-1/CREB-w | ACCCGTGCCCGGGGCCGTGACGTAAGTGCGCCGCG |
| AP-1/CREB-m | ACCCGTGCCAGGGATAGTGGTGTGAGTGCGCCGCG |
| Egr-3-w | GCCGTCGCGGACCACGCCCACGCCGATCTTGTTGG |
| Egr-3-m | GCCGTCGCGGACCACTAACACTCCGATCTTGTTGG |
| NF-1-w | ACGCCGATCTTGTTGGCGCTGCCTTCGAACCCCAG |
| NF-1-m | ACGCCGATCTTGTGATAGCTGCCTTCGAACCCCAG |
Wild-type oligonucleotide sequences are indicated by a w at the end of the oligonucleotide name, and derivatives with mutated binding sites are indicated by an m at the end of the oligonucleotide name.
The mutated binding sites are underlined.
mAPE1 promoter-binding activity in cells.
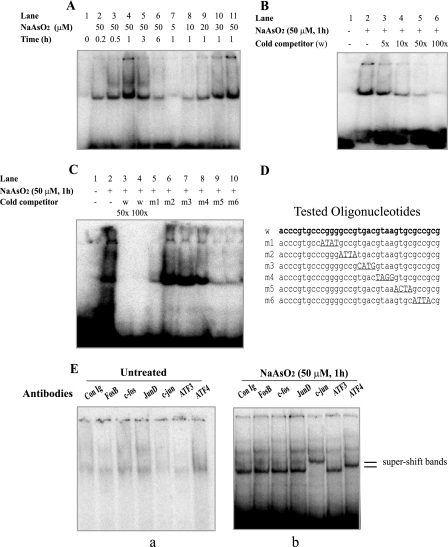
To identify DNA-binding proteins responsible for arsenite-mediated mAPE1 gene induction, EMSA reactions were carried out using whole-cell extracts and a 35-bp mAPE1 promoter fragment containing the AP-1/CREB site. A single, strong DNA-protein complex band was detected in extracts of arsenite-treated 10T½ cells (Fig. 4A). The binding activity was dose and time dependent, with an apparent maximum at 1 h of treatment with 50 μM sodium arsenite. The specificity of the complexes was tested using increasing excess amounts of cold wild-type 35-mer duplex DNA competitor, which gradually diminished the band intensity (Fig. 4B). Moreover, to map the important base pairs for binding, six altered oligonucleotide sequences m1 to m6 (Fig. 4D) were synthesized and tested in competition experiments. Sequences m1, m5, and m6 were nearly as effective as the wild-type oligonucleotide competitor (Fig. 4C, compare lanes 3 and 4 to lanes 5, 9, and 10). In contrast, m2, m3, and m4 oligonucleotides were far less effective, which suggested that the GCCGTGACGTAA sequence contains sites necessary for binding the regulatory proteins.
FIG. 4.
Identification of arsenite-activated DNA-binding activity using an EMSA. (A) Arsenite treatment of 10T½ cells activates in vitro protein binding for the 123-bp APE1 promoter fragment. The cells were treated without arsenite or with the indicated arsenite concentration for 30 min, after which the cells were washed and incubated in fresh medium for the time shown. Preparation of cell extracts and EMSA reactions were carried out as described in Materials and Methods. (B) Competition with excess (5- to 100-fold) concentrations of unlabeled wild-type DNA sequence (w). The presence (+) or absence (−) of sodium arsenite or cold competitor is shown. (C) Competition with oligonucleotides containing altered factor binding sites. Competitor oligonucleotides were used at a 50-fold (wild type [w] only) or 100-fold molar excess. Following electrophoresis, the gels were autoradiographed for 16 h. (D) Competitor oligonucleotide sequences. The altered sites are underlined. (E) EMSA supershift only by ATF4 or c-Jun antibodies. Extracts prepared from 10T½ cells treated for 30 min with or without 50 μM sodium arsenite were used in EMSA reactions. Supershift analysis using rabbit immunoglobulin G control (Con Ig) or antibodies directed against the indicated transcription factors was carried out as described in Materials and Methods. The gels were autoradiographed for 16 to 48 h. Only the top portion of the autoradiograms is shown. Separate panels are shown for control (a) and arsenite-treated cells (b).
To further identify specific binding proteins for the mAPE1 promoter as a potential arsenite-responsive transcription factor(s), “supershift” reactions were performed using commercially available antibodies. The antibodies against FosB, c-Fos, JunB, JunD, Fra-1, Fra-2, CREB-1, ATF1, ATF2, and ATF3 had no detectable effect (Fig. 4E and data not shown). However, antibodies against c-Jun and ATF4 clearly shifted the arsenite-induced complexes (Fig. 4E). These transcription factors are therefore likely to be binding the AP-1/CREB sequence GCCGTGACGTAA, (−282 bp to −271 bp in the mAPE1 promoter) to help mediate the response to arsenite.
Knockdown of c-Jun or ATF4 suppresses the mAPE1 transcriptional response to arsenite.
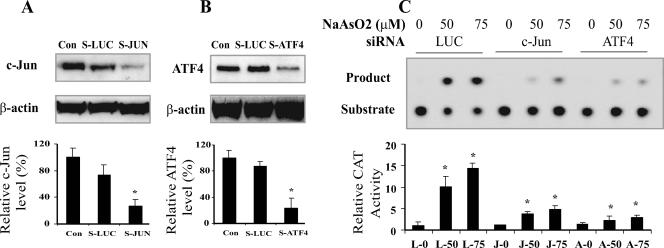
To test whether arsenite-induced mAPE1 gene expression is dependent on c-Jun and ATF4, we suppressed c-Jun or ATF4 in 10T½ cells by expressing specific siRNAs via a retroviral vector (7). Immunoblotting (Fig. 5A and B) demonstrated that the endogenous c-Jun and ATF4 protein levels were significantly down-regulated (by 83% and 76%, respectively) in 10T½ cells infected with the specific siRNA vectors, while the control siRNA against luciferase (sLUC) had little effect. These siRNA-expressing cells were subjected to the full-length mAPE1 promoter-CAT reporter assay as described above for Fig. 2. Arsenite activation of the mAPE1 promoter was suppressed in the 10T½ cells deficient in either c-Jun (64% reduced expression) or ATF4 (76% reduced), whereas the control sLUC had no effect on the activation of the mAPE1 promoter (Fig. 5C). Thus, mAPE1 promoter activation by arsenite in 10T½ cells depends on both c-Jun and ATF4.
FIG. 5.
ATF4-deficient or c-Jun-deficient 10T½ cells are defective for arsenite-induced mAPE1 promoter activation. (A and B) Down-regulation of c-Jun (A) and ATF4 (B) proteins by siRNA (indicated by S-JUN and S-ATF4, respectively, with S-LUC as the control). Cells were infected with retrovirus producing the indicated siRNA, and puromycin-resistant cells were expanded (for 7 or 8 days for S-ATF4 and 10 to 12 days for S-JUN) for treatment and analysis. Cell-free lysates were generated for protein analysis by immunoblotting. Con, control with no siRNA treatment. (C) Defective mAPE1 response to sodium arsenite in c-Jun-deficient or ATF4-deficient cells. Following treatment as shown in panels A and B to suppress c-Jun or ATF4 expression, cells were treated with the indicated concentration of sodium arsenite for 30 min, followed by a 6-h incubation in arsenite-free medium before harvesting and assay. CAT reporter assays were carried out as described in the legend to Fig. 2. The data shown are based on at least three independent experiments. Standard deviations are indicated by error bars. Values that were significantly different (P < 0.0) from the value for the control are indicated by an asterisk. The designations below the bars indicate the siRNA and arsenite treatments (e.g., L-0 corresponds to LUC siRNA and no arsenite, J-50 corresponds to c-JUN siRNA and 50 μM arsenite treatment, and A-0 corresponds to ATF4 siRNA and no arsenite).
mAPE1 induction via an oxidative mechanism.
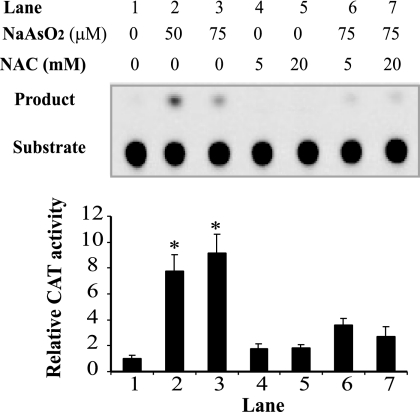
Arsenite frequently damages cells via an oxidative mechanism (6, 39, 55). Several previous studies also demonstrated that oxidative stress, such as exposure to H2O2, can directly activate mAPE1 transcription (20, 51). To determine whether oxidative stress is an underlying mechanism in arsenite-induced mAPE1 induction, 10T½ cells were first pretreated with the antioxidant NAC for 1 hour and then exposed (in the continued presence of NAC) to 50 to 75 μM sodium arsenite. NAC at a concentration of 20 mM efficiently suppressed arsenite-stimulated mAPE1 promoter activity in a full-length mAPE1 promoter-CAT reporter assay (Fig. 6). These results show that arsenite-mediated transcriptional activation of mAPE1 occurs via an oxidative mechanism.
FIG. 6.
Arsenite activation of the mAPE1 promoter via an oxidative mechanism. Confluent 10T½ cells cultured in 10-cm dishes were pretreated with freshly prepared 5 or 20 mM N-acetyl-l-cysteine (NAC) for 1 h. Following the initial incubation, 50 to 75 μM sodium arsenite was added and incubation continued for 30 min. The cells were washed twice and incubated in fresh medium for 6 h and then collected for CAT assays. The top panel shows a representative autoradiogram of one set of CAT assays; three independent experiments were run to generate the data for quantification shown in the lower panel. Standard deviations are indicated by error bars. Values that were significantly different (P < 0.05) from the value for the untreated control are indicated by an asterisk.
ATF4-deficient cells.
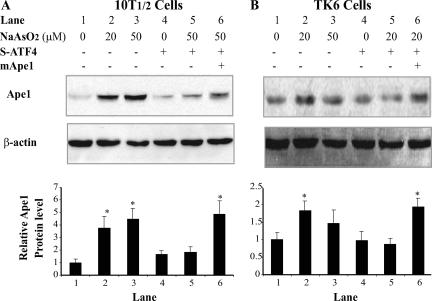
The foregoing results indicate that the induction of mAPE1 by arsenite is dependent on an ATF4/c-Jun heterodimer. Thus, inhibiting ATF4 or c-Jun should disrupt arsenite-induced mAPE1 expression. ATF4 is an important transcription factor in response to a variety of cell stresses (58). It is likely that mAPE1 is not the only gene regulated by ATF4 in response to arsenite, but the information collected about ATF4 so far has mostly focused on endoplasmic reticulum stress, and little is known about its role in DNA repair and antimutagenesis. To address the possible role of ATF4-dependent mAPE1 induction in resistance to arsenite-induced genotoxicity, an ATF4-deficient cell model was developed using RNA interference. In addition to 10T½ cells, the human lymphoblast cell line TK6 was used because of its utility for TK and HPRT mutation assay and apoptosis assay employing annexin V-propidium iodide double staining. Previous studies showed that ATF4 deficiency could affect cell growth (23); to circumvent this problem, the cell culture was supplemented with nonessential amino acids and the reducing agent β-mercaptoethanol. Under these conditions, no growth defect was observed in either type of ATF4-deficient cell (data not shown). Immunoblotting analysis of mApe1 in 10T½ cells showed that suppression of ATF4 did not affect the basal level of mApe1 expression (Fig. 7). However, ATF4 deficiency in both cell lines strongly diminished the induction of Ape1 in response to arsenite (Fig. 7A). As seen for 10T½ cells, Ape1 induction after sodium arsenite treatment was also observed in TK6 cells. Although the extent of increase of Ape1 was lower in TK6 (less than twofold), expression of siRNA against ATF4 completely blunted the hApe1 induction (Fig. 7B), which indicates that human TK6 cells shared this transcriptional regulation mechanism with murine 10T½ cells.
FIG. 7.
Changes of Ape1 levels in ATF4-deficient cells. The cells infected with a retrovirus expressing siRNA against ATF4 (S-ATF4) or with an mApe1 retroviral expression vector were cultured to confluence under the appropriate selection conditions (puromycin for the siRNA vectors and hygromycin for the mApe1 expression vector). The cells were then treated with 20 to 50 μM sodium arsenite for 30 min. The cells were washed twice, and the incubation in fresh medium continued for 4 h. The Ape1 protein levels were determined by immunoblotting and phosphorimaging and normalized to the β-actin signal. (A) 10T½ cells. (B) TK6 cells. The top panel in each frame shows a representative immunoblot, and the graph shows the quantification from three independent experiments. Standard deviations are indicated by error bars. Values that were significantly different (P < 0.05) from the value for the control are indicated by an asterisk.
Ape1 induction and cellular resistance to arsenite.
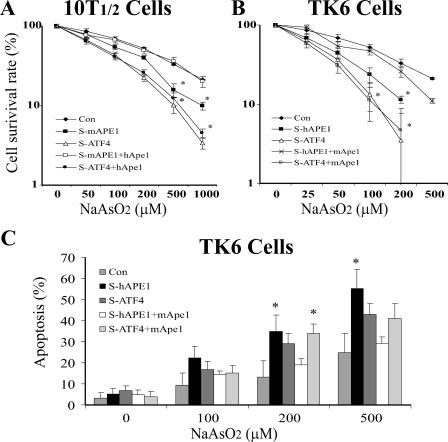
The toxicity of arsenite exposure could involve DNA damage as a significant component. Therefore, we first examined the role of mApe1 in arsenite-induced cell killing in 10T½ and TK6 cells. The cells were infected with the indicated retroviral siRNA vector, incubated for 48 h, then selected in puromycin for an additional 5 to 7 days, and finally treated with increasing amounts of sodium arsenite for 30 min. The arsenite-induced cytotoxicity was determined by the cell survival assay. This experiment included a group of Ape1-deficient cells infected with a specific siRNA retroviral vector. The infection resulted in a partial (60 to 70%) reduction of cellular Ape1 but did not affect cell spontaneous death or cell proliferation (data not shown). In 10T½ cells, the lowest survival rate upon arsenite challenge was in ATF4-deficient cells, and to a lesser extent, in mApe1-deficient cells. To determine whether arsenite toxicity was affected by Ape1, we conducted a rescue experiment by expression of hApe1 in ATF4-depleted or mApe1-depleted 10T½ cells. The hAPE1 mRNA is immune to siRNA directed against mAPE1 because of six mismatched bases within the target sequence (data not shown). The expression of hApe1 (to a level comparable to that induced by arsenite in ATF4-proficient cells) efficiently reduced the apoptosis to control levels in mApe1-depleted 10T½ cells but had little effect in the ATF4-depleted counterpart (Fig. 8A). A similar result was observed in TK6 cells (Fig. 8B), despite lower Ape1 induction by arsenite in this type of cell. Although the cell death endpoint is complex and the comparison involves two different cell types, this result may suggest that the role of Ape1 against arsenite toxicity may involve mechanisms other than protein induction, such as posttranslational modification or intracellular localization (4, 14, 28).
FIG. 8.
Arsenite-induced apoptosis and cytotoxicity in ATF4-deficient cells. Cells (107) preinfected with siRNA to LUC (control [Con]), mAPE1(S-mAPE1), hAPE1 (S-hAPE1), ATF4 (S-ATF4), or hApe1 or mApe1 retroviral expression vectors (hApe1 and mApe1, respectively) were challenged with the indicated concentration of sodium arsenite for 30 min. The cells were washed twice, and the incubation was continued in fresh medium for 4 h. 10T½ (A) or TK6 (B) cells were then reseeded into 96-well plates (103 cells per well) and incubated with fresh medium for 96 h. The cell number was determined by using Cell Counting kit 8. The results were confirmed via a parallel traditional colony-forming assay for 10T½ cells or trypan blue exclusion for TK6 cells (data not shown). (C) Apoptosis determined by the annexin V-FITC-propidium iodide assay using flow cytometry. Arsenite-challenged cells were incubated for 24 h in fresh medium before sampling for the apoptosis assay. The data were quantified from three independent experiments. Standard deviations are indicated by error bars. Values that were significantly different (P < 0.0) from the value for the control are indicated by an asterisk.
Because arsenite-induced cell killing often has an apoptotic component, apoptosis was also examined using an annexin V-propidium iodide double staining assay. Since the arsenite sensitivity of TK6 cells was somewhat higher than that of 10T½ cells, the apoptosis assay was performed in TK6 cells. As shown in Fig. 8C, apoptosis was observed in an arsenite dose-dependent manner. Notably, in each arsenite dose group, the severity of apoptosis induction was higher in hAPE1-depleted TK6 cells than in ATF4-depleted TK6 cells. This result diverges somewhat from the results of the cell survival assay, where arsenite-induced cell killing was higher under ATF4 deficiency than under hAPE1 deficiency (Fig. 8B). As seen in the cell survival assays, expressing mApe1 (resistant to suppression by the hAPE1-specific siRNA) successfully rescued cells with hAPE1 deficiency but not those with hATF4 deficiency from arsenite-induced apoptosis.
Ape1 induction and DNA damage.
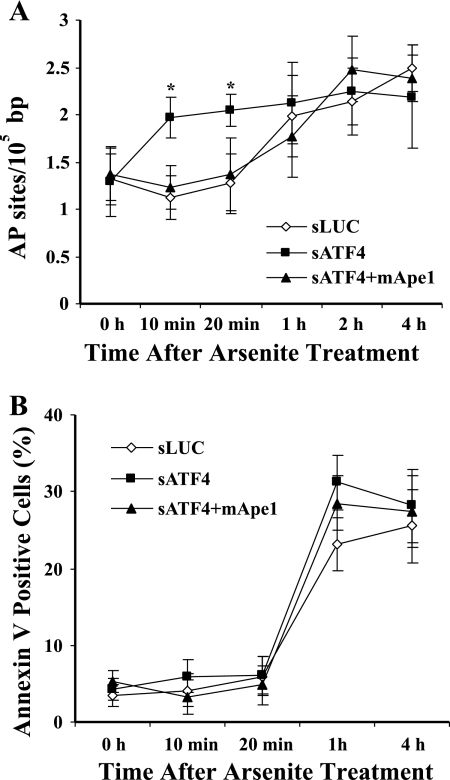
Ape1 is important in the repair of AP sites (10, 18). The arsenite hypersensitivity of cells deficient in Ape1 or ATF4 suggested that they might accumulate unrepaired AP sites. Therefore, we measured abasic sites in cellular DNA. As shown in Fig. 9A, the basal level of AP sites was unchanged as a function of the ATF4 status. Shortly (10 to 20 min) after arsenite treatment, an increase in the level of AP sites in ATF4-depleted 10T½ cells was observed. However, 1 h later, the AP sites in the control cells had rapidly climbed to the same level as in the ATF4-depleted cells, and no further increase was seen for the latter. The number of AP sites from all groups then stayed at the elevated level through 4 h after arsenite exposure. Expression of mApe1 in ATF4-depleted 10T½ cells efficiently suppressed the AP site level during the first 10 to 20 min after arsenite exposure, suggesting that Ape1 is responsible for the initial changes. Since apoptosis, especially in the later phase, involves DNA fragmentation, cell disintegration, and necrosis-like effects, which often release reactive oxygen species (68), it seemed possible that such effects might add to the signals detected in the AP site assay. We therefore measured early apoptotic changes using an annexin V-propidium iodide double staining assay in arsenite-treated 10T½ cells transfected with siRNA against LUC (control) or ATF4 or with pQHIX-mApe1 (Fig. 9B). A significant level of annexin V-positive cells could be detected 1 h after arsenite treatment, and it stayed at this level for the next 3 h. Although the activation of apoptosis might account for some of the abasic damage observed at later times, that appears not to be the case during the first 10 to 20 min after arsenite exposure. Instead, it appears that Ape1 induction dependent on ATF4 contributes importantly to initiate BER of arsenite-induced damage to cellular DNA. Nevertheless, the early (≤4-h) apoptosis found in this experiment contrasts with the later (24-h) apoptosis we observed in Fig. 8. These differences may reflect cell cycle distribution or involve different mechanisms, although Ape1 seems to have no effect on early apoptosis (Fig. 9B).
FIG. 9.
Abasic DNA damage and apoptosis in ATF4-deficient cells. The 10T½ cells preinfected with vectors expressing siRNA against luciferase (sLUC) or ATF4 (sATF4) or with sATF4 and an mApe1 retroviral expression vector (sATF4+mApe1) were cultured to confluence. The cells were then treated with 50 μM sodium arsenite for 30 min. The cells were washed twice and incubated in fresh medium for the indicated times. The cells were subjected to DNA extraction to determine abasic damage using an aldehyde-reactive probe assay (A) or collected to determine apoptosis by annexin V-FITC-propidium iodide staining and flow cytometry (B). The data were quantified from three independent experiments. Standard deviations are indicated by error bars. Values that were significantly different (P < 0.05) from the value for the sLUC control are indicated by an asterisk.
Ape1 in arsenite-induced mutations.
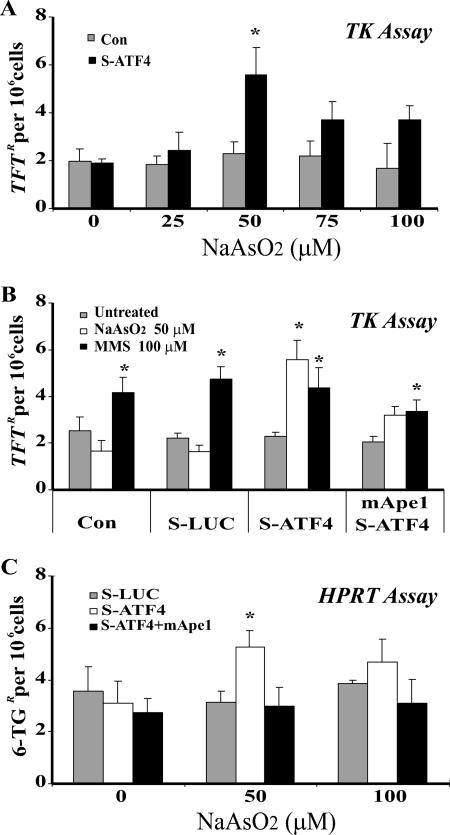
Since AP sites can be highly mutagenic (2, 22), accumulation of AP sites in ATF4-deficient cells could increase the mutation rate. We therefore examined arsenite-induced mutagenesis in TK6 cells. Mutation in TK or HPRT genes can be positively selected using trifluorothymidine or 6-thioguanine, respectively, as each is present in TK6 cells in the haploid state (44, 46, 74). The TK mutation frequency (MF) increased 2.4-fold (P = 0.007) in ATF4-deficient cells exposed to up to 50 μM arsenite (Fig. 10A). The decrease in MF at higher arsenite challenges coincided with increasing arsenite-induced cell killing (Fig. 8B). The MF following arsenite treatment was unchanged in cells with the control siRNA (sLUC) or no siRNA (Fig. 10B). Thus, arsenite mutagenesis is specific for ATF4-deficient cells. When mApe1 was expressed in ATF4-depleted cells, the mutagenesis was suppressed (Fig. 10B); evidently, Ape1 plays a key role in the ATF4-dependent antimutation response. The HPRT assay showed a similar result (Fig. 10C), further supporting this conclusion. These data, taken together, indicate that arsenite at modest levels does possess a mutagenic potential, which is offset by an antimutation response. This cellular response includes an ATF4-mediated pathway in which Ape1 is a key component.
FIG. 10.
Arsenite-induced mutation in the TK and HPRT genes in ATF4-deficient TK6 cells. TK6 cells were infected with vectors as described in the legend to Fig. 9 and treated with sodium arsenite at the indicated doses for 30 min. The medium was then replaced, and the cells were subjected to mutation assays as described in Materials and Methods. (A) TK mutations as a function of the arsenite dose. TFTR, trifluorothymidine-resistant; Con, control, S-ATF4, siRNA from cells expressing siRNA against ATF4. (B) TK mutations after a 50 μM arsenite challenge with or without reexpression of mApe1. Methyl methane sulfonate (MMS) treatment (24 h) was included as a control not involving APE1 induction or an oxidative mechanism. (C) Effects of ATF4 and mApe1 on arsenite-induced HPRT mutations. 6TGR, 6-thioguanine-resistant. The data were quantified from three independent experiments. Standard deviations are indicated by error bars. Values that were significantly different (P < 0.05) from the value for the control are indicated by an asterisk.
DISCUSSION
The carcinogenicity of arsenic in humans is well established, but the mechanisms of arsenic carcinogenesis remain elusive. Typically, chemically induced cancer development involves a gene mutagenesis step (47), but many attempts have failed to detect mutagenesis by the important arsenic form arsenite. For instance, sodium arsenite was barely mutagenic (at 10 μM) in the L5178Y/TK+/− mouse lymphoma assay (43) or (at 15 μM) in the TK assay in Chinese hamster ovary G12 cells (37). There is, however, accumulating evidence for DNA damage in arsenite-exposed cells. An improved comet assay using Fpg glycosylase as a probe provided new evidence that arsenite levels equivalent to those associated with clinical pathology (1 to 500 nM) were indeed able to cause rapid (1- to 6-h) formation of DNA lesions, such as 8-oxoguanine and AP sites (55, 59, 69). For arsenic mutagenesis and tumorigenesis, Mure et al. (45) showed that human osteosarcoma TE85 cells cultured with 100 nM sodium arsenite for 20 generations (3 weeks) increased the mutation frequency at the HPRT locus three- to fivefold, and after 30 generations (5 weeks), phenotypically transformed cells could be detected. Mutations were induced by arsenite but not by methyl arsenic derivates (45). These studies provide important evidence that genotoxicity and mutagenesis can play a role in arsenic carcinogenesis. However, the long exposures used in the Mure et al. work suggests possible indirect effects to account for mutagenesis in those studies. Note that a large number of previous studies using shorter treatments with 1 nM to 100 μM sodium arsenite failed to show mutagenesis (36, 38, 54, 55). Some explanation emerges from our current studies. We have presented evidence that arsenite rapidly activates a cellular DNA repair and antimutation response, in which the BER enzyme Ape1 is up-regulated. This induction is controlled by the ATF4 and c-Jun transcription factors. Suppressing ATF4 promotes arsenite-induced mutagenesis, while ectopically expressing Ape1 diminished the mutagenesis. In addition, arsenite mutagenesis in both mouse embryonic fibroblasts and human lymphoblastoid cells was relatively inefficient, with cytotoxic effects dominating at the higher doses. An interesting finding here is that Ape1 induction by arsenite was observed only in confluent 10T½ cells but not in actively proliferating 10T½ cells. In this regard, our previous study (17) showed that Ape1 is induced during S phase in murine fibroblasts, which might mask or supersede the arsenite-mediated activation.
ATF4-mediated resistance to arsenite genotoxicity.
ATF4, also called cyclic AMP response element-binding protein 2, is a member of the large family of ATF/CREB transcription factors. Through a leucine zipper domain, ATF4 is able to form a homodimer or various heterodimers with members of the AP-1 family, such as c-Fos, c-Jun, and JunD. ATF4 interacting with c-Jun to form an ATF4/c-Jun heterodimer has been demonstrated previously (33). ATF4 reacts differently in response to different types of cellular stress. Under endoplasmic reticulum stress, the ATF4 homodimer serves as a downstream component of eIF-2α kinases, regulating cell proliferation through the transcriptional activation of CCAAT/enhancer-binding protein (23). ATF4 by itself appeared to be sufficient for arsenite-induced expression of the vascular endothelial growth factor promoter for neovascularization (56). Here we report an additional function of ATF4, the formation of a heterodimer with c-Jun to activate APE1 in response to arsenite-induced stress. ATF4-deficient cells are impaired in amino acid metabolism and hypersensitive to oxidative stress (23). Our data showed that ATF4 deficiency causes a defect in resistance to arsenite-induced DNA damage and mutagenesis. Although Ape1 is an important component of this response, there are likely also other factors involved. Moreover, other ATF4-independent antimutation pathways may also act to limit arsenite-induced mutagenesis. ATF4 has been reported to account for resistance to another DNA-damaging agent, the cancer treatment drug cisplatin (64), which is consistent with a broader role of ATF4 in responses to genotoxicity.
Several studies (42, 66) show that ATF4 homozygous knockout mice suffered growth defects, anemia, delayed hair growth, and eye abnormalities, including microphthalmia. Tanaka also reported that ATF4−/− mice were born in a sub-Mendelian ratio (66), but this finding was not confirmed in two other studies (15, 42). The phenotypic changes in ATF4−/− mice may reflect a requirement for ATF4 for cellular proliferation in the affected tissues, particularly in the developing lens and hair follicles, as well as possible cell-specific functions. It is possible that some growth defects are linked to deficiency in the ATF4-dependent induction of Ape1, but disrupting both APE1 alleles causes a more severe phenotype: embryonic death at the 5.5-day stage (73). A severe growth defect was also observed in Ape1-null zebra fish, with dysmorphic hearts, pericardial edema, few erythrocytes, small eyes, and abnormal notochords (70). However, some other features of ATF4 mice are apparently Ape1 independent. For example, the death of APE1-null zebra fish was suggested to be due to failure in heart development, which was absent in ATF4-null mice.
Activation of APE1 expression via an oxidative mechanism.
Direct exposure of cells to the oxidant H2O2 can activate the hAPE1 promoter (20, 21). In the APE1 promoter-CAT reporter experiments reported here, the antioxidant NAC blocked arsenite-induced APE1 transcription in 10T½ cells, which indicates that a redox reaction(s) is critical for c-Jun/ATF4-mediated APE1 transcription. Interestingly, a previous study identified a CREB binding site in the human APE1 promoter as necessary for the response of a transgene to H2O2 in Chinese hamster ovary cells (20). Comparison of hAPE1 and mAPE1 (from −1500 bp to 2800 bp) shows high sequence similarity in the coding region (87% identity at the DNA level) but a much lower similarity in the promoter region (<25% identity). However, the promoter contains a small fragment (<120 bp) of high (>85%) sequence identity. This shared segment is located between bp −563 and −445 in the human gene and between bp −295 and −177 of the mouse gene (see Fig. S3 in the supplemental material). The mAPE1 AP-1/CREB binding site (starting at bp −282) is within this conserved segment and matches the previously reported human CREB site (starting at bp −550) (21) with >90% sequence identity. The high conservation of this region in the hAPE1 and mAPE1 promoters highlights its likely importance in oxidative stress-induced transcription activation. In this context, recent evidence shows that the entire BER pathway may be activated by some type of oxidative stress (57).
Ape1 is essential in repair of oxidative DNA damage.
Ape1 was initially recognized as an AP endonuclease enzyme (32). Subsequently, other DNA repair activities, such as 3′ repair diesterase, 3′-phosphatase, and 3′→5′ exonuclease activities, were discovered (62). These activities are critical for repair of oxidative DNA damage. Oxidative DNA damage involves several types of substrates for Ape1 (9, 31, 48): 3′-phosphoglycolates at oxidative strand breaks; oxidized abasic sites, such as deoxyribonolactones; AP sites generated by DNA glycosylases acting on base lesions; and perhaps 3′-deoxyribose (unsaturated) generated by DNA glycosylases with lyase activity. AP sites are also formed by hydrolytic decay of DNA, which is estimated to produce around 10,000 AP sites per day in each mammalian cell (3). AP sites are cytotoxic (35, 50), and as they lack genetic information, mutagenic (2, 22, 40).
The contribution of Ape1 to cellular arsenite resistance is consistent with these proposed DNA repair roles. The induction of this key BER protein under oxidative stress suggests that Ape1 activity may become limiting for efficient DNA repair in some circumstances. In this context, the fact that APE1 induction in response to arsenite was restricted to nondividing cells may relate to the lower basal Ape1 levels found in the G1 phase (17). On the other hand, the failure of Ape1 to restore arsenite resistance in ATF4-deficient cells (Fig. 8) suggests that there are important repair-independent mechanisms for survival.
Ape1 induction and human cancer.
There are multiple reports of Ape1 levels in cancer cells being above the background level. For example, compared to healthy tissues, Ape1 expression was increased in gliomas, in prostate and cervical cancers (5, 13, 34, 53, 75), as well as in chronically inflamed ulcerative colitis (26). Because cancer cells are often accompanied by inflammation and oxidative stress (41), these results are consistent with APE1 induction driven by oxidative stress. In addition, various research groups have reported that Ape1 translocates from the cytoplasm to the nucleus: for example, after exposure to H2O2 in human B lymphocytes (67) and HeLa cells (14). With this dynamic behavior, Ape1 may play an important part in maintaining genetic integrity in the face of environmental carcinogens or the inflammatory effects of chronic infections. However, we note that the initial results from the Cancer Genome Project for the APE1 coding sequence from 41 tumors (most from lung, breast, skin and kidney cancers) failed to detect any somatic mutations (http://www.sanger.ac.uk/perl/genetics/CGP/cosmic?action=gene&ln=APEX1). These results imply that, in most cancer cases, Ape1 functions (including the response to oxidative stress) may be still intact.
In conclusion, arsenite exposure triggers an ATF4-mediated cellular response that efficiently reduces the mutagenic effects of arsenite. Ape1 plays an important role in this response. Deficiencies in the ATF4 response and the induction of Ape1 may contribute to genomic instability and promote certain types of cancer development.
Supplementary Material
Acknowledgments
We are grateful to Vanessa Lopez-Pajares and Jonathan Wing for help with some initial experiments, to John B. Little for cell lines, to Amy Imrich and Lester Kobzik of the Harvard-NIEHS Center for Environmental Health for flow cytometry experiments, and to members of the Demple laboratory for helpful discussions. We appreciate helpful comments on the manuscript from Zhi-Min Yuan and Nurten Saydam.
This study was supported by NIH grant GM40000 to B.D., the Superfund Basic Sciences Research Program at the Harvard School of Public Health, and NIH grant U19-AI067773 (to D. J. Brenner, Columbia University).
Footnotes
Published ahead of print on 15 October 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Atamna, H., I. Cheung, and B. N. Ames. 2000. A method for detecting abasic sites in living cells: age-dependent changes in base excision repair. Proc. Natl. Acad. Sci. USA 97:686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auerbach, P., R. A. Bennett, E. A. Bailey, H. E. Krokan, and B. Demple. 2005. Mutagenic specificity of endogenously generated abasic sites in Saccharomyces cerevisiae chromosomal DNA. Proc. Natl. Acad. Sci. USA 102:17711-17716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes, D. E., and T. Lindahl. 2004. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 38:445-476. [DOI] [PubMed] [Google Scholar]
- 4.Bhakat, K. K., T. Izumi, S. H. Yang, T. K. Hazra, and S. Mitra. 2003. Role of acetylated human AP-endonuclease (APE1/Ref-1) in regulation of the parathyroid hormone gene. EMBO J. 22:6299-6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bobola, M. S., A. Blank, M. S. Berger, B. A. Stevens, and J. R. Silber. 2001. Apurinic/apyrimidinic endonuclease activity is elevated in human adult gliomas. Clin. Cancer Res. 7:3510-3518. [PubMed] [Google Scholar]
- 6.Bower, J. J., S. S. Leonard, F. Chen, and X. Shi. 2006. As(III) transcriptionally activates the gadd45a gene via the formation of H2O2. Free Radic. Biol. Med. 41:285-294. [DOI] [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Danaee, H., H. H. Nelson, H. Liber, J. B. Little, and K. T. Kelsey. 2004. Low dose exposure to sodium arsenite synergistically interacts with UV radiation to induce mutations and alter DNA repair in human cells. Mutagenesis 19:143-148. [DOI] [PubMed] [Google Scholar]
- 9.Demple, B., J. Halbrook, and S. Linn. 1983. Escherichia coli xth mutants are hypersensitive to hydrogen peroxide. J. Bacteriol. 153:1079-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demple, B., and L. Harrison. 1994. Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem. 63:915-948. [DOI] [PubMed] [Google Scholar]
- 11.Demple, B., and J. S. Sung. 2005. Molecular and biological roles of Ape1 protein in mammalian base excision repair. DNA Repair (Amsterdam) 4:1442-1449. [DOI] [PubMed] [Google Scholar]
- 12.Eferl, R., and E. F. Wagner. 2003. AP-1: a double-edged sword in tumorigenesis. Nat. Rev. Cancer 3:859-868. [DOI] [PubMed] [Google Scholar]
- 13.Evans, A. R., M. Limp-Foster, and M. R. Kelley. 2000. Going APE over ref-1. Mutat. Res. 461:83-108. [DOI] [PubMed] [Google Scholar]
- 14.Fan, Z., P. J. Beresford, D. Zhang, Z. Xu, C. D. Novina, A. Yoshida, Y. Pommier, and J. Lieberman. 2003. Cleaving the oxidative repair protein Ape1 enhances cell death mediated by granzyme A. Nat. Immunol. 4:145-153. [DOI] [PubMed] [Google Scholar]
- 15.Fischer, C., J. Johnson, B. Stillwell, J. Conner, Z. Cerovac, J. Wilson-Rawls, and A. Rawls. 2004. Activating transcription factor 4 is required for the differentiation of the lamina propria layer of the vas deferens. Biol. Reprod. 70:371-378. [DOI] [PubMed] [Google Scholar]
- 16.Florenes, V. A., G. M. Maelandsmo, A. Forus, A. Andreassen, O. Myklebost, and O. Fodstad. 1994. MDM2 gene amplification and transcript levels in human sarcomas: relationship to TP53 gene status. J. Natl. Cancer Inst. 86:1297-1302. [DOI] [PubMed] [Google Scholar]
- 17.Fung, H., R. A. Bennett, and B. Demple. 2001. Key role of a downstream specificity protein 1 site in cell cycle-regulated transcription of the AP endonuclease gene APE1/APEX in NIH 3T3 cells. J. Biol. Chem. 276:42011-42017. [DOI] [PubMed] [Google Scholar]
- 18.Fung, H., and B. Demple. 2005. A vital role for Ape1/Ref1 protein in repairing spontaneous DNA damage in human cells. Mol. Cell 17:463-470. [DOI] [PubMed] [Google Scholar]
- 19.Fung, H., Y. W. Kow, B. Van Houten, D. J. Taatjes, Z. Hatahet, Y. M. Janssen, P. Vacek, S. P. Faux, and B. T. Mossman. 1998. Asbestos increases mammalian AP-endonuclease gene expression, protein levels, and enzyme activity in mesothelial cells. Cancer Res. 58:189-194. [PubMed] [Google Scholar]
- 20.Grosch, S., G. Fritz, and B. Kaina. 1998. Apurinic endonuclease (Ref-1) is induced in mammalian cells by oxidative stress and involved in clastogenic adaptation. Cancer Res. 58:4410-4416. [PubMed] [Google Scholar]
- 21.Grosch, S., and B. Kaina. 1999. Transcriptional activation of apurinic/apyrimidinic endonuclease (Ape, Ref-1) by oxidative stress requires CREB. Biochem. Biophys. Res. Commun. 261:859-863. [DOI] [PubMed] [Google Scholar]
- 22.Haracska, L., I. Unk, R. E. Johnson, E. Johansson, P. M. Burgers, S. Prakash, and L. Prakash. 2001. Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites. Genes Dev. 15:945-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harding, H. P., Y. Zhang, H. Zeng, I. Novoa, P. D. Lu, M. Calfon, N. Sadri, C. Yun, B. Popko, R. Paules, D. F. Stojdl, J. C. Bell, T. Hettmann, J. M. Leiden, and D. Ron. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11:619-633. [DOI] [PubMed] [Google Scholar]
- 24.Harrison, L., A. G. Ascione, Y. Takiguchi, D. M. Wilson III, D. J. Chen, and B. Demple. 1997. Comparison of the promoters of the mouse (APEX) and human (APE) apurinic endonuclease genes. Mutat. Res. 385:159-172. [DOI] [PubMed] [Google Scholar]
- 25.Harrison, L., A. G. Ascione, D. M. Wilson III, and B. Demple. 1995. Characterization of the promoter region of the human apurinic endonuclease gene (APE). J. Biol. Chem. 270:5556-5564. [DOI] [PubMed] [Google Scholar]
- 26.Hofseth, L. J., M. A. Khan, M. Ambrose, O. Nikolayeva, M. Xu-Welliver, M. Kartalou, S. P. Hussain, R. B. Roth, X. Zhou, L. E. Mechanic, I. Zurer, V. Rotter, L. D. Samson, and C. C. Harris. 2003. The adaptive imbalance in base excision-repair enzymes generates microsatellite instability in chronic inflammation. J. Clin. Investig. 112:1887-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Honma, M., M. Hayashi, and T. Sofuni. 1997. Cytotoxic and mutagenic responses to X-rays and chemical mutagens in normal and p53-mutated human lymphoblastoid cells. Mutat. Res. 374:89-98. [DOI] [PubMed] [Google Scholar]
- 28.Hsieh, M. M., V. Hegde, M. R. Kelley, and W. A. Deutsch. 2001. Activation of APE/Ref-1 redox activity is mediated by reactive oxygen species and PKC phosphorylation. Nucleic Acids Res. 29:3116-3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu, Y., X. Jin, and E. T. Snow. 2002. Effect of arsenic on transcription factor AP-1 and NF-kappaB DNA binding activity and related gene expression. Toxicol. Lett. 133:33-45. [DOI] [PubMed] [Google Scholar]
- 30.Izumi, T., D. B. Brown, C. V. Naidu, K. K. Bhakat, M. A. Macinnes, H. Saito, D. J. Chen, and S. Mitra. 2005. Two essential but distinct functions of the mammalian abasic endonuclease. Proc. Natl. Acad. Sci. USA 102:5739-5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Izumi, T., T. K. Hazra, I. Boldogh, A. E. Tomkinson, M. S. Park, S. Ikeda, and S. Mitra. 2000. Requirement for human AP endonuclease 1 for repair of 3′-blocking damage at DNA single-strand breaks induced by reactive oxygen species. Carcinogenesis 21:1329-1334. [PubMed] [Google Scholar]
- 32.Kane, C. M., and S. Linn. 1981. Purification and characterization of an apurinic/apyrimidinic endonuclease from HeLa cells. J. Biol. Chem. 256:3405-3414. [PubMed] [Google Scholar]
- 33.Kato, Y., Y. Koike, K. Tomizawa, S. Ogawa, K. Hosaka, S. Tanaka, and T. Kato. 1999. Presence of activating transcription factor 4 (ATF4) in the porcine anterior pituitary. Mol. Cell. Endocrinol. 154:151-159. [DOI] [PubMed] [Google Scholar]
- 34.Kelley, M. R., L. Cheng, R. Foster, R. Tritt, J. Jiang, J. Broshears, and M. Koch. 2001. Elevated and altered expression of the multifunctional DNA base excision repair and redox enzyme Ape1/ref-1 in prostate cancer. Clin. Cancer Res. 7:824-830. [PubMed] [Google Scholar]
- 35.Kingma, P. S., and N. Osheroff. 1997. Spontaneous DNA damage stimulates topoisomerase II-mediated DNA cleavage. J. Biol. Chem. 272:7488-7493. [DOI] [PubMed] [Google Scholar]
- 36.Lee, T. C., M. Oshimura, and J. C. Barrett. 1985. Comparison of arsenic-induced cell transformation, cytotoxicity, mutation and cytogenetic effects in Syrian hamster embryo cells in culture. Carcinogenesis 6:1421-1426. [DOI] [PubMed] [Google Scholar]
- 37.Li, J. H., and T. G. Rossman. 1991. Comutagenesis of sodium arsenite with ultraviolet radiation in Chinese hamster V79 cells. Biol. Metals 4:197-200. [DOI] [PubMed] [Google Scholar]
- 38.Li, J. H., and T. G. Rossman. 1989. Mechanism of comutagenesis of sodium arsenite with n-methyl-n-nitrosourea. Biol. Trace Elem. Res. 21:373-381. [DOI] [PubMed] [Google Scholar]
- 39.Liu, S. X., M. Athar, I. Lippai, C. Waldren, and T. K. Hei. 2001. Induction of oxyradicals by arsenic: implication for mechanism of genotoxicity. Proc. Natl. Acad. Sci. USA 98:1643-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loeb, L. A., and B. D. Preston. 1986. Mutagenesis by apurinic/apyrimidinic sites. Annu. Rev. Genet. 20:201-230. [DOI] [PubMed] [Google Scholar]
- 41.Marx, J. 2004. Inflammation and cancer: the link grows stronger. Science 306:966-968. [DOI] [PubMed] [Google Scholar]
- 42.Masuoka, H. C., and T. M. Townes. 2002. Targeted disruption of the activating transcription factor 4 gene results in severe fetal anemia in mice. Blood 99:736-745. [DOI] [PubMed] [Google Scholar]
- 43.Moore, M. M., K. Harrington-Brock, and C. L. Doerr. 1997. Relative genotoxic potency of arsenic and its methylated metabolites. Mutat. Res. 386:279-290. [DOI] [PubMed] [Google Scholar]
- 44.Morris, S. M., O. E. Domon, L. J. McGarrity, J. J. Chen, and D. A. Casciano. 1995. Programmed cell death and mutation induction in AHH-1 human lymphoblastoid cells exposed to m-amsa. Mutat. Res. 329:79-96. [DOI] [PubMed] [Google Scholar]
- 45.Mure, K., A. N. Uddin, L. C. Lopez, M. Styblo, and T. G. Rossman. 2003. Arsenite induces delayed mutagenesis and transformation in human osteosarcoma cells at extremely low concentrations. Environ. Mol. Mutagen. 41:322-331. [DOI] [PubMed] [Google Scholar]
- 46.Nohmi, T., T. Suzuki, and K. Masumura. 2000. Recent advances in the protocols of transgenic mouse mutation assays. Mutat. Res. 455:191-215. [DOI] [PubMed] [Google Scholar]
- 47.Owens, D. M., S. Wei, and R. C. Smart. 1999. A multihit, multistage model of chemical carcinogenesis. Carcinogenesis 20:1837-1844. [DOI] [PubMed] [Google Scholar]
- 48.Parsons, J. L., I. I. Dianova, and G. L. Dianov. 2004. APE1 is the major 3′-phosphoglycolate activity in human cell extracts. Nucleic Acids Res. 32:3531-3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pi, J., Y. Kumagai, G. Sun, H. Yamauchi, T. Yoshida, H. Iso, A. Endo, L. Yu, K. Yuki, T. Miyauchi, and N. Shimojo. 2000. Decreased serum concentrations of nitric oxide metabolites among Chinese in an endemic area of chronic arsenic poisoning in inner Mongolia. Free Radic. Biol. Med. 28:1137-1142. [DOI] [PubMed] [Google Scholar]
- 50.Pourquier, P., L. M. Ueng, G. Kohlhagen, A. Mazumder, M. Gupta, K. W. Kohn, and Y. Pommier. 1997. Effects of uracil incorporation, DNA mismatches, and abasic sites on cleavage and religation activities of mammalian topoisomerase I. J. Biol. Chem. 272:7792-7796. [DOI] [PubMed] [Google Scholar]
- 51.Ramana, C. V., I. Boldogh, T. Izumi, and S. Mitra. 1998. Activation of apurinic/apyrimidinic endonuclease in human cells by reactive oxygen species and its correlation with their adaptive response to genotoxicity of free radicals. Proc. Natl. Acad. Sci. USA 95:5061-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramotar, D., C. Kim, R. Lillis, and B. Demple. 1993. Intracellular localization of the Apn1 DNA repair enzyme of Saccharomyces cerevisiae. Nuclear transport signals and biological role. J. Biol. Chem. 268:20533-20539. [PubMed] [Google Scholar]
- 53.Robertson, K. A., H. A. Bullock, Y. Xu, R. Tritt, E. Zimmerman, T. M. Ulbright, R. S. Foster, L. H. Einhorn, and M. R. Kelley. 2001. Altered expression of Ape1/ref-1 in germ cell tumors and overexpression in NT2 cells confers resistance to bleomycin and radiation. Cancer Res. 61:2220-2225. [PubMed] [Google Scholar]
- 54.Rossman, T. G. 1981. Effect of metals on mutagenesis and DNA repair. Environ. Health Perspect. 40:189-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rossman, T. G. 2003. Mechanism of arsenic carcinogenesis: an integrated approach. Mutat. Res. 533:37-65. [DOI] [PubMed] [Google Scholar]
- 56.Roybal, C. N., L. A. Hunsaker, O. Barbash, D. L. Vander Jagt, and S. F. Abcouwer. 2005. The oxidative stressor arsenite activates vascular endothelial growth factor mRNA transcription by an ATF4-dependent mechanism. J. Biol. Chem. 280:20331-20339. [DOI] [PubMed] [Google Scholar]
- 57.Rusyn, I., S. Asakura, B. Pachkowski, B. U. Bradford, M. F. Denissenko, J. M. Peters, S. M. Holland, J. K. Reddy, M. L. Cunningham, and J. A. Swenberg. 2004. Expression of base excision DNA repair genes is a sensitive biomarker for in vivo detection of chemical-induced chronic oxidative stress: identification of the molecular source of radicals responsible for DNA damage by peroxisome proliferators. Cancer Res. 64:1050-1057. [DOI] [PubMed] [Google Scholar]
- 58.Rutkowski, D. T., and R. J. Kaufman. 2003. All roads lead to ATF4. Dev. Cell 4:442-444. [DOI] [PubMed] [Google Scholar]
- 59.Schwerdtle, T., I. Walter, I. Mackiw, and A. Hartwig. 2003. Induction of oxidative DNA damage by arsenite and its trivalent and pentavalent methylated metabolites in cultured human cells and isolated DNA. Carcinogenesis 24:967-974. [DOI] [PubMed] [Google Scholar]
- 60.Shen, Z. X., G. Q. Chen, J. H. Ni, X. S. Li, S. M. Xiong, Q. Y. Qiu, J. Zhu, W. Tang, G. L. Sun, K. Q. Yang, Y. Chen, L. Zhou, Z. W. Fang, Y. T. Wang, J. Ma, P. Zhang, T. D. Zhang, S. J. Chen, Z. Chen, and Z. Y. Wang. 1997. Use of arsenic trioxide (As2O3) in the treatment of acute promyelocytic leukemia (APL). II. Clinical efficacy and pharmacokinetics in relapsed patients. Blood 89:3354-3360. [PubMed] [Google Scholar]
- 61.Soignet, S. L., S. R. Frankel, D. Douer, M. S. Tallman, H. Kantarjian, E. Calleja, R. M. Stone, M. Kalaycio, D. A. Scheinberg, P. Steinherz, E. L. Sievers, S. Coutre, S. Dahlberg, R. Ellison, and R. P. Warrell, Jr. 2001. United States multicenter study of arsenic trioxide in relapsed acute promyelocytic leukemia. J. Clin. Oncol. 19:3852-3860. [DOI] [PubMed] [Google Scholar]
- 62.Sung, J. S., and B. Demple. 2006. Roles of base excision repair subpathways in correcting oxidized abasic sites in DNA. FEBS J. 273:1620-1629. [DOI] [PubMed] [Google Scholar]
- 63.Syljuasen, R. G., B. Krolewski, and J. B. Little. 1999. Loss of normal G1 checkpoint control is an early step in carcinogenesis, independent of p53 status. Cancer Res. 59:1008-1014. [PubMed] [Google Scholar]
- 64.Tanabe, M., H. Izumi, T. Ise, S. Higuchi, T. Yamori, K. Yasumoto, and K. Kohno. 2003. Activating transcription factor 4 increases the cisplatin resistance of human cancer cell lines. Cancer Res. 63:8592-8595. [PubMed] [Google Scholar]
- 65.Tanaka, M., J. S. Lai, and W. Herr. 1992. Promoter-selective activation domains in Oct-1 and Oct-2 direct differential activation of an snRNA and mRNA promoter. Cell 68:755-767. [DOI] [PubMed] [Google Scholar]
- 66.Tanaka, T., T. Tsujimura, K. Takeda, A. Sugihara, A. Maekawa, N. Terada, N. Yoshida, and S. Akira. 1998. Targeted disruption of ATF4 discloses its essential role in the formation of eye lens fibres. Genes Cells 3:801-810. [DOI] [PubMed] [Google Scholar]
- 67.Tell, G., A. Zecca, L. Pellizzari, P. Spessotto, A. Colombatti, M. R. Kelley, G. Damante, and C. Pucillo. 2000. An ‘environment to nucleus’ signaling system operates in B lymphocytes: redox status modulates BSAP/Pax-5 activation through Ref-1 nuclear translocation. Nucleic Acids Res. 28:1099-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vermeulen, K., D. R. Van Bockstaele, and Z. N. Berneman. 2005. Apoptosis: mechanisms and relevance in cancer. Ann. Hematol. 84:627-639. [DOI] [PubMed] [Google Scholar]
- 69.Wang, T. S., T. Y. Hsu, C. H. Chung, A. S. Wang, D. T. Bau, and K. Y. Jan. 2001. Arsenite induces oxidative DNA adducts and DNA-protein cross-links in mammalian cells. Free Radic. Biol. Med. 31:321-330. [DOI] [PubMed] [Google Scholar]
- 70.Wang, Y., C. C. Shupenko, L. F. Melo, and P. R. Strauss. 2006. DNA repair protein involved in heart and blood development. Mol. Cell. Biol. 26:9083-9093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wiederhold, L., J. B. Leppard, P. Kedar, F. Karimi-Busheri, A. Rasouli-Nia, M. Weinfeld, A. E. Tomkinson, T. Izumi, R. Prasad, S. H. Wilson, S. Mitra, and T. K. Hazra. 2004. AP endonuclease-independent DNA base excision repair in human cells. Mol. Cell 15:209-220. [DOI] [PubMed] [Google Scholar]
- 72.Wilson, D. M., III, M. Takeshita, A. P. Grollman, and B. Demple. 1995. Incision activity of human apurinic endonuclease (Ape) at abasic site analogs in DNA. J. Biol. Chem. 270:16002-16007. [DOI] [PubMed] [Google Scholar]
- 73.Xanthoudakis, S., R. J. Smeyne, J. D. Wallace, and T. Curran. 1996. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc. Natl. Acad. Sci. USA 93:8919-8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xia, F., S. A. Amundson, J. A. Nickoloff, and H. L. Liber. 1994. Different capacities for recombination in closely related human lymphoblastoid cell lines with different mutational responses to X-irradiation. Mol. Cell. Biol. 14:5850-5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu, Y., D. H. Moore, J. Broshears, L. Liu, T. M. Wilson, and M. R. Kelley. 1997. The apurinic/apyrimidinic endonuclease (APE/ref-1) DNA repair enzyme is elevated in premalignant and malignant cervical cancer. Anticancer Res. 17:3713-3719. [PubMed] [Google Scholar]
- 76.Yu, Y., and J. B. Little. 1998. p53 is involved in but not required for ionizing radiation-induced caspase-3 activation and apoptosis in human lymphoblast cell lines. Cancer Res. 58:4277-4281. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.