Abstract
Oxidative lesions account for much of the spontaneously occurring DNA damage in normal cells and, left unrepaired, can be mutagenic or cytotoxic. We have investigated the capacity of purified human enzymes to initiate the base excision repair (BER) of oxidative lesions in model nucleosomes. In a construct where the minor groove of a thymine glycol lesion faced outward from the histone octamer, the human DNA glycosylase NTH1 (hNTH1) processed the lesion with nearly the same efficiency as in naked DNA. The hNTH1 reaction did not generate free DNA, indicating that the first step in BER occurred without irreversibly disrupting nucleosomes. Instead, lesion processing entailed the formation of nucleosome-hNTH1 ternary complexes that could be visualized in a gel mobility shift assay. These complexes contained both processed and unprocessed DNA. hNTH1 processing of lesions whose minor groove faced toward the histone octamer was poor at low hNTH1 concentrations but increased substantially as hNTH1 concentrations increased to nearly physiological levels. Additionally, an inward-facing lesion near the nucleosome edge was more efficiently processed than one closer to the nucleosome dyad. These observations suggest that access to sterically occluded lesions entails the partial, reversible unwrapping of DNA from the histone octamer, allowing hNTH1 to capture its DNA substrate when it is in an unwound state.
DNA in eukaryotes is packaged in chromatin, which consists predominantly of nucleosome arrays, punctuated by protein-DNA complexes that carry out such processes as transcription, DNA replication and repair, and chromosome segregation (for reviews, see references 11, 20, 49, and 52). Nucleosomes influence these and other DNA transactions to various extents. For example, nucleosomes are partially or fully disrupted ahead of the moving replication forks (28; reviewed in reference 14), and although newly replicated DNA is quickly packaged into new nucleosomes, correction of replication errors by proofreading activities of replicative DNA polymerases probably occurs in a quasi-nucleosome-free region. PCNA-mediated recruitment of mismatch repair machinery to replication foci (19) may permit it to act in a nucleosome-free zone as well, although it appears that only a small fraction of mismatch repair activity is replication associated. Nucleotide excision repair (NER) of UV photoproducts and bulky chemical adducts requires at least 30 proteins and 100 bp of DNA to either side of the DNA lesion and occurs independently of genomic replication. This suggests that NER requires the transient disruption of one or more nucleosomes. Nucleosome disruption may be facilitated by the reduced stability and increased mobility of nucleosomes in transcribed regions (reviewed in references 22, 37, and 45) in cases where NER enzymes are recruited to transcription elongation complexes stalled at sites of DNA damage (15, 48). Additionally, some, but not all, bulky lesions subject to NER have been reported to occur preferentially in linker regions (e.g., UV-induced 6,4 photoproducts but not cyclobutane pyrimidine dimers) (reviewed in reference 38), suggesting either that nucleosomes suppress the formation of such lesions or that bulky lesions destabilize or induce the migration of histone octamers to nearby undamaged DNA.
Oxidative lesions account for much of the spontaneously occurring DNA damage in normal cells and are efficiently repaired by enzymes in the base excision repair (BER) pathway (reviewed in references 7, 50, and 51). DNA lesions that result from oxidative damage or ionizing radiation generally are small and relatively nondistorting and occur both within and outside of nucleosomes, although the pattern of oxidative damage within nucleosomes may sometimes exhibit a periodicity that reflects the helical periodicity of nucleosomal DNA (for details, see references 10, 21, 26, 33, and 41). This makes it important to determine which step(s) in BER is nucleosome limited and to investigate mechanisms that cells use to circumvent these limits. BER is initiated by either mono- or bifunctional DNA glycosylases that remove the damaged base from its deoxyribose sugar. Monofunctional DNA glycosylases leave an apurinic (AP) site that is a substrate for AP endonuclease (APE1), which cleaves 5′ of the AP site, leaving a 5′ blocked terminus that must be removed by a 5′ deoxyribose phosphatase, an activity intrinsic to DNA polymerase β (31). Bifunctional DNA glycosylases contain a lyase activity that cleaves 3′ of the AP site, such that subsequent action by APE1 produces a repairable gap. In short-patch BER, the resulting gap can then be filled by DNA polymerase β and sealed by ligase III, in concert with XRCC1 (5, 8).
The capacity of human monofunctional DNA glycosylases to act on lesions in nucleosomes has been studied in vitro by two groups. Using model nucleosomes assembled with glucocorticoid hormone receptor response element (GRE)-containing DNA, Smerdon and colleagues found a substantial reduction in both the rate and extent of removal of uracil residues from nucleosomes by human uracil DNA glycosylase (UDG), with the extent of inhibition varying with the helical orientation of the uracil residues relative to the underlying histone octamer (3). Using model nucleosomes assembled with the Lytechinus variegatus 5S rRNA gene, Nilsen et al. (32) found a similar reduction in the efficiency of removal of uracil residues from nucleosomes by either human UNG2 or human SMUG1. However, this second group reported that the efficiency of uracil removal was essentially uniform, irrespective of the rotational position of the uracil relative to the histone octamer. Further studies indicated that DNA polymerase β, which acts at a later step in BER, was partially inhibited by the L. variegatus 5S DNA-containing nucleosome but completely inhibited by the GRE-containing nucleosomes (3, 32). Interestingly, however, both ligase I and FEN I, which act in long-patch BER, were able to act on nucleosomal substrates with nearly the same efficiency as seen in linker DNA (6, 17). The ligase I result was particularly surprising since structural studies indicate that it virtually encircles its DNA substrate (35).
The above-described studies suggested that early steps in BER can occur on nucleosomes but that some degree of perturbation, remodeling, or disruption of nucleosomes is required to complete BER. It is still not clear, however, how DNA glycosylases gain access to sterically occluded lesions. Possibly, DNA glycosylases are able to capture such lesions because of spontaneous partial unwrapping of DNA from the histone octamer, a phenomenon that may also help transcription and other factors bind to nucleosomal targets (12, 23, 24, 39, 40, 46). To test this hypothesis and to extend the above-described studies to bifunctional DNA glycosylases, we examined the capacity of purified human NTH1 (hNTH1) to act on a series of thymine glycol-containing model nucleosomes in vitro. We report that hNTH1 can process thymine glycol residues that face away from the histone octamer with an efficiency approaching that observed for naked DNA. Processing occurs without the discernible disruption of nucleosomes (as visualized by native gel electrophoresis). Under substrate excess conditions, processing of inward-facing lesions predicted to be sterically occluded was substantially inhibited. However, as concentrations of hNTH1 were increased to nearly physiological levels, the efficiency of cleavage of inward-facing lesions increased substantially. As with outward-facing lesions, cleavage at inward-facing lesions occurred without nucleosome disruption. The efficiency of cleavage of an inward-facing lesion closer to the center (dyad axis) of the nucleosome was lower than that of a similarly oriented lesion closer to the nucleosome edge. These observations support the idea that repair of inward-facing lesions involves the capture of lesions by hNTH1 during cycles of spontaneous, reversible, partial DNA unwrapping from the histone octamer.
MATERIALS AND METHODS
Preparation of DNA substrates for nucleosome assembly.
Partially complementary DNA oligomers (The Midland Certified Reagent Co.) were annealed and extended with exo− Klenow enzyme (NEB) to produce a 171-bp DNA fragment that included the L. variegatus 5S ribosomal DNA (rDNA) nucleosome-positioning sequence (44) flanked by KpnI and BamHI sites at one end and EcoRV and XbaI sites at the other end. This fragment was gel purified, cleaved with KpnI and XbaI, and ligated into pBlueScribe (pBS), and the resulting plasmids were transformed into DH5α cells. Positive clones were selected, and insertion of the desired fragment was confirmed by PCR analyses and DNA sequencing. One such plasmid was designated pBS-5SLv and used in the preparation of model nucleosomes. DNA for lesion-free nucleosomes was excised from pBS-5SLv by cleavage at EcoRI and HindIII sites that flanked the 5S rDNA insert. The excised DNA was gel purified and labeled at both ends with [α-32P]dATP (NEN) and exo− Klenow enzyme (NEB). The labeled DNA was then digested with either BamHI or XbaI and rendered blunt ended with exo− Klenow enzyme (NEB) and unlabeled deoxynucleoside triphosphates to produce uniquely end-labeled 177- and 171-bp fragments that were used to reconstitute nucleosomes. DNA for lesion-containing nucleosomes was prepared by digesting pBS-5SLv with KpnI and HindIII to liberate a fragment with a bottom strand that was 8 nucleotides (nt) longer than the top strand. The longer bottom strand was gel purified and used as a template in primer extension reactions with DNA oligomers (The Midland Certified Reagent Co.) that each contained a single discretely positioned thymine glycol residue (at sites indicated in Fig. 1). The lesion-containing oligomers were gel purified, quantified by spectrophotometry, 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase (NEB), annealed (in an ∼1.5-fold molar excess) to the unlabeled template strand, and extended with exo− Klenow enzyme (NEB) and unlabeled deoxynucleoside triphosphates at 25°C. The resulting full-length double-stranded DNA fragment was gel purified, quantified by liquid scintillation counting, and used for the reconstitution of nucleosomes.
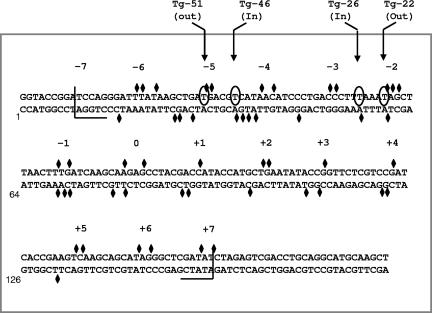
FIG. 1.
Sequence of the 5S rDNA-containing fragment used to assemble lesion-containing positioned nucleosomes. The sequence is numbered from the first base of a KpnI site that marks one end of the DNA used for reconstitution of lesion-containing nucleosomes. Diamonds indicate sites of DNase I cleavage within the reconstituted nucleosome, based on data from gels such as those shown in Fig. 2C and 3A. The cleavage sites occur in clusters, indicated by numbers −7, −6...0...+6, and +7, where 0 denotes the inferred center (dyad axis) of the nucleosome. These clusters are spaced at roughly 10-bp intervals, indicative of a rotationally positioned nucleosome. Half brackets indicate nucleosome boundaries that were also inferred from the DNase I data and from earlier studies by Simpson and Stafford (44). Circled bases indicate sites of substitution by thymine glycol to form nucleosomes Tg-51, Tg-46, Tg-26, and Tg-22, where the number in each name refers to the approximate distance between the thymine glycol and the dyad axis and where in or out refers to the approximate orientation of the lesion's minor groove relative to the histone octamer.
Preparation of donor chromatin for nucleosome assembly.
Donor chromatin was prepared by a modified version of standard methods (42, 43). Chicken red blood cells were obtained from citrated chicken blood (Pel-Freeze Biologicals), washed twice with prechilled buffer A (150 mM NaCl, 2.5 mM EGTA, 10 mM glucose, and 10 mM morpholinemethanesulfonic acid [pH 6.0] with 2 mM dithiothreitol [DTT], 0.25 mM phenylmethylsulfonyl fluoride [PMSF], and 1 mM benzamidine hydrochloride hydrate added just before use). The washed erythrocytes were suspended in cold buffer B (100 mM potassium acetate, 2.5 mM MgCl2, and 20 mM HEPES [pH 7.6] with 2 mM DTT, 0.25 mM PMSF, 1 mM benzamidine hydrochloride hydrate, 0.5 mM EGTA, 0.5 mM spermidine HCl, and 0.05 mM tosylsulfonyl phenylalanyl chloromethyl ketone [TPCK] added just before use), and lysed while stirring by the dropwise addition of 10% NP-40 to a final concentration of 0.1%. Nuclei were recovered from the lysate by centrifugation, washed twice with buffer B plus 0.1% NP-40, suspended in buffer C (100 mM potassium acetate, 2.5 mM MgCl2, 20 mM HEPES [pH 7.6], 2 mM DTT, 1 mM CaCl2), and incubated with 4,000 (Worthington) U of micrococcal nuclease (Sigma) at 37°C for 15 min. Digestion was terminated by adding EDTA to 10 mM and chilling the reaction mixture in wet ice. The digested nuclei were collected by centrifugation and suspended in cold 0.5 mM EDTA (pH 7.6) containing freshly added 0.25 mM PMSF and 0.05 mM TPCK to elute nucleosome oligomers. After an ∼20-min elution, nuclei were collected and suspended for a second ∼20-min elution. The eluates were combined and cleared by centrifugation, and the soluble chromatin was stripped of its linker histones by the dropwise addition, while stirring, of cold, sixfold-concentrated buffer D (600 mM NaCl, 50 mM Tris [pH 8.0], 1 mM EDTA, 1 mM DTT [final concentrations]). The stripped chromatin was then separated from linker histones and other free proteins by Sephacryl S500 chromatography in buffer D. Column fractions were collected and assayed by absorbance at 260 nm, and DNA and proteins in alternate fractions were analyzed by gel electrophoresis. Fractions devoid of linker histones and containing high levels of core histones were pooled and concentrated with Centricon Plus-20 centrifugal filters (Millipore). The NaCl concentration of the pooled chromatin was reduced to about 70 mM by multiple rounds of dilution with buffer E0 (25 mM HEPES [pH 8.0], 1 mM EDTA, 1 mM freshly added DTT), followed by reconcentration, after which the donor chromatin was made 15% in glycerol, divided into small aliquots, and frozen at −80°C.
Reconstitution and characterization of nucleosomes.
Nucleosomes were reconstituted by high-salt-mediated octamer transfer (13, 43). Generally, a 250-fold molar excess of chicken erythrocyte donor chromatin was mixed with end-labeled, nucleosome length or longer DNA, and NaCl was added to a final concentration of 1 M in buffer E0. After 30 min of incubation at 37°C, the mixture was shifted to 30°C and diluted to a final NaCl concentration of 0.1 M over a 2-h period by the stepwise addition of buffer E0 containing 0.05% NP-40. Nucleosome assembly was monitored by electrophoresis through native 4% polyacrylamide gels (in 45 mM Tris base-45 mM boric acid-1 mM EDTA). Gels were fixed in 10% acetic acid and 20% methanol and dried, and the background-adjusted counts in the nucleosome and naked-DNA bands were quantified by phosphorimaging. Reconstitution efficiencies, calculated as nucleosome counts/counts in naked DNA plus nucleosome (after correction for the background), generally exceeded 90%. To assess the helical orientation of DNA relative to the underlying histone octamer, end-labeled nucleosomes and naked-DNA controls in buffer E100 (i.e., buffer E0 with 100 mM NaCl) plus 0.05% NP-40 were mixed with 0.25 volume of buffer F (25 mM HEPES [pH 8.0], 25 mM MgCl2, 12.5 mM CaCl2, 50 μg/ml bovine serum albumin [NEB]) containing freshly diluted DNase I (Gibco/BRL; 0.25 and 5.0 U/ml for naked DNA and nucleosomes, respectively) and incubated for various times (24 and 60 s for naked-DNA substrates and 1, 3, and 9 min for nucleosome substrates) at room temperature (∼23°C; nota bene, Simpson and colleagues [44] obtained identical DNase I cleavage patterns in 37°C reactions). Digestion reactions were stopped by addition of one-third volume of 50 mM HNa3EDTA-0.4% sodium dodecyl sulfate (SDS)-0.2 mg/ml proteinase K, followed by incubation at 50°C for 1 h. DNase I digestion products were further purified by extraction with phenol and CHCl3 and fractionated on 8% sequencing gels. Gels were fixed in 10% acetic acid and 20% methanol, dried, and exposed to X-ray film. Cleavage data were taken from gel lanes in which the average DNA strand had been cleaved just once or less, as judged by the fraction of input DNA that remained intact (and assuming that cleavage frequencies follow a Poisson distribution). The translational position of lesion-containing DNA relative to the histone octamer was assessed by using data from both the DNase I assays described above and restriction enzyme assays conducted as follows. Reconstituted nucleosomes and naked-DNA control substrates in buffer E100 containing donor chromatin were made 10 mM in MgCl2 and incubated for 1 h at 37°C with a nominal 50-fold unit excess of the selected restriction enzyme. DNA from these reactions was fractionated on sequencing gels and visualized by autoradiography.
Preparation and assay of hNTH1.
hNTH1 cDNA was cloned into pTYB2 to generate the plasmid pTYB2-hNTH1, allowing the expression of hNTH1 as a C-terminal intein fusion protein. ER2566 (fpg−) cells were transformed with pTYB2-hNTH1, grown to an A600 of 0.5, and induced by addition of isopropyl-β-d-thiogalactopyranoside to 1 mM. After overnight induction at 16°C, cells were collected, flash-frozen in liquid nitrogen, and lysed by sonication in buffer F (50 mM Tris-HCl [pH 8.0], 500 mM NaCl, 1 mM EDTA) containing freshly added 1 mM PMSF and 10 mM benzamidine HCl. Cell lysates were clarified by centrifugation and loaded onto a chitin column in buffer F. Self-cleavage of the column-bound intein was induced by incubating the column for 18 h at 4°C in buffer F containing 50 mM DTT. hNTH1 was then eluted in buffer F and further purified through a HiTrap SP-FF column, which was eluted with 20 mM HEPES (pH 7.6)-1 M NaCl-5 mM β-mercaptoethanol-10% glycerol. Purified hNTH1 was dialyzed into 0.1 M NaCl, 50 mM Tris-HCl (pH 8.0), 1 mM DTT, and 25% glycerol and then made 50% in glycerol for storage. Protein concentrations were determined by the Bradford assay, and the fraction of active enzyme (20 to 30%, on average) was determined with a Schiff base assay (4) (nota bene, total enzyme concentrations are indicated in the figures). Glycosylase reactions with lesion-containing nucleosomes and their naked-DNA counterparts were carried out in buffer E100 containing 0.05% NP-40 at the substrate and enzyme concentrations specified in the figure legends. For the naked-DNA reactions, equivalent amounts of the donor chromatin were added just prior to enzyme addition to match the nucleosome reaction conditions. Reactions were generally conducted at 37°C for various times of up to 60 min. To measure DNA glycosylase-limited reaction rates, reactions were halted by the addition of NaOH to 0.1 N. Samples were boiled for 3 min, mixed with an equal volume of 2× formamide loading buffer, and fractionated in an 8% sequencing gel. To measure lyase-limited reaction rates, reactions were halted by the direct addition of an equal volume of 2× formamide loading buffer. Samples were boiled briefly and loaded onto sequencing gels. Gels were fixed and dried, and the substrate and reaction products were quantified by phosphorimaging.
Quantification of enzyme rate data.
Using data obtained by phosphorimaging, we first calculated the reaction extent for naked-DNA substrates as a function of time (equal to the counts per minute in the product band divided by the sum of the counts per minute in the substrate and product bands, after correction for the background). To obtain true rate data from the nucleosome reactions, we next subtracted from the apparent nucleosome rates that portion of the signal contributed by the small amount of contaminating naked-DNA substrate. Specifically, N-truet = [N-apparentt − (fxn DNA in nuc · Dt)]/(1 − fxn DNA in nuc), where N-truet and N-apparentt are the true and apparent reaction extents, respectively, for the nucleosome sample at time t, Dt is the true reaction extent for the naked DNA sample at time t, and fxn DNA in nuc is equal to 1 minus the nucleosome reconstitution efficiency, measured as described above. Data from experiments where reconstitution efficiencies varied from 85 to 95%, when corrected in this fashion, gave essentially identical results. For the substrate excess reactions (see Fig. 4), corrected data were normalized to the maximum reaction extent observed for naked DNA; this generally was limited to ∼60% of the total substrate by the slow turnover of hNTH1 in the absence of other BER enzymes. The normalized results from multiple independent experiments were then averaged and plotted as first-order exponential reaction curves (with Prism software from GraphPad). Corrected data from the enzyme excess reactions (see Fig. 6) were also normalized to the maximum reaction extents for their respective naked DNAs (∼77% and ∼98%, respectively, for the Tg-46 and Tg-51 DNA substrates) prior to their being plotted as hyperbolic curves. Because Escherichia coli Nei glycosylase was able to cleave equivalent substrates to completion, the failure of excess hNTH1 to digest the Tg-46 and Tg-51 substrates to completion was probably due to contamination by the nonpreferred 5S stereoisomer of thymine glycol (18); it is highly unlikely that such contamination would influence the results of the studies described in this paper.
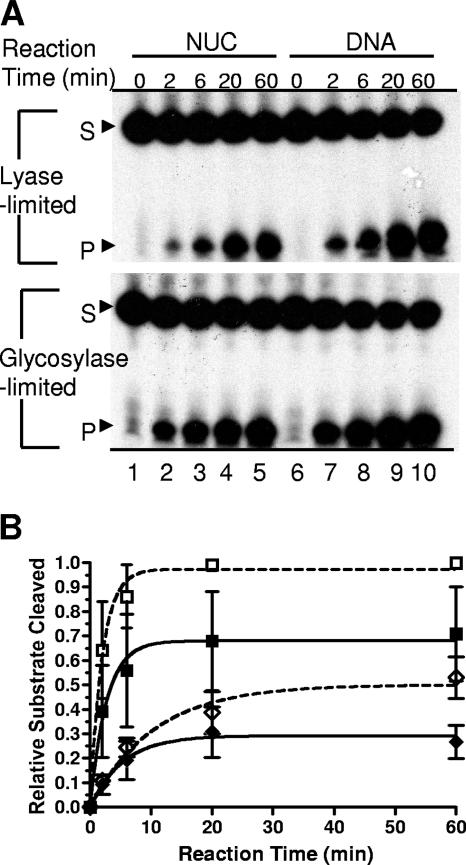
FIG. 4.
Activity of hNTH1 toward thymine glycol-containing nucleosomes. (A) Tg-51 nucleosomes (NUC, minor grove of Tg facing away from the histone octamer) and the corresponding naked DNAs were incubated for various times at 37°C at substrate and active hNTH1 concentrations of approximately 0.5 and 0.4 nM, respectively. Shown are denaturing gels used to measure glycosylase-limited (bottom gel) and lyase-limited (top gel) activities of hNTH1 (see the text for details). S, substrate; P, product. (B) Glycosylase-limited reaction curves for the Tg-51 nucleosome (solid squares) and the corresponding naked DNA (open squares) and lyase-limited reaction curves for the same nucleosome (closed diamonds) and naked-DNA samples (open diamonds). Points are average values from three independent experiments.
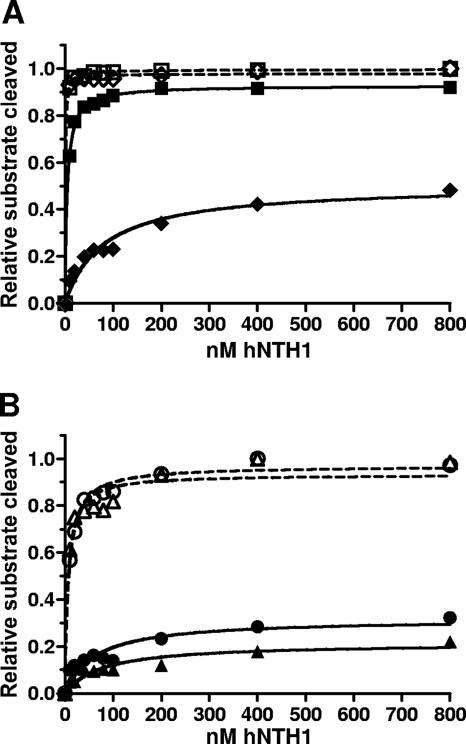
FIG. 6.
Increasing concentrations of hNTH1 increase the capacity of hNTH1 to process sterically occluded lesions. The glycosylase-limited activity of hNTH1 toward Tg-51, Tg-46, Tg-22, and Tg-26 nucleosomal and naked-DNA substrates was measured as a function of enzyme concentration. (A) The activity of hNTH1 toward Tg-51 and Tg-46 nucleosomes (solid squares and diamonds, respectively) is compared with its activity toward Tg-51 and Tg-46 naked DNAs (open squares and diamonds, respectively). (B) The activity of hNTH1 toward Tg-22 and Tg-26 nucleosomes (solid circles and triangles, respectively) is compared with its activity toward Tg-22 and Tg-26 naked DNAs (open circles and triangles, respectively). On the basis of a sign test, the probability that curves for Tg-22 and Tg-26 are identical is <0.004, indicating that hNTH1 is indeed more active toward the outward-facing Tg-22 lesion than toward the inward-facing Tg-26 lesion.
RESULTS
Assembly and characterization of model nucleosomes.
To determine if hNTH1 can act on lesions in nucleosomes, we assembled model nucleosomes containing the sea urchin 5S rDNA nucleosome-positioning sequence (Fig. 1). This DNA segment adopts a preferred rotational and translational position relative to the underlying histone octamer (44), making it possible to investigate the efficiency of BER as a function of a lesion's position in a nucleosome. Nucleosomes were reconstituted by high-salt-catalyzed transfer of histone octamers from donor chromatin isolated from chicken erythrocyte nuclei, as described in Materials and Methods. Gel electrophoresis of DNA, after limited cleavage of the donor chromatin by micrococcal nuclease and removal of linker histones by fractionation through an S500 column in 0.6 M NaCl, revealed a characteristic oligomeric repeat pattern indicative of short nucleosomal arrays (Fig. 2A); gel electrophoresis of proteins from the same fractions revealed intact core histones, as well as the stripped linker histones H1 and H5 in the trailing fractions (Fig. 2A). Gel electrophoresis also revealed highly efficient reconstitution of 5S rDNA nucleosomes (Fig. 2B) and indicated that the electrophoretic mobility of the reconstituted nucleosomes was similar to that of ethidium bromide-stained mononucleosomes from the donor chromatin population (not shown). The efficiency of nucleosome reconstitution was quantified by phosphorimaging (see Materials and Methods) and found generally to exceed 90%.
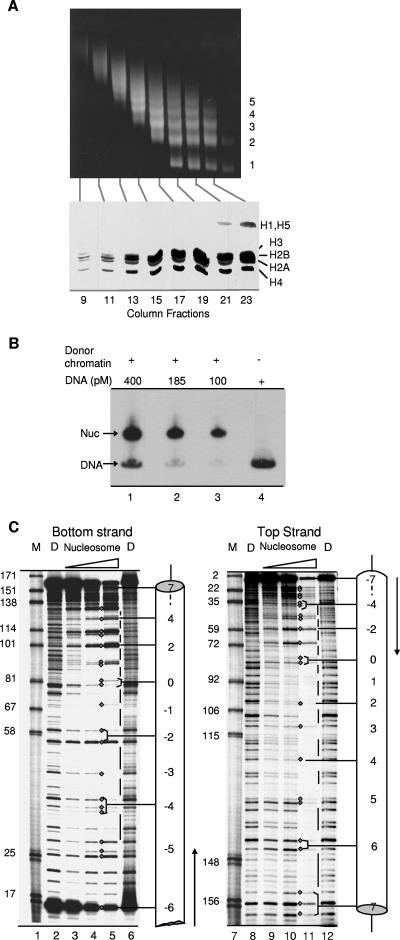
FIG. 2.
Analyses of donor chromatin and reconstituted nucleosomes. (A) DNA and total protein from size-fractionated donor chromatin were assayed by electrophoresis through 1.4% agarose and 20% SDS gels, respectively. The oligomeric series of DNA fragments (denoted by numbers on the right of the gel in the upper part) is the characteristic result of the partial micrococcal nuclease cleavage used to release chromatin from nuclei. The lower part shows intact core histones free of linker histones and other proteins in column fractions 13 to 19 that were pooled for reconstitutions. (B) Gel mobility shift analyses illustrating efficient nucleosome (Nuc) assembly, using fixed amounts of donor chromatin and various amounts of labeled recipient DNA. The mobility of the reconstituted nucleosomes is similar to that of ethidium bromide-stained mononucleosomes in the donor chromatin population (data not shown). (C) Reconstituted nucleosomes and naked-DNA controls (lanes D) were treated with DNase I for various times (see Materials and Methods), and DNA cleavage products were fractionated on 8% sequencing gels. Cleavage sites in the bottom (171-nt) strand of the reconstituted nucleosomes and naked-DNA controls were visualized and mapped in relation to a 32P-end label at an EcoRI site that is located 12 bp upstream of the KpnI site (Fig. 1). Cleavage sites in the top (177-nt) strand were visualized and mapped in relation to a 32P-end label at a HindIII site (position 179 in Fig. 1). Diamonds denote sites of cleavage in the nucleosome, while lines indicate regions that were protected from cleavage in the nucleosome. Lanes M contained Klenow end-labeled pBR322 DNA digested with MspI. Numbers adjacent to the marker fragments correspond to the labeling scheme in Fig. 1.
To assess the helical orientation of DNA in the reconstituted nucleosomes, 3′-end-labeled nucleosomes and their corresponding naked DNAs were treated with DNase I to similar extents (see Materials and Methods) and the cleavage products were separated on sequencing gels. DNase I cleavage sites in the lesion-free parental nucleosomes tended to cluster, forming cleavage maxima spaced at approximately 10-bp intervals (indicated by diamonds between lanes 4 and 5 and between lanes 10 and 11 in Fig. 2C). The cleavage maxima were separated by DNA segments that were protected in the nucleosome but vulnerable to DNase I in naked DNA (indicated by vertical lines between lanes 5 and 6, and lanes 11 and 12 in Fig. 2C). Using size markers (lanes M), we were able to map most of the DNase I cleavage sites at single-base resolution (shown as diamonds in Fig. 1). The positions of the cleavage maxima indicated that the helical orientation of the nucleosomal DNA was the same as that first mapped by Simpson and colleagues (44). Variation in pattern among different cleavage maxima suggested that the predominant translational position of DNA in the reconstituted nucleosome also matched that reported by Simpson and colleagues. This was confirmed by restriction enzyme accessibility studies described below.
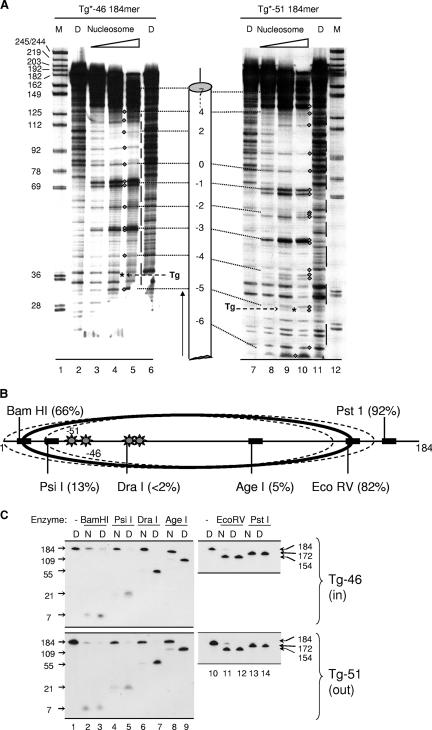
The above-described DNase I data were used to design four nucleosomes for the studies described in this paper. The first pair of nucleosomes each contained a single thymine glycol (Tg) residue positioned so that the minor groove of the Tg-A base pair would either face away from or toward the underlying histone octamer (nucleosomes Tg-51 and Tg-46, respectively, where 51 or 46 refers to the number of base pairs between the lesion and the dyad axis of the nucleosome). Tg residues in the second pair of nucleosomes also faced either away from or toward the histone octamer but were located closer to the dyad axis (nucleosomes Tg-22 and Tg-26, respectively). 5′-end-labeled, lesion-containing DNAs were prepared as described in Materials and Methods and assembled into nucleosomes. Gel mobility shift analyses indicated that inclusion of a single thymine glycol lesion did not affect reconstitution efficiency. To rule out the possibility that inclusion of the lesion had altered the rotational position of the nucleosomal DNA, we treated lesion-containing nucleosomes with DNase I as before. Figure 3A shows that, apart from a slightly reduced frequency of DNase I cleavage at the bond immediately 3′ of the Tg lesion in nucleosome Tg-51, the cleavage patterns for the lesion-containing nucleosomes were virtually identical to that of the parental nucleosome, indicating that the rotational setting was not affected by the Tg lesion. It remained possible that introduction of lesions induced DNA shifts of one or more helical turns relative to the dyad axis (thus preserving the original helical orientation), a phenomenon seen even in the absence of DNA lesions (9, 16, 36, 47). To address this possibility, we treated lesion-containing nucleosomes and corresponding naked-DNA controls with the restriction enzymes depicted in Fig. 3B, as described in Materials and Methods. As shown in Fig. 3C, the resulting cleavage patterns were virtually identical for nucleosomes Tg-51 and Tg-46. DraI and AgeI sites within the central wrap of the nucleosome were fully protected from cleavage (compare lanes 6 and 8 to lanes 7 and 9 in Fig. 3C), whereas BamHI and EcoRV sites, located 145 bp apart and close to the entry and exit points of the nucleosome, were, for the most part, vulnerable to cleavage (compare lanes 2 and 11 to lanes 3 and 12 in Fig. 3C). A PsiI site about midway between the BamHI site and the lesion in nucleosome Tg-51 was cleaved in about 13% of the molecules. This result might reflect a population of nucleosomes in which the histone octamer is shifted 10 or 20 bp to partially or fully expose the PsiI site but could also reflect transient exposure of the PsiI site because of spontaneous partial unwrapping of DNA near the edge of the histone octamer, a phenomenon that is discussed in greater detail below. Collectively, these results indicated that the DNA in most of the Tg-51 and Tg-46 nucleosomes was positioned with a single dominant helical and translational position but did not exclude the existence of minor translational variants. However, the key point, insofar as this study is concerned, is that a translational shift large enough to expose the lesions in either nucleosome Tg-51 or Tg-46 (20 to 30 bp or more) would occlude the PstI site, which was cleaved almost quantitatively. Thus, virtually all of the lesions in each of the model nucleosomes must reside within the nucleosome, with a discrete helical orientation.
FIG. 3.
Helical and translational positioning of lesion-containing nucleosomes. (A) Uniquely end-labeled Tg-46 and Tg-51 nucleosomes and their corresponding naked DNAs (lanes D) were incubated with DNase I, and their cleavage products were fractionated on 8% sequencing gels as described in Materials and Methods. Diamonds denote bands that make up the regularly spaced cleavage maxima in nucleosomes; intervening, nuclease-protected DNA segments that are vulnerable to DNase I cleavage in naked DNA are indicated by vertical lines. Asterisks denote the positions of thymine glycol residues. Size markers in lanes marked M are numbered to match the numbering scheme in Fig. 1. (B) Restriction enzyme sites used to map the translational position(s) of nucleosomes containing thymine glycol lesions (denoted by asterisks). Each percentage in parentheses is the fraction of nucleosomal DNA cleaved relative to naked DNA by a given enzyme, calculated from data shown in panel C. The predominant translational position is indicated by a solid ellipse, while two probable minor translational variants are depicted by dashed ellipses. In nucleosome Tg-51, the thymine glycol lesion would lie outside the nucleosome only in translational variants that block digestion by PstI; such variants represent, at most, ∼8% of the total Tg-51 population. (C) Uniquely end-labeled nucleosomes containing inward- and outward-facing lesions (nucleosomes Tg-46 and Tg-51, respectively) and the corresponding naked DNAs (lanes N and D, respectively) were incubated with restriction enzymes as described in Materials and Methods. To allow adequate resolution of substrate and product bands, DNA fragments produced by BamHI, PsiI, DraI, and AgeI were separated on 12% sequencing gels while the EcoRV and PstI products were separated on 6% sequencing gels. Band intensities were quantified by phosphorimaging and cleavage extents were calculated as outlined in the Materials and Methods section entitled “Quantification of enzyme rate data.”
Efficient processing of outward-facing Tg lesions by hNTH1.
Having determined that the DNA in nucleosomes Tg-51 and Tg-46 was positioned as expected, we measured the capacity of hNTH1 to remove lesions from these nucleosomes and their corresponding naked DNAs. Previous studies indicated that the activity of hNTH1 on naked-DNA substrates is lyase rather than glycosylase limited (29, 30, 34). To determine if its activity is also lyase limited on nucleosomal substrates, we added hNTH1 to molar excess amounts of nucleosome Tg-51 (lesion facing out) and corresponding naked DNAs, in identical buffers, containing equal amounts of donor chromatin. Aliquots from these reaction mixtures were collected at various times and divided into two equal parts. Half of each aliquot was loaded directly onto sequencing gels to assess the overall reaction rate (i.e., glycosylase plus lyase). To assess the glycosylase-only reaction rate, the second half aliquot was treated with NaOH prior to electrophoresis. Figure 4 shows that the lyase activity of hNTH1 was significantly lower than its glycosylase activity on nucleosomes, as well as on naked DNA. Parallel experiments with the Tg-46 (lesion facing in) nucleosome and naked-DNA substrate revealed similarly reduced lyase activity (data not shown). To quantify these results, we quantified substrate and cleavage products in Fig. 4A by phosphorimaging and calculated reaction extents as a function of time as described in Materials and Methods. These calculations included a correction for the small amounts of naked substrate DNA that contaminated our nucleosome preparations. We normalized the resulting data and averaged with data from other experiments to generate the reaction curves shown in Fig. 4B. Comparison of the initial slopes of the reaction curves indicated that the lyase activity of hNTH1 is about fivefold lower than its glycosylase activity. This result indicated that glycosylase-limited reaction rates would best reflect the impact of nucleosomes on lesion recognition by hNTH1. Hence, in later experiments, we generally assayed only the glycosylase-limited reaction rates.
Comparison of the initial slopes of the glycosylase-limited reaction curves in Fig. 4B indicated that the activity of hNTH1 toward the outward-facing lesion in the Tg-51 nucleosome is only slightly reduced (∼1.6-fold) compared to naked DNA. We observed a similar reduction in reaction extent (i.e., the overall fraction of substrate cleaved within the 60-min time frame of the reaction). These results suggested that the modest reduction in the observed reaction rate is due to a reduction in substrate availability rather than a slower catalytic rate. This in turn may partly reflect a moderate degree of substrate heterogeneity, which could be the result of minor rotational or translational variants. However, as described above, it is unlikely that there were translational shifts of sufficient magnitude to expose the lesion in nucleosome Tg-51. Further evidence against translational shifts large enough to expose the lesion in nucleosome Tg-51 (i.e., >20 bp) came from control experiments in which the ability of E. coli Udg to remove an inward-facing uracil residue at a site even closer to the edge of the 5S rDNA nucleosome was substantially lower than its ability to remove a uracil located at the outward-facing −51 site (A. Prasad, S. S. Wallace, and D. S. Pederson, unpublished data). Minor rotational variants within the Tg-51 nucleosome population might also affect the accessibility of lesions. Specifically, while our DNase I analyses indicate that DNA in most of the model nucleosomes has a single preferred helical orientation, neither DNase I nor hydroxy radical footprinting assays are precise enough to rule out helical configurations that differ from the dominant position by ±1 bp. The orientation of a Tg residue in a 1-bp rotational variant would differ from that of the dominant variant by ∼35° and might substantially reduce the efficiency of hNTH1 binding. Regardless of the exact explanation, these studies indicated that hNTH1 can efficiently recognize appropriately oriented DNA lesions in nucleosomes.
Lesion processing by hNTH1 occurs without irreversible nucleosome disruption.
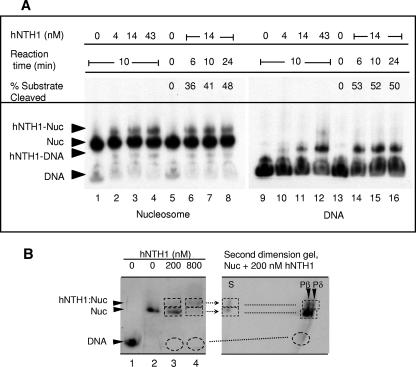
To determine if lesion processing by hNTH1 is accompanied by nucleosome disruption, we incubated thymine glycol-containing Tg-51 nucleosomes with a molar excess of hNTH1 and then electrophoretically separated free enzyme from nucleosomes. Increasing concentrations of hNTH1 led to increasing amounts of supershifted complexes that formed with both Tg-51 nucleosomes and their corresponding naked DNAs (lanes 2 to 4 and 10 to 12 in Fig. 5A). The fraction of nucleosomes that formed supershifted complexes in the presence of 14 nM hNTH1 remained relatively constant (at ∼11%) over a 6- to 24-min range of incubation times, even as the amount of lesion processed in the Tg-51 nucleosome increased to nearly 50% (Fig. 5A, lanes 6 to 8). At no point did we observe the accumulation of free naked DNA. This result indicated that hNTH1 can act on lesions without causing or requiring irreversible nucleosome disruption. These results also suggested that lesion processing occurs within the supershifted complex and that once processing is complete, the enzyme is free to dissociate. To further test this idea, nucleosomes and supershifted complexes were electrophoretically fractionated as before and the DNA from these complexes was released into and electrophoresed in a second-dimension sequencing gel. Figure 5B shows that hNTH1-processed DNA was present in both the supershifted and nucleosomal particles, which provided further evidence that lesion processing can indeed take place in the hNTH1-nucleosome complex.
FIG. 5.
Gel mobility shift analyses of complexes formed by addition of hNTH1 to lesion-containing nucleosomes and naked DNA. (A) Preformed Tg-51 nucleosomes (lanes 1 to 8) and the corresponding naked DNAs (lanes 9 to 16) were incubated with increasing concentrations of hNTH1 (lanes 1 to 4 and 9 to 12) or with 14 nM hNTH1 for increasing time periods (lanes 6 to 8 and 14 to 16) at 37°C. Arrowheads indicate naked DNA (DNA), nucleosomes (NUC), and complexes that form upon the addition of hNTH1 (hNTH1-NUC and hNTH1-DNA). The percent substrate processed by the hNTH1 glycosylase with increasing time is provided. Note the absence of any increase in naked-DNA amounts even after hNTH1 has processed nearly half of the lesion-containing nucleosomes (lane 8). (B) Nucleosomes (Nuc) were incubated with 200 or 800 nM hNTH1 for 30 min at 25°C, and the resulting complexes were fractionated as described above. The unfixed wet gels were exposed to X-ray film, and gel strips containing the fractionated complexes were excised, incubated at 50°C for 1 h in 0.1% SDS-12.5 mM HNa3EDTA-100 μg/ml proteinase K, boiled for ∼5 min in 0.1 N NaOH, soaked in formamide loading buffer, and loaded onto a second-dimension 8% sequencing gel. The first- and second-dimension gels were aligned to show the path of migration into the second-dimension gel and that hNTH1-processed DNA was present in both the supershifted and nucleosomal particles. S denotes unprocessed DNA, while Pβ and Pδ denote the β and δ elimination products produced, respectively, by hNTH1 and NaOH treatment.
Inward-facing lesions become amenable to repair by hNTH1 at high hNTH1 concentrations.
While hNTH1 efficiently cleaved the outward-facing Tg residue in nucleosome Tg-51, steric considerations suggested that hNTH1 would be far less active toward the inward-facing Tg in nucleosome Tg-46. This proved true in substrate excess reactions where the concentration of active hNTH1 was approximately 0.4 nM but much less so as hNTH1 concentrations were increased to nearly physiological levels (Fig. 6A and data not shown). Specifically, the concentration of hNTH1 in the nucleus has been estimated to be ∼2.3 μM (27). To investigate the effect of similarly high concentrations of hNTH1 on repair of nucleosomal lesions, we incubated Tg-51 and Tg-46 nucleosomes and the corresponding naked DNAs for 30 min with increasing concentrations of hNTH1. As in the substrate excess experiments described above, the outward-facing Tg-51 lesion was processed nearly as efficiently as lesions in naked DNA (Fig. 6A). At a relatively low hNTH1 concentration (10 nM), processing of the inward-facing Tg-46 lesion was about 10-fold reduced relative to that of naked DNA. However, with increasing concentrations of hNTH1, the amount of inward-facing lesion processed progressively increased, reaching ∼48% cleavage relative to that of naked DNA at the maximum enzyme concentration tested (800 nM total enzyme; Fig. 6A). This result was consistent with the hypothesis that processing of inward-facing lesions requires partial unwrapping of DNA from the histone octamer core and that high concentrations of hNTH1 are required for efficient binding in the brief interval during which DNA unwrapping exposes the target lesion (12, 23, 24, 39, 40, 46).
The above-described DNA-unwrapping hypothesis predicted that the frequency or duration of exposure of inward-facing lesions would be lower for lesions near the center of the nucleosome than for lesions near the nucleosome edge. To test this, we constructed nucleosomes carrying a single inward- or outward-facing thymine glycol located closer to the dyad axis of the nucleosome (nucleosomes Tg-26 and Tg-22, respectively). Figure 6B shows that the inward-facing Tg-26 lesion was processed only about 22% as efficiently as the naked DNA at the highest concentration of enzyme tested, compared to ∼48% for the inward-facing Tg-46 lesion, which is located 20 bp closer to the nucleosome edge. This result supported the unwrapping hypothesis.
In nucleosome Tg-22, the thymine glycol lesion was even closer to the dyad axis but its minor groove faced outward from the histone octamer and proved to be somewhat more accessible to hNTH1 than the lesion in nucleosome Tg-26, as would be expected if hNTH1 were able to bind directly to the Tg-22 lesion. It is important to note, however, that the Tg-22 lesion was far less accessible than the outward-facing lesion in nucleosome Tg-51, with excision of the Tg-22 lesion reduced about threefold relative to that of naked DNA at the highest concentration tested (Fig. 6B). Because the lesions in nucleosomes Tg-22 and Tg-51 are separated by just 29 bp (i.e., <3.0 helical turns), the reduced cleavage of the Tg-22 lesion might be due to both a suboptimal helical orientation and its more central location in the nucleosome.
Excision of sterically occluded inward-facing lesions by hNTH1 does not require nucleosome disruption.
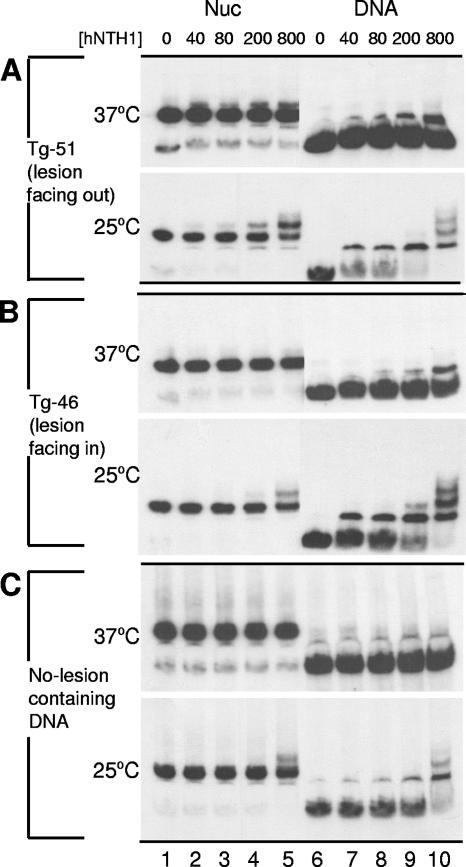
As was the case for nucleosomes with relatively accessible lesions, the processing of sterically occluded lesions occurred without generating naked DNA (Fig. 7). However, we were unable to detect a discrete supershifted complex when nucleosomes with inward-facing lesions were incubated with up to 43 nM hNTH1 at 37°C (not shown). To enhance our ability to detect such complexes, we repeated these studies with increased hNTH1 concentrations. We also reduced the hNTH1 reaction temperature from 37°C to 25°C, reasoning that this would reduce turnover of the hNTH1-nucleosome and hNTH1-DNA complexes, making them easier to detect. Figure 7A shows that Tg-51 nucleosomes with outward-facing lesions readily formed supershifted complexes when incubated with the same high levels of hNTH1 used in Fig. 6 and that greater amounts of the supershifted complexes formed when hNTH1 was incubated with nucleosomes at 25°C rather than at 37°C. As before, addition of enzyme did not lead to the release of naked DNA, indicating that processing occurred without irreversible disruption of nucleosomes. As shown in Fig. 7B, incubation of the Tg-46 nucleosome with a 200 or 800 nM concentration of hNTH1 produced a supershifted complex similar to the complex that formed with the Tg-51 nucleosome at lower hNTH1 concentrations. Importantly, however, at the highest enzyme concentration tested, a supershifted complex also formed with a lesion-free nucleosome (Fig. 7C). This result is in accord with the observation that incubation of the Tg-51 and Tg-46 naked-DNA substrates with 800 nM hNTH1 at 25°C produced not just the complex that probably reflects direct binding of thymine glycol by hNTH1 but also a slower-moving complex that might reflect nonspecific binding to the same DNA fragment (rightmost lanes in Fig. 7A and B). Given these results, it is risky to conclude that the hNTH1 complex that forms with nucleosomes containing inward-facing lesions is lesion specific. The problem, however, has little to do with differences in lesion-specific and nonspecific binding constants, since a comparison of hNTH1 binding to naked DNA indicates a high degree of specificity for thymine glycol-containing DNA (compare lanes 6 to 10 in Fig. 7A and B to the equivalent lanes in panel C). Rather, we think that the problem in distinguishing between lesion-specific and nonspecific complexes is that hNTH1 binding to inward-facing lesions is limited by the frequency and duration of transient unwrapping of DNA from the histone octamer, and the capacity of hNTH1 to capture the lesion during episodes of unwrapping (see reference 23 for a general discussion). The matter is further complicated by the dimerization of hNTH1 at high concentrations (which appears to enhance hNTH1 turnover) (27) and the fact that the supershifted complexes are quasistable intermediates rather than stable endpoint complexes and as such provide only an indirect measure of enzyme binding affinities. Although there is not enough information to evaluate the contribution of each of these variables, our data strongly support the basic conclusion from these studies: that transient DNA unwrapping facilitates the first step in BER of sterically occluded lesions.
FIG. 7.
Gel mobility shift analyses of nucleosomes (Nuc) or naked DNA incubated with large amounts of hNTH1. Tg-51 and Tg-46 nucleosomes and the corresponding naked DNAs, as well as lesion-free nucleosomes and naked DNAs (177-mer) (A, B, and C, respectively), were incubated at either 37°C or 25°C with increasing concentrations of hNTH1 as indicated (except for lanes 7, 8, and 9 in the 37°C samples in panel A, where the hNTH1 concentrations were 10, 40, and 80 nM, respectively).
DISCUSSION
Efficiency of BER of lesions in nucleosomes varies with the lesion's helical and translational position relative to the histone octamer.
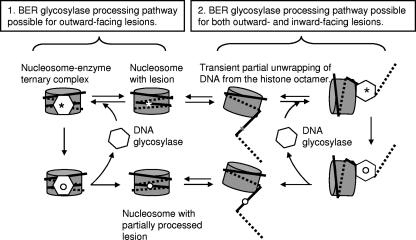
This study has investigated the ability of the bifunctional DNA glycosylase hNTH1 to process thymine glycol lesions at discrete sites in model nucleosomes. We show that the efficiency of cleavage of an outward-facing thymine glycol residue located approximately 20 nt in from the nucleosome edge (Tg-51) was nearly as high as that observed for naked DNA and substantially higher than it was for an inward-facing lesion located just 5 nt further in from the nucleosome edge (Tg-46). Cleavage of an inward-facing thymine glycol residue positioned closer to the dyad axis (Tg-26) was much reduced compared to that observed for an inward-facing residue closer to the edge of the nucleosome (Tg-46), consistent with the hypothesis that access to sterically occluded lesions is facilitated by spontaneous transient partial unwrapping of DNA from the edge of the histone octamer, as depicted in the right half of Fig. 8. In addition, cleavage of inward-facing lesions increased significantly as a function of enzyme concentration. These observations suggest that lesions within partially unwrapped nucleosomal DNA can be captured by hNTH1, thereby driving an increasing fraction of lesion-containing nucleosomes into a partially unwrapped state.
FIG. 8.
Model of how hNTH1-mediated processing of lesions occurs in nucleosomes. Optimally oriented lesions may be bound directly by hNTH1 with little or no perturbation of the nucleosome (diagram on the left). Processing of inward-facing lesions likely requires spontaneous partial unwrapping of DNA from the histone octamer to allow enzyme access to the lesion (diagram on the right). High concentrations of hNTH1 may be needed to efficiently capture lesions in the transiently unwound DNA. See the text for further discussion.
Of the two nucleosomes in which the minor groove of the thymine glycol faced outward from the histone octamer, only one (Tg-51) was processed with an efficiency approaching that seen for naked DNA. The lesion in the second of these two nucleosomes (Tg-22) was ∼2.9 helical turns (i.e., 29 bp) closer to the dyad axis of the nucleosome. Hence, while the reduced processing efficiency of the Tg-22 lesion may be due partly to its more dyad-proximal position, the highly efficient processing of the Tg-51 lesion may be an exceptional case. In other words, there may be only a small number of sites in nucleosomes where a lesion's helical orientation would permit direct binding by hNTH1. If this is correct, it means that access to most lesions in nucleosomes will depend on transient partial unwrapping of nucleosomal DNA (or on nucleosome remodeling agents, as discussed below).
Regardless of lesion orientation, processing by hNTH1 did not result in irreversible nucleosome disruption (as defined by release of DNA from preformed nucleosomes in a gel mobility shift assay). Instead, addition of hNTH1 to a nucleosome with a relatively accessible (outward-facing) lesion revealed a supershifted complex that contained partially processed DNA. This result suggested that processing occurs in an enzyme-nucleosome ternary complex, as depicted in the left half of Fig. 8. (Although we think this inference is the most straightforward, we have yet to definitively rule out alternative interpretations, for example, that enzyme processing might occur in a complex that is too short-lived to readily detect and that the supershifted complex reflects the rebinding of hNTH1 to already-processed DNA.)
As indicated in the introduction, both Nilsen et al. (32) and Smerdon and colleagues (3) investigated the capacity of monofunctional DNA glycosylases to excise uracil residues from positioned nucleosomes in vitro. Differences in how these groups calculated reaction rates make it difficult to compare their initial velocity data with ours. Hence, for comparative purposes, we focused on the extent of cleavage observed within a standard time frame. Specifically, we measured cleavage extents in 30-min reaction mixtures where the concentration of active hNTH1 exceeded the total lesion concentration (and equaled or exceeded the apparent Km of hNTH1 for naked DNA; data not shown). At 10 nM hNTH1, the cleavage extents observed for inward-facing lesions in nucleosomes Tg-46 and Tg-26 were, respectively, approximately 10- and 13-fold reduced relative to those of the corresponding naked DNAs. The cleavage extents observed for the outward-facing lesions in nucleosomes Tg-51 and Tg-22 were, respectively, reduced approximately 1.5- and 6-fold relative to those of the corresponding naked DNAs. Beard et al. (3) observed an ∼10-fold suppression of the capacity of the monofunctional DNA glycosylase UDG to excise an inward-facing uracil residue near the dyad axis (i.e., center) of the nucleosome, compared to an ∼3-fold reduction for the excision of an outward-facing uracil residue, also close to the dyad axis. Thus, while our results differed in magnitude from those of Smerdon and colleagues, we both observed differences in the extent of suppression as a function of helical orientation. The levels of activity reported by Smerdon and colleagues (3) were generally higher than those we observed, probably because of the inclusion of APE1, which was not used in the studies reported here.
The above-described sensitivity to the helical orientation of a target lesion or uracil residue is in accord with expected steric constraints imposed by the histone octamer. Hence, it was surprising that Nilsen et al. (32) reported virtually no difference in the capacity of UNG2 to excise differently oriented uracil residues from nucleosomes (i.e., the authors reported three- and ninefold reductions in the activities of UNG2 and SMUG1, respectively, toward nucleosomes carrying a single uracil [U:A] compared with the corresponding naked DNA). The explanation for these different results cannot be DNA sequence differences since we used the same 5S rDNA-positioning element as Nilsen et al. (32). These differences are also unlikely to be to due to our use of thymine glycol rather than uracil since, in our experiments, the efficiency of removal of uracil by E. coli Udg is even more sensitive to the rotational context than is processing of thymine glycol residues by hNTH1 (Prasad et al., unpublished). Control experiments also indicated that differences between our results and those of Nilsen et al. are not due to the presence or absence of Mg2+ and ATP in enzyme reaction buffers (data not shown). As shown in Fig. 6 and discussed below, differences due to helical orientation were diminished in reactions containing very high enzyme concentrations. Beard et al. (3) also noted that, at high concentrations of UDG and APE1, the efficiency of processing of both outward- and inward-facing uracil residues approached that observed for naked DNA.
Transient, partial unwrapping of DNA from the histone octamer enables hNTH1 to process sterically occluded lesions.
Nucleosomes have been shown to be conformationally dynamic in vitro, with spontaneous partial unwrapping of nucleosomal DNA transiently exposing otherwise buried DNA sites (1, 2, 23, 24). In vivo competition studies suggested that partial unwrapping of nucleosomal DNA may also facilitate the binding of certain transcription factors to their cognate sequences in cells (12). Generally, and also in the case of initiation of BER by hNTH1, we hypothesize that access to an inward-facing lesion requires partial unwrapping of the DNA from the histone octamer core. Since DNA unwrapping would begin at the nucleosome edge, an inward-facing lesion close to the nucleosome dyad would be less accessible to repair than an inward-facing lesion closer to the edge. Consistent with this prediction, we found much less cleavage of the Tg-26 (closer to the dyad) lesion than of the Tg-46 (closer to the edge) lesion on the nucleosome. These results are reminiscent of the observation that the glucocorticoid receptor was able to bind (albeit inefficiently) to its target (GRE) sequence in DNA even when key determinants in the GRE faced into the histone octamer, provided the GRE was located ∼40 bp away from the dyad axis; no binding to a similarly oriented GRE near the dyad axis of the nucleosome occurred (25). Our results also are consistent with “relative site exposure” measurements by Widom and colleagues, who found that access to sites in nucleosomal DNA decreased progressively from the edge of the nucleosome toward the dyad (2, 39).
The transient-DNA-unwrapping hypothesis predicts that the concentration of inward-facing lesions available for hNTH1 binding is much less than the total lesion concentration and, accordingly, that efficient binding of such lesions will require much greater amounts of enzyme than does binding to lesions in naked DNA. Consistent with this prediction, we found that hNTH1 cleaved progressively more of the inward-facing lesion in nucleosome Tg-46 as a function of enzyme concentration, reaching ∼50% of the cleavage seen on naked DNA at 200 to 800 nM hNTH1; such concentrations appear to be physiologically relevant in that the hNTH1 concentration in the nucleus is estimated to be ∼2.3 μM (∼0.65 μM in the cytosol) (27). While elevated concentrations that promote hNTH1 dimerization may have contributed to this increase in cleavage efficiency (27), the increase was far more substantial for inward-facing than for outward-facing lesions. This suggests that the principal mechanism for the increase in cleavage of the inward-facing lesion at high hNTH1 concentrations is enhanced efficiency of lesion capturing that drives the equilibrium toward the partially unwrapped state, as proposed by Widom and colleagues (23, 24).
In summary, hNTH1 can carry out the first step in BER in nucleosomes without inducing irreversible nucleosome disruption. While processing of an outward-facing lesion positioned within two to three helical turns of the nucleosome edge occurred without the aid of any other accessory BER factor, differently oriented lesions and lesions closer to the dyad axis were cleaved less efficiently. Thus, we suspect that lesion processing in vivo requires both BER accessory proteins and nucleosome remodeling and/or modifying factors. Our present study has established a reliable model system that will enable us to investigate factors that ensure efficient BER in cells.
Acknowledgments
We thank Alicia Holmes for purified hNTH1, Jeffrey Bond and Jeff Blaisdell for advice on data analyses, Anne MacLeod for help with manuscript preparation, and members of the Structure, Function, and Evolution of DNA Repair Enzymes Program Project Group for helpful comments and discussion.
This study was funded by a grant from the NCI (P01-CA098993).
Footnotes
Published ahead of print on 8 October 2007.
REFERENCES
- 1.Anderson, J., A. Thastrom, and J. Widom. 2002. Spontaneous access of proteins to buried nucleosomal DNA target sites occurs via a mechanism that is distinct from nucleosome translocation. Mol. Cell. Biol. 22:7147-7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, J., and J. Widom. 2000. Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J. Mol. Biol. 296:979-987. [DOI] [PubMed] [Google Scholar]
- 3.Beard, B. C., S. H. Wilson, and M. J. Smerdon. 2003. Suppressed catalytic activity of base excision repair enzymes on rotationally positioned uracil in nucleosomes. Proc. Natl. Acad. Sci. USA 100:7465-7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaisdell, J. O., and S. S. Wallace. 2007. Rapid determination of the active fraction of DNA repair glycosylases: a novel fluorescence assay for trapped intermediates. Nucleic Acids Res. 35:1601-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cappelli, E., R. Taylor, M. Cevasco, A. Abbondandolo, K. Caldecott, and G. Frosina. 1997. Involvement of XRCC1 and DNA ligase III gene products in DNA base excision repair. J. Biol. Chem. 272:23970-23975. [DOI] [PubMed] [Google Scholar]
- 6.Chafin, D. R., J. M. Vitolo, L. A. Henricksen, R. A. Bambara, and J. J. Hayes. 2000. Human DNA ligase I efficiently seals nicks in nucleosomes. EMBO J. 19:5492-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.David, S. S., and S. D. Williams. 1998. Chemistry of glycosylases and endonucleases involved in base-excision repair. Chem. Rev. 98:1221-1262. [DOI] [PubMed] [Google Scholar]
- 8.Dianova, I. I., K. M. Sleeth, S. L. Allinson, J. L. Parsons, C. Breslin, K. W. Caldecott, and G. L. Dianov. 2004. XRCC1-DNA polymerase beta interaction is required for efficient base excision repair. Nucleic Acids Res. 32:2550-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong, F., J. Hansen, and K. van Holde. 1990. DNA and protein determinants of nucleosome positioning on sea urchin 5S rRNA gene sequences in vitro. Proc. Natl. Acad. Sci. USA 87:5724-5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enright, H. U., W. J. Miller, and R. P. Hebbel. 1992. Nucleosomal histone protein protects DNA from iron-mediated damage. Nucleic Acids Res. 20:3341-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felsenfeld, G., and M. Groudine. 2003. Controlling the double helix. Nature 421:448-453. [DOI] [PubMed] [Google Scholar]
- 12.Geraghty, D. S., H. B. Sucic, J. Chen, and D. S. Pederson. 1998. Evidence that partial unwrapping of DNA from nucleosomes facilitates the binding of heat shock factor following DNA replication in yeast. J. Biol. Chem. 273:20463-20472. [DOI] [PubMed] [Google Scholar]
- 13.Godde, J. S., and A. P. Wolffe. 1995. Disruption of reconstituted nucleosomes. The effect of particle concentration, MgCl2 and KCl concentration, the histone tails, and temperature. J. Biol. Chem. 270:27399-27402. [DOI] [PubMed] [Google Scholar]
- 14.Groth, A., W. Rocha, A. Verreault, and G. Almouzni. 2007. Chromatin challenges during DNA replication and repair. Cell 128:721-733. [DOI] [PubMed] [Google Scholar]
- 15.Hanawalt, P., and G. Sivak. 1999. Transcription-coupled DNA repair, p. 169-179. In D. A. Karakaya (ed.), Advances in DNA damage and repair. Plenum Publishers, New York, NY.
- 16.Howe, L., and J. Ausio. 1998. Nucleosome translational position, not histone acetylation, determines TFIIIA binding to nucleosomal Xenopus laevis 5S rRNA genes. Mol. Cell. Biol. 18:1156-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huggins, C. F., D. R. Chafin, S. Aoyagi, L. A. Henricksen, R. A. Bambara, and J. J. Hayes. 2002. Flap endonuclease 1 efficiently cleaves base excision repair and DNA replication intermediates assembled into nucleosomes. Mol. Cell 10:1201-1211. [DOI] [PubMed] [Google Scholar]
- 18.Katafuchi, A., T. Nakano, A. Masaoka, H. Terato, S. Iwai, F. Hanaoka, and H. Ide. 2004. Differential specificity of human and Escherichia coli endonuclease III and VIII homologues for oxidative base lesions. J. Biol. Chem. 279:14464-14471. [DOI] [PubMed] [Google Scholar]
- 19.Kleczkowska, H. E., G. Marra, T. Lettieri, and J. Jiricny. 2001. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes Dev. 15:724-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kornberg, R. D., and Y. Lorch. 1999. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98:285-294. [DOI] [PubMed] [Google Scholar]
- 21.Lett, J. T. 1990. Damage to DNA and chromatin structure from ionizing radiations, and the radiation sensitivities of mammalian cells. Prog. Nucleic Acid Res. Mol. Biol. 39:305-352. [DOI] [PubMed] [Google Scholar]
- 22.Li, B., M. Carey, and J. L. Workman. 2007. The role of chromatin during transcription. Cell 128:707-719. [DOI] [PubMed] [Google Scholar]
- 23.Li, G., M. Levitus, C. Bustamante, and J. Widom. 2005. Rapid spontaneous accessibility of nucleosomal DNA. Nat. Struct. Mol. Biol. 12:46-53. [DOI] [PubMed] [Google Scholar]
- 24.Li, G., and J. Widom. 2004. Nucleosomes facilitate their own invasion. Nat. Struct. Mol. Biol. 11:763-769. [DOI] [PubMed] [Google Scholar]
- 25.Li, Q., and O. Wrange. 1995. Accessibility of a glucocorticoid response element in a nucleosome depends on its rotational positioning. Mol. Cell. Biol. 15:4375-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang, Q., and P. C. Dedon. 2001. Cu(II)/H2O2-induced DNA damage is enhanced by packaging of DNA as a nucleosome. Chem. Res. Toxicol. 14:416-422. [DOI] [PubMed] [Google Scholar]
- 27.Liu, X., S. Choudhury, and R. Roy. 2003. In vitro and in vivo dimerization of human endonuclease III stimulates its activity. J. Biol. Chem. 278:50061-50069. [DOI] [PubMed] [Google Scholar]
- 28.Lucchini, R., R. E. Wellinger, and J. M. Sogo. 2001. Nucleosome positioning at the replication fork. EMBO J. 20:7294-7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marenstein, D. R., M. K. Chan, A. Altamirano, A. K. Basu, R. J. Boorstein, R. P. Cunningham, and G. W. Teebor. 2003. Substrate specificity of human endonuclease III (hNTH1). Effect of human APE1 on hNTH1 activity. J. Biol. Chem. 278:9005-9012. [DOI] [PubMed] [Google Scholar]
- 30.Marenstein, D. R., M. T. Ocampo, M. K. Chan, A. Altamirano, A. K. Basu, R. J. Boorstein, R. P. Cunningham, and G. W. Teebor. 2001. Stimulation of human endonuclease III by Y box-binding protein 1 (DNA-binding protein B). Interaction between a base excision repair enzyme and a transcription factor. J. Biol. Chem. 276:21242-21249. [DOI] [PubMed] [Google Scholar]
- 31.Matsumoto, Y., and K. Kim. 1995. Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science 269:699-702. [DOI] [PubMed] [Google Scholar]
- 32.Nilsen, H., T. Lindahl, and A. Verreault. 2002. DNA base excision repair of uracil residues in reconstituted nucleosome core particles. EMBO J. 21:5943-5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Núñez, M. E., K. T. Noyes, and J. K. Barton. 2002. Oxidative charge transport through DNA in nucleosome core particles. Chem. Biol. 9:403-415. [DOI] [PubMed] [Google Scholar]
- 34.Ocampo-Hafalla, M. T., A. Altamirano, A. K. Basu, M. K. Chan, J. E. A. Ocampo, A. Cummings, Jr., R. J. Boorstein, R. P. Cunningham, and G. W. Teebor. 2006. Repair of thymine glycol by hNth1 and hNeil1 is modulated by base pairing and cis-trans epimerization. DNA Repair 5:444-454. [DOI] [PubMed] [Google Scholar]
- 35.Pascal, J. M., P. J. O'Brien, A. E. Tomkinson, and T. Ellenberger. 2004. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature 432:473-478. [DOI] [PubMed] [Google Scholar]
- 36.Pennings, S., G. Meersseman, and E. M. Bradbury. 1991. Mobility of positioned nucleosomes on 5S rDNA. J. Mol. Biol. 220:101-110. [DOI] [PubMed] [Google Scholar]
- 37.Peterson, C. L., and J. Cote. 2004. Cellular machineries for chromosomal DNA repair. Genes Dev. 18:602-616. [DOI] [PubMed] [Google Scholar]
- 38.Pfeifer, G. P. 1997. Formation and processing of UV photoproducts: effects of DNA sequence and chromatin environment. Photochem. Photobiol. 65:270-283. [DOI] [PubMed] [Google Scholar]
- 39.Polach, K. J., and J. Widom. 1995. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J. Mol. Biol. 254:130-149. [DOI] [PubMed] [Google Scholar]
- 40.Protacio, R. U., K. J. Polach, and J. Widom. 1997. Coupled-enzymatic assays for the rate and mechanism of DNA site exposure in a nucleosome. J. Mol. Biol. 274:708-721. [DOI] [PubMed] [Google Scholar]
- 41.Rydberg, B. 2001. Radiation-induced DNA damage and chromatin structure. Acta Oncol. 40:682-685. [DOI] [PubMed] [Google Scholar]
- 42.Simon, R. H., and G. Felsenfeld. 1979. A new procedure for purifying histone pairs H2A + H2B and H3 + H4 from chromatin using hydroxylapatite. Nucleic Acids Res. 6:689-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simpson, R. T., and P. Kunzler. 1979. Chromatin and core particles formed from the inner histones and synthetic polydeoxyribonucleotides of defined sequence. Nucleic Acids Res. 6:1387-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simpson, R. T., and D. W. Stafford. 1983. Structural features of a phased nucleosome core particle. Proc. Natl. Acad. Sci. USA 80:51-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Studitsky, V. M., W. Walter, M. Kireeva, M. Kashlev, and G. Felsenfeld. 2004. Chromatin remodeling by RNA polymerases. Trends Biochem. Sci. 29:127-135. [DOI] [PubMed] [Google Scholar]
- 46.Tomschik, M., H. Zheng, K. van Holde, J. Zlatanova, and S. H. Leuba. 2005. Fast, long-range, reversible conformational fluctuations in nucleosomes revealed by single-pair fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. USA 102:3278-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ura, K., J. J. Hayes, and A. P. Wolffe. 1995. A positive role for nucleosome mobility in the transcriptional activity of chromatin templates: restriction by linker histones. EMBO J. 14:3752-3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Hoffen, A., A. S. Balajee, A. A. van Zeeland, and L. H. Mullenders. 2003. Nucleotide excision repair and its interplay with transcription. Toxicology 193:79-90. [DOI] [PubMed] [Google Scholar]
- 49.van Holde, K. E. 1989. Chromatin. Springer Verlag, New York, NY.
- 50.Wallace, S. S. 2002. Biological consequences of free radical-damaged DNA bases. Free Radic. Biol. Med. 33:1-14. [DOI] [PubMed] [Google Scholar]
- 51.Wallace, S. S. 1998. Enzymatic processing of radiation-induced free radical damage in DNA. Radiat. Res. 150:S60-S79. [PubMed] [Google Scholar]
- 52.Wolffe, A. 1998. Chromatin structure and function, p. 240-341. Academic Press, San Diego, CA.