Abstract
Mice lacking the zinc finger transcription factor specificity protein 3 (Sp3) die prenatally in the C57BL/6 background. To elucidate the cause of mortality we analyzed the potential role of Sp3 in embryonic heart development. Sp3 null hearts display defective looping at embryonic day 10.5 (E10.5), and at E14.5 the Sp3 null mutants have developed a range of severe cardiac malformations. In an attempt to position Sp3 in the cardiac developmental hierarchy, we analyzed the expression patterns of >15 marker genes in Sp3 null hearts. Expression of cardiac ankyrin repeat protein (Carp) was downregulated prematurely after E12.5, while expression of the other marker genes was not affected. Chromatin immunoprecipitation analysis revealed that Sp3 is bound to the Carp promoter region in vivo. Microarray analysis indicates that small-molecule metabolism and cell-cell interactions are the most significantly affected biological processes in E12.5 Sp3 null myocardium. Since the epicardium showed distension from the myocardium, we studied expression of Wt1, a marker for epicardial cells. Wt1 expression was diminished in epicardium-derived cells in the myocardium of Sp3 null hearts. We conclude that Sp3 is required for normal cardiac development and suggest that it has a crucial role in myocardial differentiation.
Specificity protein 3 (Sp3) is a member of the Sp/KLF (specificity protein/Krüppel-like factor) transcription factor family, which is characterized by three conserved Cys2-His2 zinc fingers located in the C termini of the proteins (49). The three fingers form the DNA binding domain recognizing the GC (ggGGCGGgg) box and the related GT (ggGGTGTgg) box in the regulatory regions of many housekeeping and tissue-specific genes. The Sp subclass members Sp1, Sp3, and Sp4 bind with the same specificity and affinity to GC and GT boxes, and they all contain two glutamine-rich activation domains in their N termini. Sp1 and Sp3 are widely expressed, whereas Sp4 is most prominently expressed in neuronal tissues (7, 41). Sp1−/− embryos are severely growth retarded and die after embryonic day 10.5 (E10.5) (36). Although Sp3 is similar to Sp1 in terms of expression pattern and protein sequence, its biological function is different from that of Sp1. In a mixed C57BL/6 and 129/Ola genetic background, Sp3−/− mice invariably die immediately after birth (6). The immediate postpartum lethality could result from respiratory failure. However, only minor morphological abnormalities were observed in Sp3−/− lungs, and the expression of essential lung-specific genes, such as surfactant proteins, is unaltered (6). Sp3−/− fetuses have a defect in tooth development, reflected by the absence of ameloblast-specific transcripts, and their skeletal ossification is impaired (21). Furthermore, absence of Sp3 results in cell-autonomous differentiation defects in the erythroid and myeloid cell lineages, indicating that Sp3 also has a function in hematopoiesis (52). Sp4−/− mice initially have a reduced body weight and display an increased incidence of early postnatal mortality (20, 48), which is related to disturbed function of the cardiac conduction system (39).
We found that Sp3−/− fetuses in the C57BL/6 background display a high prenatal mortality rate. The majority of these fetuses die in utero before E18.5, suggesting that lung failure is not the main cause of the previously described immediate postnatal lethality of Sp3−/− fetuses (6). Congenital heart defects are a common cause of pre- and perinatal mortality. Early during embryogenesis, cells of the lateral mesoderm become committed to the cardiac fate and start to express cardiac-specific genes (16). The newly formed cardiac cells assemble into a beating linear heart tube that undergoes rightward looping. This process is essential to position the in- and outflow tracts in close proximity to the developing four heart chambers that acquire their own specific morphological characteristics. A complex transcriptional network controls these processes (12, 16).
To investigate the potential cause of the high prenatal mortality rate of Sp3−/− fetuses, we performed a detailed morphological study of heart development. This was combined with a study of the expression of over 15 cardiac marker genes in the Sp3 null hearts at different stages of development and with microarray analysis of gene expression at E12.5. Collectively, our results demonstrate that Sp3 is required for normal cardiac development and suggest a crucial role for Sp3 in myocardial differentiation, possibly resulting from disturbed cell-cell interactions.
MATERIALS AND METHODS
Mice.
Sp3+/− mice were bred for more than 10 generations with C57BL/6 mice. Embryos were derived from timed matings of Sp3+/− × Sp3+/− mice. Genotyping was performed by PCR (6). E9.5 to E14.5 embryos were immersion fixed in a 4% paraformaldehyde solution in phosphate-buffered saline at 4°C overnight, E16.5 fetuses were first perfused with the paraformaldehyde solution via the liver and then immersion fixed at 4°C overnight. The embryos were embedded in paraffin, and transverse serial sections (5 μm) were made.
In situ hybridization.
The probes for Tgfβ1, Tgfβ2, and Tgfβ3 have been described previously (37); the probe for Mlc2v was a gift from A. F. Moorman. Other probe sequences were generated via PCR and subsequently cloned using the pGEM-T-Easy vector system I A1360 (Promega, Madison, WI) and sequenced. Riboprobes were labeled with 35S-UTP (SJ603; 1,000 Ci/mmol) (Amersham, Little Chalfont, United Kingdom), and hybridization was performed as described previously (25). PCR primers used to generate the probes were as follows (s, sense; as, antisense): Anf, 5′-GACAGCAAACATCAGATCGT-3′ (s) and 5′-CTCTGGGCTCCAATCCTGTC-3′ (as) (461-bp fragment); Carp, 5′-AGGAGCTGGTAACAGGCAAA-3′ (s) and 5′-TTCAGGACATCTGCGTTTCC-3′ (as) (489-bp fragment); eHAND, 5′-TGCAACTACCCACTAGGATC-3′ (s) and 5′-TTCAGCAACGAATGGGAACG-3′ (as) (579-bp fragment); Gata4, 5′-AATGCCTGTGGCCTCTATCA-3′ (s) and 5′-CGCTGATTACGCGGTGATTA-3′ (as) (621-bp fragment); Gata5, 5′-GACTTTGCCTTCACCTCCT-3′ (s) and 5′-AGTCCTGCGTCTGTAAGCAA-3′ (as) (541-bp fragment); Irx4, 5′-ATGCTGGCAAAGACGACAAG-3′ (s) and 5′-GGTGGCCCAGGCCTGGTTCA-3′ (as) (623-bp fragment); Mlc2a, 5′-GCACAACGTGGCTCTTCTAA-3′ (s) and 5′-GTGGGTGATGATGTAGCAGA-3′ (as) (455-bp fragment); Sp3, 5′-TTGGCTTCTGCACAGTTAGG-3′ (s) and 5′-CATTGTCTGAGAACTTCCCG-3′ (570-bp fragment); and Tbx5, 5′-AAGACACCTTCTATCGCTCG-3′ (s) and 5′-TATTCTCACTCCACTCTGGC-3′ (as) (504-bp fragment).
Immunohistochemistry.
The following primary antibodies were used: actin HHF35 (Dako, Glostrup, Denmark); Mlc2a (a kind gift from S. W. Kubalak); and Nkx2.5 (sc-8697), Sp3 (sc-13018), Wt1 (sc-192), and E-cadherin (sc-7870) (all from Santa Cruz Biotechnology, Santa Cruz, CA). As secondary antibodies, we used biotinylated goat anti-rabbit BA-1000 for Mlc2a, Sp3, Wt1, and E-cadherin; biotinylated rabbit anti-goat PK-6105 (Vector Laboratories, Burlingame, CA) for Nkx2.5; and rabbit anti-mouse P0260 (Dako, Glostrup, Denmark) and goat anti-rabbit and rabbit peroxidase anti-peroxidase (Nordic Immunological Laboratories, Tilburg, The Netherlands) for actin. The Vectastain ABC kit HRP PK6100 (Vector Laboratories, Burlingame, CA) was used as the third step for biotinylated secondary antibodies. For visualization, sections were incubated with 400 μg/ml 3,3′ diaminobenzidine tetrahydrocholoride (D5637; Sigma-Aldrich, St. Louis, MO) in 0.05 mM Tris-maleate buffer, pH 7.6, and finally counterstained with Mayer's hematoxylin.
Morphometry and 3D reconstruction.
Myocardial sampling and volume estimation for 15 wild-type (WT) and 16 Sp3−/− hearts were performed as described previously (23). For the study of looping, micrographs were made of E10.5 Sp3+/+ and Sp3−/− hearts. The 3D reconstructions were made as described previously (29) using the AMIRA software package (Template Graphics Software, San Diego, CA). Statistical analysis was performed with an independent sample t test using the SPSS 11.0 software program (SPSS Inc, Chicago, IL).
Gel retardation analysis.
The sequences of the oligonucleotides used are as follows (the sense strand is given): C1, 5′-ACCCCTGCCCCCACCAGTGGC-3′; C2, 5′-TAAGAACCCCCACCCCACTTCA-3′; C3, 5′-TTCGCTCCACCCACGATGCGT-3′; C4, 5′-TTGGCTCCACCCATAAGAAGC-3′; C5, 5′-ACCTTTCCCCACCCAGGTGAT-3′; and Sp, 5′-TTATGGGCGGAGTTAGGGGCGGGACTAT-3′.
Samples were incubated for 30 min at room temperature, loaded on a 4% polyacrylamide-0.5× Tris-borate-EDTA gel, and run at 250 V for 2 h at room temperature. Antibodies against Sp1 and Sp3 were used at a 1:16 dilution in the binding reactions (6). Nuclear protein isolation was performed as described previously (1).
ChIP assays.
Hearts were isolated from E14.5 WT embryos and collected in Dulbecco's modified Eagle's medium containing 10% fetal calf serum, followed by homogenization with a 1-ml glass homogenizer (pestle B). Chromatin immunoprecipitation (ChIP) was performed as described previously (42) using either Sp3 (sc-644), Nkx2.5 (sc-8697), or preimmune immunoglobulin G (IgG) antibody (all from Santa Cruz Biotechnology, Santa Cruz, CA). PCR was performed on an MJ Research Opticon 2 PCR machine, using SYBR green to measure amplification products (Bio-Rad Laboratories, Hercules, CA). Primers were used to detect the upstream region of the Carp promoter (5′-ATCACCTGGGTGGGGAAAGGT-3′ and 5′-TTGGCTCCACCCATAAGAAGC-3′) or an area of the Gata2 gene that is not though to interact with either Sp3 or Nkx2.5 (5′-CCGGGCAGATAACGATTGG-3′ and 5′-TTCATCTCGGCCGGCTAAT-3′). The enrichment of Carp sequences was calculated relative to the amplification efficiency of the Gata2 sequences, normalized to the ratio observed before the ChIP reactions.
Quantitative RT-PCR analysis.
Total RNA was isolated from E14.5 WT and Sp3−/− hearts and subjected to quantitative reverse transcription-PCR (RT-PCR) analysis for the quantitation of gene expression (13). The primers used were 5′-CGACTCTTGATGACCTTCGG-3′ and 5′-ATTGCTTTGGTTCCACTCTG-3′ for Carp and 5′-TCACCATTTCCGACTGTGGAC-3′ and 5′-ACAGGACATTGCGAGCAGATG-3′ for cyclophilin A (Ppia), which was used as an internal standard to normalize for the amount of template used in the reactions.
Microarray analysis.
Hearts were dissected from E12.5 WT and Sp3−/− embryos, and RNA was isolated from individual hearts. Total RNA (5 μg) was used for labeling and hybridization to 430 2.0 Gene Chips (Affymetrix, Santa Clara, CA). Data extraction and normalization were done as described previously (51) using Affymetrix Gene Chip Operating Software (GCOS) version 1.4. The overall intensity value of each Gene Chip was scaled to an average value of 200 according to the method of global scaling provided by GCOS. Intensity values of between 0 and 30 were set to 30. An unsupervised correlation analysis using all the probe sets expressed above the detection level in at least one of the samples was performed with the OmniViz correlation tool (OmniViz, Maynard, MA); this measures correlation patterns by use of Pearson's correlation coefficient. We used robust multichip average normalization to obtain a list of differentially expressed probe sets (27), using a fold change of >1.5 and false discovery rate of 0.01 as criteria. This list was used for cluster analysis by standard hierarchical clustering methods with similarity measured by Euclidean distance, provided by OmniViz software. Potentially relevant biological processes were determined with Ingenuity Pathways Analysis software (Ingenuity, Redwood, CA). Functional annotation was manually curated using the Mouse Genome Informatics website from The Jackson Laboratory (Bar Harbor, ME) (http://www.informatics.jax.org); The Online Mendelian Inheritance in Man McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University (Baltimore, MD); the National Center for Biotechnology Information, National Library of Medicine (Bethesda, MD) (http://www.ncbi.nlm.nih.gov/omim/); and the Ensembl genome database at the European Bioinformatics Institute (Hinxton, United Kingdom) (http://www.ensembl.org/) (July 2007).
Microarray accession number.
Microarray data have been deposited at http://www.ncbi.nlm.nih.gov/geo (GSE9124).
RESULTS
Sp3−/− embryos display severe cardiac malformations.
To maintain the Sp3 knockout line in a defined genetic background, we backcrossed Sp3+/− animals with C57BL/6 animals for more than 10 generations. Timed matings of C57BL/6 Sp3+/− mice revealed that in this background the majority of Sp3−/− fetuses die before birth. To establish the approximate time of in utero mortality, we recovered embryos at different developmental time points. At E18.5, 1 out of 30 embryos (3%) was homozygous for the Sp3 knockout, significantly below the expected frequency of 25% (P < 0.01). In contrast, at E14.5 30/145 embryos (21%) were homozygous, close to the expected frequency. We conclude that the C57BL/6 Sp3−/− embryos die between E14.5 and E18.5 in utero.
To find developmental defects that might cause this late embryonic lethality, we studied the Sp3−/− embryos in more detail. In addition to the previously reported growth retardation of Sp3−/− embryos (6), we often observed nuchal edema in the mutants at E14.5 (Fig. 1a and d). This indicates that cardiac dysfunction may contribute to the prenatal mortality of the Sp3−/− embryos. Consistent with this notion, histological sections of E14.5 Sp3−/− hearts revealed an array of severe cardiac defects. In 15/17 Sp3−/− hearts the aorta and the pulmonary trunk were both positioned side by side (Fig. 1b and e) with the aorta connecting to the right ventricle (not shown). In all cases the arterial trunk had become septated, so no persistent truncus arteriosus was observed. Atrioventricular canal malformations were found in 11/17 cases showing a common atrioventricular valve, an atrioventricular septal defect (Fig. 1c, f, and g), or a straddling tricuspid valve (not shown).
FIG. 1.
Sp3−/− embryos display cardiac abnormalities. (a and d) The majority of the Sp3−/− embryos are edematous (arrows in panel d) at E14.5, which is suggestive of cardiac dysfunction. (b, c, and e to j) E14.5 sections of the heart stained for actin. In a WT heart (Sp3+/+) the pulmonary trunk (Pt) arises from the right ventricle (RV) (b), while in the mutant (e) the Pt and aorta (Ao) are side by side. At the level of the AVC, an atrioventricular septal defect (asterisk) occurs with a free hanging underrim of the atrial septum (AS) (f). The atrioventricular septal defect has a left dominance but is also connected to the RV, as is evident from a more posterior section of the same heart (g). In Sp3−/− hearts the myocardium is spongy with perforations (g and h) allowing for direct subendocardial-to-subepicardial contact, indicated by arrows (h). (i and j) Normal epicardial (EP) covering (i) compared to epicardial blebbing (EPB) in a mutant heart (j). LA, left atrium; LV, left ventricle; RA, right atrium. Bars; 100 μm (b, c, and e to h) and 50 μm (i and j).
In addition to these malformations, the structure of the myocardial wall was highly abnormal in Sp3−/− hearts at E12.5 and E14.5. Myocardial volume measurements showed a significant decrease in the volume of the mutant hearts; this was already observed at E10.5 (Table 1). The ventricular compact myocardium and the ventricular septum were hypoplastic and had a spongy structure (Fig. 1g and h) compared to those of WT littermates (Fig. 1c). Furthermore, we observed perforations of the atrial and ventricular myocardium in about 50% of the E14.5 Sp3−/− hearts (Fig. 1g to i). The perforations allowed for a direct contact of subendocardial and subepicardial layers. However, there was no pericardial hemorrhage, as the lining epicardium itself was intact.
TABLE 1.
Myocardial volumes of Sp3−/− hearts during development
| E | Myocardial vol (mm3)
|
t test P value | |||
|---|---|---|---|---|---|
| WT
|
Sp3−/−
|
||||
| Mean (n) | SD | Mean (n) | SD | ||
| 10.5 | 0.048 (5) | 0.0092 | 0.021 (5) | 0.0074 | 0.033 |
| 12.5 | 0.20 (5) | 0.032 | 0.14 (6) | 0.030 | 0.011 |
| 14.5 | 0.84 (5) | 0.098 | 0.37 (5) | 0.080 | 0.036 |
Morphological analysis of Sp3−/− hearts.
For the evaluation of cardiac morphology, we studied serial sections of E10.5 (n = 15), E12.5 (n = 12), and E14.5 (n = 17) Sp3−/− embryos and compared these to sections from WT littermates. We found that abnormal looping of the heart tube, with a wide inner curvature and separation of atrioventricular cushions (AVC) and outflow tract cushions, was already apparent at E10.5. Three-dimensional reconstructions of representative E10.5 hearts are shown in Fig. 2a to d.
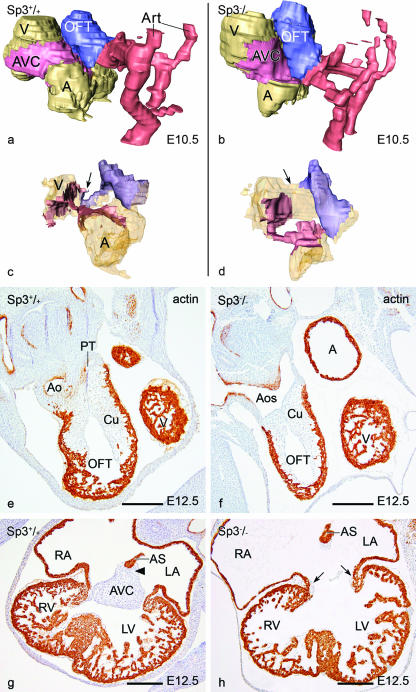
FIG. 2.
Morphological analysis of Sp3−/− embryonic hearts. (a to d) 3D reconstructions of a WT (a and c) and an Sp3−/− (b and d) embryonic heart at E10.5. Color codes: light brown, atria (A) and ventricles (V); pink, AVC; blue, outflow tract cushions (OFT); red, arteries (Art). The outer surface of the heart (a and b) shows the different relative positions of the cardiac segments in the Sp3−/− embryo. In the WT heart, the AVC and OFT cushions meet at the inner curvature (arrow in panel c), while in the knockout there is a wide inner curvature and separation of the cushions (arrow in panel d). (e to h) Sections of a WT (e and g) and an Sp3−/− (f and h) heart, stained for actin, show at E12.5 that in the Sp3−/− embryo the septation of the aortic sac (Aos) is not yet complete (f) compared to in the normal embryo (e), where the aorta (Ao) and pulmonary trunk (Pt) are already separated. At the atrioventricular canal level (g) in the WT, the AVC tissue is already fused with the cushion tissue (arrowhead) at the base of the atrial septum (AS). In the mutant heart (h), a common valve is shown. Small lateral cushions are present (arrows), and the cushion tissue at the base of the AS is not connected to the remaining AVC tissue (small patch in the middle). Bars, 200 μm.
At E12.5, we observed an incomplete outflow tract septation at the aortic sac level in 4/12 cases (Fig. 2e and f). In the remaining 8/12 cases, the aorta and pulmonary trunk were side by side, still abutting completely from the right ventricle. The atrioventricular canal region was still mainly connected to the left ventricle, while one case showed a common valve with no fusion of the AVC with the cushion ridge at the base of the atrial septum (Fig. 2g and h). Furthermore, the development of the ventricular septum was deficient (Fig. 2g and h).
Sp3 expression in the developing heart.
Since the expression pattern of Sp3 in the heart might provide clues to explain the phenotype of the Sp3 null mutation, we analyzed expression of the Sp3 protein during cardiac development. Nuclear staining of Sp3 was already detected at E9.5 in the myocardium and endocardium as well as in the cells of the endocardial cushion tissue (Fig. 3a to c). This persisted in later stages in the Anlage of the semilunar and atrioventricular valves (Fig. 3d, e, g, and h to j). A somewhat mosaic positivity of myocardial cells was observed in the atria and ventricles through E12.5 and E14.5 (Fig. 3i and k). At these stages there was a clearly marked expression in the neural crest-derived mesenchyme (Fig. 3h), the epicardium (Fig. 3i and j) and the smooth muscle cells of the vascular wall (Fig. 3d, e, and h). Thus, while there appears to be some variation in expression levels, we conclude that Sp3 is expressed ubiquitously in the developing heart between E9.5 and E14.5. The expression pattern of Sp3 therefore does not provide an indication of the cell types directly affected by Sp3 deficiency during heart development.
FIG. 3.
Sp3 expression during cardiac development. Sections show Sp3 expression (a and c to k) and actin (b) in E9.5 to E14.5 WT hearts. At E10.5 not only the myocardium but also the endocardium and the endocardial cushion cells (Cu) of the outflow tract (OFT) are positive. This persists in the OFT at E12.5 and E14.5 (asterisks in panels d and e) as well as in the AVC (g and j). The mostly positive myocardium shows a mosaic pattern of positive and negative cells in some regions (arrows in panel k). Furthermore, there is a marked expression in the epicardium (EP) (j), the neural crest-derived condensed mesenchyme (CM) (h), and the smooth muscle cells of the aorta (Ao) (e and h). LA, left atrium; LV, left ventricle; RA, right atrium; RV, right ventricle; V, ventricle. Bars, 100 μm (a to i) and 200 μm (j and k). The nuclear Sp3 staining is negative in Sp3−/− hearts (not shown).
Expression analysis of cardiac marker genes.
The cardiac phenotype of Sp3−/− embryos shows abnormalities in looping and remodeling of the heart tube and myocardial anomalies. We therefore analyzed the expression of a number of genes that are essential for myocardial differentiation during heart development. We compared the expression patterns observed in Sp3−/− hearts with those found in WT hearts at different developmental stages.
Nkx2.5 is the first marker of mesodermal cells fated to cardiac development (5), regulating the expression of many structural cardiac muscle genes (47). Gata4 was studied because of its close interactions with Nkx2.5 (34). Tbx5 is involved in atrial and ventricular septation and is a regulator of several cardiac-specific genes (8, 17). We used atrial natriuretic factor (Anf) (26) and cardiac ankyrin repeat protein (Carp; Ankrd1) (58) as markers for late differentiation-related cardiac genes. Anf and Carp are expressed in the proliferating and developing myocardium. To differentiate between atrial and ventricular specification, we used the iroquois homeobox gene 4 (Irx4). This transcription factor regulates the chamber-specific myosin isoforms by activating the ventricular myosin heavy-chain 1 gene (Vmhc1) and inhibiting the atrial myosin heavy-chain 1 (Amhc1) in the ventricles (2). Finally, we used the myosin light-chain 2 (Mlc2a) (32) and the ventricular light-chain 2 (Mlc2v) genes (40) as markers for atrial specification. The contribution of neural crest cells in myocardialization and outflow tract formation was studied using the expression of Msx2, which is a marker for neural crest-derived mesenchymal cells (10). Finally, we examined the expression of the growth factors transforming growth factor β1 (TGFβ1) to -3, since TGFβ2 is known to directly influence cardiac development (3, 45).
This analysis demonstrated that the expression of most of these genes did not appear to be different between WT and the Sp3−/− hearts at E10.5, E12.5, and E14.5. As an example, the expression pattern of the early cardiac gene Nkx2.5 at E12.5 and E14.5 is shown in Fig. 4a, b, e, and f. The only gene analyzed that showed a changed expression pattern was the Carp gene (Fig. 4c, d, g, and h). Here we found abnormal downregulation in Sp3−/− hearts after E12.5, specifically in the compact layer of the ventricular myocardium. At E14.5, Carp is still present in the right and left atria of Sp3−/− hearts, but expression of Carp is almost completely lost from the ventricular myocardium, with the exception of an area in the ventricular septum correlating with the position of the future ventricular conduction system. The aberrant downregulation of Carp expression after E12.5 occurs relatively late, when the Sp3−/− hearts already display severe abnormalities, and could be due to a secondary effect of the knockout phenotype. We therefore decided to investigate whether the Carp gene could be directly regulated by Sp3.
FIG. 4.
Expression of markers of cardiac development. In the myocardia of E12.5 and E14.5 Sp3−/− hearts (a, b, e, and f), Nkx2.5 remains expressed. Carp is downregulated after E12.5 in the absence of Sp3, as shown by in situ hybridization (c, d, g, and h). At E14.5, Carp expression is almost completely absent in the Sp3−/− heart. The remaining expression site at E14.5 correlates with the position of the future atrioventricular conduction system bridging the ventricular septum. Ao, aorta; LA, left atrium; RA, right atrium; RV, right ventricle. Bars, 100 μm.
Sp3 binds to the Carp promoter in vitro and in vivo.
To assess the potential regulation of the Carp gene by Sp3, we first studied the possible binding of Sp3 to the Carp promoter. We searched the 2.5-kb upstream promoter region of the Carp gene for the presence of GC and GT boxes. This region was previously found to be sufficient to drive correct Carp expression (33). We found that this regulatory region contains five putative Sp binding sites (Fig. 5a). To test whether these putative Sp binding sites are functional, we performed gel mobility shift assays. The results show that four of the five putative Sp binding sites are capable of binding Sp3 and Sp1 (Fig. 5b). We were interested to know if Sp3 interacts with this region of the Carp promoter in vivo. To investigate this, we performed ChIP reactions on formaldehyde-cross-linked chromatin isolated from E14.5 WT ventricles, using antibodies against Sp3, Nkx2.5, and a control preimmune IgG. We observed a seven- to eightfold increase in the amplification efficiency of the Carp fragments when the Sp3 and Nkx2.5 antibodies were used (Fig. 5c). In contrast, ChIP reactions with the control preimmune IgG did not result in any enrichment of Carp sequences (Fig. 5c). Finally, we used real time RT-PCR to quantitate Carp expression. As could be expected, we found that Carp expression is reduced, but not completely absent, in E14.5 Sp3−/− hearts (Fig. 5d). Collectively, our data strongly support the notion that Sp3 is a direct regulator of the Carp gene.
FIG. 5.
Sp3 binds to the mouse Carp promoter in vitro and in vivo. (a) The Carp promoter region. The five Sp motifs (Sp C1 to Sp C5) are indicated, as are the Nkx2.5 and GATA motifs. (b) Gel retardation assay. The specificity of binding to the Sp C1 to Sp C5 oligonucleotides was analyzed through the addition of a 50-fold molar excess of a competitor oligonucleotide containing two Sp sites (18). Supershifts were performed to analyze the protein complexes with antibodies against Sp1 or Sp3. The arrows point to the different protein complexes formed; note that there are several isoforms of Sp3 (46). (c) ChIP reactions were performed on formaldehyde-cross-linked chromatin isolated from E14.5 ventricles, using the antibodies indicated. Enrichment for Carp promoter sequences was determined by real-time quantitative PCR, using amplification of an unrelated fragment of the Gata2 gene as a standard to normalize for the amount of template DNA in the reactions. (d) Real-time PCR analysis of Carp mRNA expression in E14.5 hearts of WT and Sp3 knockout (Sp3−/−) embryos. Expression of cyclophilin A mRNA was used to normalize for the amount of RNA. Error bars indicate standard errors of the means. The average Carp expression level in WT hearts was set to 100.
Analysis of epicardial cells in Sp3−/− hearts.
The dysregulation of Carp expression occurs relatively late and is therefore not causally related to the myocardial defects displayed by Sp3−/− embryos. For proper differentiation and development, the myocardium needs signals from the epicardium and epicardium-derived cells (EPDCs) (19), which are formed by epithelial-to-mesenchymal transformation (EMT) (53). Since myocardial development was severely affected in Sp3−/− hearts, we examined the histology of the epicardium in more detail. We used expression of the Wilms' tumor suppressor gene (Wt1) as a marker for the epicardial layer and EPDCs (38) and found that there is still epicardial outgrowth in the mutant hearts (Fig. 6d, e, j, and k), similar to that observed in WT hearts (Fig. 6a, b, g, and h). However, already at E12.5 the epicardial layer is abnormally attached to the myocardium and forms bleb-like structures (Fig. 1b, e, i, and j). In the mutant hearts we found fewer Wt1-positive epicardial cells and EPDCs in the subepicardial and intramyocardial regions, compared to WT (Fig. 6g, h, j, and k). To assess whether EMT might be impaired in the EPDCs, we studied the expression of the cell adhesion molecule E-cadherin (Cdh1) (24). It has been suggested that Wt1 downregulates E-cadherin during normal EMT in human mammary epithelial cells (9). We found that E-cadherin expression was upregulated in Sp3−/− hearts compared to WT hearts (Fig. 6c, f, i, and l), suggesting that EMT of epicardial cells is impaired in Sp3−/− hearts.
FIG. 6.
Expression of epicardial markers in Sp3−/− hearts. Sections of E12.5 (a to f) and E14.5 (g to l) hearts stained for Wt1 and E-cadherin. AVC region of an E12.5 WT heart (a) with detail of the left ventricle (LV) (b) showing the epicardial (EP) covering and the EPDCs in the subepicardial layer (arrows). The latter are missing in the mutant (e) taken from a similar position in a heart (d) displaying a common atrioventricular orifice, a thin spongy myocardium, and a deficient ventricular septum (VS). At E14.5 Wt1-stained EPDCs (arrows in panel g) are missing around the developing coronary vascular system (j). In the compact layer of the myocardium, fewer Wt1-stained EPDCs (arrow) are found in the mutant (k) than in the WT heart (h). In the Sp3−/− hearts E-cadherin (f and l) is upregulated compared to that in WT hearts (c and i), where EPDCs (arrows) are also located in the subepicardial region. Ao, aorta; LA, left atrium; RV, right ventricle. Bars, 60 μm (a, d, g, and j) and 30 μm (b, c, e, f, h, i, k, and l).
Genome-wide expression analysis of Sp3−/− hearts.
Myocardial development is severely compromised in Sp3−/− embryos, but our in situ analysis of the expression of >15 marker genes of cardiac development provided surprisingly few clues on the genes involved. As an independent approach to find such genes, we performed a microarray analysis of E12.5 hearts. RNA was isolated from individual WT and Sp3−/− hearts, labeled, and used for hybridization to Affymetrix 430 2.0 Gene Chips. An unsupervised correlation analysis, which included all the genes expressed at above the detection level in at least one of the samples (15,506 probe sets), did not divide the WT and Sp3−/− samples into distinct subgroups (data not shown). This indicates that the number of genes displaying deregulated expression in Sp3−/− hearts is relatively small. To analyze this in more detail, we performed a supervised analysis designed to discover genes differentially expressed in the WT and Sp3−/− samples, using a >1.5-fold change in the mean expression level (WT versus Sp3−/−) and a false discovery rate of 0.01 as the cutoffs. This resulted in a list of 116 probe sets representing 103 genes, which clearly distinguished WT from Sp3−/− samples in a hierarchical cluster analysis (Fig. 7a). Of these 103 genes, 9 were upregulated and 94 (including Sp3) were downregulated in the Sp3−/− samples. We then used this gene list for pathway analysis, to identify biological processes that might be affected in the Sp3−/− hearts. We found that two broad categories are overrepresented (Fig. 7b). The first category, represented by 29/103 genes, is small-molecule metabolism (lipid, carbohydrate, amino acid, nucleotide, and redox) and includes a relatively large number of mitochondrial proteins (8/29). The relevance of this category is not clear at the moment, but it is possible that a general metabolic deficiency contributes to the Sp3−/− phenotype. The second category, represented by 45/103 genes, is cell-cell interactions. From this category, 27/45 genes are associated with the membrane, 11/45 with the extracellular compartment, and 7/45 with signal transduction. This is consistent with the notion that cell-cell interactions are affected in Sp3−/− myocardial development and that these abnormal interactions contribute to the phenotype. At present this list of 103 genes does not point to a known developmental pathway that might be disturbed in Sp3−/− embryos. However, it does provide leads for investigations aimed at unraveling the molecular mechanism underlying the severe cardiac phenotype of Sp3−/− embryos.
FIG. 7.
Gene expression profiling of E12.5 Sp3−/− hearts. RNA was isolated from E12.5 WT (n = 3) and Sp3−/− (n = 3) hearts, and gene expression profiles were determined using Affymetrix 430 2.0 Gene Chips. (a) Cluster analysis of the 116 probe sets representing 103 differentially expressed genes. Blue represents donwregulation and red upregulation in Sp3−/− hearts. (b) Biological processes represented by the 103 differentially expressed genes. ME, metabolism. Green, amino acid; yellow, carbohydrate; orange, lipid; purple, nucleotide; plum, redox; white, other. MB, membrane; ES, extracellular space; ST, signal transduction; NU, nuclear; MS, miscellaneous. Genes in gray boxes encode mitochondrial proteins. FC, change (n-fold). Blue and red are as defined for panel a.
DISCUSSION
Previous studies on Sp3−/− mice in a C57BL/6 and 129/Ola mixed genetic background failed to identify the cause of the observed perinatal mortality (6, 21). Our current results, obtained with the Sp3 null mutation in the C57BL/6 background, provide ample evidence that severe cardiac malformations are the primary cause of death. We show that Sp3 is expressed in all components of the heart, including the myocardium, the neural crest-derived condensed mesenchyme, the endocardium, the endocardial cushions, the epicardium, and EPDCs. Thus, several of the complex interactions between these components of the heart may be disturbed in Sp3-deficient embryos, in addition to cell-intrinsic defects that may occur in any of the cell lineages contributing to the developing heart. The analysis of the expression patterns of over 15 marker genes relevant for cardiac development and genome-wide expression profiling of E12.5 hearts provided limited insight in the molecular mechanism of the cardiac phenotype. The interpretation of the results will therefore be based primarily on morphological analysis of Sp3−/− hearts.
The early defect in cardiac looping, resulting in an abnormal wide inner curvature at E10.5, points at a myocardial problem that might relate to defective myocardial-epicardial interactions (19). Remodeling of the primitive heart through looping is absolutely necessary to properly establish the definitive atrioventricular and ventriculoarterial connections. The observed inflow and outflow abnormalities can occur as direct consequences of defective cardiac looping. Furthermore, the observed common atrioventricular valve and atrioventricular septal defects also suggest that the development and fusion of the atrioventricular canal are abnormal. As we did not observe cases with persistent truncus arteriosus, outflow tract septation has been accomplished normally, indicating that neural crest cell migration to this area has been achieved (55). This is supported by the normal Msx2 expression pattern in the outflow tract (10). Evidence for abnormal myocardial differentiation is obtained from the ventricular myocardium, which failed to form a consistent compact layer in the Sp3−/− hearts. Furthermore, the ventricular wall was very thin, presenting myocardial perforations and fistulae after E12.5 in the majority of cases. This phenomenon might be due to an abnormal contribution of EPDCs to the heart (15, 19, 28, 35).
Wt1 is expressed in the epicardium and temporarily in EPDCs after EMT (38). Comparing WT and Sp3−/− hearts, we found that epicardial outgrowth had taken place normally but that EMT and subsequent differentiation of EPDCs might be abnormal. This is suggested from diminished Wt1 expression in the epicardium and the EPDCs in the compact myocardium of the ventricular wall. Wt1 is thought to stimulate the formation of EPDCs by activation of EMT (38). Wt1−/− embryos show epicardial defects with myocardial hypoplasia (30), which may be caused by disturbed formation of the subepicardial mesenchymal cells (i.e., EPDCs) by impaired EMT (54). To further substantiate disturbed EMT in Sp3−/− hearts we have used the cell-adhesion molecule E-cadherin. This marker should be downregulated during normal EMT (9). We found that E-cadherin expression was upregulated in the epicardium of the Sp3−/− hearts. This is consistent with diminished EMT and EPDC formation. Deregulated E-cadherin expression might explain the abnormal morphology and blebbing of the epicardial surface as well. We postulate that an abnormal cross talk between myocardium and EPDCs contributes to the myocardial pathology in Sp3−/− hearts. The role of Sp3 in regulation of Wt1 expression has not yet been described; however, there is evidence for a role of Sp1 (highly homologous to Sp3) in downregulation of Wt1 (11). Sp3 is expressed in both EPDCs and myocardium, and we observe changes in the expression of both Wt1 (EPDC specific) and Carp (myocardium specific). Further studies are necessary to unravel whether this is a concomitant “dual hit” or whether the primary defect resides in either the myocardial or EPDC compartment.
Sp3−/− versus other knockout models.
Several mouse knockout models present with cardiac and myocardial phenotypes that are comparable to those displayed by Sp3−/− embryos. For instance, TGFβ2−/− embryos have malformations in the inflow and outflow tracts that are very similar to those observed in Sp3−/− embryos, but the severe thinning of the myocardium and the perforations are missing (3). Another example is provided by RxRα−/− embryos, which also show great variability in outflow and inflow malformations (22). Furthermore, RxRα−/− embryos display a serious epicardial insufficiency, leading to an abnormal cross talk of myocardium and epicardium and resulting in myocardial deficiency (28). Elevated levels of Mlc2a (14) and TGFβ2 (31) have been observed in RxRα−/− embryos. We did not detect changes in the expression of these genes in the Sp3−/− hearts. Remarkably, the RxRα−/− embryos display a dramatic downregulation of Carp expression at E10.5 (43). The timing of this aberrant downregulation of Carp is different from that in Sp3−/− hearts, where it sets in after E12.5. Thus, although the phenotypes of several previously described knockouts display similarities to the cardiac phenotype of the Sp3 null mutants, none of those appear to recapitulate the Sp3 knockout phenotype completely.
Sp3 deficiency affects expression of the Carp gene.
Carp transcription is mediated by the cooperative action of Gata4 and Nkx2.5 (33), which are not disturbed in their expression in the Sp3−/− heart. The specific loss of Carp expression in the ventricles can therefore not be directly attributed to these genes. The identification of four Sp binding sites in the 2.5-kb upstream regulating region of the Carp gene, a region essential for Carp expression (33), suggests that the Carp gene is a direct target of Sp3-mediated transcription. This is strongly supported by the ChIP assays demonstrating that Sp3 binds to this region of the Carp gene. Interestingly, we found that Nkx2.5 also interacts with this region in vivo. Thus, a model emerges in which Nkx2.5 and Gata4 initially activate the Carp gene. After this initial activation, Sp3 is required, most likely in conjunction with Nkx2.5 and Gata4, to maintain the appropriate expression pattern of Carp in the ventricles from E12.5 onwards. This is reminiscent of the role of the related family member Sp8 during early limb development, in which it is required to maintain, but not to initiate, Wnt/β-catenin-dependent fibroblast growth factor-, sonic hedgehog, and bone morphogenetic protein-mediated signaling (4, 50).
Defining the molecular role of Sp3 in heart development.
The observed dysregulation of Carp expression occurs at a developmental time point when the Sp3−/− embryos already display major myocardial defects. The genome-wide gene expression profiling of E12.5 Sp3−/− hearts shows that, despite the severe myocardial phenotype, the expression of a relatively small number of genes is affected. In a bird's eye view, these data suggest that impaired small-molecule metabolism and abnormal cell-cell interactions may contribute to the Sp3−/− myocardial phenotype. However, this analysis does not point to a specific developmental pathway that is disturbed in Sp3−/− embryos. Several of the 103 differentially expressed genes are currently associated with cardiovascular phenotypes. P2rx4 encodes the ATP-gated P2X4 ion channel; it has a key role in the response of endothelial cells to changes in blood flow (56). Evc encodes a protein that is mutated in Ellis-van Creveld syndrome (44). About 60% of patients with Ellis-van Creveld syndrome have congenital cardiac defects, usually manifest as a common atrium. Itg4a encodes integrin α4; Itg4a knockout mice fail to form an epicardium and display cardiac hemorrhage (57). These genes provide leads for future studies of the Sp3−/− heart phenotype aimed at accurate positioning of Sp3 in the pathway of cardiac development. Since Sp3 is widely expressed, the Sp3−/− phenotype could be the result of improper functioning and interaction of several cell types that interact with myocardial development. A conditional knockout strategy should help to reveal which cell types contribute to the spectrum of the Sp3−/− heart phenotype.
Acknowledgments
This work was supported by the Dutch scientific organization NWO (grants DN 82-94 and 901-08-092 to P.F.v.L. and S.P.).
We thank Jan Lens (LUMC Department of Anatomy and Embryology) for preparation of the figures and Wilfred van IJcken (Erasmus MC Department of Biomics) and Peter van der Spek (Erasmus MC Department of Bioinformatics) for microarray hybridizations and bioinformatics support, respectively.
Footnotes
Published ahead of print on 8 October 2007.
REFERENCES
- 1.Andrews, N. C., and D. V. Faller. 1991. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19:2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bao, Z. Z., B. G. Bruneau, J. G. Seidman, C. E. Seidman, and C. L. Cepko. 1999. Regulation of chamber-specific gene expression in the developing heart by Irx4. Science 283:1161-1164. [DOI] [PubMed] [Google Scholar]
- 3.Bartram, U., D. G. Molin, L. J. Wisse, A. Mohamad, L. P. Sanford, T. Doetschman, C. P. Speer, R. E. Poelmann, and A. C. Gittenberger-de Groot. 2001. Double-outlet right ventricle and overriding tricuspid valve reflect disturbances of looping, myocardialization, endocardial cushion differentiation, and apoptosis in TGF-beta(2)-knockout mice. Circulation 103:2745-2752. [DOI] [PubMed] [Google Scholar]
- 4.Bell, S. M., C. M. Schreiner, R. R. Waclaw, K. Campbell, S. S. Potter, and W. J. Scott. 2003. Sp8 is crucial for limb outgrowth and neuropore closure. Proc. Natl. Acad. Sci. USA 100:12195-12200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodmer, R. 1993. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development 118:719-729. [DOI] [PubMed] [Google Scholar]
- 6.Bouwman, P., H. Gollner, H. P. Elsasser, G. Eckhoff, A. Karis, F. Grosveld, S. Philipsen, and G. Suske. 2000. Transcription factor Sp3 is essential for post-natal survival and late tooth development. EMBO J. 19:655-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouwman, P., and S. Philipsen. 2002. Regulation of the activity of Sp1-related transcription factors. Mol. Cell Endocrinol. 195:27-38. [DOI] [PubMed] [Google Scholar]
- 8.Bruneau, B. G., G. Nemer, J. P. Schmitt, F. Charron, L. Robitaille, S. Caron, D. A. Conner, M. Gessler, M. Nemer, C. E. Seidman, and J. G. Seidman. 2001. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell 106:709-721. [DOI] [PubMed] [Google Scholar]
- 9.Burwell, E. A., G. P. McCarty, L. A. Simpson, K. A. Thompson, and D. M. Loeb. 2007. Isoforms of Wilms' tumor suppressor gene (WT1) have distinct effects on mammary epithelial cells. Oncogene 26:3423-3430. [DOI] [PubMed] [Google Scholar]
- 10.Chan-Thomas, P. S., R. P. Thompson, B. Robert, M. H. Yacoub, and P. J. Barton. 1993. Expression of homeobox genes Msx-1 (Hox-7) and Msx-2 (Hox-8) during cardiac development in the chick. Dev. Dyn. 197:203-216. [DOI] [PubMed] [Google Scholar]
- 11.Cohen, H. T., S. A. Bossone, G. Zhu, G. A. McDonald, and V. P. Sukhatme. 1997. Sp1 is a critical regulator of the Wilms' tumor-1 gene. J. Biol. Chem. 272:2901-2913. [DOI] [PubMed] [Google Scholar]
- 12.Cripps, R. M., and E. N. Olson. 2002. Control of cardiac development by an evolutionarily conserved transcriptional network. Dev. Biol. 246:14-28. [DOI] [PubMed] [Google Scholar]
- 13.Drissen, R., M. von Lindern, A. Kolbus, S. Driegen, P. Steinlein, H. Beug, F. Grosveld, and S. Philipsen. 2005. The erythroid phenotype of EKLF-null mice: defects in hemoglobin metabolism and membrane stability. Mol. Cell. Biol. 25:5205-5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dyson, E., H. M. Sucov, S. W. Kubalak, G. W. Schmid-Schonbein, F. A. DeLano, R. M. Evans, J. Ross, Jr., and K. R. Chien. 1995. Atrial-like phenotype is associated with embryonic ventricular failure in retinoid X receptor alpha−/− mice. Proc. Natl. Acad. Sci. USA 92:7386-7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eralp, I., H. Lie-Venema, M. C. DeRuiter, N. M. van den Akker, A. J. Bogers, M. M. Mentink, R. E. Poelmann, and A. C. Gittenberger-de Groot. 2005. Coronary artery and orifice development is associated with proper timing of epicardial outgrowth and correlated Fas-ligand-associated apoptosis patterns. Circ. Res. 96:526-534. [DOI] [PubMed] [Google Scholar]
- 16.Fishman, M. C., and K. R. Chien. 1997. Fashioning the vertebrate heart: earliest embryonic decisions. Development 124:2099-2117. [DOI] [PubMed] [Google Scholar]
- 17.Garg, V., I. S. Kathiriya, R. Barnes, M. K. Schluterman, I. N. King, C. A. Butler, C. R. Rothrock, R. S. Eapen, K. Hirayama-Yamada, K. Joo, R. Matsuoka, J. C. Cohen, and D. Srivastava. 2003. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 424:443-447. [DOI] [PubMed] [Google Scholar]
- 18.Gillemans, N., R. Tewari, F. Lindeboom, R. Rottier, T. de Wit, M. Wijgerde, F. Grosveld, and S. Philipsen. 1998. Altered DNA-binding specificity mutants of EKLF and Sp1 show that EKLF is an activator of the beta-globin locus control region in vivo. Genes Dev. 12:2863-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gittenberger-de Groot, A. C., M. P. Vrancken Peeters, M. Bergwerff, M. M. Mentink, and R. E. Poelmann. 2000. Epicardial outgrowth inhibition leads to compensatory mesothelial outflow tract collar and abnormal cardiac septation and coronary formation. Circ. Res. 87:969-971. [DOI] [PubMed] [Google Scholar]
- 20.Gollner, H., P. Bouwman, M. Mangold, A. Karis, H. Braun, I. Rohner, A. Del Rey, H. O. Besedovsky, A. Meinhardt, M. van den Broek, T. Cutforth, F. Grosveld, S. Philipsen, and G. Suske. 2001. Complex phenotype of mice homozygous for a null mutation in the Sp4 transcription factor gene. Genes Cells 6:689-697. [DOI] [PubMed] [Google Scholar]
- 21.Gollner, H., C. Dani, B. Phillips, S. Philipsen, and G. Suske. 2001. Impaired ossification in mice lacking the transcription factor Sp3. Mech. Dev. 106: 77-83. [DOI] [PubMed] [Google Scholar]
- 22.Gruber, P. J., S. W. Kubalak, T. Pexieder, H. M. Sucov, R. M. Evans, and K. R. Chien. 1996. RXR alpha deficiency confers genetic susceptibility for aortic sac, conotruncal, atrioventricular cushion, and ventricular muscle defects in mice. J. Clin. Investig. 98:1332-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gundersen, H. J., and E. B. Jensen. 1987. The efficiency of systematic sampling in stereology and its prediction. J. Microsc. 147:229-263. [DOI] [PubMed] [Google Scholar]
- 24.Hay, E. D. 2005. The mesenchymal cell, its role in the embryo, and the remarkable signaling mechanisms that create it. Dev. Dyn. 233:706-720. [DOI] [PubMed] [Google Scholar]
- 25.Hierck, B. P., R. E. Poelmann, L. van Iperen, A. Brouwer, and A. C. Gittenberger-de Groot. 1996. Differential expression of alpha-6 and other subunits of laminin binding integrins during development of the murine heart. Dev. Dyn. 206:100-111. [DOI] [PubMed] [Google Scholar]
- 26.Houweling, A. C., S. Somi, M. J. Van Den Hoff, A. F. Moorman, and V. M. Christoffels. 2002. Developmental pattern of ANF gene expression reveals a strict localization of cardiac chamber formation in chicken. Anat. Rec. 266:93-102. [DOI] [PubMed] [Google Scholar]
- 27.Irizarry, R. A., B. M. Bolstad, F. Collin, L. M. Cope, B. Hobbs, and T. P. Speed. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jenkins, S. J., D. R. Hutson, and S. W. Kubalak. 2005. Analysis of the proepicardium-epicardium transition during the malformation of the RXRalpha−/− epicardium. Dev. Dyn. 233:1091-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jongbloed, M. R., M. J. Schalij, R. E. Poelmann, N. A. Blom, M. L. Fekkes, Z. Wang, G. I. Fishman, and A. C. Gittenberger-De Groot. 2004. Embryonic conduction tissue: a spatial correlation with adult arrhythmogenic areas. J. Cardiovasc. Electrophysiol. 15:349-355. [DOI] [PubMed] [Google Scholar]
- 30.Kreidberg, J. A., H. Sariola, J. M. Loring, M. Maeda, J. Pelletier, D. Housman, and R. Jaenisch. 1993. WT-1 is required for early kidney development. Cell 74:679-691. [DOI] [PubMed] [Google Scholar]
- 31.Kubalak, S. W., D. R. Hutson, K. K. Scott, and R. A. Shannon. 2002. Elevated transforming growth factor beta2 enhances apoptosis and contributes to abnormal outflow tract and aortic sac development in retinoic X receptor alpha knockout embryos. Development 129:733-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kubalak, S. W., W. C. Miller-Hance, T. X. O'Brien, E. Dyson, and K. R. Chien. 1994. Chamber specification of atrial myosin light chain-2 expression precedes septation during murine cardiogenesis. J. Biol. Chem. 269:16961-16970. [PubMed] [Google Scholar]
- 33.Kuo, H., J. Chen, P. Ruiz-Lozano, Y. Zou, M. Nemer, and K. R. Chien. 1999. Control of segmental expression of the cardiac-restricted ankyrin repeat protein gene by distinct regulatory pathways in murine cardiogenesis. Development 126:4223-4234. [DOI] [PubMed] [Google Scholar]
- 34.Lien, C. L., C. Wu, B. Mercer, R. Webb, J. A. Richardson, and E. N. Olson. 1999. Control of early cardiac-specific transcription of Nkx2-5 by a GATA-dependent enhancer. Development 126:75-84. [DOI] [PubMed] [Google Scholar]
- 35.Lie-Venema, H., A. C. Gittenberger-de Groot, L. J. van Empel, M. J. Boot, H. Kerkdijk, E. de Kant, and M. C. DeRuiter. 2003. Ets-1 and Ets-2 transcription factors are essential for normal coronary and myocardial development in chicken embryos. Circ. Res. 92:749-756. [DOI] [PubMed] [Google Scholar]
- 36.Marin, M., A. Karis, P. Visser, F. Grosveld, and S. Philipsen. 1997. Transcription factor Sp1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell 89:619-628. [DOI] [PubMed] [Google Scholar]
- 37.Molin, D. G., U. Bartram, K. Van der Heiden, L. Van Iperen, C. P. Speer, B. P. Hierck, R. E. Poelmann, and A. C. Gittenberger-de-Groot. 2003. Expression patterns of Tgfbeta1-3 associate with myocardialisation of the outflow tract and the development of the epicardium and the fibrous heart skeleton. Dev. Dyn. 227:431-444. [DOI] [PubMed] [Google Scholar]
- 38.Moore, A. W., L. McInnes, J. Kreidberg, N. D. Hastie, and A. Schedl. 1999. YAC complementation shows a requirement for Wt1 in the development of epicardium, adrenal gland and throughout nephrogenesis. Development 126:1845-1857. [DOI] [PubMed] [Google Scholar]
- 39.Nguyen-Tran, V. T., S. W. Kubalak, S. Minamisawa, C. Fiset, K. C. Wollert, A. B. Brown, P. Ruiz-Lozano, S. Barrere-Lemaire, R. Kondo, L. W. Norman, R. G. Gourdie, M. M. Rahme, G. K. Feld, R. B. Clark, W. R. Giles, and K. R. Chien. 2000. A novel genetic pathway for sudden cardiac death via defects in the transition between ventricular and conduction system cell lineages. Cell 102:671-682. [DOI] [PubMed] [Google Scholar]
- 40.O'Brien, T. X., K. J. Lee, and K. R. Chien. 1993. Positional specification of ventricular myosin light chain 2 expression in the primitive murine heart tube. Proc. Natl. Acad. Sci. USA 90:5157-5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Philipsen, S., and G. Suske. 1999. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 27:2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez, P., E. Bonte, J. Krijgsveld, K. E. Kolodziej, B. Guyot, A. J. Heck, P. Vyas, E. de Boer, F. Grosveld, and J. Strouboulis. 2005. GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J. 24:2354-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruiz-Lozano, P., S. M. Smith, G. Perkins, S. W. Kubalak, G. R. Boss, H. M. Sucov, R. M. Evans, and K. R. Chien. 1998. Energy deprivation and a deficiency in downstream metabolic target genes during the onset of embryonic heart failure in RXRalpha−/− embryos. Development 125:533-544. [DOI] [PubMed] [Google Scholar]
- 44.Ruiz-Perez, V. L., S. E. Ide, T. M. Strom, B. Lorenz, D. Wilson, K. Woods, L. King, C. Francomano, P. Freisinger, S. Spranger, B. Marino, B. Dallapiccola, M. Wright, T. Meitinger, M. H. Polymeropoulos, and J. Goodship. 2000. Mutations in a new gene in Ellis-van Creveld syndrome and Weyers acrodental dysostosis. Nat. Genet. 24:283-286. [DOI] [PubMed] [Google Scholar]
- 45.Sanford, L. P., I. Ormsby, A. C. Gittenberger-de Groot, H. Sariola, R. Friedman, G. P. Boivin, E. L. Cardell, and T. Doetschman. 1997. TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development 124:2659-2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sapetschnig, A., F. Koch, G. Rischitor, T. Mennenga, and G. Suske. 2004. Complexity of translationally controlled transcription factor Sp3 isoform expression. J. Biol. Chem. 279:42095-42105. [DOI] [PubMed] [Google Scholar]
- 47.Small, E. M., and P. A. Krieg. 2004. Molecular regulation of cardiac chamber-specific gene expression. Trends Cardiovasc. Med. 14:13-18. [DOI] [PubMed] [Google Scholar]
- 48.Supp, D. M., D. P. Witte, W. W. Branford, E. P. Smith, and S. S. Potter. 1996. Sp4, a member of the Sp1-family of zinc finger transcription factors, is required for normal murine growth, viability, and male fertility. Dev. Biol. 176:284-299. [DOI] [PubMed] [Google Scholar]
- 49.Suske, G., E. Bruford, and S. Philipsen. 2005. Mammalian SP/KLF transcription factors: bring in the family. Genomics 85:551-556. [DOI] [PubMed] [Google Scholar]
- 50.Treichel, D., F. Schock, H. Jackle, P. Gruss, and A. Mansouri. 2003. mBtd is required to maintain signaling during murine limb development. Genes Dev. 17:2630-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Valk, P. J., R. G. Verhaak, M. A. Beijen, C. A. Erpelinck, S. Barjesteh van Waalwijk van Doorn-Khosrovani, J. M. Boer, H. B. Beverloo, M. J. Moorhouse, P. J. van der Spek, B. Lowenberg, and R. Delwel. 2004. Prognostically useful gene-expression profiles in acute myeloid leukemia. N. Engl. J. Med. 350:1617-1628. [DOI] [PubMed] [Google Scholar]
- 52.Van Loo, P. F., P. Bouwman, K. W. Ling, S. Middendorp, G. Suske, F. Grosveld, E. Dzierzak, S. Philipsen, and R. W. Hendriks. 2003. Impaired hematopoiesis in mice lacking the transcription factor Sp3. Blood 102:858-866. [DOI] [PubMed] [Google Scholar]
- 53.Vrancken Peeters, M. P., A. C. Gittenberger-de Groot, M. M. Mentink, and R. E. Poelmann. 1999. Smooth muscle cells and fibroblasts of the coronary arteries derive from epithelial-mesenchymal transformation of the epicardium. Anat. Embryol. (Berlin) 199:367-378. [DOI] [PubMed] [Google Scholar]
- 54.Wagner, N., K. D. Wagner, H. Scholz, K. M. Kirschner, and A. Schedl. 2006. Intermediate filament protein nestin is expressed in developing kidney and heart and might be regulated by the Wilms' tumor suppressor Wt1. Am. J. Physiol. Regul. Integr. Comp. Physiol. 291:R779-R787. [DOI] [PubMed] [Google Scholar]
- 55.Waldo, K., S. Miyagawa-Tomita, D. Kumiski, and M. L. Kirby. 1998. Cardiac neural crest cells provide new insight into septation of the cardiac outflow tract: aortic sac to ventricular septal closure. Dev. Biol. 196:129-144. [DOI] [PubMed] [Google Scholar]
- 56.Yamamoto, K., T. Sokabe, T. Matsumoto, K. Yoshimura, M. Shibata, N. Ohura, T. Fukuda, T. Sato, K. Sekine, S. Kato, M. Isshiki, T. Fujita, M. Kobayashi, K. Kawamura, H. Masuda, A. Kamiya, and J. Ando. 2006. Impaired flow-dependent control of vascular tone and remodeling in P2X4-deficient mice. Nat. Med. 12:133-137. [DOI] [PubMed] [Google Scholar]
- 57.Yang, J. T., H. Rayburn, and R. O. Hynes. 1995. Cell adhesion events mediated by alpha 4 integrins are essential in placental and cardiac development. Development 121:549-560. [DOI] [PubMed] [Google Scholar]
- 58.Zou, Y., S. Evans, J. Chen, H. C. Kuo, R. P. Harvey, and K. R. Chien. 1997. CARP, a cardiac ankyrin repeat protein, is downstream in the Nkx2-5 homeobox gene pathway. Development 124:793-804. [DOI] [PubMed] [Google Scholar]