Abstract
HMGN1, an abundant nucleosomal binding protein, can affect both the chromatin higher order structure and the modification of nucleosomal histones, but it alters the expression of only a subset of genes. We investigated specific gene targeting by HMGN1 in the context of estrogen induction of gene expression. Knockdown and overexpression experiments indicated that HMGN1 limits the induction of several estrogen-regulated genes, including TFF1 and FOS, which are induced by estrogen through entirely distinct mechanisms. HMGN1 specifically interacts with estrogen receptor α (ERα), both in vitro and in vivo. At the TFF1 promoter, estrogen increases HMGN1 association through recruitment by the ERα. HMGN1 S20E/S24E, although deficient in binding nucleosomal DNA, still interacts with ERα and, strikingly, still represses estrogen-driven activation of the TFF1 gene. On the FOS promoter, which lacks the ERα binding sites, constitutively bound serum response factor (SRF) mediates estrogen stimulation. HMGN1 also interacts specifically with SRF, but HMGN1 S20E/S24E does not. Consistent with the protein interactions, only wild-type HMGN1 significantly inhibits the estrogen-driven activation of the FOS gene. Mechanistically, the inhibition of estrogen induction of several ERα-associated genes, including TFF1, by HMGN1 correlates with decreased levels of acetylation of Lys9 on histone H3. Together, these findings indicate that HMGN1 regulates the expression of particular genes via specific protein-protein interactions with transcription factors at target gene regulatory regions.
The HMGN family of proteins modulates the chromatin structure in vertebrates. These small, highly charged, abundant proteins bind with higher affinity to nucleosomes than to double-stranded DNA. Nucleosomal binding is due to both direct interactions with nucleosomal DNA and interactions of HMGN proteins with histones (10, 18, 24, 46, 66, 71, 75). The binding of HMGN to chromatin relaxes the higher order chromatin structure. In particular, HMGN1 counteracts the ability of histone H1 to compact chromatin through competition for H1 binding sites on the chromatin. Such structural mechanisms result in the general transcription stimulation by HMGN proteins on chromatin templates in vitro (20, 74). The binding of HMGN proteins to nucleosomes also alters the posttranslational modification of nucleosomal histones at several distinct sites, which can result in either transcriptional stimulation or inhibition (42, 43, 77).
Despite the seemingly indiscriminate effects of HMGN on chromatin compactness and histone modifications, the overexpression or knockout of HMGN proteins modulates the expression of only a small subset of genes, some of which are activated and others are inhibited by HMGNs (42, 63, 79). These results raised the question as to why HMGN proteins would affect transcription only at specific promoters. HMGN proteins exhibit no nucleosome binding preference based on the underlying DNA sequences in the nucleosomes (72). Thus, it is unlikely that the targeting of HMGNs to gene regulatory regions can occur through the recognition of specific DNA sequences. Alternatively, the targeting of HMGN proteins could be mediated by specific protein-protein interactions (34). Consistent with such a hypothesis, HMGN1/2 exist in multiple complexes in HeLa cells (41); however, the prominently associated proteins in this case were hnRNPs and annexin II, which sheds little light on how the specificity of HMGN1/2 could be obtained at chromatin targets.
We have investigated the hypothesis that HMGN proteins are recruited to gene regulatory regions through direct interactions with specific DNA-binding transcription factors. A link between HMGN1 in particular and the estrogen receptor was suggested by early experiments in which HMGN1 was selectively modified in response to estrogen in the uteri of guinea pigs. This modification was connected to transcription activity, as an estrogen receptor antagonist prevented this acetylation of HMGN1 (5, 54).
Estrogen-regulated gene expression in breast cancer cells provides an extensively studied model system for examining multiple modes of gene induction (51, 65). First, in the canonical genomic estrogen response pathway, the association of estrogen with nuclear estrogen receptor α (ERα) induces its binding to estrogen response elements (EREs) within the regulatory regions of target genes. One well-characterized gene that is regulated in this manner is TFF1 (11, 47, 67, 68). Upon the stimulation of MCF-7 cells with estrogen, the binding of ERα to the ERE within the TFF1 promoter induces a cyclic recruitment to the promoter of an array of both positive transcription cofactors (including histone acetyltransferases [HATs], histone methyltransferases [HMTs], p68 RNA helicase, p160 coactivators, Mediator, and the SWI/SNF ATP-dependent nucleosome-remodeling complex) and negative transcription cofactors, including histone deacetylases (HDACs) and the AAA proteins independent of O0S (APIS) 19S proteasome subunit (47, 69). The concomitant cyclic changes in chromatin modification and organization of the nucleosomes on the promoter promotes transcription activation and at the same time allows the transcription rate of the gene to respond rapidly to different stimuli by restricting the duration of activation. Second, estrogen activates the transcription from gene regulatory regions through the protein-protein interaction between ERα and other promoter-bound DNA-binding transcription factors, such as Sp1 (64). In such cases, ERα need not interact directly with the DNA. This type of regulation has been suggested to occur on the IGFBP4 gene promoter (59). Indeed, Sp1 is critical for the induction of this gene by estrogen, although Sp1 binding to this promoter is at a very low constitutive level (44). Finally, gene expression is activated downstream of estrogen interaction with plasma membrane-associated ERα via “nongenomic” signal transduction pathways (40). The extracellular signal-regulated kinase pathway, one of the major targets of estrogen stimulation (9, 40, 61), impacts gene expression by multiple mechanisms, including rapid activation of the serum response factor (SRF)/Elk-1 complex. Consequences include stimulation of the transcription of FOS (22, 23), a highly characterized SRF target gene.
We have focused our study of the effects of HMGN1 on the induction of two estrogen-responsive genes, TFF1 and FOS, which are regulated through an ERE and a serum response element, respectively. Using these two particular genes, we tested the hypothesis that HMGN1 targets genes for regulation through interaction with DNA-binding transcription factors. We demonstrate, first, that HMGN1 inhibits the degree of estrogen induction at both genes. In addition, HMGN1 interacts specifically with both ERα and SRF, which regulate the responses of TFF1 and FOS to estrogen, respectively. Upon estrogen treatment, HMGN1 is recruited to the TFF1 gene regulatory region, but not to genomic regions lacking EREs, in parallel with the binding of ERα. Unexpectedly, although the regulation of the TFF1 gene expression by HMGN1 requires binding to specific transcription factors, it does not require high-affinity nucleosomal DNA-binding activity of HMGN1. Taken together, these results indicate that HMGN1 is targeted to specific gene regulatory regions through protein-protein interactions with transcription factors and that such interactions are required for HMGN1 to modulate transcriptional regulation. Regarding the mechanism of gene regulation, HMGN1 reduces the level of acetylation of Lys9 on histone H3 (AcLys9H3) at ERE-containing genes, such as TFF1, in which HMGN1 inhibits induction in response to estrogen. Thus, HMGN1 displays another means for negative feedback regulation to limit the extent of induction by estrogen, which is functionally analogous to, although mechanistically distinct from, the negative coregulators recruited to the promoter in an alternating manner with coactivators.
MATERIALS AND METHODS
Plasmid constructs.
Bacterial expression pGEX-2TK constructs expressing either wild-type or truncated HMGN1 proteins were constructed by subcloning PCR-amplified fragments of HMGN1 cDNA from pQE-HMGN1 (20) into BamHI and EcoRI sites of the pGEX-2TK vector (GE Healthcare, Piscataway, NJ). pGEX-4T-ERα and corresponding constructs expressing truncated ERα proteins were from R. Karas (45); pcDNA3.1-ERα was from M. Brown (26). Full-length wild-type and mutant HMGN1 cDNAs expressing either untagged or N-terminally Flag-tagged HMGN1 were cloned into the retroviral vector LZRSpBMN-linker-IRES-EGFP (31). The HMGN1 S20E/S24E cDNA was generated by site-directed mutagenesis. Retroviral small hairpin RNA (shRNA) constructs were generated by ligating annealed, double-stranded oligonucleotides into a pSIREN-RetroQ vector (Clontech, Mountain View, CA). Oligonucleotide sequences for HMGN1shRNA were GATCCGTCATTTGCTGGTTGTTTATTTCAAGAGAATAAACAACCAGCAAATGACTTTTTTGCTAGCG and AATTCGCTAGCAAAAAAGTCATTTGCTGGTTGTTTATTCTCTTGAAATAAACAACCAGCAAATGACG. The control shRNA against luciferase was provided by the manufacturer.
Antibodies.
Antipeptide HMGN1 antisera were generated separately against an N-terminal peptide (VSSAEGAAKEEPKC) and a C-terminal peptide (PASDEAGEKEAKSDC) of HMGN1. Peptides synthesized by the Tufts Medical School Core Facility (Boston, MA) were conjugated to maleimide-activated keyhole limpet hemocyanin (Pierce, Rockford, IL). Rabbit polyclonal antisera were generated by Covance Research Products, Inc. (Denver, PA). Antiserum was purified through a protein A column (GE Healthcare, Piscataway, NJ), followed by affinity purification using the respective peptide coupled to SulfoLink coupling gel (Pierce, Rockford, IL).
Antibodies for chromatin immunoprecipitation also included anti-ERα antibody (HC20), anti-SRF antibody (G-20) (Santa Cruz Biotechnology, Santa Cruz, CA), and anti-AcLys9H3 (Millipore, Billerica, MA). Immunoprecipitation and chromatin immunoprecipitation of Flag-tagged proteins were performed using anti-Flag-M2 beads (Sigma-Aldrich, St. Louis, MO). Antibodies for immunoblotting included anti-ERα (Ab15; Lab Vision, Fremont, CA) and anti-β-actin antibody (Sigma-Aldrich, St. Louis, MO). Anti-HMGN2 antibody was a gift from Michael Bustin.
Cell culture and retrovirus transduction.
MCF-7 cells were cultured in Dulbecco's modified essential medium (DMEM) with 2 mM l-glutamine and 10% fetal bovine serum (FBS) (HyClone, Logan, UT) at 37°C in 5% CO2. For estrogen stimulation experiments, MCF-7 cells were incubated in phenol red-free DMEM (Cellgro, Herndon, VA) with 10% charcoal-stripped FBS for two days prior to induction with 100 nM 17β-estradiol (Sigma-Aldrich, St. Louis, MO).
Retrovirus was produced in BOSC cells transfected by the LZRSpBMN-linker-IRES-EGFP (NotI-), the LZRSpBMN-linker-IRES-EGFP (NotI-)-HMGN1, or the pSIREN-RetroQ construct together with pVpack-VSV-G (Stratagene, La Jolla, CA) and pVpack-GP (Stratagene, La Jolla, CA) using Fugene 6 (Roche, Indianapolis, IN). Supernatants containing virus were collected at 48 and 72 h after transfection and were used to transduce MCF-7 cells in the presence of 4 μg/ml Polybrene (Sigma-Aldrich, St. Louis, MO). The transduction efficiency was greater than 90% judging from green fluorescent protein (GFP) expression. There were no effects observed for overexpression of either the wild-type or the mutant HMGN1 on cell growth or survival.
Transient and stable knockdown of HMGN1.
To transiently knockdown HMGN1, the small interfering RNA (siRNA) sequence CCAAGAAACUAAAGAAGAC was used. RNA oligonucleotides (Dharmacon, Lafayette, CO) were annealed to form double-stranded siRNA. Control siRNA oligonucleotides were obtained from Ambion (catalog no. AM4611) (Austin, TX). The siRNA oligonucleotides were transfected into MCF-7 cells in 24-well plates, using 1 μl of Lipofectamine 2000 (Invitrogen, Carlsbad, CA) and 50 pmol of siRNA oligonucleotides in serum-free DMEM. After 6 h of incubation, the medium was replaced, using phenol red-free DMEM plus 10% charcoal-stripped FBS.
To generate stable HMGN1 knockdown cell lines, MCF-7 cells were transduced with retroviral supernatants capable of expressing HMGN1 or control shRNA. Transduced cells were selected by the addition of 1 to 2 μg/ml puromycin in the culture medium for 1 week. Single clones were isolated by the limiting dilution method. Well contents initially containing only a single cell were propagated further and used in subsequent experiments.
Cell extracts.
Cells were washed with phosphate-buffered saline before cell extracts were prepared. TLB buffer lysates were prepared by lysing cells on ice for 1 h in 10 mM HEPES, pH 7.4, 137 mM NaCl, 1% Triton X-100 with Complete Miniprotease inhibitor cocktail tablets (Roche, Indianapolis, IN) and centrifuging at 20,800 × g for 20 min. Protein concentration was determined by the Bradford assay (Bio-Rad). TD buffer lysates were prepared with 50 mM HEPES, pH 7.4, 250 mM NaCl, 50 mM NaF, 5 mM EDTA, 1% Triton X-100 with Complete Miniprotease inhibitor cocktail tablets. Following Dounce homogenization and a 20-min incubation at 4°C, cell debris was removed by centrifugation. Before immunoprecipitation, the lysate was diluted onefold with 20% glycerol and 1% Triton X-100. Whole-cell sodium dodecyl sulfate (SDS) lysates were prepared by lysing cells in 50 mM Tris-Cl, pH 6.7, 2% SDS, 5% glycerol. Protein concentration was determined by bicinchoninic acid assay (Pierce, Rockford, IL).
Immunoblotting.
Following SDS-polyacrylamide gel electrophoresis (PAGE), gels were electrophoretically transferred to polyvinylidene fluoride membranes (GE Healthcare, Piscataway, NJ). Membranes were blocked in 5% nonfat dry milk in TBST buffer (10 mM Tris-Cl, pH 7.4, 150 mM NaCl, 0.1% Tween 10). Affinity-purified anti-HMGN1-N and anti-HMGN1-C antibody were each diluted 1:1,000; anti-ERα, anti-SRF, and anti-AcLys9H3 antibodies were diluted as recommended by the manufacturer. Following incubation with horseradish peroxidase-conjugated goat secondary antibodies (Bio-Rad), protein was visualized using enhanced chemiluminescent substrate (Pierce, Rockford, IL) and Biomax XAR film (Perkin Elmer, Waltham, MA).
In vitro protein-protein interactions.
ERα was translated in vitro in the presence of l-[35S]methionine (37 TBq/mmol; Perkin Elmer, Waltham, MA) with TNT Quick Coupled transcription/translation systems (Promega, Madison, WI), using pcDNA3.1-ERα as template.
Glutathione S transferase (GST)-HMGN1 fusion proteins were first purified using glutathione resin (GE Healthcare, Piscataway, NJ). For GST pulldown assays, 20 μg of purified GST-HMGN1 fusion protein was incubated with glutathione Sepharose 4B beads (GE Healthcare, Piscataway, NJ) in phosphate-buffered saline, 10% glycerol, 0.1% Triton X-100, followed by a 1-h incubation at 4°C with in vitro-translated ERα. Bound proteins were resolved by SDS-PAGE through an 8% gel and visualized by phosphorimaging (Storm system; GE Healthcare, Piscataway, NJ) and analysis with ImageQuant 5.2 software (GE Healthcare, Piscataway, NJ).
For GST pulldown assays using GST-ERα fusions, proteins expressed in 6 ml of bacterial culture were isolated on a glutathione resin, followed by incubation with 1 to 1.5 mg of MCF-7 TLB lysate overnight at 4°C. Bound proteins were resolved by SDS-PAGE through a 15% gel. HMGN1 proteins were detected by immunoblotting.
Coimmunoprecipitation experiments.
MCF-7 TD lysate was immunoprecipitated with 2.5 μg of a mixture of the two anti-HMGN1 antibodies or normal rabbit immunoglobulin G (Santa Cruz Biotechnology, Santa Cruz, CA). Immunocomplexes were collected on protein A beads (Upstate, Lake Placid, NY) and washed five times with 50 mM Tris-HCl, pH 7.4, 150 mM NaCl. Coimmunoprecipitation involving Flag-tagged HMGN1 protein was performed by using anti-Flag-M2 beads (Sigma-Aldrich, St. Louis, MO) and by washing with 50 mM HEPES, pH 7.5, 7 mM MgCl2, 2 mM EDTA. Proteins were resolved by SDS-PAGE through either 8% or 15% polyacrylamide gels for analysis of ERα or HMGN1, respectively.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation (ChIP) experiments were performed essentially as described previously (67). Chromatin-containing extracts from cross-linked MCF-7 cells (4 × 106 to 5 × 106 cells) sheared by sonication to a length of 100 to 500 bp were immunoprecipitated with their respective antibodies. Precipitated chromatin was eluted; cross-linking was reversed, and the DNA was purified using a gel extraction column (Qiagen, Valencia CA). Immunoprecipitated DNA was analyzed by real-time PCR using the TFF1 promoter P1 primers (CACCCCGTGAGCCACTGT and CTGCAGAAGTGATTCATAGTGAGAGAT) (14), the TFF1 coding region primers G1 (GTCACGCCCTCCCAGTGT and CGAACGGTGTCGTCGAAAC) and G2 (same as those used for mRNA primers, shown below), the FOS promoter primers P1 (CAGACACCCCCTTCAAATGTCT and AGATAAACACTGTGCAAAACCTACGT) and P2 (GCGAGCAGTTCCCGTCAAT and CCTAATATGGACATCCTGTGTAAGGG) (76), the FOS coding region G1 primers (GGACCTTATCTGTGCGTGAAACA and CCGGAAGAGGTAAGGACTTGAG), the IGFBP4 promoter primers (GGCGGGAGAGGTTGCAA and GGTAAAGCGTACAGGCACAGCTA), the IGFBP4 coding region primer (same as that used for the mRNA primers), the XBP1 primers as described previously (14), the XBP1 coding region primer (same as the heterogeneous nuclear RNA [hnRNA] primers), and the ACTB promoter primers P1 (CACTCCACCCAGTTCAACGTT and TGTTGATGAGGGCTGGTTCTG) and P2 (GACTCCCCCAACACCACACT and TCACTAGGGAGAGACCTGGGC).
RNA analysis.
Total RNA was prepared using Trizol (Invitrogen, Carlsbad, CA). One microgram of RNA was transcribed with Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA) and random hexamers (Applied Biosystems, Foster City, CA). The cDNA was subjected to triplicate real-time PCR analyses, using SYBR green PCR master mix (Applied Biosystems, Foster City, CA) and an ABI Prism 7900 sequence detection system (Applied Biosystems, Foster City, CA). All primer sets amplified linearly when characterized by serial dilution, and all generated unique products as determined by the dissociation curves. Primers included those for TFF1 hnRNA (TTGGAGAAGGAAGCTGGATGG and ACCACAATTCTGTCTTTCACGG), for TFF1 mRNA (ACCGGACACCTCAGACACG and CTGTGTTGTGAGCCGAGGC), for FOS mRNA, as described previously (76), for IGFBP4 mRNA (AACAACAGCTTCAGCCCCTGTA and CCATTGACCTTCATCTTGCCC), for XBP1 hnRNA (CAGTGTGGTATGTTCCTTCTCTTCC and GCTGCTACTCTGTTTTTCAGTTTCC), for GREB1 mRNA (CAGATGACAATGGCCACAATG and CAACACAATTCCCAGAGAAACCA), for MYC mRNA (CGAGACCTTCATCAAAAACATCAT and CCAGCTTCTCTGAGACGAGCTT), for CTSD hnRNA (GGTGCTCAAGAACTACATGGACG and TTGCAGCAGGGCTAAGACCT), and for 18S rRNA (GTAACCCGTTGAACCCCATT and CCATCCAATCGGTAGTAGCG).
RESULTS
HMGN1 inhibits estrogen-mediated transcriptional induction.
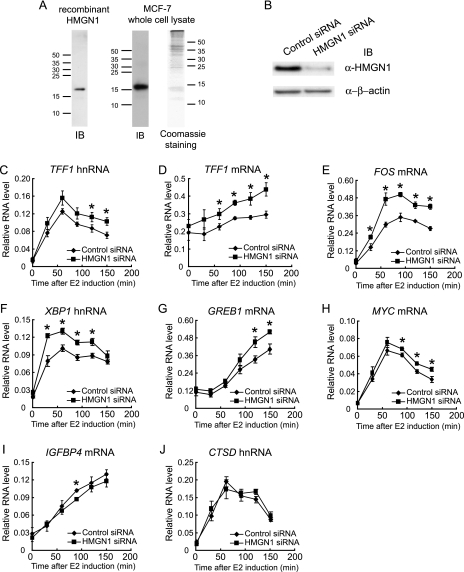
To test whether HMGN1 influences estrogen-mediated transcription, we used MCF-7 cells, an estrogen receptor-positive breast cancer cell line, as the model system. The role of HMGN1 was determined initially by diminishing the endogenous levels of HMGN1 with siRNA and examining the effect on estrogen-mediated transcriptional induction. In order to assay levels of HMGN1, we generated antipeptide antibodies against both the N- and the C-terminal peptides. These anti-peptide HMGN1 antibodies are highly specific for HMGN1, recognizing only a single band of endogenous HMGN1 in an MCF-7 whole-cell extract by immunoblotting, which migrates to the same position as that of recombinant HMGN1 (Fig. 1A). The expression of HMGN1 was then assayed by immunoblotting with a combination of both antibodies with MCF-7 cells transfected either with siRNA against HMGN1 or with a control siRNA. Immunoblotting confirmed that siRNA against HMGN1 knocked down the endogenous protein level by approximately 80% (Fig. 1B). To monitor the estrogen induction of gene expression, MCF-7 cells transfected with either control siRNA or HMGN1 siRNA were depleted of estrogen for 2 days, followed by the addition of 17β-estradiol. The RNA levels of several target genes, including the highly characterized TFF1 and FOS genes that are activated by distinct mechanisms, were monitored over a 2.5-h time frame. Although mRNA levels for most of these genes were rapidly induced by estrogen, this was not the case for mRNA levels for the TFF1, XBP1, and CTSD genes. Thus, for these three genes, the rapid induction of hnRNA was assayed in order to determine, in the most accurate way, the effect of HMGN1 on the rate of transcription. In all experiments, similar results were obtained with both hnRNA and mRNA; results of both for TFF1 are shown (Fig. 1C and D).
FIG. 1.
Knockdown of HMGN1 levels in MCF-7 cells enhances estrogen-mediated transcription of several but not all genes. (A) Characterization of anti-HMGN1 antibodies. Either recombinant HMGN1 or MCF-7 whole-cell SDS lysate was resolved by SDS-PAGE and immunoblotted (IB) with a mixture of antibodies against HMGN1 N-terminal and C-terminal peptides. Coomassie blue staining of the MCF-7 whole-cell extract is also shown. Migrations of molecular weight markers are indicated by tick marks at the side of each gel. (B) Immunoblotting of whole-cell lysates of MCF-7 cells transfected either with siRNA against HMGN1 or with control siRNA. (Upper panel) Immunoblotting with antibodies against HMGN1 (α-HMGN1). (Lower panel) Immunoblotting with antibodies against β-actin. (C to J) MCF-7 cells were transfected either with siRNA against HMGN1 (filled squares) or control siRNA (filled diamonds). After transfection, cells were cultured under estrogen-deprived conditions. The time courses of induction of TFF1 hnRNA, TFF1 mRNA, FOS mRNA, XBP1 hnRNA, GREB1 mRNA, MYC mRNA, IGFBP4 mRNA, and CTSD hnRNA are indicated following the addition of 100 nM 17β-estradiol. RNA levels were analyzed as described in Materials and Methods; RNA levels from each gene are expressed relative to levels of 18S rRNA in the same sample. Representative data from three independent experiments are shown, with experiments performed in triplicate. Error bars indicate standard deviations. Asterisks indicate points that are statistically significant by a two-tailed t test, assuming unequal variance (P < 0.05) between control siRNA versus HMGN1 siRNA samples.
Diminished levels of HMGN1 reproducibly enhanced the induction of TFF1 and FOS expression by estrogen but not that of either IGFBP4 or CTSD (Fig. 1, panels C to G and I to J). Although CTSD was considered to be another classical estrogen-responsive gene regulated by direct ERα binding to the promoter ERE (3, 49, 69), others reported that an Sp1 site was important (17, 36, 78), and recent genome-wide surveys of ERα binding sites in the genome failed to detect binding to the promoter-proximal EREs (15, 44). Thus, the mechanism of activation of CTSD by estrogen is questionable. Similar enhancements were also observed for the induction of other target genes, such as XBP1, GREB1, and MYC (Fig. 1F to H), for which the mechanisms of estrogen regulation have been less extensively studied. The regulation of XBP1 and GREB1 involves the long-range action of ERE-containing enhancer sequences (14, 15, 19, 73). In the case of MYC, a complex of β-catenin, an enhancer of zeste homolog 2 (EZH2), and ERα apparently mediates estrogen activation (70). The degree of increase in estrogen-induced expression of these genes varies from around 50% for FOS to 30% for TFF1, XBP1, and GREB1 to 20% for MYC. This degree of modulation of gene expression is comparable to effects observed for HMGN1 with other inducible genes (42, 43).
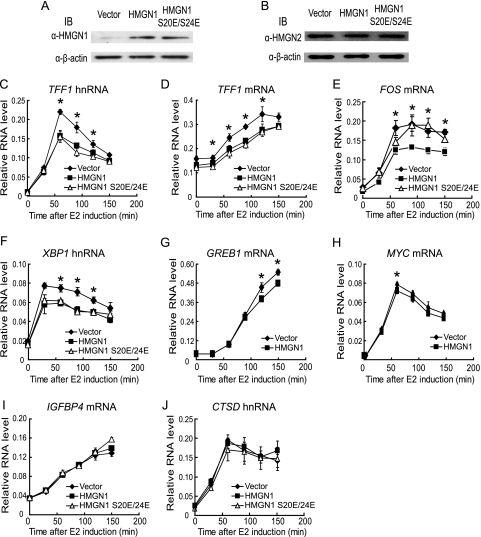
The impact of HMGN1 on estrogen-mediated transcription activation was further examined by overexpression in MCF-7 cells, comparing gene induction of the same panel of genes following infection with either a control retrovirus or a retrovirus expressing HMGN1. Exogenous HMGN1 was expressed at sevenfold-higher levels than endogenous protein (Fig. 2A), without observable changes in the levels of the paralog HMGN2 (Fig. 2B). Consistent with the siRNA studies, the overexpression of HMGN1 reproducibly reduced the estrogen-induced expression of TFF1, FOS, XBP1, GREB1, and MYC but not that of IGFBP4 or CTSD (Fig. 2, panels C to J). The extent of inhibition in different experiments ranged from 30 to 50% for TFF1, 30 to 35% for FOS and XBP1, and 10 to 20% for GREB1 and MYC. Although the effects of HMGN1 overexpression on the induction of GREB1 and MYC, in particular, were very modest, with fewer time points that were statistically different than that of the control, the trend toward decreased transcription activation by estrogen upon overexpression of HMGN1, even for these two genes, was in agreement with the increased induction upon knockdown of HMGN1. Together, these results demonstrated that HMGN1 inhibits several estrogen-induced genes in MCF-7 cells. However, the differing extents of modulation and the lack of impact on every gene that is transcriptionally induced by estrogen (i.e., IGFBP4 and CTSD) indicate that HMGN1 does not target the general transcription machinery but rather more specifically some component required for activation of these genes.
FIG. 2.
Overexpression of HMGN1 in MCF-7 cells represses estrogen-mediated transcription of several but not all genes. MCF-7 cells were transduced with retrovirus expressing the wild-type HMGN1, the HMGN1 S20E/S24E mutant, or the parental retroviral vector as a control. (A and B) Immunoblotting (IB) of whole-cell lysates of MCF-7 cells transduced with the indicated retroviruses. (Upper panel) Levels of HMGN1 (and HMGN1 S20E/S24E) (A) or HMGN2 (B) as monitored with antibodies against HMGN1 or HMGN2 (α-HMGN1 or α-HMGN2, respectively). (Lower panel) Levels of β-actin. (C to J) Transduced cells were deprived of estrogen and then subjected to 100 nM 17β-estradiol for up to 2.5 h. Time courses of induction of the indicated genes after stimulation are shown. Filled diamonds, RNA from control cells; filled squares, RNA from HMGN1-expressing cells; open triangles, RNA from HMGN1 S20E/S24E-expressing cells. Averages of data from experiments performed in triplicate are shown, which are representative of five independent experiments with HMGN1 and two independent experiments with HMGN1 S20E/S24E using MCF-7 cells transduced with different preparations of retroviruses. Error bars indicate standard deviations. Asterisks indicate points that are statistically significant by a two-tailed t test, assuming unequal variance (P < 0.05) between vector versus HMGN1 samples. Points that are statistically significant by t test between vector and HMGN1 S20E/24E samples are not shown on the graph, to prevent confusion, but include all the same time points as that between vector and HMGN1, plus the 30-min time point of XBP1 hnRNA.
High-affinity nucleosome-binding of HMGN1 is not required for the modulation of estrogen induction of TFF1.
The overexpression experiments afforded the opportunity to test the mutant HMGN1 proteins for their abilities to impact estrogen induction of gene expression. Of considerable interest were the consequences of the effects of HMGN1 S20E/S24E, assayed in parallel with wild-type HMGN1. The two substitution mutations lie within the nucleosomal DNA-binding domain (NDB) of HMGN1; reports indicate that the mutant is unable to bind nucleosomes in vitro and it exhibits a lower residence time on chromatin in vivo (56, 58). Furthermore, this mutant is unable to rescue a number of phenotypes associated with hmgn1−/− mouse embryonic fibroblasts (MEFs) (6, 7, 42, 43, 63), a finding which previously suggested that the two substitution mutations abolish the biological activity of HMGN1. Unexpectedly therefore, the effect of the HMGN1 S20E/S24E mutant upon estrogen stimulation differed depending on the target gene. Whereas the mutant was significantly reduced in its ability to inhibit the induction of FOS expression (Fig. 2E), it retained the ability to inhibit the induction of expression of TFF1 and XBP1 (Fig. 2C, D, and F). As with wild-type HMGN1, the induction of IGFBP4 and CTSD remained the same with or without the expression of HMGN1 S20E/S24E. Definitive results of the consequences of the S20E/24E mutations on the gene expression from GREB1 and MYC could not be obtained due to the modest effect of overexpression of wild-type HMGN1 on these two genes.
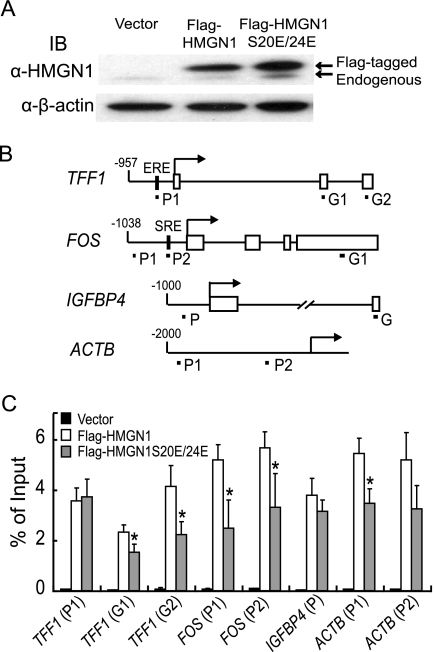
Given the remaining biological activity of HMGN1 S20E/S24E, particularly with the extensively studied TFF1 gene, we determined whether the mutant protein was indeed diminished in binding chromatin in vivo. Flag-tagged wild-type and mutant HMGN1 samples were overexpressed with MCF-7 cells to comparable levels (Fig. 3A); binding of the proteins to chromatin was then monitored by ChIP assays using antibodies against the Flag epitope. Binding was analyzed at representative estrogen-responsive genes that were affected differentially by HMGN1 and at the ACT8 gene, which is not an estrogen-responsive gene. (Fig. 3B). As expected, and consistent with previous fluorescence loss in photobleaching (FLIP) and fluorescence recovery after photobleaching (FRAP) data (56, 58), the mutant exhibited a general reduction (an average of twofold) in its occupancy on chromatin compared to that of wild-type HMGN1 at a number of different chromosomal locations (Fig. 3C). The remaining binding of the HMGN1 mutant to chromatin is more likely to result from the protein-protein interactions of HMGN1 with nucleosomal histones than from the binding of HMGN1 to nucleosomal DNA (see Discussion). Despite the general decrease in chromatin occupancy, similar levels of Flag-tagged wild-type and HMGN1 S20E/S24E were observed for the TFF1 promoter. (It should be noted that these experiments were performed using normal media, in which the estrogen signaling pathway is active. Under these conditions, ChIP assays demonstrate that ERα is associated with the TFF1 promoter [data not shown].) As discussed below, this suggested that HMGN1 can be targeted to the TFF1 promoter by a second mechanism, distinct from its NDB activity.
FIG. 3.
The S20E/S24E mutations diminish the association of HMGN1 at most chromosomal locations but not at the TFF1 promoter. (A) Immunoblotting (IB) of whole-cell SDS lysates from MCF-7 cells transduced with control retrovirus (vector) or with retroviruses expressing either Flag-tagged HMGN1 or Flag-tagged HMGN1 S20E/S24E. (Upper panel) Endogenous and Flag-tagged HMGN1 (wild-type or S20E/S24E mutant), as indicated, were detected with antibodies against HMGN1 (α-HMGN1). (Lower panel) Immunoblotting with antibodies against β-actin. (B) Locations of the primers for various promoter (P) or gene coding regions (G). Transcription start sites are indicated by arrows, exons are shown as open boxes, and primers are shown as short lines. The scale for each diagram is proportional to the length in nucleotides. (C) ChIP assay comparing the binding of HMGN1 versus that of HMGN1 S20E/S24E to different chromosomal regions. MCF-7 cells were transduced with control retrovirus (vector), Flag-HMGN1-expressing virus (white bars), or Flag-HMGN1 S20E/S24E-expressing virus (gray bars). Exponentially growing cells were treated with formaldehyde and harvested, and chromatin was immunoprecipitated with anti-Flag antibody coupled to beads. Locations of the primers are indicated in panel B. The binding level of protein to the indicated promoter or gene coding region is expressed as the percentage of DNA immunoprecipitated compared to the total amount of chromosomal DNA before immunoprecipitation. Error bars represents standard deviations from triplicate samples. Asterisks indicate values that were statistically different, as calculated by a two-tailed t test assuming unequal variance (P < 0.05).
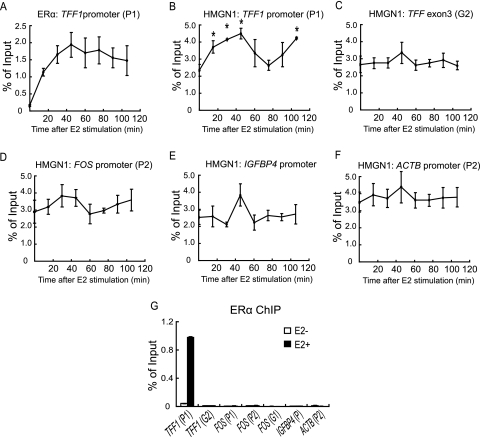
Recruitment of HMGN1 to the TFF1 promoter upon estrogen stimulation.
To investigate the mechanism whereby HMGN1 affects the induction of specific estrogen-regulated genes, we examined the binding of endogenous HMGN1 in vivo to promoters of a subset of estrogen-induced genes. Appropriate estrogen stimulation of the cells was verified by performing ChIP assays with antibodies against ERα. As expected, the binding of ERα to the TFF1 promoter was dramatically elevated by the hormone (Fig. 4A). The association of HMGN1 was tested using the anti-peptide HMGN1 antibodies characterized in Fig. 1A. A number of genomic locations were measured for HMGN1 association, within both the promoter and the coding regions of representative genes with differing responses to HMGN1 (Fig. 3B, locations of probes are indicated). As shown in Fig. 4B, HMGN1 binding to the TFF1 promoter was reproducibly and specifically elevated upon estrogen induction (Fig. 4B, asterisks indicate data points that are statistically different from that at time zero; P < 0.05). At 30 to 45 min, levels were increased twofold above the already high levels of occupancy, characteristic of this abundant nucleosome-binding protein. In contrast, binding was unaltered within the TFF1 coding region (Fig. 4C). Moreover, the level of HMGN1 at the TFF1 promoter did not remain constant after recruitment but rather decreased to the baseline level and then increased again (Fig. 4B). This cyclic binding of HMGN1 resembles that of a number of cofactors recruited to the TFF1 promoter after estrogen stimulation (47, 69). Although the trend was also toward enhanced binding on the FOS promoter, no statistically significant alteration in the level of HMGN1 was demonstrable (Fig. 4D). There was random variability but no statistically significant changes in the association of HMGN1 on either the IGFBP4 inducible promoter (Fig. 4E) or on the control β-actin promoter (Fig. 4F); analysis of variance of HMGN1 binding indicated no statistically significant changes in the level of binding over time except for that on the TFF1 promoter. This significant alteration in the level of HMGN1 only on the TFF1 promoter correlates with the robust, estrogen-mediated recruitment of ERα only to this promoter (Fig. 4G).
FIG. 4.
Association of HMGN1 with the TFF1 promoter increases upon estrogen induction. MCF-7 cells were cultured under estrogen-deprived conditions and then incubated with 17β-estradiol for up to 105 min. ChIP assays were performed using antibodies against either ERα (A and G) or HMGN1 (B to F). Input DNA and DNA isolated by the ChIP assays were quantified by real-time PCR. (A to F) The binding level of protein to the indicated promoter or gene coding region is expressed as indicated in the legend to Fig. 3C. Error bars represent standard errors of the means of three independent experiments. By analysis of variance, the association of HMGN1 only at the TFF1 promoter (P = 0.03) was statistically altered by the addition of estrogen. Pairwise t tests (two-tailed, assuming unequal variance) indicated the specific data points (indicated by asterisks) for which the level of HMGN1 binding was statistically different (P < 0.05) from that at the 0 time point. (G) Association of ERα to the indicated promoter or gene coding regions prior to (white bars) or after (black bars) a 45-min treatment with 17β-estradiol. The means from duplicate samples are shown; error bars represent standard deviations.
HMGN1 interacts with ERα both in vitro and in vivo.
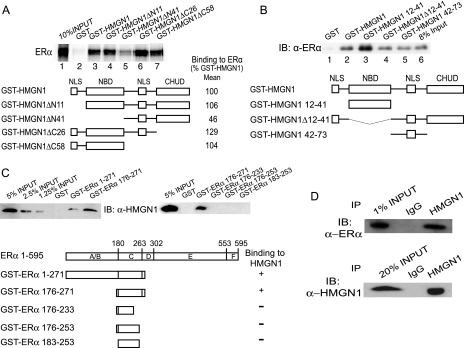
The increased binding of HMGN1 specifically at the TFF1 promoter suggested directed recruitment to this promoter, potentially through protein-protein interactions. Thus, we tested the hypothesis that HMGN1 interacts with ERα. In vitro protein-protein interaction studies with purified GST-HMGN1 and in vitro-translated ERα demonstrated such a specific interaction (Fig. 5A, compare lanes labeled GST versus GST-HMGN1). We mapped the region(s) of HMGN1 important for this interaction, using fusion proteins linking GST to different truncated portions of HMGN1. The only region whose removal diminished the degree of interaction in these studies was the NBD (Fig. 5A, compare GST-HMGN1 versus GST-HMGN1 ΔN41). These data suggested that a major source of the interaction lies within the NBD of HMGN1. Consistent with this interpretation, a GST fusion with HMGN2, another family member whose largest region of conservation with HMGN1 is the NBD, also interacted with ERα in the same assay (data not shown). In order to prove that the NBD is sufficient for interaction with ERα, we measured the ability of a purified GST fusion protein containing only the NBD (GST-HMGN1 12-41) to bind to purified, recombinant ERα. The association between ERα and the NBD of HMGN1 is at least as robust as its binding to full-length HMGN1 (Fig. 5B, lanes 1 to 3), confirming that the NBD is both sufficient for and a major source of the protein-protein interaction between HMGN1 and ERα. Importantly, the latter results also demonstrated that the interaction between HMGN1 and ERα is direct, since purified components were used in this experiment.
FIG. 5.
HMGN1 interacts with ERα both in vitro and in vivo. (A) GST pulldown assay between purified GST fusion proteins to wild-type or truncated HMGN1 and in vitro-translated 35S-labeled ERα (see Fig. S1 in the supplemental material for visualization of the GST fusion proteins). (Upper panel) Lanes 2 to 7, autoradiograph of the amount of ERα bound to the indicated GST fusion proteins; 1, amount of ERα in 10% of the input applied to each resin. (Lower panel) Schematic of the HMGN1 truncation proteins shown with the means of quantification of the data from two independent experiments; data are normalized to the amount of ERα binding to the wild-type HMGN1. (B) GST pulldown assay between purified GST fusion proteins to wild-type or truncated HMGN1 and purified recombinant ERα (see Fig. S1 in the supplemental material for visualization of the GST fusion proteins). (Upper panel) Lanes 1 to 5, immunoblot (IB) of the amount of ERα bound to the indicated GST fusion proteins; 6, amount of ERα in 8% of the input applied to each resin. α-ERα, anti-ERα antibody. (Lower panel) Schematic of the HMGN1 truncation proteins. (C) GST pulldown assay between GST fusion proteins to truncated ERα and MCF-7 cell extract (see Fig. S1 in the supplemental material for visualization of the GST fusion proteins). Bound proteins were immunoblotted for HMGN1. The leftmost three lanes and one lane in the left and right upper panels, respectively, contain the indicated percentages of the input extract that was used for each assay. The remaining lanes show the amounts of cellular HMGN1 bound to the indicated GST fusion proteins. (Lower panel) Schematic of the ERα truncations, along with a qualitative indication of whether the binding to HMGN1 was observed. Full-length ERα is drawn at the top, showing locations of domains A through F; domain C is the DNA-binding domain. (D) Coimmunoprecipitation assay between the endogenous HMGN1 and the endogenous ERα in MCF-7 cells. Cell lysate was immunoprecipitated with either anti-HMGN1 antibody or control immunoglobulin G. Immunoprecipitates were resolved by SDS-PAGE through either 8% or 15% gels, for the detection by immunoblotting of ERα (top panel) or HMGN1 (bottom panel), respectively. The leftmost lanes contained the indicated amounts of the extract used for the immunoprecipitation (IP).
However, binding of ERα to the initial truncation mutants of HMGN1 also suggested that regions other than the NBD of HMGN1 are capable of interacting with ERα (Fig. 5A). Also, the retention of purified ERα to resin containing the internal deletion mutant GST-HMGN1 Δ12-41, which specifically lacks the NBD, confirmed this interpretation (Fig. 5B). Moreover, upon expression in MCF-7 cells (in an experiment similar to that shown in Fig. 2), HMGN1 Δ12-41 retained the ability to specifically inhibit the induction of TFF1 expression in response to estrogen. This gene regulatory activity was observed even though HMGN1 Δ12-41 could not be highly expressed, maximal levels were comparable only to that of endogenous HMGN1 (data not shown). In vitro, this second ERα interaction region, although less robust than the NBD, was further mapped to residues 42 to 73 (Fig. 5B). This ability of HMGN1 to interact specifically with ERα through multiple regions prevents the generation of a stable mutant of HMGN1 that does not interact with ERα.
To map the region in ERα that is required for the interaction with HMGN1, reciprocal protein-protein interaction experiments were performed using GST-ERα truncation proteins combined with MCF-7 cell lysate as a source of HMGN1 (Fig. 5C). Again, a highly specific protein-protein interaction between HMGN1 and ERα was observed (Fig. 5C, left panel, compare GST versus GST-ERα 1-271). The truncation mutant of ERα containing only residues 176 to 271 was sufficient to specifically bind HMGN1, but further deletion into this region (i.e., GST-ERα 176-253 [Fig. 5C, right panel]) abolished the interaction between the two proteins. The region of ERα sufficient to interact with HMGN1 includes the double zinc finger DNA-binding domain (region C; residues 180 to 263), plus a few amino acids of flanking sequences.
Finally, to test whether endogenous HMGN1 and ERα proteins interact in vivo, MCF-7 cell extracts were immunoprecipitated using anti-HMGN1 antibody, and the complexes were immunoblotted for ERα. The significant amount of ERα coimmunoprecipitating with HMGN1 (Fig. 5D) verified the ability of these two proteins to associate in vivo.
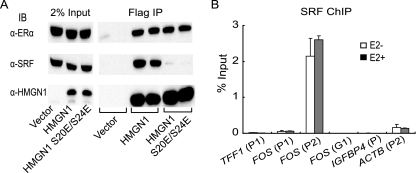
These interaction studies supported the supposition that the estrogen-inducible binding of ERα to the TFF1 promoter is what directly recruits HMGN1, leading to the moderation of gene induction. Since HMGN1 S20E/S24E also, surprisingly, fully repressed transcriptional induction of the TFF1 gene (Fig. 2C and D), even though it is deficient in binding nucleosomal DNA (58), we determined whether this double-amino-acid substitution mutant HMGN1 could still associate with ERα in cells. Flag-tagged HMGN1 S20E/S24E or wild-type HMGN1 was expressed with MCF-7 cells via retroviral transduction. The exogenous HMGN1 was then immunoprecipitated from cellular extract with anti-Flag antibody. As predicted, the amounts of ERα associating with the wild-type and the mutant HMGN1s were equivalent, as detected by immunoblotting (Fig. 6A, top panel). These data demonstrate that the S20E/S24E mutations do not significantly affect the interaction of HMGN1 with ERα.
FIG. 6.
S20E/S24E mutations in HMGN1 reduce specific interaction with SRF but not with ERα. (A) MCF-7 cells were transduced with control, parental retrovirus (vector), or retroviruses expressing either Flag-tagged HMGN1 or Flag-tagged HMGN1 S20E/S24E. TD buffer lysates were subjected to immunoprecipitation (IP) with anti-Flag M2 beads. Bound proteins were separated by SDS-PAGE before immunoblotting (IB) with antibodies against ERα (α-ERα) (upper panel), SRF (middle panel), or HMGN1 (bottom panel); the immunoprecipitations were performed in duplicate as indicated. In each case, the three lanes on the left contain 2% of the amount of input cell extracts used in the immunoprecipitation from each sets of transduced cells. (B) ChIP analysis for SRF binding to chromosomal sites in cells treated without (open bars) or with (dark gray bars) estrogen. MCF-7 cells were deprived of estrogen, and half were stimulated with 100 nM 17β-estradiol for 45 min. Both sets of cells were subjected to ChIP analysis with anti-SRF antibody. The binding level of the protein to the indicated promoter or gene region is expressed as indicated in the legend to Fig. 3C; the primers are shown in Fig. 3B. The means of duplicate samples are shown. Error bars represent standard deviations.
SRF interaction with HMGN1 is diminished by NBD mutations.
Although estrogen induction of the TFF1 gene is mediated by the binding of ERα to this promoter, estrogen induction of the FOS gene is instead mediated through SRF binding sites (22, 23). In order to test whether the ability of HMGN1 to moderate the induction of FOS gene expression is due to interactions with SRF, we first tested whether HMGN1 specifically associates with SRF in MCF-7 cells. Immunoprecipitates from cells exogenously expressing Flag-tagged wild-type or HMGN1 S20E/S24E were immunoblotted with antibodies against SRF (Fig. 6A, middle panel). Specific interaction between wild-type HMGN1 and SRF was, indeed, observed, consistent with the model. However, in contrast to ERα, the SRF-HMGN1 interaction was diminished by the HMGN1 S20E/S24E mutations (Fig. 6A, middle panel). Based on this result, the S20E/S24E mutations would be expected to be less efficient at inhibiting the estrogen induction of the FOS gene expression, which is what is observed (Fig. 2E).
Finally, if the HMGN1-SRF interaction mediates the transcriptional consequences of HMGN1 at the FOS promoter, then the association of HMGN1 with the promoter should mirror that of SRF. This was tested by measuring the binding of SRF to the FOS promoter by ChIP, in the absence and presence of estrogen treatment (Fig. 6B). In contrast to the inducible binding of ERα on the TFF1 promoter (Fig. 4A and G), SRF associates constitutively with the FOS promoter, as is also observed for other cell types (32). This is consistent with the constant association of HMGN1 with the FOS promoter (Fig. 4D), in contrast to its inducible association with the TFF1 promoter (Fig. 4B).
HMGN1 diminishes the level of histone H3 Lys9 acetylation on the TFF1 gene.
Several possible mechanisms could be envisioned to explain how HMGN1 might repress estrogen-mediated gene activation at specific target genes, such as the TFF1 gene: the abundant HMGN1 could decrease productive binding of ERα to the promoter through squelching, HMGN1 could alter higher order chromatin structure, or it could alter specific nucleosomal histone modifications. Since we have demonstrated protein-protein interactions between HMGN1 and ERα, we initially examined whether HMGN1 squelches ERα binding to the TFF1 promoter region, both in vitro and in vivo. Binding of ERα to a consensus ERE, as measured by electrophoretic mobility shift assay, was unaffected by increasing amounts of HMGN1, up to a 40-fold molar ratio (see Fig. S2A in the supplemental material). Similarly, the overexpression of either HMGN1 or HMGN1 S20E/24E did not alter the extent of binding of ERα to the TFF1 promoter, as measured by ChIP, assaying either the basal or the stimulated level of the ERα association (see Fig. S2B in the supplemental material). Therefore, squelching of ERα binding is not how HMGN1 inhibits induction of the TFF1 gene by estrogen.
Alterations in higher order chromatin structure by HMGN1 are mediated by its C-terminal chromatin unfolding domain. Thus, the involvement of higher order chromatin structure in repression by HMGN1 of estrogen-mediated gene activation was tested by monitoring the activity of HMGN1 ΔC26, which lacks this domain. This mutant robustly repressed the induction of TFF1 and FOS as effectively as the wild-type HMGN1 did (see Fig. S3 in the supplemental material), demonstrating that direct alterations in higher order chromatin structure by HMGN1 are not required.
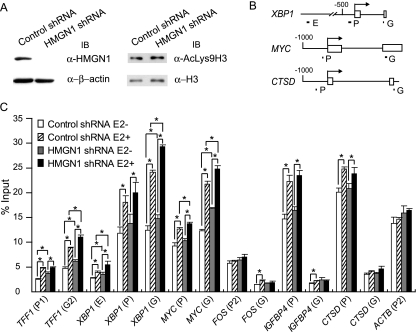
Finally, we investigated the consequences of the effects of HMGN1 on the levels of specific nucleosomal histone modifications at its target genes before and after estrogen induction. HMGN1 expression was previously linked to alterations in the overall cellular levels of several nucleosomal histone modifications (see Discussion). HMGN1 might repress gene induction by inhibiting the phosphorylation of Ser10 on histone H3 (pSer10H3) or AcLys9H3, both of which are associated with gene activation, or by enhancing the methylation of Lys9 on histone H3 (MeLys9H3), which is associated with gene repression. In order to measure histone modification states at specific genes, we established MCF-7 cell lines stably expressing either shRNA against HMGN1 or control shRNA. The expression of HMGN1 is dramatically diminished in these lines (Fig. 7A, left panel), without reproducible effects on the expression levels of HMGN2 (data not shown). In a comparison of cell lines with or without diminished HMGN1 expression, we were unable to observe a change in the levels of pSer10H3 or of MeLys9H3 at specific gene sites, as monitored by ChIP (data not shown). The total cellular levels of AcLys9H3 were also unaltered (Fig. 7A, right panel). However, upon examination of the level of AcLys9H3 associated with specific genomic regions, differences were observed. A previous study showed that the amount of AcLys9H3 increases on the TFF1 promoter upon estrogen stimulation (28). In both the control and the HMGN1 knockdown MCF-7 cell lines, lysine 9 acetylation on histone H3 also increased 1.3- to 2-fold at several inducible promoters with estrogen treatment, including the TFF1, XBP1, MYC, IGFBP4, and CTSD genes, but not at the FOS or ACTB promoters (Fig. 7C). Increases in acetylation were observed not only at the promoters listed above but also at the XBP1 enhancer containing the major ERα binding site and within the coding regions for the TFF1, XBP1, and MYC genes.
FIG. 7.
Loss of HMGN1 enhances the acetylation of histone H3 Lys9 at specific genes. (A) Immunoblotting (IB) of whole-cell lysates of a stable HMGN1 shRNA knockdown cell line or a control shRNA cell line (see Materials and Methods). (Upper left panel) Levels of HMGN1; (lower left panel) levels of β-actin; (upper right panel) levels of acetylated Lys9 H3; (lower right panel) levels of histone H3. α-HMGN1, anti-HMGN1 antibody. (B) Locations of the primers for various enhancer (E), promoter (P), or gene coding regions (G) are shown, as described in the legend to Fig. 3B. (C) ChIP assay of stable knockdown cell lines performed using anti-AcLys9H3 antibody. Similar results were observed for two independent HMGN1 stable knockdown cell lines; results from one comparison are shown here. The level of AcLys9H3 on the indicated enhancer, promoter, or gene region is expressed as indicated in the legend to Fig. 3C. The means of triplicate samples are shown. Error bars represent standard deviations. Asterisks show the values that are statistically significantly different by a two-tailed t test assuming unequal variance (P < 0.05) (values being compared are indicated by the brackets).
In the absence of HMGN1, the basal levels of AcLys9H3 (after the incubation of cells for 2 days in the absence of estrogen; Fig. 7C, compare open and gray bars) showed statistically significant, albeit subtle, increases only at the TFF1 promoter and its coding regions (30 and 50% increases, respectively), the XBP1 enhancer and its coding region (30 and 20% increases, respectively), and the MYC coding region (30% increase); these are the genes whose induction is inhibited by HMGN1. In addition, in the absence of HMGN1, there were increases in the stimulated levels of AcLys9H3 (after the addition of estrogen) (Fig. 7C, compare slashed and black bars) in the TFF1 coding region (25%), the XBP1 enhancer and the coding region (40 and 20%, respectively), and the MYC promoter and the coding regions (10 and 14%). In contrast, no differences in the levels of acetylation at Lys9 of histone H3 were observed between the control and the HMGN1 knockdown cells at the IGFBP4 or CTSD promoter, both of which responded to estrogen with enhanced levels of this histone modification. This result is consistent with the enhancement in the transcriptional induction of TFF1, but not of either IGFBP4 or CTSD, in the HMGN1 knockdown cells versus the control cells. These results, taken together, indicate that the acetylation of Lys9 on histone H3 correlates fully with HMGN1-mediated inhibition of the induction of genes via the canonical ERα-ERE activation pathway (TFF1 and XBP1), as well as with specific other pathways involving ERα recruitment in the absence of an ERE (e.g., MYC).
On the FOS gene, there was no appreciable modulation of AcLys9H3 levels in response to estrogen induction. Furthermore, the reduction of HMGN1 did not alter the extent of this modification on FOS. Thus, estrogen induction of FOS expression was inhibited by HMGN1 without the involvement of the AcLys9H3 modification state. This is not surprising, as regulation of FOS by HMGN1 occurs even in the absence of estrogen signaling; reduction in cellular HMGN1 levels also enhances the extent of the stimulation of FOS expression upon growth factor stimulation of resting T98G glioblastoma cells (data not shown). We propose that HMGN1 reduces the recruitment of distinct cofactors by SRF, which may be required either for the formation of transcriptional competent complexes at the FOS gene or for distinct histone modifications.
DISCUSSION
Given the ubiquitous presence, abundance, and lack of affinity for specific DNA sequences of HMGN1, it was initially surprising that its effect on transcription is limited to a small subset of genes (43, 63, 79). Our studies regarding the effects of HMGN1 on the estrogen-mediated induction of gene expression provide the first molecular basis for understanding such gene-specific effects, that HMGN1 is recruited to particular gene regulatory regions through specific protein-protein interactions with DNA-binding transcription factors. In support of this model, we demonstrated the following. First, HMGN1 inhibits estrogen-mediated transcriptional activation of genes (e.g., TFF1 and FOS) through a specific mechanism, as it does not influence estrogen induction of other genes (e.g., IGFBP4 and CTSD). Second, HMGN1 specifically interacts with the DNA-binding transcription factors responsible for estrogen induction at its target promoters (ERα and SRF). Third, the association of HMGN1 with these promoter regions mirrors that of the respective transcription factor, ERα or SRF. Fourth, the ability of HMGN1 to alter the regulation of a specific target gene depends on its ability to interact with the respective transcription factor. These data also provide the first demonstration that HMGN1 proteins lacking intrinsic nucleosomal DNA-binding activity retain biological phenotypes. All previous studies have linked the function of HMGN1 with its ability to bind nucleosomal DNA.
Target gene specificity of HMGN1.
Previous studies with various tissues or cell types have shown that HMGN proteins regulate mRNA levels of only a small fraction of expressed genes. In monitoring the HMGN effects in a rapidly regulated system, the transcriptional induction of genes by estrogen in breast cancer cells, we demonstrated similarly that the regulation of some genes, including TFF1, XBP1, and FOS, is reciprocally affected by the knockdown versus the overexpression of HMGN1, whereas the regulation of other genes, such as IGFBP4 and CTSD, remains unaffected. Such specificity is also consistent with previous studies of inducible, immediate-early gene expression in hmgn1−/− MEFs treated with anisomycin, in which the induction of only 4 out of 18 genes was affected by HMGN1 (42). However, in all previous studies, the molecular cause of the target gene specificity was not identified.
The binding of HMGN1 to a variety of chromosomal loci in multiple cell types has been assayed by chromatin immunoprecipitation. Such studies uniformly demonstrate both an overall association of the highly abundant HMGN1 with all chromatin, as well as its enrichment at specific chromatin locations (8, 27). Upon induction of immediate-early genes with stress stimuli, levels of HMGN1 specifically at the FosB promoter diminished rapidly. This is apparently due to the phosphorylation of HMGN1 associated with the promoter, which reduces HMGN1's affinity to nucleosomes in general (42). We also observed HMGN1 at basal levels at a number of chromosomal regions in MCF-7 cells. More importantly, we demonstrated for the first time the rapid, specific recruitment of HMGN1 to a regulatory region in response to a stimulus, the TFF1 promoter, mirroring the specific recruitment of ERα to this location. Even more strikingly, the HMGN1 S20E/S24E mutant, which has been demonstrated to lack nucleosomal DNA-binding ability in vitro (35) and which shows diminished occupancy on chromatin at most genomic locations (Fig. 3C), nonetheless retained its association with the TFF1 promoter and was also fully competent in inhibiting TFF1 transcriptional induction. As we have shown here, a plausible explanation for this apparent paradox is that HMGN1 S20E/S24E can be recruited to the TFF1 promoter through protein-protein interaction with ERα (see Discussion below). The residual interaction with chromatin in general is likely to be due to interactions of the N- and C-terminal regions of HMGN1 with nucleosomal histones (75), which should be retained in the mutant HMGN1. The functionality of a nucleosomal DNA-binding HMGN1 mutant is unprecedented and argues strongly for the ability of HMGN1 to mediate biological effects independent of its normal interaction with nucleosomal DNA, although perhaps still through other interactions with nucleosomes.
Specific protein-protein interactions between HMGN1 and DNA-binding transcription factors.
The only previous report of a specific interaction between HMGN proteins and the transcription factors was the initial identification of one member of the HMGN family of proteins, HMGN3a (Trip 7), through its interaction with thyroid hormone receptor in a yeast two-hybrid screen (37). HMGN3 augments transcriptional activation by TR-RXR heterodimers on reporter gene constructs in transient transfection assays both in Xenopus laevis (2) and in mammalian cells (H.-F. Ding and U. Hansen, unpublished observations). However, the functional consequences of this interaction on endogenous genes have remained elusive. In support of the hypothesis that specific protein-protein interactions drive the specificity of HMGN effects on gene expression, we demonstrated that HMGN1 and the transcription factor ERα directly interact in vitro and that these two proteins form, at endogenous levels, a complex in vivo. Furthermore, both the HMGN1 S20E/S24E NBD mutant and the HMGN1 Δ12-41 mutant, which deletes the entire NBD but still interacts with ERα, are fully competent to inhibit estrogen-mediated induction of TFF1 and other genes whose induction is mediated by ERα binding directly to an ERE. Unfortunately, given that two distinct regions in HMGN1 can interact independently with ERα, it is not possible to construct a stable HMGN1 mutant that no longer binds ERα. Nonetheless, these results support the conclusion that the functional consequences of HMGN1 at TFF1 and the genes similarly regulated are mediated by the HMGN1-ERα interaction, rather than by the general binding to nucleosomes.
With other genes, where estrogen induction is not mediated by ERα-ERE binding, the binding of distinct transcription factors is required for estrogen induction. In particular, FOS lacks a functional ERE and does not interact significantly with ERα upon estrogen stimulation (Fig. 4G). The regulation of FOS expression by estrogen in MCF-7 cells is mediated instead through the nongenomic pathway of estrogen receptor, via the activation of the mitogen-activated protein kinase- and phosphoinositide-3 kinase signaling pathways that target Elk-1 and SRF, respectively, at the serum response element on this promoter (22, 23). Although a role for Sp1 in tethering ERα onto this promoter was also previously suggested (21), the association of ERα was not detectable in our experiments, either before or after induction. Instead, SRF is constitutively bound and can interact with HMGN1 in vivo, consistent with the diminution of FOS transcriptional stimulation by HMGN1. Furthermore, the reduced interaction of HMGN1 S20E/S24E with SRF in vivo explains the mutant's limited ability to inhibit expression from the FOS promoter, in contrast to that of wild-type HMGN1.
In summary, we demonstrated that the recruitment of HMGN1 to specific promoters through the interaction with transcription factors is consistent with the underlying mechanism of the differential rather than the indiscriminative effects of this protein on transcription. Given that other family members, HMGN3 and HMGN1, can interact with thyroid hormone receptor (2) and that HMGN2 can interact with ERα in vitro (data not shown), it is likely that binding to specific transcription factors is a general property of all HMGN proteins. Such interactions either may be complementary to the well-characterized abilities of HMGNs to influence chromatin structure and modifications, in simply getting these proteins to the correct chromosomal locations at the correct time, or under other circumstances, may provide distinct biological functions from the chromatin-modifying roles of HMGNs.
The issue arises as to how such a small protein (only 99 amino acids) could interact with such a wide variety of other proteins, especially since it even contains two distinct domains for binding to ERα. The answer is likely to lie with the highly flexible structure of HMGN1. In solution, it adopts an unstructured, random coil configuration (1, 16). This allows it to conform to the partner proteins (or nucleosomes), with which it interacts specifically by an induced fit model, and would permit a wider range of protein-protein interactions. Such flexibility has been proposed for another distinct type of HMG (small and highly charged) proteins, the HMGA family (60).
A mechanism for the inhibition of estrogen-mediated transcription activation by HMGN1.
Previous studies identified two general mechanisms that underlie the ability of HMGN1 to affect transcription by affecting nucleosomal histone modification (42, 43, 57, 77) and by counteracting the ability of histone H1 to compact the chromatin fiber, mediated through the C-terminal CHUD domain (20, 74). Our finding that the abundant HMGN1 interacts with specific DNA-binding transcription factors raised another possibility, that HMGN1 may squelch transcription factor activities. We tested and excluded the latter two mechanisms as explanations for how HMGN1 regulates estrogen-mediated gene expression.
The remaining mechanism known for regulation by HMGN1 involves the alteration of various nucleosomal histone modifications leading to either facilitation or inhibition of transcription. Specifically, HMGN1 decreases cellular levels of pSer10H3, AcLys9H3, and MeLys9H3, while it increases that of AcLys14H3 (42). Previous studies with HMGN1−/− MEFs linked alterations in the levels of pSer10H3 and AcLys14H3 with the ability of HMGN1 to limit the induction of Fosb and enhance the induction of Jund, respectively, in response to stress stimuli (42, 43). We examined the involvement of pSer10H3, AcLys9H3, and MeLys9H3 in HMGN1-mediated transcription inhibition of estrogen-responsive genes. Although modest increases (approximately 1.5-fold) in the level of pSer10H3 on the TFF1 promoter have been reported after estrogen stimulation of MCF-7 cells (25), we did not detect such changes in pSer10H3 levels at the promoter in our cell line (data not shown). Such differences in specific gene regulatory properties are not unanticipated, given that MCF-7 cells have been independently propagated in multiple laboratories over decades, readily driving production of variant, although related, cell lines. Estrogen also decreases both di- and trimethylation of Lys9 in histone H3 on the TFF1 promoter (13, 28). We confirmed this result; however, neither the basal nor the diminished estrogen-treated levels of MeLys9H3 were affected by the loss of HMGN1 in the knockdown cells (data not shown). Only levels of AcLys9H3 at specific genes responded to changes in HMGN1 protein levels. We noted that the overall cellular levels of AcLys9H3 were unchanged in our HMGN1 knockdown MCF-7 cell lines, which differs from the previous report of an approximately 1.7-fold increase in the cellular level of AcLys9H3 in HMGN1−/− MEFs. This distinction may be due either to the shRNA-mediated reduction versus removal of HMGN1 or to cell type-specific differences.
Estrogen stimulation induces acetylation at Lys9 of H3 at the TFF1 promoter (13). We first expanded upon this observation, demonstrating enhanced acetylation at Lys9 of H3 at several estrogen-induced genes, both in the regulatory regions that bind ERα (promoters and enhancers, including the TFF1 promoter) and in the coding regions in a subset of these genes. However, levels of AcLys9H3 did not appreciably increase on the promoters of either FOS, which is stimulated by estrogen through the nongenomic pathway, or on ACTB, which is not responsive to estrogen (Fig. 7C). Next, we compared the levels of AcLys9H3 at the same genomic locations in cells with greatly diminished levels of HMGN1. The reduction of cellular HMGN1 led to increases in both the basal and the stimulated levels of AcLys9H3 on a subset of the genes, including TFF1, XBP1, and MYC, but not on the IGFBP4 and CTSD promoters, even though H3 Lys9 acetylation increased at these two promoters upon estrogen stimulation also. In general, the subset of genes on which the loss of HMGN1 enhances levels of AcLys9H3 coincides with the subset of estrogen-responsive genes on which the RNA induction is inhibited by HMGN1, and conversely, the subset of estrogen-responsive genes at which levels of AcLys9H3 are the same irrespective of HMGN1 levels coincides with the subset whose expression is also unaffected by HMGN1. The only outlier to this generalization is FOS, whose activation by estrogen does not require nuclear ERα and whose regulation by HMGN1 must therefore occur through a distinct mechanism.
In any mechanistic model of the molecular action of HMGN1, three aspects of the regulation of AcLys9H3 levels must be incorporated: first, the regulation of both basal and stimulated levels of this histone modification; second, the regulation only at specific estrogen-stimulated genes; and third, at these HMGN1 target genes, the regulation of the histone modification both within the regulatory and the coding regions. The impact of HMGN1 on both the basal and the stimulated levels of AcLys9H3 is reminiscent of previous observations in which HMGN affected both the basal and the stimulated levels of pSer10H3 and AcLys14H3 modifications on specific promoters (42, 43). Furthermore, it is consistent with the trend (although not statistically significant) toward an increase in basal TFF1 mRNA levels upon transient diminishment of HMGN1 levels with siRNA (Fig. 1D) and toward a decrease in basal TFF1 levels upon overexpression of HMGN1 (Fig. 2D). Of all the genes studied, TFF1 exhibits the highest basal level of mRNA, making it the easiest on which to visualize this effect. Because both basal and estrogen-stimulated levels of H3K9Ac at specific genes are elevated in HMGN1 knockdown cells, it is likely that the stimulated chromatin modification state is maintained over the 2-day period in which the MCF-7 cells are incubated in the absence of estrogen. MCF-7 cells require estrogen for cell growth and survival and can therefore be propagated in the absence of estrogen for only relatively short periods of time.
Two types of models could explain the gene target specificity of HMGN1 action. The simpler model would invoke specific recruitment of HMGN1 to specific locations by protein-protein interaction with DNA-binding transcription factors, coupled with direct alteration of histone modification by HMGN1, either through steric hindrance via interaction of HMGN1 with histone tails or through the modulation of histone-modifying enzymes. One example of the latter mechanism is the ability of HMGN to facilitate acetylation of Lys14 on H3 by p/CAF (43). Alternatively, our data support the model in which HMGN1 inhibits recruitment of a specific HAT by ERα or, alternatively, facilitates the subsequent recruitment of an HDAC. In the presence of estrogen, liganded ERα recruits a number of HATs to the TFF1 promoter and to other gene regulatory regions. Almost all of the recruited HATs can acetylate Lys9 on histone H3 (38, 62). Then, in an alternating kinetic pattern, HDACs are recruited. Further studies are required to pinpoint which specific HAT (or HDAC) may be dysregulated by HMGN1, resulting in altered recruitment or activity at estrogen-responsive genes. In considering why only some estrogen-responsive genes are inhibited by HMGN1, we propose that the ERα coregulator targeted by HMGN1 is essential for the induction of TFF1, XBP1, and MYC but not for the induction of IGFBP4 and CTSD. In fact, our findings indicate that the mechanism of estrogen-mediated stimulation of IGFBP4 and CTSD expression must be distinct from that of TFF1, XBP1, and MYC, because AcLys9H3 acetylation is not induced by estrogen in the coding regions of IGFBP4 and CTSD, unlike the subset of high-affinity ERE-containing genes (Fig. 7C).
Finally, the models above address how levels of acetylated H3 could be altered by HMGN1 only at promoter and enhancer regions, where ERα binds. Given that HMGN1 is not recruited above its constitutive levels of association at coding regions, the increase in AcK9H3 levels within gene coding regions upon the reduction of cellular HMGN1 suggests that the altered activity of coregulators at the promoter is propagated into the coding region. A precedent for this hypothesis is the targeting and spreading of the GCN5-containing SAGA complex to the GAL1 coding region, which is dependent on the preinitiation complex assembly and transcription (30).
The above discussion of HMGN1-mediated regulation deals with canonical ERα target genes. Despite different specific mechanisms on these genes compared with the FOS gene, the general mode of regulation by HMGN1 may nonetheless be similar. We propose that in both instances, HMGN1 exerts its regulatory effects through interaction with other components of the transcription machinery recruited after the application of stimuli.
Biological relevance of recruitment of HMGN by DNA-binding transcription factors.
Since HMGN1 is ubiquitous and abundant and binds chromatin at basal levels throughout the genome, the advantage of protein-protein interactions to recruit it to a specific location is not inherently obvious. FRAP experiments indicate that HMGN1/2 move rapidly throughout the nucleus by diffusion, although they are slowed by transient binding to chromatin (29, 55, 56). We suggest that this general binding of HMGN1 keeps the chromatin in a generally accessible state (8, 20). In contrast to this dynamic low-affinity binding of HMGN to chromatin, the interaction of HMGN with specific DNA-bound transcription factors stabilizes its interaction at particular sites. As a consequence, HMGN1 either specifically potentiates or represses the expression of particular genes to which it associates with higher affinity. The biological function of repressing the rapid gene induction by particular stimuli, such as estrogen, although initially counterintuitive, would be to limit the degree and duration of the activation. This is in keeping with many other negative feedback mechanisms, which prevent uncontrolled activation and ensure rapid downregulation as well as the upregulation of transcription in response to the application and removal of cellular stimuli. Through its effect on chromatin-related events, including transcription and DNA repair, as described here and previously (reviewed in references 12 and 34), HMGN plays a role in a number of physiological processes. These include tumorigenesis (6), development (8, 35, 48), and differentiation, in which its level is tightly regulated and progressively diminished (4, 7, 33, 39, 50, 52, 53). Its effect on estrogen-mediated transcriptional regulation suggests that HMGN may also be involved in the in vivo physiology of estrogen action, which requires further exploration.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grant R01 GM54808 to U.H. and a Boston University SPRInG award to U.H.
We thank Richard Karas for the generous gifts of pGEX-4T-ERα and corresponding constructs expressing truncated ERα proteins, Myles Brown for pcDNA3.1-ERα, and Michael Bustin for antibody against HMGN2. We also thank Geoffrey Cooper for critical suggestions on the manuscript.
Footnotes
Published ahead of print on 15 October 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abercrombie, B. D., G. G. Kneale, C. Crane-Robinson, E. M. Bradbury, G. H. Goodwin, J. M. Walker, and E. W. Johns. 1978. Studies on the conformational properties of the high-mobility-group chromosomal protein HMG 17 and its interaction with DNA. Eur. J. Biochem. 84:173-177. [DOI] [PubMed] [Google Scholar]
- 2.Amano, T., K. Leu, K. Yoshizato, and Y. B. Shi. 2002. Thyroid hormone regulation of a transcriptional coactivator in Xenopus laevis: implication for a role in postembryonic tissue remodeling. Dev. Dyn. 223:526-535. [DOI] [PubMed] [Google Scholar]
- 3.Augereau, P., F. Miralles, V. Cavailles, C. Gaudelet, M. Parker, and H. Rochefort. 1994. Characterization of the proximal estrogen-responsive element of human cathepsin D gene. Mol. Endocrinol. 8:693-703. [DOI] [PubMed] [Google Scholar]
- 4.Begum, N., J. M. Pash, and J. S. Bhorjee. 1990. Expression and synthesis of high mobility group chromosomal proteins in different rat skeletal cell lines during myogenesis. J. Biol. Chem. 265:11936-11941. [PubMed] [Google Scholar]
- 5.Bertrand-Mercat, P., and J. R. Pasqualini. 1991. Antagonistic effect of the antiestrogen 4-hydroxytamoxifen on estradiol-stimulated acetylation of nuclear high mobility group (HMG) proteins in the uterus of newborn guinea pigs. Life Sci. 48:2081-2087. [DOI] [PubMed] [Google Scholar]
- 6.Birger, Y., F. Catez, T. Furusawa, J. H. Lim, M. Prymakowska-Bosak, K. L. West, Y. V. Postnikov, D. C. Haines, and M. Bustin. 2005. Increased tumorigenicity and sensitivity to ionizing radiation upon loss of chromosomal protein HMGN1. Cancer Res. 65:6711-6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birger, Y., J. Davis, T. Furusawa, E. Rand, J. Piatigorsky, and M. Bustin. 2006. A role for chromosomal protein HMGN1 in corneal maturation. Differentiation 74:19-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birger, Y., K. L. West, Y. V. Postnikov, J. H. Lim, T. Furusawa, J. P. Wagner, C. S. Laufer, K. H. Kraemer, and M. Bustin. 2003. Chromosomal protein HMGN1 enhances the rate of DNA repair in chromatin. EMBO J. 22:1665-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Björnström, L., and M. Sjöberg. 2005. Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol. Endocrinol. 19:833-842. [DOI] [PubMed] [Google Scholar]
- 10.Brawley, J. V., and H. G. Martinson. 1992. HMG proteins 14 and 17 become cross-linked to the globular domain of histone H3 near the nucleosome core particle dyad. Biochemistry 31:364-370. [DOI] [PubMed] [Google Scholar]
- 11.Brown, A. M., J. M. Jeltsch, M. Roberts, and P. Chambon. 1984. Activation of pS2 gene transcription is a primary response to estrogen in the human breast cancer cell line MCF-7. Proc. Natl. Acad. Sci. USA 81:6344-6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bustin, M. 2001. Chromatin unfolding and activation by HMGN* chromosomal proteins. Trends Biochem. Sci. 26:431-437. [DOI] [PubMed] [Google Scholar]
- 13.Carling, T., K. C. Kim, X. H. Yang, J. Gu, X. K. Zhang, and S. Huang. 2004. A histone methyltransferase is required for maximal response to female sex hormones. Mol. Cell. Biol. 24:7032-7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carroll, J. S., X. S. Liu, A. S. Brodsky, W. Li, C. A. Meyer, A. J. Szary, J. Eeckhoute, W. Shao, E. V. Hestermann, T. R. Geistlinger, E. A. Fox, P. A. Silver, and M. Brown. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122:33-43. [DOI] [PubMed] [Google Scholar]
- 15.Carroll, J. S., C. A. Meyer, J. Song, W. Li, T. R. Geistlinger, J. Eeckhoute, A. S. Brodsky, E. K. Keeton, K. C. Fertuck, G. F. Hall, Q. Wang, S. Bekiranov, V. Sementchenko, E. A. Fox, P. A. Silver, T. R. Gingeras, X. S. Liu, and M. Brown. 2006. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 38:1289-1297. [DOI] [PubMed] [Google Scholar]
- 16.Cary, P. D., D. S. King, C. Crane-Robinson, E. M. Bradbury, A. Rabbani, G. H. Goodwin, and E. W. Johns. 1980. Structural studies on two high-mobility-group proteins from calf thymus, HMG-14 and HMG-20 (ubiquitin), and their interaction with DNA. Eur. J. Biochem. 112:577-580. [DOI] [PubMed] [Google Scholar]
- 17.Cavailles, V., P. Augereau, and H. Rochefort. 1993. Cathepsin D gene is controlled by a mixed promoter, and estrogens stimulate only TATA-dependent transcription in breast cancer cells. Proc. Natl. Acad. Sci. USA 90:203-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cook, G. R., P. Yau, H. Yasuda, R. R. Traut, and E. M. Bradbury. 1986. High mobility group protein 17 cross-links primarily to histone H2A in the reconstituted HMG 17-nucleosome core particle complex. J. Biol. Chem. 261:16185-16190. [PubMed] [Google Scholar]
- 19.Deschenes, J., V. Bourdeau, J. H. White, and S. Mader. 2007. Regulation of GREB1 transcription by estrogen receptor α through a multipartite enhancer spread over 20 kb of upstream flanking sequences. J. Biol. Chem. 282:17335-17339. [DOI] [PubMed] [Google Scholar]
- 20.Ding, H.-F., M. Bustin, and U. Hansen. 1997. Alleviation of histone H1-mediated transcriptional repression and chromatin compaction by the acidic activation region in chromosomal protein HMG-14. Mol. Cell. Biol. 17:5843-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duan, R., W. Porter, and S. Safe. 1998. Estrogen-induced c-fos protooncogene expression in MCF-7 human breast cancer cells: role of estrogen receptor Sp1 complex formation. Endocrinology 139:1981-1990. [DOI] [PubMed] [Google Scholar]
- 22.Duan, R., W. Xie, R. C. Burghardt, and S. Safe. 2001. Estrogen receptor-mediated activation of the serum response element in MCF-7 cells through MAPK-dependent phosphorylation of Elk-1. J. Biol. Chem. 276:11590-11598. [DOI] [PubMed] [Google Scholar]
- 23.Duan, R., W. Xie, X. Li, A. McDougal, and S. Safe. 2002. Estrogen regulation of c-fos gene expression through phosphatidylinositol-3-kinase-dependent activation of serum response factor in MCF-7 breast cancer cells. Biochem. Biophys. Res. Commun. 294:384-394. [DOI] [PubMed] [Google Scholar]
- 24.Espel, E., J. Bernués, J. A. Pérez-Pons, and E. Querol. 1985. Binding of HMG14 non-histone protein to histones H2A, H2B, H1 and DNA in reconstituted chromatin. Biochem. Biophys. Res. Commun. 132:1031-1037. [DOI] [PubMed] [Google Scholar]
- 25.Espino, P. S., L. Li, S. He, J. Yu, and J. R. Davie. 2006. Chromatin modification of the trefoil factor 1 gene in human breast cancer cells by the Ras/mitogen-activated protein kinase pathway. Cancer Res. 66:4610-4616. [DOI] [PubMed] [Google Scholar]
- 26.Font de Mora, J., and M. Brown. 2000. AIB1 is a conduit for kinase-mediated growth factor signaling to the estrogen receptor. Mol. Cell. Biol. 20:5041-5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furusawa, T., J. H. Lim, F. Catez, Y. Birger, S. Mackem, and M. Bustin. 2006. Down-regulation of nucleosomal binding protein HMGN1 expression during embryogenesis modulates Sox9 expression in chondrocytes. Mol. Cell. Biol. 26:592-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Bassets, I., Y. S. Kwon, F. Telese, G. G. Prefontaine, K. R. Hutt, C. S. Cheng, B. G. Ju, K. A. Ohgi, J. Wang, L. Escoubet-Lozach, D. W. Rose, C. K. Glass, X. D. Fu, and M. G. Rosenfeld. 2007. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell 128:505-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorski, S. A., M. Dundr, and T. Misteli. 2006. The road much traveled: trafficking in the cell nucleus. Curr. Opin. Cell Biol. 18:284-290. [DOI] [PubMed] [Google Scholar]
- 30.Govind, C. K., F. Zhang, H. Qiu, K. Hofmeyer, and A. G. Hinnebusch. 2007. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol. Cell 25:31-42. [DOI] [PubMed] [Google Scholar]
- 31.Heemskerk, M. H., B. Blom, G. Nolan, A. P. Stegmann, A. Q. Bakker, K. Weijer, P. C. Res, and H. Spits. 1997. Inhibition of T cell and promotion of natural killer cell development by the dominant negative helix loop helix factor Id3. J. Exp. Med. 186:1597-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herrera, R. E., P. E. Shaw, and A. Nordheim. 1989. Occupation of the c-fos serum response element in vivo by a multi-protein complex is unaltered by growth factor induction. Nature 340:68-70. [DOI] [PubMed] [Google Scholar]
- 33.Hill, D. A., and A. N. Imbalzano. 2006. HMGN1 is dispensable for myogenesis and adipogenesis. Gene 371:59-67. [DOI] [PubMed] [Google Scholar]
- 34.Hock, R., T. Furusawa, T. Ueda, and M. Bustin. 2007. HMG chromosomal proteins in development and disease. Trends Cell Biol. 17:72-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korner, U., M. Bustin, U. Scheer, and R. Hock. 2003. Developmental role of HMGN proteins in Xenopus laevis. Mech. Dev. 120:1177-1192. [DOI] [PubMed] [Google Scholar]
- 36.Krishnan, V., X. Wang, and S. Safe. 1994. Estrogen receptor-Sp1 complexes mediate estrogen-induced cathepsin D gene expression in MCF-7 human breast cancer cells. J. Biol. Chem. 269:15912-15917. [PubMed] [Google Scholar]
- 37.Lee, J. W., H. S. Choi, J. Gyuris, R. Brent, and D. D. Moore. 1995. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol. Endocrinol. 9:243-254. [DOI] [PubMed] [Google Scholar]
- 38.Lee, K. K., and J. L. Workman. 2007. Histone acetyltransferase complexes: one size doesn't fit all. Nat. Rev. Mol. Cell. Biol. 8:284-295. [DOI] [PubMed] [Google Scholar]
- 39.Lehtonen, S., V. M. Olkkonen, M. Stapleton, M. Zerial, and E. Lehtonen. 1998. HMG-17, a chromosomal non-histone protein, shows developmental regulation during organogenesis. Int. J. Dev. Biol 42:775-782. [PubMed] [Google Scholar]
- 40.Levin, E. R. 2005. Integration of the extranuclear and nuclear actions of estrogen. Mol. Endocrinol. 19:1951-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim, J. H., M. Bustin, V. V. Ogryzko, and Y. V. Postnikov. 2002. Metastable macromolecular complexes containing high mobility group nucleosome-binding chromosomal proteins in HeLa nuclei. J. Biol. Chem. 277:20774-20782. [DOI] [PubMed] [Google Scholar]
- 42.Lim, J. H., F. Catez, Y. Birger, K. L. West, M. Prymakowska-Bosak, Y. V. Postnikov, and M. Bustin. 2004. Chromosomal protein HMGN1 modulates histone H3 phosphorylation. Mol. Cell 15:573-584. [DOI] [PubMed] [Google Scholar]
- 43.Lim, J. H., K. L. West, Y. Rubinstein, M. Bergel, Y. V. Postnikov, and M. Bustin. 2005. Chromosomal protein HMGN1 enhances the acetylation of lysine 14 in histone H3. EMBO J. 24:3038-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin, C. Y., V. B. Vega, J. S. Thomsen, T. Zhang, S. L. Kong, M. Xie, K. P. Chiu, L. Lipovich, D. H. Barnett, F. Stossi, A. Yeo, J. George, V. A. Kuznetsov, Y. K. Lee, T. H. Charn, N. Palanisamy, L. D. Miller, E. Cheung, B. S. Katzenellenbogen, Y. Ruan, G. Bourque, C. L. Wei, and E. T. Liu. 2007. Whole-genome cartography of estrogen receptor α binding sites. PLoS Genet. 3:e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu, Q., D. C. Pallas, H. K. Surks, W. E. Baur, M. E. Mendelsohn, and R. H. Karas. 2004. Striatin assembles a membrane signaling complex necessary for rapid, nongenomic activation of endothelial NO synthase by estrogen receptor alpha. Proc. Natl. Acad. Sci. USA 101:17126-17131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mardian, J. K., A. E. Paton, G. J. Bunick, and D. E. Olins. 1980. Nucleosome cores have two specific binding sites for nonhistone chromosomal proteins HMG 14 and HMG 17. Science 209:1534-1536. [DOI] [PubMed] [Google Scholar]
- 47.Métivier, R., G. Penot, M. R. Hübner, G. Reid, H. Brand, M. Koš, and F. Gannon. 2003. Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115:751-763. [DOI] [PubMed] [Google Scholar]
- 48.Mohamed, O. A., M. Bustin, and H. J. Clarke. 2001. High-mobility group proteins 14 and 17 maintain the timing of early embryonic development in the mouse. Dev. Biol. 229:237-249. [DOI] [PubMed] [Google Scholar]
- 49.Morisset, M., F. Capony, and H. Rochefort. 1986. The 52-kDa estrogen-induced protein secreted by MCF7 cells is a lysosomal acidic protease. Biochem. Biophys. Res. Commun. 138:102-109. [DOI] [PubMed] [Google Scholar]
- 50.Norman, B., J. Davis, and J. Piatigorsky. 2004. Postnatal gene expression in the normal mouse cornea by SAGE. Investig. Ophthalmol. Vis. Sci. 45:429-440. [DOI] [PubMed] [Google Scholar]
- 51.O'Lone, R., M. C. Frith, E. K. Karlsson, and U. Hansen. 2004. Genomic targets of nuclear estrogen receptors. Mol. Endocrinol. 18:1859-1875. [DOI] [PubMed] [Google Scholar]
- 52.Pash, J. M., P. J. Alfonso, and M. Bustin. 1993. Aberrant expression of high mobility group chromosomal protein 14 affects cellular differentiation. J. Biol. Chem. 268:13632-13638. [PubMed] [Google Scholar]
- 53.Pash, J. M., J. S. Bhorjee, B. M. Patterson, and M. Bustin. 1990. Persistence of chromosomal proteins HMG-14/-17 in myotubes following differentiation-dependent reduction of HMG mRNA. J. Biol. Chem. 265:4197-4199. [PubMed] [Google Scholar]
- 54.Pasqualini, J. R., R. Sterner, P. Mercat, and V. G. Allfrey. 1989. Estradiol enhanced acetylation of nuclear high mobility group proteins of the uterus of newborn guinea pigs. Biochem. Biophys. Res. Commun. 161:1260-1266. [DOI] [PubMed] [Google Scholar]
- 55.Phair, R. D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404:604-609. [DOI] [PubMed] [Google Scholar]
- 56.Phair, R. D., P. Scaffidi, C. Elbi, J. Vecerova, A. Dey, K. Ozato, D. T. Brown, G. Hager, M. Bustin, and T. Misteli. 2004. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 24:6393-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Postnikov, Y. V., G. I. Belova, J. H. Lim, and M. Bustin. 2006. Chromosomal protein HMGN1 modulates the phosphorylation of serine 1 in histone H2A. Biochemistry 45:15092-15099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prymakowska-Bosak, M., T. Misteli, J. E. Herrera, H. Shirakawa, Y. Birger, S. Garfield, and M. Bustin. 2001. Mitotic phosphorylation prevents the binding of HMGN proteins to chromatin. Mol. Cell. Biol. 21:5169-5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qin, C., P. Singh, and S. Safe. 1999. Transcriptional activation of insulin-like growth factor-binding protein-4 by 17β-estradiol in MCF-7 cells: role of estrogen receptor-Sp1 complexes. Endocrinology 140:2501-2508. [DOI] [PubMed] [Google Scholar]
- 60.Reeves, R. 2003. HMGA proteins: flexibility finds a nuclear niche? Biochem. Cell. Biol. 81:185-195. [DOI] [PubMed] [Google Scholar]
- 61.Rollerova, E., and M. Urbancikova. 2000. Intracellular estrogen receptors, their characterization and function. Endocr. Regul. 34:203-218. [PubMed] [Google Scholar]
- 62.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70:81-120. [DOI] [PubMed] [Google Scholar]
- 63.Rubinstein, Y. R., T. Furusawa, J. H. Lim, Y. V. Postnikov, K. L. West, Y. Birger, S. Lee, P. Nguyen, J. B. Trepel, and M. Bustin. 2005. Chromosomal protein HMGN1 modulates the expression of N-cadherin. FEBS J. 272:5853-5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Safe, S. 2001. Transcriptional activation of genes by 17β-estradiol through estrogen receptor-Sp1 interactions. Vitam. Horm. 62:231-252. [DOI] [PubMed] [Google Scholar]
- 65.Sanchez, R., D. Nguyen, W. Rocha, J. H. White, and S. Mader. 2002. Diversity in the mechanisms of gene regulation by estrogen receptors. Bioessays 24:244-254. [DOI] [PubMed] [Google Scholar]
- 66.Sandeen, G., W. I. Wood, and G. Felsenfeld. 1980. The interaction of high mobility proteins HMG14 and 17 with nucleosomes. Nucleic Acids Res. 8:3757-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sewack, G. F., T. W. Ellis, and U. Hansen. 2001. Binding of TATA binding protein to a naturally positioned nucleosome is facilitated by histone acetylation. Mol. Cell. Biol. 21:1404-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sewack, G. F., and U. Hansen. 1997. Nucleosome positioning and transcription-associated chromatin alterations on the human estrogen-responsive pS2 promoter. J. Biol. Chem. 272:31118-31129. [DOI] [PubMed] [Google Scholar]
- 69.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843-852. [DOI] [PubMed] [Google Scholar]
- 70.Shi, B., J. Liang, X. Yang, Y. Wang, Y. Zhao, H. Wu, L. Sun, Y. Zhang, Y. Chen, R. Li, M. Hong, and Y. Shang. 2007. Integration of estrogen and Wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Mol. Cell. Biol. 27:5105-5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shick, V. V., A. V. Belyavsky, and A. D. Mirzabekov. 1985. Primary organization of nucleosomes. Interaction of non-histone high mobility group proteins 14 and 17 with nucleosomes, as revealed by DNA-protein crosslinking and immunoaffinity isolation. J. Mol. Biol. 185:329-339. [DOI] [PubMed] [Google Scholar]
- 72.Shirakawa, H., J. E. Herrera, M. Bustin, and Y. Postnikov. 2000. Targeting of high mobility group-14/-17 proteins in chromatin is independent of DNA sequence. J. Biol. Chem. 275:37937-37944. [DOI] [PubMed] [Google Scholar]
- 73.Sun, J., Z. Nawaz, and J. J. Slingerland. 2007. Long-range activation of Greb1 by estrogen receptor via three distal consensus estrogen-responsive elements in breast cancer cells. Mol. Endocrinol. 21:2651-2662. [DOI] [PubMed] [Google Scholar]
- 74.Trieschmann, L., P. J. Alfonso, M. P. Crippa, A. P. Wolffe, and M. Bustin. 1995. Incorporation of chromosomal proteins HMG-14/HMG-17 into nascent nucleosomes induces an extended chromatin conformation and enhances the utilization of active transcription complexes. EMBO J. 14:1478-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Trieschmann, L., B. Martin, and M. Bustin. 1998. The chromatin unfolding domain of chromosomal protein HMG-14 targets the N-terminal tail of histone H3 in nucleosomes. Proc. Natl. Acad. Sci. USA 95:5468-5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tullai, J. W., M. E. Schaffer, S. Mullenbrock, S. Kasif, and G. M. Cooper. 2004. Identification of transcription factor binding sites upstream of human genes regulated by the phosphatidylinositol 3-kinase and MEK/ERK signaling pathways. J. Biol. Chem. 279:20167-20177. [DOI] [PubMed] [Google Scholar]
- 77.Ueda, T., Y. V. Postnikov, and M. Bustin. 2006. Distinct domains in high mobility group N variants modulate specific chromatin modifications. J. Biol. Chem. 281:10182-10187. [DOI] [PubMed] [Google Scholar]
- 78.Wang, F., W. Porter, W. Xing, T. K. Archer, and S. Safe. 1997. Identification of a functional imperfect estrogen-responsive element in the 5′-promoter region of the human cathepsin D gene. Biochemistry 36:7793-7801. [DOI] [PubMed] [Google Scholar]
- 79.West, K. L., M. A. Castellini, M. K. Duncan, and M. Bustin. 2004. Chromosomal proteins HMGN3a and HMGN3b regulate the expression of glycine transporter 1. Mol. Cell. Biol. 24:3747-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.