Abstract
DNA methylation plays an important role in gene silencing in mammals. Two de novo methyltransferases, Dnmt3a and Dnmt3b, are required for the establishment of genomic methylation patterns in development. However, little is known about their coordinate function in the silencing of genes critical for embryonic development and how their activity is regulated. Here we show that Dnmt3a and Dnmt3b are the major components of a native complex purified from embryonic stem cells. The two enzymes directly interact and mutually stimulate each other both in vitro and in vivo. The stimulatory effect is independent of the catalytic activity of the enzyme. In differentiating embryonic carcinoma or embryonic stem cells and mouse postimplantation embryos, they function synergistically to methylate the promoters of the Oct4 and Nanog genes. Inadequate methylation caused by ablating Dnmt3a and Dnmt3b is associated with dysregulated expression of Oct4 and Nanog during the differentiation of pluripotent cells and mouse embryonic development. These results suggest that Dnmt3a and Dnmt3b form a complex through direct contact in living cells and cooperate in the methylation of the promoters of Oct4 and Nanog during cell differentiation. The physical and functional interaction between Dnmt3a and Dnmt3b represents a novel regulatory mechanism to ensure the proper establishment of genomic methylation patterns for gene silencing in development.
DNA methylation is an epigenetic modification indispensable for multiple cellular processes, including transcriptional repression of tissue-specific genes, genomic imprinting, X chromosome inactivation, and transposon silencing (6, 9, 36). Aberrant methylation contributes to tumorigenesis and other diseases (30).
Methylation occurs on ∼5% of the cytosines in the genomic DNA of mammals. The distribution of methylcytosines forms specific patterns in the genome. Methylation patterns are propagated in mitotic cells by maintenance methylation that takes place in concert with DNA replication. The methyltransferase Dnmt1 performs the maintenance function (4, 37). Consistent with its role, Dnmt1 has a substrate preference for hemimethylated DNA in vitro and is usually associated with replication foci in the S phase of dividing cells (18, 35).
As DNA methylation patterns are stably maintained in differentiated mitotic cells, new patterns arise in embryonic cell differentiation and germ line specification in development. Dnmt3a and Dnmt3b, the two closely related de novo methyltransferases, are required for this process (5). Inactivation of both genes caused a complete failure to establish genome-wide methylation. Inactivation of either gene alone led to hypomethylation at a subset of sequences in the genome, but most sequences are unaffected (40). Dnmt3a is specifically required for the methylation of imprinted genes in germ cells (27, 31) and the Xist gene on the X chromosome (14). Dnmt3b has an irreplaceable function in the methylation of centromeric minor satellite repeats (40, 51). Thus, most genomic targets can be methylated by both enzymes, and sequences requiring a particular enzyme for methylation might be exceptions. This suggests a possibility that Dnmt3a and Dnmt3b may independently establish methylation for the bulk of genomic sequences during development.
Structurally, Dnmt3a and Dnmt3b have a similar organization, with three conserved domains: a long N-terminal region contains a PWWP domain, which targets the enzyme to chromatin (13, 24), a cysteine-rich PHD zinc finger domain, which interacts with transcriptional repressors, histone deacetylases, and histone methyltransferases generating inactive chromatin marks (2, 22, 23), and a C-terminal catalytic domain resembling bacterial cytosine methyltransferases (41).
Isolated recombinant Dnmt3a and Dnmt3b show very low enzymatic activity in comparison to other methyltransferases, including Dnmt1 (33, 41). The mere existence of the enzymes at high levels in embryonic cells does not result in an increase in genomic methylation. Genomic regions devoid of methylation remain unmethylated faithfully during cell proliferation (7). These observations suggest that the methylation activity is tightly regulated by factors which directly or indirectly affect the function of the two de novo methyltransferases. For example, Dnmt3L, a short paralog of Dnmt3a and -3b that lacks critical residues found in the active center of all cytosine methyltransferases, is required for the methylation of imprinted genes and some repetitive sequences in germ cells (10, 11, 27). Biochemical studies have demonstrated that Dnmt3L, albeit inactive per se, stimulates the methylation activity of both Dnmt3a and Dnmt3b by interacting with them through the conserved C-terminal domain (12, 16, 17, 25). However, in Dnmt3L mutant mice, methylation of most sequences occurs normally and no developmental defects are apparent except in the germ cells. Therefore, other mechanisms may exist to regulate the activity of Dnmt3a and Dnmt3b for the methylation of various genomic sequences during somatic development.
In this study, we sought to identify regulatory factors for Dnmt3a and Dnmt3b by purifying their associated proteins from mouse embryonic cells. To our surprise, Dnmt3a and Dnmt3b were the two major components present in a stable complex. In vitro, their collective enzymatic activity is much higher than the total activities of the two individual proteins combined. In vivo, the two enzymes contribute synergistically to the methylation of Oct4 and Nanog during mouse embryonic cell differentiation. Thus, our studies have revealed a physical association and functional cooperation between the two de novo methyltransferases which provides a framework for future understanding of the regulation of DNA methylation during development and pathogenesis.
MATERIALS AND METHODS
Affinity purification of methyltransferase-associated proteins from embryonic stem (ES) and P19 cells.
For isolation of Dnmt3b-associated proteins, chromatin extract was prepared from approximately 1 × 108 ES cells as described previously (28), except that 0.25 M (NH4)2SO4 was replaced with 500 mM NaCl in the extraction buffer. The extract was then precleared by incubating with 200 μl of protein G-agarose suspension (Roche) in a total volume of 5 ml at 4°C for 3 h. The precleared supernatant was incubated with 10 μg of anti-Dnmt3b antibody (mouse monoclonal; Shanghai Wolwo Biotech Co. Ltd) for 3 h. Two hundred microliters of protein G-agarose slurry was then added and incubated overnight. The agarose beads were washed with DNase I digestion buffer (28) containing 500 mM NaCl. Finally, the beads were resuspended in 100 μl of 1× sodium dodecyl sulfate loading buffer and heated to 100°C for 3 min to denature proteins.
For isolation of Dnmt3a-associated proteins, tandem-affinity purification (TAP) was carried out using the previous procedure (44) with slight modifications. Chromatin extract was prepared as described above from about 7 × 108 P19 cells stably expressing TAP-tagged Dnmt3a and dialyzed into immunoglobulin G (IgG)-Sepharose binding buffer (20 mM Tris-HCl, pH 7.9, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 μg/ml pepstatin). The dialyzed extract was then incubated at 4°C with 100 μl of IgG Sepharose slurry (GE Healthcare). After washing, bound proteins were released from beads by treatment with 15 U/μl AcTEV protease (Invitrogen) in 100 μl of tobacco etch virus cleavage buffer. The sample was subsequently incubated with 50 μl of calmodulin resin (GE Healthcare) to bind CBP-containing proteins, and the bound proteins were finally eluted with 100 μl of 50 mM NH4HCO3, pH 8.0, and 25 mM EGTA.
The protein samples were resolved on 7% denaturing polyacrylamide gel and visualized by silver staining. Specific bands were excised, and the protein sequence was determined by using electrospray ionization LTQ tandem mass spectrometry at the Research Center for Proteome Analysis, Shanghai Institutes for Biological Sciences.
Gel filtration chromatography.
A Superose 6 column (GL 10/300; GE Healthcare) was equilibrated with buffer containing 50 mM Tris-HCl, pH 7.5, 400 mM NaCl, and 1 mM EDTA. Five hundred microliters of chromatin extract was applied to the column, the chromatography was carried out at a flow rate of 0.2 ml/min at 4°C, and 0.5-ml fractions were collected.
In vitro methylation assay.
The assay was performed as described previously (45). A biotinylated 30-bp DNA oligonucleotide duplex (5′ biotin-GAAGCTGGGACTTCCGGGAGGAGAGTGCAA-3′ and 5′-TTGCACTCTCCTCCCGGAAGTCCCAGCTTC-3′) containing a single CG site (45) was used as a substrate. The DNA substrate (400 nM) was incubated with full-length Dnmt3a or Dnmt3b (20 nM each) or their catalytic domains (50 nM each) alone or in equimolar combination in methylation buffer containing 20 mM HEPES, pH 7.5, 15 mM NaCl, 1 mM EDTA, 20 μg/ml bovine serum albumin (BSA), and 0.25 μM S-[methyl-3H]AdoMet (80 Ci/mmol; GE Healthcare) in a total volume of 50 μl at 37°C for 30 min. Scintillation counts were then measured to determine the incorporation of methyl-3H into the substrate DNA.
In vivo DNA methylation assay.
Genomic DNA (30 to 300 ng) was restricted with EcoRV and treated with sodium bisulfite as previously described (42). Treated DNA was subjected to nested PCR (information available upon request). For combined bisulfite restriction analysis (COBRA), about 500 ng of PCR products were restricted with TaqI (Takara), followed by electrophoresis in a 2% agarose gel, and quantified by TotalLab1.10 (Nonlinear Dynamics Ltd.). For sequencing analysis, the PCR products were cloned into T-vectors (Takara) and individual clones sequenced by Invitrogen Ltd.
Whole-mount in situ hybridization.
Digoxigenin-labeled riboprobes (Oct4 and Nanog cDNA, sense and antisense) were generated using T7 and Sp6 polymerases from pSPT19 (Roche) containing full-length Oct4 or Nanog cDNA using the DIG RNA labeling kit (Roche). Embryos were processed according to the standard procedure (38), and in situ hybridization was carried out as described previously (21). Embryos were stored in PBT (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4, 1% Tween 20) containing 70% glycerol and photographed using a digital camera (DC300F; Leica) on a microscope (MZ FLIII; Leica).
Quantitative PCR.
Total cell RNA was reverse transcribed and assayed by quantitative real-time PCR using SYBR Green incorporation. The expression of genes under investigation was normalized to that of β-actin. Primer information is available upon request.
RESULTS
Dnmt3a and Dnmt3b form a common complex in ES and P19 cells.
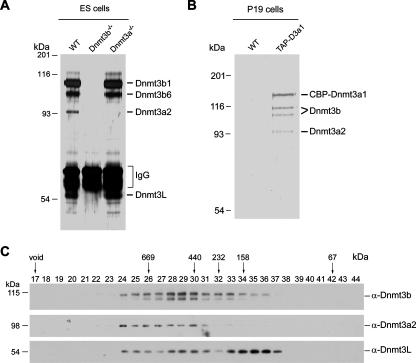
Mouse ES cells express high levels of Dnmt3a2 and Dnmt3b (15, 41) and are capable of conferring de novo methylation on retroviral elements (34) and certain endogenous genes upon cell differentiation (39). ES cells may retain the methylation activity of the inner cell mass of a mouse blastocyst prior to the onset of genome-wide methylation during embryonic development. In order to investigate the regulation of DNA methylation, we tried to identify proteins associated with the de novo DNA methyltransferases in ES cells. Affinity purification using a monoclonal antibody specific for Dnmt3b revealed four prominent proteins in the wild-type ES cells, which could not be purified from the Dnmt3b-deficient ES cells (Fig. 1A, lanes 1 and 2). Mass spectrometric analysis (data not shown) indicated that the two upper bands are Dnmt3b1 and Dnmt3b6, two isoforms of Dnmt3b (14). The third but less abundant one is Dnmt3a2, the Dnmt3a isoform predominantly expressed in ES cells (15). Consistently, Dnmt3a2 is absent in the fraction purified from the Dnmt3a knockout ES cells (Fig. 1A, lane 3). The fourth copurified major protein is Dnmt3L, a factor known to interact with Dnmt3a and Dnmt3b and stimulate their activity (25). Notably, copurification of Dnmt3L with Dnmt3b was also achieved in the Dnmt3a knockout ES cells (Fig. 1A, lane 3). These data suggest that the two methyltransferases and their regulator may form a complex in ES cells. Importantly, the association between the endogenous enzymes remains stable in differentiating ES cells as shown by coimmunoprecipitation (data not shown).
FIG. 1.
Affinity purification of Dnmt3a- and Dnmt3b-associated proteins from ES and EC cells. (A) Silver staining of copurified Dnmt3a and Dnmt3b proteins from ES cells. Affinity purification was performed with an anti-Dnmt3b monoclonal antibody, followed by protein identification by matrix-assisted laser desorption ionization (MALDI) mass spectrometry. WT, wild type. (B) Silver staining of copurified Dnmt3a and Dnmt3b proteins from P19 cells. Affinity purification was performed using a TAP-tagging procedure through stably expressed TAP-tagged Dnmt3a, followed by protein identification by MALDI mass spectrometry. The protein identification was also confirmed by Western blotting using specific antibodies (data not shown). (C) Western detection of Dnmt3a, Dnmt3b, and Dnmt3L in the fractions separated by gel filtration using a Sepharose 6 column. The input was chromatin extract prepared from ES cells. Antibodies used are shown at the right (α-Dnmt3b, -Dnmt3a2, and -Dnmt3L, anti-Dnmt3b, -Dnmt3a2, and -Dnmt3L, respectively). All three blots were from the same gel. The elution profile of the protein markers is indicated at the top.
Since Dnmt3b appears to be associated with Dnmt3L in ES cells and Dnmt3L is known to interact with Dnmt3a (27), we asked whether the complex formation between Dnmt3a2 and Dnmt3b depends on Dnmt3L. P19 is a teratocarcinoma cell line expressing Dnmt3a and Dnmt3b but not Dnmt3L (data not shown). We established a stable P19 cell line which expresses TAP-tagged Dnmt3a1 with an expression level similar to that of the endogenous protein. Tandem affinity purification revealed three major proteins copurified with CBP-Dnmt3a1 (Fig. 1B). These proteins appear in a similar molar ratio and are absent in the fraction from the wild-type P19 cells lacking TAP-tagged Dnmt3a1. Mass spectrometric analysis indicated that the copurified proteins are two isoforms of Dnmt3b (likely Dnmt3b1 and -3b6) and Dnmt3a2, respectively (data not shown). This result confirms the association between Dnmt3a and Dnmt3b, which is independent of Dnmt3L. Moreover, the Dnmt3a and Dnmt3b isoforms appear to be close to equimolar in the fraction. Comparison with the purification result from ES cells suggests that most Dnmt3a2 in ES cells associates with Dnmt3b while a small amount of Dnmt3b associates with Dnmt3a2.
To further characterize the methyltransferase complex(es), we applied the chromatin fraction of ES cells to a gel filtration chromatography column to explore the relationship of the major components. As shown in Fig. 1C, Dnmt3a2 and the two Dnmt3b isoforms (Dnmt3b1 and -3b6) coeluted in the fractions with molecular masses ranging from 750 to 350 kDa (Fig. 1C), suggesting that they may exist in a multisubunit complex. Consistent with the detection of the Dnmt3b-Dnmt3L complex in Dnmt3a knockout cells (Fig. 1A, lane 3), the elution profiles of two components overlapped in a wider range of fractions, even when Dnmt3a2 was hardly detected after the 232-kDa marker (Fig. 1C). In sum, the purification and characterization of proteins associated with Dnmt3a and Dnmt3b from both ES and P19 cells unequivocally indicates that the two methyltransferases exist in a stable complex in vivo, though each of them alone might also be associated with Dnmt3L.
Direct association of Dnmt3a and Dnmt3b.
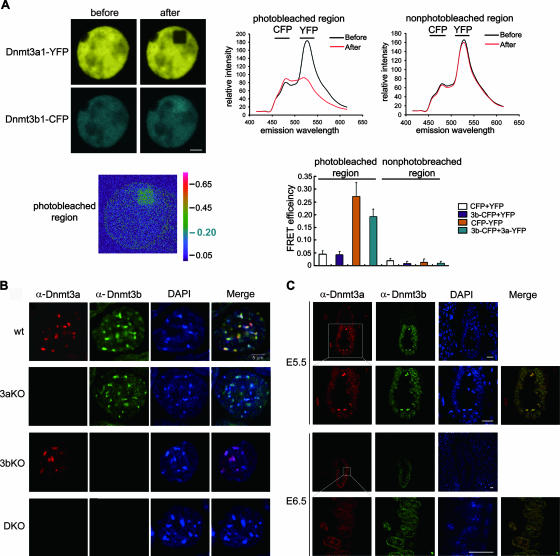
The copurification of Dnmt3a and Dnmt3b did not address whether the association of the two enzymes occurs directly or is mediated by any unknown proteins. We therefore performed a fluorescence resonance energy transfer (FRET) assay to test the spatial relationship between Dnmt3a and Dnmt3b in living cells (Fig. 2A). Dnmt3a1-yellow fluorescent protein (YFP) and Dnmt3b1-cyan-fluorescent protein (CFP) fusion proteins were coexpressed in 293T cells as acceptor and donor fluorophores, respectively. Upon photobleaching of YFP, the energy of CFP increased by about 20% in cells coexpressing Dnmt3a1-YFP and Dnmt3b1-CFP (Fig. 2A). Such an efficiency of energy transfer indicates a <10-nm distance between two fluorescent partners (19), suggesting a direct contact between Dnmt3a and Dnmt3b in transfected cells.
FIG. 2.
Association between Dnmt3a and Dnmt3b in vivo. (A) FRET analysis of Dnmt3a1-YFP and Dnmt3b1-CFP fusion proteins expressed in 293T cells. Scale bar, 3 μm. The line chart to the right illustrates emission spectra of Dnmt3b1-CFP and Dnmt3a1-YFP before (black) or after (red) photobleaching. The bar chart below shows the averaged FRET efficiency for coexpression of CFP and YFP (CFP+YFP) (negative control; n = 25), Dnmt3b1-CFP and -YFP (3b-CFP+YFP) (n = 25), Dnmt3b1-CFP and Dnmt3a1-YFP (3b-CFP+3a-YFP) (n = 30), or CFP-YFP fusion protein (positive control; n = 25). Data are representative of at least three independent experiments. (B) Colocalization of endogenous Dnmt3a and Dnmt3b in the interphase nuclei of ES cells. Heterochromatin domains are brightly stained with 4′,6′-diamidino-2-phenylindole (DAPI). A similar staining pattern was observed in differentiating ES cells treated with RA for 2 days. Antibodies used did not distinguish different isoforms. wt, wild type; 3aKO, Dnmt3a knockout; 3bKO, Dnmt3b knockout; DKO, double knockout; α-Dnmt3a and -Dnmt3b, anti-Dnmt3a and -Dnmt3b antibodies. (C) Colocalization of Dnmt3a and Dnmt3b in mouse embryos. Embryos were frozen and sliced, followed by immunostaining with the antibodies indicated at the top. The squared epiblast regions are shown at higher magnification below. Scale bar, 20 μm.
To further characterize the relationship of Dnmt3a and Dnmt3b, we examined their subcellular distribution in ES cells by immunostaining for the endogenous proteins. In wild-type ES cells, Dnmt3a2 and Dnmt3b are dispersed throughout the nucleus with colocalization of speckles in heterochromatin domains in the interphase (Fig. 2B, upper row). The heterochromatin association is independent of the complex formation, because enrichment of one enzyme at the heterochromatin foci still occurs in cells lacking the other enzyme (Fig. 2B, two middle rows). The similar distribution patterns of Dnmt3a2 and Dnmt3b in ES cells is consistent with their presence as a complex revealed by affinity purification.
Methylation patterns are reprogrammed genome-wide during postimplantation embryonic development. We next investigated whether these two enzymes are present together in embryonic cells undergoing de novo methylation by using fluorescence immunostaining of embryo sections. Both enzymes were detected in the nuclei of epiblast cells in the 5.5- and 6.5-day-postcoitum (dpc) embryos (Fig. 2C). Subcellular colocalization of the two proteins was again observed in heterochromatin regions including the nuclear peripherals (Fig. 2C, second and fourth rows), consistent with the need for heavy methylation of the centromeric repeats. Altogether, the results provide further support for interaction between Dnmt3a and Dnmt3b and their presence within a common complex in embryonic cells.
Dnmt3a and Dnmt3b stimulate methylation activity in vitro.
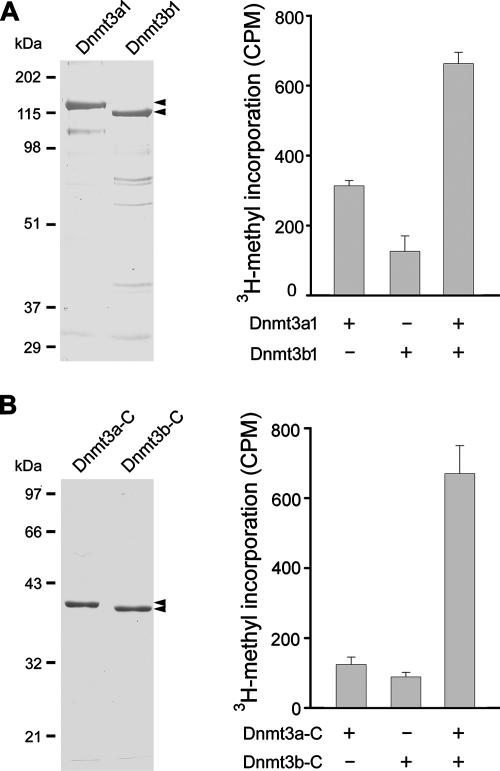
The isolated Dnmt3a and Dnmt3b recombinant proteins display substantially lower de novo methylation activity than other methyltransferases in vitro (5). We surmised that association between the two enzymes may modulate the enzymatic activity, particularly given the fact that the interaction involves the catalytic domain of each enzyme, as we have shown (data not shown). An in vitro methylation assay with purified recombinant proteins did show a strong stimulation between the two enzymes (Fig. 3A); the methyl transfer in the presence of both enzymes exceeded by at least 40% the sum of methyl transfer by the two corresponding enzymes acting alone. The stimulation was even more striking between the two catalytic domains (Fig. 3B). The activity in the presence of both exceeded by 200% the combined activities of two individual enzymes acting alone. These results clearly demonstrate that each of the two enzymes stimulates the methylation activity of the other in vitro. It remains to be seen whether the stimulation between the Dnmt3a and Dnmt3b catalytic domains occurs through a mechanism similar to that of Dnmt3L, which promotes the binding of Dnmt3a to DNA and the cofactor AdoMet (25).
FIG. 3.
Stimulation of methyltransferase activity by Dnmt3a-Dnmt3b interaction in vitro. (A) Stimulation of methyltransferase activity between full-length enzymes. Six-His-tagged Dnmt3a1 and Dnmt3b1 purified from Escherichia coli were visualized by Coomassie blue staining in a 10% sodium dodecyl sulfate-polyacrylamide gel (left panel). Methyltransferase activity of Dnmt3a1 or Dnmt3b1 alone or in the equimolar presence of both was measured (right panel). Data for incorporation in cpm are averages from three independent experiments ± standard deviations. (B) Stimulation of methyltransferase activity between the catalytic domains. Six-His-tagged catalytic domains of Dnmt3a (Dnmt3a-C) and Dnmt3b (Dnmt3b-C) purified from E. coli were visualized by Coomassie blue staining (left panel). Their methyltransferase activities were examined alone or in equimolar combination (right panel).
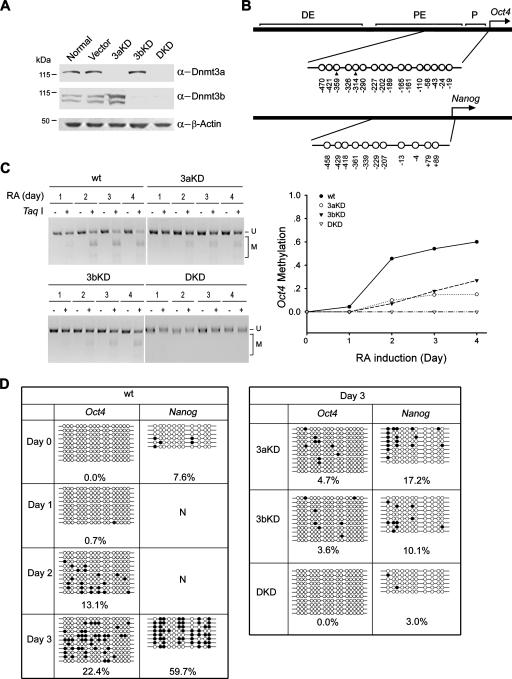
Cooperation of Dnmt3a and Dnmt3b in methylation of Oct4 and Nanog promoters during differentiation of P19 cells upon RA induction.
The results presented in the previous sections demonstrate that Dnmt3a and Dnmt3b associate in a complex to enhance each other's activity. We anticipated that their physical and functional interaction may contribute to the regulation of gene expression in vivo. We next chose cis-regulatory elements of Oct4 and Nanog, the genes which control pluripotency in development, to further explore the in vivo biological relevance of the interaction of Dnmt3a and Dnmt3b. The promoter of Oct4 undergoes progressive methylation in association with transcriptional silencing in embryonic carcinoma (EC) cells after the onset of differentiation induced by the addition of retinoic acid (RA) to the growth medium (3). To determine the involvement of Dnmt3a and Dnmt3b in the methylation of the Oct4 and Nanog promoters, we knocked down the expression of Dnmt3a and Dnmt3b individually or in combination by stably transfecting small interfering RNA into P19 EC cells (Fig. 4A) and analyzed the methylation pattern of a regulatory regions upstream of Oct4 and Nanog (Fig. 4B) upon induction. Consistent with a previous report (20), COBRA and bisulfite sequencing analyses showed no methylation in uninduced proliferating P19 cells (Fig. 4C and D). During 4 days of induction with RA, the methylation level of the Oct4 promoter increased steadily in wild-type P19 cells, indicating a process of de novo methylation accompanying gene silencing during cell differentiation. However, when Dnmt3a or Dnmt3b was knocked down, methylation levels were much lower than those in wild-type cells starting from day 2 (Fig. 4C). Bisulfite sequencing revealed that less than 5% of the scored CpG sites were methylated in these cells after 3 days of RA induction (Fig. 4D, right), while the methylation level in wild-type cells had reached 22.4% by this time (Fig. 4D, left). In Dnmt3a and Dnmt3b double-knockdown cells, no single CpG site could be methylated after RA induction (Fig. 4D), indicating that Dnmt3a and Dnmt3b are required and sufficient for the establishment of methylation of the Oct4 promoter. Notably, the sum of methylated CpG sites of the two single knockdown cell lines is much smaller than that of wild-type cells (4.7% plus 3.6% versus 22.4%) (Fig. 4D, right), indicating a functional cooperation between Dnmt3a and Dnmt3b for the methylation of Oct4. The synergistic effect of Dnmt3a and Dnmt3b is even more evident for the methylation of the promoter of Nanog (17.2% plus 10.1% versus 59.7%) (Fig. 4D).
FIG. 4.
Depletion of either Dnmt3a or Dnmt3b affects methylation of the Oct4 promoter in P19 cells upon differentiation. (A) Depletion of Dnmt3a and Dnmt3b in stably transfected P19 cell lines expressing small interfering RNA. The expression level of Dnmt3a and Dnmt3b in knockdown (KD) cell lines was examined by Western analysis of total cell lysates using specific antibodies (α-Dnmt3a, -Dnmt3b, and -β-actin, anti-Dnmt3a, -Dnmt3b, and -β-actin). Equal loading was verified by the detection of β-actin. (B) Schematic representation of the regulatory region upstream of Oct4 and Nanog. The distribution of CpG dinucleotides (circles), analyzed by bisulfite sequencing in panel D, is shown below. Two (filled triangles) of the 16 CpGs in the Oct4 upstream region are present in the TaqI restriction sites used for the COBRA methylation assay. DE, distal enhancer; PE, proximal enhancer; P, promoter. (C) COBRA methylation analysis of Oct4 upon cell differentiation induced with RA. Genomic DNA was isolated from wild-type (wt) and Dnmt3 knockdown cells at different days with RA treatment. The methylation status of two CpG sites was assessed by TaqI digestion of the PCR products derived from bisulfite-converted genomic DNA (left) and quantified by measuring the ratio of fully cleaved DNA to total DNA (right). The ratio from the complete digestion corresponding to full methylation at the TaqI sites was set to 1. U, uncut fragments derived from unmethylated DNA; M, cleaved fragments derived from methylated DNA. (D) Bisulfite sequencing analysis. PCR products amplified from bisulfite-treated genomic DNA were cloned and sequenced to reveal the methylation statuses of individual CpG sites. Percentages of the methylated CpG sites (filled circles) among all scored sites are indicated.
Cooperation of Dnmt3a and Dnmt3b in the methylation of Oct4 and Nanog promoters during ES cell differentiation and embryonic development.
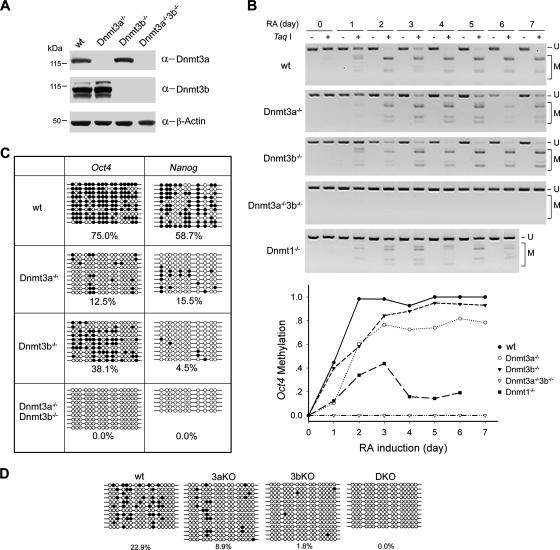
While P19 cells may represent embryonic cells from a postimplantation embryo around dpc 6.5, mouse ES cells are generally believed to have a developmental potential similar to that of an earlier embryo at 3.5 and 4.5 dpc. Dnmt3L, one of the proteins stably associated with Dnmt3a and Dnmt3b, is expressed in ES but not in P19 cells (Fig. 1; also data not shown). To assess the functional cooperation between Dnmt3a and Dnmt3b in the presence of different cell line-specific factors, we extended the investigation of the methylation process of Oct4 to ES cells (Fig. 5). In wild-type ES cells, the methylation of Oct4 was rapidly established upon RA induction and reached a plateau by day 2 (Fig. 5B). When either Dnmt3a or Dnmt3b was knocked out, Oct4 methylation was substantially reduced in the first 2 days. Though methylation gradually increased in the following days, there was still a considerable gap compared to the wild-type level (Fig. 5B). Quantification of bisulfite sequencing data revealed that by day 3, only 12.5% and 38.1% of CpG sites were methylated in Dnmt3a and Dnmt3b knockout ES cells, respectively (Fig. 5C). The combined methylation levels of the two single-knockout ES cell lines (50.6%) are much lower than that of wild-type cells (75.0%), indicating that a lack of either paralog affects the de novo methylation activity at Oct4 at this time point. A similar result was seen in the analysis of methylation at the promoter of Nanog (15.5% plus 4.5% is <58.7%) (Fig. 5C). Methylation did not occur at all in the absence of the two Dnmt3 enzymes, because no single sites were found to be methylated in the double-knockout ES cells (Fig. 5C). In contrast, the lack of the Dnmt1 methyltransferase does not prevent the establishment of methylation (Fig. 5B). The reduction in Oct4 methylation in Dnmt1-deficient cells was nevertheless more drastic than in cells without Dnmt3a or Dnmt3b, presumably as a consequence of the complete failure to maintain freshly established methylation patterns in dividing cells. Interestingly, the methylation level in Dnmt1−/− cells stopped rising at day 3 and then started to drop. This is likely caused by a passive demethylation during cell divisions in the absence of Dnmt1 plus insufficient de novo methylation due to the down-regulation of Dnmt3b and Dnmt3L after day 3 in the course of RA induction (data not shown).
FIG. 5.
Deficiency in either Dnmt3a or Dnmt3b reduces DNA methylation at the Oct4 promoter in ES cells and embryos. (A) Confirmation of wild-type (wt) and knockout ES cell lines by Western blotting. (B) COBRA methylation analysis of the Oct4 promoter. Wild-type and knockout ES cells were induced with RA for up to 7 days, and the methylation levels of Oct4 at different time points were assessed by TaqI restriction assay of the PCR products derived from bisulfite-treated genomic DNA and quantified (bottom). Labeling of the digested fragments is as in Fig. 4C. (C) Bisulfite sequencing analysis of Oct4 and Nanog at day 3 upon RA treatment. Percentages of the methylated CpG sites (filled circles) are indicated. (D) Bisulfite sequencing analysis of Oct4 in mouse embryos. Genomic DNA was from wild-type (wt), single knockout (3aKO and 3bKO), or double knockout (DKO) embryos collected at E9.5 (three for each genotype).
The colocalization of Dnmt3a and Dnmt3b in postimplantation embryos (Fig. 2C) suggests a functional cooperation of the two enzymes in de novo methylation of Oct4 during development. We next assessed their contribution to Oct4 methylation by analyzing knockout embryos (Fig. 5D). In wild-type embryos, the methylation level reached 22.9% at embryonic day 9.5 (E9.5), similar to a previous observation by Gu et al. (26). In Dnmt3a or Dnmt3b knockout embryos, the methylation levels dropped to 8.9% and 1.8%, respectively. In double-knockout embryos, not a single methylated site could be detected. Similar to the situation in P19 and ES cells, far more extensive methylation occurs in wild-type embryos than in single-knockout embryos. The methylation level in wild-type embryos exceeded by >100% the sum of methylation found in singly knocked out embryos (compare 22.9% to 8.9% plus 1.8%). These results indicate cooperation by the two methyltransferases in the establishment of methylation patterns on Oct4 during embryonic development. Interestingly, reduction of methylation in the Dnmt3b-deficient embryo is more drastic than that in the Dnmt3a-deficient embryo.
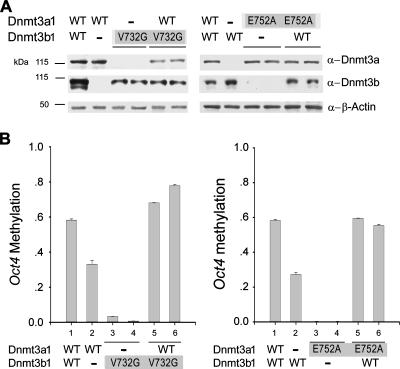
Dnmt3a and Dnmt3b mutually stimulate de novo methylation at the Oct4 promoter in ES cells.
We have shown above that the presence of both Dnmt3a and Dnmt3b increases their collective methylation activity in vitro and in vivo. To further characterize the relationship between their physical and functional interactions, we introduced a catalytically inactive mutant of Dnmt3b (V732G) (50) into Dnmt3b−/− ES cells and examined whether the mutant can stimulate de novo methylation at Oct4 by Dnmt3a. The Dnmt3b mutant was expressed at a lower level than the endogenous protein in wild-type ES cells (Fig. 6A) but lacked activity on its own because <5% of methylation could be detected in DKO cells expressing the mutant (Fig. 6B, left graph, bars 3 and 4). However, the introduction of this Dnmt3b mutant into Dnmt3b−/− cells increased the methylation activity of the endogenous Dnmt3a by 25% (from 50% to 75%) (compare bars 5 and 6 to bar 2). An inactive mutant of Dnmt3a (E752A) stimulated Dnmt3b-mediated methylation at the Oct4 promoter as efficiently as the endogenous wild-type Dnmt3a (Fig. 6B, right graph). Consistent with their capacity for enzymatic stimulation, both the Dnmt3a and Dnmt3b mutant proteins are able to interact with their respective paralog in a coimmunoprecipitation assay (data not shown). Taken together, we conclude that protein interaction and reciprocal stimulation occur between the two methyltransferases in the methylation of the Oct4 promoter during ES cell differentiation and that this stimulation is not dependent on the methyltransferase activity of the stimulator.
FIG. 6.
Dnmt3a and Dnmt3b mutually stimulate de novo methylation at the Oct4 promoter in differentiating ES cells. (A) Expression of ectopic (with gray-shadowed background) and endogenous methyltransferases in stably established ES cell lines by Western blotting. Catalytically inactive mutant Dnmt3b1 V732G, Dnmt3a1 E752A, or wild-type (WT) counterparts were stably introduced into Dnmt3a−/−, Dnmt3b−/−, or Dnmt3a−/−3b−/− ES cells. (B) Analysis of Oct4 methylation 3 days upon RA induction by COBRA. Each bar represents an individual stable cell line.
DNA methylation is required for normal progression of Oct4 and Nanog inactivation.
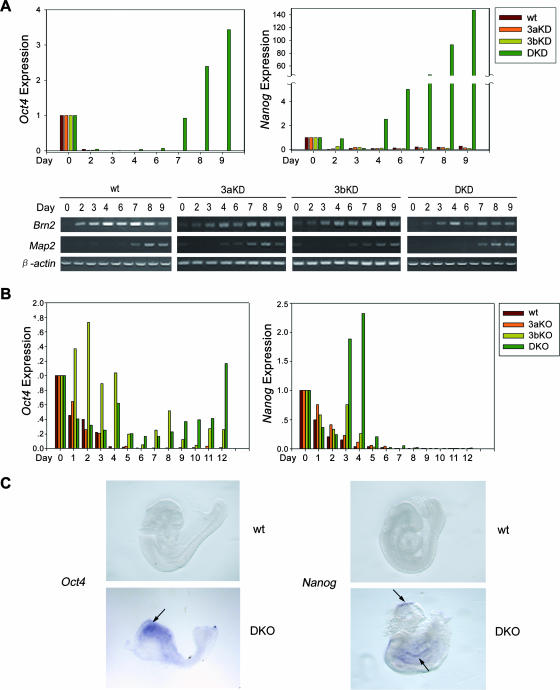
Inactivation of Oct4 and other early embryonic genes in development is a multistep process involving the action of transcription repressors, histone deacetylation and methylation, and chromatin remodeling in addition to DNA methylation. However, little is known about the precise role of DNA methylation in the inactivation of these target genes. We analyzed their expression in cells and embryos with inadequate DNA methylation resulting from genetic ablation of the de novo methyltransferases. In P19 cells, Oct4 and Nanog are inactivated in response to treatment with the differentiation inducer, retinoic acid (Fig. 7A). In cells depleted of both Dnmt3a and Dnmt3b, transcription of both genes was reactivated (Fig. 7A, DKD [double knockdown] panel) after 3 days of down-regulation. The increase in the transcription level is unlikely to be due to an increase in the percentage of surviving multipotent cells unresponsive to RA amid differentiating and dying cells. First, transcription in methylation-deficient cells started to rise at day 4 (Fig. 7A), but severe cell death was not observed until day 5 (data not shown). Second, some progression of neuronal differentiation occurred because the expression of the neural stem cell marker gene Brn2 and the neuron marker gene Map2 was detectable by reverse transcription-PCR in the cell population (Fig. 7A). Moreover, immunostaining for another neuron marker, Tuj1, revealed a similar expression pattern in normal and double-knockdown P19 cells 7 days after RA induction (data not shown).
FIG. 7.
Promoter hypomethylation leads to misregulation of expression of Oct4 and Nanog in differentiating cells and developing embryos. (A) Expression of Oct4 and Nanog in P19 cells at various time points upon RA induction, measured by quantitative PCR analyses. The expression of differentiation markers Brn2 and Map2 was examined by reverse transcription-PCR (lower panels). Labels of cell lines are as in Fig. 4. (B) Expression of Oct4 and Nanog in ES cells at different time points upon RA induction measured by quantitative PCR analyses. Cell lines used are as in Fig. 5A. (C) Whole-mount in situ hybridization of Oct4 and Nanog in E9.5 wild-type and double knockout (DKO) embryos.
Transcriptional repression was also compromised during the differentiation of ES cells deficient in both Dnmt3a and Dnmt3b (Fig. 7B). Oct4 was reactivated at day 10 following earlier repression. Though Nanog was silenced after day 6, its reactivation was obvious at days 2 and 4. Notably, depletion of either enzyme altered the progression course of silencing of the two genes in both P19 and ES cells, with delayed or relatively inefficient inactivation at certain time points. The timing and degree of reactivation could also be influenced by the presence of epigenetic modifications other than DNA methylation or a change in available transcription factors upon differentiation.
We then wanted to know whether deficiency in methyltransferases and inadequate promoter methylation could bring about misexpression of Oct4 and Nanog in mouse embryos. By whole-mount in situ hybridization, we observed abnormal expression of Oct4 in the anterior part and Nanog in two different regions of E9.5 mutant embryos lacking both Dnmt3a and Dnmt3b, while normal silencing of both target genes occurred in wild-type embryos from the same litter (Fig. 7C). In single-knockout embryos, no significant upregulation of the two genes was observed (data not shown). These data, along with the observations for P19 and ES cells, indicate that aberrant DNA methylation can cause dysregulation of the expression of the two pluripotency genes in development.
DISCUSSION
DNA methylation is an epigenetic modification responsible for transcriptional silencing of gene expression. Methylation patterns are introduced onto the genome by the action of two de novo methyltransferases, Dnmt3a and Dnmt3b, in embryonic development and are then stably maintained in cell divisions. Alterations of both genome-wide and gene-specific methylation are incompatible with normal development. For example, aberrant hypermethylation of tumor suppressor genes cause their inactivation, promoting carcinogenesis. The activity of de novo methylation must therefore be tightly regulated. In this work, we demonstrate that the two de novo methyltransferases form a complex in mammalian cells. The two enzymes interact directly both in vitro and in vivo. The absence of either leads to decreased methylation, which would not be the case if they were functionally redundant. More importantly, the two enzymes cooperate in the methylation of the Oct4 and Nanog genes during embryonic cell differentiation; disruption of either gene leads to a drastic reduction in the methylation level and misexpression of Oct4 and Nanog. We propose that the synergy derived from the interaction and stimulation between Dnmt3a and Dnmt3b is a molecular mechanism for the regulation of methyltransferase activity in development.
Complex formation between Dnmt3a and Dnmt3b and regulation of cellular methylation activity.
The expression of Dnmt3a and Dnmt3b is developmentally regulated. Both genes are expressed at a high level in undifferentiated cells, including embryonic stem cells, and undergo down-regulation upon cell differentiation (15, 49). In adult tissues, both enzymes are undetectable or exist at a low level, in agreement with the notion that there is no need for de novo methylation in differentiated cells. Though the tissue-specific expression patterns of the two methyltransferase genes are not fully identical, both are expressed in somatic and germ cells undergoing de novo methylation. Indeed, our data show that both proteins colocalize in embryonic cells and form a complex by direct protein interaction. This allows the regulatory potential for methylation activity to be extended through the selective expression of isoforms from the two paralogous genes in a given tissue. It is known that there are two isoforms of Dnmt3a, Dnmt3a1 and Dnmt3a2, transcribed from alternative promoters (15). Both isoforms are enzymatically active, but they may have different properties. Dnmt3a2 lacks the N-terminal 219 amino acids and tends to associate with euchromatin. Dnmt3b has six isoforms that result from alternative splicing of three exons. Only the full-length isoform 3b1 and a short isoform, 3b6, are present in ES cells (14, 15). Dnmt3b6 lacks part of motif IX (63 amino acids) in the catalytic domain and is enzymatically inactive. This short form shows reduced association with small nuclear foci and accumulation in pericentric heterochromatin (48). However, the absence of sequences in the shorter Dnmt3a and Dnmt3b isoforms should not influence interaction between the two paralogous proteins (48) (data not shown). Indeed, our work demonstrates the presence of all ES isoforms in copurified fractions (Fig. 1). Such a combinational property of the methyltransferase complex(es) may add another layer to regulation of methylation activity in development, for example, through selective expression of isoforms with various catalytic activities and subcellular targeting abilities mentioned above. We anticipate that the characterization of the precise composition along with that of less stably associated proteins and the determination of stoichiometry will facilitate understanding of the regulatory mechanism of DNA methylation.
Most recently, Dnmt3L was reported to form a complex with Dnmt3a and to direct DNA methylation through binding to unmethylated lysine 4 on the histone H3 tail (29, 43). Since Dnmt3a and Dnmt3b are coexpressed with Dnmt3L in pluripotent and germ line cells, the relationship between these three proteins in complexes needs to be characterized to understand the mechanism of de novo methylation.
Significance of functional interaction between Dnmt3a and Dnmt3b in development.
Cell differentiation during early embryonic development depends on the transcriptional activation of lineage-specific genes as well as inactivation of genes that specify proliferative characteristics of embryonic stem cells (46). Oct4 and Nanog are the two most important genes for the self-renewal and maintenance of the undifferentiated state of ES cells and are down-regulated with the initiation of cell differentiation. The inactivation of Oct4 is a multistep program involving recruitment of transcriptional repressors, alterations in chromatin structure, acquisition of histone H3K9 methylation, and DNA methylation at their promoters (20, 26). The detailed mechanism and the relative contribution of DNA methylation to the regulation of Oct4 and Nanog are not understood. We have addressed these points by analyzing promoter methylation and transcription of Oct4 and Nanog genes in the differentiation of cultured EC cells and ES cells and in the development of early mouse embryos which are deficient in DNA methylation. One major finding is a clear physical and functional cooperation between the two de novo methyltransferases for the proper methylation of Oct4 and Nanog in early development. Though methylation still proceeds with one enzyme, the efficiency is much reduced, resulting in partial or delayed methylation. The cooperative action between the two enzymes may not be restricted to Oct4 and Nanog. It may rather extend to the methylation of other genes or sequences, including those in the heterochromatin, since both enzymes are expressed and show a colocalized subcellular distribution in early postimplantation embryos known to undergo genome-wide de novo methylation (Fig. 2C). The functional interdependency required to achieve full methylation may provide a mechanistic explanation for why both methyltransferase genes are required for normal development in mice and humans (40, 51).
Our second finding is the role of DNA methylation in the transcriptional control of Oct4 and Nanog during mouse embryonic development. While localized overexpression is evident in mutant embryos deficient in both methyltransferases, up-regulation does not occurs when one of the enzymes is absent (Fig. 7C), irrespective of the defective methylation state of their promoters (Fig. 5D). This baffling inconsistency might be explained by overcompensation through a drastic increase in the expression of the remaining paralogous enzyme and Dnmt3L in the mutant embryos. Indeed, Dnmt3L expression increases by fivefold in Dnmt3b knockout embryos (unpublished data). The recruitment of these up-regulated factors to the promoters may exert transcriptional repression independently of cytosine methylation (1, 2). Overcompensation of transcriptional regulation can also arise through epigenetic mechanisms other than DNA methylation, such as histone methylation in the embryonic tissues. We propose that a subtle change, including up- and down-regulation of developmentally important genes such as Oct4 and Nanog in early embryonic stages, may have a long-lasting impact on development. This possibility could be of particular relevance for the interpretation of the late lethality of Dnmt3a null mice. The mutant animals can develop to term but die a few weeks after birth, with no detectable hypomethylation in the genome (40). Thus, some of the developmental defects seen in the adult mice could be a result of misexpression of pluripotency genes in early stages of embryonic development in a manner similar to that in EC and ES cells as we have observed.
Consistent with the importance of DNA methylation in embryos, derepression of Oct4 in association with promoter hypomethylation has been found in the in vitro differentiation of pluripotent cells deficient in the histone methyltransferase G9a and the orphan receptor GCNF (20, 26). All these observations suggest that DNA methylation is essential for establishing a spatially and temporally correct expression pattern for the target genes crucial for normal development. This is particularly important in somatic cloning, which requires timely reactivation and subsequent resilencing of Oct4 and Nanog (8, 47). In light of the essential function of DNA methylation in transcriptional repression of Oct4 and Nanog and the well-established roles of these two genes in specifying toti-/pluripotency (32), we suggest that the state of DNA methylation is an important determinant and indicator of cellular developmental potential.
Acknowledgments
We thank Y.-G. Hu for Dnmt3b antibodies and C. P. Walsh (University of Ulster) for critical review of the manuscript.
This work was supported by grants from the Max-Planck-Gesellschaft of Germany, the Ministry of Science and Technology of China (grants 2005CB522400 and 2006CB943900), the National Science Foundation of China, and Shanghai Municipal Commission for Science and Technology to G.-L.X.
Footnotes
Published ahead of print on 15 October 2007.
REFERENCES
- 1.Aapola, U., I. Liiv, and P. Peterson. 2002. Imprinting regulator DNMT3L is a transcriptional repressor associated with histone deacetylase activity. Nucleic Acids Res. 30:3602-3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachman, K. E., M. R. Rountree, and S. B. Baylin. 2001. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J. Biol. Chem. 276:32282-32287. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Shushan, E., E. Pikarsky, A. Klar, and Y. Bergman. 1993. Extinction of Oct-3/4 gene expression in embryonal carcinoma x fibroblast somatic cell hybrids is accompanied by changes in the methylation status, chromatin structure, and transcriptional activity of the Oct-3/4 upstream region. Mol. Cell. Biol. 13:891-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bestor, T., A. Laudano, R. Mattaliano, and V. Ingram. 1988. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J. Mol. Biol. 203:971-983. [DOI] [PubMed] [Google Scholar]
- 5.Bestor, T. H. 2000. The DNA methyltransferases of mammals. Hum. Mol. Genet. 9:2395-2402. [DOI] [PubMed] [Google Scholar]
- 6.Bestor, T. H., and D. Bourc'his. 2004. Transposon silencing and imprint establishment in mammalian germ cells. Cold Spring Harb. Symp. Quant. Biol. 69:381-387. [DOI] [PubMed] [Google Scholar]
- 7.Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6-21. [DOI] [PubMed] [Google Scholar]
- 8.Boiani, M., S. Eckardt, H. R. Scholer, and K. J. McLaughlin. 2002. Oct4 distribution and level in mouse clones: consequences for pluripotency. Genes Dev. 16:1209-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bortvin, A., K. Eggan, H. Skaletsky, H. Akutsu, D. L. Berry, R. Yanagimachi, D. C. Page, and R. Jaenisch. 2003. Incomplete reactivation of Oct4-related genes in mouse embryos cloned from somatic nuclei. Development 130:1673-1680. [DOI] [PubMed] [Google Scholar]
- 10.Bourc'his, D., F. Ledeist, and E. Viegas-Pequignot. 2003. DNMT3B and the immunodeficiency-centromeric instability-facial anomalies syndrome, p. 776-779. In C. J. Epstein, R. P. Erickson, and A. Wynshaw-Boris (ed.), Inborn errors of development: the molecular basis of clinical disorders of morphogenesis. Oxford University Press, New York, NY.
- 11.Bourc'his, D., G. L. Xu, C. S. Lin, B. Bollman, and T. H. Bestor. 2001. Dnmt3L and the establishment of maternal genomic imprints. Science 294:2536-2539. [DOI] [PubMed] [Google Scholar]
- 12.Chedin, F., M. R. Lieber, and C. L. Hsieh. 2002. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc. Natl. Acad. Sci. USA 99:16916-16921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, T., N. Tsujimoto, and E. Li. 2004. The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol. Cell. Biol. 24:9048-9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, T., Y. Ueda, J. E. Dodge, Z. Wang, and E. Li. 2003. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell. Biol. 23:5594-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, T., Y. Ueda, S. Xie, and E. Li. 2002. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J. Biol. Chem. 277:38746-38754. [DOI] [PubMed] [Google Scholar]
- 16.Chen, Z. X., J. R. Mann, C. L. Hsieh, A. D. Riggs, and F. Chedin. 2005. Physical and functional interactions between the human DNMT3L protein and members of the de novo methyltransferase family. J. Cell Biochem. 95:902-917. [DOI] [PubMed] [Google Scholar]
- 17.Chen, Z. X., and A. D. Riggs. 2005. Maintenance and regulation of DNA methylation patterns in mammals. Biochem. Cell Biol. 83:438-448. [DOI] [PubMed] [Google Scholar]
- 18.Chuang, L. S., H. I. Ian, T. W. Koh, H. H. Ng, G. Xu, and B. F. Li. 1997. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science 277:1996-2000. [DOI] [PubMed] [Google Scholar]
- 19.Day, R. N., and F. Schaufele. 2005. Imaging molecular interactions in living cells. Mol. Endocrinol. 19:1675-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feldman, N., A. Gerson, J. Fang, E. Li, Y. Zhang, Y. Shinkai, H. Cedar, and Y. Bergman. 2006. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat. Cell Biol. 8:188-194. [DOI] [PubMed] [Google Scholar]
- 21.Fuhrmann, G., A. C. Chung, K. J. Jackson, G. Hummelke, A. Baniahmad, J. Sutter, I. Sylvester, H. R. Scholer, and A. J. Cooney. 2001. Mouse germline restriction of Oct4 expression by germ cell nuclear factor. Dev. Cell 1:377-387. [DOI] [PubMed] [Google Scholar]
- 22.Fuks, F., W. A. Burgers, N. Godin, M. Kasai, and T. Kouzarides. 2001. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J. 20:2536-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuks, F., P. J. Hurd, R. Deplus, and T. Kouzarides. 2003. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 31:2305-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ge, Y. Z., M. T. Pu, H. Gowher, H. P. Wu, J. P. Ding, A. Jeltsch, and G. L. Xu. 2004. Chromatin targeting of de novo DNA methyltransferases by the PWWP domain. J. Biol. Chem. 279:25447-25454. [DOI] [PubMed] [Google Scholar]
- 25.Gowher, H., K. Liebert, A. Hermann, G. Xu, and A. Jeltsch. 2005. Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J. Biol. Chem. 280:13341-13348. [DOI] [PubMed] [Google Scholar]
- 26.Gu, P., D. Le Menuet, A. C. Chung, and A. J. Cooney. 2006. Differential recruitment of methylated CpG binding domains by the orphan receptor GCNF initiates the repression and silencing of Oct4 expression. Mol. Cell. Biol. 26:9471-9483. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Hata, K., M. Okano, H. Lei, and E. Li. 2002. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129:1983-1993. [DOI] [PubMed] [Google Scholar]
- 28.He, D. C., J. A. Nickerson, and S. Penman. 1990. Core filaments of the nuclear matrix. J. Cell Biol. 110:569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia, D., R. Z. Jurkowska, X. Zhang, A. Jeltsch, and X. Cheng. 2007. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature 449:248-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones, P. A., and S. B. Baylin. 2002. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 3:415-428. [DOI] [PubMed] [Google Scholar]
- 31.Kaneda, M., M. Okano, K. Hata, T. Sado, N. Tsujimoto, E. Li, and H. Sasaki. 2004. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429:900-903. [DOI] [PubMed] [Google Scholar]
- 32.Kehler, J., E. Tolkunova, B. Koschorz, M. Pesce, L. Gentile, M. Boiani, H. Lomeli, A. Nagy, K. J. McLaughlin, H. R. Scholer, and A. Tomilin. 2004. Oct4 is required for primordial germ cell survival. EMBO Rep. 5:1078-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim, G. D., J. Ni, N. Kelesoglu, R. J. Roberts, and S. Pradhan. 2002. Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. EMBO J. 21:4183-4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lei, H., S. P. Oh, M. Okano, R. Juttermann, K. A. Goss, R. Jaenisch, and E. Li. 1996. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 122:3195-3205. [DOI] [PubMed] [Google Scholar]
- 35.Leonhardt, H., A. W. Page, H. U. Weier, and T. H. Bestor. 1992. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71:865-873. [DOI] [PubMed] [Google Scholar]
- 36.Li, E. 2002. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 3:662-673. [DOI] [PubMed] [Google Scholar]
- 37.Li, E., T. H. Bestor, and R. Jaenisch. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69:915-926. [DOI] [PubMed] [Google Scholar]
- 38.Loosli, F., R. W. Koster, M. Carl, A. Krone, and J. Wittbrodt. 1998. Six3, a medaka homologue of the Drosophila homeobox gene sine oculis is expressed in the anterior embryonic shield and the developing eye. Mech. Dev. 74:159-164. [DOI] [PubMed] [Google Scholar]
- 39.Oka, M., A. M. Meacham, T. Hamazaki, N. Rodic, L. J. Chang, and N. Terada. 2005. De novo DNA methyltransferases Dnmt3a and Dnmt3b primarily mediate the cytotoxic effect of 5-aza-2′-deoxycytidine. Oncogene 24:3091-3099. [DOI] [PubMed] [Google Scholar]
- 40.Okano, M., D. W. Bell, D. A. Haber, and E. Li. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247-257. [DOI] [PubMed] [Google Scholar]
- 41.Okano, M., S. Xie, and E. Li. 1998. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 19:219-220. [DOI] [PubMed] [Google Scholar]
- 42.Olek, A., J. Oswald, and J. Walter. 1996. A modified and improved method for bisulphite based cytosine methylation analysis. Nucleic Acids Res. 24:5064-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ooi, S. K., C. Qiu, E. Bernstein, K. Li, D. Jia, Z. Yang, H. Erdjument-Bromage, P. Tempst, S. P. Lin, C. D. Allis, X. Cheng, and T. H. Bestor. 2007. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448:714-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 45.Roth, M., and A. Jeltsch. 2000. Biotin-avidin microplate assay for the quantitative analysis of enzymatic methylation of DNA by DNA methyltransferases. Biol. Chem. 381:269-272. [DOI] [PubMed] [Google Scholar]
- 46.Simonsson, S., and J. Gurdon. 2004. DNA demethylation is necessary for the epigenetic reprogramming of somatic cell nuclei. Nat. Cell Biol. 6:984-990. [DOI] [PubMed] [Google Scholar]
- 47.Takahashi, K., and S. Yamanaka. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663-676. [DOI] [PubMed] [Google Scholar]
- 48.Ueda, Y., M. Okano, C. Williams, T. Chen, K. Georgopoulos, and E. Li. 2006. Roles for Dnmt3b in mammalian development: a mouse model for the ICF syndrome. Development 133:1183-1192. [DOI] [PubMed] [Google Scholar]
- 49.Watanabe, D., I. Suetake, T. Tada, and S. Tajima. 2002. Stage- and cell-specific expression of Dnmt3a and Dnmt3b during embryogenesis. Mech. Dev. 118:187-190. [DOI] [PubMed] [Google Scholar]
- 50.Xie, Z. H., Y. N. Huang, Z. X. Chen, A. D. Riggs, J. P. Ding, H. Gowher, A. Jeltsch, H. Sasaki, K. Hata, and G. L. Xu. 2006. Mutations in DNA methyltransferase DNMT3B in ICF syndrome affect its regulation by DNMT3L. Hum. Mol. Genet. 15:1375-1385. [DOI] [PubMed] [Google Scholar]
- 51.Xu, G. L., T. H. Bestor, D. Bourc'his, C. L. Hsieh, N. Tommerup, M. Bugge, M. Hulten, X. Qu, J. J. Russo, and E. Viegas-Pequignot. 1999. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402:187-191. [DOI] [PubMed] [Google Scholar]