Abstract
Many pre-mRNAs are alternatively spliced in a tissue-specific manner in multicellular organisms. The Fox-1 family of RNA-binding proteins regulate alternative splicing by either activating or repressing exon inclusion through specific binding to UGCAUG stretches. However, the precise cellular contexts that determine the action of the Fox-1 family in vivo remain to be elucidated. We have recently demonstrated that ASD-1 and FOX-1, members of the Fox-1 family in Caenorhabditis elegans, regulate tissue-specific alternative splicing of the fibroblast growth factor receptor gene, egl-15, which eventually determines the ligand specificity of the receptor in vivo. Here we report that another RNA-binding protein, SUP-12, coregulates the egl-15 alternative splicing. By screening for mutants defective in the muscle-specific expression of our alternative splicing reporter, we identified the muscle-specific RNA-binding protein SUP-12. We identified juxtaposed conserved stretches as the cis elements responsible for the regulation. The Fox-1 family and the SUP-12 proteins form a stable complex with egl-15 RNA, depending on the cis elements. Furthermore, the asd-1; sup-12 double mutant is defective in sex myoblast migration, phenocopying the isoform-specific egl-15(5A) mutant. These results establish an in vivo model that coordination of the two families of RNA-binding proteins regulates tissue-specific alternative splicing of a specific target gene.
Tissue-specific alternative splicing of pre-mRNAs is considered to be a major source of proteomic complexity in metazoans (7, 21). Consequences of the alternative splicing for protein structure and function as well as for cellular processes have been discussed (45). Recent global studies demonstrated that as many as two-thirds of human genes have multiple isoforms of mature mRNAs (28, 39). Consistent with the functional significance of the fine regulation of the pre-mRNA processing, it is estimated that at least 15% of human hereditary diseases are caused by defects in posttranscriptional processing (15, 19). Utilization of alternative splicing microarrays revealed that many alternative splicing events are controlled in a tissue- and cell-type- and/or developmental stage-dependent manner, raising the novel question of how so many genes are regulated by a limited number of regulatory factors (8, 34).
Studies on multiple model genes established a view that a complex interplay among both positive and negative functions of various trans-acting factors and multiple exonic and intronic cis-acting elements affects tissue-specific alternative splicing. SR protein family members and heterogeneous nuclear ribonucleoproteins (hnRNPs) that bind to the exonic splicing enhancers and silencers, respectively, have been shown to regulate constitutive and alternative splicing of several model genes (6, 33). Conditional knockout and transgenic models demonstrated that balanced expression of these ubiquitous factors regulates a specific subset of target genes in a tissue-specific manner in vivo (44, 53). In other cases, tissue-specific RNA-binding proteins regulate tissue-specific alternative splicing by binding to their specific target sequences (6). The function of the Nova family, consisting of brain-specific KH-type RNA-biding proteins (11, 54), has been the most thoroughly studied. They bind to YCAY clusters (11, 47) and regulate a specific subset of brain-specific alternative splicing in vivo to modulate synaptic functions (48). Recent computational comparison of the intronic sequences among alternatively spliced genes predicted many other uncharacterized candidate cis elements (46, 55). Further experimental identification of the trans factors and elucidation of rules for their functional coordination would contribute to deciphering the “cellular codes” underlying tissue-specific and developmentally regulated alternative splicing in living organisms (8, 33, 34).
Members of the Fox-1 family of RNA-binding proteins are tissue-specific alternative splicing regulators that specifically bind to (U)GCAUG. Experimental studies had demonstrated that the (U)GCAUG stretches are involved in cell-type-dependent regulation of well-studied model exons from alternatively spliced genes (16, 22, 26, 29, 32, 38). Computational analysis of the brain-specific alternative cassette exons demonstrated that GCAUG pentanucleotide and UGCAUG hexanucleotide elements are the most overrepresented elements in the downstream introns (10). The UGCAUG stretch is phylogenetically and spatially conserved in the flanking introns of brain-enriched exons among multiple orthologous vertebrate genes (37). Jin et al. first demonstrated that zebrafish (zFox-1) and murine (mFox-1) homologs of Caenorhabditis elegans FOX-1 specifically bind to the (U)GCAUG stretch in vitro, and they provided evidence that expression of the vertebrate Fox-1 proteins in cultured cells promotes the inclusion of the fibronectin EIIIB exon, mimics the muscle-specific skipping of the mitochondrial ATP synthase F1γ, and affects the choice of mutually exclusive exons of the α-actinin pre-mRNA (27). Since this discovery, other laboratories have also reported that closely related members of the Fox-1 family of RNA-binding proteins regulate cell-type-specific alternative splicing of several genes through UGCAUG stretches in mammalian cells (3, 41, 43, 49, 56). It is yet to be elucidated, however, what precisely determines whether the Fox-1 family proteins enhance or inhibit inclusion of the alternatively spliced exons.
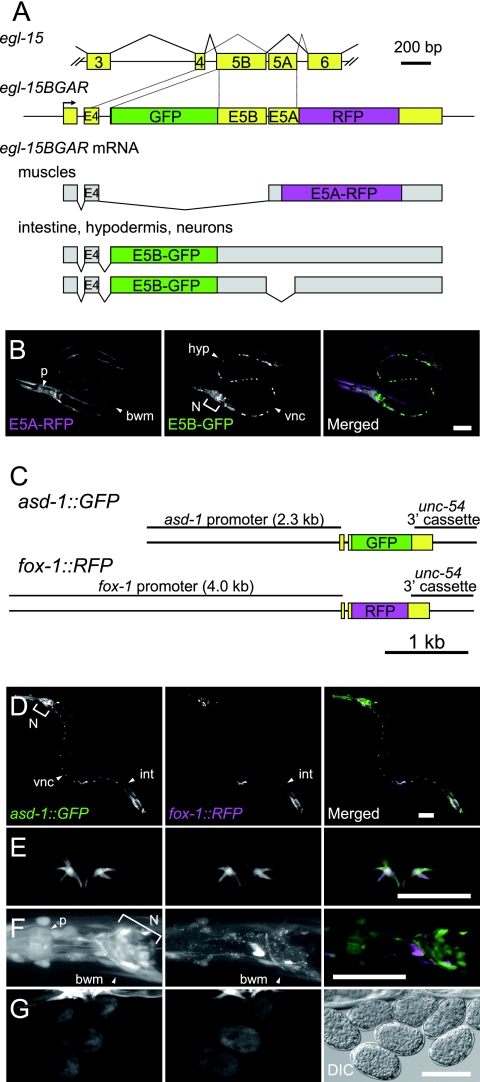
C. elegans is an excellent model organism to study mechanisms of regulation of alternative splicing in vivo. At least 5% of the total genes in C. elegans are alternatively spliced in manners similar to those for vertebrates (30, 40). We have recently developed a transgenic reporter worm system that can monitor the expression profiles of alternatively spliced exons at single-cell resolution in vivo by utilizing the egl-15 gene as a model (31). The egl-15 gene encodes the sole homolog of the fibroblast growth factor receptors in C. elegans (17), and selection of its mutually exclusive alternative exon 5A and exon 5B confers a functional difference (5). Stern and colleagues provided genetic evidence that the egl-15(5A) isoform is required for the migration of the sex myoblast towards the center of the body, where its specific ligand, EGL-17, is expressed and secreted, while the egl-15(5B) isoform exerts functions essential in embryonic and larval development (13, 14, 20). A functional difference between the egl-15 isoforms has also been reported in axon outgrowth and maintenance in the nervous system (12). In a previous study, we constructed a reporter minigene, egl-15BGAR, so that we could monitor the selection of the egl-15 exons 5B and 5A through expression of green fluorescent protein (GFP) and red fluorescent protein (RFP), respectively (Fig. 1A) (31). We have demonstrated that the pre-mRNA from the egl-15BGAR minigene is alternatively spliced into three isoforms, yielding the two fluorescent proteins in a tissue-specific manner (Fig. 1A and B) (31). In muscle tissues, such as egg-laying vulval muscle, body wall muscles, pharyngeal muscles, and an anal muscle, egl-15BGAR pre-mRNA is predominantly spliced to form the E5A-RFP isoform (Fig. 1A and B, left panel), while in epidermal tissues such as intestine and hypodermis and in neurons, the E5B-GFP isoform is exclusively or predominantly expressed (Fig. 1A and B, middle panel).
FIG. 1.
Expression patterns of the egl-15 alternative splicing reporter and the Fox-1 family. (A) The egl-15BGAR reporter minigene. Top, schematic drawing of the genomic fragment spanning exons 3 through 6 of the egl-15 gene. Boxes indicate exons, and horizontal lines indicate introns. Middle, schematic drawing of the egl-15BGAR minigene. The GFP cDNA was inserted into exon 5B (E5B), and the RFP cDNA was connected in frame to exon 5A (E5A). Bottom, expression and structures of the mRNAs derived from the egl-15BGAR minigene. Putative open reading frames are colored in magenta for E5A-RFP fusion proteins and in green for E5B-GFP fusion proteins. Note that repression of E5B results in splicing between exon 4 (E4) and E5A to produce E5A-RFP, while inclusion of E5B results in production of either of the two isoforms producing the GFP protein (E5B-GFP). (B) Tissue-dependent expression patterns of the egl-15BGAR reporter. Expression patterns of E5A-RFP (left) and E5B-GFP (middle) and a merged image (right) are shown. (C) Schematic drawing of the asd-1 and fox-1 transcriptional fusion constructs. Genomic fragments 5′ flanking the initiation codons of the asd-1 and fox-1 genes were used to drive expression of the GFP and RFP cDNAs, respectively. (D to G) Expression patterns of asd-1::GFP (left column) and fox-1::RFP (middle column) and merged views (right column). (D) Lateral view of an adult worm. (E) Enlarged lateral view of an adult vulva. (F) Enlarged lateral view of an adult head. (G) Embryos. A differential interference contrast (DIC) image is shown instead of a merged image in the right panel of panel G. Bars, 50 μm. bwm, body wall muscles; hyp, hypodermis; int, intestine; N, neurons in the head ganglia; p, pharynx; vnc, ventral nerve cord.
We have also provided evidence that the Fox-1 family of RNA-binding proteins regulates the tissue-specific alternative splicing of the egl-15 gene through the UGCAUG stretch in intron 4 (31). Through screening for mutants defective in E5A-RFP expression from the egl-15BGAR reporter in the body wall muscles, we isolated mutants with a characteristic color phenotype, “chimera,” and identified the alternative-splicing-defective-1 (asd-1) gene, a member of the Fox-1 family (31). Our genetic evidence led to a model that ASD-1 and FOX-1 redundantly regulate the tissue-specific selection of the alternative exons of the egl-15 reporter and the endogenous egl-15 gene by binding to the UGCAUG stretch in intron 4 to repress inclusion of the upstream exon 5B and to allow inclusion of the downstream exon 5A. This work was the first demonstration that Fox-1- and UGCAUG-mediated tissue-specific alternative splicing is evolutionarily conserved in nematodes as in vertebrates, reinforcing the feasibility of genetic studies with C. elegans to decipher complex codes of alternative splicing in vivo.
In the present study, we demonstrate that the expression of the asd-1 and fox-1 is observed beyond muscular tissues and is insufficient to explain the tissue-dependent profiles of the egl-15 reporter expression. We therefore screened for further mutants defective in muscle-specific alternative splicing of the egl-15BGAR reporter. We provide genetic evidence that another, muscle-specific RNA-binding protein, SUP-12, regulates the muscle-specific alternative splicing of the endogenous egl-15 gene by cooperatively binding with the Fox-1 family proteins to the juxtaposed cis elements in intron 4. This study provides the first genetic evidence that two families of tissue-specific factors coregulate tissue-specific alternative splicing of a target gene in vivo through cooperative binding to their juxtaposed cis elements in a specific subset of tissues where expression of the two regulator families overlaps.
MATERIALS AND METHODS
Worm culture, mutant screening, and mapping.
We cultured worms by standard methods. We prepared transgenic lines as described previously (31). We mutagenized the transgenic reporter allele ybIs733 [myo-3::EGL-15BGAR lin-15 (+)] with ethyl methanesulfonate as described previously (23, 31). Since the completely “green” mutants were sterile, we randomly isolated F1 worms with a fluorescence-assisted worm sorter (Copas Biosort; Union Biometrica) and screened the F2 worms for siblings with color phenotypes. We eventually found that all the green sup-12 mutants are sterile when they are homozygous for the transgenic reporter allele, while they are fertile when heterozygous for the reporter allele. We can therefore maintain the homozygous sup-12 alleles as reporter heterozygotes. We performed single-nucleotide polymorphism-based mapping as described previously (31, 51).
Minigene construction.
We constructed the promoter vectors as Gateway vectors (Invitrogen). We amplified the asd-1 promoter fragment with PCR primers GCAGGTACCGCACTGACTGGGATGATGAGC and TATGGTACCCGCCGTTGTGATTTGTAGAGA (underlining indicates KpnI digestion sites), digested the fragment with KpnI, and subcloned the fragment into pDEST-PL (31) to construct pDEST-asd-1p. We amplified the fox-1 promoter fragment with TGATTACGCCAAGCTTCGACACGTGCATTAGGCACA and CCTTTGGCCAATCCCTACAGGGCTTGGATGGGCAGA and subcloned the fragment into pDEST-PL (31) to construct pDEST-fox-1p. The nucleotide sequences of these vectors are available in the C. elegans promoter database (http://www.shigen.nig.ac.jp/c.elegans/promoter/index.jsp). We mutagenized the myo-3::egl-15BGAR minigene (31) to construct the myo-3::egl-15BGARctc minigene by utilizing QuikChange (Stratagene) and mutagenizing oligonucleotide DNAs TTCCATGCATGGTCTCCTTTGTTTTCAGA and TCTGAAAACAAAGGAGACCATGCATGGAA.
Microscopy.
We used confocal microscopes (Fluoview FV500 and FV1000; Olympus), for image scanning and processed the acquired images with Metamorph (Molecular Devices).
Sequence alignment.
We aligned the amino acid sequences of the RNA recognition motif (RRM) domains of the SUP-12 family of RNA-binding proteins by the Clustal W method using MegAlign (DNASTAR). The accession numbers of the sequences used are NP_508674 (SUP-12), NP_059965 (Homo sapiens Rnpc1), CAG31969 (Gallus gallus SEB4), AAP42281 (Xenopus laevis SEB4), CAB96420 (Xenopus laevis MTG-1a), BAD12194 (Danio rerio seb-4), AAH81649 (Danio rerio Rnpc1), AAM61541 (Arabidopsis thaliana SEB-4), and NP_665898 (Homo sapiens A2BP1/Fox-1).
In vitro binding assay.
We prepared His-tagged and FLAG-tagged recombinant proteins by using Ni-nitrilotriacetic acid Sepharose beads (QIAGEN) and M2 agarose beads (Sigma), respectively, according to standard protocols. We prepared the bacterial expression constructs as Gateway expression vectors (Invitrogen) by modifying a cold-induction vector (pCold II; Takara). We synthesized RNA probes by in vitro transcription with [α-32P]UTP and T7 RNA polymerase. We performed all the in vitro binding experiments with RNA binding buffer consisting of 100 mM KCl, 5% glycerol, and 1% Triton X-100 in HEPES-KOH (pH 7.9) supplemented with 1 mM dithiothreitol and 0.1 mM phenylmethylsulfonyl fluoride). We performed UV cross-linking essentially as described previously (31) in the presence of 130 ng/μl Escherichia coli tRNA. We performed electrophoretic mobility shift assay (EMSA) essentially as described previously (31) in the presence of 130 ng/μl E. coli tRNA and 50 ng/μl bovine serum albumin. We performed pull-down experiments with FLAG-tagged recombinant proteins bound to M2 agarose beads (Sigma) and purified His-tagged recombinant proteins in a total volume of 1 ml in the presence of 100 ng/μl yeast tRNA and 50 ng/μl bovine serum albumin. The egl-15 RNA, AUUUCUUCCAUGCAUGGUGUGCUUUG, was chemically synthesized (Hokkaido System Science).
Reverse transcription-PCR (RT-PCR).
We prepared total RNAs and amplified the endogenous egl-15 and inf-1 cDNA fragments as previously described (31).
RESULTS
Expression of asd-1 and fox-1 is not restricted to muscle.
In order to investigate whether expression of either asd-1 or fox-1 is correlated with the tissue-dependent expression of the E5A-RFP isoform from the egl-15BGAR reporter, we analyzed the expression profiles of the asd-1 and fox-1 genes. We prepared promoter fusion constructs (Fig. 1C) and generated transgenic worms by coinjecting the two minigenes. We found that both asd-1 and fox-1 are expressed in a relatively broad range of tissues (Fig. 1D to G). Consistent with our previous observation that asd-1 and fox-1 are redundantly required for the expression of the E5A isoform in muscle (Fig. 1B) (31), they are robustly expressed in vulval muscles (Fig. 1E) and modestly expressed in body wall muscles and in the pharynx (Fig. 1D and F). However, we also observed expression of both asd-1 and fox-1 in the intestine (Fig. 1D) and expression of asd-1 in the nervous system (Fig. 1D and F), where the E5B isoform predominated (Fig. 1B) (31). Since fox-1 was originally identified as a regulator of sex determination in C. elegans (24, 42), we investigated the expression of these genes in embryogenesis and found that they become widely expressed in the early embryos before morphogenesis (Fig. 1G). These expression profiles raised a possibility that mere expression of the Fox-1 family proteins is insufficient to regulate the tissue-specific alternative splicing of the egl-15 reporter in vivo.
sup-12 mutants are also defective in egl-15 reporter expression.
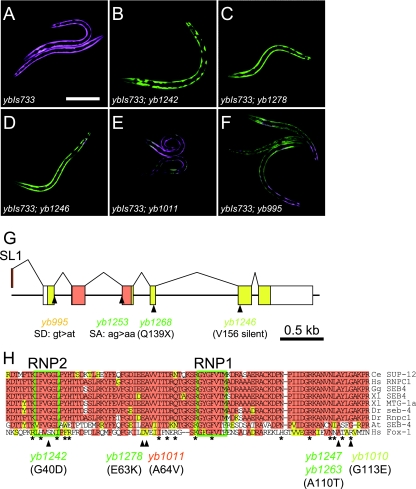
In order to search for other factors involved in the regulation of tissue-specific expression of the egl-15 reporter, we screened for further color mutants. The parental transgenic reporter allele, ybIs733, predominantly expressed E5A-RFP in body wall muscles (Fig. 2A). In the screening, we randomly isolated many F1 animals and searched the next generation for color mutants so that we could obtain sterile mutants (see Materials and Methods for details). We isolated several novel green (Fig. 2B and C) and chimera (Fig. 2D) mutants as well as chimera, “orange,” and other mutants similar to those we have reported previously (Fig. 2E and F) (31). Although all the green mutants were sterile in the reporter homozygous background, they were fertile in the reporter heterozygous background, which allowed us to maintain the homozygous mutant alleles. Most of the chimera mutants were indistinguishable from the asd-1 (yb978) mutants (31) and turned out to carry new alleles of the asd-1 gene, including both nonsense and missense mutations (see Wormbase at http://www.wormbase.org/ for further allele information), demonstrating reproducibility of the color mutant screening and the asd-1 color phenotype.
FIG. 2.
sup-12 mutants defective in tissue-specific expression of the egl-15 alternative splicing reporter. (A) The parent allele, ybIs733, expressing the egl-15BGAR minigene in the body wall muscles, used for the mutant screening. (B to F) sup-12 mutant alleles identified in the screening. (B and C) Green mutants yb1242 (B) and yb1278 (C). (D to F) chimera mutants, yb1246 (D), yb1011 (E), and yb995 (F). Bar, 100 μm. (G) Schematic representation of the sup-12 gene and the mutations identified in the screening. Open reading frames are colored. The RRM is orange. (H) Alignment of the RRM amino acid sequences among the SUP-12 family and comparison to that of the human Fox-1. Identical residues are shaded in orange, and residues with similar properties are shaded in yellow. Gaps were introduced to maximize the alignment. Conserved RNP1 and RNP2 motifs are boxed in green. Positions and allele names of the missense mutations identified within the RRM are indicated. Allele names are colored to reflect the severity of the color phenotype of the egl-15 reporter expression. Asterisks indicate residues of human Fox-1 involved in recognition of the UGCAUG RNA (2). Ce, C. elegans; Hs, Homo sapiens (human); Gg, Gallus gallus (chick); Xl, Xenopus laevis (frog); Dr, Danio rerio (zebrafish); At, Arabidopsis thaliana (plant). Panel F is reproduced from reference 31.
Single-nucleotide polymorphism-based chromosome mapping and sequencing of the mutant genome revealed that the green mutants (Fig. 2B and C) and unidentified chimera mutants (Fig. 2D to F) carry alleles of the sup-12 gene (Fig. 2G and H). The sup-12 gene was originally cloned as a gene responsible for the genetic suppression of unc-60B mutants (1). sup-12 encodes an evolutionarily conserved RNA-binding protein specifically expressed in body wall muscles, pharynx, vulval muscles, and an anal depressor muscle (1, 18, 35), consistent with our previous observation that the E5A-RFP form of the egl-15 reporter was predominantly expressed in the muscle (Fig. 1B) (31). SUP-12 protein has a highly conserved N-terminal RRM (Fig. 2H) and a C-terminal AQ-rich domain (1). The yb1253 allele is likely to be a null allele, since it has a mutation in the splice acceptor of exon 3 corresponding to the C-terminal half of the RRM (Fig. 2G). This leads to cryptic splicing causing a one-base deletion in the mature sup-12 mRNA (data not shown). Although other green mutants with missense mutations within the RRM are indistinguishable from yb1253 mutants in the color phenotype, they may be reduction-of-function (rf) alleles, since the yb1278(E63K) allele (Fig. 2C) has a less severe growth phenotype than the yb1253 allele in the asd-1 or fox-1 mutant background (see below) and the recombinant SUP-12(E63K) protein has a trace of RNA-binding activity (see below). All the missense mutations affected residues within the RRM, which is well conserved among the family members, and caused various color phenotypes ranging from green to almost red chimera (Fig. 2H), suggesting that they are rf alleles with variable RNA-binding properties.
We also mapped the genes responsible for the orange phenotype (31) and found that the phenotype is caused by mutation in the smg-1 (see Fig. S1 in the supplemental material) and smg-2 (see Fig. S2 in the supplemental material) genes. We conclude that the E5B-GFP isoforms of the mRNAs derived from the egl-15BGAR reporter (Fig. 1A) are substrates for nonsense-mediated mRNA decay (see the supplemental material).
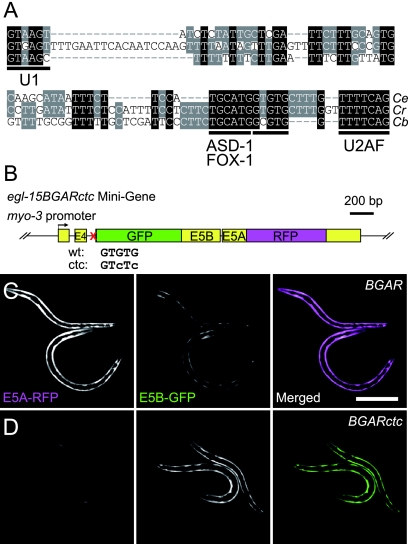
A GUGUG stretch is an essential cis element for egl-15 reporter regulation.
As the sup-12 gene encodes an RNA-binding protein, we searched for a cis element involved in regulation by the SUP-12 protein. We have previously reported that the conserved cis element for the ASD-1 and FOX-1 proteins, UGCAUG, resides in intron 4 of the egl-15 pre-mRNA (Fig. 3A) (31). Further comparison of the nucleotide sequences of egl-15 intron 4 among three related nematodes revealed that another short stretch, GYGUG, is conserved just downstream of the UGCAUG stretch (Fig. 3A). Anyanful et al. have reported that SUP-12 preferentially binds to a (UG)4 repeat in vitro (1), consistent with the sequence of the short stretch. In order to investigate whether the GYGUG stretch is involved in the tissue-specific expression of the egl-15BGAR reporter, we constructed a mutant minigene in which the GUGUG stretch is mutagenized to GUCUC (Fig. 3B). We found that the disruption of the GUGUG stretch resulted in the green phenotype (Fig. 3C and D), as did the disruption of the UGCAUG stretch (31). This result indicates that the GUGUG stretch is also required for the muscle-specific expression of the egl-15 reporter in C. elegans.
FIG. 3.
cis elements for the tissue-specific alternative splicing of the egl-15 gene. (A) Alignment of the nucleotide sequence of egl-15 intron 4 among three related nematodes. Nucleotides conserved among all three species are shaded in black, and those conserved in two of the three species are in gray. Conserved stretches are underlined, and the known binding factors are indicated. Ce, C. elegans; Cr, C. remanei; Cb, C. briggsae. (B) Schematic representation of the modified reporter minigene, egl-15BGARctc. wt, wild type. (C and D) Expression of E5A-RFP (left panels) and E5B-GFP (middle panels) and merged views (right panels) of the egl-15BGAR reporter (C) and the egl-15BGARctc reporter (D) in the body wall muscles. Bar, 100 μm.
SUP-12 specifically binds to the GUGUG stretch in egl-15 intron 4 in vitro.
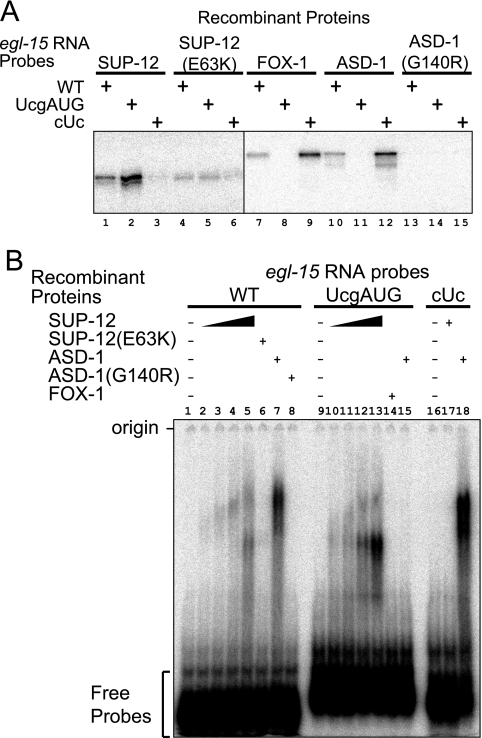
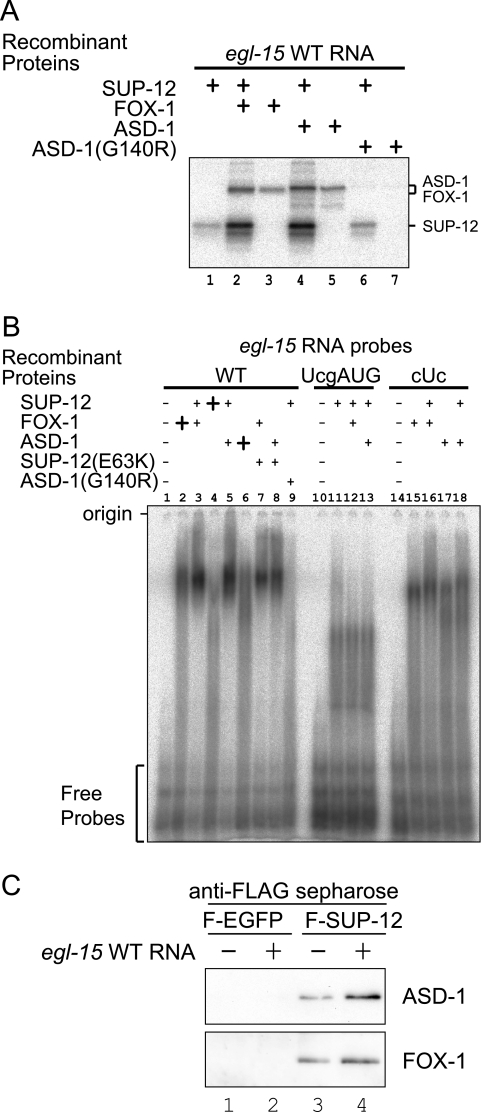
In order to characterize the RNA-binding property of the SUP-12 protein, we performed UV cross-linking and EMSA. We prepared a radioisotope-labeled RNA probe from the wild-type egl-15 intron 4 and two mutant probes, UcgAUG and cUc, in which the UGCAUG and GUGUG stretches were mutagenized, respectively (see Fig. S3A and B in the supplemental material). We also prepared recombinant full-length proteins (see Fig. S3C in the supplemental material). UV cross-linking experiments (Fig. 4A) revealed that the SUP-12 protein binds to the wild-type (lane 1) and the UcgAUG (lane 2) probes but not to the cUc probe (lane 3). The mutant SUP-12(E63K) protein binds only weakly to these probes (lanes 4 to 6). On the other hand, ASD-1 and FOX-1 bind to the wild-type and the cUc probes but not to the UcgAUG probe (lanes 7 to 12), while the mutant ASD-1(G140R) protein did not bind to any of these probes (lanes 13 to 15), confirming the sequence specificity of the Fox-1 family RNA-binding proteins (31). EMSA revealed the same RNA-binding properties (Fig. 4B). SUP-12 shifted the mobility of the wild-type (lanes 2 to 5) and the UcgAUG (lanes 10 to 13) probes in a dose-dependent manner but not that of the cUc probe (lane 17). SUP-12(E63K) has little effect on the mobility of the wild-type probe (lane 6). These results indicate that SUP-12 specifically binds to the GUGUG stretch of egl-15 intron 4.
FIG. 4.
Specific binding of SUP-12 to the GUGUG stretch in egl-15 intron 4. (A) UV cross-linking assays with single recombinant proteins. Recombinant proteins and RNA probes used in the assay are indicated. WT, wild type. (B) EMSA with single recombinant proteins. Recombinant proteins and RNA probes used in the assay are indicated. Twofold dilution series of SUP-12 proteins were used in lanes 2 to 5 and 10 to 13.
ASD-1/FOX-1 and SUP-12 cooperatively bind to egl-15 intron 4.
We next investigated whether interaction between the Fox-1 family and SUP-12 affects the RNA-binding properties of each other (Fig. 5A). SUP-12 enhanced the UV cross-linking of the FOX-1 and ASD-1 proteins to the wild-type RNA probe by 1.7- and 1.9-fold, respectively (lanes 2, 3, 4, and 5). In the same way, FOX-1 and ASD-1 enhanced the UV cross-linking of the SUP-12 protein to the wild-type probe by 3.0- and 3.2-fold, respectively (lanes 1, 2, and 4), but the ASD-1(G140R) mutant protein had little effect (1.2-fold) (lanes 1, 6, and 7). EMSA also revealed cooperative binding (Fig. 5B). Coincubation of the wild-type RNA probe with both the Fox-1 family and the SUP-12 proteins promoted formation of more stable complexes (lanes 3 and 5) than did incubation with any of the single proteins (lanes 2, 4, and 6). The stable complex was not formed when the ASD-1(G140R) protein (lane 9) or either of the mutant probes (lanes 10 to 13 and 14 to 18) was used, indicating that formation of the stable complex requires the specific binding of both Fox-1 family members and SUP-12 to the egl-15 RNA. Unexpectedly, SUP-12(E63K), which binds only weakly to egl-15 RNA, can also form the stable complex with the Fox-1 family proteins and the egl-15 RNA (lanes 7 and 8), suggesting a direct interaction between SUP-12 and the Fox-1 family proteins. We therefore investigated the physical interaction between the two families of RNA-binding proteins in vitro by pull-down experiments using purified recombinant proteins (Fig. 5C). SUP-12 (lane 3) but not GFP (lanes 1 and 2) bound to ASD-1 and FOX-1 proteins even in the absence of the egl-15 RNA, and egl-15 RNA enhanced the binding of SUP-12 to ASD-1 and FOX-1 by 2.9- and 1.9-fold, respectively (lane 4).
FIG. 5.
Cooperative binding of the Fox-1 family and SUP-12 to the juxtaposed cis elements in egl-15 intron 4. (A) UV cross-linking with combinations of recombinant proteins. WT, wild type. (B) EMSA with combinations of recombinant proteins. Large plus signs indicate twice the amount of the recombinant proteins compared to small plus signs. (C) Pull-down assay. Purified FLAG-tagged proteins and a synthesized RNA used in the pull-down assay are indicated.
sup-12 and the Fox-1 family cooperatively regulate the endogenous egl-15 gene.
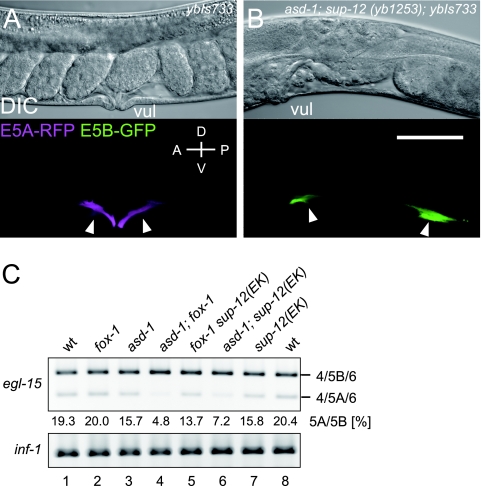
We investigated whether sup-12 is involved in the alternative splicing regulation of the endogenous egl-15 in vivo. Since the putative null allele of the sup-12 mutant, yb1253, is normal in egg-laying behavior (data not shown), we analyzed the genetic interaction between sup-12 and the fox-1 family. The asd-1; sup-12 (yb1253) double mutant grew to sterile adults in which the vulval muscles differentiated in the posterior of the body (Fig. 6A and B), phenocopying the aberrant migration of the sex myoblasts in the egl-15(5A) isoform-specific mutant (20) and the asd-1; fox-1 double mutant (31). The fox-1 sup-12 (yb1253) double mutant arrested as larvae, and we could not investigate the vulval muscle phenotype. On the other hand, asd-1; sup-12 (E63K) and fox-1 sup-12 (E63K) double mutants were fertile and their vulval muscles were normally formed (data not shown), indicating that sup-12 function is more seriously affected by the yb1253 allele. We then compared the amounts of the egl-15 mRNA isoforms in the single mutants and the fertile double mutants by RT-PCR (Fig. 6C). Although the amounts of the egl-15(5A) isoform in the single mutants (lanes 2, 3, and 7) were comparable to that in the wild type (lanes 1 and 8), we found a ∼2.8-fold reduction in the asd-1; sup-12 (E63K) double mutant (lane 6) as in the asd-1; fox-1 double mutant (>4-fold) (lane 4). These results indicate that SUP-12 functions as a coregulator of the muscle-specific alternative splicing of the endogenous egl-15 gene in vivo.
FIG. 6.
Regulation of the endogenous egl-15 gene by sup-12. (A and B) Lateral views of adult bodies expressing the egl-15BGAR reporter in the wild-type background (A) and in the asd-1; sup-12 (yb1253) background (B). Top, differential interference contrast (DIC) images. Bottom, merged confocal views. vul, vulva. Arrowheads indicate vulval muscles. Anterior is to the left and dorsal is to the top. Bar, 50 μm. (C) RT-PCR analysis of the endogenous egl-15 gene. The amount of the egl-15(5A) isoform relative to that of the egl-15(5B) isoform in each strain is indicated. inf-1 cDNA was also amplified as a loading control. The genotypes of the worms used are indicated. wt, wild type.
DISCUSSION
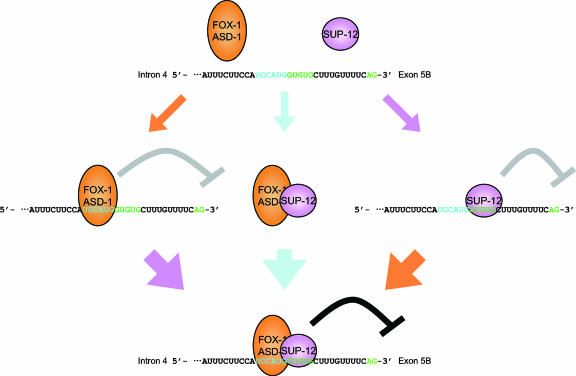
We have provided genetic and biochemical evidence that the muscle-specific and evolutionarily conserved RNA-binding protein SUP-12 cooperatively regulates, with ASD-1 and FOX-1, the tissue-specific alternative splicing of the fibroblast growth factor receptor gene egl-15. Figure 7 summarizes the coordinated binding of the Fox-1 family and SUP-12 to the egl-15 RNA. The Fox-1 family and SUP-12 can specifically bind alone to the egl-15 RNA through the UGCAUG and the GUGUG stretches, respectively (left and right panels). The Fox-1 family and SUP-12 can directly bind to each other even in the absence of the target RNA (middle panel). When the three components meet together, they form a more stable ternary complex through their sequence-specific binding properties (bottom panel). This scheme also explains the mode of the egl-15 alternative splicing regulation. The two families of RNA-binding proteins form a stable complex on intron 4 to repress selection of E5B (bottom panel), which eventually leads to the selection of the downstream E5A in muscle. Disruption of either of the UGCAUG or the GUGUG stretches or mutation in the asd-1, fox-1, or sup-12 gene (left and right panels) also leads to derepression of E5B. The molecular mechanisms of E5B repression by the Fox-1 family/SUP-12/egl-15 intron 4 ternary complex formation remain to be elucidated. Although C. elegans does not have a recognizable branch point consensus or a polypyrimidine tract (25), the putative branch point is the A at position −20 in the UGCAUG element, which is the first A upstream of the 3′ splice site for E5B (Fig. 3A) and is close to the positions where the putative branch point A is frequently found (9). It is reasonable to suggest that the SUP-12/Fox-1 complex directly competes with the branch point binding proteins or that the complex interferes with the recruitment of U2 snRNP by U2AF. Mutation in any one of the asd-1, fox-1, and sup-12 genes did not cause an egg-laying (Egl) phenotype, suggesting that the Egl phenotype is the binary readout of the amount of the endogenous egl-15(5A) isoform in the sex myoblasts. On the other hand, asd-1 and sup-12 single mutants showed reproducible color phenotypes in the body wall muscles (Fig. 2) (31), suggesting that the color phenotype is quantitative and directly reflects the net amounts of the mature reporter mRNA isoforms in that tissue. Our approach with the alternative splicing reporter worm system thus allowed us to identify the trans-acting factors involved in the tissue-specific regulation and led us to the conclusion that the endogenous egl-15 gene is coordinately regulated by these three RNA-binding proteins in vivo.
FIG. 7.
Schematic representation of the in vitro binding of ASD-1, FOX-1, and SUP-12 to the egl-15 pre-mRNA. See text for details.
Previous work suggested that SUP-12 is a muscle-specific splicing regulator of the unc-60 gene (1). The unc-60 gene, encoding two related isoforms of actin-depolymerizing factor (also known as cofilin), has a common first exon and two separate series of downstream exons (36). Exons 2A through 5A of the unc-60A isoform are selected in nonmuscle tissues, while exons 2B through 5B of the unc-60B isoform reside downstream of the polyadenylation signal of unc-60A and are selected only in muscle tissues. Genetic evidence indicated that SUP-12 represses expression of unc-60A in muscle, allowing selection of downstream unc-60B (1). SUP-12 has been shown to bind to UG-rich stretches in intron 1 and intron 2A of the unc-60 pre-mRNA in vitro (1). Therefore, the situations of the sup-12-dependent muscle specific exon repression are quite similar for unc-60 and egl-15, although the Fox-1 family RNA-binding consensus, (U)GCAUG, is absent from the unc-60 pre-mRNA. It is not yet clear whether other trans factors and cis elements are involved in regulation of the unc-60 gene or whether SUP-12 functions as the sole repressor. As the SUP-12 family of RNA-binding proteins are evolutionarily conserved among metazoans and plants (Fig. 3F) (1), it is likely that the family is involved in the regulation of tissue-specific alternative splicing in other multicellular organisms.
A remarkable feature of the Fox-1 family RNA-binding proteins is that their single evolutionarily conserved RRM binds to a highly specific hexameric nucleotide stretch (2, 27). In contrast, other tissue-specific alternative splicing regulator families usually have multiple copies of RNA-binding domains or form dimers. For instance, the Nova family has three KH motifs (54), the CELF/Bruno-like family has three RRMs (4), PTB and its paralogue nPTB have four RRMs (50), and the GSG/STAR family forms homodimers (52). Although the target sequence of the Fox-1 family, UGCAUG, is the most highly overrepresented hexanucleotide in the downstream introns of tissue-specific alternative exons (10, 37), it is unlikely that all the UGCAUG-containing pre-mRNAs are spliced in a common way depending merely on the expression of the Fox-1 family proteins, because such binary regulation mechanisms would reduce diversity in gene expression. In fact, mammalian pre-mRNAs regulated by the Fox-1 family usually have multiple copies of other cis elements involved in alternative splicing regulation. Furthermore, the UGCAUG stretch may function as both an enhancer and a silencer of the cassette exons (27, 49). These characteristic features of the Fox-1 family proteins raise a question about what other tissue-specific regulators or components of the general splicing apparatus mediate tissue-specific and context-dependent alternative splicing regulation by the Fox-1 family. Our present study provides the first example that two families of RNA-binding proteins, each of which has only one RNA-binding domain, cooperatively form a stable complex on a target pre-mRNA that has a tandem cluster of the cis elements for each of the families. The two Fox-1 family proteins, ASD-1 and FOX-1, are coexpressed in a broad range of tissues, redundantly function through high sequence specificity, and may contribute to confer robustness to egl-15 regulation, while their muscle-specific partner protein, SUP-12, may contribute to confer tissue specificity and also to specify the targets to a smaller subset. This kind of coordinated regulation of alternative splicing by the Fox-1 family may be evolutionarily conserved in higher eukaryotes, since (i) the mammalian Fox-1 family members are coexpressed in neurons and muscles (41, 49) and the target pre-mRNAs usually have multiple copies of the UGCAUG stretches (3, 27, 41, 43, 49, 56), allowing robust regulation, and (ii) the domain composition of the Fox-1 family is well conserved (27), suggesting conservation of the physical interaction with other regulators.
Multiple cis elements are usually involved in cell-type-specific regulation of alternative splicing. Functional antagonism between juxtaposed enhancer and silencer elements via positive and negative regulatory factors have been reported in both exonic and intronic regions from multiple model genes (34). The functional coordination of the Fox-1 family and SUP-12 presented in this study suggest the presence of more orchestrated modes of splicing regulation. Recent global analyses of tissue-specific splicing patterns and comparison of the genomic sequences predicted many uncharacterized candidate cis elements (10, 46, 55). Further identification of trans-acting factors and characterization of their RNA-binding properties, as in the present study, would lead to understanding of the tissue-specific splicing regulation and to prediction of the effect of nucleotide polymorphisms on gene expression.
Supplementary Material
Acknowledgments
We thank T. Nojima, and A. Takeuchi at TMDU for discussion. We thank Y. Ogawa and N. Kataoka at TMDU for critically reading the paper. We thank the Caenorhabditis Genetics Center for materials. We thank M. Hagiwara and N. Sekine for technical assistance.
This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to H.K. and M.H.).
H.K. contributed to the overall experiments. G.O. contributed to the mutant analyses. S.M. contributed to the mutant screening. H.K. and M.H. organized this work. All authors discussed the results and commented on the manuscript.
Footnotes
Published ahead of print on 8 October 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Anyanful, A., K. Ono, R. C. Johnsen, H. Ly, V. Jensen, D. L. Baillie, and S. Ono. 2004. The RNA-binding protein SUP-12 controls muscle-specific splicing of the ADF/cofilin pre-mRNA in C. elegans. J. Cell Biol. 167:639-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auweter, S. D., R. Fasan, L. Reymond, J. G. Underwood, D. L. Black, S. Pitsch, and F. H. Allain. 2006. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J. 25:163-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baraniak, A. P., J. R. Chen, and M. A. Garcia-Blanco. 2006. Fox-2 mediates epithelial cell-specific fibroblast growth factor receptor 2 exon choice. Mol. Cell. Biol. 26:1209-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barreau, C., L. Paillard, A. Mereau, and H. B. Osborne. 2006. Mammalian CELF/Bruno-like RNA-binding proteins: molecular characteristics and biological functions. Biochimie 88:515-525. [DOI] [PubMed] [Google Scholar]
- 5.Birnbaum, D., C. Popovici, and R. Roubin. 2005. A pair as a minimum: the two fibroblast growth factors of the nematode Caenorhabditis elegans. Dev. Dyn. 232:247-255. [DOI] [PubMed] [Google Scholar]
- 6.Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72:291-336. [DOI] [PubMed] [Google Scholar]
- 7.Black, D. L. 2000. Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell 103:367-370. [DOI] [PubMed] [Google Scholar]
- 8.Blencowe, B. J. 2006. Alternative splicing: new insights from global analyses. Cell 126:37-47. [DOI] [PubMed] [Google Scholar]
- 9.Blumenthal, T., and K. Steward. 1997. RNA processing and gene structure, p. 117-145. In D. Riddle, T. Blumenthal, B. Meyer, and J. Priess (ed.), C. elegans II. Cold Spring Harbor Laboratory Press, Woodbury, NY. [PubMed]
- 10.Brudno, M., M. S. Gelfand, S. Spengler, M. Zorn, I. Dubchak, and J. G. Conboy. 2001. Computational analysis of candidate intron regulatory elements for tissue-specific alternative pre-mRNA splicing. Nucleic Acids Res. 29:2338-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buckanovich, R. J., and R. B. Darnell. 1997. The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo. Mol. Cell. Biol. 17:3194-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bulow, H. E., T. Boulin, and O. Hobert. 2004. Differential functions of the C. elegans FGF receptor in axon outgrowth and maintenance of axon position. Neuron 42:367-374. [DOI] [PubMed] [Google Scholar]
- 13.Burdine, R. D., C. S. Branda, and M. J. Stern. 1998. EGL-17(FGF) expression coordinates the attraction of the migrating sex myoblasts with vulval induction in C. elegans. Development 125:1083-1093. [DOI] [PubMed] [Google Scholar]
- 14.Burdine, R. D., E. B. Chen, S. F. Kwok, and M. J. Stern. 1997. egl-17 encodes an invertebrate fibroblast growth factor family member required specifically for sex myoblast migration in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 94:2433-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caceres, J. F., and A. R. Kornblihtt. 2002. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 18:186-193. [DOI] [PubMed] [Google Scholar]
- 16.Deguillien, M., S. C. Huang, M. Moriniere, N. Dreumont, E. J. Benz, Jr., and F. Baklouti. 2001. Multiple cis elements regulate an alternative splicing event at 4.1R pre-mRNA during erythroid differentiation. Blood 98:3809-3816. [DOI] [PubMed] [Google Scholar]
- 17.DeVore, D. L., H. R. Horvitz, and M. J. Stern. 1995. An FGF receptor signaling pathway is required for the normal cell migrations of the sex myoblasts in C. elegans hermaphrodites. Cell 83:611-620. [DOI] [PubMed] [Google Scholar]
- 18.Dupuy, D., N. Bertin, C. A. Hidalgo, K. Venkatesan, D. Tu, D. Lee, J. Rosenberg, N. Svrzikapa, A. Blanc, A. Carnec, A. R. Carvunis, R. Pulak, J. Shingles, J. Reece-Hoyes, R. Hunt-Newbury, R. Viveiros, W. A. Mohler, M. Tasan, F. P. Roth, C. Le Peuch, I. A. Hope, R. Johnsen, D. G. Moerman, A. L. Barabasi, D. Baillie, and M. Vidal. 2007. Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nat. Biotechnol. 25:663-668. [DOI] [PubMed] [Google Scholar]
- 19.Faustino, N. A., and T. A. Cooper. 2003. Pre-mRNA splicing and human disease. Genes Dev. 17:419-437. [DOI] [PubMed] [Google Scholar]
- 20.Goodman, S. J., C. S. Branda, M. K. Robinson, R. D. Burdine, and M. J. Stern. 2003. Alternative splicing affecting a novel domain in the C. elegans EGL-15 FGF receptor confers functional specificity. Development 130:3757-3766. [DOI] [PubMed] [Google Scholar]
- 21.Graveley, B. R. 2001. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 17:100-107. [DOI] [PubMed] [Google Scholar]
- 22.Hedjran, F., J. M. Yeakley, G. S. Huh, R. O. Hynes, and M. G. Rosenfeld. 1997. Control of alternative pre-mRNA splicing by distributed pentameric repeats. Proc. Natl. Acad. Sci. USA 94:12343-12347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hodgkin, J. 1999. Conventional genetics, p. 245-270. In I. A. Hope (ed.), C. elegans. A practical approach. Oxford, New York, NY.
- 24.Hodgkin, J., J. D. Zellan, and D. G. Albertson. 1994. Identification of a candidate primary sex determination locus, fox-1, on the X chromosome of Caenorhabditis elegans. Development 120:3681-3689. [DOI] [PubMed] [Google Scholar]
- 25.Hollins, C., D. A. Zorio, M. MacMorris, and T. Blumenthal. 2005. U2AF binding selects for the high conservation of the C. elegans 3′ splice site. RNA 11:248-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu, Y., Z. Zhou, X. Huang, M. Xu, L. Lu, Z. Xu, J. Li, and J. Sha. 2004. Expression of a novel DnaJA1 alternative splicing in human testis and sperm. Int J. Androl. 27:343-349. [DOI] [PubMed] [Google Scholar]
- 27.Jin, Y., H. Suzuki, S. Maegawa, H. Endo, S. Sugano, K. Hashimoto, K. Yasuda, and K. Inoue. 2003. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 22:905-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson, J. M., J. Castle, P. Garrett-Engele, Z. Kan, P. M. Loerch, C. D. Armour, R. Santos, E. E. Schadt, R. Stoughton, and D. D. Shoemaker. 2003. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302:2141-2144. [DOI] [PubMed] [Google Scholar]
- 29.Kawamoto, S. 1996. Neuron-specific alternative splicing of nonmuscle myosin II heavy chain-B pre-mRNA requires a cis-acting intron sequence. J. Biol. Chem. 271:17613-17616. [PubMed] [Google Scholar]
- 30.Kim, E., A. Magen, and G. Ast. 2007. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 35:125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuroyanagi, H., T. Kobayashi, S. Mitani, and M. Hagiwara. 2006. Transgenic alternative-splicing reporters reveal tissue-specific expression profiles and regulation mechanisms in vivo. Nat. Methods 3:909-915. [DOI] [PubMed] [Google Scholar]
- 32.Lim, L. P., and P. A. Sharp. 1998. Alternative splicing of the fibronectin EIIIB exon depends on specific TGCATG repeats. Mol. Cell. Biol. 18:3900-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maniatis, T., and B. Tasic. 2002. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 418:236-243. [DOI] [PubMed] [Google Scholar]
- 34.Matlin, A. J., F. Clark, and C. W. Smith. 2005. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell. Biol. 6:386-398. [DOI] [PubMed] [Google Scholar]
- 35.McKay, S. J., R. Johnsen, J. Khattra, J. Asano, D. L. Baillie, S. Chan, N. Dube, L. Fang, B. Goszczynski, E. Ha, E. Halfnight, R. Hollebakken, P. Huang, K. Hung, V. Jensen, S. J. Jones, H. Kai, D. Li, A. Mah, M. Marra, J. McGhee, R. Newbury, A. Pouzyrev, D. L. Riddle, E. Sonnhammer, H. Tian, D. Tu, J. R. Tyson, G. Vatcher, A. Warner, K. Wong, Z. Zhao, and D. G. Moerman. 2003. Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harbor Symp. Quant. Biol. 68:159-169. [DOI] [PubMed] [Google Scholar]
- 36.McKim, K. S., C. Matheson, M. A. Marra, M. F. Wakarchuk, and D. L. Baillie. 1994. The Caenorhabditis elegans unc-60 gene encodes proteins homologous to a family of actin-binding proteins. Mol. Gen. Genet. 242:346-357. [DOI] [PubMed] [Google Scholar]
- 37.Minovitsky, S., S. L. Gee, S. Schokrpur, I. Dubchak, and J. G. Conboy. 2005. The splicing regulatory element, UGCAUG, is phylogenetically and spatially conserved in introns that flank tissue-specific alternative exons. Nucleic Acids Res. 33:714-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Modafferi, E. F., and D. L. Black. 1997. A complex intronic splicing enhancer from the c-src pre-mRNA activates inclusion of a heterologous exon. Mol. Cell. Biol. 17:6537-6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Modrek, B., and C. Lee. 2002. A genomic view of alternative splicing. Nat. Genet. 30:13-19. [DOI] [PubMed] [Google Scholar]
- 40.Nagasaki, H., M. Arita, T. Nishizawa, M. Suwa, and O. Gotoh. 2005. Species-specific variation of alternative splicing and transcriptional initiation in six eukaryotes. Gene 364:53-62. [DOI] [PubMed] [Google Scholar]
- 41.Nakahata, S., and S. Kawamoto. 2005. Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res. 33:2078-2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicoll, M., C. C. Akerib, and B. J. Meyer. 1997. X-chromosome-counting mechanisms that determine nematode sex. Nature 388:200-204. [DOI] [PubMed] [Google Scholar]
- 43.Ponthier, J. L., C. Schluepen, W. Chen, R. A. Lersch, S. L. Gee, V. C. Hou, A. J. Lo, S. A. Short, J. A. Chasis, J. C. Winkelmann, and J. G. Conboy. 2006. Fox-2 splicing factor binds to a conserved intron motif to promote inclusion of protein 4.1R alternative exon 16. J. Biol. Chem. 281:12468-12474. [DOI] [PubMed] [Google Scholar]
- 44.Qi, J., S. Su, M. E. McGuffin, and W. Mattox. 2006. Concentration dependent selection of targets by an SR splicing regulator results in tissue-specific RNA processing. Nucleic Acids Res. 34:6256-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stamm, S., S. Ben-Ari, I. Rafalska, Y. Tang, Z. Zhang, D. Toiber, T. A. Thanaraj, and H. Soreq. 2005. Function of alternative splicing. Gene 344:1-20. [DOI] [PubMed] [Google Scholar]
- 46.Sugnet, C. W., K. Srinivasan, T. A. Clark, G. O'Brien, M. S. Cline, H. Wang, A. Williams, D. Kulp, J. E. Blume, D. Haussler, and M. Ares, Jr. 2006. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput. Biol. 2:e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ule, J., K. B. Jensen, M. Ruggiu, A. Mele, A. Ule, and R. B. Darnell. 2003. CLIP identifies Nova-regulated RNA networks in the brain. Science 302:1212-1215. [DOI] [PubMed] [Google Scholar]
- 48.Ule, J., A. Ule, J. Spencer, A. Williams, J. S. Hu, M. Cline, H. Wang, T. Clark, C. Fraser, M. Ruggiu, B. R. Zeeberg, D. Kane, J. N. Weinstein, J. Blume, and R. B. Darnell. 2005. Nova regulates brain-specific splicing to shape the synapse. Nat. Genet. 37:844-852. [DOI] [PubMed] [Google Scholar]
- 49.Underwood, J. G., P. L. Boutz, J. D. Dougherty, P. Stoilov, and D. L. Black. 2005. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol. Cell. Biol. 25:10005-10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner, E. J., and M. A. Garcia-Blanco. 2001. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 21:3281-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wicks, S. R., R. T. Yeh, W. R. Gish, R. H. Waterston, and R. H. Plasterk. 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28:160-164. [DOI] [PubMed] [Google Scholar]
- 52.Wu, J., L. Zhou, K. Tonissen, R. Tee, and K. Artzt. 1999. The quaking I-5 protein (QKI-5) has a novel nuclear localization signal and shuttles between the nucleus and the cytoplasm. J. Biol. Chem. 274:29202-29210. [DOI] [PubMed] [Google Scholar]
- 53.Xu, X., D. Yang, J. H. Ding, W. Wang, P. H. Chu, N. D. Dalton, H. Y. Wang, J. R. Bermingham, Jr., Z. Ye, F. Liu, M. G. Rosenfeld, J. L. Manley, J. Ross, Jr., J. Chen, R. P. Xiao, H. Cheng, and X. D. Fu. 2005. ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell 120:59-72. [DOI] [PubMed] [Google Scholar]
- 54.Yang, Y. Y., G. L. Yin, and R. B. Darnell. 1998. The neuronal RNA-binding protein Nova-2 is implicated as the autoantigen targeted in POMA patients with dementia. Proc. Natl. Acad. Sci. USA 95:13254-13259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yeo, G. W., E. L. Nostrand, and T. Y. Liang. 2007. Discovery and analysis of evolutionarily conserved intronic splicing regulatory elements. PLoS Genet. 3:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou, H. L., A. P. Baraniak, and H. Lou. 2007. Role for Fox-1/Fox-2 in mediating the neuronal pathway of calcitonin/calcitonin gene-related peptide alternative RNA processing. Mol. Cell. Biol. 27:830-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.