Abstract
Eukaryotic pre-mRNA 3′-end formation is catalyzed by a complex set of factors that must be intricately regulated. In this study, we have discovered a novel role for the small ubiquitin-like modifier SUMO in the regulation of mammalian 3′-end processing. We identified symplekin, a factor involved in complex assembly, and CPSF-73, an endonuclease, as SUMO modification substrates. The major sites of sumoylation in symplekin and CPSF-73 were determined and found to be highly conserved across species. A sumoylation-deficient mutant was defective in rescuing cell viability in symplekin small interfering RNA (siRNA)-treated cells, supporting the importance of this modification in symplekin function. We also analyzed the involvement of sumoylation in 3′-end processing by altering the sumoylation status of nuclear extracts. This was done by the addition of a SUMO protease, which we show interacts with both symplekin and CPSF-73, or by siRNA-mediated depletion of ubc9, the SUMO E2-conjugating enzyme. Both treatments resulted in a marked inhibition of processing. The assembly of a functional polyadenylation complex was also impaired by the SUMO protease. Our identification of two key polyadenylation factors as SUMO targets and of the role of SUMO in enhancing the assembly and activity of the 3′-end-processing complex together reveal an important function for SUMO in the processing of mRNA precursors.
The poly(A) tail at the 3′ end of nearly all eukaryotic mRNAs is essential for the stability of the transcript, for its transport into the cytoplasm, and for translation initiation. The 3′ ends of pre-mRNAs are formed in a two-step process, with an endonucleolytic cleavage generating a 3′ OH end followed by the synthesis of a poly(A) tail (reviewed in references 10 and 47). This apparently simple reaction requires a surprisingly complex set of factors. The multisubunit cleavage/polyadenylation specificity factor (CPSF) and cleavage stimulatory factor (CstF) define the poly(A) site by binding cooperatively to the conserved AAUAAA and GU-rich sequence elements upstream and downstream, respectively, of the cleavage site (32, 43, 55). Cleavage factors I and II help in the complex assembly and in the first step (7, 15, 58). Poly(A) polymerase (PAP) catalyzes poly(A) addition and is also, in most cases, required in vitro for the cleavage reaction (48). The C-terminal domain of the largest subunit of RNA polymerase II (CTD) participates in the 3′-end-processing reaction and plays a critical stimulatory role (25, 39).
CPSF-73, one of the subunits of the CPSF complex, has generated considerable interest recently as evidence has accumulated that it is the endonuclease that catalyzes the cleavage reaction. This was suggested first by its identification as a member of the metallo-β-lactamase family of Zn-dependent hydrolytic enzymes (8, 49). More conclusively, recent structural and biochemical studies carried out with purified CPSF-73 provided unequivocal evidence that CPSF-73 indeed possesses endonucleolytic activity (38).
In addition to the factors described above, symplekin, a protein sharing similarity with the yeast (Saccharomyces cerevisiae) polyadenylation factor Pta1, was identified as a CstF-64-interacting factor and was found to associate with both CstF and CPSF (55). Symplekin was also found to colocalize with CPSF both in nuclei and in cytoplasmic extracts (27). Later studies showed the involvement of symplekin as an essential factor participating in the related processes of histone pre-mRNA 3′-end processing (34) and cytoplasmic polyadenylation (4). In the above-mentioned processes, symplekin appears to function as a scaffold, making contact with multiple factors in these complexes.
3′-end processing constitutes an important step at which gene expression can be regulated. For instance, several examples indicate that 3′ processing is regulated by alterations in the levels of basal polyadenylation factors (3). When B cells differentiate, CstF-64 levels increase, and high levels of CstF-64 favor the use of a weaker, proximal poly(A) site, thus mediating at least in part the switch from the membrane-bound to the secreted form of immunoglobulin heavy chain in plasma cells (56, 59). Similarly, the stimulation of macrophages by lipopolysaccharides elevates CstF-64 levels, thus influencing gene expression and alternative polyadenylation (53). Human immunodeficiency virus type 1 Tat protein upregulates CPSF-73 mRNA levels in infected cells, and this appears to manipulate cellular transcription and processing to further viral replication (9).
The regulation by posttranslational modification of basal polyadenylation factors in mammals is restricted to the single known example of PAP. PAP possesses several sites for phosphorylation by cyclin-dependent kinases in its CTD and is known to be phosphorylated throughout the cell cycle (2, 11). During mitosis and meiotic progression, PAP is hyperphosphorylated by cdc2-cyclin B (11, 12). This results in the downregulation of PAP enzymatic activity, which in turn appears to be important for normal cell growth (11, 65). Alterations in PAP phosphorylation and activity have also been reported in human immunodeficiency virus-infected cells (42).
Posttranslational modification by the small ubiquitin-like modifier (SUMO) has gained prominence as a regulator of protein stability, subcellular localization, and interaction (23, 30). Similar to ubiquitin, the multienzymatic cascade of SUMO conjugation utilizes an E1-activating-enzyme heterodimer (SAE2/SAE1), a conjugation enzyme (ubc9), and several E3 ligases to attach SUMO via an isopeptide bond to the target lysine in the substrate. Of the SUMO isoforms found in mammals, SUMO-2 and -3 share 95% identity and can form polymeric chains, while SUMO-1 shares 40% identity with SUMO-2/3 and can only be added as a monomer (52, 60). Various SUMO proteases, members of the SENP family, catalyze the deconjugation of SUMO from substrates (30). Proteins that can interact with SUMO or sumoylated proteins noncovalently often possess SUMO interaction motifs, or SIMs (24).
The growing list of cellular processes influenced by sumoylation includes transcriptional regulation, DNA repair, maintenance of genomic integrity, subcellular transport, and signal transduction (23, 30). Proteomic analysis showed clustering of sumoylation substrates in functional protein complexes, suggesting a role for sumoylation in regulating the functions of these complexes (63). In keeping with this finding, there are now examples of functional complexes with more than one component modified by SUMO, such as PML bodies, the Smc5-6 complex, and PcG bodies (1, 28, 31). Three independent proteomic efforts in yeast identified the polyadenylation factors Ysh1 (CPSF-73 homolog) and Cft2p (CPSF-100 homolog) as potential targets for modification by SUMO (21, 45, 63). Additionally, an in vitro-expression cloning approach has identified symplekin as a potential SUMO-1 target (19). These studies suggest a possible role for sumoylation in polyadenylation.
In this study, we have analyzed the role of sumoylation in 3′-end processing in mammals. We identified CPSF-73 and symplekin as targets for SUMO-2/3 modification in vivo. Sumoylation of symplekin was found to be essential for a function required for cell viability. By reducing the levels of sumoylation in cell extracts, either by SUMO protease addition or by prior ubc9 RNA interference (RNAi)-mediated depletion, we provided evidence that sumoylation is necessary for efficient 3′-end formation. The SUMO protease directly interacted with the two SUMO targets in nuclear extracts of HeLa cells (NEs), consistent with the idea that desumoylation of these factors may at least partly account for the observed effect on 3′ processing. Our results suggest a novel role for SUMO modification in enhancing the assembly and activity of the pre-mRNA 3′-end-processing complex.
MATERIALS AND METHODS
Plasmids, siRNAs, and antibodies.
pcDNA3-Flag symplekin was generated by reverse transcription-PCR from HeLa total RNA, and the PCR product inserted downstream of the Flag epitope between EcoRI and XbaI in vector pcDNA3.1. CPSF-73 was subcloned into pCMV14 upstream of the three-Flag tag. Plasmids expressing SUMO-2GG and SUMO-3GG were gifts from R. T. Hay, plasmids expressing ubc9 and SUMO-1 were gifts from D. Wotton, and pQE-his SENP2 was a gift from M. Matunis. Anti-SUMO-3, anti-SUMO-1 (Invitrogen), antisymplekin (BD Biosciences), antiactin, anti-ubc9 (Santa Cruz Biotechnology), anti-Flag (Stratagene), and anti-GST (Molecular Probes) were purchased from the indicated companies. Anti-CstF-64 was described earlier (57). Polyclonal antisymplekin and anti-CPSF-73 were gifts from Orit Rosen. The small interfering RNAs (siRNAs) used were as follows: ubc9 siRNA1 (CAATGAACCTGATGAACTG), ubc9 siRNA2 (ubc9 Smartpool; M-004910-00), symplekin siRNA 1 (CCACACTACTGGACAACTT in 5′UTR), symplekin siRNA 2 (TCATCGCATTCCAAGCAGA in 5′UTR), and nontargeting control siRNA (D-001210). All siRNAs were purchased from Dharmacon.
Site-directed mutagenesis.
Lysine-to-arginine mutations in pcDNA3-symplekin, pCMV14-CPSF-73, and R577L, K578M pQE-HisSENP2 were generated by PCR-based mutagenesis, using appropriate pairs of site-specific mutagenic primers. Double and triple mutations were generated by subsequent mutagenesis. Mutations were confirmed by sequence analysis.
Cell culture and transfections.
HeLa and 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum. NIH 3T3 cells were cultured in DMEM with 10% bovine calf serum. Small-scale plasmid transfections in HeLa cells were carried out in six-well plates with 4 μg of total DNA using Lipofectamine 2000, following the manufacturer's instructions. For immunoprecipation (IP) experiments, 293T cells were grown to 50% confluence in 10-cm plates and 12 μg DNA was transfected by the calcium phosphate method. For large-scale siRNA transfections, HeLa cells were grown to 50% confluence in 10-cm plates, and 250 pmol of siRNAs was transfected with 40 μl Lipofectamine 2000. Forty-eight hours after transfections, cells were harvested and NEs were prepared as described below. siRNA-plasmid cotransfections in HeLa cells were carried out in six-well plates with 50 pmol siRNAs and 1 μg plasmid using 8 μl of Lipofectamine 2000 per well. Cells were harvested 48 to 60 h after the siRNA transfections or as indicated in the Fig. 4 legend. RNAiFECT transfection reagent (QIAGEN) was used for siRNA transfections in NIH 3T3 cells. Cells were grown to 40% confluence in six-well plates, and 100 pmol of siRNAs was transfected with 8 μl reagent.
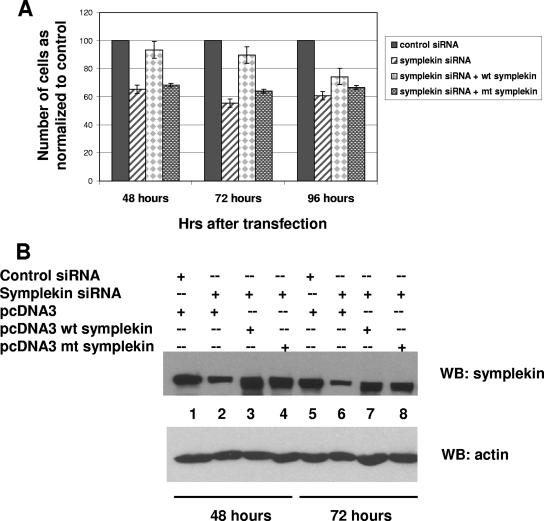
FIG. 4.
Cells expressing a sumoylation-deficient mutant of symplekin are inviable. (A) Symplekin siRNA1 or control siRNAs were cotransfected with pcDNA3 empty vector, pcDNA3-wt symplekin, or pcDNA3-mt symplekin with Lipofectamine 2000 in six-well plates as described in Materials and Methods. Adherent cells were harvested 48 h, 72 h, and 96 h after transfection from three independent experiments and counted, and the mean value plotted as a percentage of cells in the control transfection (control siRNA plus empty vector). Standard error bars are indicated. (B) Western blot analysis with antisymplekin antibodies (top panel) and antiactin (bottom panel) of cells harvested at 48 h (lanes 1 through 4) and 72 h (lanes 5 through 8) after transfection. mt, mutant; WB, Western blot; +, present; −, absent.
NE preparation and IPs.
HeLa NEs were prepared as described earlier (33). For assessing the sumoylation of endogenous proteins, buffers used in NE preparations contained 20 mM N-ethyl maleimide (NEM). Extracts were diluted in 50 mM Tris, pH 8.0, 250 mM NaCl, 5 mM EDTA, 0.5% NP-40 and precleared with protein A-Sepharose beads for 1 h. Antibody-bound Sepharose beads were incubated with precleared extracts, washed three times with lysis buffer containing 500 mM NaCl, and eluted by boiling in sodium dodecyl sulfate (SDS) sample buffer. The inputs and bound proteins were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotted with the antibodies indicated in Fig. 1 and 2.
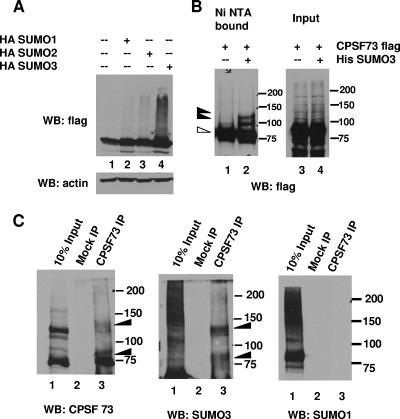
FIG. 1.
Symplekin is modified by SUMO-2/3 in HeLa cells. (A) HeLa cells were cotransfected with a plasmid encoding Flag-tagged symplekin along with empty vector (lane 1) or vectors encoding HA-SUMO-1, -2 or -3 (lanes 2, 3, and 4). Cell lysates were subjected to Western blot analysis with anti-Flag or antiactin antibodies. Higher-molecular-weight forms and the unmodified form of symplekin are indicated by closed and open arrowheads, respectively. (B) 293 cells were cotransfected with a plasmid encoding His-tagged SUMO-3 (lanes 1 and 4) or with Flag-tagged symplekin plus empty vector (lanes 2 and 5) or vectors encoding His-tagged SUMO-3 (lanes 3 and 6). Lysates were prepared under denaturing conditions and subjected to Ni-NTA chromatography. Inputs (lanes 1, 2, and 3) and the Ni-NTA-bound proteins (lanes 4, 5, and 6) were analyzed by Western blotting with anti-Flag antibodies. The unmodified (open arrowhead) and SUMO-modified (bracket) forms of symplekin are indicated. (C) Symplekin immunoglobulin G or control immunoglobulin G was used to immunoprecipitate proteins from HeLa NE, and IPs were analyzed by Western blotting with antisymplekin (left panel), anti-SUMO-2/3 (middle panel), and anti-SUMO-1 (right panel) antibodies. The closed arrowheads indicate the slower-migrating form of symplekin. Positions of molecular weight markers are indicated. WB, Western blot; +, present; −, absent.
FIG. 2.
CPSF-73 is modified by SUMO-2/3. (A) HeLa cells were transfected with a plasmid encoding Flag-tagged CPSF-73 along with empty vector (lane 1) or plasmids encoding HA-SUMO-1, -2 or -3 (lanes 2, 3, and 4). Whole-cell lysates were subjected to Western blotting with anti-Flag and antiactin antibodies. (B) HeLa cells were transfected with a plasmid encoding Flag-tagged CPSF-73 along with empty vector (lanes 1 and 3) or a plasmid encoding His-tagged SUMO-3 (lanes 2 and 4), and cell lysates were partially purified under denaturing conditions by Ni-NTA chromatography as described in Materials and Methods. Inputs (lanes 3 and 4) and bound fractions (lanes 1 and 2) were subjected to Western blotting with anti-Flag antibodies. Higher-molecular-weight forms of CPSF-73 are indicated by closed arrowheads, and unmodified CPSF-73 is indicated by an open arrowhead. (C) Antibodies to CPSF-73 or control immunoglobulin Gs were used to immunoprecipitate proteins from HeLa NEs, and IPs were analyzed by Western blotting with anti-CPSF-73 (left panel), anti-SUMO-2/3 (middle panel), and anti-SUMO-1 (right panel) antibodies. Higher-molecular-weight forms of CPSF-73 are indicated by closed arrowheads. Positions of molecular weight markers are indicated. WB, Western blot; +, present; −, absent.
In vivo sumoylation and denaturing purification.
293 cells were grown to 50% confluence in 10-cm plates and transfected with 6 μg CPSF-73 expression vector and 6 μg empty vector or His-SUMO-3 vector using the calcium phosphate method. Forty-eight hours after transfection, cells were lysed in 6 M guanidine-HCl, 50 mM NaH2PO4 (pH 8.0), 10 mM Tris-HCl (pH 8.0), and 100 mM NaCl. Six-His-tagged proteins were bound to nickel-nitrilotriacetic acid (Ni-NTA)-agarose (QIAGEN) by rocking for 3 h. Beads were washed several times sequentially in 8 M urea, 50 mM NaH2PO4, 100 mM NaCl, pH 8.0 (buffer A), buffer A with 10 mM imidazole, and buffer A with the pH adjusted to 6.3. Bound proteins were eluted by boiling in 1% SDS, 100 mM dithiothreitol, 50 mM Tris, pH 6.8, 10% glycerol containing 100 mM imidazole. The inputs and the Ni-NTA-bound fractions were resolved by SDS-PAGE and immunoblotted with the antibodies indicated below.
3′-processing assays.
32P-labeled simian virus 40 late (SVL) substrate, SVL precleaved substrate, and L3 substrates were prepared as described previously (50). For 3′ cleavage assays, the reaction mixtures consisted of 0.2 to 0.5 ng labeled RNA, 250 ng tRNA, 0.25 U RNasin (Promega), 8 mM Tris, pH 7.9, 10% glycerol, 20 mM creatine phosphate, 120 mM NaCl, 0.2 mM dithiothreitol, 2.5% polyvinyl alcohol, 2 mM EDTA, and 0.2 mM phenylmethylsulfonyl fluoride. Specific polyadenylation assays with precleaved SVL substrate and cleavage/polyadenylation reaction mixtures with L3 contained 1 mM MgCl2 and 1 mM ATP instead of EDTA. In all cases, the reaction mixtures were preincubated with the indicated proteins for 15 min at 30°C before the addition of substrate RNA.
Complex assembly assays.
Cleavage/polyadenylation reactions with wt SVL or SVL AAAAAA mutant substrates were performed as described above. The indicated proteins were preincubated with reaction mixtures prior to the addition of substrate for 15 min at 30°C, and the reaction mixture was further incubated for 20 min with the substrate at 30°C. Heparin was added to a final concentration of 5 μg/μl, and the complexes resolved on a 1.5% low-melting-point agarose gel in buffer containing 50 mM Tris and 50 mM glycine. The gel was fixed, dried, and subjected to autoradiography.
RESULTS
Symplekin is modified by SUMO-3 in HeLa cells.
We began our analysis of the possible role of sumoylation in pre-mRNA polyadenylation by examining symplekin, which is a potential SUMO-1 substrate found by expression screening (19). We first tested whether symplekin can be modified by any of the three isoforms in vivo by coexpressing Flag-tagged symplekin with hemagglutinin (HA)-tagged SUMO-1, -2, and -3 in HeLa cells. Immunoblotting of whole-cell lysates with anti-Flag antibodies revealed slower-migrating forms (Fig. 1A, lane 4) that were detected strongly and reproducibly with SUMO-3, but not with SUMO-1. Interestingly, however, cotransfection with SUMO-1 resulted in a significant decrease in symplekin levels, even though no SUMO-1-conjugated forms of symplekin were detected (Fig. 1A, lane 2). In order to confirm that the slower-migrating species were SUMO-modified forms of symplekin, we carried out a similar coexpression experiment with Flag-symplekin and His-tagged SUMO-3. His-SUMO-3 conjugates were partially purified under denaturing conditions by being bound to Ni-NTA-agarose beads. As shown in Fig. 1B, lanes 3 and 6, the higher-molecular-weight species of symplekin detected by anti-Flag antibodies were enriched in the Ni-NTA-bound fraction. In addition, the higher-molecular-weight species were recognized by SUMO-3 antibodies following IP with anti-Flag antibodies (see Fig. S1A, bottom panel, in the supplemental material). The results with SUMO-2 coexpression were inconclusive since HA-SUMO-2 was only poorly expressed (results not shown). These experiments reveal that symplekin is specifically modified by SUMO-3 in vivo.
We next wished to examine whether endogenous symplekin is naturally sumoylated. To this end, we prepared NEs utilizing conditions that minimize desumoylation activity, i.e., in the presence of the cysteine protease inhibitor NEM. We immunoprecipitated endogenous symplekin from these extracts and analyzed the IPs by Western blotting with symplekin, SUMO-1, and SUMO-2/3 antibodies (Fig. 1C). In addition to apparently unmodified symplekin, a higher-molecular-weight form was detected with the antisymplekin antibody (left panel, lane 3). This species was recognized by anti-SUMO-2/3 (middle panel, lane 3), but not anti-SUMO-1 (right panel, lane 3) antibodies. To confirm that endogenous symplekin can be modified by SUMO-3, HeLa cells were transfected with plasmids encoding SUMO-1, -2, or -3. Antisymplekin antibodies detected higher-molecular-weight species in cells that overexpressed SUMO-3 (see Fig. S1B in the supplemental material). We conclude that symplekin is targeted for modification by SUMO-2/3 in HeLa cells.
CPSF-73 is modified by SUMO-2/3.
We next sought to investigate the possible sumoylation of CPSF-73 and CPSF-100 in mammalian cells. First, all three SUMO isoforms were coexpressed with human Flag-tagged CPSF-73 and CPSF-100 in HeLa cells. With CPSF-100, no changes in the expression levels or any appearance of higher-molecular-weight modified forms was observed (data not shown). Coexpression of SUMO with CPSF-73, however, resulted in an increase in expression levels, an effect that was most obvious with SUMO-3 coexpression (Fig. 2A, lane 4). In this experiment, specific SUMO-3-conjugated isoforms were not visible in Western blot analysis of cell lysates. In order to detect possible SUMO-conjugated forms, Flag-tagged CPSF-73 was coexpressed with His-SUMO-3 and His-tagged SUMO-3 conjugates were partially purified by being bound to Ni-NTA-agarose under denaturing conditions. Immunoblotting with anti-Flag antibodies revealed the presence of distinct higher-molecular-weight species that were enriched in the Ni-NTA-bound fraction (Fig. 2B, lane 2). To provide additional evidence that these higher-molecular-weight species were indeed SUMO-modified forms, Flag-tagged CPSF-73 and HA-tagged SUMO-3 were coexpressed. Higher-molecular-weight species were detected by anti-HA antibody in the anti-Flag immunoprecipitates (see Fig. S2 in the supplemental material).
We next investigated whether endogenous CPSF-73 is modified by SUMO in HeLa cells. To this end, NE was prepared as described above, and IP with anti-CPSF-73 antibodies carried out. Figure 2C shows that two higher-molecular-mass forms, in addition to the 73-kDa isoform, were immunoprecipitated (left panel, lane 3). Furthermore, both higher-molecular-weight forms were recognized by the SUMO-2/3 antibody (middle panel, lane 3), but not the SUMO-1 antibody (right panel, lane 3), a result consistent with the results of the initial coexpression experiment. Therefore both exogenously expressed and endogenous CPSF-73, like symplekin, are targeted for modification by the SUMO-2/3 isoform.
The major sites of SUMO modification in symplekin and CPSF-73 are highly conserved.
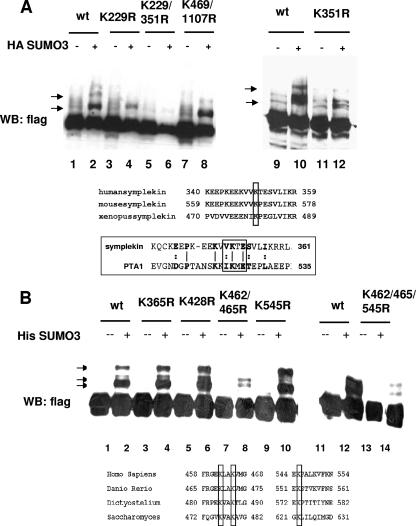
The above experiments establish that CPSF-73 and symplekin are targets for SUMO-2/3 modification. We next wished to identify the SUMO acceptor lysines in these proteins. Symplekin has six lysines that lie within the sumoylation consensus, ΨKXE/D, where Ψ represents a hydrophobic residue (30). As the mutation of lysine to arginine abolishes sumoylation, all six lysines were individually mutated to arginine. In vivo sumoylation assays were then carried out by coexpressing the Flag-tagged mutant proteins with SUMO-3 in transient transfection experiments, followed by Western blot analysis of the lysates with anti-Flag antibodies. Mutations at lysines 229 and 351 caused changes in the sumoylation pattern (Fig. 3A, lanes 4 and 12), and no sumoylated forms were detected with the appropriate double mutant (Fig. 3A, lane 6). The sumoylation site at lysine 351 is highly conserved across species and is present in the yeast homolog Pta1, as shown in the amino acid alignment in Fig. 3A (bottom panel). This is especially notable because Pta1 shares only 23% similarity with human symplekin (55).
FIG. 3.
The major sites of sumoylation in symplekin and CPSF-73 are conserved among species. (A) A plasmid encoding HA-SUMO-3 was cotransfected with plasmids encoding Flag-tagged wt symplekin or the indicated mutants into 293T cells, and lysates subjected to immunoblotting with anti-Flag antibodies. The sumoylated forms of symplekin are indicated by arrows. The panel below is a CLUSTALW alignment of human, mouse, and Xenopus symplekin. The bottom panel shows an amino acid alignment of human symplekin and yeast Pta1. Conserved lysines are boxed. (B) 293T cells were cotransfected with plasmids encoding His-SUMO-3 and Flag-tagged wt CPSF-73 (lanes 2 and 12) or the indicated mutants. Lysates were prepared and subjected to immunoblotting with anti-Flag antibodies. Sumoylated forms of CPSF-73 are indicated by arrows. The bottom panel shows a CLUSTALW amino acid alignment of human, zebrafish, D. discoideum and S. cerevisiae homologs of CPSF-73. Conserved SUMO acceptor lysines are boxed. WB, Western blot; +, present; −, absent.
CPSF-73 has two sumoylation consensus sites, lysine 197 (IKPD) and lysine 630 (VKDD). However, when both sites were simultaneously mutated in Flag-CPSF-73, the sumoylated forms of the proteins produced in transfected HeLa cells were unchanged (results not shown), indicating that the acceptor lysines are in nonconsensus sites. While many SUMO substrates are commonly modified at consensus lysines, there are many examples of proteins that are modified at lysines in sites not conforming to the consensus motif (30), and several proteins are modified at nonconsensus lysines despite the presence of consensus sites (46, 51). In such cases, it has been postulated that secondary-structure elements may contribute to the recognition of the SUMO attachment site or that the consensus motifs are inaccessible to the sumoylation machinery (46).
CPSF-73 contains 42 lysines. We first created lysine-to-arginine mutations in 14 lysines that are conserved in Ysh1. Additionally, lysines 410, 474, 604, and 214, predicted as potential SUMO sites by the program SUMOplot, were mutated (see Materials and Methods). The simultaneous mutation of conserved lysines at 462 and 465 changed the pattern of sumoylated species (Fig. 3B, lane 8), as did mutation of lysine 545 (Fig. 3B, lane 10), while mutation of other lysines had no effect. Mutating all three sites together significantly reduced overall sumoylation levels (Fig. 3B, lane 14). Faint bands were still detected with this mutant, suggesting that there may be alternative, less-preferred lysines at which SUMO can be attached. However, the three lysines identified are highly conserved across species, as shown by the alignment in Fig. 3B, and they are not in consensus sites in any of the species examined. The conservation of SUMO attachment sites for both proteins suggests that sumoylation of these proteins may also be conserved across species.
Cells expressing a sumoylation-defective symplekin are unable to grow.
A key question is whether sumoylation of symplekin and/or CPSF-73 is important for function in vivo. However, a difficulty in addressing this problem is that very little is known about how these proteins function in vivo, and assays to measure the roles of either protein in vivo have not been developed. For example, although we assume both proteins are essential for cell viability, this has not been addressed experimentally. To do so, we determined the effect of siRNA-mediated depletion of each protein on the viability of HeLa cells. Surprisingly, two different siRNAs directed against CPSF-73 had no apparent effects on growth for periods of up to 96 h (results not shown). Although both siRNAs depleted CPSF-73 effectively, we assume the remaining protein was sufficient to allow cell viability for the time period analyzed. Two different antisymplekin siRNAs, however, each had a significant effect on cell growth and viability. A large decrease (40%) in cell number compared to the number of control siRNA-treated cells was observed as early as 48 h following transfection (Fig. 4A).
The above results indicate that symplekin is required for normal cell growth. Although whether this reflects symplekin's role in polyadenylation is unknown, these results provide an assay for evaluating the requirement of sumoylation for symplekin function in vivo. Specifically, we first wished to determine whether cell growth could be rescued by cotransfection of a plasmid expressing wild-type (wt) symplekin, and if so, to ask if a sumoylation-deficient mutant could also rescue viability. The strategy was to express symplekin together with siRNA in a cotransfection experiment. To avoid targeting the exogenously expressed protein, the siRNA used was directed against the 5′ UTR and exogenous symplekin was expressed from a construct lacking this region. wt symplekin was indeed able to rescue the growth phenotype, increasing cell numbers by up to 90% of the number of control siRNA-treated cells (Fig. 4A). However, the expression of the sumoylation-deficient K229/351R mutant symplekin was unable to rescue the growth phenotype (Fig. 4A). Western blot analysis indicates that both proteins were expressed at comparable levels (Fig. 4B). We conclude from these experiments that symplekin is required for cell growth and that a sumoylation-defective symplekin mutant is unable to perform the essential function(s).
Sumoylation in HeLa NE is decreased by ubc9 siRNA and SENP2 protease.
We next wished to determine whether sumoylation has an effect on 3′-processing activity. With transcription factors, the wt and sumoylation-deficient mutants are commonly cotransfected with reporter constructs to examine transcription activity in reporter gene assays. However, a similar approach to examine the activity of basal polyadenylation factors is not feasible, as changing the activity of a single factor introduced exogenously by transfection would likely not alter the polyadenylation of a reporter substrate. Furthermore, neither factor can be tested individually in assays since both form part of larger complexes that participate both in steps of cleavage and polyadenylation. Therefore, we by necessity took a more indirect approach, which was to examine 3′ cleavage and polyadenylation in NEs whose sumoylation status has been altered.
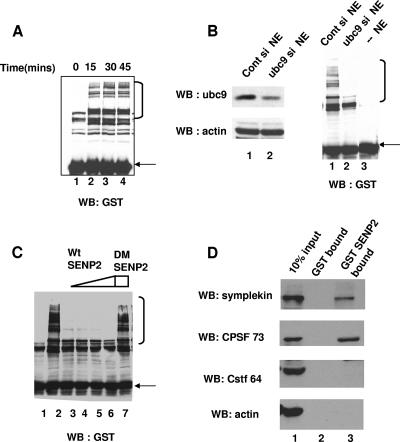
We first wished to confirm that sumoylation enzymes are active in NE. To test this, GST-SUMO-3 was added to NE, the mixture was incubated at 30°C under conditions optimal for cleavage and polyadenylation, and proteins were resolved by SDS-PAGE. Several higher-molecular-weight GST-SUMO conjugates, which formed in a time-dependent manner, were detected by Western blotting with an anti-GST antibody (Fig. 5A). The GST-SUMO conjugates increased for up to 30 min, and their levels remained constant thereafter, suggesting that a balance of sumoylation and desumoylation is maintained in these extracts. We next sought to determine whether downregulating the SUMO pathway in vivo could affect sumoylation in vitro. NEs prepared from cells depleted of ubc9 by siRNA were in fact deficient in the formation of high-molecular-weight GST-SUMO conjugates (Fig. 5B, right panel, lane 3). The addition of the SUMO protease SENP2 (64) also resulted in loss of the high-molecular-weight GST-SUMO conjugates, while SENP2 with a catalytically inactive double mutant with mutations R577L and K578M (DM SENP2) had no effect (Fig. 5C).
FIG. 5.
Sumoylation in HeLa NE is downregulated by ubc9 siRNA and SENP2 protease, which interacts specifically with symplekin and CPSF-73. (A) GST-SUMO-3 was added to reaction mixtures containing HeLa NE under polyadenylation conditions. Reaction products were analyzed by Western blotting with anti-GST antibodies. (B) HeLa cells were transfected with control or ubc9 siRNAs (si), and NEs were subjected to Western blotting with anti-ubc9 and antiactin antibodies (left panels). Control siRNA NE, ubc9 siRNA NE, or buffer (right panel, lanes 1 to 3, respectively) was added to the reaction mixtures described above. (C) Reactions were performed as described for panel A in the presence of buffer (lane 2); 50 ng, 200 ng, 500 ng, and 1 μg of WT SENP2 (lanes, 3, 4, 5, and 6); or 1 μg of DM SENP2 (lane 7) and analyzed by Western blotting with GST antibodies. Lane 1 is a negative control without NE. In all panels, free GST-SUMO-3 is indicated by a closed arrow and higher-molecular-weight GST-SUMO-3 conjugates are indicated by an open bracket. (D) HeLa NE was incubated overnight with 8 μg GST or GST-SENP2 proteins bound to glutathione beads. Ten percent of input NE, bound proteins from GST, and GST-SENP2 beads were subjected to Western blot analysis with anti-CPSF-73, antisymplekin, anti-CstF-64, and antiactin antibodies as indicated. WB, Western blot.
SENP2 interacts with both symplekin and CPSF-73.
We also wished to test whether SENP2 can target symplekin and/or CPSF-73 in the NEs under polyadenylation conditions. Since only a small fraction of these proteins are sumoylated (see above), it would be difficult to directly observe the effect of SENP2 activity on the two proteins. However, a specific interaction of SENP2 with CPSF-73 or symplekin would provide evidence that SENP2 can target the protein for desumoylation. Therefore, GST SENP2 or GST alone was incubated with NE and bound proteins analyzed by Western blotting (Fig. 5D). Both CPSF-73 and symplekin bound to GST SENP2, but not to GST (top and second panel). SENP2 did not interact with either CstF-64, another core component of the polyadenylation machinery (lane 3, third panel from top), or with actin (lane 3, bottom panel). These data suggest that SENP2 associates specifically with both SUMO targets in NE. SENP2 can target symplekin for desumoylation in vivo, as cotransfection of SENP2 with symplekin and SUMO-3 abrogated the formation of higher-molecular-weight forms of symplekin (see Fig. S3A in the supplemental material).
NE prepared from cells depleted of ubc9 is deficient in cleavage and polyadenylation.
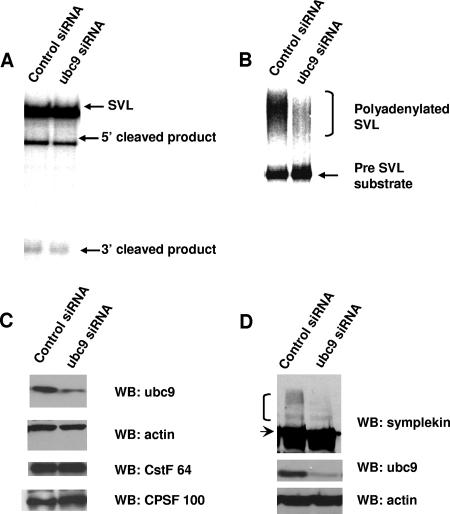
We next used the NEs prepared from ubc9 siRNA or control siRNA-treated HeLa cells in 3′ cleavage and polyadenylation assays. For these, the substrate, a 32P-labeled SVL RNA, undergoes cleavage but is not polyadenylated because the reaction mixture contains EDTA instead of a divalent cation (58). Significantly, NEs made from cells transfected with ubc9 siRNA displayed reduced cleavage activity in comparison to the activity in NEs from cells transfected with the control siRNA (Fig. 6A). Polyadenylation was assayed in both NEs by using a precleaved SVL substrate, which when incubated with ATP and MgCl2 undergoes AAUAAA-dependent polyadenylation (50). Polyadenylation in the ubc9-depleted NE was markedly reduced compared to the level in control extracts (Fig. 6B). To rule out nonspecific effects of the ubc9 siRNA used, we designed a second siRNA to deplete ubc9. NEs prepared from these cells showed a similar impairment of both cleavage and polyadenylation (data not shown). Western blot analysis of the CstF and CPSF subunits showed similar levels in NEs from control and ubc9 siRNA-treated cells (Fig. 6C), suggesting that the accumulation of these factors in the nucleus was not significantly affected. Importantly, the accumulation of sumoylated forms of symplekin was greatly reduced in NEs from the ubc9 siRNA-treated cells, indicating that sumoylation of symplekin was reduced by ubc9 siRNA treatment (Fig. 6D). Sumoylated forms of CPSF-73 were not detectable in the NEs (results not shown), and therefore, changes in CPSF-73 sumoylation could not be ascertained.
FIG. 6.
Knockdown of ubc9 by RNAi results in loss of cleavage and polyadenylation activity. (A) 32P-labeled pG3SVL-A pre-RNA was incubated under standard cleavage conditions in NEs prepared from control siRNA-treated HeLa cells or ubc9 siRNA-treated cells. RNAs were resolved by denaturing PAGE. The unprocessed pre-RNA (SVL) and the 5′ and 3′ cleavage products are indicated. (B) 32P-labeled pG3SVL-A precleaved pre-mRNA substrate (pre-SVL) was incubated as described for panel A under polyadenylation conditions, and RNAs were resolved by denaturing PAGE. The pre-mRNA substrate (precleaved SVL) and the polyadenylated products are indicated. (C) NEs made from control siRNA-treated cells (lane 1) or ubc9 siRNA-treated cells (lane 2) were analyzed by Western blotting with the antibodies indicated on the right. (D) HeLa cells were transfected with control siRNA (lane 1) or ubc9 siRNA (lane 2) and harvested, and extracts analyzed by Western blotting with antisymplekin (top panel), anti-ubc9 (middle panel), or antiactin (bottom panel) antibodies. The higher-molecular-weight forms of symplekin and the unmodified form are indicated by a bracket and an arrow, respectively. WB, Western blot.
The above data provide evidence that ubc9-dependent sumoylation, perhaps of CPSF-73 and/or symplekin, is necessary for efficient 3′ processing in NE. In the case of cleavage, at least, this must reflect a loss of sumoylation in vivo rather than of ongoing SUMO addition in the NE, because sumoylation would not be expected to occur in the presence of EDTA.
Desumoylation of NEs by SENP2 also inhibits 3′-end processing.
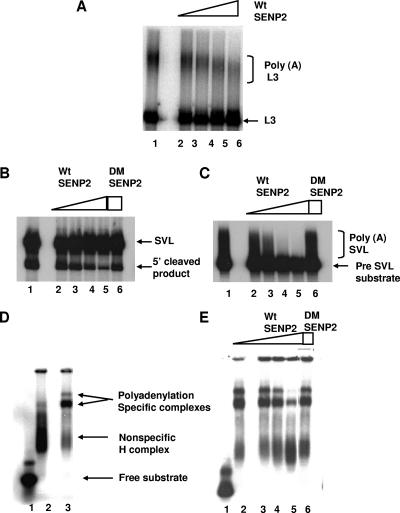
We next wished to determine whether the desumoylation of proteins in NE was sufficient to block 3′ processing in vitro. To this end, we preincubated NEs with increasing amounts of SENP2 and carried out polyadenylation reactions. Following a 15-min preincubation, 32P-labeled adenovirus L3 RNA (similar results were obtained with SVL RNA; results not shown) was added and reaction mixtures incubated for a further 90 min at 30°C. The addition of SENP2 resulted in a significant reduction in the formation of polyadenylated RNA (Fig. 7A, lanes 2 to 5). To extend this result, separate cleavage and polyadenylation reactions were carried out as described above. The addition of SENP2 to cleavage reaction mixtures resulted in a concentration-dependent decrease in the formation of the cleaved product (Fig. 7B, lanes 2 to 5). Similarly, the addition of SENP2 resulted in a strong decrease in the polyadenylation of the precleaved SVL substrate (Fig. 7C, lanes 2 to 5). The addition of DM SENP2 to these reaction mixtures had no effect on cleavage (Fig. 7B, lane 6) or polyadenylation (Fig. 7C, lane 6), indicating that the desumoylation activity of SENP2 was necessary for the effects observed. Taken together, these results show that the desumoylation of proteins in NEs compromises 3′ processing at both the cleavage and polyadenylation steps. We confirmed that SENP2 incubation does not result in the proteolysis of proteins in the NE by Western blotting of the reaction products with antisymplekin and CPSF-73 antibodies (see Fig. S3B in the supplemental material).
FIG. 7.
Desumoylation of NE by the SUMO protease SENP2 inhibits 3′ processing and blocks the formation of polyadenylation-specific complexes. (A) 32P-labeled pG3L3-pre-RNA was incubated under cleavage/polyadenylation conditions following preincubation with buffer (lane 1) or 50 ng, 100 ng, 500 ng, and 1 μg wt SENP2 (lanes 2, 3, 4, and 5, respectively). RNAs were resolved by denaturing PAGE. (B) 32P-labeled pG3SVL-A pre-RNA (SVL) was incubated under cleavage conditions following preincubation with buffer (lane 1); 50 ng, 100 ng, 500 ng, and 1 μg wt SENP2 (lanes 2, 3, 4, and 5, respectively); or 1 μg DM SENP2 (lane 6). RNAs were resolved by denaturing PAGE. (C) 32P-labeled pG3SVL-A precleaved pre-mRNA (pre-SVL) was incubated under polyadenylation conditions after preincubation as described above. Pre-RNA, 5′ cleaved products, and the polyadenylated products are indicated on the right in all panels. (D) Standard cleavage/polyadenylation reaction mixtures using 32P-labeled pG3SVL-A pre-RNA were assembled with buffer (lane 1) or NE (lane 3). Reaction mixtures using 32P-labeled pG3SVL-AAAAAA mutant RNA were assembled with NE (lane 2). Following incubation, complexes were resolved by nondenaturing PAGE. Positions of free pG3SVL-A pre-RNA substrate polyadenylation-specific complexes and nonspecific complexes are indicated. (E) Standard reaction mixtures were assembled with NE as above, and preincubation was carried out with buffer (lane 1); 50 ng, 100 ng, 500 ng, and 1 μg wt SENP2 (lanes 2, 3, 4, and 5, respectively); or 1 μg DM SENP2 (lane 6) before the addition of 32P-labeled pG3SVL-A pre-RNA. Complexes were analyzed as described for panel D.
SENP2 impairs the formation of specific polyadenylation complexes.
Sumoylation is known to have significant effects on protein-protein interactions. It is thus conceivable that interactions that occur during the assembly of the polyadenylation complex are affected by sumoylation. To address this possibility, the complex assembly on 32P-labeled SVL RNA in NE was monitored. Polyadenylation-specific complexes A and B (40) were distinguished from nonspecific (H) complexes by incubating SVL RNA and a poly(A) site mutant (SVL AAAAAA) with NE and resolving the complexes by native gel electrophoresis. Complexes A and B were formed with the wt but not the mutant substrate (Fig. 7D). NE was then preincubated with SENP2 or DM SENP2 before assembly with wt SVL RNA. After 15 min of incubation, the complexes were analyzed by native gel electrophoresis. The addition of increasing amounts of SENP2 resulted in the retention of the substrate in H complexes, with poor formation of A and B complexes (Fig. 7E, lanes 3 to 5). DM SENP2 had no effect on complex formation (Fig. 7E, lane 6). Significantly, the preincubation of NE with SENP2 was necessary for the inhibition, and the addition of SENP2 had no effect on preformed complexes (data not shown). Thus, desumoylation indeed disrupts the productive interactions necessary for the formation of specific complexes, providing a mechanism by which SENP2 inhibits the cleavage/polyadenylation reactions.
DISCUSSION
The work described here has provided the first evidence for a regulatory role of sumoylation in the processing of mRNA precursors. We demonstrated that symplekin and CPSF-73 are modified specifically by the SUMO-2/3 isoform in vivo and identified evolutionarily conserved sites of modification in both proteins. We also showed that reducing sumoylation in NEs diminished the capacity of the extracts to cleave and polyadenylate RNA substrates, at least in part by inhibiting the assembly of processing complexes. Our observations that SENP2 directly interacts with symplekin and CPSF-73 and that a sumoylation-deficient mutant of symplekin was unable to provide a function necessary for cell viability support the idea that the desumoylation of these substrates is involved in the inhibition of 3′ processing. However, further work is required to establish the specific functional roles of symplekin and CPSF-73 sumoylation.
Our data have indicated that both symplekin and CPSF-73 are specifically targeted by the SUMO-2/3 isoforms. Several previous studies have indicated that certain substrates are indeed preferentially conjugated to SUMO-1 or SUMO-2/3 (22, 26, 52, 62), which is consistent with our data showing that SUMO-2/3 specifically modifies symplekin and CPSF-73 in vivo. However, we also found that SUMO-1 overexpression had a significant effect on symplekin, as the coexpression of SUMO-1 with symplekin reduced the levels of symplekin, and that the reduction occurred in a proteasome-dependent manner (unpublished data). This is not the first example of SUMO-1 inducing the degradation of a substrate. For example, SUMO-1 overexpression has been shown to induce the proteasome-mediated degradation of the p53-related protein p63 and of the glucocorticoid receptor (17, 35). Notably however, we were unable to detect SUMO-1 modification of symplekin in vivo. This could indicate that SUMO-1-modified symplekin is so unstable that it cannot be detected or that the effect of SUMO-1 on symplekin is indirect. In any event, our data point to a role for SUMO-1 that is distinct from the role of SUMO-2/3 in affecting symplekin function.
The lysines at modification sites in symplekin and CPSF-73 were found to be conserved across species, an indication that the sumoylation of 3′-processing factors may also be conserved in different species. In keeping with this, Ysh1, the yeast homolog of CPSF-73, is known to be sumoylated (21, 45). Yeast PAP was shown to interact with the SUMO E1, Uba2, and although PAP was not shown to be sumoylated, the depletion of Uba2 resulted in increased polyadenylation activity in yeast extracts (13). This would appear to contrast with our findings, but it would not be the first instance in which yeast 3′ processing differs from mammalian 3′ processing. For example, yeast PAP lacks the regulatory domain of mammalian PAP that is important for the cell cycle-dependent phosphorylation and regulation of PAP activity (11, 12, 66). In addition, although we have recently discovered that mammalian PAP is sumoylated, the SUMO acceptor lysines are not conserved in yeast PAP (unpublished data).
Symplekin was first suggested to be a scaffolding protein when it was found to interact with purified CstF-64 and to associate with both CPSF and CstF as part of a higher-molecular-weight complex (55). Symplekin was also observed to associate with CPSF subunits in enucleated oocytes (27) and to form an essential scaffold upon which the cytoplasmic polyadenylation components CPSF and CPEB were assembled, together with the GLD-2 PAP (4). Similarly, in histone mRNA 3′-end processing, symplekin functions as a key component of a multisubunit complex required for the reaction and was found to be necessary for the functional integrity of the complex (34). These multiple functions are consistent with our discovery that symplekin is required for cell viability, and the fact that a sumoylation-deficient mutant was unable to rescue viability supports the involvement of sumoylation in symplekin function in one or more aspects of 3′ processing.
We have shown that the SUMO protease SENP2 interacts with both CPSF-73 and symplekin and also inhibits 3′ processing in NE. We utilized SENP2 in our experiments because it efficiently desumoylates both SUMO-1- and SUMO-2/3-conjugated substrates in vitro (44, 64). However, whether SENP2 influences 3′ processing in vivo is unclear. The substrate specificities of SUMO proteases are often determined by their subcellular localization. For example, the SENP2 gene produces three alternatively spliced isoforms, SENP2, SMT3IP2, and SuPR-1, which localize to the nucleoplasmic side of the nuclear pore, cytoplasm, and nuclear bodies, respectively (30, 44, 64). While the localization makes it possible that SENP2 or one of the forms generated by alternative splicing may target the 3′-processing complex in vivo, it is also possible that another SUMO protease, such as a SUMO-2/3-specific protease like SENP3 and SENP5 (16, 20), functions in this capacity in vivo.
While there are numerous examples of transcription factors that are modified by sumoylation, there are very few studies examining SUMO modification of proteins potentially involved in RNA processing. Several hnRNPs were found in proteomic analysis to be modified by SUMO, and SUMO modification of the hnRNP C and M proteins was reported to reduce RNA binding by both proteins (36, 61). The RNA-editing enzyme ADAR1 is also sumoylated, and SUMO modification was found to reduce editing activity (14). Ours, however, is the first study demonstrating a role for sumoylation in an RNA-processing event carried out by a large complex. The negative effects of hnRNP and ADAR1 sumoylation on activity contrast with the positive role that sumoylation plays in 3′-end formation. As discussed below, this likely reflects the fact that multiple subunits of the complex polyadenylation machinery may be SUMO targets and is also consistent with the idea that SUMO-mediated interactions may enhance the assembly of the polyadenylation complex. While CPSF-73 and/or symplekin is likely involved in mediating this assembly, the sumoylation of other factors may also be important.
Several examples of sumoylation modifying multiple factors in a functional protein complex and affecting its assembly have been described. For example, PML nuclear bodies, thought to be storage depots for nuclear factors or sites of assembly of transcription factors, contain several sumoylation targets, such as PML, Daxx, and SP100 (28). The integrity of nuclear bodies depends on maintaining the sumoylation of all these components, and the overexpression of the SUMO protease Su-PR1, which localizes to PML bodies, disassembles this nuclear substructure (5). Recently, PML and Daxx were found to possess SUMO binding motifs, and the interaction between the SIM and the sumoylated components promoted the complex assembly (37, 54). An attractive possibility is that the sumoylation of components of the 3′-processing complex functions in a similar manner. Supporting this idea, we identified a 30-amino-acid stretch in symplekin (amino acids 368 to 398) that has a striking resemblance to SIMs found in PIASy and PIASxα (24). Preliminary experiments have revealed a weak interaction between symplekin and SUMO (unpublished data), and we suggest that the presence of SUMO moieties in polyadenylation factors may augment their interactions with symplekin, thereby enhancing its role as a scaffolding factor and facilitating the assembly of the polyadenylation complex.
The cleavage/polyadenylation reaction has been reconstituted with highly enriched or purified fractions of NE (e.g., see references 6 and 58). It is doubtful that sumoylated forms of proteins are retained through such extensive purification processes. It thus may be that the activity detected may reflect a low, SUMO-independent activity, perhaps requiring higher concentrations of factors than would be necessary if the proteins were sumoylated. However, it is also possible that the role of sumoylation in facilitating assembly is more important in the nucleus or in a crude environment such as HeLa NE, where other factors may interfere with the formation of a functional complex due to inhibitory interactions with the polyadenylation factors. Sumoylation may be necessary in such an environment to regulate such interactions and facilitate the proper assembly of the functional 3′-processing complex. While additional work is required to establish the specific roles of CPSF-73 and symplekin sumoylation in 3′-end formation, our data have both extended the potential regulatory roles of sumoylation to RNA processing and also highlighted the complexity of the 3′-processing reaction.
Supplementary Material
Acknowledgments
We thank R. T. Hay, D. Wotton, M. Matunis, and O. Rosen for generous gifts of plasmids and antibodies, N. Rao and D. di Giammartino for technical assistance, Y. Shi and C. Lima for helpful discussions and suggestions, and I. Boluk for help with preparing the manuscript.
The work was supported by a grant from the NIH.
Footnotes
Published ahead of print on 8 October 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Andrews, E. A., J. Palecek, J. Sergeant, E. Taylor, A. R. Lehmann, and F. Z. Watts. 2005. Nse2, a component of the Smc5-6 complex, is a SUMO ligase required for the response to DNA damage. Mol. Cell. Biol. 25:185-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballantyne, S., A. Bilger, J. Astrom, A. Virtanen, and M. Wickens. 1995. Poly (A) polymerases in the nucleus and cytoplasm of frog oocytes: dynamic changes during oocyte maturation and early development. RNA 1:64-78. [PMC free article] [PubMed] [Google Scholar]
- 3.Barabino, S. M., and W. Keller. 1999. Last but not least: regulated poly(A) tail formation. Cell 99:9-11. [DOI] [PubMed] [Google Scholar]
- 4.Barnard, D. C., K. Ryan, J. L. Manley, and J. D. Richter. 2004. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell 119:641-651. [DOI] [PubMed] [Google Scholar]
- 5.Best, J. L., S. Ganiatsas, S. Agarwal, A. Changou, P. Salomoni, O. Shirihai, P. B. Meluh, P. P. Pandolfi, and L. I. Zon. 2002. SUMO-1 protease-1 regulates gene transcription through PML. Mol. Cell 10:843-855. [DOI] [PubMed] [Google Scholar]
- 6.Bienroth, S., G. Christofori, K. M. Lang, E. Wahle, and W. Keller. 1990. Components involved in 3′ processing of precursors to polyadenylated messenger RNA. Mol. Biol. Rep. 14:197. [DOI] [PubMed] [Google Scholar]
- 7.Brown, K. M., and G. M. Gilmartin. 2003. A mechanism for the regulation of pre-mRNA 3′ processing by human cleavage factor Im. Mol. Cell 12:1467-1476. [DOI] [PubMed] [Google Scholar]
- 8.Callebaut, I., D. Moshous, J. P. Mornon, and J. P. de Villartay. 2002. Metallo-beta-lactamase fold within nucleic acids processing enzymes: the beta-CASP family. Nucleic Acids Res. 30:3592-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calzado, M. A., R. Sancho, and E. Munoz. 2004. Human immunodeficiency virus type 1 Tat increases the expression of cleavage and polyadenylation specificity factor 73-kilodalton subunit modulating cellular and viral expression. J. Virol. 78:6846-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colgan, D. F., and J. L. Manley. 1997. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 11:2755-2766. [DOI] [PubMed] [Google Scholar]
- 11.Colgan, D. F., K. G. Murthy, C. Prives, and J. L. Manley. 1996. Cell-cycle related regulation of poly(A) polymerase by phosphorylation. Nature 384:282-285. [DOI] [PubMed] [Google Scholar]
- 12.Colgan, D. F., K. G. Murthy, W. Zhao, C. Prives, and J. L. Manley. 1998. Inhibition of poly(A) polymerase requires p34cdc2/cyclin B phosphorylation of multiple consensus and non-consensus sites. EMBO J. 17:1053-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.del Olmo, M., N. Mizrahi, S. Gross, and C. L. Moore. 1997. The Uba2 and Ufd1 proteins of Saccharomyces cerevisiae interact with poly(A) polymerase and affect the polyadenylation activity of cell extracts. Mol. Gen. Genet. 255:209-218. [DOI] [PubMed] [Google Scholar]
- 14.Desterro, J. M., L. P. Keegan, E. Jaffray, R. T. Hay, M. A. O'Connell, and M. Carmo-Fonseca. 2005. SUMO-1 modification alters ADAR1 editing activity. Mol. Biol. Cell 16:5115-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Vries, H., U. Ruegsegger, W. Hubner, A. Friedlein, H. Langen, and W. Keller. 2000. Human pre-mRNA cleavage factor II(m) contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J. 19:5895-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Bacco, A., J. Ouyang, H. Y. Lee, A. Catic, H. Ploegh, and G. Gill. 2006. The SUMO-specific protease SENP5 is required for cell division. Mol. Cell. Biol. 26:4489-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghioni, P., Y. D'Alessandra, G. Mansueto, E. Jaffray, R. T. Hay, G. La Mantia, and L. Guerrini. 2005. The protein stability and transcriptional activity of p63alpha are regulated by SUMO-1 conjugation. Cell Cycle 4:183-190. [DOI] [PubMed] [Google Scholar]
- 18.Reference deleted.
- 19.Gocke, C. B., H. Yu, and J. Kang. 2005. Systematic identification and analysis of mammalian small ubiquitin-like modifier substrates. J. Biol. Chem. 280:5004-5012. [DOI] [PubMed] [Google Scholar]
- 20.Gong, L., and E. T. Yeh. 2006. Characterization of a family of nucleolar SUMO-specific proteases with preference for SUMO-2 or SUMO-3. J. Biol. Chem. 281:15869-15877. [DOI] [PubMed] [Google Scholar]
- 21.Hannich, J. T., A. Lewis, M. B. Kroetz, S. J. Li, H. Heide, A. Emili, and M. Hochstrasser. 2005. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J. Biol. Chem. 280:4102-4110. [DOI] [PubMed] [Google Scholar]
- 22.Hardeland, U., R. Steinacher, J. Jiricny, and P. Schar. 2002. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 21:1456-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hay, R. T. 2005. SUMO: a history of modification. Mol. Cell 18:1-12. [DOI] [PubMed] [Google Scholar]
- 24.Hecker, C. M., M. Rabiller, K. Haglund, P. Bayer, and I. Dikic. 2006. Specification of SUMO1- and SUMO2-interacting motifs. J. Biol. Chem. 281:16117-16127. [DOI] [PubMed] [Google Scholar]
- 25.Hirose, Y., and J. L. Manley. 1998. RNA polymerase II is an essential mRNA polyadenylation factor. Nature 395:93-96. [DOI] [PubMed] [Google Scholar]
- 26.Hofmann, H., S. Floss, and T. Stamminger. 2000. Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J. Virol. 74:2510-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hofmann, I., M. Schnolzer, I. Kaufmann, and W. W. Franke. 2002. Symplekin, a constitutive protein of karyo- and cytoplasmic particles involved in mRNA biogenesis in Xenopus laevis oocytes. Mol. Biol. Cell 13:1665-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. Yeh, J. F. Strauss III, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reference deleted.
- 30.Johnson, E. S. 2004. Protein modification by SUMO. Annu. Rev. Biochem. 73:355-382. [DOI] [PubMed] [Google Scholar]
- 31.Kagey, M. H., T. A. Melhuish, and D. Wotton. 2003. The polycomb protein Pc2 is a SUMO E3. Cell 113:127-137. [DOI] [PubMed] [Google Scholar]
- 32.Kaufmann, I., G. Martin, A. Friedlein, H. Langen, and W. Keller. 2004. Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J. 23:616-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kleiman, F. E., and J. L. Manley. 2001. The BARD1-CstF-50 interaction links mRNA 3′ end formation to DNA damage and tumor suppression. Cell 104:743-753. [DOI] [PubMed] [Google Scholar]
- 34.Kolev, N. G., and J. A. Steitz. 2005. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Dev. 19:2583-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le Drean, Y., N. Mincheneau, P. Le Goff, and D. Michel. 2002. Potentiation of glucocorticoid receptor transcriptional activity by sumoylation. Endocrinology 143:3482-3489. [DOI] [PubMed] [Google Scholar]
- 36.Li, T., E. Evdokimov, R. F. Shen, C. C. Chao, E. Tekle, T. Wang, E. R. Stadtman, D. C. Yang, and P. B. Chock. 2004. Sumoylation of heterogeneous nuclear ribonucleoproteins, zinc finger proteins, and nuclear pore complex proteins: a proteomic analysis. Proc. Natl. Acad. Sci. USA 101:8551-8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin, D. Y., Y. S. Huang, J. C. Jeng, H. Y. Kuo, C. C. Chang, T. T. Chao, C. C. Ho, Y. C. Chen, T. P. Lin, H. I. Fang, C. C. Hung, C. S. Suen, M. J. Hwang, K. S. Chang, G. G. Maul, and H. M. Shih. 2006. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell 24:341-354. [DOI] [PubMed] [Google Scholar]
- 38.Mandel, C. R., S. Kaneko, H. Zhang, D. Gebauer, V. Vethantham, J. L. Manley, and L. Tong. 2006. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 444:953-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCracken, S., N. Fong, K. Yankulov, S. Ballantyne, G. Pan, J. Greenblatt, S. D. Patterson, M. Wickens, and D. L. Bentley. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385:357-361. [DOI] [PubMed] [Google Scholar]
- 40.McLauchlan, J., C. L. Moore, S. Simpson, and J. B. Clements. 1988. Components required for in vitro cleavage and polyadenylation of eukaryotic mRNA. Nucleic Acids Res. 16:5323-5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reference deleted.
- 42.Mouland, A. J., M. Coady, X. J. Yao, and E. A. Cohen. 2002. Hypophosphorylation of poly(A) polymerase and increased polyadenylation activity are associated with human immunodeficiency virus type 1 Vpr expression. Virology 292:321-330. [DOI] [PubMed] [Google Scholar]
- 43.Murthy, K. G., and J. L. Manley. 1995. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev. 9:2672-2683. [DOI] [PubMed] [Google Scholar]
- 44.Nishida, T., H. Tanaka, and H. Yasuda. 2000. A novel mammalian Smt3-specific isopeptidase 1 (SMT3IP1) localized in the nucleolus at interphase. Eur. J. Biochem. 267:6423-6427. [DOI] [PubMed] [Google Scholar]
- 45.Panse, V. G., U. Hardeland, T. Werner, B. Kuster, and E. Hurt. 2004. A proteome-wide approach identifies sumoylated substrate proteins in yeast. J. Biol. Chem. 279:41346-41351. [DOI] [PubMed] [Google Scholar]
- 46.Pichler, A., P. Knipscheer, E. Oberhofer, W. J. van Dijk, R. Korner, J. V. Olsen, S. Jentsch, F. Melchior, and T. K. Sixma. 2005. SUMO modification of the ubiquitin-conjugating enzyme E2-25K. Nat. Struct. Mol. Biol. 12:264-269. [DOI] [PubMed] [Google Scholar]
- 47.Proudfoot, N., and J. O'Sullivan. 2002. Polyadenylation: a tail of two complexes. Curr. Biol. 12:R855-R857. [DOI] [PubMed] [Google Scholar]
- 48.Raabe, T., F. J. Bollum, and J. L. Manley. 1991. Primary structure and expression of bovine poly(A) polymerase. Nature 353:229-234. [DOI] [PubMed] [Google Scholar]
- 49.Ryan, K., O. Calvo, and J. L. Manley. 2004. Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA 10:565-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ryner, L. C., Y. Takagaki, and J. L. Manley. 1989. Multiple forms of poly(A) polymerases purified from HeLa cells function in specific mRNA 3′-end formation. Mol. Cell. Biol. 9:4229-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sacher, M., B. Pfander, C. Hoege, and S. Jentsch. 2006. Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nat. Cell Biol. 8:1284-1290. [DOI] [PubMed] [Google Scholar]
- 52.Saitoh, H., and J. Hinchey. 2000. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 275:6252-6258. [DOI] [PubMed] [Google Scholar]
- 53.Shell, S. A., C. Hesse, S. M. Morris, Jr., and C. Milcarek. 2005. Elevated levels of the 64-kDa cleavage stimulatory factor (CstF-64) in lipopolysaccharide-stimulated macrophages influence gene expression and induce alternative poly(A) site selection. J. Biol. Chem. 280:39950-39961. [DOI] [PubMed] [Google Scholar]
- 54.Shen, T. H., H. K. Lin, P. P. Scaglioni, T. M. Yung, and P. P. Pandolfi. 2006. The mechanisms of PML-nuclear body formation. Mol. Cell 24:331-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takagaki, Y., and J. L. Manley. 2000. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol. Cell. Biol. 20:1515-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takagaki, Y., and J. L. Manley. 1998. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol. Cell 2:761-771. [DOI] [PubMed] [Google Scholar]
- 57.Takagaki, Y., J. L. Manley, C. C. MacDonald, J. Wilusz, and T. Shenk. 1990. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes Dev. 4:2112-2120. [DOI] [PubMed] [Google Scholar]
- 58.Takagaki, Y., L. C. Ryner, and J. L. Manley. 1989. Four factors are required for 3′-end cleavage of pre-mRNAs. Genes Dev. 3:1711-1724. [DOI] [PubMed] [Google Scholar]
- 59.Takagaki, Y., R. L. Seipelt, M. L. Peterson, and J. L. Manley. 1996. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 87:941-952. [DOI] [PubMed] [Google Scholar]
- 60.Tatham, M. H., E. Jaffray, O. A. Vaughan, J. M. Desterro, C. H. Botting, J. H. Naismith, and R. T. Hay. 2001. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276:35368-35374. [DOI] [PubMed] [Google Scholar]
- 61.Vassileva, M. T., and M. J. Matunis. 2004. SUMO modification of heterogeneous nuclear ribonucleoproteins. Mol. Cell. Biol. 24:3623-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vertegaal, A. C., J. S. Andersen, S. C. Ogg, R. T. Hay, M. Mann, and A. I. Lamond. 2006. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol. Cell Proteomics 5:2298-2310. [DOI] [PubMed] [Google Scholar]
- 63.Wohlschlegel, J. A., E. S. Johnson, S. I. Reed, and J. R. Yates III. 2004. Global analysis of protein sumoylation in Saccharomyces cerevisiae. J. Biol. Chem. 279:45662-45668. [DOI] [PubMed] [Google Scholar]
- 64.Zhang, H., H. Saitoh, and M. J. Matunis. 2002. Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol. Cell. Biol. 22:6498-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao, W., and J. L. Manley. 1998. Deregulation of poly(A) polymerase interferes with cell growth. Mol. Cell. Biol. 18:5010-5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhelkovsky, A. M., M. M. Kessler, and C. L. Moore. 1995. Structure-function relationships in the Saccharomyces cerevisiae poly(A) polymerase. Identification of a novel RNA binding site and a domain that interacts with specificity factor(s). J. Biol. Chem. 270:26715-26720. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.