Abstract
Transcription factor E1AF is widely known to play critical roles in tumor metastasis via directly binding to the promoters of genes involved in tumor migration and invasion. Here, we report for the first time E1AF as a novel binding partner for ubiquitously expressed Sp1 transcription factor. E1AF forms a complex with Sp1, contributes to Sp1 phosphorylation and transcriptional activity, and functions as a mediator between epidermal growth factor and Sp1 phosphorylation and activity. Sp1 functions as a carrier bringing E1AF to the promoter region, thus activating transcription of glioma-related gene for β1,4-galactosyltransferase V (GalT V; EC 2.4.1.38). Biologically, E1AF functions as a positive invasion regulator in glioma in cooperation with Sp1 partly via up-regulation of GalT V. This report describes a new mechanism of glioma invasion involving a cooperative effort between E1AF and Sp1 transcription factors.
E1AF, a member of a subfamily of ETS domain transcription factors, is capable of regulating transcription by binding to the Ets-binding site (EBS) in the promoter of its target genes (39) and is involved in a number of processes, including neuronal pathfinding (23) and mammary gland development and male sexual function (22, 25). Pathologically, E1AF plays an important role in HER2/Neu-mediated mammary oncogenesis and hepatocyte growth factor-induced cancer invasiveness and metastasis via directly binding to the promoters of genes involved in tumor migration and invasion (17, 18, 22, 29, 30, 38, 39, 41), suggesting the contribution of E1AF to various malignant phenotypes of cancer cells. However, the mechanisms of E1AF-induced tumor metastasis remain to be discovered.
Sp1 is a well-known DNA-binding nuclear protein that is widely expressed in tissues (2). It binds to GC box motifs in promoters of numerous genes involved in cell growth regulation and cancer (7), including p21 (14), caspase-8 (28), cyclin D1, and GalT V (35, 47), which effectively galactosylates the GlcNAcβ1,6 branch of N-glycans and functions as a positive regulator in glioma invasion (9, 16, 20). Biologically, Sp1 plays important roles in a wide variety of physiological processes, including the cell cycle, hormonal activation, apoptosis, angiogenesis, oncogenesis, etc. (10). Sp1 phosphorylation is tied to functional changes in DNA binding and promoter activation, contributes to the regulation of cell physiology, and functions as a link between various pathophysiological signals and transcription of their target genes (6).
Here, we found that E1AF physically and functionally interacted with Sp1 through a glutamine-rich (Gln-rich) domain and contributed to Sp1 phosphorylation and transcriptional activity. Sp1 functioned as a carrier bringing E1AF to the region of the glioma-related gene GalT V promoter, thus activating its transcription. Furthermore, E1AF functioned as a positive invasion regulator in glioma in cooperation with Sp1. This report describes new mechanisms of glioma invasion involving cooperative efforts of E1AF and Sp1 transcription factors and E1AF-induced tumor invasion, providing a novel model of invasion-associated transcription regulation in glioma.
MATERIALS AND METHODS
Antibodies and reagents.
G418, phenylmethylsulfonyl fluoride, aprotinin, pepstatin, epidermal growth factor (EGF), ethidium bromide (EtBr), mithramycin A, antiphosphoserine antibody, and antiphosphothreonine antibody were from Sigma Chemical Co. 32PO4 and an enhanced chemiluminescence assay kit were from Amersham Pharmacia Biotech. Antihemagglutinin (anti-HA) antibody, anti-E1AF antibody, anti-Sp1 antibody, anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH), and TransCruz Gel Supershift reagent anti-E1AF and anti-Sp1 antibodies were purchased from Santa Cruz Biotechnology. Anti-poly(ADP-ribose) polymerase (anti-PARP) antibody and anti-EGF receptor (anti-EGFR) antibody were purchased from Cell Signaling. Anti-green fluorescent protein (anti-GFP) antibody was purchased from Roche Applied Science. Anti-myc antibody was purchased from Invitrogen. Normal human brain tissues and glioma tissues were obtained from Huashan Hospital, China. Other reagents were commercially available in China.
Plasmids.
Expression constructs for HA-pcDNA3.0, pcDNA3.1-myc, pcDNA3.0-E1AF, pGL3-Basic, pRL-CMV, GalT V promoter construct GalT V-Luc, and M(Sp1) have been described previously (20, 49, 50). The Sp1-Luc, mSp1-Luc, CDK2-Luc, mCDK2-Luc, hSR-Luc, mhSR-Luc, Ap2γ-Luc, and mAp2γ-Luc vectors were constructed as previously described (27, 31, 40, 44). PEVR2-Sp1 vector was kindly provided by Guntram Suske (Marburg, Germany). Myc-tagged E1AF plasmid was constructed by inserting E1AF coding sequence into the HindIII/XhoI site of pcDNA3.1/myc (−) vector by use of pcDNA3.0-E1AF as the template. The deletion mutants of E1AF have been designated Δ148-244 (representing the deletion of amino acids [aa] 148 to 244) and 148-244 (aa 148 to 244). HA-tagged Sp1 plasmid was constructed by inserting Sp1 coding sequence using the EcoRI/XhoI site of pcDNA3.0-HA vector and pEVR2-Sp1 vector as templates. The deletion mutants of Sp1 were designated Δ138-232, Δ352-500, 138-232, and 352-500. Mutagenesis was carried out using a TakaRa MutanBEST mutagenesis kit. Mutated constructs were sequenced, and the correct ones were selected for further experiments. The mutagenic primers used were R397/400K sense (5′-TCGCTCAAATACTATTAT-3′) and R397/400K antisense (5′-TTTGCTCAGCTTGTCGTA-3′); S59A sense (5′-GCACCTTTGGCTCTGCTGGCA-3′) and S59A antisense (5′-TGGCTGGGACTCCTGCCCTC-3′); S131A sense (5′-GCAAATGGCAGTGAGTCTTCCAAGA-3′) and S131A antisense (5′-GCCATTGGTACTGCTGCCACTCTGT-3′); T355A sense (5′-GCACCCCAGAGGGTCAGTGG-3′) and T355A antisense (5′-CTGGCCTTGAGAGTTGGTCCCTGAT-3′); T453A sense (5′-GCACCAACAGTGGGGCCCAATG-3′) and T453A antisense (5′-CCGGATGATGATGGGACCAGAGTT-3′); T579A sense (5′-GCAGCAGGTGGAGAGGAAGGAGAA-3′) and T579A antisense (5′-GTCATCATGTATTCCATCACCACCA-3′); T739A sense (5′-GCACCTTCAGCCCTTATTACCACCA-3′) and T739 antisense (5′-GGCAGTGCCACTGCCTTCTGAAC-3′); C658S sense (5′-AGTACCTGGTCATACTG-3′) and C658S antisense (5′-CATAAATGGCCTCTCGC-3′); and C688S sense (5′-AGCCCTGAGTGTCCTAAG-3′) and C688S antisense (5′-GGCAAATTTCTTCTCACC-3′).
Construction of E1AF RNA interference (RNAi) or Sp1 RNAi was performed using a siRNA construction kit (KCsiRNA) according to the manufacturer's suggestions (50). The sequence of the E1AF mRNA target oligonucleotide was as follows: AGGATCTAAGTCACTTCCA (annealed and cloned into pSilencer-2.0 vector). The sequence of the Sp1 mRNA target oligonucleotide was as follows: GGAACAGAGTGGCAACAGT.
Cell culture and transfection.
Human glioma cell lines U251 and SHG44 have been described previously (20). Cell transfection was performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. For stable transfection, the original medium was replaced after 48 h with G418-containing medium and individual clones were picked and analyzed.
Invasion and migration analysis.
A wound healing assay was performed as described previously (50). A Boyden chamber invasion assay was performed basically as described previously by Albini et al. (1). Cells were added to the upper compartment of the chamber, and 800 μl medium (containing 0.1% bovine serum albumin) was added into the lower chamber. Cells were incubated and allowed to migrate for 24 h. After removal of nonmigrated cells, cells that had migrated through the filter were counted under a microscope in five fields at a magnification of ×400.
Dual-luciferase assay, gel shift assay, and DNA affinity precipitation assay.
A dual-luciferase assay was performed by the method used in our previous study (50). Nuclear proteins were isolated according to the method of Schreiber et al. (36), and a gel shift assay was performed according to the method of our previous study (50). The association of Sp1 with chromatin DNA in SHG44 cells was confirmed using a chromatin immunoprecipitation (ChIP) assay kit (Upstate Biotechnology) with anti-Sp1 antibody as described by the manufacturer. Normal anti-rabbit immunoglobulin G (IgG) was used as a negative control. The GalT V promoter region (−200 to +1) was amplified by conventional PCR (with forward primer 5′-AAGACTGGTGGGGGAATTTCATGG-3′ and reverse primer 5′-CAGGCGGCCGCTAGAGA-3′). DNA affinity precipitation assays were performed as previously reported (34). Oligonucleotides containing biotin on the 5′ nucleotide of the sense strand were used in the assays. The sequences of the oligonucleotides were as follows: for the wild-type (WT) oligonucleotide, 5′-CTGGCCCCGCCTCCCGCGCGTGCGCC, which corresponded to bp −82 to −57 of the human GalT V promoter; and for the M (Sp1) oligonucleotide, 5′-CTGGCCCAAACTCCCGCGCGTGCGCC, which contained the mutation of the Sp1-binding site (underlined).
Immunoblotting and immunoprecipitation assays.
Immunoblotting and immunoprecipitation assays were performed as previously described (13). Immunoblot analysis was performed with anti-E1AF, anti-GAPDH, anti-Sp1, anti-EGFR, and anti-GFP. Lysates of nuclear extract were also subjected to immunoprecipitation with anti-Sp1 or control IgG, and the immune complex was analyzed by immunoblotting using anti-Sp1. In some experiments the precipitated complexes were treated with EtBr prior to elution to test specific dependence on DNA structural integrity as previous described (15, 24). EtBr was added (50 to 400 μg/ml), and the lysates were incubated on ice for 30 min. Precipitates were removed by centrifugation in a microcentrifuge, and the supernatant was transferred to a fresh tube. The resulting lysate was then ready for immunoprecipitation.
In vivo labeling and Western blot analysis.
Cells were incubated in phosphate-free medium for 2 h prior to being labeled in phosphate-free medium containing 7.5 to 15 mCi/ml 32PO4 for 2 h as previous described (4). Cells were rinsed with phosphate-buffered saline and lysed directly in boiling 10 mM Tris-HCl (pH = 7.2)-1% sodium dodecyl sulfate and reboiled, and DNA was sheared. Following addition of 2.2 volumes of ice-cold 15 mM Tris-HCl (pH = 7.2)-7.5 mM EDTA-150 mM sodium fluoride-230 mM NaCl-1.5% Triton X-100-0.75% Nonidet P-40-100 mM β-glycerophosphate-15 mM sodium pyrophosphate-400 mM Na2VO3-2 mM phenylmethylsulfonyl fluoride-20 mM leupeptin-10 mg/ml aprotinin, particulate material was removed by centrifugation. Supernatants were precleared with normal rabbit IgG and protein A-Sepharose, and Sp1 was immunoprecipitated with anti-Sp1 antibody (PEP2; Santa Cruz) and protein A-Sepharose. Immunoprecipitates were washed four times with radioimmunoprecipitation assay buffer, separated by 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and subjected to autoradiography.
Statistics and presentation of data.
All experiments were repeated three times. All numerical data are expressed as means ± standard deviations. Data were analyzed using the two-tailed t test.
RESULTS
Interaction between E1AF and Sp1 in glioma.
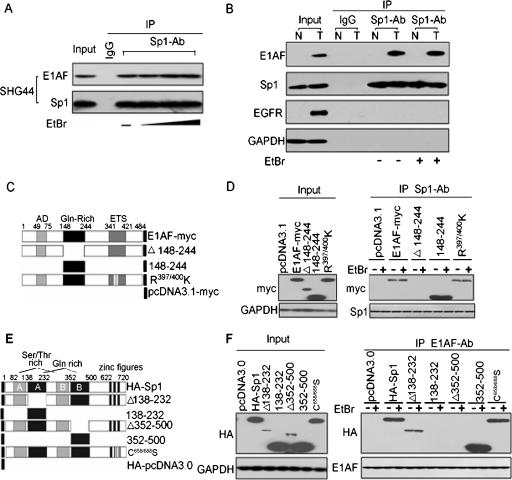
The association between E1AF and Sp1 was first demonstrated by coimmunoprecipitation-Western blot analysis using the glioma cell line SHG44. E1AF was detected in anti-Sp1 immunoprecipitates in SHG44 cells (Fig. 1A). To investigate the DNA dependence of E1AF-Sp1 association, cell lysates were treated prior to immunoprecipitation with EtBr, which distorted DNA structure and disrupted DNA-protein interaction, as previously described (24). Addition of increasing amounts of EtBr did not affect the interaction between E1AF and Sp1 (Fig. 1A). To further ensure the physical association of E1AF and Sp1 in glioma, the interaction of these two proteins was determined in glioma tissues and normal brain tissues. The interaction of Sp1 and E1AF was observed in glioma tissues but not in normal brain tissues in the rare cases of expression of E1AF in normal brain tissues (Fig. 1B). Also, the interaction between E1AF and Sp1 in glioma tissues was not sensitive to the presence of EtBr (Fig. 1B). These data indicated that E1AF might physically interact with Sp1 transcription factor in a DNA-independent manner.
FIG. 1.
Identification E1AF as a Sp1 binding protein. (A) In vivo association of E1AF with Sp1 determined using cells of the glioma SHG44 cell line and a coimmunoprecipitation assay. Lysates from SHG44 cells were immunoprecipitated (IP) with anti-Sp1 antibody (Ab) or control IgG in the absence or presence of EtBr (50 μg/ml, 200 μg/mln or 400 μg/ml) and sequentially immunoblotted with anti-E1AF or anti-Sp1 antibody. (B) Sp1 IP of glioma tissue (T) and normal brain tissue (N) lysates in the absence or presence of EtBr (50 μg/ml) probed with anti-E1AF, anti-Sp1, anti-EGFR, or anti-GAPDH antibodies. Expression of GAPDH served as a loading control. (C) Schematic representations of E1AF and myc-tagged E1AF mutants used in a coimmunoprecipitation assay. The ETS domain and acidic domain (AD) are shown as gray boxes. The Gln-rich domain is shown as a black box. Amino acid numbers mark the N and C termini and the deletion breakpoints. (D) SHG44 cells were transfected with constructs for expression of control or myc-tagged E1AF mutants and harvested 48 h after transfection. The results of Sp1 IP of these cell lysates in the absence or presence of EtBr (50 μg/ml) blotted with anti-myc or anti-Sp1 antibodies are shown. (E) The structural domains of Sp1 and HA-tagged Sp1 mutants in this work are diagrammed. (F) SHG44 cells were transfected with expression constructs for control or HA-tagged Sp1 or its mutants and harvested 48 h after transfection. The results of E1AF IP of these cell lysates in the absence or presence of EtBr (50 μg/ml) blotted with anti-HA or anti-E1AF antibodies are shown.
E1AF has a functional acidic domain, a Gln-rich domain, and ETS domain (39). To identify the binding surface(s) of Sp1 on E1AF, a fusion protein consisting of WT or various truncated forms of E1AF fused at the C terminus of myc were constructed and utilized in immunoprecipitation assays (Fig. 1C). Both myc-tagged full-length E1AF and E1AF(148-244) containing the Gln-rich domain were sufficient for binding to Sp1 that was not sensitive to the presence of EtBr, whereas deletion of the Gln-rich domain of E1AF abolished interaction with Sp1 (Fig. 1D). The ETS domain of E1AF protein contains two conserved residues (R397/400) which are important for the DNA binding ability of Ets family members (5, 8, 19). To investigate the role of the contribution of these residues in DNA binding and the interaction with Sp1, the myc-tagged mutant of E1AF (R397/400K) was constructed and utilized in immunoprecipitation assays. The R397/400K mutant abolished the DNA binding ability of E1AF (data not shown) and did not affect its interaction with Sp1 (Fig. 1D).
We next turned to mapping the domain(s) of Sp1 that was required for the interaction with E1AF. Sp1 contains a DNA-binding domain consisting of three C2H2-type zinc fingers close to the C terminus, two serine/threonine stretches (A and B) in the N-terminal part, and two glutamine-rich activation domains (A and B) contributing to the interaction with its partners (10). To address the role of Gln-rich domains in the interaction with E1AF, HA-tagged mutants of Sp1 with or without the Gln-rich domain were constructed and utilized in immunoprecipitation assays (Fig. 1E). As shown in Fig. 1F, the presence of HA-tagged full-length Sp1(Δ138-232) or Sp1(353-500) containing the Gln-rich domain B was sufficient for binding to E1AF that was not sensitive to the presence of EtBr, whereas deletion of the Gln-rich B domain of Sp1 abolished the interaction between E1AF and Sp1 (Fig. 1F). In addition, a mutant of Sp1 harboring a two-cysteine mutation in the zinc finger domain that impaired its ability to bind to DNA (26) (data not shown) did not affect its interaction with E1AF (Fig. 1F).
E1AF increases transactivation by Sp1.
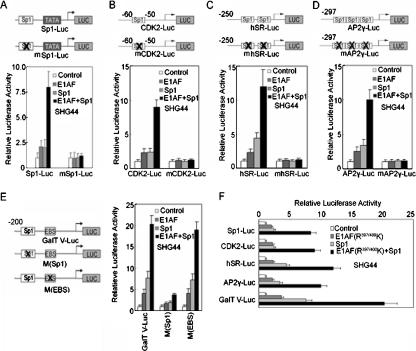
To elucidate whether E1AF had an effect on the transcription capacity of Sp1, we performed transient cotransfection assays using SHG44 cells and a reporter construct containing one Sp1 consensus binding site and the Sp1 expression vector. E1AF overexpression enhanced transactivation by Sp1 that was dependent on which Sp1-binding site was used (Fig. 2A). Similarly, E1AF overexpression induced the activity of a reporter construct containing the promoter region of the CDK2, hSR, or hAP2γ gene which contained identified Sp1-binding site(s) and lacked any potential ETS binding site (Fig. 2B to D) (27, 31, 44).
FIG. 2.
Activation of Sp1 transcription potential by E1AF. (A to D) PcDNA3.0 and/or E1AF and/or Sp1 expression vector was transiently cotransfected into SHG44 cells with a pSp1-Luc, pmSp1-Luc, CDK2-Luc, mCDK2-Luc, hSR-Luc, mhSR-Luc, AP2γ-Luc, or mAP2γ-Luc construct. The luciferase activity was determined as described in Materials and Methods. (E) PcDNA3.0 and/or E1AF and/or Sp1 expression vectors were transiently cotransfected into SHG44 cells with GalT V-Luc, M(Sp1), or M(EBS). The luciferase activity was determined as described in Materials and Methods. (F) PcDNA3.0 and/or E1AF(R397/400K) and/or Sp1 expression vector was transiently cotransfected into SHG44 cells with a pSp1-Luc, CDK2-Luc, hSR-Luc, AP2γ-Luc, or GalT V-Luc construct. The luciferase activity was determined as previously described.
To study whether the enhancing effect of E1AF on Sp1 activity played a role in the activation of Sp1-targeting glioma-related genes, we studied the contribution of E1AF to the transcription of GalT V, which functions in a positive role in glioma growth (20, 35). We transfected SHG44 cells with a luciferase reporter gene driven by a −200-to-+120 fragment of the GalT V promoter (GalT V-Luc) with or without mutation of the Sp1-binding site or Ets-binding site together with a combination of expression constructs for E1AF and Sp1. As shown in Fig. 2E, E1AF activated the GalT V promoter in cooperation with Sp1, and mutation of Sp1-binding site but not that of the Ets-binding site abolished the positive effect of E1AF on the activity of the GalT V promoter. In addition, inhibition of Sp1 binding by its inhibitor mithramycin A or Sp1 RNAi dramatically inhibited the activation of the GalT V promoter mediated by E1AF (data not shown).
These data indicated that E1AF might control gene expression through the Sp1-binding site without direct binding to DNA. To address this point, the DNA binding-defective mutant of E1AF (R397/400K) was transiently cotransfected into SHG44 cells with a Sp1 expression vector and reporter construct containing an Sp1-binding site. As depicted in Fig. 2F, overexpression of R397/400K increased the activity of the reporter construct in cooperation with Sp1. These data indicated that E1AF regulated gene expression in cooperation with Sp1.
The E1AF/Sp1 complex binds to the GalT V promoter in vitro and in vivo.
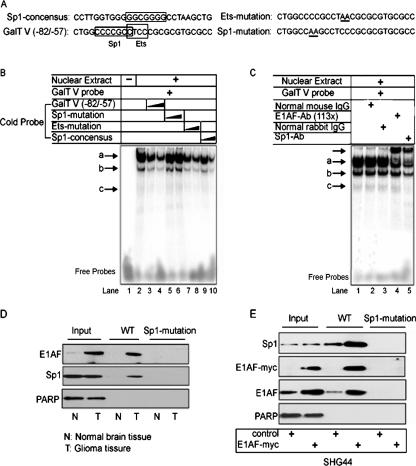
Next, an electrophoretic mobility shift assay was performed to determine whether E1AF/Sp1 recognized the GalT V promoter. Nuclear extracts from SHG44 cells were incubated with labeled double-stranded oligonucleotides spanning the region between nucleotides −82 and −57 of the GalT V promoter containing one Sp1-binding site and one Ets-binding site with or without competition (indicated in Fig. 3A). Three main DNA-protein complexes were detected (Fig. 3B, lane 2) which were gradually competed with by excess unlabeled GalT V (−82 to −57) oligonucleotides or the unlabeled Ets mutation oligonucleotides (Fig. 3B, lanes 3, 4, 7, and 8). In contrast to this finding, the unlabeled Sp1-binding site mutation oligonucleotides failed to compete with the binding (Fig. 3B, lanes 5 and 6), indicating that the Sp1-binding site was important for the formation of three complexes. Consistently, unlabeled Sp1 consensus oligonucleotides significantly inhibited complexes a, b, and c (Fig. 3B, lanes 9 and 10). To identify specific proteins that bound to the Sp1 binding site, we used TransCruz Gel supershift antibodies against E1AF or Sp1 or control IgG. It was found that antibody against E1AF supershifted complex a without changing other complexes and that antibody against Sp1 supershifted complex a and decreased the levels of complexes b and c (Fig. 3C), indicating the contribution of E1AF and Sp1 to the formation of complex a and the binding of E1AF and Sp1 to the GalT V promoter.
FIG. 3.
Analysis of E1AF/Sp1 complex binding to the GalT V promoter in glioma cell and glioma tissue in vitro. (A) Oligonucleotides used in an electrophoretic mobility shift assay. The putative Sp1 and Ets binding sites are indicated with boxes. The mutated nucleotides are underlined. (B) An electrophoretic mobility shift assay was performed using nuclear proteins of SHG44 cells and a human GalT V promoter sequence (−82 to −57) double-stranded radiolabeled probe. Competition assays were carried out with a 10- to 20-fold excess of GalT V promoter sequence (−82 to −57) oligonucleotides with or without the Ets-binding site or Sp1-binding site mutated or Sp1 consensus oligonucleotides. The protein-DNA complexes (arrows a to c) and free DNA are indicated. (C) E1AF/Sp1 bound to a GC box site within a human GalT V promoter. Nuclear extracts from SHG44 cells were incubated with 32P-labeled double-stranded oligonucleotides spanning the GC box and an Ets-binding site within the GalT V promoter in the presence or absence of control IgG or an antibody specific to Sp1 or E1AF. The unlabeled arrow indicates the protein-DNA-antibody complex. (D) The same amounts of nuclear extracts from glioma tissues or normal brain tissues were incubated with biotin-labeled oligonucleotides as described in Materials and Methods. Proteins bound to these nucleotides were isolated with streptavidin-agarose, and E1AF or Sp1 was detected by immunoblotting. PARP expression served as a loading control. (E) The same nuclear extracts from SHG44 cells transiently transfected with control or E1AF-myc plasmids incubated with biotin-labeled oligonucleotides as described in Materials and Methods. Proteins bound to these nucleotides were isolated with streptavidin-agarose, and E1AF, Sp1, or myc was detected using immunoblotting. PARP expression served as a loading control.
To accurately determine whether E1AF and Sp1 could bind to the GalT V promoter in glioma tissues and normal brain tissues, we performed a DNA affinity precipitation assay using WT oligonucleotides spanning the region between nucleotides −82 to −57 of the human GalT V promoter and Mut oligonucleotide containing the mutation of the Sp1-binding site. The results showed that the activity of Sp1 or E1AF binding to GalT V promoter in glioma tissues was dependent on which Sp1 binding site was used (Fig. 3D). To see the effect of E1AF overexpression on Sp1 binding to the GalT V promoter, the same amount of nuclear protein extracted from SHG44 cells transfected with control or myc-tagged E1AF plasmid was used in a DNA affinity precipitation assay. As depicted in Fig. 3E, binding of Sp1 and endogenous and exogenous E1AF to the GalT V promoter was dependent on the Sp1 binding site and E1AF overexpression induced the binding of Sp1 to the GalT V promoter (Fig. 3E).
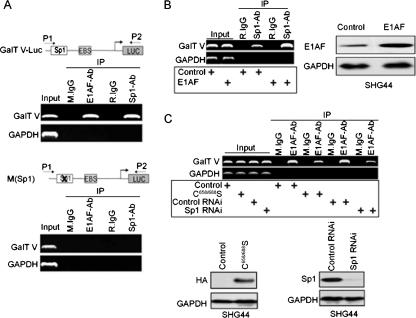
To further demonstrate that the two proteins actually cooccupied the GalT V promoter, we carried out a series of ChIP assays. First, we investigated the recruitment of E1AF and Sp1 to the GalT V-Luc reporter gene with or without the mutation of Sp1-binding site. An obvious level of E1AF and Sp1 binding to GalT V promoter region was seen (Fig. 4A, upper panel). However, mutation of the Sp1-binding site within the GalT V-Luc reporter gene resulted in no E1AF or Sp1 binding (Fig. 4A, lower panel). Next, we determined whether E1AF and Sp1 could be detected at the promoter of endogenous GalT V in SHG44 cells by use of a ChIP assay. In contrast to control IgG results, E1AF and Sp1 were detected in the promoter region of GalT V gene, and no binding to the GAPDH gene was observed (Fig. 4B and C). Furthermore, E1AF overexpression increased the ability of Sp1 to bind to the GalT V promoter (Fig. 4B). In addition, to address the Sp1 dependence of E1AF binding to GalT V promoter, C658/688S, a mutant of Sp1 harboring a two-cysteine mutation in the zinc finger domain, or Sp1 RNAi was transiently transfected into SHG44 cells and subsequently subjected to ChIP analysis using anti-E1AF antibody. As depicted in Fig. 4C, decreasing Sp1 DNA binding or down-regulation of Sp1 expression decreased the binding of E1AF to the GalT V promoter (Fig. 4C).
FIG. 4.
Analysis of E1AF/Sp1 complex binding to the GalT V promoter in glioma cell in vivo. (A) ChIP assay of endogenous E1AF or Sp1 on GalT V-Luc (upper panel) or M(Sp1) (lower panel) constructs in SHG44 cells. Immunoprecipitations were carried out with rabbit IgG (R.IgG), mouse IgG (M.IgG), anti-E1AF antibody (E1AF-Ab), or anti-Sp1 antibody (Sp1-Ab). Coprecipitating DNA was revealed by PCR with the indicated primers. Input DNA was diluted 10-fold before amplification. (B) A ChIP assay was performed using SHG44 cells transfected with control or E1AF expression vector and control IgG or an antibody against Sp1. PCR primers for the GalT V promoter or the GAPDH promoter were used to detect promoter fragments in immunoprecipitates (left panel). The presence of transfected E1AF is indicated (right panel). (C) A ChIP assay was performed using SHG44 cells transfected with control, C658/688S, control RNAi, or Sp1 RNAi vector and control IgG or an antibody against E1AF. PCR primers for the GalT V promoter or the GAPDH promoter were used to detect promoter fragments in immunoprecipitates (upper panel). The presence of endogenous Sp1 expression or C658/688S is indicated (lower panel).
E1AF overexpression induces Sp1 phosphorylation activity.
As Sp1 DNA binding and transcription activity could be regulated by its phosphorylation (10), we examined its phosphorylation status in response to E1AF overexpression. Whole-cell extracts of vector- or E1AF-transfected cells previously treated with EtBr were immunoprecipitated by Sp1 antibody and were subsequently subjected to Western blot analysis utilizing a panel of antibodies, recognizing a distinct pattern of serine/threonine-phosphorylated proteins (32). As shown in Fig. 4A, the forced expression of E1AF increased relative amounts of serine/threonine-phosphorylated Sp1 transcription factor without changing Sp1 expression in a DNA-independent manner (Fig. 5A). Furthermore, immunoprecipitation assays were also performed using E1AF-transfected cells labeled with [32P]orthophosphate to investigate in intact cells whether expression of E1AF altered Sp1 phosphorylation. As shown in Fig. 5B, the incorporation of [32P]orthophosphate into immunoprecipitated wild-type Sp1 increased in response to E1AF overexpression.
FIG. 5.
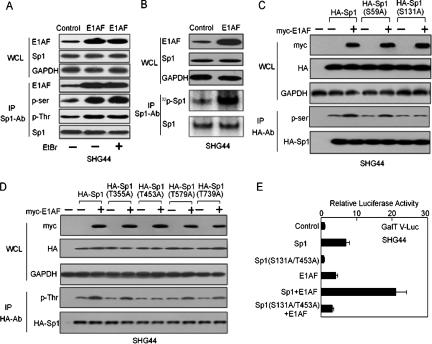
E1AF was associated with phosphorylation of Sp1. (A, WCL panels) Whole-cell lysates from SHG44 cells transfected with control or E1AF expression vectors in the absence or presence of EtBr (50 μg/ml) were loaded onto a 8% denatured polyacrylamide gel, and E1AF and Sp1 protein levels were determined by Western blotting using anti-E1AF or anti-Sp1 antibody (Sp1-Ab). (IP panels) The results of Sp1 immunoprecipitation of the lysates of SHG44 cells transfected with control or E1AF expression vectors in the absence of EtBr or in the presence of EtBr (50 μg/ml) blotted with the indicated antibodies are shown. (B) Whole-cell lysates from SHG44 cells transfected with control or E1AF expression vectors labeled with 32PO4 for 2 h prior to harvesting and the levels of 32P labeling of Sp1 were determined as described in Materials and Methods. (C, WCL panels) Whole-cell lysates from SHG44 cells transfected with control or myc-E1AF expression vector and pcDNA3.0-HA, HA-Sp1, S59A, or S131A were loaded onto an 8% denatured polyacrylamide gel, and E1AF and HA-Sp1 protein levels were determined by Western blotting using anti-myc or anti-HA antibody. (IP panels) The results of Sp1 IP of the lysates of SHG44 cells transfected with control or myc-E1AF expression vector and pcDNA3.0-HA, HA-Sp1, S59A, or S131A blotted with the indicated antibodies are shown. (D, WCL panels) Whole-cell lysates from SHG44 cells transfected with the indicated constructs were loaded onto an 8% denatured polyacrylamide gel, and E1AF and HA-Sp1 protein levels were determined by Western blotting using anti-myc or anti-HA antibody. (IP panels) The results of Sp1 IP of the lysates of SHG44 cells transfected with the indicated constructs blotted with the indicated antibodies are shown. (E) PcDNA3.0 and/or E1AF and/or Sp1 and/or Sp1 (S131A/T453A) expression vectors and GalT V-Luc were transiently cotransfected into SHG44 cells. The luciferase activity was determined as described in Materials and Methods.
Six SP1 phosphorylation sites, Ser59, Ser131, Thr355, Thr453, Thr579, and Thr739, have been confirmed (10). To further demonstrate the effect of E1AF overexpression on Sp1 phosphorylation, point mutations of potential phosphorylation sites in Sp1 were constructed and transiently cotransfected into SHG44 cells with control or myc-E1AF expression vector and subsequently subjected to Western blot analysis. As depicted in Fig. 5C, mutation of Ser131 of Sp1 abolished E1AF-induced Sp1 phosphorylation at serine residues. In addition, mutation of Thr453 of Sp1 abolished E1AF-induced phosphorylation at threonine residues (Fig. 5D). To investigate the role of Sp1 phosphorylation in E1AF-induced GalT V promoter activity, the mutant of Sp1 (S131A/T453A) was transiently cotransfected into SHG44 cells with E1AF expression vector and GalT V-Luc construct. Compared to wild-type Sp1 results, mutation of Sp1 residues S131 and T453 reduced E1AF-induced GalT V promoter activity and abolished the effect of cooperation of the GalT V promoter with E1AF (Fig. 5E).
E1AF expression is important for EGF-induced GalT V promoter activity.
Previously, we reported that EGF could activate GalT V transactivation in a Sp1-binding site-dependent manner (20). To address whether E1AF was important during such activation, we generated an E1AF RNAi construct and transiently cotransfected the construct into the SHG44 glioma cell line with GalT V-Luc treated with EGF. As shown in Fig. 6A, decreasing E1AF expression inhibited EGF-induced activation of GalT V promoter, indicating that E1AF functioned in an essential role in EGF-induced GalT V transcription. Supporting this point, EGF induced GalT V transcription in an E1AF-dependent manner and overexpression of E1AF and Sp1 significantly increased EGF-induced GalT V transcription in HEK293 cells (data not shown).
FIG. 6.
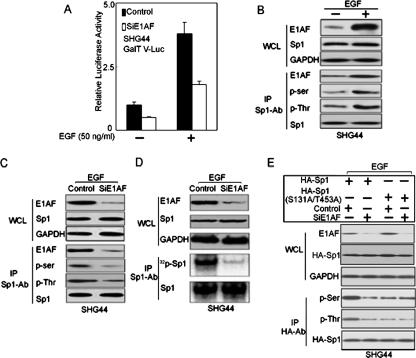
The contribution of E1AF to EGF-induced GalT V promoter activity. (A) SHG44 cells transiently cotransfected with GalT V-Luc and control RNAi or E1AF RNAi construct were treated with EGF (50 ng/ml) for 24 h or left untreated, and the luciferase activity was assayed as described in Materials and Methods. (B) EGF increased E1AF expression and enhanced the interaction between E1AF and Sp1. (WCL panels) Expression of E1AF and Sp1 in SHG44 cells left untreated or treated with EGF (50 ng/ml) for 24 h was studied by immunoblot analysis using the indicated antibodies. (IP panels) The results of Sp1 immunoprecipitation of the lysates of these cells blotted with the indicated antibodies are shown. (C) The role of E1AF in the phosphorylation of Sp1 induced by EGF. (WCL panels) Whole-cell lysates from SHG44 cells transfected with control or E1AF RNAi construct in the presence of EGF (50 ng/ml) for 24 h were determined by Western blotting using anti-E1AF or anti-Sp1 antibody. (IP panels) The results of Sp1 immunoprecipitation of the lysates of these cells blotted with the indicated antibodies are shown. (D, WCL panels) Whole-cell lysates from SHG44 cells transfected with control or E1AF RNAi construct in presence of EGF (50 ng/ml) for 24 h and after being labeled with 32PO4 for 2 h were investigated by Western blotting using anti-E1AF or anti-Sp1 antibody. (IP panels) Sp1 immunoprecipitation of the lysates of these cells transfected with control or E1AF RNAi construct in the presence of EGF (50 ng/ml) for 24 h were labeled with 32PO4 for 2 h prior to harvesting, and the levels of 32P labeling of Sp1 were determined as described in Materials and Methods. (E, WCL panels) Whole-cell lysates from SHG44 cells cotransfected with control or E1AF RNAi and HA-Sp1 or HA-Sp1(S131A/T453A) construct in presence of EGF (50 ng/ml) for 24 h were subjected to Western blot analysis using anti-E1AF, anti-HA, and anti-GAPDH antibody. (IP panels) Results of HA IP of the lysates of these cells blotted with the indicated antibodies are shown.
To clarity the mechanism involved, we investigated the effect of EGF on E1AF expression and the interaction of E1AF and Sp1. As shown in Fig. 6B, EGF increased expression of E1AF and phosphorylation of Sp1 and enhanced the interaction of Sp1 and E1AF proteins in SHG44 cells. Also, decreasing expression of E1AF reduced EGF-induced Sp1 phosphorylation without changing Sp1 expression levels (Fig. 6C) and inhibited EGF-induced incorporation of [32P]orthophosphate into immunoprecipitated wild-type Sp1 (Fig. 6D). Furthermore, decreasing expression of E1AF reduced EGF-induced exogenous Sp1 phosphorylation that was dependent on the presence of S131 and T453 (Fig. 6E). Together, the data reported above suggested that E1AF might act as a mediator between EGF signaling and Sp1.
Positive correlation between levels of E1AF and GalT V in glioma.
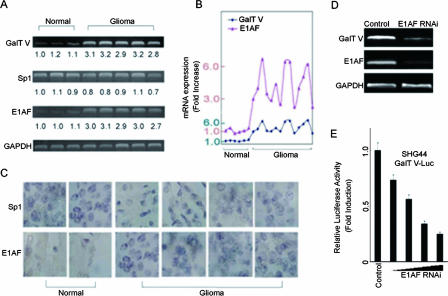
We have thus far established a positive role for E1AF and Sp1 in GalT V transcription in the cell culture system. Our previous study showed that GalT V was highly expressed in glioma (20, 45, 46). To address the mechanism of the high level of expression of GalT V in glioma, we investigated expression of E1AF and Sp1 by use of glioma and normal brain tissues and a reverse transcription-PCR (RT-PCR) assay. A positive correlation was found between the levels of E1AF and GalT V; in contrast, we did not find a correlation between the levels of Sp1 and GalT V (Fig. 7A and B). High expression of E1AF in glioma was confirmed using a series of glioma and normal brain tissue samples and immunohistochemistry (Fig. 7C); the results indicated that E1AF was highly expressed in glioma. The same results were obtained using Western blot analysis (data not shown).
FIG. 7.
Positive correlation between levels of E1AF and GalT V in glioma. (A) RT-PCR analysis of Sp1, E1AF, and GalT V mRNA expression in normal brain tissues (n = 3) and glioma tissues (n = 5). The relative quantities of mRNA expression compared to that of the first sample are indicated. (B) The correlation of expression of E1AF and GalT V mRNA in normal brain tissues (n = 6) and glioma tissues (n = 14). The values are presented as upregulation compared to the results seen with the first sample. (C) Immunohistochemical analysis of Sp1 and E1AF protein expression was performed with normal brain tissues (n = 2) and glioma tissues (n = 4). Scale bar, 10 μm. (D) RT-PCR analysis of GalT V and E1AF mRNA expression levels in SHG44 cells transfected with control or E1AF RNAi construct. GAPDH mRNA expression served as a loading control. (E) Control RNAi or increasing amounts of E1AF RNAi construct were transiently cotransfected into SHG44 cells with GalT V-Luc. The luciferase activity was measured as described in Materials and Methods.
To address the effect of E1AF on GalT V expression, control RNAi or E1AF RNAi was transiently transfected into SHG44 cells. As depicted in Fig. 7D and E, downregulation of E1AF expression by E1AF RNAi reduced GalT V mRNA expression and GalT V promoter activity in a dose-dependent manner.
E1AF promotes glioma invasion in cooperation with Sp1.
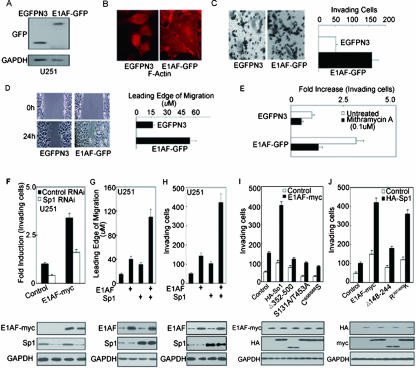
To evaluate the relationship between E1AF overexpression and tumor behavior, cells from the glioma cell line U251 were stably transfected with the E1AF-GFP construct (Fig. 8A). As shown in Fig. 8B, actin clustered in long filaments along the edges of the GFP-E1AF-transfected cells but not in the control cells. E1AF overexpression resulted in an almost threefold increase in in vitro invasion through a reconstituted material basement membrane (Fig. 8C), a striking increase of cell migration (Fig. 8D). Taken together, these results indicated that E1AF functioned as a positive regulator in glioma invasion.
FIG. 8.
E1AF promotes glioma migration and invasion in cooperation with Sp1. (A) Western blot assay demonstrating E1AF-GFP expression using U251 cells stably transfected with EGFPN3 or E1AF-GFP construct and anti-GFP antibody. (B) Control cells and E1AF-GFP-transfected U251 cells were subjected to actin staining by fluorescein isothiocyanate-phalloidin. Scale bar, 10 μm. (C) E1AF overexpression in U251 cells increased invasive ability as assayed in a modified Boyden chamber (P < 0.05; n = 3). Photomicrographs of the bottom of a Transwell filter (8-μm pores) are shown (upper panel). Scale bar, 100 μm. For quantification of invasion assays, the numbers of invading cells in 10 photographic fields from three separate experiments were counted. The values represent n-fold activation compared to control cell results. Data represent the means ± standard deviations of the results of three independent experiments (lower panel). (D) Cell migration assay of control or E1AF-GFP-transfected U251 cells. A wound-healing assay was prepared as described in Material and Methods. Scale bar, 50 μm. For quantification of migration assays, the wound-induced migration of cells was measured after 24 h. (E) U251 cells stably transfected with EGFPN3 or E1AF-GFP construct were plated on top of gels. Mithramycin A (0.1 μM) was added to the medium 1 day later. Quantification of the invasion assay was performed as described in Materials and Methods. (F) Control and/or Sp1 RNAi and/or myc-E1AF expression vectors were cotransfected into U251 cells. After 24 h, these cells were subjected to invasion assays. The values represent activation compared to control cell results (upper panel). Expression of myc-E1AF and Sp1 is indicated; expression of GAPDH served as a loading control (lower panel). (G) PcDNA3.0 and/or Sp1 and/or E1AF expression vector was cotransfected into U251 cells. After 24 h, these cells were subjected to wound-healing assays (upper panel). Expression of E1AF and Sp1 is indicated; expression of GAPDH served as a loading control (lower panel). (H) PcDNA3.0 and/or Sp1 and/or E1AF expression vector was cotransfected into U251 cells. After 24 h, these cells were subjected to invasion assays. The values represent activation compared to control cell results (upper panel). Expression of E1AF and Sp1 is indicated; expression of GAPDH served as a loading control (lower panel). (I) Control Sp1 or HA-Sp1 or its mutant was transiently cotransfected into U251 cells with control or myc-tagged E1AF expression vector. After 24 h, these cells were subjected to invasion assays. The values represent activation compared to control cell results (upper panel). Expression of myc-E1AF and HA is indicated; expression of GAPDH served as a loading control (lower panel). (J) Control, Myc-E1AF, R397/400K, or 148-244 expression vector was transiently cotransfected into U251 cells with control or HA-Sp1 expression vector. After 24 h, these cells were subjected to invasion assays. The values represent activation compared to control cell results (upper panel). Expression of myc and HA-Sp1 is indicated; expression of GAPDH served as a loading control (lower panel).
We next examined the role of Sp1 in E1AF-mediated glioma invasion. As shown in Fig. 8E and F, inhibition of Sp1 activity by mithramycin A or Sp1 expression by Sp1 RNAi dramatically inhibited E1AF-induced glioma invasion. To determine whether E1AF could collaborate with Sp1 to induce migration and invasion of glioma cells, U251 cells transiently transfected with E1AF or/and Sp1 constructs were subjected to invasion and migration assays. A significant synergistic effect on U251 cells that was induced by the presence of E1AF and Sp1 was found with respect to migration and invasion (Fig. 8G and H). Our data reported above suggested the presence of a cooperative effect of E1AF and Sp1 on tumor behaviors.
To examine whether E1AF-Sp1 interaction was essential for promotion of cell invasion by E1AF, U251 cells transiently transfected with myc-E1AF and Sp1 full-length construct or its mutants were subjected to invasion assays. As expected, deletion of Gln-rich domain B of Sp1 abolished the effect on glioma invasion of its cooperation with E1AF. In addition, mutation of Sp1 residues S131 and T453 or the Sp1 DNA-binding domain inhibited glioma cell invasion and abrogated the effect on glioma invasion of its cooperation with E1AF (Fig. 8I). Consistent with this, wild-type E1AF, but not its Gln-rich domain deletion mutation, cooperated with Sp1 to induce glioma cell invasion and E1AF mutation (R397/400K), which resulted in a deficiency in DNA binding-induced glioma cell invasion in cooperation with Sp1 (Fig. 8G), indicating that E1AF promoted glioma cell invasion at least partly via its interaction with Sp1.
E1AF is required for EGF-induced glioma cell migration and invasion.
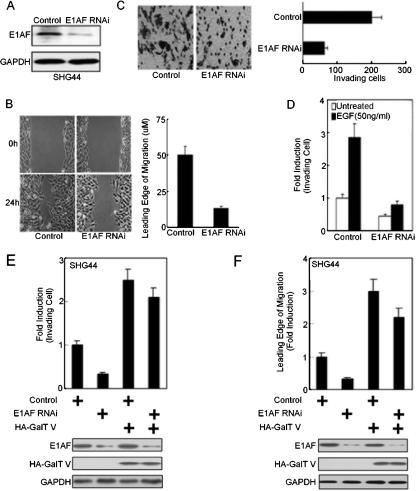
To further elucidate the biological significance of E1AF in glioma, the E1AF RNAi construct was stably transfected into SHG44 cells and stable clones were selected (Fig. 9A). Compared to the control cell results, inhibition of E1AF expression downregulated GalT V mRNA expression, decreased the activity of the GalT V promoter in a Sp1-binding site-dependent manner, and reduced the content of the β1,4-galactosidase branch in the cell surface glycoconjugates (data not shown).
FIG. 9.
Downregulation of E1AF expression decreases the invasion of glioma left untreated or treated with EGF. (A) A Western blot assay demonstrated E1AF expression by use of SHG44 cells stably transfected with control RNAi or E1AF RNAi construct and anti-E1AF antibody. (B) Cell migration assay of control or E1AF RNAi-transfected SHG44 cells (left panel). A wound-healing assay was prepared as described in Material and Methods. Scale bar, 50 μm. The wound-induced migration of cells was measured after 24 h (right panel). (C) Decreasing E1AF expression in SHG44 cells inhibited invasive ability as assayed in a modified Boyden chamber (P < 0.05, n = 3) (left panel). Scale bar, 100 μm. Quantification of invasion assays is shown in the right panel. (D) SHG44 cells stably transfected with control or E1AF RNAi vector was plated on the top of gels, and EGF (50 ng/ml) was added to the medium 1 day later. The numbers of invading cells in 10 photographic fields from three separate experiments were counted. The values represent activation compared to levels observed in untreated cells. (E) Control and/or E1AF RNAi and/or HA-GalT V expression vector was cotransfected into SHG44 cells. After 24 h, these cells were subjected to invasion assays. The values represent activation compared to control cell results (upper panel). Expression of E1AF and HA-GalT V is indicated; expression of GAPDH served as a loading control (lower panel). (F) Control and/or E1AF RNAi and/or HA-GalT V expression vector was cotransfected into SHG44 cells. After 24 h, these cells were subjected to wound-healing assays (upper panel). Expression of E1AF and HA-GalT V is indicated; expression of GAPDH served as a loading control (lower panel).
In culture, decreasing E1AF expression levels resulted in a significant decrease in cell migration in vitro (Fig. 9B) and decreased the ability of cells to migrate through material-coated 8-μm-pore-size membranes (Fig. 9C). Furthermore, reducing E1AF expression inhibited EGF-induced glioma invasion (Fig. 9D). To address the effect of GalT V on E1AF-induced glioma cell invasion, SHG44 cells transiently transfected with E1AF RNAi or/and HA-GalT V constructs were subjected to invasion and migration assays. As shown in Fig. 9E and F, GalT V overexpression inhibited the negative effect of downregulation of E1AF on glioma cell migration and invasion. Taking all the results together, involvement of E1AF in EGF-induced glioma invasion was at least partly via upregulation of GalT V expression.
DISCUSSION
Gliomas are the most common intracranial tumors (11). In the US, approximately 15,000 patients die of glioblastoma per year. One of the most important hallmarks of the malignant glioma is its invasive behavior (43). EGF has been reported to participate in glioma invasion with largely unknown mechanisms of action (3). Here, we demonstrated a new mechanism of EGF-induced glioma invasion: the direct interaction of E1AF and Sp1 enhanced by EGF cooperatively activated transcription of glioma-related gene GalT V and glioma invasion.
Members of the Ets family of transcription factors characterized by an evolutionarily conserved DNA-binding domain regulate viral and cellular gene expression by binding to a purine-rich GGAA/T core sequence in cooperation with other transcriptional factors and cofactors (37). Numerous forms of regulation involving Ets family members and Sp1 have been documented, but in all cases, the two proteins interact directly with DNA on their respective binding sites (EBS and GC box) (12, 21, 28, 34, 48). Here, we found that the Ets family member E1AF could functionally and physically interact with Sp1 without direct DNA binding, as suggested by previously determined quantitative evidence (1). E1AF cooperated with Sp1 to transactivate the GalT V promoter (2). Inhibition of Sp1 activity by the presence of mithramycin A or Sp1 RNAi reduced the binding of E1AF to the GalT V promoter and E1AF-induced GalT V promoter activity (3). E1AF physically interacted with the Sp1 protein in vivo and colocalized with Sp1 in the nucleus (data not shown) (4). E1AF could bind to the GalT V promoter via interaction with Sp1. E1AF has a functional acidic domain, a Gln-rich domain, and an ETS domain. E1AF could functionally and physically interact with Sp1 through the glutamine-rich domain. These observations were consistent with the theory that Gln-rich domains tend to interact with each other (33, 42). We also investigated whether other glutamine-rich transcription factors act as E1AF binding partners. We found that E1AF interacted with itself, TFIIB, and CBP (data not shown), indicating that interaction with transcription factors containing a Gln-rich domain might contribute to E1AF functions, a topic that needs further investigation.
Interaction with other protein factors may be one of the most important activities of Sp1 (10), but the functions and mechanisms of such interactions have not been studied in detail. Phosphorylation of Sp1 is tied to functional changes in DNA binding and promoter activation and is involved in cell growth (7). Here, we provide evidence that E1AF-induced Sp1 target gene transcription may be conveyed via increased Sp1 phosphorylation as a way to enhance Sp1 transactivating activity. As a result, E1AF overexpression could enhance the binding of Sp1 to the promoter of the GalT V gene. Although the evidence presented here is largely indirect, an increase in the transcriptional potential of Sp1, along with previously established mechanisms of Sp1-site-dependent transcription, owing to changes in its phosphorylation state may point to a rational and likely conclusion concerning the mechanism by which E1AF conveys induction of Sp1 target gene transcription. Biologically, E1AF and Sp1 cooperatively regulate glioma migration and invasion. To our knowledge, this is the first report of the contribution of E1AF or Sp1 to glioma invasion. Overexpression of E1AF increases invasion of glioma cell U251 in a Sp1- and GalT V-dependent manner (data not shown), suggesting that E1AF might be involved in glioma cell metastasis phenotypes through association with other transcription factors.
To date, many signals of divergent natures involved in regulation of gene transcription via altering Sp1 transcriptional activity have been identified (7, 10). EGF is widely known to induce gene transcription via alteration of Sp1 phosphorylation (10). Here, we provide evidence that E1AF might act as a new mediator between EGF signaling and Sp1. EGF could increase E1AF expression in glioma cells and enhance the association of E1AF and Sp1. In addition, decreasing expression of E1AF inhibited EGF-induced phosphorylation of Sp1, transcription activity, and glioma invasion. The data reported above suggest that E1AF might act as a mediator between EGF signaling and Sp1. Taken together, the data indicate that E1AF links the EGF signaling and Sp1 transcription factor in the system of Sp1-target gene transcription, which expands our knowledge of EGF signal pathways.
In summary, the transcriptional complex consisting of E1AF and Sp1 may potentially represent a class of transcription factors that functionally interact and participate in transcription regulation and tumor behavior. The findings of the current study advance our knowledge of EGF-induced glioma invasion and establish a functional link between EGF signaling and transcription of its target genes.
Acknowledgments
This work was supported by 863 Program of China (grant 2001AA234031), National Natural Scientific Foundation of China (30330320 and 30700132), National Basic Research Program (2002CB512803), and a grant from the Development of Science and Technology of Shanghai (02DJ14002) and sponsored by Shanghai Educational Development Foundation.
We thank Guntram Suske (Marburg, Germany) for providing the PEVR2-Sp1 (human). We thank Liangfu Zhou (Nurosurgery, Huaashang Hospital, China) and Aiguo Shen (Laboratory of Neurobiology, Nantong University) for providing normal human brain tissues and glioma tissues.
Footnotes
Published ahead of print on 15 October 2007.
REFERENCES
- 1.Albini, A., Y. Iwamoto, H. K. Kleinman, G. R. Martin, S. A. Aaronson, J. M. Kozlowski, and R. N. McEwan. 1987. A rapid in vitro assay for quantitating the invasive potential of tumor cells. Cancer Res. 47:3239-3245. [PubMed] [Google Scholar]
- 2.Azizkhan, J. C., D. E. Jensen, A. J. Pierce, and M. Wade. 1993. Transcription from TATA-less promoters: dihydrofolate reductase as a model. Crit. Rev. Eukaryot. Gene Expr. 3:229-254. [PubMed] [Google Scholar]
- 3.Bachoo, R. M., E. A. Maher, K. L. Ligon, N. E. Sharpless, S. S. Chan, M. J. You, Y. Tang, J. DeFrances, E. Stover, R. Weissleder, D. H. Rowitch, D. N. Louis, and R. A. DePinho. 2002. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell 1:269-277. [DOI] [PubMed] [Google Scholar]
- 4.Baert, J. L., C. Beaudoin, L. Coutte, and Y. de Launoit. 2002. ERM transactivation is up-regulated by the repression of DNA binding after the PKA phosphorylation of a consensus site at the edge of the ETS domain. J. Biol. Chem. 277:1002-1012. [DOI] [PubMed] [Google Scholar]
- 5.Bailly, R. A., R. Bosselut, J. Zucman, F. Cormier, O. Delattre, M. Roussel, G. Thomas, and J. Ghysdael. 1994. DNA-binding and transcriptional activation properties of the EWS-FLI-1 fusion protein resulting from the t(11;22) translocation in Ewing sarcoma. Mol. Cell. Biol. 14:3230-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black, A. R., J. D. Black, and J. Azizkhan-Clifford. 2001. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J. Cell Physiol. 188:143-160. [DOI] [PubMed] [Google Scholar]
- 7.Black, A. R., D. Jensen, S. Y. Lin, and J. C. Azizkhan. 1999. Growth/cell cycle regulation of Sp1 phosphorylation. J. Biol. Chem. 274:1207-1215. [DOI] [PubMed] [Google Scholar]
- 8.Bosselut, R., J. Levin, E. Adjadj, and J. Ghysdael. 1993. A single amino-acid substitution in the Ets domain alters core DNA binding specificity of Ets1 to that of the related transcription factors Elf1 and E74. Nucleic Acids Res. 21:5184-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, X., J. Jiang, J. Yang, C. Chen, M. Sun, Y. Wei, X. Guang, and J. Gu. 2006. Down-regulation of the expression of β1,4-galactosyltransferase V promotes integrin β1 maturation. Biochem. Biophys. Res. Commun. 343:910-916. [DOI] [PubMed] [Google Scholar]
- 10.Chu, S., and T. J. Ferro. 2005. Sp1: regulation of gene expression by phosphorylation. Gene 348:1-11. [DOI] [PubMed] [Google Scholar]
- 11.DeAngelis, L. M. 2001. Brain tumors. N. Engl. J. Med. 344:114-123. [DOI] [PubMed] [Google Scholar]
- 12.Dittmer, J., A. Gegonne, S. D. Gitlin, J. Ghysdael, and J. N. Brady. 1994. Regulation of parathyroid hormone-related protein (PTHrP) gene expression. Sp1 binds through an inverted CACCC motif and regulates promoter activity in cooperation with Ets1. J. Biol. Chem. 269:21428-21434. [PubMed] [Google Scholar]
- 13.Firlej, V., B. Bocquet, X. Desbiens, Y. de Launoit, and A. Chotteau-Lelievre. 2005. Pea3 transcription factor cooperates with USF-1 in regulation of the murine bax transcription without binding to an Ets-binding site. J. Biol. Chem. 280:887-898. [DOI] [PubMed] [Google Scholar]
- 14.Gartel, A. L., E. Goufman, F. Najmabadi, and A. L. Tyner. 2000. Sp1 and Sp3 activate p21 (WAF1/CIP1) gene transcription in the Caco-2 colon adenocarcinoma cell line. Oncogene 19:5182-5188. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez, M. I., and D. M. Robins. 2001. Oct-1 preferentially interacts with androgen receptor in a DNA-dependent manner that facilitates recruitment of SRC-1. J. Biol. Chem. 276:6420-6428. [DOI] [PubMed] [Google Scholar]
- 16.Guo, S., T. Sato, K. Shirane, and K. Furukawa. 2001. Galactosylation of N-linked oligosaccharides by human beta-1,4-galactosyltransferases I, II, III, IV, V, and VI expressed in Sf-9 cells. Glycobiology 11:813-820. [DOI] [PubMed] [Google Scholar]
- 17.Hakuma, N., I. Kinoshita, Y. Shimizu, K. Yamazaki, K. Yoshida, M. Nishimura, and H. Dosaka-Akita. 2005. E1AF/PEA3 activates the Rho/Rho-associated kinase pathway to increase the malignancy potential of non-small-cell lung cancer cells. Cancer Res. 65:10776-10782. [DOI] [PubMed] [Google Scholar]
- 18.Hanzawa, M., M. Shindoh, F. Higashino, M. Yasuda, N. Inoue, K. Hida, M. Ono, T. Kohgo, M. Nakamura, K. Notani, H. Fukuda, Y. Totsuka, K. Yoshida, and K. Fujinaga. 2000. Hepatocyte growth factor upregulates E1AF that induces oral squamous cell carcinoma cell invasion by activating matrix metalloproteinase genes. Carcinogenesis 21:1079-1085. [PubMed] [Google Scholar]
- 19.Jaishankar, S., J. Zhang, M. F. Roussel, and S. J. Baker. 1999. Transforming activity of EWS/FLI is not strictly dependent upon DNA-binding activity. Oncogene 18:5592-5597. [DOI] [PubMed] [Google Scholar]
- 20.Jiang, J., X. Chen, J. Shen, Y. Wei, T. Wu, Y. Yang, H. Wang, H. Zong, J. Yang, S. Zhang, J. Xie, X. Kong, W. Liu, and J. Gu. 2006. β1,4-Galactosyltransferase V functions as a positive growth regulator in glioma. J. Biol. Chem. 281:9482-9489. [DOI] [PubMed] [Google Scholar]
- 21.Kavurma, M. M., Y. Bobryshev, and L. M. Khachigian. 2002. Ets-1 positively regulates Fas ligand transcription via cooperative interactions with Sp1. J. Biol. Chem. 277:36244-36252. [DOI] [PubMed] [Google Scholar]
- 22.Kurpios, N. A., N. A. Sabolic, T. G. Shepherd, G. M. Fidalgo, and J. A. Hassell. 2003. Function of PEA3 Ets transcription factors in mammary gland development and oncogenesis. J. Mammary Gland Biol. Neoplasia 8:177-190. [DOI] [PubMed] [Google Scholar]
- 23.Ladle, D. R., and E. Frank. 2002. The role of the ETS gene PEA3 in the development of motor and sensory neurons. Physiol. Behav. 77:571-576. [DOI] [PubMed] [Google Scholar]
- 24.Lai, J. S., and W. Herr. 1992. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl. Acad. Sci. USA 89:6958-6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laing, M. A., S. Coonrod, B. T. Hinton, J. W. Downie, R. Tozer, M. A. Rudnicki, and J. A. Hassell. 2000. Male sexual dysfunction in mice bearing targeted mutant alleles of the PEA3 ets gene. Mol. Cell. Biol. 20:9337-9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, J. A., D. C. Suh, J. E. Kang, M. H. Kim, H. Park, M. N. Lee, J. M. Kim, B. N. Jeon, H. E. Roh, M. Y. Yu, K. Y. Choi, K. Y. Kim, and M. W. Hur. 2005. Transcriptional activity of Sp1 is regulated by molecular interactions between the zinc finger DNA binding domain and the inhibitory domain with corepressors, and this interaction is modulated by MEK. J. Biol. Chem. 280:28061-28071. [DOI] [PubMed] [Google Scholar]
- 27.Li, M., and R. E. Kellems. 2003. Sp1 and Sp3 are important regulators of AP-2γ gene transcription. Biol. Reprod. 69:1220-1230. [DOI] [PubMed] [Google Scholar]
- 28.Liedtke, C., N. Groger, M. P. Manns, and C. Trautwein. 2003. The human caspase-8 promoter sustains basal activity through SP1 and ETS-like transcription factors and can be up-regulated by a p53-dependent mechanism. J. Biol. Chem. 278:27593-27604. [DOI] [PubMed] [Google Scholar]
- 29.O'Hagan, R. C., and J. A. Hassell. 1998. The PEA3 Ets transcription factor is a downstream target of the HER2/Neu receptor tyrosine kinase. Oncogene 16:301-310. [DOI] [PubMed] [Google Scholar]
- 30.O'Hagan, R. C., R. G. Tozer, M. Symons, F. McCormick, and J. A. Hassell. 1996. The activity of the Ets transcription factor PEA3 is regulated by two distinct MAPK cascades. Oncogene 13:1323-1333. [PubMed] [Google Scholar]
- 31.Pang, R. T., L. T. Lee, S. S. Ng, W. H. Yung, and B. K. Chow. 2004. CpG methylation and transcription factors Sp1 and Sp3 regulate the expression of the human secretin receptor gene. Mol. Endocrinol. 18:471-483. [DOI] [PubMed] [Google Scholar]
- 32.Pore, N., S. Liu, H. K. Shu, B. Li, D. Haas-Kogan, D. Stokoe, J. Milanini-Mongiat, G. Pages, D. M. O'Rourke, E. Bernhard, and A. Maity. 2004. Sp1 is involved in Akt-mediated induction of VEGF expression through an HIF-1-independent mechanism. Mol. Biol. Cell 15:4841-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saluja, D., M. F. Vassallo, and N. Tanese. 1998. Distinct subdomains of human TAFII130 are required for interactions with glutamine-rich transcriptional activators. Mol. Cell. Biol. 18:5734-5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santiago, F. S., and L. M. Khachigian. 2004. Ets-1 stimulates platelet-derived growth factor A-chain gene transcription and vascular smooth muscle cell growth via cooperative interactions with Sp1. Circ. Res. 95:479-487. [DOI] [PubMed] [Google Scholar]
- 35.Sato, T., and K. Furukawa. 2004. Transcriptional regulation of the human beta-1,4-galactosyltransferase V gene in cancer cells: essential role of transcription factor Sp1. J. Biol. Chem. 279:39574-39583. [DOI] [PubMed] [Google Scholar]
- 36.Schreiber, E., P. Matthias, M. M. Muller, and W. Schaffner. 1989. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 17:6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharrocks, A. D. 2001. The ETS-domain transcription factor family. Nat. Rev. Mol. Cell Biol. 2:827-837. [DOI] [PubMed] [Google Scholar]
- 38.Shepherd, T. G., L. Kockeritz, M. R. Szrajber, W. J. Muller, and J. A. Hassell. 2001. The pea3 subfamily ets genes are required for HER2/Neu-mediated mammary oncogenesis. Curr. Biol. 11:1739-1748. [DOI] [PubMed] [Google Scholar]
- 39.Shindoh, M., F. Higashino, and T. Kohgo. 2004. E1AF, an ets-oncogene family transcription factor. Cancer Lett. 216:1-8. [DOI] [PubMed] [Google Scholar]
- 40.Sowa, Y., T. Orita, S. Minamikawa, K. Nakano, T. Mizuno, H. Nomura, and T. Sakai. 1997. Histone deacetylase inhibitor activates the WAF1/Cip1 gene promoter through the Sp1 sites. Biochem. Biophys. Res. Commun. 241:142-150. [DOI] [PubMed] [Google Scholar]
- 41.Subbaramaiah, K., L. Norton, W. Gerald, and A. J. Dannenberg. 2002. Cyclooxygenase-2 is overexpressed in HER-2/neu-positive breast cancer: evidence for involvement of AP-1 and PEA3. J. Biol. Chem. 277:18649-18657. [DOI] [PubMed] [Google Scholar]
- 42.Subramanian, C., and E. S. Robertson. 2002. The metastatic suppressor Nm23-H1 interacts with EBNA3C at sequences located between the glutamine- and proline-rich domains and can cooperate in activation of transcription. J. Virol. 76:8702-8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tonn, J. C., and R. Goldbrunner. 2003. Mechanisms of glioma cell invasion. Acta Neurochir. Suppl. 88:163-167. [DOI] [PubMed] [Google Scholar]
- 44.Xie, R. L., S. Gupta, A. Miele, D. Shiffman, J. L. Stein, G. S. Stein, and A. J. van Wijnen. 2003. The tumor suppressor interferon regulatory factor 1 interferes with SP1 activation to repress the human CDK2 promoter. J. Biol. Chem. 278:26589-26596. [DOI] [PubMed] [Google Scholar]
- 45.Xu, S., S. Zhang, C. Chen, J. Yan, M. Cai, X. Zhu, and J. Gu. 2002. Over-expression of beta-1,4-galactosyltransferase V increases the growth of astrocytoma cell line. J. Exp. Clin. Cancer Res. 21:409-414. [PubMed] [Google Scholar]
- 46.Xu, S., X. Zhu, S. Zhang, S. Yin, L. Zhou, C. Chen, and J. Gu. 2001. Over-expression of beta-1,4-galactosyltransferase I, II, and V in human astrocytoma. J. Cancer Res. Clin. Oncol. 127:502-506. [DOI] [PubMed] [Google Scholar]
- 47.Yan, G. Z., and E. B. Ziff. 1997. Nerve growth factor induces transcription of the p21 WAF1/CIP1 and cyclin D1 genes in PC12 cells by activating the Sp1 transcription factor. J. Neurosci. 17:6122-6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan, S., I. M. Berquin, B. R. Troen, and B. F. Sloane. 2000. Transcription of human cathepsin B is mediated by Sp1 and Ets family factors in glioma. DNA Cell Biol. 19:79-91. [DOI] [PubMed] [Google Scholar]
- 49.Zhu, X., S. Chen, X. Yin, A. Shen, S. Ji, Z. Shen, and J. Gu. 2003. Constitutively active PKB/Akt inhibited apoptosis and down-regulated beta1,4-galactosyltransferase 1 in hepatocarcinoma cells. Biochem. Biophys. Res. Commun. 309:279-285. [DOI] [PubMed] [Google Scholar]
- 50.Zhu, X., J. Jiang, H. Shen, H. Wang, H. Zong, Z. Li, Y. Yang, Z. Niu, W. Liu, X. Chen, Y. Hu, and J. Gu. 2005. Elevated β1,4-galactosyltransferase I in highly metastatic human lung cancer cells. Identification of E1AF as important transcription activator. J. Biol. Chem. 280:12503-12516. [DOI] [PubMed] [Google Scholar]