Abstract
It has been a long-standing dogma in life sciences that only eukaryotic organisms possess a cytoskeleton. Recently, this belief was questioned by the finding that the bacterial cell division protein FtsZ resembles tubulin in sequence and structure and, thus, may be the progenitor of this major eukaryotic cytoskeletal element. Here, we report two nuclear-encoded plant ftsZ genes which are highly conserved in coding sequence and intron structure. Both their encoded proteins are imported into plastids and there, like in bacteria, they act on the division process in a dose-dependent manner. Whereas in bacteria FtsZ only transiently polymerizes to a ring-like structure, in chloroplasts we identified persistent, highly organized filamentous scaffolds that are most likely involved in the maintenance of plastid integrity and in plastid division. As these networks resemble the eukaryotic cytoskeleton in form and function, we suggest the term “plastoskeleton” for this newly described subcellular structure.
Keywords: FtsZ, plastid division, plastoskeleton, cytoskeleton, Physcomitrella
Introduction
The functional complexity of eukaryotic cells is guaranteed at the structural level by different cell compartments; e.g., most eukaryotes possess mitochondria, and all plant cells possess plastids. These organelles are remnants of free-living prokaryotes which lost their autonomy during evolution by establishing an endosymbiosis with their host cells. In the course of this coevolution, the majority of prokaryotic genes were lost or transferred to the eukaryotic nucleus. Thus, recent eukaryotic cells are mosaics of at least two (animals and fungi) or three (plants) different genetic systems and their encoded structures (Gray 1992; Martin and Herrmann 1998; Gray et al. 1999).
It has been a long-standing canon in life sciences that a major difference between prokaryotes and eukaryotes is the cytoskeleton, a complex network of actin, tubulin, and intermediate filaments spanning the eukaryotic cytoplasm. This view was questioned by analysis of FtsZ, a protein which initiates bacterial cell division by polymerizing to a ring at the division site (Lutkenhaus 1993; Ma et al. 1996) and thus forming the scaffold for assembly of the complex cell division machinery (Rothfield et al. 1999). This ring clearly represents a prokaryotic cytoskeletal structure even though it could be detected only transiently (Bi and Lutkenhaus 1991; Lutkenhaus and Addinall 1997). Eukaryotic tubulin and prokaryotic FtsZ share several biochemical and structural features (de Boer et al. 1992; Raychaudhuri and Park 1992; Mukherjee and Lutkenhaus 1994; Löwe and Amos 1998) identifying FtsZ as an ancestral tubulin and leading to the hypothesis that the eukaryotic cytoskeleton has evolved from prokaryotic cytoskeletal elements (Bork et al. 1992; Erickson 1997; Faguy and Doolittle 1998).
In plants, FtsZ has been retained, encoded by a small nuclear gene family whose proteins are re-imported to their evolutionary origins, plastids and mitochondria (Osteryoung and Vierling 1995; Beech et al. 2000). In a functional conservation of prokaryotic cell– and eukaryotic organelle division, plant FtsZ is involved in chloroplast division (Osteryoung et al. 1998; Strepp et al. 1998). From this evidence, it was suggested that the solely morphologically described plastid dividing ring might be a simple reminiscence of the bacterial FtsZ ring (Lutkenhaus 1998; Osteryoung and Pyke 1998).
Here, we promote a novel, more complex view. As bacteria are kept in shape by their cell walls, they may only transiently need cytoskeletal elements in the course of their division. To facilitate endosymbiotic metabolite exchange, however, bacterial cell walls had to be reduced, concomitantly enhancing the need for internal structures that maintain the integrity and shape of the organelle. Thus, the eukaryotic cell may have been forced to generate structures based on preexisting bacterial compounds that keep their organelles in shape. Such an as yet undescribed structure has now been identified inside chloroplasts in this work and is termed the plastoskeleton.
Materials and Methods
Plant Material
Physcomitrella patens (Hedw.) B.S.G. was cultivated as described previously (Strepp et al. 1998).
Isolation of PpftsZ2
A cDNA of PpftsZ1 has been described previously (Strepp et al. 1998). To identify homologues of PpftsZ1, a Physcomitrella λZAP library (Strepp et al. 1998) was screened as described previously (Reski et al. 1998). Clones were sequenced from both ends with standard T7 and T3 primers as well as with insert-specific primers with the Taq Dye–Primer Cycle sequencing kit on an automatic sequencer (model 373A; PE Biosystems, PerkinElmer), and sequences were analyzed with DNASIS (Amersham Pharmacia Biotech).
Phylogenetic Tree Reconstruction
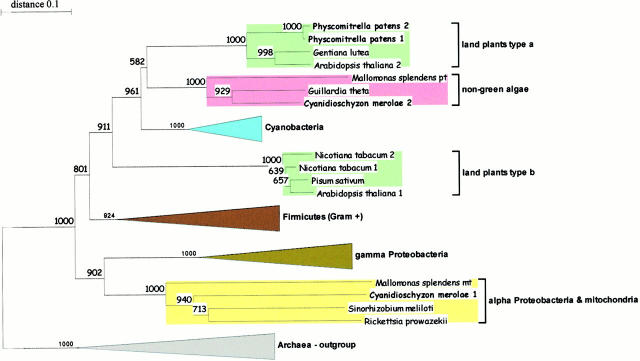
40 full-length FtsZ amino acid sequences were aligned (CLUSTAL W v1.8.; Thompson et al. 1994), and initial trees were reconstructed. Subsequently, sequence alignment was reduced to 317 positions of the protein core. As all but one of the five available red algae sequences clustered with the cryptophyte (Guillardia theta) and the chromophyte (Mallomonas splendens), the two divergent sequences from Cyanidioschyzon merolae were selected to represent the Rhodophyta. In monophyletic clades of Archaea (outgroup), γ Proteobacteria, Gram+ Eubacteria, and Cyanobacteria, the number of sequences was reduced to 3 per cluster, leading to 27 sequences (accession numbers upon request) for final tree construction. Trees were reconstructed using TREECON (van de Peer and de Wachter 1993) with Tajima and Nei's correction (insertions and/or deletions were taken into account) and neighbor joining (Saitou and Nei 1987). Bootstrap resampling (Felsenstein 1985) of the original data set with 1,000 replicates was carried out before tree reconstruction and building of the consensus tree.
PpftsZ1 and PpftsZ2 Gene Analysis
To analyze the genomic structure of the two ftsZ genes, different sets of primers were synthesized on the basis of the respective cDNA sequences. Genomic Physcomitrella DNA was extracted as described previously (Reski et al. 1994) and used as template for PCR amplification. The resulting PCR products were subcloned into pCR® 2.1-TOPO (Invitrogen) and sequenced in both directions with appropriate overlaps by primer walking.
Cloning of PpftsZ Green Fluorescent Protein Fusions and Transfection Assays
The coding region of PpftsZ1 was PCR-amplified with primers F1 (5′-GCGGGGATCCGTATGGCTAGCGGTACCGCGTTGTTTAGTGG-GTGCTCGGG-3′) and R1 (5′-GCGGTCGACCCCGGGATGACGTGTCTGGCCTCGCTTCCTTAAG-3′) introducing BamHI and SmaI restriction sites 5′ and 3′ of the cDNA, respectively. The PCR product was cleaved and ligated into the BamHI-SmaI–digested green fluorescent protein (GFP) reporter plasmid pMAV4 (Kircher et al. 1999). PpFtsZ2-GFP was constructed in essentially the same way using primers F2 (5′-GCGGAGATCTGTATGGCTAGCGGTACCGCACTGTTAGGCAGTCGC TCG-3′) and R2 (5′-GCGGTCGACGATATCCCTTTGCCCTCGCTTT CG-3′), adding BglII and EcoRV restriction sites to the PCR product. The plasmid pFtsZ1(1–35)-GFP contains the first 105 base pairs of the ftsZ1 cDNA. This fragment was amplified by PCR with primers pF1 (5′-TTTGTCGACATGGCGTTGTTTAGTGGGTGC-3′) and pR1 (5′-TTTAGATCTATGCATGCTGCAGTGAACAACG-3′) introducing SalI and BglII restriction sites to the PCR product which was inserted into pMAV4. pFtsZ1(1–93)-GFP contains a 297-bp ftsZ1 cDNA fragment coding for the FtsZ1 chloroplast transit peptide. The fragment was amplified with primers pF1 and pR2 (5′-TTTAGATCTGTCTCCGCTTCCTTCAAATGC-3′). The DNA for transfection was prepared with the Jetstar Maxiprep kit (Genomed). 3 × 105 protoplasts were transiently transfected with 25 μg of circular plasmid DNA. Isolation and regeneration of protoplasts in the absence of any antibiotic drug was carried out as described previously (Rother et al. 1994).
Confocal Laser Scanning Microscopy
Localization of FtsZ-GFP was analyzed in transfected protoplasts by confocal laser scanning microscopy (CLSM) (TCS 4D; Leica) using 488-nm excitation and two-channel measurement of emission from 510–580 nm (green/GFP) as well as >590 nm (red/chlorophyll).
Results and Discussion
Isolation of PpftsZ2
Recently, a role of eukaryotic PpFtsZ1 in plastid division had been demonstrated in the moss Physcomitrella patens (Strepp et al. 1998). As reported for Arabidopsis thaliana, seed plants may have more than one ftsZ gene functional in plastid division (Osteryoung et al. 1998) unlike most Eubacteria where only one ftsZ gene per species has been identified (Rothfield et al. 1999). To evaluate the evolutionary implications of these findings, a second FtsZ was isolated as a full-length cDNA of 1,395 bp from the moss (PpftsZ2) and sequenced on both strands (sequence data available from EMBL/GenBank/DDBJ under accession no. AJ249140).
Genomic Sequences of the Two PpftsZ Genes
To investigate the evolutionary relationship of the two different plant ftsZ genes, genomic fragments of PpftsZ were PCR-amplified, cloned, and subsequently sequenced. Sequence assembly yielded two loci 3,088 and 3,459 bp in length (sequence data available from EMBL/GenBank/DDBJ under accession nos. AJ249138 and AJ249139). Each locus contains six introns ranging in size from 78 to 461 bp.
The two ftsZ loci are highly conserved: (a) all six introns are in exactly the same position within the coding sequence of the two genes, and (b) five out of the six pairs of introns show only small differences in length (1–16 bp). The remaining pair of introns (intron 6) differs in length by 180 bp. The precise conservation of intron positions and their nearly identical lengths indicate that a single copy of the prokaryotic ftsZ was transferred to the nucleus during establishment of endosymbiosis and acquired eukaryotic features like introns before duplication and subsequent divergence occurred.
Phylogeny of FtsZ
27 deduced FtsZ protein sequences were compared with each other, 14 of them from photosynthetic eukaryotes and 13 from photosynthetically inactive bacteria, mitochondria, and Archaea (Fig. 1). Archaea, Gram+ Eubacteria, Cyanobacteria, nongreen algae, as well as α and γ Proteobacteria form monophyletic clusters. As recently described, the mitochondrial Mallomonas sequence clusters within the α Proteobacteria (Beech et al. 2000).
Figure 1.
Consensus phylogenetic tree of FtsZ amino acid sequences. Branch numbers represent bootstrap values (1,000 replicates). Accession nos. of the sequences are available on request.
In addition, one Cyanidioschyzon sequence is placed in that clade as well, identifying a second eukaryote with an α Proteobacterium–related FtsZ that, therefore, most likely is located in mitochondria. Interestingly, Mallomonas and Cyanidioschyzon are nongreen algae. Despite huge sequencing projects, a mitochondrial FtsZ has never been identified from any organism other than the nongreen algae. Thus, only their mitochondria might divide by an ancient FtsZ-dependent mechanism whereas in yeast, chlorobionts, and higher eukaryotes, mitochondria may be divided by dynamin (Erickson 2000).
Surprisingly, the land plant sequences formed two monophyletic clusters designated types a and b. Both Physcomitrella sequences cluster in clade a, whereas the two divergent Arabidopsis ftsZ genes, whose products are putatively located in different cell compartments (Osteryoung et al. 1998), group in different clades. AtFtsZ2, which is supposed to remain cytoplasmic (Osteryoung et al. 1998), clusters with the two Physcomitrella proteins in clade a.
Both FtsZ Proteins Are Imported into Plastids
The existence of two distinct monophyletic FtsZ clusters in land plants indicates differences in function or localization, respectively, between these two protein classes. Arabidopsis possesses at least two different ftsZ genes, the products of which were suggested to be localized in different cell compartments: in vitro studies demonstrated that AtFtsZ1 was imported into plastids (Osteryoung and Vierling 1995), whereas the same could not be demonstrated for AtFtsZ2, revealing that AtFtsZ2 might be localized in the cytoplasm (Osteryoung et al. 1998; Colletti et al. 2000). Both Physcomitrella sequences cluster with AtFtsZ2 and thus may be localized in the cytoplasm as well. However, sequence analysis (Center for Biological Sequences Analysis, available at http://www.cbs.dtu.dk/services/ChloroP/) identified transit signal sequences indicating that both Physcomitrella FtsZ proteins are imported into plastids.
Because in vitro as well as in silico studies are indirect and may easily generate contradictory and inconclusive data, we analyzed subcellular localization of both Physcomitrella FtsZ proteins in vivo as COOH-terminal GFP fusion proteins. Physcomitrella protoplasts were transiently transfected with expression plasmids PpFtsZ1-GFP or PpFtsZ2-GFP, respectively, and GFP fluorescence was visualized by CLSM. 2 d after transfection, GFP signals could be solely detected within plastids (Fig. 2, a–g). The absence of GFP fluorescence in all other cell compartments, including the plastid outer membrane, was confirmed by series of optical sections through protoplasts and single plastids (Fig. 2, d–g), respectively.
Figure 2.
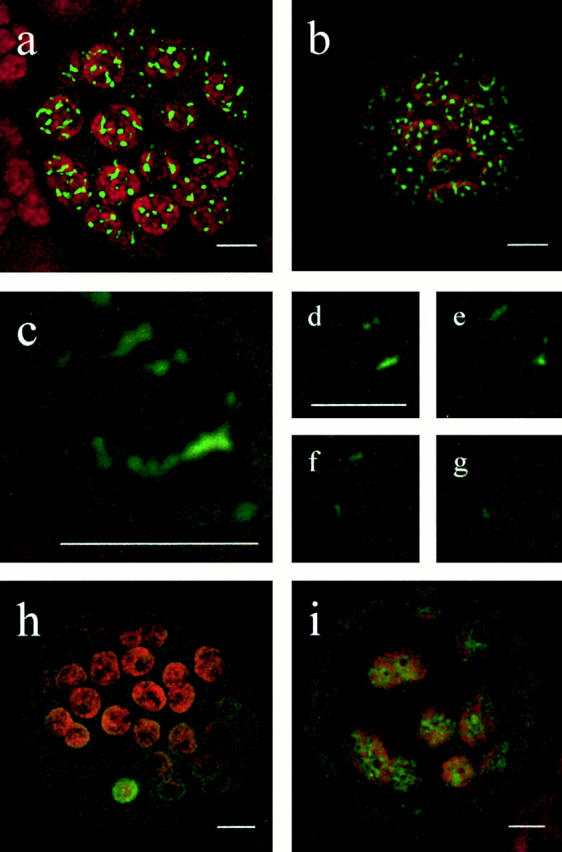
Subcellular localization of FtsZ-GFP fusion proteins. Physcomitrella protoplasts were transiently transfected with the corresponding expression plasmid and GFP fluorescence was visualized by CLSM 2 d after transfection. (a) Protoplast transfected with PpFtsZ1-GFP. (b) Protoplast transfected with PpFtsZ2-GFP. (c) Single chloroplast (detail of a). (d–g) Sections of the chloroplast shown in c demonstrating the localization of PpFtsZ1 solely within chloroplasts. (h) Protoplast transfected with pFtsZ1(1–35)-GFP (incomplete transit peptide) showing cytoplasmic distribution of fusion protein. (i) Protoplast transfected with pFtsZ1(1–93)-GFP (complete transit peptide) showing plastidic distribution of fusion protein. a–c, and h and i, represent overlays of all sections of the object. Chlorophyll fluorescence is shown in red; GFP signals are shown in green. Bars, 5 μm.
As controls, a COOH-terminal fusion of GFP to a short NH2-terminal fragment of PpFtsZ1, designated pFtsZ1(1–35)-GFP, accumulated in the cytoplasm (Fig. 2 h), whereas pFtsZ1(1–93)-GFP accumulated exclusively in the plastids (Fig. 2 i), revealing that the NH2-terminal 93 amino acid residues, but not the first 35, contain the functional transit peptide of PpFtsZ1.
Thus, in vivo both Physcomitrella FtsZ proteins are exclusively imported into plastids.
FtsZ-GFP Fusion Proteins Are Functional in Plastid Division
In bacteria, FtsZ acts together with a variety of associated proteins in forming the cell division apparatus. The molar ratios of some of its constituents, such as FtsZ, FtsA, and ZipA, were shown to be critical for its functionality: a slight overexpression of FtsZ enhanced division frequency, whereas high levels of FtsZ inhibited bacterial cell division (Ward and Lutkenhaus 1985).
Like their bacterial progenitors, both Physcomitrella proteins acted in a dose-dependent manner on the division process despite the presence of the COOH-terminal GFP fusion domain. Low FtsZ-GFP levels (indicated by weak GFP fluorescence) led to an enhancement of the division process and resulted in plastids that were significantly reduced in size (Fig. 3). In contrast, high FtsZ levels (indicated by strong GFP fluorescence) blocked plastid division (Fig. 4, a–d), leading to undivided, abnormally large plastids closely resembling the PpftsZ1-knockout phenotype (Strepp et al. 1998). The reduction of plastid size in cells overexpressing FtsZ-GFP indicates that the recombinant proteins are functional in division.
Figure 3.
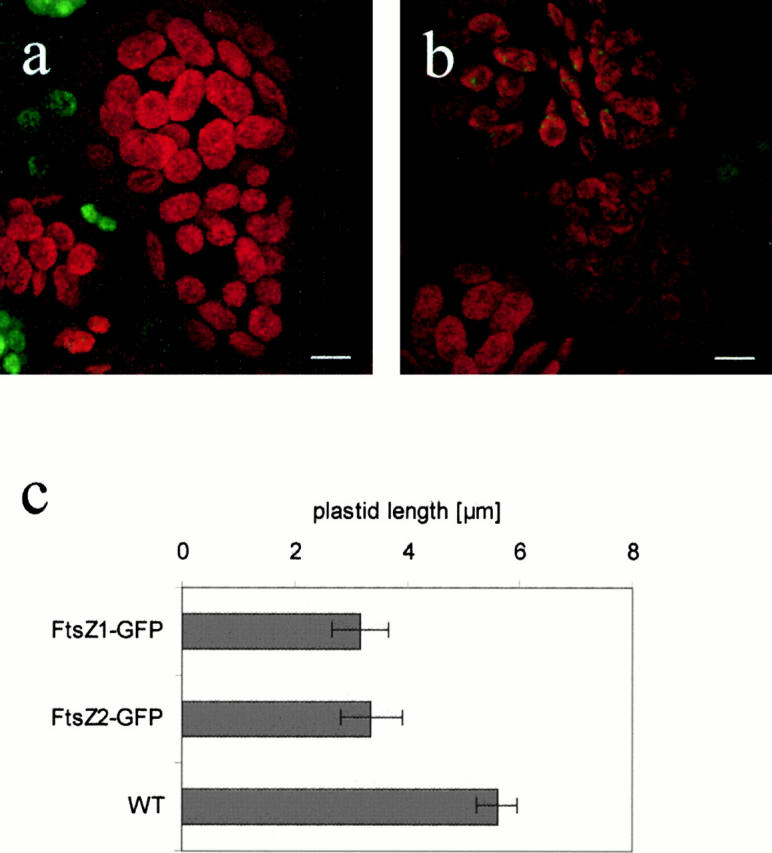
Reduction of chloroplast size by slight overexpression of FtsZ. (a) Nontransfected cells; (b) FtsZ2-GFP–expressing cells with reduced chloroplast size. Chlorophyll fluorescence is shown in red; GFP signals are shown in green. (c) Chloroplast size of FtsZ-overexpressing cells. Moss protoplasts were analyzed after their first cell division. Within each analyzed cell, the length of 20 chloroplasts was determined. Average plastid length and standard deviation are shown for two protoplasts expressing FtsZ1-GFP, for five expressing FtsZ2-GFP, and for three nontransformed protoplasts (WT), respectively. Bars, 5 μm.
Figure 4.
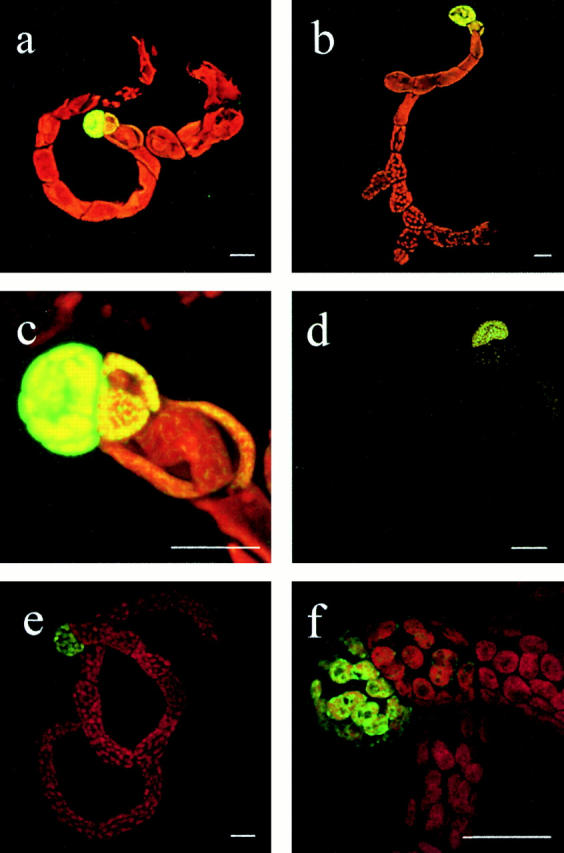
Inhibition of chloroplast division by high overexpression of FtsZ-GFP. Moss protonemata were analyzed by CLSM 10 d after protoplast transfection with expression plasmids FtsZ1-GFP (a and c), FtsZ2-GFP (b and d), or pFtsZ1(1–93)-GFP (e and f); (c) detail of a; (f) detail of e. a–c, and e and f, represent overlays of chlorophyll (red) and GFP (green) channel, in d only GFP channel is shown. Bars, 20 μm.
The correlation between FtsZ-GFP dosage and plastid division was most strikingly evident in regenerating Physcomitrella plants: the initial cell, the protoplast, regenerates into a protonema. As only apical cells divide, the protonema filament represents a cell lineage from the initial protoplast to the dividing tip cell. In our transfection experiments, cells immediately after the initial protoplast showed strong GFP fluorescence in one giant plastid (Fig. 4, a–d). In subsequent cells, GFP fluorescence as well as plastid sizes decreased, finally resulting in cells with wild-type–like plastid number and size (Fig. 4, a, b, and d). As a control, pFtsZ(1–93)-GFP had no influence on plastid division but exhibited the same gradient in fluorescence intensities as FtsZ-GFPs (Fig. 4e and Fig. f).
As both PpFtsZ proteins are independently acting on plastid division, they are—although highly conserved—not redundant in function.
FtsZ Proteins Contribute to a Filamentous Plastoskeleton
It has been long believed that cytoskeletal filaments occur solely in the cytoplasm of eukaryotes. Nonetheless, in the late 1960s, some authors by means of electron microscopy detected microtubule-like structures within plastids of several plant species (Sprey 1968; Lawrence and Possingham 1984). These structures were suggested to play a role in altering the shape of plastids and plastid division (Sprey 1968). Such reports, however, were not widely accepted. Recent analyses have now identified bacterial FtsZ as an ancestral tubulin (Löwe and Amos 1998), thus supporting the idea that cytoskeletal elements in eukaryotes may have arisen from bacterial progenitors (Erickson 1995; Faguy and Doolittle 1998). In this work, we could extend this hypothesis by visualizing a proteinaceous scaffold inside a eukaryotic organelle, the chloroplast.
In vivo localization by CLSM showed that both PpFtsZ-GFP fusions form organized, branched filamentous structures within Physcomitrella plastids (Fig. 5). This polymerization is due to the FtsZ part of the fusion protein, since the truncated pFtsZ(1–93)-GFP is distributed more diffusely within the organelle (see Fig. 2 i) compared with the complete FtsZ-GFP fusions (see Fig. 2, a and b). Both FtsZ scaffolds span the entire plastids and thus resemble the cytoskeleton in eukaryotic cytoplasm. We therefore suggest the term “plastoskeleton” to describe these structures. Additionally, these networks represent the first report to date on eukaryotic FtsZ filamentation in vivo. As FtsZ1-GFP and FtsZ2-GFP independently influence plastid division and even in low concentration are functional in division, we postulate that the scaffolds built by these fusion proteins mimic a naturally occurring state.
Figure 5.
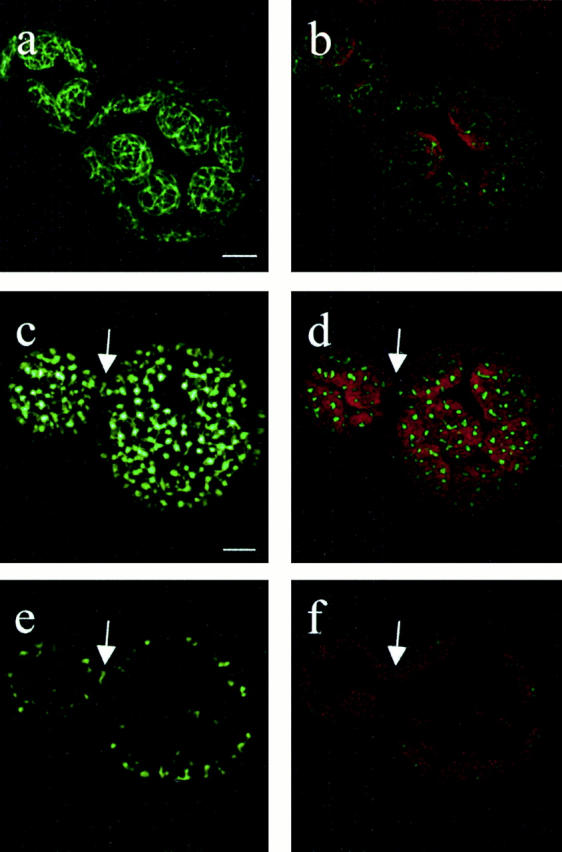
In vivo filamentation of PpFtsZ1 and PpFtsZ2. FtsZ1-expressing cells (a and b) were analyzed by CLSM 2 d after transfection with FtsZ1-GFP plasmid. FtsZ2-expressing cells (c and d) were analyzed by CLSM 2 d after transfection with FtsZ2-GFP plasmid. An arrow points to the S-shaped structure, which may represent a plastid division ring. (e and f) Single section of the overlay shown in c and d visualizing the S-shaped structure in more detail. Bars, 5 μm.
Although PpFtsZ1 and PpFtsZ2 share 84% identity at the peptide sequence level, they could be discriminated according to their spatial organization in plastids. FtsZ1-GFP formed highly regular networks (Fig. 5, a and b), whereas FtsZ2-GFP networks were not as well organized (Fig. 5c and Fig. d). In both cases, the filaments were connected by nodes that seemed to anchor the skeleton to the inner chloroplast membrane.
Although bacterial and eukaryotic FtsZ proteins are highly conserved in molecular terms, our results indicate that there are striking differences between them: in E. coli, structures formed by FtsZ are only transiently detected as Z rings during cell division (Bi and Lutkenhaus 1991). In plastids, however, we detected highly organized filamentous networks spanning the whole organelle. As bacteria are single cells surrounded by a rigid cell wall, they may not need cytoskeletal structures except for division. During establishment of endosymbiosis, bacteria were engulfed by the host cell and subsequently lost components of their cell walls in order to establish an endosymbiotic metabolism (Gray 1992; Vellai et al. 1998). To maintain organelle integrity after loss of cell wall components, a novel structure may have evolved. Here, we suggest that this novel structure is the FtsZ-based plastoskeleton.
This striking difference between bacteria and plastids may be due to differences in the FtsZ molecule itself. Bacterial and eukaryotic FtsZ proteins are highly conserved in the NH2-terminal region, which is responsible for FtsZ polymerization to protofilaments (Löwe and Amos 1999) and ring formation (Ma et al. 1996). However, they are more variable in the COOH terminus, which is involved in heterologous protein–protein interactions (Ma and Margolin 1999; van den Ent et al. 1999). Thus, it seems more likely that structural distinctions are due to different interactions with FtsZ-associated proteins. As FtsZ networks have as yet not been identified in prokaryotes, the plastoskeleton may represent a chimera of prokaryotic-derived FtsZ and associated proteins recruited from the host cell.
In addition to these networks, we occasionally identified an S-shaped structure at the constriction site of dividing chloroplasts in regenerating protoplasts transfected with FtsZ2-GFP (Fig. 5, c–f; arrows). This S-shaped structure may represent a PpFtsZ2-based plastid division ring, indicating that the eukaryotic PpFtsZ2 has maintained a conserved function besides polymerizing into filamentous networks.
In this study, we identified two types of plant FtsZ proteins. Our phylogenetic analysis revealed that both Physcomitrella proteins group in land plant clade a. In contrast, the two Arabidopsis proteins group in different clades: AtFtsZ1, which was shown to be imported into plastids (Osteryoung and Vierling 1995), clusters in clade b. In contrast, AtFtsZ2 which seemed to be localized in the cytoplasm (Osteryoung et al. 1998) groups in the same clade as both Physcomitrella proteins. However, by in vivo analysis we demonstrated that both PpFtsZs are imported into plastids and also are involved in plastid division. In addition to their role in plastid division, they contribute to a novel structure here named the plastoskeleton.
The existence of two clearly distinct clades of land plant FtsZ, both involved in plastid division, strongly suggests differences in localization or function, respectively, between both protein groups. The existing data indicate that type b may represent a basal FtsZ type that is involved in the ancient function of plastid division, whereas type a may be a more evolved type that is additionally involved in the maintenance of plastid shape and flexibility via the newly described plastoskeleton.
Acknowledgments
We thank William Martin, Randall Cassada, Gunther Neuhaus, and Eberhard Schäfer for helpful discussions.
Financial support by the Deutsche Forschungsgemeinschaft (Re 837/4) is gratefully acknowledged.
Footnotes
Abbreviations used in this paper: CLSM, confocal laser scanning microscope; GFP, green fluorescent protein.
References
- Beech P.L., Nheu T., Schultz T., Herbert S., Lithgow T., Gilson P.R., McFadden G.I. Mitochondrial FtsZ in a chromophyte alga. Science. 2000;287:1276–1279. doi: 10.1126/science.287.5456.1276. [DOI] [PubMed] [Google Scholar]
- Bi E.F., Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli . Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Bork P., Sander C., Valencia A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. USA. 1992;89:7290–7294. doi: 10.1073/pnas.89.16.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colletti K.S., Tattersall E.A., Pyke K.A., Froelich J.E., Stokes K.D., Osteryoung K.W. A homologue of the bacterial cell division site-determining factor MinD mediates placement of the chloroplast division apparatus. Curr. Biol. 2000;10:507–516. doi: 10.1016/s0960-9822(00)00466-8. [DOI] [PubMed] [Google Scholar]
- de Boer P., Crossley R., Rothfield L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature. 1992;359:254–256. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- Erickson H.P. FtsZ, a prokaryotic homolog of tubulin? Cell. 1995;80:367–370. doi: 10.1016/0092-8674(95)90486-7. [DOI] [PubMed] [Google Scholar]
- Erickson H.P. FtsZ, a tubulin homologue in prokaryote cell division. Trends Cell Biol. 1997;7:362–367. doi: 10.1016/S0962-8924(97)01108-2. [DOI] [PubMed] [Google Scholar]
- Erickson H.P. Dynamin and FtsZ. Missing links in mitochondrial and bacterial division. J. Cell Biol. 2000;148:1103–1105. doi: 10.1083/jcb.148.6.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faguy D.M., Doolittle W.F. Cytoskeletal proteinsthe evolution of cell division. Curr. Biol. 1998;8:R338–R341. doi: 10.1016/s0960-9822(98)70216-7. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogeniesan approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Gray M.W. The endosymbiont hypothesis revisited. Int. Rev. Cytol. 1992;141:233–357. doi: 10.1016/s0074-7696(08)62068-9. [DOI] [PubMed] [Google Scholar]
- Gray M.W., Burger G., Lang F. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- Kircher S., Wellmer F., Nick P., Rugner A., Schafer E., Harter K. Nuclear import of the parsley bZIP transcription factor CPRF2 is regulated by phytochrome photoreceptors. J. Cell Biol. 1999;144:201–211. doi: 10.1083/jcb.144.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M.E., Possingham J.V. Observations of microtubule-like structures within spinach plastids. Biol. Cell. 1984;52:77–82. [Google Scholar]
- Löwe J., Amos L.A. Crystal structure of the bacterial cell-division protein FtsZ. Nature. 1998;391:203–206. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- Löwe J., Amos L.A. Tubulin-like protofilaments in Ca2+-induced FtsZ sheets. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:2364–2371. doi: 10.1093/emboj/18.9.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J.F. FtsZ ring in bacterial cytokinesis. Mol. Microbiol. 1993;9:403–409. doi: 10.1111/j.1365-2958.1993.tb01701.x. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J. Organelle divisionfrom coli to chloroplasts. Curr. Biol. 1998;8:R619–R621. doi: 10.1016/s0960-9822(98)70391-4. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J., Addinall S.G. Bacterial cell division and the Z ring. Annu. Rev. Biochem. 1997;66:93–116. doi: 10.1146/annurev.biochem.66.1.93. [DOI] [PubMed] [Google Scholar]
- Ma X., Margolin W. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol. 1999;181:7531–7544. doi: 10.1128/jb.181.24.7531-7544.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Ehrhardt D.W., Margolin W. Co-localization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli . Proc. Natl. Acad. Sci. USA. 1996;93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W., Herrmann R.G. Gene transfer from organelles to the nucleushow much, what happens, and why? Plant Physiol. 1998;118:9–17. doi: 10.1104/pp.118.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A., Lutkenhaus J. Guanine nucleotide-dependent assembly of FtsZ into filaments. J. Bacteriol. 1994;176:2754–2758. doi: 10.1128/jb.176.9.2754-2758.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung K.W., Vierling E. Conserved cell and organelle division. Nature. 1995;376:473–474. doi: 10.1038/376473b0. [DOI] [PubMed] [Google Scholar]
- Osteryoung K.W., Pyke K.A. Plastid divisionevidence for a prokaryotically derived mechanism. Curr. Opin. Plant Biol. 1998;1:475–479. doi: 10.1016/s1369-5266(98)80038-1. [DOI] [PubMed] [Google Scholar]
- Osteryoung K.W., Stokes K.D., Rutherford S.M., Percival A.L., Lee W.Y. Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ . Plant Cell. 1998;10:1991–2004. doi: 10.1105/tpc.10.12.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri D., Park J.T. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature. 1992;359:251–254. doi: 10.1038/359251a0. [DOI] [PubMed] [Google Scholar]
- Reski R., Faust M., Wang X.H., Wehe M., Abel W.O. Genome analysis of the moss Physcomitrella patens (Hedw.) B.S.G. Mol. Gen. Genet. 1994;244:352–359. doi: 10.1007/BF00286686. [DOI] [PubMed] [Google Scholar]
- Reski R., Reynolds S., Wehe M., Kleber-Janke T., Kruse S. Moss (Physcomitrella patens) expressed sequence tags include several sequences which are novel for plants. Bot. Acta. 1998;111:143–149. [Google Scholar]
- Rother S., Hadeler B., Orsini J.M., Abel W.O., Reski R. Fate of a mutant macrochloroplast in somatic hybrids. J. Plant Physiol. 1994;143:72–77. [Google Scholar]
- Rothfield L., Justice S., Garcia-Lara J. Bacterial cell division. Annu. Rev. Genet. 1999;33:423–448. doi: 10.1146/annurev.genet.33.1.423. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbour-joining methoda new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sprey B. Zur feinstruktur des plastidenstromas von Hordeum vulgare L. Protoplasma. 1968;66:469–479. [Google Scholar]
- Strepp R., Scholz S., Kruse S., Speth V., Reski R. Plant nuclear gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin. Proc. Natl. Acad. Sci. USA. 1998;95:4368–4373. doi: 10.1073/pnas.95.8.4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL Wimproving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ent F., Lockhart A., Kendrick-Jones J., Löwe J. Crystal structure of the NH2-terminal domain of MukBa protein involved in chromosome partitioning. Structure Fold Des. 1999;7:1181–1187. doi: 10.1016/s0969-2126(00)80052-0. [DOI] [PubMed] [Google Scholar]
- van de Peer Y., de Wachter R. TREECONA software package for the construction and drawing of evolutionary trees. Comput. Appl. Biosci. 1993;9:177–182. doi: 10.1093/bioinformatics/9.2.177. [DOI] [PubMed] [Google Scholar]
- Vellai T., Takacs K., Vida G. A new aspect to the origin and evolution of eukaryotes. J. Mol. Evol. 1998;46:499–507. doi: 10.1007/pl00006331. [DOI] [PubMed] [Google Scholar]
- Ward J.E., Jr., Lutkenhaus J. Overproduction of FtsZ induces minicell formation in E. coli . Cell. 1985;42:941–949. doi: 10.1016/0092-8674(85)90290-9. [DOI] [PubMed] [Google Scholar]