Abstract
We describe here a novel sarcomeric 145-kD protein, myopalladin, which tethers together the COOH-terminal Src homology 3 domains of nebulin and nebulette with the EF hand motifs of α-actinin in vertebrate Z-lines. Myopalladin's nebulin/nebulette and α-actinin–binding sites are contained in two distinct regions within its COOH-terminal 90-kD domain. Both sites are highly homologous with those found in palladin, a protein described recently required for actin cytoskeletal assembly (Parast, M.M., and C.A. Otey. 2000. J. Cell Biol. 150:643–656). This suggests that palladin and myopalladin may have conserved roles in stress fiber and Z-line assembly. The NH2-terminal region of myopalladin specifically binds to the cardiac ankyrin repeat protein (CARP), a nuclear protein involved in control of muscle gene expression. Immunofluorescence and immunoelectron microscopy studies revealed that myopalladin also colocalized with CARP in the central I-band of striated muscle sarcomeres. Overexpression of myopalladin's NH2-terminal CARP-binding region in live cardiac myocytes resulted in severe disruption of all sarcomeric components studied, suggesting that the myopalladin–CARP complex in the central I-band may have an important regulatory role in maintaining sarcomeric integrity. Our data also suggest that myopalladin may link regulatory mechanisms involved in Z-line structure (via α-actinin and nebulin/nebulette) to those involved in muscle gene expression (via CARP).
Keywords: α-actinin, nebulin, palladin, myopalladin, CARP
Introduction
The precise organization of Z-lines, the borders of individual sarcomeres in vertebrate striated muscle, is a remarkable example of supramolecular assembly in eukaryotic cells. Z-lines contain the barbed ends of actin thin filaments, the NH2-terminal ends of titin filaments, the COOH-terminal ends of nebulin filaments (skeletal muscle), and nebulette (cardiac muscle), as well as a variety of other regulatory and structural proteins. In addition to being a boundary between successive sarcomeres, Z-lines are responsible for transmitting tension generated by individual sarcomeres along the length of the myofibril, allowing for efficient contractile activity (for discussion see Vigoreaux 1994; Squire 1997). Z-line–associated proteins also appear to be crucial for early stages of myofibril assembly since I-Z-I structures (i.e., Z-line precursors) form the earliest identifiable protein assemblies observed during muscle differentiation (Epstein and Fischman 1991; Holtzer et al. 1997; Ehler et al. 1999; Ojima et al. 1999; Gregorio and Antin 2000). Thus, deciphering the molecular interactions of proteins forming Z-lines is pivotal for understanding the regulation of myofibril assembly, sarcomeric organization, and mechanical properties of striated muscle.
Detailed ultrastructural and biochemical investigations of the Z-disc and its various components have yielded valuable information concerning its structural architecture. The width of the Z-line can vary from ∼30 nm in fish skeletal muscle, up to >1 μm in patients with certain forms of nemaline myopathy (Franzini-Armstrong 1973; Rowe 1973; Yamaguchi et al. 1985; Vigoreaux 1994). The thin filaments from adjacent sarcomeres fully overlap within the Z-line and are cross-linked by the direct interaction of actin filaments and the actin filament barbed end–capping protein, CapZ, with α-actinin (Goll et al. 1972; Papa et al. 1999). Both the NH2-terminal region of titin and the COOH-terminal region of nebulin are also integral components of the Z-line lattice. Titin filaments from adjacent sarcomeres fully overlap in the Z-lines (Gregorio et al. 1998; Young et al. 1998). In vitro studies have identified two distinct α-actinin–binding sites within titin's NH2-terminal, 80-kD, Z-disc integral segment. These binding sites (the Z-repeats and titin's NH2-terminal sequences adjacent to the Ig-repeat Z4), may link together titin and α-actinin filaments both inside the Z-line and at its periphery (Ohtsuka et al. 1997; Sorimachi et al. 1997; Gregorio et al. 1998; Young et al. 1998). The ∼50-kD COOH-terminal region of nebulin also extends into the Z-line lattice in skeletal muscle: the Src homology (SH) 3 domain of nebulin is localized ∼25 nm inside the Z-line, whereas the more NH2-terminal repeating modules of nebulin are located at the periphery of the Z-line (Millevoi et al. 1998). Although cardiac muscle does not appear to contain nebulin, it contains a smaller, nebulin-related protein, nebulette (Moncman and Wang 1995). Nebulette is highly homologous to the COOH-terminal region of nebulin and it also extends into the Z-line. It has been suggested that this homology between nebulin and nebulette reflects conserved roles for their COOH-terminal ends in Z-line integration (Moncman and Wang 1995; Millevoi et al. 1998).
To date, the molecular interactions that are responsible for anchoring nebulin and nebulette filaments inside Z-lines are unknown, although previous models have proposed potential direct interactions between nebulin/nebulette and α-actinin (Nave et al. 1990; Moncman and Wang 1999). Here, we have used the yeast two-hybrid system to search for components that interact with the COOH-terminal (intra–Z-line) region of nebulin and thereby anchor it within the sarcomere. This approach identified a direct interaction between the Z-line peripheral region of nebulin and the intermediate filament protein, desmin; this link may contribute to the lateral connections between adjacent Z-lines. We also identified a novel 145-kD protein, myopalladin, that interacts with both the intra–Z-line, SH3 domain of nebulin and with α-actinin's EF hand region. Our in vitro binding data predict that myopalladin tethers the nebulin (directly), titin, and thin filaments (indirectly) via α-actinin filaments, thereby forming intra–Z-line meshworks. Additionally, we found that the NH2-terminal region of myopalladin interacts with the cardiac ankyrin repeat protein (CARP) (Chu et al. 1995; Baumeister et al. 1997; Jeyaseelan et al. 1997; Zou et al. 1997) within the I-band, an inducible protein implicated in the control of muscle gene expression (Zou et al. 1997). Our binding, immunolocalization, and cellular targeting data, together with the severe disruption of all sarcomeric components observed by the overexpression of NH2-terminal myopalladin fragments in live cardiac myocytes, suggest that myopalladin has important regulatory roles in the maintenance of sarcomere structure.
Materials and Methods
Yeast Two-Hybrid Interaction Studies
For a survey of potential nebulin and myopalladin interactions, cDNA fragments were amplified from rabbit psoas and human skeletal muscle cDNA by PCR (Saiki et al. 1985). The nebulin M160-M185+Ser+SH3 cDNA sequence was amplified with the primer pair 5′-tttgaattc CCT GAC ACC CCT CAG ATC CTC CTG GCA AAG and 3′-tttggatccc TCC TGC AGA CAG GTG TAA TAC TTG TAA GTA. For details on PCR conditions, see Centner et al. 2000. Amplified PCR fragments were inserted into pAS2-1 (Matchmaker system II; CLONTECH Laboratories, Inc.) or BTM117c (Wanker et al. 1997) vectors to obtain GAL4-BD or LexA-BD fusions, respectively. For screening, bait constructs were transformed into Saccharomyces cerevisiae, strain CG1945 (pAS2-1 bait) or L40c (BTM117c bait). Subsequently, the cells were transformed with a human skeletal muscle cDNA library in the pGAD10 prey vector (HL4010AB; CLONTECH Laboratories, Inc.) essentially as described by the manufacturer. Cells were plated onto SD/Leu-/Trp-/His-plates and incubated at 30°C until colonies appeared (after ∼5 d). In screens where the pAS2-1 bait vector was used, the plates were supplemented with 1,5 mM 3-amino-1,2,4-triazole (3-AT; Sigma-Aldrich).
Transformants were picked, restreaked onto SD/Leu-/Trp-/His-plates, and screened for β-galactosidase activity. β-Galactosidase activity of the cells was measured either by colony lift filter assays using X-gal or in liquid culture using chlorophenol red-β-d-galactopyranoside (cPRG) as described by the manufacturer (CLONTECH Laboratories, Inc.). β-Galactosidase–positive colonies were processed to loosen the bait plasmid, and prey clones were recovered by electroporation of yeast DNA in Escherichia coli and sequenced. To further confirm binding, transformants were retransformed into yeast with either the bait or the vector. In addition, the inserts of the bait and the prey vector were swapped and cotransformed into yeast.
For α-actinin–myopalladin interaction studies, a previously described α–actinin-2 deletion series in the pGAD424 vector was used (Sorimachi et al. 1997). Furthermore, the COOH-terminal part of α–actinin-3 containing two 4-EF hands cloned in pGAD10 was used (provided by Alan Beggs, Children's Hospital, Boston, MA). The sequence of all constructs was verified by sequencing.
cDNA Cloning and Sequence Analysis
The identified myopalladin prey cDNA corresponded to a 780-bp partial cDNA clone. The partial myopalladin cDNA was labeled randomly with 32P (Optiprime kit; Stratagene) and hybridized to a human heart cDNA library (number 936208; Stratagene). From a total of 400,000 screened clones, ∼80 clones hybridized to the probe. 24 myopalladin-positive phages were randomly picked and characterized by PCR, using combinations of specific internal primers and vector arm–derived primers. Clones extending 1–2 kb into the 5′ and 3′ directions were selected for sequencing. In total, three fragments extending towards the 5′ end, and one fragment extending towards the 3′ end provided a 5,696-bp cDNA. The presence of 5′ and 3′ untranslated regions, a putative start ATG at the 5′ end, and polyadenylation at the 3′ end indicated that the 5.7 kb cDNA corresponded to the complete myopalladin message (see Fig. 4 A). The complete myopalladin coding sequence was also amplified from a human skeletal muscle cDNA library by PCR (HL4010AB; CLONTECH Laboratories, Inc.) and sequenced. No sequence differences between cardiac and skeletal myopalladin were observed. All myopalladin fragments were sequenced on both DNA strands. Reads were edited and assembled using Geneskipper v1.2 software (Christian Schwager, European Molecular Biology Laboratory). For sequence analysis, Ig-I repeats were aligned using ClustalX (Higgins et al. 1996). Insertions were introduced to optimize the alignments. The complete myopalladin cDNA sequence from human heart and skeletal muscle is available from EMBL/GenBank/DDBJ under accession number AF328296.
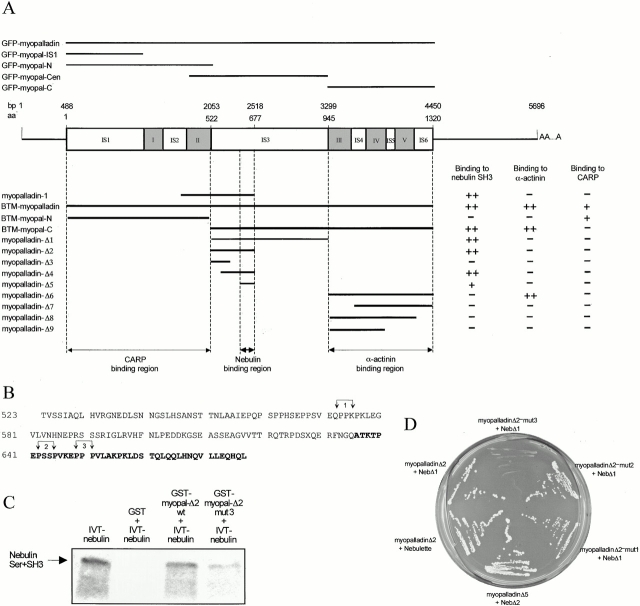
Figure 4.
Yeast two-hybrid screens reveal that myopalladin's nebulin-binding and α-actinin–binding sites are located in two distinct domains within its COOH-terminal 90-kD region. (A) Schematic structure of human full-length myopalladin cDNA. Numbers indicate nucleotide residue numbers (top, labeled bp) and amino acid residue numbers (below the nucleotide number, labeled aa). AAA indicates a poly(A+) tail. The Ig domains are shown in gray and are numbered I–V. Unique sequences are labeled IS1–6. Lines above the myopalladin domain structure indicate the myopalladin regions that were expressed as GFP fusion proteins in live cardiac myocytes (full-length myopalladin, two NH2-terminal fragments, one COOH-terminal fragment, and one internal fragment). Lines below indicate the partial myopalladin bait constructs that were generated for yeast two-hybrid screens to test for binding to the nebulin SH3 domain, α-actinin, and CARP. (+) and (−) denote the presence or absence of the growth of yeast colonies on SD/Trp-/Leu-/His-plates supplemented with 1.5 mM 3-AT. Myopalladin's nebulin-binding region was mapped to a part of IS3, whereas myopalladin's α-actinin–binding site was mapped to its COOH-terminal 375 residues. The CARP-binding site was assigned to myopalladin's NH2-terminal 522 residues. (B) The IS3 sequence of myopalladin contains three proline residue–rich motifs (labeled 1, 2, and 3). In each motif, pairs of proline residues (arrows) were mutated pairwise to glycine residues to obtain the bait constructs myopalladin-Δ2-mut1, 2, and 3. (C) Myopalladin interacted with nebulin in GST pull-down assays. In vitro–translated Ser+SH3 nebulin fragment (IVT-nebulin, lane 1) bound to glutathione-Sepharose 4B beads in the presence of GST-myopal-Δ2 wild-type (wt) fusion peptide (lane 3); a weaker binding was observed when proline residues 649 and 651 were mutated (lane 4; GST-myopal-Δ2-mut3 in B). As a negative control, binding of GST and nebulin Ser+SH3 to beads was tested (lane 2). (D) Interaction of myopalladin and nebulin/nebulette in the yeast two-hybrid system. Partial myopalladin cDNAs were cotransformed with partial nebulin/nebulette cDNAs into S. cerevisiae (see Fig. 3 A). The partial myopalladin sequence, myopalladin-Δ5 (IS3), was sufficient for binding to the SH3 domain of nebulin (Neb-Δ2). The myopalladin-Δ2-mut1,2 bait, but not the myopalladin-Δ2-mut3 bait interacted with nebulin preys, indicating that the PPP motif in myopalladin (residues 649–651) is involved in the interaction with nebulin. Additionally, myopalladin also bound to the SH3 domain of nebulette.
Protein Expression and Antibody Production
For obtaining specific antibodies against myopalladin and palladin, residues 264–430 and 1168–1320 of myopalladin and residues 301–385 of palladin (EMBL/GenBank/DDBJ accession number AB023209; see Fig. 4 for location of epitopes) were expressed as His6-tagged fusion proteins in the pET9D vector (Studier and Moffatt 1986). For generating antibodies against CARP, the NH2-terminal 93 residues of CARP were fused to His6 in the pET9D vector. Protein expression and purification were performed essentially as described (Studier et al. 1990). Polyclonal antibodies were raised in rabbits by Eurogentec and the specific IgG fraction was isolated by affinity chromatography. Western blot analysis was used to verify the specificity of the antibodies (see below).
In Vitro Transcription/Translation and GST Pull-down Experiments
In vitro transcription and translation were carried out in the presence of [35S]methionine (Amersham Pharmacia Biotech) using a TNT T7-coupled reticulocyte lysate system according to the manufacturer's instructions (Promega). His6–glutathione S-transferase (GST) double-tagged fusion proteins were obtained by cloning into a modified pET9D vector. The constructs were transformed into BL21-DE3 (CLONTECH Laboratories, Inc.) cells. Whole cell lysates in coating buffer (2× PBS, 1% Triton X-100) were prepared as described previously (Studier et al. 1990). The GST fusion protein was immobilized on glutathione-Sepharose 4B beads (Amersham Pharmacia Biotech) by incubating ∼200 μl of the lysate with 50 μl of beads (50% slurry) for 1 h at 4°C. The beads were washed three times with coating buffer and resuspended in binding buffer (20 mM Tris, pH 7.4, 100 mM KCl, 1 mM EDTA, 1% Triton X-100, plus protease inhibitors). 5 μl of in vitro–translated 35S-labeled proteins were added to 50 μl of beads coated with 20 μg of bound GST fusion proteins in 300 μl binding buffer. The mixture was incubated for 1.5 h at 4°C, washed three times with binding buffer, and resuspended in SDS sample buffer. The protein complexes were fractionated by SDS-PAGE using 15% gels. The gels were fixed (20% methanol, 10% acetic acid), stained with Coomassie blue, treated with Amplify (Amersham Pharmacia Biotech), dried, and exposed using BioMax MR-1 film (Eastman Kodak Co.). The results of the Coomassie blue staining confirmed that equal amounts of each GST protein were bound to the beads in the different samples (data not shown).
Northern and Western Blot Analysis
For determining myopalladin and palladin's tissue-specific transcription, the full-length coding sequence of myopalladin and the subfragment bp 1289–4172 of palladin (EMBL/GenBank/DDBJ accession number AB023209) was randomly labeled with 32P using the Prime-It RmT kit (Stratagene) and purified on a NucTrap column (Stratagene). The myopalladin-specific probe was hybridized to a multiple tissue Northern blot (CLONTECH Laboratories, Inc.) as described by the manufacturer. For protein expression analysis, Western blots were probed with rabbit affinity-purified antimyopalladin, antipalladin, and anti-CARP polyclonal antibodies essentially as described previously (Gregorio et al. 1995). Snap-frozen heart, skeletal, or smooth muscle was ground in liquid nitrogen into a fine powder using a mortar and pestle. The muscle powder was solubilized in 2× SDS sample buffer, boiled for 5 min, and sonicated. Solubilized proteins were fractionated by SDS-PAGE using 8% gels and transferred to a nitrocellulose membrane. 0.5 μg/ml of affinity-purified rabbit antimyopalladin-1 IgG was used to probe rabbit heart, soleus, and psoas muscle lysates. 2.0 μg/ml of affinity-purified rabbit antipalladin IgG was used to probe rat heart, psoas, and small intestine (source of smooth muscle) lysates. 1.0 μg/ml of affinity-purified rabbit anti-CARP IgG was used to probe rat heart lysates. Primary antibody incubations were followed by HRP-conjugated goat anti–rabbit IgG (1:25,000; Jackson ImmunoResearch Laboratories). Both primary and secondary antibody incubations were for 1 h at 4°C. The blots were incubated in Super Signal chemiluminescent substrate (Pierce Chemical Co.) according to the manufacturer's instructions and exposed to Biomax MR film (Eastman Kodak Co.).
Cell Culture and Transfection Procedures
For myocyte expression studies, the entire myopalladin open reading frame (bp 488–4450) and subfragments 488–1327, 488–2254, 1789–3299, and 3300–4450 (see Fig. 3) were amplified by PCR and cloned into pEGFP-C1 (CLONTECH Laboratories, Inc.). Recombinant pEGFP-C1 constructs were purified using QIAGEN columns before transfection into myocytes. Plasmids were verified by sequencing. To rule out any potential artifacts resulting from the green fluorescent protein (GFP) tag, pCMVmyc-myopalladin constructs were also generated (as in Gregorio et al. 1998) and tested in overexpression studies: identical results were obtained (data not shown).
Figure 3.
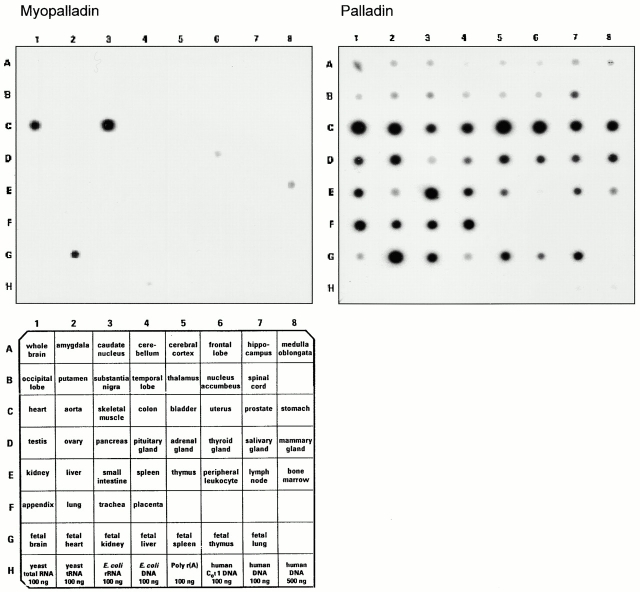
Northern blot analysis reveals that detectable myopalladin gene expression is restricted to striated muscle. Myopalladin- and palladin-specific cDNA probes were hybridized to human RNA master blots (CLONTECH Laboratories, Inc.). Left, myopalladin gene expression is restricted to skeletal muscle (C3), fetal heart (G2), and adult heart (C1). Right, palladin transcripts are detected in all human tissues tested and are particularly abundant in striated and smooth muscle tissues (e.g., bladder, uterus, prostate, and stomach, C5–C8, respectively).
Cardiac myocytes were prepared from day 6 embryonic chick hearts and cultured as described previously (Gregorio and Fowler 1995). Isolated cells were plated in 35-mm tissue culture dishes containing 12-mm round coverslips (106 cells/dish). 15–20% of the cells in our primary cultures are fibroblasts. 24 h after plating, cultured myocytes were washed two times in OptiMEM, placed in 800 μl fresh OptiMEM, and returned to the incubator while DNA liposome complexes were prepared. DNA liposome complexes were prepared by combining 0.5 or 1 μg plasmid with 4 μl LipofectAmine and 6 μl PLUS Reagent in 200 μl serum-free OptiMEM. After 15 min, the complexes were added dropwise to the culture dish. 3 h later, 1 ml of MEM/10% FBS (Hyclone Laboratories, Inc.) was added to the dish. 3–6 d later, cells were gently washed with PBS and fixed with 2% formaldehyde in PBS for 10 min. Coverslips were washed and stored in PBS at 4°C until staining. Over 200 transfected cells per construct were analyzed. Our transfection efficiencies ranged from 10 to 50%. All tissue culture reagents (except where noted) were purchased from Life Technologies.
Immunoelectron Microscopy
Cardiac muscles were dissected from the left ventricular wall of hearts rapidly excised from 8–9-wk-old mice (Balb/C). Muscle strips were skinned in relaxing solution containing 1% vol/vol Triton X-100 overnight at 4°C. After extensive washing with relaxing solutions, the muscles were fixed in 3% formaldehyde/PBS solution, blocked for 1 h with PBS supplemented with 1% BSA, and labeled with the primary antibodies (typically 50 μg/ml), followed by washing. Affinity-purified Fab′ fragments, raised in goat against the whole rabbit IgG molecule, were used as secondary antibodies (no. 2004; Nanoprobes, Inc.). Secondary antibodies were conjugated with 1.4-nm gold particles and were further intensified with HQ silver developer for 4 min. For more details, see Trombitás et al. 1995 and Gregorio et al. 1998. Z-disc–to-epitope distances were measured from electron micrographs after high-resolution scanning (UC-1260; UMAX Data System, Inc.) as described in Gregorio et al. 1998.
Indirect Immunofluorescence Microscopy
Rat cardiac and skeletal muscle myofibrils were isolated and washed according to Knight and Trinick 1982, as modified by Fowler et al. 1993. Primary cultures of rat cardiac myocytes were isolated and maintained as described (Gustafson et al. 1987). Myofibrils attached to poly-d-lysine (Sigma-Aldrich) coated coverslips were fixed in 2% formaldehyde/PBS for 10 min, washed in PBS, and permeabilized in 0.2% Triton X-100/PBS for 15 min. The coverslips were preincubated in 2% BSA, 1% normal donkey serum/PBS for 1 h to minimize nonspecific binding of antibodies. Isolated myofibrils and cells were incubated with affinity-purified rabbit polyclonal antibodies specific to myopalladin, palladin, or CARP (5–10 μg/ml1), followed by Cy-2–conjugated goat anti–rabbit IgG (1:600). For double-labeling protocols, myofibrils and cells were also incubated with antimyomesin monoclonal antibodies (1:20; provided by Perriard, J.-C., and E. Ehler (Institute for Cell Biology, Zurich, Switzerland; Grove et al. 1985), followed by Texas red–conjugated goat anti–mouse IgG+IgM (1:600). To confirm the location of precursor I-Z-I (i.e., precursor Z-lines/stress-fiber–like) structures in cultured cardiac myocytes, cells were also double-stained with Texas red–conjugated phalloidin (data not shown).
Transfected cardiac myocytes were essentially stained as described (Gregorio et al. 1998). Six well-characterized antibodies specific to various sarcomeric components and phalloidin were used to analyze sarcomeres in transfected cells. The transfected cells were stained with monoclonal sarcomeric anti–α-actinin antibodies (1:1,500, EA-53; Sigma-Aldrich) or rabbit polyclonal α-actinin antibodies (1:2,000; provided by Dr. S. Craig, Johns Hopkins University, Baltimore, MD), antititin T11 monoclonal antibodies (1:1,000; Sigma-Aldrich), monoclonal antimyomesin B4 antibodies (1:20; see above), or Texas red–conjugated phalloidin (Molecular Probes, Inc.) for 1 h and washed in PBS. Subsequently, the cells were stained with Texas red–conjugated goat anti–mouse IgG+IgM (1:600) or Texas red–conjugated donkey anti–rabbit antibodies (1:700) for 45 min. All coverslips were mounted on slides using Aqua Poly/Mount (Polysciences, Inc.) and subsequently analyzed on an Axiovert microscope (ZEISS) using 63× (NA 1.4) or 100× (NA 1.3) objectives, and micrographs were recorded as digital images on a SenSys cooled HCCD (Photometrics). Images were processed for presentation using Adobe Photoshop® 5.0. All secondary antibodies were purchased from Jackson ImmunoResearch Laboratories, except the Cy2-conjugated antibodies that were purchased from Pierce Chemical Co.
Results
Interaction of the COOH-terminal Region of Nebulin with Desmin and Myopalladin
To search for proteins interacting with the Z-disc region of nebulin, a fragment corresponding to its COOH-terminal 2,687 kb was amplified from rabbit psoas skeletal muscle and inserted into the yeast two-hybrid bait vector pAS2-1. The bait, referred to as pAS2-M160-M185+Ser+SH3, included the nebulin repeats, M160–M176 and M182–M185, the Ser-rich region, and the SH3 domain (Fig. 1). The nebulin repeats M177–M181 are absent from rabbit psoas muscle nebulin (Millevoi et al. 1998). When screening 700,000 clones from a human skeletal library with pAS2-M160-M185+Ser+SH3, >1,000 potentially interacting clones were identified. From these, 100 clones were randomly picked and analyzed for β-galactosidase expression. 12 clones with the highest levels of β-galactosidase expression were sequenced. One clone corresponded to a partial cDNA, which encoded one Ig repeat and was derived from a palladin-homologous gene (Parast and Otey 2000) described here as “myopalladin.” The full-length cDNA sequence of myopalladin was obtained by the screening of both human skeletal and heart muscle cDNA libraries and performing extensions to reach the 5′ and 3′ end. This identified a 5.7-kb full-length cDNA for myopalladin (see Materials and Methods), which contains a single open reading frame and encodes a protein of 1,320 amino acids with a predicted molecular mass of 145 kD (see Fig. 4 A). We found no evidence for differentially spliced myopalladin isoforms in heart and skeletal muscle (not shown).
Figure 1.
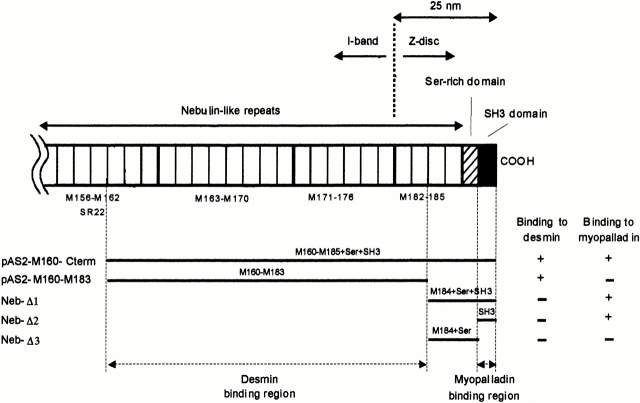
Yeast two-hybrid screens identify myopalladin and desmin as Z-line nebulin-binding proteins. Schematic structure of the COOH-terminal region of nebulin and its location within the sarcomere is shown based on previous immunoelectron microscopy studies (Millevoi et al. 1998). The nebulin COOH-terminal region constructs pAS2-M160-M185+Ser+SH3C and pAS2-M160-M183 identified myopalladin and desmin prey clones in yeast two-hybrid screens. Nebulin bait deletion constructs (NebΔ1–NebΔ3) allowed us to further map the myopalladin-binding region on nebulin to within the nebulin SH3 domain. The desmin-binding site on nebulin was found to be located within the nebulin M160-M183 modules. (+) and (−) denote the presence or absence of the growth of yeast colonies on SD/Trp-/Leu-/His-plates supplemented with 1.5 mM 3-AT.
Another clone corresponded to the intermediate filament protein, desmin (Vicart et al. 1996). The remaining 10 prey clones were devoid of open reading frames, or corresponded to ribosomal and nuclear proteins. Therefore, we assumed that a large amount of prey clones identified in this screen corresponded to false positive clones.
Subsequently, we performed yeast two-hybrid screens with subfragments of pAS2-M160-M185+Ser+SH3. pAS2-M160-M183 (Fig. 1) identified 23 prey clones from the screening of 800,000 clones. 21 of the prey clones corresponded to desmin (Vicart et al. 1996), thus confirming the interaction identified in the initial screen with pAS2-M160-M185+Ser+SH3 (Fig. 1). Further analysis of desmin and nebulin's interaction will be presented elsewhere (Bang, M.-L., and S. Labeit, manuscript in preparation). In screenings performed with pAS2-M184-M185+Ser+SH3, as well as pAS2-SH3 (Fig. 1), a high frequency of prey clones was identified, suggesting that the presence of the nebulin SH3 domain in the bait was responsible for the high occurrence of false positives in the first screen. Therefore, our clones, in particular those interacting with pAS2-SH3 in the two-hybrid system, were analyzed in detail by GST pull-down assays. The interaction of nebulin Ser-SH3 with myopalladin was confirmed by GST pull-down assays (see Fig. 4 D).
In summary, we conclude that the COOH-terminal end of nebulin, which is located ∼25 nm inside the Z-line lattice, specifically interacted with the novel 145-kD sarcomeric protein, myopalladin. Studies in progress are focused on characterizing further the interaction of nebulin M160–M183, shown previously to be located in the periphery of the Z-line (Millevoi et al. 1998), with desmin.
Analysis of Myopalladin's Primary Structure and Its Expression Patterns
Analysis of the primary structure of myopalladin revealed the presence of five Ig domains from the Ig-I subset (Harpaz and Chothia 1994; designated I–V in Fig. 2 A) in its 1,320-residue sequence. Six interdomain insertions (designated IS1–IS6) separate the five Ig repeats. To search for proteins homologous to myopalladin, a BLAST search was performed with its 145-kD peptide sequence (BLAST2 from the National Center for Biotechnology Information server). From known proteins, Ig repeats from palladin (human: EMBL/GenBank/DDBJ under accession number AB023209; mouse: Parast and Otey 2000) (68% identity), the Z-disc protein myotilin (Salmikangas et al. 1999) (50% identity), and titin's central I-band N2B region (48% identity) were most related to myopalladin (Fig. 2 C). The two NH2-terminal Ig domains in myopalladin are most homologous to Ig domains from the N2B region of titin in the I-band region. Myopalladin's three COOH-terminal Ig domains are most related to palladin's Ig domains, whereas the two most COOH-terminal Ig domains also share high homology to myotilin's Ig domains (Fig. 2 D).
Figure 2.
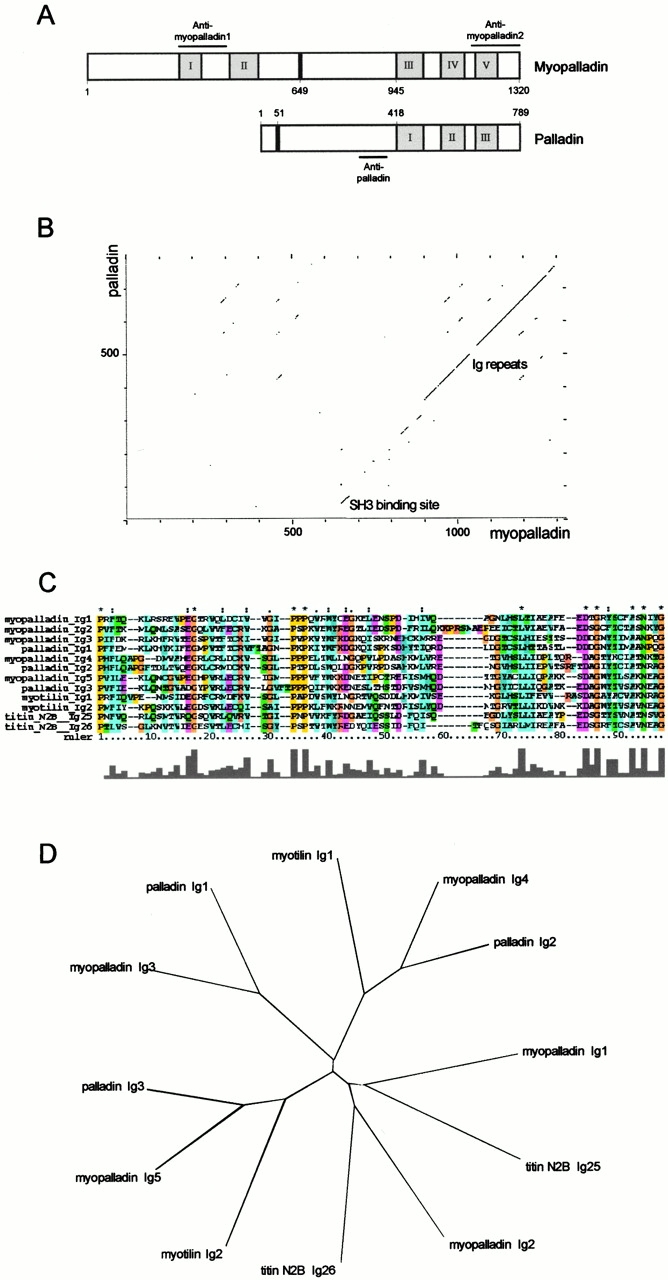
Myopalladin is a novel 145-kD protein with homology to palladin. (A) Comparison of the domain architecture of myopalladin and the recently described palladin protein (Parast and Otey 2000). The Ig-I repeats are shown in gray, the unique sequences are shown in white, and the proline-rich PPP motif regions are shown in black. The bars above the myopalladin and below the palladin schematics indicate the recombinant fragments of the proteins that were used as antigens to generate specific antibodies (see Fig. 7 for the characterization of the antibodies). (B) Dot plot matrix sequence comparison of myopalladin and palladin. The regions of myopalladin and palladin that bind to the nebulin SH3 domain and to α-actinin are the most conserved regions between the two proteins. (C) Peptide sequence alignment of myopalladin's five Ig-I domains and comparison with Ig-I domains from myotilin (EMBL/GenBank/DDBJ accession number AF144477), titin N2B (accession number X90568), and palladin (accession number AB023209). All sequences are from humans. Identical residues are indicated by asterisks. (D) Phylogenetic tree based on the comparison of myopalladin's Ig-I repeats and the most related Ig-I repeats from myotilin, titin, and palladin.
The sequence homology of myopalladin to the recently described 90–92-kD palladin protein from mouse is most significant, because palladin's entire sequence, including its COOH-terminal three Ig repeats and its unique NH2-terminal domain, is colinear with and conserved to myopalladin's COOH-terminal 92 kD (Fig. 2 B). In particular, the COOH-terminal part of palladin containing three Ig domains is 63% identical to myopalladin's COOH-terminal region (Fig. 2A and Fig. B). Also highly conserved to palladin is a 15-residue box in myopalladin (residues 643–658; Fig. 2 B), which corresponds to myopalladin's SH3-binding site (see below). Palladin's PPPPP domains and Ser-rich domains, noted by Parast and Otey 2000, are not found in myopalladin (corresponding to gaps in Fig. 2 B).
By searching the human genome database with myopalladin and palladin cDNAs, the myopalladin gene was shown to be derived from a single locus, located on chromosome 10q21.1 between the markers D10S207 and D10S561. Palladin was mapped to a single locus on chromosome 4q31.3 between the markers D4S1596 and D4S2910. To determine the expression pattern of the myopalladin gene, a specific probe corresponding to its full-length cDNA sequence was hybridized to a multiple tissue RNA blot. Myopalladin gene expression was found to be restricted in adult to striated muscle tissues (Fig. 3). Consistent with the results of Parast and Otey 2000, the palladin gene was found to be ubiquitously expressed in all tested tissues. The highest levels of palladin expression were noted in cardiac muscle and in tissues with a high content of smooth muscle (Fig. 3).
In conclusion, myopalladin and palladin are members of the same gene family which have conserved COOH-terminal and SH3-binding domains. Palladin is encoded by a gene located on chromosome 4q31.3 and is ubiquitously expressed, whereas the myopalladin gene on chromosome 10q21.1 encodes a 145-kD protein, whose expression is restricted to striated muscle tissues (explaining the name given to this protein).
Myopalladin's Central IS3 Domain Interacts with Nebulin and Nebulette's SH3 Domain
To identify more precisely the sequences responsible for the nebulin–myopalladin interaction, nebulin and myopalladin subfragments were cotransformed into yeast (Fig. 1 and Fig. 4 A). Since the nebulin bait, pAS2-M160-M185+Ser+SH3, was partly autoactivating, nebulin truncations were made in the prey vector and myopalladin truncations in the bait vector. Truncations of pAS2-M160-M185+Ser+SH3 indicated that nebulin's SH3 domain alone is sufficient for the interaction with myopalladin (Fig. 1). The additional presence of the Ser-rich domain, however, further enhanced yeast growth and β-galactosidase activity and may therefore be responsible for strengthening the interaction (Fig. 1).
Truncation analysis of myopalladin indicated that a 42-residue proline-rich stretch within IS3 is sufficient for the interaction with nebulin's SH3 domain (Fig. 4 A). Binding sites for SH3 domains are typically proline-rich stretches of ∼10 amino acids, which are capable of adopting a polyproline II helix conformation (Musacchio et al. 1992). Within myopalladin's IS3 domain, three such proline-rich motifs are present. To determine if these motifs are responsible for SH3-binding, pairs of proline residues in myopalladin-Δ2 (Fig. 4 A), within three predicted potential SH3-binding motifs, were mutated to glycines (myopalladin-Δ2-mut1,2,3; Fig. 4 B). Cotransformation of myopalladin-Δ2-mut3 with nebulin indicated that myopalladin's proline residue triplet (PPP), residues 649–651, are required for binding, since this mutation abolished the interaction. The other two mutants, myopalladin-Δ2-mut1,2 (residues 573–576 and 642–645) did not affect nebulin binding (Fig. 4 D). The interaction of nebulin M184+ Ser+SH3 and myopalladin-Δ2/myopalladin-Δ2-mut3 fusion peptides was also tested by GST pull-down assays. Nebulin's COOH-terminal region coprecipitated with the wild-type GST–myopalladin-Δ2 but not with GST alone and only weakly with the GST–myopalladin-Δ2-mut3 peptide (Fig. 4 C). This confirmed that the nebulin SH3 domain specifically binds to myopalladin, involving the PPP motif, residues 649–651, which is located within myopalladin's central IS3 domain (Fig. 4 A).
Finally, as nebulette is thought to be a functional homologue of the Z-disc part of nebulin in the heart (Moncman and Wang 1995; Millevoi et al. 1998), the Ser+SH3 domains of nebulette from human heart were cotransformed with myopalladin-Δ2. This experiment demonstrated that the SH3 domain of nebulette also interacts with myopalladin (Fig. 4 D). Therefore, both the SH3 domains of skeletal muscle nebulin and cardiac muscle nebulette specifically interact with myopalladin.
Myopalladin's COOH-terminal Region Interacts with the COOH-terminal, EF Hand Region of α-Actinin
To search for other myopalladin binding partners, we screened a human skeletal muscle cDNA library with a bait containing myopalladin's full-length coding sequence (BTM-myopalladin; Fig. 4 A). By screening of ∼1.2 × 106 colonies, 700 prey clones were identified, all of which were confirmed in the β-galactosidase assay. 65 β-galactosidase–expressing clones were randomly chosen and sequenced. Three of the prey clones were derived from the COOH-terminal part of nebulin, confirming again the nebulin SH3–myopalladin interaction. 13 of the prey clones contained parts of the ACTN2 gene, which encodes the α–actinin-2 isoform (Beggs et al. 1992). When hybridizing α–actinin-2 probes to the 700 yeast colonies from the myopalladin screen, 155 clones (22%) hybridized to ACTN2. Sequence analysis of 12 ACTN2-positive clones showed that they all share a region of overlap in the COOH-terminal region of sarcomeric α–actinin-2. To further narrow down the myopalladin-binding site within α–actinin-2, the interaction of myopalladin with various deletion constructs of α–actinin-2 (Sorimachi et al. 1997) was tested (Fig. 5 A). This analysis showed that α-actinin's binding site for myopalladin is located in the COOH-terminal part of α–actinin-2, from just after the spectrin repeats (before the two EF hands) up to the extreme COOH terminus. This interaction was confirmed by GST pull-down assays (Fig. 5 B).
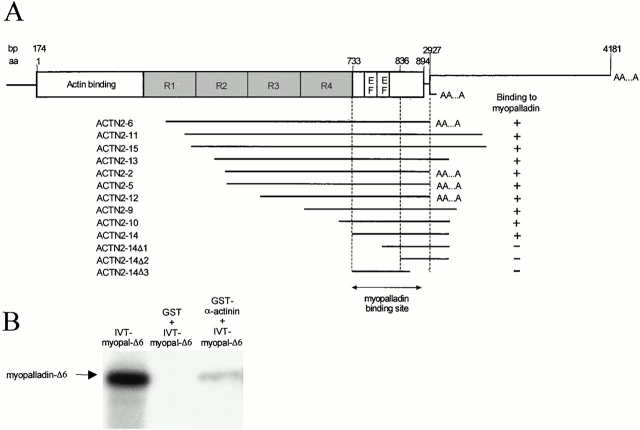
Figure 5.
Myopalladin interacts with the EF hand region of α–actinin-2. (A) The schematic structure of α–actinin-2 (ACTN2) with its four central rod domains, R1–R4, and its unique terminal sequence is shown. Numbers indicate the nucleotide or amino acid residue numbers deduced from the human full-length cDNA sequence. AAA indicates a poly(A+) tail. A series of ACTN2 deletion constructs (below) were tested for interaction with full-length myopalladin in the yeast two-hybrid system. This assigned the myopalladin-binding domain within α-actinin to its COOH-terminal region containing the two EF hands. (+) and (−) denote the presence or absence of the growth of yeast colonies on SD-/Trp-/Leu-/His-plates supplemented with 1.5 mM 3-AT. (B) Interaction of myopalladin with α-actinin in GST pull-down assays. Myopalladin's COOH-terminal 376 residues were translated in vitro (IVT-myopal-Δ6, left). When incubated together with expressed ACTN14-GST fusion peptides, their binding to glutathione-Sepharose 4B beads was observed (right). Middle lane, negative control pull-down performed without the addition of the α-actinin fragment (for details, see Materials and Methods).
Our two-hybrid screens isolated only α–actinin-2 isoforms. To test if the interaction with myopalladin was specific to the α–actinin-2 isoform, we also tested α–actinin-3 prey constructs, encoding a skeletal muscle–specific isoform of α-actinin (Beggs et al. 1992). This showed that α–actinin-3 also interacted with myopalladin (data not shown).
To determine the sequences in myopalladin that are required for binding to α-actinin, truncated myopalladin constructs were tested (Fig. 4 A). Using this approach, myopalladin's α-actinin–binding site was assigned to the COOH-terminal region of myopalladin. The results indicated that all three Ig domains, III+IV+V within the COOH-terminal region of myopalladin, are required for binding to α–actinin-2. We conclude that myopalladin's COOH-terminal 43 kD and α-actinin's COOH-terminal EF hand region specifically interact.
Myopalladin's NH2-terminal Domain Interacts with Sarcomeric CARP
To search for proteins interacting with myopalladin's NH2-terminal domain, the NH2-terminal 1,605 kb was cloned into the yeast two-hybrid bait vector BTM117c and used to screen a human skeletal muscle cDNA library. By screening of ∼900,000 clones, 100 clones were identified. 88 were confirmed in the β-galactosidase assay, and 20 were analyzed by sequencing. Six of the sequenced clones encoded the cardiac ankyrin repeat protein, CARP (Chu et al. 1995). All six CARP clones corresponded to full-length cDNAs. Also, when 5′ and 3′ deletions were introduced into CARP, the interaction with myopalladin's NH2-terminal domain was ablated, suggesting that a full-length version of CARP is required for its interaction with myopalladin (data not shown). The specific interaction of myopalladin's NH2-terminal region with a full-length CARP protein was confirmed by GST pull-down experiments (Fig. 6 A).
Figure 6.
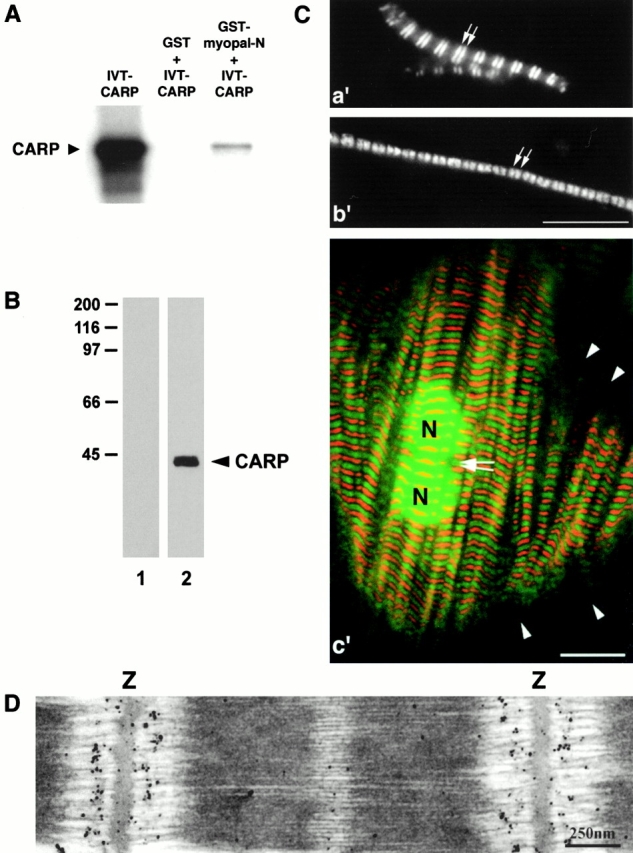
Identification of CARP as a sarcomeric component. (A) Interaction of myopalladin with CARP in GST pull-down assays. Full-length CARP was translated in vitro (left). When incubated together with GST-myopalladin fusion peptide (fragment myopal-N; see Fig. 4 A), binding to glutathione-Sepharose 4B beads was observed (right). Middle lane, negative control pull-down performed with GST and CARP peptides. (B) Specificity of affinity-purified anti-CARP antibodies. Western blot analyses of rat heart muscle lysates reveal that polyclonal antibodies raised to CARP react with a single 40-kD band (lane 2). No bands are detected in the control lane, which was incubated with secondary antibody alone (lane 1). (C) Immunofluorescence staining of CARP in washed isolated myofibrils from rat heart (a′) and skeletal (b′) muscle, as well as in primary cultures of rat cardiac myocytes (c′; CARP staining in green, myomesin staining in red), demonstrating that CARP is a sarcomeric component. Isolated myofibrils and cardiac myocytes were labeled with affinity-purified anti-CARP antibodies, followed by Cy2-conjugated secondary antibodies, and with antimyomesin antibodies (data not shown in a′ and b′) followed by Texas red–conjugated secondary antibodies. The merged images revealed that CARP is localized as a doublet within the I-band (in close proximity to the Z-line) in both heart and skeletal myofibrils and in isolated cardiac myocytes. Double arrows in a′ and b′ mark I-band staining. Arrowheads (in c′) indicate the absence of detectable CARP (and myomesin) staining in I-Z-I bodies, located at the edges of cultured cardiac myocytes. Note the additional staining of CARP in the nucleus in c′. N, nucleus. (D) Immunoelectron microscopy with CARP-specific antibodies demonstrates that CARP is present in the central I-band in cardiac myofibrils from mouse left ventricle. Z, Z-disc. Bars: (C) 10 μm; (D) 250 nm.
Endogenous CARP has been detected previously in the nucleus and no sarcomeric forms have been reported (e.g., Zou et al. 1997). Therefore, to test if endogenous CARP is indeed present in the sarcomere, a novel antibody was raised to an NH2-terminal subfragment of CARP that appears to be specific for CARP. On Western blots, this antibody recognized a single 40-kD band which corresponded to the previously reported (Jeyaseelan et al. 1997; Zou et al. 1997) molecular mass of CARP protein (Fig. 6 B). To ascertain the cellular localization of CARP in striated muscle, we stained isolated myofibrils from rat heart and skeletal muscle, as well as isolated rat cardiac myocytes with our anti-CARP–specific antibody. Consistent with earlier data on the nuclear targeting of CARP, endogenous CARP was detected within the nucleus of rat cardiac myocytes (Fig. 6 C, c′; CARP staining in green, myomesin staining in red). However, endogenous CARP protein was also detected in a striated pattern in both isolated myofibrils and in cardiac myocytes, indicating that CARP is a myofibrillar component. Costaining with a marker of the M-line, the myosin-associated protein, myomesin, revealed that CARP is distributed in the I-band, as a doublet in close proximity to the Z-line in both heart and skeletal muscle (Fig. 6 C, a′ and b′, myomesin staining not shown). Notably, CARP was not detected in I-Z-I (i.e., precursor Z-line/stress-fiber–like) structures at the edges of cardiac myocytes in culture (Fig. 6 C, c′, arrowheads). To determine more precisely the localization of sarcomeric CARP, immunoelectron microscopy was performed on mouse cardiac myofibrils. This experiment confirmed that a CARP protein is present in the central I-band region of the sarcomere (Fig. 6 D).
Localization and Assembly of Myopalladin and Palladin in Striated Muscle
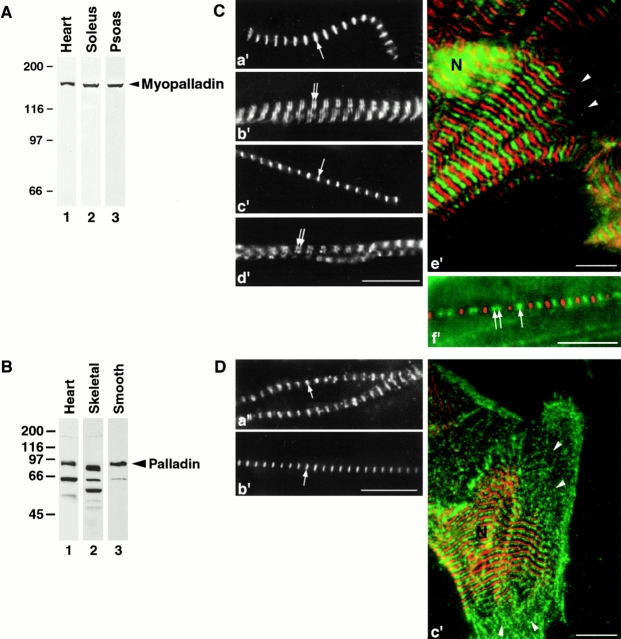
To study the endogenous localization of myopalladin and palladin, specific antibodies to expressed myopalladin and palladin fragments were raised (Fig. 2 A). Western blot analysis of different tissues using our affinity-purified antimyopalladin-1 antibodies detected a single band of ∼155 kD in rabbit cardiac, soleus, and psoas skeletal muscle (Fig. 7 A). This slightly slower mobility of myopalladin observed on gels than expected from its predicted molecular mass raises the possibility of posttranslational modifications (e.g., differential phosphorylation). For palladin, a single, prominent band at ∼92 kD was detected with our polyclonal antipalladin antibodies in smooth muscle (Parast and Otey 2000), whereas bands at ∼90–92 and ∼60 kD in heart, as well as a band at ∼55 kD in skeletal muscle, were observed (Fig. 7 B). With longer exposure times, reactivity to a faint band at ∼155 kD was also detected in cardiac and skeletal muscle. This could result from the cross-reactivity of our polyclonal antipalladin antibodies with the homologous myopalladin protein. However, since the palladin antibodies were raised to a region which is not homologous to myopalladin, it is not likely that the antipalladin antibodies cross-react with myopalladin. Additionally, by immunofluorescence staining the antimyopalladin and antipalladin antibodies appear not to cross-react, since both antibodies demonstrated differences in their staining patterns (Fig. 7). The results from our Western blot analysis are consistent with those reported in adult tissues using monoclonal antipalladin antibodies in Parast and Otey 2000, with the exception that our polyclonal antipalladin antibodies appeared to recognize more palladin-related proteins (although the smaller proteins we detected may not have been resolved on their gel system).
Figure 7.
Characterization of endogenous myopalladins and palladins in striated muscle. (A) Specificity of affinity-purified antimyopalladin antibodies. Antimyopalladin antibodies recognize a band at ∼155 kD in rabbit heart (lane 1), soleus (lane 2), and psoas muscle (lane 3) by Western blot analysis. Note, on some blots an ∼35-kD band was also detected; the detection of this band was variable and its significance is unknown. (B) Specificity of affinity-purified antipalladin antibodies. Antipalladin antibodies recognize a 92-kD band in rat smooth muscle from the small intestine (lane 3), as well as multiple bands in heart (lane 1) and skeletal muscle (lane 2). (C) Immunofluorescence staining of myopalladin in washed, isolated myofibrils from rat heart (a′ and b′) and skeletal (c′ and d′) muscle, as well as in primary cultures of rat cardiac myocytes (e′; myopalladin staining in green, myomesin staining in red) demonstrating that myopalladin can be detected as a single striation at the Z-line (a′ and c′, arrows) and as a doublet within the I-band (in close proximity to the Z-line) (b′ and d′, double arrows). Isolated myofibrils and cardiac myocytes were labeled with affinity-purified antimyopalladin-1 antibodies, followed by Cy2-conjugated secondary antibodies, and with antimyomesin antibodies (data not shown in a′–d′) followed by Texas red–conjugated secondary antibodies. Note the additional staining of myopalladin in the nucleus in e′. Arrowheads in e′ mark the absence of detectable myopalladin (and myomesin) staining in I-Z-I bodies, located at the edges of cultured cardiac myocytes. (f′) Immunofluorescence image demonstrating the targeting of expressed GFP-myopalladin to the Z-line and to the I-band in primary cultures of chick cardiac myocytes. Cardiomyocytes expressing GFP–full-length myopalladin were fixed 3–5 d after transfection and stained with antimyomesin antibodies followed by Texas red–conjugated secondary antibodies and analyzed by immunofluorescence microscopy. Single arrows mark Z-line staining, whereas double arrows mark I-band staining. Significant variability in the relative labeling intensities of myopalladin at the Z-line vs. the I-band was observed. N, nucleus. (D) Immunofluorescence staining of palladin in washed, isolated myofibrils from rat heart (a′) and skeletal (b′) muscle, as well as in primary cultures of rat cardiac myocytes (c′; palladin staining in green, myomesin staining in red), demonstrating that palladin is localized at the Z-line. Isolated myofibrils and cardiac myocytes were labeled with affinity-purified antipalladin antibodies, followed by Cy2-conjugated secondary antibodies, and with antimyomesin antibodies (data not shown in a′ and b′) followed by Texas red–conjugated secondary antibodies. Arrowheads in c′ mark the presence of palladin (but not myomesin) staining in I-Z-I bodies, located at the edges of cultured cardiac myocytes. Note that palladin staining was not detected in the nucleus in c′. N, nucleus. Bars, 10 μm.
To ascertain the cellular localization of myopalladin in striated muscle, we performed immunofluorescence staining with our antimyopalladin-1–specific antibodies on isolated rat heart and skeletal myofibrils. Costaining with a marker of the M-line, the myosin-associated protein myomesin, revealed that myopalladin is localized at the Z-line region in both heart and skeletal myofibrils (Fig. 7 C, a′ and c′); however, in some sarcomeres, a doublet of myopalladin staining is detected, indicating that this protein is also localized within the I-band (Fig. 7 C, b′ and d′; staining with myomesin not shown). In isolated rat cardiac myocytes, myopalladin is also detected (with varying intensities) within the nucleus of ∼75% cells (Fig. 7 C, e′; myopalladin staining in green, myomesin staining in red). Palladin was also detected at the Z-line (but not within the I-band) in isolated rat heart and skeletal myofibrils (Fig. 7 D, a′ and b′; staining for myomesin not shown). No staining for palladin was detected in the nucleus in rat cardiac myocytes (Fig. 7 C, c′; palladin staining in green, myomesin in red). Furthermore, palladin (Fig. 7 D, c′) but not myopalladin (Fig. 7 C, c′) was detected in I-Z-I bodies at the edges of the cardiac myocytes.
To determine the cellular localization of myopalladin when being expressed in primary cultures of chick cardiac myocytes, we transfected cells with a GFP fusion construct encoding full-length myopalladin. Full-length GFP-myopalladin was mainly targeted to Z-lines (single band in green), with some targeting to I-bands (double band in green), as demonstrated by costaining for myomesin (shown in red) (Fig. 7 C, f′).
Consistent with the immunofluorescence data, immunoelectron microscopy studies with myopalladin-specific antibodies detected myopalladin both within the periphery of the Z-line (Fig. 8 A) and in the central I-band (Fig. 8, A–C). In fact, measurements of the CARP and myopalladin epitopes from the Z-disc revealed that the two proteins colocalized within the central I-band (Fig. 8 D). Interestingly, for both I-band–bound myopalladin and CARP the distances of their epitopes from the Z-line were dependent on stretch (i.e., sarcomere length). In contrast, the Z-line–bound myopalladin remained in a fixed position. This raises the possibility that CARP and the I-band–bound myopalladin are attached to the elastic titin filament, whereas the Z-line–bound myopalladin is an integral component within Z-lines.
Figure 8.
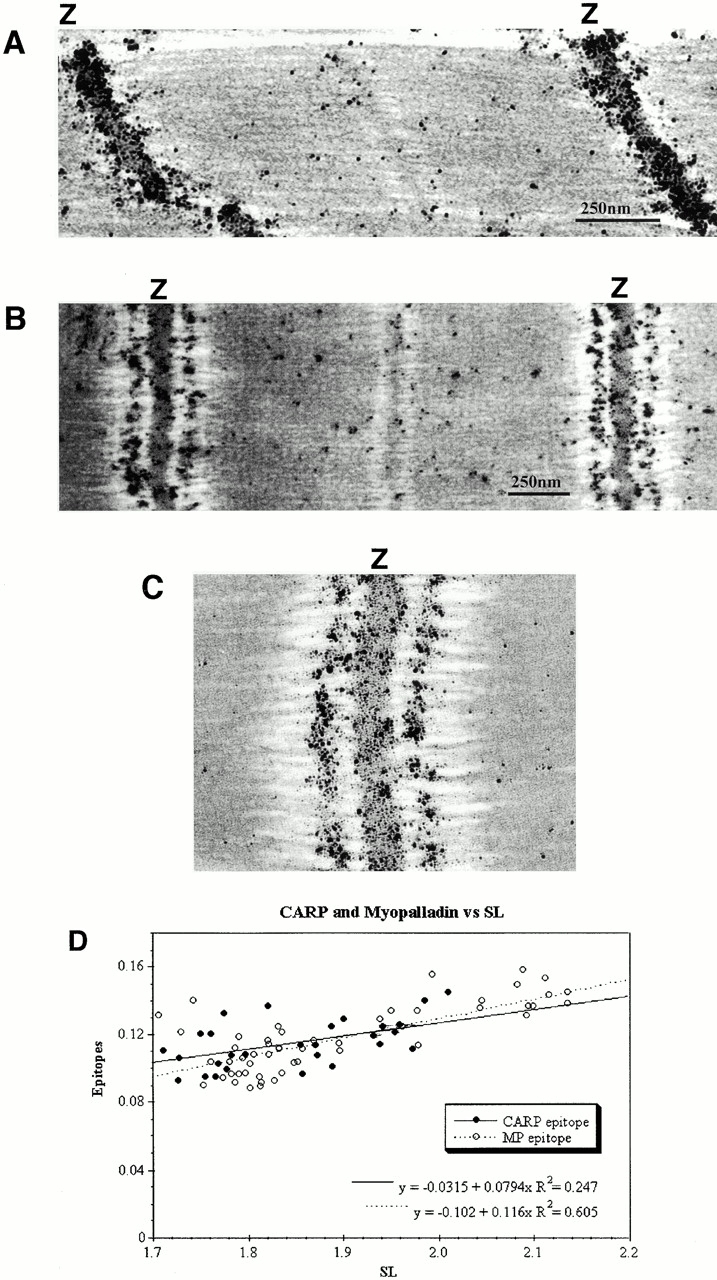
Immunoelectron microscopy reveals colocalization of myopalladin and CARP in mouse cardiac myofibrils. (A–C) Isolated myofibrils from mouse were prepared and stretched according to Trombitás et al. 1995; immunoelectron microscopy with affinity-purified antimyopalladin antibodies reveals that myopalladin is detected both within the periphery of the Z-line (A) and in the central I-band region (A–C). In stretched sarcomeres (B), the Z-line to I-band distance increases. A, unstretched; B, slightly stretched; C, high magnification view of Z-disc in stretched myofibrils. (D) The distances of I-band–bound CARP and myopalladin from the Z-disk were measured from immunoelectron micrographs at different sarcomere lengths (29 micrographs were analyzed for CARP; 54 micrographs for myopalladin). Both CARP and myopalladin colocalize in the central I-band and their epitope distances (in μm) from the Z-disc were dependent on sarcomeric length (SL).
Effects of Overexpression of Myopalladin on Z-Line Integrity
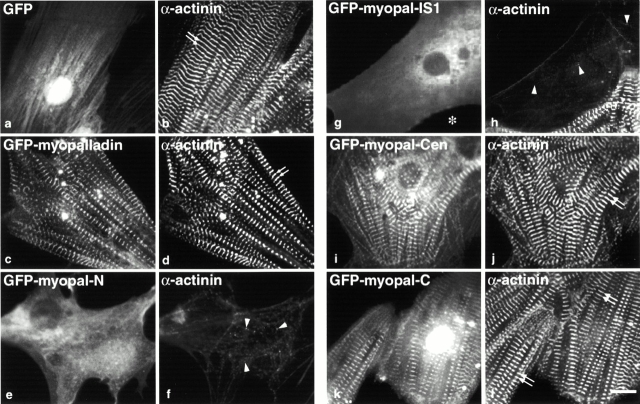
To investigate the functional significance of the individual domains of myopalladin in Z-line organization, we transfected chick cardiac myocytes with constructs encoding GFP fusions with full-length myopalladin, as well as four myopalladin subfragments (Fig. 4 A). Two fragments corresponded to myopalladin's NH2-terminal CARP-binding region, one to its middle nebulette/nebulin-binding region, and one to its COOH-terminal α-actinin–binding region. We observed the effects of overexpressing these domains on the organization of α-actinin, a marker for Z-line structure, using immunofluorescence microscopy (Fig. 9). Control cells expressing GFP alone exhibited a diffuse staining pattern for GFP (Fig. 9 a) with mature, repeating α-actinin staining (Fig. 9 b). When full-length GFP-myopalladin was overexpressed in cells, it mainly assembled in a striated pattern at the Z-line (Fig. 9 c), colocalizing with α-actinin, which was assembled in a mature striated pattern (Fig. 9 d), whereas some assembled at the I-band (Fig. 7 C, f′). In cells overexpressing either the middle region (Fig. 9 i) or the COOH-terminal region (Fig. 9 k) of myopalladin, no perturbation of Z-line structure was observed (Fig. 9j and Fig. l, respectively). In contrast, in cells overexpressing either of two myopalladin NH2-terminal domain GFP fusion proteins (Fig. 9e and Fig. g), an extensively disrupted Z-line structure (i.e., α-actinin staining) in 80–85% of all transfected myocytes was observed (Fig. 9f and Fig. h).
Figure 9.
Overexpression of the NH2-terminal region of myopalladin in cardiac myocytes results in marked disruption of Z-line architecture as indicated by α-actinin staining. Cardiac myocytes expressing GFP alone (GFP; a), GFP–full-length myopalladin (GFP-myopalladin; c), GFP-myopalladin NH2-terminal region (GFP–myopal-N; e), a smaller GFP–NH2-terminal myopalladin IS1 region (GFP–myopal-IS1; g), GFP-myopalladin central region (GFP–myopal-Cen; i), and GFP-myopalladin COOH-terminal region (GFP–myopal-C; k) were fixed 3–5 d after transfection and stained with anti–α-actinin antibodies followed by Texas red–conjugated secondary antibodies (b, d, f, h, j, and l). Arrows point to the typical striated staining pattern of α-actinin in transfected cardiac myocytes (b, d, j, and l); arrowheads point to disrupted α-actinin staining pattern in transfected myocytes (f and h). Bar, 10 μm.
Effects of Overexpression of Myopalladin on Sarcomeric Integrity
Since the Z-lines of cells overexpressing the NH2-terminal region of myopalladin were disrupted, we next investigated the effects of overexpressing this region on overall sarcomeric integrity by observing effects on the thin, thick, and titin filament systems. Thin filament integrity was examined by staining transfected cells for actin using Texas red–labeled phalloidin, whereas thick filament organization was investigated using two markers, antimyosin and antimyomesin antibodies. Antibodies specific for a region close to the NH2-terminal (Z-line) region of titin (T11) and the COOH-terminal M-line region of titin (T114) were used to analyze the effects of the overexpression of the NH2-terminal region of myopalladin on the titin third filament system. In cells expressing GFP alone (Fig. 10, a, g, and m), actin filaments (Fig. 10 b), titin T11 (Fig. 10 h), titin T114 (data not shown), myomesin (Fig. 10 n), and myosin (data not shown) appeared in their respective typical striated pattern. Additionally, myocytes overexpressing full-length GFP-myopalladin demonstrated a striated pattern (Fig. 10c, Fig. i, and Fig. o) and typically assembled actin (Fig. 10 d), titin T11 (Fig. 10 j), titin T114 (data not shown), myomesin (Fig. 10 r), and myosin (data not shown). In contrast, myocytes overexpressing the GFP–NH2-terminal region of myopalladin (Fig. 10e, Fig. k, and Fig. q), exhibited a dramatic sarcomeric disruption, owing to the disrupted thin (Fig. 10 f), titin (Fig. 10 l), and thick (Fig. 10 p) filament systems in 80–85% of transfected cells. Similar results were obtained in myocytes overexpressing GFP–myopalladin-IS1, a smaller fragment of the NH2-terminal region of myopalladin that also contains the CARP-binding site (data not shown). The most likely explanation for this phenomenon is that a dominant negative phenotype occurred. That is, overexpression of the NH2-terminal region of myopalladin competed for the CARP-binding site on endogenous myopalladin. These data indicate a critical role for the NH2 terminus of myopalladin containing the CARP-binding domain for sarcomeric structure.
Figure 10.
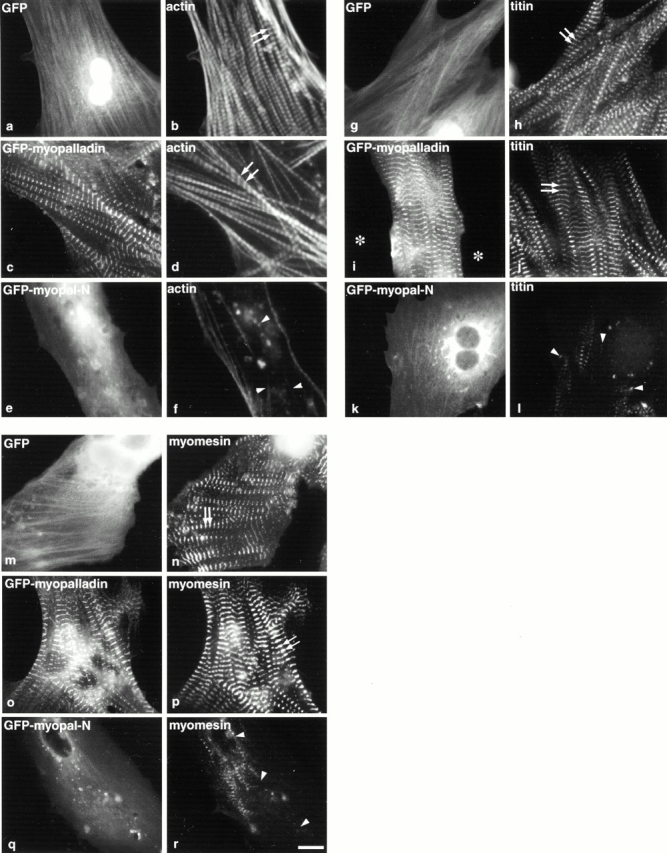
Overexpression of the NH2-terminal region of myopalladin in cardiac myocytes results in disruption of thin, titin, and thick filaments. Chick cardiac myocytes expressing GFP alone (GFP; a, g, and m), GFP–full-length myopalladin (GFP-myopalladin; c, i, and o), and GFP–NH2-terminal myopalladin region (GFP–myopalladin-N; e, k and p) were fixed 3–5 d after transfection and stained with Texas red–conjugated phalloidin (b, d, and f), antititin T11 antibodies followed by Texas red–conjugated secondary antibodies (h, j, and l), or antimyomesin antibodies followed by Texas red–conjugated secondary antibodies (n, p, and r). Arrows point to typical striated staining pattern of actin (b and d), titin (h and j), and myomesin (n and p); arrowheads point to disrupted actin (f), titin (l), and myomesin (r). Bar, 10 μm.
Discussion
In this study, we aimed to further dissect the molecular components required for the coordinated organization of Z-line components into regular, hexagonal lattices. We took the approach of searching for protein interactions that anchor the COOH-terminal region of nebulin within the sarcomere. This allowed us to identify the specific binding of the nebulin modules M160–M183, from the peripheral (I-band side) region of the Z-line (Millevoi et al. 1998), with the intermediate filament protein, desmin. Studies are currently under investigation to determine whether desmin and nebulin indeed participate in a coordinated cytoskeletal network that could provide lateral linkages in the Z-line perimyofibrillar space, allowing for efficient force transmission and mechanochemical signaling.
This study has also led to the identification of a novel nebulin-binding protein, myopalladin, which we named based on its striking homology with the recently described ubiquitously expressed protein, palladin (Parast and Otey 2000). The homology of palladin and myopalladin suggests that they are members of the same gene family. Palladin colocalizes with α-actinin in focal adhesions, stress fibers, cell–cell junctions, and Z-lines, and is critical for the organization of the actin cytoskeleton and focal adhesions (Parast and Otey 2000). The 63% sequence identity of myopalladin to palladin within its α-actinin–binding regions raises the intriguing possibility that the assembly of nonmuscle stress fibers and sarcomeres share common regulatory mechanisms involving myopalladin and palladin.
Previous studies have demonstrated that both the nebulette and nebulin SH3 domains target to vertebrate Z-lines when expressed in avian cardiac and skeletal myocytes, respectively (Moncman and Wang 1999; Ojima et al. 2000). However, the molecular basis that allows for the targeting of the SH3 domain to Z-lines has been unclear; that is, could a direct interaction of nebulin/nebulette with α-actinin account for the targeting of the nebulin/nebulette SH3 fragments to the Z-line (Nave et al. 1990; Moncman and Wang 1999)? Based on our immunoelectron microscopy (this report and Millevoi et al. 1998), cellular targeting, and biochemical studies, we predict that the proline residue–rich motif (the triple proline PPP motif, residues 649–651) in the central IS3 domain of myopalladin interacts with the SH3 domains of nebulin and nebulette, thereby anchoring them ∼25 nm inside the Z-line. Also, myopalladin's PPP motif, and its flanking residues, are highly conserved in palladin (Fig. 2 B). This strongly suggests that palladin's PPP motif can also interact with the SH3 domains of nebulin and nebulette in skeletal and cardiac muscle, respectively.
Myopalladin's COOH-terminal region, in turn, specifically binds to the EF hand motifs of α-actinin. By extrapolating our data on the properties of myopalladin to what is known about palladin, we would predict that palladin's COOH-terminal three Ig domains interact with α-actinin in muscle Z-lines and in nonmuscle cells. Therefore, the data presented here, together with previous reports demonstrating the association of α-actinin with titin Z-repeats (Ohtsuka et al. 1997; Sorimachi et al. 1997; Gregorio et al. 1998; Young et al. 1998), predict that palladin/myopalladin and α-actinin may form a linking system, tethering the barbed ends of the actin-thin filaments, the NH2-terminal ends of titin filaments, and the COOH-terminal ends of nebulin filaments (skeletal muscle) and nebulette (cardiac muscle) within the Z-line. Therefore, we speculate that the palladin/myopalladin–α-actinin complexes are major players in assembling I-Z-I bodies in striated muscle (Epstein and Fischman 1991; Holtzer et al. 1997; Ehler et al. 1999; Ojima et al. 1999; Gregorio and Antin 2000). The palladin/myopalladin–α-actinin interaction could provide the anchor to stabilize these structures during development, similar to palladin's proposed role in stabilizing stress fibers in nonmuscle cells (Parast and Otey 2000). It will be interesting to see if the PPP motif from nonmuscle palladin also interacts with other SH3 domain–containing proteins in nonmuscle cells, which are involved in regulation of stress fiber assembly.
Although myopalladin and palladin are highly homologous and are coexpressed and localized in Z-lines in mature sarcomeres (Fig. 7), these two molecules appear to have very different cellular properties. Palladin, but not myopalladin, is detected in the earliest I-Z-I bodies, structures which are precursor Z-lines (for cardiac myocytes, Fig. 7; for myopalladin in skeletal myogenic cells, Holtzer, H., personal communication). Possibly during myofibril assembly, some palladin complexes can be replaced by myopalladin Z-line complexes. Furthermore, myopalladin, in contrast to palladin, also localizes to the I-band and interacts with CARP (Fig. 4 and Fig. 7). Also, the myopalladin NH2-terminal domain is rich in potential phosphorylation sites and is efficiently phosphorylated by muscle extracts (data not shown), whereas palladin is missing these sites. Finally, palladin is extensively differentially spliced, accounting for the multiple bands detected on Western blots (this study and Parast and Otey 2000). In contrast, blots probed for myopalladin did not display multiple bands (Fig. 7). Interestingly, we identified a specific splice isoform of palladin that is missing the exon encoding the COOH-terminal Ig repeat that is predicted to be involved in binding to α-actinin (data not shown). In summary, it appears that although myopalladin and palladin may interact with SH3 domain–containing proteins via their conserved binding sites, the two proteins are likely to be regulated by different mechanisms. A molecular understanding of how the palladin/myopalladin–α-actinin complex assembly is regulated is likely to improve our general understanding of Z-line assembly and maintenance.
Finally, myopalladin's NH2-terminal domain binds CARP. The CARP protein contains a nuclear localization signal, has been reported to be exclusively localized in the nucleus, and has been shown to negatively regulate the expression of cardiac genes, including the ventricular-specific myosin light chain-2, natriuretic factor, and cardiac troponin-C genes (Jeyaseelan et al. 1997; Zou et al. 1997). CARP is expressed abundantly and specifically in the early embryonic heart (E8.5) and is downstream in the Nkx2-5 pathway that defines the early heart field (Zou et al. 1997). In the adult heart, CARP is developmentally downregulated, but can be dramatically induced as part of the onset of an embryonic gene program during cardiac hypertrophy (Arber et al. 1997; Kuo et al. 1999). Furthermore, CARP expression is induced during denervation of skeletal muscle (Baumeister et al. 1997) and by stimuli such as the muscle toxin adriamycin (doxorubicin) (Jeyaseelan et al. 1997). In summary, these studies reveal that CARP is involved in the regulation of muscle gene expression, physiologically during development and pathologically in response to various cellular stresses.
Here, using a novel affinity-purified CARP-specific antibody (Fig. 6 B), an intranuclear localization of CARP was confirmed. However, high levels of CARP staining were also detected in the cytoplasm, as a sarcomeric bound form in the central I-band. The discovery of this large proportion of CARP that is associated with sarcomeric I-bands is likely to provide insights into understanding regulation of CARP-based signaling, since free CARP presumably is rapidly degraded by PEST-directed proteases (Baumeister et al. 1997) whereas I-band bound CARP may be more stable. Interestingly, in some myocytes, we detected only sarcomeric CARP. In other myocytes nuclear CARP was predominant, whereas in still others both localization patterns were detected. This suggests that CARP may be dynamically redistributed between sarcomeres and nuclei. In this respect, our immunolocalization results for CARP are similar to those obtained for myopalladin: myopalladin was also detected in variable amounts inside the nucleus, but also as a sarcomeric bound form both at the Z-line periphery and in the central I-band region where CARP is colocalized. Myopalladin contains a potential nuclear import signal in its sequence (KKPR, residues 486–489). It remains to be seen if this motif is involved in nuclear import, or if myopalladin can be imported into nuclei as a complex with the nuclear form of CARP. Interestingly, our overexpression data demonstrated that the NH2-terminal region of myopalladin containing the CARP-binding region, but not the central region (containing the nebulin-binding region) or the COOH-terminal region (containing the α-actinin–binding region), caused disruption of all sarcomeric proteins studied (Fig. 9 and Fig. 10). These data suggest that the interaction of myopalladin and CARP is critical for sarcomeric structure in cardiac myocytes. Clearly, future studies are required to investigate the molecular basis for the targeting of myopalladin and CARP to multiple cellular sites, and how these dynamic redistributions are involved in myofibril assembly, muscle structure, and muscle gene expression.
The further biochemical characterization of myopalladin, that is, its interaction with molecules crucially involved in Z-line assembly (α-actinin and nebulin/nebulette) and in muscle gene expression regulation (CARP), together with an understanding of its multiple cellular localizations, may provide insights into how myofibril assembly/disassembly and gene expression regulatory mechanisms are linked.
Acknowledgments
The authors would like to thank Mark McNabb for excellent technical assistance and Dr. J. Bahl for providing us with rat cardiac myocytes.
This work was supported by grants from the Human Frontier Science Program (S. Labeit, H. Sorimachi, and C. Gregorio), CREST grant 11B-1 from the Ministry of Health and Welfare (to H. Sorimachi), National Institutes of Health grants HL57461 and HL03985 (C. Gregorio), HL07249 (A.S. McElhinny), and HL61497 and HL62881 (H. Granzier), and the Deutsche Forschungsgemeinschaft (La668/3-3 and 4-2) to S. Labeit.
Footnotes
C. Gregorio and S. Labeit contributed equally to this work.
Abbreviations used in this paper: CARP, cardiac ankyrin repeat protein; GFP, green fluorescent protein; GST, glutathione S-transferase; SH, Src homology.
References
- Arber S., Hunter J.J., Ross J., Jr., Hongo M., Sansig G., Borg J., Perriard J.C., Chien K.R., Caroni P. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell. 1997;88:393–403. doi: 10.1016/s0092-8674(00)81878-4. [DOI] [PubMed] [Google Scholar]
- Baumeister A., Arber S., Caroni P. Accumulation of muscle ankyrin repeat protein transcript reveals local activation of primary myotube endcompartments during muscle morphogenesis. J. Cell Biol. 1997;139:1231–1242. doi: 10.1083/jcb.139.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggs A.H., Byers T.J., Knoll J.H., Boyce F.M., Bruns G.A., Kunkel L.M. Cloning and characterization of two human skeletal muscle alpha-actinin genes located on chromosomes 1 and 11. J. Biol. Chem. 1992;267:9281–9288. [PubMed] [Google Scholar]
- Centner T., Fougerousse F., Freiburg A., Witt C., Beckmann J.S., Granzier H., Trombitas K., Gregorio C.C., Labeit S. Molecular tools for the study of titin's differential expression. Adv. Exp. Med. Biol. 2000;481:35–49. doi: 10.1007/978-1-4615-4267-4_3. [DOI] [PubMed] [Google Scholar]
- Chu W., Burns D.K., Swerlick R.A., Presky D.H. Identification and characterization of a novel cytokine-inducible nuclear protein from human endothelial cells. J. Biol. Chem. 1995;270:10236–10245. doi: 10.1074/jbc.270.17.10236. [DOI] [PubMed] [Google Scholar]
- Ehler E., Rothen B.M., Hammerle S.P., Komiyama M., Perriard J.C. Myofibrillogenesis in the developing chicken heartassembly of Z-disk, M-line and the thick filaments. J. Cell Sci. 1999;112:1529–1539. doi: 10.1242/jcs.112.10.1529. [DOI] [PubMed] [Google Scholar]
- Epstein H.F., Fischman D.A. Molecular analysis of protein assembly in muscle development. Science. 1991;251:1039–1144. doi: 10.1126/science.1998120. [DOI] [PubMed] [Google Scholar]
- Franzini-Armstrong C. The structure of a simple Z-line. J. Cell Biol. 1973;58:630–642. doi: 10.1083/jcb.58.3.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler V.M., Sussmann M.A., Miller P.G., Flucher B.E., Daniels M.P. Tropomodulin is associated with the free (pointed) ends of the thin filaments in rat skeletal muscle. J. Cell Biol. 1993;120:411–420. doi: 10.1083/jcb.120.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll D.E., Suzuki A., Temple J., Holmes G.R. Studies on purified α-actinin. I. Effect of temperature and tropomyosin on the α-actinin/F-actin interaction. J. Mol. Biol. 1972;67:469–488. doi: 10.1016/0022-2836(72)90464-0. [DOI] [PubMed] [Google Scholar]
- Gregorio C.C., Antin P.B. To the heart of myofibril assembly. Trends Cell Biol. 2000;10:355–362. doi: 10.1016/s0962-8924(00)01793-1. [DOI] [PubMed] [Google Scholar]
- Gregorio C.C., Fowler V.M. Mechanisms of thin filament assembly in embryonic chick cardiac myocytestropomodulin requires tropomyosin for assembly. J. Cell Biol. 1995;129:683–695. doi: 10.1083/jcb.129.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorio C.C., Weber A., Bondad M., Pennise C.R., Fowler V.M. Requirement of pointed end capping by tropomodulin to maintain actin filament length in embryonic chick cardiac myocytes. Nature. 1995;377:83–86. doi: 10.1038/377083a0. [DOI] [PubMed] [Google Scholar]
- Gregorio C.C., Trombitás K., Center T., Kolmerer B., Stier G., Kunkel K., Suzuki K., Obermayr F., Herrmann B., Granzier H. The NH2 terminus of titin spans the Z-discits interaction with a novel 19-kD ligand (T-cap) is required for sarcomeric integrity. J. Cell Biol. 1998;143:1013–1027. doi: 10.1083/jcb.143.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove B.K., Cerny L., Perriard J.C., Eppenberger H.M. Myomesin and M proteinexpression of two M-band proteins in pectoral muscle and heart during development. J. Cell Biol. 1985;101:1413–1421. doi: 10.1083/jcb.101.4.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson T.A., Bahl J.J, Markham B.E., Roeske W.R., Morkin E. Hormonal regulation of myosin heavy chain and alpha-actin gene expression in cultured fetal rat heart myocytes. J. Biol. Chem. 1987;262:13316–13322. [PubMed] [Google Scholar]
- Harpaz Y., Chothia C. Many of the immunoglobulin superfamily domains in cell adhesion molecules and surface receptors belong to a new structural set which is close to that containing variable domains. J. Mol. Biol. 1994;238:528–539. doi: 10.1006/jmbi.1994.1312. [DOI] [PubMed] [Google Scholar]
- Higgins D.G., Thompson J.D., Gibson T.J. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- Holtzer H., Hijikata T., Lin Z.X., Zhang Z.Q., Holtzer S., Protasi F., Franzini-Armstrong C., Sweeney H.L. Independent assembly of 1.6 microns long bipolar MHC filaments and I-Z-I bodies. Cell Struct. Funct. 1997;22:83–93. doi: 10.1247/csf.22.83. [DOI] [PubMed] [Google Scholar]
- Jeyaseelan R., Poizat C., Baker R.K., Abdishoo S., Isterabadi L.B., Lyons G.E., Kedes L. A novel cardiac-restricted target for doxorubicin. J. Biol. Chem. 1997;272:22800–22808. doi: 10.1074/jbc.272.36.22800. [DOI] [PubMed] [Google Scholar]
- Knight P.J., Trinick J.A. Preparation of myofibrils. Methods Enzymol. 1982;85:9–12. doi: 10.1016/0076-6879(82)85004-0. [DOI] [PubMed] [Google Scholar]
- Kuo H.-C., Chen J., Ruiz-Lozano P., Zou Y., Nemer M., Chien K.R. Control of segmental expression of the cardiac-restricted ankyrin repeat protein gene by distinct regulatory pathways in murine cardiogenesis. Development. 1999;126:4223–4234. doi: 10.1242/dev.126.19.4223. [DOI] [PubMed] [Google Scholar]
- Millevoi S., Trombitas K., Kolmerer B., Kostin S., Schaper J., Pelin K., Granzier B.H., Labeit S. Characterization of nebulette and nebulin and emerging concepts of their roles for vertebrate Z-discs. J. Mol. Biol. 1998;282:111–123. doi: 10.1006/jmbi.1998.1999. [DOI] [PubMed] [Google Scholar]
- Moncman C.L., Wang K. Nebulettea 107 kD nebulin-like protein in cardiac muscle. Cell Motil. Cytoskeleton. 1995;32:205–225. doi: 10.1002/cm.970320305. [DOI] [PubMed] [Google Scholar]
- Moncman C.L., Wang K. Functional dissection of nebulette demonstrates actin binding of nebulin-like repeats and Z-line targeting of SH3 and linker domains. Cell Motil. Cytoskeleton. 1999;44:1–22. doi: 10.1002/(SICI)1097-0169(199909)44:1<1::AID-CM1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Musacchio A., Gibson T., Lehto V.P., Saraste M. SH3 - an abundant protein domain in search of a function. FEBS Lett. 1992;307:55–61. doi: 10.1016/0014-5793(92)80901-r. [DOI] [PubMed] [Google Scholar]
- Ohtsuka H., Yajima H., Maruyama K., Kimura S. Binding of the N-terminal 63 kDa portion of connectin/titin to alpha-actinin as revealed by the yeast two-hybrid system. FEBS Lett. 1997;401:65–67. doi: 10.1016/s0014-5793(96)01432-9. [DOI] [PubMed] [Google Scholar]
- Ojima K., Lin Z.X., Zhang Z.Q., Hijikata T., Holtzer S., Labeit S., Sweeney H.L., Holtzer H. Initiation and maturation of I-Z-I bodies in the growth tips of transfected myotubes. J. Cell Sci. 1999;112:4101–4112. doi: 10.1242/jcs.112.22.4101. [DOI] [PubMed] [Google Scholar]
- Ojima K., Lin Z.X., Bang M.-L., Holtzer S., Matsuda R., Labeit S., Sweeney H.L., Holtzer H. Distinct families of Z-line targeting modules in the C-terminal region of nebulin. J. Cell Biol. 2000;150:553–566. doi: 10.1083/jcb.150.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa I., Astier C., Kwiatek O., Raynaud F., Bonnal C., Lebart M.C., Roustan C., Benyamin Y. Alpha-actinin-CapZ, an anchoring complex for thin filaments in Z-line. J. Muscle Res. Cell Motil. 1999;20:187–197. doi: 10.1023/a:1005489319058. [DOI] [PubMed] [Google Scholar]
- Parast M.M., Otey C.A. Characterization of palladin, a novel protein localized to stress fibers and cell adhesions. J. Cell Biol. 2000;150:643–656. doi: 10.1083/jcb.150.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe R.W. The ultrastructure of Z disks from white, intermediate, and red fibers of mammalian striated muscles. J. Cell Biol. 1973;57:261–277. doi: 10.1083/jcb.57.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R.K., Scharf S., Faloona F., Mullis K.B., Horn G.T., Erlich H.A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Salmikangas P., Mykkanen O.M., Gronholm M., Heiska L., Kere J., Carpen O. Myotilin, a novel sarcomeric protein with two Ig-like domains, is encoded by a candidate gene for limb-girdle muscular dystrophy. Hum. Mol. Genet. 1999;8:1329–1336. doi: 10.1093/hmg/8.7.1329. [DOI] [PubMed] [Google Scholar]
- Sorimachi H., Freiburg A., Kolmerer B., Ishiura S., Stier G., Gregorio C.C., Labeit D., Linke W.A., Suzuki K., Labeit S. Tissue-specific expression and alpha-actinin binding properties of the Z-disc titinimplications for the nature of vertebrate Z-discs. J. Mol. Biol. 1997;270:688–695. doi: 10.1006/jmbi.1997.1145. [DOI] [PubMed] [Google Scholar]
- Squire J.M. Architecture and function in the muscle sarcomere. Curr. Opin. Struct. Biol. 1997;7:247–257. doi: 10.1016/s0959-440x(97)80033-4. [DOI] [PubMed] [Google Scholar]
- Studier F.W., Moffatt B.A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Studier F.W., Rosenberg A.H., Dunn J.J., Dubendorff J.W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Trombitás K., Jin J.P., Granzier H. The mechanically active domain of titin in cardiac muscle. Circ. Res. 1995;77:856–861. doi: 10.1161/01.res.77.4.856. [DOI] [PubMed] [Google Scholar]
- Vicart P., Dupret J.M., Hazan J., Li Z., Gyapay G., Krishnamoorthy R., Weissenbach J., Fardeau M., Paulin D. Human desmin genecDNA sequence, regional localization and exclusion of the locus in a familial desmin-related myopathy. Hum. Genet. 1996;98:422–429. doi: 10.1007/s004390050233. [DOI] [PubMed] [Google Scholar]
- Vigoreaux J.O. The muscle Z-bandlessons in stress management. J. Muscle Res. Cell Motil. 1994;15:237–255. doi: 10.1007/BF00123477. [DOI] [PubMed] [Google Scholar]
- Wanker E.E., Rovira C., Scherzinger E., Hasenbank R., Walter S., Tait D., Colicelli J., Lehrach H. HIP-Ia huntingtin interacting protein isolated by the yeast two-hybrid system. Hum. Mol. Genet. 1997;6:487–495. doi: 10.1093/hmg/6.3.487. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Izumimoto M., Robson R.M., Stromer M.H. Fine structure of wide and narrow vertebrate muscle Z-lines. A proposed model and computer simulation of Z-line architecture. J. Mol. Biol. 1985;184:621–644. doi: 10.1016/0022-2836(85)90308-0. [DOI] [PubMed] [Google Scholar]
- Young P., Ferguson C., Bañuelos S., Gautel M. Molecular structure of the sarcomeric Z-disktwo types of titin interactions lead to an asymmetrical sorting of alpha-actinin. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:1614–1624. doi: 10.1093/emboj/17.6.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y., Evans S., Chen J., Kuo H.-C., Harvey R.P., Chien K.R. CARP, a cardiac ankyrin repeat protein, is downstream in the Nkx2-5 homeobox gene pathway. Development. 1997;124:793–804. doi: 10.1242/dev.124.4.793. [DOI] [PubMed] [Google Scholar]