Abstract
The first step in the assembly of new chromatin is the cell cycle–regulated synthesis and nuclear import of core histones. The core histones include H2A and H2B, which are assembled into nucleosomes as heterodimers. We show here that the import of histone H2A and H2B is mediated by several members of the karyopherin (Kap; importin) family. An abundant complex of H2A, H2B, and Kap114p was detected in cytosol. In addition, two other Kaps, Kap121p and Kap123p, and the histone chaperone Nap1p were isolated with H2A and H2B. Nap1p is not necessary for the formation of the Kap114p-H2A/H2B complex or for import of H2A and H2B. We demonstrate that both histones contain a nuclear localization sequence (NLS) in the amino-terminal tail. Fusions of the NLSs to green fluorescent protein were specifically mislocalized to the cytoplasm in kap mutant strains. In addition, we detected a specific mislocalization in a kap95 temperature-sensitive strain, suggesting that this Kap is also involved in the import of H2A and H2B in vivo. Importantly, we show that Kap114p, Kap121p, and Kap95 interact directly with both histone NLSs and that RanGTP inhibits this association. These data suggest that the import of H2A and H2B is mediated by a network of Kaps, in which Kap114p may play the major role.
Keywords: yeast, nuclear import, karyopherin, histone, nuclear localization signal
Introduction
The eukaryotic chromosome is comprised of DNA and its major protein component, histones. This group of proteins includes the core histones H2A, H2B, H3, and H4 as well as the linker histone H1 (Kornberg and Lorch 1999). Two copies of each core histone are assembled into an octamer, which has ∼146 base pairs of DNA wrapped around it, to form a nucleosome core (Luger et al. 1997). The four core histones have a similar overall structure, consisting of basic amino and carboxy termini and a hydrophobic, globular internal region that forms the histone-fold domain (Luger et al. 1997). Posttranslational modifications of the positively charged histone amino-terminal tails modulate the structure of chromatin and play an important role in many DNA template-dependent processes such as replication, recombination, repair, and transcription (Strahl and Allis 2000). They may also function in histone deposition and nucleosome assembly (Strahl and Allis 2000).
Before their assembly onto the nucleosome, histones are complexed with specific chaperones, such as Nap1p, which are believed to play an important role in directing histone deposition, although the specific function of these factors is not well understood (Adams and Kamakaka 1999). Nap1p forms a complex with histones H2A and H2B in the cytoplasm, and has been postulated to play a role in their import into the nucleus (Ishimi et al. 1984; Kellogg and Murray 1995; Chang et al. 1997). In higher eukaryotes, Nap1 has been shown to localize to the nucleus in a cell cycle–dependent manner (Ito et al. 1996). This dynamic nuclear localization in S phase correlates with the synthesis of histones, which also peaks in S phase (Adams and Kamakaka 1999). In yeast, additional functions have been ascribed to Nap1p, as it has been shown to interact with B-type cyclins and may play a role in mitosis (Kellogg et al. 1995).
One of the first steps of nucleosome assembly is the import of newly synthesized histones into the nucleus. Nucleocytoplasmic transport of proteins and RNA is mediated through the nuclear pore complex (NPC), a large protein complex embedded in the nuclear envelope (reviewed in Nakielny and Dreyfuss 1999; Gorlich and Kutay 1999). The NPC serves as the site of interaction for soluble factors involved in nuclear transport (Rout et al. 2000). Transport is mediated by a family of evolutionarily conserved, structurally similar, soluble transport receptors called karyopherin βs (Kaps, also called importins, exportins, and transportins) (Görlich et al. 1997; Pemberton et al. 1998; Wozniak et al. 1998). Members of this family mediate the import and export of cargo through the NPC and are able to dock at specific sites on the NPC (Nakielny and Dreyfuss 1999; Gorlich and Kutay 1999). Most Kaps bind directly to nuclear localization sequences (NLS) within their cargo. The NLSs recognized by most Kaps are largely undefined, and those identified so far suggest that they are very diverse (Rosenblum et al. 1998; Senger et al. 1998; Nakielny and Dreyfuss 1999; Lee and Aitchison 1999). Kaps can also interact with a subset of NPC proteins and the small GTPase Ran, which acts as a molecular switch and terminates transport cycles (Rexach and Blobel 1995; Izaurralde et al. 1997; Moore 1998). One member of the Kap family, Kapβ1 (Kap95p in yeast; Enenkel et al. 1995), can also bind its cargo via an adaptor protein, Kapα (Kap60p in yeast). Kapα binds to “classical” or “basic” NLSs, exemplified by that found in SV40 large T antigen (SV40) and nucleoplasmin, which include several basic amino acids (Gorlich and Kutay 1999; Nakielny and Dreyfuss 1999).
The Saccharomyces cerevisiae genome encodes 14 Kaps, and most of these appear to have one if not several human homologues (Görlich et al. 1997; Pemberton et al. 1998; Wozniak et al. 1998). 10 Kaps have been shown to be involved in nuclear import, three have been demonstrated to be export receptors, and one has been shown to function in both import and export (Görlich et al. 1997; Pemberton et al. 1998; Wozniak et al. 1998; Yoshida and Blobel 2001). To date, there are only one or two cargoes known for most Kaps. This is likely far from complete, as there are ∼1,000–2,000 nuclear or nucleocytoplasmic shuttling proteins and 10 import Kaps in yeast, suggesting that each Kap has many cargoes. How each Kap can interact with several NLSs and distinguish between distinct NLSs is not known. The import Kaps, with the exception of Kap95p and Kap121p, are encoded by nonessential genes (Pemberton et al. 1998; Wozniak et al. 1998). However, nonessential Kaps function in the import of essential proteins, such as TFIIA and the TATA binding protein (TBP), suggesting that the members of the Kap family may have partially overlapping functions (Pemberton et al. 1999; Titov and Blobel 1999). This notion, coupled with the fact that they probably recognize several if not hundreds of cargoes, suggests that their coordination may be very complex.
The import of histone H1 has been previously shown to be a receptor-mediated process, even though H1 is small enough to diffuse through the NPC (Breeuwer and Goldfarb 1990; Kurz et al. 1997), and recently the mammalian importin β1/importin 7 heterodimer was shown to be able to mediate H1 import in vitro (Jakel et al. 1999). Of the core histones, only the histone H2B NLS has been studied in any detail (Moreland et al. 1987; Marchetti et al. 2000). Amino acids 22–33 of yeast H2B were shown to be sufficient to localize a reporter protein to the nucleus (Moreland et al. 1987). Since this domain of the H2B amino terminal tail is positively charged and resembles the SV40 NLS, it was assumed that H2B import is mediated by a Kapα-dependent pathway (Moreland et al. 1987). Contradictory reports in different systems have suggested that the import of H2A/H2B is both Kapα/Kapβ1 independent and that H2A/H2B binds directly to Kapβ1 (Johnson-Saliba et al. 2000; Langer 2000).
We have previously shown that Kap114p imports TBP into the nucleus (Pemberton et al. 1999). We were able to show that while Kap114p mediates the major import pathway for TBP, two other Kaps, Kap121p and Kap123p, may also function in TBP import (Pemberton et al. 1999). Here we show that Kap114p also mediates the nuclear import of histones H2A and H2B, suggesting an important role for this Kap in the assembly of new nucleosomes. We show for the first time that H2A and H2B each have distinct NLSs in their amino terminal tails, both of which are recognized by Kap114p. We also demonstrate that the chaperone Nap1p is not necessary for the formation of a competent transport complex in vivo. The import of H2A and H2B is mediated by a network of Kaps, including Kap114p, Kap121p, Kap123p, and Kap95p, several of which we have shown to bind directly to the amino terminal tails of H2A and H2B. This suggests a new model for the organization of a subfamily of import Kaps and their essential cargoes.
Materials and Methods
Yeast Strains
All yeast strains (except where noted as W303) were derived from DF5 and the procedures for yeast manipulation were as described (Sherman et al. 1986; Pemberton et al. 1997). Construction of all Kap mutant and deletion strains has been described previously: Δkap104 (Aitchison et al. 1996), Δkap119 (nmd5; Albertini et al. 1998), Δkap108 (sxm1; Rosenblum et al. 1997), Δkap111 (mtr10; Pemberton et al. 1997), Δkap114 (Pemberton et al. 1999), Δkap123 (Rout et al. 1997), Δkap142 (msn5; Yoshida and Blobel 2001), kap95ts (in W303; Koepp et al. 1996), kap121ts (pse1-1 in W303; Seedorf and Silver 1997), srp1-31 (in W303; Loeb et al. 1995), except for Δkap120, which was a gift of Susana Chaves and Günter Blobel (The Rockefeller University, New York, NY). The kap114/pse1-1 and Kap114-protein A (PrA)/Δnap1 strains were created by mating, sporulating, and dissecting the relevant haploid strains. Histones in yeast are each encoded by two separate genes, strains expressing the Htb1-PrA (H2B-PrA) and Hta1-PrA (H2A-PrA) were constructed by integrative transformation of the coding sequence of four and a half IgG binding repeats of Staphylococcus aureus PrA immediately upstream of the relevant stop codon, as described (Aitchison et al. 1995). H2A-PrA/Δkap114 and H2B-PrA/Δkap114 were created by integration of the PrA sequences into haploid Δkap114 strains.
Plasmids
Green fluorescent protein (GFP) reporter constructs were based on pGFP-C-FUS and pGFP-N-FUS as previously described (Niedenthal et al. 1996). The sequence of a second GFP was cloned into these vectors to create the GFP2 vectors. All H2A and H2B constructs were made using sequences from the HTA1 (H2A) and HTB1 (H2B) genes, and cloned into the relevant GFP2 vectors. For the in vitro translation experiments Kap114, Kap121, Kap108, Kap95 was expressed from pET21a (Invitrogen). The NLS domains of H2A (amino acids 1–46) and H2B (amino acids 1–52) were expressed as glutathione-S-transferase (GST) fusions in pGEX-4T1 (Amersham Pharmacia Biotech). Kap ORFs were expressed as GST fusions in pGEX-5X1 (Kap114), pGEX-4T1 (Kap121), or pGEX-2TK (Kap95).
Cell Culture and Microscopy
Conventional immunofluorescence microscopy on yeast spheroplasts was done as previously described (Pemberton et al. 1997). Strains harboring plasmids based on pGFP-C-FUS and pGFP-N-FUS were grown in SC-ura, transferred to SC-ura-met, and incubation was continued for 2 h to induce the reporter. Cells were cultured at 23° or 30°C as described. All microscopy was performed on a Nikon Microphot-SA microscope using a 60× (in figures) or 100× (for quantitation) objective. Images were captured with digital camera into the OpenLab 2.0.6 software. All images (in figures) were taken at 200 ms (except for wild type, which were taken at 100 ms because the nuclear fluorescence was so bright) and manipulated identically in Adobe Photoshop. For quantitation, 10–20 random images containing multiple cells were captured for each cell type in 12-bit format, avoiding saturation of maximum pixel value. Mean fluorescent intensity in a 36-pixel box (0.72 × 0.72 μm) was measured in the nuclear and cytoplasmic compartment (excluding vacuoles) of multiple cells. Mean pixel values were used to determine the nuclear:cytoplasmic (N:C) ratio of fluorescence intensity. The mean N:C ratio was determined from 50 cells for each cell type and reporter. Images shown in figures were manipulated using Adobe Photoshop.
Protein Purification
Postnuclear, postribosomal cytosol was prepared from 2 liters of the Kap114-PrA and H2B-PrA strains and 1 liter of the other PrA-tagged strains, grown to an OD600 of 1.6 as described (Aitchison et al. 1996). Kap114-PrA, H2A-PrA or H2B-PrA and associated proteins were immunoisolated by overnight incubation of cytosol with rabbit IgG Sepharose as described (Aitchison et al. 1996; Pemberton et al. 1997). After washing in TB (20 mM Hepes, pH 7.5, 110 mM KOAc, 2 mM MgCl2, 1 mM DTT, 0.1% Tween-20), proteins were eluted from the Sepharose with a step gradient of MgCl2 and precipitated with methanol before analysis by SDS-PAGE. Coomassie blue staining bands were excised and prepared for analysis by matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) mass spectrometry (MS), and by peptide sequencing as described (Fernandez et al. 1994; Gharahdaghi et al. 1996). For Western blotting, proteins were transferred from SDS-PAGE gels onto PVDF membrane and probed with antibodies as noted. The Nap1p (Santa Cruz Biotechnology, Inc.), Kap121p, and Kap123p antibodies have been previously described (Rout et al. 1997; Marelli et al. 1998). Antibody interaction was visualized by labeling with HRP-conjugated secondary antibodies followed by ECL (Amersham Pharmacia Biotech).
Nano-HPLC Microelectrospray Ionization Mass Spectrometry Analysis and Database Searching
For analysis of the entire interacting fraction, the Sepharose and associated proteins were washed extensively in TB and 50 mM MgCl2. The associated proteins were then eluted with 1 M MgCl2. This fraction was buffer exchanged five times with 100 mM ammonium acetate, pH 8, 0.05% CHAPS, and concentrated in a Minicon. An aliquot of the sample was digested with modified trypsin (Promega) at room temperature for ∼18 h. The digest was frozen at −35°C until analyzed. An aliquot of the digested sample was acidified, and then analyzed by nano-HPLC microelectrospray ionization (μESI) mass spectrometry. All mass spectrometric analyses were performed on an LCQ ion trap mass spectrometer (Finnigan). Briefly, nano-HPLC columns were constructed from 360 × 75-μm fused silica and packed with 7-cm C18 beads (YMC ODS-AQ; Waters). The HPLC gradient was 0–60% B in 70 min, and 60–100% B in 15 min. Solvents A and B were 0.1 M acetic acid in water and 0.1 M acetic acid in 70% acetonitrile, respectively. To determine amino acid sequence, the mass spectrometer was operated in a data-dependent mode in which six scan events were cycled continuously. Scan event 1 was an MS scan from m/z 300 to 2,000, and scan events 2–6 were MS/MS scans on the five most abundant ions detected in scan event 1. The m/z ratio for the ions that had been selected for fragmentation were then dynamically excluded for 1 min from further fragmentation repeat count 1 (pre-exclude time, 30 s). Filtering software was employed to reduce the number of low quality MS/MS spectra and to determine the charge state of the peptides subjected to MS/MS. The filtered MS/MS spectra were searched against S. cerevisiae database using SEQUEST (Eng et al. 1994; Yates et al. 1995). Peptides from the protein databases are scored and ranked based on the similarity between the experimental and theoretical spectra. Peptides with cross correlation scores >2 were manually confirmed. Peptides were synthesized by the solid phase method using standard fmoc chemistry, purified by reverse phase HPLC, and analyzed for sequence.
Recombinant Protein Expression and Binding Studies
Kaps and histone NLS constructs were expressed as GST fusions and purified from bacteria using glutathione-Sepharose. The GST tag was cleaved from purified Kaps using factor Xa (Kap114) or thrombin (Kap121 and Kap95). For binding studies, approximately equal amounts of purified GST-NLS fusion protein were incubated with 20 μl of glutathione-sepharose at 4°C. The glutathione-Sepharose was preblocked with 10% BSA in TB/0.1% Tween-20 (binding buffer). After washing briefly with binding buffer, 16 μl of a TNT (Promega) reaction (see below) in binding buffer was added to the Sepharose and incubated at 4°C for 1 h. For binding studies with bacterially expressed Kap, 1 μg of Kap was substituted for each assay. Where indicated, purified His6-tagged human RanGTP Q69L (a gift from Ian Macara, University of Virginia, Charlottesville, VA) was added to a final concentration of 20 μM. (Human Q69L Ran is much easier to produce in active form than its yeast counterpart. It has previously been shown to bind to yeast Kaps and been used in similar experiments; Schlenstedt et al. 1997.) The Sepharose was then washed three times in binding buffer and once in PBS. The Sepharose was boiled in sample buffer and half the sample (for TNT synthesized Kaps) or the entire sample (for recombinant Kaps) was loaded on an SDS-PAGE gel. The TNT (Promega) reaction was carried out as per manufacturer's instructions using T7 polymerase. 50-μl reactions contained 2 μg of DNA (Kap114, Kap121, Kap108, or Kap95 in pET21a) and 20 μCi of translabel 35S-methionine/cysteine (ICN Biomedicals). 1 μl (2%) of the reaction was removed before histone binding and loaded on gel as “input.”
Results
Kap114p Interacts with Histone H2A and H2B
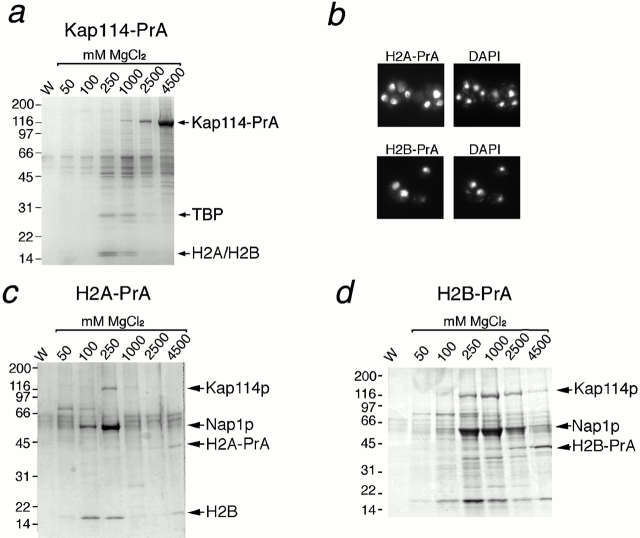
Kap114p was genomically tagged with an in-frame carboxy-terminal fusion of the IgG binding domain of PrA. Kap114-PrA and its associated proteins were isolated by incubation of a post-ribosomal cytosol with IgG-Sepharose and elution with a step gradient of MgCl2 (ranging from 0.05 to 4.5 M). The eluted fractions were analyzed by SDS-PAGE and Coomassie blue staining. Kap114-PrA was eluted at high concentrations of MgCl2 (1–4.5 M). Several other bands were visible at lower MgCl2 concentrations. A few faint bands were also seen in every fraction, including the final wash, suggesting that they may be nonspecific. We have recently shown that the import of TBP into the nucleus is mediated by Kap114p, and TBP was identified among a group of proteins that copurify with Kap114-PrA (Pemberton et al. 1999). In addition, a previously uncharacterized doublet was visible that eluted at the same concentrations as TBP (0.25–1 M) and migrated at ∼16 kD (Fig. 1 a). Each band from this doublet was excised individually and subjected to analysis by MALDI-TOF MS and Edman sequencing. Both methods demonstrated that the bands contained histones H2A and H2B (Edman sequenced residues: H2A amino acids 84–90, H2B amino acids 105–112), suggesting that H2A and H2B may be additional import cargoes for Kap114p.
Figure 1.
Kap114 binds to histones H2A and H2B in the cytoplasm. (a) Kap114-PrA and associated proteins were isolated from cytosol by IgG-Sepharose, eluted with a MgCl2 gradient, separated by SDS-PAGE, and visualized by Coomassie blue staining. The bands representing Kap114-PrA, TBP and histones H2A and H2B are indicated. (b) Cells expressing either H2A-PrA or H2B-PrA were fixed and the PrA moiety detected by indirect immunofluorescence. The coincident DAPI staining is shown. (c) H2A-PrA and associated proteins were isolated from cytosol. Bands representing Kap114p, Nap1p, H2A-PrA, and H2B are indicated. (d) H2B-PrA and associated proteins were isolated as described. Bands representing Kap114p, Nap1p, and H2B-PrA are indicated. Positions of molecular mass standards in kilodaltons are shown.
H2A-PrA and H2B-PrA Localize to the Nucleus
To identify the proteins that may potentially mediate H2A and H2B import, we searched for proteins that interacted with H2A and H2B in cytosol. H2A and H2B are each encoded by two separate genes (Wallis et al. 1980; Choe et al. 1982), and one copy of H2A and H2B were genomically tagged with PrA. We determined where the tagged proteins were localized by visualization of the PrA tags using indirect immunofluorescence on fixed cells. H2A-PrA and H2B-PrA both localized to the nucleus (Fig. 1 b). This nuclear staining pattern suggested that the PrA-tagged copies of the histones were at least functional for nuclear import and were able to interact with the import machinery.
H2A-PrA and H2B-PrA Interact with Kap114p
In separate experiments, either H2A-PrA and interacting proteins or H2B-PrA and interacting proteins were isolated from cytosol as before. The PrA-tagged H2A and H2B both predominantly eluted in 4.5 M MgCl2 fraction as expected (Fig. 1c and Fig. d), and Western blotting confirmed that these bands indeed contained PrA-tagged histone (data not shown). The amount of H2A-PrA and H2B-PrA visible by Coomassie staining appeared low, which may be due to low solubility after MgCl2 elution and methanol-chloroform precipitation (Fig. 1c and Fig. d; note H2B pullout was performed with twice as much cytosol). Consistent with this notion, much larger amounts of histone were obtained by directly solubilizing the proteins bound to the IgG Sepharose in sample buffer (data not shown).
In both cases, the most abundant interacting proteins, migrating at ∼116, 60, and 16 kD, were excised from the gel and analyzed by MS as described (one band of ∼70 kD was not examined thus far). This analysis identified the H2A-interacting proteins as Kap114p, Nap1p, and H2B, respectively, and the H2B-PrA–interacting proteins as Kap114p and Nap1p. In addition, a band was visible that migrated with the same mobility as H2A that has yet to be analyzed. This data correlated with what we observed with Kap114-PrA, and suggested that Kap114p formed a cytoplasmic complex with H2A and H2B. It was not possible from this experiment to determine whether H2A or H2B bound directly to Kap114p as in both cases, both histones were present. Interestingly, with H2A-PrA, H2B appeared to elute in the 100–250-mM and the 4.5-M MgCl2 fractions. Its presence in the 4.5-M fraction suggested that H2A-PrA and H2B were forming a tight heterodimer that was resistant to high concentrations of MgCl2. The presence of H2B in fractions eluting at lower concentrations of MgCl2 suggested that some H2B may be bound less tightly or may interact with H2A via another protein.
H2A and H2B both Contain a Functional NLS
These experiments suggested that both H2A and H2B may be import cargoes for the karyopherin, Kap114p. As these histones are imported as a heterodimer, we wished to determine whether one or both histones contained a functional NLS. Previous studies have predicted that H2B contains a basic NLS in the amino terminus, from residues 22–33 (Moreland et al. 1987). However, H2B lacking these amino acids was also imported, possibly via dimerization with H2A (Moreland et al. 1987). We expressed H2A and H2B fragments that did not include their dimerization domains to determine the precise location of the NLSs (the dimerization domain is located primarily in the alpha-2 helix, see schematic representations in Fig. 2, a and b; Luger et al. 1997). Full-length H2A and H2B were expressed as fusions with two GFP moieties (GFP2) and were strictly nuclear (shown in Fig. 3 b, below). In addition, GFP2 alone, with no NLS, appeared to be mainly cytoplasmic, with no detectable concentration in the nucleus (Fig. 2 a).
Figure 2.
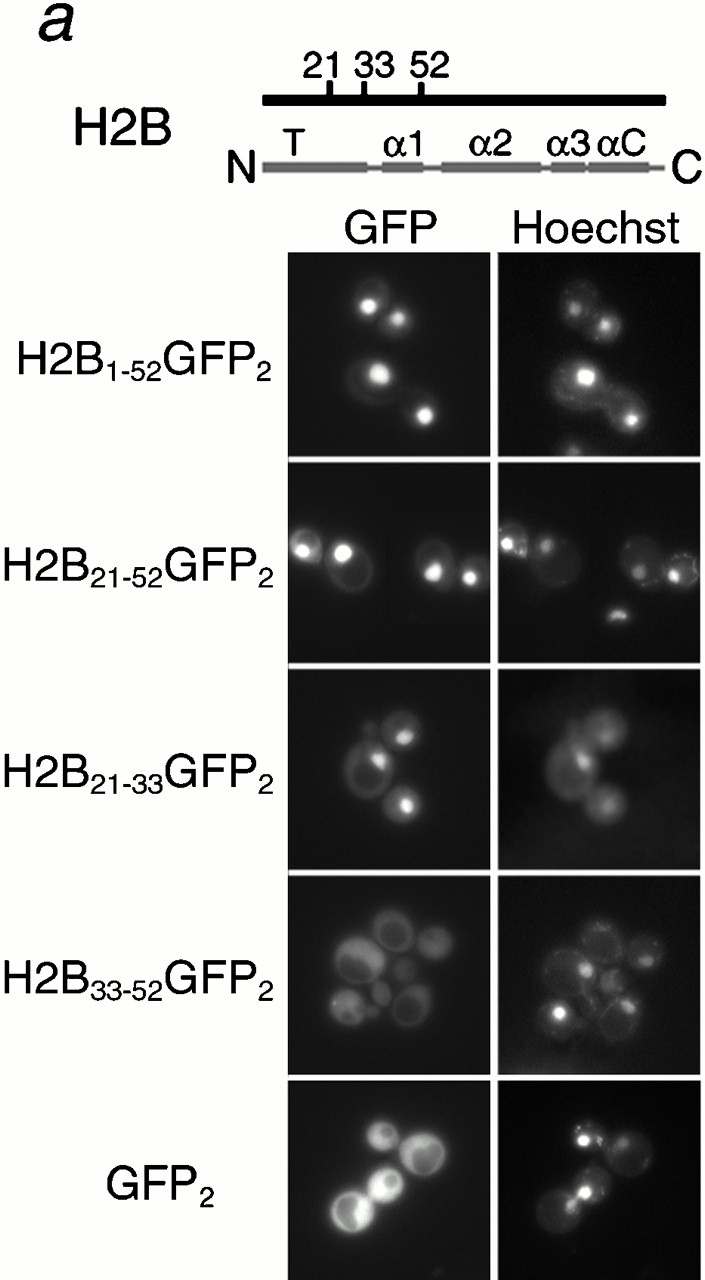
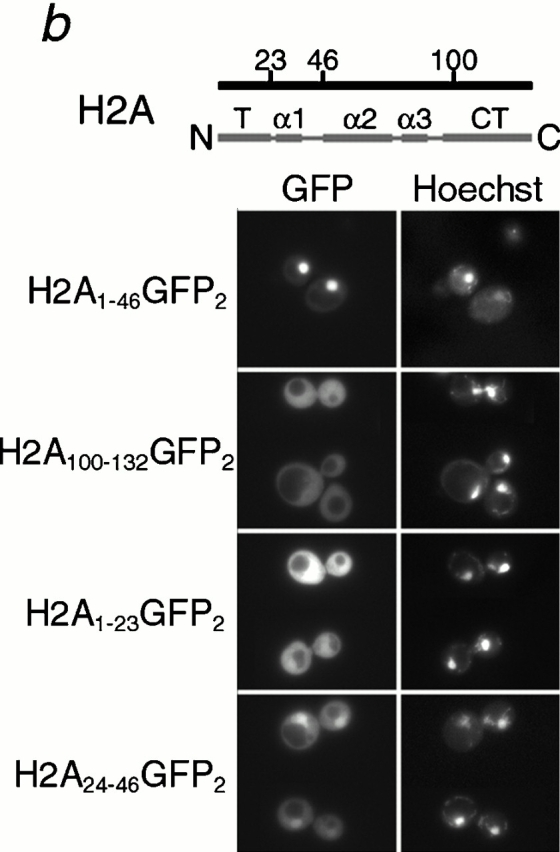
Histone H2A and H2B both contain distinct NLSs. Different fragments of H2B (a) and H2A (b) (as indicated by amino acid number) were expressed as GFP2 fusions in wild-type cells, and the GFP moiety detected by fluorescence imaging. The coincident Hoechst staining is shown. Schematic of H2A and H2B drawn to scale; T, amino-terminal tail; α, alpha helix; αC, carboxy-terminal alpha-helix; CT, carboxy-terminal tail.
Figure 3.
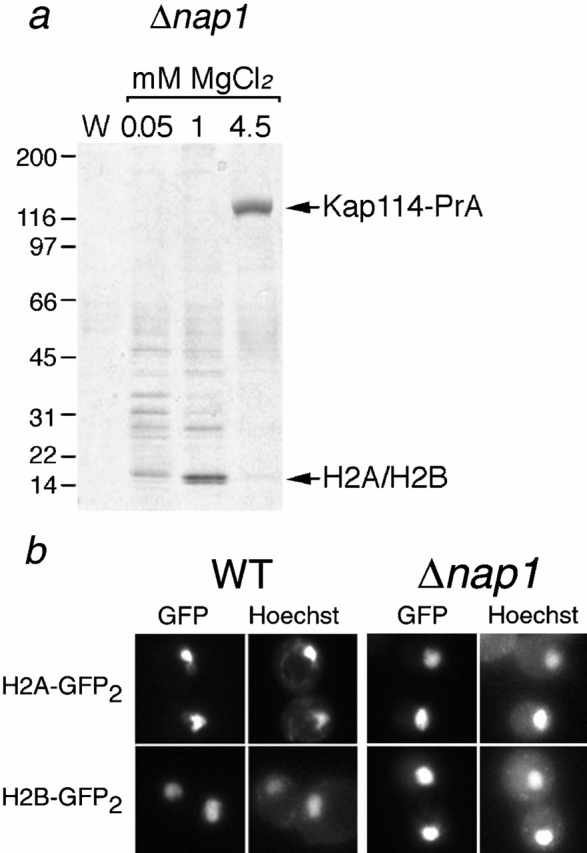
Nap1p is not required for the formation of a functional Kap114p-H2A/H2B complex. (a) Kap114-PrA was expressed in a Δnap1 strain. Kap114-PrA and associated proteins were isolated as described and bands representing Kap114-PrA and H2A/H2B are indicated. Positions of molecular mass standards in kilodaltons are shown. (b) H2A-GFP2 or H2B-GFP2 were expressed in wild-type (WT) and Δnap1 strains and detected by fluorescence imaging. The coincident Hoechst staining is shown.
To determine which domain of H2B contained the NLS, amino acids 1–52, 21–52, and 21–33 of H2B were fused to GFP2 and shown to be sufficient to direct the reporter into the nucleus (Fig. 2 a). In all cases, the fluorescent signal was concentrated in the nucleus and relatively low levels of cytoplasmic signal were visible. In contrast, residues 33–52 of H2B were insufficient to mediate nuclear accumulation of the reporter and the GFP signal was visible throughout the entire cell (Fig. 2 a). Western blotting of these reporters with an anti–GFP antibody showed that the H2B1-52GFP2 and H2B21-33GFP2 fusion proteins were expressed at their expected sizes (data not shown). The H2B NLS we mapped was in good agreement with that determined by Moreland et al. 1987. However, as H2B1-52 was more nuclear than H2B21-33, it is possible that flanking sequences around amino acids 21–33 were also important. In addition, as the predicted H2A–H2B interaction domain was not present in these reporters, it was unlikely they were dimerizing with endogenous H2A (Luger et al. 1997).
Similar experiments were carried out to determine whether H2A had an NLS, and where it was located in the protein. H2A has both an amino and a carboxy terminal tail outside of the main histone fold domain, and we postulated that the putative H2A NLS may be localized there. Amino acids 1–46 and 100–132 were expressed as amino- and carboxy-terminal GFP2 fusions, respectively, in wild-type cells. H2A1-46 was localized to the nucleus while H2A100-132 appeared to be throughout the cell (Fig. 2 b), suggesting that H2A contained an NLS localized in the amino-terminal tail. We also expressed H2A1-23 and H2A24-46 as GFP2 fusions. H2A1-23 was cytoplasmic, whereas for H2A24-46 the nucleus was clearly visible (Fig. 2 b). As the shorter NLS (amino acids 24–46) was significantly more cytoplasmic than the longer one (amino acids 1–46), it suggested that determinants amino terminal to this domain were also important (Fig. 2 b). Thus, both H2A and H2B contain NLSs in their amino-terminal tails.
Nap1p Is Necessary Neither for the Interaction between Kap114p and H2A/H2B, Nor for the Nuclear Localization of H2A and H2B
We identified Nap1p in coprecipitation experiments using both H2A-PrA and H2B-PrA. In addition, we also identified several peptides from Nap1p by liquid chromatography/tandem mass spectrometry of a Kap114-PrA interacting fraction, and by immunoblotting Kap114-PrA interacting proteins with an anti–Nap1p antibody (data not shown). These experiments suggested that Nap1p may be bridging the interaction between the histones and Kap114p, or mediating their import into the nucleus in some other way. To test this, we PrA-tagged Kap114p in a haploid nap1 deletion strain. Western blotting of a whole-cell lysate from this strain with an anti–Nap1p antibody confirmed the absence of Nap1p (data not shown). Kap114-PrA was isolated from cytosol made from this strain and the interacting proteins separated by SDS-PAGE. Consistent with earlier experiments, we detected two Kap114-PrA–interacting proteins that migrated at ∼16 kD (Fig. 3 a). MS analysis of these bands determined that they contained histones H2A and H2B. These results suggest that Nap1p is not required for the interaction of H2A and H2B with Kap114p.
We determined whether Nap1p was required for the nuclear localization of H2A and H2B. The localization of H2A-GFP2 and H2B-GFP2 reporters was examined in wild-type cells and cells lacking Nap1p. We determined that H2A and H2B were localized to the nucleus with virtually no cytoplasmic signal in both wild-type and nap1 deletion strains (Fig. 3 b). These results suggested that Nap1p is not required for the localization of H2A and H2B to the nucleus.
H2A and H2B Interact with other Kaps in the Absence of Kap114p
Our data suggested that H2A and H2B form an abundant cytoplasmic complex with Kap114p, suggesting Kap114p may represent their cognate import factor. Since Kap114p is not essential, whereas histones have an essential nuclear function, we speculated that other Kaps may also participate in the import of H2A and H2B. H2A and H2B were genomically tagged with PrA as before in kap114 deletion strains. After immunoisolation of H2A-PrA or H2B-PrA and separation of their associated proteins by SDS-PAGE from these strains, the visible bands appeared similar to those observed from strains expressing Kap114p (Fig. 4, a and b, compare with Fig. 1c and Fig. d). However, in this case a major band in the Kap range (∼120 kD) was not visible.
Figure 4.
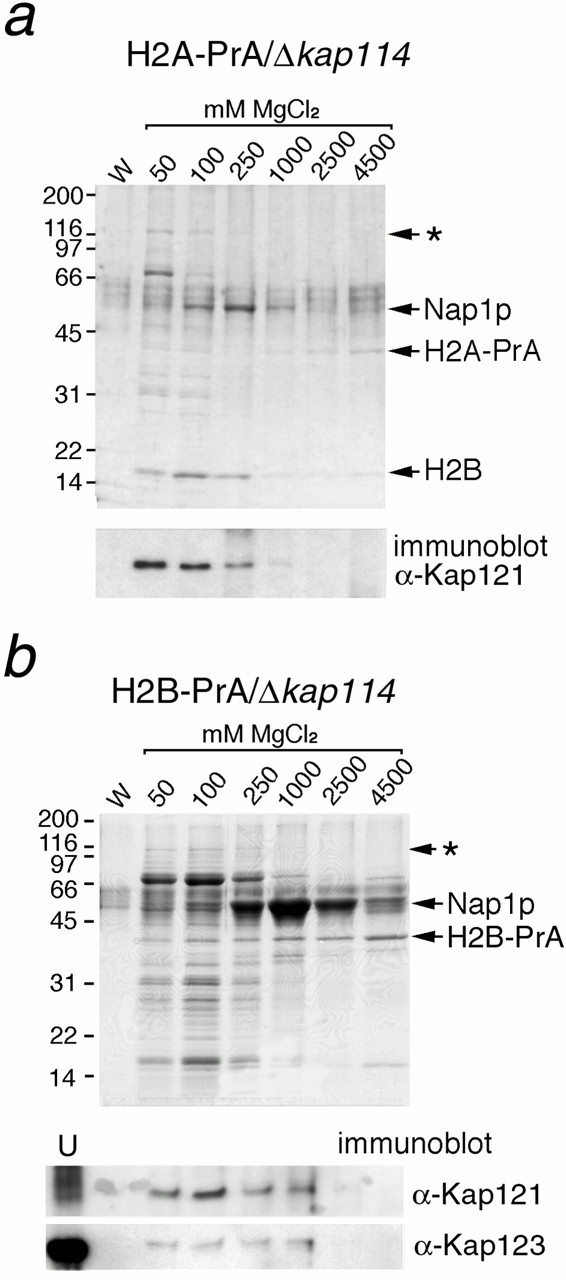
H2A and H2B interact with additional Kaps in the absence of Kap114p. (a) H2A-PrA was expressed in a Δkap114 strain. H2A-PrA and associated proteins were isolated as described. Bands representing potential Kaps (*), Nap1p, H2A-PrA, and H2B are indicated. Identical fractions were Western blotted with an antibody to Kap121p, shown below. (b) H2B-PrA was expressed in a Δkap114 strain. H2B-PrA and associated proteins were isolated as described. Bands representing potential Kaps (*), Nap1p, and H2B-PrA are indicated. The fractions were Western blotted with antibodies to Kap121p and Kap123p as indicated below. (U) Unbound supernatant fraction. Positions of molecular mass standards in kilodaltons are shown.
Bands with the expected mobility of Nap1p were observed (Fig. 4, a and b), and confirmed by Western blotting (data not shown) and MS. Faint bands migrated in the expected position for Kap family members. To determine the interacting proteins more comprehensively, we devised an approach to analyze them all in one eluate. H2A–PrA interacting proteins were eluted by incubation with 1 M MgCl2, enzymatically digested, and the resulting peptides were analyzed by liquid chromatography/tandem mass spectrometry. For comparison, we first analyzed interacting fractions from H2A–PrA strains expressing Kap114p; this fraction contained multiple peptides from H2B, Nap1p, and Kap114p, consistent with our original data. The presence of these proteins was further confirmed by comparison of the MS/MS spectra with those of synthetic peptides. Interacting fractions from H2A-PrA/Δkap114 strains contained no peptides corresponding to Kap114p, as expected, but several peptides corresponding to H2B and Nap1p were detected. In addition, peptides corresponding to karyopherins, Kap121p and Kap123p, were detected. We were also able to confirm the presence of Kap121p in the fraction by Western blotting with anti–Kap121p antibodies (Fig. 4 a). Interestingly, the Kap121p appeared to elute at lower (50–100 mM) MgCl2 concentrations than Kap114p, which eluted at 250 mM MgCl2 (compare Fig. 1 c and 4 a). These experiments suggested that in cytosol H2A-PrA is complexed, directly or indirectly, with H2B, Kap114p, Nap1p, Kap121p, and Kap123p.
We determined whether Kap121p and Kap123p were also present in H2B-PrA/Δkap114 interacting fractions by Western blotting with anti–Kap121p and anti–Kap123 antibodies. This demonstrated that Kap121p and Kap123p were also isolated with H2B-PrA (Fig. 4 b). These data suggested that as with H2A, H2B interacted with Kap114p, Nap1p, Kap121p, and Kap123p.
Several Kaps, Including Kap114p and Kap121p, Import H2A and H2B
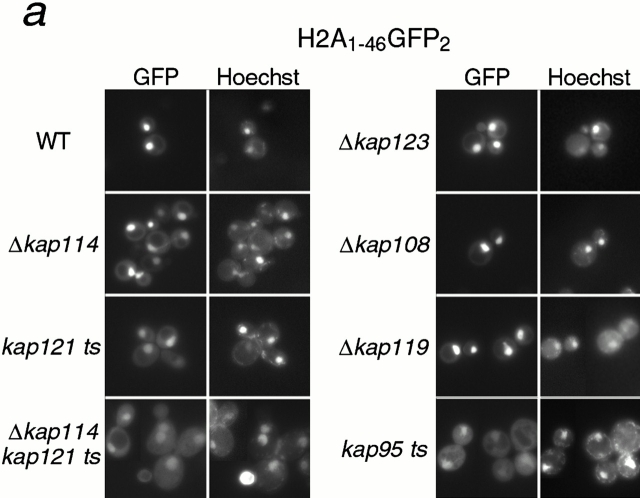
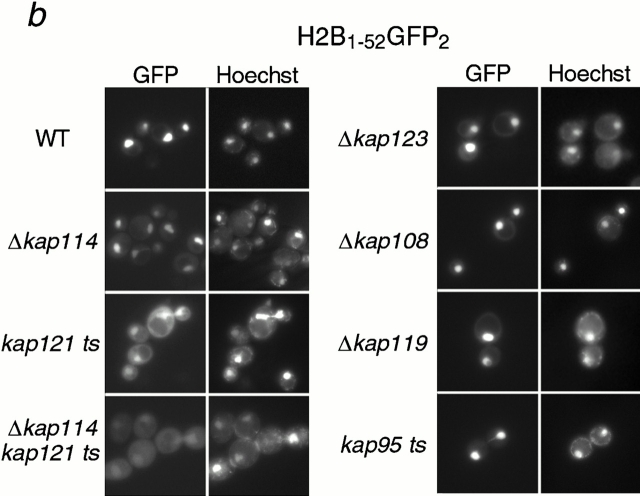
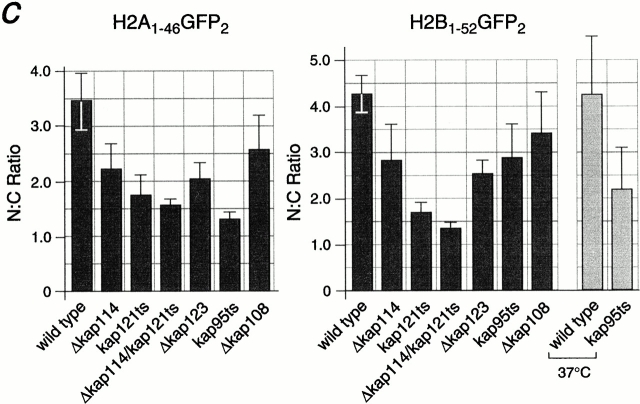
It was possible that Kap114p interacted with either the NLS of H2A or H2B, whereas another Kap may recognize the other NLS. Hence, heterodimerization of H2A and H2B in the cytosol would result in the coisolation of both histones with Kap114-PrA. In addition, either histone would be able to coisolate all of the Kaps involved. To determine the direct role of Kap114p and the additional Kaps in the import of both histones in vivo, we analyzed the localization of the NLS-GFP2 constructs in strains bearing deletions or mutations in the KAP genes. The longer NLS constructs, H2A1-46GFP2 and H2B1-52GFP2, were used for these studies as they were the most nuclear in wild-type cells, but were unlikely to dimerize with endogenous histones. In addition, we quantitated the mean N:C ratios of GFP fluorescence intensity for both reporters in the different strains. In wild-type cells, these reporters were strictly nuclear with an N:C ratio of ∼3.5 (H2A) and ∼4.3 (H2B) (Fig. 5 c). In Δkap114 cells, both reporters were present in the nucleus, although there was more cytoplasmic signal than detected in wild-type cells. This correlated with the observed decrease in the N:C ratio compared with wild type (Fig. 5 c). Due to its coisolation with H2A and H2B, we were particularly interested in the role of Kap121p in histone import. In a kap121ts (pse1-1) strain, which has a temperature-sensitive allele of KAP121, significant mislocalization of both H2A and H2B NLS reporters was observed (Fig. 5, a and b), which correlated with a decrease in the N:C ratio (Fig. 5 c). This decrease was larger than that observed with Δkap114 and more pronounced with H2B. In addition, in a kap121ts/Δkap114 double-mutant strain, mislocalization of both reporters was even greater than in the single mutants, with much of the GFP signal localized to the cytoplasm (Fig. 5 a). This mislocalization was reflected in a further statistically significant decrease in the N:C ratio from that observed in the kap121ts strain. We also determined the effect of shifting these strains to 37°C for 1 h; however, the GFP localization appeared similar to unshifted cells (data not shown). These results show that both Kap121p and Kap114p function in the import of both H2A and H2B in vivo.
Figure 5.
Kap114p, Kap121p, Kap123p, and Kap95p participate in the nuclear import of H2A and H2B. H2A1-46GFP2 (a) or H2B1-52GFP2 (b) were expressed in wild-type (WT) and kap mutant strains (as indicated) and the GFP moiety detected by fluorescence imaging. The coincident Hoechst staining is shown. Strains were grown at 30°C, except for strains with temperature-sensitive alleles (kap121ts and kap95ts), which were grown at room temperature. Images were identically manipulated in Photoshop. (c) Images of the NLS reporter-bearing strains, grown as above or as indicated, were captured by fluorescence imaging. The mean fluorescence intensity of a defined pixel area was measured in the nucleus (N) and cytoplasm (C), and used to calculate the N:C ratio of mean fluorescence intensity. The mean ratio for 50 cells is shown (columns), as well as the SD.
In addition, we had detected a biochemical interaction of H2A and H2B with Kap123p. We observed a slight mislocalization of both reporters to the cytoplasm in the Δkap123 strain, with a concomitant decrease in the N:C ratio, similar to that observed with Δkap114 (Fig. 5, a–c). We observed no mislocalization in the following deletion strains: Δkap119 (nmd5) and Δkap108 (sxm1) (Fig. 5, a and b), as well as Δkap104, Δkap111 (mtr10), Δkap120, and Δkap142 (msn5) (data not shown). However, when we quantitated the N:C ratio of the NLS reporters in the control Δkap108 strain, we did observe a small decrease that was more apparent with H2A. At present, we cannot rule out a minor role in import or nonspecific effects from the mutation. We also tested a kap95ts strain (Koepp et al. 1996). In cells grown at room temperature, we detected significant cytoplasmic mislocalization of the H2A1-46GFP2 reporter (Fig. 5 a). This was reflected in the lowest N:C ratio observed in these experiments (Fig. 5 c). Significantly less mislocalization of H2B1-52GFP2 was observed (Fig. 5 b), although after shifting the cells to 37°C for 3 h the reporter became more mislocalized (data not shown). Additionally, the N:C ratio decreased in contrast to similarly treated wild-type cells (Fig. 5 c). To determine whether this effect was via the Kap60p/Kap95p heterodimer, we expressed the H2A1-46GFP2 and H2B1-52GFP2 fusion proteins in the kap60ts strain, srp1-31 (Loeb et al. 1995). After shifting the temperature to 37°C for 3 h, many of the cells were dead, although cells with intact nuclei still expressed the GFP2 reporter in the nucleus (data not shown).
These results suggested that Kap114p, Kap121p, Kap123p, and Kap95p play a role in the import of histones H2A and H2B. In addition, the role of Kap114p, Kap121p, and Kap123p has been suggested by the isolation of endogenous proteins in a complex with H2A and H2B.
Kap114p, Kap121p, and Kap95p Interact Directly with the NLSs of H2A and H2B
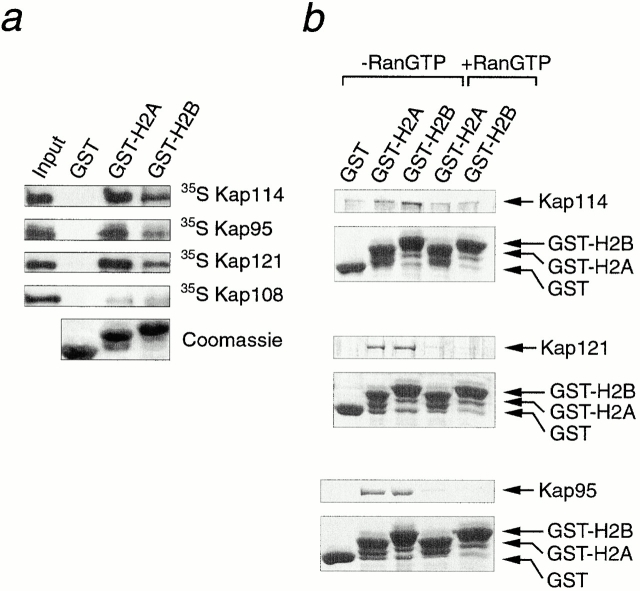
The observed mislocalization of H2A and H2B NLS reporters in various Kap mutant strains suggested that several Kaps may interact with these domains. The primary sequences of the H2A and H2B NLSs are not significantly similar, although they both contain several basic residues, characteristic of histone tails. We therefore used in vitro binding studies to determine whether Kap114p, as well as other Kaps, could interact with recombinant NLS domains from H2A and H2B. Approximately equal amounts of GST-H2A1-46, GST-H2B1-52, and GST alone were bound to glutathione-Sepharose. The Sepharose was incubated with reticulocyte lysate containing 35S-methionine-labeled Kap114p, Kap95p, Kap121p, or the control Kap, Kap108p. After washing and separating the bound proteins by SDS-PAGE, the gel was fluorographed. Kap114p bound to both the H2A and H2B NLSs, but not to GST alone (Fig. 6 a). Interestingly, Kap95p and Kap121p also bound to the H2A and H2B NLSs, but not to GST (Fig. 6 a), with more Kap95p binding to the H2A NLS than the H2B NLS. These results were consistent with the observed cytoplasmic mislocalization of the NLS-GFP fusions in the mutant strains (Fig. 5). Only low levels of binding to Kap108p could be detected, which is consistent with our biochemical and in vivo experiments, which do not implicate this Kap as a major import factor for either H2A or H2B.
Figure 6.
Kap114p, Kap121p, and Kap95p bind directly to the H2A and H2B NLSs. (a) Reticulocyte lysate containing 35S-methionine–labeled Kap114p, Kap95p, Kap121p, or Kap108p was incubated with Sepharose-bound GST (GST), GST-H2A amino acids 1–46 (GST-H2A), or GST-H2B amino acids 1–52 (GST-H2B). The bound material was separated by SDS-PAGE. Kaps were visualized by fluorography of the gels (top four panels). The reticulocyte lysate input (2% of reaction) is also shown. The GST and GST-fusion proteins for a representative experiment, visualized by Coomassie blue staining, are shown (bottom). (b) 1 μg of purified, bacterially expressed Kap114p, Kap121p, or Kap95p was incubated with Sepharose-bound GST (GST), GST-H2A amino acids 1–46 (GST-H2A), or GST-H2B amino acids 1–52 (GST-H2B). Where indicated, the Kap was preincubated with 20 μM human RanGTP Q69L. The bound material was separated by SDS-PAGE and visualized by Coomassie blue staining. Positions of GST-fusion proteins are indicated; additional lower bands are probably degradation products.
We wished to determine whether the binding observed for Kap114p, Kap121p, and Kap95p to the histone NLSs was mediated by direct interaction. It was possible that proteins present in the reticulocyte lysate were bridging the interaction. These Kaps were expressed and purified from bacteria as GST fusions. After removal of the GST moiety, binding assays were performed with purified protein as above. Analysis of the bound fractions revealed that recombinant Kap114p bound to the H2A and H2B NLSs (Fig. 6 b). Kap114p was difficult to purify in a cleaved and active form, and some binding was observed between Kap114p and GST, although the relative amount was less than that bound to either histone NLS. Similar binding assays were performed with recombinant Kap121p and Kap95p, and both of these Kaps bound specifically to both histone NLSs and not GST alone (Fig. 6 b). A maltose binding protein–Kap123p fusion also bound to the H2A and H2B NLSs and not to GST alone (data not shown). To show these Kap-NLS interactions were specific, each Kap was preincubated with a Ran mutant that was deficient in GTP hydrolysis, and consequently remains GTP bound. The binding of Kap121p and Kap95p to the histone NLSs was nearly completely prevented in the presence of RanGTP. The level of Kap114p binding was also reduced, although as this Kap also bound nonspecifically to GST it was not possible to determine how much binding was inhibited. These experiments suggest that Kap121p, Kap95p, and Kap114p bind directly to the NH2-terminal tails of H2A and H2B.
Discussion
Here we show for the first time that histones H2A and H2B both contain distinct NLSs in their amino-terminal tails. We demonstrate that the import factor Kap114p forms a cytoplasmic complex with the H2A/H2B heterodimer, via direct interactions with both NLSs. H2A and H2B also interact with other Kaps, including Kap121p, Kap95p, and Kap123p. Experiments in vivo suggest that each of these Kaps participates in H2A/H2B import and we propose that the import of abundant, essential proteins such as histones is mediated by a network of Kaps.
Histones H2A and H2B are imported into the nucleus as a heterodimer. We have demonstrated that both H2A and H2B have a functional NLS in their amino termini. Amino acids 21–33 of H2B represents a “minimal” NLS domain, which correlates with the NLS previously reported by Moreland et al. 1987 in fixed cells. H2B1-52 was more nuclear, suggesting that the amino acids flanking the minimal domain are also important. We identified an NLS domain in H2A consisting of amino acids 1–46, although residues 24–46 gave some nuclear accumulation. In vitro binding experiments showed that amino acids H2A1-46 and H2B1-52 were sufficient to confer Kap114p binding. In vivo, it remains to be determined whether one or both NLSs in the H2A/H2B heterodimer simultaneously bind to Kap114p. Comparison of these two amino-terminal NLS sequences does not reveal any obvious similarities. Since the amino-terminal domains of histones have been shown to be substrates for several posttranslational modifications (Strahl and Allis 2000), it is possible that these modifications may also play a role in NLS recognition. Similarly, posttranslational modifications of the transcription factor Pho4p regulate its interaction with its import Kap (Kaffman et al. 1998).
Kap114p also imports TBP into the nucleus, and this Kap may have several cargoes (Pemberton et al. 1999). It is intriguing that the same Kap imports the H2A/H2B histone dimer and TBP. These proteins are fundamental for all aspects of gene regulation, but are generally perceived to have opposing effects. Sequence comparison of the NLSs of H2A and H2B with TBP does not reveal any obvious similarities at the amino acid level. Kap114p interacts directly with the NLSs of H2A, H2B, and with TBP (Pemberton et al. 1999). In vivo, we do not know whether the Kap114p-TBP and Kap114p-H2A/H2B complexes are assembled independently or on the same Kap. How three apparently different NLSs could interact with the same domain on Kap114p is unclear, and structural determination of Kap114-cargo interactions should elucidate the mechanism of binding.
The NLS of H2B is rich in basic amino acids, suggesting it may represent a “classical” NLS (Moreland et al. 1987). However, we have no evidence of Kapα binding to either the H2A or H2B NLS by PrA coimmunoprecipitation or by localization of the reporters in the Kapα ts strain, srp1-31. This data correlates with experiments in mammalian cells, where H2B import was not blocked by addition of excess SV40 NLS peptide in a permeabilized cell system (Langer 2000), suggesting that histones are imported by an α-independent pathway.
The biochemical coisolation of endogenous Kap114p, Kap121p, and Kap123p with H2A-PrA and H2B-PrA suggested that these Kaps mediate the import of H2A and H2B. Kap114p may play a more major role in H2A/H2B import, suggested by the fact that Kap114p was the predominantly isolated Kap. Kap114p is less abundant than Kap121p, Kap123p, and Kap95p (Morehouse et al. 1999; Pemberton et al. 1999). It is intriguing that when H2A-PrA, H2B-PrA, and TBP-PrA cytosolic complexes were purified, Kap121p and Kap123p were only readily detectable in the absence of Kap114p, and these Kaps were eluted from histones with lower concentrations of MgCl2. This suggests that these cargoes preferentially bind Kap114p before Kap121p and Kap123p.
To determine the role of these Kaps in vivo, we investigated the localization of H2A NLS-GFP and H2B NLS-GFP reporters in the corresponding Kap mutant strains. The H2A and H2B NLS-GFP reporters were mislocalized in strains with mutations in kap114, kap121, kap123, and kap95. Quantitation of fluorescence showed a significant decrease in the N:C ratios between wild-type and these mutant strains. In addition, an additive effect was measured in the Δkap114/kap121 ts mutant, with significantly more cytoplasmic mislocalization of both reporters than was detected in either single mutant. This demonstrates that both these Kaps function in histone import in vivo. We do not know why the effect was greater in the kap121ts strain than the Δkap114 strain, as H2A and H2B appear to preferentially bind Kap114p. It is possible that there are other pleiotropic defects in nuclear transport in this temperature-sensitive strain.
These experiments also suggested that Kap95p functioned in H2A import and to a lesser extent H2B import. We did not detect Kap95p by MS in our immunoprecipitation experiments using endogenous proteins. We could, however, show binding of Kap95p to the H2A and H2B NLSs using recombinant proteins. In addition, published studies using recombinant mammalian proteins in ELISAs suggest that Kapβ1 (human Kap95) may directly bind H2A/H2B in vitro (Johnson-Saliba et al. 2000). We do not know whether additional Kaps can import histones H2A and H2B. Surprisingly, we did detect a small decrease in the N:C ratio of our “control” Kap, Kap108, that was not obvious from the fluorescent images. This decrease was less than that observed for kap114 and kap123 deletions. We cannot rule out the possibility that Kap108p plays a very minor role in import, or that this a pleiotropic defect of the mutant strain. Together, our in vivo data, both the isolation of endogenous protein complexes and the localization of NLS reporters in mutant strains, suggest a major role for Kap114p and Kap121p, and Kap123p and Kap95p in H2A and H2B import.
Our in vitro binding studies using recombinant NLSs and Kaps expressed in reticulocyte lysate or purified from bacteria confirmed that the same NLSs are recognized by Kap114p, Kap121p, Kap95p, and Kap123p. We were able to show that these interactions were specific as they were blocked by preincubation of the Kaps with RanGTP. Future experiments should determine whether additional factors stimulate the RanGTP-mediated dissociation of histones from Kaps, as was observed with Kap114p and TBP (Pemberton et al. 1999). Although both H2A and H2B interact similarly with Kap114p, our results suggest that there maybe some subtle differences between the interactions of H2A and H2B with the other Kaps. We have evidence that more Kap121p and Kap123p are isolated with H2B. In fact, Kap123p was detected in the H2A-PrA coisolation by MS, but not detected by Western blotting, suggesting that it was present at lower levels than observed with H2B-PrA. In addition, we observed a more dramatic decrease in the N:C ratio of the H2B NLS-GFP, than of H2A NLS-GFP, in the kap121ts strain. In contrast, evidence that Kap95p appears to function preferentially with H2A comes from both the in vivo localization and in vitro binding experiments. It is interesting that these Kaps, which have low apparent sequence homology, bound the same NLSs in H2A and H2B.
In addition to the import of TBP, which is mediated by the same subgroup of Kaps discussed here (Pemberton et al. 1999), the only other published examples of multiple Kaps importing a single cargo are in the Kap123p- and Kap121p-mediated import of yeast ribosomal proteins. There, Kap123p mediates the primary route into the nucleus (Rout et al. 1997). Multiple Kaps have also been shown to import mammalian ribosomal proteins into the nucleus in vitro (Jakel and Gorlich 1998). However, in these experiments, it is not possible to compare the specific in vivo requirement for, and the contribution by, each Kap. Further examples of networks of import Kaps need to be characterized to determine their importance and hierarchy in vivo. We suggest that the import of essential proteins by several Kaps will represent a broadly relevant model.
It is probable that Nap1p interacts with the Kap114p-histone complex via one of the histones. The majority of cytosolic Nap1p is not complexed with Kap114p (Pemberton, L.F., unpublished data) and may be interacting with a pool of free histone monomers or dimers. One model is that Nap1p binds to histones as they are synthesized and, upon the interaction of Kap114p with the histone dimer, some fraction of Nap1p remains bound to the Kap114p-histone complex. This may be the route of Nap1p into the nucleus, where it carries out chromatin assembly functions. Immunofluorescence studies have demonstrated that the majority of Nap1p remains in the cytoplasm, and distinct cytoplasmic functions in mitosis have been determined (Kellogg et al. 1995; Kellogg and Murray 1995; Ito et al. 1996). We have not detected any Nap1p in a complex with Kap123-PrA or Kap121-PrA in the presence of Kap114p (Pemberton, L.F., unpublished data). This may be due to the fact that there is little histone bound to these Kaps in the presence of Kap114p, or that Nap1p may be functionally linked to the Kap114p-mediated import pathway. It is possible that Nap1p may help regulate the interaction between histones and Kap114p.
We propose a model where histone H2A/H2B is synthesized and heterodimerizes in the cytoplasm, and is bound by the chaperone Nap1p. The majority of this complex is imported into the nucleus by Kap114p. However, a fraction of H2A/H2B is imported by Kap121p or Kap123p, preferentially binding via H2B, and some is imported by Kap95p, binding via H2A. Future experiments will determine what factors regulate which Kap pathway is used, and whether different pathways are used under different conditions.
H2A and H2B are synthesized in S phase and must rapidly enter the nucleus as DNA is replicated. The production and import of H2A and H2B needs to also be coordinated with that of H3 and H4. Because of the importance of having the correct amounts of histones in the nucleus, it is not surprising that the import of these proteins is mediated by several independent transport factors. We propose that all essential proteins may have more than one route into the nucleus, explaining why the Kap family members are functionally overlapping.
Acknowledgments
We thank Günter Blobel, in whose laboratory this work was initiated, for his kind help and support. We thank Robert Moreland (Abbott Labs) for sharing his unpublished results and reagents. We thank the following for reagents and strains: Susana Chaves, Rick Wozniak, John Aitchison, Douglas Kellogg, Pam Silver, Kimi Yoshida, and Ian Macara. We thank Judson Frye for technical help. We acknowledge the Rockefeller DNA Technology Center for MALDI MS and Edman sequencing. We thank Jonathan Rosenblum and David Wotton for helpful comments on the manuscript.
N. Mosammaparast is supported by the National Institutes of Health (NIH) Medical Scientist Training Program. This work was supported in part by a grant from the United States Public Health Service to D.F. Hunt (NIH GM-37537).
Footnotes
Abbreviations used in this paper: GFP, green fluorescent protein; GST, glutathione-S-transferase; MALDI-TOF, matrix-assisted laser desorption/ionization time of flight; MS, mass spectrometry; N:C, nuclear:cytoplasmic; NLS, nuclear localization sequence; NPC, nuclear pore complex; PrA, protein A; TBP, TATA binding protein.
References
- Adams C.R., Kamakaka R.T. Chromatin assemblybiochemical identities and genetic redundancy. Curr. Opin. Genet. Dev. 1999;9:185–190. doi: 10.1016/S0959-437X(99)80028-8. [DOI] [PubMed] [Google Scholar]
- Aitchison J.D., Blobel G., Rout M.P. Kap104pa karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- Aitchison J.D., Rout M.P., Marelli M., Blobel G., Wozniak R.W. Two novel related yeast nucleoporins Nup170p and Nup157pcomplementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J. Cell Biol. 1995;131:1133–1148. doi: 10.1083/jcb.131.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertini M., Pemberton L.F., Rosenblum J.S., Blobel G. A novel nuclear import pathway for the transcription factor TFIIS. J. Cell Biol. 1998;143:1447–1455. doi: 10.1083/jcb.143.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeuwer M., Goldfarb D.S. Facilitated nuclear transport of histone H1 and other small nucleophilic proteins. Cell. 1990;60:999–1008. doi: 10.1016/0092-8674(90)90348-i. [DOI] [PubMed] [Google Scholar]
- Chang L., Loranger S.S., Mizzen C., Ernst S.G., Allis C.D., Annunziato A.T. Histones in transitcytosolic histone complexes and diacetylation of H4 during nucleosome assembly in human cells. Biochemistry. 1997;36:469–480. doi: 10.1021/bi962069i. [DOI] [PubMed] [Google Scholar]
- Choe J., Kolodrubetz D., Grunstein M. The two yeast histone H2A genes encode similar protein subtypes. Proc. Natl. Acad. Sci. USA. 1982;79:1484–1487. doi: 10.1073/pnas.79.5.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enenkel C., Blobel G., Rexach M. Identification of a yeast karyopherin heterodimer that targets import substrate to mammalian nuclear pore complexes. J. Biol. Chem. 1995;270:16499–16502. doi: 10.1074/jbc.270.28.16499. [DOI] [PubMed] [Google Scholar]
- Eng J., McCormack A.L., Yates J.R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Fernandez J., Andrews L., Mische S.M. An improved procedure for enzymatic digestion of polyvinylidene difluoride-bound proteins for internal sequence analysis. Anal. Biochem. 1994;218:112–117. doi: 10.1006/abio.1994.1148. [DOI] [PubMed] [Google Scholar]
- Gharahdaghi F., Kirchner M., Fernandez J., Mische S.M. Peptide-mass profiles of polyvinylidene difluoride-bound proteins by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in the presence of nonionic detergents. Anal. Biochem. 1996;233:94–99. doi: 10.1006/abio.1996.0012. [DOI] [PubMed] [Google Scholar]
- Görlich D., Dabrowski M., Bischoff F.R., Kutay U., Bork P., Hartmann E., Prehn S., Izaurralde E. A novel class of RanGTP binding proteins. J. Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D., Kutay U. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell. Dev. Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- Ishimi Y., Hirosumi J., Sato W., Sugasawa K., Yokota S., Hanaoka F., Yamada M. Purification and initial characterization of a protein which facilitates assembly of nucleosome-like structure from mammalian cells. Eur. J. Biochem. 1984;142:431–439. doi: 10.1111/j.1432-1033.1984.tb08305.x. [DOI] [PubMed] [Google Scholar]
- Ito T., Bulger M., Kobayashi R., Kadonaga J.T. Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Mol. Cell. Biol. 1996;16:3112–3124. doi: 10.1128/mcb.16.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E., Kutay U., von Kobbe C., Mattaj I.W., Gorlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakel S., Albig W., Kutay U., Bischoff F.R., Schwamborn K., Doenecke D., Gorlich D. The importin beta/importin 7 heterodimer is a functional nuclear import receptor for histone H1. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:2411–2423. doi: 10.1093/emboj/18.9.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakel S., Gorlich D. Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Saliba M., Siddon N.A., Clarkson M.J., Tremethick D.J., Jans D.A. Distinct importin recognition properties of histones and chromatin assembly factors. FEBS Lett. 2000;467:169–174. doi: 10.1016/s0014-5793(00)01142-x. [DOI] [PubMed] [Google Scholar]
- Kaffman A., Rank N.M., O'Shea E.K. Phosphorylation regulates association of the transcription factor Pho4 with its import receptor Pse1/Kap121. Genes Dev. 1998;12:2673–2683. doi: 10.1101/gad.12.17.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg D.R., Kikuchi A., Fujii-Nakata T., Turck C.W., Murray A.W. Members of the NAP/SET family of proteins interact specifically with B-type cyclins. J. Cell Biol. 1995;130:661–673. doi: 10.1083/jcb.130.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg D.R., Murray A.W. NAP1 acts with Clb1 to perform mitotic functions and to suppress polar bud growth in budding yeast. J. Cell Biol. 1995;130:675–685. doi: 10.1083/jcb.130.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepp D.M., Wong D.H., Corbett A.H., Silver P.A. Dynamic localization of the nuclear import receptor and its interactions with transport factors. J. Cell Biol. 1996;133:1163–1176. doi: 10.1083/jcb.133.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R.D., Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- Kurz M., Doenecke D., Albig W. Nuclear transport of H1 histones meets the criteria of a nuclear localization signal-mediated process. J. Cell. Biochem. 1997;64:573–578. [PubMed] [Google Scholar]
- Langer T. Nuclear transport of histone 2b in mammalian cells is signal- and energy-dependent and different from the importin alpha/beta-mediated process. Histochem. Cell Biol. 2000;113:455–465. doi: 10.1007/s004180000147. [DOI] [PubMed] [Google Scholar]
- Lee D.C., Aitchison J.D. Kap104p-mediated nuclear import. Nuclear localization signals in mRNA-binding proteins and the role of Ran and Rna. J. Biol. Chem. 1999;274:29031–29037. doi: 10.1074/jbc.274.41.29031. [DOI] [PubMed] [Google Scholar]
- Loeb J.D., Schlenstedt G., Pellman D., Kornitzer D., Silver P.A., Fink G.R. The yeast nuclear import receptor is required for mitosis. Proc. Natl. Acad. Sci. USA. 1995;92:7647–7651. doi: 10.1073/pnas.92.17.7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K., Mader A.W., Richmond R.K., Sargent D.F., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Marchetti M.A., Tschudi C., Kwon H., Wolin S.L., Ullu E. Import of proteins into the trypanosome nucleus and their distribution at karyokinesis. J. Cell Sci. 2000;113:899–906. doi: 10.1242/jcs.113.5.899. [DOI] [PubMed] [Google Scholar]
- Marelli M., Aitchison J.D., Wozniak R.W. Specific binding of the karyopherin Kap121p to a subunit of the nuclear pore complex containing Nup53p, Nup59p, and Nup170p. J. Cell Biol. 1998;143:1813–1830. doi: 10.1083/jcb.143.7.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M.S. Ran and nuclear transport. J. Biol. Chem. 1998;273:22857–22860. doi: 10.1074/jbc.273.36.22857. [DOI] [PubMed] [Google Scholar]
- Morehouse H., Buratowski R.M., Silver P.A., Buratowski S. The importin/karyopherin Kap114 mediates the nuclear import of TATA-binding protein. Proc. Natl. Acad. Sci. USA. 1999;96:12542–12547. doi: 10.1073/pnas.96.22.12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreland R.B., Langevin G.L., Singer R.H., Garcea R.L., Hereford L.M. Amino acid sequences that determine the nuclear localization of yeast histone 2B. Mol. Cell. Biol. 1987;7:4048–4057. doi: 10.1128/mcb.7.11.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S., Dreyfuss G. Transport of proteins and RNAs in and out of the nucleus. Cell. 1999;99:677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- Niedenthal R.K., Riles L., Johnston M., Hegemann J.H. Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast. 1996;12:773–786. doi: 10.1002/(SICI)1097-0061(19960630)12:8%3C773::AID-YEA972%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Pemberton L.F., Blobel G., Rosenblum J.S. Transport routes through the nuclear pore complex. Curr. Opin. Cell Biol. 1998;10:392–399. doi: 10.1016/s0955-0674(98)80016-1. [DOI] [PubMed] [Google Scholar]
- Pemberton L.F., Rosenblum J.S., Blobel G. A distinct and parallel pathway for the nuclear import of an mRNA-binding protein. J. Cell Biol. 1997;139:1645–1653. doi: 10.1083/jcb.139.7.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton L.F., Rosenblum J.S., Blobel G. Nuclear import of the TATA-binding proteinmediation by the karyopherin Kap114p and a possible mechanism for intranuclear targeting. J. Cell Biol. 1999;145:1407–1417. doi: 10.1083/jcb.145.7.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach M., Blobel G. Protein import into nucleiassociation and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- Rosenblum J.S., Pemberton L.F., Blobel G. A nuclear import pathway for a protein involved in tRNA maturation. J. Cell Biol. 1997;139:1655–1661. doi: 10.1083/jcb.139.7.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum J.S., Pemberton L.F., Bonifaci N., Blobel G. Nuclear import and the evolution of a multifunctional RNA-binding protein. J. Cell Biol. 1998;143:887–899. doi: 10.1083/jcb.143.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout M.P., Aitchison J.D., Suprapto A., Hjertaas K., Zhao Y., Chait B.T. The yeast nuclear pore complexcomposition, architecture, and transport mechanism. J. Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout M.P., Blobel G., Aitchison J.D. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- Schlenstedt G., Smirnova E., Deane R., Solsbacher J., Kutay U., Görlich D., Ponstingl H., Bischoff F.R. Yrb4p, a yeast ran-GTP-binding protein involved in import of ribosomal protein L25 into the nucleus. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:6237–6249. doi: 10.1093/emboj/16.20.6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seedorf M., Silver P.A. Importin/karyopherin protein family members required for mRNA export from the nucleus. Proc. Natl. Acad. Sci. USA. 1997;94:8590–8595. doi: 10.1073/pnas.94.16.8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger B., Simos G., Bischoff F.R., Podtelejnikov A., Mann M., Hurt E. Mtr10p functions as a nuclear import receptor for the mRNA-binding protein Npl3p. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:2196–2207. doi: 10.1093/emboj/17.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Fink G.R., Hicks J.B. Methods in yeast genetics. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1986. [Google Scholar]
- Strahl B.D., Allis C.D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Titov A.A., Blobel G. The karyopherin Kap122p/Pdr6p imports both subunits of the transcription factor IIA into the nucleus. J. Cell Biol. 1999;147:235–246. doi: 10.1083/jcb.147.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis J.W., Hereford L., Grunstein M. Histone H2B genes of yeast encode two different proteins. Cell. 1980;22:799–805. doi: 10.1016/0092-8674(80)90556-5. [DOI] [PubMed] [Google Scholar]
- Wozniak R.W., Rout M.P., Aitchison J.D. Karyopherins and kissing cousins. Trends Cell Biol. 1998;8:184–188. doi: 10.1016/s0962-8924(98)01248-3. [DOI] [PubMed] [Google Scholar]
- Yates J.R., III, Eng J.K., McCormack A.L., Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 1995;67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- Yoshida K., Blobel G. The karyopherin Kap142p/Msn5p mediates nuclear import and nuclear export of different cargo proteins. J. Cell Biol. 2001;152:729–740. doi: 10.1083/jcb.152.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]