Abstract
During anaphase, mitotic spindles elongate up to five times their metaphase length. This process, known as anaphase B, is essential for correct segregation of chromosomes. Here, we examine the control of spindle length during anaphase in the budding yeast Saccharomyces cerevisiae. We show that microtubule stabilization during anaphase requires the microtubule-associated protein Stu2. We further show that the activity of Stu2 is opposed by the activity of the kinesin-related protein Kip3. Reexamination of the kinesin homology tree suggests that KIP3 is the S. cerevisiae orthologue of the microtubule-destabilizing subfamily of kinesins (Kin I). We conclude that a balance of activity between evolutionally conserved microtubule-stabilizing and microtubule-destabilizing factors is essential for correct spindle elongation during anaphase B.
Keywords: stu2, checkpoint, yeast, spindle, anaphase
Introduction
At metaphase, sister chromatids are joined together and attached to the microtubules of the metaphase spindle (for reviews see Hyman and Karsenti 1996; Nicklas 1997; Saunders 1999). After the metaphase-to-anaphase transition, sister chromatids separate, and chromosomes segregate apart from one another. Chromosome segregation occurs in two phases: chromosomes move towards spindle poles during anaphase A, and spindle poles separate from each other during anaphase B. Although anaphase A requires that the chromosome-to-pole microtubules shorten, anaphase B requires that pole-to-pole microtubules elongate. Analysis of anaphase B in Saccharomyces cerevisiae (Straight et al. 1997) suggests that spindle elongation proceeds in two stages. Initially overlapping microtubules of the metaphase spindle slide apart, probably as a consequence of the action of bipolar microtubule-based motors. Subsequent elongation is driven by coupling the polymerization of microtubule plus ends to the sliding of opposing spindle pole microtubules. This sequence of events has been best analyzed during the elongation of isolated diatom spindles in vitro. These spindles will elongate after addition of ATP. However, once they elongate past the spindle overlap zone, spindles then break into two halves unless tubulin is added to the reaction (Baskin and Cande 1990). Although it seems likely that microtubule motors drive spindle sliding, the mechanisms by which microtubule growth is coupled to anaphase B spindle elongation are unknown.
The Dis1 family of microtubule-associated proteins plays a central role in the regulation of microtubule polymerization in diverse eukaryotic species. Members of this protein family have been shown to promote microtubule growth from yeast to mammals (Charrasse et al. 1998; Matthews et al. 1998; Cullen et al. 1999; Tournebize et al. 2000). Removal of the Xenopus homologue XMAP215 from cytoplasmic extracts prevents microtubule growth, whereas embryos with mutations in the Caenorhabditis elegans homologue zyg-9 have short microtubules. Work in Xenopus extracts has shown that the stabilizing activity of XMAP215 opposes the destabilizing activity of a member of the Kin I subfamily of kinesins, XKCM1 (Tournebize et al. 2000). However, neither the role of XKCM1–XMAP215 pathway during anaphase nor its relationship to control of microtubule dynamics in organisms other then Xenopus is known. The central role of the Dis1 family proteins in regulation of microtubule dynamics suggested to us that the function of this protein family may be important for microtubule stabilization during anaphase B.
The metaphase spindle of a haploid yeast S. cerevisiae is 1.5–2 μm long, but elongates to 7–10 μm by the end of anaphase. This fact, combined with the powerful molecular genetics of S. cerevisiae, makes budding yeast a very good experimental organism to study factors involved in spindle elongation. The roles of S. cerevisiae kinesins in spindle function were revealed by the works of Saunders and Hoyt 1992, Saunders and Hoyt 1997 and Hoyt 1994. We used this system to analyze the coupling of microtubule growth to spindle elongation during anaphase. Given the central role of Dis1 family of proteins in regulation of microtubule dynamics throughout eukaryotes, we focused on the role of this class of microtubule-associated proteins during anaphase B. The orthologue of dis1 + in S. cerevisiae is STU2, initially identified as a suppressor of a β-tubulin mutation. STU2 is an essential gene and its product is localized to the spindle and spindle poles (Wang and Huffaker 1997). However, its detailed role in spindle elongation and regulation of microtubule dynamics during anaphase has not been analyzed. To examine the role of Stu2 in spindle elongation, we have characterized the phenotypic consequences arising from STU2 inhibition. A temperature-sensitive allele of stu2 arrests at metaphase by engaging the Mad2-dependent spindle checkpoint. Combining a mad2Δ deletion with the inactivation of stu2 results in exit from mitosis in the absence of spindle elongation. Remarkably, spindle elongation can be restored in these double mutants by introducing a deletion of the kinesin-related protein Kip3. These results show that stabilization of microtubules during spindle elongation requires the opposed activities of Stu2 and Kip3, and that Kip3 may be the functional orthologue of the Kin I family of microtubule-destabilizing kinesins in S. cerevisiae.
Materials and Methods
The strains used in this study are all derivatives of W303 and are shown below. Media have been described (Rose et al. 1990). Yeast extract with peptone (YEP) YEP medium was supplemented with either 2% raffinose or 2% glucose (YEPD). To obtain synchronous cultures, cells were grown at 25°C in YEP–rafinose, and unbudded cells were isolated by centrifugal elutriation (Schwob and Nasmyth 1993). These cells were then inoculated into YEPD to a density of 5 × 106 and incubated at 34°C for 210 min.
Chromosomes were visualized by a tetracycline repressor (tetR)–green fluorescent protein (GFP) fusion protein binding to an array of tetracycline operators (tetOs) integrated at ura locus near the centromere V (Michaelis et al. 1997), referred to as CENV-GFP. To observe the GFP, cells from the elutriations were fixed in 100% ethanol and then washed in water. At least 100 cells were scored for each time point.
A FACScan® was used as described (Epstein and Cross 1992). Cells were prepared for indirect immunofluorescence (Piatti et al. 1996). Spindles were detected with a mouse monoclonal antitubulin antibody and a rhodamine-conjugated secondary antibody. Photographs were taken with Hamamatsu C4742-95 camera mounted on a ZEISS Axioplan 2 microscope. To estimate the percentage of long spindles, ≥100 cells were counted for each time point. Spindles that were longer than the diameter of the mother cell were scored as long. For real-time imaging of spindle elongation, cells were transformed with GFP-tub1::HIS3. Cycling cells were mounted in a growth chamber and filmed as described (Maddox et al. 2000). A heated 63× objective lens was used and the images were acquired every 20 s.
stu2-10 mutant was a gift of Zarina Alam (Cornell University, Ithaca, NY). GFP–tub1 plasmid was a gift of Aaron Straight (Harvard Medical School, Boston, MA).
Yeast Strains
The following strains with relevant genotypes were used in this study: TH452 (tetR-GFP:LEU2, TetOs:URA3), TH859 (stu2-10, tetR-GFP:LEU2, TetOs:URA3), TH865 (stu2-10, kip3::HIS, tetR-GFP:LEU2, TetOs:URA3), TH866 (stu2-10, mad2::HIS, tetR-GFP:LEU2, TetOs:URA3), TH867 (stu2-10, mad2::HIS, kip3::HIS, tetR-GFP:LEU2, TetOs:URA3), TH644 (tetR-GFP:LEU2, TetOs:URA3, GFP-Tub1::HIS), and TH891 (stu2-10, mad2::HIS, kip3::HIS, tetR-GFP:LEU2, TetOs:URA3, GFP-Tub1::HIS).
Sequence Alignment
Sequences used for construction of the multiple sequence alignment were extracted from the nonredundant database of the National Center for Biotechnology Information. The kinesin motor domain of each sequence was located using the program SMART (Schultz et al. 1998). Kinesin motor domain sequences were loaded into the program ClustalX (Thompson et al. 1997) and aligned using either the Blosum or the Gonnet weight matrix. After construction of the alignment, sequences that produced low scoring segments were discarded, as well as those producing gaps in highly conserved sequence blocks of the kinesin motor domain superfamily. A neighbor-joining tree (Saitou and Nei 1987) was calculated from the resulting alignment using the Blosum matrix, whereby positions with gaps were excluded and the tree was corrected for multiple substitutions. For calculation of the phylogenetic tree, the bootstrap option was used with default settings (random number generator seed of 111/1,000 bootstrap trials). The tree was displayed as radial tree using the program TreeView and edited for publication using a graphics program (Freehand8). Accession numbers of sequences used for multiple sequence alignment and the phylogenetic tree can be found at http://www.proweb.org/kinesin/ MotorSeqTable.html.
Results and Discussion
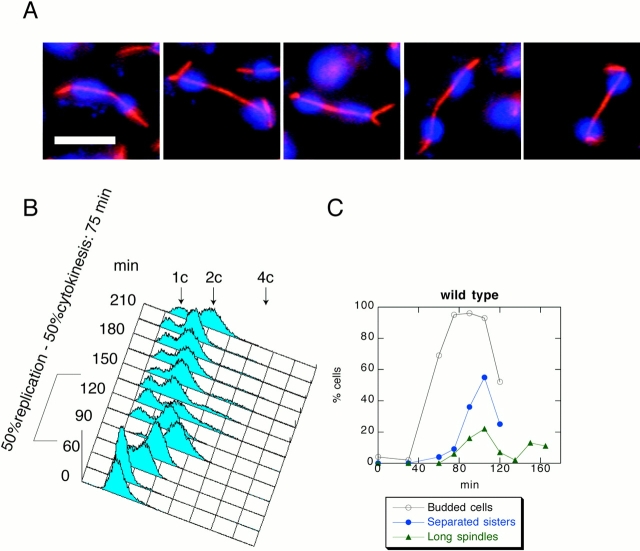
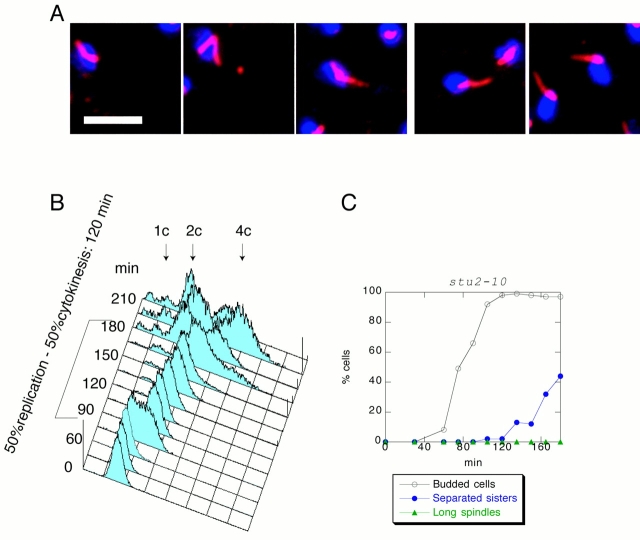
We examined the role of Stu2 during anaphase by using the temperature-sensitive allele stu2-10. We obtained a population of stu2-10 G1 cells at room temperature using centrifugal elutriation and released them into fresh medium at 37°C. The cells arrested with short spindles, 2C DNA, and nonseparated sister chromatids, showing that mutants in stu2-10 generate a metaphase arrest (data not shown). To minimize complications arising from destabilization of spindles at this high temperature, we determined the minimal restrictive temperature for stu2-10 by shifting a cycling population of stu2-10 cells to various temperatures. This analysis showed that the minimal nonpermissive temperature for stu2-10 cells was 34°C (data not shown). To confirm this result, we grew stu2-10 cells at room temperature, obtained a population of G1 cells by centrifugal elutriation, and released them at 34°C. Although control cells produced long spindles by 105 min after the release (Fig. 1A and Fig. C), stu2-10 cells never elongated their spindles (Fig. 2A and Fig. C). To quantify spindle elongation, we counted the percentage of spindles that had completed anaphase B (long spindles) throughout the release. Spindles of a length greater than the diameter of the mother cell were counted as long. As shown in Fig. 2 C, at 90 min after the release, <1% of total number of stu2-10 cells had long spindles, whereas in the population of the control cells the proportion of the cells with long spindles is 22% (Fig. 1 C). FACS® analysis showed that stu2-10 cells delay cytokinesis (Fig. 2 B) by 45 min compared with the control cells (Fig. 1 B), suggesting that stu2-10 delays progression from metaphase to anaphase. To further analyze the cell cycle state in stu2-10 mutants, we analyzed sister chromatid separation throughout the release by introducing a CENV–GFP construct (Michaelis et al. 1997) into all the yeast strains described in this study. Although control cells start to separate sister chromatids 45 min after budding (Fig. 1 C), stu2-10 mutants start sister chromatid separation at ∼90 min after budding (Fig. 2 C), confirming the existence of a significant metaphase delay in stu2-10.
Figure 1.
The control G1 cells released at 34°C. (A) Photomicrographs taken 105 min after release. Microtubules were detected by indirect immunofluorescence and are shown in red. DNA was visualized by DAPI and shown in blue. (B) FACS® profile shows that for these cells the time between DNA replication and cytokinesis is ∼75 min. (C) Sister chromatids separate 30 min after budding. 22% of the cells have long spindles in anaphase. Bar, 5 μm.
Figure 2.
stu2-10 cells do not elongate their spindles and delay in metaphase. stu2-10 cells were grown at 25°C, and then elutriated and released at 34°C. (A) Photomicrographs taken 135 min after release. Microtubules were detected by indirect immunofluorescence and are shown in red. DNA was visualized by DAPI and is shown in blue. (B) FACS® profile shows that the cells delay with 2C DNA and start to cytokinese with a 45-min delay relative to the wild type. (C) Sister chromatids separate 90 min after budding. Less than 1% of stu2-10 cells elongate their spindles. Bar, 5 μm.
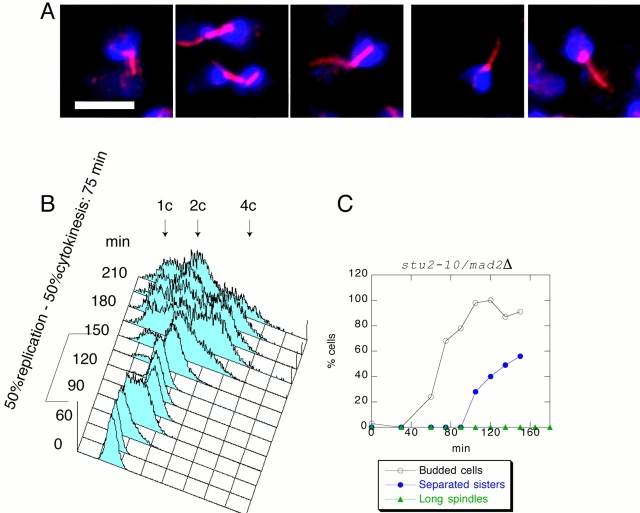
The metaphase delay of stu2-10 limits analysis of its role in anaphase. Previous studies have shown that malformed spindles are monitored by the spindle checkpoint pathway, a key member of which is MAD2 (Li and Murray 1991; Alexandru et al. 1999). To check whether the stu2-10 arrest was dependent on MAD2, we constructed a double mad2Δ/stu2-10 mutant. We obtained a population of mad2Δ/stu2-10 G1 cells using centrifugal elutriation, released them at 34°C, and monitored cell cycle state by FACS analysis and separation of GFP–CEN dots. Fig. 3 shows that deletion of MAD2 abolishes the stu2-10 arrest in metaphase, and the timing of sister chromatid separation is indistinguishable from wild type. (Compare Fig. 1 C with Fig. 3 C. Use time of the budding as a time reference for cell cycle position.) We therefore conclude that stu2-10 mutant cells arrest in metaphase because they engage the spindle checkpoint. To analyze the role of Stu2 during anaphase, we monitored spindle length in stu2-10/mad2Δ double mutants. Although the double mutant cells progressed through mitosis, no spindle elongation was seen (Fig. 3A and Fig. C). From these results we conclude that Stu2 is required for spindle elongation during anaphase.
Figure 3.
stu2-10/mad2Δ cells do not elongate spindles but otherwise progress normally through the cell cycle at 34°C. stu2-10/mad2Δ cells were grown at 25°C, and then elutriated and released at 34°C. (A) Microphotographs taken 135 min after the release. Microtubules were detected by indirect immunofluorescence and are shown in red. DNA was visualized by DAPI and is shown in blue. (B) FACS® profile shows that cytokinesis happens with the wild-type timing. (C) Sister chromatids separate 90 min after budding. Less than 1% of stu2-10/mad2Δ cells elongate their spindles. Bar, 5 μm.
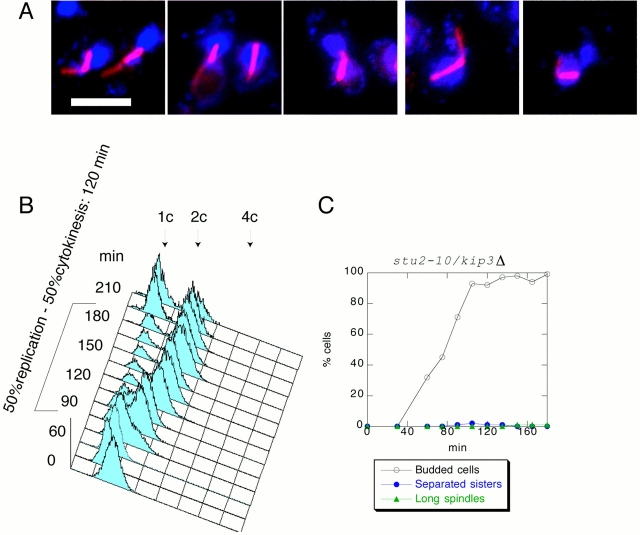
Homologues of Stu2 are required for microtubule stabilization in other eukaryotic species. Therefore, it seemed likely that stu2 mutants are unable to elongate their spindles because they cannot stabilize spindle microtubules. However, other possibilities exist, such as a role for Stu2 in spindle pole body structure or kinetochore function. In Xenopus egg extracts, it has been shown that activity of the Stu2 homologue XMAP215 is opposed by XKCM1, a member of the Kin I subfamily of microtubule-destabilizing kinesins. Removal of XMAP215 from a Xenopus extract results in microtubule destabilization, but this microtubule destabilization can be rescued by concomitant removal of XKCM1 (Tournebize et al. 2000). Extrapolating from the Xenopus experiments, we reasoned that if anaphase fails in stu2-10 because of a defect in microtubule stabilization, then removal of a microtubule-destabilizing factor in stu2-10 would stabilize spindles during anaphase. No orthologue of XKCM1 has been identified in S. cerevisiae. However, two kinesins, KAR3 and KIP3, have been genetically implicated in microtubule destabilization in S. cerevisiae (Cottingham et al. 1999). A double mutant kar3Δ /stu2-10 was synthetically lethal (data not shown). However, we were able to construct a kip3Δ/stu2-10 double mutant. To test whether kip3Δ/stu2-10 cells elongate their spindles at the restrictive temperature (34°C), we analyzed the double mutant by centrifugal elutriation and found that it arrests with short spindles (Fig. 4), as is seen for the stu2-10 single mutant. This arrest corresponds to metaphase: the DNA content is 2C, and the sister chromatids are not separated (Fig. 4B and Fig. C). Interestingly, the arrest of the double mutant is tighter than that of the single stu2-10 mutant, whereas spindle structure is indistinguishable by light microscopy. To examine anaphase spindle structure in the stu2-10/kip3Δ double mutant, we deleted MAD2 and analyzed the release of G1 cells of a triple mutant stu2-10/kip3Δ/mad2Δ at 34°C. Fig. 5 shows that the stu2-10/kip3Δ/mad2Δ triple mutant does not delay in metaphase, and sister chromatids separate with similar timing as wild-type cells. Analysis of spindle length in the triple mutant showed that many long spindles are seen during anaphase. The morphology of the elongated spindles appears to be similar to that of wild-type spindles as judged by light microscopy (Fig. 5 A). To illustrate these data, we reploted spindle lengths of all the elutriation–release experiments in this paper on the same graph (Fig. 5 D). This analysis shows that the percentage of long spindles in the triple mutant reaches 75% of the wild-type level (Fig. 5 C).
Figure 4.
stu2-10/kip3Δ cells do not elongate their spindles and delay in metaphase. stu2-10/kip3Δ cells were grown at 25°C, and then elutriated and released at 34°C. (A) Photomicrographs taken 135 min after the release. Microtubules were detected by indirect immunofluorescence and are shown in red. DNA was visualized by DAPI and is shown in blue. (B) FACS® profile shows that the cells delay with 2C DNA and start to undergo cytokinesis late in the release. (C) Sister chromatids separate 120 min after budding. Less than 1% of stu2-10/kip3Δ cells elongate their spindles. Bar, 5 μm.
Figure 5.
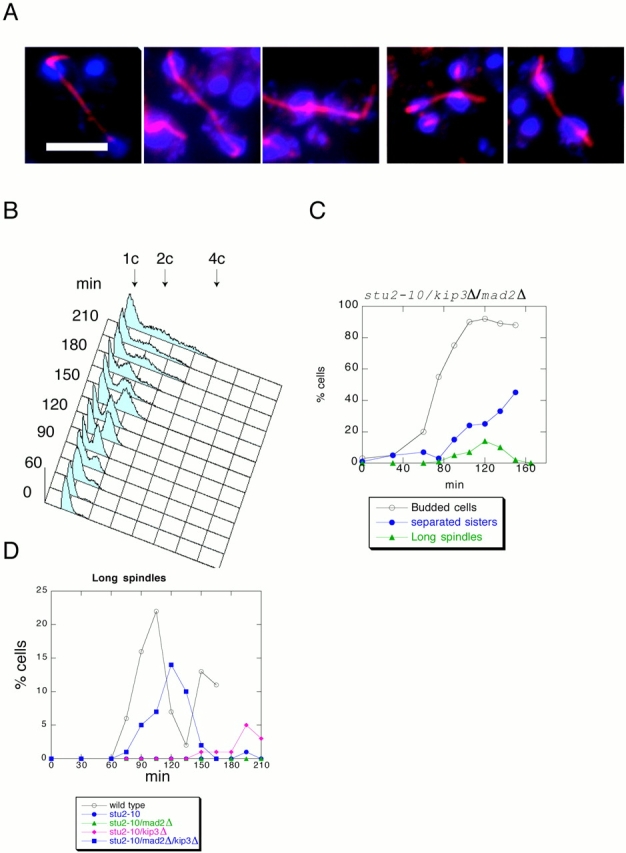
stu2-10/kip3Δ/mad2Δ cells do elongate their spindles and do not delay in metaphase. stu2-10/kip3Δ/mad2Δ l cells were grown at 25°C, and then elutriated and released at 34°C. (A) Photomicrographs taken 135 min after the release. Microtubules were detected by indirect immunofluorescence and are shown in red. DNA was visualized by DAPI and is shown in blue. (B) A FACS® profile shows that there were many dead cells that failed to replicate. The dead cells obscure the FACS® profile and do not allow the determination of the exact time point of cytokinesis. (C) Sister chromatids separate without any delay. (Only those cells that showed GFP–CEN signal were scored for budding. This allowed us to exclude dead cells from the count.) stu2-10/kip3Δ/mad2Δ cells elongate their spindles similar to the wild type. (D) The kinetics of long spindles accumulation in all of the analyzed strains. Bar, 5 μm.
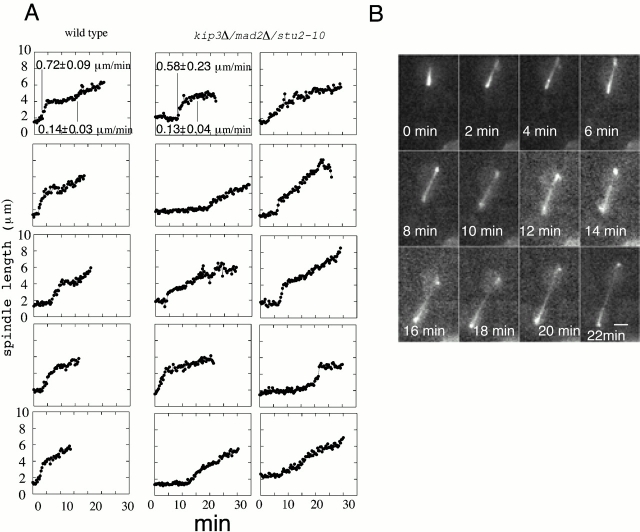
To compare the kinetics of spindle elongation in wild-type cells and stu2-10/kip3Δ/mad2Δ mutant, we introduced a GFP–tubulin construct into the cells and filmed spindle elongation in real time at 34°C. Fig. 6 A shows tracings of spindle elongation from wild-type and mutant cells. Fig. 6 B shows a typical mutant spindle elongating. Spindles from both wild-type and mutant cells show biphasic kinetics consistent with the data reported by Straight et al. 1997. The rates of spindle elongation between the wild-type and the mutant cells are similar at ∼0.6 and 0.14 μm/min. Therefore, we conclude that, indeed, spindles in the triple mutant elongate with similar kinetics to spindles in wild type.
Figure 6.
Real-time imaging of anaphase spindle elongation. (A) The kinetics of spindle elongation of five control and 10 stu2-10/kip3Δ/mad2Δ cells. (B) Anaphase video sequence of a stu2-10/kip3Δ/mad2Δ cell. Bar, 2 μm.
Interestingly, in 3 out of 10 of the triple mutant cells the GFP–centromere dots failed to segregate correctly, consistent with DAPI staining of fixed cells (Fig. 5 A). There are several possible reasons for the segregation defect in mad2Δ/kip3Δ/stu2-10 cells. One possibility is that Stu2 is involved in the connection between kinetochore microtubules and the spindle poles (Desai and Mitchison 1995). Another possibility is that Stu2 plays a role in chromosome attachment to microtubules at the kinetochores. Although members of the Dis1 family do not seem to have an obvious kinetochore localization (Charrasse et al. 1998; Matthews et al. 1998; Cullen et al. 1999; Graf et al. 2000; Tournebize et al. 2000), evidence for a kinetochore function comes from the analysis of centromere movement in dis1 mutants, where centromeres cease motility despite presence of relatively normal-looking spindles (Nabeshima et al. 1998). Further evidence comes from the similarity of the chromosomal missegregation phenotype of mad2Δ/kip3Δ/stu2-10 cells to that seen in mutants of ipl1, a gene that has been implicated in kinetochore function (Biggins et al. 1999).
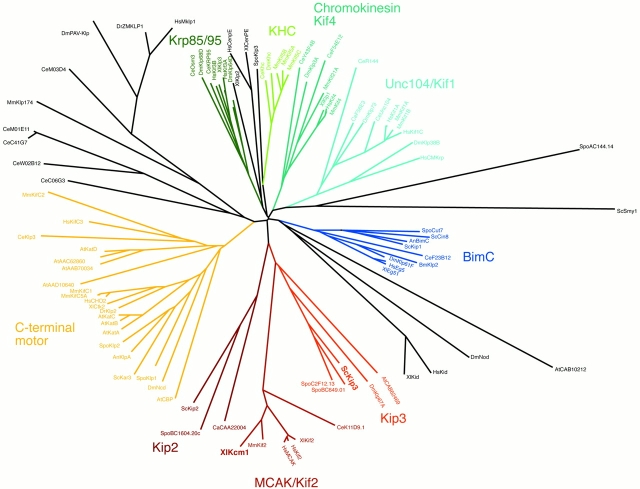
In summary, we conclude that stabilization of the central spindle during anaphase B depends on a balance between the opposing activities of Stu2 and Kip3. Our data further suggests that KIP3 is the functional orthologue of the Kin I subfamily of proteins. Members of this family have been found in all organisms examined (Desai et al. 1999) except yeast. Our discovery of the effect of Kip3 in opposing Stu2 suggests either that the Kin I family evolved an orthologous function in metazoans, or that the bioinformatics analysis has missed the similarities in sequence. On the basis of our observations, we reanalyzed the relationship of XKCM1 to the kinesin-related proteins in yeast. We recalculated a phylogenetic tree from a multiple sequence alignment of the kinesin superfamily using only the sequence of the kinesin motor domain. In the resulting phylogenetic tree (Fig. 7), the Kip3 family of kinesin motor domain proteins did congregate with the MCAK/Kif2 (Kin I) subfamily, which is in accordance with our experimental data. The resulting tree is opposed to a recently published phylogenetic tree (available at kinesin homepage, http://www.proweb.org/kinesin/ KinesinTree.html), in which the two subfamilies are found on two separate branches of the phylogenetic tree. The observed disagreement is most likely due to the fact that the kinesin domain from Kip3 we used for building the multiple sequence alignment differs in length from the one at kinesin homepage. In the case of S. cerevisiae KIP3, the sequence was 97 amino acids longer than the one used for the tree at the kinesin homepage. This results in a 56–amino acid gap produced by ScKip3, yet the conservation at the very NH2 terminus of the kinesin domains, which includes a V-x-[V,I]-R-x-R-P-[V,I,L,F]-x(3)-[E,N,Q,D] motif in this part of the kinesin domain that is present in most of the family members, justifies the inclusion of this part of Kip3 family members. This motif in the first conserved part of the kinesin domain furthermore aligns to a conserved β-sheet (β-1) (data not shown). In addition, we excluded gap columns, and only conserved alignment blocks were used for calculating the phylogenetic tree. Sequences that produced gaps in otherwise conserved alignment blocks were excluded from the analysis. Two additional phylogenetic trees have been published before (Goodson et al. 1994; Moore and Endow 1996), none of which contain the Kip3 subfamily of kinesin motor domain proteins and therefore make comparison to our results impossible. Interestingly, MCAK, a member of Kin I subfamily of kinesins, is localized to kinetochores in mammalian cells (Wordeman and Mitchison 1995). Since it now appears to be part of this subfamily, perhaps Kip3 is a kinetochore component in S. cerevisiae as well, thus explaining the chromosome missegregation.
Figure 7.
Unrooted phylogenetic tree of the kinesin motor domains. A multiple sequence alignment for the kinesin motor domain sequences was constructed using the program ClustalX, a bootstrapped phylogenetic tree (neighbor-joining tree), was calculated using the same program and displayed as a radial tree using the program TreeView. Congruent with biological data, the Kip3 subfamily is found on the same branch as the MCAK/Kif2 subfamily, suggesting close evolutionary relationship between the members of these subfamilies. Only the bootstrap values for the Kip3 and MCAK/Kif2 subbranches are shown (538 and 528 for the branch leading to Kip2, Kip3, MACK/Kif2, and Kip3, MACK/Kif2, respectively).
Our results have shown that anaphase spindle elongation in S. cerevisiae requires the coordinated action of Stu2 and Kip3. However, we do not know whether Stu2 and Kip3 are constitutively active throughout the cell cycle, or if Stu2 or Kip3 are directly regulated during the metaphase to anaphase transition. Evidence for direct control of microtubule dynamics using this pathway comes from analysis of XMAP215, the Xenopus homologue. In vitro hyperphosphorylation of XMAP215 by Cdc2 decreases its ability to stimulate microtubule elongation (Gard and Kirschner 1987), and phosphorylation of XMAP215 significantly increases during mitosis. Analysis of Stu2 phosphorylation may help determine its role in the switch in microtubule dynamics that occurs at the metaphase–anaphase transition.
Acknowledgments
We thank Arshad Desai for critical reading of the manuscript, Kerry Bloom and Kim Nasmyth for the helpful discussion, and Aliona Bogdanova for helping us to construct the strains.
Footnotes
Abbreviations used in this paper: GFP, green fluorescent protein; tetO, tetracycline operator; tetR, tetracycline repressor; YEP, yeast extract with peptone.
References
- Alexandru G., Zachariae W., Schleiffer A., Nasmyth K. Sister chromatid separation and chromosome re-duplication are regulated by different mechanisms in response to spindle damage. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:2707–2721. doi: 10.1093/emboj/18.10.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin T.I., Cande W.Z. Kinetic analysis of mitotic spindle elongation in vitro. J. Cell Sci. 1990;97:79–89. doi: 10.1242/jcs.97.1.79. [DOI] [PubMed] [Google Scholar]
- Biggins S., Severin F.F., Bhalla N., Sassoon I., Hyman A.A., Murray A.W. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrasse S., Schroeder M., Gauthier-Rouviere C., Ango F., Cassimeris L., Gard D.L., Larroque C. The TOGp protein is a new human microtubule-associated protein homologous to the Xenopus XMAP215. J. Cell Sci. 1998;111:1371–1383. doi: 10.1242/jcs.111.10.1371. [DOI] [PubMed] [Google Scholar]
- Cottingham F.R., Gheber L., Miller D.L., Hoyt M.A. Novel roles for Saccharomyces cerevisiae mitotic spindle motors. J. Cell Biol. 1999;147:335–350. doi: 10.1083/jcb.147.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen C.F., Deak P., Glover D.M., Ohkura H. mini spindlesa gene encoding a conserved microtubule-associated protein required for the integrity of the mitotic spindle in Drosophila . J. Cell Biol. 1999;146:1005–1018. doi: 10.1083/jcb.146.5.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Mitchison T.J. A new role for motor proteins as couplers to depolymerizing microtubules. J. Cell Biol. 1995;128:1–4. doi: 10.1083/jcb.128.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Verma S., Mitchison T.J., Walczak C.E. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- Epstein C.B., Cross F.R. CLB5a novel B cyclin from budding yeast with a role in S phase. Genes Dev. 1992;6:1695–1706. doi: 10.1101/gad.6.9.1695. [DOI] [PubMed] [Google Scholar]
- Gard D.L., Kirschner M.W. A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plus-end. J. Cell Biol. 1987;105:2203–2215. doi: 10.1083/jcb.105.5.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson H.V., Kang S.J., Endow S.A. Molecular phylogeny of the kinesin family of microtubule motor proteins. J. Cell Sci. 1994;107:1875–1884. doi: 10.1242/jcs.107.7.1875. [DOI] [PubMed] [Google Scholar]
- Graf R., Daunderer C., Schliwa M. Dictyostelium DdCP224 is a microtubule-associated protein and a permanent centrosomal resident involved in centrosome duplication. J. Cell Sci. 2000;113:1747–1758. doi: 10.1242/jcs.113.10.1747. [DOI] [PubMed] [Google Scholar]
- Hoyt M.A. Cellular roles of kinesin and related proteins. Curr. Opin. Cell Biol. 1994;6:63–68. doi: 10.1016/0955-0674(94)90117-1. [DOI] [PubMed] [Google Scholar]
- Hyman A.A., Karsenti E. Morphogenetic properties of microtubules and mitotic spindle assembly. Cell. 1996;84:401–410. doi: 10.1016/s0092-8674(00)81285-4. [DOI] [PubMed] [Google Scholar]
- Li R., Murray A.W. Feedback control of mitosis in budding yeast. 1994 Cell 66 1991. 519 531(erratum published 79:388). [DOI] [PubMed] [Google Scholar]
- Maddox P.S., Bloom K.S., Salmon E.D. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae . Nat. Cell Biol. 2000;2:36–41. doi: 10.1038/71357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews L.R., Carter P., Thierry-Mieg D., Kemphues K. ZYG-9, a Caenorhabditis elegans protein required for microtubule organization and function, is a component of meiotic and mitotic spindle poles. J. Cell Biol. 1998;141:1159–1168. doi: 10.1083/jcb.141.5.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C., Ciosk R., Nasmyth K. Cohesinschromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Moore J.D., Endow S.A. Kinesin proteinsa phylum of motors for microtubule-based motility. Bioessays. 1996;18:207–219. doi: 10.1002/bies.950180308. [DOI] [PubMed] [Google Scholar]
- Nabeshima K., Nakagawa T., Straight A.F., Murray A., Chikashige Y., Yamashita Y.M., Hiraoka Y., Yanagida M. Dynamics of centromeres during metaphase-anaphase transition in fission yeastDis1 is implicated in force balance in metaphase bipolar spindle. Mol. Biol. Cell. 1998;9:3211–3225. doi: 10.1091/mbc.9.11.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas R.B. How cells get the right chromosomes. Science. 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- Piatti S., Bohm T., Cocker J.H., Diffley J.F., Nasmyth K. Activation of S-phase-promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- Rose M.D., Winston F., Hieter P. Methods in Yeast Genetics 1990. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: pp. 198 [Google Scholar]
- Saitou N., Nei M. The neighbor-joining methoda new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Saunders W.S. Action at the ends of microtubules. Curr. Opin. Cell Biol. 1999;11:129–133. doi: 10.1016/s0955-0674(99)80016-7. [DOI] [PubMed] [Google Scholar]
- Saunders W.S., Hoyt M.A. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- Saunders W., Hoyt M.A. Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol. Biol. Cell. 1997;8:1025–1033. doi: 10.1091/mbc.8.6.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J., Milpetz F., Bork P., Ponting C.P. SMART, a simple modular architecture research toolidentification of signaling domains. Proc. Natl. Acad. Sci. USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob E., Nasmyth K. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae . Genes Dev. 1993;7:1160–1175. doi: 10.1101/gad.7.7a.1160. [DOI] [PubMed] [Google Scholar]
- Straight A.F., Marshall W.F., Sedat J.W., Murray A.W. Mitosis in living budding yeastanaphase A but no metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The CLUSTAL_X windows interfaceflexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournebize R., Popov A., Kinoshita K., Ashford A.J., Rybina S., Pozniakovsky A., Mayer T.U., Walczak C.E., Karsenti E., Hyman A.A. Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat. Cell Biol. 2000;2:13–19. doi: 10.1038/71330. [DOI] [PubMed] [Google Scholar]
- Wang P.J., Huffaker T.C. Stu2pa microtubule-binding protein that is an essential component of the yeast spindle pole body. J. Cell Biol. 1997;139:1271–1280. doi: 10.1083/jcb.139.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wordeman L., Mitchison T.J. Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J. Cell Biol. 1995;128:95–104. doi: 10.1083/jcb.128.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]