Abstract
The loss of chromosomal integrity from DNA double-strand breaks introduced into mammalian cells by ionizing radiation results in the specific phosphorylation of histone H2AX on serine residue 139, yielding a specific modified form named γ-H2AX. An antibody prepared to the unique region of human γ-H2AX shows that H2AX homologues are phosphorylated not only in irradiated mammalian cells but also in irradiated cells from other species, including Xenopus laevis, Drosophila melanogaster, and Saccharomyces cerevisiae. The antibody reveals that γ-H2AX appears as discrete nuclear foci within 1 min after exposure of cells to ionizing radiation. The numbers of these foci are comparable to the numbers of induced DNA double-strand breaks. When DNA double-strand breaks are introduced into specific partial nuclear volumes of cells by means of a pulsed microbeam laser, γ-H2AX foci form at these sites. In mitotic cells from cultures exposed to nonlethal amounts of ionizing radiation, γ-H2AX foci form band-like structures on chromosome arms and on the end of broken arms. These results offer direct visual confirmation that γ-H2AX forms en masse at chromosomal sites of DNA double-strand breaks. The results further suggest the possible existence of units of higher order chromatin structure involved in monitoring DNA integrity.
Keywords: ionizing radiation, histone H2AX, DNA double-strand breaks, phosphorylation, indirect immunofluorescence
The universal presence of ionizing radiation poses a constant threat to the chromosomal integrity of living organisms. One of the most serious cellular lesions induced by radiation is the random DNA double-strand break. This lesion jeopardizes a chromosome's physical integrity, which is essential for its correct segregation during mitosis and meiosis, as well as its informational redundancy, which is critical for maintaining accurate encoding of cellular components. Thus, it is not surprising that multiple mechanisms exist for dealing with this serious lesion, including nonhomologous end-joining, homologous recombination, and apoptosis (Jeggo 1998; Kanaar et al. 1998; Kerr 1998; Rathmell and Chu 1998). On the other hand, DNA double-strand cleavages are necessary steps in several important cellular processes, including recombination during meiosis and mitosis, V(D)J recombination during immune system development (Jeggo et al. 1995), and mating type switching in Saccharomyces cerevisiae. It is also relevant to note that DNA double-strand breaks introduced by ionizing radiation and radiomimetic procedures are accompanied by multiple random single-strand breaks and other lesions, resulting in locally multiply damaged sites (Ward 1990), whereas those introduced as part of cellular processes presumably are not. However, in spite of these differences, the resultant DNA ends appear to be rejoined by similar mechanisms in all of these situations.
In mammals, nonhomologous end-joining appears to be the major mechanism of rejoining DNA ends. This process involves a DNA-activated protein kinase complex with three subunits, a catalytic subunit, DNA-PKcs (Anderson 1994), and two subunits with DNA end-binding capabilities, KU70 and KU80 (Li et al. 1995). A fourth protein, XRCC4, may be an accessory factor for DNA ligase IV (Grawunder et al. 1997; Jeggo 1998; Modesti et al. 1999). Homologues of all of these proteins, except DNA-PKcs, have been reported in S. cerevisiae. These proteins are involved in the end-joining of exogenously as well as endogenously-induced DNA double-strand breaks. In contrast to higher eucaryotes, homologous recombination appears to be the major mechanism for DNA double-strand break rejoining in S. cerevisiae (Nickoloff and Hoekstra 1998). This pathway involves several genes in the RAD52 epistasis group (Game 1993; Bai and Symington 1996); one subgroup comprising RAD50, MRE11, and XRS2 (NBS1 in human) is also implicated in nonhomologous end-joining (Johzuka and Ogawa 1995).
Whereas much is known about the rejoining of DNA double-strand breaks, much less is known about how these breaks are initially recognized. Our group reported previously that mammalian cells and mice respond to agents that introduce DNA double-strand breaks with the immediate and substantial phosphorylation of histone H2AX (Rogakou et al. 1998). H2AX, one of the three types of conserved histone H2A protein species, differs from the other two by the presence of a conserved motif SQ(E/D)-(I/L/Y) at the COOH terminus (Mannironi et al. 1989). It is the phosphorylation of the serine in this motif, residue 139 in mammals, that yields the modified form named γ-H2AX (Rogakou et al. 1998). H2AX is rapidly γ-phosphorylated after exposure of cells to ionizing radiation, with half-maximal amounts reached at 1–3 min. At the maximum, 10–30 min after irradiation, the stoichiometry suggests that hundreds to several thousand γ-H2AX molecules are present per DNA double-strand break in mammals. In this study, we show that the H2AX molecules are γ-phosphorylated en masse at the sites of DNA double-strand breaks.
Materials and Methods
Cell Culture
Cell lines were obtained from the American Type Culture Collection unless otherwise noted. IMR90 normal human fibroblast cells were grown in MEM Alpha (No. 12561; GIBCO BRL) containing 10% FBS. MCF7 human breast cancer and SF268 human astrocytoma cells (National Cancer Institute, Division of Cancer Treatment and Diagnosis, Developmental Therapeutics Program, National Institutes of Health) were grown in RPMI 1640 (No. 11875; GIBCO BRL) containing 10% FBS. Indian muntjac, Muntiacus muntjak, normal skin fibroblasts were grown in F-10 Ham's nutrient mixture (No. 11550; GIBCO BRL) containing 20% FBS. Xenopus laevis A6 normal kidney cells were grown in medium NCTC-109 (No. 21340; GIBCO BRL) containing 15% deionized water and 10% FBS. Cultures were maintained at room temperature, ∼24°C, in an atmosphere of 5% CO2. Drosophila melanogaster epithelial cells, a gift of C. Wu (National Cancer Institute, Division of Basic Sciences, Laboratory of Molecular Cell Biology, National Institutes of Health, Bethesda, MD), were grown in Schneider's Drosophila medium (No. 11720; GIBCO BRL) containing 10% heat-inactivated FBS at room temperature.
Antibody Production
Anti-γ was prepared by Genosys Biotechnologies Inc. The peptide CKATQAS(PO4)QEY was synthesized, conjugated to keyhole limpet hemocyanin, and injected into rabbits. The immune serum from the third bleed was passed through a column containing immobilized CKATQASQEY to absorb antibodies to unphosphorylated H2AX.
Ionizing Radiation
Cells growing in 10-cm dishes, on Labtek II slides (Nalge Nunc International), or on coverslips were exposed to the indicated amount of ionizing radiation from a 137Cs source in a Mark I irradiator (J.L. Shepherd and Associates). Doses >20 Gy were given at a rate of 15.7 Gy/min. Doses of 2 and 0.6 Gy were given in 1 min.
Laser-introduced DNA Double-Strand Breaks
Except for the source of UVA irradiation, the method of Limoli and Ward 1993 was followed. MCF7 cells, grown in the presence or absence of 0.4 μM bromo deoxyuridine (BrdU)1 and 2.4 μM thymidine for 3 d, were subcultured onto No. 1 1/2 coverslips that had been gently scribed with lines by a diamond pencil. After growth for 24 h, the cells were incubated with Hoechst dye 33258 for 5 min. The coverslips were mounted on a glass slide with a 0.5-mm-thick silicone gasket (Electron Microscopy Sciences) to form a chamber which was filled with PBS. The slides were kept on ice until placed on the stage of the microscope fitted with a LaserScissors™ Module 390/20 (Cell Robotics, Inc.). This laser emits at 390 nm, a wavelength at which the Hoescht dye has substantial absorption. An image of the chosen field of cells containing an inscribed line was recorded and printed. A proposed path of the laser was traced on the print. The laser was operated at various power outputs (100% = 20 μj/pulse) and focused through a 100× objective to a 0.5-μm-diameter circle in the focal plane of the cells with the pulse rate set at 10 pulses/s. The laser was guided by means of a joystick along the traced path at a maximum rate of 8 μm/s. After irradiation, the coverslips were transferred to a culture dish with growth media for 30 min at 37°C before fixation. After processing for laser scanning confocal microscopy, the coverslips were mounted on slides and the irradiated cell groupings were found with the aid of the inscribed lines.
Laser Scanning Confocal Microscopy
Cells were grown on Labtek II slides or coverslips. After irradiation and recovery at 37°C, the cell preparations were fixed in 2% paraformaldehyde in PBS for 5 min, washed in PBS, permeabilized in 100% methanol at −20°C for 5 min, washed, blocked with 8% BSA for 1 h, incubated with the γ-H2AX first antibody at 800-fold dilution for 2 h, washed, incubated with a Cy2-conjugated goat anti–rabbit second antibody (Jackson Immunolabs) at 200-fold dilution for 1 h, washed, mounted with or without propidium iodide, and viewed with a PCM2000 laser scanning confocal microscope (Nikon Inc.) using a 100× objective. Optical sections (0.5 μm) through the thickness of the sample were imaged and combined in a maximum projection with Simple32 software (Compix Inc.) so that all of the visible foci and bands in a nucleus or mitotic figure were recorded. The projection was saved as a BMP file and brought into Paint Shop Pro 5 (Jasc Software, Inc.) and Powerpoint (Microsoft Corp.) for presentation.
Immunoblotting
Polyvinylidene difluoride (PVDF) membranes containing transferred proteins were blocked with 1% dried nonfat milk for 1 h, incubated with the γ-H2AX first antibody at 12,000-fold dilution for 2 h, washed, incubated with peroxidase goat anti–rabbit second IgG (Calbiochem-Novabiochem Corp.) at 3,000-fold dilution for 1.5 h and washed. Anti-γ binding was visualized by chemiluminescence (ECL RPN 2209; Amersham Pharmacia Biotech).
Results
Antibody to γ-H2AX COOH Terminus
To examine the spatial distribution of γ-H2AX in the chromatin of irradiated cells, a polyclonal antibody (anti-γ) was raised in rabbits against a synthetic phosphorylated peptide containing the mammalian γ-H2AX COOH-terminal sequence. On immunoblots of total protein extracts from irradiated MCF7 cells, anti-γ detected one band at the position expected for γ-H2AX (Fig. 1 A). No binding was detected in irradiated samples when the immunizing peptide was present as competitor (Fig. 1 A, P-pep) or with preimmune serum (Fig. 1 A, pre). Although anti-γ binding was not apparent in unirradiated MCF7 samples at film exposures optimal for γ-H2AX detection (Fig. 1 A, 0 Gy), small amounts of binding were detectable on highly exposed immunoblots. Results from our laboratory show that γ-H2AX is present in apoptotic cells with fragmented DNA (Rogakou, E.P., W. Nieves-Neira, C. Boon, Y. Pommier, and W.M. Bonner, manuscript submitted for publication), indicating a possible source of γ-H2AX in unirradiated cultures. Another possibility is that anti-γ cross-reacts slightly with unmodified H2AX. However, two-dimension gels which separate these two protein species (Fig. 1 B) showed that anti-γ bound only γ-H2AX (Fig. 1 C, solid line) with no detectable cross-reaction to unmodified H2AX. This result indicates that the binding of anti-γ in extracts of unirradiated cells is due to the presence of γ-H2AX in some of the cells in those cultures.
Figure 1.
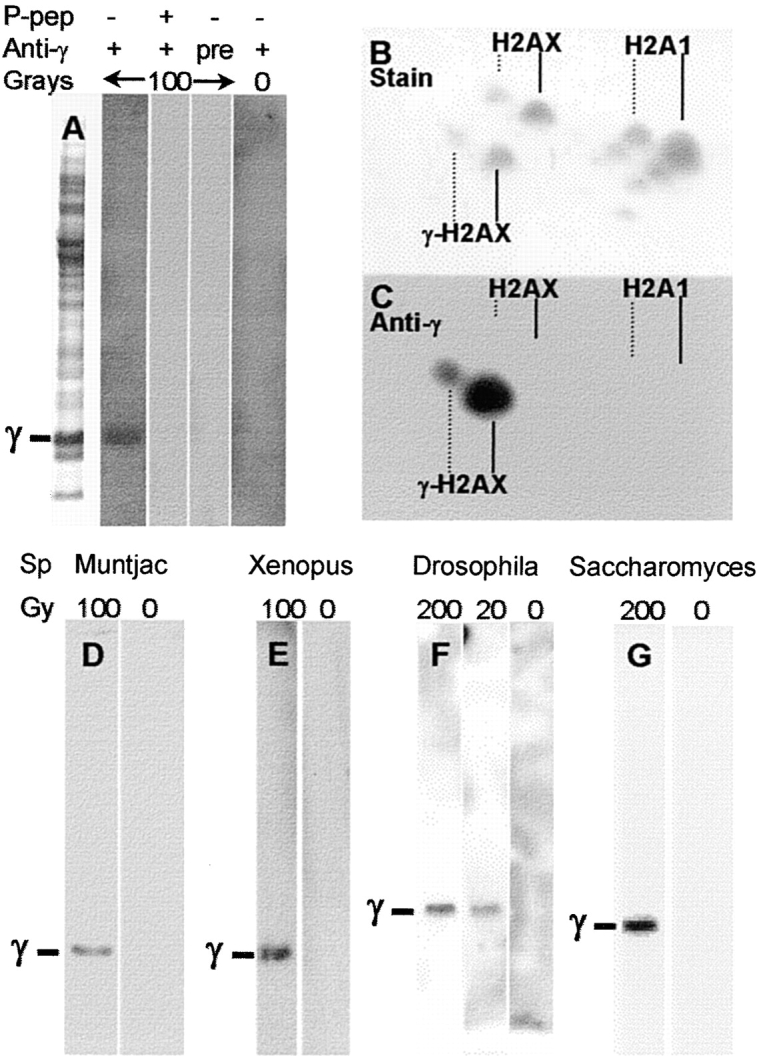
Immunoblots. After exposure to the indicated amount of ionizing radiation and a 30-min recovery, cells were harvested. Extracts were prepared and analyzed by gel electrophoresis and immunoblotting on polyvinylidene difluoride (PVDF) membranes as described in Materials and Methods. (A) Human MCF7 breast cancer cells. Blots of fractionated total SDS extracts were probed as indicated with anti-γ preimmune serum (pre) or with anti-γ containing 1 μM immunizing peptide (P-pep). The left-most lane shows the protein staining pattern on SDS gels. (B and C) Human SF268 astrocytoma cells. Cultures were irradiated with 100 Gy and analyzed on high-resolution two-dimension acetic acid gels. (D–G) Other eucaryotes. The position of migration of the respective H2AX homologues, H2AX in M. muntjak (D) and X. laevis (E), H2AvD in D. melanogaster (F), and H2A in S. cerevisiae (G) are indicated by γ. Cultures of S. cerevisiae strain BY4733 were irradiated and allowed to recover for 30 min at 30°C. Nuclei were prepared from spheroplasts (Celis 1998), and histones were extracted as described (Ueda and Tanaka 1995). For MCF7, M. muntjak, X. laevis, and S. cerevisiae extracts were prepared with SDS and fractionated on 12% NuPage SDS gels (Novex Novel Technology). For D. melanogaster, extracts were prepared with 0.5 N HCl and fractionated on 12% acetic acid–urea–Triton X-100 gels.
H2AX as well as H2A1, the most plentiful of the H2A species in mammals, can be phosphorylated on serine residue 1 and acetylated on lysine residue 5 (Fig. 1 B, dotted lines). H2AX molecules modified in the NH2-terminal region bound anti-γ only when they were γ-modified (Fig. 1 C, dotted line). Also noticeable was the lack of anti-γ binding to H2A1, whose sequence, except for the COOH-terminal motif, is almost identical to the H2AX.
Since the H2AX COOH terminus is highly conserved, immunoblots were prepared from irradiated cell cultures of various species to examine whether anti-γ could detect γ-H2AX homologues. Anti-γ detected one band that migrated as expected for the appropriate γ-H2AX homologue not only in other examined mammalian species such as the mouse, rat, hamster, (data not shown), and M. muntjak (Fig. 1 D), but also in X. laevis (Fig. 1 E), D. melanogaster (Fig. 1 F), and S. cerevisiae (Fig. 1 G). These experiments show that not only H2AX itself, but also its phosphorylation in response to ionizing radiation, have been highly conserved during evolution.
γ-H2AX Foci in Cells Subjected to Ionizing Radiation
With the demonstration that anti-γ is specific for γ-H2AX, the distribution of γ-H2AX in irradiated cells was examined. Cells of the normal human fibroblast line IMR90 and the human breast cancer line MCF7 both responded to ionizing radiation with the formation of discrete foci containing γ-H2AX throughout the nuclei (Fig. 2). Some cells of both lines contained foci in the absence of irradiation; most of the unirradiated MCF7 cells contained one to two foci (Fig. 2 I), whereas fewer of the IMR90 cells did (Fig. 2 A). The amount of γ-H2AX present in these foci in unirradiated cells is evidently below the level of detection of the immunoblots shown in Fig. 1 A, but may account for the signal seen on highly exposed immunoblots. No foci were apparent in unirradiated or irradiated cells when 1 μM immunizing peptide was included in the first antibody solution (data not presented). The relationship between the presence of γ-H2AX in unirradiated and irradiated cells is being examined.
Figure 2.
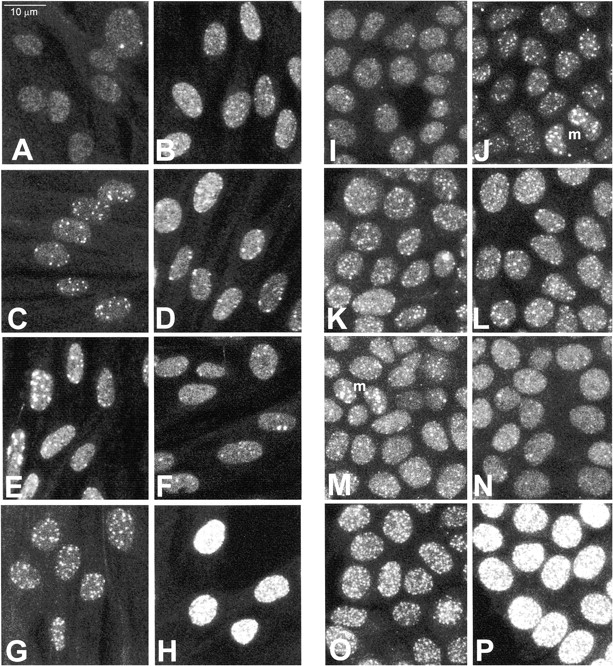
γ-H2AX foci formation in human cells after irradiation (maximum projections showing all recorded foci). IMR90 normal human fibroblasts (A–H) and MCF7 human breast cancer cells (I–P) were exposed to various amounts of ionizing radiation and permitted to recover for various lengths of time. (A and I) Unirradiated control, (B and J) 3 min after 0.6 Gy, (C and K) 15 min after 0.6 Gy, (D and L) 30 min after 0.6 Gy, (E and M) 60 min after 0.6 Gy, (F and N) 180 min after 0.6 Gy, (G and O) 15 min after 2 Gy, (H and P) 15 min after 22 Gy. m indicates mitotic MCF7 cells in J and M. Cells were processed for laser scanning confocal microscopy as described in Materials and Methods.
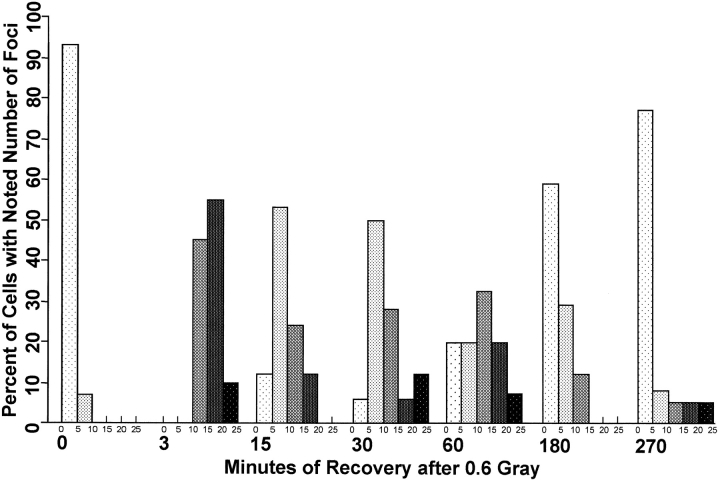
With IMR90 cells, foci were apparent 3 min after irradiation with 0.6 Gy (Fig. 2 B), persisted at 15–60 min (Fig. 2, C–E), then decreased in number at 180 min (Fig. 2 F). With MCF7 cells, the time course of foci appearance and disappearance was similar (Fig. 2, I–N). A more detailed analysis of IMR90 cells is presented in Fig. 3, in which individual nuclei in fields of cells were scored for the number of foci. Compared with the unirradiated control cells, which contained an average of 1.5 ± 2.4 (14 nuclei) foci per nucleus, all the IMR90 cells 3 min after exposure to 0.6 Gy contained numerous small foci, with an average of 16.3 ± 3.6 (11 nuclei) foci per nucleus. The foci became fewer in number but better defined after 15 min, 10.1 ± 3.9 (17 nuclei) foci per nucleus; 30 min, 11.6 ± 5.3 (18 nuclei) foci per nucleus; and 60 min, 11.4 ± 6.1 (15 nuclei) foci per nucleus. After 180 min recovery, the number of foci again decreased to 4.8 ± 3.3 (17 nuclei) foci per nucleus, and at 270 min there were 4.5 ± 5.3 (26 nuclei) foci per nucleus. In addition, after 270 min recovery, 2 of the 26 scored nuclei appeared to be free of foci, possibly suggesting that in these two cells all of the introduced DNA double-strand breaks had been rejoined. This time course obtained by counting foci is very similar to that obtained by Rogakou et al. 1998, in which mass formation of γ-H2AX was measured after CHO cells had been irradiated with 200 Gy. This similarity suggests that the processes of γ-H2AX appearance and disappearance are the same at these two very different amounts of ionizing radiation.
Figure 3.
Numbers of foci in IMR90 cells. Maximum projections of fields of IMR90 cells, similar to those shown in Fig. 2A–F, were analyzed by eye for numbers of foci per nucleus. Nuclei were scored as containing 0–5, 6–10, 11–15, 16–20, or 20–25 foci. All whole nuclei in a field were included, none of which contained >25 foci.
Larger amounts of radiation resulted in larger numbers of foci in both IMR90 (Fig. 2G and Fig. H) and MCF7 (Fig. 2O) cultures. Compared with IMR90 cultures 15 min after 0.6 Gy, with 10.1 to ± 3.9 (17 nuclei) foci per nucleus, IMR90 cultures 15 min after 2 Gy contained 24 ± 5.7 (10 nuclei) foci per nucleus. For MCF7 cultures, the comparable numbers are 12.2 ± 5.7 (26 nuclei) foci per nucleus 15 min after 0.6 Gy, and 27.1 ± 10.8 (24 nuclei) foci per nucleus 15 min after 2 Gy. For both IMR90 and MCF7 cultures, the values for 2 Gy are somewhat less than expected for linear proportionality with respect to the amount of radiation. This discrepancy may be at least partly explained if two foci at different levels in the nuclei overlap in the maximum projection and are scored as a single focus. This overlapping is more likely as the number of foci increases. In addition, it is relevant to mention that the SD values are presented as a means of displaying the range of the values in different nuclei, not as a measure of reproducibility. DNA breakage by ionizing radiation is a stochastic process, and thus the values of foci per nucleus are expected to follow a Poisson distribution. The distribution of values shown in Fig. 3 for each time point as well as the calculated SDs are consistent with those of Poisson distributions.
Each Gy introduces one double-strand break per 0.2 × 109 bp DNA when irradiated cells are analyzed before any repair can take place (Ruiz de Almodovar et al. 1994; Cedervall et al. 1995). In typical mammalian cells with a diploid DNA content between 6 × 109 bp in G1 and 12 × 109 bp in G2, this number corresponds to 30–60 DNA initial double-strand breaks per Gy depending on the cell cycle position of the cell. About 60% of these initial breaks are transient with rejoining half-lives on the order of minutes, whereas the other 40% are much more persistent with rejoining half-lives on the order of hours (Ward 1988, Ward 1990; Kodym and Hörth 1995; Nunez et al. 1995). The transient fraction may be composed of multiply nicked DNA molecules that maintain a continuous double helix in vivo but which are pulled apart under the in vitro assay conditions (Ward 1988, Ward 1990).
Thus, depending on their position in the cell cycle, IMR90 cells are predicted to contain 18–36 initial and 7–14 persistent DNA double-strand breaks after exposure to 0.6 Gy. At 3 min after irradiation, most of the IMR90 cells displayed between 10 and 20 foci (Fig. 3), with an average of 16.3 ± 3.6 foci per nucleus, values near the low number of expected initial breaks (18–36) and the high number of expected persistent breaks (7–14). At 15 and 30 min after irradiation, most of the IMR90 cells displayed between 5 and 15 foci (Fig. 3) with averages of 10.1 ± 3.9 and 11.6 ± 5.3 foci per nucleus, respectively. These values agree well with the number of expected persistent breaks (7–14). As there is likely to be a wide range in the severity of the DNA damage at locally multiply damaged sites (Ward 1990), perhaps some of the small γ-H2AX foci seen at 3 min after irradiation cease growing or disappear because those DNA double-strand breaks were rejoined very quickly. These observations show that there is a close correlation between the numbers of γ-H2AX foci and the numbers of expected DNA double-strand breaks, leading to the conclusion that each γ-H2AX focus may represent a DNA double-strand break in vivo.
Laser-induced DNA Double-Strand Breaks
If each γ-H2AX focus identifies a DNA double-strand break, then the two should coincide. To determine this, advantage was taken of the finding that γ-H2AX was formed when DNA double-strand breaks were introduced into cells by the BrdU dye–UVA light procedure of Limoli and Ward 1994 (Rogakou et al. 1998). A UVA pulsed laser (390 nm) commonly used in LaserScissors™ devices can be substituted for the UVA fluorescent light source (365 nm), and has the advantage of permitting the illumination of specific partial nuclear volumes. When MCF7 cells with BrdU-containing DNA were exposed to the laser in the presence of the dye, those nuclear regions traversed by the laser at 1, 10, and 30% relative power contained γ-H2AX foci (Fig. 4 A). γ-H2AX formation was dependent on the presence of BrdU; when BrdU was absent but dye still present, γ-H2AX foci were consistently found only in the cells traversed with the laser at 30% relative power (Fig. 4 B). This experiment demonstrates that γ-H2AX foci form at the sites of DNA double-strand breaks.
Figure 4.
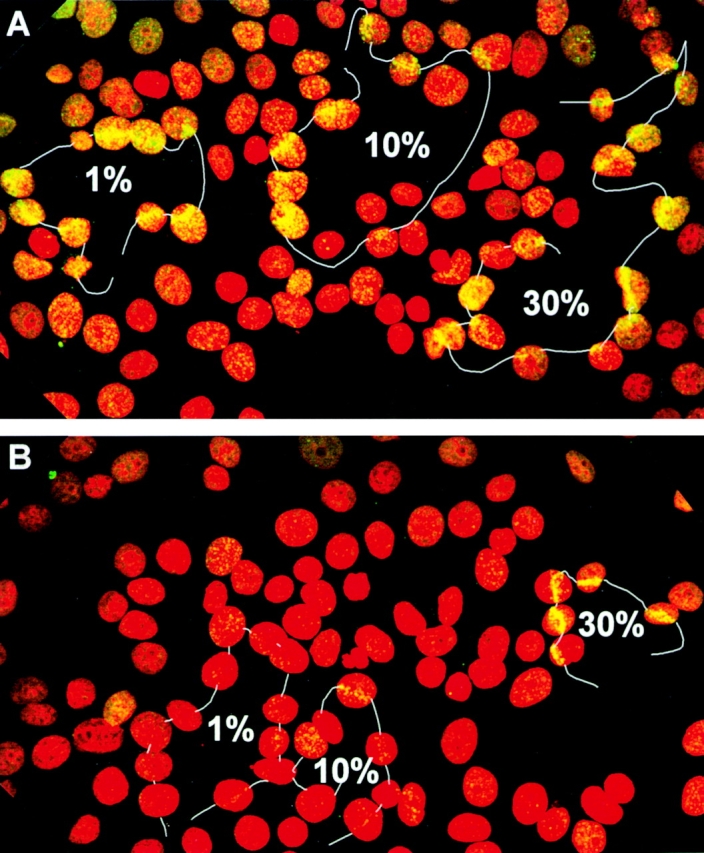
Laser-directed DNA double-strand breaks in MCF7 cells. UVA light was delivered by a 390-nm laser as described in Materials and Methods. The white lines trace the path of the laser as guided with a joystick. The percentages refer to the relative laser energy used in each transit. (A) Cells grown with BrdU. (B) Cells grown without BrdU. In both cases, cells were incubated with Hoechst dye 33258 as described in Materials and Methods.
Notably, there appears to be no difference in the efficacy of 1, 10, and 30% relative power in the formation of γ-H2AX foci (Fig. 4 A), possibly because even at 1% the exposure was more than sufficient to lead to the complete conversion of H2AX in the path of the laser to γ-H2AX. Limoli and Ward 1994 used exposures of up to 3 kJ/m2, whereas Rogakou et al. 1998 used 10 kJ/m2. The LaserScissors™ at 1% relative power produces 0.2 μj/pulse in a focused region ∼0.5 μm in diameter, or 1,000 kJ/m2 per pulse, values at least 100 times more than those used in the earlier studies with UVA fluorescent bulbs. Thus, it is quite possible that extensive formation of γ-H2AX occurs in the beam path at 1% relative power and that more is not formed at the higher power settings. It is relevant to mention that cells are generally quite transparent to these UVA wavelengths, thus most of the laser radiation probably passes completely through the cell, inflicting little if any damage except for that absorbed by the dye.
It is also notable that there are more γ-H2AX foci in the regions adjacent to the path of the microbeam laser in the cells grown with BrdU than in those without (compare Fig. 4 A and Fig. 4 B), probably because the BrdU and dye present in the former make the whole nucleus very sensitive to any UVA light that might be scattered from the laser beam. Thus, to limit DNA damage to more defined regions, it may be more appropriate to omit BrdU and use higher laser power settings.
Foci in Irradiated Mitotic Cells
Mitotic MCF7 cells were present in some of the cultures analyzed in Fig. 2. The mitotic cell noted in Fig. 2 J (m) is of particular interest because this culture was fixed only 3 min after irradiation, a result indicating that γ-H2AX foci form on mitotic chromosomes as well as on interphase chromatin. When fields of cells from MCF7 cultures containing both mitotic and interphase cells were analyzed for γ-H2AX foci 15 and 225 min after exposure to 0.6 Gy, the number of foci was decreased at the latter time in both interphase and mitotic cells (Fig. 5, A–C). These results show that the kinetics of γ-H2AX foci appearance and disappearance are similar whether cells are interphase or mitotic.
Figure 5.
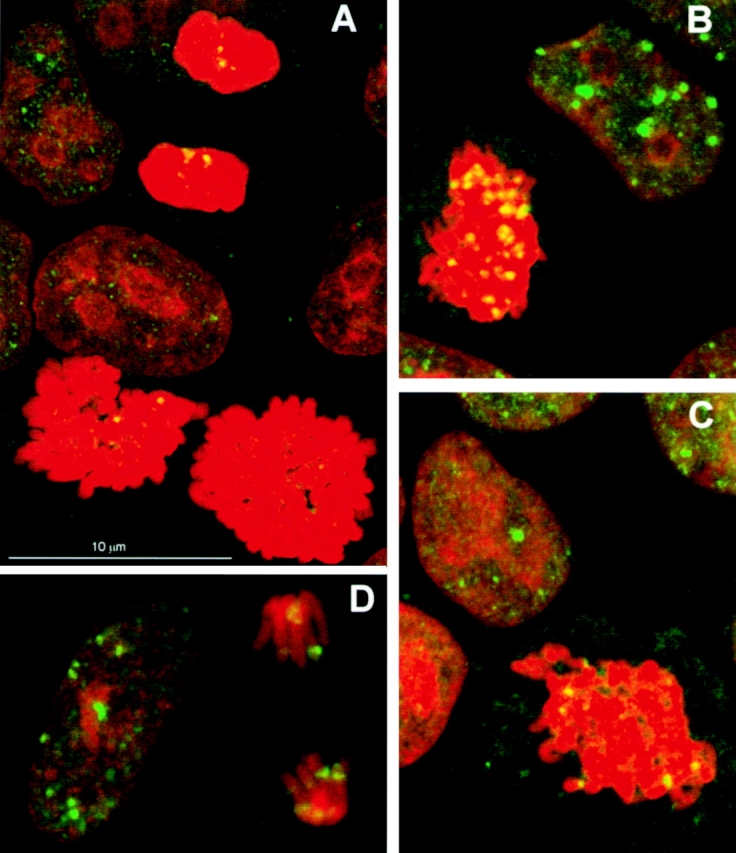
Interphase and mitotic cells from human MCF7 and M. muntjak cultures. (A–C) MCF7 cells from an unirradiated culture (A) or from cultures exposed to 0.6 Gy and allowed to recover for 15 (B) or 225 (C) min before fixation. (D) M. muntjak cells from a culture exposed to 0.6 Gy and allowed to recover for 15 min before fixation.
Because of the large number of small chromosomes in human mitotic cells, it is not possible to consistently visualize γ-H2AX foci on individual chromosomal arms. However, individual chromosome arms can be easily visualized in fibroblasts of the Indian muntjac, M. muntjak, a small deer with the typical mammalian DNA complement divided into 6 chromosomes instead of the 46 in humans (Wurster and Benirschke 1970). Interphase nuclei and condensed chromosomes from muntjac cells exposed to 0.6 Gy were found to show distinct γ-H2AX foci similar in number and intensity to those seen in the MCF7 cells (Fig. 5 D). In addition, it is possible to see the relationship of γ-H2AX foci to the chromosome arms.
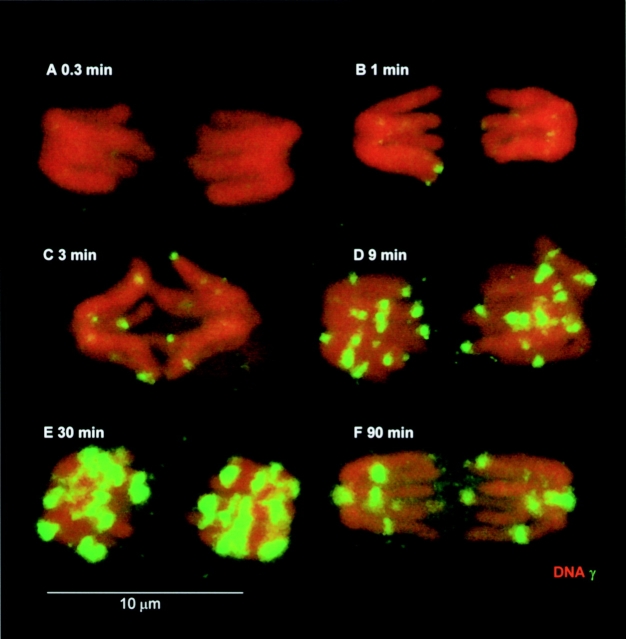
A study was performed with muntjac cell cultures exposed to 0.6 Gy and permitted to recover for various times (Fig. 6). The fields were searched for mitotic cells, which were imaged. A mitotic figure with discretely visible arms taken from each recovery time period is presented in Fig. 6. γ-H2AX foci, although not detectable after 0.3 min (Fig. 6 A), were detectable as small punctate foci after 1 min and continued to grow in number and size until 30 min (Fig. 6, B–E). At 90 min, the γ-H2AX foci were fewer in number but similar in size to those seen at 30 min. Since muntjac cells contain ∼90% of the DNA per cell as human cells (Levy et al. 1993), similar numbers of γ-H2AX foci per Gy would be expected in the two. If each half mitotic figure contains the G1 complement of DNA, one would expect ∼6 initial and 6 persistent DNA double-strand breaks in each. The number of γ-H2AX foci visible in each half of the 9- and 30-min mitotic figures (Fig. 6D and Fig. E) is higher than that found in the IMR90 cells (Fig. 2C and Fig. D, and Fig. 3), and nearer the expected value for initial rather than persistent DNA double-strand breaks. This difference might reflect a greater sensitivity of detection of small γ-H2AX foci in mitotic cells due to the greater compaction of the chromatin; mitotic cells often display more distinct foci than do interphase cells (compare the mitotic and interphase cells in Fig. 2J and Fig. M). The difference could also be due to differences in DNA double-strand break detection and rejoining between interphase and mitotic cells, or to differences between human and muntjac cell metabolism. The IMR90 cells contained more foci at 3 min after 0.6 Gy than later (Fig. 2B and Fig. C), whereas the opposite was the case for the muntjac mitotic cells (Fig. 6C and Fig. D). These findings indicate that these types of differences do exist, but whether they are due to differences in detection sensitivity, DNA compaction, species metabolism, or other factors requires further study.
Figure 6.
γ-H2AX foci developing on M. muntjak mitotic chromosomes (maximum projections). M. muntjak cell cultures were exposed to 0.6 Gy on ice, covered with growth media at 37°C, and allowed to recover for 0.3 (A), 1 (B), 3 (C), 9 (D), 30 (E), and 90 min (F) before fixation. Fields were scanned by eye using phase optics for mitotic cells. Selected cells were optically sectioned at 0.5-μm intervals. The sections were recombined into a maximum projection. The green channel is amplified to the same extent in all samples to visualize the nascent foci at 1 and 3 min; however, this results in some overexposed foci in the 9-, 30-, and 90-min samples.
These findings show that DNA double-strand breaks are rapidly detected and marked in condensed chromosomes as well as in interphase chromatin. The mitotic cells imaged in Fig. 6 appear to have been dividing normally when fixed. However, in cultures fixed at 90 min after irradiation, defective mitotic cells could be found (Fig. 7, A–C). Three mitotic cells containing six isolated chromosomal arm fragments are shown, each with a large γ-H2AX focus at one end (indicated by green arrows). In addition, no isolated chromosomal arms lacking a terminal γ-H2AX focus were found in any mitotic figure. These results provide direct visual confirmation that γ-H2AX forms en masse at the sites of DNA double-strand breaks.
Figure 7.
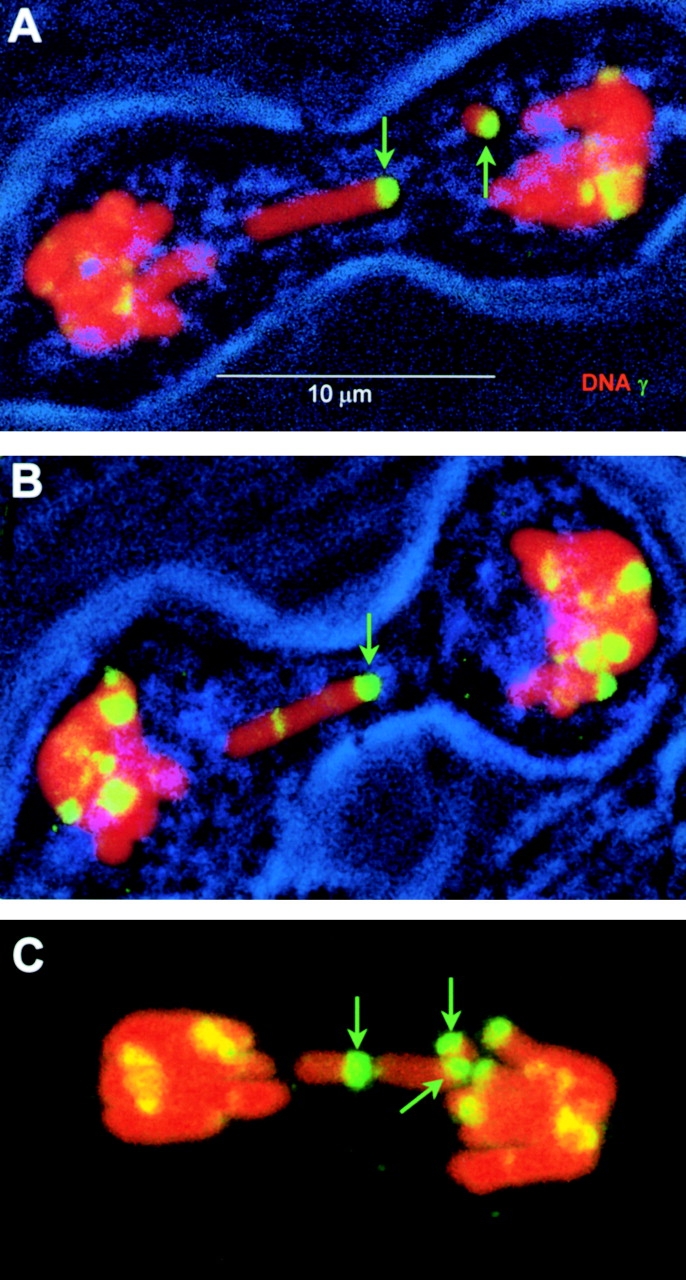
γ-H2AX foci on defective M. muntjak mitotic figures (maximum projections). Selected mitotic figures were imaged in M. muntjak cell cultures that had been exposed to 0.6 Gy on ice and covered with growth media at 37°C for 90 min before fixation. Green arrows point to ends of isolated chromosome arms with γ-H2AX foci. In A and B, transmitted light was collected to show the outline of the cell membrane.
Ionizing radiation introduces a delay in cell cycle progression in early G2. Kimler et al. 1981 determined that in CHO cells, the duration of the G2 delay could be calculated as 1 h per Gy for cells that were irradiated before prophase. The arrest point was calculated to be 17 min before metaphase and 49 min before completion of cytokinesis. Similar values had been found for mouse leukemic cells (Doida and Okada 1969), indicating that these values may apply to mammalian cells in general. Thus, the M. muntjak cells exposed to 0.6 Gy (Fig. 7) could have arrested in early G2 for ∼30 min, then entered mitosis and progressed toward cytokinesis, but were unable to complete cytokinesis because chromosome arms abandoned during anaphase blocked the constriction of the cleavage furrow.
Discussion
Collectively, these data reveal the presence of an immediate, substantial, and evolutionarily conserved response of cells to the introduction of DNA double-strand breaks. This response involves the formation of γ-H2AX on chromosomal regions encompassing megabase lengths of DNA adjacent to break sites. No mammalian cell line, normal or repair-defective, has been found that lacks the ability to form γ-H2AX when exposed to ionizing radiation. Cell lines from patients with ataxia telangiectasia, Werner's syndrome, Bloom's syndrome, and Nijmegen breakage syndrome all form γ-H2AX after exposure to ionizing radiation (Rogakou et al. 1998; Rogakou, E.P., C. Boon, and W.M. Bonner, unpublished results). M059J human, scid mouse, and V3 hamster cells that lack the DNA-activated protein kinase catalytic subunit (DNA-PKcs) form γ-H2AX by gel analysis, and show γ-H2AX foci after exposure to ionizing radiation (Rogakou, E.P., C. Boon, and W.M. Bonner, unpublished results). A KU70 knockout cell line (Nussenzweig et al. 1995) also forms γ-H2AX. The lack of mutant cell lines unable to form γ-H2AX indicates that it may fulfill essential functions in organisms throughout the evolutionary scale.
It has been reported that when DNA double-strand breaks were introduced into regions of fibroblast nuclei by partial volume irradiation, MRE11, one of the proteins involved in DNA double-strand break rejoining, was found in these regions 30 min after irradiation (Nelms et al. 1998). Maximal amounts of γ-H2AX are formed in irradiated human fibroblasts by 10 min, amounts corresponding to regions of chromatin containing about 2 × 106 bp of DNA and 2,000 γ-H2AX molecules (Rogakou et al. 1998). One possible role of these γ-H2AX foci at the sites of DNA double-strand breaks could be to serve in recruiting proteins that are involved in rejoining DNA ends such as MRE11 or RAD50 to those sites, either directly, through binding to the γ-H2AX COOH terminus, or indirectly, through an altered regional chromatin structure. With the γ-H2AX antibody, regions of chromatin containing DNA double-strand breaks may be isolated and components that interact with those regions may be characterized.
The γ-H2AX domains seen on chromosome arms are similar in appearance to chromosome bands and also appear to stop enlarging after 30 min, even though some DNA double-strand breaks are still present. Although other explanations are possible, these findings may suggest the existence of units of higher-order chromatin structures that are involved in monitoring DNA integrity.
Acknowledgments
We thank Dr. Kurt Kohn (National Cancer Institute, Division of Basic Sciences, Laboratory of Molecular Pharmacology, National Institutes of Health) for his continuous support of this work, Dr. Kenneth Yamada (National Institute of Dental and Craniofacial Research, Craniofacial Developmental Biology and Regeneration Branch, National Institutes of Health) for the use of the LaserScissors™ instrument, and Dr. Robert F. Bonner (National Institute of Child Health and Human Development, Laboratory of Integrative and Medical Biophysics, National Institutes of Health) for instruction on the LaserScissors™.
Footnotes
1.used in this paper: BrdU, bromo deoxyuridine
References
- Anderson C.W. Protein kinases and the response to DNA damage. Semin. Cell Biol. 1994;5:427–436. doi: 10.1006/scel.1994.1050. [DOI] [PubMed] [Google Scholar]
- Bai Y., Symington L.S. A RAD52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae . Genes Dev. 1996;10:2025–2037. doi: 10.1101/gad.10.16.2025. [DOI] [PubMed] [Google Scholar]
- Cedervall B., Wong R., Albright N., Dynlacht J., Lambin P., Dewey W.C. Methods for the quantification of DNA double-strand breaks determined from the distribution of DNA fragment sizes measured by pulsed-field gel electrophoresis. Radiat. Res. 1995;143:8–16. [PubMed] [Google Scholar]
- Celis J.E. Cell Biology. Vol. 2. 2nd ed Academic Press 1998. 533 San Diego; CA: pp [Google Scholar]
- Doida Y., Okada S. Radiation-induced mitotic delay in cultured mammalian cells (L5178Y) Radiat. Res. 1969;38:513–529. [PubMed] [Google Scholar]
- Game J.C. DNA double-strand breaks and the RAD50-RAD57 genes in Saccharomyces . Semin. Cancer Biol. 1993;4:73–83. [PubMed] [Google Scholar]
- Grawunder U., Wilm M., Wu X., Kulesza P., Wilson T.E., Mann M., Lieber M.R. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- Jeggo P.A. Identification of genes involved in repair of DNA double-strand breaks in mammalian cells. Radiat. Res. 1998;150:S80–S91. [PubMed] [Google Scholar]
- Jeggo P.A., Taccioli G.E., Jackson S.P. Menage a troisdouble strand break repair, V(D)J recombination and DNA-PK. Bioessays. 1995;17:949–957. doi: 10.1002/bies.950171108. [DOI] [PubMed] [Google Scholar]
- Johzuka K., Ogawa H. Interaction of Mre11 and Rad50two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae . Genetics. 1995;139:1521–1532. doi: 10.1093/genetics/139.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaar R., Heijmakers J.H.J., van Gent D.C. Molecular mechanisms of DNA double-strand break repair. Trends Cell Biol. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- Kerr J. An overview of apoptosis with emphasis on its biodefence functions. In: Ozawa T., Tatsumi K., Hori T., editors. Biodefence Mechanisms against Environmental Stress. Springer-Verlag; Heidelberg, Germany: 1998. pp. 69–75. [Google Scholar]
- Kimler B.F., Leeper D.B., Schneiderman M.H. Radiation-induced division delay in Chinese hamster ovary fibroblast and carcinoma cellsdose effect and ploidy. Radiat. Res. 1981;74:430–438. [PubMed] [Google Scholar]
- Kodym R., Hörth E. Determination of radiation-induced DNA strand breaks in individual cells by non-radioactive labeling of 3′ OH ends. Int. J. Radiat. Biol. 1995;68:133–139. doi: 10.1080/09553009514551031. [DOI] [PubMed] [Google Scholar]
- Levy H.P., Schultz R.A., Ordonez J.V., Cohen M.M. DNA content measurements and an improved idiogram for the Indian muntjac. Cytometry. 1993;14:362–368. doi: 10.1002/cyto.990140404. [DOI] [PubMed] [Google Scholar]
- Li Z., Otevrel T., Gao Y., Cheng H.L., Seed B., Stamato T.D., Taccioli G.E., Alt F.W. The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell. 1995;83:1079–1089. doi: 10.1016/0092-8674(95)90135-3. [DOI] [PubMed] [Google Scholar]
- Limoli C.L., Ward J.F. A new method for introducing double-strand breaks into cellular DNA. Radiat. Res. 1993;134:160–169. [PubMed] [Google Scholar]
- Limoli C.L., Ward J.F. Response of bromo-deoxyuridine–substituted Chinese hamster cells to UVA light exposure in the presence of Hoechst dye #33258survival and DNA repair studies. Radiat. Res. 1994;138:312–319. [PubMed] [Google Scholar]
- Mannironi C., Bonner W.M., Hatch C.L. H2AX histone isoprotein with a conserved C-terminal sequence, is encoded by a novel mRNA with both DNA replication type and poly A 3′ processing signals. Nucleic Acids Res. 1989;17:9113–9126. doi: 10.1093/nar/17.22.9113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modesti M., Hesse J.E., Gellert M. DNA binding of Xrcc4 protein is associated with V(D)J recombination but not with stimulation of DNA ligase IV activity. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:2008–2018. doi: 10.1093/emboj/18.7.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelms B.E., Maser R.S., MacKay J.F., Lagally M.G., Petrini J.H. In situ visualization of DNA double-strand break repair in human fibroblasts. Science. 1998;280:590–592. doi: 10.1126/science.280.5363.590. [DOI] [PubMed] [Google Scholar]
- Nickoloff J.A., Hoekstra M.F. Double-strand break and recombinational repair in Saccharomyces cerevisiae . In: Nickoloff J.A., Hoekstra M.F., editors. DNA Damage and Repair. Vol. 1. Humana Press; Totowa, NJ: 1998. pp. 335–362. [Google Scholar]
- Nunez M.I., Villalobos M., Olea N., Valenzuela M.T., Pedraza V., McMillan T.J., Ruiz de Almodovar J.M. Radiation-induced DNA double-strand break rejoining in human tumor cells. Br. J. Cancer. 1995;71:311–316. doi: 10.1038/bjc.1995.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussenzweig A., Chen C., da Costa Soares V., Sanchez M., Sokol K., Nussenzweig M.C., Li G.C. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature. 1995;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- Rathmell W.K., Chu G. Mechanisms for DNA double-strand break repair in eucaryotes. In: Nickoloff J.A., Hoekstra M.F., editors. DNA Damage and Repair. Vol. 2. Humana Press; Totowa, NJ: 1998. pp. 299–316. [Google Scholar]
- Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Ruiz de Almodovar J.M., Steel G.G., Whitaker S.J., McMillan T.J. A comparison of methods for calculating DNA double-strand break induction frequency in mammalian cells by pulsed-field gel electrophoresis. Int. J. Radiat. Biol. 1994;6:641–649. doi: 10.1080/09553009414550751. [DOI] [PubMed] [Google Scholar]
- Ueda K., Tanaka I. The appearance of male gamete-specific histones gH2B and gH3 during pollen development in Lilium longiflorum . Dev. Biol. 1995;169:210–217. doi: 10.1006/dbio.1995.1138. [DOI] [PubMed] [Google Scholar]
- Ward J.F. DNA damage produced by ionizing radiation in mammalian cellsidentities, mechanisms of formation and repairability. Prog. Nucleic Acid Res. Mol. Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- Ward J.F. DNA damage and repair. In: Glass W.A., Varma M.N., editors. Physical and Chemical Mechanisms in Molecular Radiation Biology. Plenum Press; New York: 1990. pp. 403–421. [Google Scholar]
- Wurster D.H., Benirschke K. Indian muntjac, Muntiacus muntjaka deer with a low diploid chromosome number. Science. 1970;168:1364–1366. doi: 10.1126/science.168.3937.1364. [DOI] [PubMed] [Google Scholar]